Abstract
Eukaryotic translation initiation factor 2 (eIF2) is a heterotrimeric GTPase, which plays a critical role in protein synthesis regulation. eIF2-GTP binds Met-tRNAi to form the eIF2-GTP•Met-tRNAi ternary complex (TC), which is recruited to the 40S ribosomal subunit. Following GTP hydrolysis, eIF2-GDP is recycled back to TC by its guanine nucleotide exchange factor (GEF), eIF2B. Phosphorylation of the eIF2α subunit in response to various cellular stresses converts eIF2 into a competitive inhibitor of eIF2B, which triggers the integrated stress response (ISR). Dysregulation of eIF2B activity is associated with a number of pathologies, including neurodegenerative diseases, metabolic disorders, and cancer. However, despite decades of research, the underlying molecular mechanisms of eIF2B action and regulation remain unknown. Here we employ a combination of NMR, fluorescence spectroscopy, site-directed mutagenesis, and thermodynamics to elucidate the mechanisms of eIF2B action and its regulation by phosphorylation of the substrate eIF2. We present: (i) a novel mechanism for the inhibition of eIF2B activity, whereby eIF2α phosphorylation destabilizes an autoregulatory intramolecular interaction within eIF2α; and (ii) the first structural model for the complex of eIF2B with its substrate, eIF2-GDP, reaction intermediates, apo-eIF2 and eIF2-GTP, and product, TC, with direct implications for the eIF2B catalytic mechanism.
INTRODUCTION
Eukaryotic translation initiation factor 2 (eIF2) is a heterotrimeric GTPase, which in its active GTP-bound state is responsible for recruiting the initiator methionyl tRNA (Met-tRNAi) to the 40S ribosomal subunit as part of the eIF2-GTP•Met-tRNAi ternary complex (TC). Upon start codon selection and GTP hydrolysis, eIF2-GDP dissociates from the ribosome in complex with eIF5, its GTPase-activating protein (GAP). eIF2 is re-activated by recycling its bound GDP for GTP and binds another Met-tRNAi. Both nucleotide exchange and Met-tRNAi recruitment are performed by the guanine nucleotide exchange factor (GEF) eIF2B, resulting in a new TC capable of initiating another round of translation (reviewed in (1–3)).
The GEF eIF2B consists of five subunits, α-ϵ, recently shown by us and others to assemble into a ∼600 kDa decamer (4–8), making it one of the largest and most complex GEFs characterized to date. eIF2B is the target of multiple pathways regulating protein synthesis and the cellular stress response. The catalytic activity of eIF2B is regulated by phosphorylation of both its substrate eIF2 and eIF2B itself, as well as by small molecules, such as nucleotides and sugars, binding to eIF2B. eIF2 consists of α, β and γ subunits, with eIF2γ being the actual GTPase, and eIF2α and β serving accessory functions. eIF2α is phosphorylated at S51 by four kinases in what is collectively known as the integrated stress response (ISR): Protein Kinase R (PKR), in response to viral infection; PKR-like Endoplasmic Reticulum Kinase (PERK), in response to ER stress, oxidative stress, and other stimuli; General Control Nonderepressible 2 Kinase (GCN2), in response to amino-acid starvation; and Heme-Regulated Inhibitor Kinase (HRI), in response to heme deficiency. Of all the layers of regulation of eIF2B activity, regulation by phosphorylation of its substrate eIF2 has attracted the most attention since it has wide-ranging effects on the cell and is at the core of ISR.
eIF2α phosphorylation converts eIF2-GDP (eIF2(α-P)-GDP) into a competitive inhibitor of eIF2B, thereby slowing down protein synthesis, while at the same time inducing the production of a set of transcription factors, including Activating Transcription Factor 4 (ATF4) in mammals, and GCN4 in Saccharomyces cerevisiae. The result is the activation of both pro-survival pathways aimed at restoring homeostasis and pro-apoptotic pathways. The fate of the cell depends largely on the magnitude and duration of the stress response, as well as other factors. Since the stressors themselves can cause cell death, either an insufficient or overly aggressive stress response can lead to apoptosis (reviewed in (3,9–13)). While apoptosis can be the desired outcome in some cases, such as viral infection, in many cases it occurs as the result of uncontrolled ISR and is a causative factor in the pathology of a number of diseases. It was recently reported that PERK activation mediates neuronal cell death in prion disease, Alzheimer's, and other neurodegenerative disorders (reviewed in (14)). Accordingly, PERK inhibitors, as well as an eIF2B activator called ISR inhibitor (ISRIB), were shown to be neuroprotective in animal models of these diseases (15,16). However, the mechanisms of TC regeneration by eIF2B and its regulation still remain poorly understood.
Mutations in all five eIF2B subunits cause a genetic neurodegenerative disorder known as childhood ataxia with CNS hypomyelination (CACH) or leukoencephalopathy with vanishing white matter (VWM) (reviewed in (12,17,18)). The molecular mechanisms of CACH/VWM are not well understood, but the mutations appear to affect eIF2B complex assembly, stability, and/or activity (4,6–8,19).
The eIF2Bϵ subunit contains the catalytic domain, and together with the eIF2Bγ subunit forms the catalytic subcomplex (eIF2Bcat). eIF2Bα, β and δ form the regulatory subcomplex (eIF2Breg). Four of the five eIF2B subunits are essential in yeast, except eIF2Bα, which is required for inhibition by eIF2α phosphorylation: its deletion has a General control nonderepressible (Gcn−) phenotype, characterized by an inability to induce ISR under conditions of amino acid starvation (reviewed in (10,11)). In S. cerevisiae, Gcn− mutations have been found in the eIF2Bα, β, and δ subunits (reviewed in (10,11)). The eIF2Bβ, γ, and ϵ subunits are essential for nucleotide exchange, whereas eIF2Bδ has been proposed to be important for Met-tRNAi binding to eIF2-GTP (20). Therefore, at least the β, γ, δ, and ϵ subunits of eIF2B must contribute to catalysis. Mutations that lower eIF2B activity and mimic the effects of eIF2α phosphorylation, thereby causing constitutively induced ISR in the absence of amino acid starvation (General control derepressed, Gcd−), have been reported in all five eIF2B subunits, including eIF2Bα. Thus, even eIF2Bα appears to play a role in catalysis, albeit not an essential one, supported by the fact that combining eIF2Bα deletion with certain Gcn− mutations causes a Gcd− phenotype (21). The vast amount of genetic and biochemical data indicates that eIF2 interacts simultaneously with the catalytic and regulatory subcomplexes of eIF2B, and that eIF2α phosphorylation causes the interaction with the regulatory subcomplex to become inhibitory (10,11,20).
The past two years have marked a radical change in our understanding of the structure of eIF2B and the nature of the eIF2B•eIF2 interaction. First, we and others reported that eIF2B is in fact a decamer with a hexameric regulatory subcomplex, rather than a pentamer as had been widely assumed (4,5,7,8). Next, the crystal structure of the nearly complete Schizosaccharomyces pombe eIF2B decamer was reported (6). The only eIF2B segments missing from the structure are the C-terminal domain (CTD) of eIF2Bϵ (the catalytic domain) and the long eIF2Bδ N-terminal tail (NTT). The regulatory subcomplex, eIF2Breg, is composed of an eIF2Bα2 homodimer and two eIF2Bβδ heterodimers (eIF2Bα2(βδ)2), packed against each other with a pseudo-three-fold symmetry. The three dimers are organized in such a way that the CTDs of the subunits form the core of the hexamer, while the N-terminal domains (NTDs) form two pockets. Two eIF2Bγϵ heterodimers (eIF2Bcat) are bound on opposite sides of the eIF2Breg core, with the ϵ subunit forming a large contact surface with the β subunit and the γ subunit contacting the δ-subunit (6). Cross-linking data showed that eIF2α binds in the eIF2Breg pocket formed by the NTDs of the eIF2Bα, β, and δ subunits, whereas eIF2γ binds to a platform on eIF2Bcat formed by eIF2Bγ and ϵ (6). Based solely on their cross-linking data, Kashiwagi and co-authors proposed a model for the orientation of eIF2α-NTD in the eIF2Breg pocket, where it contacts all three eIF2Breg subunits, though it primarily lies in the groove between eIF2Bβ and δ. As simultaneous binding of eIF2α in the pocket and of eIF2γ to eIF2Bcat appeared to be impossible, given their orientation of eIF2α-NTD in the eIF2Breg pocket, the authors proposed that binding of eIF2α-NTD into the pocket precludes the productive eIF2Bcat/eIF2γ interaction necessary for nucleotide exchange, and vice versa. They thus suggested that phosphorylation of eIF2α would favor the former interaction and disfavor the latter, leading to inhibition of nucleotide exchange (6). However, the idea of two mutually exclusive one-site interactions is in apparent contradiction with the vast amounts of data indicating that eIF2α-NTD interactions with the eIF2Breg pocket play a role in catalysis, and not just in eIF2B inhibition by phosphorylated eIF2-GDP (eIF2(α-P)-GDP) (10,11,20).
It is commonly assumed that the primary mechanism responsible for the increased affinity of eIF2B for eIF2(α-P)-GDP over unphosphorylated eIF2-GDP is the direct effect of phosphorylation on the affinity of the eIF2α phosphorylation loop (P-loop) for a corresponding surface on eIF2B. It has been shown in yeast (22,23) and in mammals (24) that the loss of eIF2Bα results in loss of eIF2B inhibition by eIF2(α-P)-GDP. Therefore, the ‘direct effect’ is widely believed to be mediated by eIF2Bα. Work by Hinnebusch and co-authors (25), however, suggests the mechanism may be more complex. While phosphorylated eIF2α (eIF2α-P) could pull down eIF2B and unphosphorylated eIF2α could not, a ∼100 amino acid C-terminal deletion in eIF2α resulted in an equally effective pull-down regardless of eIF2α’s phosphorylation state. From the structure of eIF2α (26), it becomes clear that the C-terminal deletion of eIF2α that abrogates eIF2B’s preference for eIF2α-P over eIF2α corresponds to an effective deletion of most of eIF2α-CTD. This suggests an autoregulatory role for eIF2α-CTD in eIF2α binding to eIF2B. We hypothesized that in addition to a potential ‘direct effect,’ there is an ‘indirect effect’ whereby phosphorylation of S51 disrupts an intramolecular interaction between eIF2α-NTD and -CTD. This disruption exposes an eIF2B-binding surface on eIF2α-NTD that is otherwise obstructed by eIF2α-CTD. The contribution of this new ‘indirect effect’ to the increase in affinity of eIF2B for eIF2α-P over eIF2α can be at least as important as the addition of the negatively charged phosphate group, if not more so.
In this work, we demonstrate for the first time that the two domains of eIF2α interact with each other and that their interaction is destabilized by phosphorylation of S51. We further demonstrate that this intramolecular binding interface in eIF2α overlaps with the eIF2α binding surfaces for eIF2Bβ, and possibly also eIF2Bδ, in support of the hypothesis for an indirect effect of eIF2α phosphorylation on eIF2B•eIF2 binding. We show that while eIF2α phosphorylation has a modest direct effect on eIF2B binding, this direct effect is too small to account for the much greater eIF2B affinity for eIF2(α-P)-GDP, compared to unphosphorylated eIF2-GDP. We also show that eIF2 can bind simultaneously in the eIF2Breg pocket (via eIF2α-NTD) and to eIF2Bcat (via eIF2γ), in an ‘extended’ conformation in which the eIF2α-NTD/-CTD intramolecular interaction is disrupted. We propose a model wherein the eIF2B•eIF2 complex is in equilibrium between a ‘closed’ eIF2B•apo-eIF2 state in which eIF2 only contacts eIF2Bcat, and an ‘extended’ nucleotide-bound state in which eIF2 contacts both eIF2Bcat and eIF2Breg simultaneously. When eIF2α is not phosphorylated, eIF2B prefers the apo-state, thus promoting GDP release, whereas when bound to eIF2(α-P)-GDP, eIF2B has no preference for the apo- or GDP-bound state and thus has no effect on nucleotide exchange.
MATERIALS AND METHODS
Nuclear magnetic resonance (NMR) spectroscopy
NMR spectra were recorded at 298 K on a Bruker 500 MHz (Boston University School of Medicine) or on a Bruker 800 MHz (Brandeis University) magnet, each equipped with a cryoprobe. Backbone resonance assignments of eIF2α-NTD, eIF2α-NTDS51D, and eIF2α-CTD were obtained using standard triple-resonance experiments on 2H/15N/13C-labeled samples at 300 mM NaCl.
Samples for NMR measurements were prepared in Buffer A150 (10 mM Na-phosphate (pH 7), 150 mM NaCl, 0.01% NaN3, 2 mM DTT, 1 mM EDTA, 0.1 mM AEBSF) with 5% 2H2O. Transverse relaxation optimized 1H–15N-heteronuclear single-quantum coherence (TROSY-HSQC) spectra of 2H/15N-labeled eIF2α, eIF2αS51D, eIF2α-NTD and eIF2α-NTDS51D were collected in the absence and presence of excess eIF2Bα or eIF2Bβ and compared. TROSY-HSQC spectra of 2H/15N/13C-labeled eIF2α-CTD were collected in the presence and absence of excess eIF2Bα or eIF2Bβ and compared. HSQC spectra of 2H/15N-labeled eIF2αS51D both free and in the presence of eIF2Bα were also collected.
NMR chemical shift perturbation (CSP) assays were performed in combination with deletion analysis to map the intramolecular surface, and in combination with site-directed mutagenesis to determine the effects of phosphorylation, as previously described (27–30). When comparing spectra of the various constructs in the absence and presence of excess eIF2Bα or eIF2Bβ, a pattern of selective signal loss in peaks belonging to interface residues was observed upon binding. The selective signal loss is due to cross-relaxation between the free and bound state, a phenomenon observed in cases with a weak interaction in which the labeled protein is in fast equilibrium between a free state and a complex that is too large to observe by NMR (31). A statistical analysis of both chemical shift perturbation and signal loss was performed by calculating average change and standard deviation for all peaks on the protein, as described previously (27). Peak movement, or decrease in intensity, by more than two standard deviations from the average was considered statistically significant. Changes between one and two standard deviations were also mapped on the protein structures.
Fluorescence anisotropy
eIF2α-NTD and eIF2α-NTDS51D were labeled in a 1:5 ratio with fluorescein-5-malemeide (F5M) in Buffer B300 (10 mM Na-phosphate (pH 7), 300 mM NaCl, 0.1 mM AEBSF, 1 mM EDTA, and 0.1 mM TCEP), degassed and deoxygenated by bubbling N2. Size-exclusion chromatography was used to separate free F5M and also to exchange the labeled protein back into Buffer A150.
Fluorescence anisotropy measurements were performed on a QuantaMaster QM4 fluorescent spectrometer (PTI), equipped with polarizers and dual monochromators. Proteins of interest were titrated into F5M-labeled eIF2α-NTD or eIF2α-NTDS51D; eIF2Bα and eIF2Bβ were titrated up to their solubility limit. Increases in fluorescence anisotropy as a function of protein concentration were recorded and used to fit the KD of the interaction in MATLAB.
Vectors, protein expression, and purification
Human eIF2Bα was cloned in a pET21a-derivative vector with a C-terminal His6-tag (eIF2Bα-H). An eIF2BαI301E/L305D mutant was derived from the WT eIF2Bα-H plasmid using site-directed mutagenesis. WT eIF2Bα has a tendency to self-associate at high concentrations, which is reduced in the eIF2BαI301E/L305D mutant. The mutation did not cause visible difference in any binding experiments compared to WT eIF2Bα (data not shown). Therefore, the eIF2BαI301E/L305D mutant was used in this work, unless otherwise stated, and designated as eIF2Bα. Human eIF2Bβ and eIF2Bδ were cloned in a pET21a-derivative vector with an N-terminal IgG-binding domain 1 (GB1) tag, followed by a His6-tag and a TEV protease cleavage site (GH-eIF2Bβ and GH-eIF2Bδ).
Human full length eIF2α was cloned in a pET30a-derivative vector with a C-terminal His6-tag (eIF2α-H). The N-terminal domain (NTD) of eIF2α (aa 1–184) was derived from the WT eIF2α-H plasmid using site-directed mutagenesis to insert a stop codon and a C-terminal His6-tag (eIF2α-NTD-H). Phosphomimetic variants of eIF2α and eIF2α-NTD, containing the S51D mutation, were derived from WT eIF2α-H and eIF2α-NTD-H, respectively, via site-directed mutagenesis. The C-terminal domain (CTD) of eIF2α (aa 185–314) was obtained by subcloning the C-terminal portion of eIF2α-H into a pET21a-derivative vector, incorporating the same N-terminal GB1-His6-TEV protease cleavage site sequence as detailed above for eIF2Bβ.
eIF2Bα constructs were expressed in Rosetta2(DE3) cells at 20°C overnight. eIF2Bβ was expressed in Rosetta2(DE3)-pLysS cells at 37°C for 3 hours. eIF2α-NTD and eIF2α-CTD were expressed in Rosetta2(DE3) cells at 20°C overnight. Full length eIF2α and eIF2αS51D were expressed in BL21(DE3) cells at 20°C overnight, co-expressed with the GroEL-GroES and trigger factor (Tf) chaperones, contained on a pG-Tf2 plasmid (Clontech). Induction of chaperone expression was initiated at least one hour before induction of eIF2α or eIF2αS51D by addition of 0.5 ng/ml anhydrotetracycline. 15N and 13C labeling of eIF2α constructs were performed by expression in minimal media, supplemented with 15NH4Cl and 13C6-d-glucose as the sole nitrogen and carbon source, respectively. Partial 2H labeling was achieved by replacing the H2O in the minimal media with 2H2O.
Following expression, cells were collected, spun down, and re-suspended in lysis buffer (10 mM Na-phosphate buffer (pH 7), 150 mM NaCl, 0.01% NaN3, 7 mM β-mercaptoethanol, 0.1 mM AEBSF, a Complete EDTA-free tablet (Roche Diagnostics) per 50 ml and 1 mg/ml lysozyme). Cells were then subjected to sonication. Proteins were purified using His-tag affinity chromatography on TALON CellThru resin (Clontech), followed by Gel Filtration Chromatography (GFC) on either a Superdex 75 (GE Healthcare) or an ENrich SEC 650 (BioRad) column in 10 mM Na-phosphate buffer (pH 7), 150 mM NaCl, 0.01% NaN3, 2 mM DTT, 0.1 mM AEBSF and 1 mM EDTA. Analytical GFC was performed on a Superdex 75 (GE Healthcare), as above, loading 300 μl of eIF2α at 15 μM concentration. When necessary, eIF2Bβ and eIF2α-CTD were separated from their GB1-His6-tags by cleavage with TEV protease. Tag was removed by using His-tag affinity chromatography on TALON CellThru resin (after exchanging into buffer lacking DTT and EDTA), by using GB1-affinity chromatography on IgG resin (GE Healthcare), or by size-exclusion chromatography in the manner previously described. eIF2α-NTD and eIF2α-NTDS51D purifications were performed in buffers containing 300 mM NaCl. GH-eIF2Bδ was expressed in Rosetta2(DE3) cells at 37°C for 3 h. as inclusion bodies. Following expression, cells were collected, spun down, and re-suspended in lysis buffer. Cells were then subjected to sonication. The insoluble fraction was washed with lysis buffer and dissolved in 8M urea. Proteins were purified using His-tag affinity chromatography on TALON CellThru resin (ClonTech) under denaturing conditions. Refolding was done by serial dialysis into Buffer A150, supplemented with 20% glycerol. eIF2Bδ was then mixed with eIF2Bα and eIF2Bβ, followed by size-exclusion chromatography on an ENrich SEC 650 (BioRad) column in Buffer A150, to separate eIF2Breg from free subunits. PKR was purchased from SignalChem and used to phosphorylate eIF2α following the manufacturer's instructions.
Molecular docking
Closed conformation of eIF2α
The model for the closed conformation of eIF2α (Figure 1E), used to generate the model shown in Figure 6D, was generated from the structure of human eIF2α (1Q8K.pdb) (26), based on the intramolecular contact interface mapped using CSPs from the NMR deletion analysis (Figure 1), comparing spectra of full-length eIF2α with those of its individual domains, eIF2α-NTD and -CTD. Modeling was done interactively in Molmol by allowing the two domains to rotate with respect to each other as rigid bodies along the flexible linker between them.
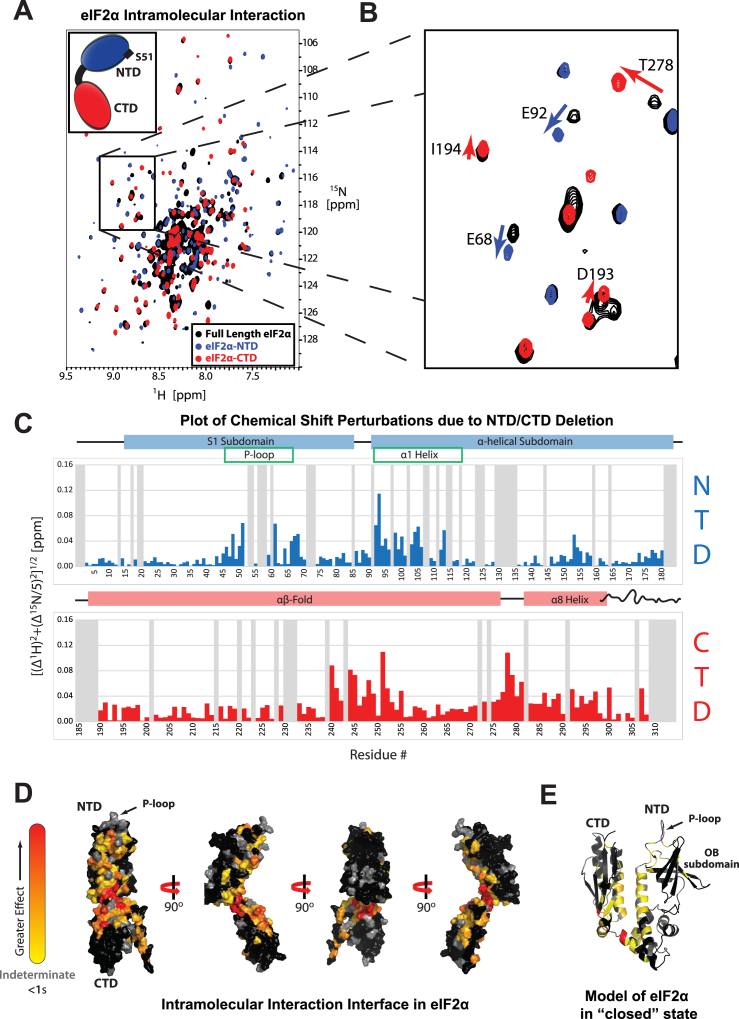
Figure 1.
Intramolecular interaction in eIF2α. (A) TROSY-HSQC spectra of 2H/15N-labeled eIF2α (black), eIF2α-NTD (blue), and 2H/15N/13C-labeled eIF2α-CTD (red). (B) Boxed area in (A). The arrows indicate movement of peaks away from their full length WT positions. (C) Plot of chemical shift perturbations (CSP) between eIF2α and its individual domains. Gray bars represent indeterminate residues for which no analysis could be performed. Key structural features are shown above the plot for reference, with the extreme end (aa 302–314) of the C-terminal tail (CTT) shown as a wavy line. (D) Residues affected by the intramolecular interaction mapped onto the structure of eIF2α, colored from yellow (>1 standard deviation (σ)) to red (>3σ). Indeterminate residues are colored gray. Residues <1σ are colored black. Residues 302–314 were absent in the NMR structure and are thus not displayed here. (E) Model for the closed conformation of eIF2α shown in ribbon and colored as in panel D.
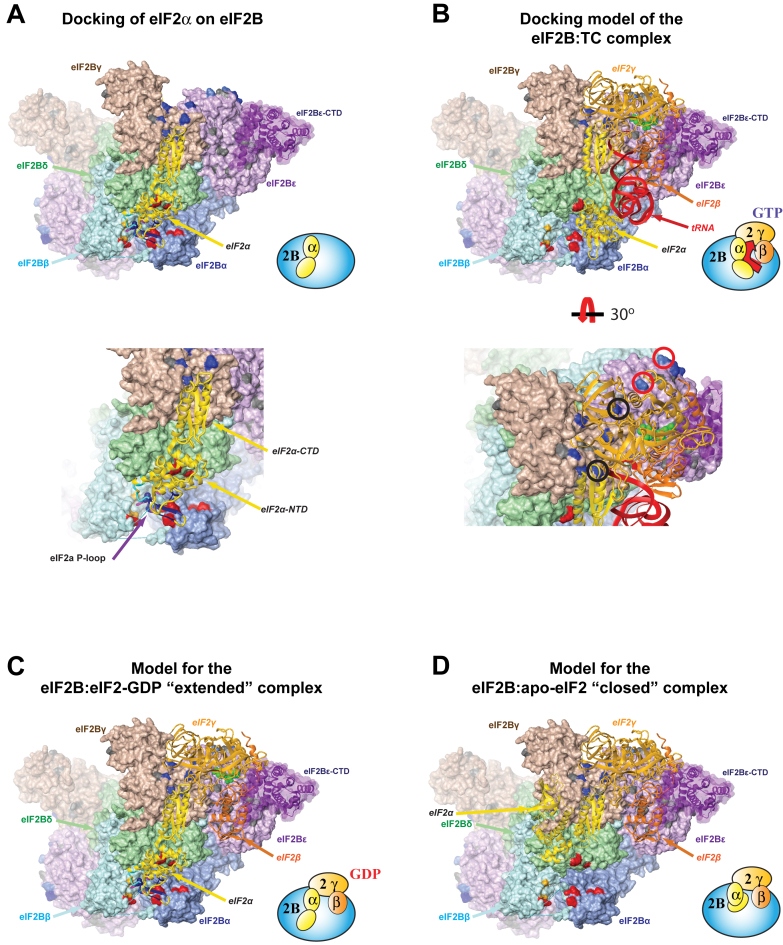
Figure 6.
Structural model for the eIF2B/eIF2 interaction. (A) Model of the eIF2B•eIF2α interaction, where eIF2α-NTD is docked into the eIF2B regulatory subcomplex pocket (6). eIF2B subunits are shown in surface representation; eIF2α is shown as ribbon. eIF2Bα/β/δ residues shown to cross-link to both phosphorylated and unphosphorylated eIF2α are red; residues in eIF2Bβ that cross-link only to unphosphorylated eIF2α are orange (6). eIF2Bγ/ϵ residues shown to cross-link to eIF2γ in eIF2B•apo-eIF2 complexes are navy, except the two eIF2Bϵ residues with lower efficiency of cross-linking to eIF2(α-P)-GDP than to apo-eIF2 (6), which are light blue. The sites of CACH/VWM mutations in eIF2Bγϵ are gray. Residues in eIF2α are colored according to the following scheme: (i) residues in the P-loop are purple, unless colored as detailed below; (ii) residues affected by eIF2Bα binding (see Figure 3) are navy; (iii) residues affected by eIF2Bβ binding (see Figure 4) are cyan and (iv) residues affected by both are blue. Inset: zoomed-in view of the eIF2Breg pocket, with eIF2α-NTD, -CTD, and P-loop labeled. (B) Model of the eIF2B•TC complex, oriented and aligned using cross-linking data (6), as well as a proposed docking of the eIF2Bϵ-CTD catalytic domain (purple, semi-transparent). eIF2B coloring is as in panel (A). TC is shown as ribbon. The nucleotide is displayed in green. Inset: zoomed-in view of the eIF2γ-ibnding surface of eIF2Bcat, rotated 30 degrees as shown. The two eIF2Bγ/ϵ residues shown to cross-link equally well to both apo-eIF2 and eIF2(α-P)-GDP (6) are navy and marked with black circles. The two eIF2Bϵ residues with lower efficiency of cross-linking to eIF2(α-P)-GDP than to apo-eIF2 (6) are light blue and marked with red circles. (C) Model of the eIF2B complex with nucleotide-bound eIF2 in ‘extended’ conformation, obtained by merging the model of the eIF2B•eIF2α complex shown in (A) and the model of the eIF2B•TC complex shown in (B). eIF2B and eIF2α coloring is as in panel (A). (D) Model of the eIF2B•apo-eIF2 complex, in which eIF2β/γ remain docked as in the eIF2B•TC complex shown in (B), but eIF2α is ‘closed’ as per the intramolecular interaction displayed in Figure 1. eIF2B coloring is as in panel (A). Cartoon representation of the complexes is shown on the bottom right of each panel.
eIF2α binding in the eIF2Breg regulatory subcomplex pocket
The structure of human eIF2α (1Q8K.pdb) (26) was docked in the eIF2Breg pocket of the structure of S. pombe eIF2B (5B04.pdb) (6) guided by our NMR interaction mapping results for human eIF2α with eIF2Bα (Figure 3) and eIF2Bβ (Figure 4), as well as by published cross-linking data between S. pombe eIF2α and eIF2Bα, β and δ (6). The proteins were docked as rigid bodies, allowing some side-chain, but no backbone steric clashes.
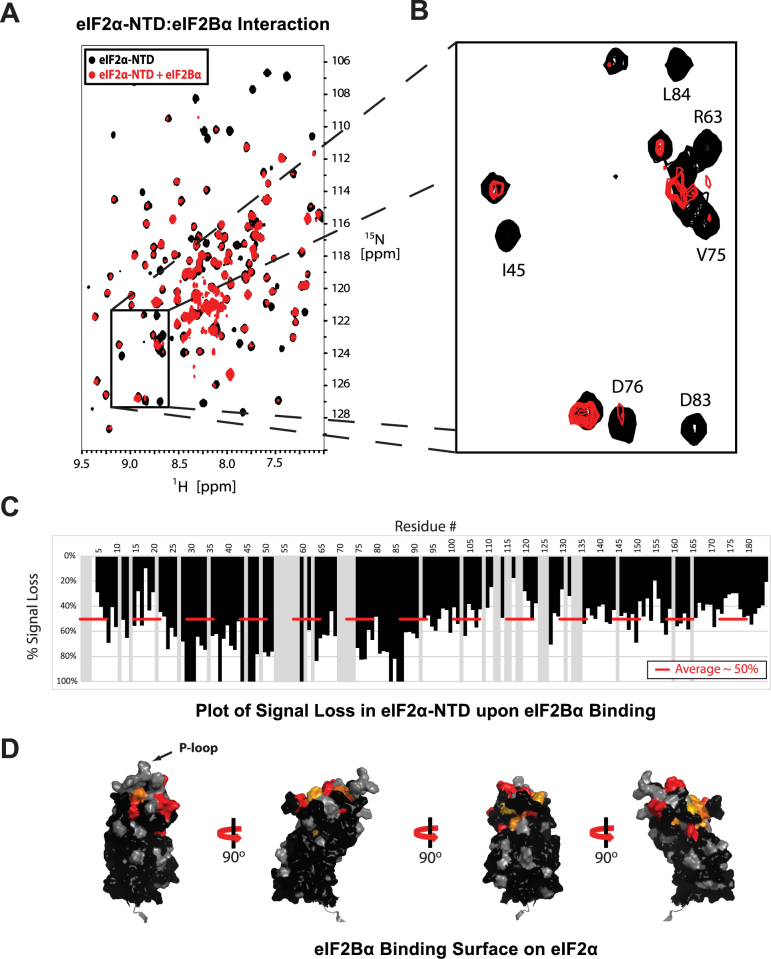
Figure 3.
eIF2α-NTD surfaces affected by binding to eIF2Bα. (A) TROSY-HSQC spectra of 2H/15N-labeled eIF2α-NTD, free (black) and in the presence of excess unlabeled eIF2Bα (red). (B) The boxed area in (A). Some of the peaks experiencing selective signal loss are labeled. (C) Plot of signal loss for residues in eIF2α-NTD. Gray bars represent indeterminate residues for which no analysis could be performed. (D) Residues experiencing selective signal loss mapped onto the structure of eIF2α-NTD, colored from yellow (>1σ) to red (>3σ). Indeterminate residues are colored gray. Residues <1σ are colored black.
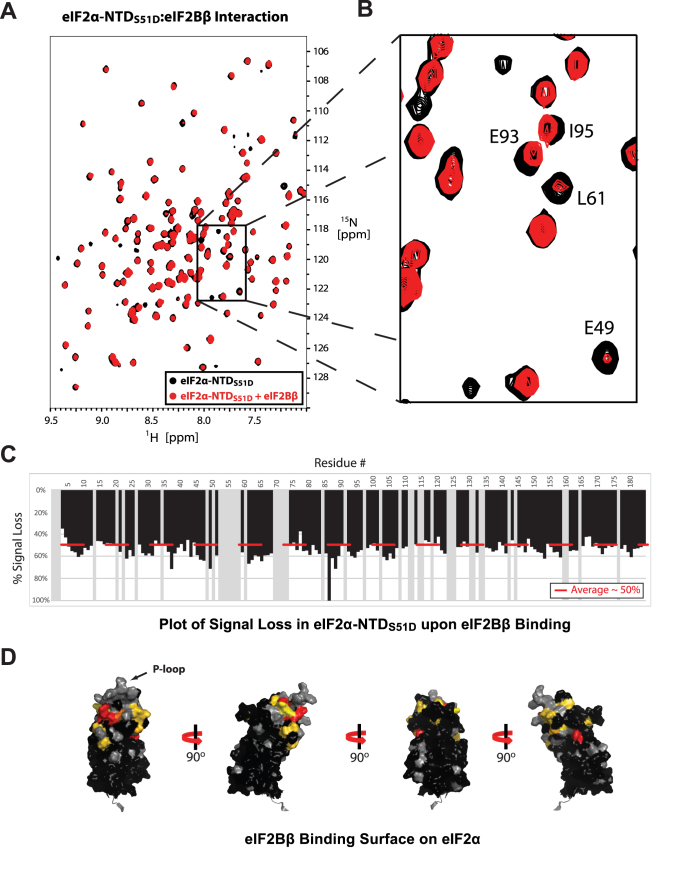
Figure 4.
eIF2α-NTD surfaces affected by binding to eIF2Bβ. (A) TROSY-HSQC spectra of 2H/15N-labeled eIF2α-NTDS51D, free (black) and in the presence of excess unlabeled eIF2Bβ (red). (B) The boxed area in (A). Some of the peaks experiencing selective signal loss and/or chemical shift perturbations are labeled. (C) Plot of signal loss for residues in eIF2α-NTDS51D. Gray bars represent indeterminate residues for which no analysis could be performed. (D) Residues experiencing selective signal loss mapped onto the structure of eIF2α-NTD, colored from yellow (>1σ) to red (>2σ). Residues experiencing chemical shift perturbations of >0.01ppm, but without significant selective signal loss (<1σ), are colored in gold. Indeterminate residues are colored grey. Residues <1σ are colored black. Comparison to the intramolecular surface in Figure 1D reveals overlap between the eIF2Bβ binding surface and the intramolecular interface.
Distance restraints for NMR mapping effects were set to 5 Å, with distances up to 10 Å considered allowed, to account for uncertainties due to the fact that we were docking human eIF2α to S. pombe eIF2B as rigid bodies. Distances >10 Å were considered violations. Since the observed NMR effects can be either due to direct contact or indirect, distance violations for residues on the periphery of, or away from, the main contact surface could be due to indirect effects. Distance violations for residues at the main contact surfaces were not allowed. Distance restraints for cross-links were set to 10 Å, based on the length of the cross-linker pBpa (6). Distances up to 15 Å were considered allowed, to account for uncertainties, as above. The docking model in (6) was also analyzed and used as a control. No distance restraints derived from cross-linking were violated (>15 Å) in either docking model (see Supplementary Table S1).
eIF2B complex with TC
We used the TC structure from the S. cerevisiae 48S pre-initiation complex (3JAP.pdb) (32), which is very similar to the structure of archaeal aIF2-TC (3V11.pdb) (33), but more complete. eIF2B and TC were docked as rigid bodies, using the published cross-linking data between Komagataella pastoris eIF2α and S. pombe eIF2Bγϵ (6). TC orientation was guided by the mapped interaction of eIF2α with the eIF2Breg pocket described above, as well as by maximizing contacts and surface complementarity. Cross-links between S. cerevisiae eIF2 and eIF2B (5) and charge complementarity (electrostatic surface potential) were not considered for the docking, but were used for validation of the resulting model. Since eIF2Bϵ-CTD is attached to the rest of the protein through a flexible linker and is not resolved in the structure of S. pombe eIF2B (5B04.pdb) (6), we docked the structure of human eIF2Bϵ-CTD (3JUI.pdb) (34) onto the eIF2B•TC complex, on a surface of eIF2Bϵ adjacent to the binding site for the eIF2γ G-domain and rich in solvent-exposed VWM/CACH mutations. eIF2Bϵ-CTD was oriented with the residues near its N-terminus, known to be important for catalysis (35), facing eIF2γ. The molecules were docked as rigid bodies, allowing some side-chain, but no backbone steric clashes.
eIF2B complex with eIF2 in an ‘extended’ conformation
Starting from the complexes of eIF2B with TC and with eIF2α-NTD docked in the eIF2Breg pocket, we held in place the entire TC and the eIF2α-NTD, while eIF2α-CTD was rotated toward the orientation of eIF2α-CTD in the eIF2B•TC complex, using the flexible linker between the NTD and CTD as a hinge. The eIF2γ binding surface of eIF2α-CTD was readily able to reach the corresponding eIF2α-CTD binding surface on eIF2γ. For simplicity, in Figure 6A, eIF2α-CTD is shown already in the same conformation as in the eIF2B•eIF2 complex in ‘extended’ conformation (Figure 6C).
eIF2B complex with eIF2 in ‘closed’ conformation
Starting from the complex of eIF2B with eIF2 in an ‘extended’ conformation, we replaced the structure of eIF2α with the model for eIF2α in a ‘closed’ conformation (Figure 1E) by aligning their CTDs. In the resulting model, eIF2α-NTD clashed with eIF2Bγ. We rotated eIF2α away from the eIF2Bγ surface to eliminate steric clashes, while maintaining the contact between eIF2α and eIF2γ.
Potential ligand-binding sites in eIF2B subunits
To model the potential ligand-binding sites in the interdomain pockets of eIF2Bα, β and δ, we aligned the structure of the ribose-1,5-bisphosphate isomerase from T. kodakarensis KOD1 (tkRBPI) in complex with α-d-ribose-1,5-bisphosphate (PRPP) (3VM6.pdb) (36), to the hexameric eIF2Bα2(βδ)2 regulatory subcomplex in the S. pombe eIF2B structure (6). To model the potential ligand-binding sites in eIF2Bγ and ϵ, we aligned the structure of the pyrophosphorylase domain of the bifunctional enzyme GlmU from Escherichia coli in complex with Uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) bound in the pyrophosphorylase active site (2OI7.pdb) (37), to the homologous domain of each of the eIF2Bγ and ϵ subunits in the S. pombe eIF2B structure (6).
RESULTS
eIF2α-NTD and -CTD interact with each other
The structure of human eIF2α (1Q8K.pdb) (26) reveals the presence of two folded domains, an NTD (aa 1–183) and a CTD (aa 188–280), connected to each other by a flexible linker, followed by a C-terminal tail (CTT). eIF2α-NTD consists of two subdomains: 1) an oligonucleotide/oligosaccharide-binding (OB)-fold subdomain formed by residues 18–85, and 2) an α-helical subdomain formed by residues 91–183. The OB-fold subdomain has an extended B3/B4 loop (numbering of secondary structure elements in eIF2α is from (26)), called the phosphorylation loop (P-loop) since it contains the regulatory phosphorylation site. This stretch of residues (His46 to Arg66) is highly conserved among eukaryotes and is highly positively charged. Most of eIF2α-CTT is intrinsically disordered, except for an α-helical region (residues 281–288).
In order to test whether eIF2α-NTD and -CTD interact with each other, we combined NMR chemical shift perturbation (CSP) assays (27,29,30) with deletion analysis, comparing peak movements between spectra of full length eIF2α and spectra of eIF2α-NTD and -CTD alone. This approach is effective for observing dynamic interactions (28,38). We collected transverse relaxation optimized spectroscopy heteronuclear single-quantum coherence (TROSY-HSQC) NMR spectra of 2H/15N-labeled full length eIF2α and its individual domains. Comparison of the spectra of the individual domains with that of full length eIF2α showed a large number of moving peaks in both domains, confirming the presence of an intramolecular interaction (Figure 1ABC). We next mapped the affected residues on the structure of eIF2α, revealing large contiguous surfaces on both eIF2α-NTD and eIF2α-CTD that included the P-loop as well as the CTT (Figure 1D) We then oriented the two domains based on the observed CSPs, yielding a model for eIF2α in the ‘closed’ conformation (Figure 1E). The ‘closed’ eIF2α conformation is fully compatible with the structure of archaeal aIF2-TC (3V11.pdb) (33), with no steric clashes between eIF2α-NTD and the rest of eIF2 (Supplementary Figure S3A). Furthermore, the ‘closed’ eIF2α conformation places eIF2α-NTD and eIF2γ in proximity to each other, which is consistent with reported crosslinking between them (5,39).
Phosphorylation destabilizes the eIF2α intramolecular interaction
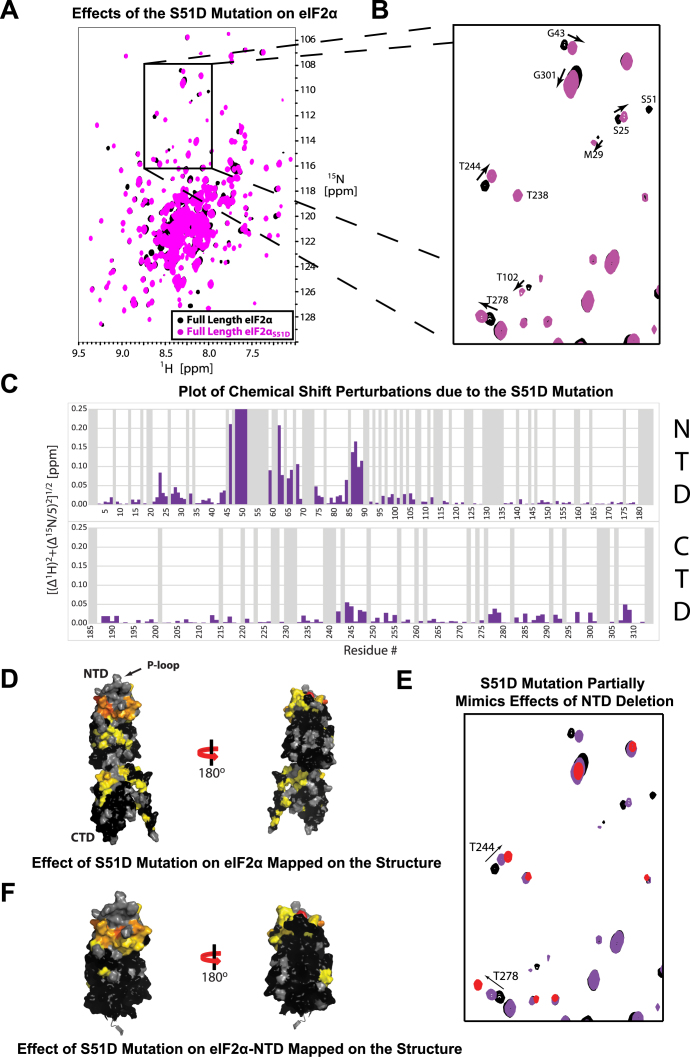
We next sought to test whether phosphorylation affects the intramolecular interaction. Once again we used a CSP assay, this time comparing spectra of 2H/15N-labeled WT eIF2α and a phosphomimetic mutant, eIF2αS51D (Figure 2A and B). The S51D mutation in mammalian eIF2α has been extensively studied and shown to cause the same in vivo effects as eIF2α phosphorylation (see e.g. (40–42)). A number of residues in the CTD were affected by the phosphomimetic mutation (Figure 2C). Mapping these residues on the structure of eIF2α revealed that the same surface on eIF2α-CTD is affected by the S51D mutation (Figure 2D) as by deleting the NTD (Figure 1D), but with a smaller magnitude of chemical shift changes. The mapped surfaces in Figures 1D and 2D are shown side-by-side in Supplementary Figure S1A and B, respectively. Residues throughout the NTD were affected by the S51D mutation, with the largest chemical shift perturbations occurring in the OB-fold and extending into the first half of the α1 helix (Figure 2D). CSP deletion analysis of eIF2αS51D identified the same intramolecular contact interface in eIF2αS51D (Supplementary Figure S1C) as in WT eIF2α (Supplementary Figure S1A), but with a smaller magnitude of chemical shift changes. Therefore, eIF2αS51D appears intermediate between WT eIF2α (closed state) and the free domains (open state), indicating that the S51D mutation destabilizes the intramolecular interaction between eIF2α-NTD and CTD.
Figure 2.
Effects of the phosphomimetic S51D mutation in eIF2α. (A) TROSY-HSQC spectra of 2H/15N-labeled eIF2α (black) and a phosphomimetic mutant, eIF2αS51D (purple). (B) The boxed area in (A). The arrows indicate movement of peaks away from their WT positions. (C) Plot of chemical shift perturbations between eIF2α and eIF2αS51D. Gray bars represent indeterminate residues for which no analysis could be performed. (D) Residues affected by the phosphomimetic mutation mapped onto the structure of eIF2α, colored from yellow (>1σ) to red (>3σ). Indeterminate residues are colored gray. Residues <1σ are colored black. Comparison to Figure 1D reveals that the effects of the phosphomimetic mutation overlap substantially with the intramolecular surface. (E) The same boxed area as in (B), but with the addition of a third overlain spectrum of 2H/15N/13C-labeled eIF2α-CTD (red). Peaks corresponding to residues affected by the phosphomimetic mutation displayed a consistent pattern of movement, where they are located halfway between the peak positions in the eIF2α spectrum (‘bound’ state) and the eIF2α-CTD spectrum (‘free’ state). Arrows indicate movement of peaks away from their full length WT positions. (F) Residues affected by the phosphomimetic mutation mapped onto the structure of eIF2α-NTD. Coloring scheme is the same as in (D).
We next looked to understand whether phosphorylation does indeed destabilize and/or modify the contact interface between eIF2α-NTD and CTD. Since CSPs in eIF2α-NTD can be both a direct result of the S51D mutation and an indirect result of changes in the interaction between the NTD and CTD, we used eIF2α-CTD to analyze the effects of phosphorylation on the intramolecular interaction. Peaks in the eIF2α-CTD spectrum served as a reference for the ‘free’ state, while peaks in the full-length WT eIF2α spectrum served as a reference for the ‘bound’ state. When comparing residues in the CTD of eIF2αS51D, we observed a pattern of peak movement along a line from ‘bound’ to ‘free.’ Analysis of moving peaks in eIF2α-CTD showed that the peaks in the eIF2αS51D mutant were approximately halfway between the ‘free’ and ‘bound’ states, indicating that the S51D mutation destabilizes the intramolecular interaction by inducing eIF2α to spend more time open (Figure 2E, Supplementary Figure S2). Some of these peaks lay slightly off the connecting line, indicating that the S51D mutation might modulate the nature of the interaction, in addition to simply destabilizing it. No specific surface in eIF2α-CTD appears differentially affected by the S51D mutation, since no correlation could be found between any one surface on eIF2α-CTD and the degree of deviation from the line connecting the ‘free’ and ‘bound’ states. Consistent with our conclusion that the S51D mutation destabilizes the interaction between eIF2α-NTD and -CTD, eIF2αS51D has larger apparent MW than WT eIF2α, indicative of a change in the average shape of the molecule (Supplementary Figure S3B).
To test whether the effects of the S51D mutation on the intramolecular interaction in eIF2α are the same as those of S51 phosphorylation, we phosphorylated 2H/15N-labeled WT eIF2α in vitro with PKR and collected a TROSY-HSQC spectrum. While protease contamination in the PKR stock interfered with the quality of the NMR spectra, the results showed that phosphorylation causes the same CSP patterns in eIF2α-CTD as the S51D mutation, even causing the peaks to move slightly further toward their positions in free eIF2α-CTD (Supplementary Figure S2E).
In order to isolate the direct effect of phosphorylation on eIF2α-NTD from the secondary effect of modulation of the intramolecular interaction, we compared TROSY-HSQC spectra of 2H/15N-labeled WT eIF2α-NTD and phosphomimetic eIF2α-NTDS51D (Supplementary Figure S2B). Consistent with our results from the comparison of full length WT and phosphomimetic eIF2α, we noticed substantial perturbations throughout the OB-fold and extending into the α1 helix (Figure 2F). It is clear that the S51D mutation affects a large part of the surface on eIF2α-NTD responsible for the intramolecular interaction (compare Figures 1D and 2F). Therefore, it appears that phosphorylation not only affects the residues in the immediate vicinity of S51, but also changes the P-loop conformation, and likely also the P-loop's interactions with the rest of eIF2α-NTD, potentially magnifying its effect on the eIF2α-NTD/CTD intramolecular interaction.
eIF2Bα and eIF2Bβ bind to adjacent surfaces on eIF2α-NTD
As described above, eIF2Breg presents a pocket formed by the NTDs of its constituent subunits, eIF2Bα, β and δ. Cross-linking data indicate that eIF2α-NTD binds in this pocket (6), though its orientation and contact interfaces remain unknown. In order to unambiguously determine the orientation, we sought to map the eIF2Bα and eIF2Bβ binding surfaces on the structure of eIF2α. We were unable to study binding to eIF2Bδ by NMR due to its insufficient solubility. HSQC spectra were initially obtained for 15N-labeled eIF2αS51D, free and mixed with excess eIF2Bα. Addition of eIF2Bα caused nearly complete loss of signal due to relaxation, consistent with formation of a large molecular weight complex (data not shown). This result confirmed that eIF2αS51D binds to eIF2Bα, but made any mapping impossible.
When the experiment was repeated using 2H/15N-labeled eIF2α-NTD and collecting TROSY-HSQC spectra, we were able to observe the spectrum of eIF2α-NTD in the presence of eIF2Bα (Figure 3AB). However, rather than detecting a change in chemical shift positions, we observed a selective loss in signal intensity for certain peaks compared to the domain as a whole, ranging from partial to complete loss of signal (Figure 3C). The selective loss in signal is due to cross-relaxation between the free and bound states of eIF2α. The phenomenon is seen in cases like this one, with a weak interaction, where the labeled protein is in fast equilibrium between a free state and a complex that is too large to observe by NMR, even with deuteration and TROSY (31). The spectrum corresponds to the free state of the protein, but the signal is reduced because the protein spends part of the time in a large slowly-tumbling complex characterized by fast relaxation (leading to loss of signal). Peaks that correspond to residues experiencing different environments in the free and bound states, and therefore having different chemical shifts, demonstrate stronger cross-relaxation effects (i.e. signal intensity loss) (31). We mapped those peaks with the largest drops in relative signal intensity on the structure of eIF2α, revealing the eIF2Bα-binding surface on eIF2α-NTD (Figure 3D). There was little overlap between this intermolecular surface and the previously identified intramolecular contact surface with eIF2α-CTD, except for the P-loop, which lies on the periphery.
The experiment was repeated for 2H/15N-labeled eIF2α-NTDS51D as well as for both 2H/15N-labeled full length WT eIF2α and eIF2αS51D (Supplementary Figure S4). Both the degree of binding and the surface were similar among all four constructs, indicating that neither the S51D mutation, nor the presence of eIF2α-CTD affected the observed eIF2α/eIF2Bα interaction (Supplementary Figure S4), consistent with the lack of significant overlap between the eIF2α/eIF2Bα interface and eIF2α-NTD/CTD interface. The similar nature of the interactions of WT and phosphomimetic eIF2α with eIF2Bα indicates that the eIF2α-NTD/eIF2Bα interaction does not mediate any direct effects of eIF2α phosphorylation.
We next turned to mapping the eIF2Bβ contact surface on eIF2α-NTD. Once again TROSY-HSQC spectra were collected for 2H/15N-labeled WT eIF2α-NTD and eIF2α-NTDS51D, both free and mixed with excess eIF2Bβ. The same phenomenon of selective signal loss was observed as with eIF2Bα, however in this case we also observed CSP effects in a number of peaks. CSPs tended to be more prevalent at lower ratios of eIF2Bβ to eIF2α-NTDS51D, whereas signal loss was more prominent at higher eIF2Bβ concentrations (data not shown), consistent with eIF2α-NTDS51D spending more time in a large complex with faster relaxation. The eIF2Bβ contact surface on eIF2α-NTD is adjacent to the eIF2Bα contact surface (compare Figures 3D and 4D) and overlaps with the intramolecular eIF2α-NTD/CTD interface (compare Figures 1D and 4D). Once again, binding of eIF2Bβ to eIF2α-NTD and eIF2α-NTDS51D caused similar changes in peak intensities (compare Supplementary Figures S5AB and S4AB). This suggests that, as was the case for the eIF2α-NTD–eIF2Bα interaction, phosphorylation seems to have no direct effect on the eIF2α-NTD–eIF2Bβ interaction. Unlike for eIF2Bα, however, we were unable to observe significant binding of eIF2Bβ to either full length WT or phosphomimetic eIF2α (Supplementary Figure S5CD). This indicates that eIF2α-CTD must be interfering with eIF2α-NTD binding to eIF2Bβ, as would be expected from the overlap between the intramolecular eIF2α-NTD/CTD contact surface and the eIF2α-NTD/eIF2Bβ contact surface.
In order to test for possible direct binding of eIF2α-CTD to eIF2Bα or eIF2Bβ, we collected TROSY-HSQC spectra of 2H/15N/13C-labeled eIF2α-CTD both free and in the presence of excess eIF2Bα, as well as HSQC spectra of 15N-labeled eIF2α-CTD both free and in the presence of excess eIF2Bβ. No binding was observed between eIF2α-CTD and either eIF2Bα or eIF2Bβ (data not shown).
eIF2Bα, eIF2Bβ and eIF2Breg show no significant preference for phosphomimetic over WT eIF2α-NTD
eIF2B missing the eIF2Bα subunit can no longer be inhibited by eIF2(α-P)-GDP (22–24). It is therefore a commonly held (and logical) belief that the tight, non-productive binding of eIF2(α-P)-GDP to eIF2B is mediated, at least in part, by a ‘direct effect’ characterized by a greater affinity of eIF2Bα for the phosphorylated P-loop. Our NMR data suggest, however, that eIF2Bα binding to eIF2α-NTD and eIF2α-NTDS51D is at least qualitatively similar and certainly not sufficiently different to explain the substantial increase in affinity expected for competitive inhibition.
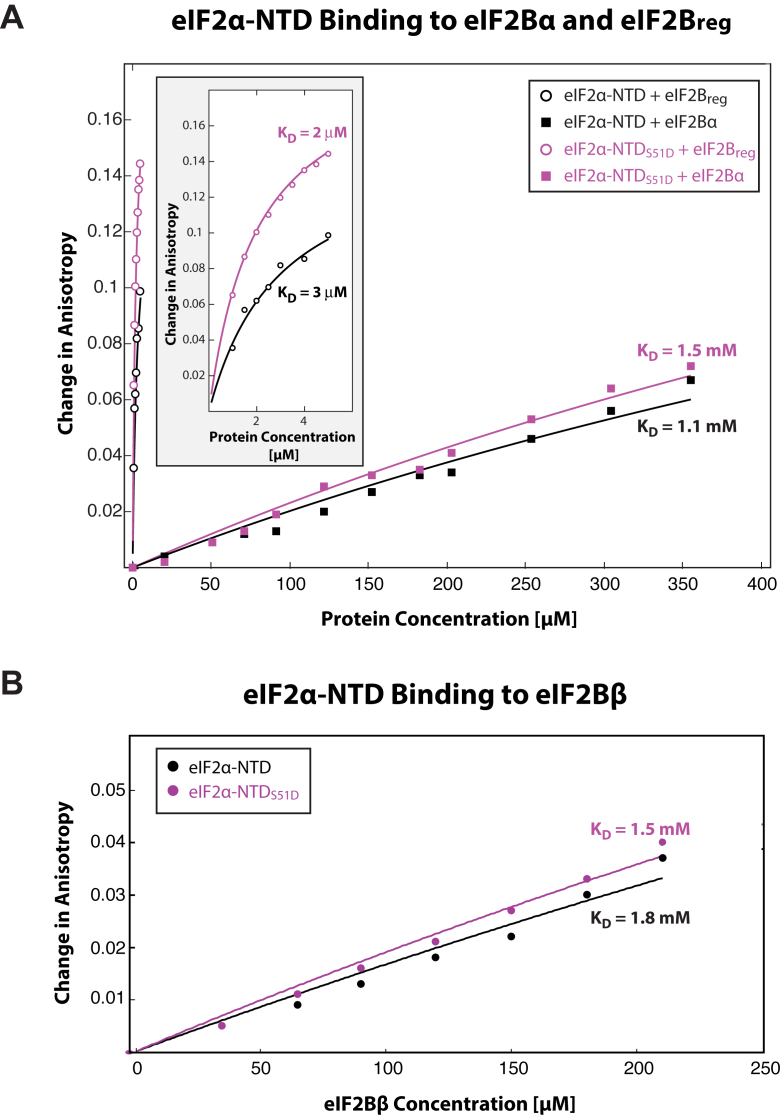
In order to more directly test whether eIF2Bα does in fact bind eIF2α-NTDS51D more tightly than WT eIF2α-NTD, we sought to obtain KDs of these interactions using fluorescence anisotropy. eIF2α-NTD and eIF2α-NTDS51D were labeled with fluorescein-5-maleimide using existing surface exposed cysteines and unlabeled eIF2Bα was titrated in up to the maximum achievable concentration. Confirming our NMR data, we found that eIF2Bα showed no preference for phosphomimetic over WT eIF2α-NTD (Figure 5A).
Figure 5.
eIF2Bα, eIF2Bβ, and eIF2Breg bind eIF2α-NTD and eIF2α-NTDS51D with similar affinities. Plot of the changes in fluorescence anisotropy of fluorescein-labeled eIF2α-NTD (black) and eIF2α-NTDS51D (purple) in the presence of increasing concentrations of unlabeled eIF2Bα (solid squares) and eIF2Breg (circles) (A), and eIF2Bβ (B). Calculated KDs are shown next to the corresponding titration curves. Zoomed-in eIF2Breg titration curves are shown in the inset of panel A. No substantive difference in affinity was observed for eIF2Bα, eIF2Bβ, or eIF2Breg as a function of the phosphorylation state of eIF2α-NTD.
We next sought to determine whether the ‘direct effect’ of phosphorylation may be mediated by eIF2Bβ, rather than by eIF2Bα. Titrations were performed in the same manner as for eIF2Bα. We observed no substantive preference of eIF2Bβ for phosphomimetic over WT eIF2α-NTD either (Figure 5B). From this data it can be concluded that, if it does exist, any ‘direct effect’ of the phosphate group on the binding of eIF2α-P to eIF2Breg would have to be mediated either by eIF2Bδ or by an aggregate interaction with two or more of the eIF2B regulatory subunits.
In order to definitively test for the presence of a direct effect of phosphorylation on increasing the affinity of eIF2α binding to the intact eIF2Breg pocket, we compared eIF2α-NTDS51D and WT eIF2α-NTD binding to eIF2Breg reconstituted from individual subunits (Figure 5A, Supplementary Figure S5E). While eIF2α-NTDS51D had slightly higher affinity than WT eIF2α-NTD, the difference was less than two-fold, indicating that while there is some direct effect of eIF2α phosphorylation on binding to eIF2B, the indirect effect is predominant and accounts for most of the increase in affinity.
Orientation of eIF2α in the eIF2Breg pocket
The eIF2α contact surfaces for eIF2Bα and eIF2Bβ identified here by NMR are consistent with most of the cross-linking data in reference (6), but not compatible with the model for the eIF2α/eIF2B interaction, which suggested that eIF2α-NTD enters the eIF2Breg pocket from the side through the groove between eIF2Bβ and eIF2Bδ. While, as expected, no violations were observed in that model for distance restraints from cross-linking, between one-third (eIF2Bα) and one-half (eIF2Bβ) of our NMR distance restraints were violated (see Materials and Methods for description of the docking and analysis). Docking eIF2α-NTD into the eIF2Breg pocket using our NMR data in addition to cross-linking indicated that it is more centrally positioned in the eIF2Breg pocket than suggested in reference (6) (Figure 6A). The resulting complex is compatible with the cross-linking data, yielding an equally good fit to the cross-linking data as the model in reference (6): no distance restraint violations and the same average of all distances as the model in reference (6). However, the new docking model yielded a far superior fit to our NMR mapping results (Supplementary Table S1). The only distance violations were for several eIF2α residues affected by eIF2Bβ binding, which are located away from the main contact surface. Thus, these residues could be affected indirectly by eIF2Bβ binding. Another possible explanation for some of these effects is that they could be due to contacts with the long loop in eIF2Bβ-NTD, where Gcn− mutations have been found (21). A portion of this loop is seen contacting eIF2Bα, whereas the rest of it remains disordered in the eIF2B crystal structure (6).
In the new orientation, the eIF2α P-loop faces the two eIF2Bβ residues that cross-link only to unphosphorylated eIF2α (shown in orange in Figure 6A). Therefore, our results indicate that the loss of cross-linking at these positions upon eIF2α phosphorylation is due to the ‘relocation’ of the P-loop upon S51 phosphorylation, whereas the overall eIF2α orientation remains similar.
Architecture of the eIF2B•eIF2 complex
The most important implication of the eIF2α orientation determined here is that eIF2α-CTD is directed toward the more proximal eIF2Bcat dimer in eIF2B and ends up in proximity to the eIF2Bγ surface cross-linked to eIF2γ. This observation indicates that eIF2 can simultaneously contact eIF2Breg (through eIF2α-NTD) and eIF2Bcat (through eIF2γ), in a two-site interaction, consistent with a wealth of existing data.
We proceeded to model the position of eIF2 on eIF2B. The structure of eIF2 is rather dynamic (see e.g. (43)), which could compromise the quality of docking. We reasoned that the eIF2-GTP•Met-tRNAi ternary complex (TC) is the best candidate for docking to eIF2B as: (i) it is the true product of the eIF2B-catalyzed process (44,45) and is bound rather tightly to eIF2B ((45), reviewed in (3)); (ii) it is more rigid than free eIF2 (43) and (iii) by being larger and more complex, it provides a greater number of steric constraints. The most complete TC structure to date is that from the Cryo-EM structure of the S. cerevisiae pre-initiation complex (3JAP.pdb) (32). The TC was docked to the eIF2B surface shown in Supplementary Figure S6A, between the eIF2α- and eIF2γ-binding sites. First, the TC was positioned with eIF2γ facing the eIF2Bγϵ surfaces, where it was shown to cross-link (6), and oriented such that eIF2α faced in the direction of the eIF2Breg pocket, where eIF2α-NTD binds. The eIF2B/TC contacts were then optimized to yield the complex shown in Supplementary Figure S6B. The resulting complex displays a remarkable degree of surface complementarity, especially considering that there must be at least some differences between the structures of TC when bound to the ribosome and when bound to eIF2B, as well as between S. pombe and S. cerevisiae. The eIF2B•TC complex also shows good charge complementarity, as shown in Supplementary Figure S7A, with Met-tRNAi contacting positively charged surfaces. Finally, to further validate our complex, we compared it to recently reported cross-links between eIF2 and eIF2B (5). As can be seen in Supplementary Figure S7B, the positions of the cross-links are fully consistent with the docking model. Therefore, it is safe to conclude that we have identified the correct eIF2B/TC interface and that TC binds to eIF2B in roughly the orientation shown in Supplementary Figure S6B.
We then wondered whether we could identify the position of eIF2Bϵ-CTD (the catalytic domain) within the eIF2B•TC complex. eIF2Bϵ-CTD is connected to the rest of eIF2Bϵ by a flexible linker and is not visible in the S. pombe eIF2B crystal structure (6). However, it is known to contact eIF2γ (46), and if eIF2γ is docked onto the eIF2Bγϵ surface, it is likely that eIF2Bϵ-CTD also occupies a specific position in the eIF2B•eIF2 complex. We noticed that while the vast majority of the surface-exposed mutations in eIF2B subunits known to cause the CACH/VWM neurodegenerative disorder are now known to affect residues buried at intersubunit interfaces or at the interface with eIF2γ, one surface on eIF2Bϵ with a high density of CACH/VWM mutations remains unaccounted for. Remarkably, this surface is adjacent to the eIF2γ-binding surface and faces the GTPase domain (G-domain) of eIF2γ. Therefore, we docked eIF2Bϵ-CTD onto the eIF2Bϵ surface, with its N-terminal portion (where critically important residues have been identified (35)) facing the G-domain of eIF2γ, yielding the model for the eIF2B•TC complex shown in Figure 6B and Supplementary Figure S8A. eIF2Bϵ-CTD is shown semi-transparent in both Figure 6 and Supplementary Figure S8, to emphasize that its orientation is purely speculative.
A comparison of Figure 6A and B illustrates that eIF2α-NTD can easily bind in the eIF2Breg pocket (as in Figure 6A), while the rest of eIF2 is bound to eIF2B in the orientation shown in Figure 6B, forming a simultaneous two-site interaction. To obtain a model for the structure of the eIF2B•eIF2 complex with a two-site interaction, we merged the model for eIF2α binding in the eIF2Breg pocket (Figure 6A) with the model of the eIF2B•TC complex (Figure 6B), by aligning eIF2α-CTD from the eIF2B•eIF2α model to eIF2α-CTD from the eIF2B•TC model, as shown in Supplementary Figure S8B. This yielded the model shown in Figure 6C. Here, eIF2α is in an ‘extended’ conformation, where the intramolecular contacts between the NTD and the CTD are disrupted, both in order to reach the eIF2Breg pocket and to fit in it.
Complexes of eIF2B with inhibitor (eIF2(α-P)-GDP), substrate (eIF2-GDP) and reaction intermediates (apo-eIF2 and eIF2-GTP)
Having built a model for the enzyme-product complex, eIF2B•TC (Figure 6B), and shown that eIF2 can bind to eIF2B in a two-site interaction (Figure 6C), we proceeded to explore the architectures of the enzyme-inhibitor (eIF2B•eIF2(α-P)-GDP), enzyme-substrate (eIF2B•eIF2-GDP), and enzyme-reaction intermediates (eIF2B•apo-eIF2 and eIF2B•eIF2-GTP) complexes. It has been assumed that the eIF2B complexes with substrate and inhibitor must be somehow different, as a way to rationalize that one of them is nonproductive (20). However, there is no experimental evidence to support this assumption. In fact, the cross-linking of phosphorylated and unphosphorylated eIF2α to eIF2Breg yields very similar results, with most cross-links being identical and only two cross-links lost upon phosphorylation (6). Remarkably, no additional cross-links specific for eIF2α-P were observed. In our docking model of eIF2α in the eIF2Breg pocket, the two cross-links specific for unphosphorylated eIF2α face the P-loop, indicating that the loss of these cross-links is due to changes in the P-loop upon phosphorylation, while the rest of the complex remains the same. Therefore, we postulate that the complexes of eIF2B with the inhibitor, eIF2(α-P)-GDP and with the substrate, eIF2-GDP, are similar to each other, and likely also similar to the complex with eIF2-GTP (Figure 6C). Since eIF2α phosphorylation both destabilizes its closed conformation and stabilizes the eIF2B•eIF2(α-P)-GDP complex, we conclude that in the eIF2B•apo-eIF2 complex, eIF2α is in a closed conformation and does not bind in the eIF2Breg pocket.
eIF2γ cross-links to eIF2Bcat in both the eIF2B•apo-eIF2 and eIF2B•eIF2(α-P)-GDP complexes, but a subset of the cross-links are weaker in the eIF2B•eIF2(α-P)-GDP complex (6). This observation indicates that the conformation of eIF2 and its interface with eIF2B are different between the two complexes. A comparison of our eIF2B•TC docking model with the cross-linking results in (6) indicates that eIF2B residues that cross-link better in the eIF2B•apo-eIF2 complex face eIF2γ domain 1 (the G-domain) and are the ones distal from eIF2Breg (see Figure 6B, inset, and Supplementary Figure S6A). Therefore, in the eIF2B•apo-eIF2 complex, the eIF2γ G-domain likely moves away from domain 2 and toward these sites of cross-links in eIF2Bϵ. The domain 2 orientation could also change, causing a different eIF2α-CTD orientation.
To build a model for the complex of eIF2B with apo-eIF2, we used eIF2 with the eIF2α subunit in a ‘closed’ conformation, obtained using the NMR data reported here (Figure 1E). Modeling eIF2α in the ‘closed’ conformation onto the structure shown in Figure 6C would cause eIF2α-NTD to clash with eIF2B. To eliminate the steric clash, we rotated eIF2α slightly in a way that causes eIF2α-NTD to rotate away from eIF2B, thus yielding the model shown in Figure 6D. It should be noted that in building the model for the architecture of the eIF2B•apo-eIF2 complex, we kept the eIF2γ conformation unchanged because we do not have enough restraints to model the apo-eIF2γ conformation reliably. However, the differences in conformation between eIF2-GDP and apo-eIF2 in the eIF2B•eIF2 complex are likely what causes the apo-state of eIF2 to be compatible with the closed, but not the extended eIF2 conformation.
DISCUSSION
Implications of the eIF2α intramolecular interaction
In this work we show that eIF2α-NTD and -CTD interact with each other (Figure 1) and that eIF2α phosphorylation destabilizes this intramolecular interaction (Figure 2). The intramolecular interface in eIF2α overlaps significantly with the eIF2α/eIF2Bβ interface (Figure 4), and, based on the docking of eIF2α in the eIF2Breg pocket (Figure 6A), possibly also with the eIF2α/eIF2Bδ interface. This intramolecular interface must be disrupted in order for eIF2α both to reach and to bind into the eIF2Breg pocket (Figure 6C). Therefore, eIF2α phosphorylation increases the affinity of eIF2 for eIF2B mostly by destabilizing the autoregulatory intramolecular interaction within eIF2α and increasing the fraction of time eIF2α spends in an ‘extended’ conformation able to bind in the eIF2Breg pocket. While eIF2α phosphorylation has some direct effect on the binding to eIF2Breg, as is currently believed, such an effect appears to be no greater than 2-fold (Figure 5).
The eIF2α NMR solution structure (26) was solved using a solubility-enhanced mutant protein and at high salt, 350 mM Na2SO4 (47). One of the mutated residues in the construct used previously (L46) is affected by the intramolecular interaction. The deleted N-terminus is also at the interface, while the other two point mutations (A27Q and V71K) target residues in the direct vicinity of the intramolecular interface. Even under those conditions, the S51D phosphomimetic mutation had some effect on chemical shifts in the CTD (26), although far fewer residues were affected, and to a lesser degree than what we observed with the WT eIF2α at physiological salt, 150 mM NaCl (Figure 2). Therefore, even at high salt and with a solubility-enhanced mutant eIF2α, the intramolecular interaction was not fully disrupted.
It was reported by Dever et al. (48) that PKR binding induces conformational changes in eIF2α, exposing the P-loop, which is a prerequisite for S51 phosphorylation. Mutations in the P-loop that increased its mobility made it easier for PKR to phosphorylate S51, even when its affinity for eIF2α was compromised. The phosphomimetic S51D mutation causes similar effects on eIF2α to these mutations (compare Figure 2 with Supplementary Figure S5 from (48)), indicating that eIF2α phosphorylation likely stabilizes the exposed P-loop conformation imposed by PKR binding.
Structural mechanism of eIF2B catalysis
The eIF2B•eIF2 complex is in equilibrium between an eIF2B•apo-eIF2 state and a nucleotide-bound state. The newly identified orientation of eIF2α in the eIF2Breg pocket allows for simultaneous binding of both eIF2α-NTD to eIF2Breg, and eIF2γ to eIF2Bcat (two-site interaction) (Figure 6C). This simultaneous interaction is only possible when eIF2α is in the ‘extended’ conformation; in the ‘closed’ conformation, eIF2 would not be able to reach both binding sites on eIF2B simultaneously (one-site interaction mediated only by eIF2γ binding to eIF2Bcat) (Figure 6D). Our results indicate that the nucleotide-bound eIF2B•eIF2 state is in an extended conformation (Figure 6C), whereas the eIF2B•apo-eIF2 state is in a closed conformation (Figure 6D). Binding to eIF2B lowers the affinity of eIF2 for GDP at least 100-fold (49) and accordingly eIF2B has at least a 100-fold higher affinity for apo-eIF2 than for eIF2-GDP (49–51). Thus, eIF2B shifts the equilibrium toward the ‘closed’ apo-eIF2 complex at least 100-fold. Since eIF2B favors apo-eIF2 over eIF2-GDP, the interaction of eIF2γ with eIF2Bcat must be much stronger in the eIF2B•apo-eIF2 complex than in the eIF2B•eIF2-GDP complex, in order to compensate for the loss of the eIF2α•eIF2Breg contacts and account for this ∼100-fold difference in affinity. At physiological nucleotide concentrations (∼500 μM for GTP and ∼50 μM for GDP), the equilibrium is still strongly in favor of the eIF2B•eIF2 complex bound to GTP or GDP. This conclusion is consistent with the observation that the eIF2γ - eIF2Bcat cross-linking patterns are different between the eIF2B•apo-eIF2 and eIF2B•eIF2(α-P)-GDP complexes (6), which indicates different interaction interfaces. A subset of the cross-links observed in eIF2B•apo-eIF2 are weaker in the eIF2B•eIF2(α-P)-GDP complex (6) (see Figure 6B, inset, and Supplementary Figure S6A). In addition to eIF2γ binding to the eIF2Bcat platform, the interaction between eIF2γ and the catalytic eIF2Bϵ-CTD must have major contribution to shifting the equilibrium toward apo-eIF2 upon binding to eIF2B, because eIF2Bϵ-CTD has some GDP dissociating activity on its own (52). If as we propose here, the eIF2B•eIF2 complexes with GTP and GDP are similar, with eIF2α-NTD bound in the eIF2Breg pocket (Figure 6C), then Met-tRNAi binding is accompanied by eIF2α-NTD leaving the pocket and binding to the tRNA instead, as in the structure of the TC (Figure 6B), leading to lower eIF2B affinity for the TC than for eIF2-GTP, a prerequisite for TC release from the enzyme.
It was recently reported by Jennings and co-authors that eIF2B has the same affinity for apo-eIF2, eIF2-GDP, and eIF2-GTP, based on affinity pull-down data (53). However, their result contradicts existing data for both eIF2B (49,50,54) and all known GEFs (reviewed in (55,56)), and is thermodynamically impossible. Enzymes stabilize the transition state by having higher affinity for it than for the substrate and product. eIF2B lowers the affinity of eIF2 for GDP at least 100-fold (49); therefore, GDP must lower the affinity of eIF2 for eIF2B at least 100-fold, since these processes are thermodynamically coupled; accordingly, the affinity of eIF2B for eIF2-GDP has been reported to be much lower than its affinity for apo-eIF2 (49,50,54). This discrepancy is likely due to the use of affinity pull-down titration in reference (53), which is not a standard method for KD determination, and/or to other issues with their assay. Jennings and co-authors also suggested that eIF2B and Met-tRNAi compete for binding to eIF2 (53). However, their results show that eIF2B and Met-tRNAi only weaken, but do not prevent, each other's binding to eIF2, and are therefore, fully consistent with previous work showing the existence of an eIF2B•TC complex, and reports that eIF2B has lower affinity for the TC than for eIF2-GTP, which allows release of the TC from eIF2B (44,45).
Structural mechanism of eIF2B inhibition by eIF2(α-P)-GDP
Our results presented here, combined with the available information in the field, indicate that the nucleotide-bound state (Figure 6C) is similar between the eIF2B•eIF2-GDP and eIF2(α-P)-GDP complexes. How then does eIF2(α-P)-GDP inhibit eIF2B activity? An enzyme has higher affinity for the transition state (in this case apo-eIF2) than for the substrate (in this case eIF2-GDP), which lowers the activation barrier and accelerates the reaction. eIF2 phosphorylation increases the relative affinity of eIF2B for eIF2(α-P)-GDP compared to apo-eIF2(α-P). This leads to eIF2B having the same affinity for eIF2(α-P)-GDP as for the transition state apo-eIF2(α-P), effectively preventing eIF2B from catalyzing GDP dissociation from eIF2(α-P). The results described in this work (Figures 1 and 2) offer insights into the underlying mechanism: eIF2α phosphorylation promotes the extended conformation by destabilizing the intramolecular interaction between eIF2α-NTD and -CTD. Of course, phosphorylation could also directly affect the affinity of eIF2α-NTD for the pocket as currently thought, since the two effects are not mutually exclusive. However, based on our results with eIF2α-NTD binding to eIF2Breg (Figure 5), the direct effect appears to be no more than 2-fold; therefore, the indirect effect must be predominant.
The affinities of eIF2(α-P)-GDP and apo-eIF2(α-P) for eIF2B are approximately the same (50); therefore, eIF2B does not prefer the apo- form of eIF2(α-P). Accordingly, eIF2B neither promotes nor inhibits GDP release from eIF2(α-P) (51). Apo-eIF2(α-P) has been reported to have ∼10-fold lower affinity for eIF2B than unphosphorylated apo-eIF2 (50), while eIF2α-P has a few-fold higher affinity for eIF2B than does unphosphorylated eIF2α (6). Therefore, the shift from a ∼100-fold preference of eIF2B for apo-eIF2 over eIF2-GDP to no preference between apo-eIF2(α-P) and eIF2(α-P)-GDP appears to result from a ∼10-fold destabilization of the apo- state and a ∼10-fold stabilization of the GDP-bound state. Given the intramolecular eIF2α-NTD/CTD interaction reported here, eIF2α phosphorylation likely increases the affinity of eIF2B for the ‘extended’ eIF2-GDP state, in which eIF2α-NTD is bound in the eIF2Breg pocket, at least in part by destabilizing the intramolecular interaction within eIF2α. Phosphorylation also lowers the affinity of eIF2B for the ‘closed’ apo-eIF2 state, at least in part by destabilizing the intramolecular interaction within eIF2α. It should be noted that if phosphorylation instead stabilizes the ‘extended’ state 100-fold with no effect on the ‘closed’ state, the final effect on eIF2B activity would be the same. The tight binding of eIF2B to eIF2(α-P)-GDP, in the absence of conversion to eIF2(α-P)-GTP and eIF2(α-P) TC, results in eIF2B remaining trapped in an unproductive eIF2B•eIF2(α-P)-GDP complex, thus explaining the observed competitive inhibition.
It is important to compare the mechanism described here with previously proposed models. Based on extensive genetic and biochemical data, it has been generally accepted that eIF2 simultaneously contacts both the eIF2B catalytic and regulatory subcomplexes and does so in catalysis as well as in inhibition by eIF2α phosphorylation. However, the active and inhibited complexes are thought to be somehow different, in order to account for the latter being non-productive (see e.g. (20)). Thus it came as a surprise that the recent results of Kashiwagi and co-authors seemed to suggest that eIF2 binding to the two sites on eIF2B is mutually exclusive and therefore that the eIF2B•eIF2 complex exists in one of two alternative one-site interaction states (bound either to eIF2Breg or to eIF2Bcat (6)). As described above, our results, placed in the context of the report by Kashiwagi and co-authors, show that eIF2 can in fact contact both eIF2B subcomplexes at the same time as previously believed, thus resolving this discrepancy. Furthermore, the different relative intensities of cross-links between eIF2γ and eIF2Bcat in eIF2B•apo-eIF2 and eIF2B•eIF2(α-P)-GDP also support the conclusion that eIF2γ and eIF2Bcat contact each other in eIF2B•eIF2(α-P)-GDP. In contrast, mutually exclusive binding, as proposed in (6), would cause uniform weakening or complete loss of cross-linking over the entire eIF2γ/eIF2Bcat interface in the complex with eIF2(α-P)-GDP.
However, in contrast to previous two-site interaction models, the eIF2B•eIF2 complex is in equilibrium between an ‘extended’ two-site interaction state and a ‘closed’ one-site interaction state, with this equilibrium playing a central role in catalysis by eIF2B. This conclusion is also supported by cross-linking data in (6) that phosphorylated eIF2α shows essentially the same pattern of cross-linking to eIF2Breg as does unphosphorylated eIF2α and has only a few-fold higher affinity. If, as we propose here, the complexes of eIF2B with eIF2-GTP, eIF2-GDP, and eIF2(α-P)-GDP are similar, then any mutation or modification that affects eIF2B’s affinity for eIF2-GDP should have a similar effect on its affinity for eIF2-GTP and vice versa. The same should also be true for binding to eIF2(α-P)-GDP, possibly with the exception of mutations in the eIF2α P-loop or in the respective contact surfaces in eIF2B, where a direct effect of phosphorylation could play a role.
Implications for the regulation of eIF2B activity by small molecules
eIF2B is allosterically regulated by small molecules, such as nucleotides, dinucleotides and phosphosugars (57–59), and all eIF2B subunits are homologs of metabolic enzymes (reviewed in (3)). While the eIF2B subunits have most likely lost the enzymatic activity of their distant ancestors, the inter-domain active site pockets are highly conserved and are the most likely binding sites for these allosteric regulators (reviewed in (3)). In the model for the eIF2B•eIF2 complex (Figure 6), the positions of the ligand-binding sites in all eIF2B subunits are such that it is possible, and even likely, that ligand binding would affect the eIF2B/eIF2 interactions (Supplementary Figure S9). The ligand-binding sites in eIF2Bγ and ϵ lie underneath the eIF2γ-binding surface (compare Supplementary Figure S9A and B). Based on the structures of the hexameric enzyme ribose 1,5-bisphosphate isomerase from Thermococcus kodakarensis (tkRBPI) bound to substrate, product and in ligand-free form (36), it was proposed by Kuhle and co-authors that the eIF2Breg pocket undergoes conformational changes upon eIF2α binding (7). The tkRBPI active sites alternate between a closed and an open state, with the latter characterized by the NTDs of its subunits moving away from the CTDs and closer to each other (36). If eIF2Breg undergoes similar conformational changes, the open state would result in a ‘narrowing’ of the eIF2α-binding pocket. The authors proposed that ligand binding to the inter-domain pockets of eIF2Bα, β and/or δ could modulate the conformation of the eIF2α-binding pocket and thus its affinity for eIF2α (7). In the eIF2B crystal structure (6), eIF2Bα and δ are in an open (‘narrowed’) conformation, whereas eIF2Bβ is in a closed (‘widened’) conformation (Supplementary Figure S9A). If eIF2Bα were in an open conformation, it would be fully compatible with the contact surfaces we report here and with the cross-linking data of Kashiwagi and co-authors (6). A closed conformation for eIF2Bβ does not appear favorable as it would pull the eIF2α contact surface on eIF2Bβ away from those on eIF2Bα and eIF2Bδ. An open conformation for eIF2Bδ also appears unfavorable as it would make the pocket too narrow for eIF2α to fit. Therefore, conformational changes in any of the eIF2Bα, β or δ subunits could have an effect on eIF2α binding and thus eIF2B activity: stabilization of eIF2α-NTD binding in the pocket could stabilize the ‘extended’ state, while destabilization of eIF2α-NTD in the pocket could favor the ‘closed’ state.
In summary, we have proposed a novel model for the structural and thermodynamic basis of the eIF2B/eIF2 and eIF2B/eIF2(α-P) interactions, the mechanism of catalysis, and the inhibition of eIF2B activity by eIF2α phosphorylation. First, catalysis involves an equilibrium between an ‘extended’ nucleotide-bound conformation of eIF2, in which eIF2 binds eIF2Bregvia eIF2α-NTD, and eIF2Bcatvia eIF2γ simultaneously, and a ‘closed’ apo- conformation, in which eIF2 only binds eIF2Bcat (via eIF2γ). Second, eIF2α phosphorylation exerts its inhibitory effects by shifting the equilibrium of the eIF2B•eIF2 complex toward the ‘extended’ GDP-bound state, at least in part by destabilizing an intramolecular interaction between eIF2α-NTD and -CTD. This work offers a wide array of testable predictions that will guide future research.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Alan Hinnebusch and Boriana Marintcheva for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM095720 to A.M. and Shared Instrument Grant, 1S10-OD11941 to C.J. McKnight]. Funding for open access charge: Research grant.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hinnebusch A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014; 83:779–812. [DOI] [PubMed] [Google Scholar]
- 2. Jackson R.J., Hellen C.U., Pestova T.V.. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2010; 11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marintchev A., Wagner G.. Translation initiation: structures, mechanisms and evolution. Q. Rev. Biophys. 2004; 37:197–284. [DOI] [PubMed] [Google Scholar]
- 4. Bogorad A.M., Xia B., Sandor D.G., Mamonov A.B., Cafarella T.R., Jehle S., Vajda S., Kozakov D., Marintchev A.. Insights into the architecture of the eIF2Balpha/beta/delta regulatory subcomplex. Biochemistry. 2014; 53:3432–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gordiyenko Y., Schmidt C., Jennings M.D., Matak-Vinkovic D., Pavitt G.D., Robinson C.V.. eIF2B is a decameric guanine nucleotide exchange factor with a gamma2epsilon2 tetrameric core. Nat. Commun. 2014; 5:3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kashiwagi K., Takahashi M., Nishimoto M., Hiyama T.B., Higo T., Umehara T., Sakamoto K., Ito T., Yokoyama S.. Crystal structure of eukaryotic translation initiation factor 2B. Nature. 2016; 531:122–125. [DOI] [PubMed] [Google Scholar]
- 7. Kuhle B., Eulig N.K., Ficner R.. Architecture of the eIF2B regulatory subcomplex and its implications for the regulation of guanine nucleotide exchange on eIF2. Nucleic Acids Res. 2015; 43:9994–10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wortham N.C., Martinez M., Gordiyenko Y., Robinson C.V., Proud C.G.. Analysis of the subunit organization of the eIF2B complex reveals new insights into its structure and regulation. FASEB J. 2014; 28:2225–2237. [DOI] [PubMed] [Google Scholar]
- 9. Dever T.E., Dar A.C., Sicheri F.. Mathews MB, Sonenberg N, Hershey JWB. Translational Control in Biology and Medicine. 2007; NY: Cold Spring Harbor Laboratory Press; 319–344. [Google Scholar]
- 10. Hinnebusch A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005; 59:407–450. [DOI] [PubMed] [Google Scholar]
- 11. Hinnebusch A.G., Dever T.E., Asano K.. Mathews MB, Sonenberg N, Hershey JWB. Translational Control in Biology and Medicine. 2007; NY: Cold Spring Harbor Laboratory Press; 225–268. [Google Scholar]
- 12. Ron D., Harding H.P.. Mathews MB, Sonenberg N, Hershey JWB. Translational Control in Biology and Medicine. 2007; NY: Cold Spring Harbor Laboratory Press; 345–368. [Google Scholar]
- 13. Wek R.C., Jiang H.Y., Anthony T.G.. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006; 34:7–11. [DOI] [PubMed] [Google Scholar]
- 14. Smith H.L., Mallucci G.R.. The unfolded protein response: mechanisms and therapy of neurodegeneration. Brain. 2016; 139:2113–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Halliday M., Radford H., Sekine Y., Moreno J., Verity N., le Quesne J., Ortori C.A., Barrett D.A., Fromont C., Fischer P.M. et al. . Partial restoration of protein synthesis rates by the small molecule ISRIB prevents neurodegeneration without pancreatic toxicity. Cell Death Dis. 2015; 6:e1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moreno J.A., Halliday M., Molloy C., Radford H., Verity N., Axten J.M., Ortori C.A., Willis A.E., Fischer P.M., Barrett D.A. et al. . Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci. Transl. Med. 2013; 5:206ra138. [DOI] [PubMed] [Google Scholar]
- 17. Bugiani M., Boor I., Powers J.M., Scheper G.C., van der Knaap M.S.. Leukoencephalopathy with vanishing white matter: a review. J. Neuropathol. Exp. Neurol. 2010; 69:987–996. [DOI] [PubMed] [Google Scholar]
- 18. Fogli A., Boespflug-Tanguy O.. The large spectrum of eIF2B-related diseases. Biochem. Soc. Trans. 2006; 34:22–29. [DOI] [PubMed] [Google Scholar]
- 19. Richardson J.P., Mohammad S.S., Pavitt G.D.. Mutations causing childhood ataxia with central nervous system hypomyelination reduce eukaryotic initiation factor 2B complex formation and activity. Mol. Cell. Biol. 2004; 24:2352–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dev K., Qiu H., Dong J., Zhang F., Barthlme D., Hinnebusch A.G.. The beta/Gcd7 subunit of eukaryotic translation initiation factor 2B (eIF2B), a guanine nucleotide exchange factor, is crucial for binding eIF2 in vivo. Mol. Cell. Biol. 2010; 30:5218–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pavitt G.D., Yang W., Hinnebusch A.G.. Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol. Cell. Biol. 1997; 17:1298–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hannig E.M., Hinnebusch A.G.. Molecular analysis of GCN3, a translational activator of GCN4: evidence for posttranslational control of GCN3 regulatory function. Mol. Cell. Biol. 1988; 8:4808–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hinnebusch A.G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev. 1988; 52:248–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fabian J.R., Kimball S.R., Jefferson L.S.. Reconstitution and purification of eukaryotic initiation factor 2B (eIF2B) expressed in Sf21 insect cells. Protein Expr. Purif. 1998; 13:16–22. [DOI] [PubMed] [Google Scholar]
- 25. Krishnamoorthy T., Pavitt G.D., Zhang F., Dever T.E., Hinnebusch A.G.. Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2alpha) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell. Biol. 2001; 21:5018–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ito T., Marintchev A., Wagner G.. Solution structure of human initiation factor eIF2alpha reveals homology to the elongation factor eEF1B. Structure. 2004; 12:1693–1704. [DOI] [PubMed] [Google Scholar]
- 27. Marintchev A., Frueh D., Wagner G.. NMR methods for studying protein-protein interactions involved in translation initiation. Methods Enzymol. 2007; 430:283–331. [DOI] [PubMed] [Google Scholar]
- 28. Nag N., Lin K.Y., Edmonds K.A., Yu J., Nadkarni D., Marintcheva B., Marintchev A.. eIF1A/eIF5B interaction network and its functions in translation initiation complex assembly and remodeling. Nucleic Acids Res. 2016; 44:7441–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qin J., Vinogradova O., Gronenborn A.M.. Protein-protein interactions probed by nuclear magnetic resonance spectroscopy. Methods Enzymol. 2001; 339:377–389. [DOI] [PubMed] [Google Scholar]
- 30. Zuiderweg E.R. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002; 41:1–7. [DOI] [PubMed] [Google Scholar]
- 31. Anthis N.J., Clore G.M.. Visualizing transient dark states by NMR spectroscopy. Q. Rev. Biophys. 2015; 48:35–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Llacer J.L., Hussain T., Marler L., Aitken C.E., Thakur A., Lorsch J.R., Hinnebusch A.G., Ramakrishnan V.. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol. Cell. 2015; 59:399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmitt E., Panvert M., Lazennec-Schurdevin C., Coureux P.D., Perez J., Thompson A., Mechulam Y.. Structure of the ternary initiation complex aIF2-GDPNP-methionylated initiator tRNA. Nat. Struct. Mol. Biol. 2012; 19:450–454. [DOI] [PubMed] [Google Scholar]
- 34. Wei J., Jia M., Zhang C., Wang M., Gao F., Xu H., Gong W.. Crystal structure of the C-terminal domain of the varepsilon subunit of human translation initiation factor eIF2B. Protein Cell. 2010; 1:595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mohammad-Qureshi S.S., Haddad R., Hemingway E.J., Richardson J.P., Pavitt G.D.. Critical contacts between the eukaryotic initiation factor 2B (eIF2B) catalytic domain and both eIF2beta and -2gamma mediate guanine nucleotide exchange. Mol. Cell. Biol. 2007; 27:5225–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakamura A., Fujihashi M., Aono R., Sato T., Nishiba Y., Yoshida S., Yano A., Atomi H., Imanaka T., Miki K.. Dynamic, ligand-dependent conformational change triggers reaction of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1. J. Biol. Chem. 2012; 287:20784–20796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olsen L.R., Vetting M.W., Roderick S.L.. Structure of the E. coli bifunctional GlmU acetyltransferase active site with substrates and products. Protein Sci. 2007; 16:1230–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhuravleva A., Clerico E.M., Gierasch L.M.. An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell. 2012; 151:1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Beilsten-Edmands V., Gordiyenko Y., Kung J.C., Mohammed S., Schmidt C., Robinson C.V.. eIF2 interactions with initiator tRNA and eIF2B are regulated by post-translational modifications and conformational dynamics. Cell Discov. 2015; 1:15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Perkins D.J., Barber G.N.. Defects in translational regulation mediated by the alpha subunit of eukaryotic initiation factor 2 inhibit antiviral activity and facilitate the malignant transformation of human fibroblasts. Mol. Cell. Biol. 2004; 24:2025–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sidrauski C., Acosta-Alvear D., Khoutorsky A., Vedantham P., Hearn B.R., Li H., Gamache K., Gallagher C.M., Ang K.K., Wilson C. et al. . Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife. 2013; 2:e00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Srivastava S.P., Kumar K.U., Kaufman R.J.. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem. 1998; 273:2416–2423. [DOI] [PubMed] [Google Scholar]
- 43. Yatime L., Mechulam Y., Blanquet S., Schmitt E.. Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:18445–18450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gross M., Rubino M.S., Hessefort S.M.. The conversion of eIF-2.GDP to eIF-2.GTP by eIF-2B requires Met-tRNA(fMet). Biochem. Biophys. Res. Commun. 1991; 181:1500–1507. [DOI] [PubMed] [Google Scholar]
- 45. Salimans M., Goumans H., Amesz H., Benne R., Voorma H.O.. Regulation of protein synthesis in eukaryotes. Mode of action of eRF, an eIF-2-recycling factor from rabbit reticulocytes involved in GDP/GTP exchange. Eur. J. Biochem. 1984; 145:91–98. [DOI] [PubMed] [Google Scholar]
- 46. Alone P.V., Dever T.E.. Direct binding of translation initiation factor eIF2gamma-G domain to its GTPase-activating and GDP-GTP exchange factors eIF5 and eIF2B epsilon. J. Biol. Chem. 2006; 281:12636–12644. [DOI] [PubMed] [Google Scholar]
- 47. Ito T., Wagner G.. Using codon optimization, chaperone co-expression, and rational mutagenesis for production and NMR assignments of human eIF2 alpha. J. Biomol. NMR. 2004; 28:357–367. [DOI] [PubMed] [Google Scholar]
- 48. Dey M., Velyvis A., Li J.J., Chiu E., Chiovitti D., Kay L.E., Sicheri F., Dever T.E.. Requirement for kinase-induced conformational change in eukaryotic initiation factor 2alpha (eIF2alpha) restricts phosphorylation of Ser51. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:4316–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Panniers R., Rowlands A.G., Henshaw E.C.. The effect of Mg2+ and guanine nucleotide exchange factor on the binding of guanine nucleotides to eukaryotic initiation factor 2. J. Biol. Chem. 1988; 263:5519–5525. [PubMed] [Google Scholar]
- 50. Goss D.J., Parkhurst L.J., Mehta H.B., Woodley C.L., Wahba A.J.. Studies on the role of eukaryotic nucleotide exchange factor in polypeptide chain initiation. J. Biol. Chem. 1984; 259:7374–7377. [PubMed] [Google Scholar]
- 51. Jennings M.D., Zhou Y., Mohammad-Qureshi S.S., Bennett D., Pavitt G.D.. eIF2B promotes eIF5 dissociation from eIF2*GDP to facilitate guanine nucleotide exchange for translation initiation. Genes Dev. 2013; 27:2696–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gomez E., Mohammad S.S., Pavitt G.D.. Characterization of the minimal catalytic domain within eIF2B: the guanine-nucleotide exchange factor for translation initiation. EMBO J. 2002; 21:5292–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jennings M.D., Kershaw C.J., Adomavicius T., Pavitt G.D.. Fail-safe control of translation initiation by dissociation of eIF2alpha phosphorylated ternary complexes. Elife. 2017; 6:e24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rowlands A.G., Panniers R., Henshaw E.C.. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J. Biol. Chem. 1988; 263:5526–5533. [PubMed] [Google Scholar]
- 55. Bos J.L., Rehmann H., Wittinghofer A.. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007; 129:865–877. [DOI] [PubMed] [Google Scholar]
- 56. Sprang S.R., Coleman D.E.. Invasion of the nucleotide snatchers: structural insights into the mechanism of G protein GEFs. Cell. 1998; 95:155–158. [DOI] [PubMed] [Google Scholar]
- 57. Dholakia J.N., Mueser T.C., Woodley C.L., Parkhurst L.J., Wahba A.J.. The association of NADPH with the guanine nucleotide exchange factor from rabbit reticulocytes: a role of pyridine dinucleotides in eukaryotic polypeptide chain initiation. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:6746–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kimball S.R., Jefferson L.S.. Allosteric regulation of eukaryotic initiation factor eIF-2B by adenine nucleotides. Biochem. Biophys. Res. Commun. 1995; 212:1074–1081. [DOI] [PubMed] [Google Scholar]
- 59. Webb B.L., Proud C.G.. Eukaryotic initiation factor 2B (eIF2B). Int. J. Biochem. Cell. Biol. 1997; 29:1127–1131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.