Abstract
Purpose
How to optimally detect bilateral mammographic asymmetry and improve risk prediction accuracy remains a difficult and unsolved issue. Our aim was to find an effective mammographic density segmentation method to improve accuracy of breast cancer risk prediction.
Methods
A dataset including 168 negative mammography screening cases was used. We applied a mutual threshold to bilateral mammograms of left and right breasts to segment the dense breast regions. The mutual threshold was determined by the median grayscale value of all pixels in both left and right breast regions. For each case, we then computed three types of image features representing asymmetry, mean and the maximum of the image features, respectively. A two-stage classification scheme was developed to fuse three types of features. The risk prediction performance was tested using a leave-one-case-out cross-validation method.
Results
By using the new density segmentation method, the computed area under the receiver operating characteristic curve was 0.830±0.033 and overall prediction accuracy was 81.0%, significantly higher than those of 0.633±0.043 and 57.1% achieved by using the previous density segmentation method (p < 0.01, t-test).
Conclusions
A new mammographic density segmentation method based on a bilateral mutual threshold can be used to more effectively detect bilateral mammographic density asymmetry and help significantly improve accuracy of near-term breast cancer risk prediction.
Keywords: Breast, Cancer, Computer-aided detection, Mammographic density segmentation, Risk stratification
I. INTRODUCTION
Due to the potential harmful effects of cumulative radiation exposure [1], unnecessary biopsies [2], high cost and limited healthcare resources [3], the current population-based mammography screening paradigm remains quite controversial [4]. In order to solve this clinical dilemma, developing an optimal breast cancer screening paradigm to detect early cancers has attracted extensive research interest recently [5].
The success of establishing an optimal personalized breast cancer screening paradigm depends on developing a reliable risk prediction model. Although many epidemiology study-based breast cancer risk models, such as Gail, Claus, and Tyrer-Cuzick model [6] have been developed, they typically aim to assess the risk of a woman developing breast cancer in a long term or lifetime. Thus, it is required to develop new models that have higher discriminatory power in predicting the risk of individual women developing breast cancer in the near-term [7]. Based on the computed quantitative image features, several research groups have developed and tested a number of new risk stratification models to predict breast cancer risk [8–12]. These image features could be divided into two major categories: unilateral features (depicting the information of every single breast mammogram), and concurrent features (describing the asymmetry of bilateral mammograms of left and right breasts) [13]. Using unilateral features, including mammographic texture and density features, has shown the value in breast cancer risk assessment [14]. A recent study also demonstrated a moderately high positive association between the risk prediction scores generated by the mammographic density related image feature analysis and the actual risk of women having an image-detectable breast cancer in the next subsequent examinations [10]. Despite that mammographic density has the highest discriminatory power besides women’s age in the existing epidemiology-based risk models [6], its discriminatory power at the individual level remains low and controversial [15,16].
In our studies, we hypothesized that bilateral mammographic density asymmetry may be an important indicator of breast cancer development [4]. To test this hypothesis, we have preliminarily investigated computerized methods to detect bilateral mammographic density asymmetry features and the potential of using asymmetry scores to predict near-term breast cancer risk [13, 17, 18]. However, how to optimally detect bilateral mammographic asymmetry and improve risk prediction accuracy remains a difficult and unsolved issue.
In this study, we investigated a new bilateral mammographic asymmetry detecting approach and tested whether it can help significantly improving the performance of near-term breast cancer risk prediction. Specifically, we proposed to use a single “mutual” threshold to bilateral mammograms of left and right breasts to segment dense breast regions and compute relevant image features, and developed a unique two-stage classification scheme to fuse the image features for breast cancer risk prediction.
II. Materials
We retrospectively assembled an image dataset, which includes two sequential mammographic screening examinations acquired from 168 women. Each examination includes two full-field digital mammography (FFDM) images representing the craniocaudal (CC) view of the left and right breasts. All images were acquired using Hologic Selenia FFDM systems (Hologic Inc., Bedford, MA, USA).
For each case (woman), the first (“prior”) examination and the next (“current”) examination were acquired within a time lag of 12–36 months. All the “prior” examinations were interpreted by radiologists as “negative”. Among the “current” examinations, 83 were positive cases with cancer verified and 85 were negative (cancer-free) cases. The average ages and standard deviations were 57.5±11.8 and 52.5±10.9 years old for the positive and negative groups of women, respectively. In this study, only “prior” screening images were used for computing features and building prediction models, which were divided into two different classes based on their status in the “current” examinations.
In this study, we only focused on predicting whether women will develop breast cancers in the next sequential screening after the first negative screening. It was not validated whether the “negative” cases remained in the “cancer-free” status in the third sequential examinations.
III. Methods
The new risk model was developed using four steps including (1) segmenting breast regions, (2) creating local pixel value fluctuation maps, (3) computing image features, and (4) optimizing classification scheme.
A. Breast Region Segmentation
For each case, a pair of bilateral CC view images were analyzed. We applied an automatic segmentation scheme on each image to extract the whole breast region as described in [19, 20]. In brief, a gray level histogram of the image was plotted and an iterative searching method was used to detect the smoothest curvature between the breast tissue and background or air region. The pixels in the background were discarded and the skin region was removed by a morphological erosion operation (Fig.1). In addition, we also segmented the dense breast region from each image. In the previous studies, the dense breast region of each image (left or right mammographic image) was defined as the region that encompasses the pixel values above the median value of the whole breast region[7,21–22]. In these studies, dense regions of left and right mammographic images were segmented using their respective medians as thresholds. It was found that some individual features computed for the dense breast areas showed good correlation with the radiologists’ ratings [21] and some texture features computed on the dense breast regions are most effective for breast cancer risk prediction [7]. In this study, we also used median value for thresholding to take its advantage, but in a different way. It is possible that the thresholding method used in previous studies may reduce the capability of detecting bilateral density asymmetry. In this study, we used a single “mutual” threshold, instead of two different thresholds, to segment dense regions from left and right mammographic images (Fig.1). The “mutual” threshold was defined as median grayscale value of all pixels in the whole breast regions of both left and right mammographic images. This was done to investigate a new approach aiming to more accurately detect bilateral mammographic density or tissue asymmetry and test whether it can help further improve the performance of applying quantitative image feature analysis methods to predict near-term breast cancer risk.
Fig. 1.
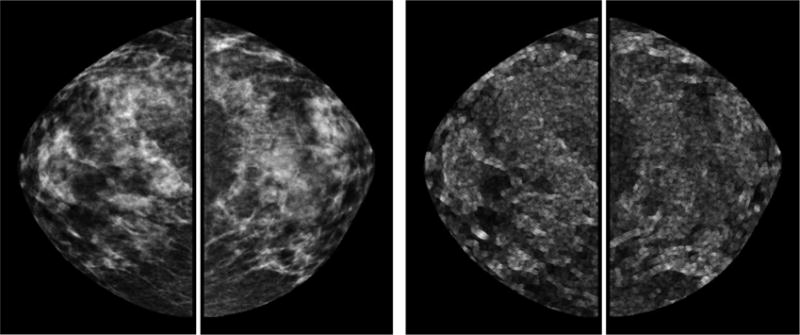
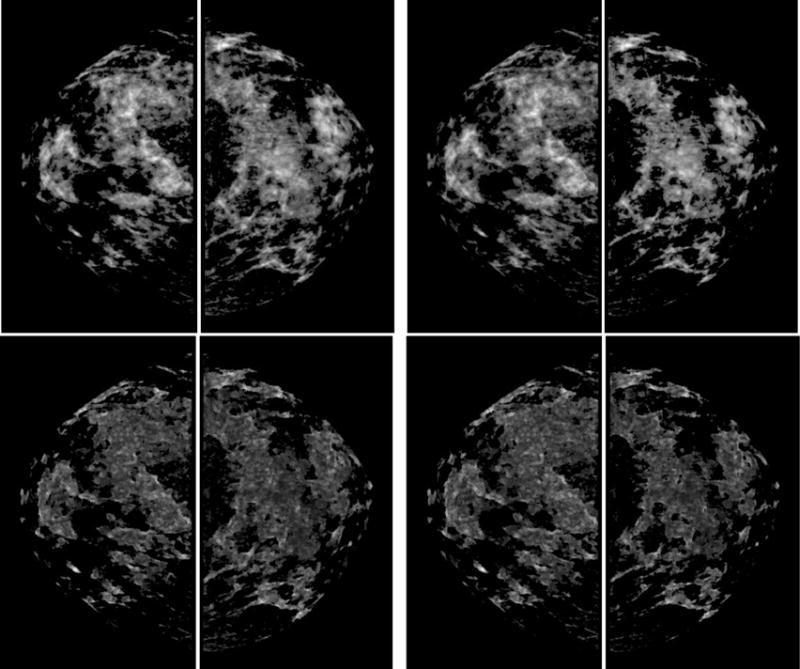
Example of a positive case acquired from the “prior” screening examination. It shows the segmented whole breast regions of original images (top left) and local pixel value fluctuation maps (top right), dense breast regions of original images segmented with the new segmentation method (middle left) and previous segmentation method (middle right), dense breast regions of local pixel value fluctuation maps segmented with the new segmentation method (bottom left) and previous segmentation method (bottom right).
B. Local Pixel Value Fluctuation Map Generation
In order to compute image features of higher predictive power, we generated local pixel value fluctuation (LPVF) maps for each image by applying a 5×5 square convolution kernel to scan the image with the segmented whole breast region and dense breast region, respectively (Fig.1). The absolute pixel value differences between the center pixel and each of the other pixels inside the kernel were computed. The maximum difference value computed inside the kernel was used to replace the original pixel value in the map (center pixel). As a result, unlike the original mammogram, the generated LPVF map can better show the image features related to the pixel value fluctuation and/or the local density variation, which may provide useful and complementary information to the image features acquired or computed from the original mammogram. Similar image conversion method was also used in [4, 18].
C. Image Feature Computation
Various studies have analyzed the correlation of mammographic image features with breast cancer risk [23–26]. In this study, we computed some features that have been proposed in the literature, as well as redefined some existing features and explored some new features that have never been examined for breast cancer risk prediction.
For each case, after reducing the gray level range of the original mammographic images from 4,096 to 256 gray levels, we computed the same 220 features from (1) whole breast regions and (2) dense breast regions in the original bilateral CC view images, which are named as WBR-ORG and DBR-ORG images, respectively, (3) whole breast regions and (4) dense breast regions in the bilateral LPVF maps, which are named as WBR-LPVF and DBR-LPVF maps, respectively. In the following, these features will be name as NFi (i = 1,2,…,220×4). For comparison, we also computed the same 220×4 features from the original mammographic images and the LPVF maps in which the dense breast regions were segmented by applying two different thresholds separately to two bilateral mammograms (previous segmentation method). These features will be name as PFi (i = 1,2,…,220×4).
Each group of 220 features were divided into three distinct types namely: (1) 76 asymmetry features, which were represented by the absolute subtraction of two matched bilateral feature values computed from the left and right images; (2) 72 mean features, which were computed by taking average value of two matched bilateral feature values; and (3) 72 maximum features, which were represented by the greater one of two matched bilateral feature values.
1) Asymmetry Features
The asymmetry features can be divided into five subgroups. The first subgroup include 13 image statistics based features (NFi and PFi, i = 1,2,…,13) [17], which are average local value fluctuation of gray level histogram, mean of gray level histogram values, standard deviation of gray level histogram values, statistics based features computed from the pixel value distributions of the ROI (region of interest) including mean value, variance, standard deviation, skewness, kurtosis, energy, and entropy, statistics based features computed from the local pixel value fluctuation map of the ROI including mean value, standard deviation, and skewness.
The second subgroup includes 4 fractal dimension related breast tissue composition features (NFi and PFi, i = 14,15,16,17) [21], which are estimated with variation method, mathematical morphology, two slopes of fitting lines using textural edgeness and Gaussian subtraction.
The third subgroup has 39 texture related image features (NFi and PFi, i = 18,19,…,56) [27], which include 8 Gray-level co-occurrence matrix based features, 13 Gray-level run-length matrix based features, 13 Gray-level size zone matrix based features, and 5 Neighborhood gray-tone difference matrix based features.
The fourth subgroup includes 16 features (NFi and PFi, i = 57,58,…,72) modified from those used in [17] to compute a variety of percentage of mammographic density (PMD) for mimicking Breast Imaging Reporting and Data System (BIRADS) used in clinical practice. These features are defined and computed as following: NF57 (or PF57) = NHA/NU, NF58 (or PF58) = NHA/NU, NF59 (or PF59) = NHA/NB and NF60 (or PF60) = NHA/NB. Where, NU is the total number of pixels in the ROI of left or right breast; NB is the total number of pixels in the ROIs of both left and right breasts. For NF57 and NF59, NHA is the number of pixels in the ROI of left or right breast with gray value larger than the average value of all pixels in the ROIs of both left and right breasts. For PF57 and PF59 NHA is the number of pixels in the ROI of left or right breast with gray value larger than the average value of all pixels in the ROI. For NF58 and NF60, NHM is the number of pixels in the ROI of left or right breast with gray value larger than the median value of all pixels in the ROIs of both left and right breasts. For PF58 and PF60, NHM is the number of pixels in the ROI of left or right breast with gray value larger than the median value of all pixels in the ROI. These 4 features are redefined from a feature used in Ref 25.
Next, to mimic BIRADS, we computed 3 features (NFi (or PFi) = IK/IS,i = 61,62,63). Specifically, IS is the average gray value of all breast tissue pixels in one image and IK are the average gray values of the pixels whose gray values are under the threshold of K = 25%, 50%, 75% of the maximum breast tissue pixel value in the image, respectively. In addition, we also computed other 3 features (NFi(or PFi) = NK/NS,i = 64,65,66), where NS is the total number of breast tissue pixels in a single image and NK are numbers of pixels whose gray values are under the threshold of K = 25%, 50%, 75% of the maximum pixel value in the image, respectively. Similarly, we also computed another set of 6 features combining all pixels in two bilateral images (NFi(or PFi) = IK/IB,i = 67,68,69) and (NFi(or PFi) = NK/NB,i = 70,71,72), where IB is the average pixel value of total breast tissue pixels (NB) of both left and right mammograms.
The last subgroup includes 4 newly explored features (NFi or PFi, i = 73,74,75,76) which are defined and computed as following:
, where and are the maximum breast tissue pixel values in the left and right images, respectively; GVh is the number of gray levels in the ROI of left or right breast, in which the maximum pixel value is higher. Specifically, if , GVh refers to the number of gray levels in the ROI of the left breast; otherwise GVh refers to the number of gray levels in the ROI of the right breast.
Where, Ng is the number of pixels in the ROI of higher maximum pixel value with gray values higher than the maximum pixel value of another ROI; Nh is the total number of pixels in the ROI of higher maximum pixel value. Specifically, if , Ng refers to the number of pixels in the ROI of the left breast with gray values higher than and Nh refers to the number of pixels in the ROI of the left breast; otherwise Ng refers to the number of pixels in the ROI of the right breast with gray values higher than and Nh refers to the number of pixels in the ROI of the right breast.
From each pair of matched ROIs, the first 72 asymmetry features were independently computed for the left and right breasts. Each asymmetry feature was computed by the absolute difference of two feature values computed from the two matched ROIs. The last 4 asymmetry features were computed directly from each pair of matched ROIs.
2) Mean Features
This type includes 72 features (NF77 to NF148 (or PF77 to PF148)), which were similar to the first 72 features in type 1 (NF1 to NF72 (or PF1 to PF72)). From each of two matched ROIs of left and right breasts, each of the 72 features was independently computed. Then, the average of two values of the same feature was used to represent the mean feature.
3) Maximum Features
This type includes another set of 72 features (NF149 to NF220 (or PF149 to PF220)), which were similar to the features in type 1 (NF1 to NF72 (or PF1 to PF72)). From each of two matched ROIs of left and right breasts, each of the 72 features was also independently computed. Then, the greater one of the two values of the same feature was chosen to represent the maximum feature.
D. Risk Prediction and Performance Assessment
The mean and standard deviation of values of different features may vary widely. In order to compare different features, a normalization process was applied for each feature to normalize the values to the range of 0 to 1. Similar normalization process was applied in [17, 18]. We used two analysis approaches to assess discriminatory power of these features to predict near-term breast cancer risk. In the first approach, we split the feature data 168 times and aggregated the validation performance of all splits. In each split, data of 167 cases were used for “optimal” feature selection (redundant and uninformative features may downgrade the classification accuracy) and data of the left out 1 case was used to test discriminatory power of the “optimal” features. In the feature selection step, the cross-validation method was carried out by using a publicly available WEKA data mining and machine learning software package [28]. In the validation step, the “optimal” features of the test case were fused with 3 simple methods (taking the average, maximum, and minimum value) [29] to generate 3 new classification scores, which were used for validating the prediction performance. The feature selection and performance validation protocol is characterized by a ‘double-cross-validation-loop’: an inner selection loop and an outer validation loop [30,31]. Prediction performance of the new classification scores of all 168 cases was assessed with an area under a receiver operating characteristic curve (AUC). The AUCs were computed applying a publically available ROC curve fitting program (ROCKIT, http://www-radiology.uchicago.edu/krl/).
In the second analysis approach, we trained a “two-stage” classification scheme using WEKA. As shown in Fig. 2. In the first stage, 3 ANNs (Artificial Neural Networks) were independently trained using the 76×4 = 304 asymmetry features, 72×4 = 288 mean features, and 72×4 = 288 maximum features, respectively. In the second stage, the 3 sets of scores produced by the 3 ANNs was further fused by another ANN to derive the final risk prediction scores. For comparison, we also trained another “one-stage” ANN, which was built using the mixed pool of all three types of features.
Fig. 2.
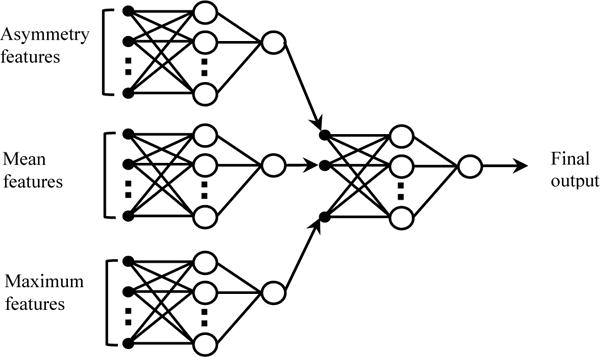
A two-stage “scoring fusion” RBFN-based ANN classification scheme, whereby the final classification score is derived by optimally fusing the prediction scores produced by 3 ANNs trained using bilateral asymmetrical features, mean features, and the maximum features, respectively. In the ANNs, the circles and black dots represent artificial neurons analogous to human neural units, the connecting lines represent links between adjacent artificial neurons analogous to neural axons.
In using WEKA, we chose the classifier of “AttributeSelectedClassifier” to integrate the normalized Gaussian Radial Basis Function Network (RBFN) as a base classifier, the wrapper subset evaluator (WSE) as a feature evaluator, and the “BestFirst” (searches the space of feature subsets by greedy hillclimbing augmented with a backtracking facility) as a feature search method. RBFN has drawn much attention due to its good generalization ability and simple network structure [32]. Past research has shown that any nonlinear function over a compact set with arbitrary accuracy can be approximated by RBFN [33]. In this study, the ANNs were trained using a leave-one-case-out (LOCO) based cross-validation method [34]. In each of LOCO training/testing iterations, 1 case was selected as an independent testing case and the remaining 167 cases were used as a training dataset. Feature selection was performed on the training dataset (redundant and uninformative features may downgrade the classification accuracy) and the selected features were then used to train a RBFN based classifier. The trained classifier was applied to the testing case to generate a risk prediction score ranging from 0 to 1. According to this validation method, in each training and testing iteration cycle, different image features may be selected from the initial feature pool and used to build a corresponding classifier.
We used AUC and prediction accuracy to compare performance of the asymmetry feature based classifier, mean feature based classifier, the maximum feature based classifier, the one-stage classification scheme, and the two-stage classification scheme. We compared the prediction accuracy on “positive”, “negative”, and all cases by applying a threshold of 0.5 on the classification scores to divide the 168 testing cases into two predicted groups.
IV. RESULTS
Table 1 compares two sets of AUCs yielded by 8 schemes. Using the previous segmentation method, AUCs ranged from 0.603±0.043 to 0.641±0.042. Using the new segmentation method, AUCs ranged from 0.603±0.043 to 0.830±0.033, which yields a significantly higher maximum AUC value (p < 0.01, t-test). Fig. 3 shows 2 sets of ROC curves generated using the classification scores yielded by 5 ANN-based classification schemes. Fig. 4 compares two ROC curves generated using the classification scores yielded by the two-stage classification scheme. By using the new segmentation methods, AUC was significantly increased from 0.633±0.043 to 0.830±0.033 (p < 0.01, t-test).
TABLE 1.
Comparison of AUC values and corresponding standard deviations of 8 different prediction schemes by using 2 dense breast region segmentation methods.
| Feature fusion method | New dense breast region segmentation method | Previous dense breast region segmentation method |
|---|---|---|
| Scheme 1 (Taking maximum value of the selected features) | 0.603±0.043 | 0.603±0.043 |
| Scheme 2 (Taking minimum value of the selected features) | 0.690±0.040 | 0.606±0.043 |
| Scheme 3 (Taking mean value of the selected features) | 0.640±0.043 | 0.621±0.043 |
| Scheme 4 (one-stage classification scheme) | 0.813±0.034 | 0.641±0.042 |
| Scheme 5 (asymmetry feature based classifier) | 0.807±0.035 | 0.623±0.043 |
| Scheme 6 (mean feature based classifier) | 0.657±0.043 | 0.603±0.043 |
| Scheme 7 (maximum feature based classifier) | 0.704±0.040 | 0.616±0.043 |
| Scheme 8 (two-stage classification scheme) | 0.830±0.033 | 0.633±0.043 |
Fig. 3.
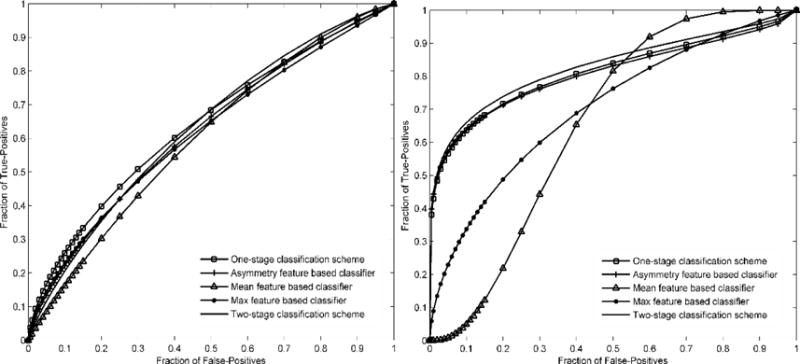
Comparison of receiver operating characteristic (ROC) curves generated by 5 breast cancer risk prediction schemes. Left and right correspond to the previous and new dense breast region segmentation methods, respectively
Fig. 4.
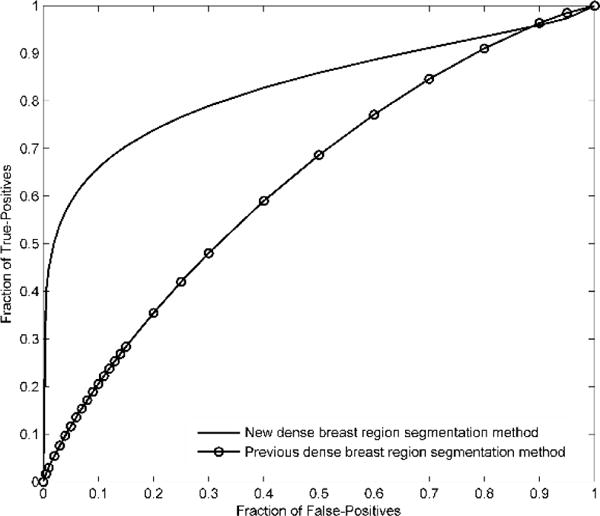
Comparison of receiver operating characteristic (ROC) curves generated by the proposed two-stage classification scheme. Two curves correspond to the previous and new dense breast region segmentation methods, respectively
Table 2 summarizes the prediction accuracy values yielded by 5 classification schemes. The right column shows 5 groups of prediction accuracy values yielded by using the previous dense breast tissue segmentation method, which are comparable with relatively low overall prediction accuracy (i.e., ≤ 60%). When using the new dense breast tissue segmentation method, the overall prediction accuracy was significantly increased (p < 0.01, t-test). For example, using the “two-stage” ANN-based classifier, it reached 81%. Table 2 also shows that the prediction accuracy for the positive cases is substantially higher than that for the negative cases (95.2% versus 67.1%).
TABLE 2.
Comparison of the overall prediction accuracy, prediction accuracy for the case groups with and without breast cancer of 5 different prediction schemes by using 2 dense breast region segmentation methods.
| Feature fusion method | Overall prediction accuracy, prediction accuracy for cases with cancer, and prediction accuracy for cases without cancer (%) | |
|---|---|---|
| New dense breast region segmentation method | Previous dense breast region segmentation method | |
| Scheme 4 (one-stage classification scheme) | 59.5 | 55.9 |
| 42.2 | 38.6 | |
| 76.5 | 72.9 | |
| Scheme 5 (asymmetry feature based classifier) | 54.2 | 60.7 |
| 36.1 | 42.2 | |
| 71.8 | 78.8 | |
| Scheme 6 (mean feature based classifier) | 50.0 | 54.2 |
| 62.7 | 38.6 | |
| 37.6 | 69.4 | |
| Scheme 7 (maximum feature based classifier) | 69.0 | 57.1 |
| 60.2 | 38.6 | |
| 77.6 | 75.3 | |
| Scheme 8 (two-stage classification scheme) | 81.0 | 57.1 |
| 95.2 | 60.2 | |
| 67.1 | 54.1 | |
V. DISCUSION
The contribution of this study is that we successfully demonstrated that a “mutual” threshold approach could play a surprisingly important role to significantly improve performance of near-term breast cancer risk prediction.
It was demonstrated that bilateral mammographic density asymmetry was a significantly stronger breast cancer risk factor than woman’s age, and mammographic density rated or assessed based on single images by either radiologists (BIRADS) or computerized schemes [18]. Thus, this study aims to investigate a new bilateral mammographic asymmetry detecting approach with and an assumption that it can help improving near-term breast cancer prediction. The assumption is based on several underlying scientific evidences and validated experimental observations: (1) mammographic tissue asymmetry is an important radiographic image phenotype related to the biological processes [35], (2) radiologists routinely examine the bilateral mammographic tissue asymmetry pattern and change over time when making clinical decisions [36].
We observed in the literature that image features computed from the dense mammographic regions had higher discriminatory power than those computed from the whole mammographic regions. However, how to optimally segment dense mammographic regions is important, which determines the subsequently computed image features. In this study, we developed a new mammographic image segmentation approach, in which the dense breast regions of bilateral mammograms were segmented using a “mutual” threshold instead of two individual thresholds. The threshold was determined according to the pixel grayscale value distribution of the CC view images of both left and right breasts.
From our study, we can make the following observations. First, by using the new “mutual” threshold segmentation method, bilateral mammographic density asymmetry related image features can be more sensitively detected and the discriminatory power of these features can be increased.
Second, previous study reported that using the maximum features to train an ANN yielded higher performance than the ANN trained using the bilateral asymmetrical features. However, we observed in this study, by using our new segmentation approach, the ANN trained using asymmetry features yielded much higher performance than the ANNs trained using the maximum features (Table 1). Therefore, defining an optimal dense mammographic region segmentation method is important for developing an optimal near-term breast cancer risk model.
Third, we observed that asymmetry, mean, and the maximum features of two bilateral images might contain complementary information. As a result, comparing to training one large integrated ANN using all three types of features, developing a two-stage ANN based classifier that fuses the prediction outcomes of 3 ANNs, which were separately trained with 3 types of features, yielded much higher prediction performance (AUC=0.830±0.033 vs AUC=0.813±0.034). The AUC is much higher than that of 0.633±0.043 achieved by using the previous density segmentation method and those of 0.761±0.025 and 0.725±0.018 reported in [18] and [21] achieved by using image features related to bilateral mammographic asymmetry for breast cancer prediction.
Last, we also observed that by using the proposed two-stage classification scheme, the prediction accuracy for the “positive” cases was substantially higher than that for the “negative” cases (95.2% versus 67.1%). This observation indicates high sensitivity and lower specificity, which is a potentially weakness of applying this risk prediction model in the real screening environment. Thus, improving specificity is important to improve efficacy of mammography screening. This is our goal in the future.
Despite the promising results and new observations, this study is with a number of limitations. First, the size of the dataset used is small and the ratio between positive and negative cases also does not represent the actual cancer prevalence ratio. Hence, the clinical utility of our new risk model needs to be eventually tested in future large prospective studies. Second, studies have shown that age is an important breast cancer risk factor [18] and association between mammographic density and breast cancer may be weaker in women with larger breasts [37]. In this study, we didn`t incorporate age and breast size as features in our new risk model or check their influence on the prediction accuracy. We will take them into account in the future study work. Third, only RBFN based ANN was trained and tested. Other machine learning methods can be developed for this purpose too.
In summary, we tested and demonstrated a new quantitative image segmentation and feature analysis approach to more accurately detect bilateral mammographic density asymmetry, as well as to improve performance in predicting near-term breast cancer risk. The initial testing results are promising. However, the robustness of this new approach and risk prediction model needs to be further evaluated before it can be clinically acceptable to help establish a new optimal and personalized breast cancer screening paradigm.
Acknowledgments
Funding: This study was supported in part by Grant R01 CA160205 and CA197150 from the National Cancer Institute, National Institutes of Health, USA.
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
References
- 1.Yaffe MJ, Mainprize JG. Risk of radiation-induced breast cancer from mammographic screening. Radiology. 2011;258:98–105. doi: 10.1148/radiol.10100655. [DOI] [PubMed] [Google Scholar]
- 2.Hubbard RA, Kerlikowske K, Flowers CI, Yankaskas BC, Zhu W, Miglioretti DL. Cumulative probability of false-positive recall or biopsy recommendation after 10 years of screening mammography: a cohort study. Ann Intern Med. 2011;155:481–492. doi: 10.1059/0003-4819-155-8-201110180-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buist DS, Anderson ML, Haneuse SJ, Sickles EA, Smith RA, Carney PA, Taplin SH, Rosenberg RD, Geller BM, Onega TL, Monsees BS, Bassett LW, Yankaskas BC, Elmore JG, Kerlikowske K, Miglioretti DL. Influence of annual interpretive volume on screening mammography performance in the United States. Radiology. 2011;259:72–84. doi: 10.1148/radiol.10101698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng B, Tan M, Ramalingam P, Gur D. Association between Computed Tissue Density Asymmetry in Bilateral Mammograms and Near-term Breast Cancer Risk. The Breast Journal. 2014;20:249–257. doi: 10.1111/tbj.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brawley OW. Risk-based mammography screening: an effort to maximize the benefits and minimize the harms. Ann Intern Med. 2012;156:662–663. doi: 10.7326/0003-4819-156-9-201205010-00012. [DOI] [PubMed] [Google Scholar]
- 6.Amir E, Freedman OC, Seruga B, Evans DG. Assessing women at high risk of breast cancer: a review of risk assessment models. J Natl Cancer Inst. 2010;102:680–691. doi: 10.1093/jnci/djq088. [DOI] [PubMed] [Google Scholar]
- 7.Tan M, Pu J, Cheng S, Liu H, Zheng B. Assessment of a Four-View Mammographic Image Feature Based Fusion Model to Predict Near-Term Breast Cancer Risk. Annals of Biomedical Engineering. 2015;40:2416–2428. doi: 10.1007/s10439-015-1316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen M, Karemore G, Loog M, Raundahl J, Karssemeijer N, Otten JD, Karsdal MA, Vachon CM, Christiansen C. A novel and automatic mammographic texture resemblance marker is an independent risk factor for breast cancer. Cancer Epidemiol. 2011;35:381–387. doi: 10.1016/j.canep.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Haberle L, Wagner F, Fasching PA, Jud SM, Heusinger K, Loehberg CR, Hein A, Bayer CM, Hack CC, Lux MP, Binder K, Elter M, Münzenmayer C, Schulz-Wendtland R, Meier-Meitinger M, Adamietz BR, Uder M, Beckmann MW, Wittenberg T. Characterizing mammographic images by using generic texture features. Breast Cancer Res. 2012;14:R59. doi: 10.1186/bcr3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun W, Qian W, Zhang J, Saltzstein EC, Zheng B, Lure F, Yu H, Zhou S. Using multi-scale texture and density features for near-term breast cancer risk analysis. Med Phys. 2015;42:2853–2862. doi: 10.1118/1.4919772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan M, Qian W, Pu J, Liu H, Zheng B. A new approach to develop computer-aided detection schemes of digital mammograms. Phys Med Biol. 2015;60:4413–4427. doi: 10.1088/0031-9155/60/11/4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian W, Sun W, Zheng B. Improving the efficacy of mammography screening: the potential and challenge of developing new computer-aided detection approaches. Expert Rev Med Devices. 2015;12:497–499. doi: 10.1586/17434440.2015.1068115. [DOI] [PubMed] [Google Scholar]
- 13.Tan M, Zheng B, Ramalingam P, Gur D. Prediction of near-term risk of developing breast cancer using computerized features from bilateral mammograms. Academic Radiology. 2013;20:1542–1550. doi: 10.1016/j.acra.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ray S, Keller BM, Kontos D. Application of computer-extracted breast tissue texture features in predicting false-positive recalls from screening mammography. Spie Medical Imaging. 2014;9035(1):90351X. [Google Scholar]
- 15.Gail MH, Mai PL. Comparing breast cancer risk assessment models. J Natl Cancer Inst. 2010;102:665–668. doi: 10.1093/jnci/djq141. [DOI] [PubMed] [Google Scholar]
- 16.Passaperuma K, Warner E, Hill KA, Gunasekara A, Yaffe MJ. Is mammographic breast density a breast cancer risk factor in women with BRCA mutations? J Clin Oncol. 2010;28:3779–3783. doi: 10.1200/JCO.2009.27.5933. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Lederman D, Tan J, Wang X, Zheng B. Computerized prediction of risk for developing breast cancer based on bilateral mammographic breast tissue asymmetry. Medical Engineering & Physics. 2011;33:934–942. doi: 10.1016/j.medengphy.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng B, Sumkin JH, Zuley ML, Wang X, Klym AH, Gur D. Bilateral mammographic density asymmetry and breast cancer risk: a preliminary assessment. European Journal of Radiology. 2012;81:3222–3228. doi: 10.1016/j.ejrad.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan M, Pu J, Zheng B. Reduction of false-positive recalls using a computerized mammographic image feature analysis scheme. Phys Med Biol. 2014;59:4357–4373. doi: 10.1088/0031-9155/59/15/4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng B, Sumkin JH, Zuley ML, Lederman D, Wang X, Gur D. Computer-aided detection of breast masses depicted on full-field digital mammograms: a performance assessment. Br J Radiol. 2012;85:e153–e161. doi: 10.1259/bjr/51461617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang YH, Wang XH, Hardesty LA, Chang TS, Poller WR. Computerized assessment of tissue composition on digitized mammograms. Acad Radiol. 2002;9:898–905. doi: 10.1016/s1076-6332(03)80459-2. [DOI] [PubMed] [Google Scholar]
- 22.Tan M, Zheng B, Leader JK, Gur D. Association between Changes in Mammographic Image Features and Risk for Near-term Breast Cancer Development. IEEE Trans Med Imaging. 2016;35:1719–1728. doi: 10.1109/TMI.2016.2527619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gierach GL, Li H, Loud JT, Greene MH, Chow CK. Relationships between computer-extracted mammographic texture pattern features and BRCA1/2 mutation status: a cross-sectional study. Breast Cancer Res. 2014;16:1–16. doi: 10.1186/s13058-014-0424-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertrand KA, Tamimi RM, Scott CG, Jensen MR, Pankratz V, Visscher D, Norman A, Couch F, Shepherd J, Fan B, Chen YY, Ma L, Beck AH, Cummings SR, Kerlikowske K, Vachon CM. Mammographic density and risk of breast cancer by age and tumor characteristics. Breast Cancer Res. 2013;15:R104–R104. doi: 10.1186/bcr3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heine JJ, Scott CG, Sellers TA, Brandt KR, Serie DJ, Wu FF, Morton MJ, Schueler BA, Couch FJ, Olson JE, Pankratz VS, Vachon CM. A novel automated mammographic density measure and breast cancer risk. J Natl Cancer Inst. 2012;104:1028–1037. doi: 10.1093/jnci/djs254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Szekely L, Eriksson L, Heddson B, Sundbom A. High-throughput mammographic-density measurement: a tool for risk prediction of breast cancer. Breast Cancer Res. 2012;14:R114. doi: 10.1186/bcr3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vallières M, Freeman CR, Skamene SR, EI Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Physics in Medicine & Biology. 2015;60:5471–5496. doi: 10.1088/0031-9155/60/14/5471. [DOI] [PubMed] [Google Scholar]
- 28.Witten I, Frank E, Hall MA. Data Mining: Practical Machine Learning Tools and Techniques. Elsevier; New York: 2011. [Google Scholar]
- 29.Lederman D, Zheng B, Wang X, Wang XH, Gur D. Improving breast cancer risk stratification using resonance-frequency electrical impedance spectroscopy through fusion of multiple classifiers. Ann Biomed Eng. 2011;39:931–945. doi: 10.1007/s10439-010-0210-4. [DOI] [PubMed] [Google Scholar]
- 30.Bøvelstad HM, Nygård S, Størvold HL, Aldrin M, Borgan Ø, Frigessi A, Lingjaerde OC. Predicting survival from microarray data-a comparative study. Bioinformatics. 2007;23:2080–2087. doi: 10.1093/bioinformatics/btm305. [DOI] [PubMed] [Google Scholar]
- 31.Wessels LF, Reinders MJ, Hart AA, Veenman CJ, Dai H, He YD, van’t Veer LJ. A protocol for building and evaluating predictors of disease state based on microarray data. Bioinformatics. 2005;21:3755–3762. doi: 10.1093/bioinformatics/bti429. [DOI] [PubMed] [Google Scholar]
- 32.Fang Y, Fei J, Ma K. Model reference adaptive sliding mode control using RBF neural network for active power filter. Electrical Power and Energy Systems. 2015;73:249–258. [Google Scholar]
- 33.Liu D, Wang D, Wu J, Wang Y, Wang L. A risk assessment method based on RBF artificial neural network-cloud model for urban water hazard. Journal of Intelligent & Fuzzy Systems. 2014;27:2409–2416. [Google Scholar]
- 34.Li Q, Doi K. Reduction of bias and variance of evaluation of computer aided diagnostic schemes. Med Phys. 2006;33:868–875. doi: 10.1118/1.2179750. [DOI] [PubMed] [Google Scholar]
- 35.Scutt D, Manning JT, Whitehouse GH, Leinster SJ, Massey CP. The relationship between breast asymmetry, breast size and the occurrence of breast cancer. British Journal of Radiology. 1997;70:1017–1021. doi: 10.1259/bjr.70.838.9404205. [DOI] [PubMed] [Google Scholar]
- 36.Blanks RG, Wallis MG, Given-Wilson RM. Observer variability in cancer detection during routine repeat (incident) mammographic screening in a study of two versus one view mammography. Journal of Medical Screening. 1999;6:152–158. doi: 10.1136/jms.6.3.152. [DOI] [PubMed] [Google Scholar]
- 37.Stuedal A, Ma H, Bernstein L, Pike MC, Ursin G. Does breast size modify the association between mammographic density and breast cancer risk? Cancer Epidemiol Biomarkers Prev. 2008;17(3):621–627. doi: 10.1158/1055-9965.EPI-07-2554. [DOI] [PubMed] [Google Scholar]


