Stacked paper skiving paves the way for industrial manufacturing of paper-based analytical devices for barcode assays.
Abstract
This paper shows that stacked sheets of paper preincubated with different biological reagents and skiving them into uniform test paper sheets allow mass manufacturing of multiplexed immunoassay devices and simultaneous detection of multiplex targets that can be read out by a barcode scanner. The thickness of one sheet of paper can form the width of a module for the barcode; when stacked, these sheets of paper can form a series of barcodes representing the targets, depending on the color contrast provided by a colored precipitate of an immunoassay. The uniform thickness of sheets of paper allows high-quality signal readout. The manufacturing method allows highly efficient fabrication of the materials and substrates for a straightforward assay of targets that range from drugs of abuse to biomarkers of blood-transmitted infections. In addition, as a novel alternative to the conventional point-of-care testing method, the paper-based barcode assay system can provide highly efficient, accurate, and objective diagnoses.
INTRODUCTION
This article describes a rapid and efficient mass manufacturing method to fabricate paper-based barcode chips (PBCs) for multiplexed immunoassays. The edges of paper sheets with constant thickness comprise the modules of the barcodes, displaying superiority over complicated processing technologies in the precise fabrication of barcodes or reaction channels. When stacked together, the edges of the paper form the reaction regions, without the need for manual operation to create hydrophilic channels for immunoreaction. Stacked sheets of paper form a series of barcodes, representing the information of multiplex targets. Hundreds of PBCs can be fabricated just by skiving glued stacked sheets of paper in minutes, without requiring precise instruments and professionals, as does nanoskiving (1, 2). We expect the rapid, efficient, and straightforward paper-based mass manufacturing method and immunoassay system can be widely used and further developed in the field of clinical diagnosis and point-of-care testing (POCT).
Disease control, food safety, and environmental monitoring require affordable point-of-care diagnostic devices that are sensitive, specific, rapid, portable, automated, and easy to manufacture and operate (3–8). Recently, paper-based analytical devices (PADs) have demonstrated huge potential in rapid diagnosis and POCT because of their simplicity of operation, rapidity of fabrication, and cost-effectiveness (9–12). PADs have become attractive alternatives to conventional microfluidics (13–16). The physical and chemical properties of paper make it an extremely versatile material for fabricating PADs. Simple operations enable processing of the functional regions in PADs to store reagents, manipulate fluid, permit reactions, and recover waste (17). Because liquid transport relies entirely on capillary wicking in PADs, all the functional regions can be fabricated by creating hydrophilic channels in hydrophobic paper. To process hydrophilic channels and hydrophobic barriers, techniques including wax printing, photolithography, screen printing, inkjet printing, and plasma etching are applied in PAD fabrication (18–20). The process of the fabrication is dependent not only on sophisticated instruments but also on professionals, resulting in increases in costs and time. Further, for most PADs, the results are mainly read out with the naked eye or by small, inexpensive equipment (21) and have to be analyzed manually one by one, making it an inefficient process. Our group and others have pioneered the use of a barcode scanner and smartphone application to read out the barcode results of multiplexed assays (22, 23). In the assay, we used barcodes to encode the information of multiplex targets and designed microfluidic barcoded chips by using “bar” and “space” to correspond to the “positive” and “negative” information in diagnosis. This works because microfluidic channels fabricated with precise sizes can comprise the wide modules and narrow modules in barcodes. Nonexperts, such as cashiers in a supermarket, can decode the information using a barcode scanner, making the readout and analysis of the results a rapid and efficient process. However, the repeating operations of introducing samples, incubating, and washing in the microfluidic channels are not only time-consuming but also complex to nonprofessionals, leading to restricted applications of microfluidic barcoded chips in POCT fields.
Here, we present a paper-based barcode assay system (PBAS) that allows simultaneous detection of multiplex targets in a single-step within 10 min. We use the PBAS in the detection of drugs of abuse and biomarkers of blood-transmitted infections. As in a lateral flow immunoassay (LFIA) system (24, 25), we use gold nanoparticles (AuNPs) to generate colorimetric signals in the test regions. After the completion of the detection, we directly decipher the barcode results of multiplex biomarkers by a barcode scanner. Compared with the LFIA that relies on visual readout, the readout based on barcode scanning is more objective and accurate (17, 26). Analysis and recording using a barcode scanner connected to a computer are efficient and can avoid occasional mistakes that exist in keyboard input of the results.
RESULTS
Design of PBAS
We use the Codabar code and the constant thickness of papers to realize the barcode-based immunoassay. A typical Codabar barcode consists of a Start character, data of the message, a Stop character, and intercharacter spaces. One character in a Codabar barcode comprises seven modules, four bars, and three spaces. Every module can be either narrow or wide. The ratio of the wide module and the narrow module can be set between 2:1 and 3:1. We use three sheets of paper to achieve the wide module, one sheet of paper to achieve the narrow module, and one sheet of paper as the intercharacter space. We choose character “A” as the Start Code and the Stop Code, and characters “1” and “:” as the data of the message. Characters “1” and “:” can be transformed from one to another by interchanging two of the narrow bars with wide ones (Fig. 1A). When the left two sheets of paper beside the first and fourth bars in character “1” have AuNPs aggregated and appear to be red, they form two wide bars with the right one sheet of red paper, and character “1” transforms to become character “:”; otherwise, when the left two sheets of paper are white, the first and fourth bars are narrow bars, and character “1” remains character “1.” We define these two sheets of paper as variable regions and other sheets of paper as constant regions. When the pair of variable regions in the same character is red, the character is “:”; otherwise, the character is “1.” We can obtain barcodes that contain the information of multiplex biomarkers through colorimetric reaction in the variable regions.
Fig. 1. The design and fabrication of PBCs and the assay system.
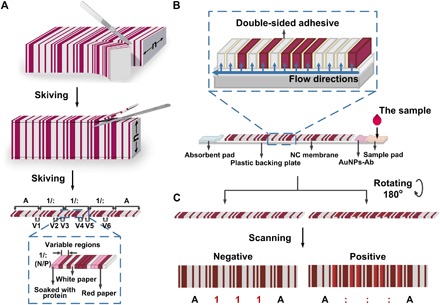
(A) Fabrication of PBCs. We fabricated the PBCs by gluing the white papers, the red papers, and the papers soaked with capture probes together according to the Codabar rules and skived the combined papers into a small size (6 mm × 0.5 mm). V1 and V2, V3 and V4, and V5 and V6 are variable regions used for target 1, target 2, and target 3. N, negative; P, positive. (B) Assay system of the PBCs. We placed the PBCs on the NC membrane of the LFTSs and pressed it with an external force during the assay. After the detection, we rotated the PBC 180° for the readout of the results. Ab, antibody. (C) Readout of the results. In an indirect or sandwich assay, when the targets are all positive, the result is read as “A:::A;” otherwise, the result is read as “A111A.”
The barcode assay system that we present here consists of two major parts: the PBCs and the lateral flow test strips (LFTSs) (Fig. 1B). The PBCs play the role of immobilizing capture probes, conducting immunoassays, and supporting the readout of the barcode results. We choose chromatography paper for the fabrication of the PBCs. The most commonly used substrates in PADs are filter paper and chromatography paper (17). The thickness, pore size, and strength and deformation after saturation are important parameters in fabricating the PADs. The paper used for the PBCs should be homogeneous and of constant thickness. The paper thickness also influences its strength (20). The pore size relates to the transporting speed of AuNPs in paper and the amount of immunoreactive product aggregating on the edge of the paper. Paper strength and deformation after saturation affect the generation of the immunoreaction on the edge of the paper and the readout of the results. We choose two kinds of chromatography paper and one kind of filter paper as candidates and characterize the surface morphology, pore size, dry tensile strength, and wet tensile strength of each [Whatman Grade chromatography (Chr), Grade 1 Chr, and Grade 1]. The fiber structure of Grade 3MM Chr is more compact than the fiber structure of Grade 1 Chr and Grade 1. The pore size of Grade 3MM Chr is smaller than the other kinds of paper and the nitrocellulose (NC) membrane (fig. S1). In addition, the Grade 3MM Chr has higher dry tensile strength and wet tensile strength compared with the other kinds of paper (fig. S2). The Grade 3MM Chr paper is the optimal one for the fabrication of the PBCs.
According to the Chinese standard for Codabar barcode (GB/T 12907-2008), the deviation of the width of every module should not be higher than the maximum permissible deviation to guarantee the accurate readout of the barcode, which is represented as “t.” The maximum permissible deviation is dependent on the ratio of the wide module and the narrow module and the width of the narrow module
| (1) |
where N is the ratio between the wide module and the narrow module and X is the width of the narrow module. In our system, N is 3 and X is 340 μm, so t is ±119 μm. We characterize the width of the papers in the PBCs and measure the widths of the four different modules by ImageJ (Fig. 2A). The widths of the narrow bar (one sheet of red paper), the narrow space (one sheet of white paper), the wide bar (three sheets of red paper), and the wide space (three sheets of white paper) are 342.7, 338.7, 1057.7, and 1007.7 μm (Fig. 2B). The deviations are all lower than the maximum permissible deviation (Fig. 2C). Once they become wet, the papers become swollen and the widths of the papers change. To investigate whether the change of the widths affects the readout of the barcodes, we characterize the widths of the modules in the wet PBCs (fig. S3). The widths of the narrow bar, the narrow space, the wide bar, and the wide space are 410.3, 393.9, 1215.6, and 1195 μm (fig. S3C). According to Eq. 1, the maximum permissible deviation is around ±141 μm, and all the deviations are lower than the maximum permissible deviation.
Fig. 2. Characterization of the widths of the modules in the PBCs.
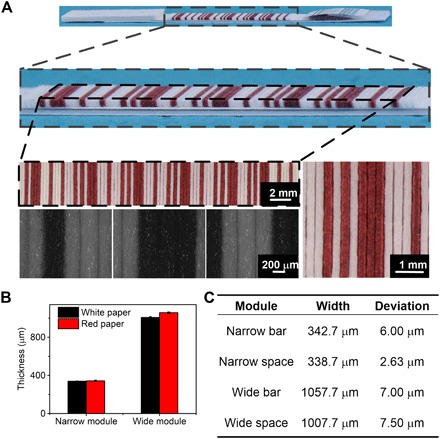
(A) Pictures of the PBCs taken by cameras and microscopes. (B) The thickness of the narrow and wide modules consists of papers. (C) Table of the widths and deviations of the narrow bar, the narrow space, the wide bar, and the wide space.
To further illustrate the transport of the liquid during the detection, we characterize the hydrophilicity of the double-sided adhesive, the white paper, the red paper, and the paper soaked with protein by water contact angle analysis. The contact angle of the double-sided adhesive is around 108°, showing the hydrophobicity of the adhesive. The contact angle of the red paper is around 104° in the beginning, and the water droplets are absorbed by the red paper within 130 s, meaning that the red paper is still hydrophilic even after soaking with hydrophobic ink. The water droplets appear to have spherical shapes on the white paper and the paper soaked with protein within 1 s (fig. S4), demonstrating high hydrophilicity of the papers.
The LFTSs are used to transport reagents and samples during the detection, and contain five components, namely, the plastic backing plate, the sample pad, the conjugate pad, the NC membrane, and the absorbent pad. As in an LFIA, the plastic backing plate is the substrate to assemble all the other components, and the sample pad, the conjugate pad, and the absorbent pad play the role of introducing the sample, immobilizing AuNP-conjugated detection probes, and absorbing excess solution. The NC membrane transports the fluid to the PBC and facilitates the accumulation of AuNPs. We do not need to coat capture probes on the surface of the NC membrane but just assemble the PBCs on it. When the sample is introduced onto the sample pad of the LFTS, the liquid flows through the conjugate pad and the NC membrane and is finally absorbed by the absorbent pad. There are two directions of liquid flow. The first direction is the same as that of a typical LFTS. In the second direction, in the PBCs, the liquid flows from the NC membrane to the stacked paper (Fig. 1B, blue arrows). Because the double-sided adhesive in the stacked paper is hydrophobic (fig. S4), the liquid can only transport to every sheet of paper from the NC membrane but cannot transport between neighboring sheets of paper (Fig. 1B). This design avoids smears between different sheets of paper to get rid of interference.
In a sandwich immunoassay or an indirect immunoassay, when we introduce the positive sample on the sample pad, the fluid migrates to the conjugate pad and dissolves the AuNP-conjugated detection probes. The target reacts with the AuNP-conjugated detection probes and generates complexes of the targets and AuNPs. While the complexes migrate to the PBC, the fluid flows along the vertical direction of the PBC, and the complexes migrate through the chromatography paper and react with the capture probes immobilized in the paper. At the same time, the complexes bind on the edges of the chromatography paper through the immunoreaction. When more and more fluid migrates through the paper, AuNPs accumulate and the edge of the paper gradually changes from white to red. When the sample is negative, no immunoreaction is generated, and the edge of the paper remains white. The results indicate that blank samples do not appear to contribute to nonspecific reading, suggesting that AuNPs trapped in the pores in the fibers of paper do not affect the results of the assay (fig. S5). These results agree with the study of Zhang et al. (27). After the detection, we rotate the PBCs and expose the edges participating in the reaction to the scanner for barcode reading (Fig. 1C). The results can be read out by the barcode scanner and recorded in any kind of word-processing software (we used Notepad in the experiment). When the three kinds of targets are all positive, the result is read as “A:::A.” When all the targets are negative, the result is read as “A111A.” In a competitive immunoassay, white and red in the variable regions are inverted. The result is read as “A:::A” for the negative sample and “A111A” for the positive sample. The cutoff value of the color intensity is dependent on the algorithm of the application installed in the barcode scanner. The device captures the images, measures the average gray values of the bars and spaces, calculates the cutoff values for the bars and spaces, and gives the results. For a particular detection, the cutoff value of the color intensity for a positive signal or a negative signal can just be determined by the barcode scanner.
Immunoassay of drugs of abuse
We carry out the simultaneous detection of cocaine (COC), morphine (MOR), and methamphetamine (mAMP) to investigate the applicability of PBAS. Rapid testing of illegal drugs is important for the control of drugs and criminal investigations (28, 29). Because the targets of interest are all small organic molecules, we use competitive immunoassays. In the cases where the samples are positive, COC, MOR, and mAMP react with the detection probes and cover all the immunocompetent domains on the antibodies that bound on the surface of the AuNPs. When the complexes of the targets and the AuNP-conjugated detection probes migrate to the PBCs, no more antibodies can react with the antigens immobilized on the papers; thus, no more AuNPs accumulate and the papers remain white. The variable regions all remain white after detection. The barcode is read as “A111A” by the barcode scanner (Fig. 3A). When the samples are negative, the immunocompetent domains on the antibodies remain exposed. AuNP-conjugated detection probes react with antigens immobilized on the papers, and AuNPs accumulate on the edges of the papers. The edges of the papers change from white to dark red. The barcode is read as “A:::A” by the barcode scanner (Fig. 3A).
Fig. 3. Simultaneous detection of drugs of abuse using the PBAS.
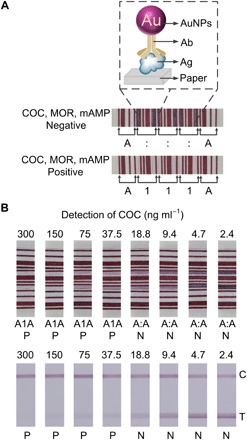
(A) Negative and positive results of COC, MOR, and mAMP. Ag, antigen. (B) Comparison of the LOD of COC using the PBCs and LFTSs. C, control line; T, test line.
To determine the limit of detection (LOD) of COC by the PBAS, we test COC with the concentration from 300 to 2.4 ng ml−1 on the PBAS and read out the results using the barcode scanner. The amount of AuNPs aggregating on the edge of the paper decreases as the COC concentration increases (fig. S5). The color intensity of the variable regions decreases as the concentration of COC increases. The barcodes of COC with the concentrations of 300, 150, 75, 37.5, 18.8, 9.4, 4.7, and 2.4 ng ml−1 are read as “A1A,” “A1A,” “A1A,” “A1A,” “A:A,” “A:A,” “A:A,” and “A:A” by the barcode scanner after the detection (Fig. 3B). The result indicates that the LOD of the immunoassay of COC by the PBAS is 37.5 ng ml−1. The threshold values of the commercially available kits for the qualitative detection of COC, MOR, and mAMP are 300, 300, and 1000 ng ml−1. The above results show the PBAS can satisfy the demand of simultaneous detection of COC, MOR, and mAMP.
We further compare the LOD of COC by the PBAS with the LFIA system. The minimum concentration that gives very weak color intensity in the test line is defined as the visual LOD in a qualitative evaluation (25). We judge that the LOD of COC by the LFIA system is also around 37.5 ng ml−1, which is almost at the same level as the PBAS (Fig. 3B). However, the judgment dependent on the naked eye is subjective and differs among individuals. In the PBAS, the readout of the results is objective since it depends on the barcode scanner. Additionally, we can improve the LOD of the PBAS by amplification strategies after the assay (30, 31). Decreasing the cutoff value of the gray value in the barcode reading by soaking the papers for the constant bars with light-colored ink can also improve the LOD.
To investigate the repeatability of the PBAS, we detect three groups of COC with concentrations from 300 to 2.4 ng ml−1. The barcodes of the three groups are read as “A1A,” “A1A,” “A1A,” “A1A,” “A:A,” “A:A,” “A:A,” and “A:A,” indicating that the LOD is around 37.5 ng ml−1, and the PBAS has good repeatability (fig. S6). To further evaluate the stability of the PBAS, we detect the positive sample and the negative sample of COC with the PBCs stored from 1 to 60 days. All the positive samples are read as “A1A” and all the negative samples are read as “A:A,” indicating that the PBCs are stable during storage for several months (table S1).
Immunoassay of infections among blood donors
To further determine the feasibility of the PBAS, we perform detection of the hepatitis B virus (HBV) surface antigen and human anti–hepatitis C virus (HCV), HIV-1, and Treponema pallidum (TP) antibodies from patient blood by the sandwich immunoassay. The mandated blood screenings in China are serological tests for the HBV surface antigen and antibodies for HCV, HIV, and TP. In the detection of the positive sample, the edges of the papers in touch with the NC membrane change from white to red, and the result is read as “A::::A” (Fig. 4A). When the sample is negative, the papers remain white, and the result is read as “A1111A.” Next, we demonstrate the PBAS with the serum samples of patients. The serum samples of patients are collected from a local hospital. All the results can be read out by the barcode scanner and are consistent with the results detected in the laboratory by enzyme-linked immunosorbent assay and polymerase chain reaction (Fig. 4B).
Fig. 4. Simultaneous detection of the infections of HBV, HCV, HIV-1, and TP using the PBAS.
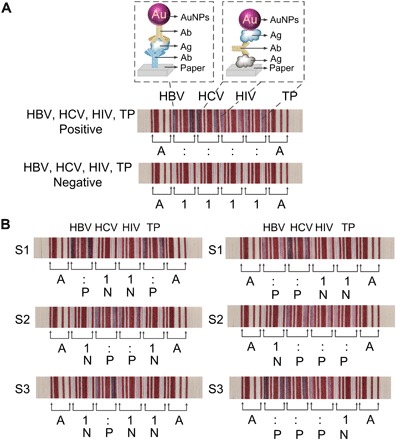
(A) Positive and negative results of the simultaneous detection of HBV surface antigen and human anti-HCV, HIV-1, and TP antibodies. (B) Detection of six samples from patients infected by HBV, HCV, HIV-1, and TP.
DISCUSSION
We present a strategy to fabricate precise and complex barcode-based PADs with high efficiency and convenience, independent of sophisticated facilities and professional operations required in a conventional clinical laboratory (32). We provide an integrated assay system with the advantages of rapidity, low cost, and robustness, making it extremely suitable for application in developing countries, low-resource regions, and private homes. In particular, the barcode-based encoding and decoding modes provide efficient, accurate, and objective readout and analysis of a large number of multiplex results. Combined with the methods for amplification and advanced hardware/software, we expect that the PBAS will have improved analytical performance, such as sensitivity, linear range, and throughput, broadening its applications. The manufacturing of an old material (paper, which is thousands of years old) can bring about novel functions, and we hope that approaches like this will allow researchers to explore totally new manufacturing methods and novel applications of old materials.
MATERIALS AND METHODS
Fabrication of PBCs
To fabricate the PBCs, we used three kinds of paper: the white paper, the red paper, and the paper for immobilizing protein. The three kinds of paper used in our experiment were all from the chromatography paper of Whatman (Grade 3MM Chr, 340 μm, white). We obtained the red paper by soaking the paper with red ink and drying and the paper for immobilizing proteins by soaking the paper with certain antibodies or antigens that can recognize corresponding targets, forming the variable regions. We dried the wet paper soaked with proteins at 37°C and glued (with 3M’s double-sided adhesive tape) them with the red paper and the white paper according to the regulations of the Codabar codes. Finally, we skived the combined papers into a small size (6 mm × 0.5 mm) with an electric paper cutter (X-920, Guoli) and obtained the PBCs.
Characterization of the papers and the PBCs
We characterized the surface morphology, pore size, and the reaction regions on paper after the immunoassay by a scanning electron microscope (S4800, Hitachi), and the widths of the modules in the dry and wet PBCs by a phase contrast microscope (Axio Imager 2, Zeiss) and a single-lens reflex camera (D90, Nikon). We took all the results of the PBCs with the single-lens reflex camera (D90, Nikon). We characterized the dry tensile strength and the wet tensile strength of the papers by a tensile test machine (Tensile Tester, Lorentzen & Wettre). We evaluated the hydrophilicity of the double-sided adhesive, the white paper, the red paper, and the paper soaked with protein with a water contact angle analysis system (DSA100). We dripped the water droplets with a volume of 10 μl separately onto the surface of the double-sided adhesive and the papers. Each group consisted of five tests. The contact angles were the average values with SDs.
Immunoassay of drugs of abuse
The detection of drugs of abuse was carried out on the PBC with competitive immunoassay. The targets were COC, MOR, and mAMP. The procedures were as follows:
1.) For the fabrication of PBCs for immunoassay of COC, MOR, and mAMP, we soaked four sheets of chromatography paper with conjugates of haptens (drugs of abuse or their metabolites) and bovine serum albumin (BSA) at concentrations of 1 mg ml−1 in 0.01 M phosphate-buffered saline (PBS) (pH 7.4) and dried them at 37°C. We glued the dried chromatography papers with red papers and white papers in the order described in Fig. 1A and skived the combined papers into a small size (6 mm × 0.5 mm).
2.) For the detection of COC, MOR, and mAMP, we fabricated the blank LFTSs by fixing the NC membrane, the absorbent pad, the sample pad, and the conjugate pad on the plastic backing plates and cut the strips into a width of 8 mm. To ensure that the edges of every sheet of paper in the PBCs touched the NC membrane completely, we added a layer of double-sided foam tape under the NC membrane. To prepare the conjugate pad, we mixed the three kinds of AuNP-labeled antibodies (anti-COC, anti-MOR, and anti-mAMP antibodies) at equal ratios, dispensed the mixture onto the glass fiber, and freeze dried it. In the assay, we placed the PBC on the NC membrane of the LFTS, pressed the PBC with an external force on the top, and dropped the solution of the sample on the sample pad. We diluted all the positive samples from the standard samples and used the solution of PBS (0.01 M) as the negative control. We detected the positive sample of COC, MOR, and mAMP with the concentration of 300 ng ml−1. To determine the LOD of COC, we detected COC with concentrations of 300, 150, 75, 37.5, 18.8, 9.4, 4.7, and 2.4 ng ml−1 with the PBCs and the LFTSs. Finally, we read out the results using a barcode scanner after the detection. The whole process of every detection was completed within 10 min.
3.) For the repeatability and stability of the PBAS, we detected three groups of COC with the concentration from 300 to 2.4 ng ml−1 on the PBCs and four groups of the positive (300 ng ml−1) and negative samples (0 ng ml−1) of COC with the PBCs stored for 1, 7, 30, and 60 days in dry conditions and at room temperature.
Immunoassay of infections among blood donors
We detected the infections caused by HBV, HCV, HIV, and TP by sandwich immunoassay based on the PBCs. The target contains HBV surface antigen and human anti-HCV, human anti–HIV-1, and human anti-TP antibodies. We detected the HBV surface antigen by double-antibody sandwich immunoassay and the other three by double-antigen sandwich immunoassay. The procedures were similar to the immunoassay of drugs of abuse, except for soaking the chromatography papers with mouse anti-HBV surface antibody and HCV, HIV-1, and TP antigens with a concentration of 2 mg ml−1 and conjugating AuNPs with mouse anti-HBV surface antibody and HCV, HIV-1, and TP antigens. We mixed the four kinds of AuNP-labeled antibodies and antigens (mouse anti-HBV surface antibody and HCV, HIV-1, and TP antigens) at equal ratios, dispensed the mixture onto the glass fiber, and freeze dried it to fabricate the conjugate pad. The positive sample was the mixture of the positive standard samples of HBV, HCV, HIV, and TP, and the negative control was the solution of BSA (1 mg ml−1). The serum samples of patients were collected from the Chinese PLA General Hospital. All the experiments were performed in compliance with the hospital guidelines (The Ethics Guidelines for Research Involving Human Subjects or Human Tissue from the Chinese PLA General Hospital).We read out the results using the barcode scanner after the detection.
Barcode reading
We read out the results of the immunoassay using a compact laser handheld barcode scanner (BL-N70UBE, Keyence). First, we connected the barcode scanner with a computer through the universal serial bus interface. Next, we opened any kind of word-processing software, such as Notepad, Microsoft Office Word, or Microsoft Office Excel, pressed on the switch and scanned the barcodes. Correspondingly, the laser was emitted, the green status light-emitting diode lighted up, a buzzer sounded, and the data were sent to the computer and automatically recorded by the word-processing software.
Supplementary Material
Acknowledgments
We thank R. Cha and S. Shi for providing technical assistance. We thank Q. Niu in the Guoli Machinery Limited Liability Company for providing us with the electric paper cutter. Funding: We thank the Ministry of Science and Technology of China (2013YQ190467), the Chinese Academy of Sciences (XDA09030305), and the National Science Foundation of China (81361140345, 51373043, and 21535001) for financial support. Author contributions: M.Y. and W. Zhang conceived and initiated the PBAS. M.Y., B.H., and W. Zheng designed the PBCs. M.Y. and J.Y. characterized the papers. M.Y. fabricated the PBCs. M.Y., F.C., and Y.C. fabricated the LFTSs. M.Y. completed the detection of the drugs of abuse and biomarkers of blood-transmitted infections and the analysis of the results. M.Y. and X.J. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Date and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/12/eaao4862/DC1
fig. S1. Characterization of the three kinds of paper (Grade 3MM Chr, Grade 1 Chr, and Grade 1).
fig. S2. The dry tensile strength and wet tensile strength of the three kinds of paper (Grade 3MM Chr, Grade 1 Chr, and Grade 1).
fig. S3. Characterization of the widths of the modules in the wet PBCs.
fig. S4. Wetting behavior of the double-sided adhesive, the white paper, the red paper, and the paper soaked with protein.
fig. S5. Characterization of the reaction regions on paper after the immunoassay.
fig. S6. The repeatability of the PBAS.
table S1. The detection of COC with the PBCs stored for 1 to 60 days.
REFERENCES AND NOTES
- 1.Xu Q., Rioux R. M., Whitesides G. M., Fabrication of complex metallic nanostructures by nanoskiving. ACS Nano 1, 215–227 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Xu Q., Bao J., Rioux R. M., Perez-Castillejos R., Capasso F., Whitesides G. M., Fabrication of large-area patterned nanostructures for optical applications by nanoskiving. Nano Lett. 7, 2800–2805 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Fraikin J. L., Teesalu T., McKenney C. M., Ruoslahti E., Cleland A. N., A high-throughput label-free nanoparticle analyser. Nat. Nanotechnol. 6, 308–313 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Sun J., Xianyu Y., Jiang X., Point-of-care biochemical assays using gold nanoparticle-implemented microfluidics. Chem. Soc. Rev. 43, 6239–6253 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Chen Y., Xianyu Y., Wang Y., Zhang X., Cha R., Sun J., Jiang X., One-step detection of pathogens and viruses: Combining magnetic relaxation switching and magnetic separation. ACS Nano 9, 3184–3191 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Xianyu Y., Xie Y., Wang N., Wang Z., Jiang X., A dispersion-dominated chromogenic strategy for colorimetric sensing of glutathione at the nanomolar level using gold nanoparticles. Small 11, 5510–5514 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Liu D., Chen W., Wei J., Li X., Wang Z., Jiang X., A highly sensitive, dual-readout assay based on gold nanoparticles for organophosphorus and carbamate pesticides. Anal. Chem. 84, 4185–4191 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Zhu Z., Guan Z., Liu D., Jia S., Li J., Lei Z., Lin S., Ji T., Tian Z., Yang C. J., Translating molecular recognition into a pressure signal to enable rapid, sensitive, and portable biomedical analysis. Angew. Chem. Int. Ed. 54, 10448–10453 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Fang X., Wei S., Kong J., Paper-based microfluidics with high resolution, cut on a glass fiber membrane for bioassays. Lab Chip 14, 911–915 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Cunningham J. C., Brenes N. J., Crooks R. M., Paper electrochemical device for detection of DNA and thrombin by target-induced conformational switching. Anal. Chem. 86, 6166–6170 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Guan L., Tian J., Cao R., Li M., Cai Z., Shen W., Barcode-like paper sensor for smartphone diagnostics: An application of blood typing. Anal. Chem. 86, 11362–11367 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Yu J., Ge L., Huang J., Wang S., Ge S., Microfluidic paper-based chemiluminescence biosensor for simultaneous determination of glucose and uric acid. Lab Chip 11, 1286–1291 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Martinez A. W., Phillips S. T., Butte M. J., Whitesides G. M., Patterned paper as a platform for inexpensive, low-volume, portable bioassays. Angew. Chem. Int. Ed. 46, 1318–1320 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renault C., Anderson M. J., Crooks R. M., Electrochemistry in hollow-channel paper analytical devices. J. Am. Chem. Soc. 136, 4616–4623 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Li M., Tian J., Al-Tamimi M., Shen W., Paper-based blood typing device that reports patient’s blood type “in writing”. Angew. Chem. Int. Ed. 51, 5497–5501 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Badu-Tawiah A. K., Lathwal S., Kaastrup K., Al-Sayah M., Christodouleas D. C., Smith B. S., Whitesides G. M., Sikes H. D., Polymerization-based signal amplification for paper-based immunoassays. Lab Chip 15, 655–659 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Yamada K., Henares T. G., Suzuki K., Citterio D., Paper-based inkjet-printed microfluidic analytical devices. Angew. Chem. Int. Ed. 54, 5294–5310 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Liu M., Hui C. Y., Zhang Q., Gu J., Kannan B., Jahanshahi-Anbuhi S., Filipe C. D. M., Brennan J. D., Li Y., Target-induced and equipment-free DNA amplification with a simple paper device. Angew. Chem. Int. Ed. 55, 2709–2713 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Yu L., Shi Z. Z., Microfluidic paper-based analytical devices fabricated by low-cost photolithography and embossing of Parafilm®. Lab Chip 15, 1642–1645 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Yetisen A. K., Akram M. S., Lowe C. R., Paper-based microfluidic point-of-care diagnostic devices. Lab Chip 13, 2210–2251 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Parolo C., Merkoçi A., Paper-based nanobiosensors for diagnostics. Chem. Soc. Rev. 42, 450–457 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Sun J., Zou Y., Chen W., Zhang W., Xi J. J., Jiang X., Barcoded microchips for biomolecular assays. Anal. Chem. 87, 900–906 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Wong J. X. H., Li X., Liu F. S. F., Yu H.-Z., Direct reading of bona fide barcode assays for diagnostics with smartphone apps. Sci. Rep. 5, 11727 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin Z., Chan W. C. W., Boulware D. R., Akkin T., Butler E. K., Bischof J. C., Significantly improved analytical sensitivity of lateral flow immunoassays by using thermal contrast. Angew. Chem. Int. Ed. 51, 4358–4361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song S., Liu N., Zhao Z., Njumbe Ediage E., Wu S., Sun C., De Saeger S., Wu A., Multiplex lateral flow immunoassay for mycotoxin determination. Anal. Chem. 86, 4995–5001 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Chen J., Zhou S., Wen J., Disposable strip biosensor for visual detection of Hg2+ based on Hg2+-triggered toehold binding and exonuclease III-assisted signal amplification. Anal. Chem. 86, 3108–3114 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Zhang C., Zhang Y., Wang S., Development of multianalyte flow-through and lateral-flow assays using gold particles and horseradish peroxidase as tracers for the rapid determination of carbaryl and endosulfan in agricultural products. J. Agric. Food Chem. 54, 2502–2507 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Zhang C.-Y., Johnson L. W., Single quantum-dot-based aptameric nanosensor for cocaine. Anal. Chem. 81, 3051–3055 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawano R., Osaki T., Sasaki H., Takinoue M., Yoshizawa S., Takeuchi S., Rapid detection of a cocaine-binding aptamer using biological nanopores on a chip. J. Am. Chem. Soc. 133, 8474–8477 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Qu W., Liu Y., Liu D., Wang Z., Jiang X., Copper-mediated amplification allows readout of immunoassay by the naked eye. Angew. Chem. Int. Ed. 50, 3442–3445 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Xianyu Y., Wang Z., Jiang X., A plasmonic nanosensor for immunoassay via enzyme-triggered click chemistry. ACS Nano 8, 12741–12747 (2014). [DOI] [PubMed] [Google Scholar]
- 32.T. C. Tisone, B. O’Farrell, Manufacturing the next generation of highly sensitive and reproducible lateral flow immunoassay, in Lateral Flow Immunoassay, R. C. Wong, H. Y. Tse, Eds. (Humana Press, 2009), pp. 131–156. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/12/eaao4862/DC1
fig. S1. Characterization of the three kinds of paper (Grade 3MM Chr, Grade 1 Chr, and Grade 1).
fig. S2. The dry tensile strength and wet tensile strength of the three kinds of paper (Grade 3MM Chr, Grade 1 Chr, and Grade 1).
fig. S3. Characterization of the widths of the modules in the wet PBCs.
fig. S4. Wetting behavior of the double-sided adhesive, the white paper, the red paper, and the paper soaked with protein.
fig. S5. Characterization of the reaction regions on paper after the immunoassay.
fig. S6. The repeatability of the PBAS.
table S1. The detection of COC with the PBCs stored for 1 to 60 days.


