Abstract
The human cytidine deaminase APOBEC3G (A3G) is a potent inhibitor of the HIV-1 virus in the absence of viral infectivity factor (Vif). The molecular mechanism of A3G antiviral activity is primarily attributed to deamination of single-stranded DNA (ssDNA); however, the nondeamination mechanism also contributes to HIV-1 restriction. The interaction of A3G with ssDNA and RNA is required for its antiviral activity. Here we used atomic force microscopy to directly visualize A3G–RNA and A3G–ssDNA complexes and compare them to each other. Our results showed that A3G in A3G–RNA complexes exists primarily in monomeric–dimeric states, similar to its stoichiometry in complexes with ssDNA. New A3G–RNA complexes in which A3G binds to two RNA molecules were identified. These data suggest the existence of two separate RNA binding sites on A3G. Such complexes were not observed with ssDNA substrates. Time-lapse high-speed atomic force microscopy was applied to characterize the dynamics of the complexes. The data revealed that the two RNA binding sites have different affinities for A3G. On the basis of the obtained results, a model for the interaction of A3G with RNA is proposed.
Graphical Abstract
APOBEC3G (A3G) belongs to the APOBEC3 family of DNA-editing cytidine deaminases that are responsible for deami-nation of HIV-1 cDNA during reverse transcription in the absence of viral infectivity factor (Vif).1,2 A3G consists of two zinc-coordinating domains,3 the N-terminal domain (NTD) that is incapable of catalysis but responsible for binding ssDNA and RNA4,5 and the C-terminal domain (CTD) that catalyzes ssDNA deamination.6,7 The primary antiviral activity of A3G is deamination of the viral ssDNA and the most studied mechanism of action.7–10 However, there is evidence that deaminase-independent mechanisms are also involved in the antiviral activity of A3G.4,11,12 One is the so-called “road block” mechanism, in which A3G physically blocks the elongation of reverse transcribed proviral ssDNA,12 and the other includes RNA-dependent incorporation of newly synthesized A3G into viral particles.13–16 It was shown that A3G binds to diverse cellular RNA,14,17–19 but during the encapsidation process, A3G forms a complex with viral RNA.16 These deaminase-independent activities include A3G binding capabilities that are specific to ssDNA and RNA and involve both A3G domains. It was shown that the extents of most deaminase-independent functions of A3G decrease after the introduction of mutations into the NTD,4 but mutations in the CTD do not completely abolish the antiviral A3G activity against HIV.20,21
Considerable biochemical and biophysical data are available for A3G–RNA complexes.2 It was shown that RNA inhibits A3G catalytic activity by binding to the CTD of A3G. This suggests a mechanistic explanation for the different modes of binding of A3G22 to ssDNA and RNA. However, no studies have directly characterized A3G–RNA complexes or compared them with A3G–ssDNA complexes. It is currently unclear whether RNA shares the A3G binding site with ssDNA or if RNA interacts with a different region of A3G. Currently, there is a lack of direct data about the stoichiometry of A3G–RNA complexes. It is also not clear whether the A3G stoichiometry of A3G–RNA complexes differs from the stoichiometry of A3G–ssDNA complexes.
To address these issues, we characterized properties of A3G–RNA complexes and compared them with the properties of A3G–ssDNA complexes. We, for the first time, directly visualized the A3G–RNA complexes with AFM and estimated the stoichiometry of A3G in these complexes. From experiments performed for both A3G–ssDNA and A3G–RNA complexes, we conclude that A3G appears in similar oligomeric states on both substrates. However, a remarkable difference in assemblies of ssDNA and RNA complexed with A3G was observed. Similar to the case for A3G–ssDNA complexes, when one RNA molecule bound to A3G, we also observed two RNA molecules bound to A3G. Complexes containing two molecules of ssDNA bound to A3G were never observed. By analyzing the data for each A3G domain independently, we show that RNA forms complexes with both the sNTD and CTD-191–384-E259A, while ssDNA is not able to form such complexes. Together, these results indicate the existence of an additional region on A3G different from ssDNA that is capable of binding RNA. The dynamic behavior of the A3G–RNA complexes suggests that the two binding regions of A3G have different affinities for RNA. On the basis of these results, a model for binding of RNA to A3G is discussed.
MATERIALS AND METHODS
RNA Hybrid Substrate Preparation
The hybrid RNA substrate was assembled like the hybrid DNA substrate as described in ref 23. The schematic of the procedure is shown in Figure S1. First, the DNA–RNA hybrid consisting of a 21-nucleotide ssDNA and a 69-nucleotide RNA strand was synthesized (Integrated DNA Technology, Coralville, IA). Second, phosphorylated 18-mer ssDNA was annealed in a 1:1 ratio with a 90-nucleotide hybrid ssDNA–RNA strand to create an 18 bp dsDNA region with three sticky ends and a 69-nucleotide RNA tail. Third, the strand formed after annealing 18 bp dsDNA with a 69-nucleotide RNA tail was ligated with a previously gel-purified 124 bp dsDNA fragment with three-nucleotide sticky ends. As a result, the hybrid RNA product consists of 145 bp dsDNA and a 69-nucleotide RNA tail. The ligated product was immediately treated with phenol, precipitated with ethanol, and stored at –80 °C. To protect the ssRNA from degradation, RNAsecure inactivation reagent (Thermo Fisher Scientific) was added at each step of the hybrid RNA assembly process. Specifically, all DEPC-treated buffers with 1× RNAsecure inactivation reagent were heated to 60 °C for 10 min and then cooled to room temperature.
The gel electrophoresis of the ligated RNA hybrid product is shown in Figure S2A, and the estimated yield of the ligated product was 78%. The final product of the RNA hybrid has 145 bp dsDNA fused with 69-nucleotide RNA, as chematically shown in Figure S3B.
DNA Hybrid Substrate Preparation
The hybrid DNA substrate was assembled as described in detail23 and is similar to the assembly of the RNA hybrid, described above. Briefly, the 89-nucleotide sequence was annealed at a 1:1 ratio with phosphorylated 20-mer followed by ligation overnight at 16 °C with a previously gel-purified 356 bp dsDNA fragment with three-nucleotide sticky ends. The ligated product containing a 69-nucleotide ssDNA region attached to the 376 bp dsDNA was immediately treated with phenol, precipitated with ethanol, and stored at –20 °C. The gel electrophoresis for the ligated ssDNA hybrid product is shown in Figure S2B. The yield of the ligated product was 90%. The final product of the ssDNA hybrid has 376 bp dsDNA fused with 69-nucleotide ssDNA, as shown in Figure S3A.
Preparation of Complexes of A3G with RNA and DNA
Full length human A3G was purified as described in refs 23–25. The A3G–ssDNA and A3G–RNA complexes were formed at a 4:1 protein:DNA ratio in reaction buffer containing 50 mM HEPES (pH 7.5), 100 mM NaCl, 5 mM MgCl2, and 1 mM DTT and incubated for 15 min at 37 °C. All components of the buffer were prepared in DEPC-treated deionized water (AquaMax) and treated with RNAsecure inactivation reagent, as described above. To ensure that treatment does not affect the activity of A3G, the A3G–ssDNA complex was prepared at a 4:1 ratio. The yield of the A3G–ssDNA complex (~83%) was similar to the yield of the A3G–ssDNA complexes obtained without treatment,26 which shows that RNAsecure inactivation reagent-treated A3G maintains its binding activity. The schematic for formation of the A3G complex between the ssDNA hybrid and the RNA hybrid is shown in Figure S3. The protein, depicted as green spheres, forms complexes that are seen at the end of the 376 bp dsDNA for the A3G–ssDNA complex (C) and the 145 bp RNA (D).
Purification of sNTD and CTD-191–384-E259A
For the domain experiments, we used two soluble variants of the NTD and CTD of human A3G, sNTD27 and inactive CTD-191–384-E259A,28,29 respectively. Despite sequence variability (see the sequences of the sNTD and CTD-191–384-E259A in Figure S4), previous studies suggested that the sNTD and CTD-191–384-E259A are structurally similar to the wild-type NTD and CTD.29 The pGEX6P-1 expression vector carrying either the sNTD or the CTD-191–384-E259A (inactive variant) genes fused with an N-terminal GST tag was transformed into BL21 (DE3) cells (Invitrogen).27,28 The amino acid sequences for the sNTD and CTD-191–384-E259A are shown in Figure S4. Cells were grown in LB medium at 37 °C until they reached an optical density of 0.5–0.6 at 600 nm. Then the temperature was changed to 17 °C, and cells were induced by adding 0.2 mM isopropyl β-D-1-thiogalactopyranoside at an optical density of 0.6–0.8. Cells were further grown overnight at 17 °C.
All steps for protein purification were performed at 4 °C, unless specified. Escherichia coli cells were harvested by centrifugation and resuspended in lysis buffer [50 mM sodium phosphate (pH 7.3), 150 mM NaCl, 25 mM ZnCl2, 2 mM DTT, and 0.002% Tween 20] and protease inhibitor (Roche, Basel, Switzerland). The suspended cells were disrupted by sonication, and then cells debris was separated by centrifugation at 20000 rpm for 30 min. The supernatant containing the desired protein was applied to Glutathione-Sepharose (GE Healthcare Life Science) beads, pre-equilibrated with lysis buffer, and agitated for ~2 h. The beads were washed with Pre-Scission Protease cleavage buffer [50 mM sodium phosphate (pH 7.5), 100 mM NaCl, 10 mM ZnCl2, 2 mM DTT, and 0.002% Tween 20] and incubated with Pre-Scission protease (GE Healthcare Life Science) overnight. GST free proteins were separated from beads by centrifugation. The purity and concentration of the proteins were measured by gel electrophoresis and UV spectroscopy.
Preparation of Complexes of RNA with the sNTD and CTD-191–384-E259A
Complexes of each domain and RNA were prepared in the treated binding buffer [50 mM HEPES (pH 7.5), 100 mM NaCl, 5 mM MgCl2, and 1 mM DTT] and incubated at 37 °C for 15 min, as described above. The protein:RNA or protein:ssDNA ratio was 48:1 for both the CTD-191–384-E259A and the sNTD.
Sample Preparation and AFM Imaging in Air
The AFM experiments were performed with the use of an APS-functionalized mica surface as described in detail previously.30 Specifically, 5 μL of the complexes was deposited onto a mica surface for 2 min, washed with DEPC-treated, deionized (AquaMax) water, and dried with argon gas. AFM images were acquired in tapping mode in air with a Multimode IIIa system (Bruker, Santa Barbara, CA). Silicon TESP probes from Bruker with a spring constant of around 42 pN/nm and resonance frequencies around 320 kHz were used.
Sample Preparation and HS-AFM Imaging in Liquid
The methodology for the sample preparation for high-speed AFM (HS-AFM) imaging is described in ref 30. Briefly, a 1.5 mm diameter APS-treated mica disk was mounted on the top of the cylinder stage that was partially submerged in binding buffer [50 mM HEPES (pH 7.5), 100 mM NaCl, 5 mM MgCl2, and 1 mM DTT]. Two microliters of the complexes was deposit onto the APS mica disc for 2 min to allow the complexes to attach to the mica surface. The surface was then washed with binding buffer, and the cylinder stage with the attached sample was immediately immersed in binding buffer to prevent the sample from drying. The data were acquired by continuous scanning of the selected area (200 nm × 200 nm) with a scan rate of 398 ms/frame. The tips for imaging were grown under an electron beam using short cantilevers (BL-AC10DS-A2, Olympus) with a spring constant between 0.1 and 0.2 N/m and a resonance frequency of 400–1000 kHz.
Data Analysis
For each type of complex, the protein volume was obtained using the cross-section feature of the FemtoScan Online software (Advance Technologies Center, Moscow, Russia) as described in detail in refs 23 and 31. The data for protein volume were assembled as a histogram using Origin 6.0 (OriginLab, Northampton, MA).
RESULTS
Characterization of Complexes of A3G with RNA
AFM studies of A3G–RNA complexes were performed using the hybrid substrate approach described in refs 23, 25, and 32. In this approach, RNA or ssDNA is spliced with the DNA duplex that functions as a tag. The substrate is shown schematically in Figure 1A, in which the DNA duplex is depicted with a black line and the RNA tail is shown as a gray line (see Figure S3B,D for specifics of substrate assembly). During formation of the complex, A3G binds RNA and appeares at the end of the dsDNA tag. In our setup, the RNA hybrid substrate consists of a 145 bp dsDNA tag and a 69-nucleotide RNA tail (see Materials and Methods and Figure S3 for details).
Figure 1.
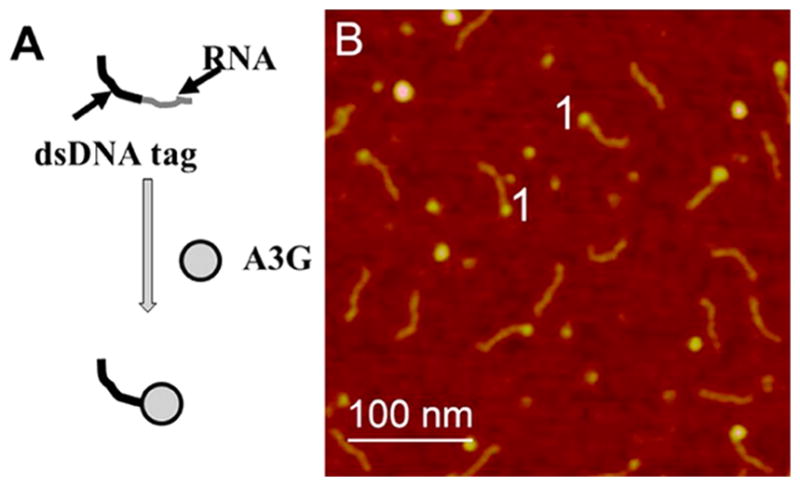
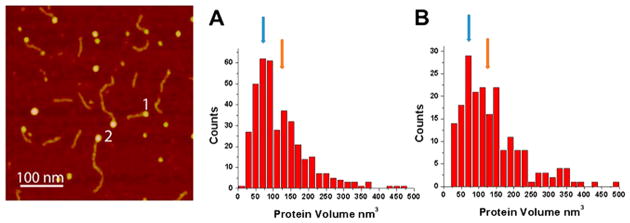
AFM images of A3G–RNA complexes. (A) Hybrid RNA substrate consisting of the dsDNA tag (black line) and the RNA tail (gray line) forming the complex with A3G (imaged by AFM). (B) Image of the hybrid RNA substrate complexed with A3G protein. Complexes appear as bright blobs clearly seen at the end of the dsDNA tag. The complexes are marked with the number 1.
A3G–RNA complexes were assembled at a 4:1 protein:RNA ratio, and examples of typical AFM images for the complexes are shown in Figure 1B. A3G–RNA complexes appear as bright globular features (blobs) at the end of the dsDNA tag and are marked with the number 1. This approach allows us to directly visualize the A3G–RNA complexes and confirm that A3G efficiently binds RNA. Note that the brightness and size of the blobs vary, suggesting that different oligomeric states of A3G exist when it is complexed with RNA. To compare the oligomerization states of A3G in complex with RNA and ssDNA, we characterized the protein stoichiometry of A3G–RNA and A3G–ssDNA complexes.
In these experiments, we used a hybrid DNA substrate with the same length of ssDNA as RNA (69 nucleotides) and a much longer DNA duplex tag (376 bp). Because of the large difference in the length of the dsDNA tags, we were able to distinguish and characterize complexes of A3G with RNA and ssDNA in the mixture of both substrates. Such an experimental setup eliminates the potential concern about the variability of the experimental conditions when the complexes are prepared and visualized with each substrate separately. As in the previous experiment, RNA and ssDNA substrates were mixed with A3G at 4:1 protein:substrate ratio. Figure 2 shows a typical AFM image for the mixed complexes. As seen in the images, A3G complexes appear as bright blobs at the ends of both short (RNA) and long (ssDNA) substrates. In this image, the complexes formed with the RNA hybrid are marked with the number 1 and complexes formed with the ssDNA hybrid with the number 2. We estimated the stoichiometry of both types of complexes by measuring the protein volumes from AFM images. The histograms for the protein volumes were obtained from more than 150 molecules for both A3G–RNA and A3G–ssDNA complexes and are shown in panels A and B of Figure 2, respectively. The protein volumes in the images were measured and translated into protein molecular weights, as described in refs 23 and 30. The two distributions are similar to each other and show a majority of A3G for both substrates as monomers and dimers with a mixture of larger oligomers.
Figure 2.
Determination of the stoichiometry of A3G–RNA and A3G–ssDNA complexes. AFM image of A3G complexes with RNA (1) and ssDNA (2) substrates. The distributions of the protein volume for more than 150 complexes of (A) A3G with RNA and (B) A3G with ssDNA were calculated on the basis of the differences in the length of the dsDNA parts for RNA and ssDNA substrates (see Figure S3 for schematics). The blue and brown arrows indicate the volume values for the A3G monomer and dimer, respectively.
Characterization of Complexes of A3G with Two RNA Molecules
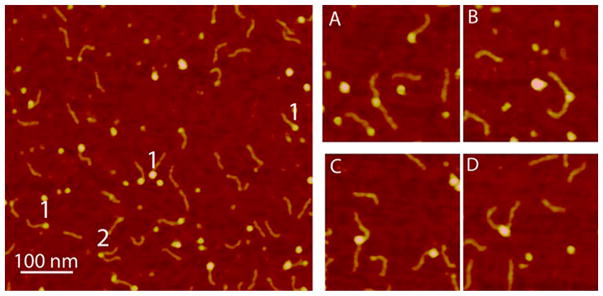
During the analysis of AFM images of A3G with the RNA substrate, we found complexes composed of two RNA molecules attached to A3G. Figure 3 shows the example of this type of complex labeled with the number 2, along with a regular A3G–RNA complex labeld with the number 1. The yield of complexes, labeled with the number 2, is ~10% of all the A3G–RNA complexes. The gallery of selected images in which A3G binds two RNA substrates is shown in Figure 3A–D. The sizes of A3G in these complexes varied. Two RNA molecules can be bound to the A3G monomer (Figure 3A,B) or dimer (Figure 3C,D). Note that complexes with two ssDNA molecules attached to A3G were not found, suggesting that the formation of complexes with two RNA molecules is a unique feature of binding of RNA to A3G.
Figure 3.
AFM images of A3G–RNA complexes. The number 1 marks the regular A3G–RNA complexes, and the number 2 marks complexes with two RNA molecules bound to A3G. At the right is the gallery of AFM images that show the two RNA molecules attached to the (A and B) A3G monomer and (C and D) A3G dimer. The image size is 200 nm.
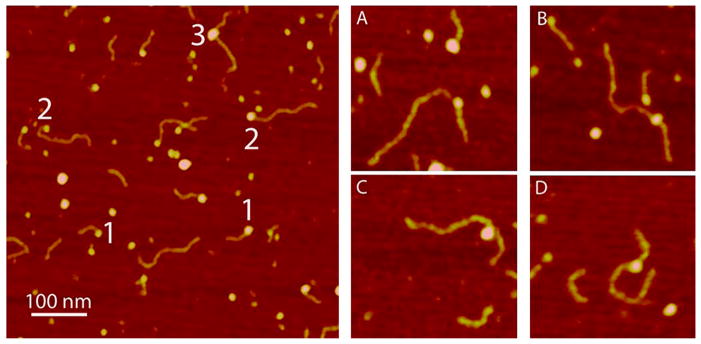
Next, we imaged complexes formed by A3G with the mixture of ssDNA and RNA substrates, and the typical AFM image is presented in Figure 4 at the left. The AFM image shows that in addition to complexes of A3G with RNA (marked with the number 1) and ssDNA (marked with the number 2), there are complexes that contain both ssDNA and RNA substrates bound to the same A3G protein (marked with the number 3). However, the yield of these chimeric complexes is quite low, ~2% of all the complexes. The gallery of chimeric complexes is shown in Figure 4A–D. The sizes of A3G vary, suggesting that in chimera A3G complexes exist as a monomer (Figure 4A,B), as well as a dimer (Figure 4C,D).
Figure 4.
AFM image of complexes of A3G with the RNA/ssDNA mixture. The number 1 marks A3G–RNA complexes; the number 2 marks A3G–ssDNA complexes, and the number 3 marks A3G–RNA–ssDNA complexes. The gallery of AFM images (at the right) shows chimeric ssDNA and RNA complexes bound to the (A and B) A3G monomer and (C and D) dimer. The image size is 200 nm.
Characterization of Complexes of RNA with the sNTD and CTD-191–384-E259A
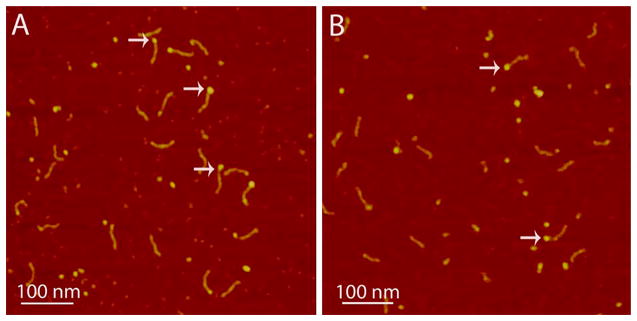
To determine which domain contributes to A3G–RNA interactions, we assembled complexes of RNA with the isolated CTD-191–384-E259A or the sNTD. Typical AFM images of CTD-191–384-E259A–RNA and sNTD–RNA complexes are presented in panels A and B of Figure 5, respectively. As in the images obtained for the full size A3G (Figures 1–4), complexes with individual domains are identified as blobs located at the end of the dsDNA tag, as indicated with arrows in Figure 5. However, the sizes of the blobs for both domains are smaller that the sizes obtained with the full size protein, which reflect the smaller sizes of the individual domains compared to full length A3G. Note that RNA binds less efficiently to the individual the CTD-2K3A-E259A and sNTD than to full size A3G; therefore, it was necessary to use protein:substrate molar ratio as large as 48:1 to obtain a reasonable yield of complexes.
Figure 5.
Complexes of RNA with the (A) CTD-2K3A-E259A and (B) sNTD. A few typical complexes are indicated with arrows. The yield of RNA–CTD-2K3A-E259A complexes is 42%, and the yield of RNA–sNTD complexes is 22%.
Yields of complexes for both domains were 22 and 42% for sNTD–RNA and CTD-191–384-E259A–RNA complexes, respectively. Our observation of 2-fold lower yield of the sNTD–RNA complex does not correlate with previous studies that suggest that the NTD plays a major role in RNA binding.33 The 2-fold lower yield of the sNTD–RNA complex is most likely due to the mutation introduced into the sNTD variant to make it soluble.27 However, the fact that the CTD-2K3A-E259A forms complexes with RNA suggests that the CTD also contribute to RNA binding with the full-size A3G.
Under our experimental conditions, we do not observe any formation of a complex of ssDNA with either the CTD-191–384-E259A or the sNTD (see Figure S5). The fact that no complex formed between the CTD-191–384-E259A and ssDNA is consistent with previous observations by Xiao et al.,34 who showed that the hA3G–CTD complex produces no shift or a very weak band shift in a gel shift assay with ssDNA, even at significantly higher hA3G–CTD complex concentrations.34 However, the lack of complex formation between the sNTD and ssDNA could be a reflection of mutations introduced into the sNTD. Therefore, our observations suggest that both the CTD and the NTD are required to achieve high-affinity binding to ssDNA.
Dynamics of A3G–RNA Complexes
To follow the dynamics of A3G–RNA complexes, we used time-lapse HS-AFM. In this AFM mode, imaging is performed in solution, which eliminates the potential concern of sample drying. Additionally, time-lapse imaging over the same area allows one to follow the dynamics of the complex. The acquisition of data during HS-AFM experiments is 1000 times faster than in regular time-lapse AFM, which allows us to follow the dynamics of the complex on the millisecond time scale.
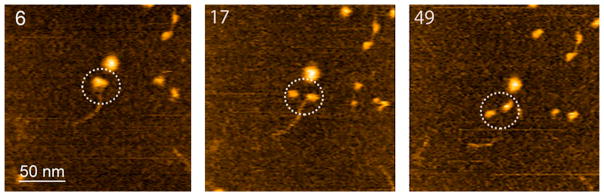
Figure 6 shows three frames of 52 from Movie 1 that demonstrate the dissociation process of the A3G–RNA complex. The initial A3G–RNA complex in frame 6 appears as a globular feature. Over time, one monomer dissociates (frame 17) while the other remains attached to RNA, suggesting that A3G was initially a dimer. The remaining monomeric complex was stable and dissociated only at frame 49. This two-step dissociation pattern was observed in seven different movies, suggesting that this is a common dissociation pathway for dimeric A3G–RNA complexes. A similar pattern was observed in our experiments with A3G–ssDNA complexes.30 The two-step dissociation pattern suggests that free protein, dissociated from the complex, is mostly monomeric. Indeed, the measured volumes of free protein from dry AFM images of complexes of A3G with RNA and ssDNA substrates show that A3G exists primarily in the monomeric state (Figure S6), thereby supporting our hypothesis.
Figure 6.
HS-AFM images of RNA–A3G complexes. Selected frames of 52 assembled as Movie 1. The circles indicate the complex undergoing dissociation. The scanning rate corresponds to 398 ms/frame.
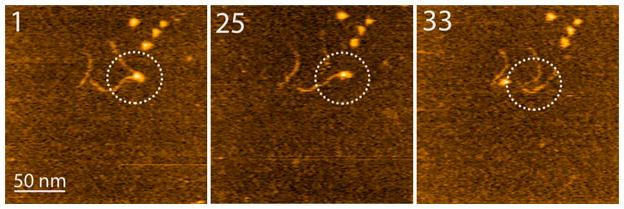
Next, we used time-lapse HS-AFM to characterize the dynamics of complexes in which A3G binds two RNA molecules. The three frames in Figure 7 illustrate the dynamics of the complex formed by the A3G monomer. The entire dynamics can be seen in Movie 2. Initially, the A3G monomer forms a complex with two RNA molecules (frame 1). Then, one of the RNA molecules dissociates (frame 25), while another RNA molecule remains bound to the A3G monomer. This RNA eventually dissociates, so the fully dissociated complex is shown in frame 33. This dissociation pattern was observed in five different movies, but simultaneous dissociation of two RNA molecules was never observed. This suggests that the efficiency of binding of RNA to two A3G sites within these complexes is different.
Figure 7.
HS-AFM images of the dissociation of the A3G monomer complexed with RNA. Selected frames of 36 assembled as Movie 2. The complexes undergoing dissociation are circled. The scanning rate corresponds to 398 ms/frame.
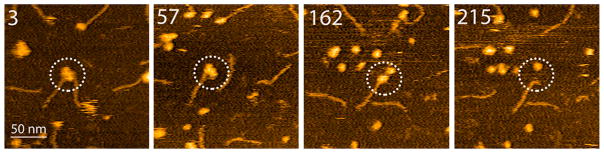
Four frames in Figure 8 show the dissociation process for the complexes of two RNA molecules bound to the A3G dimer (frame 3), and the entire process can be seen in Movie 3. In frame 57, one RNA molecule dissociates, leaving the A3G dimer attached to the second RNA molecule. This pattern is similar to that in Figure 6 in which one RNA molecule binds to the A3G dimer. In frame 162, the A3G monomer dissociates from the complex, demonstrating a two-step dissociation pattern. Finally, in frame 215, the A3G monomer dissociates from RNA, suggesting stronger interaction of RNA with one A3G region compared to the strength of the monomer–monomer bond in the A3G dimer.
Figure 8.
HS-AFM images of the dissociation of the A3G dimer complexed with RNA. Selected frames of 226 assembled as Movie 3. The circles indicate the complex undergoing dissociation. The scanning rate corresponds to 398 ms/frame.
DISCUSSION
Here we present the first direct visualization and dynamic behavior of A3G–RNA complexes and reveal a number of novel properties that might have important biological implications.
Specificity and Stoichiometry of A3G–RNA Complexes
Our results show that A3G binds RNA with the same binding efficiency as ssDNA, despite the lack of deaminase activity for RNA. Moreover, the stoichiometries of A3G in complex with RNA and ssDNA as determined from the volume measurements are very similar to one another. Importantly, these measurements were taken from the samples in which two different substrates were mixed together, so potential concerns about the variability of the reaction conditions have been eliminated. The primary species in these complexes were monomers and dimers, with a very low percentage of higher-order oligomers. Although there was a discussion in the literature regarding the stimulation of A3G oligomerization upon binding to RNA,14,35,36 our results primarily show the existence of monomeric and dimeric A3G states bound to RNA. These results are in line with the data presented in ref 37 that showed the capability of A3G monomers to bind to RNA
Time-lapse HS-AFM data allowed us to follow the dynamics of the A3G–RNA complexes (Figure 6). The data demonstrate that the dissociation of the complex containing the A3G dimer is a two-step process in which one monomer dissociates prior to the dissociation of the A3G–RNA complex. These findings suggest that in these complexes, A3G–RNA interactions are stronger than interprotein interactions. This is a property similar to one we observed for complexes of A3G dimers with ssDNA.30
Two RNA Molecules Binding to A3G
One of our novel findings is the formation of complexes in which two RNA molecules are bound to A3G. This property of RNA is entirely different from that of ssDNA, which never binds to A3G in pairs. Moreover, the appearance of the chimera complexes that contain one ssDNA and one RNA bound to A3G strongly indicate an extra binding region on A3G for RNA only. Therefore, our data demonstrate that while ssDNA has only one binding site on A3G, RNA has an additional binding region on A3G. This finding is consistent with the data presented in ref 22 that revealed an additional binding region for RNA located at the N-terminal domain at amino acids 15–29, 41–52, and 83–99.
Using HS-AFM (Figure 7), we also were able to characterize the dynamics of such complexes. The time-lapse studies show that dissociation of RNA molecules from two RNA–A3G complexes is a two-step process, suggesting that the efficiency of binding of RNA to two A3G sites is different. The dissociation of the complex when the A3G monomer bound to two RNA molecules suggests that one RNA binding region of A3G interacts stronger with RNA than a second does. This effect is even more pronounced when two RNA molecules are bound to the A3G dimer (see Movie 3 and selected frames in Figure 8). In this case, one RNA molecule dissociates first, leaving the A3G dimer still attached to the other RNA molecule. These data confirm that the two A3G sites have different binding efficiencies with RNA and indicate that the interaction between RNA and one of the A3G binding sites is weaker than protein–protein interaction in the A3G dimer.
Complexes of RNA and ssDNA with the NTD and CTD
The existence of two regions on A3G that are capable of binding to RNA is fully supported by the results from experiments with RNA in complex with the sNTD and CTD-191–384-E259A. Indeed, our results clearly show that RNA is capable of binding the CTD-191–384-E259A or sNTD, but no such complexes were observed with ssDNA. Our observation of chimeric complexes in which one ssDNA molecule and one RNA molecule are bound to A3G also confirms that RNA has an additional binding site on A3G that is separate from the one that binds to ssDNA.
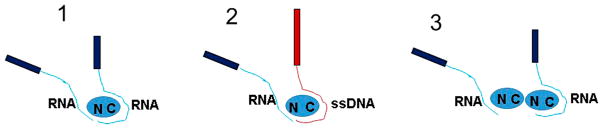
Model for A3G–RNA Complexes
Our results clearly demonstrate that A3G has two separate regions for binding RNA, but only one for ssDNA. The presence of an additional RNA binding site is directly supported by the appearance of two RNA molecules bound to A3G and chimeric complexes consisting of one ssDNA and one RNA molecule bound to A3G simultaneously. From the dynamic behavior of two A3G–RNA complexes, we showed that these two sites have a different binding affinity for A3G. Together, these data allow us to propose a model for binding of RNA to A3G, which is illustrated in Figure 9. The RNA hybrid on the cartoon consists of two parts, a short thick blue bar representing the dsDNA tag and a thin blue line representing the RNA tail; the ssDNA hybrid has a long red thick bar representing the dsDNA tag with a thin red line representing the ssDNA tail. The model in which two RNA molecules are bound to two A3G sites is indicated with the number 1. We suggest that the site, shown as a long blue line on the right, is the strongest RNA binding site on A3G and includes both the CTD and the NTD, similar to A3G–ssDNA complexes as described in refs 30 and 37 and shown by the red thin line in the cartoon (marked with the number 2 on the right). Indeed, the majority of the A3G–RNA complexes contain one RNA bound to A3G. This model is also supported by our data showing that no ssDNA complexes are formed with either the CTD-191–384-E259A or the sNTD, even at a very high protein:substrate ratio.
Figure 9.
Model of the A3G–RNA complex. Proposed model of A3G in complex with two RNA molecules and with RNA and ssDNA. The blue and red thin lines represent RNA and ssDNA, respectively. The bars represent the dsDNA tags for RNA (short blue bar) and dsDNA (long red bar) (see the schematic for the substrate design in Figure S3): (1) complex with two RNA molecules attached to the A3G monomer, (2) complex with one ssDNA and one RNA attached to the A3G monomer, and (3) complex with two RNA molecules attached to the hypothesized assembly of the A3G dimer.
We suggest that the second, additional RNA binding site on A3G is located on the NTD, as illustrated by the thin blue line on the left and marked with the number 1. The model for this site on A3G is derived from the fact that RNA may form complexes with the sNTD. The data obtained from the dynamic behavior of two RNA molecules bound to a monomer or dimer of A3G confirm that one of the RNA binding sites on A3G is less efficient than the other. The existence of chimeric complexes in which one RNA and one ssDNA are bound to A3G also provides additional support for this model.
Model 2, depicted in the cartoon in Figure 9, illustrates the chimeric complex with ssDNA and RNA attached to A3G. The ssDNA is shown as a thin red line covering the CTD and the NTD on A3G (see the number 2). The RNA molecule is shown as a blue line that is in contact with only the NTD (see the number 2), which is the additional binding site for RNA within A3G. We suggest that the existence of A3G complexes containing two RNA molecules or one RNA and one ssDNA attached to A3G is more rare than the existence of regular RNA–A3G complexes because RNA binds less efficiently to the NTD than to both the CTD and the NTD.
Model 3 shows complexes consisting of two RNA molecules interacting with the A3G dimer. The dynamic behavior of complexes illustrated in Figure 8 (Movie 3) shows that one RNA molecule dissociates first, suggesting that this RNA was bound to the weaker binding site. Moreover, it shows that protein–protein interactions within the A3G dimer are stronger than the interactions between RNA and A3G at this binding site.
Note that the data obtained from complexes consisting of two RNA molecules or one RNA and one ssDNA molecule bound to the A3G monomer allow us to speculate about the assembly of A3G into dimers. On the basis of the proposed model for A3G monomer complexes, the most probable arrangement for dimeric A3G is a head-to-tail structure. Indeed, when the A3G dimer has a head-to-tail structure, two RNA molecules or one ssDNA and one RNA molecule have a binding pattern similar to the pattern associated with A3G monomeric complexes. Recently, the head-to-tail dimer conformation was suggested as the most probable structural model of full length A3G.38
In summary, single-molecule AFM imaging and HS-AFM approaches allowed us to directly visualize RNA–A3G complexes and follow their dynamic behavior. The hybrid design in which dsDNA is used as a ruler to unambiguously identify each individual ssDNA and RNA complex with A3G allowed us to directly compare the A3G stoichiometry in the complexes without the ambiguity of separate sample preparations. The analysis of the data shows similar oligomerization states of A3G bound to RNA and ssDNA. Moreover, new, previously unknown, A3G–RNA complexes have been discovered. Two separate RNA binding regions on A3G have been observed and characterized. Finally, we proposed a model for different mechanisms for binding of RNA and ssDNA to A3G.
Supplementary Material
Acknowledgments
Funding
This work was supported by grants from the National Institute of General Medical Sciences (R01-GM118006, GM096039, and GM100156 to Y.L.L. and P01-GM091743 to R.S.H.) and from the National Science Foundation (MCB 1515346 to Y.L.L.). R.S.H. is an Investigator of the Howard Hughes Medical Institute. For T.K., A.M. and H.M., this project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
We thank Lyubchenko’s lab members for very helpful discussion.
ABBREVIATIONS
- A3G
APOBEC3G
- AFM
atomic force microscopy
- HS-AFM
high-speed AFM
- APS
1-(3-aminopropyl)silatrane
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
Footnotes
Notes
The authors declare the following competing financial interest(s): R.S.H. is a co-founder, shareholder, and consultant of ApoGen Biotechnologies, Inc. The other authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.6b01189.
Figures S1–S6 (PDF)
Movie 1 (AVI)
Movie 2 (AVI)
Movie 3 (AVI)
References
- 1.Harris RS, Dudley JP. APOBECs and virus restriction. Virology. 2015;479–480:131–145. doi: 10.1016/j.virol.2015.03.012. and references therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salter JD, Bennett RP, Smith HC. The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem Sci. 2016;41:578–594. doi: 10.1016/j.tibs.2016.05.001. and references therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 4.Belanger K, Savoie M, Rosales Gerpe MC, Couture JF, Langlois MA. Binding of RNA by APOBEC3G controls deamination-independent restriction of retroviruses. Nucleic Acids Res. 2013;41:7438–7452. doi: 10.1093/nar/gkt527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belanger K, Langlois MA. RNA-binding residues in the N-terminus of APOBEC3G influence its DNA sequence specificity and retrovirus restriction efficiency. Virology. 2015;483:141–148. doi: 10.1016/j.virol.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 6.Hache G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J Biol Chem. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- 7.Chelico L, Pham P, Goodman MF. Mechanisms of APOBEC3G-catalyzed processive deamination of deoxycytidine on single-stranded DNA. Nat Struct Mol Biol. 2009;16:454–455. doi: 10.1038/nsmb0509-454. author reply 455–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 9.Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3′ –> 5′ on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y, Baig TT, Love RP, Chelico L. Suppression of APOBEC3-mediated restriction of HIV-1 by Vif. Front Microbiol. 2014;5:450. doi: 10.3389/fmicb.2014.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaurasiya KR, McCauley MJ, Wang W, Qualley DF, Wu T, Kitamura S, Geertsema H, Chan DS, Hertz A, Iwatani Y, Levin JG, Musier-Forsyth K, Rouzina I, Williams MC. Oligomerization transforms human APOBEC3G from an efficient enzyme to a slowly dissociating nucleic acid-binding protein. Nat Chem. 2014;6:28–33. doi: 10.1038/nchem.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan MA, Kao S, Miyagi E, Takeuchi H, Goila-Gaur R, Opi S, Gipson CL, Parslow TG, Ly H, Strebel K. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. Journal of virology. 2005;79:5870–5874. doi: 10.1128/JVI.79.9.5870-5874.2005. and references therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strebel K, Khan MA. APOBEC3G encapsidation into HIV-1 virions: which RNA is it? Retrovirology. 2008;5:55. doi: 10.1186/1742-4690-5-55. and references therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shindo K, Li M, Gross PJ, Brown WL, Harjes E, Lu Y, Matsuo H, Harris RS. A Comparison of Two Single-Stranded DNA Binding Models by Mutational Analysis of APOBEC3G. Biology. 2012;1:260–276. doi: 10.3390/biology1020260. and references therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prohaska KM, Bennett RP, Salter JD, Smith HC. The multifaceted roles of RNA binding in APOBEC cytidine deaminase functions. Wiley interdisciplinary reviews RNA. 2014;5:493–508. doi: 10.1002/wrna.1226. and references therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDougall WM, Smith HC. Direct evidence that RNA inhibits APOBEC3G ssDNA cytidine deaminase activity. Biochem Biophys Res Commun. 2011;412:612–617. doi: 10.1016/j.bbrc.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Apolonia L, Schulz R, Curk T, Rocha P, Swanson CM, Schaller T, Ule J, Malim MH. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog. 2015;11:e1004609. doi: 10.1371/journal.ppat.1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.York A, Kutluay SB, Errando M, Bieniasz PD. The RNA Binding Specificity of Human APOBEC3 Proteins Resembles That of HIV-1 Nucleocapsid. PLoS Pathog. 2016;12:e1005833. doi: 10.1371/journal.ppat.1005833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 21.Imahashi M, Nakashima M, Iwatani Y. Antiviral Mechanism and Biochemical Basis of the Human APOBEC3 Family. Front Microbiol. 2012;3:250. doi: 10.3389/fmicb.2012.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polevoda B, McDougall WM, Tun BN, Cheung M, Salter JD, Friedman AE, Smith HC. RNA binding to APOBEC3G induces the disassembly of functional deaminase complexes by displacing single-stranded DNA substrates. Nucleic Acids Res. 2015;43:9434–9445. doi: 10.1093/nar/gkv970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shlyakhtenko LS, Lushnikov AY, Li M, Lackey L, Harris RS, Lyubchenko YL. Atomic force microscopy studies provide direct evidence for dimerization of the HIV restriction factor APOBEC3G. J Biol Chem. 2011;286:3387–3395. doi: 10.1074/jbc.M110.195685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Shandilya SM, Carpenter MA, Rathore A, Brown WL, Perkins AL, Harki DA, Solberg J, Hook DJ, Pandey KK, Parniak MA, Johnson JR, Krogan NJ, Somasundaran M, Ali A, Schiffer CA, Harris RS. First-in-class small molecule inhibitors of the single-strand DNA cytosine deaminase APOBEC3G. ACS Chem Biol. 2012;7:506–517. doi: 10.1021/cb200440y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shlyakhtenko LS, Lushnikov AY, Miyagi A, Lyubchenko YL. Specificity of binding of single-stranded DNA-binding protein to its target. Biochemistry. 2012;51:1500–1509. doi: 10.1021/bi201863z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shlyakhtenko LS, Dutta S, Banga J, Li M, Harris RS, Lyubchenko YL. APOBEC3G Interacts with ssDNA by Two Modes: AFM Studies. Sci Rep. 2015;5:15648. doi: 10.1038/srep15648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouno T, Luengas EM, Shigematsu M, Shandilya SM, Zhang J, Chen L, Hara M, Schiffer CA, Harris RS, Matsuo H. Structure of the Vif-binding domain of the antiviral enzyme APOBEC3G. Nat Struct Mol Biol. 2015;22:485–491. doi: 10.1038/nsmb.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen KM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, Harris RS, Matsuo H. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 29.Shandilya SM, Nalam MN, Nalivaika EA, Gross PJ, Valesano JC, Shindo K, Li M, Munson M, Royer WE, Harjes E, Kono T, Matsuo H, Harris RS, Somasundaran M, Schiffer CA. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010;18:28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shlyakhtenko LS, Lushnikov AY, Miyagi A, Li M, Harris RS, Lyubchenko YL. Atomic force microscopy studies of APOBEC3G oligomerization and dynamics. J Struct Biol. 2013;184:217–225. doi: 10.1016/j.jsb.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shlyakhtenko LS, Gilmore J, Kriatchko AN, Kumar S, Swanson PC, Lyubchenko YL. Molecular mechanism underlying RAG1/RAG2 synaptic complex formation. J Biol Chem. 2009;284:20956–20965. doi: 10.1074/jbc.M109.028977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shlyakhtenko LS, Lushnikov AJ, Li M, Harris RS, Lyubchenko YL. Interaction of APOBEC3A with DNA assessed by atomic force microscopy. PLoS One. 2014;9:e99354. doi: 10.1371/journal.pone.0099354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao X, Li SX, Yang H, Chen XS. Crystal structures of APOBEC3G N-domain alone and its complex with DNA. Nat Commun. 2016;7:12193. doi: 10.1038/ncomms12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Chen Y, Li M, Carpenter MA, McDougle RM, Luengas EM, Macdonald PJ, Harris RS, Mueller JD. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. J Mol Biol. 2014;426:1296–1307. doi: 10.1016/j.jmb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 2009;5:e1000330. doi: 10.1371/journal.ppat.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chelico L, Prochnow C, Erie DA, Chen XS, Goodman MF. Structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. J Biol Chem. 2010;285:16195–16205. doi: 10.1074/jbc.M110.107987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu X, Zhang T, Xu Z, Liu S, Zhao B, Lan W, Wang C, Ding J, Cao C. Crystal Structure of DNA Cytidine Deaminase ABOBEC3G Catalytic Deamination Domain Suggests a Binding Mode of Full-length Enzyme to Single-stranded DNA. J Biol Chem. 2015;290:4010–4021. doi: 10.1074/jbc.M114.624262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.