Abstract
Eukaryotic cells consist of a complex network of thousands of proteins present in different organelles where organelle-specific cellular processes occur. Identification of the subcellular localization of a protein is important for understanding its potential biochemical functions. In the post-genomic era, localization of unknown proteins is achieved using multiple tools including a fluorescent-tagged protein approach. Several fluorescent-tagged protein organelle markers have been introduced into dicot plants, but its use is still limited in monocot plants. Here, we generated a set of multicolored organelle markers (fluorescent-tagged proteins) based on well-established targeting sequences. We used a series of pGWBs binary vectors to ameliorate localization and co-localization experiments using monocot plants. We constructed different fluorescent-tagged markers to visualize rice cell organelles, i.e., nucleus, plastids, mitochondria, peroxisomes, golgi body, endoplasmic reticulum, plasma membrane, and tonoplast, with four different fluorescent proteins (FPs) (G3GFP, mRFP, YFP, and CFP). Visualization of FP-tagged markers in their respective compartments has been reported for dicot and monocot plants. The comparative localization of the nucleus marker with a nucleus localizing sequence, and the similar, characteristic morphology of mCherry-tagged Arabidopsis organelle markers and our generated organelle markers in onion cells, provide further evidence for the correct subcellular localization of the Oryza sativa (rice) organelle marker. The set of eight different rice organelle markers with four different FPs provides a valuable resource for determining the sub-cellular localization of newly identified proteins, conducting co-localization assays, and generating stable transgenic localization in monocot plants.
Keywords: biolistic bombardment, cell organelle markers, co-localization, fluorescent proteins, EYFP, ECFP, GFP, mRFP, subcellular localization, rice
INTRODUCTION
Eukaryotic cells consist of distinct membrane-bound organelles with specific biochemical functions. Plant cells also contain many membrane-bound cellular structures that perform specific functions required for survival and normal functioning of the cells. Marker proteins/enzymes present in a particular compartment define the biochemical function of that organelle; therefore, determining the subcellular localization of unknown proteins is important for understanding their biological functions and the organization of cellular activity. Based on this concept, several computational and experimental approaches to determine the subcellular localization of proteins have been developed (Lunn, 2007; Tanz et al., 2013).
The computational approach identifies a short nucleic acid sequence of a gene known as a targeting sequence (Li et al., 2006). The specific protein recognized by the targeting sequence helps to sort the protein into a specific organelle. Most bioinformatics tools aim to identify the signal sequence at the amino (N)-terminus that targets the protein either to plastids, mitochondria, or the endomembrane system [endoplasmic reticulum, plasmalemma, vacuoles] and apoplast (Lunn, 2007). In general, this approach might be useful for prediction; however, in some cases, it gives false negative results (Hezlewoaod et al., 2004; Richly and Leister, 2004). Though the computational analysis can likely identify the specific compartment of an unknown protein, eventually these results need further confirmation by experimental analysis. Traditionally, cell fractionation and immunohistochemistry were commonly used experimental approaches. Cell fractionation relies on fractionation of the cell lysate by differential centrifugation, followed by measuring the activity of the unknown protein using a known marker protein for each compartment (Lunn, 2007). These studies are usually carried out on spinach leaves, castor bean seedlings, and cell suspension cultures, and although information obtained from these models is valid for comparable tissues, there is a high probability of contamination with different organelles (Lunn, 2007). Additionally, immunohistochemistry is based on specific antibodies that are labeled with fluorescence, heavy atom, or enzyme markers (Tobin and Yamaya, 2001). Generating specific antibodies is time-consuming, expensive, and unspecific, as in the case of closely related gene family members. Further, proteomic approaches, combined with traditional cell fractionation methods, are used to specify subcellular compartments (Lunn, 2007). Since data obtained using this approach are dependent on the purity of the original organelle preparation and the level of contamination with other cellular proteins (Dunkley et al., 2006; Komatsu et al., 2007; Lilley and Dupree, 2006), this method is not an effective means of locating unknown proteins.
Recently, the simple and highly efficient fluorescent-tagged protein (FTP) approach has been used to visualize protein localization in cells. Fluorescent proteins (FPs) are generated using marine invertebrates e.g., green fluorescent protein (GFP) from jelly fish (Aequorea victoria) and red fluorescent protein (RFP) from sea anemone, Discosoma striata (Chalfie et al., 1994; Matz et al., 1999). Remarkable progress has been made in the development and availability of FPs (Day and Davidson, 2009), and they have been applied in protein localization studies in different organisms, including yeast, Drosophila, Caenorhabditis elegans, bacteria such as Streptococcus pneumonia, and plants (Henriques et al., 2013; O’Rourke et al., 2005; Stadler et al., 2013). In plant studies, most organelle markers have been generated for Arabidopsis thaliana, Medicago truncatula, maize, and algae (Nannochloropsis oceanica) (Cutler et al., 2000; Luo and Nakata, 2012; Moog et al., 2015; Nelson et al., 2007; Wu et al., 2013).
Rice (Oryza sativa), the monocot model plant, is a major world crop containing thousands of genes with unknown biological functions. It is crucial to define where novel genes are located in order to determine their functional significance. Determining the subcellular compartment of a gene using traditional approaches is more applicable to dicot than monocot plants (Komatsu et al., 2007). Several FTP organelle markers have been developed for dicot plants, but marker development has been limited in rice. At present, a mCherry-tagged gene expression system and GFP-tagged organelle markers have been constructed for subcellular localization and protein secretion analysis in rice cells (Wang et al., 2013; Wu et al., 2016). Here, we present information on the generation of fluorescent markers for several organelles in rice. All markers were constructed by fusing four FPs [EYFP (enhanced yellow fluorescent protein), ECFP (enhanced cyan fluorescent protein), G3GFP (G3 green fluorescent protein), and mRFP (monomeric red fluorescent protein)] at the amino (N)-terminal using a series of Gateway binary vectors (pGWBs) (Nakagawa et al., 2007). These organelle markers can be used for both in vitro and in vivo localization studies and are compatible with both transient and stable expression systems. Using these organelle markers, it will be easy to locate plasma membrane, nucleus, endoplasmic reticulum, tonoplast, mitochondria, golgi bodies, plastids, and peroxisomes in rice cells. This set of organelle markers could provide an additional powerful resource for rice and the monocot crop research community for localization and functional studies.
MATERIALS AND METHODS
Plant material and growth conditions
The rice wild-type cultivar Nipponbare was used for all localization experiments. Plants were placed in a growth chamber under 60% humidity and 16 h light and 8 h dark conditions at 28°C for 5–6 weeks. The wild-type rice seeds were obtained from the National Institute of Crop Science (http://www.nics.go.kr).
Generation of binary constructs of organelle markers
All targeting signal peptide and protein sequences required to generate organelle markers were selected on the basis of previously reported organelle markers in Arabidopsis (Nelson et al., 2007). The selected organelle markers were first amplified from the rice cDNA library with gene-specific primers containing attB1 and attB2 sites, as mentioned in the primer table (Supplementary Table S1). The amplified PCR products were further used as a template for the second PCR using attB1 and attB2 primers. The second PCR product was then sub-cloned into the pDONR™201 entry vector using BP clonase (Invitrogen) to create entry clones. The entry clones were recombined into Gateway binary vectors (pGWBs) containing four different FP tags: EYFP (pGWB542); ECFP (pGWB545); G3GFP (pGWB552), and mRFP (pGWB555). This was achieved using LR clonase (Invitrogen). A similar procedure was used for the construction of mRFP-fused nuclear localization sequence (NLS), GFP-fused OsMEKK25, and mRFP-fused OsMEKK24. All cloned marker plasmids were finally confirmed by sequencing (Macrogen).
Subcellular localization and co-localization assay
Rice leaf sheath preparation, onion tissue preparation, DNA preparation, and biolistic bombardment were performed as described previously (Singh et al., 2012; 2014; Wang et al., 2013). Briefly, 20 μg of DNA was coated with tungsten M-17 particles and delivered into rice epidermal cells placed on half-strength Murashige and Skoog medium plates (MS media), prepared from 4–5-week-old seedlings using biolistic bombardment (Bio-Rad, Biolistic®-PDS-1000/He Particle Delivery System), as described previously (Singh et al., 2012; Wang et al., 2013). Similarly, 8 μg of DNA coated with tungsten M-17 particles was delivered into onion epidermal cells placed on MS media using biolistic bombardment. The bombarded samples of both rice and onion were incubated in the dark at 25°C for 48 h and 12–24 h, respectively. For the co-localization assay, 8 μg of mRFP-fused NLS and 8 μg of G3GFP-fused nucleus marker were co-transformed in onion epidermal cells and 20 μg of both DNAs were co-transformed in rice cells using biolistic bombardment. The bombarded samples of both rice and onion were incubated in the dark at 25°C for 48 h and 12–24 h, respectively. Similar procedures were applied for co-expression of mRFP-fused OsMEKK24 with GFP-fused ER marker and GFP-fused Os-MEKK25 with mRFP-fused plasma membrane marker in rice and onion epidermal cells.
Microscopic analysis
After incubation, the subcellular localization and co-localization of each expressed protein was visualized using a confocal microscope (Leica, TCS SP5). Images of transformed onion and rice cells were captured using a 20× objective lens and a 40× oil immersion objective lens, respectively, using bright field, YFP (Ex/Em: 514/520–554 nm wavelength), GFP (Ex/Em: 488/498–548 nm), RFP (Ex/Em: 543/592–627 nm), CFP (Ex/Em: 458/467–511 nm), mCherry (Ex/Em: 543/560–631 nm), and DAPI (Ex/Em: 405/421–523 nm) filters. Additionally, to prevent autofluorescence, an extra filter was used in the autofluorescence range for each FP. Chlorophyll autofluorescence was distinguished from real fluorescence with emission wavelengths >600 nm when excited with light of wavelengths between 420 and 460 nm. Images were taken in sequential line mode (Multitracking mode) to prevent crosstalk with chlorophyll fluorescence.
Accession numbers
Sequence data from this article can be found on the Rice Genome Project website (http://rice.plantbiology.msu.edu/) under the following accession numbers: OsSPP1, Os02g 02530; OsManI, Os04g51690; OsTIP, Os03g05290; OsALDH2a, Os02g49720; OsPIP2.1, Os02g41860; OsTAF2H, Os09g26180; OsAPX3, Os04g14680; OsERD1, Os02g32520; OsMEKK24, Os04g56530.1; OsMEKK25, Os02g38080.
RESULTS
Short targeting sequences and full-length sequences were selected for multicolored organelle markers
The localization of the well-studied short targeting and full-length sequences to the specific subcellular compartments provides the basis for visualizing the rice organelles using the fluorescent markers generated in this study. Since these markers contain exact subcellular organelle targeting sequences, they can localize directly to the respective compartment without altering cellular native function. Initially, we selected short targeting and full-length protein sequences that had been used successfully for localization assays in Arabidopsis as a reference to search for orthologs (https://www.ncbi.nlm.nih.gov/UniGene/) or similar proteins in rice; these were subsequently used to generate multicolored fluorescence markers (Nelson et al., 2007; Saint-Jore-Dupas et al., 2006).
The first 99 amino acids (aa) of the rice mitochondrial aldehyde dehydrogenase, OsALDH2a, were used as the targeting sequence for mitochondria (Nakazono et al., 2000). Additionally, the cytoplasmic tail, transmembrane domain, and luminal stalk (first 49 aa) of the rice alpha-1–2 mannosidase I (OsManI), an ortholog of Glycine max GmManI, which localizes in the Golgi body, were used to generate the Golgi body marker (Saint-Jore-Dupas et al., 2006). In the present study, only mitochondrial and golgi body markers were based on fusion of short targeting signals to the FP and the remaining markers were based on full-length protein fusions.
The full-length rice signal peptide peptidase (OsSPP1) sequence, which has an endoplasmic reticulum targeting sequence (first 26 aa) and an endoplasmic reticulum retrieval motif (last 4 aa), was used to generate the endoplasmic reticulum marker (Tamura et al., 2009). For the tonoplast marker, we used the rice tonoplast intrinsic protein (OsTIP), which is an ortholog of the Arabidopsis tonoplast targeting protein [gamma-tonoplast intrinsic protein (γ-TIP)] (Saito et al., 2002). The plasma membrane marker was constructed using a full-length sequence of the rice plasma membrane aquaporin (PIP2.1), which is an ortholog of Arabidopsis AtPIP2A, whereas the peroxisome targeting sequence was based on the previously reported rice peroxisome-bound ascorbate peroxidase (OsAPX3) (Li et al., 2011; Teixeira et al., 2006). Furthermore, the rice transcription factor TFIID subunit 10, OsTAF2H, was found to be localized in the nucleus (Singh et al., 2012). We constructed the nuclear marker using the full-length sequence of OsTAF2H. For the plastid marker, the chloroplast precursor protein (OsERD1), an ortholog of Arabidopsis ClpC-like protein (AtERD1), was used (Weaver et al., 1999). Essential information on selected organelle markers with references from other plant species is summarized in Table 1.
Table 1.
List of organelle markers used in this study
| Organelle | Binary plasmids used | Signal peptide or fused protein | Reference | |||||
|---|---|---|---|---|---|---|---|---|
| G3GFP | EYFP | mRFP | ECFP | Bacterial Selection Marker | Plant selection marker | |||
| Endoplasmic reticulumn (ER) | pGWB552 | pGWB542 | pGWB555 | pGWB545 | Spectinomycin | Hygromycin | Signal Peptide Peptidase OsPP-1 (AK061815) | Tamura et al., 2009 |
| Nucleus (NU) | pGWB552 | pGWB542 | pGWB555 | pGWB545 | Rice transcription initiation factor TFIID subunit 10, OsTAF2H (AK103717) | Singh et al., 2012 | ||
| Mitochondria (MT) | pGWB552 | pGWB542 | pGWB555 | pGWB545 | First 99 aa of the rice mitochondrial aldehyde dehydrogenase, OsALDH2a | Nakazono et al., 2000 | ||
| Tonoplast (TP) | pGWB552 | pGWB542 | pGWB555 | pGWB545 | Rice tonoplast intrinsic protein, OsTIP which is ortholog to the Arabidopsis tonoplast targeting protein, gamma-tonoplast intrinsic protein (γ-TIP) | Saito et al., 2002 | ||
| Plasma membrane (PM) | pGWB552 | pGWB542 | pGWB555 | pGWB545 | Full length sequence of rice aquaporin PIP2.1, a plasma membrane aquaporin which is ortholog to Arabidopsis AtPIP2A | Li et al., 2011 | ||
| Peroxisome (PR) | pGWB552 | pGWB542 | pGWB555 | pGWB545 | Full length sequence of rice peroxisome-bound ascorbate peroxidase (OsAPX3) which is previously reported to localized in the peroxisome in rice | Teixeira et al., 2006 | ||
| Chloroplast (CHL) | pGWB552 | pGWB542 | pGWB555 | pGWB545 | Full length coding region of OsERD1 as a chloroplast marker | Weaver et al., 1999 | ||
Cloning of organelle targeting sequences into multicolor expression vectors
After selection of the respective organelle targeting sequences, all were cloned to the Gateway binary vectors pGWBs series, containing four different FPs tagged at the N-terminal: EYFP (pGWB542); ECFP (pGWB545); G3GFP (pGWB552); and mRFP (pGWB555) (Nakagawa et al., 2007). These pGWBs series binary vectors carry a cauliflower mosaic virus (CaMV) 35S promoter for constitutive expression in plants and a hygromycin selection marker for Agro-bacterium tumefaciens-mediated plant transformation (Fang et al., 1989).
Localization of fluorescent-tagged organelle markers to respective organelles
Peroxisomes are a group of organelles found throughout the cytoplasm, roughly spherical in size, and bounded by a single membrane. Peroxisomes are about 0.1 to 1 μm in diameter, although size can vary according to biochemical changes within the cell. They contain oxidative enzymes such as catalase and peroxidase, required to neutralize toxic entities within the cell (Muench and Mullen, 2003). The fluorescence of peroxisomes marker, OsAPX3 appeared as dot-like structures distributed throughout the periphery of the cytosol (Figs. 1A and 2A; Supplementary Fig. S1), and in some cases appeared as clusters (Fig. 2A). The GFP-fused OsAPX3 exhibited a dispersed fluorescence pattern around the cytosol as well as some reticulated fluorescence around the nucleus of BY-2 cells (Teixeira et al., 2006); however, in the present study, only the dispersed fluorescence around the cytosol was found.
Fig. 1. Subcellular localization of eight G3GFP-tagged organelle markers in rice epidermal cells.
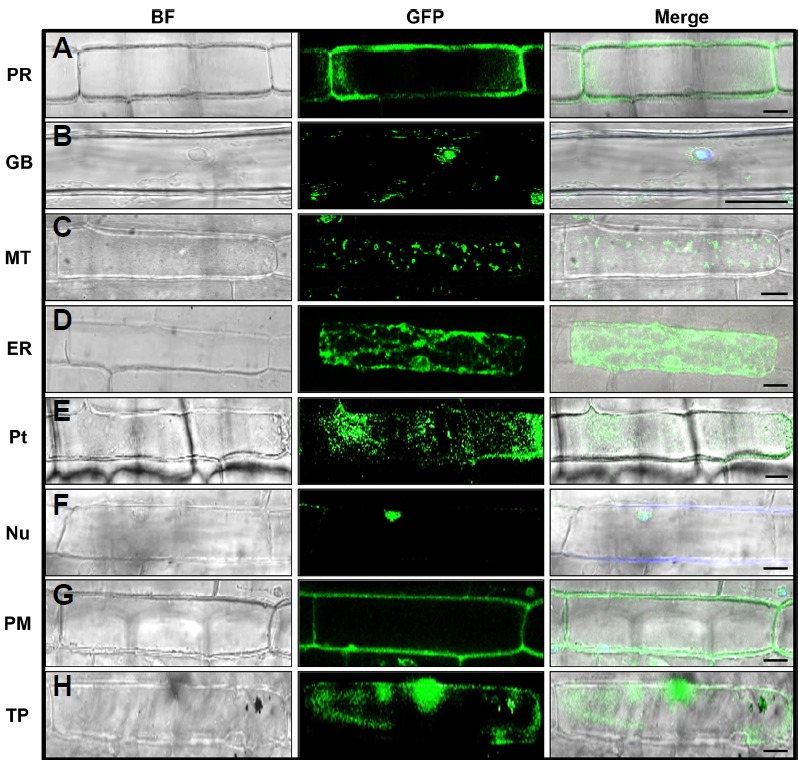
The organelle targeting marker constructs were transiently expressed in rice epidermal cells by biolistic bombardment. The subcellular localization of peroxisome (PR), golgi body (GB), mitochondria (MT), endoplasmic reticulum (ER), plastid (Pt), nucleus (NU), plasma membrane (PM), and tonoplast (TP) is shown (A–H). The microscopic images were taken with a confocal microscope (Leica, TCS SP5) using bright field and GFP filters. BF Bright Field. Scale bars 20 μm.
Fig. 2. Subcellular localization of seven organelle markers tagged with four different fluorescent proteins (FPs) in onion epidermal cells.
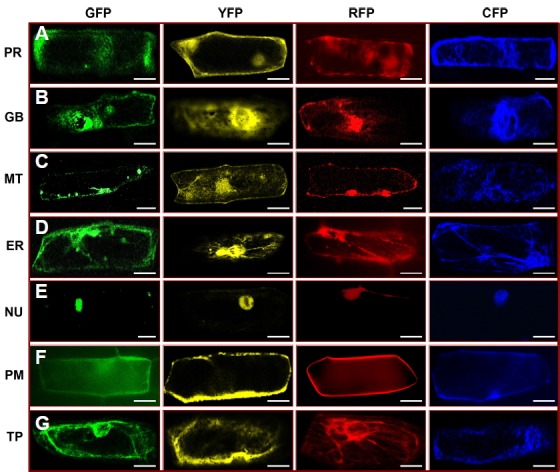
The organelle targeting marker constructs were transiently expressed in onion epidermal cells by biolistic bombardment. The sub-cellular localization of peroxisome (PR), golgi body (GB), mitochondria (MT), endoplasmic reticulum (ER), nucleus (NU), plasma membrane (PM), and tonoplast (TP) tagged with four different FPs (G3GFP, EYFP, mRFP, and ECFP) is shown (A–G). The microscopic images were taken with a confocal microscope (Leica, TCS SP5) using bright field, GFP, YFP, RFP, and CFP filters. Scale bars 20 μm.
Golgi bodies consist of individual stacks of membrane-bound sacs surrounded by vesicles. Mostly, golgi bodies are small, round, spot-like structures (Fig. 1B), but in some cases, they appeared as short lines or disc-like structures (Fig. 2B). We also co-bombarded the G3GFP-tagged rice golgi body marker with the mCherry-tagged Arabidopsis golgi body marker obtained from The Arabidopsis Information Resource (TAIR) (Nelson et al., 2007), to verify localization in the same organelle (Supplementary Fig. S2). In a previous study, ManI proteins localized in both cis Golgi bodies and the endoplasmic reticulum, but the first 49 aa sequence, which includes the cytosolic tail (29 aa), a transmembrane domain (16 aa), and a luminal part (4 aa), shifted localization more towards the trans half of the golgi stacks; however, it exhibited a weak signal for the endoplasmic reticulum. This signal was distinguished from endoplasmic reticulum after treating with brefeldin A (BFA), which relocates the marker protein into the endoplasmic reticulum by aggregating golgi bodies (Saint-Jore-Dupas et al., 2006).
Mitochondria are round, small structures (0.5 to 1 μm in diameter) that vary widely in number according to the organism, tissue, and cell type. The shape and number of mitochondria vary from cell to cell depending on its energy status (Detmer and Chan, 2007). Here, we found the mitochondrial marker as a small spot-like structure in some cells, while in other cells it appeared as a longer, worm-like structure (Figs. 1C and 2C).
The endoplasmic reticulum is the inner core of cytoplasm that is continuously connected with the outer membrane of the nuclear envelope. It forms an interconnected network throughout the cytoplasm. The FP tagged with the endoplasmic reticulum marker (OsSPP1) exhibited prominent fluorescence around the nucleus, forming an extensive network throughout the cytoplasm (Figs. 1D and 2D; Supplementary Fig. S3). The stippled fluorescent signal of OsSPP1 in the present study is similar to that previously reported for the endoplasmic reticulum marker AtWAK2 (Arabidopsis thaliana wall-associated kinase-2) and the endoplasmic reticulum retention signal (His-Asp-Glu-Leu at its N- and C-terminal) in Arabidopsis and Medicago truncatula, respectively (Luo and Nakata 2012; Nelson et al., 2007).
The plastid is a double membrane-bound organelle and the site of photosynthesis. They are generally distributed homogeneously throughout the cytoplasm. Mostly, plastids appear as small circular spots within cells, roughly 5 μm in diameter, although they occasionally appear as long tubular branches, called stromules (Nelson et al., 2007). OsERD1 appeared as small dots of uniform diameter scattered throughout the cytoplasm (Fig. 1E; Supplementary Fig. S4), similar to the previously reported Arabidopsis plastid marker (Nelson et al., 2007) and the Arabidopsis Whirly protein (AtWhy1) (Krause et al., 2005).
The protein localized in the nucleus exhibited a spherical fluorescent pattern. In this study, the nucleus marker (OsTAF2H) exhibited strong fluorescence within the nucleus, giving a roughly spherical shape (Figs. 1F and 2E). To evaluate the accuracy of the nucleus marker, we also examined the co-expression of OsTAF2H with the NLS (Kalderon et al., 1984). The G3GFP-fused nucleus marker was co-expressed with mRFP-fused NLS in both rice and onion epidermal cells. The co-localization of the nucleus marker with NLS further confirms that our nucleus marker can be used for subcellular localization studies (Fig. 3A; Supplementary Fig. S5).
Fig. 3. Application of rice organelle markers in a co-localization assay.
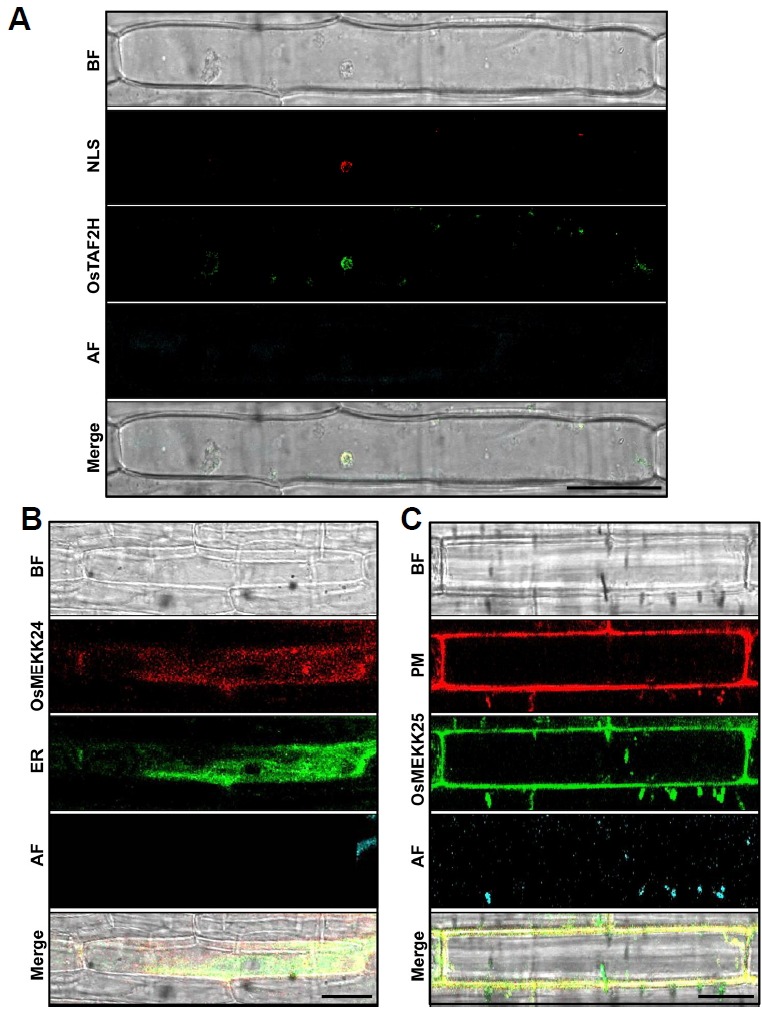
To verify the application of rice organelle markers in the co-localization assay, mRFP-fused NLS (35S:mRFP:NLS), uncharacterized rice MAP3K OsMEKK24 (35S:mRFP:OsMEKK24), and G3GFP-fused OsMEKK25 (35S:G3GFP:OsMEKK25) were transiently co-expressed with a G3GFP-fused nucleus marker (35S:G3GFP:OsTAF2H), an endoplasmic reticulum (ER) marker (35S:G3GFP:OsSPP1), and a mRFP-fused plasma membrane (PM) marker (35S:G3GFP:OsOsPIP2.1) in rice epidermal cells. Merged images of both GFP and RFP show clear co-localization of NLS (A), OsMEKK24 (B), and OsMEKK25 (C) with rice Nucleus, ER, and PM markers. The microscopic images were taken with a confocal microscope (Leica, TCS SP5) using the bright field (BF), GFP, and RFP filters. AF autofluorescence. Scale bars 20 μm.
The plasma membrane is the outer membrane of a cell that surrounds the cytoplasm and the tonoplast is the membrane that surrounds the vacuole. The fluorescence patterns of the plasma membrane and tonoplast markers are similar, since both almost overlap around the periphery of the cell; however, on close inspection, the plasma membrane signal is found only at the periphery of the cell (Figs. 1G and 2F), whereas the tonoplast signal is found in the interior of the cell (Fig. 1H and 2G). In the present study, the tonoplast marker exhibited a uniform distribution of fluorescence around the tonoplast in some cells (Fig. 2G), although in some cases the signal was not uniform around the membrane (Fig. 1H). This could be explained by the fact that, due to the low pH of the vacuole, the tonoplast aquaporin (Os-TIP) may not label all of the vacuolar membrane (Jauh, 1999). We also co-bombarded G3GFP-tagged OsPIP2.1 and OsTIP with the reported mCherry-tagged Arabidopsis plasma membrane and tonoplast markers (TAIR) to confirm localization in the same organelle (Supplementary Fig. S2). The signal obtained in the present study is similar to that reported previously for the tonoplast marker protein of Arabidopsis and maize (Nelson et al., 2007; Wu et al., 2013).
Application of organelle markers in the co-localization assay
We selected two rice MAP3K proteins (OsMEKK24 and Os-MEKK25), based on their predicted subcellular localization (https://psort.hgc.jp) and subcellular distribution in cells (Singh et al., 2014), to illustrate the application of rice organelle markers in the co-localization assay. To detect a co-localization image, green and red fluorescence were employed, since their overlap results in a different color (yellow) that can easily be detected.
The rice MAP3K, OsMEKK24, exhibits putative nucleotide-binding and kinase activity and is predicted to localize to the endoplasmic reticulum (https://psort.hgc.jp). We tagged OsMEKK24 with mRFP and co-expressed it with the G3GFP-tagged endoplasmic reticulum marker in rice and onion cells. Similarly, OsMEKK25 possesses kinase activity and its fluorescence was distributed around the plasma membrane as predicted (Singh et al., 2014). Thus, we co-expressed G3GFP-fused OsMEKK25 with the mRFP-fused plasma membrane marker in rice and onion cells. The mRFP-fused OsMEKK24 and G3GFP-fused endoplasmic reticulum markers, as well as the G3GFP-fused OsMEKK25 and mRFP-fused plasma membrane markers, were perfectly merged (Figs. 3B and 3C; Supplementary Fig. S6), suggesting that the constructed organelle markers can be used for co-localization studies of newly identified proteins.
Conducting fluorescence-based analysis in rice is difficult due to the existing high level of non-specific autofluorescence of molecules such as chlorophyll. To exclude this non-specific fluorescence, we used autofluorescence fields consisting of a different wavelength of the emission states of the fluorescent markers (labeled as autofluorescence in the figures) to differentiate between real fluorescence and autofluorescence (Fig. 3; Supplementary Figs. S1, S3, S4 and S5). In detail, we added one more field along with bright field, GFP, RFP YFP, or CFP, to distinguish chlorophyll autofluorescence from real fluorescence with emission wavelengths >600 nm and excited wavelengths between 420 and 460 nm. We took images of fluorescent markers in combination with chlorophyll autofluorescence in multitrack mode instead of classical simultaneous mode to prevent any crosstalk with the autofluorescence signal. This technique significantly reduces the level of false signal visualization.
DISCUSSION
In this study, we generated G3GFP-, mRFP-, ECFP-, and EYFP-tagged rice cell organelle markers. As reported for Arabidopsis, the morphology of rice cell organelles can be divided into two general groups. The first group includes plastids, peroxisomes, mitochondria, and Golgi bodies, which share a characteristic punctuate fluorescence pattern due to their small size and number per cell. The second group includes the endoplasmic reticulum, tonoplast, and plasma membrane, exhibiting an extended, continuous, and sheet-like morphology (Nelson et al., 2007). The fluorescence morphology of peroxisomes, mitochondria, and Golgi bodies was similar, visualized as small, round, spot-like structures (Figs. 1A–1C, 2A and 2C). Golgi bodies also exhibited heterogeneity in their morphology: in addition to the round small structures, they also appeared as disc-shaped structures (Fig. 2B). The endoplasmic reticulum showed an extensive network in the cell cytoplasm (Fig. 1D and 2D; Supplementary Fig. S3), whereas the morphology of the tonoplast and plasma membrane exhibited a uniform distribution of fluorescence (Figs. 1G, 1H, 2F and 2G). This is the general localization morphology observed in all plant species (Luo and Nakata, 2012; Nelson et al., 2007; Wu et al., 2016; Wu et al., 2013).
Fluorescent signals are difficult to visualize in a rice cell because of its small size and multilayered composition; therefore, most localization studies of rice proteins use a heterologous expression system in onion epidermal cells, and to a lesser extent, in rice protoplasts (Singh et al., 2014; Wu et al., 2016; Xia et al., 2013; Zhang et al., 2011). Signal sequences of proteins from monocots may not be conserved in heterologous systems, which might lead to mis-targeting of fusion proteins (Collings, 2013). In the present work, we found similar expression patterns of organelle markers in both onion and rice cells (Figs. 1 and 2; Supplementary Figs. S1, S3, and S4). Moreover, the organelle targeting sequence of OsManI, OsTIP, OsPIP2.1, and OsERD1 is based on the heterologous plants Glycine max and Arabidopsis (Nelson et al., 2007; Saint-Jore-Dupas et al., 2006). The homologs Os-Man1, OsTIP, OsPIP2.1, and OsERD1 were found to successfully localize in respective organelles in Arabidopsis (Nelson et al., 2007). We compared the localization patterns of Os-Man1, OsTIP, and OsPIP2.1 with mCherry-tagged Arabidopsis golgi body, tonoplast, and plasma membrane markers (TAIR): the similarity of subcellular localization confirmed that these homologs are not paralogs in rice (Supplementary Fig. S2).
We also compared the localization patterns of our rice organelle markers with those of mCherry-tagged Arabidopsis organelle markers (TAIR). The Arabidopsis organelle markers for peroxisomes, plasma membrane, golgi bodies, mitochondria, endoplasmic reticulum, plastids, and tonoplast were expressed in onion epidermal cells by biolistic bombardment. The resulting microscopic morphology of mCherry-tagged Arabidopsis organelle marker proteins was similar to that of our rice organelle marker proteins (Supplementary Fig. S7), providing further support for the usefulness of our rice cell organelle markers in subcellular localization studies.
Although construction of many FP-fused organelle targeting proteins has been reported for plants (Tanz et al., 2013), only a set of GFP-fused cell organelle markers have been reported for rice (Wu et al., 2016). Wu et al. (2016) used the same constructs for golgi bodies and tonoplast from previous report (Nelson et al., 2007) and confirmed their localization in rice cells. However, we generated a set of organelle markers using rice endogenous proteins tagged with four different FPs, which are relevant for protein localization, are usually targeted by biolistic bombardment, and are easily accessible to fluorescence microscope. The use of endogenous rice proteins eliminates the risk of cross species mislocalization. Visualization of each organelle marker and comparison with other systems showed that each organelle marker was targeted to the correct intracellular location. This suggests that the markers can be used as comparative standards in determining organelle distribution and investigating organelle dynamics and cell fractionation. No previous study in rice has confirmed organelle dynamics of more than one protein, whereas the present study confirmed organelle dynamics of two newly identified proteins and NLS with our newly generated markers (Fig. 3; Supplementary Figs. S5 and S6). These findings provide further evidence for the biological significance of the present study. Since we constructed the organelle markers tagged with four different FPs (G3GFP, mRFP, YFP, and CFP), they can also be used for co-localization assays by choosing two different channels with no crosstalk in emission and excitation wavelengths (Fig. 3; Supplementary Figs. S2, S5 and S6). Additionally, researchers can use these organelle markers for generating transgenic lines with stable localization markers. These constructs are available to the scientific community on request.
Supplementary data
ACKNOWLEDGMENTS
SD, RS, and NSJ conceived the study. SD and RS generated the necessary plasmids, and SD, RS, and YC performed the experiments. SD, RS, and NSJ analyzed the data and wrote the manuscript. All authors read and approved the final manuscript. This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1D1A1A09918756).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Chalfie M., Tu Y., Euskirchen G., Ward W.W., Prasher D.C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Collings D.A. Subcellular localization of transiently expressed fluorescent fusion proteins. Methods Mol Biol. 2013;1069:227–258. doi: 10.1007/978-1-62703-613-9_16. [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Ehrhardt D.W., Griffitts J.S., Somerville C.R. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day R.N., Davidson M.W. The fluorescent protein palette: tools for cellular imaging. Chem Soc Rev. 2009;38:2887–2921. doi: 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmer S.A., Chan D.C. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–900. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- Dunkley T.P., Hester S., Shadforth I.P., Runions J., Weimar T., Hanton S.L., Griffin J.L., Bessant C., Brandizzi F., Hawes C., et al. Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA. 2006;103:6518–6523. doi: 10.1073/pnas.0506958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R.X., Nagy F., Sivasubramaniam S., Chua N.H. Multiple cis regulatory elements for maximal expression of the cauliflower mosaic virus 35S promoter in transgenic plants. Plant Cell. 1989;1:141–50. doi: 10.1105/tpc.1.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques M.X., Catalão M.J., Fi gueiredo J., Gomes J.P., Filipe S.R. Construction of improved tools for protein localization studies in Streptococcus pneumoniae. PLoS One. 2013;8:e55049. doi: 10.1371/journal.pone.0055049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezlewoaod J.L., Tonti-Filippini J.S., Gout A.M., Day D.A., Whelan J., Millar A.H. Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell. 2004;16:241–256. doi: 10.1105/tpc.016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauh G.Y., Phillips T.E., Rogers J.C. Tonoplast intrinsic protein isoforms as markers for vacuolar functions. Plant Cell. 1999;11:1867–1882. doi: 10.1105/tpc.11.10.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D., Roberts B.L., Richardson W.D., Smith A.E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Krause K., Kilbienski I., Mulisch M., Rödiger A., Schäfer A., Krupinska K. DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS Lett. 2005;17:3707–3712. doi: 10.1016/j.febslet.2005.05.059. [DOI] [PubMed] [Google Scholar]
- Komatsu S., Konishi H., Hashimoto M. The proteomics of plant cell membranes. J Exp Bot. 2007;58:103–112. doi: 10.1093/jxb/erj209. [DOI] [PubMed] [Google Scholar]
- Li S., Ehrhardt D.W., Rhee S.Y. Systematic analysis of Arabidopsis organelles and a protein localization database for facilitating fluorescent tagging of full-length Arabidopsis proteins. Plant Physiol. 2006;141:527–539. doi: 10.1104/pp.106.078881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang X., Yang Y., Li R., He Q., Fang X., Luu D.T., Maurel C., Lin J. Single-molecule analysis of PIP2.1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell. 2011;23:3780–3797. doi: 10.1105/tpc.111.091454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley K.S., Dupree P. Methods of quantitative proteomics and their application to plant organelle characterization. J Exp Bot. 2006;57:11493–11499. doi: 10.1093/jxb/erj141. [DOI] [PubMed] [Google Scholar]
- Lunn J.E. Compartmentation in plant metabolism. J Exp Bot. 2007;58:35–47. doi: 10.1093/jxb/erl134. [DOI] [PubMed] [Google Scholar]
- Luo B., Nakata P.A. A set of GFP organelle marker lines for intracellular localization studies in Medicago truncatula. Plant Sci. 2012;188–189:19–24. doi: 10.1016/j.plantsci.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Matz M.V., Fradkov A.F., Labas Y.A., Savitsky A.P., Zaraisky A.G., Markelov M.L., Lukyanov S.A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- Moog D., Stork S., Reislöhner S., Grosche C., Maier U.G. In vivo localization studies in the stramenopile alga Nannochloropsis oceanica. Protist. 2015;166:161–171. doi: 10.1016/j.protis.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Muench D.G., Mullen R.T. Peroxisome dynamics in plant cells: a role for the cytoskeleton. Plant Sci. 2003;64:307–315. [Google Scholar]
- Nakagawa T., Suzuki T., murata S., Nakamura S., Hino T., Maeo K., Tabata R., Kawai T., Tanaka K., Niwa Y., et al. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem. 2007;71:2095–2100. doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- Nakazono M., Tsuji H., Li Y., Saisho D., Arimura S., Tsutsumi N., Hirai A. Expression of a gene encoding mitochondrial aldehyde dehydrogenase in rice increases under submerged conditions. Plant Physiol. 2000;124:587–598. doi: 10.1104/pp.124.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- O’Rourke N.A., Meyer T., Chandy G. Protein localization studies in the age of ‘Omics’. Curr Opin Chem Biol. 2005;9:82–87. doi: 10.1016/j.cbpa.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Richly E., Leister D. An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene. 2004;329:11–16. doi: 10.1016/j.gene.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Saint-Jore-Dupas C., Nebenführ A., Boulaflous A., Follet-Gueye M.L., Plasson C., Hawes C., Driouich A., Faye L., Gomord V. Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell. 2006;18:3182–3200. doi: 10.1105/tpc.105.036400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito C., Ueda T., Abe H., Wada Y., Kuroiwa T., Hisada A., Furuya M., Nakano A. A complex and mobile structure forms a distinct subregion within the continuous vacuolar membrane in young cotyledons of Arabidopsis. Plant J. 2002;29:245–255. doi: 10.1046/j.0960-7412.2001.01189.x. [DOI] [PubMed] [Google Scholar]
- Singh R., Lee J.E., Dangol S., Choi J., Yoo R.H., Moon J.S., Shim J.K., Rakwal R., Agrawal G.K., Jwa N.S. Protein interactome analysis of 12 mitogen-activated protein kinase kinase kinase in rice using a yeast two-hybrid system. Proteomics. 2014;14:105–115. doi: 10.1002/pmic.201300125. [DOI] [PubMed] [Google Scholar]
- Singh R., Lee M.O., Lee J.E., Choi J., Park J.H., Kim E.H., Yoo R.H., Cho J.I., Jeon J.S., Rakwal R., et al. Rice mitogen-activated protein kinase interactome analysis using the yeast two-hybrid system. Plant Physiol. 2012;160:477–487. doi: 10.1104/pp.112.200071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler C., Rexhepaj E., Singan V.R., Murphy R.F., Pepperkok R., Uhlén M., Simpson J.C., Lundberg E. Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat Methods. 2013;10:315–323. doi: 10.1038/nmeth.2377. [DOI] [PubMed] [Google Scholar]
- Tamura T., Kuroda M., Oikawa T., Kyozuka J., Terauchi K., Ishimaru Y., Abe K., Asakura T. Signal peptide peptidases are expressed in the shoot apex of rice, localized to the endoplasmic reticulum. Plant Cell Rep. 2009;28:1615–1621. doi: 10.1007/s00299-009-0760-9. [DOI] [PubMed] [Google Scholar]
- Tanz SK, Castleden I, Small ID, Millar AH. Fluorescent protein tagging as a tool to define the subcellular distribution of proteins in plants. Front Plant Sci. 2013;4:214. doi: 10.3389/fpls.2013.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira F.K., Menezes-Benavente L., Galvão V.C., Margis R., Margis-Pinheiro M. Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta. 2006;224:300–314. doi: 10.1007/s00425-005-0214-8. [DOI] [PubMed] [Google Scholar]
- Tobin A.K., Yamaya T. Cellular compartmentation of ammonium assimilation in rice and barley. J Exp Bot. 2001;52:591–594. [PubMed] [Google Scholar]
- Wang Y., Wu J., Kim S.G., Kim S.T., Kang K.Y. A transient gene expression protocol for subcellular protein localization and protein secretion analyses in rice. Protocol Exch. 2013:064. [Google Scholar]
- Weaver L.M., Froehlich J.E., Amasino R.M. Chloroplast-targeted ERD1 protein declines but its mRNA increases during senescence in Arabidopsis. Plant Physiol. 1999;119:1209–1216. doi: 10.1104/pp.119.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Luo A., Zadrozny T., Sylvester A., Jackson D. Fluorescent protein marker lines in maize: generation and applications. Int J Dev Biol. 2013;57:535–543. doi: 10.1387/ijdb.130240qw. [DOI] [PubMed] [Google Scholar]
- Wu T.M., Lin K.C., Liau W.S., Chao Y.Y., Yang L.H., Chen S.Y., Lu C.A., Hong C.Y. A set of GFP-based organelle marker lines combined with DsRed-based gateway vectors for subcellular localization study in rice (Oryza sativa L.) Plant Mol Biol. 2016;90:107–115. doi: 10.1007/s11103-015-0397-8. [DOI] [PubMed] [Google Scholar]
- Xia J., Yamaji N., Ma J.F. A plasma membrane-localized small peptide is involved in rice aluminum tolerance. Plant J. 2013;76:345–355. doi: 10.1111/tpj.12296. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., Wang P., Li Y., Liu B., Feng D., et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods. 2011;7:30. doi: 10.1186/1746-4811-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


