Abstract
Fructose-1,6-bisphosphate (FBP) aldolase, a glycolytic enzyme, catalyzes the reversible and stereospecific aldol addition of dihydroxyacetone phosphate (DHAP) and d-glyceraldehyde 3-phosphate (d-G3P) by an unresolved mechanism. To afford insight into the molecular determinants of FBP aldolase stereospecificity during aldol addition, a key ternary complex formed by DHAP and d-G3P, comprising 2% of the equilibrium population at physiological pH, was cryotrapped in the active site of Toxoplasma gondii aldolase crystals to high resolution. The growth of T. gondii aldolase crystals in acidic conditions enabled trapping of the ternary complex as a dominant population. The obligate 3(S)-4(R) stereochemistry at the nascent C3–C4 bond of FBP requires a si-face attack by the covalent DHAP nucleophile on the d-G3P aldehyde si-face in the active site. The cis-isomer of the d-G3P aldehyde, representing the dominant population trapped in the ternary complex, would lead to re-face attack on the aldehyde and yield tagatose 1,6-bisphosphate, a competitive inhibitor of the enzyme. We propose that unhindered rotational isomerization by the d-G3P aldehyde moiety in the ternary complex generates the active trans-isomer competent for carbonyl bond activation by active-site residues, thereby enabling si-face attack by the DHAP enamine. C–C bond formation by the cis-isomer is suppressed by hydrogen bonding of the cis-aldehyde carbonyl with the DHAP enamine phosphate dianion through a tetrahedrally coordinated water molecule. The active site geometry further suppresses C–C bond formation with the l-G3P enantiomer of d-G3P. Understanding C–C formation is of fundamental importance in biological reactions and has considerable relevance to biosynthetic reactions in organic chemistry.
Keywords: crystal structure, enzyme catalysis, enzyme mechanism, glycolysis, stereoselectivity
Introduction
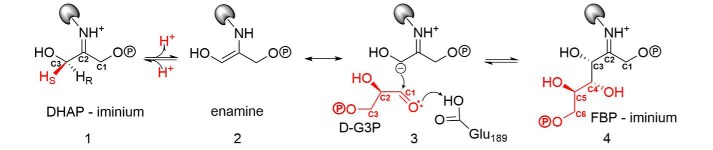
Aldolases are central enzymes that catalyze stereospecific carbon–carbon bond formation. Their role is best known in glycolysis, where fructose 1,6-bisphosphate (FBP)3 aldolase (EC 4.1.2.13) reversibly cleaves FBP into the triose-phosphates, d-glyceraldehyde-3-phosphate (d-G3P) and dihydroxyacetone phosphate (DHAP). The catalytic mechanism has been extensively investigated in class I aldolases from rabbit muscle, namely for its ease of purification and high sequence identity to the human counterpart. Class I aldolases make use of a covalent mechanism implicating Schiff base formation between a lysine residue and a ketose substrate (1).
The thermodynamically favored aldol addition involves formation of a Schiff base with the keto triose-P (DHAP), shown as intermediate 1 in Scheme 1. Stereospecific abstraction of the pro(S) C3 proton on DHAP ensures the development of a carbanionic character at C3 of DHAP, which is stabilized by resonance with the enamine form (intermediate 2). The DHAP-enamine condenses with d-G3P (intermediate 3) to form the C3–C4 bond, generating the ketimine intermediate (intermediate 4), which undergoes hydrolysis to release FBP. The ordered Uni Bi reaction mechanism is a consequence of structural changes at the molecular level, whereby DHAP binding stabilizes a conformational change with respect to the free enzyme that narrows the active site cleft (2). The narrowing is responsible for ordered binding, as it positions side chains at the P6-phosphate binding locus such that they are now able to interact with d-G3P or with the nascent aldehyde moiety of FBP, as FBP binding mimics the structural changes induced by DHAP (3). In the absence of these structural changes, stabilized by attachment at the P1-phosphate binding locus, d-G3P binding is unlikely or so weak that it has not been observed.
Scheme 1.
Intermediates of the catalytic mechanism in class I aldolases.
Studies with substrates and analogues have contributed immensely to our understanding of the active-site architecture and catalytic mechanism in aldolases (4–9). Class I aldolases are specific for DHAP but can accept diverse aldehydes (8). Selectivity studies have explored the stereochemistry of substrate analogs at every position, including the C5 position of FBP. Conflicting reports regarding the discrimination of diastereomers at the C5 position have been reported, notably regarding the l-isomer of G3P. First reports indicated that aldolase catalyzed the condensation of DHAP and l-G3P, producing l-sorbose 1,6-bisphosphate (SBP), albeit at a much lower rate than with incorporation of the d-isomer (10, 11). Cleavage of SBP by FBP aldolase was equally reduced compared with FBP. However, subsequent experiments by fluorescence quenching (4) and labeling studies (12) demonstrated that aldolase is rather progressively inactivated by l-G3P and that l-G3P is not a substrate for aldolase (4).
The prototypical class I aldolase used for mechanistic studies has long been the muscle isoform from rabbit aldolase. Cryocrystallography has enabled active-site trapping of a number of reaction intermediates that were determined to high resolution and, in complement with site-directed mutagenesis, delineated precise roles for several key residues, including Lys-229, the Schiff base–forming residue, and Glu-187, which mediates various proton transfer steps (2, 3, 13), notably carbinolamine formation and proton transfer during C–C bond cleavage.
Crystal structures emanating from previous studies involved long soaking times (≥3 min) and high substrate concentrations (10 mm FBP) (2) before flash-freezing. Therefore, the cryogenic structures trapped only the highly populated intermediates, consistent with biochemical determination of active-site populations near equilibrium (14). This has precluded the trapping of the elusive ternary enzymatic complex, aldolase·enamine·d-G3P, consistent with incipient aldol addition. Under neutral equilibrium conditions, this complex comprises ∼2% of the total bound intermediates (6, 15). Crystallographic characterization of this low-abundance active-site population would afford insight into the structural constraints imposed by the active-site architecture upon the formation of the ternary complex that ensures 3(S)-4(R) stereospecificity in C–C bond formation in FBP aldolases.
In class I aldolases, several interconversions of reaction intermediates are pH-dependent in the catalytic mechanism. We reasoned that crystallization conditions favoring acidic pH would provide an opportunity to stabilize reaction intermediates that would otherwise be of low occupancy in human and rabbit muscle aldolase crystals, which are typically grown at neutral pH. Specifically, a key proton transfer step, mediated by Glu-187, is required for both aldol addition and cleavage in mammalian class I aldolases and has a pKa of 5.2 in the muscle isozyme (16). We reasoned that acidic crystallization conditions would make proton transfer at Glu-187 a rate-limiting step. Crystals suitable for detailed structural analysis in acidic conditions (pH 4.5–5.5) were grown for a class I aldolase from Toxoplasma gondii (TgALD), a unicellular protozoan, that shares a 57% overall sequence identity (363 residues) and 100% conservation of active-site residues with human muscle aldolase. Additionally, acid pH stabilizes Schiff base formation rather than enamine formation (6), and together these considerations would argue in favor of cryotrapping intermediates with reaction geometries conducive for incipient aldolization in the direction of triose-P condensation.
Crystals for TgALD were soaked with FBP substrate for various times and then flash-frozen in liquid nitrogen to cryotrap enzymatic intermediates. The acid crystallization conditions yielded a series of high-resolution structures of enzymatic intermediates that corresponded to a series of snapshots delineating the progress of central reaction intermediates along the reaction pathway in TgALD. Notably, a ternary intermediate complex was successfully cryotrapped as a dominant population that could be shown to be consistent with incipient aldol addition. The active-site configuration of the intermediate provided intimate detail of the stereo control determinants during aldol addition. Further analyses of these structures revealed the structural basis by which glycolytic aldolases discriminate against the l-G3P enantiomer to ensure aldehyde stereospecificity.
Results and discussion
Ternary complex
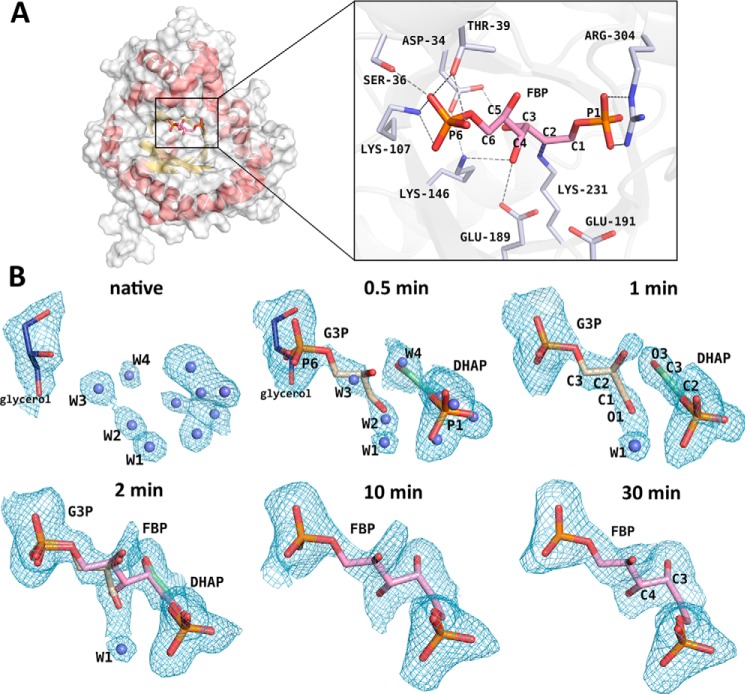
TgALD crystallized with one tetrameric subunit in the asymmetric unit of space group P42212, adopting the classic (α/β)8-barrel fold, characteristic of other class I aldolases (Fig. 1A, left). The TgALD quaternary structure corresponds to a tetramer and has 222-fold symmetry from the crystallographic packing. Data collection and refinement statistics for all crystal structures are shown in Table 1. All crystal structures were determined to high resolution (1.75–2.09 Å) and had comparable data collection and refinement statistics with Rfree values close to or inferior to 20%. Flash freezing of class I TgALD crystals previously soaked in the presence of substrate for various time intervals ranging between 0 and 30 min trapped a series of enzymatic intermediates. The intermediates included d-G3P and covalently bound DHAP simultaneously trapped before the aldol reaction step, leading to product formation (Fig. 1).
Figure 1.
Progress of the aldol addition reaction in a class I aldolase. A, overall view of the subunit globular TIM-barrel structure (helices in red; sheets in yellow) and of the active-site pocket in TgALD (structure shown corresponds to 30-min soaking with FBP in crystallization buffer). In the 30-min structure, FBP is covalently bound to the Schiff base–forming Lys-231. Key active-site residues that participate in hydrogen bonding (gray dashes) with substrate and in catalytic activity are depicted in the close-up (right inset). Noteworthy is the hydrogen bond made between FBP O4 and Glu-189 and that with Lys-146. The FBP phosphates (P1 and P6) participate in extensive contacts with adjacent residues (some omitted). Residue numbering is shown for TgALD. B, crystal structures obtained corresponding to different soaking times of native TgALD crystals with FBP in the crystallization buffer and the resultant intermediates that were trapped at the incremental time points by flash-freezing soaked crystals in liquid nitrogen. Difference electron density maps were calculated from simulated-annealing Fo − Fc omit maps encompassing the substrate-binding site and are contoured at a 2.1σ level. Where appropriate, the covalently bound intermediates, DHAP-enamine (green) or FBP-Schiff base (pink), are modeled into the active site as well as the aldehyde triose-P product d-G3P (wheat). Water molecules (blue spheres) were also modeled where appropriate. Ligands juxtaposed with water molecules at the 0.5-min structure correspond to partially occupied configurations. Soaking for at least 10 min is required for FBP to fully populate the TgALD active site.
Table 1.
Data collection and refinement statistics
| Crystal structure |
||||||
|---|---|---|---|---|---|---|
| Native | 0.5 min | 1 min | 2 min | 10 min | 30 min | |
| PDB code | 5TJS | 5TK3 | 5TKC | 5TKL | 5TKN | 5TKP |
| Data collection | ||||||
| Resolution range (Å) | 38.97–1.78 (1.84–1.78)a | 49.03–1.83 (1.90–1.83) | 38.82–1.78 (1.84–1.78) | 49.02–1.75 (1.81–1.75) | 27.27–1.92 (1.99–1.92) | 48.97–2.09 (2.165–2.09) |
| Space group | P 42 21 2 | P 42 21 2 | P 42 21 2 | P 42 21 2 | P 42 21 2 | P 42 21 2 |
| No. in asymmetric unit | 1 | 1 | 1 | 1 | 1 | 1 |
| Unit cell a (Å), c (Å), degrees | 110.2, 54.9, 90 | 109.8, 54.8, 90 | 109.8, 54.9, 90 | 109.6, 55.0, 90 | 109.1, 54.7, 90 | 109.12, 54.8, 90 |
| Total reflections | 426,298 (25,526) | 319,675 (24,753) | 404,145 (23,443) | 414,630 (34,281) | 169,255 (5154) | 154,210 (4866) |
| Unique reflections | 32,599 (2922) | 29,287 (2843) | 31,953 (2659) | 34,191 (3285) | 21,046 (1229) | 19,150 (1602) |
| Multiplicity | 13.1 (8.9) | 10.9 (8.7) | 12.7 (9.0) | 12.1 (7.5) | 8.0 (4.3) | 8.1 (3.7) |
| Completeness | 0.99 (0.89) | 0.97 (0.97) | 0.97 (0.82) | 1.00 | 0.82 (0.48) | 0.87 (0.67) |
| Average I/σ(I) | 20.8 (1.5) | 23.5 (2.5) | 26.2 (2.7) | 26.0 (1.9) | 20.0 (1.4) | 17.0 (1.3) |
| Wilson B-factor | 24.6 | 26.2 | 22.6 | 23.9 | 29.6 | 30.5 |
| Rmergeb | 0.085 (1.243) | 0.061 (0.909) | 0.059 (0.713) | 0.064 (0.896) | 0.067 (0.717) | 0.084 (0.929) |
| Rmeasc | 0.089 (1.322) | 0.064 (0.966) | 0.062 (0.754) | 0.067 (0.962) | 0.071 (0.800) | 0.089 (1.055) |
| CC* | 1 (0.863) | 1 (0.949) | 1 (0.958) | 1 (0.914) | 1 (0.886) | 1 (0.837) |
| Refinement | ||||||
| Rworkd | 0.155 (0.284) | 0.161 (0.259) | 0.152 (0.235) | 0.149 (0.270) | 0.150 (0.303) | 0.161 (0.239) |
| Rfreee | 0.200 (0.335) | 0.191 (0.301) | 0.192 (0.272) | 0.176 (0.292) | 0.199 (0.340) | 0.213 (0.295) |
| No. of non-H atoms | 2991 | 2972 | 3016 | 3066 | 2932 | 2882 |
| Macromolecules | 2653 | 2656 | 2670 | 2698 | 2677 | 2657 |
| Ligands | 6 | 25 | 25 | 38 | 19 | 19 |
| r.m.s. deviation (bond length) (Å) | 0.010 | 0.006 | 0.008 | 0.007 | 0.005 | 0.006 |
| r.m.s. deviation (angles) (degrees) | 1.20 | 0.72 | 0.83 | 0.88 | 0.92 | 0.93 |
| Ramachandran favored (%) | 97 | 97 | 97 | 97 | 98 | 98 |
| Average B-factor (Å2) | 33.91 | 35.09 | 30.16 | 30.66 | 34.86 | 35.90 |
a All values in parentheses are given for the highest resolution shell.
b Rmerge = ΣhklΣi|Ii(hkl) − Īi(hkl)|/ΣhklΣiIi(hkl), with i running over the number of independent observations of reflection hkl.
c Rmeas = Σhkl Σi(|Ii − Imean|)/ΣhklΣi(Ii).
d Rwork = Σhkl‖Io(hkl)|−|Ic(hkl)‖/Σhkl|Io(hkl)|.
e Rfree = ΣhklϵT‖Io(hkl) − |Ic(hkl)‖/ΣhklϵT|Io(hkl)|, where T is a test data set randomly selected from the observed reflections before refinement. The test data set was not used throughout refinement and contains a minimum of 2000 unique reflections (or 5%) (smaller value is selected).
Active-site residues in TgALD, shown in Fig. 1A (inset), are identical to rabbit muscle aldolase except for a serine residue in the rabbit homolog, which becomes Thr-39 in TgALD. At the structural level, there is complete conservation of the subunit β-barrel fold. The TgALD structure at 30-min soaking time (Fig. 1A) corresponds to an active site fully occupied by FBP that superposes identically with the FBP-bound structure of the rabbit homolog (PDB entry 1ZAI) (r.m.s. deviation = 0.41 Å based on equivalent Cα atoms) (shown in supplemental Fig. S1). Surrounding active-site residues of the same intermediates also superpose identically (r.m.s. deviation = 0.14 Å based on equivalent Cα atoms of active-site residues). In the homology alignment of class I aldolases from diverse species shown in supplemental Fig. S2, key active-site residues, shown in Fig. 1A, are colored according to their biochemical properties. The alignment underscores the high level of sequence conservation of active-site residues in class I aldolases. The blue color scheme emphasizes the level of similarity in residues flanking the active site. The active-site similarity in both sequence and structure makes TgALD a potential candidate for studying reaction mechanisms in class I aldolases.
Catalytic activity
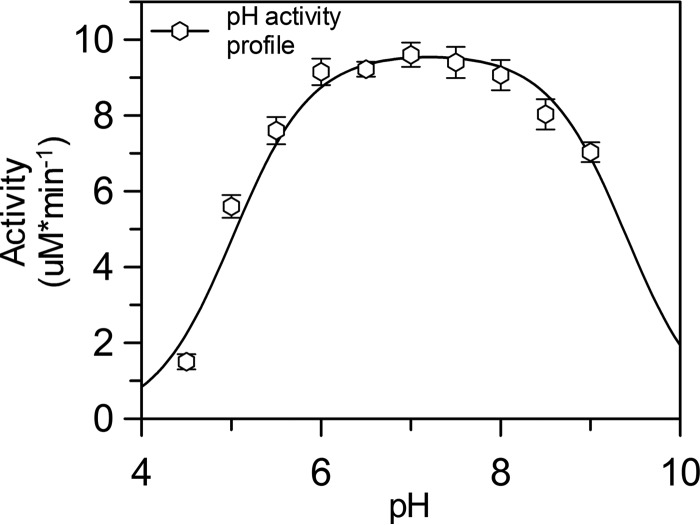
To validate TgALD as a suitable model for studying the reaction mechanism of FBP aldolase, kinetic parameters were determined (Table 2). Values were comparable with cited values for rabbit muscle aldolase (13). Likewise, a bell-shaped pH activity profile for TgALD with pKa estimates of 5.0 ± 0.1 for the acidic limb and 9.4 ± 0.2 for the basic limb (Fig. 2) were comparable with values obtained previously for rabbit muscle aldolase (16). The active-site residues, which are evolutionarily conserved in both TgALD and rabbit muscle aldolase, together with activity profiles are consistent with Glu-189 of TgALD, which hydrogen-bonds to the C4 hydroxyl of FBP in the 30-min structure (Fig. 1A, inset), as the titrated residue responsible for proton transfer in aldol and reverse aldol reactions. In the mammalian enzyme, the homologous residue Glu-187 has been shown by structural and site-directed mutagenesis to be responsible for proton abstraction at the C4 hydroxyl of FBP and constitutes the acidic limb of the pH profile (2, 16). As indicated previously, the basic limb corresponds most likely to a proton transfer step involving the carbinolamine intermediate (17, 18). Empirical pKa calculations of TgALD using the Propka and H++ programs support pKa estimate ∼5 for Glu-189 (19–23). The kinetic data imply that TgALD active-site functionality is typical of class I FBP aldolases and is stereospecific.
Table 2.
Kinetic parameters of the FBP cleavage reaction for recombinant native aldolases from T. gondii and rabbit muscle
Values are shown ± S.E.
Figure 2.
pH activity profile of the FBP cleavage reaction for TgALD. Saturating concentrations of FBP were used to assay activity at pH 4–9 for every 0.5 pH unit. A double pKa curve was fitted against the kinetic data using Equation 6 (see “Experimental procedures”) in GraFit (version 6.0.12). The acidic limb of the profile has a pKa of 5.0, consistent with the pKa of a glutamate residue. Error bars, S.E. (n ≥ 3 for each point).
Aldol addition
Active-site electron density consistent with potential reaction intermediates was carefully analyzed at each time point with simulated-annealing and iterative-build electron density omit maps to reduce model bias and to assess the fidelity of the intermediate modeling. In the native structure (Fig. 1B), the active site is solvated by numerous water molecules that extend to two and three hydration shells. The tightly bound water molecules in the first hydration shell of the active site (average B-factor of 35 ± 3 Å2 versus overall solvent B-factor of 44 ± 12 Å2) superimpose with the ligand binding loci. Water molecules W2 and W3 overlap with the d-G3P–binding site, whereas W4 overlaps with the DHAP O3 atom position. In addition, the P1-phosphate site is occupied by several water molecules. The P6-phosphate–binding site is bound by the glycerol cryoprotectant that can be seen in the 0.5min structure, where refinement indicated partial occupancies of triose-P, glycerol, and water molecules. Refinement of the active-site binding loci with water molecules revealed only residual electron density in the difference density maps, notably around water molecules whose positions coincided with those of FBP or triose-P atoms bound under saturating conditions. Diffusion of FBP into the crystal from the mother liquor results in an initial concentration gradient that eventually dissipates over time; however, before equilibration, the interior of the crystal lattice will be undersaturated with respect to the exterior substrate concentration used for crystal soaking.
To assess occupancies of various active-site entities, a linear combination of three active-site populations was refined and consisted of water molecules as positioned in the native structure, FBP as found in the fully equilibrated structures, and triose-Ps as these became evident in the difference electron density map. Densities corresponding to other active-site entities were not evident. Populations having partial occupancies of <0.1 at any given time point were excluded from further refinement. At 0.5 min, the refined occupancies of DHAP and d-G3P were 0.44 and 0.25, respectively (Table 3), whereas that of water molecules was 0.68 for W2, 0.72 for W3, and 0.62 for W4, yielding combined occupancies near 1.00, as expected. Upon 1 min of soaking with FBP, the populations increased to 0.72 and 0.66 for DHAP and d-G3P, respectively. The first sign of FBP occurs at 2 min, with a refined occupancy of 0.46 and 0.52/0.51 for DHAP/d-G3P. Longer soaking times (10 min) sufficed to fully populate the TgALD active site with FBP, where trace amounts of triose-Ps were rejected from the refinement. No changes were noted in the FBP population beyond 10 min (occupancy of 1.00 at 30 min). These results indicate that the TgALD conformer has demonstrable catalytic activity in the crystal state.
Table 3.
Progress of populations of active-site species in TgALD crystals soaked with FBP
| Structure | Refined substrate occupancy |
||
|---|---|---|---|
| DHAP | d-G3P | FBP | |
| Native | 0 | 0 | 0 |
| 0.5 min | 0.44 | 0.25 | 0 |
| 1 min | 0.72 | 0.66 | 0 |
| 2 min | 0.52 | 0.51 | 0.46 |
| 10 min | 0 | 0 | 1 |
| 30 min | 0 | 0 | 1 |
Substrate diffusion into catalytically competent crystals
Surprisingly, the first distinguishable ligands observed in the active site as FBP diffused into the crystals were the products of the aldolase forward reaction, namely the triose-Ps, DHAP and d-G3P. This would appear contrary to the thermodynamic equilibrium governing the aldolase reaction, whose Keq for the cleavage reaction is ∼10−4 to 10−5 m, favoring aldol addition (24, 25). However, diffusion by FBP into the crystal lattice establishes a transient concentration gradient of FBP, before its equilibration, providing an explanation for these observations. At low FBP concentrations, as would occur at the diffusion front inside the crystal, the thermodynamic equilibrium is not in favor of FBP formation but promotes triose-P formation (Table 4). Therefore, FBP that is bound in the active sites at the diffusion front is transformed into triose-Ps. For example, for a Keq = 10−4 m, at 1 μm FBP, almost 100% of the reactant is cleaved into product. In the pre-equilibrium state, at 0.5 min for instance, a snapshot would thus show that FBP has been cleaved into triose-Ps, which is consistent with the chemical species observed in our crystal structures. Furthermore, the occupancies will be inferior to 1.0, as equilibrium has not been attained, as was observed.
Table 4.
Populations of the FBP cleavage product (triose-Ps) at varying local FBP concentrations in class I aldolase
| [FBP] | Percentage of product as triose-Pa |
|
|---|---|---|
| Keq = 10−4 mb | Keq = 10−5 m | |
| μm | % | % |
| 1 | 99.0 | 91.6 |
| 10 | 91.6 | 61.8 |
| 100 | 61.8 | 27.0 |
| 1000 | 27.0 | 9.5 |
a Theoretical populations were calculated for the Keq range (10−4 to 10−5 m) reported in the literature.
b Keq, equilibrium constant for the aldolase cleavage reaction. Equation 3 (see “Experimental procedures”) was used to calculate the population of species.
In addition, the triose-P products, released from the active sites at the periphery of the crystal lattice, due to their smaller mass would diffuse more rapidly, populating the crystal lattice ahead of the advancing FBP concentration front. Even at low micromolar concentrations, the KD for DHAP of 2.4 μm (26) at pH 6.0 would favor binding and even more so if its KD is corrected for the desolvation of DHAP (K′D = 1.2 μm) (27). Binding for d-G3P is weaker (KD = 12.5 μm) (4) but more favorable when the desolvated form is considered, yielding an approximate micromolar binding for the nascent d-G3P of K′D = 4.3 μm (28, 29). As the local concentration of FBP increases throughout the crystal to its maximal concentration (1 mm in mother liquor), the chemical equilibrium shifts decidedly toward FBP, and the population of FBP rises in the active site. The shortest time interval (0.5 min) reflects the initial concentration gradient produced by substrate as it diffuses into the crystal lattice; DHAP and d-G3P have similar active-site occupancies (Table 3) as do water molecules W1, W2, and W3. After 1 min, the population of the triose-Ps approaches 70% with a coincidental reduction in water occupancies. As the local concentration of FBP increases, equilibrium favors condensation with the appearance of the FBP-iminium, which becomes clearly recognizable (2-min structure; Fig. 1B), whereas after 10 min, FBP becomes the predominant species (10-min structure; Fig. 1B). Upon full equilibration with the 1 mm FBP, triose-P populations drop below 10% and are rejected by the refinement, consistent with the reported Keq of 10−5 m.
Geometry of aldol addition
The refined reaction intermediates revealed a d-G3P·enamine ternary complex trapped in TgALD crystals with a population amenable for structural analysis (Fig. 1B, 1- and 2-min structures). A ternary complex competent for incipient aldol addition in FBP aldolases requires that the DHAP C3 carbanion be capable of making a si-face attack on the aldehyde si-face, generating the 3(S)-4(R) stereochemistry of FBP about the C3–C4 bond. Attack on the re-face of the aldehyde would yield the 3(S)-4(S) stereochemistry, a stereoisomer of FBP. However, class I FBP aldolases are highly discriminatory toward substrates that do not have the cognate 3(S)-4(R) stereochemistry. Accordingly, FBP aldolases do not cleave d-ribulose-1,5-bisphosphate (6) or d-tagatose 1,6-bisphosphate (TBP) (5), which have 3(R)-4(R) and 3(S)-4(S) stereochemistry. Rather, these compounds act as competitive inhibitors of FBP aldolase. To confirm this behavior against TgALD, the TBP cleavage activity was measured for TgALD at neutral (pH 7.5) and acidic pH (pH 5.5) (data not shown). No cleavage activity was observed at either pH, indicating that TgALD exhibits the same stereospecificity as other members of the FBP aldolase family.
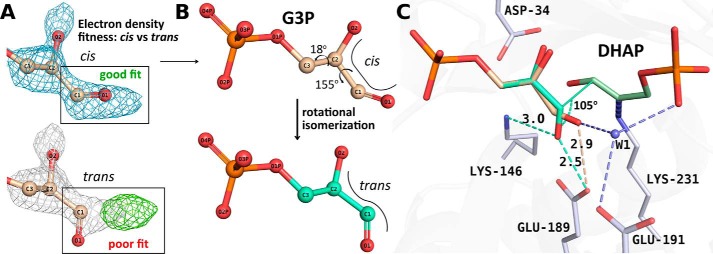
In the ternary complex of the early structures showing triose-Ps, such as the 1-min structure, the orientation of the d-G3P aldehyde, however, was not the expected trans-configuration requisite for si-face attack. Rather, we observed a cis-configuration (Fig. 3A) that upon addition would yield the incorrect chirality (3(S)-4(S)) about the C3–C4 bond. Close inspection of the difference density in simulated-annealing omit maps was consistent with discernment of only the cis-configuration for the bound aldehyde (illustrated in Fig. 3A). To validate the fit of the aldehyde to the electron density, we conducted several refinements using the trans-configuration as a starting model (0.5-, 1-, and 2-min structures), yet refinement invariably provoked a rotational isomerization back to the cis-configuration (see details under “Experimental Procedures”). This observation clearly indicates that the trans-configuration can undergo unhindered rotational isomerization about the C1–C2 aldehyde bond (Fig. 3B) in the ternary complex.
Figure 3.
Reaction geometry associated with aldol addition implicating putative cis-trans rotational isomerization. A, active-site difference density calculated from a simulated-annealing Fo − Fc omit map is illustrated encompassing the substrate-binding site surrounding the cis-d-G3P aldehyde group in the 1-min structure and contoured at a 2.1σ level. Unambiguous modeling of the cis-configuration was possible. Refinement with the trans-configuration afforded a poor fit, as shown by the residual difference density (green) calculated from a Fo − Fc map (contour at 2.1σ). The corresponding 2Fo − Fc map (contoured at 1σ) encompassing the trans-configuration is colored gray. Refinement with the trans-configuration modeled into the 0.5-, 1-, and 2-min structures, reverted to the cis-form in all cases (see “Experimental procedures” for more details). B, the 1-min structure is shown with d-G3P bound in the unproductive cis-form (wheat). The 3(S)-4(R) stereochemistry of the condensation product, FBP, requires prior rotational isomerization about the d-G3P C1–C2 bond, yielding a putative d-G3P trans-configuration (model shown in cyan). The position of trans-d-G3P in the active site was energy-minimized using geometric restraints generated by refmac5 (67). C, superposition of the reported d-G3P configurations shows a hydrogen bond with Glu-189 (dotted cyan line) in the modeled trans-configuration, which, together with Lys-146, orients the carbonyl O1 to promote efficient proton transfer to the polarized d-G3P carbonyl that would otherwise be unproductive in the cis-form.
The ternary complex structures offer several structural clues that help to understand the preferential stabilization of the cis-configuration. First, in the 1-min structure, d-G3P carbonyl is stabilized by hydrogen bonding to the presumably protonated form of Glu-189 OE2 (2.9 Å), shown in Fig. 3C, and a nearby bridging water molecule, W1 (2.8 Å). W1 is highly ordered (fully occupied), and hydrogen-bonding consistency requires that it donate one hydrogen bond to the d-G3P O1 carbonyl and a second one to the Glu-191. Consequently, it must accept a third hydrogen bond from the P1-phosphate oxyanion (2.6 Å) and a fourth from a water molecule (2.8 Å) in the second hydration shell. The Glu-191 carboxylate is expected to have a pKa close to the theoretical value of 4.3, as it is solvent-exposed, and is in proximity of Arg-148 that would stabilize the conjugate base form, supporting a proton acceptor role at pH 5.5 with regard to W1. Proton donation to W1 by the P1-phosphate oxyanion is supported by an experimental pKa of 6.45 for the P1-phosphate dianion (30) and would thus be largely protonated at pH 5.5. The tetrahedrally coordinated hydrogen-bonding pattern stabilizing W1 would thus ensure trapping of the unproductive cis-configuration of the aldehyde phosphate in the active site.
Proton transfer would be unlikely in the cis-configuration, as the nearest basic residue, Lys-146, is not proximal (>4.2 Å) to efficiently polarize the aldehyde carbonyl, and the configuration is least favorable for in-line proton transfer, as the transfer geometry bisects the sp2 orbitals of d-G3P O1 (31). Although the cis-configuration is unproductive with respect to aldol addition, isomerization to a minor population (<0.1 in occupancy) corresponding to the trans-configuration of the aldehyde, competent for proton transfer, is not hindered in the ternary complex. The cryotrapping of the cis-configuration can be attributed to the acidic pH of the crystallization buffer that has stabilized in acid form a significant fraction of Glu-189 in the crystallization buffer (pH 5.5), allowing it to participate in a hydrogen-bonding interaction with the d-G3P aldehyde. At neutral pH, a slight positional rearrangement of the cis-configuration could relieve the close contact of d-G3P O1 with the acidic population of Glu-189, seen in Fig. 3C, and allow the cis-configuration of d-G3P to persist in the active site. Furthermore, stabilization of the cis-configuration by W1 interaction would be maintained at pH 7, as a sizable fraction of the DHAP monobasic phosphate anion remains protonated and thus competent to hydrogen-bond with W1. This strongly supports the interpretation that the hydrogen-bonding pattern made by the phosphate oxyanion of the DHAP enamine with d-G3P is responsible for stereospecific suppression of re-face attack to the aldehyde in glycolytic FBP aldolases and indicates that aldolase stereospecificity is in fact enforced by its proper reactant.
Binding of the cis-d-G3P configuration is also consistent with computational and experimental studies on the solution stability of its glycoaldehyde analogue (32). The most stable glycoaldehyde complexes have cis-configurations with respect to the water molecules that solvate glycolaldehyde. The cis-configuration optimizes bridging hydrogen bonds by water molecules with the hydroxyl and carbonyl groups of glycolaldehyde. This lends support to the notion that d-G3P bound in the aldolase active site would possess a predominant cis-configuration upon attachment.
d-G3P interacts extensively via its P6-phosphate site with residues Lys-107, Ser-36, and Thr-39 (5 hydrogen bonds) and ordered water molecules (6 hydrogen bonds), some of which interact with active-site residues, thereby firmly anchoring d-G3P phosphate in the active site. However, there are fewer structural restraints that would reduce the rotational degrees of freedom of the aldehyde moiety (2 hydrogen bonds by ordered water molecules interacting with the O1 hydroxyl and O2 carbonyl respectively), allowing for considerable configurational rearrangement and consistent with the observations that aldolase can accept structurally diverse aldehydes (8).
To attain the competent reaction geometry for incipient C3–C4 bond formation by the bound d-G3P, the observed cis-configuration would have to undergo a rearrangement (Fig. 3B) whereby the carbonyl O1 rotates about the d-G3P C1–C2 bond by ∼155° and undergoes a slight torsional rotation about its C2–C3 bond (∼18°). This places the carbonyl oxygen within 2.5 Å of the Glu-189 carboxylate and parallel to the enamine plane (Fig. 3B, trans-configuration). In trans, the d-G3P carbonyl O1 is able to make a charged interaction with the Lys-146, thereby polarizing the d-G3P carbonyl. Subsequent proton transfer from Glu-189 then initiates aldol addition. Configurational isomerization by C–C bonds during nucleophilic attack has also been noted for Schiff base formation in transaldolases (33).
In the trans-configuration model, the plane comprising the d-G3P-carbonyl defined by atoms C2, C1, and O1 would indicate that the DHAP C3 follows an approach angle of 105° during the aldol reaction that is consistent with the geometry of incipient aldol addition adhering to the Bürgi–Dunitz angle of 105 ± 5° (34, 35). In enzyme catalysis, adherence to the Bürgi–Dunitz trajectory has been shown in Schiff base formation by transaldolases for the angle of attack by the Lys nucleophile with respect to the ketone plane of the acyclic substrate (33). The trapping of the ternary complex in TgALD, as seen in Fig. 3, provides experimental corroboration of an aldol addition reaction adhering to the Bürgi–Dunitz trajectory during enzyme-catalyzed C–C bond formation.
Finally, the putative trans-geometry of the aldehyde affords a pericyclic transition state (36, 37), as was pointed out in the crystal structure determination of the Schiff base intermediate formed between FBP and rabbit muscle aldolase (2). In TgALD, the atoms of Lys-231 Nζ, FBP C2, C3, C4, and O4 as well as a Glu-189 carboxylate oxygen form a nearly identical chairlike structure that mimics the spatial arrangement of reactants for a pericyclic transition state proposed in non-enzymatic aldol additions with preformed enolates. Comparison of the atomic positions in TgALD intermediate with the previously demonstrated chairlike conformation in rabbit muscle aldolase gives an r.m.s. deviation of 0.45 Å. The cis-form of the aldehyde could not participate in such a transition state, further reinforcing our interpretation of the trans-configuration as the catalytically productive form.
The formation of the C3–C4 bond between trans-G3P and DHAP-enamine is consistent with the least atomic motion during the aldol addition. The motions require a slight hinge movement of the aldehyde carbonyl pivoting about the P6-phosphate binding locus (supplemental Fig. S3), by <1 Å, enabling the aldehyde C1 carbon to approach the DHAP C2 carbon, thereby forming a hydrogen bond (2.8 Å) interaction between the aldehyde O1 and Lys-146 Nζ (shown in supplemental Fig. S3).
Aldehyde stereospecificity
Notwithstanding the promiscuity of aldolase toward aldehydes (8), not all aldehydes condense with DHAP, most notable of which is the l-enantiomer of G3P. Inactivation of aldolase by l-G3P has been reported in the literature (12, 38), whereas other reports have shown the putative reaction product of DHAP and l-G3P, SBP, to be a poor substrate for aldolase cleavage (10, 11). The active-site constraints imposed on aldehyde binding for productive condensation provide a basis for understanding the enantiomeric selectivity at the aldehyde O2 position.
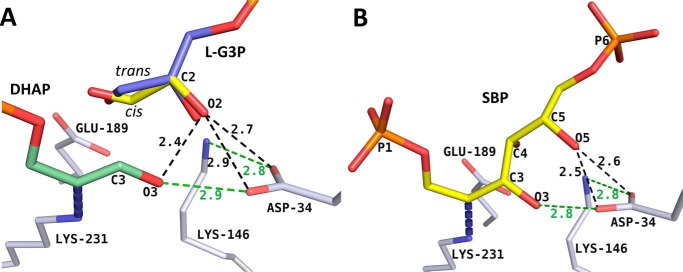
The l-enantiomer of G3P superposed and geometrically optimized upon the cis-form of d-G3P in the 1-min structure introduces steric clashes in the active site. In d-G3P, the C2 hydroxyl is stabilized by hydrogen-bonding with an active-site water molecule (3.0 Å) weakly coordinated to the Ala-32 backbone carbonyl oxygen (3.2 Å). Altering the chirality at the C2 carbon while maintaining a geometry competent for incipient aldol addition is destabilizing by +2.08 kcal/mol compared with the d-form, as per binding energy calculations performed with AutoDock version 4.2 (39). The principal factors include destabilizing van der Waals interactions and the interaction of O2 hydroxyl oxygen with Asp-34 carboxylate oxygens as well as clashes with the DHAP O3 oxygen. The O2 hydroxyl group in the l-enantiomer is positioned opposite the conserved Asp-34 carboxylate oxygens (2.7 Å/2.9 Å) and makes a steric clash with the DHAP O3 oxygen (2.4 Å) (Fig. 4A, dark dashes). This symmetrical positioning of the l-G3P O2 oxygen opposite the Asp-34 carboxylate oxygens is non-ideal due to unfavorable bifurcated hydrogen bonding (i.e. acute C=O···H angles) (40, 41) and results in a repulsive interaction between oxygens, thereby disfavoring binding by the cis-configuration of the l-G3P. This clash cannot be relieved by hydrogen bond donation of the DHAP O3 hydroxyl to l-G3P O2 oxygen, as it maintains a tight linear syn hydrogen bond with the Asp-34 carboxylate (Fig. 4A, green dash). The trans-configuration (cyan) for l-enantiomer based on the trans-d-G3P model, when conformationally optimized, exacerbates the repulsive effect, due to the close contact by the O2 hydroxyl with the Asp-34 carboxylate oxygens and DHAP O3. For both cis/trans-isomers of l-G3P, energy minimization with the surrounding residues relieved the repulsive interactions at the O2 hydroxyl but caused a displacement of the aldehyde away from the aldehyde binding pocket, precluding the potential for aldol addition.
Figure 4.
l-G3P model reveals basis for enantiomeric discrimination in class I aldolase. A, the l-enantiomer of the bound d-G3P (yellow) was superimposed on the d-G3P geometry from the 1-min structure. Favorable linear syn hydrogen bonds are depicted as green dashes. The steric repulsion resulting from proximity of l-G3P O2 to surrounding oxygens (DHAP O3 oxygen, Asp-34 carboxylate oxygens) is depicted as dark dashes. The trans-configuration (blue) for l-enantiomer modeled using the trans-d-G3P model yields repulsions comparable with those observed for the cis-configuration. B, SBP was modeled into the active site using the FBP Schiff base intermediate as a template. Clashes (dark dashes) and favorable hydrogen bonds (green dashes) are illustrated.
The overlay of SBP onto the active-site binding mode of FBP rationalizes the poor substrate specificity for the hexose SBP. Superposition results in the same unfavorable steric interaction as above between the C5 hydroxyl and Asp-34 (Fig. 4B, dark dashes) and would entail an unproductive binding mode for proton transfer between the SBP C4 hydroxyl and Glu-189. Its minimization to relieve repulsive effects induced distortions at the O4 hydroxyl that disrupt the electrostatic stabilization afforded by Lys-146, which is necessary for cleavage of the C3–C4 bond.
Racemic aldol addition products with respect to the configuration at C4 have been noted in a number of class I aldolases. In both 2-keto-4-hydroxyglutarate aldolase (42) and l-fuculose-1-phosphate aldolase (43), racemization occurs at C4 of the condensation products, resulting from isomerization of the aldehyde during stereofacial attack and from the chemical nature of the acceptor aldehyde (44). In the class I TBP aldolase from Staphylococcus aureus and Streptococcus pyogenes, configuration at both C3 and C4 is not retained by the reaction mechanism, as aldol addition yields a mixture of sorbose-P2, psicose-P2, fructose-P2, and tagatose-P2 (45, 46). Although class I FBP aldolases maintain strict stereospecificity regarding the configuration at C3 and C4, the class I aldolase reaction mechanism in general does not appear to preclude active-site binding of diverse isomers (rotamers, diastereomers, and enantiomers) or preclude the existence of such enzymatic intermediate forms. Rather the active-site geometry in glycolytic class I FBP aldolases has evolved to suppress specific isomers by disfavoring their activation to ensure strict stereospecificity in aldol addition. The ternary complex intermediate promoting the chemistry of bond formation is thus promiscuous to attachment by diverse isomers, as ternary complex formation does not entail stereospecific discrimination. Consequently, from an evolutionary perspective, different active sites may give rise to similar ternary complexes, and the differential activation of these complexes would then provide the evolutionary mechanism by which to generate stereospecificity or its lack thereof. Finally, interest in understanding C–C formation is not only of fundamental importance in biological reactions but also has considerable relevance to biosynthetic reactions in organic chemistry dependent on C–C bond formation.
Experimental procedures
Materials
Fru(1,6)P2, glycerol-3-phosphate dehydrogenase and triose-phosphate isomerase were purchased from Sigma-Aldrich. NADH was from Roche Diagnostics. All other chemicals and materials were of analytical grade and were obtained from Sigma-Aldrich, Fisher, Bioshop Canada, and GE Healthcare.
Purification and crystallization
Recombinant TgALD was cloned and prepared as described previously (47). The TgALD gene was then subcloned into a T7-inducible pE-SUMO AMP vector available from LifeSensors Inc. (SUMOpro® gene fusion technology); this was generously provided by Dr. David Sibley (Washington University, St. Louis, MO). The resulting plasmid was transformed into Escherichia coli BL21-StarTM (DE3) cells (Invitrogen). Overexpressing clones were identified from a mini-lysis test using a mixture of sucrose (25%), lysozyme, and Triton X-100 (0.3%) for cell lysis. The overexpressing recombinant strain grown to A600 = 0.6 in LB medium was induced with 1 mm isopropyl β-d-1-thiogalactopyranoside at 37 °C for 4 h. Cells were harvested by centrifugation and frozen at −20 °C overnight. The bacterial pellet was lysed by mechanical disruption with type A-5 aluminum oxide using a mortar and pestle. Cells were resuspended in a lysis buffer (20 mm Tris-HCl, pH 7.4, 0.5 m NaCl, 20 mm imidazole, 10 mm MgCl2, 0.2 mm PMSF) containing 1 mg/g RNase and recombinant DNase. The clear lysate obtained after centrifugation was dialyzed with binding buffer (20 mm Tris-HCl, pH 7.4, 0.5 m NaCl, 20 mm imidazole) and was purified by affinity purification using a HisTrapTM HP 5-ml column (Ni2+-immobilized) connected to an ÄKTA purification system (GE Healthcare). The N-terminal His6-SUMO-TgALD was eluted using elution buffer (20 mm Tris-HCl, pH 7.4, 0.5 m NaCl, 0.5 m imidazole) applied on a linear gradient. The His-SUMO tag from eluted protein was cleaved by incubation for 4 h at 30 °C with SUMO protease after overnight dialysis in SUMO buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 10% glycerol). Before reinjection on the HisTrap column, samples were dialyzed with binding buffer. Fractions from the flow-through containing the protein, as determined by SDS-PAGE and enzyme activity, were pooled and precipitated overnight with precipitation buffer (100 mm Tris-HCl, pH 7.6, 1 mm EDTA, 70% ammonium sulfate, 5 mm DTT). Precipitated protein was pelleted, resuspended in gel filtration buffer (20 mm Tris-HCl, pH 7.4, 1 mm EDTA, 50 mm NaCl), and loaded onto a HiLoadTM 16/60 SuperdexTM, preparation grade (GE Healthcare). TgALD eluted from the gel filtration column at an estimated molecular mass of ∼160 kDa, consistent with tetramerization of TgALD. Purity was assessed by SDS-PAGE and mass spectrometry. Peak fractions were pooled, dialyzed with a minimal buffer (20 mm Tris-HCl, pH 7.0), and concentrated to 5–10 mg/ml before crystallization trials.
Crystallographic data collection and processing
Native TgALD crystals were identified from the Clear StrategyTM Screen I (Molecular Dimensions) and were optimized to the following conditions: 0.1 m sodium acetate, pH 5.5, 0.2 m lithium sulfate, 3.5% (w/v) PEG 8000, 10% (w/v) PEG 1000. Crystals were grown at 20 °C by hanging-drop vapor diffusion using a 1:1 mixture of protein/precipitant. TgALD crystals were first washed by three serial transfers in fresh precipitant solution to remove soluble protein and then transferred to a soaking solution (precipitant + ligand) containing FBP (mother liquor with 1 mm FBP) for incremental time intervals (0.5, 1, 2, 10, and 30 min). To improve reproducibility, crystals of discernably similar lengths (∼100 μm in length) with rodlike morphology were selected for all diffraction experiments. A minimum of two crystals were selected for each time point, and crystal structures were compared to validate the precision of the soaking method. Before flash-freezing in liquid nitrogen, single crystals were cryoprotected by briefly soaking them (5 s) in the ligand buffer and 20% (v/v) glycerol. The data were collected at beamlines X25 and X29 of the National Synchrotron Light Source at Brookhaven National Laboratories. The diffraction data were processed and scaled with the HKL package (48), and the final results are listed in Table 1. Duplicates had comparable collection and refinement statistics (not shown), and only the structures with lowest R-factors were retained for final analysis.
Structure determination and refinement
Structures were solved by molecular replacement with Phaser-MR (49) in the PHENIX software (50) using subunit A from the native rabbit aldolase homotetramer structure as a reference model (PDB entry 1ZAH) for which there is 57.1% sequence homology (51) (Oryctolagus cuniculus aldolase (Uniprot ID P00883) and T. gondii aldolase (Uniprot ID Q8l8l2)). A TgALD crystal structure was solved previously independently from this study and is available in the PDB (entry 4TU1) (52). TgALD crystals have space group P 42212 with one tetrameric TgALD subunit in the asymmetric unit. Refinement and model building were performed using the refinement module in Phenix (53) and Coot (54), respectively. Data quality was assessed and cut-off using correlation coefficient–based criteria, CC½ (55). Ligand fitting and interpretation was performed using a combination of simulated-annealing Fo − Fc omit maps and feature-enhanced σ-A–weighted 2Fo − Fc maps (56) that were calculated in the final round of refinement. All difference density maps (Fo − Fc) shown in the paper correspond to simulated-annealed Fo − Fc omit maps. Ligand coordinates and restraints were generated using Phenix eLBOW (57). Final model statistics were calculated with MolProbity (58) and are shown in Table 1. The coordinates and structure factors have been deposited in the Protein Data Bank. All figures were prepared using PyMOL graphics software (59).
Active-site intermediates
The identification of whether substrate and/or product species was bound in the active site for a given soaking condition (snapshot) was derived from inspection of electron density features characteristic of the ligands' chemical features. The features included continuous electron density, extending beyond Lys-231 Nζ, indicative of formation of a stable covalent bond with the C2 carbon. In the 1-min structure, discrimination between DHAP-enamine and DHAP-iminium intermediate is feasible by considering the geometry of the electron density observed about the C2 carbon (supplemental Fig. S4A). The planar shape observed in the 1-min structure indicates trigonal hybridization, whereas non-planar shape of the electron density about the Lys-231 Nζ atom would suggest tetrahedral hybridization and is consistent with trapping of a DHAP-enamine intermediate, as was observed previously with DHAP covalently bound in rabbit muscle aldolase (3). Another indication of DHAP-enamine trapping is the presence of an additional albeit weak hydrogen bond between Ser-301 hydroxyl and Lys-231 Nζ (3.1 Å in the 0.5-min structure; 3.3 Å in the 1-min structure), which contributes to stabilize the enamine intermediate (shown in supplemental Fig. S4A). This stabilization is not the DHAP-carbanion, the DHAP-iminium, and FBP-iminium intermediates, which all have formal sp2 hybridization at Lys-231 Nζ and no longer contact the Ser-301 hydroxyl (e.g. 3.7 Å in the 10-min structure, which has predominantly FBP covalently bound). Further, electron density developing between the DHAP C3 and d-G3P C1 atoms in the 2-min structure and not present in the 1-min structure suggests C–C bond formation and the presence of both the ternary complex (aldolase· enamine·d-G3P) and covalently bound FBP intermediate in the active site (Fig. 1). Indeed, nascent C–C bond formation was the clearest indication of the FBP-iminium intermediate progressively populating the active site. The iminium geometry was consistent with similar geometries observed for previously reported aldolase and transaldolase structures where FBP was trapped in the active site (2, 33).
Active-site occupancy
Occupancies of substrates in each model were determined by introducing systematic differences in their starting occupancies (e.g. model 1, FBP 0.40 and triose-Ps 0.60; model 2, FBP 0.60 and triose-Ps 0.40; model 3, FBP 0.80 and triose-Ps 0.20; etc.), creating for each structure an ensemble of five models that were refined for 10 cycles each with identical refinement protocols until convergence of occupancies was observed to a common value (60). Further confirmation of model fitness affording distinction between various intermediates was accomplished by assessment of real-space correlation coefficients (CCs) in Phenix (53). Phenix performs bulk-solvent correction and scaling on the data and calculates a likelihood-weighted 2mFo − DFc map. This is compared with a map calculated from the model alone, and CCs for each residue are obtained. Comparison of CC values for substrate modeled at different occupancies allowed cross-validation of the convergence test performed above. Because CC scores are best interpreted as relative scores (53), they provided a corroborative indication of the above methods for active-site discernment of intermediate species. This analysis was conducted at every time point, and the final models shown in Fig. 1B were consistent with the highest CC scores. For example, in a preaddition state (1-min structure), as there was no density between the nascent C–C bond, modeling of the FBP Schiff base complex in the 1-min structure yielded a lower CC score (CC = 0.85) than modeling of the DHAP·enamine·d-G3P ternary complex (CC = 0.96). Discernment of DHAP-enamine from DHAP-iminium (a minor structural difference) yielded a higher CC for the DHAP-enamine (0.97) than for the DHAP-iminium (0.92). Likewise, slightly higher CCs were determined for cis-modeled d-G3P versus trans-modeled d-G3P in the 0.5-min (0.92 versus 0.87), 1-min (0.93 versus 0.87), and 2-min (0.92 versus 0.89) structures, corroborating the structural interpretation from omit maps and validating the retention of cis-d-G3P in our models.
Equilibrium triose-P concentrations as a function of initial substrate concentration and thermodynamic equilibrium constant
The concentrations of reactant and product after equilibration can be calculated by solving a quadratic equation (Equation 3), where S0 is the initial FBP concentration (pre-equilibrium), x is the concentration of triose-Ps after equilibration, and Keq is thermodynamic equilibrium constant.
| (Eq. 1) |
| (Eq. 2) |
then
| (Eq. 3) |
For S0 ≪ Keq, from Equation 3, and by Taylor expansion,
| (Eq. 4) |
and then substituting back into Equation 3, we have x ≈ [S0]. For S0 ≪ Keq, dividing Equation 3 by [S0], we have the following,
| (Eq. 5) |
by keeping the largest term. Because Keq/[S0] ≪ 1, [x] becomes negligible with respect to [S0].
Accordingly, when S0 ≪ Keq, x will be greater than [FBP] after equilibrium. As FBP diffused into the crystal, we should expect conditions where S0 will be low, which confirms the high proportion of triose-Ps observed at early time points. As the S0 ≫ Keq, the ratio of [FBP]/[x] will shift in favor of FBP, which is what should occur after several minutes of equilibrating our TgALD crystals with FBP. Table 4 was calculated using the above quadratic equation (Equation 3) and shows precisely this.
Activity assay
Aldolase cleavage activity was determined at neutral pH (pH 7.5) using a previously described coupled assay system involving triose-phosphate isomerase and glycerol-3-phosphate dehydrogenase (TIM/GDH) (16, 61). Conversion of DHAP to glycerol 3-phosphate consumes equimolar NADH (absorption at 340 nm) and was assayed using a Varian Cary 300 UV-visible spectrophotometer thermostated at 25 °C. Enzyme activity was calculated using standard Michaelis–Menten kinetics. pKa estimates were obtained by modeling the following equation for double pKa,
| (Eq. 6) |
where y represents measured catalytic activity, and Limit is highest activity. All data were processed with GraFit Data Analysis Software version 6.0.12.
Binding energy calculations
AutoDock version 4.2.6, which considers van der Waals, hydrogen-bonding, and electrostatic energies, was used to estimate binding free energies and perform a detailed ligand intramolecular energy analysis of stereochemically related aldehydes with aldolase (39). The crystal structure with FBP soaked for 1 min was used to generate atomic energy grids. Customary preparation of the protein, including the addition of polar hydrogens, Kollman charges, and solvation parameters, was undertaken with AutoDockTools version 1.5.6 (http://mgltools. scripps.edu/),4 which uses a modified version of Open Babel (62). The fully refined model with d-G3P was used for the scoring function, from which Gasteiger charges and all hydrogens were computed using an input charge of −2 for d-G3P. To optimize the orientation of the O2 hydroxyl hydrogen with respect to localized interactions in the active site, d-G3P and l-G3P were optimized using an Amber force field (63) in the Antechamber module (64) from USCF Chimera (65) for assignment of ligand parameters (bond length, charge, etc.).
PROPKA version 3.1 (19, 20) and H++ version 3.1 (http://biophysics.cs.vt.edu/)4 (21–23) were used to determine the pKa of active-site residues. Structures for pKa calculations were prepared using the PDB2PQR server in which a PARSE force field was used to assign charges (66).
Author contributions
P. H. and J. S. designed the study, P. H. conducted the experiments, P. H. and J. S. analyzed the results, and P. H. wrote most of the paper with critical revisions by J. S. Both authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We are grateful to Dr. David Sibley and Bang Shen (University of Washington, St. Louis) for providing T. gondii aldolase expression vector. This research used resources of beamlines X25 and X29A of the National Synchrotron Light Source, a United States Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under contract DE-AC02-98CH10886.
This work was supported by a Discovery grant from the National Science and Engineering Research Council of Canada (NSERC) (to J. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S4.
The atomic coordinates and structure factors (codes 5TJS, 5TK3, 5TKC, 5TKL, 5TKN, and 5TKP) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- FBP
- fructose 1,6-bisphosphate
- DHAP
- dihydroxyacetone phosphate
- G3P
- glyceraldehyde 3-phosphate
- TgALD
- T. gondii aldolase
- TBP
- d-tagatose-1,6-bisphosphate
- SBP
- l-sorbose 1,6-bisphosphate
- triose-P
- triose-phosphate
- PDB
- Protein Data Bank
- r.m.s.
- root mean square
- CC
- correlation coefficient.
References
- 1. Grazi E., Rowley P. T., Cheng T., Tchola O., and Horecker B. L. (1962) The mechanism of action of aldolases. III. Schiff base formation with lysine. Biochem. Biophys. Res. Commun. 9, 38–43 [DOI] [PubMed] [Google Scholar]
- 2. St-Jean M., Lafrance-Vanasse J., Liotard B., and Sygusch J. (2005) High resolution reaction intermediates of rabbit muscle fructose-1,6-bisphosphate aldolase: substrate cleavage and induced fit. J. Biol. Chem. 280, 27262–27270 [DOI] [PubMed] [Google Scholar]
- 3. St-Jean M., and Sygusch J. (2007) Stereospecific proton transfer by a mobile catalyst in mammalian fructose-1,6-bisphosphate aldolase. J. Biol. Chem. 282, 31028–31037 [DOI] [PubMed] [Google Scholar]
- 4. Rose I. A., and O'Connell E. L. (1969) Studies on the interaction of aldolase with substrate analogues. J. Biol. Chem. 244, 126–134 [PubMed] [Google Scholar]
- 5. Hartman F. C., and Barker R. (1965) An exploration of the active site of aldolase using structural analogs of fructose diphosphate. Biochemistry 4, 1068–1075 [DOI] [PubMed] [Google Scholar]
- 6. Rose I. A., and Warms J. V. B. (1985) Complexes of muscle aldolase in equilibrium with fructose 1,6-bisphosphate. Biochemistry 24, 3952–3957 [DOI] [PubMed] [Google Scholar]
- 7. Kuo D. J., and Rose I. A. (1985) Chemical trapping of complexes of dihydroxyacetone phosphate with muscle fructose-1,6-bisphosphate aldolase. Biochemistry 24, 3947–3952 [DOI] [PubMed] [Google Scholar]
- 8. Bednarski M. D., Simon E. S., Bischofberger N., Fessner W. D., Kim M. J., Lees W., Saito T., Waldmann H., and Whitesides G. M. (1989) Rabbit muscle aldolase as a catalyst in organic synthesis. J. Am. Chem. Soc. 111, 627–635 [Google Scholar]
- 9. Gefflaut T., Blonski C., Perie J., and Willson M. (1995) Class I aldolases: substrate specificity, mechanism, inhibitors and structural aspects. Prog. Biophys. Mol. Biol. 63, 301–340 [DOI] [PubMed] [Google Scholar]
- 10. Richards O. C., and Rutter W. J. (1961) Comparative properties of yeast and muscle aldolase. J. Biol. Chem. 236, 3185–3192 [PubMed] [Google Scholar]
- 11. Tung T. C., Ling K. H., Byrne W. L., and Lardy H. A. (1954) Substrate specificity of muscle aldolase. Biochim. Biophys. Acta. 14, 488–494 [DOI] [PubMed] [Google Scholar]
- 12. Lai C. Y., Martinez-de Dretz G., Bacila M., Marinello E., and Horecker B. L. (1968) Labeling of the active site of aldolase with glyceraldehyde 3-phosphate and erythrose 4-phosphate. Biochem. Biophys. Res. Commun. 30, 665–672 [DOI] [PubMed] [Google Scholar]
- 13. St-Jean M., Blonski C., and Sygusch J. (2009) Charge stabilization and entropy reduction of central lysine residues in fructose-bisphosphate aldolase. Biochemistry 48, 4528–4537 [DOI] [PubMed] [Google Scholar]
- 14. Rose I. A., Warms J. V., and Kuo D. J. (1987) Concentration and partitioning of intermediates in the fructose bisphosphate aldolase reaction: comparison of the muscle and liver enzymes. J. Biol. Chem. 262, 692–701 [PubMed] [Google Scholar]
- 15. Rose I. A., and O'Connell E. L. (1977) Specificity of fructose-1,6-P2 aldolase (muscle) and partition of the enzyme among catalytic intermediates in the steady state. J. Biol. Chem. 252, 479–482 [PubMed] [Google Scholar]
- 16. Maurady A., Zdanov A., de Moissac D., Beaudry D., and Sygusch J. (2002) A conserved glutamate residue exhibits multifunctional catalytic roles in d-fructose-1,6-bisphosphate aldolases. J. Biol. Chem. 277, 9474–9483 [DOI] [PubMed] [Google Scholar]
- 17. Cordes E. H., and Jencks W. P. (1962) On the mechanism of Schiff base formation and hydrolysis. J. Am. Chem. Soc. 84, 832–837 [Google Scholar]
- 18. Sayer J. M., Pinsky B., Schonbrunn A., and Washtien W. (1974) Mechanism of carbinolamine formation. J. Am. Chem. Soc. 96, 7998–8009 [Google Scholar]
- 19. Søndergaard C. R., Olsson M. H. M., Rostkowski M., and Jensen J. H. (2011) Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 7, 2284–2295 [DOI] [PubMed] [Google Scholar]
- 20. Olsson M. H. M., Søndergaard C. R., Rostkowski M., and Jensen J. H. (2011) PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 [DOI] [PubMed] [Google Scholar]
- 21. Myers J., Grothaus G., Narayanan S., and Onufriev A. (2006) A simple clustering algorithm can be accurate enough for use in calculations of pKs in macromolecules. Proteins 63, 928–938 [DOI] [PubMed] [Google Scholar]
- 22. Gordon J. C., Myers J. B., Folta T., Shoja V., Heath L. S., and Onufriev A. (2005) H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 33, W368–W371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Anandakrishnan R., Aguilar B., and Onufriev A. V. (2012) H++ 3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 40, W537–W541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Connett R. J. (1985) In vivo glycolytic equilibria in dog gracilis muscle. J. Biol. Chem. 260, 3314–3320 [PubMed] [Google Scholar]
- 25. Veech R. L., Raijman L., Dalziel K., and Krebs H. A. (1969) Disequilibrium in the triose phosphate isomerase system in rat liver. Biochem. J. 115, 837–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grazi E., Sivieri-Pecorari C., Gagliano R., and Trombetta G. (1973) Complexes of fructose diphosphate aldolase with dihydroxyacetone phosphate and dihydroxyacetone sulfate. Biochemistry 12, 2583–2590 [DOI] [PubMed] [Google Scholar]
- 27. Reynolds S. J., Yates D. W., and Pogson C. I. (1971) Dihydroxyacetone phosphate: its structure and reactivity with α-glycerophosphate dehydrogenase, aldolase and triose phosphate isomerase and some possible metabolic implications. Biochem. J. 122, 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trentham D. R., McMurray C. H., and Pogson C. I. (1969) The active chemical state of d-glyceraldehyde 3-phosphate in its reactions with d-glyceraldehyde 3-phosphate dehydrogenase, aldolase and triose phosphate isomerase. Biochem. J. 114, 19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peczon B. D., and Spivey H. O. (1972) Catalytic sites in rabbit muscle glyceraldehyde-3-phosphate dehydrogenase. Their number and their kinetic and spectral properties. Biochemistry 11, 2209–2217 [DOI] [PubMed] [Google Scholar]
- 30. Grazi E. (1974) Quantitative evaluation of the carbanion intermediate in the aldolase reaction. Biochem. Biophys. Res. Commun. 56, 106–111 [DOI] [PubMed] [Google Scholar]
- 31. Duan X., and Scheiner S. (1992) Energetics, proton transfer rates, and kinetic isotope effects in bent hydrogen bonds. J. Am. Chem. Soc. 114, 5849–5856 [Google Scholar]
- 32. Azofra L. M., Quesada-Moreno M. M., Alkorta I., Avilés-Moreno J. R., Elguero J., and López-González J. J. (2015) Understanding the aldo-enediolate tautomerism of glycolaldehyde in basic aqueous solutions. ChemPhysChem 16, 2226–2236 [DOI] [PubMed] [Google Scholar]
- 33. Light S. H., Minasov G., Duban M.-E., and Anderson W. F. (2014) Adherence to Bürgi–Dunitz stereochemical principles requires significant structural rearrangements in Schiff-base formation: insights from transaldolase complexes. Acta Crystallogr. D Biol. Crystallogr. 70, 544–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bürgi H. B., Dunitz J. D., Lehn J. M., and Wipff G. (1974) Stereochemistry of reaction paths at carbonyl centres. Tetrahedron 30, 1563–1572 [Google Scholar]
- 35. Fleming I. (2010) Molecular Orbitals and Organic Chemical Reactions: Reference Edition, John Wiley & Sons, Inc., New York [Google Scholar]
- 36. Zimmerman H. E., and Traxler M. D. (1957) The stereochemistry of the Ivanov and Reformatsky reactions. I. J. Am. Chem. Soc. 79, 1920–1923 [Google Scholar]
- 37. Dubois J.-E., and Fort J.-F. (1972) Dynamic stereochemistry of aldolization—XX. Tetrahedron 28, 1653–1663 [Google Scholar]
- 38. Rose I. A., and O'Connell E. L. (1969) Inactivation and labeling of triose phosphate isomerase and enolase by glycidol phosphate. J. Biol. Chem. 244, 6548–6550 [PubMed] [Google Scholar]
- 39. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., and Olson A. J. (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McDonald I. K., and Thornton J. M. (1994) Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238, 777–793 [DOI] [PubMed] [Google Scholar]
- 41. Görbitz C. H., and Etter M. C. (1992) Hydrogen bonds to carboxylate groups: the question of three-centre interactions. J. Chem. Soc. Perkin Trans. 2 1, 131–135 [Google Scholar]
- 42. Kobes R. D., and Dekker E. E. (1969) 2-Keto-4-hydroxyglutarate aldolase of bovine liver purification, criteria of purity, and general properties. J. Biol. Chem. 244, 1919–1925 [PubMed] [Google Scholar]
- 43. Espelt L., Bujons J., Parella T., Calveras J., Joglar J., Delgado A., and Clapés P. (2005) Aldol additions of dihydroxyacetone phosphate to N-Cbz-amino aldehydes catalyzed by l-fuculose-1-phosphate aldolase in emulsion systems: inversion of stereoselectivity as a function of the acceptor aldehyde. Chemistry 11, 1392–1401 [DOI] [PubMed] [Google Scholar]
- 44. Meloche H. P., and Mehler L. (1973) The stereochemistry at carbon 3 of pyruvate lyase condensation products aldolases forming condensation products racemic at carbon 4. J. Biol. Chem. 248, 6333–6338 [PubMed] [Google Scholar]
- 45. Bissett D. L., and Anderson R. L. (1980) Lactose and d-galactose metabolism in Staphylococcus aureus. III. Purification and properties of d-tagatose-6-phosphate kinase. J. Biol. Chem. 255, 8745–8749 [PubMed] [Google Scholar]
- 46. LowKam C., Liotard B., and Sygusch J. (2010) Structure of a Class I tagatose-1,6-bisphosphate aldolase: investigation into an apparent loss of stereospecificity. J. Biol. Chem. 285, 21143–21152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jewett T. J., and Sibley L. D. (2003) Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites. Mol. Cell. 11, 885–894 [DOI] [PubMed] [Google Scholar]
- 48. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 49. Adams P. D., Grosse-Kunstleve R. W., Hung L.-W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 50. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Söding J. (2005) Protein homology detection by HMM–HMM comparison. Bioinformatics 21, 951–960 [DOI] [PubMed] [Google Scholar]
- 52. Boucher L. E., and Bosch J. (2014) Structure of Toxoplasma gondii fructose-1,6-bisphosphate aldolase. Acta Crystallogr. Sect. F Struct. Biol. Commun. 70, 1186–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with <phenix.refine>. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Karplus P. A., and Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Afonine P. V., Moriarty N. W., Mustyakimov M., Sobolev O. V., Terwilliger T. C., Turk D., Urzhumtsev A., and Adams P. D. (2015) FEM: feature-enhanced map. Acta Crystallogr. D Biol. Crystallogr. 71, 646–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moriarty N. W., Grosse-Kunstleve R. W., and Adams P. D. (2009) electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 65, 1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. DeLano W. L. (2014) The PyMol Molecular Graphics System, version 1.7.4, Schrödinger, LLC, New York [Google Scholar]
- 60. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Adams P. D., Moriarty N. W., Zwart P., Read R. J., Turk D., and Hung L.-W. (2007) Interpretation of ensembles created by multiple iterative rebuilding of macromolecular models. Acta Crystallogr. D Biol. Crystallogr. 63, 597–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Racker E. (1947) Spectrophotometric measurement of hexokinase and phosphohexokinase activity. J. Biol. Chem. 167, 843–854 [PubMed] [Google Scholar]
- 62. O'Boyle N. M., Banck M., James C. A., Morley C., Vandermeersch T., and Hutchison G. R. (2011) Open Babel: an open chemical toolbox. J. Cheminformatics 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang J., Wolf R. M., Caldwell J. W., Kollman P. A., and Case D. A. (2004) Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 [DOI] [PubMed] [Google Scholar]
- 64. Wang J., Wang W., Kollman P. A., and Case D. A. (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 25, 247–260 [DOI] [PubMed] [Google Scholar]
- 65. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 66. Dolinsky T. J., Nielsen J. E., McCammon J. A., and Baker N. A. (2004) PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.