Abstract
Background
Metacercariae of Diplostomum are important fish pathogens, but reliable data on their diversity in natural fish populations are virtually lacking. This study was conducted to explore the species diversity and host-parasite association patterns of Diplostomum spp. in a large riverine system in Europe, using molecular and morphological data.
Methods
Twenty-eight species of fish of nine families were sampled in the River Danube at Nyergesújfalu in Hungary in 2012 and Štúrovo in Slovakia in 2015. Isolates of Diplostomum spp. were characterised morphologically and molecularly. Partial sequences of the ‘barcode’ region of the cytochrome c oxidase subunit 1 (cox1) and complete sequences of the nicotinamide adenine dinucleotide dehydrogenase subunit 3 (nad3) mitochondrial genes were amplified for 76 and 30 isolates, respectively. The partial cox1 sequences were used for molecular identification of the isolates and an assessment of haplotype diversity and possible host-associated structuring of the most prevalent parasite species. New primers were designed for amplification of the mitochondrial nad3 gene.
Results
Only lens-infecting Diplostomum spp. were recovered in 16 fish species of five families. Barcoding of representative isolates provided molecular identification for three species/species-level genetic lineages, D. spathaceum, D. pseudospathaceum and ‘D. mergi Lineage 2’, and three single isolates potentially representing distinct species. Molecular data helped to elucidate partially the life-cycle of ‘D. mergi Lineage 2’. Many of the haplotypes of D. spathaceum (16 in total), D. pseudospathaceum (15 in total) and ‘D. mergi Lineage 2’ (7 in total) were shared by a number of fish hosts and there was no indication of genetic structuring associated with the second intermediate host. The most frequent Diplostomum spp. exhibited a low host-specificity, predominantly infecting a wide range of cyprinid fishes, but also species of distant fish families such as the Acipenseridae, Lotidae, Percidae and Siluridae. The nad3 gene exhibited distinctly higher levels of interspecific divergence in comparison with the cox1 gene.
Conclusions
This first exploration of the species diversity and host ranges of Diplostomum spp., in natural fish populations in the River Danube, provided novel molecular, morphological and host-use data which will advance further ecological studies on the distribution and host ranges of these important fish parasites in Europe. Our results also indicate that the nad3 gene is a good candidate marker for multi-gene approaches to systematic estimates within the genus.
Electronic supplementary material
The online version of this article (10.1186/s13071-017-2518-5) contains supplementary material, which is available to authorized users.
Keywords: Diplostomum, Diplostomidae, Metacercariae, Freshwater fishes, Barcodes, cox1, nad3, River Danube, Europe
Background
Metacercariae of the genus Diplostomum von Nordmann, 1832 (Digenea: Diplostomidae) are important fish pathogens [1–3] and represent a case study illustrating the difficulties of species identification based solely on morphological data. The recent use of molecular markers proved to be a valuable and efficient approach to species delimitation and identification, especially for the larval stages of Diplostomum spp. which lack reliable distinguishing morphological characters. Recent intensive molecular studies, following the publication of the genus-specific primers for the ‘barcode’ region of the cytochrome c oxidase subunit 1 (cox1) gene [4], resulted in the generation of sequence libraries for the North American [5, 6] and European species [3, 7–12] of the genus. Thus providing a sound basis for molecular identification and provisional species delineation. These libraries provide a foundation that will allow identification of life-cycle stages and ensure an increased taxonomic resolution in epidemiological and ecological studies of these important fish parasites (e.g. Locke et al. [13]; Désilets et al. [14]; Pérez-del-Olmo et al. [3]) as well as for further exploration of species host and geographical ranges [6].
To date, molecular data for a total of 19 species/species-level genetic lineages of Diplostomum exist from North America including three named species, i.e. Diplostomum baeri Dubois, 1937, Diplostomum huronense (La Rue, 1927) and Diplostomum indistinctum (Guberlet, 1923), and 16 otherwise unidentified species or species-level lineages [4–6, 15]. Extensive studies carried out in Europe recently revealed a total of 12 species/species-level genetic lineages including two species complexes: D. spathaceum (Rudolphi, 1819); D. pseudospathaceum Niewiadomska, 1984; D. parviventosum Dubois, 1932; three species-level lineages within the “D. baeri” species complex (Diplostomum sp. ‘Lineages 3–5’ sensu Blasco-Costa et al., 2014 [9]); three species-level lineages within the “D. mergi” species complex (Diplostomum sp. ‘Lineages 2–4’ sensu Georgieva et al., 2013 [7] and Selbach et al., 2015 [10]); Diplostomum sp. ‘Clade Q’ sensu Georgieva et al., 2013 [7]; and Diplostomum sp. ‘Lineages 2 and 6’ sensu Blasco-Costa et al., 2014 [9] (see [3, 7, 9, 10, 12, 16]).
However, although molecular data for metacercariae of Diplostomum spp. in fishes from European freshwater ecosystems have accumulated recently, most of the sequences originate from fish populations sampled in ponds and lakes in central and northern Europe (Germany, Iceland, Norway), and also predominantly from salmonid fishes. A single study provided molecular and morphological data for metacercariae of three species of Diplostomum spp. in endemic and invasive fish host species in Spain, at the southern distributional range of Diplostomum spp. in Europe [3]. However, no molecular data exist on species diversity and host ranges of these fish pathogens in large river systems in Europe.
Our study is the first to explore species diversity and host-parasite association patterns of Diplostomum spp. in a large riverine system in Europe. Here we extend the cox1 ‘barcode’ reference library for Diplostomum spp. based on an extensive sampling of metacercariae from a broad range of fish hosts collected at two localities in the middle section of the River Danube. We provide molecular identification based on the cox1 gene in association with a thorough morphological characterisation of the metacercariae. Further, we provide primers and the first assessment of the usefulness of the mitochondrial nicotinamide adenine dinucleotide dehydrogenase subunit 3 (nad3) gene for species delineation within Diplostomum spp.
Methods
Sample collection and processing
A total of 174 fish belonging to 28 species of 9 families were sampled in the River Danube near Nyergesújfalu (47.7658N, 18.5417E) in Hungary in 2012 and at Štúrovo (47.8197N, 18.7286E) in Slovakia in 2015. As a part of a complete helminthological examination, fish eyes and brains were isolated and examined for the presence of metacercariae of Diplostomum spp. The eyes were dissected and lens, vitreous humour and retina were placed in 0.9% saline solution and examined under a dissecting microscope. All metacercariae were collected and counted. Representative subsamples were selected for DNA isolation and sequencing.
Morphological examination
The morphology of the metacercariae selected for sequencing was initially studied in live parasites; these were then transferred to molecular grade ethanol and re-examined. A series of photomicrographs was made for each isolate (live and fixed) using a digital camera of an Olympus BX51 microscope (Olympus Corporation, Tokyo, Japan). Measurements for each isolate were taken from the digital images with the aid of Quick Photo Camera 2.3 image analysis software. All measurements in the descriptions and tables are in micrometres and are presented as the range, followed by the mean in parentheses.
Fourteen morphometric variables were measured from the digital images of live and fixed metacercariae and the number of excretory concretions was recorded from live material. The following abbreviations for variables were used: BL, body length; BW, body width; HL, hindbody length; OSL, oral sucker length; OSW, oral sucker width; PSL, pseudosucker length; PSW, pseudosucker width; VSL, ventral sucker length; VSW, ventral sucker width; PHL, pharynx length; PHW, pharynx width; HOL, holdfast organ length; HOW, holdfast organ width; AVS, distance from anterior extremity of body to ventral sucker.
Sequence generation
Genomic DNA (gDNA) was isolated from single metacercariae using the E.Z.N.A. Tissue DNA Kit (Omega Bio-tek, Norcross, USA) following the manufacturer’s instructions. Amplification of the mitochondrial (mt) cox1 gene was performed with the forward primer Plat-diploCOX1F (5′-CGT TTR AAT TAT ACG GAT CC-3′) and the reverse primer Plat-diploCOX1R (5′-AGC ATA GTA ATM GCA GCA GC-3′) [4]. A pair of newly designed primers was used for amplification of the complete nad3 mt gene: forward Diplo-nad3F (5′-ATG TGA AAG TGG TGT TTG TT-3′) and reverse Diplo-nad3R (5′-ATG CGC TTA TGA TCT AAC GT-3′). PCR amplifications for both genes were performed in a total volume of 20 μl (8 pmol of each primer) with c.50 ng of gDNA and 10 μl of 2× MyFi™ DNA Polymerase mix (Bioline Inc., Taunton, USA). Thermocycling started with an initial DNA denaturation for 2 min at 94 °C followed by 35 cycles with 30 s DNA denaturation at 94 °C, 30 s primer annealing at 50 °C for cox1 (57 °C for nad3), and 60 s at 72 °C for primer extension, followed by a final extension step of 10 min at 72 °C. PCR amplicons were purified using a QIAquick PCR purification kit (Qiagen Ltd., Hilden, Germany). Cycle sequencing of purified DNA was carried out using ABI Big Dye™ chemistry (ABI Perkin-Elmer, London, UK) on an Applied Biosystems 3730xl DNA Analyser following the manufacturer’s recommendations, using the primers used for PCR amplification. Contiguous sequences were assembled with MEGA v6 [17] and submitted to GenBank under accession numbers KY653961–KY654066.
Unique cox1 haplotypes were identified with DnaSP [18] against all published sequences for a given species/lineage. Unrooted statistical parsimony haplotype networks were constructed for D. spathaceum and D. pseudospathaceum using TCS 1.21 [19] with plausible branch connections between the haplotypes at a connection limit of 95% [20].
Phylogenetic analyses
Sequences were aligned using MUSCLE implemented in MEGA v6. Two alignments were analysed. The cox1 alignment (410 nt) comprised 76 newly generated sequences and 31 sequences for Diplostomum spp. retrieved from GenBank; Tylodelphys clavata (von Nordmann, 1832) was used as the outgroup. The nad3 alignment (357 nt) comprised 30 newly generated sequences and two published sequences, D. pseudospathaceum and D. spathaceum. Both alignments included no insertions or deletions and were aligned with reference to the amino acid translation, using the echinoderm and flatworm mitochondrial code [21]. Distance-based neighbour-joining (NJ) and model-based Bayesian inference (BI) algorithms were conducted to identify and explore relationships among the species/isolates. Neighbour-joining analyses of Kimura 2-parameter distances were carried out using MEGA v6; nodal support was estimated using 1000 bootstrap resamplings. Bayesian inference analysis was performed for the cox1 dataset using MrBayes version 3.2.3 [22]. Prior to BI analysis, the best-fit nucleotide substitution model was selected in jModelTest 2.1.1 [23] using the Akaike Information Criterion (AIC). This was the general time reversible model, with estimates of invariant sites and gamma distributed among-site rate variation (GTR + I + Г). BI analysis was run with the following nucleotide substitution model settings: lset nst = 6, rates = invgamma, samplefreq = 100, ncat = 4, shape = estimate, inferrates = yes and basefreq = empirical. Markov chain Monte Carlo (MCMC) chains were run for 10,000,000 generations, log-likelihood scores were plotted and only the final 75% of trees were used to produce the consensus trees by setting the ‘burn-in’ parameter at 2500. Results were visualised in Tracer v.1.6 (http://tree.bio.ed.ac.uk/software/tracer/) to assess convergence and proper sampling and to identify the ‘burn-in’ period.
Distance matrices (uncorrected p-distance model) were calculated with MEGA v6. The nomenclature of Georgieva et al. [7] for the lineages of Diplostomum spp. was applied for consistency with previous records.
Results
General observations
A total of 174 fish individuals belonging to 28 species and 9 families were examined for the presence of metacercariae of Diplostomum spp. in the eyes and brain. Only lens-infecting metacercariae were found in 16 fish species of 5 families: 12 cyprinids, one acipenserid, one lotid, one percid and one silurid (Table 1). The overall Diplostomum spp. intensity of infection was low (1–15 metacercariae per fish) with two exceptions: Abramis brama (25–43, four fishes) and Blicca bjoerkna (27, one fish). The overall Diplostomum spp. prevalence appeared rather high in five cyprinids (Leuciscus aspius: 89%; Vimba vimba: 89%; A. brama: 83%; B. bjoerkna: 77%; and Alburnus alburnus: 57%) but reliable estimates for prevalence could be obtained only for the sample of A. brama. In this sample, the prevalence of three species/lineages identified in our study (see below) was high: D. spathaceum: 75%; ‘D. mergi Lineage 2’: 58%; D. pseudospathaceum: 50%. Twelve species of fish, for which fewer specimens were examined, were not infected.
Table 1.
Summary data for the fish species examined/infected with Diplostomum spp.
| Host species | No. examined | No. infected | Diplostomum spp. |
|---|---|---|---|
| Acipenseridae | |||
| Acipenser ruthenus L. | 1 | 1 | D. spathaceum |
| Anguillidae | |||
| Anguilla anguilla (L.) | 1 | – | – |
| Centrarchidae | |||
| Lepomis gibbosus (L.) | 11 | – | – |
| Cyprinidae | |||
| Abramis brama (L.) | 41 | 34 | D. spathaceum, D. pseudospathaceum, ‘D. mergi Lineage 2’ |
| Alburnus alburnus (L.) | 7 | 4 | ‘D. mergi Lineage 2’ |
| Ballerus sapa (Pallas) | 9 | 2 | D. pseudospathaceum, ‘D. mergi Lineage 2’ |
| Blicca bjoerkna (L.) | 13 | 10 | D. spathaceum, D. pseudospathaceum, ‘D. mergi Lineage 2’, Diplostomum sp. A |
| Carassius gibelio (Bloch) | 6 | 1 | Diplostomum sp. B |
| Chondrostoma nasus (L.) | 11 | 4 | D. spathaceum, ‘D. mergi Lineage 2’ |
| Cyprinus carpio L. | 3 | 1 | D. pseudospathaceum |
| Leuciscus aspius (L.) | 9 | 8 | D. spathaceum, D. pseudospathaceum |
| Leuciscus idus (L.) | 4 | 1 | D. pseudospathaceum |
| Rutilus pigus (Lacépède) | 3 | 2 | D. spathaceum |
| Rutilus rutilus (L.) | 9 | 4 | D. spathaceum, Diplostomum sp. C |
| Vimba vimba (L.) | 9 | 8 | D. spathaceum, D. pseudospathaceum, ‘D. mergi Lineage 2’ |
| Barbus barbus (L.) | 2 | – | – |
| Gobio gobio (L.) | 6 | – | – |
| Esocidae | |||
| Esox lucius L. | 3 | – | – |
| Gobiidae | |||
| Neogobius melanostomus (Pallas) | 8 | – | – |
| Ponticola kessleri (Günther) | 2 | – | – |
| Lotidae | |||
| Lota lota (L.) | 2 | 1 | D. pseudospathaceum |
| Percidae | |||
| Gymnocephalus schraetser (L.) | 5 | 1 | D. pseudospathaceum |
| Perca fluviatilis L. | 3 | – | – |
| Sander lucioperca (L.) | 1 | – | – |
| Sander volgensis (Gmelin) | 2 | – | – |
| Zingel zingel (L.) | 1 | – | – |
| Zingel streber (Siebold) | 1 | – | – |
| Siluridae | |||
| Silurus glanis L. | 1 | 1 | D. spathaceum |
Molecular identification, haplotype diversity and host-use
We generated partial cox1 sequences (410 nt) for 76 isolates of Diplostomum spp. recovered from fishes of the River Danube (Table 2). These sequences were analysed together with 31 sequences for 10 Diplostomum species/species-level genetic lineages retrieved from the GenBank database (see Additional file 1: Table S1 for details). All lens-infecting species/lineages of Diplostomum (7) reported in Europe were included in analyses: D. parviventosum, D. pseudospathaceum, D. spathaceum, ‘D. mergi Lineage 2’, ‘D. mergi Lineage 3’, ‘D. mergi Lineage 4’, ‘Diplostomum sp. Clade Q’ sensu Georgieva et al., 2013 [7]. We also included sequences for D. huronense (a species believed to have a Holarctic distribution; see [24]) and two representatives of non-lens infecting species of the “D. baeri” complex. The branch topologies of the trees resulting from both, NJ and BI analyses, were in consensus in depicting species/species-level genetic lineages (Figs. 1, 2). The newly generated sequences clustered within three well-supported clades representing D. pseudospathaceum, D. spathaceum and ‘D. mergi Lineage 2’ except for three singletons which may potentially represent distinct species (labelled as Diplostomum sp. A, B and C in Fig. 2). Two of these (Diplostomum sp. A and B) were resolved as basal to the clade representing the “D. mergi” species complex, whereas Diplostomum sp. C appeared associated with ‘Clade Q’; however, these relationships were not supported.
Table 2.
Summary data for the isolates of Diplostomum spp. used for generation of the cox1 and nad3 sequences
| Species | Host | Country | Isolate | Haplotype (cox1) | GenBank ID | |
|---|---|---|---|---|---|---|
| cox1 | nad3 | |||||
| D. spathaceum | Abramis brama | S | ABD1 | H11 | KY653961 | KY654037 |
| D. spathaceum | Abramis brama | S | ABD2 | H1 | KY653962 | |
| D. spathaceum | Abramis brama | S | ABD3 | H1 | KY653963 | |
| D. spathaceum | Abramis brama | S | ABD4 | H5 | KY653964 | |
| D. spathaceum | Abramis brama | S | ABD5 | H9 | KY653965 | |
| D. spathaceum | Abramis brama | S | ABD6 | H12 | KY653966 | KY654038 |
| D. spathaceum | Abramis brama | S | ABD7 | H10 | KY653967 | |
| D. spathaceum | Abramis brama | S | ABD8 | H2 | KY653968 | |
| D. spathaceum | Abramis brama | S | ABD9 | H3 | KY653969 | KY654039 |
| D. spathaceum | Acipenser ruthenus | S | ARD | H4 | KY653970 | |
| D. spathaceum | Blicca bjoerkna | S | BBD1 | H6 | KY653971 | |
| D. spathaceum | Blicca bjoerkna | S | BBD2 | H4 | KY653972 | KY654040 |
| D. spathaceum | Blicca bjoerkna | H | BBD3 | H14 | KY653973 | |
| D. spathaceum | Chondrostoma nasus | S | CND1 | H7 | KY653974 | KY654041 |
| D. spathaceum | Chondrostoma nasus | H | CND2 | H15 | KY653975 | |
| D. spathaceum | Leuciscus aspius | H | LAD1 | H13 | KY653976 | KY654042 |
| D. spathaceum | Leuciscus aspius | S | LAD2 | H2 | KY653977 | |
| D. spathaceum | Rutilus pigus | S | RPD1 | H5 | KY653978 | |
| D. spathaceum | Rutilus pigus | S | RPD2 | H2 | KY653979 | KY654043 |
| D. spathaceum | Rutilus pigus | S | RPD3 | H8 | KY653980 | |
| D. spathaceum | Rutilus pigus | S | RPD4 | H3 | KY653981 | KY654044 |
| D. spathaceum | Rutilus rutilus | S | RRD1 | H1 | KY653982 | KY654045 |
| D. spathaceum | Rutilus rutilus | H | RRD2 | H16 | KY653983 | |
| D. spathaceum | Silurus glanis | S | SGD | H3 | KY653984 | KY654046 |
| D. spathaceum | Vimba vimba | S | VVD1 | H1 | KY653985 | |
| D. spathaceum | Vimba vimba | S | VVD2 | H1 | KY653986 | |
| D. pseudospathaceum | Abramis brama | S | ABD10 | H1 | KY653987 | KY654047 |
| D. pseudospathaceum | Abramis brama | S | ABD11 | H1 | KY653988 | |
| D. pseudospathaceum | Abramis brama | S | ABD12 | H2 | KY653989 | KY654048 |
| D. pseudospathaceum | Abramis brama | S | ABD13 | H14 | KY653990 | |
| D. pseudospathaceum | Abramis brama | S | ABD14 | H15 | KY653991 | |
| D. pseudospathaceum | Ballerus sapa | S | BSD1 | H1 | KY653992 | KY654049 |
| D. pseudospathaceum | Ballerus sapa | S | BSD2 | H3 | KY653993 | KY654050 |
| D. pseudospathaceum | Ballerus sapa | S | BSD3 | H3 | KY653994 | |
| D. pseudospathaceum | Ballerus sapa | S | BSD4 | H2 | KY653995 | |
| D. pseudospathaceum | Blicca bjoerkna | H | BBD4 | H1 | KY653996 | |
| D. pseudospathaceum | Blicca bjoerkna | S | BBD5 | H7 | KY653997 | KY654051 |
| D. pseudospathaceum | Blicca bjoerkna | S | BBD6 | H8 | KY653998 | KY654052 |
| D. pseudospathaceum | Blicca bjoerkna | S | BBD7 | H10 | KY653999 | |
| D. pseudospathaceum | Blicca bjoerkna | S | BBD8 | H11 | KY654000 | |
| D. pseudospathaceum | Blicca bjoerkna | H | BBD9 | H4 | KY654001 | |
| D. pseudospathaceum | Blicca bjoerkna | S | BBD10 | H9 | KY654002 | |
| D. pseudospathaceum | Cyprinus carpio | S | CCD | H1 | KY654003 | KY654053 |
| D. pseudospathaceum | Gymnocephalus schraetser | H | GSD | H4 | KY654004 | |
| D. pseudospathaceum | Leuciscus aspius | S | LAD3 | H13 | KY654005 | |
| D. pseudospathaceum | Leuciscus aspius | S | LAD4 | H1 | KY654006 | |
| D. pseudospathaceum | Leuciscus aspius | S | LAD5 | H2 | KY654007 | |
| D. pseudospathaceum | Leuciscus aspius | S | LAD6 | H6 | KY654008 | |
| D. pseudospathaceum | Leuciscus aspius | S | LAD7 | H5 | KY654009 | KY654054 |
| D. pseudospathaceum | Leuciscus aspius | S | LAD8 | H5 | KY654010 | |
| D. pseudospathaceum | Leuciscus aspius | H | LAD9 | H4 | KY654011 | |
| D. pseudospathaceum | Leuciscus idus | S | LID1 | H1 | KY654012 | KY654055 |
| D. pseudospathaceum | Leuciscus idus | S | LID2 | H12 | KY654013 | |
| D. pseudospathaceum | Lota lota | H | LLD | H3 | KY654014 | |
| D. pseudospathaceum | Vimba vimba | S | VVD3 | H1 | KY654015 | KY654056 |
| D. pseudospathaceum | Vimba vimba | H | VVD4 | H1 | KY654016 | |
| ‘D. mergi Lineage 2’ | Abramis brama | S | ABD15 | H2 | KY654017 | |
| ‘D. mergi Lineage 2’ | Abramis brama | S | ABD16 | H4 | KY654018 | KY654057 |
| ‘D. mergi Lineage 2’ | Abramis brama | S | ABD17 | H1 | KY654019 | KY654058 |
| ‘D. mergi Lineage 2’ | Abramis brama | S | ABD18 | H2 | KY654020 | KY654059 |
| ‘D. mergi Lineage 2’ | Alburnus alburnus | H | AAD1 | H2 | KY654021 | |
| ‘D. mergi Lineage 2’ | Alburnus alburnus | S | AAD2 | H5 | KY654022 | KY654060 |
| ‘D. mergi Lineage 2’ | Alburnus alburnus | H | AAD3 | H1 | KY654023 | KY654061 |
| ‘D. mergi Lineage 2’ | Alburnus alburnus | H | AAD4 | H1 | KY654024 | |
| ‘D. mergi Lineage 2’ | Alburnus alburnus | H | AAD5 | H1 | KY654025 | |
| ‘D. mergi Lineage 2’ | Alburnus alburnus | H | AAD6 | H1 | KY654026 | |
| ‘D. mergi Lineage 2’ | Ballerus sapa | H | BSD5 | H7 | KY654027 | KY654062 |
| ‘D. mergi Lineage 2’ | Blicca bjoerkna | S | BBD11 | H3 | KY654028 | KY654063 |
| ‘D. mergi Lineage 2’ | Blicca bjoerkna | S | BBD12 | H1 | KY654029 | KY654064 |
| ‘D. mergi Lineage 2’ | Blicca bjoerkna | H | BBD13 | H1 | KY654030 | |
| ‘D. mergi Lineage 2’ | Chondrostoma nasus | S | CND3 | H1 | KY654031 | KY654065 |
| ‘D. mergi Lineage 2’ | Vimba vimba | H | VVD5 | H6 | KY654032 | |
| ‘D. mergi Lineage 2’ | Vimba vimba | H | VVD6 | H1 | KY654033 | KY654066 |
| Diplostomum sp. A | Blicca bjoerkna | S | BBD14 | – | KY654034 | |
| Diplostomum sp. B | Carassius gibelio | S | CGD | – | KY654035 | |
| Diplostomum sp. C | Rutilus rutilus | S | RRD3 | – | KY654036 | |
Abbreviations: H Hungary, S Slovakia
Fig. 1.
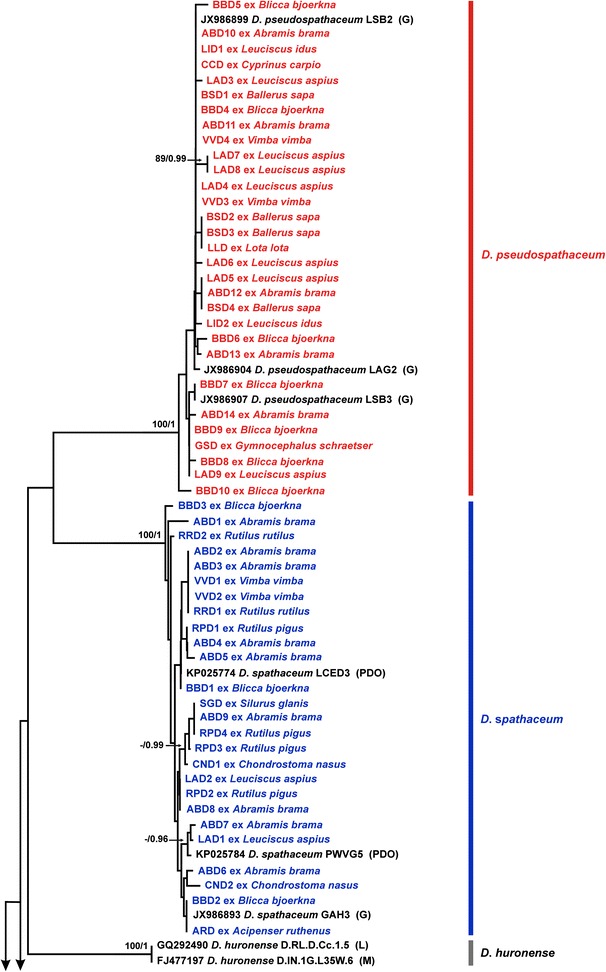
Neighbour-joining (NJ) phylogram for Diplostomum spp. reconstructed using 76 newly generated and 31 cox1 sequences retrieved from GenBank. Outgroup: Tylodelphys clavata. Nodal support from NJ and Bayesian inference (BI) analyses are indicated as NJ/BI; only values > 70% (NJ) and > 0.95 (BI) are shown. The scale-bar indicates the expected number of substitutions per site. Codes for the newly sequenced isolates are provided in Table 2. Sequence identification is as in GenBank, followed by a letter: G, Georgieva et al. [7]; L, Locke et al. [5]; M, Moszczynska et al. [4]; PDO, Pérez-del-Olmo et al. [3]
Fig. 2.
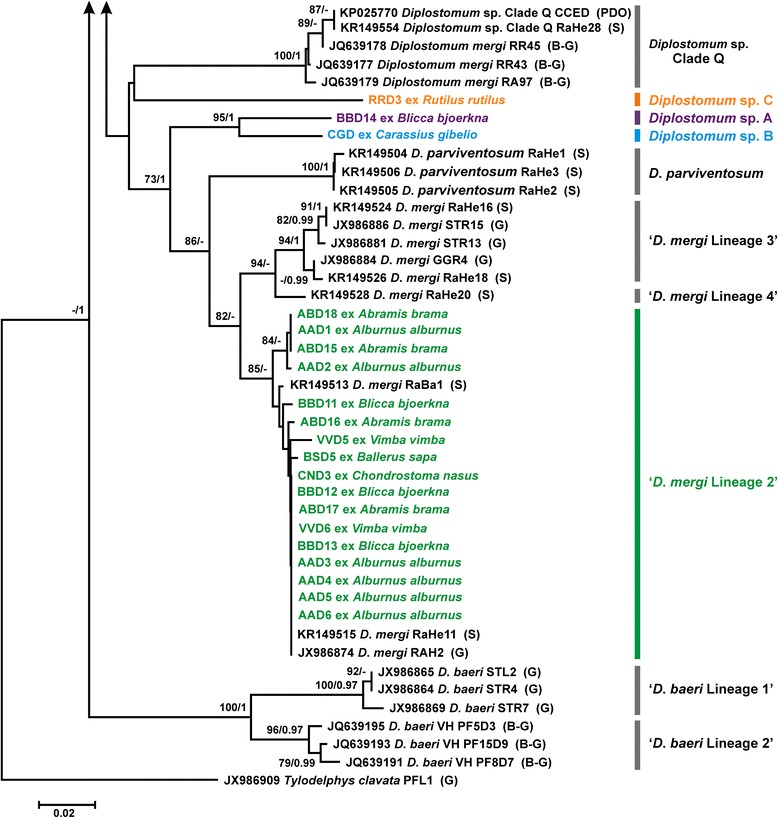
Neighbour-joining (NJ) phylogram for Diplostomum spp. reconstructed using 76 newly generated and 31 cox1 sequences retrieved from GenBank; continuation of Fig. 1. Nodal support from NJ and Bayesian inference (BI) analyses are indicated as NJ/BI; only values > 70% (NJ) and > 0.95 (BI) are shown. The scale-bar indicates the expected number of substitutions per site. Codes for the newly sequenced isolates are provided in Table 2. Sequence identification is as in GenBank, followed by a letter: B-G, Behrmann-Godel [8]; G, Georgieva et al. [7]; PDO, Pérez-del-Olmo et al. [3]; S, Selbach et al. [10]
The intraspecific divergence (uncorrected p-distance range), observed within the newly generated cox1 sequences, ranged between 0 and 1.71% (mean 0.56%) for D. pseudospathaceum, 0–1.95% (mean 0.82%) for D. spathaceum and 0–1.71% (mean 0.47%) for ‘D. mergi Lineage 2’. The three singletons exhibited high levels of divergence compared with the isolates of Diplostomum spp. included in the analyses: 7.1–15.6% for Diplostomum sp. A; 5.6–15.9% for Diplostomum sp. B; and 11.5–15.0% for Diplostomum sp. C.
The newly generated sequences for the three Diplostomum spp. were collapsed into 16 haplotypes for D. spathaceum, 15 haplotypes for D. pseudospathaceum and 7 haplotypes for ‘D. mergi Lineage 2’. Of these, D. spathaceum and D. pseudospathaceum had 7 unique haplotypes each (H1, H8, H9, H11, H14, H15, H16 and H3, H6, H8, H9, H11, H13, H14, respectively); and ‘D. mergi Lineage 2’ had 4 unique haplotypes (H3, H4, H5, H6).
Nine haplotypes of D. spathaceum were shared among isolates studied here and previously published sequences, predominantly generated in studies carried out in Europe (Germany, Iceland and Spain; see Georgieva et al. [7]; Pérez-del-Olmo et al. [3]; Selbach et al. [10]) (see Table 3 for details). Notably, four haplotypes (H2, H5, H6 and H10) were shared between isolates from all three hosts in the species life-cycle (first intermediate hosts: Radix auricularia (L.) and Radix peregra (Müller); definitive hosts: Larus argentatus (s.l.) and L. ridibundus; second intermediate host: a number of fish species). Due to the geographical coverage of the previous studies, most of the shared haplotypes originate from Europe; however, sequence matches for isolates from Asia [6] indicate a wider distribution of six haplotypes (Iraq: H2, H5, H7 and H10; China: H2, H13) (Table 3). It is also worth noting that four of the haplotypes were shared with haplotypes implicated in a case of diplostomiasis in aquaculture of Pseudochondrostoma willkommii (Steindachner) [3].
Table 3.
Details for the hosts, localities and GenBank accession numbers for the shared haplotypes of Diplostomum spp. identified in fishes from the River Danube
| Species/Haplotype | Present study | Published isolates with matching sequences | ||||
|---|---|---|---|---|---|---|
| Isolate codea | Host | GenBank ID | Host | Origin | Reference | |
| Diplostomum spathaceum | ||||||
| H2 | ABD8; LAD2; RPD2 | A. brama; L. aspius; R. pigus | JX986889; KR149550; KR149553; JX986888; KJ726433, KJ726434; KR271463; KR271451; KR271426; KR271430; JX986887 | Snails: Radix auricularia
Fishes: Abramis brama; Acanthobrama marmid; Barbus luteus; Cyprinion macrostomum; Gasterosteus aculeatus Birds: Larus cachinnans |
China; Czech Republic; Germany; Iceland; Iraq | [6, 7, 9, 10] |
| H3 | ABD9; RPD4; SGD | A. brama; R. pigus; S. glanis | JX986894; KR271417 | Fishes: Gasterosteus aculeatus; Perca fluviatilis | Germany; Italy | [6, 7] |
| H4 | ARD; BBD2 | A. ruthenus; B. bjoerkna | JX986893; KP025775; KP025785; KJ726438; KR271462 | Fishes: Gasterosteus aculeatus; Pseudochondrostoma willkommii; Salvelinus alpinus; Silurus glanis
Birds: Larus ridibundus |
Germany; Iceland; Romania; Spain | [3, 6, 7, 9] |
| H5 | ABD4; RPD1 | A. brama; R. pigus | JX986892; KR149551; KR271422, KR271429; KP025783; KP025772 | Snails: Radix auricularia.
Fishes: Cyprinion macrostomum; Pseudochondrostoma willkommii Birds: Larus argentatus; L. argentatus michahellis |
Germany; Iraq; Poland; Spain | [3, 6, 7, 10] |
| H6 | BBD1 | B. bjoerkna | KR149547, KR149548; KP025781; KP025778; KP025774; KJ726435, KJ726436; KR271431 | Snails: Radix auricularia; Radix peregra
Fishes: Gasterosteus aculeatus; Misgurnus anguillicaudatus; Pseudochondrostoma willkommii Birds: Larus argentatus michahellis |
Germany; Iceland; Spain | [3, 6, 9, 10] |
| H7 | CND1 | C. nasus | JX986891; KR149552; JX986890; KP025786, KP025782; KR271452; KR271423 | Snails: Radix auricularia
Fishes: Acanthobrama marmid; Cyprinion macrostomum; Gasterosteus aculeatus; Pseudochondrostoma willkommii |
Germany; Iraq; Spain | [3, 6, 7, 10] |
| H10 | ABD7 | A. brama | KR149549; KP025779; KR271428; JX986895 | Snails: Radix auricularia
Fishes: Barbus luteus; Misgurnus anguillicaudatus Birds: Larus cachinnans |
Germany; Iraq; Poland; Spain | [3, 6, 7, 10] |
| H12 | ABD6 | A. brama | KR271420 | Fishes: Perca fluviatilis | Italy | [6] |
| H13 | LAD1 | L. aspius | KR271459 | Fishes: Abramis brama | China | [6] |
| Diplostomum pseudospathaceum | ||||||
| H1 | ABD10; ABD11; BBD4; BSD1; CCD; LAD4; LID1; VVD3; VVD4 | A. brama; B. bjoerkna; B. sapa; C. carpio; L. aspius; L. idus; V. vimba | JX986899; JX986900; KR149529; KR149535; KR149536; KR271088; JX986901; KR271090; KR271091 | Snails: Lymnaea stagnalis; Stagnicola palustris
Fishes: Silurus glanis |
Germany; Romania | [6, 7, 10] |
| H2 | ABD12; BSD4; LAD5 | A. brama; B. sapa; L. aspius | JX986897; KR149534; KR149533; KR149532; KR149530; JX986898; KR149541; KR271093; JX986896 | Snails: Lymnaea stagnalis; Stagnicola palustris
Fishes: Cyprinus carpio Birds: Larus cachinnans |
Czech Republic; Germany; Romania | [6, 7, 10] |
| H4 | BBD9; GSD; LAD9 | B. bjoerkna; G. schraetsor; L. aspius | KR149546 | Snails: Stagnicola palustris | Germany | [10] |
| H5 | LAD7; LAD8 | L. aspius | JX986902; JX986903 | Fishes: Gasterosteus aculeatus | Germany | [7] |
| H7 | BBD5 | B. bjoerkna | KR149542 | Snails: Stagnicola palustris | Germany | [10] |
| H10 | BBD7 | B. bjoerkna | JX986907 | Snails: Lymnaea stagnalis | Germany | [7] |
| H12 | LID2 | L. idus | KR149531 | Snails: Lymnaea stagnalis | Germany | [10] |
| H15 | ABD14 | A. brama | KR149537 | Snails: Stagnicola palustris | Germany | [10] |
| ‘Diplostomum mergi Lineage 2’ | ||||||
| H1 | AAD3; AAD4; AAD5; AAD6; ABD17; BBD12; BBD13; CND3; VVD6 | A. alburnus; A. brama; B. bjoerkna; C. nasus; V. vimba | JX986874; JX986875; JX986876; KR149522; KR149521; KR149520; KR149518; KR149517; KR149515; KR149514 | Snails: Radix auricularia | Germany | [7, 10] |
| H2 | AAD1; ABD15; ABD18 | A. alburnus; A. brama | KR149523; KR149519; KR149516 | Snails: Radix auricularia | Germany | [10] |
| H7 | BSD5 | B. sapa | KR271082 | Fishes: Abramis brama | China | [6] |
aSee Table 2 for details
Of the 15 haplotypes of D. pseudospathaceum, 8 were shared with previously reported isolates, predominantly from the first intermediate hosts, Lymnaea stagnalis (L.) and Stagnicola palustris (Müller), from the Czech Republic, Germany and Romania [6, 7, 10]; among these, a single haplotype (H2) was shared between isolates from all three hosts in the species life-cycle (Table 3). Finally, three haplotypes of ‘D. mergi Lineage 2’ were shared with isolates from the snail host R. auricularia in Germany (H1 and H2) and one with a metacercaria from A. brama in China (H7, see Table 3).
The cox1 haplotype networks for D. spathaceum and D. pseudospathaceum, generated by statistical parsimony analysis, are presented in Figs. 3 and 4, respectively. For both species, haplotypes identified in the present material were sampled from 9 fish host species and there was no indication of genetic structuring associated with the host. The ancestral haplotype (H1) of D. spathaceum was recovered as unique and represented by isolates from 3 cyprinid hosts (A. brama, R. rutilus and V. vimba). Two other haplotypes (H2 and H3) were shared by isolates from 3 fish hosts each (A. brama, L. aspius and R. pigus and A. brama, R. pigus and S. glanis, respectively) (Fig. 3a). The cyprinid A. brama was the host with the largest haplotype diversity (8 haplotypes; 2 unique).
Fig. 3.
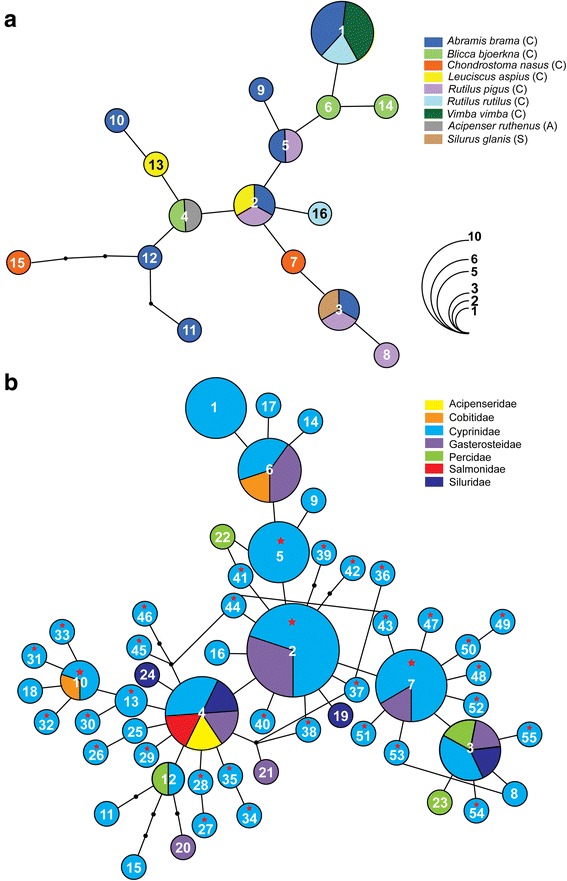
Haplotype networks for Diplostomum spathaceum: (a) based on the novel cox1 sequences from metacercarial isolates sampled from nine fish species in the River Danube; (b) based on all currently published cox1 sequences from metacercarial isolates sampled from fishes in Europe and Asia. Numbers indicate the haplotype code number (see Table 2 and Additional file 2: Table S2 for details). Black dots represent inferred unsampled intermediate haplotypes and connective lines represent one mutational step. Pie chart size is proportional to the number of isolates sharing a haplotype; haplotype frequency is indicated by colourless semicircles. Hosts reported in this study (a) and host families (b) are colour-indicated; stars indicate haplotypes recovered in Asia. Abbreviations: A, Acipenseridae; C, Cyprinidae; S, Siluridae
Fig. 4.
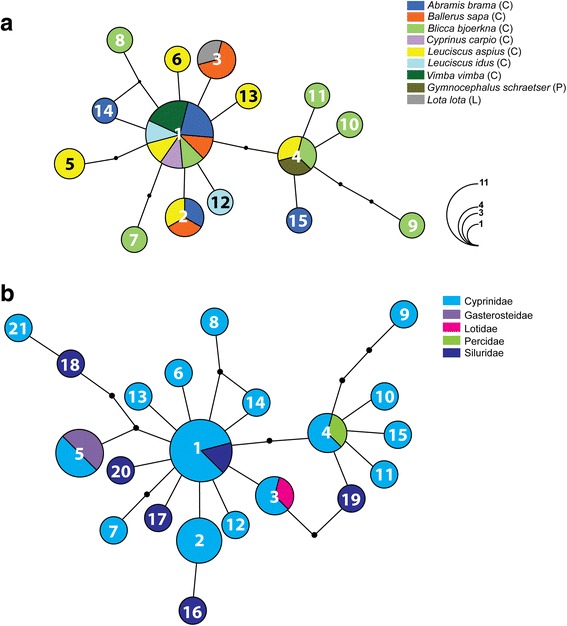
Haplotype networks for Diplostomum pseudospathaceum: (a) based on the novel cox1 sequences from metacercarial isolates sampled from nine fish species in the River Danube; (b) based on all currently published cox1 sequences from metacercarial isolates sampled from fishes in Europe. Numbers indicate the haplotype code number (see Table 2 and Additional file 2: Table S2 for details). Black dots represent inferred unsampled intermediate haplotypes and connective lines represent one mutational step. Pie chart size is proportional to the number of isolates sharing a haplotype; haplotype frequency is indicated by colourless semicircles. Hosts reported in this study (a) and host families (b) are colour-indicated. Abbreviations: C, Cyprinidae; L, Lotidae; P, Percidae
Figure 3b illustrates a haplotype network including all available sequence data for D. spathaceum from fish hosts in Europe and Asia. A total of 68 sequences was added for isolates from 12 fish species of five families: Cyprinidae (7 species; Locke et al. [6], Pérez-del-Olmo et al. [3]); Gasterosteidae (1 species; Georgieva et al. [7], Blasco-Costa et al. [9]); Cobitidae (1 species; Pérez-del-Olmo et al. [3]); Percidae (1 species; Locke et al. [6]); Salmonidae (1 species; Blasco-Costa et al. [9]) and Siluridae (1 species; Locke et al. [6]) (see Additional file 2: Table S2 for details). This expanded dataset comprising 94 sequences (trimmed to 402 nt) for isolates from 17 fish host species of 7 families revealed a much higher haplotype diversity (55 haplotypes) and a generally similar pattern for the most common haplotypes. However, a large number of haplotypes were represented by singletons (45 haplotypes: H8, H9, H11, H14-H55, see Additional file 2: Table S2) and H2 was the most common haplotype in the expanded network. A total of 30 haplotypes was identified in isolates sampled recently in China (n = 4) and Iraq (n = 26) by Locke et al. [6], and five haplotypes (H2, H5, H7, H10 and H13) were shared by isolates from Europe and Asia (Fig. 3b; Table 3). Notably, three of the five major haplotypes (H2-H4) recovered from different host species in the River Danube (Fig. 3a) also exhibited low host-specificity at the level of host family (associated with fish hosts of 2–5 families, see Fig. 3b) whereas haplotypes H1 and H5 appear to be restricted to the Cyprinidae based on the currently available data.
Diplostomum pseudospathaceum exhibited a marked contrast in haplotype network structure (star-shaped network, indicative of range expansion, see Fig. 4a) compared to the more complex network for D. spathaceum. The ancestral haplotype (H1) was shared among isolates from 7 of the 9 fish hosts (all cyprinids). The largest haplotype diversity was also found in cyprinid fishes: B. bjoerkna (7 haplotypes; 3 unique) followed by L. aspius (6 haplotypes, 2 unique). The haplotype network, including all available sequence data for D. pseudospathaceum from fish hosts in Europe (Fig. 4b) (12 host species of 5 families), includes 11 additional sequences for isolates from 3 fish species of 3 families: Cyprinidae (2 species; Locke et al. [6]); Gasterosteidae (1 species; Georgieva et al. [7]); and Siluridae (1 species; Locke et al. [6]) (see Additional file 2: Table S2 for details). This resulted in adding 6 new haplotypes (all singletons) to the dataset (41 sequences, trimmed to 402 nt; 21 haplotypes, see Additional file 2: Table S2). The haplotype network (Fig. 4b) closely resembled that for fishes sampled in the River Danube (Fig. 4a). Three of the four haplotypes identified in isolates from different fish species in the River Danube were also recovered in non-cyprinid fishes (Fig. 4b) (H1: Siluridae; H3: Lotidae; and H4: Percidae) and one haplotype (H5) was also identified in isolates from G. aculeatus (Gasterosteidae) (Georgieva et al. [7]).
To aid further exploration of species boundaries among the most widespread lens-infecting Diplostomum spp., the nad3 gene was selected based on its lower level of sequence conservation (83.3%) compared with the ‘barcode’ region of the cox1 gene (90.6%) (see Brabec et al. [25]). A total of 30 complete nad3 sequences (357 nt) were generated for the three species identified based on the cox1 gene subsampling (10 isolates per species; see Table 2 for details). NJ analysis of the nad3 dataset depicted three distinct well-supported monophyletic clades corresponding to the cox1 lineages (Fig. 5). The levels of the interspecific divergence for the nad3 gene was distinctly higher with minimum p-distance values well above the maximum values for cox1 (14.6–15.7 vs 9–11.2%) (Table 4). It is worth noting that the use of the newly designed primers resulted in successful amplification of nad3 in the distantly related lineage of the “D. mergi” complex of cryptic species.
Fig. 5.
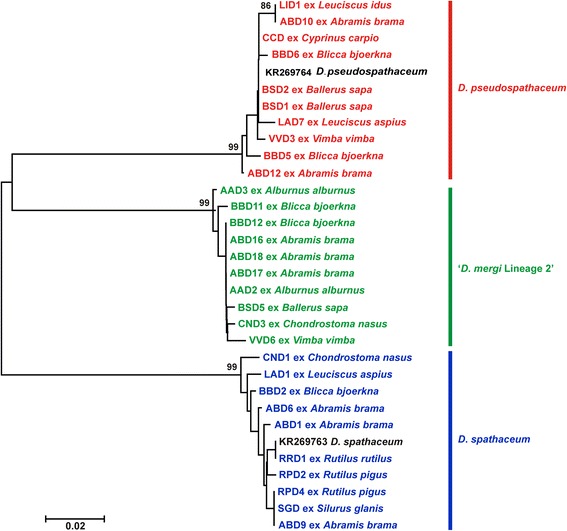
Neighbour-joining (NJ) phylogram for Diplostomum spp. reconstructed using 30 newly generated and two nad3 sequences retrieved from GenBank. The scale-bar indicates the expected number of substitutions per site. Codes for the newly sequenced isolates are provided in Table 2
Table 4.
Levels of divergence (p-distance in %) for cox1 and nad3 gene sequences in interspecific comparisons of Diplostomum spp.
| Species comparison | cox1 | nad3 |
|---|---|---|
| D. pseudospathaceum vs D. spathaceum | 9.0–10.7 | 15.7–17.4 |
| D. spathaceum vs ‘D. mergi Lineage 2’ | 10.0–11.7 | 15.4–16.8 |
| D. pseudospathaceum vs ‘D. mergi Lineage 2’ | 11.2–12.9 | 14.6–16.2 |
Descriptions of the molecular voucher material
Comparisons based on live metacercariae of the most frequent species in this study, D. spathaceum, D. pseudospathaceum and ‘D. mergi Lineage 2’ revealed that metacercariae of D. spathaceum exhibit the highest mean values for the width of the body, the length of the hindbody, and the size of the oral sucker, pseudosuckers and pharynx. Live metacercariae of D. pseudospathaceum were characterised by the lowest mean values for the size of the body, pseudosuckers and holdfast organ whereas those of ‘D. mergi Lineage 2’ exhibited the highest mean values for the length of the body and the size of the ventral sucker and holdfast organ. Surprisingly, fixed metacercariae of ‘D. mergi Lineage 2’ demonstrated the highest mean values for the size of the body, pseudosuckers, ventral sucker, holdfast organ and hindbody whereas the dimensions of specimens of D. spathaceum and D. pseudospathaceum were rather similar (see Tables 5, 6). We have therefore provided morphological and morphometric characterisation based on both live and fixed material.
Table 5.
Comparative metrical data for metacercariae of Diplostomum spathaceum
| Host Source |
Multiple hostsa
Present study |
Gasterosteus aculeatus L.; Salvelinus alpinus (L.) Faltýnková et al. [16] |
Cyprinus carpio L. Pérez-del-Olmo et al. [3] |
|||||
|---|---|---|---|---|---|---|---|---|
| Fixed | Live | Fixed | Fixed | |||||
| Variable | Range (n = 21) | Mean | Range | Mean | Range | Mean | Range | Mean |
| BL | 288–415 | 346 | 360–570 | 498 | 262–574 | 376 | 277–453 | 376 |
| BW | 241–333 | 288 | 252–332 | 286 | 171–313 | 235 | 198–295 | 248 |
| HL | 17 | 17 | 36–80 | 53 | 22–67 | 41 | 10–26 | 16 |
| PSL | 46–61 | 53 | – | – | 35–40 | 37 | 44–55 | 48 |
| PSW | 24–36 | 29 | – | – | – | – | 22–30 | 26 |
| OSL | 40–54 | 47 | 44–65 | 57 | 44–64 | 52 | 40–57 | 45 |
| OSW | 37–52 | 46 | 44–72 | 60 | 41–72 | 50 | 36–41 | 39 |
| PHL | 30–42 | 38 | 36–51 | 42 | 29–45 | 35 | 29–43 | 37 |
| PHW | 16–26 | 21 | 20–32 | 26 | 16–19 | 17 | 19–26 | 23 |
| VSL | 38–51 | 45 | 35–55 | 45 | 40–56 | 49 | 30–43 | 38 |
| VSW | 48–61 | 54 | 38–62 | 50 | 34–53 | 43 | 33–48 | 43 |
| AVS | 135–248 | 181 | – | – | – | – | – | – |
| HOL | 67–99 | 84 | 78–131 | 104 | 72–82 | 77 | 63–89 | 75 |
| HOW | 92–130 | 112 | 83–181 | 131 | 63–95 | 81 | 59–90 | 80 |
Abbreviations: BL body length, BW body width, HL hindbody length, PSL pseudosucker length, PSW pseudosucker width, OSL oral sucker length, OSW oral sucker width, PHL pharynx length, PHW pharynx width, VSL ventral sucker length, VSW ventral sucker width, AVS distance from anterior extremity of body to ventral sucker, HOL holdfast organ length, HOW holdfast organ width
a Acipenser ruthenus L.; Abramis brama (L.); Blicca bjoerkna (L.); Chondrostoma nasus (L.); Leuciscus aspius (L.); Rutilus pigus (Lacépède); Rutilus rutilus (L.); Vimba vimba (L.); Silurus glanis L.
Table 6.
Comparative metrical data for metacercariae of Diplostomum spp.
| Species | Diplostomum pseudospathaceum | Diplostomum pseudospathaceum | ‘Diplostomum mergi Lineage 2’ | Diplostomum sp. A | Diplostomum sp. B | Diplostomum sp. C | |||
|---|---|---|---|---|---|---|---|---|---|
| Host | Multiple hostsa | Cyprinus carpio L. | Multiple hostsb | Blicca bjoerkna (L.) | Carassius gibelio (Bloch) | Rutilus rutilus (L.) | |||
| Source | Present study | Niewiadomska [26] | Present study | Present study | Present study | Present study | |||
| Fixed | Fixed | Fixed | Fixed | Fixed | Fixed | ||||
| Variable | Range (n = 24) | Mean | Range | Mean | Range (n = 18) | Mean | n = 1 | n = 1 | n = 1 |
| BL | 288–447 | 364 | 347–458 | 381 | 362–485 | 420 | 338 | 426 | 381 |
| BW | 234–301 | 264 | 162–296 | 201 | 242–338 | 287 | 242 | 304 | 278 |
| HL | 19–19 | 19 | – | – | 14–45 | 26 | 20 | 19 | 16 |
| PSL | 40–65 | 52 | – | – | 52–68 | 60 | 47–52 | 56–58 | 61–67 |
| PSW | 25–35 | 30 | – | – | 31–36 | 34 | – | – | – |
| OSL | 39–56 | 47 | 42–52 | 45.8 | 41–53 | 47 | 37 | 46 | 47 |
| OSW | 36–53 | 44 | 30–51 | 37.7 | 34–49 | 43 | 44 | 41 | 47 |
| PHL | 32–45 | 38 | 28–35 | 31.8 | 30–45 | 38 | 30 | 41 | 30 |
| PHW | 19–25 | 21 | 17–30 | 20.4 | 19–23 | 22 | 20 | 22 | – |
| VSL | 33–53 | 42 | 34–42 | 38.9 | 40–62 | 51 | 51 | 51 | 43 |
| VSW | 43–56 | 51 | 35–51 | 42.2 | 49–70 | 61 | 64 | 59 | 49 |
| AVS | 158–243 | 191 | – | – | 174–261 | 208 | 143 | 215 | 174 |
| HOL | 68–96 | 82 | 62–81 | 67.5 | 95–115 | 104 | 65 | 115 | – |
| HOW | 79–126 | 99 | 54–76 | 61.7 | 102–187 | 136 | 106 | 136 | – |
Abbreviations: BL body length, BW body width, HL hindbody length, PSL pseudosucker length, PSW pseudosucker width, OSL oral sucker length, OSW oral sucker width, PHL pharynx length, PHW pharynx width, VSL ventral sucker length, VSW ventral sucker width, AVS distance from anterior extremity of body to ventral sucker, HOL holdfast organ length, HOW holdfast organ width
a Abramis brama (L.); Ballerus sapa (Pallas); Blicca bjoerkna (L.); Cyprinus carpio L.; Leuciscus aspius (L.); L. idus (L.); Vimba vimba (L.); Lota lota (L.); Gymnocephalus schraetser (L.)
b Abramis brama (L.); Alburnus alburnus (L.); Ballerus sapa (Pallas); Blicca bjoerkna (L.); Chondrostoma nasus (L.); Vimba vimba (L.)
Unfortunately, the single metacercariae of Diplostomum sp. A, Diplostomum sp. B and Diplostomum sp. C were fixed in the field and their descriptions are based on fixed material. Nevertheless, comparisons based on fixed metacercariae of the six forms recovered in the present study indicate that the sucker ratios and the number and relative size of the excretory concretions are the most prominent characters that can be used for their discrimination. Diplostomum sp. A and B exhibited the largest values for the sucker width ratio and were characterised by having large excretory concretions, similar to those observed in D. spathaceum. However, the metacercaria of Diplostomum sp. B is much larger (426 × 304 vs a mean of 346 × 288 μm for D. spathaceum) and the excretory concretions in the metacercaria of Diplostomum sp. A also appear larger than in the metacercaria of D. spathaceum (Fig. 6). The metacercaria of Diplostomum sp. C can be distinguished from the other five forms in having the largest number of excretory concretions (482 vs a maximum of 254, 360, 440 in D. spathaceum, D. pseudospathaceum and ‘Diplostomum mergi Lineage 2’, respectively, and 154 and 261 in Diplostomum sp. A and Diplostomum sp. B, respectively) (see also Fig. 6).
Fig. 6.
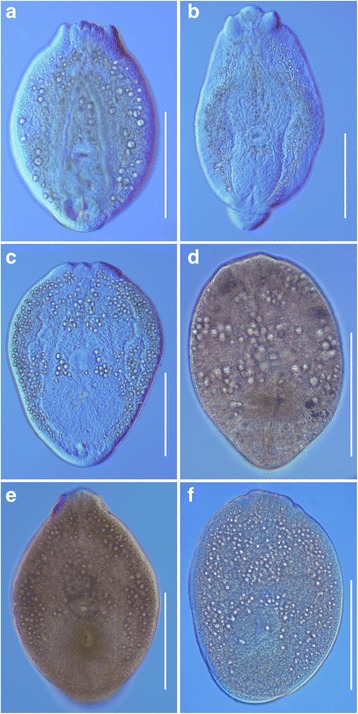
Metacercariae of Diplostomum spp. (a-c, live; d-f, fixed). a D. spathaceum from the eye lens of Rutilus pigus (hologenophore; GenBank KY653979 and KY654043). b D. pseudospathaceum from the eye lens of Abramis brama (hologenophore; GenBank KY653989 and KY654048). c ‘D. mergi Lineage 2’ from the eye lens of Abramis brama (hologenophore; GenBank KY654020 and KY654059). d Diplostomum sp. A from the eye lens of Blicca bjoerkna (hologenophore; GenBank KY654034). e Diplostomum sp. B from the eye lens of Carassius gibelio (hologenophore; GenBank KY654035). f Diplostomum sp. C from the eye lens of Rutilus rutilus (hologenophore; GenBank KY654036). Scale-bars: 200 μm
Diplostomum spathaceum (Rudolphi, 1819)
Hosts : Acipenser ruthenus L. (Chondrostei: Acipenseridae), Abramis brama (L.), Blicca bjoerkna (L.), Chondrostoma nasus (L.), Leuciscus aspius (L.), Rutilus pigus (Lacépède), Rutilus rutilus (L.), Vimba vimba (L.) (Teleostei: Cyprinidae); Silurus glanis L. (Teleostei: Siluridae).
Prevalence : A. ruthenus: 1/1 (Slovakia, S); A. brama: 75% (29/40, S); B. bjoerkna: 1/5 (Hungary, H), 1/8 (S); C. nasus: 2/7 (H), 1/5 (S); L. aspius: 3/6 (H), 1/4 (S); R. pigus: 2/3 (S); R. rutilus: 1/1 (H), 2/8 (S); V. vimba: 2/4 (S); S. glanis: 1/1 (S).
Representative DNA sequences : KY653961–KY653986 (cox1); KY654037–KY654046 (nad3).
Description
[Based on 20 live metacercariae. Metrical data for fixed material are provided in Table 5; Fig. 6a.] Body oval, 349–601 × 265–442 (474 × 341), with maximum width just anterior to ventral sucker. Oral sucker elongate-oval, 51–80 × 46–69 (62 × 57). Pseudosuckers strongly muscular, elongate-oval, 58–90 × 31–51 (76 × 41). Oral opening terminal; prepharynx absent; pharynx elongate-oval, 32–47 × 20–39 (40 × 28); oesophagus short, bifurcates close posterior to pharynx; caeca long, narrow, reach posterior to holdfast organ. Ventral sucker transversely oval, 34–64 × 38–66 (50 × 56), smaller or equal to oral sucker (sucker width ratio 1:0.83–1.19 (1:1.01), posterior to mid-body length. Distance from anterior extremity of body to ventral sucker 191–365 (262). Holdfast organ relatively small, transversely oval, bipartite, contiguous with ventral sucker, 71–153 × 78–180 (108 × 124). Excretory vesicle small, V-shaped; reserve excretory system of diplostomid type; excretory concretions relatively large, 171–346 (246) in number, grouped into 2 lateral extracaecal [106–254 (179) excretory concretions] and 1 median [39–109 (67) excretory concretions] fields. Hindbody 34–59 (44) long.
Remarks
The morphology of the present metacercariae of D. spathaceum (Fig. 6a) agrees with the descriptions of metacercariae of D. spathaceum by Faltýnková et al. [16] and Pérez-del-Olmo et al. [3] with some variations. The present live specimens differ from the live material described by Faltýnková et al. [16] by having on average shorter and wider body, somewhat larger pseudosuckers and ventral sucker, narrower holdfast organ and a different sucker width ratio (mean 1:1.01 vs 1:0.84) (also see Table 5). Similarly, the present fixed specimens differ from the fixed material described by Faltýnková et al. [16] and Pérez-del-Olmo et al. [3] in having on average shorter and wider body and larger pseudosuckers and ventral sucker and a distinctly wider holdfast organ. The number of excretory concretions in D. spathaceum falls within the range provided by Shigin [1] but the mean is distinctly higher: 171–346 (246) vs 117–401 (143).
Our study adds 8 fish species to the hosts of D. spathaceum in Europe confirmed by molecular evidence. Previous records include Gasterosteus aculeatus L. in Germany [7]; G. aculetaus and Salvelinus alpinus (L.) in Iceland [9]; Misgurnus anguillicaudatus (Cantor), S. glanis and P. willkommii in Spain [3]; and Perca fluviatilis L. in Italy and S. glanis in Romania [6]. Among these hosts, cyprinids predominate (7 species) and are more diverse; a very high prevalence (75%) was also registered in a cyprinid (A. brama; present study).
Diplostomum pseudospathaceum Niewiadomska, 1984
Hosts : Abramis brama (L.), Ballerus sapa (Pallas), Blicca bjoerkna (L.), Cyprinus carpio L., Leuciscus aspius (L.), L. idus (L.), Vimba vimba (L.) (Teleostei: Cyprinidae); Lota lota (L.) (Teleostei: Lotidae), Gymnocephalus schraetser (L.) (Teleostei: Percidae).
Prevalence : A. brama: 50% (20/40, S); B. sapa: 1/1 (S); B. bjoerkna: 3/5 (H), 5/8 (S); C. carpio: 1/3 (S); L. aspius: 2/5 (H), 3/4 (S); L. idus: 1/1 (S); V. vimba: 1/5 (H), 1/4 (S); L. lota: 1/2 (H); G. schraetser: 1/5 (H).
Representative DNA sequences : KY653987–KY654016 (cox1); KY654047–KY654056 (nad3).
Description
[Based on 18 live metacercariae. Metrical data for fixed material are provided in Table 6; Fig. 6b.] Body elongate-oval, 325–490 × 234–410 (406 × 306), with maximum width just anterior to ventral sucker. Oral sucker elongate-oval, 48–65 × 43–58 (55 × 50). Pseudosuckers strongly muscular, elongate-oval, 42–73 × 26–43 (54 × 33). Oral opening terminal; prepharynx short or absent; pharynx elongate-oval, 31–52 × 19–37 (38 × 24); oesophagus short, bifurcates close posterior to pharynx; caeca long, narrow, reach posterior to holdfast organ. Ventral sucker transversely oval, 37–56 × 45–66 (47 × 55), smaller or larger than oral sucker [sucker width ratio 1:0.93–1.35 (1:1.11)], slightly posterior to mid-body length. Distance from anterior extremity of body to ventral sucker 177–279 (216). Holdfast organ relatively small, transversely oval, bipartite, contiguous with ventral sucker, 69–111 × 88–170 (90 × 115). Excretory vesicle small, V-shaped; reserve excretory system of diplostomid type; excretory concretions small, 185–360 (241) in number, grouped into 2 lateral extracaecal [122–244 (164) excretory concretions] and 1 median [57–116 (77) excretory concretions] fields. Hindbody 19–47 (31) long.
Remarks
The present metacercariae were identified as D. pseudospathaceum based on molecular data. The metrical data for the present material (fixed specimens) exhibit overlapping ranges with the data for experimentally developed metacercariae of D. pseudospathaceum described by Niewiadomska [26] but differ in the possesion of on average shorter and wider body, wider suckers and distinctly wider holdfast organ (Table 6). Shigin [1] reported 151–309 (234) excretory concretions for D. pseudospathaceum (as D. chromatophorum); these values agree very well with our observations, i.e. 185–360 (241).
Our study reports nine fish hosts for D. pseudospathaceum in Europe confrmed by sequencing. Previous molecularly identified records in fishes are few: G. aculeatus in Germany [7] and C. carpio and S. glanis in Romania [6]. Among the hosts studied here, cyprinids predominated (7 species) with a high prevalence in A. brama (50%).
‘Diplostomum mergi Lineage 2’ sensu Georgieva et al. (2013)
Hosts : Abramis brama (L.), Alburnus alburnus (L.), Ballerus sapa (Pallas), Blicca bjoerkna (L.), Chondrostoma nasus (L.), Vimba vimba (L.) (Teleostei: Cyprinidae).
Prevalence : A. brama: 58% (23/40, S); A. alburnus: 3/5 (H), 1/3 (S); B. sapa: 1/8 (H), 1/1 (S); B. bjoerkna: 1/5 (H), 2/8 (S); C. nasus: 1/4 (S); V. vimba: 4/5 (H).
Representative DNA sequences : KY654017–KY654033 (cox1); KY654057–KY654066 (nad3).
Description
[Based on 8 live metacercariae. Metrical data for fixed material are provided in Table 6; Fig. 6c.] Body elongate-oval, 456–529 × 256–382 (490 × 328), with maximum width just anterior to ventral sucker. Oral sucker subspherical, 48–57 × 46–61 (52 × 53). Pseudosuckers elongate-oval, 69–73 × 32–40 (67 × 36). Oral opening terminal; prepharynx short; pharynx elongate-oval, 29–40 × 23–34 (35 × 26); oesophagus short, bifurcates close posterior to pharynx; caeca long, narrow, reach posterior to holdfast organ. Ventral sucker transversely oval, 54–61 × 64–71 (57 × 67), distinctly larger than oral sucker (sucker width ratio 1:1.14–1.31 (1:1.25), at mid-body length. Distance from anterior extremity of body to ventral sucker 205–265 (237). Holdfast organ large, transversely oval, bipartite, contiguous with ventral sucker, 120–158 × 152–205 (134 × 174). Excretory vesicle small, V-shaped; reserve excretory system of diplostomid type; excretory concretions predominantly medium-sized, 316–440 (372) in number, grouped into 2 lateral extracaecal [229–360 (285) excretory concretions] and 1 median [58–122 (87) excretory concretions] fields.
Remarks
Shigin [1] suggested that the large size and number [702–854 (772)] of the excretory concretions in the metacercariae of D. mergi (sensu lato) clearly distinguish this species from all lens-infecting forms. However, molecular analyses by Georgieva et al. [7] and Selbach et al. [10] revealed the presence of at least four cryptic species within this complex. The present material is characterised by a distinctly smaller number of excretory concretions, i.e. 316–443 (372) thus adding morphological evidence to the genetic differentiation of ‘D. mergi Lineage 2’.
To date, ‘D. mergi Lineage 2’ has only been recorded/sequenced in Europe from snails in Germany: three cercarial isolates from R. auricularia from Hengsteysee [7] and 13 cercarial isolates from the same host in Baldeneysee, Hengsteysee and Sorpetalsperre [10]. Our study, therefore partially elucidates the life-cycle of this species, providing the first data for the second intermediate hosts in Europe comprising six new host records, all cyprinids. Similarly to the other two Diplostomum spp. reported here, high prevalence of infection (58%) was detected in A. brama. It is worth noting that a single metacercarial isolate has been sequenced from A. brama in China [6].
Diplostomum sp. A
Host : Blicca bjoerkna (L.) (Teleostei: Cyprinidae).
Prevalence : 1/8 (Slovakia).
Representative DNA sequence : KY654034 (cox1).
Description
[Based on 1 fixed metacercaria; see also Table 6, Fig. 6d.] Body elongate-oval, 338 × 242, with maximum width at level of ventral sucker. Oral sucker transversely oval, 37 × 44. Pseudosuckers distinct, muscular, 47–52 long. Oral opening terminal; prepharynx absent; pharynx elongate-oval, 30 × 20; oesophagus short. Ventral sucker transversely oval, 51 × 64, larger than oral sucker (sucker width ratio 1:1.45), located at mid-body length. Distance from anterior extremity of body to ventral sucker 143. Holdfast organ small, transversely oval, bipartite, contiguous with ventral sucker, 65 × 106. Excretory vesicle small, V-shaped; reserve excretory system of diplostomid type; excretory concretions very large, 154 in number, grouped into 2 lateral extracaecal (107 excretory concretions) and 1 median (47 excretory concretions) fields. Hindbody 20 long.
Diplostomum sp. B
Host : Carassius gibelio (Bloch) (Teleostei: Cyprinidae).
Prevalence : 1/6 (Slovakia).
Representative DNA sequence : KY654035 (cox1).
Description
[Based on 1 fixed metacercaria; see also Table 6, Fig. 6e.] Body elongate-oval, 426 × 304, with maximum width at level of ventral sucker. Oral sucker elongate-oval, 46 × 41. Pseudosuckers muscular, 56–58 long. Oral opening terminal; prepharynx short; pharynx elongate-oval, 41 × 22; oesophagus short, bifurcates close posterior to pharynx; caeca long, narrow, reach posterior to holdfast organ. Ventral sucker transversely oval, 51 × 59, larger than oral sucker (sucker width ratio 1:1.44), located at mid-body length. Distance from anterior extremity of body to ventral sucker 215. Holdfast organ large, transversely oval, bipartite, contiguous with ventral sucker, 115 × 136. Excretory vesicle small, V-shaped; reserve excretory system of diplostomid type; excretory concretions predominantly large, 261 in number, grouped into 2 lateral extracaecal (168 excretory concretions) and 1 median (93 excretory concretions) fields. Hindbody 19 long.
Diplostomum sp. C
Host : Rutilus rutilus (L.) (Teleostei: Cyprinidae).
Prevalence : 1/8 (Slovakia).
Representative DNA sequence : KY654036 (cox1).
Description
[Based on 1 fixed metacercariae. Metrical data for the isolate are provided in Table 6; Fig. 6f.] Body oval, 381 × 278, with maximum width at level of ventral sucker. Oral sucker spherical, 47 × 47. Pseudosuckers strongly muscular, 61–67 long. Oral opening terminal; prepharynx short; pharynx 30 long. Ventral sucker transversely oval, 43 × 49, similar in size to oral sucker (sucker width ratio 1:1.04), located at mid-body length. Distance from anterior extremity of body to ventral sucker 174. Holdfast organ transversely oval, bipartite, contiguous with ventral sucker. Excretory vesicle small, V-shaped; reserve excretory system of diplostomid type; excretory concretions predominantly small, 482 in number, grouped into 2 lateral extracaecal (334 excretory concretions) and 1 median (148 excretory concretions) fields. Hindbody 16 long.
Discussion
Parasite diversity in fishes from the River Danube has been studied extensively in the past (see Moravec [27]). However, remarkably little is known about the actual species diversity of the metacercariae of the genus Diplostomum. These have been typically reported as D. spathaceum, without any morphological evidence confirming species identification, or left unidentified (see Moravec [27] for details of the records). Due to the failure in achieving species identification of the metacercariae based on morphology, this practice is observed in a number of recent ecological studies of fish parasites from the River Danube (e.g. [28–32]). Recently, a single cox1 sequence for D. pseudospathaceum has been published from S. glanis in the River Danube in Romania [6].
The present study is the first taxonomically broad screening of fish hosts to provide data on the diversity of Diplostomum spp. from the River Danube applying molecular identification methods. The analyses based on the newly generated and published cox1 sequences demonstrated the presence of three species/species-level genetic lineages of Diplostomum, i.e. D. spathaceum, D. pseudospathaceum and ‘D. mergi Lineage 2’, and three single isolates potentially representing distinct species, i.e. Diplostomum spp. A-C. Our approach ensured a refined taxonomic resolution and allowed an assessment of the host ranges of the three most frequent Diplostomum spp. and to partly elucidate the life-cycle of one species. The large number of isolates from a wide range of hosts examined led to the detection of the somewhat higher level of mean intraspecific divergence for D. spathaceum and ‘D. mergi Lineage 2’ compared with previous data: 0.82 vs 0.43% [7] and 0.53% [10], and 0.47 vs 0% [7] and 0.30% [10], respectively.
Our novel data for host ranges of D. spathaceum, D. pseudospathaceum and ‘D. mergi Lineage 2’, based on molecular identification of the metacercariae, indicate that the transmission of these species in the River Danube is primarily associated with cyprinid fishes as second intermediate hosts. Twelve out of fourteen cyprinid species were infected with at least one species of Diplostomum; the largest number of species/lineages (4 out of 6) was detected in B. bjoerkna. Diplostomum spathaceum was also found in A. ruthenus (Acipenseridae) and S. glanis (Siluridae) and D. pseudospathaceum was recovered in G. schraetser (Percidae) and Lota lota (Lotidae). All three species of Diplostomum exhibited remarkably high prevalence in A. brama, the most well-sampled species. Although the lack of infections with Diplostomum spp. in 12 out of the 28 species of fish examined may be due to the small sample sizes, infections were detected in a large number of similarly under-sampled species, i.e. the acipenserid A. ruthenus (D. spathaceum), the cyprinids A. alburnus (‘D. mergi Lineage 2’), B. sapa (D. pseudospathaceum and ‘D. mergi Lineage 2’), C. gibelio (Diplostomum sp. B), C. nasus (D. spathaceum and ‘D. mergi Lineage 2’), C. carpio (D. pseudospathaceum), L. aspius (D. spathaceum and D. pseudospathaceum), L. idus (D. pseudospathaceum), R. pigus (D. spathaceum), R. rutilus (D. spathaceum and Diplostomum sp. C), V. vimba (D. spathaceum, D. pseudospathaceum and ‘D. mergi Lineage 2’), the lotid L. lota (D. pseudospathaceum), the percid G. schraetser (D. pseudospathaceum) and the silurid S. glanis (D. spathaceum). These data indicate that the species/lineages reported here may parasitise a wide range of hosts. The lack of specific host-related pattern of genetic structuring, illustrated by the haplotype networks for D. spathaceum and D. pseudospathaceum, based on the novel data and the pattern of shared haplotypes with isolates from fish hosts of the Cobitidae, Gasterosteidae, Percidae, Salmonidae and Siluridae (detailed in Table 3), also tend to support this suggestion. Furthermore, the apparent lack of host-specificity for D. spathaceum and D. pseudospathaceum is confirmed by the wide host ranges (17 fish species of 7 families and 12 host species of 5 families, respectively) in the expanded datasets comprising the cox1 sequences available to date (Figs. 3b, 4b; Additional file 2: Table S2). The most common haplotypes exhibited low host-specificity at the level of both host species (our novel data) and host family (expanded datasets).
Regarding the geographical distribution, the present comparisons with all published sequences revealed haplotypes with a wide Palaearctic distribution for two of the species, reported from Iraq and China by Locke et al. [6], i.e. D. spathaceum (H2: Iraq, China; H5, H7 and H10: Iraq; H13: China); ‘D. mergi Lineage 2’ (H7: China); a number of haplotypes of D. spathaceum (n = 30) are currently known from Asia only (see Locke et al. [6]; Additional file 2: Table S2).
Our study represents the first record of ‘D. mergi Lineage 2’ in a fish host in Europe and is the first to provide a morphological description of the metacercaria. The new isolates clustered together, and exhibited additional shared haplotypes, with cercarial isolates sequenced by Georgieva et al. [7] and Selbach et al. [10]. Thus, the life-cycle of this lineage was partially elucidated using molecular data, with the pulmonate snail R. auricularia acting as the first intermediate host and six cyprinid fishes (A. alburnus, A. brama, B. bjoerkna, B. sapa, C. nasus and V. vimba) acting as second intermediate hosts. The cercaria of ‘D. mergi Lineage 2’ was described in detail by Selbach et al. [10] who differentiated it from the cercaria of D. mergi sensu Niewiadomska & Kiselienė, 1994 [33] by having furcae longer than the tail stem and by morphometry, and from the cercariae of the four species within the “D. mergi” species complex by five unique morphometric features (see Selbach et al. [10] for details). The present metacercariae exhibited markedly smaller number of excretory concretions in comparison with the metacercariae of D. mergi (sensu lato) (mean 372 vs 772; see [1]) and showed morphometric differences from the metacercariae of the other lens-infecting species, D. spathaceum and D. pseudospathaceum. These data, in association with the genetic evidence, support the distinct species status of ‘D. mergi Lineage 2’; however, formal description of the species would require the discovery of the adult stage. The distribution of this species-level genetic lineage is apparently wider, and not restricted to Europe, since Locke et al. [6] reported a single sequence from a metacercaria in the cyprinid A. brama from China. Further studies would add to our knowledge of haplotype diversity, host ranges and geographical distribution of this lineage.
Brabec et al. [25] characterised the complete mitochondrial genomes of the two closely related species, D. spathaceum and D. pseudospathaceum and carried out a comparative genome assessment. These authors revealed that the cox1 gene and its ‘barcode’ region, currently applied for molecular identification, represent the most conserved protein-coding regions of the mitochondrial genome of Diplostomum spp. and identified nad4 and nad5 genes as most promising molecular diagnostic markers. In the pilot nad gene sequencing carried out here, we opted for nad3 gene, a slightly more conserved in comparison to the nad4 and nad5 genes, because the identification based on cox1 revealed the presence of a lineage of the “D. mergi” species complex that was shown to be rather distant to the two sibling species studied by Brabec et al. [25] (e.g. [7, 10]). Our results indicate that the newly designed primers can be used for successful amplification of nad3 within the “D. mergi” complex and possibly in other distantly related lineages of Diplostomum; the markedly higher levels of interspecific divergence compared to cox1 indicate that the nad3 gene is a good candidate marker for multi-gene approaches to systematic estimates within the genus.
Conclusions
The first exploration of the species diversity and host ranges of Diplostomum spp., based on molecular and morphological evidence from a broad range of fish hosts in the River Danube (Hungary and Slovakia), revealed the presence of three species/species-level genetic lineages of Diplostomum, i.e. D. spathaceum, D. pseudospathaceum and ‘D. mergi Lineage 2’, and three single isolates potentially representing distinct species. The most frequently found Diplostomum spp. exhibited a low host-specificity, predominantly infecting a wide range of cyprinid fishes but also species of distant fish families such as the Acipenseridae, Lotidae, Percidae and Siluridae. Our study provided the first cox1 and nad3 sequences associated with a morphological characterisation for metacercariae of ‘D. mergi Lineage 2’ in a fish host in Europe and partially elucidated the life-cycle of this species using molecular data. The novel sequence data will advance further ecological studies on the distribution and host ranges of these important fish parasites in Europe.
Additional files
Summary data for the sequences from isolates of Diplostomum spp. isolates retrieved from the GenBank database and used in the phylogenetic analyses. (DOC 67 kb)
Summary data for the sequences for Diplostomum spathaceum and D. pseudospathaceum from metacercarial isolates used in the expanded haplotype networks. (DOCX 31 kb)
Acknowledgements
We are grateful to Jan Brabec and Roman Kuchta (Institute of Parasitology, Biology Centre of the Czech Academy of Sciences) and Tibor Eros (Balaton Limnological Institute, Hungarian Academy of Sciences) for their invaluable help during material collection. We thank the three anonymous reviewers for their constructive comments and suggestions.
Funding
This research was partially supported by the Czech Science Foundation, grants 15-14198S (SG and AK) and ECIP P505/12/G112 (OK); the Research & Development Operational Programme funded by the ERDF (code ITMS: 26220120022) (0.3) (MO). SG benefited from a postdoctoral fellowship of the Czech Academy of Sciences. This is contribution number 214 from the NWU-Water Research Group.
Availability of data and materials
The data supporting the conclusions of this article are included within the article and its additional files. The newly generated sequences are submitted to the GenBank database under the accession numbers KY653961–KY654066.
Authors’ contributions
SG and MO: obtained the samples, undertook the identification and morphological characterisation of the isolates. OK and SG: carried out the morphological analysis, sequencing, performed the phylogenetic analyses and drafted the MS. AK: conceived and coordinated the study, discussed the results and helped draft the manuscript. All authors read and approved the final manuscript.
Ethics approval
All applicable institutional, national and international guidelines for the care and use of animals were followed.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13071-017-2518-5) contains supplementary material, which is available to authorized users.
Contributor Information
Olena Kudlai, Email: olena.kudlai@gmail.com.
Mikuláš Oros, Email: oros@saske.sk.
Aneta Kostadinova, Email: aneta.kostadinova@gmail.com.
Simona Georgieva, Email: simona.georgieva@gmail.com.
References
- 1.Shigin AA. [Trematode fauna of the USSR. Genus Diplostomum. Metacercariae.] Moscow: Nauka; 1986 (In Russian).
- 2.Chappell LH, Hardie LJ, Secombes CJ. Diplostomiasis: the disease and host-parasite interactions. In: Pike AW, Lewis JW, editors. Parasitic diseases of fish. Tresaith, Dyfed, UK: Samara Publishing Ltd.; 1994. pp. 59–86. [Google Scholar]
- 3.Pérez-del-Olmo A, Georgieva S, Pula HJ, Kostadinova A. Molecular and morphological evidence for three species of Diplostomum (Digenea: Diplostomidae), parasites of fishes and fish-eating birds in Spain. Parasit Vectors. 2014;7:502. doi: 10.1186/s13071-014-0502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moszczynska A, Locke SA, McLaughlin JD, Marcogliese DJ, Crease TJ. Development of primers for the mitochondrial cytochrome c oxidase I gene in digenetic trematodes illustrates the challenge of barcoding parasitic helminths. Mol Ecol Resour. 2009;9:75–82. doi: 10.1111/j.1755-0998.2009.02634.x. [DOI] [PubMed] [Google Scholar]
- 5.Locke SA, McLaughlin JD, Dayanandan S, Marcogliese DJ. Diversity, specificity and evidence of hybridization in Diplostomum spp. metacercariae in freshwater fishes is revealed by DNA barcodes and ITS sequences. Int J Parasitol. 2010;40:333–343. doi: 10.1016/j.ijpara.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Locke SA, Al-Nasiri FS, Caffara M, Drago F, Kalbe M, Lapierre AR, et al. Diversity, specificity and speciation in larval Diplostomidae (Platyhelminthes: Digenea) in the eyes of freshwater fish, as revealed by DNA barcodes. Int J Parasitol. 2015;45:841–855. doi: 10.1016/j.ijpara.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Georgieva S, Soldánová M, Pérez-del-Olmo A, Dangel RD, Sitko J, Sures B, et al. Molecular prospecting for European Diplostomum (Digenea: Diplostomidae) reveals cryptic diversity. Int J Parasitol. 2013;43:57–72. doi: 10.1016/j.ijpara.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 8.Behrmann-Godel J. Parasite identification, succession and infection pathways in perch fry (Perca fluviatilis): new insights through a combined morphological and genetic approach. Parasitology. 2013;140:509–520. doi: 10.1017/S0031182012001989. [DOI] [PubMed] [Google Scholar]
- 9.Blasco-Costa I, Faltýnková A, Georgieva S, Skírnisson K, Scholz T, Kostadinova A. Fish pathogens near the Arctic circle: molecular, morphological and ecological evidence for unexpected diversity of Diplostomum (Digenea: Diplostomidae) in Iceland. Int J Parasitol. 2014;44:703–715. doi: 10.1016/j.ijpara.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Selbach C, Soldánová M, Georgieva S, Kostadinova A, Sures B. Integrative taxonomic approach to the cryptic diversity of Diplostomum spp. in lymnaeid snails from Europe with a focus on the ‘Diplostomum mergi’ species complex. Parasit Vectors. 2015;8:300. doi: 10.1186/s13071-015-0904-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn JA, Kristoffersen R, Knudsen R, Jakobsen J, Marcogliese DJ, Locke SA, et al. Parasite communities of two three-spined stickleback populations in subarctic Norway - effects of a small spatial-scale host introduction. Parasitol Res. 2015;114:1327–1339. doi: 10.1007/s00436-015-4309-2. [DOI] [PubMed] [Google Scholar]
- 12.Soldánová M, Georgieva S, Roháčová J, Knudsen R, Kuhn JA, Henriksen EH, et al. Molecular analyses reveal high species diversity of trematodes in a sub-Arctic lake. Int J Parasitol. 2017;47:327–345. doi: 10.1016/j.ijpara.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Locke SA, McLaughlin JD, Marcogliese DJ. Predicting the similarity of parasite communities in freshwater fishes using the phylogeny, ecology and proximity of hosts. Oikos. 2013;122:73–83. doi: 10.1111/j.1600-0706.2012.20211.x. [DOI] [Google Scholar]
- 14.Désilets HD, Locke SA, McLaughlin JD, Marcogliese DJ. Community structure of Diplostomum spp. (Digenea: Diplostomidae) in eyes of fish: main determinants and potential interspecific interactions. Int J Parasitol. 2013;43:929–939. doi: 10.1016/j.ijpara.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Galazzo DE, Dayanandan S, Marcogliese DJ, McLaughlin JD. Molecular systematics of some north American species of Diplostomum (Digenea) based on rDNA-sequence data and comparisons with European congeners. Can J Zool. 2002;80:2207–2217. doi: 10.1139/z02-198. [DOI] [Google Scholar]
- 16.Faltýnková A, Georgieva S, Kostadinova A, Blasco-Costa I, Scholz T, Skírnisson K. Diplostomum von Nordmann, 1832 (Digenea: Diplostomidae) in the sub-Arctic: descriptions of the larval stages of six species discovered recently in Iceland. Syst Parasitol. 2014;89:195–213. [DOI] [PubMed]
- 17.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysisversion 6.0. Mol Biol Evol. 2013;30:2725–9. [DOI] [PMC free article] [PubMed]
- 18.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by thecoalescent and other methods. Bioinformatics. 2003;19:2496–7. [DOI] [PubMed]
- 19.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 20.Templeton AR, Crandall KA, Sing CFA. Cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Telford MJ, Herniou EA, Russell RB, Littlewood DTJ. Changes in mitochondrial genetic codes as phylogenetic characters: two examples from the flatworms. Proc Natl Acad Sci USA. 2000;97:11359–64. [DOI] [PMC free article] [PubMed]
- 22.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shigin AA. [Trematodes of the fauna of Russia and neighbouring regions. Genus Diplostomum. Adults]. Moscow: Nauka; 1993 (In Russian).
- 25.Brabec J, Kostadinova A, Scholz T, Littlewood DT. Complete mitochondrial genomes and nuclear ribosomal RNA operons of two species of Diplostomum (Platyhelminthes: Trematoda): a molecular resource for taxonomy and molecular epidemiology of important fish pathogens. Parasit Vectors. 2015;8:336. doi: 10.1186/s13071-015-0949-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niewiadomska K. Verification of the life-cycles of Diplostomum spathaceum (Rudolphi, 1819) and D. pseudospathaceum Niewiadomska, 1984 (Trematoda, Diplostomidae) Syst Parasitol. 1986;8:23–31. doi: 10.1007/BF00010306. [DOI] [Google Scholar]
- 27.Moravec F. Checklist of the metazoan parasites of fishes of the Czech Republic and the Slovak Republic (1873–2000) Prague: Academia; 2001. [Google Scholar]
- 28.Ondračková M, Dávidová M, Pečínková M, Blažek R, Gelnar M, Valová Z, et al. Metazoan parasites of Neogobius fishes in the Slovak section of the river Danube. J Appl Ichthyol. 2005;21:345–349. doi: 10.1111/j.1439-0426.2005.00682.x. [DOI] [Google Scholar]
- 29.Ondračková M, Slováčková I, Trichkova T, Polačik M, Jurajda P. Shoreline distribution and parasite infection of black-striped pipefish Syngnathus abaster Risso, 1827 in the lower river Danube. J Appl Ichthyol. 2012;28:590–596. doi: 10.1111/j.1439-0426.2012.01967.x. [DOI] [Google Scholar]
- 30.Müehlegger JM, Jirsa F, Konecny R, Frank C. Parasites of Apollonia melanostoma (Pallas 1814) and Neogobius kessleri (Guenther 1861) (Osteichthyes, Gobiidae) from the Danube River in Austria. J Helminthol. 2010;84:87–92. doi: 10.1017/S0022149X09990095. [DOI] [PubMed] [Google Scholar]
- 31.Jirsa F, Konecny R, Frank C, Sures B. The parasite community of the nase Chondrostoma nasus (L. 1758) from Austrian rivers. J Helminthol. 2011;85:255–262. doi: 10.1017/S0022149X10000490. [DOI] [PubMed] [Google Scholar]
- 32.Francová K, Ondračková M, Polačik M, Jurajda P. Parasite fauna of native and non-native populations of Neogobius melanostomus (Pallas, 1814) (Gobiidae) in the longitudinal profile of the Danube River. J Appl Ichthyol. 2011;27:879–886. doi: 10.1111/j.1439-0426.2010.01582.x. [DOI] [Google Scholar]
- 33.Niewiadomska K, Kiselienė V. Diplostomum cercariae (Digenea) in snails from Lithuania. II. Survey of species. Acta Zool Lituan. 1994;39:179–86.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary data for the sequences from isolates of Diplostomum spp. isolates retrieved from the GenBank database and used in the phylogenetic analyses. (DOC 67 kb)
Summary data for the sequences for Diplostomum spathaceum and D. pseudospathaceum from metacercarial isolates used in the expanded haplotype networks. (DOCX 31 kb)
Data Availability Statement
The data supporting the conclusions of this article are included within the article and its additional files. The newly generated sequences are submitted to the GenBank database under the accession numbers KY653961–KY654066.


