Abstract
One of the major biological functions of the retinal pigmented epithelium (RPE) is the clearance of shed photoreceptor outer segments (POS) through a multistep process resembling phagocytosis. RPE phagocytosis helps maintain the viability of photoreceptors that otherwise could succumb to the high metabolic flux and photo-oxidative stress associated with visual processing. The regulatory mechanisms underlying phagocytosis in the RPE are not fully understood, although dysfunction of this process contributes to the pathogenesis of multiple human retinal degenerative disorders, including age-related macular degeneration. Here, we present an integrated transcriptomic, proteomic, and phosphoproteomic analysis of phagocytosing RPE cells, utilizing three different experimental models: the human-derived RPE-like cell line ARPE-19, cultured murine primary RPE cells, and RPE samples from live mice. Our combined results indicated that early stages of phagocytosis in the RPE are mainly characterized by pronounced changes in the protein phosphorylation level. Global phosphoprotein enrichment analysis revealed involvement of PI3K/Akt, mechanistic target of rapamycin (mTOR), and MEK/ERK pathways in the regulation of RPE phagocytosis, confirmed by immunoblot analyses and in vitro phagocytosis assays. Most strikingly, phagocytosis of POS by cultured RPE cells was almost completely blocked by pharmacological inhibition of phosphorylation of Akt. Our findings, along with those of previous studies, indicate that these phosphorylation events allow the RPE to integrate multiple signals instigated by shed POS at different stages of the phagocytic process.
Keywords: eye, receptor recycling, retina, retinal degeneration, retinal metabolism
Introduction
The retinal pigmented epithelium (RPE)4 is a specialized epithelial cell layer in the back of the eye that performs a number of critical functions to maintain the health of the neural retina and sustain vision. As a part of the blood–retina barrier, the RPE helps to maintain the immune privileged state of the eye as well as regulate the transport of oxygen and needed nutrients from the underlying vasculature into the retina (1). Its essential role in the retinoid (visual) cycle is well documented, with perturbations resulting in a wide range of retinal dystrophies (2, 3). Moreover, the continuous removal of the most distal portions of photoreceptor outer segments (POS) by a phagocytic mechanism maintains the health of postmitotic photoreceptor cells that contain the light-sensing pigments required for vision (4, 5). This circadian regulated process removes toxic compounds formed in the high-oxygen, high-lighting conditions present in the retina that threaten cellular health. Although this process was first described more than 5 decades ago, our understanding of the molecular events that regulate POS engulfment remains limited.
Phagocytosis by professional phagocytes, like monocytes or macrophages, involves four temporally distinct steps, namely recognition, binding, engulfment, and subsequent degradation of ingested material, which is then either recycled or sequestered. Similar to many other phagocytic cell types, RPE cells recognize phosphatidylserine (PtdSer), which decorates the plasma membrane of the most distal portions of the POS (6). This substrate is detected either directly by the scavenger receptor CD36 (7, 8) or through opsonins like Gas6/ProS and MFG-E8, for the Mer receptor tyrosine kinase (MERTK) (9) and integrin αvβ5 (10), respectively. Once recognized, POS material binds to the RPE plasma membrane, initiating a signal transduction pathway that results in the phosphorylation and activation of intracellular kinases, such as focal adhesion kinase and Src (11, 12), amplifying the signal and preparing the cell for engulfment. Although several other signal transduction members have been implicated in downstream signaling, including p130cas and Rac1, as well as other members of the small GTPase family (11, 13), just how initiation of the signal transduction pathway causes a large rearrangement of the underlying cytoskeleton and subsequent ingestion is poorly understood.
Once initiated, signal transduction pathways can be amplified in several ways, including post-translational modifications and second messenger generation, with protein phosphorylation representing one of the best-studied systems. From previous work, it is clear that phosphorylation is an essential component of RPE phagocytosis (14), with at least three aforementioned kinases being unequivocally linked to this process. The lack of significant scientific progress in this area stems mainly from its complexity, which involves a close interaction between two postmitotic cell types. Protocols for the isolation of pure RPE cells and POS from a number of species have facilitated the development of several ex vivo assays. Additionally, due to its circadian nature, investigators have observed a significant burst of phagocytosis shortly after sunrise, allowing time course experiments to be performed during a typical day/night cycle. Here we used an unbiased approach to probe phosphorylation events that occur during engulfment of POS in three different biological settings. Thus, mass spectrometry was employed to quantitatively analyze the phosphoproteome of the human-derived RPE-like cell line, ARPE-19, as well as the phosphoproteome of cultured primary RPE cells and RPE samples from live animals taken at different times during a 24-h period that matched the high and low levels of RPE phagocytosis in vivo. We then created a catalogue of phosphosites that revealed a change in phosphorylation state during RPE phagocytosis. Validation of several targets with phospho-specific antibodies and pharmacological inhibitors provided evidence for the circadian role of three interconnected signal transduction pathways, the MAPK, PI3K/Akt, and non-canonical Wnt pathways, which previously had not been appreciably associated with this process.
Results
Quantitative proteomic and phosphoproteomic analyses of ARPE-19 cells
To explore novel and important regulatory mechanisms related to RPE phagocytosis, we designed transcriptomic, proteomic, and phosphoproteomic experiments using three different sources of RPE cells: human-derived RPE-like ARPE-19 cells that retain the ability to phagocytose POS (15), cultured primary RPE cells, and native RPE cells isolated directly from mouse eyes.
Our initial analysis involved ARPE-19 cultured cells, which allow good control over experimental conditions and use of the stable isotope labeling by amino acids in cell culture (SILAC) method for unbiased, highly reproducible quantitative proteome and phosphoproteome profiling. To extensively characterize changes induced by POS stimulation, SILAC-labeled and unlabeled cells were exposed to POS for different time periods (15, 30, 60, 90, and 120 min) or left unchallenged (0 min, control conditions). Labeled cell lysates were pooled and spiked into each unlabeled cell lysate, resulting in a total of 18 samples (n = 3) used for further LC-MS/MS analysis (supplemental Fig. S1). For each sample, an unenriched and phosphopeptide-enriched fraction was analyzed with a 4-h gradient run on a Q-Exactive mass spectrometer, resulting in a total of 36 runs.
Pairwise Pearson's correlation analyses of 18 measurements on both the ARPE-19 proteome and phosphoproteome showed good reproducibility between biological replicates (supplemental Fig. S2A). Both data sets also exhibited good clustering of the biological replicates as well as a separation between treated and control (0-min) samples in principal component analysis (supplemental Fig. S2B). Using a false discovery rate (FDR) of 1%, we identified a total of 2571 protein groups in the proteomic analysis (Fig. 1A), among which 2307 exhibited a minimum of two peptide ratio counts, and 6918 phosphorylation events on 1990 phosphoproteins. For further downstream bioinformatic analyses, we selected a data set containing only accurately quantified proteins and phosphorylation events. This resulted in more stringent, filtered data sets of 1762 proteins (referred to as the ARPE-19 proteome, supplemental Data Set S1) and 2546 class I phosphorylation sites (referred to as the ARPE-19 phosphoproteome, characterized by a localization probability score >0.75, supplemental Data Set S2). These accurately quantified phosphosites included 2307 pSer (90.6%), 223 pThr (8.8%), and 16 pTyr (0.6%) residues, exhibiting an abundance ratio similar to that of previously published phosphoproteome data sets (16).
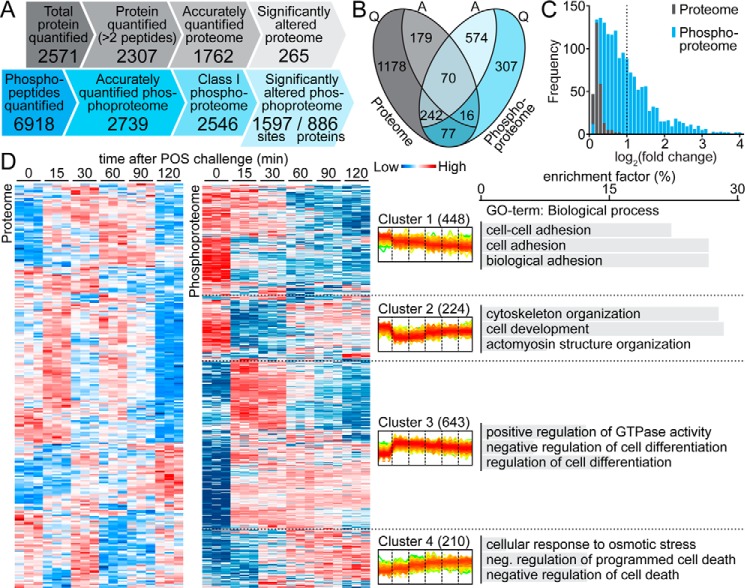
Figure 1.
Overview of the POS-stimulated ARPE-19 proteome and phosphoproteome. A, comparison of the SILAC-based quantification of ARPE-19 proteome and phosphoproteome. Only 265 of 1762 (15%) accurately quantified proteins responded to POS stimulation, whereas almost 63% of class I phosphorylation events (1597 of 2546) were significantly altered under the same conditions. B, relationships between accurately quantified (Q) and significantly altered (A) proteome (gray) and phosphoproteome (blue). C, abundance profiles of significantly altered proteome (gray) and phosphoproteome (blue). D, hierarchical clustering analysis of significantly altered proteins/phosphosites (in rows), according to their abundance profile over time (in columns). Relative abundance levels are represented with color-coded boxes (blue, low; red, high). Quantification for three biological repetitions is presented for each time point. In the center, superimposed diagrams of phosphorylation level changes over time in four clusters of phosphosites are shown. The four typical responses to POS challenge were as follows: a slow constant phosphorylation decrease (Cluster 1), a significant phosphorylation decrease followed by a slow increase (Cluster 2), a significant phosphorylation increase followed by a slow decrease (Cluster 3), and a slow constant phosphorylation increase over the experimental time period (Cluster 4). The chart on the right shows the top three enriched biological process gene ontology terms (BP-FAT) within each cluster of the POS-stimulated ARPE-19 phosphoproteome. Cluster 3 exhibited specific enrichment in the regulation of the GTPase activity biological process GO term.
As shown in Fig. 1A, statistical analysis with the one-way ANOVA identified 265 proteins (15.0%) and 1597 phosphosites (62.7%) which were significantly altered between at least two time points (q < 0.05). By comparing proteomic and phosphoproteomic data, we found most of the observed significant alterations to be exclusively related with one of the two data sets (Fig. 1B). Interestingly, the amplitude of phosphorylation change greatly exceeded the amplitude of protein abundance, as the mean logarithmic -fold changes of the POS-stimulated phosphoproteome and proteome were found to be 0.92 and 0.19, respectively. IGFBP3, an insulin-like growth factor-binding protein known to induce phagocytosis in ARPE-19 cells (17), was the only protein in which the abundance change was >2-fold. In contrast, over one-third of significantly altered phosphosites (557 of 1597, 34.9%) exhibited a >2-fold change in their phosphorylation levels (Fig. 1C). This much greater dynamic change in the phosphorylation status compared with the protein abundance suggests that post-translational mechanisms play a prominent role in the phagocytosis response within the time frame of our experiments.
To further characterize proteome and phosphoproteome fluctuations during POS phagocytosis progression, we performed a hierarchical clustering analysis of both significantly altered data sets. Interestingly, clustering of the phosphoproteome revealed a clear segregation into four clusters, reflecting four distinct patterns of phosphorylation progression (Fig. 1D). The observed behaviors included either a rapid significant change (increase or decrease) followed by a slow return to the initial state (clusters 2 and 3) or, conversely, a progressive and prolonged phosphorylation change (clusters 1 and 4). In fact, both pairs of clusters were mirror images of each other. In the case of 72 phosphorylation sites, we were unable to detect their respective phosphopeptides in all three untreated biological replicates, suggesting their low phosphorylation levels under control conditions. Those sites, which may possibly undergo a qualitative change of their phosphorylation status in response to POS stimulation, were moved into a separate cluster (Cluster 0; supplemental Fig. S3). In total, more than half of the phosphosites exhibited increased phosphorylation after 15 min of POS challenge (925 of 1597, 57.9%, clusters 3, 4, and 0), most of which also revealed a sharp phosphorylation peak at this time point.
To gain insight into the biological relevance of the POS-stimulated proteome and phosphoproteome identified here, we performed a gene ontology (GO) enrichment analysis using the bioinformatics resources available via DAVID. POS-stimulated proteins were selectively enriched for several GO terms, including extracellular matrix, adherens junction (cell component), cell adhesion (biological process), and cell adhesion molecule binding (molecular function). Compared with this data set, the POS-stimulated phosphoproteome showed an overrepresentation of GO terms associated with cell–cell junction, actin cytoskeleton, single-organism organelle organization, and cytoskeletal protein binding (supplemental Table S1). May et al. (18) pointed out that all phagocytic processes are driven by a finely controlled rearrangement of the actin cytoskeleton. Our analysis confirmed that phosphorylation events associated with the cell–cell junction and cytoskeletal protein binding are specifically involved in phagocytosis regulation.
Predicted kinase regulators of the POS-stimulated ARPE-19 phosphoproteome
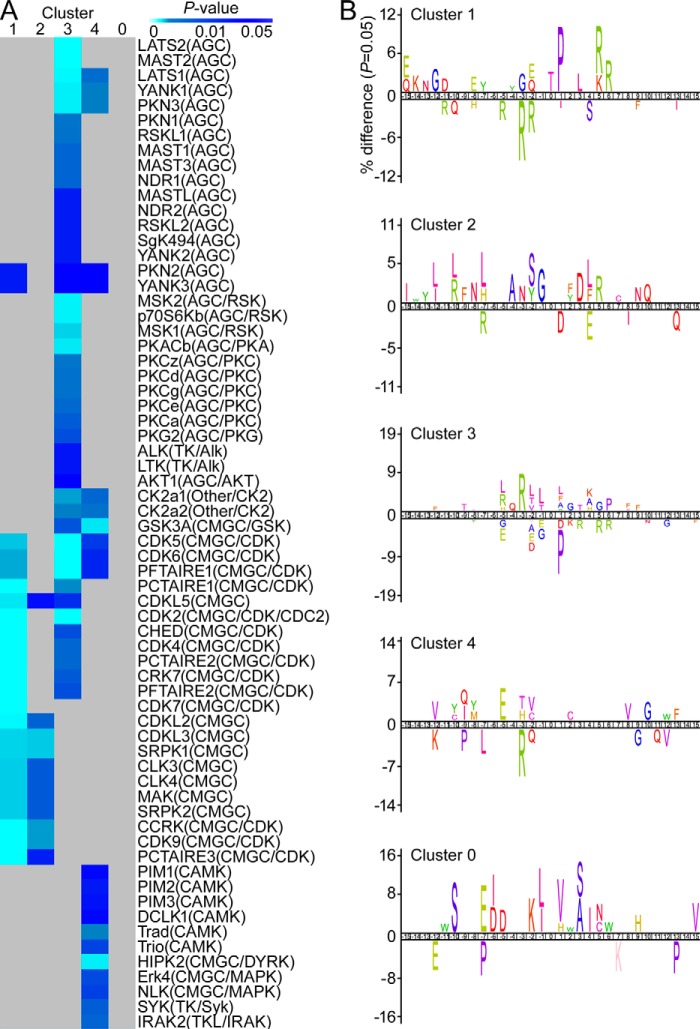
To identify putative protein kinases that might modulate POS-stimulated phosphorylation events, we employed iGPS analysis (a GPS algorithm with the interaction filter, 1.0). A total of 12,181 potential site-specific kinase-substrate relationships were predicted within a significantly altered phosphoproteome, yielding 357 protein kinases targeting 398 substrates (695 phosphopeptides) with a coverage rate of 43.7%. Next, we checked the enrichment of predicted kinases within the four previously defined phosphosite clusters (Fig. 2A). Cluster 3, which included sites markedly overphosphorylated shortly after POS challenge, showed the highest enrichment in predicted kinase regulators belonging mostly to the AGC, AGC/RSK, AGC/PKC, and CAMK groups (Fisher's exact test, p < 0.05, relative to the total POS-stimulated phosphoproteome). A detailed predicted network of interactions is shown in supplemental Fig. S4. Multiple kinases belonging to CMGC and CMGC/CDK groups were also predicted to target Cluster 1 phosphosites. Because we observed a gradual decrease in phosphorylation levels for these sites, this result could suggest which kinases are less relevant for phagocytosis regulation.
Figure 2.
Predicted kinase regulators of ARPE-19 phagocytosis-related phosphoproteome. A, heat map of enriched putative protein kinases related to the POS-stimulated phosphoproteome. Lists of enriched protein kinases were predicted from POS-stimulated phosphopeptides according to their cluster classification (in columns). B, sequence logo analysis of the POS-stimulated phosphoproteome from each cluster.
To further analyze potential regulators of phagocytosis-related phosphorylation events, we performed iceLogo analysis within the defined clusters that revealed enrichment of particular amino acids surrounding the phosphorylation sites (Fig. 2B). In accordance with our previous kinase enrichment analysis, most Cluster 1 phosphosites possessed a Pro residue at the +1 position required for phosphorylation by Pro-directed kinases belonging to the CMGC group. In contrast, the consensus sequence for Cluster 3 phosphosites showed a high avoidance for Pro in the +1 position. However, sites in Cluster 3 were significantly overrepresented (p < 0.05) with Arg-directed and Leu-containing motifs, including Arg/Leu and Arg at the −5 and −3 positions, as well as hydrophobic amino acids at the +1 position. Interestingly, when compared with the Akt-regulated substrate sequence (Arg-X-Arg-X-X-Ser/Thr-Y, where Y tends to be a hydrophobic residue (19)), Cluster 3 exhibited favorable substrate relations with Akt kinase. Moreover, when compared with the whole POS-stimulated phosphoproteome, Cluster 3 was selectively enriched for the regulation of GTPase activity GO biological process, known to be highly modulated/phosphorylated by Akt through the PI3K/Akt pathway (Fig. 1D).
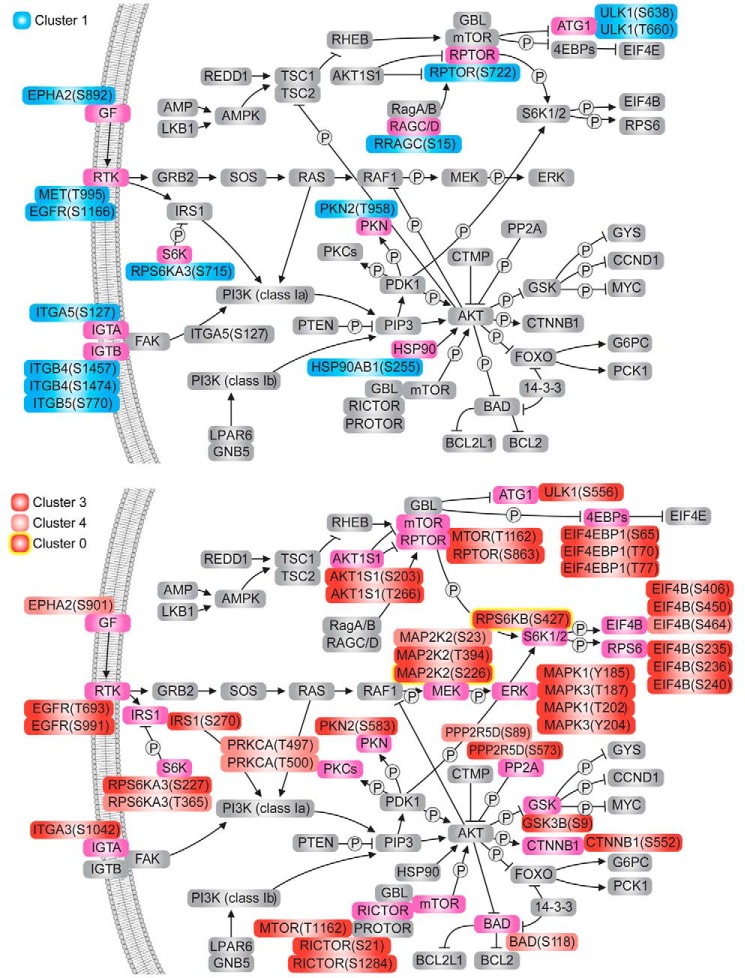
In evaluating to what extent POS-stimulated phosphorylation events may be involved in PI3K/Akt as well as the interconnected mTOR and MEK/ERK signaling pathways, we integrated our phosphoproteome data into known pathway mappings from the KEGG database (Fig. 3). Here, 54 POS-stimulated phosphorylation changes on 28 proteins were found to be related to all three pathways, most of which involved overphosphorylation events (40 of 54, 74.1%, Clusters 3, 4, and 0). Whereas the upstream proteins exhibited a well-balanced mix of increased and decreased phosphorylation, the downstream components were largely overphosphorylated, preferably achieving the maximum 15 min after POS stimulation (Cluster 3). Interestingly, the MEK-ERK axis exhibited only overphosphorylated events. Collectively, our data suggest that phosphorylation events responding to POS stimulation, especially those associated with the regulation of GTPase activity, are modulated by associated AGC kinases related to PI3K/Akt/mTOR/ERK signaling cascades.
Figure 3.
The Akt/mTOR pathway represents a major axis of phosphorylation signal transduction in response to phagocytosis. Shown is a schematic representation of Akt and mTOR signal transduction pathways with proteins undergoing significant phosphorylation level changes under POS stimulation highlighted in pink. Phosphorylation change events were divided into two categories: decreased (Clusters 1 and 2; 14 phosphosites) and increased (Clusters 3, 4, and 0; 40 phosphosites). Phosphorylation events outlined in yellow were not detectable under control conditions.
Transcriptome and phosphoproteome of cultured primary RPE cells
The biology of ARPE-19 cells differs in various aspects from that of their native counterparts (20). For this reason, we complemented the observations provided by the previous analysis with a system that would more closely mimic the in vivo process while continuing to provide the control offered by cultured cells. A previously established ex vivo phagocytosis assay with cultured primary RPE cells from mice was chosen for this analysis (21), but due to known limitations of this method (the low number of cells present in a mouse eye with poor proliferation in culture), we focused instead on a much less material-consuming transcriptome profiling, while limiting the phosphoproteomic analysis to just a qualitative registration of phosphorylation events found in POS-challenged cells. In both cases, cells were challenged with isolated POS for 90 min, a period shown in our ARPE-19 analysis to induce a sufficient change in the phosphorylation response.
Less than 2.5% of genes exhibited differential expression in response to POS challenge (supplemental Fig. S5), indicating that phagocytosing RPE cells undergo only mild transcriptome changes at early stages of the phagocytosis process. Interestingly, GO term analysis of all significantly altered genes revealed a significant overrepresentation of RNA polymerase II transcription factors and mitogen-activated protein kinase ERK1/2 signaling pathway regulators (supplemental Fig. S5D). By analyzing the phosphoproteome of POS-challenged primary RPE cells, we accurately identified 1199 phosphosites (assuming a localization probability score >0.75, supplemental Data Set S3), confirming the phosphorylation status of 678 proteins. This included 345 sites homologous to those of the Class I ARPE-19 phosphoproteome, most of which (231 phosphosites, 70%) were also homologous to the significantly altered subset of the ARPE-19 phosphoproteome. This shared data set of 231 phosphosites evidenced strong enrichment in the germ cell–Sertoli cell junction signaling pathway (supplemental Fig. S5E), confirming a high functional similarity between Sertoli and RPE cells, both known for their phagocytic capabilities. Other prominently enriched pathways included those involving cell assembly and organization, such as cell junctions and the actin cytoskeleton. This observation suggests a good mouse–human interspecies conservation of crucial phosphorylation sites related to structural rearrangements of phagocytosing RPE cells.
Quantitative proteomic and phosphoproteomic analyses of native RPE cells
To further characterize the spectrum of highly relevant phosphorylation events involved in RPE phagocytosis, we then analyzed the phosphoproteome of RPE cells sampled directly from live animals. That RPE phagocytosis is a circadian-regulated process permitted a precise collection of samples representing both high and low levels of RPE phagocytosis (cells harvested at 7:30 a.m. and 3 p.m., respectively). Proteins in each native RPE sample were extracted and enzymatically digested, and the phosphopeptide-enriched fraction was analyzed by LC-MS/MS. In parallel with the RPE phosphoproteome study, a portion of peptides released by protease digestion in each sample was used for the RPE quantitative proteomic analyses via a stable isotope dimethyl labeling strategy.
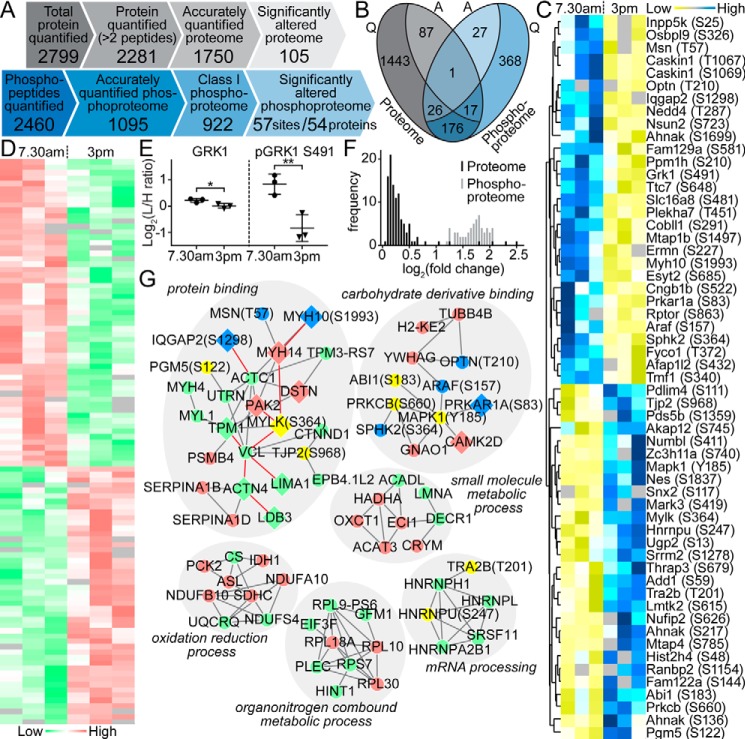
A total of 1750 proteins (proteome; supplemental Data Set S4) and 922 class I phosphorylation sites on 615 proteins (phosphoproteome; supplemental Data Set S5) were accurately quantified with a minimum of 3 of 6 MS measurements (Fig. 4A). Notably, 220 proteins were common to both data sets (Fig. 4B). Moreover, 105 proteins in the proteomic analysis (105 of 1750, 6.0%) and 57 phosphopeptides in the phosphoproteomic analysis (57 of 922, 6.2%) exhibited a significantly altered abundance between the two analyzed time points. Comparison of the significantly altered RPE phosphoproteome and proteome revealed that both data sets exhibited almost equal numbers of up- and down-regulation events (Fig. 4, C and D). The significantly altered RPE phosphoproteome displayed much higher amplitudes of change (mean of 3.1-fold) than the respective proteome data set (mean of 1.2-fold; Fig. 4F). Indeed, for the only protein that exhibited a significant fluctuation at both the protein and phosphorylation level, namely GRK1 (rhodopsin kinase), the magnitude of the phosphorylation oscillation at Ser-491 greatly exceeded the magnitude of protein abundance change (Fig. 4E). To delve into the relationship between the significantly altered proteome and phosphoproteome, we then used STRING to construct direct protein interaction networks. The top function within the largest protein interaction network was protein binding (Fig. 4G). This network included a large number of proteins and phosphorylation events involved in actin filament-based processes. Prior studies have shown that such processes are one of the modulators responsible for phagocytosis-regulated movements on a cell surface (22).
Figure 4.
Global proteomic and phosphoproteomic analysis of native RPE. A, comparison of the quantitative proteomic and phosphoproteomic analysis of native RPE cells isolated from mouse eyes sampled in the morning (7:30 a.m.) and afternoon (3 p.m.), reflecting the highest and lowest phagocytosis intensities. B, relationships between accurately quantified (Q) and significantly altered (A) proteome (gray) and phosphoproteome (blue). One protein (GRK1) exhibited significant changes in both protein abundance and phosphorylation levels. C, heat map showing phosphorylation status of 57 phosphosites (rows) that were significantly changed between the two measurement time points (7:30 a.m. versus 3 p.m., columns). D, heat map revealing the expression level of 105 proteins (rows) significantly altered between the two measurement time points (columns). E, detailed comparison of the change in expression and phosphorylation level of GRK1 indicates a greater magnitude of oscillation in phosphorylation. F, distribution of the amplitudes of -fold changes (log2 of light/intermediate intensities) for the 105 and 57 significantly altered proteins and phosphosites, respectively. G, direct physical protein–protein interaction networks of significantly expressed RPE proteins and phosphoproteins. Relative responses to POS challenge are denoted by different colors (proteome: down-regulated (green) and up-regulated (red); phosphoproteome: down-regulated (yellow) and up-regulated (blue)).
The limited change in the native RPE phosphoproteome compared with the broad response observed in ARPE-19 cell culture may be explained in part by a high in vivo cell-to-cell variability combined with the dynamics of phosphorylation signaling (23). In consequence, only 15 proteins and 16 phosphorylation sites exhibited significant changes in both experiments (supplemental Table S2). Notably, three identified phosphoproteins (mitogen-activated protein kinase 1 (MAPK1/ERK2), heterogeneous nuclear ribonucleoprotein U (HNRNPU/SAF-A), and thyroid hormone receptor–associated protein 3 (THRAP3/TRAP150)) are known modulators of circadian clock function in humans. The two latter were shown to directly influence function of BMAL1 (24, 25), a core component of the clock system, suggesting a possible link between circadian machinery and phagocytosis regulation.
Changes in the PI3K/Akt, mTOR, and MEK/ERK signaling pathways related to phagocytosis
Activation of the mTOR and PI3K/Akt pathways has been widely studied with respect to a number of physiological and pathological processes, including cell cycle regulation, apoptosis, and cancer. The role of these pathways in RPE phagocytosis has remained obscure, but recently it was shown that pharmacological inhibition of PI3K, a common upstream signaling component for both pathways, by treatment with LY294002 markedly reduced phagocytosis in cell culture (26). Our ARPE-19 phosphoproteome study revealed increased phosphorylation of multiple PI3K/Akt, mTOR, and MEK/ERK pathway components in response to POS challenge (Fig. 3). Moreover, our transcriptomic analysis showed differential expression of MEK/ERK pathway regulators shortly after phagocytosis induction in cultured mouse RPE (supplemental Fig. S5D).
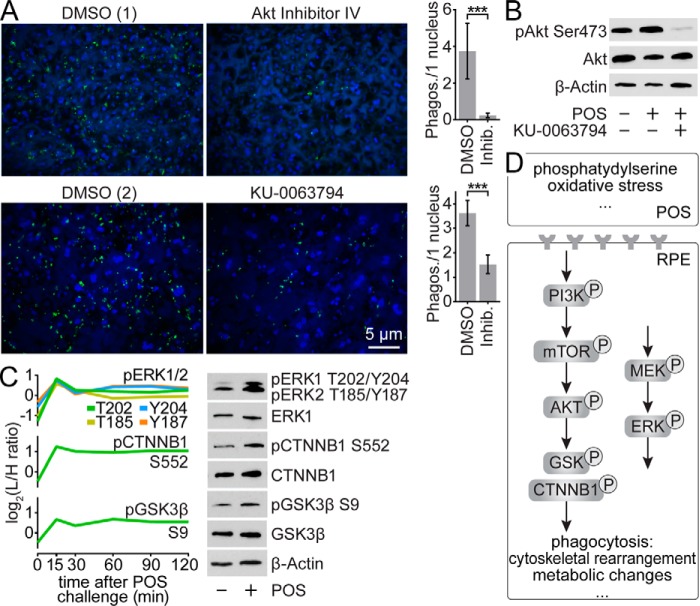
To further evaluate the role of mTOR and PI3K/Akt pathways in RPE phagocytosis, we initially focused on Akt, an important signaling node in both cascades and a downstream substrate for PI3K. Pharmacological inhibition of Akt phosphorylation with Akt inhibitor IV resulted in complete ablation of RPE phagocytosis in POS-challenged primary RPE cell culture (Fig. 5A). In contrast, we found that Ser-473 of Akt was overphosphorylated in POS-exposed RPE cells (Fig. 5B). Activation of Akt by this phosphorylation involves the direct action of mTOR kinase, known for its role in several other signaling pathways (26). To further confirm the importance of this phosphorylation event in RPE phagocytosis, we used the dual mTOR inhibitor KU-0063794 to inhibit the mTOR pathway in a similar assay. This treatment of POS-challenged primary RPE cells resulted in a significant reduction (p < 0.0001) in RPE phagocytosis (Fig. 5A) as well as nearly complete inhibition of Akt Ser-473 phosphorylation (Fig. 5B). Taken together, these observations suggest that phagocytosis initiation requires stimulation of PI3K/Akt and mTOR signaling pathways.
Figure 5.
Involvement of mTOR and PI3K/Akt signaling pathways in phagocytosis regulation. A, immunofluorescence images of primary RPE cells in culture challenged with FITC-labeled POS and showing the influence of Akt and mTOR inhibition (with Akt inhibitor IV and KU-0063794, respectively) on the number of internalized outer segments. The respective controls treated with DMSO are marked with (1) and (2). Quantification of internalized POS is shown in the bar graphs; ***, p < 0.0001 in an unpaired t test. B, immunoblots revealing phosphorylation levels of pAkt Ser-473 in cultured primary RPE cells under POS challenge and mTOR inhibition. C, influence of POS challenge on the phosphorylation level of pERK1 Thr-202/Tyr-204 and pERK2 Thr-185/Tyr-187, pβCat Ser-552, and pGSK3β Ser-9 in cultured primary RPE cells. Respective data obtained from phosphoproteomic analysis of POS-exposed ARPE-19 cells are shown in the graphs. D, overview of the phagocytosis signaling pathways in the RPE. PI3K/Akt, mTOR, and MEK/ERK pathways play important roles in the integration of multiple consecutive signals emanating from shed POS.
Multiple downstream components of the PI3K/Akt pathway showed significantly altered phosphorylation levels in our ARPE-19 phosphoproteome analysis (Fig. 3). These included increased phosphorylation of β-catenin (CTNNB1) Ser-552 and glycogen synthase kinase β (GSK3β) Ser-9 under POS challenge. To validate these results, we performed immunoblot analyses of the two proteins in cultured primary RPE cells using antibodies with specificity for their particular phosphorylation sites. In agreement with the phosphoproteomic data, we observed an increased phosphorylation in response to POS challenge in both cases (Fig. 5C). Because transcriptomic analysis showed enrichment in ERK signaling pathway regulators and ERK1/ERK2 kinases exhibited increased phosphorylation in ARPE-19 cells, we decided to validate their phosphorylation status in cultured RPE as well. Both ERK1 (MAPK3) and ERK2 (MAPK1) at residues Thr-202/Tyr-204 and Thr-185/Tyr-187, respectively, showed a positive response to POS challenge (Fig. 5C).
Discussion
Phagocytosis of POS by the RPE cell layer occurs as a highly synchronized process that bursts every morning after the onset of light. Its spatio-temporal nature requires tight coordination of multiple events that allow specific recognition, binding, internalization, and ultimate digestion of shed photoreceptor outer segments. To gain a global view of the regulatory mechanisms that underlie phagocytosis in the RPE, we used a holistic approach by evaluating the spectrum of transcriptomic, proteomic, and phosphoproteomic changes induced by POS stimulation. To ensure fully unbiased results, we employed three different experimental models: an immortalized RPE-derived cell line (ARPE-19), a primary cell culture of mouse RPE, and native RPE obtained directly from live mice.
Our transcriptomic profile of cultured mouse RPE showed only minor changes resulting from phagocytosis induction (supplemental Fig. S5), suggesting more rapid cellular responses to signals coming from shed POS. Indeed, we observed much more pronounced changes in phosphorylation than in protein abundance, both in ARPE-19 (Fig. 1C) and native RPE cells (Fig. 4F). A recent study on phosphoproteome changes in the RPE induced by light exposure indicated differential phosphorylation of 60 phosphoproteins, mainly chaperones and proteins involved in cytoskeletal function (27). However, the phosphoproteomic profile of phagocytosing RPE cells has not been reported to date.
Our study presents a comprehensive picture of the phagocytosis-related phosphosignaling network in RPE cells. We observed a limited overlap of significantly altered phosphorylation events between the ARPE-19 cell line and the native tissue, which may be explained by the inherent differences between the two experimental models. Nonetheless, we confirmed a significant activation of PI3K/Akt, mTOR, and MEK/ERK pathways during phagocytosis of POS, a physiological function of RPE cells. These interconnected signaling pathways control multiple cellular processes, including cell survival, division, and metabolism (28). Both professional and non-professional phagocytes rely on signaling through the PI3K/Akt, mTOR, and MEK/ERK pathways for their phagocytic capability (29–36). Activation of PI3K/Akt was observed previously in RPE cells under oxidative stress (37). Inhibition of PI3K but not downstream Akt inhibited phagocytosis 3 h after POS challenge of RPE-J cells (26). However, the same study showed that 7 h after POS challenge, Akt inhibition results in increased POS uptake. In contrast, our analysis of earlier events (1.5 h after POS challenge) shows a crucial stimulating role of Akt, evidenced by nearly complete ablation of phagocytosis under Akt inhibition with inhibitor IV (Fig. 5A). This result coincides with increased phosphorylation of Akt Ser-473 that peaks shortly after challenge with POS. We observed a similar response by Rictor on residues Ser-21 and Ser-1284 as well as mTOR on Thr-1162 (Fig. 3), both forming a complex (mTORC2) that directly phosphorylates Ser-473 of Akt (38), suggesting its involvement in phagocytosis. In line with this hypothesis, inhibition of mTOR with KU-0063794 significantly reduced POS uptake within 1.5 h after the challenge (Fig. 5, A and B). The opposite effect was observed 8 h after POS challenge with regard to mTORC1, another complex involving mTOR (39).
We also observed increases in phosphorylation of crucial residues within the MEK-ERK axis, shortly after POS treatment in both ARPE-19 (Fig. 3) and cultured RPE cells (Fig. 5C). A similar response in both RPE models was also demonstrated by two downstream components of the Akt pathway, β-catenin and GSK3β. Phosphorylation of β-catenin on Ser-552 promotes its dissociation from cell-to-cell contacts, nuclear accumulation, and transcriptional activity (40), whereas GSK3β phosphorylated on Ser-9 positively regulates the stability of β-catenin (41). A similar phosphorylation pattern of β-catenin and GSK3β mediated by PI3K/Akt and mTOR pathways was observed previously in mouse RPE exposed to oxidative stress, which led to detrimental cell dedifferentiation and hypertrophy (42). In addition, the ERK pathway also underwent an overphosphorylation in response to oxidative stress in ARPE-19 cells (43).
Multiple phosphorylation events reported in this study have previously been associated with oxidative stress. Indeed, long-term responses to phagocytosis or oxidative stress share common features in human RPE (44). However, an inhibitory effect of reactive oxygen intermediates on phagocytosis has also been observed (45). Considering the dynamic nature of phosphorylation events, we can speculate that the role of particular phosphorylation cascades changes as the phagocytic process progresses, possibly as different stimuli reach the phagocytosing cell. In fact, it is still not well understood at a molecular level just how phagocytosis of POS is initiated. PtdSer exposed on the surface of POS is one well-known hallmark of phagocytosis initiation. PtdSer is recognized by MERTK, which is highly abundant in RPE cell-surface membranes, through an indirect interaction involving extracellular Gas6 and Protein S (46–48). Receptor tyrosine kinases and their substrates are known activators of mTOR and PI3K/Akt pathways (49). It is therefore possible that PI3K/Akt, mTOR, and MEK/ERK comprise core signal transduction pathways that integrate multiple consecutive signals emanating from shed POS (Fig. 5D).
Previous studies have suggested a contribution of signaling through PI3K/Akt, mTOR, and MEK/ERK pathways in the pathophysiology of retinal degenerative disorders, including age-related macular degeneration (42, 50), as well as possible treatments targeting those pathways (51, 52). That these signaling cascades contribute to important physiological processes exerted by the RPE may at least partially explain the incomplete effectiveness of tested inhibitors.
Collectively, our results indicate that early stages of phagocytosis in the RPE are directed by signaling through mTOR, Akt, and MEK/ERK. Importantly, we discovered that the non-canonical Wnt pathway plays an important role in phagocytosis as well.
Experimental procedures
Chemicals
Ammonium bicarbonate (NH4HCO3), DTT, iodoacetamide, urea, triethylammonium bicarbonate buffer, acetonitrile (ACN), TFA, ammonia, formic acid, sucrose, sodium chloride (NaCl), HEPES, formaldehyde (CH2O), formaldehyde-d2 (CD2O), sodium cyanoborohydride (NaBH3CN), mTOR inhibitor, and KU-0063794 were obtained from Sigma-Aldrich. Inhibitors were dissolved in 100% DMSO to a concentration of 10 mm and used to treat cells at 10 μm unless noted otherwise. Akt inhibitor IV was purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX). The radioimmunoprecipitation assay (RIPA) buffer was purchased from Cell Signaling Technology (Danvers, MA). Lys-C was procured from Wako Chemicals (Richmond, VA), and trypsin (TPCK-treated) was from Worthington. Dulbecco's phosphate-buffered saline (DPBS) was obtained from Gibco-Invitrogen (Burlington, Canada).
Animals and cell lines
Mice were housed in the animal facility at the School of Medicine, Case Western Reserve University under a 12-h light (∼10 lux)/12-h dark cyclic environment. All animal procedures and experiments were approved by the Case Western Reserve University Animal Care Committees and conformed to recommendations of both the American Veterinary Medical Association Panel on Euthanasia and the Association for Research in Vision and Ophthalmology. C57BL/6J mice used for RPE isolation were purchased from The Jackson Laboratory (Bar Harbor, ME). ARPE-19 cells purchased from ATCC (Manassas, VA) were cultured in DMEM/F-12 medium (Invitrogen) supplemented with 10% FBS and 28 μg/ml gentamicin (Gibco-Invitrogen) at 37 °C in 5% CO2, according to the manufacturer's recommendations.
SILAC of an ARPE-19 cell line
ARPE-19 cells were cultured in customized DMEM/F-12 by AthenaES (Baltimore, MD) in which the natural arginine and lysine were replaced by heavy isotope-labeled amino acids (heavy media), 13C615N4 l-arginine (Arg-10) and 13C615N2 l-lysine (Lys-8), both from Sigma-Aldrich, at final concentrations of 147 and 91 mg/liter for arginine and lysine, respectively. The medium was supplemented with 17 mg/liter methionine (Sigma-Aldrich), 10% (v/v) dialyzed FBS (Gibco-Invitrogen), 1 mm sodium pyruvate (Gibco-Invitrogen), and 28 μg/ml gentamicin (Gibco-Invitrogen). Isotope-labeled ARPE-19 cells were generated by growth for at least 10 doublings in SILAC medium to allow complete intracellular incorporation of the isotopically labeled amino acids.
ARPE-19 cell phagocytosis assays
Both unlabeled (light) and SILAC-labeled (heavy) ARPE-19 cells were grown in 15-cm dishes to achieve confluence in about 7 days under standard growth conditions. POS membranes from bovine retinas for phagocytosis induction were isolated as described previously (53, 54); resuspended in 10 mm sodium phosphate buffer, pH 8.0, containing 150 mm NaCl and 2.5% sucrose; and kept at −80 °C before use. ARPE-19 cells were washed three times with ice-cold DPBS and challenged with POS at a density of ∼10 POS/ARPE-19 cell for the periods indicated (15, 30, 60, 90, and 120 min; n = 3). Unchallenged APRE19 cells (denoted as 0 min) were used as controls for the downstream proteomic and phosphoproteomic analyses.
Quantitative proteomic and phosphoproteomic analysis of ARPE-19 cells
POS-challenged and control ARPE-19 cells were washed twice with ice-cold DPBS and lysed by the addition of 1 ml of RIPA buffer with protease inhibitor mixture (Complete Mini, Roche Applied Science, Mississauga, Canada) and a phosphatase inhibitor tablet (PhosStop, Roche Applied Science). Lysates were ultrasonicated three times (10-s pulses each with >30 s of incubation on ice between each pulse) and centrifuged for 10 min at 16,000 × g. Five volumes of ice-cold acetone were added to lysates that were then left to precipitate overnight at −20 °C. Precipitates were pelleted by centrifugation (4500 × g, 10 min, 4 °C), washed three times with ice-cold acetone, and resolubilized in 400 μl of a 50 mm NH4HCO3 solution containing 8 m urea. Protein concentrations were determined with a Bio-Rad Bradford protein assay kit (Mississauga, Canada). Isotope-labeled samples from each time point were first pooled and then mixed with each unlabeled sample (500 μg of protein) in a 1:1 (w/w) ratio. Resulting samples (1 mg of protein each) were reduced with 5 mm DTT at 37 °C for 45 min and alkylated with 10 mm iodoacetamide in the dark at room temperature for 30 min. Next, samples were diluted with 5 volumes of 50 mm NH4HCO3, pH 8.5, to reduce the urea concentration to <2 m and then digested with Lys-C at 37 °C for 2 h (1:50 (w/w) enzyme/protein ratio). TPCK-treated trypsin at the same ratio was added to each sample before overnight incubation at 37 °C. For proteome analysis, 0.1 mg of the resulting peptides was desalted with in-house C18 desalting cartridges and dried in a SpeedVac before LC-MS analysis. For phosphoproteomic analyses, 0.9 mg of digested peptides from each sample was desalted with SepPak C18 cartridges (Waters, Mississauga, Canada), dried, and enriched for phosphopeptides by a Ti4+-IMAC bead-based procedure described previously (55). Briefly, each sample (0.9 mg of digested peptides) was resuspended in 1 ml of loading buffer (80% (v/v) ACN and 6% (v/v) TFA) with a 900-μl Ti4+-IMAC bead slurry (10 mg of beads in 1 ml of loading buffer) for 30 min at 4 °C. After centrifugation (16,000 × g, 10 min), beads were washed with 300 μl of washing buffer 1 (50% (v/v) ACN, 6% (v/v) TFA, and 200 mm NaCl) followed by two washes with 300 μl of washing buffer 2 (30% (v/v) ACN and 0.1% (v/v) TFA). Phosphopeptides were eluted from the beads by adding 500 μl of 10% (v/v) ammonia and vortexed for 15 min at 4 °C. After centrifugation (16,000 × g, 10 min), the supernatants were collected, acidified with 10% (v/v) TFA, desalted with in-house C18 desalting cartridges, and dried in a SpeedVac before LC-MS analysis.
Mouse RPE isolation
The RPE was isolated from 10–12-day-old C57BL/6J mice as described previously (56). Briefly, eyes were removed from animals and washed twice in DMEM (Thermo Fisher Scientific) supplemented with non-essential amino acids. Eyes were incubated in 2% dispase (Thermo Fisher Scientific) solution for 45 min in a 37 °C water bath with occasional tube inversion. Then eyes were washed twice in cold DMEM plus streptomycin/penicillin (Thermo Fisher Scientific), 10% FBS (Thermo Fisher Scientific), and 20 mm HEPES, pH 7.2. After eye enucleation, the cornea, lens, and iris were removed. Eye cups were incubated with DMEM plus streptomycin/penicillin, 10% FBS, and 20 mm HEPES, pH 7.2, in a 37 °C incubator for 15 min to facilitate removal of the neural retina. After neural retina removal, sheets of continuous RPE were peeled from the choroid and pipetted into a tube containing RPE growth medium (DMEM plus streptomycin/penicillin, 1× non-essential amino acids, and 10% FBS). RPE sheets were subsequently filtered with a 40-μm cell strainer (Thermo Fisher Scientific) to remove contaminating cell types. Sheets were spun at 200 × g for 3 min and then suspended in the RPE growth medium (for subsequent culturing) or placed in 1 ml of RIPA buffer (for MS analysis).
Primary RPE cell culture and phagocytosis assays
Disrupted RPE cells were seeded onto 24-well, 0.4-μm transwell permeable supports (Corning Inc., Corning, NY) with the RPE from ∼1 eye/well to allow polarization of cells. Cells were grown for 5–6 days at 37 °C in 5% CO2 before their use in phagocytosis assays. Cultured RPE cells were challenged with mouse POS membranes for 1.5 h at a density of ∼10 POS/RPE cell at 37 °C in 5% CO2. Cells were subsequently washed three times with DPBS, suspended in 100 μl of DPBS/well, and transferred to a 1.5-ml centrifuge tube. Cells then were pelleted at 1500 × g at 4 °C for 15 min. The supernatant was discarded, and cell pellets were either lysed when proceeding with an immunoblot analysis, snap frozen, and stored at −80 °C for MS analysis or suspended in RNAlater stabilization reagent (Qiagen, Valencia, CA) for RNA-seq analysis.
For inhibitor phagocytosis assays, cells were preincubated with the inhibitor in serum-free medium for at least 2 h before challenge. Then cells were challenged with either unlabeled or FITC-labeled POS, for immunochemistry or fluorescence imaging, respectively. POS membranes were covalently labeled with FITC (Thermo Fisher Scientific) according to established protocols (15). POS were suspended in DMEM plus streptomycin/penicillin, 10% FBS, and 2.5% sucrose. Then 50 μl of this mixture was added to the top of a transwell membrane, whereas 700 μl of DMEM alone plus streptomycin/penicillin and 10% FBS was added to the well of the plate. Assay mixtures were incubated in the dark at 37 °C for 1 h. Cells were then washed three times with DPBS. FITC fluorescence of externally bound photoreceptor POS was quenched by incubation with 0.2% trypan blue/DPBS (Thermo Fisher Scientific) for 10 min, after which cells were washed three times with DPBS. For imaging, cells were fixed with ice-cold methanol for 5 min at 4 °C followed by 4% paraformaldehyde/DPBS at room temperature for 10 min. Fixed cells were washed three times with DPBS. Nuclear staining was achieved by incubation with Hoechst stain (10 μm final) for 30 min at room temperature. Cells then were washed in DPBS an additional three times. Transwell membranes were removed from supports and mounted onto microscope slides with ProLong Gold antifade agent (Thermo Fisher Scientific). For immunochemistry, cells were lysed in Laemmli sample buffer (Bio-Rad).
Qualitative phosphoproteomics of cultured RPE cells
POS-challenged RPE cells were pooled together and lysed in urea lysis buffer (20 mm HEPES, 9 m urea, pH 8.0) supplemented with protease inhibitor mixture and a phosphatase inhibitor tablet. Protein content was determined with a Pierce BCA protein assay kit (Thermo Fisher Scientific). For pSer and pThr identification, a TiO2 phosphopeptide enrichment kit (Thermo Fisher Scientific) was used, whereas for pTyr identification, a PTMScan phosphotyrosine rabbit mAb (P-Tyr-1000) kit (Cell Signaling Technology) was employed. Both 280 and 500 mg of protein were used for the analyses. In both cases, protein digestion, phosphopeptide enrichment, and purification were performed based on the respective manufacturers' suggestions.
Transcriptomic analysis of cultured RPE cells
POS-challenged and control RPE cells (n = 3) were used for RNA isolation with TRIzol (Thermo Fisher Scientific), followed by poly(A) RNA fraction isolation with an Oligotex kit (Qiagen), according to the manufacturer's directions. Six cDNA libraries were generated (one for each biological replicate under each condition) and run separately, each on two lanes of the Genome Analyzer IIx (Illumina, San Diego, CA) in the Genomics Core Facility at Case Western Reserve University. Raw reads were processed and aligned with the University of California, Santa Cruz (UCSC) mouse genome assembly and transcript annotation (mm10) using the Genomic Short-read Nucleotide Alignment Program (GSNAP (57)). The Rsubread package (58) was used for read quantification. Genes with <0.5 counts/million mapped reads in at least three libraries were removed from the subsequent analyses. Additionally, data sets were corrected against potential photoreceptor contamination by removing a set of genes exhibiting very high expression in mouse photoreceptors as compared with the RPE (59). Data normalization and differential expression analyses were performed with DESeq2 (60), with a Benjamini–Hochberg adjusted p value <0.05 considered as statistically significant. DAVID software (61) was used for GO annotation.
Quantitative proteomics and phosphoproteomics of native RPE cells
RPE cells were isolated from eyes removed from mice at 7:30 a.m. and 3 p.m. (10 mice for each sample, n = 3 for each time point). Disrupted cells were resuspended in 1 ml of RIPA buffer and precipitated with 5 ml of ice-cold acetone at −20 °C overnight. Precipitates were centrifuged, washed with ice-cold acetone, resolubilized in 50 mm NH4HCO3, 8 m urea buffer, and protein concentrations were determined with the Bradford assay. Then 650 μg of protein in each sample was reduced, alkylated, digested with Lys-C and trypsin, desalted with a SepPak C18 cartridge, and dried in a SpeedVac. For quantitative analyses, stable isotope dimethyl labeling of peptides was performed (61). Briefly, samples were reconstituted in 100 mm triethylammonium bicarbonate buffer, pH 8.0, and divided into halves (325 μg of digested peptides each), which were isotopically labeled with either 0.2% CH2O and 30 mm NaBH3CN (light (L) form labeling) or 0.2% CD2O and 30 mm NaBH3CN (intermediate (M) form labeling). Ammonium solution was used to quench the reaction, and formic acid was used to acidify the samples. Next, six of the intermediate labeled peptide samples (325 μg each) were pooled together as a reference and diluted into each light sample (325 μg) with a 1:1 protein weight ratio (a total of 650 μg of peptides in each sample). The resulting samples were desalted with SepPak C18 cartridges and dried by a SpeedVac. In each case, 65 μg of peptides were used for the proteome analysis, whereas 585 μg of peptides were subjected to phosphopeptide enrichment with Ti4+-IMAC beads used for the phosphoproteome analysis.
LC-MS analysis
ARPE-19 and native RPE peptide samples were reconstituted in 100 μl of 0.1% (v/v) FA, and 4 μl of each sample was analyzed by an online reverse-phase LC-MS/MS platform consisting of an Eksigent NanoLC-Ultra 2D plus system (AB SCIEX) coupled with a Q Exactive Hybrid quadrupole-orbitrap mass spectrometer (Thermo Fisher Scientific) via a nano-electrospray source. Peptide mixtures were separated by reverse-phase chromatography on a home-packed ReproSil-Pur C18-AQ column (75-μm internal diameter × 15 cm, 1.9 μm, 200-Å pore size; Dr-Maisch GmbH, Ammerbuch, Germany) in a 4-h LC gradient of 2–80% buffer B (ACN in 0.1% (v/v) FA) at a flow rate of 300 nl/min. The Q Exactive instrument was operated in the data-dependent mode to simultaneously scan MS spectra (from m/z 400 to 2000) in the Orbitrap analyzer at resolution r = 70,000. Up to the 12 most intense peaks with a charge state ≥2 and having a signal threshold >500 counts were selected for fragmentation in the ion trap through higher-energy collisional dissociation. Dynamic exclusion was set as follows: repeat count 1, repeat duration 30 s, and an exclusion duration of 90 s. System control and data collection were carried out with Xcalibur software version 2.2 (Thermo Fisher Scientific).
Cultured RPE samples enriched in phosphopeptides were analyzed with a Dionex Ultimate 3000 nanoflow HPLC interfaced with a Finnigan Orbitrap LTQ Elite ion trap mass spectrometer system (Thermo Fisher Scientific). The HPLC system consisted of an Acclaim PepMap 100 precolumn (75-μm internal diameter × 2 cm, C18, 3 μm, 100-Å pore size) followed by an Acclaim PepMap RSLC analytical column (75-μm internal diameter × 15 cm, C18, 2 μm, 100-Å pore size). 5-μl volumes of the extract were injected, and peptides eluted from the column by an acetonitrile, 0.1% formic acid gradient at a flow rate of 0.3 μl/min were introduced into the source of the mass spectrometer online. The digest was analyzed using the data-dependent multitask capability of the instrument acquiring full-scan mass spectra to determine peptide molecular weights and product ion spectra to determine the amino acid sequence in successive instrument scans.
Mass spectrometry database search and bioinformatic analyses
Proteomic and phosphoproteomic data were analyzed and quantified with MaxQuant (version 1.3.0.5) using Andromeda as a search engine against the UniProt database restricted to human (Homo sapiens, released August 2014) and mouse (Mus musculus, released May 2013) taxonomy concatenated with a decoy of reversed sequences for ARPE-19 and RPE data, respectively. The precursor ion mass tolerance was 6 ppm, and fragment mass deviation was 0.5 Da for MS/MS spectra. The search included variable modifications of methionine oxidation, N-terminal acetylation, and Ser/Thr/Tyr phosphorylation and a fixed modification of Cys carbamidomethylation. Trypsin/P (cleavage after Lys/Arg residues, including Lys/Arg-Pro residues) was set as the cleavage specificity with two missed cleavages. For peptide and protein identification, the cutoff FDRs were both set to 0.01, and the minimum peptide length was set to 7. Identification across different replicates and adjacent fractions was achieved by enabling a match between runs within a time window of 2 min. High stringency of the analysis aimed to prevent quantification of bovine photoreceptor-derived proteins in the ARPE-19 samples. Default settings were used for all other parameters in MaxQuant. The protein group files (proteomic analysis) and phosphorylation (STY) site files (phosphoproteomic analysis) from ARPE-19 or RPE experiments were imported into Perseus (version 1.5.2.4) for their respective analyses.
One-way ANOVA was used to find the POS-stimulated proteome and phosphoproteome in the ARPE-19 data set (permutation-based FDR estimation, q < 0.05), whereas Student's t test was employed with the RPE proteome and phosphoproteome data sets to identify statistically significant (p < 0.05) differences in expression at the protein and phosphorylation levels, respectively. For the hierarchical clustering analysis, log values for the normalized L/H ratio of each protein and phosphopeptide profile were determined after z-score normalization of the data within Euclidean distances, respectively. GO annotation and pathway enrichment analyses were implemented with DAVID (61). Ingenuity Pathway Analysis (IPA) software (version 7.5; Qiagen) was used to analyze the biological functions, protein–protein interactions, and signaling pathway annotations of proteins and phosphoproteins responding to POS stimulation. Motif analysis was performed with iceLogo (62), scoring by percentage difference with a significance threshold of 0.05 for a sequence window of 15 amino acids surrounding the phosphorylated residues of POS-stimulated phosphopeptides against the class I phosphoproteome (localization probability score >0.75). Protein interaction network analyses of the significantly altered proteome and phosphoproteome in ARPE-19 and native RPE data sets were performed with STRING (63) using a high confidence (0.7) threshold. iGPS 1.0 (64) was used to predict possible site-specific kinase–substrate relationships between putative protein kinases and significantly altered phosphopeptides in the ARPE-19 phosphoproteome data set with a medium threshold.
Immunoblot analysis
Isolated proteins were resolved on 4–20% SDS-polyacrylamide gels and transferred to PVDF membranes with a Bio-Rad Mini-PROTEAN Tetra Cell. Membranes then were blocked with 5% nonfat milk for 1 h at room temperature. Primary antibodies against phospho-GSK3β (Ser-9), phospho-CTNNB1 (Ser-552), phospho-ERK1/2 (Thr-202/Tyr-204), and phospho-Akt (Ser-473) as well as anti-GSK3β, anti-ERK1/2, and anti-Akt antibodies were purchased from Cell Signaling Technology (Danvers, MA), and anti-CTNNB1 primary antibody was from Santa Cruz Biotechnology, Inc. (Dallas, TX). Primary antibodies were diluted with 5% bovine serum albumin in PBS-Tween 20 (PBST) and incubated with membranes overnight at 4 °C. Membranes were then washed in PBS three times for 10 min each and incubated with HRP-conjugated secondary antibodies (1:5000; Jackson ImmunoResearch Laboratories, West Grove, PA) in 5% nonfat milk for 1 h at room temperature. Membranes were washed again in PBS three times for 10 min each. Bands were visualized after incubation with the Thermo Fisher Scientific SuperSignal West Pico chemiluminescent substrate and subsequent exposure to CL-XPosure film (Waltham, MA).
Author contributions
B. M. K. and K. P. designed the study. B. M. K. and C.-K. C. performed the experiments: B. M. K. performed M. S., fluorescence imaging, and immunochemistry experiments, and C.-K. C. carried out the MS experiments. B. M. K., C.-K. C., and A. T. analyzed the results. D. F. and K. P. supervised the research. B. M. K. and A. T. wrote the initial draft of the manuscript, and C.-K. C., B. X., J. M., Z. N., D. F., and K. P. assisted in editing the article. All authors read and approved the final manuscript.
Supplementary Material
Acknowledgments
We thank members of the Palczewski laboratory for valuable comments regarding the manuscript. We also thank Dr. Belinda Willard and Dr. Dongmei Zhang from the Cleveland Clinic Proteomics and Metabolomics Laboratory for assistance with the MS analysis of cultured RPE.
This work was supported in part by National Institutes of Health Grants EY009339 and EY024864 (to K. P.) and EY011373 (VSRC CORE grant); the National Sciences and Engineering Research Council (NSERC) of Canada (to D. F.); the Canadian Institute for Advanced Research (CIFAR) and the Canadian Institutes of Health Research (CIHR) (to D. F.); the Arnold and Mabel Beckman Foundation (to K. P.); and the Canadian Foundation for Innovation (CFI), the Ontario Research Fund, and la Fondation J.-Louis Lévesque (to D. F.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
RNA-seq raw data reported in this work were deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE96626.
Mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifiers PXD006124, PXD006125, and PXD006144.
This article contains supplemental Tables S1 and S2, Figs. S1–S5, and Data Sets S1–S5.
- RPE
- retinal pigment epithelium
- FDR
- false discovery rate
- GO
- gene ontology
- POS
- photoreceptor outer segment
- PtdSer
- phosphatidylserine
- RIPA
- radioimmunoprecipitation assay
- RNA-seq
- RNA sequencing
- SILAC
- stable isotope labeling by amino acids in cell culture
- TPCK
- l-1-tosylamido-2-phenylethyl chloromethyl ketone
- mTOR
- mechanistic target of rapamycin
- GSK
- glycogen synthase kinase
- MERTK
- Mer receptor tyrosine kinase
- ACN
- acetonitrile
- DPBS
- Dulbecco's PBS.
References
- 1. Rizzolo L. J. (2014) Barrier properties of cultured retinal pigment epithelium. Exp. Eye Res. 126, 16–26 [DOI] [PubMed] [Google Scholar]
- 2. Kiser P. D., Golczak M., Maeda A., and Palczewski K. (2012) Key enzymes of the retinoid (visual) cycle in vertebrate retina. Biochim. Biophys. Acta 1821, 137–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kiser P. D., and Palczewski K. (2016) Retinoids and Retinal Diseases. Annu. Rev. Vis. Sci. 2, 197–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kevany B. M., and Palczewski K. (2010) Phagocytosis of retinal rod and cone photoreceptors. Physiology 25, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Young R. W., and Bok D. (1969) Participation of the retinal pigment epithelium in the rod outer segment renewal process. J. Cell Biol. 42, 392–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ruggiero L., Connor M. P., Chen J., Langen R., and Finnemann S. C. (2012) Diurnal, localized exposure of phosphatidylserine by rod outer segment tips in wild-type but not Itgb5−/− or Mfge8−/− mouse retina. Proc. Natl. Acad. Sci. U.S.A. 109, 8145–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ryeom S. W., Silverstein R. L., Scotto A., and Sparrow J. R. (1996) Binding of anionic phospholipids to retinal pigment epithelium may be mediated by the scavenger receptor CD36. J. Biol. Chem. 271, 20536–20539 [DOI] [PubMed] [Google Scholar]
- 8. Ryeom S. W., Sparrow J. R., and Silverstein R. L. (1996) CD36 participates in the phagocytosis of rod outer segments by retinal pigment epithelium. J. Cell Sci. 109, 387–395 [DOI] [PubMed] [Google Scholar]
- 9. Prasad D., Rothlin C. V., Burrola P., Burstyn-Cohen T., Lu Q., Garcia de Frutos P., and Lemke G. (2006) TAM receptor function in the retinal pigment epithelium. Mol. Cell Neurosci. 33, 96–108 [DOI] [PubMed] [Google Scholar]
- 10. Nandrot E. F., Anand M., Almeida D., Atabai K., Sheppard D., and Finnemann S. C. (2007) Essential role for MFG-E8 as ligand for αvβ5 integrin in diurnal retinal phagocytosis. Proc. Natl. Acad. Sci. U.S.A. 104, 12005–12010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Finnemann S. C. (2003) Focal adhesion kinase signaling promotes phagocytosis of integrin-bound photoreceptors. EMBO J. 22, 4143–4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Law A. L., Ling Q., Hajjar K. A., Futter C. E., Greenwood J., Adamson P., Wavre-Shapton S. T., Moss S. E., and Hayes M. J. (2009) Annexin A2 regulates phagocytosis of photoreceptor outer segments in the mouse retina. Mol. Biol. Cell 20, 3896–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mao Y., and Finnemann S. C. (2012) Essential diurnal Rac1 activation during retinal phagocytosis requires αvβ5 integrin but not tyrosine kinases focal adhesion kinase or Mer tyrosine kinase. Mol. Biol. Cell 23, 1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heth C. A., and Schmidt S. Y. (1991) Phagocytic challenge induces changes in phosphorylation of retinal pigment epithelium proteins. Curr. Eye Res. 10, 1049–1057 [DOI] [PubMed] [Google Scholar]
- 15. Finnemann S. C., Bonilha V. L., Marmorstein A. D., and Rodriguez-Boulan E. (1997) Phagocytosis of rod outer segments by retinal pigment epithelial cells requires α(v)β5 integrin for binding but not for internalization. Proc. Natl. Acad. Sci. U.S.A. 94, 12932–12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., and Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 17. Ainscough S. L., Feigl B., Malda J., and Harkin D. G. (2009) Discovery and characterization of IGFBP-mediated endocytosis in the human retinal pigment epithelial cell line ARPE-19. Exp. Eye Res. 89, 629–637 [DOI] [PubMed] [Google Scholar]
- 18. May R. C., and Machesky L. M. (2001) Phagocytosis and the actin cytoskeleton. J. Cell Sci. 114, 1061–1077 [DOI] [PubMed] [Google Scholar]
- 19. Pearce L. R., Komander D., and Alessi D. R. (2010) The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11, 9–22 [DOI] [PubMed] [Google Scholar]
- 20. Ablonczy Z., Dahrouj M., Tang P. H., Liu Y., Sambamurti K., Marmorstein A. D., and Crosson C. E. (2011) Human retinal pigment epithelium cells as functional models for the RPE in vivo. Invest. Ophthalmol. Vis. Sci. 52, 8614–8620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Palczewska G., Maeda A., Golczak M., Arai E., Dong Z., Perusek L., Kevany B., and Palczewski K. (2016) Receptor MER tyrosine kinase proto-oncogene (MERTK) is not required for transfer of bis-retinoids to the retinal pigmented epithelium. J. Biol. Chem. 291, 26937–26949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Freeman S. A., and Grinstein S. (2014) Phagocytosis: receptors, signal integration, and the cytoskeleton. Immunol. Rev. 262, 193–215 [DOI] [PubMed] [Google Scholar]
- 23. Filippi S., Barnes C. P., Kirk P. D., Kudo T., Kunida K., McMahon S. S., Tsuchiya T., Wada T., Kuroda S., and Stumpf M. P. (2016) Robustness of MEK-ERK dynamics and origins of cell-to-cell variability in MAPK signaling. Cell Rep. 15, 2524–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Onishi Y., Hanai S., Ohno T., Hara Y., and Ishida N. (2008) Rhythmic SAF-A binding underlies circadian transcription of the Bmal1 gene. Mol. Cell. Biol. 28, 3477–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lande-Diner L., Boyault C., Kim J. Y., and Weitz C. J. (2013) A positive feedback loop links circadian clock factor CLOCK-BMAL1 to the basic transcriptional machinery. Proc. Natl. Acad. Sci. U.S.A. 110, 16021–16026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bulloj A., Duan W., and Finnemann S. C. (2013) PI 3-kinase independent role for AKT in F-actin regulation during outer segment phagocytosis by RPE cells. Exp. Eye Res. 113, 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee H., Chung H., Lee S. H., and Jahng W. J. (2011) Light-induced phosphorylation of crystallins in the retinal pigment epithelium. Int. J. Biol. Macromol. 48, 194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mendoza M. C., Er E. E., and Blenis J. (2011) The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem. Sci. 36, 320–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Indik Z. K., Park J. G., Hunter S., and Schreiber A. D. (1995) The molecular dissection of Fc γ receptor mediated phagocytosis. Blood 86, 4389–4399 [PubMed] [Google Scholar]
- 30. Cox D., Tseng C. C., Bjekic G., and Greenberg S. (1999) A requirement for phosphatidylinositol 3-kinase in pseudopod extension. J. Biol. Chem. 274, 1240–1247 [DOI] [PubMed] [Google Scholar]
- 31. García-García E., Rosales R., and Rosales C. (2002) Phosphatidylinositol 3-kinase and extracellular signal-regulated kinase are recruited for Fc receptor-mediated phagocytosis during monocyte-to-macrophage differentiation. J. Leukoc. Biol. 72, 107–114 [PubMed] [Google Scholar]
- 32. Wrann C. D., Tabriz N. A., Barkhausen T., Klos A., van Griensven M., Pape H. C., Kendoff D. O., Guo R., Ward P. A., Krettek C., and Riedemann N. C. (2007) The phosphatidylinositol 3-kinase signaling pathway exerts protective effects during sepsis by controlling C5a-mediated activation of innate immune functions. J. Immunol. 178, 5940–5948 [DOI] [PubMed] [Google Scholar]
- 33. Fox R., Nhan T. Q., Law G. L., Morris D. R., Liles W. C., and Schwartz S. M. (2007) PSGL-1 and mTOR regulate translation of ROCK-1 and physiological functions of macrophages. EMBO J. 26, 505–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Parsa K. V., Butchar J. P., Rajaram M. V., Cremer T. J., and Tridandapani S. (2008) The tyrosine kinase Syk promotes phagocytosis of Francisella through the activation of Erk. Mol. Immunol. 45, 3012–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Makino-Okamura C., Niki Y., Takeuchi S., Nishigori C., Declercq L., Yaroch D. B., and Saito N. (2014) Heparin inhibits melanosome uptake and inflammatory response coupled with phagocytosis through blocking PI3k/Akt and MEK/ERK signaling pathways in human epidermal keratinocytes. Pigment Cell Melanoma Res. 27, 1063–1074 [DOI] [PubMed] [Google Scholar]
- 36. Wang X., Li M., Gao Y., Gao J., Yang W., Liang H., Ji Q., Li Y., Liu H., Huang J., Cheng T., and Yuan W. (2016) Rheb1-mTORC1 maintains macrophage differentiation and phagocytosis in mice. Exp. Cell Res. 344, 219–228 [DOI] [PubMed] [Google Scholar]
- 37. Yang P., Peairs J. J., Tano R., and Jaffe G. J. (2006) Oxidant-mediated Akt activation in human RPE cells. Invest. Ophthalmol. Vis. Sci. 47, 4598–4606 [DOI] [PubMed] [Google Scholar]
- 38. Sarbassov D. D., Guertin D. A., Ali S. M., and Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 39. Yu B., Xu P., Zhao Z., Cai J., Sternberg P., and Chen Y. (2014) Subcellular distribution and activity of mechanistic target of rapamycin in aged retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 55, 8638–8650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fang D., Hawke D., Zheng Y., Xia Y., Meisenhelder J., Nika H., Mills G. B., Kobayashi R., Hunter T., and Lu Z. (2007) Phosphorylation of β-catenin by AKT promotes β-catenin transcriptional activity. J. Biol. Chem. 282, 11221–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., and Hemmings B. A. (1995) Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378, 785–789 [DOI] [PubMed] [Google Scholar]
- 42. Zhao C., Yasumura D., Li X., Matthes M., Lloyd M., Nielsen G., Ahern K., Snyder M., Bok D., Dunaief J. L., LaVail M. M., and Vollrath D. (2011) mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J. Clin. Invest. 121, 369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garg T. K., and Chang J. Y. (2003) Oxidative stress causes ERK phosphorylation and cell death in cultured retinal pigment epithelium: prevention of cell death by AG126 and 15-deoxy-δ12,14-PGJ2. BMC Ophthalmol. 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tate D. J. Jr, Miceli M. V., and Newsome D. A. (1995) Phagocytosis and H2O2 induce catalase and metallothionein gene expression in human retinal pigment epithelial cells. Invest. Ophthalmol. Vis. Sci. 36, 1271–1279 [PubMed] [Google Scholar]
- 45. Olchawa M. M., Pilat A. K., Szewczyk G. M., and Sarna T. J. (2016) Inhibition of phagocytic activity of ARPE-19 cells by free radical mediated oxidative stress. Free Radic. Res. 50, 887–897 [DOI] [PubMed] [Google Scholar]
- 46. Hall M. O., Obin M. S., Prieto A. L., Burgess B. L., and Abrams T. A. (2002) Gas6 binding to photoreceptor outer segments requires γ-carboxyglutamic acid (Gla) and Ca2+ and is required for OS phagocytosis by RPE cells in vitro. Exp. Eye Res. 75, 391–400 [PubMed] [Google Scholar]
- 47. Ishimoto Y., Ohashi K., Mizuno K., and Nakano T. (2000) Promotion of the uptake of PS liposomes and apoptotic cells by a product of growth arrest-specific gene, gas6. J. Biochem. 127, 411–417 [DOI] [PubMed] [Google Scholar]
- 48. Nakano T., Ishimoto Y., Kishino J., Umeda M., Inoue K., Nagata K., Ohashi K., Mizuno K., and Arita H. (1997) Cell adhesion to phosphatidylserine mediated by a product of growth arrest-specific gene 6. J. Biol. Chem. 272, 29411–29414 [DOI] [PubMed] [Google Scholar]
- 49. Goruppi S., Ruaro E., Varnum B., and Schneider C. (1997) Requirement of phosphatidylinositol 3-kinase-dependent pathway and Src for Gas6-Axl mitogenic and survival activities in NIH 3T3 fibroblasts. Mol. Cell. Biol. 17, 4442–4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Glotin A. L., Calipel A., Brossas J. Y., Faussat A. M., Tréton J., and Mascarelli F. (2006) Sustained versus transient ERK1/2 signaling underlies the anti- and proapoptotic effects of oxidative stress in human RPE cells. Invest. Ophthalmol. Vis. Sci. 47, 4614–4623 [DOI] [PubMed] [Google Scholar]
- 51. Kyosseva S. V. (2016) Targeting MAPK signaling in age-related macular degeneration. Ophthalmol. Eye Dis. 8, 23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sasore T., Reynolds A. L., and Kennedy B. N. (2014) Targeting the PI3K/Akt/mTOR pathway in ocular neovascularization. Adv. Exp. Med. Biol. 801, 805–811 [DOI] [PubMed] [Google Scholar]
- 53. Liang Y., Fotiadis D., Filipek S., Saperstein D. A., Palczewski K., and Engel A. (2003) Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J. Biol. Chem. 278, 21655–21662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Papermaster D. S. (1982) Preparation of retinal rod outer segments. Methods Enzymol. 81, 48–52 [DOI] [PubMed] [Google Scholar]
- 55. Zhou H., Ye M., Dong J., Corradini E., Cristobal A., Heck A. J., Zou H., and Mohammed S. (2013) Robust phosphoproteome enrichment using monodisperse microsphere-based immobilized titanium (IV) ion affinity chromatography. Nat. Protoc. 8, 461–480 [DOI] [PubMed] [Google Scholar]
- 56. Diemer T., Gibbs D., and Williams D. S. (2008) Analysis of the rate of disk membrane digestion by cultured RPE cells. Adv. Exp. Med. Biol. 613, 321–326 [DOI] [PubMed] [Google Scholar]
- 57. Wu T. D., and Nacu S. (2010) Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liao Y., Smyth G. K., and Shi W. (2013) The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bennis A., Gorgels T. G., Ten Brink J. B., van der Spek P. J., Bossers K., Heine V. M., and Bergen A. A. (2015) Comparison of mouse and human retinal pigment epithelium gene expression profiles: potential implications for age-related macular degeneration. PLoS One 10, e0141597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 62. Colaert N., Helsens K., Martens L., Vandekerckhove J., and Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
- 63. Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., and Jensen L. J. (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Song C., Ye M., Liu Z., Cheng H., Jiang X., Han G., Songyang Z., Tan Y., Wang H., Ren J., Xue Y., and Zou H. (2012) Systematic analysis of protein phosphorylation networks from phosphoproteomic data. Mol. Cell. Proteomics 11, 1070–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.