Abstract
The anther cuticle and pollen wall function as physical barriers that protect genetic material from various environmental stresses. The anther cuticle is composed of wax and cutin, the pollen wall includes exine and intine, and the components of the outer exine are collectively called sporopollenin. Other than cuticle wax, cutin and sporopollenin are biopolymers compounds. The precise constituents and developmental mechanism of these biopolymeric are poorly understood. Here, we reported a complete male sterile mutant, male sterile6021, in maize. The mutant displayed a smooth anther surface and irregular pollen wall formation before anthesis, and its tapetum was degraded immaturely. Gas chromatography-mass spectrometry analysis revealed a severe reduction of lipid derivatives in the mutant anther. We cloned the gene by map based cloning. It encoded a fatty acyl carrier protein reductase that was localized in plastids. Expression analysis indicated that MS6021 was mainly expressed in the tapetum and microspore after the microspore was released from the tetrad. Functional complementation of the orthologous Arabidopsis mutant demonstrated that MS6021 is conserved between monocots and dicots and potentially even in flowering plants. MS6021 plays a conserved, essential role in the successful development of anther cuticle and pollen exine in maize.
Introduction
In flowering plants, male reproductive development is essential for metagenesis and genetic recombination, which is also a complex process in which cooperative interactions occur between sporophytic and gametophytic tissues1,2. After anther morphogenesis, each anther locule includes centrally localized pollen mother cells (PMC) surrounded by four somatic layers, from the surface to the interior: the epidermis (E), endothecium (En), middle layer (ML), and tapetum (T)2–4. As a secretory cell layer, the tapetum provides abundant ingredients for the anther cuticle and pollen outer wall5,6. These two rigid barriers protect the genetic material in microspores or pollen grains from various biotic and abiotic stresses7,8.
The anther cuticle is located outside of the epidermis. It seals plant anther against the environment. As the skin of the anther, the cuticle is mainly composed of cutin and cuticle wax. Cuticle wax impregnates or covers cutin9–11. Hydrophobic cutin is a polymer of hydroxylated and epoxylated fatty acids and their derivatives with chain lengths of C16 and C1812. Cuticle wax is composed of very long-chain fatty acids (VLCFA), alkanes, alkene, and fatty alcohols, among others9. The pollen wall is a multilayer, robust structure surrounding the pollen cytoplasm. The outer layer, called the exine, is principally composed of sporopollenin, highly resistant biopolymers derived from fatty acids, phenylpropanoids, and phenolic13. Although sporopollenin is commonly present in pollen grains and spores14, the fine structure of the exine is varies among species15. The durability of the exine combined with its species-specific structure enable its application in paleontological and forensic analyses16. However, the understanding of the biochemical components and biosynthesis of the exine remains largely elusive due to its high insolubility and chemical resistance.
Recent genetic and biochemical investigations of the development of Arabidopsis and rice anthers have greatly facilitated our understanding of the synthesis regulation of aliphatic biopolymers, such as anther cuticle and sporopollenin17. Arabidopsis MALE STERILITY 2 (MS2)18 and rice DEFECTIVE POLLEN WALL (DPW)19 in plastids catalyze the reduction of fatty acyl-ACP to fatty alcohols. CYP703A2 and CYP703A3 20 function as lauric acid hydrolxylase21. CYP704B1 22 and CYP704B2 7 catalyze the ω-hydroxylation of fatty acid. Both CYP703As and CYP704Bs belong to the ancient and conserved P450 gene family23. ACYL COENZYME A SYNTHETASE 5 24, two POLYKETIDE SYNTHESES, PKSA/LAP6 and PKSB/LAP5 25,26, TETRAKETIDE α-PYRONE REDUCTASE 1 (TKPR1)27,28 are proposed to function together in the synthesis of hydroxylated tetraketide α-pyrones, which are polyketides that may form the major constituent of sporopollenin29. All the above mentioned genes related to lipid-soluble precursor synthesis are predominantly expressed in tapetal cells. After the biosynthesis steps, these precursors must be secreted from the tapetum and transferred to the outside surface of microspores and anther wall surfaces to be polymerized into biopolymers of sporopollenin, and cutin30 respectively. According to recent investigations, ATP-binding cassette (ABC), lipid transfer protein (LTP), and multidrug and toxic efflux (MATE) protein may be responsible for the transport of biopolymer precursors23. OsABCG15 is believed to transfer lipid monomers for anther cuticle and exine development31, while its ortholog, AtABCG26, transports both lipid precursors and polyketides for exine formation29,32. OsC6 encodes a lipid transfer protein. It is speculated to transfer lipidic molecules from tapetal cells to other anther cells and pollen wall surfaces because the mutant displays both defective cuticle and exine development33.
Maize is one of the most important crops worldwide. Many male sterile mutants have been collected at the stock center of maize MaizeGDB (http://www.maizegdb.org/data_center/phenotype?id=24992), but nevertheless, only four genes involved in pollen exine development have been reported. MALE STERILE26 (MS26) encodes a P450 family protein, which is orthologous to CYP704B1 in Arabidopsis 34. MS45 encodes a strictosidin synthase, which serves as a vital component in seed production technology35. IRREGULAR POLLEN EXINE1 encodes a putative glucose-methanol-choline oxidoreductase36. ABNORMAL POLLEN VACUOLATION1 encodes another P450 family protein that functions in the fatty acid hydroxylation pathway37. Here, we report a complete maize male sterile mutant male sterile 6021 (ms6021), which shows defective anther cuticle and exine development. We isolated the monofactorial recessive, nuclear male sterile gene using map-based cloning. The expression pattern analysis showed that MS6021 was specifically expressed in the tapetum and microspore after meiosis, and MS6021 was mainly localized to the plastid via the N-terminal transit peptide. MS6021 could functionally complement the Arabidopsis ms2 mutant, indicating that MS6021 was the putative maize ortholog of MS2 and may also function as a fatty acyl-ACP reductase. This work would improve our understanding of anther cuticle and exine development in maize.
Results
Phenotypic and genetic analysis of the ms6021 mutant
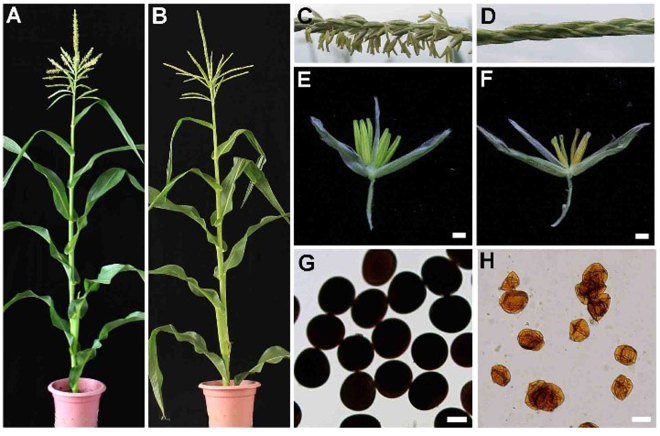
To identify maize genes that contribute to anther development, we requested a series of male sterile mutants from the stock at maizeGDB. Among these materials, ms6021 displayed normal female development but smaller anthers before flowering (Fig. 1D and F) and complete male sterility compared with the wild type (Fig. 1A,C and E). The phenotype was identical to the phenotypic description from MaizeGDB. It was first reported by Patterson E. B. in 1995. I2-KI pollen staining revealed an absence of mature pollen in ms6021 (Fig. 1H) compared with wild-type anthers (Fig. 1G).
Figure 1.
Phenotypic comparison between wild-type and the ms6021 mutant. (A,B) Wild-type (A) and ms6021 mutant (B) plants at the flowering stage. (C,D) Branches of wild-type (C) and the ms6021 mutant (D) at the flowering stage. (E,F) Spikelet of wild-type (E) and the ms6021 mutant (F) before pollen loss. (G,H) Pollen grains of wild type (G) and the ms6021 mutant (H) stained with a 1% I2-KI solution at the flowering stage. Bars = 1 mm in (E,F) and 50 µm in (G,H).
When the ms6021 plants were pollinated with wild-type (B73) pollen, all the F1 progeny displayed normal male fertility, indicating that ms6021 was a recessive mutant. The BC1F1 population was developed by crossing ms6021 mutant plants with the F1 plants. BC1F1 fertility testing showed a segregation of 76 normal and 79 mutant plants (χ2 = 0.03, P > 0.05), indicating a monofactorial recessive characteristic of ms6021. According to the information provied by maizeGDB, ms6022 (928P), ms6046 (928S) and ms6047 (928T) are allelic to ms6021 (928O). Our allelic testing confirmed the allelic relationship (Supplementary Table S1).
Defects of the ms6021 anther development
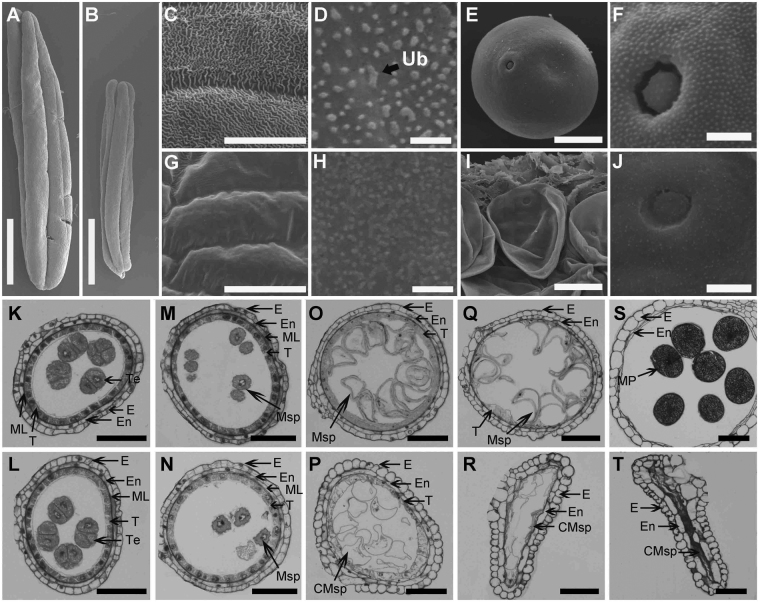
To investigate the detailed differences between the ms6021 mutant and wild type, we used scanning electron microscopy to examine the anther and pollen surfaces at different anther development stages. The anthers of ms6021 (Fig. 2B) were much smaller than those of wild-type (Fig. 2A) at the mature pollen stage. A three-dimensional reticular structure formed on wild-type epidermal cells (Fig. 2C), while the anther surface of ms6021 was glossy and smooth (Fig. 2G). At the uninuclear stage of wild type, a large number of granular Ubisch bodies were secreted from the tapetum on the inner locule surface (Fig. 2D). These sporopollenin precursor carriers would be accumulated outside of microspores to form a particulate exine pattern (Fig. 2F), and this process was critical for mature pollen formation (Fig. 2E). At the same stage, much smaller spot-like Ubisch bodies were observed on the inner side of the ms6021 tapetum (Fig. 2H), indicating unregular aliphatic component transportation to the microspore surface of the mutant. The pollen granule shrunk severely and adhered to the inside of the ms6021 anther wall (Fig. 2I), and its pollen surface was also smoother than that of wild type (Fig. 2J).
Figure 2.
Defective development of the ms6021 anther surface and pollen wall. (A,B) Anthers of wild-type (A) and ms6021 (B) at the mature pollen stage. (C,G) SEM analysis of the anther surface of wild-type (C) and ms6021 (G) at the mature pollen stage. (D,H) SEM analysis of the inner surface of wild-type (D) and ms6021 (H) at the mature pollen stage. (E,F,I,J) SEM analysis of the pollen grain (E,I) and pollen surface (F,J) of wild type (E,F) and (I,J) at the mature pollen stage. (K) to (T) Cytological comparison of anther development in wild type and ms6021 at different stages. The anthers of wild type are shown in (K,M,O,Q and S); and of the ms6021 mutant are shown in (L,N,P,R and T) tetrad stage (K,L); uninucleate stage (M,N); large vacuole stage (O,P); binucleate stage (Q,R); mature pollen stage (S,T). CMsp, collapsed microspore; E, epidermis; En, endothecium; ML, middle layer; MP, mature pollen; Msp, microspore; T, tapetum; Te, tetrad; Ub, ubisch body. Bars = 1 mm in (A,B), 20 µm in (C) to (E) and (G) to (I), 5 µm in (F,J), and 50 µm in (K) to (T).
Next, we performed a morphological analysis to identify anther developmental defects in the ms6021 mutant. Light microscopy was used to examine transverse sections of wild-type and ms6021 anthers. The tetrads formed normally in the locules of both wild type (Fig. 2K) and the ms6021 mutant (Fig. 2L). The callus surrounding the tetrads was then digested, and the microspores were easily released (Fig. 2M and N) as previously reported38. During the large vacuole stage, exine assembly was completed. The microspores rapidly inflated. Tapetal cells were squeezed, and the middle layer was almost invisible (Fig. 2O). By contrast, in the ms6021 mutant anther, a tenuous exine was formed around the irregular microspores. The tapetal cells were swollen, and the cellular outline of middle layer were still visible (Fig. 2P). The microspores then entered the trinucleate stage through two cycles of mitotic divisions in wild type. The tapetum nearly disappeared at this stage (Fig. 2Q). Mature pollen grains were formed immediately before flowering in the wild-type anther (Fig. 2S). In contrast, the ms6021 anther started to shrink (Fig. 2R) and developed into a rectangular structure (Fig. 2T).
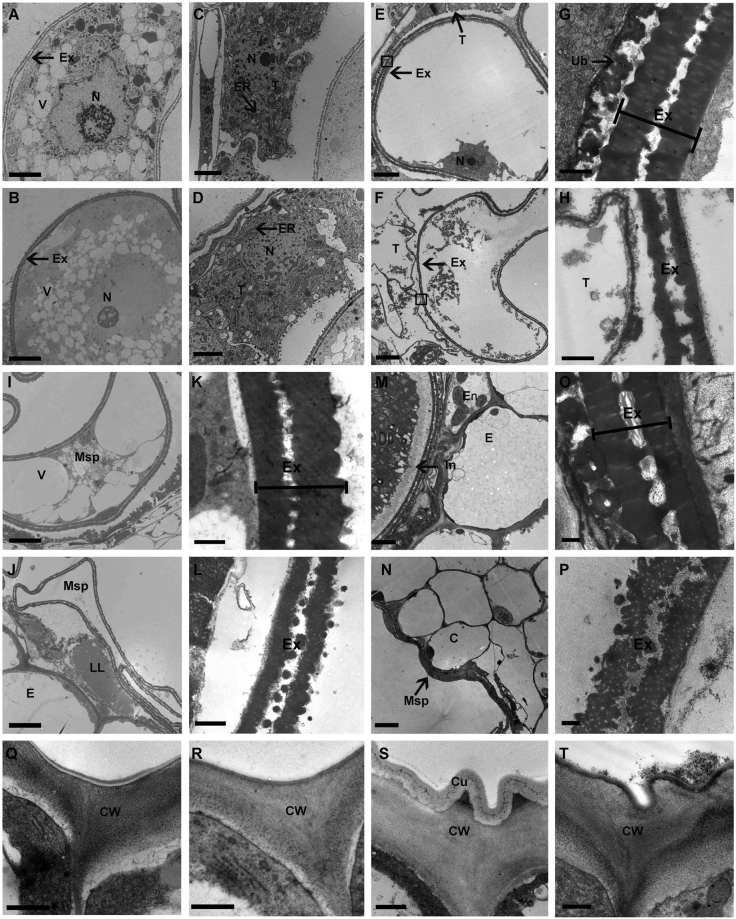
To gain more detailed insight into the defective anther development of ms6021, transmission electron microscopy (TEM) was performed. In accordance with the light microscopy results, the abnormality did not appear until the uninucleate stage. In both wild-type and ms6021 anthers, multiple small vacuoles were observed in the microspores, and the structural primexine of the microspores was clearly observed (Fig. 3A and B). The heavily stained tapetal cells revealed a vigorous metabolism (Fig. 3C and D). Subsequently, the small vacuoles merged into a central large vacuole, and the tapetum was squeezed together to form a hill-like structure (Fig. 3E) in the wild-type anther. Moreover, a large amount of Ubisch bodies were secreted out of the tapetum, which transported the sporopollenin precursor to the outside surface of the microspore. The exine rapidly thickened (Fig. 3G). By contrast, both the microspore and tapetum collapsed and the cell contour was remained in the ms6021 anther (Fig. 3F) at the vacuolated stage. In addition, we did not observe any normal Ubisch bodies in the ms6021 locule, and the exine was much thinner than that in wild type (Fig. 3H). The microspore underwent one round of mitosis and entered the binucleate stage in wild type. The large vacuole was divided, and the microspore shrunk. Programmed cell death (PCD) was launched in the tapetum (Fig. 3I) and exine morphogenesis ended (Fig. 3K). By comparison, the microspore contents of ms6021 completely disappeared. Abnormal liposome-like structures were formed in the tapetal cell (Fig. 3J). Normal exine thickening did not occur in the mutant anther (Fig. 3L). During the trinucleate stage of wild type, the tapetum and middle layer were invisible, and only the endothecium and epidermis remained in the anther wall. The intine, which was composed of polysaccharides, was accumulated inside the exine (Fig. 3M and O). The tapetum did not disappear completely in the ms6021 anther compared with wild type. The microspores further shrunk and adhered tightly to the anther wall (Fig. 3N). The intine did not form in the mutant (Fig. 3P). We also studied the anther cuticle development process. At the uninucleate stage, there was no cuticle structure outside of the epidermal cells of either wild type or the ms6021 mutant (Fig. 3Q and R). Before flowering, a hair-like cuticle formed on the surface of the wild-type epidermis (Fig. 3S). The outmost layer of ms6021 remained glossy (Fig. 3T), which was consistent with the SEM results (Fig. 2D). An identical cytological analysis was performed using the ms6047 mutant. Interestingly, the anther developmental process in this mutant was more completely disrupted compared with the ms6021 mutant (Supplementary Figure S1).
Figure 3.
TEM analysis of anthers from wild-type and ms6021. (A) to (D) Microspore (A,B) and tapetum (C,D) of anthers from wild type (A,C) and the mutant (B,D) during the uninucleate stage. (E) to (H) Microspore (E,F) and pollen exine (G,H) of anther from wild-type (E,G) and mutant (F,H) at large vacuole stage. (I) to (L) Microspore (I,J) and pollen exine (K,L) of anthers from wild type (I,K) and the mutant (J,L) at the binucleate stage. (M) to (P) Anther wall (M,N) and pollen exine (O,P) of anthers from wild type (M,O) and the mutant (N,P) at the mature pollen stage. (Q) to (T) Anther epidermal surface of wild type (Q,S) and the ms6021 mutant (R,T) at the uninucleate stage (Q,R) and mature pollen stage (S,T). C, cavity for dehiscence. Cu, cuticle; CW, cell wall; ER, endoplasmic reticulum; Ex, exine; In, intine; LL, lipidosome-like; Msp, microspore; N, nucleus; T tapetum; Ub, ubisch body; V, vacuole. G and H were zoomed form black solid boxes region in E and F respectively. Bars = 5 µm in (A,B,E,F,I,J,M and N), 2 µm in (C,D), and 0.5 µm in (G,H,K,L,O and P) and (Q) to (T).
Aliphatic alteration of ms6021 anther
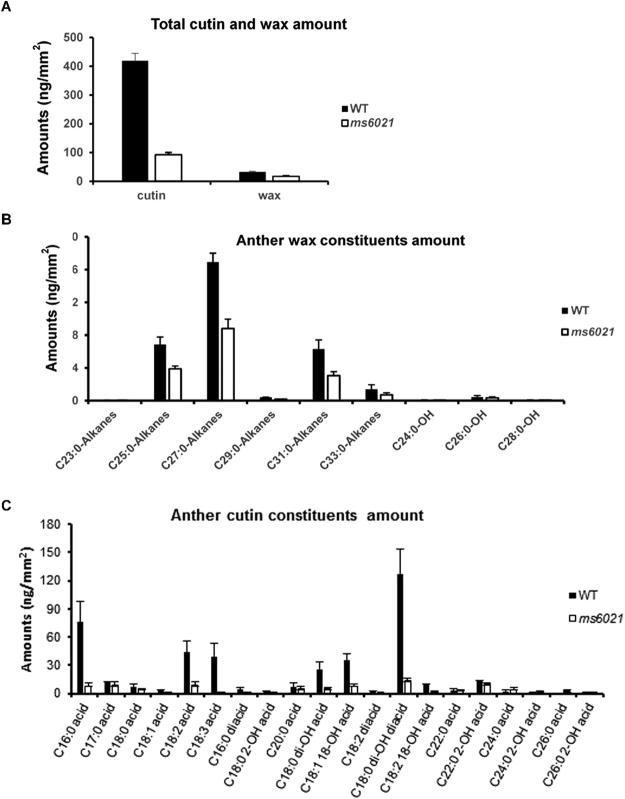
The smooth anther surface, abnormal Ubisch bodies, and defective pollen exine structure indicated a disruption of the accumulation of aliphatic components in the ms6021 anther. Next, we extracted cuticle wax, cutin and soluble fatty acid (SFA) from wild-type and ms6021 anthers step-by-step, and analyzed the composition by GC-MS39,40. The methods described by Li7 was used to plot surface area against fresh weight of corresponding samples (Supplementary Figure S2). The total wax was 32.58 ng/mm2 and 17.19 ng/mm2 in wild-type and ms6021 anthers, respectively, representing a 47.23% decrease in the mutant anther (Fig. 4A). C23, C25, C27, C29, C31, and C33 alkenes and C24 and C26 alcohols were significantly decreased in the mutant (P < 0.01) (Fig. 4B; Supplementary Table S2). The total amount of cutin in ms6021 anthers (92.93 ng/mm2) decreased by 78% compared with that of wild type (419.48 ng/mm2, P < 0.01; Fig. 4A). All monomer compositions less than 24 carbons were significantly reduced in the mutant anthers (Fig. 4C; Supplementary Table S3). The amount of total soluble fatty acids with carbon lengths ranging from 16 to 28 was 20.05 µg/mg in wild-type anthers. By contrast, in the mutant anthers, the total amount of soluble fatty acids was reduced to 2.81 µg/mg (Table 1).
Figure 4.
Analysis of anther wax and cutin monomers in wild type and the ms6021 mutant. (A) Total cutin and wax amounts per unit area (ng/mm2) in wild-type (black bars) and ms6021 (white bars) anthers. Error bars indicate the SD (n = 3). (B) Wax amounts per unit (ng/mm2) in wild type (black bars) and ms6021 (white bars) anthers. Error bars indicate the SD (n = 3). (C) Cutin amounts per unit (ng/mm2) in wild-type (black bars) and ms6021 (white bars) anthers. Error bars indicate the SD (n = 3).
Table 1.
Total soluble fatty acids of wild-type and ms6021 anthers.
| Soluble fatty acids | Wide Type ms6021 | Down | |
|---|---|---|---|
| Mean ± SD (ng/mg dry weight) | |||
| C16:1 acid | 0.001 ± 0.0004 | 0 | 100% |
| C16:0 acid | 8.838 ± 1.759 | 0.479 ± 0.086 | 94.58% |
| C18:2 acid | 4.548 ± 0.551 | 0.596 ± 0.114 | 86.90% |
| C18:3 acid | 5.241 ± 1.229 | 0.082 ± 0.015 | 98.43% |
| C18:1 acid | 0.458 ± 0.042 | 0 | 100% |
| C18:0 acid | 0.312 ± 0.036 | 0.283 ± 0.039 | 9.11% |
| C20:0 acid | 0.217 ± 0.019 | 0.381 ± 0.035 | −75.25% |
| C22:0 acid | 0.057 ± 0.010 | 0.158 ± 0.018 | −175.43% |
| C24:0 acid | 0.056 ± 0.008 | 0.227 ± 0.046 | −307.69% |
| C26:0 acid | 0.021 ± 0.003 | 0.113 ± 0.028 | −431.06% |
| C28:0 acid | 0.102 ± 0.014 | 0.301 ± 0.129 | −193.75% |
| Total | 20.055 ± 3.404 | 2.804 ± 0.417 | 86.03% |
Isolation of MS6021
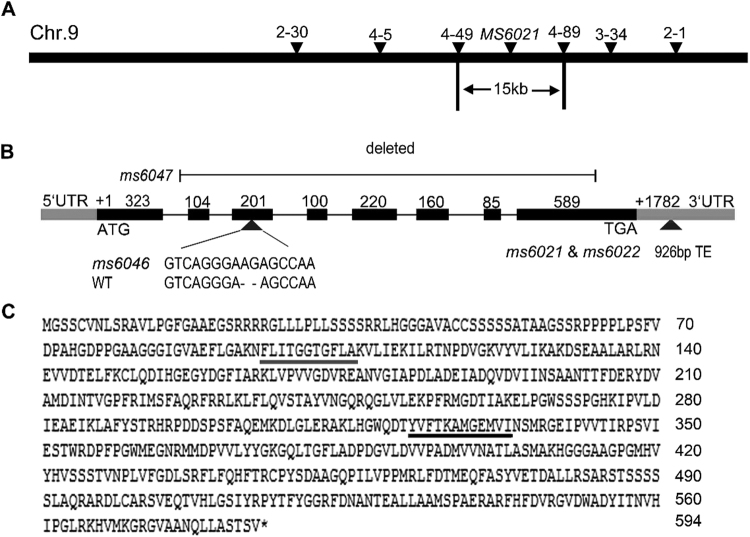
Map-based cloning was used to isolate MS6021. The gene was initially mapped to a 5.93-Mb interval on chromosome 9 between markers 2–30 and 2-1 (Fig. 5A). Then 998 individuals from the BC1F1 population were used for fine mapping. MS6021 was mapped to a 15-kb region based on the B73 reference genome. There were two complete open reading frames in this region (GRMZM2G420926 & GRMZM2G120987). The GRMZM2G420926 genomic sequence was identical between wild type and ms6021. There was a 926-bp insertion in the 3′ untranslated region (UTR) of GRMZM2G120987 in the ms6021 genome. The GRMZM2G120987 genomic sequences of the three other alleles were then analyzed. The sequencing results revealed that the genomic sequence of ms6022 was identical to that of ms6021, two residues were inserted into the third exon in ms6046 resulting in a frame shift, and a 2105-bp region from the first intron to the last exon was deleted in ms6047 (Fig. 5B). The above results indicate that variations of the GRMZM2G120987 sequence are responsible for the phenotypic defects in the mutant anther. The transcribed region of GRMZM2G120987 includes a 279-bp 5′ UTR, a 482-bp 3′ UTR, and a 1782-bp ORF encoding 593 amino acids (Fig. 5B).
Figure 5.
Molecular cloning and sequence analysis of MS6021. (A) Fine mapping of the MS6021 on chromosome 9. The location and name of the makers are indicated. The MS6021 locus was mapped to a 15-kb region between markers 4–49 and 4–89. (B) A schematic representation of the exon and intron structure of MS6021. The black boxes indicate exons, grey boxes indicate UTR region, and intervening lines indicate introns. +1 indicates the start codon (ATG); +1782 indicates the stop codon (TAA). A 926-bp fragment is inserted into the 3′UTR in ms6021 and ms6022; 2-bp is inserted into the third exon in ms6046; a 2142-bp fragment from the first intron to the last exon is deleted in ms6047. (C) The amino acid sequence of MS6021. The grey and black lines underlining indicated putative NAD-binding region and active region respectively.
MS6021 is mainly expressed in the tapetum and microspore
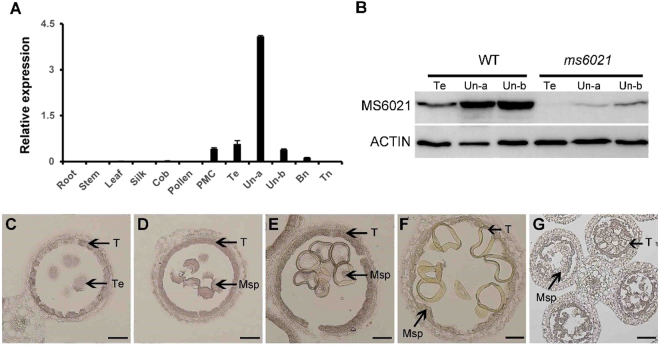
To understand the expression pattern of MS6021, qRT-PCR analysis was performed using total RNA isolated from both vegetative and reproductive organs. The results indicated that MS6021 was not expressed in vegetative and female reproductive organs. Only trace amount of MS6021 was detected in the pollen mother cell (PMC) stage and tetrad stage, peaking during the early nucleate stage and then rapidly declining (Fig. 6A). Its expression level significantly decreased in the mutant during the uninucleate stage (Supplementary Figure S3). In situ hybridization was performed to confirm the spatial and temporal expression pattern of MS6021 in wild-type anther sections. The MS6021 transcript could be detected in both microspores and tapetum from the tetrad to the binucleate stage (Fig. 6C–F); high-level expression was detected at uninucleate stage (Fig. 6D), while only background signal was detected using the sense probe during the same stage (Fig. 6G), which was consistent with the qRT-PCR results. Moreover, we produced polyclonal antibodies to perform the western blot analysis. Trace amounts of MS6021 were detected in the anthers of the ms6021 mutant (Fig. 6B), which may underlie the phenotypic differences between ms6021 and ms6047 (Fig. 2 and Supplementary Figure S1).
Figure 6.
Expression pattern of MS6021. (A) qRT- PCR analysis of MS6021 expression. Error bars indicate the SD (n = 3). (B) Protein gel blot analysis. Full-length gels are presented in Supplementary Figure S4. Te, tetrad stage; Un-a, early uninucleate stage; Un-b, late uninucleate stage. (C) to (F) RNA in situ hybridization using an anti-sense probe during the tetrad stage (C), early uninucleate stage (D), large vacuole stage (E), and binucleate stage (F). (G) RNA in situ hybridization using a sense probe during the early uninucleate stage. Msp, microspore; T, tapetum. Bars = 50 µm in (C) to (F) and 100 µm in (G).
MS6021 is localized to plastids
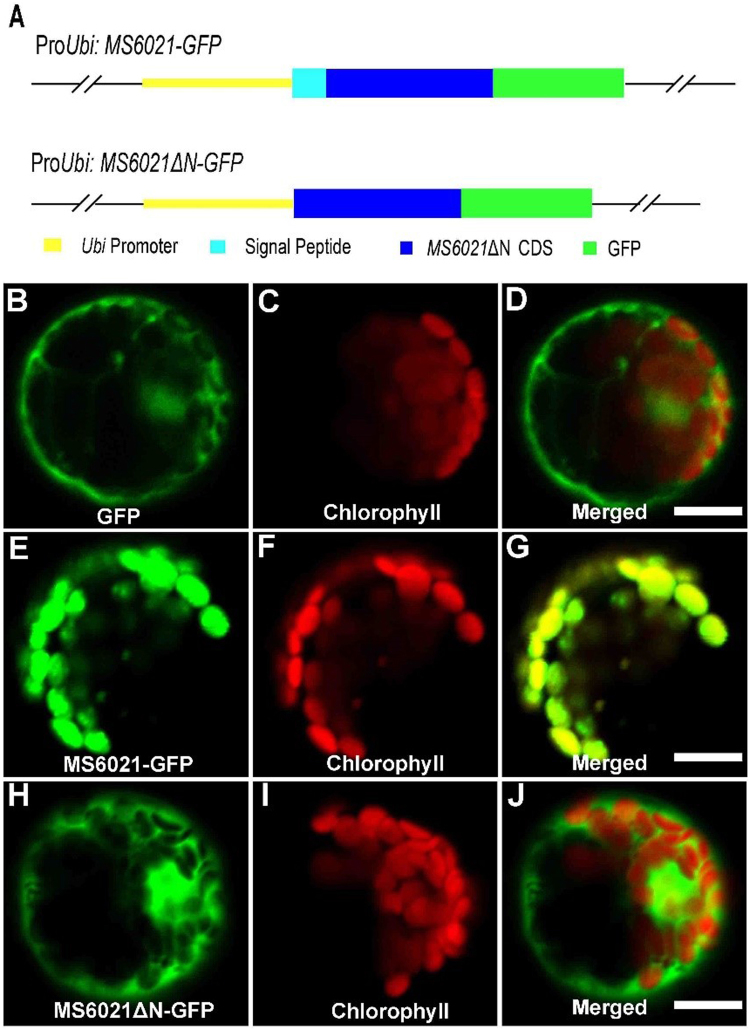
The targetP 1.1 server (http://www.cbs.dtu.dk/services/TargetP/) was used to analyze the amino acid sequence of MS6021. There was a chloroplast signal peptide predicted at the N-terminal of MS6021. To identify the subcellular localization of MS6021, we constructed plasmids containing MS6021-GFP, MS6021ΔN-GFP (the predicted signal peptide was deleted), or GFP only driven by the maize ubiquitin promoter (Fig. 7A). These plasmids were then introduced into protoplasts isolated from young maize leaves. The plastid signal was detected by chlorophyll autofluorescence. The MS6021-GFP signal co-localized with the autofluorescence of chlorophyll in the plastid (Fig. 7E–G). In contrast, the GFP alone signal (Fig. 7B–D) and the MS6021ΔN-GFP (Fig. 7H–J) signal were observed in the cytoplasm and did not co-localize with the autofluorescence of chlorophyll. This result revealed that MS6021 was localized in plastids mediated by N-terminal signal peptide.
Figure 7.
Subcellular localization analysis of MS6021. (A) Diagram of the full-length constructs of MS6021 cDNA and signal region deleted cDNA fused to GFP under the control of the maize ubiquitin promoter. (B) to (D) A maize protoplast expressing empty pJIT163-GFP showing green fluorescence (B), chlorophyll autofluorescence (C), and the merged signals (D) of (B) and (C). (E) to (G) A maize protoplast expressing fused MS6021-GFP showing green fluorescence (E), chlorophyll autofluorescence (F), and the merged signals (G) of (B) and (C). (H) to (J) A maize protoplast expressing empty fused MS6021ΔN-GFP showing green fluorescence (H), chlorophyll autofluorescence (I), and the merged signals (J) of (H) and (I). Bars = 10 µm.
Functional conservation of MS6021
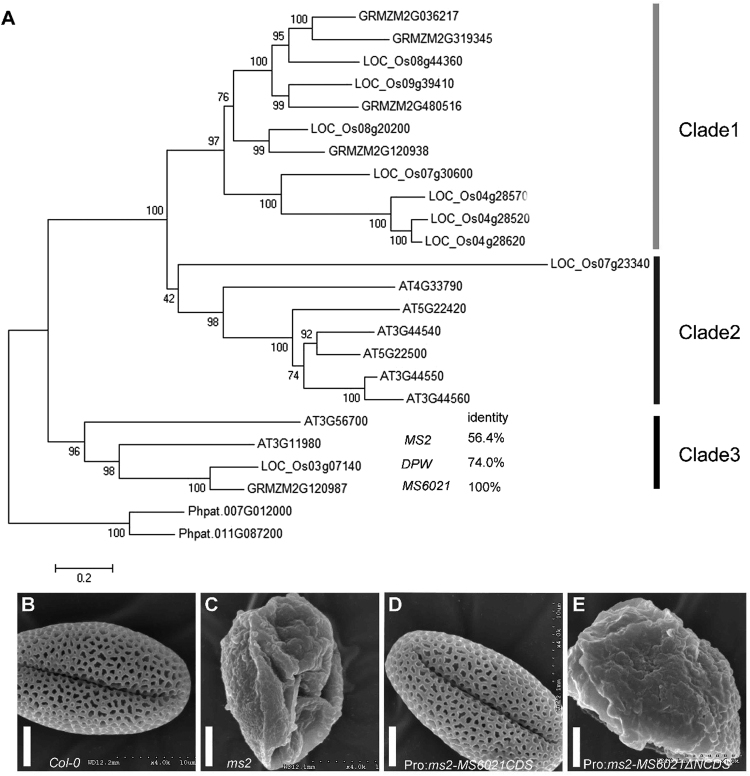
According to the phytozome11 information, MS6021 contained a NAD-binding domain and a sterile domain (Fig. 5C), and it is considered to be a member of the fatty acyl-CoA reductase family (FAR). The FAR is a cluster of reductases that catalyze the transformation of fatty acyl-CoA/ACP to the corresponding alcohol and comprises 5, 9 and 8 members in maize, rice and Arabidopsis, respectively. We compared the amino acid sequences of these 22 homologous genes (Supplementary Figure S5). A neighbor-jointing (NJ) tree was subsequently constructed with orthologous genes from Physcomitrella patens as an outgroup (Fig. 8A). The NJ tree was grouped into three clades. Clade 1 was specific to monocot, and all the genes in this clade were from maize or rice. In clade 2, all the genes were from Arabidopsis except LOC_Os07g23340 from rice. Clade 3 was composed of AT3G56700 and three male sterile genes, namely MS2, DPW and MS6021. AT3G56700 was also a plastid-localized protein that was specifically expressed in anthers. It may be functionally redundant with MS2, and it is responsible for partial fertility in the Arabidopsis ms2 mutant16,41.
Figure 8.
Functional conservation of MS6021. (A) A neighbor-joining phylogenetic tree summarizing the evolutionary relationships among FAR members in Arabidopsis, rice, and maize of (BLASTPE E-value < 1E-100). The proteins are named according to their Phytozome accession numbers. The numbers under the branches refer to the bootstrap value of the neighbor-joining phylogenetic tree. The length of the branches is proportional to the amino acid variation rates. At, Arabidopsis thaliana; Os, Oryza sativa; Zm, Zea mays. The bar indicates the estimated number of amino acid substitutions per site (for the protein alignment, see Figure S4). (B) to (E) SEM analysis of the pollen wall surface of wild-type (B), ms2 mutant (C), and transgenic ms2 mutant lines (D and E) at anthesis. Bars = 5 µm.
The phylogenetic analysis indicated that MS6021, MS2 from Arabidopsis, and DPW from rice are orthologs, and ms2 18 and dpw 19 mutants display a similar phenotype to ms6021. However, MS6021 only shares 56.4% identity with MS2 (Supplementary Figure S5). To identify the evolutionary relatedness of these two genes, a complementation experiment was performed with the MS6021 complete CDS region and the CDS region lacking the signal peptide driven by the MS2 native promoter. The two constructs were used to transform the Arabidopsis ms2 mutant. Five positive lines per transformation were selected to assess the pollen morphology by SEM. The results showed that the CDS of MS6021 was able to rescue the phenotype of ms2, whereas the CDS lacking the signal peptide failed to rescue the ms2 mutant (Fig. 8B–E). These results revealed the functional conservation of MS2/MS6021 between Arabidopsis and maize.
The ms6021 mutant exhibited abnormal expression of genes involved in aliphatic metabolism
To better understand the defects in ms6021, RNA-seq was performed with total RNA isolated from uninucleate-stage anthers of wild type and ms6021 using three biological repeats. High-throughput sequencing was performed with the Illumina Hiseq2500 platform. More than 30 million reads were generated for each sample. After filtration of the data, the clean reads were aligned to the maize genome references42. There were 23273 genes transcripts detected in wild type and ms6021 mutant. We identified 1163 differentially expressed genes (DEGs) with a threshold fold change greater than 2 and a false discovery rate less than 0.05. Among the DEGs, 594 genes were up-regulated and 569 genes were down-regulated (Supplementary Figure S6A). The RNA-seq results were confirmed by qRT-PCR (Supplementary Figure S7). The GO analysis revealed that multiple processes related to pollen maturation were impacted in ms6021 (Supplementary Figure S6B), including pollen wall assembly (GO:0010208), NADP metabolic processes (GO:0006739), oligosaccharide metabolic processes (GO:0009311), secondary metabolic processes (GO:0019748) and extracellular matrix organization (GO:0030198). Plant immune system-related processes, such as the response to stress (GO:0006950) and positive regulation of defense responses (GO:0031349), were also simulated in ms6021.
It is well known that sporopollenin is composed of fatty acids and phenolic43. The precise components of phenolic are largely unknown. They are the main products of phenylalanine metabolism, including flavonoids, stilbenes, coumarins and lignin25. The KEGG pathway analysis indicated that both the metabolisms of fatty acids and phenolic were affected in ms6021 (Supplementary Table S4). More genes involved in phenylalanine biosynthesis and flavonoid biosynthesis showed altered expression patterns in comparison to the genes involved in fatty acid metabolism, such as fatty acid elongation, fatty acid biosynthesis and fatty acid unsaturation in the mutant. In addition, the expression of ABC transporter proteins, which function as sporopollonin precursor transporters, was also affected. Taken together, these findings indicated the severely disrupted metabolism of fatty acid-derived components in the ms6021 anthers.
Discussion
Both the cuticle and pollen wall have a strategic position at the interface between the plant and environment. They must protect genetic material from various stresses. Deciphering the chemical composition of these protective tissues has encountered enormous challenges due to their extreme resistance to degradation and sophisticated fine structures. Recently, several male sterile mutants associated with defective anther cuticle or/and pollen wall development have been identified using genetic approaches, including acos5 24, cyp703a2 21, cyp704b1 22, drl1 27, lap5/6 25, ms2 18, in Arabidopsis, and cyp703a3 20, cyp704b2 7, dpw 19, osabcg15 44, osabcg26 45, tdr 6, wda1 10 in rice. In addition, some reviews have summarized the regulation network underlying cuticle and pollen wall development of these model plants4,8,23. However, only four genes, MS26 34, MS45, IPE1 36, and APV1 37, have been reported to participate in cuticle or/and pollen wall development in maize. All the four cloned genes encoded enzymes involved in aliphatic metabolism, while their exact biochemical function and substrate relationships are largely unknown.
We reported here the male sterile mutant ms6021 in maize, which also displayed defective cuticle and pollen wall development. The epidermal anther surface of ms6021 was smooth and glossy (Fig. 2G), and the TEM results showed that the cuticle layer had largely disappeared (Fig. 3T). The total amount of wax and cutin decreased by 47% and 78%, respectively (Fig. 4A; Supplementary Tables S2 and S3). The wax compositions of the mutant showed a moderate (<50%) decrease in both fatty alcohols and alkanes (Supplementary Table S2). By contrast, most of the cutin compositions decreased by more than 75% in the mutant anthers (Supplementary Table S3). This result indicated that the products of MS6021 most likely served as dominant precursors for cutin biosynthesis, not the dominant substrates for fatty acid elongation pathway. The conclusion was in accordance with KEGG analysis result that few DEGs were enriched in fatty acid elongation pathway. Metabolism analysis also showed that the proportion of total soluble fatty acids decreased to 13.97% (Table 1). The detailed constituent variation revealed a drastic reduction (up to 100%) in C16 - C18 fatty acids and a sharp increase (2-5-fold) in VLCFAs (>C20; Table 1). The results were consistent with the putative palmitic acyl ACP reductase function of MS6021 and further confirmed that the product of MS6021 was not the dominant substrate for VLCFA biosynthesis.
Aborted pollen grains were observed in the anther locule of ms6021 (Fig. 2I). Ubisch bodies, sporopollenin trafficking vehicles between the tapetum and pollen wall, disappeared from the inner surface of the mutant (Figs. 2H and 3H). Insufficient material supplementation led to failed exine thickening in ms6021 (Fig. 3L and P) after microspores release from the tetrads. Based on chemical analysis, it was assumed that sporopollenin was composed of aliphatic derivatives and a mixture of phenolic43,46,47. Phenolic is a large class of secondary metabolites48, which are mainly the products of phenylpropanoid metabolism. Flavonoids belong to a subclass of phenolic compounds. They are important for plant fertility in maize49, petunia50, and tobacco51. Among the Arabidopsis genes required for sporopollenin formation, ACYL COENME A SYNTHETASE5 24, LAP5/6 25,26, and TETRAKETIDE α-PYRONE REDUCTASE 27,28 function in the synthesis of hydroxylated tetraketide α-pyrones. TRANSPARENT TESTA4 52 and 4-COUMARATE:COENZYME A LIGASE 53 encode enzymes that participate in flavonoid biosynthesis. Both polyketides and flavonoids are believed the major composition of sporopollenin28. KEGG analysis revealed that DEGs were enriched in phenylpropanoid biosynthesis, phenylalanine metabolism, and flavonoid-related derivative biosynthesis and metabolic pathways (Supplementary Table S4). The orthologs of the above-mentioned genes (GRMZM2G108894/LAP5, GRMZM2G380650/LAP6, GRMZM2G422750/TT4, GRMZM2G004683/TKPR1) were also included in these pathways. The expression of IPE1 36, a newly reported male sterile maize gene that participated in cutin and wax biosynthesis, was influenced in ms6021 anthers as well. The expression of genes involved in maize anther cuticle and sporopollenin biosynthesis in apv1, ipe1 and ms6021 were compared. Most genes displayed different expression change (Supplementary Table S5), which was in accordance with differences of lipidomic alteration.
In Arabidopsis and rice, the ATP-binding cassette transporter has been reported to be required for sporopollenin accumulation. It has been proposed to be responsible for sporopollenin trafficking out of the tapetum29,30,32,44,54. The altered expression of multiple predicted ABC transporter genes (Supplementary Table S4) indicated abnormal sporopollenin trafficking in the mutant. Although genetic approaches provide insights into sporopollenin biosynthesis and composition, the trafficking mechanism of sporopollenin from the tapetum to the exine remains poorly understood8,55.
The FAR activity among these orthologous genes of MS6021 was conserved between monocot and dicot plants for both DPW 19 and MS6021 could functionally complement ms2 of Arabidopsis (Fig. 8D). Nevertheless, their products may not participate in the identical metabolism pathway, because mutants in different species displayed subtle differences in male sterility. The pollen of ms2 in Arabidopsis presented partial sterility, while both dpw in rice and ms6021 in maize showed complete male sterility. Furthermore, the total wax content of ms6021 was observably declined, unlike that of dpw 19 (Fig. 4A).
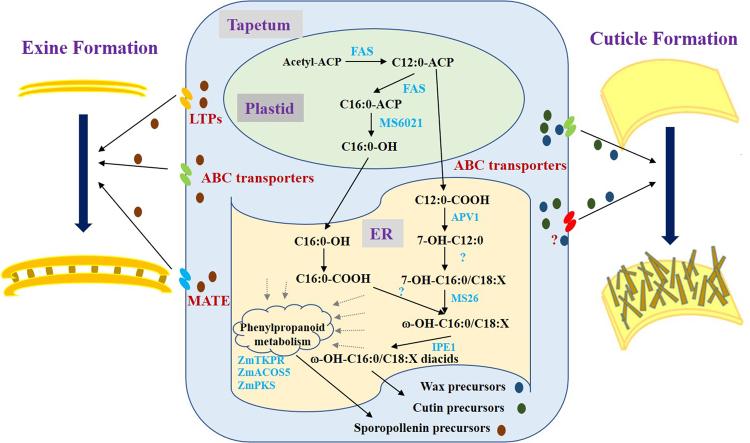
In conclusion, we proposed a working model of how MS6021 participated in maize anther cuticle and exine development (Fig. 9). MS6021 functioned as a fatty acid reductase in plastids in maize. It reduced the palmitoyl-ACP to corresponding alcohol, which would be transferred to the endoplasmic reticulum18,19. In the main organelle for fatty acid modification, the palmitoyl alcohol would influence phenylpropanoid biosynthesis, flavonoid biosynthesis, fatty acid enlongation and other fatty acid modification pathways directly or indirectly (Supplementary Table S4). These products would finally serve as precursors for cutin, wax and sporopollenin assembly8,13. The defect of MS6021 resulted in altered expression of massive genes and immature maize anther cuticle and pollen wall, while the regulation network underlying this remains unknown. More work needs to be done to decipher the complicated regulation pathway.
Figure 9.
The proposed model of MS6021 function during anther cuticle and exine development in maize. Fatty acids are de novo synthesized in the form of esterified ACP in plastids. The palmitoyl-ACP could be reduced to corresponding alcohol and then be transported from plastids to ER. Hexadecanol is converted to fatty acid19. The hydroxylated fatty acid would be oxygenated by P450 to form hydroxy fatty acid, which is further oxygenated by IPE143, or participate in VLFAs biosynthesis. The intermediate metabolites interplayed with phenylpropanoid metabolism, which functioned as supplier for sporopollenin synthesis8. Finally, the cutin, wax and sporopollenin precursors are translocated from the tapetal cells into the locule and anther epidermis by ATP binding cassette transporters29,30,32,44,54, lipid transport proteins33 and multidrug and toxic efflux transporters4.
Methods
Plant materials and growth conditions
ms6021, ms6022, ms6046, and ms6047 mutant lines were obtained from the Maize Genetics Cooperation Stock Center. ms6021 was used to generate the BC1F1 population with B73. All plants were cultivated in the experimental field of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences (IGDB, CAS) in Beijing and Hainan Province.
Phenotypic analysis of ms6021
For SEM analysis, fresh anthers from both wild type and the mutant at different stages were immersed in FAA solution (50% ethanol, 5% glacial acetic acid, 5% formalin) for 24 h at room temperature for fixation. The samples were then dehydrated in a serial of ethanol gradients (50–100%). After critical-point drying, the anthers were coated with palladium gold and then observed using a scanning electron microscope (HITACHI S-3400N).
For cytological observation, anthers were pricked and fixed in FAA solution overnight. The samples then were dehydrated using a serial of ethanol (50–100%) and embedded in spurr resin. Semi-thin sections were obtained using a Leica UE, stained with 0.05% toluidine blue and observed with an Olympus BX-53 microscope.
For TEM analysis, fresh anthers were vacuum infiltrated and prefixed in 3.5% glutaraldehyde (with 0.1 M phosphate buffer, pH7.4) followed by rinsing with 0.1 M phosphate buffer. The samples then were transferred into 1% osmium tetraoxide and rinsed with 0.1 M phosphate buffer. After fixation, the samples were dehydrated using an ethanol series from 50% to 100% and embedded in spurr resin. Ultra-thin sections were collected with a Leica EM-UC6. After double staining with uranyl acetate and lead citrate, images were obtained with a HITACHI H-7500 transmission electron microscope.
Aliphatic components analysis
To calculate the surface area, the anthers were considered as cylinders. The anther surface area was then plotted against the corresponding fresh weight7. Cuticle wax, cutin and total soluble fatty acid extraction and GC-MS analysis were performed as described previously19,43.
Isolation of MS6021
MS6021 has been reported to located on chromosome 9. For primary mapping, bulk segregation analysis (BSA) was used to identify pleomorphic markers associated to MS6021. The locus was first mapped between two simple sequence repeat (SSR) markers, 2–30 and 2-1, on the long arm of chromosome 9, at a position not far from telomere. Then, 998 individuals were used for fine mapping with pairs of primers (4–5, 4–49, 4–89, and 3–34). The primer sequences are listed in Supplementary Table 6.
RNA extraction and qRT-PCR
Total RNA was isolated from root, stem, leaf, silk, cob, pollen and different stages of anthers using the RNeasy Plant mini Kit (QIAGEN) as described by the manufacturer. The developmental stage of anthers was determined based on the semi-section morphology. One microgram of total RNA was used to synthesis cDNA using RevertAid First Strand CDNA Synthesis Kit (THERMO). qRT-PCR was performed on the Roche LightCycle480 system with SYBR Green Premix (TAKARA). All PCR reactions were conducted using 40 cycles at 98 °C for 10 s, 60 °C for 10 s, and 72 °C for 10 s, in a 20 µl reaction mixture containing 10 pmol of each primer and 2 µl of cDNA as template. All reactions were performed in triplicate, and ZmActin1 was used as the internal control for normalization. All primers used for qRT-RCR are listed in Supplementary Table 6.
In situ hybridization
Wild-type anthers at different developmental stages were fixed in FAA solution and then dehydrated in a gradient ethanol series (50%, 70%, 85%, 95% and 100%). After embedding in paraffin, 8 µm thick sections were obtained using a Leica microtome. To generate anti-sense and sense probes, a 750-bp cDNA fragment was produced by PCR amplification. RNA in-situ hybridization was performed according to a previously described protocol56.
Western blot analysis
Different developmental stages of anthers from wild type and the ms6021 mutant were grounded into a powder in liquid nitrogen. Total protein was isolated using Plant Protein Extraction Reagent (CW0885M) according to the manufacturer’s protocol (CWBIO) and quantified by the Bio-Rad protein assay. A small synthetic peptide (C-ESTWRDPFPGWMWNGNR-N) was generated to obtain the polyclonal antibody of MS6021 raised in rabbit. The small peptide and polyclonal antibody were produced by GL Biochem (Shanghai) Ltd. A protein blot was performed using the Super Signal West Femto Maximum Sensitivity Substrate Kit (THERMO).
Subcellular localization of MS6021
For the subcellular localization analysis, MS6021CDS and MS6021ΔNCDS (putative signal peptide-coding region deleted) were cloned into the pJIT-163-GFP plasmid. These constructs, as well as the empty plasmid, were introduced into maize protoplasts isolated from delicate leaves, by polyethylene glycol-mediated transformation57. The fluorescence signal were examined under a laser scanning confocal microscope (ZEISS LSM 710 NLO).
Phylogenetic analysis
The protein sequences of the FAR family members from Arabidopsis, rice and maize were aligned using the BioEdit tool. The alignment result was used to construct a neighbor-joining polygenetic tree in MEGA 6 (http://megasoftware.net/) using the following parameters: Poisson model, complete deletion, and 1,000 bootstrap replicates.
Arabidopsis transformation
To confirm the functional conservation between MS2 and MS6021, we cloned the MS2 promoter sequence from Columbia ecotype Arabidopsis, MS6021CDS, and MS6021ΔNCDS. These fragments were subcloned into the binary vector pCAMBIA1300 to generate ProMS2:MS6021CDS and ProMS2:MS6021ΔNCDS. These constructs were introduced into Agrobacterium tumefaciens GV3101 and transformed into the ms2 mutant plants by the floral dipping method58. The transformed seeds were screened using 1/2 plant MS medium containing 20 mg/L hygromycin.
Transcriptome analysis
Triplicate individuals of uninucleate-stage anthers were harvested from wild-type and ms6021 plants and were used to extract total RNA with the RNeasy Plant mini Kit (QIAGEN) according to the manufacturer’s instructions. Libraries were constructed in accordance with standard Illumina TruSeq instructions and sequenced using an Illumina Genome Analyzer (Hiseq. 2500; Illumina). The raw reads were filtered to obtain high-quality clean reads and mapped to the maize reference genome (AGPv3; MaizeSequence.org) using TopHat259 with default parameters. The gene expression level was calculated by RPKM60. The edgeR package61 was used to detect differentially expressed genes (DEGs), which were defined according to the following criteria: more than a two-fold change and FDR less than 0.05. Gene ontology (GO) enrichment and KEGG pathway enrichment analyses were performed using the Bioconductor tool62.
Electronic supplementary material
Acknowledgements
We thank Yanbao Tian (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for the SEM and TEM services, Caixia Gao (IGDB, CAS) for providing the pJIT163-GFP vector, and Zhidan Chen for the field work. This work was supported by funds from the Ministry of Agriculture of China (Grant 2016ZX0801002-001-003), and the National Natural Science Foundation of China (31601376).
Author Contributions
H.C. and Y.T. designed the experiments. Y.H. performed most experiments and wrote the manuscript. S.X. offered help on material collection and western blot. J.L. performed some of the mapping. Y.S. and H.Z. did some work about in situ hybridization and subcellular localization. M.W. constructed the NJ tree. H.Z. finished the RNA-seq analysis. L.Z. and H.C. edited the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-16930-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Scott RJ, Spielman M, Dickinson HG. Stamen structure and function. Plant Cell. 2004;16:S46–60. doi: 10.1105/tpc.017012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 2005;56:393–434. doi: 10.1146/annurev.arplant.55.031903.141717. [DOI] [PubMed] [Google Scholar]
- 3.McCormick S. Male gametophyte development. Plant cell. 1993;5:1265–1275. doi: 10.1105/tpc.5.10.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang D, Yang L. Specification of tapetum and microsporocyte cells within the anther. Curr. Opin. Plant Biol. 2014;17:49–55. doi: 10.1016/j.pbi.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg RB, Beals TP, Sanders PM. Anther development: basic principles and practical applications. Plant Cell. 1993;5:1217–1229. doi: 10.1105/tpc.5.10.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li N, et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell. 2006;18:2999–3014. doi: 10.1105/tpc.106.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, et al. Cytochrome P450 family member CYP704B2 catalyzes the {omega}-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell. 2010;22:173–190. doi: 10.1105/tpc.109.070326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ariizumi T, Toriyama K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 2011;62:437–460. doi: 10.1146/annurev-arplant-042809-112312. [DOI] [PubMed] [Google Scholar]
- 9.Kunst L, Samuels AL. Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 2003;42:51–80. doi: 10.1016/S0163-7827(02)00045-0. [DOI] [PubMed] [Google Scholar]
- 10.Jung KH, et al. Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell. 2006;18:3015–3032. doi: 10.1105/tpc.106.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeats TH, Rose JK. The formation and function of plant cuticles. Plant Physiol. 2013;163:5–20. doi: 10.1104/pp.113.222737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heredia A. Biophysical and biochemical characteristics of cutin, a plant barrier biopolymer. Biochim. Biophys. Acta. 2003;1620:1–7. doi: 10.1016/S0304-4165(02)00510-X. [DOI] [PubMed] [Google Scholar]
- 13.Ahlers, F., Lambert, J. & Wiermann, R. Acetylation and silylation of piperidine solubilized sporopollenin from pollen of typha angustifolia L. Zeitschrift für Naturforsch. C58, 10.1515/znc-2003-11-1210 (2003). [DOI] [PubMed]
- 14.Wallace S, Fleming A, Wellman CH, Beerling DJ. Evolutionary development of the plant and spore wall. AoB Plants. 2011;2011:plr027. doi: 10.1093/aobpla/plr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blackmore S, Wortley AH, Skvarla JJ, Rowley JR. Pollen wall development in flowering plants. New Phytol. 2007;174:483–498. doi: 10.1111/j.1469-8137.2007.02060.x. [DOI] [PubMed] [Google Scholar]
- 16.Wallace S, et al. Conservation of Male Sterility 2 function during spore and pollen wall development supports an evolutionarily early recruitment of a core component in the sporopollenin biosynthetic pathway. New Phytol. 2015;205:390–401. doi: 10.1111/nph.13012. [DOI] [PubMed] [Google Scholar]
- 17.Wilson ZA, Zhang DB. From Arabidopsis to rice: pathways in pollen development. J. Exp. Bot. 2009;60:1479–1492. doi: 10.1093/jxb/erp095. [DOI] [PubMed] [Google Scholar]
- 18.Chen W, et al. Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol. 2011;157:842–853. doi: 10.1104/pp.111.181693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi J, et al. Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell. 2011;23:2225–2246. doi: 10.1105/tpc.111.087528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X, et al. Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J. Integr. Plant Biol. 2014;56:979–994. doi: 10.1111/jipb.12212. [DOI] [PubMed] [Google Scholar]
- 21.Morant M, et al. CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell. 2007;19:1473–1487. doi: 10.1105/tpc.106.045948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobritsa AA, et al. CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 2009;151:574–589. doi: 10.1104/pp.109.144469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi J, Cui M, Yang L, Kim YJ, Zhang D. Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci. 2015;20:741–753. doi: 10.1016/j.tplants.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Azevedo Souza C, et al. A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell. 2009;21:507–525. doi: 10.1105/tpc.108.062513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobritsa AA, et al. LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol. 2010;153:937–955. doi: 10.1104/pp.110.157446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SS, et al. LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl alpha-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant cell. 2010;22:4045–4066. doi: 10.1105/tpc.110.080028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang LK, Chu H, Yip WK, Yeung EC, Lo C. An anther-specific dihydroflavonol 4-reductase-like gene (DRL1) is essential for male fertility in Arabidopsis. New phytol. 2009;181:576–587. doi: 10.1111/j.1469-8137.2008.02692.x. [DOI] [PubMed] [Google Scholar]
- 28.Grienenberger E, et al. Analysis of TETRAKETIDE alpha-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell. 2010;22:4067–4083. doi: 10.1105/tpc.110.080036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quilichini TD, Samuels AL, Douglas CJ. ABCG26-mediated polyketide trafficking and hydroxycinnamoyl spermidines contribute to pollen wall exine formation in arabidopsis. Plant Cell. 2014;26:4483–4498. doi: 10.1105/tpc.114.130484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu L, et al. OsABCG15 encodes a membrane protein that plays an important role in anther cuticle and pollen exine formation in rice. Plant Cell Rep. 2014;33:1881–1899. doi: 10.1007/s00299-014-1666-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu L, Shi J, Zhao G, Zhang D, Liang W. Post-meiotic deficientanther1 (PDA1) encodes an ABC transporter required for the development of anther cuticle and pollen exine in rice. J. Plant Biol. 2013;56:59–68. doi: 10.1007/s12374-013-0902-z. [DOI] [Google Scholar]
- 32.Choi H, et al. An ABCG/WBC-type ABC transporter is essential for transport of sporopollenin precursors for exine formation in developing pollen. Plant J. 2011;65:181–193. doi: 10.1111/j.1365-313X.2010.04412.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhang D, et al. OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 2010;154:149–162. doi: 10.1104/pp.110.158865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Djukanovic V, et al. Male-sterile maize plants produced by targeted mutagenesis of the cytochrome P450-like gene (MS26) using a re-designed I-CreI homing endonuclease. Plant J. 2013;76:888–899. doi: 10.1111/tpj.12335. [DOI] [PubMed] [Google Scholar]
- 35.Wu Y, et al. Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross-pollinating crops. Plant Biotechnol. J. 2016;14:1046–1054. doi: 10.1111/pbi.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, et al. IRREGULAR POLLEN EXINE1 is a novel factor in anther cuticle and pollen exine formation. Plant Physiol. 2016;173:307–325. doi: 10.1104/pp.16.00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Somaratne Y, et al. ABNORMAL POLLEN VACUOLATION1 (APV1) is required for male fertility by contributing to anther cuticle and pollen exine formation in maize. Plant J. 2017;90:96–110. doi: 10.1111/tpj.13476. [DOI] [PubMed] [Google Scholar]
- 38.Zhang D, Wilson ZA. Stamen specification and anther development in rice. Chinese Sci. Bull. 2009;54:2342–2353. doi: 10.1007/s11434-009-0348-3. [DOI] [Google Scholar]
- 39.Bonaventure G, Beisson F, Ohlrogge J, Pollard M. Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. Plant J. 2004;40:920–930. doi: 10.1111/j.1365-313X.2004.02258.x. [DOI] [PubMed] [Google Scholar]
- 40.Franke R, et al. Apoplastic polyesters in Arabidopsis surface tissues–a typical suberin and a particular cutin. Phytochemistry. 2005;66:2643–2658. doi: 10.1016/j.phytochem.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 41.Doan TT, et al. Biochemical characterization of a chloroplast localized fatty acid reductase from Arabidopsis thaliana. Biophys. Acta. 2012;1821:1244–1255. doi: 10.1016/j.bbalip.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 42.Schnable PS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 43.Guilford WJ, Schneider DM, Labovitz J, Opella SJ. High resolution solid state (13)C NMR spectroscopy of sporopollenins from different plant taxa. Plant Physiol. 1988;86:134–136. doi: 10.1104/pp.86.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin P, et al. ABCG15 encodes an ABC transporter protein, and is essential for post-meiotic anther and pollen exine development in rice. Plant Cell Physiol. 2013;54:138–154. doi: 10.1093/pcp/pcs162. [DOI] [PubMed] [Google Scholar]
- 45.Zhao G, et al. Two ATP binding cassette G transporters, rice ATP Binding Cassette G26 and ATP Binding Cassette G15, collaboratively regulate rice male reproduction. Plant Physiol. 2015;169:2064–2079. doi: 10.1104/pp.15.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rozema J, et al. UV-B absorbance and UV-B absorbing compounds (para-coumaric acid) in pollen and sporopollenin: the perspective to track historic UV-B levels. J. Photochem. Photobiol. B. 2001;62:108–117. doi: 10.1016/S1011-1344(01)00155-5. [DOI] [PubMed] [Google Scholar]
- 47.Descolas-Gros C, Scholzel C. Stable isotope ratios of carbon and nitrogen in pollen grains in order to characterize plant functional groups and photosynthetic pathway types. New Phytol. 2007;176:390–401. doi: 10.1111/j.1469-8137.2007.02176.x. [DOI] [PubMed] [Google Scholar]
- 48.Hahlbrock K, Scheel D. Physiology and molecular biology of phenylpropanoid metabolism. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1989;40:347–369. doi: 10.1146/annurev.pp.40.060189.002023. [DOI] [Google Scholar]
- 49.Edward H. C, McCormic SM, Modena SA. White pollen in mazie. J. Hered. 1981;72:318–320. doi: 10.1093/oxfordjournals.jhered.a109514. [DOI] [Google Scholar]
- 50.Napoli CA, Fahy D, Wang HY, Taylor LP. White anther: a petunia mutant that abolishes pollen flavonol accumulation, induces male sterility, and is complemented by a chalcone synthase transgene. Plant Physiol. 1999;120:615–622. doi: 10.1104/pp.120.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischer R, Budde I, Hain R. Stilbene synthase gene expression causea changes in flower colour and male sterility in tobacco. Plant J. 1997;11:489–498. doi: 10.1046/j.1365-313X.1997.11030489.x. [DOI] [Google Scholar]
- 52.Watkins JM, Hechler PJ, Muday GK. Ethylene-induced flavonol accumulation in guard cells suppresses reactive oxygen species and moderates stomatal aperture. Plant Physiol. 2014;164:1707–1717. doi: 10.1104/pp.113.233528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Kim JI, Pysh L, Chapple C. Four isoforms of Arabidopsis 4-Coumarate:CoA Ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 2015;169:2409–2421. doi: 10.1104/pp.15.00838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yadav V, et al. ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell. 2014;26:3569–3588. doi: 10.1105/tpc.114.129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quilichini TD, Grienenberger E, Douglas CJ. The biosynthesis, composition and assembly of the outer pollen wall: A tough case to crack. Phytochemistry. 2015;113:170–182. doi: 10.1016/j.phytochem.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Ding L, et al. HANABA TARANU (HAN) bridges meristem and organ primordia boundaries through PINHEAD, JAGGED, BLADE-ON-PETIOLE2 and CYTOKININ OXIDASE 3 during flower development in Arabidopsis. PLoS Genet. 2015;11:e1005479. doi: 10.1371/journal.pgen.1005479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoo S-D, Cho Y-H, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protocols. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 58.Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protocols. 2006;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 59.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics (Oxford, England) 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ali Mortazavi BAW, McCue K, Schaeffer L. Barbara Wold. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 61.Anders S, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protocols. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 62.Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ, Holmes SP. Bioconductor Workflow for Microbiome Data Analysis: from raw reads to community analyses. F1000Research. 2016;5:1492. doi: 10.12688/f1000research.8986.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.