Abstract
BRAF V600E colorectal cancers (CRC) are insensitive to RAF inhibitor monotherapy due to feedback reactivation of receptor tyrosine kinase signaling. Combined RAF and EGFR inhibition exerts a therapeutic effect, but resistance invariably develops through undefined mechanisms. In this study, we determined that CRC progression specimens invariably harbored lesions in elements of the RAS-RAF-MEK-ERK pathway. Genetic amplification of wild-type RAS was a recurrent mechanism of resistance in CRC patients that was not seen in similarly resistant melanomas. We show that wild-type RAS amplification increases receptor tyrosine kinase-dependent activation of RAS more potently in CRC than in melanoma and causes resistance only in the former. Currently approved RAF inhibitors inhibit RAF monomers but not dimers. All the drug-resistant lesions we identified activate BRAF V600E dimerization directly or by elevating RAS-GTP. Overall, our results show that mechanisms of resistance converge on formation of RAF dimers and that inhibiting EGFR and RAF dimers can effectively suppress ERK-driven growth of resistant CRC.
Keywords: BRAF, RAS amplification, BRAF inhibitor, RAF dimerization, drug resistance, colorectal cancer
Introduction
The RAS-RAF-MEK-ERK pathway is physiologically regulated by upstream signals generated by receptors and activates multiple cellular processes including proliferation. RAS activation promotes the formation of RAF homo- and hetero-dimers, which in turn induce downstream signaling(1, 2). ERK activation also induces feedback inhibition of multiple components of the pathway, which limits the duration and output of the signal. Thus, ERK inhibits activation of RAS by receptor tyrosine kinases (RTKs) by phosphorylating the SOS exchange factor and a variety of receptors and by inducing the expression of members of the Sprouty family of proteins. Active ERK also directly phosphorylates and inhibits CRAF and BRAF and induces the expression of multiple MAPK phosphatases(3, 4).
BRAF V600E is the most common mutant BRAF allele. BRAF V600 mutants are constitutively activated and, uniquely among RAF mutants, can signal as RAS-independent monomers or dimers, depending on levels of RAS activation in the tumor(5, 6). BRAF V600 mutants are therefore unaffected by upstream feedback and drive high levels of ERK signaling output, which profoundly inhibit intracellular RAS activity. Thus, in these tumors, BRAF V600E predominantly exists as a drug-sensitive monomer. Current RAF inhibitors selectively inhibit BRAF monomers and are much less potent inhibitors of RAF dimers. Accordingly, RAF inhibitors rapidly inhibit ERK signaling in BRAF V600E tumors. This relieves feedback inhibition of RAS and results in induction of both BRAF V600E and wild-type RAF dimers. These dimers are resistant to RAF inhibitors, so a rebound in ERK signaling ensues and attenuates the antitumor effects of these drugs(7). In BRAF V600E melanomas, ERK rebounds only slightly and remains significantly lower than pretreatment levels. RAF inhibitors have significant therapeutic activity in BRAF V600E melanoma, but combined inhibition of BRAF and MEK reduces the rebound and is more effective than BRAF inhibition alone(8, 9). In colorectal and thyroid carcinoma, ERK rebound after inhibition by RAF inhibitors is much greater than that observed in melanoma and can rise to pretreatment levels. RAF inhibitors have only marginal therapeutic effects in these tumors(10).
EGFR is the dominant RTK in colon and the marked rebound in ERK signaling is thought to be due predominantly to relief of feedback inhibition of this receptor(11, 12). Consistent with this idea, the rebound in ERK signaling in CRC is sensitive to EGFR inhibition and combined administration of RAF and EGFR inhibitors induces tumor regression in most patients(13–17). However, acquired resistance invariably develops, typically within 6 months(13–16). In the recent randomized trial of cetuximab and irinotecan with or without vemurafenib in BRAF V600E colorectal cancer (CRC) patients (SWOG 1406), there was an improved response rate in the triplet arm, but progression free survival in the triplet arm was only 4.4 months (17).
Materials and Methods
Genetic analysis
DNA from pre-treatment and disease progression specimens were analyzed using our custom next-generation sequencing platform, MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets). The pre-treatment samples were collected before the administration of any chemotherapy and consisted of the primary colon tumor in six cases and liver metastasis in two patients. MSK-IMPACT is a targeted exome capture assay with deep sequencing coverage. Target specific-probes for hybrid selection were designed as previously described(18, 19) to capture all protein-coding exons of 341 oncogenes, tumor suppressor genes, and components of pathways deemed actionable by targeted therapies (for full list see Cheng DT, et al)(20).
All patients were treated on BRAF inhibitor clinical trials approved by MSKCC Institutional Review Board/Privacy Board (protocols 12-131, 12-221, 14-019). Progression biopsies and collection of patient samples were conducted under appropriate Institutional Review Board/Privacy Board protocols and waivers (protocols 06-107, 12-245, 14-019). Participating patients signed written informed consent for these clinical trials and biospecimen protocols. This study was conducted in accordance with ethical guidelines in the Declaration of Helsinki.
V600E BRAF immunohistochemistry
Immunohistochemistry with an antibody specific to BRAF V600E was performed as previously described(21).
FISH
FISH analysis was performed on formalin fixed paraffin embedded (FFPE) sections. BRAF FISH was performed using a 2-color BRAF break apart probe (developed at MSKCC). BAC (bacterial artificial chromosomes) clones mapping to 5′BRAF (RP11-715H9, RP11-133N19) labeled with Red dUTP and 3′BRAF (RP11-759K14, RP11-788O6) with Green dUTP. KRAS FISH analysis was performed using a 2-color KRAS/Cen12 probe mix (developed at MSKCC). The probe mix consisted of BAC clones containing the full length KRAS gene (clones RP11-29515 and RP11-707F18; labeled with red dUTP) and a centromeric repeat plasmid for chromosome 12 served as the control (clone pa12H8; labeled with green dUTP). Probe labeling, hybridization, washing, and fluorescence detection were performed according to standard procedures. Slides were scanned using a Zeiss Axioplan 2i epifluorescence microscope equipped with a megapixel CCD camera (CV-M4+CL, JAI) controlled by Isis 5.5.9 imaging software (MetaSystems Group Inc, Waltham, MA). The entire section was scanned through 63X or 100X to assess signal pattern and select representative regions for imaging. Amplification was defined as >10 copies of each locus.
Drugs
Vemurafenib (PLX4032) and PLX4720 were obtained from Plexxikon. Cetuximab was obtained from the MSKCC hospital pharmacy. BGB659 was provided by BeiGene.
Cell culture
All cells were obtained either from the MSKCC cell collection or the American Type Culture Collection (ATCC). HT-29 and VaCo432 cells were maintained in McCoy’s medium with antibiotics and 10% FBS. A375 and SKMel28 cells were grown in DMEM medium with antibiotics and 10% FBS. Cells with inducible expression constructs were maintained in this medium with 100ug/mL Hygromycin (Invitrogen) and 0.2ug/mL Puromycin (Invitrogen). HT-29 is a mismatch repair proficient colon cancer cell line (22). Vaco432 is a mismatch repair deficient (dMMR) colon cancer cell line due to MLH1 promoter methylation and silencing (23). The four cell lines used (HT-29, Vaco432, A375, and SKMel-28) do not have any mutations in the KRAS, NRAS, or HRAS genes. All cell lines tested negative for mycoplasma. Gene alterations in cell lines from the MSKCC cell collection (SKMel-28) were confirmed by MSK-IMPACT sequencing.
Inducible expression system
Retrovirus encoding the tet regulated NRAS or KRAS gene was packaged in Phoenix-AMPHO cells obtained from ATCC. The medium containing virus was filtered with 0.45 PVDF filters followed by incubation with the target cells for six hours. After this incubation, cells were cultured in virus free medium for two days. Then the cells were selected with Puromycin (2ug/mL) or Hygromycin (250ug/mL) for three days. The positive infected cells were further sorted with GFP marker after overnight exposure to 1ug/mL doxycycline (Sigma Aldrich). The NRAS gene construct included three consecutive FLAG tags in the N-terminus, and the KRAS gene construct included one FLAG tag in the N-terminus.
Antibodies
Immunoassays were performed as previously described(24). Antibodies against phospho-ERK (T202/Y204), total ERK1/2, phospho-MEK (S217/221), phospho-EGFR (Y1068), and total EGFR were obtained from Cell Signaling; antibodies against NRAS and BRAF from Santa Cruz Biotechnology; anti-RAS from ThermoScientific; and anti-CRAF from BD Biosciences.
RAS-GTP assay
GTP-bound RAS was measured with the RAF1 RAS-binding domain (RBD) pull-down and detection kit (ThermoScientific) following the manufacturer’s instructions.
Animal model studies
Patient derived tumor models were generated by mincing about 1g of tumor tissue, mixing with matrigel (50%) and implanting orthotopically into NSG (NOD scid gamma) mice (Institutional Review Board protocols 06-107, 14-091). The growing tumor was then implanted as subcutaneous xenografts for growth experiments, and tumor measurements were performed as described(24). The patient derived xenograft (PDX) generated was sequenced to confirm the same genomic alterations present in the progressing biopsy specimen were maintained in the PDX. All studies were performed in compliance with institutional guidelines under an Institutional Animal Care and Use Committee (IACUC) approved protocol.
Results
To define the molecular basis for resistance to combined RAF/EGFR inhibition, we analyzed nine tumor samples collected from eight patients at the time of disease progression and compared the results with those obtained from matched pre-treatment tumors. The tumors of all eight patients regressed with treatment (10–100%), and all subsequently developed resistance (Table 1). DNA derived from tumor and normal DNA from blood were subjected to targeted deep sequencing of all exons and selected introns of 341 key cancer-associated genes(20) (mean tumor coverage was 649x). New alterations in genes that encode components of the RAS/RAF pathway were identified in all nine of the samples obtained at progression. These included activating mutations of KRAS or NRAS, amplification of wild-type NRAS or KRAS or mutant BRAF V600E, and an intragenic deletion of exons 2–8 in BRAF V600E (Table 1). Two separate progression samples (collected from the liver and the peritoneum) from patient 3 revealed distinct new alterations of genes in the pathway – a BRAF intragenic deletion in the former and an NRAS mutation in the latter. Amplification in the resistant cells was validated directly (Supplementary Fig. 1A, data not shown), by increased protein expression (Supplementary Fig. 1B–C), and by detection of the amplified gene in double minute chromosomes or homogenously staining regions by fluorescence in-situ hybridization (Supplementary Fig. 1D–E). Amplification or gain of wild-type RAS occurred in 3 of 9 tumors after acquisition of resistance.
Table 1.
Acquired alterations in BRAF V600E Colorectal Cancers Treated with RAF/EGFR Inhibitor Combinations
| Patient | Treatment | Best response (RECIST read) | Duration of response | Acquired ERK pathway alteration |
|---|---|---|---|---|
| 1 | Vemurafenib + Panitumumab | PR (−100%) | 40 weeks | NRAS Q61K |
| 2 | Vemurafenib + Panitumumab | PR (−64%) | 24 weeks | BRAF V600E amplification (predominantly double minute chromosomes with 10–100 copies of BRAF) |
| 3 | Encorafenib + Cetuximab + Alpelisib | PR (−62%) | 24 weeks | Peritoneal metastasis: BRAF del exons 2–8Liver metastasis: NRAS G13R |
| 4 | Vemurafenib + Cetuximab | PR (−50%) | 16 weeks | BRAF V600E amplification (predominantly clusters of 6–14 copies of BRAF) |
| 5 | Encorafenib + Cetuximab + Alpelisib | PR (−43%) | 18 weeks | KRAS G12A |
| 6 | Vemurafenib + Panitumumab | SD (−24%) | 32 weeks | KRAS gain (4 copies), MET gain (5 copies) |
| 7 | Vemurafenib + Panitumumab | SD (−20%) | 16 weeks | NRAS amplification (>10 copies) |
| 8 | Vemurafenib + Cetuximab | SD (−10%) | 24 weeks | KRAS amplification (predominantly homologous staining region type with 10 to >20 copies of KRAS) |
Our previous work showed that RAS mutation, BRAF V600E amplification, and a splice variant of BRAF V600E all cause resistance to RAF inhibitors by inducing BRAF V600E dimerization(6, 25). One of the tumor specimens from patient 3 harbored an intragenic deletion of exons 2–8 of BRAF, a region including the RAS-binding domain. This is also the domain that is deleted in the alternatively spliced isoform and in most BRAF fusion proteins in cancer(26) (Supplementary Fig. 1F). Its deletion causes RAS-independent constitutive dimerization and activation of the truncated RAF(26, 27). As demonstrated in the tumor from patient 3, different mechanisms of resistance can occur in different lesions, but, in this case, each results in RAF dimerization, suggesting that this is a common convergent mechanism underlying the progression of tumors treated with this regimen.
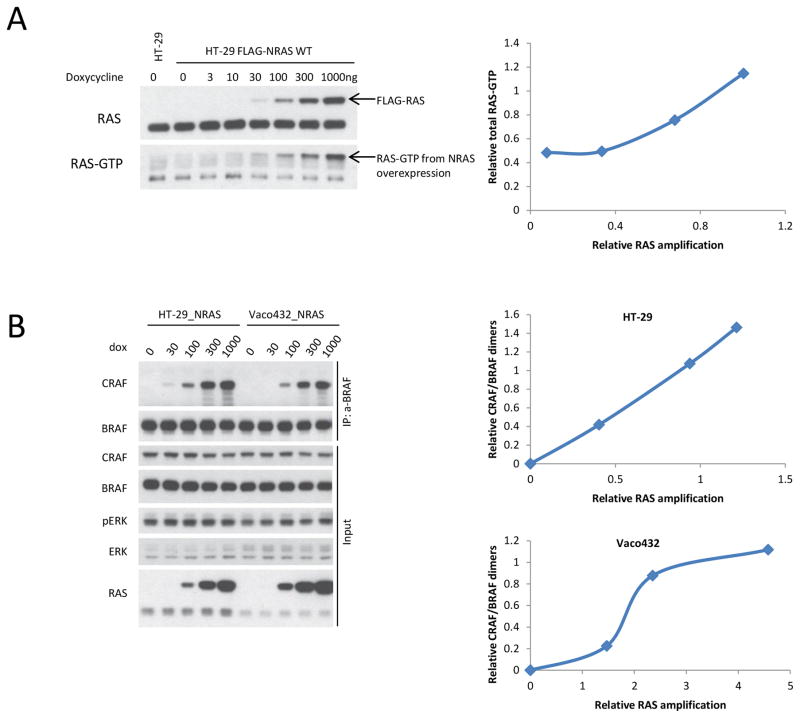
In melanoma, RAS mutation, BRAF V600E aberrant splice isoforms, and BRAF V600E amplification are the most common mechanisms for acquisition of resistance to RAF inhibitors or to RAF plus MEK inhibitors (Table 2). Amplification of wild-type RAS has not been reported as a resistance mechanism in melanoma and the biologic consequences of wild-type RAS overexpression are not clear. To investigate these issues, CRC BRAF V600E cell lines sensitive to vemurafenib/cetuximab treatment (HT-29 and Vaco432) were infected with a virus that encodes a doxycycline-inducible wild-type NRAS. Overexpression of wild-type NRAS in these cells resulted in an increase in RAS-GTP levels. In HT-29 and Vaco432 cells, an approximate doubling of wild-type NRAS expression led to just more than double the levels of the activated, GTP-bound form of RAS (Fig. 1A, Supplementary Fig. 2A). RAS overexpression resulted in an increase of co-immunoprecipitated CRAF with BRAF (Fig. 1B, Supplementary Fig. 2B). At baseline levels of RAS, BRAF/CRAF dimers could not be detected by immunoprecipitation (Supplementary Fig. 2B, Fig. 1B), but a less than doubling in RAS expression resulted in the detection of RAF hetero-dimers (Fig 1B). We observed a dose dependent relationship between levels of RAS expression and levels of BRAF/CRAF dimers (Fig. 1B). Thus, in HT-29 cells, an approximate doubling of baseline RAS expression resulted in RAS-GTP increasing approximately 2.5 times and this increase resulted in an induction of RAF hetero-dimers. These data suggest that modest increases in RAS expression, comparable to what is seen in clinical progression samples (4 to >20 copies), are enough to increase RAF dimerization.
Table 2.
Clinically Validated Mechanisms of Resistance to RAF Inhibitors in Melanoma Patients
| Reference | Treatment | Resistance Mechanisms |
|---|---|---|
| Johannessen CM et al, 2010.(34) | Vemurafenib | Increased COT expression |
| Nazarian R et al, 2010.(35) | Vemurafenib | PDGFβ upregulation NRAS mutation |
| Poulikakos P et al, 2011.(25) | Vemurafenib | Aberrantly spliced BRAF |
| Wagle H et al, 2011.(36) | Vemurafenib | MEK1 mutation |
| Villaneuva J et al, 2010.(37) | Vemurafenib | Increased IGF1R expression |
| Shi H et al, 2012.(38) | Vemurafenib | BRAF amplification BRAF truncation NRAS mutation Increased RTK expression |
| Straussman R et al, 2012.(39) | Vemurafenib or dabrafenib + trametinib | Stromal HGF secretion |
| Whittaker SR et al, 2013.(40) | Vemurafenib | NF1 loss |
| Trunzer K et al, 2013.(41) | Vemurafenib | NRAS mutation MEK1 mutation |
| Van Allen EM et al, 2014.(42) | Vemurafenib or dabrafenib | NRAS mutation BRAF amplification MEK1 mutation MEK2 mutation MITF amplification |
| Shi H, Hugo W et al, 2014.(43) | Vemurafenib or dabrafenib | NRAS mutation BRAF amplification Aberrantly spliced BRAF MEK1 mutation KRAS mutation |
| Rizos H et al, 2014 and Johnson DB et al, 2015.(44, 45) | Vemurafenib or dabrafenib | NRAS mutation Aberrantly spliced BRAF BRAF amplification MEK1 mutation MEK2 mutation KRAS mutation Increased IGF1R expression AKT1 mutation PIK3CA mutation PTEN loss DUSP4 deletion AKT3 mutation MITF amplification PDGFR upregulation |
| Sun C et al, 2014.(46) | Vemurafenib or dabrafenib or trametinib | EGFR expression |
| Wagle N et al, 2014.(47) | Dabrafenib + trametinib | MEK2 mutation BRAF amplification Aberrantly spliced BRAF |
| Villanueva J et al, 2013.(48) | Trametinib followed by dabrafenib | MEK2 mutation plus BRAF amplification |
Figure 1. RAS amplification leads to increased RAS-GTP and dimerization of RAF.
A. HT-29 cells and HT-29 FLAG-NRAS WT (wild-type) (HT-29_NRAS) cells, exposed to increasing doses of doxycycline as indicated, were harvested after 24 hours. Expression of the indicated proteins was assayed by immunoblotting. The cellular RAS-GTP was determined by the active RAS pull-down assay. Densitometric analysis of the bands was used to calculate the relative amplification of RAS and RAS-GTP.
B. HT-29_NRAS and Vaco432_NRAS cells were treated with the indicated doses of doxycycline (dox) for 16 hours. Then the cells were lysed in 0.1% NP-40 Tris-NaCl buffer. The soluble fractions were isolated and incubated with anti-BRAF antibody coupled IgG beads for 2 hours at 4°C. The immunoprecipitated protein complex and 2% input were assayed by immunoblotting with indicated antibodies. Densitometric analysis of the bands was used to calculate relative RAS amplification and CRAF/BRAF dimers. Relative levels of CRAF/BRAF dimers were normalized to levels of immunoprecipitated BRAF.
Increased expression of wild-type RAS led to increased RAS activation and induction of BRAF/CRAF dimers in a model of KRAS amplification, as well. Increasing doxycycline exposure from 100ng/mL to 1000ng/mL led to an approximate doubling of RAS expression and a large (10-fold) increase in RAS-GTP levels (Supplementary Fig. 2C). Doubling of RAS expression was again sufficient to lead to a detectable increase in CRAF/BRAF dimers (Supplementary Fig. 2D). These data suggest that moderate amplification and overexpression of either wild-type NRAS or KRAS in BRAF V600E CRC is sufficient to increase RAS activation enough to cause resistance to RAF/EGFR therapy by generating of RAF-inhibitor resistant RAF dimers.
This turned out to be the case. Overexpression of NRAS in BRAF V600E HT-29 CRC cells was sufficient to confer vemurafenib/cetuximab resistance (Supplementary Figs. 3A, 3B). A 1.7-fold amplification of RAS was found to be associated with a detectable decrease in the inhibition of phosphorylated ERK with 1-hour vemurafenib/cetuximab treatment (Supplementary Fig. 3C). RAS overexpression led to resistance in HT-29 xenografts as well (Fig. 2A). NRAS expression approximately doubled in the vehicle treated mouse and increased 3.6-fold in the mouse treated with RAF and EGFR inhibitors. These data confirm that even low level of RAS expression in colorectal cancer is sufficient to cause resistance to RAF/EGFR inhibitors in vivo and the higher level of RAS amplification in the drug treated mouse suggests a selection for RAS amplification in the tumors treated with the drug combination.
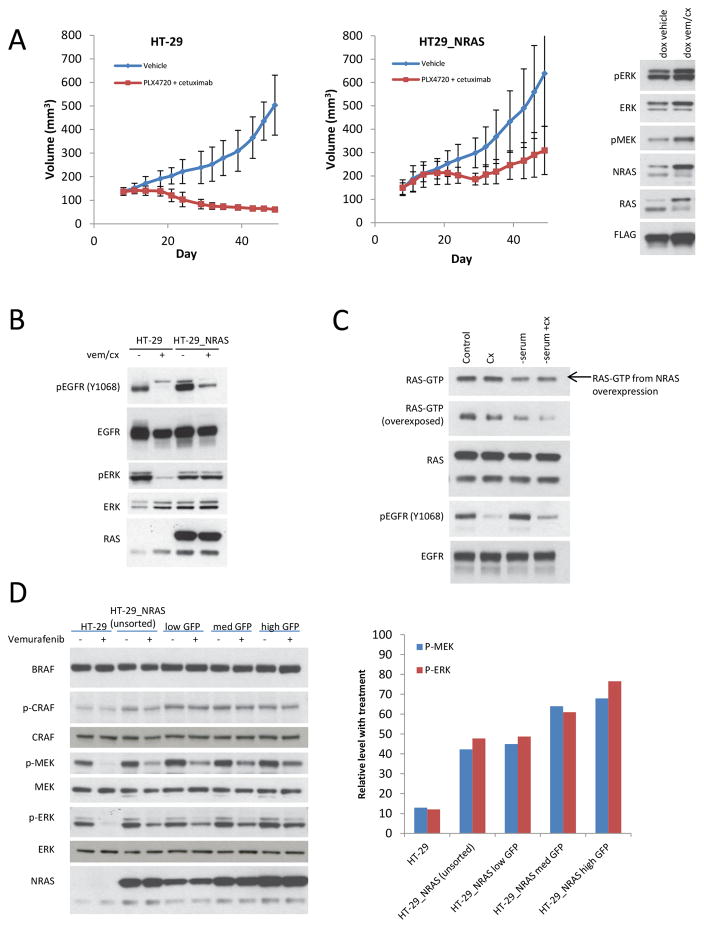
Figure 2. Increase in NRAS expression is sufficient to cause resistance to RAF/EGFR inhibition in CRC.
A. Growth curves for HT-29 and HT-29_NRAS xenografts treated with vehicle or vemurafenib (PLX4720 50 mg/kg PO twice daily) plus cetuximab (50 mg/kg intraperitoneal injection twice per week). Five mice were treated in each group, and tumor volumes (and standard deviations [SD]) are shown as a function of time on treatment. Right panel shows immunoblots from representative mice fed doxycycline (dox) and treated with vehicle control (left) or vemurafenib/cetuximab (vem/cx) (right). Tumor were collected for immunoblot analysis at the end of the growth experiment.
B. HT-29 and HT-29_NRAS, treated with doxycycline 2ug/mL for 24 hours before drug exposure, were treated with either vehicle (DMSO) or vemurafenib/cetuximab (vem/cx) for 24 hours. Expression of the indicated proteins was assayed by immunoblotting.
C. HT-29_NRAS cells, treated with doxycycline 2ug/mL, were plated for 12 hours to adhere and then serum was removed as indicated. Twelve hours later, cells were subjected to treatment with vehicle control or cetuximab (cx) for 24 hours. Cells were then collected and expression of the indicated proteins was assayed by immunoblotting. The cellular RAS-GTP was determined by the active RAS pull-down assay.
D. HT-29_NRAS cells were treated with doxycycline for 24 hours and then subjected to sorting of the cell populations by GFP expression. HT-29 cells, unsorted HT-29_NRAS cells, and HT-29 cells sorted for low, medium (med), or high GFP expression were treated with either vehicle (DMSO) or vemurafenib 1uM for one hour. Expression of the indicated proteins was assayed by immunoblotting. Densitometric analysis of the bands was used to calculate phosphorylated ERK and phosphorylated MEK levels with vemurafenib treatment at each level of RAS expression.
Inhibition of EGFR phosphorylation by vemurafenib/cetuximab treatment was unaffected by overexpression of wild-type RAS, but inhibition of ERK phosphorylation was abrogated in the HT-29_NRAS cells (Figs. 2B and Supplementary Fig. 3D). These data suggest that the sensitivity of RAS activation to EGFR inhibition is reduced in cells in which wild-type NRAS is amplified and this turned out to be the case. In the HT-29 cells, inhibition of EGFR with cetuximab alone or together with vemurafenib, inhibited endogenous levels of activated RAS (Supplementary Fig. 3D). Levels of RAS-GTP in HT-29 cells with NRAS overexpression were not sensitive to EGFR inhibition (Fig. 2C and Supplementary Fig. 3D), suggesting that amplified wild-type RAS may be activated by other RTKs. The baseline levels of RAS-GTP and the increased RAS-GTP in cells in which NRAS was overexpressed remained sensitive to serum starvation (Fig. 2C), suggesting that wild-type RAS amplification leads to increased RAS-GTP in CRC cells by amplifying upstream signaling by growth factors and may signal through multiple RTKs in addition to EGFR.
To determine if RAS amplification conferred resistance by reducing sensitivity of the pathway to the RAF inhibitor, we examined the ability of vemurafenib to inhibit ERK signaling in HT-29 cells in which different levels of NRAS expression were induced. The sensitivity of ERK signaling in these cells to one hour exposure to vemurafenib was inversely related to NRAS expression (Fig. 2D). In cells with 3.5-fold RAS overexpression, one hour exposure to vemurafenib caused a 32% decrease in phosphorylated MEK and a 23% decrease in phosphorylated ERK compared to an 88% decrease in both phosphorylated MEK and ERK levels in the absence of NRAS overexpression.
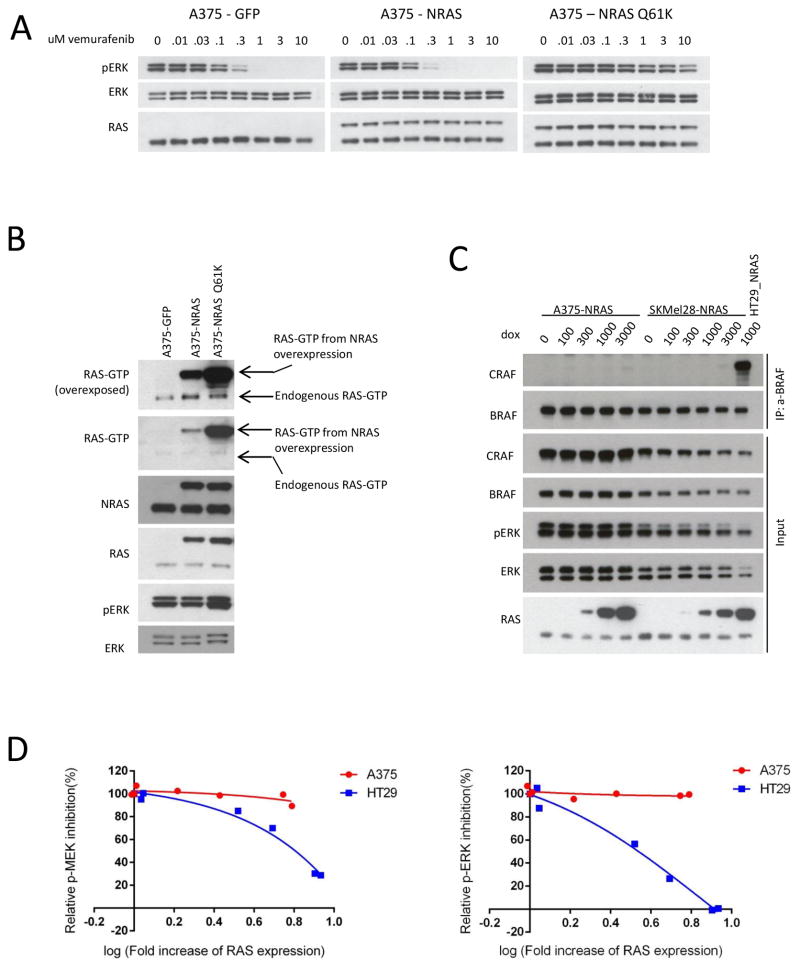
These findings suggest that wild-type RAS gene amplification causes resistance to RAF/EGFR inhibition by increasing cellular RAS-GTP levels to levels sufficient to drive BRAF V600E dimerization. To our knowledge, wild-type RAS amplification has not been described as a mechanism of resistance to RAF inhibitors in melanomas (Table 2). To evaluate if wild-type RAS amplification can cause resistance in melanoma, we inducibly expressed wild-type NRAS, Q61K mutant NRAS, or control GFP protein in A375 or SKMel-28 BRAF V600E melanoma cells. Overexpression of wild-type NRAS did not affect phospho-ERK inhibition by vemurafenib, while expression of Q61K NRAS led to insensitivity of phospho-ERK to even high concentrations of drug (Fig. 3A and Supplementary Fig. 4A). Overexpression of NRAS in A375 cells led to a small increase in RAS-GTP levels, much lower than the levels of RAS-GTP that result from Q61K mutant NRAS expression (Fig. 3B), and did not alter the growth inhibitory effect of vemurafenib treatment (Supplementary Fig. 4B). Compared to the CRC cell lines, we observed that overexpression of NRAS in A375 and SKMel-28 melanoma cells, even to high levels, resulted in a barely perceptible induction of BRAF/CRAF dimers (Fig. 3C). We compared the effect of increasing levels of NRAS expression on relative inhibition of phospho-ERK and phospho-MEK in A375 and HT-29 cells (Fig. 3D and Supplementary Fig. 4C). An approximately 6-fold increase in NRAS expression led to minimal change in the sensitivity of phospho-ERK and phospho-MEK to vemurafenib in A375 cells, but led to a greater than 50% decrease in the inhibition of phospho-ERK and phospho-MEK by vemurafenib combined with cetuximab in HT-29 cells. These data suggest that overexpression of wild-type NRAS amplifies RAS activation to a much greater degree in CRCs than in melanomas when BRAF V600E is expressed. This may have to do with the low levels of endogenous RTK activation in melanomas(28). In this context, amplification of the weak RTK signal by wild-type RAS may be insufficient to cause resistance. We speculate that this is the reason that RAS amplification has not been identified as a common cause of RAF inhibitor resistance in melanomas.
Figure 3. Increase in NRAS expression does not cause resistance to RAF inhibition in melanoma.
A and B. A375 cells expressing inducible GFP (control), wild-type NRAS, or NRAS Q61K were treated with doxycycline (2ug/mL for 24 hours) (A) followed by treatment with vemurafenib for one hour at the indicated concentrations or (B) collected for RAS-GTP analysis with the active RAS pull-down assay. Expression of the indicated proteins was assayed by immunoblotting.
C. Increasing expression of wild-type NRAS was induced into BRAF V600E mutant CRC cell lines HT-29 and VACO432 and melanoma cell lines A375 and SKMEL-28 with FLAG-NRAS by doxycycline treatment with the indicated doses for 24 hours. The cells were then collected and lysed. BRAF/CRAF heterodimers were pulled down with anti-BRAF antibody conjugated beads. The cell lysate (input) for the binding assay and the immunoprecipitated complexes were assayed by immunoblotting with the indicated antibodies.
D. A375 and HT-29 cells expressing inducible wild-type NRAS were treated with increasing doses of doxycycline for 24 hours and then treated with vemurafenib 1uM or vemurafenib 1uM plus cetuximab 50nM, respectively, for 24 hours. Expression of phospho-ERK and phospho-MEK was assayed by immunoblotting. Densitometric analysis of the bands was used to calculate the relative change in phosphorylated ERK and MEK levels with drug treatment at each level of ectopic RAS expression, where the change in phospho-MEK or phospho-ERK levels in the absence of doxycycline in each cell line was defined as 100% inhibition.
Recently, we have identified drugs that inhibit ERK signaling driven by both mutant RAF monomers and dimers(6). We hypothesized that such compounds should effectively inhibit BRAF V600E CRCs with dimer dependent acquired resistance to RAF/EGFR inhibition. We tested the effects of one of these, BGB659, in vemurafenib/cetuximab resistant colorectal tumors. This drug inhibited ERK signaling in HT-29 and HT-29_NRAS cells at similar concentrations (Fig. 4A). At later time points, BGB659 treatment was associated with a rebound in phospho-ERK levels in HT-29 cells that was largely suppressed by combination treatment with the EGFR inhibitor cetuximab (Supplementary Fig. 5A), suggesting that reactivation of RTK signaling can also reduce sensitivity to the RAF dimer inhibitor in BRAF V600E CRCs. Feedback reactivation of RTK signaling likely attenuates sensitivity to RAF dimer inhibitors due to increased RAS activation and to formation of wild-type RAF dimers. We have shown that, compared to mutant BRAF dimers, RAS driven wild-type RAF dimers are less sensitive to these drugs(6). Thus, RAF dimer inhibitors will still have to be combined with inhibitors of the dominant RTK EGFR in order to maximally inhibit both mutant and wild-type RAF. As shown in Supplementary Fig. 5B, the combination of BGB659 with cetuximab led to significantly better growth inhibition in vitro than BGB659 alone in both HT-29_NRAS and Vaco432_NRAS cells. In agreement with our model, although both the BGB659/cetuximab and vemurafenib/cetuximab combinations were able to suppress the growth of CRC cells with BRAF V600E mutation, only the former combination could effectively inhibit the growth of BRAF V600E CRC cells which also express high levels of wild-type NRAS (Fig. 4B).
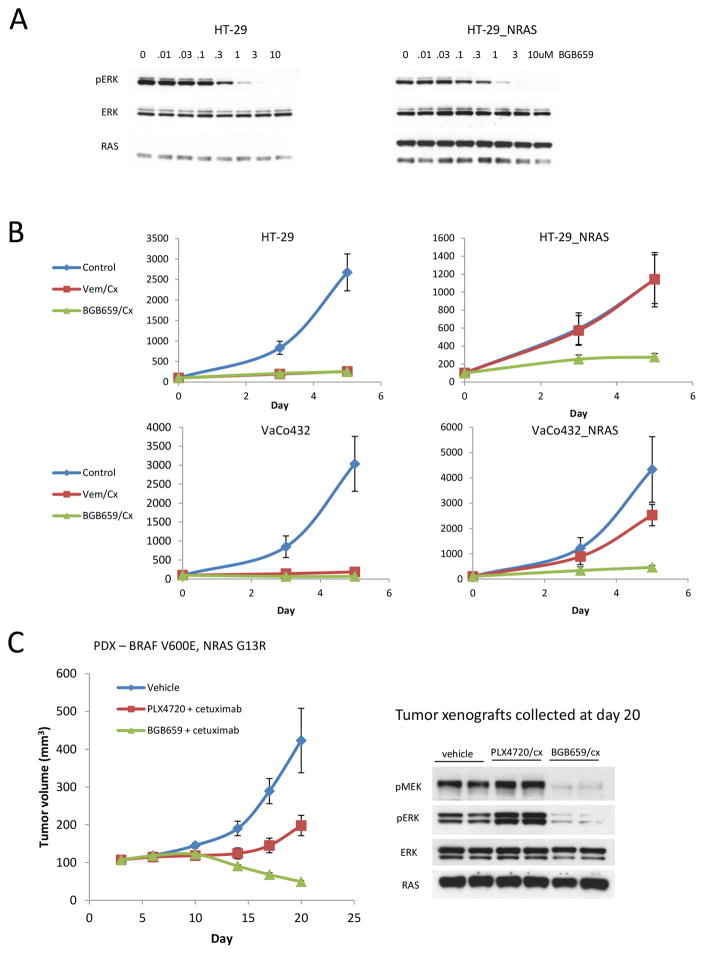
Figure 4. Combined administration of RAF dimer and EGFR inhibitors overcome resistance.
A. HT-29 and HT-29_NRAS cells (treated with 2ug/mL doxycycline for 24 hours) were treated with a range of BGB659 doses as indicated for one hour. Expression of the indicated proteins was assayed by immunoblotting.
B. Growth curves for treatment of HT-29 or Vaco432 cells with vehicle (DMSO), vemurafenib 2uM plus cetuximab 100nM (Vem/Cx), or BGB659 1uM plus cetuximab 100nM (BGB659/Cx) for 5 days. Relative cell counts were assayed by alamarBlue. For this experiment, HT-29 and Vaco432 NRAS overexpressing cells were treated with doxycycline 2ug/mL for 24 hours before drug exposure. Experiments were done in 8 replicates.
C. Patient derived xenograft (PDX) made from the progression specimen of patient 3 was expanded into mice that were treated with vehicle, vemurafenib (PLX4720 50 mg/kg PO twice daily) plus cetuximab (50 mg/kg intraperitoneal injection twice per week), or BGB659 (100mg/kg oral daily) plus cetuximab (50 mg/kg intraperitoneal injection twice per week). Tumor volumes (and SD) are shown as a function of time on treatment. Tumors were collected at day 20 and two samples from each group were lysed for immunoblotting with the indicated antibodies.
BGB659/cetuximab treatment was not only effective in the RAS amplification driven vemurafenib resistant CRC cells, but also in other models of resistance. In a patient-derived xenograft derived from the progressing liver metastasis obtained from patient 3 (NRAS G13R mutation, Table 1), treatment with PLX4720 plus cetuximab resulted in only a modest delay in tumor growth whereas BGB659/cetuximab treatment led to tumor regression in all five mice treated (Fig. 4C). In tumors treated with PLX4720/cetuximab, phospho-MEK and phospho-ERK levels increased by day 20. This is likely due to transactivation of wild-type RAF dimers with PLX4720. By contrast, BGB659/cetuximab continued to profoundly inhibit phospho-MEK and phospho-ERK on day 20. These data suggest that new RAF dimer inhibitors that equipotently inhibit both RAF mutant monomers and dimers represent a new, potentially much more effective strategy for treating BRAF V600E CRC.
Discussion
Acquired resistance to RAF/EGFR inhibitor combination therapy is mediated by multiple genetic lesions(29–31). In our work, we analyzed nine progression samples from patients and found that resistance was mediated by five different genetic aberrations. All five lesions prevent effective inhibition of RAF activation by the combination. Tumors in which resistance is mediated by parallel or downstream lesions in which RAF activity remains sensitive to the RAF inhibitors were not found. Four of the lesions we have identified have been previously shown to also cause resistance in melanoma. The exception is amplification of wild-type RAS, which causes resistance in CRC, but has not been identified as a resistance mechanism in melanoma. We have now determined that each of these lesions causes resistance by the same unifying mechanism – induction of RAF dimers which are insensitive to current RAF inhibitors. Based on this understanding, we show that combined administration of a RAF inhibitor that equipotently inhibits RAF mutant monomers and dimers and an EGFR inhibitor is able to suppress growth of resistant colorectal tumors, thus proving that induction of RAF dimers is a key event in resistance.
Amplification of wild-type RAS is often observed in carcinomas, but functional consequences are unknown. Our data suggest the possibility that high levels of wild-type RAS can amplify induction of RAS/RAF/ERK signaling by upstream inputs such as RTKs. Future studies are needed to elucidate the detailed mechanism by which amplification of RAS facilitates RAS activation by RTKs, but likely mechanisms include a simple linear increase in RAS-GTP as a function of increase in RAS expression, as seen in the CRC cell lines, or saturation of inhibitory processes involving RAS-GTP loading and hydrolysis or membrane binding.
We find that amplification of wild-type RAS in CRC, but not melanoma, is sufficient to elevate RAS-GTP to levels that induce enough dimers to cause resistance to RAF inhibitors. This result is reminiscent of the differing response of melanoma and CRC to single-agent RAF inhibitors and suggests that higher endogenous levels of RTK signaling in CRC lead to the formation of drug-resistant RAF dimers, causing adaptive resistance to RAF inhibitors and, in the presence of high RAS expression, rapid acquired resistance. This is the first example of a lineage dependent mechanism of acquired resistance and is due either to the higher level of RTK signaling in CRC than in melanoma or to the increased sensitivity of RTK signaling to ERK-dependent feedback in melanoma compared to CRC. We have recently shown that hypoactive BRAF mutants serve a similar function(32, 33). They amplify ERK signaling in a RAS-dependent manner and, in lung and colon carcinomas, cooperate with upstream RTKs to drive transformation. Interestingly, levels of RAS-GTP in melanomas are also not sufficient to cooperate with low activity RAF mutations(33). Tumors with these mutations are invariably found to coexist with NF1 inactivation or RAS mutations. Thus, both hypoactive BRAF mutants and amplified wild-type RAS significantly activate ERK signaling in carcinomas, but not melanoma. It is likely that both of these lineage dependent phenomena have the same cause—higher levels of steady state RAS-activation in lung and colon epithelial cells than in melanocytes. Further work will be required to prove this hypothesis.
We observed that RAS amplification does not significantly increase the level of phosphorylated ERK in BRAF V600E cells. The presence of high levels of RAS expression, however, affects response to vemurafenib treatment differently in melanoma and colon tumors. In BRAF V600E cells, RAS activity is almost completely feedback inhibited and ERK phosphorylation is driven by BRAF V600E monomers. In CRC cells, but not in melanomas, RTK activity is high enough to elevate RAS-GTP levels in tumor cells with high expression of wild-type RAS. This causes RAS-dependent dimerization of BRAF V600E protein, and thus reduces the sensitivity of BRAF V600E CRC to vemurafenib, since the drug does not potently inhibit these dimers. ERK phosphorylation does not go up because the activity of BRAF V600E is not regulated by RAS binding or its dimerization.
We recently reported why RAF inhibitors selectively inhibit BRAF V600E monomers at low concentrations, but require much higher concentrations to inhibit any of the RAF dimers(5). The binding of drug to one protomer of the RAF dimer reduces the affinity of the drug for the other protomer(6). This negative cooperativity leads to reduced sensitivity of RAF dimers, compared to monomers, to inhibitors and also underlies adaptive resistance to these agents in CRC.
Our work identifies a potential strategy to overcome resistance in BRAF V600E CRC, using new RAF inhibitors that are not affected by the negative cooperativity. Unlike MEK and ERK inhibitors, these drugs selectively inhibit ERK signaling that is driven by mutant BRAF(6). This provides a basis for a therapeutic index for these RAF inhibitors in the treatment of resistant tumors. Our data suggest that combining these new RAF inhibitors with an EGFR inhibitor may be useful in treating resistance and as initial targeted therapy for BRAF V600E CRC and should be explored in future clinical trials.
Supplementary Material
Acknowledgments
This research was supported by grants to N. Rosen from the National Institutes of Health (R01 CA169351 and P01 CA129243), from Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, The Center for Experimental Therapeutics at Memorial Sloan Kettering Cancer Center, and support from Mr. and Mrs. Robert A. Kramer. This work is also supported by a Career Development Award from the Conquer Cancer Foundation of the American Society of Clinical Oncology (R. Yaeger) and the NIH/NCI Cancer Center Support Grant P30 CA008748.
The authors are grateful to Yijun Gao for helpful discussions. We would like to thank Ahmet Zehir for help with figure preparation.
References
- 1.Rushworth LK, Hindley AD, O’Neill E, Kolch W. Regulation and role of Raf-1/B-Raf heterodimerization. Mol Cell Biol. 2006;26:2262–72. doi: 10.1128/MCB.26.6.2262-2272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weber CK, Slupsky JR, Kalmes HA, Rapp UR. Active Ras induces heterodimerization of cRaf and BRaf. Cancer Res. 2001;61:3595–8. [PubMed] [Google Scholar]
- 3.Dougherty MK, Muller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD, et al. Regulation of Raf-1 by direct feedback phosphorylation. Mol Cell. 2005;17:215–24. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]
- 4.Douville E, Downward J. EGF induced SOS phosphorylation in PC12 cells involves P90 RSK-2. Oncogene. 1997;15:373–83. doi: 10.1038/sj.onc.1201214. [DOI] [PubMed] [Google Scholar]
- 5.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–30. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao Z, Torres NM, Tao A, Gao Y, Luo L, Li Q, et al. BRAF Mutants Evade ERK-Dependent Feedback by Different Mechanisms that Determine Their Sensitivity to Pharmacologic Inhibition. Cancer Cell. 2015;28:370–83. doi: 10.1016/j.ccell.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell. 2012;22:668–82. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Combined BRAF and MEK Inhibition versus BRAF Inhibition Alone in Melanoma. N Engl J Med. 2014 doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 10.Kopetz S, Desai J, Chan E, Hecht JR, O’Dwyer PJ, Maru D, et al. Phase II Pilot Study of Vemurafenib in Patients With Metastatic BRAF-Mutated Colorectal Cancer. J Clin Oncol. 2015;33:4032–8. doi: 10.1200/JCO.2015.63.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer discovery. 2012;2:227–35. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–3. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 13.Atreya CE, Van Cutsem E, Bendell JC, Andre T, Schellens JH, Gordon MS, et al. Updated efficacy of the MEK inhibitor trametinib (T), BRAF inhibitor dabrafenib (D), and anti-EGFR antibody panitumumab (P) in patients (pts) with BRAF V600E mutated (BRAFm) metastatic colorectal cancer (mCRC) J Clin Oncol. 2015;33 abstract 103. [Google Scholar]
- 14.Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay JY, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med. 2015;373:726–36. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabernero J, Van Geel R, Guren TK, Yaeger R, Spreafico A, Faris JE, et al. Phase 2 results: Encorafenib (ENCO) and cetuximab (CETUX) with or without alpelisib (ALP) in patients with advanced BRAF-mutant colorectal cancer (BRAFm CRC) Journal of Clinical Oncology. 2016;34 abstract 3544. [Google Scholar]
- 16.Yaeger R, Cercek A, O’Reilly EM, Reidy DL, Kemeny NE, Wolinsky T, et al. Pilot Trial of Combined BRAF and EGFR Inhibition in BRAF Mutant Metastatic Colorectal Cancer Patients. Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-14-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kopetz S, LMS, Lenz HJ, Magliocco AM, Atreya CE, Diaz LA, et al. Randomized trial of irinotecan and cetuximab with or without vemurafenib in BRAF-mutant metastatic colorectal cancer (SWOG S1406) J Clin Oncol. 2017:35. doi: 10.1200/JCO.20.01994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nature biotechnology. 2009;27:182–9. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer discovery. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–64. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vakiani E, Yaeger R, Brooke S, Zhou Y, Klimstra DS, Shia J. Immunohistochemical Detection of the BRAF V600E Mutant Protein in Colorectal Neoplasms. Applied immunohistochemistry & molecular morphology : AIMM / official publication of the Society for Applied Immunohistochemistry. 2014 doi: 10.1097/PAI.0000000000000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmed D, Eide PW, Eilertsen IA, Danielsen SA, Eknaes M, Hektoen M, et al. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis. 2013;2:e71. doi: 10.1038/oncsis.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma AH, Xia L, Littman SJ, Swinler S, Lader G, Polinkovsky A, et al. Somatic mutation of hPMS2 as a possible cause of sporadic human colon cancer with microsatellite instability. Oncogene. 2000;19:2249–56. doi: 10.1038/sj.onc.1203568. [DOI] [PubMed] [Google Scholar]
- 24.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–62. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16:793–8. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross JS, Wang K, Chmielecki J, Gay L, Johnson A, Chudnovsky J, et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer. 2016;138:881–90. doi: 10.1002/ijc.29825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Real FX, Rettig WJ, Chesa PG, Melamed MR, Old LJ, Mendelsohn J. Expression of epidermal growth factor receptor in human cultured cells and tissues: relationship to cell lineage and stage of differentiation. Cancer Res. 1986;46:4726–31. [PubMed] [Google Scholar]
- 29.Ahronian LG, Sennott EM, Van Allen EM, Wagle N, Kwak EL, Faris JE, et al. Clinical Acquired Resistance to RAF Inhibitor Combinations in BRAF-Mutant Colorectal Cancer through MAPK Pathway Alterations. Cancer discovery. 2015;5:358–67. doi: 10.1158/2159-8290.CD-14-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oddo D, Sennott EM, Barault L, Valtorta E, Arena S, Cassingena A, et al. Molecular landscape of acquired resistance to targeted therapy combinations in BRAF mutant colorectal cancer. Cancer Res. 2016 doi: 10.1158/0008-5472.CAN-16-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pietrantonio F, Oddo D, Gloghini A, Valtorta E, Berenato R, Barault L, et al. MET-driven resistance to dual EGFR and BRAF blockade may be overcome by switching from EGFR to MET inhibition in BRAF mutated colorectal cancer. Cancer discovery. 2016 doi: 10.1158/2159-8290.CD-16-0297. [DOI] [PubMed] [Google Scholar]
- 32.Nieto P, Ambrogio C, Esteban-Burgos L, Gomez-Lopez G, Blasco MT, Yao Z, et al. A Braf kinase-inactive mutant induces lung adenocarcinoma. Nature. 2017 doi: 10.1038/nature23297. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao Z, Yaeger R, Rodrik-Outmezguine V, Tao A, Torres N, Chang MT, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature. 2017 doi: 10.1038/nature23291. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–96. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nature communications. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer discovery. 2013;3:350–62. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trunzer K, Pavlick AC, Schuchter L, Gonzalez R, McArthur GA, Hutson TE, et al. Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. J Clin Oncol. 2013;31:1767–74. doi: 10.1200/JCO.2012.44.7888. [DOI] [PubMed] [Google Scholar]
- 42.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer discovery. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer discovery. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson DB, Menzies AM, Zimmer L, Eroglu Z, Ye F, Zhao S, et al. Acquired BRAF inhibitor resistance: A multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur J Cancer. 2015;51:2792–9. doi: 10.1016/j.ejca.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rizos H, Menzies AM, Pupo GM, Carlino MS, Fung C, Hyman J, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin Cancer Res. 2014;20:1965–77. doi: 10.1158/1078-0432.CCR-13-3122. [DOI] [PubMed] [Google Scholar]
- 46.Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–22. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 47.Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer discovery. 2014;4:61–8. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villanueva J, Infante JR, Krepler C, Reyes-Uribe P, Samanta M, Chen HY, et al. Concurrent MEK2 mutation and BRAF amplification confer resistance to BRAF and MEK inhibitors in melanoma. Cell reports. 2013;4:1090–9. doi: 10.1016/j.celrep.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.