Abstract
DNA N6-methyladenine (6mA) is newly rediscovered as a potential epigenetic mark across a more diverse range of eukaryotes than previously realized. As a unicellular model organism, Tetrahymena thermophila is among the first eukaryotes reported to contain 6mA modification. However, lack of comprehensive information about 6mA distribution hinders further investigations into its function and regulatory mechanism. In this study, we provide the first genome-wide, base pair-resolution map of 6mA in Tetrahymena by applying single-molecule real-time (SMRT) sequencing. We provide evidence that 6mA occurs mostly in the AT motif of the linker DNA regions. More strikingly, these linker DNA regions with 6mA are usually flanked by well-positioned nucleosomes and/or H2A.Z-containing nucleosomes. We also find that 6mA is exclusively associated with RNA polymerase II (Pol II)-transcribed genes, but is not an unambiguous mark for active transcription. These results support that 6mA is an integral part of the chromatin landscape shaped by adenosine triphosphate (ATP)-dependent chromatin remodeling and transcription.
INTRODUCTION
DNA methylation is implicated as an epigenetic mark in various important processes in eukaryotes, including maintenance of DNA structure (1), DNA replication (2), and transcription (3). 5-methylcytosine (5mC) is the major form of DNA methylation in higher eukaryotes and consequently has been extensively studied (4). In contrast, N6-methyladenine (6mA) was largely neglected in eukaryotes until recent investigations in several model organisms (5–10). These findings not only expand our knowledge about 6mA distribution, but also substantiate its functional relevance and epigenetic roles in a wide range of eukaryotes.
Previous studies provide some important clues to the distribution of 6mA but also highlight many apparent discrepancies. In green algae Chlamydomonas reinhardtii, 6mA marks active genes with a bimodal distribution around transcription start sites (TSS) (5). In early-diverging fungi, 6mA is also enriched near the transcriptional start sites of expressed genes as dense methylated adenine clusters (11). In zebrafish, a large fraction of 6mA peaks are located in repetitive elements (8). In Xenopus laevis, 6mA is generally depleted in the gene body, especially exons (7). In mouse embryonic stem cells, 6mA is strongly enriched on young LINE-1 transposons on the X chromosome (12).
The model organism Tetrahymena thermophila is among the first eukaryotes reported to contain 6mA (13–15). 6mA is the only detectable DNA methylation in Tetrahymena (14,16), making it an ideal system to study 6mA function and regulation. Like other ciliates, Tetrahymena contains two types of nuclei in the same cytoplasmic compartment: the somatic macronucleus (MAC) and germline micronucleus (MIC) (17). 6mA is present in the transcriptionally active MAC, while absent in the transcriptionally silent MIC during vegetative growth (14,17,18). During conjugation, the sexual phase of Tetrahymena life cycle, 6mA occurs de novo in the developing new MAC which is derived from the germiline MIC (19,20). However, lack of comprehensive information about 6mA distribution prevents the molecular-level understanding of its function and regulatory mechanism.
Here, we provide the first genome-wide, base pair-resolution map of 6mA in Tetrahymena, by applying single-molecule real-time (SMRT) sequencing. We also present evidence that 6mA is associated with the linker DNA of well-positioned nucleosomes and nucleosomes containing the conserved histone variant H2A.Z in Pol II-transcribed genes. These results support that 6mA is an integral part of the chromatin landscape shaped by adenosine triphosphate (ATP)-dependent chromatin remodeling and transcription, as well as provide strong clues to the molecular mechanism for the propagation of 6mA as an epigenetic mark.
MATERIALS AND METHODS
Cell culture
Tetrahymena thermophila wild-type strains, SB210 and CU428 were obtained from the national Tetrahymena Stock Center (http://tetrahymena.vet.cornell.edu). H2A.Z-NHA strain was constructed by introducing a short sequence encoding the hemagglutinin (HA) tag to the N-terminus of the HTA3 gene (TTHERM_00143660) that encodes the histone variant H2A.Z. Cells were grown in SPP medium at 30°C (21). The cellular localization and protein size of the HA-tagged H2A.Z was validated by immunofluorescence staining and immuno-blotting, respectively (Supplementary Figure S1).
Nuclei purification, MNase-seq and ChIP-seq
Purification and digestion of MACs and MICs was carried out following established protocols (22). For MNase-seq, approximately 5 × 107 purified MACs were digested by Micrococcal Nuclease (MNase, NEB) (200U/ml, 25°C, 15 min) and mono-nucleosome sized DNA was selected by agarose gel purification (22). For ChIP-seq, MACs were purified from H2A.Z-NHA cells, with a wild-type strain (CU428) as a control. About 5 × 107 purified MACs were digested by MNase (200 U/ml, 25°C, 15 min). After extraction by Triton X-100 (0.1%) and NP-40 (0.1%), solubilized chromatin was immunoprecipitated with Pierce anti-HA magnetic beads (Thermo Scientific, I2149) at 4°C overnight (23) and HA-tagged H2A.Z was eluted with the HA peptide (Sigma, E6779).
Illumina sequencing and data processing
After sequencing by Illumina HiSeq 4000, paired-end reads were mapped back to the genome assembly of T. thermophila (SB210), using Bowtie 2 (24). The MAC genome assembly was from the Tetrahymena genome database (TGD) (http://ciliate.org) (25,26). Potential polymerase chain reaction (PCR) duplicates were removed and unique mapping results were kept by customized scripts in Perl. H2A.Z ChIP-enriched regions were identified using SICER v1.1 (W50-G150-FDR1E-2) (27).
SMRT sequencing
Genomic DNA was extracted from the SB210 strain of T. thermophila using Wizard® Genomic DNA Purification Kit (Promega, A1120). SMRT sequencing libraries were prepared according to manufacturer’s instructions and sequencing was carried out by Macrogen Inc. (Seoul, Korea). The raw data generated from eight sequencing cells (GEO accession number GSE96521) were mapped to the latest SB210 MAC genome from the TGD (http://ciliate.org) (25,26). Base modifications (6mA) were identified using RS_Modification_and_Motif_Analysis.1 protocol in the SMRT Portal v2.3.0 (Pacific Biosciences). Only identified modifications with Qv > 20 and coverage > 10 × were kept. For composite analysis and motif identification, 6mA was divided into three groups based on their methylation level (low 10–20%; intermediate 20–80%; high 80–100%).
6mA motif analysis
A total of 20 bp sequences from the upstream and downstream of each 6mA site were extracted to identify the motif. Local motifs of nearby 6mA were illustrated by WebLogo 3 (28). The genome-wide distribution of H2A.Z ChIP peaks and 6mA and its motif profiles were generated using Circos (29). The distribution of identified 6mA, H2A.Z signals and nucleosome dyads on the MAC genome was visualized using GBrowse2 (30). Phasograms were plotted to reveal the periodicity of 6mA, comparing with that of nucleosomes, based on all 6mA modifications (or nucleosome dyads) across the genome (31). Spectral density estimation (spectrum analysis) of 6mA was carried out in R.
Distribution and composite analyses of 6mA and nucleosomes
Pol III transcribed genes (including 672 tRNAs, 172 5S rRNAs, 4 U6 snRNAs and 1 snoRNA, see Supplementary Table S2) were detected in the latest version of the MAC genome using tRNAscan-SE v1.3.1, RNAmmer v1.2 and Rfam v11.0, as described in previous studies (25,32,33). For analyses related to TSS, 15 841 well-modeled genes (13 177 of which are longer than 1 kb) that are strongly supported by deep RNA-seq sequencing results are selected (34,35). Information of precise positions of nucleosomes and their positioning degrees as well as linker DNA regions are acquired from our previous study (34). Distribution heatmaps of nucleosome dyads around TSSs, including 13 485 TSSs of genes marked with 6mA and 2356 TSSs of genes without 6mA, were generated by deepTools (bin = 10 bp) (36). Composite analyses of identified 6mA with nucleosome dyads, H2A.Z ChIP signals and linker DNA regions were performed using customized Perl scripts. Comparisons of observed versus simulated (randomly picked adenines, in equal number to observed 6mA on each scaffold, were used) distributions of 6mA on the different regions across the Tetrahymena genome, and matrices of correlation among 6mA, nucleosomes, H2A.Z signals and gene expression levels, were carried out by customized scripts in Perl and R. One-factor analysis of variance (one-way ANOVA) was carried out by using SPSS v. 22.0 (37). KEGG analysis on genes without 6mA was carried out by KEGG Automatic Annotation Server (KAAS) using the bi-directional best hit (BBH) method to assign orthologs (38).
Immunofluorescence staining and imaging
Cell fixation and immuofluoresence (IF) staining were performed as described previously (6,39,40). Cells were fixed during vegetative growth or at 10 h after mixing cells of two mating types, as indicated. For the 6mA staining, fixed cells were treated with RNase A (50 μg/ml or otherwise indicated) in phosphate-buffered saline (PBS) buffer for 2 h at 37°C. Non-treated cells were either incubated at 37°C or at room temperature (RT), as indicated. Cells were then incubated with 2N HCl for 20 min at RT and neutralized with Tris–HCl (100 mM, pH 8.5) for 10 min, prior to antibody incubation. The primary antibodies are α-6mA (Synaptic Systems, 202003, 1:2000) (5,6,10), α-HA (Cell Signaling, C29F4, 1:200), α-H3K9-acetylation (Abcam, ab10812, 1:500) and α-H3K14-acetylation (Abcam, ab52946, 1:500). The primary antibodies are incubated with cells at 4°C overnight (for α-6mA) or at RT for 2 h (for other antibodies). Cells were then incubated in the secondary antibody (Goat anti-Rabbit IgG (H+L), Invitrogen, A-21428, 1:4000) at RT for 1 h. Digital images were collected using a Leica DM2500 microscope with a Leica DFC450C camera.
Immuno-blotting
The nucleic lysate was prepared from H2A.Z-NHA and CU428 (wild-type) cells, by adding the SDT buffer (4% sodium dodecyl sulphate, 100 mM Tris–HCl pH 7.6, 100 mM dithiothreitol (DTT) pH 8.0). The primary antibodies are α-HA (Cell Signaling, C29F4, 1:2000) and α-alpha-tubulin (Sigma, T6199, 1:1000). The secondary antibody is a horseradish peroxidase (HRP) -conjugated secondary Goat anti-Rabbit antibody (TransBionovo, HS101-01, 1:8000).
Dot blot assay
Genomic DNAs from purified MAC and MIC were denatured at 100°C for 10 min and spotted on Hybond + membranes (Amersham, RPN303B) in a Bio-Dot Apparatus (BIO-RAD, 1706545). After washes and blocking, membranes were incubated with an α-6mA antibody (Synaptic Systems, 202003, 1:4000) overnight at 4°C and then a HRP-conjugated secondary Goat anti-Rabbit antibody (TransBionovo, HS101-01, 1:8000) at RT for 1 h. Methylene blue hydrate (Molecular Research Center, Inc., MB119) staining was performed to determine the amount of input DNA.
DpnI/DpnII digestion and quantitative PCR (qPCR) analysis
Approximately 2 μg purified genomic DNA was treated with 40 U DpnI (NEB, R0176v) for 30 min or overnight or with 40 U DpnII (NEB, R0543L) overnight at 37°C. The samples were heat-inactivated for 20 min at 80°C (DpnI) or 65°C (DpnII) after digestion. A total of 4 ng digested (DpnI-30min, DpnI-O/N, DpnII-O/N) and non-digested DNA were subjected to quantitative PCR (qPCR) analysis using EvaGreen Express 2 × qPCR MasterMix-Low ROX (Abm, MasterMix-LR). Primers flanking selected GATC sites were used to validate methylation level at a specific position (Supplementary Table S3). Primers matched to the coding DNA sequence (CDS) of JMJ1 (TTHERM_00185640) were used as internal controls. Methylation level is reflected by normalized Ct difference (ΔCt) between digested and undigested samples. As DpnI and DpnII cut methylated and unmethylated GATC sequence respectively, the following results are expected. For highly methylated sites, ΔCtDpnI should be substantially larger than zero and ΔCtDpnII be very close to zero. For intermediately methylated sites, ΔCtDpnI should be above zero but smaller than that of highly methylated sites and ΔCtDpnII be above zero. For unmethylated sites, ΔCtDpnI should be very close to zero and ΔCtDpnII be substantially larger than zero.
RESULTS
6mA occurs in vegetative and conjugative Tetrahymena cells
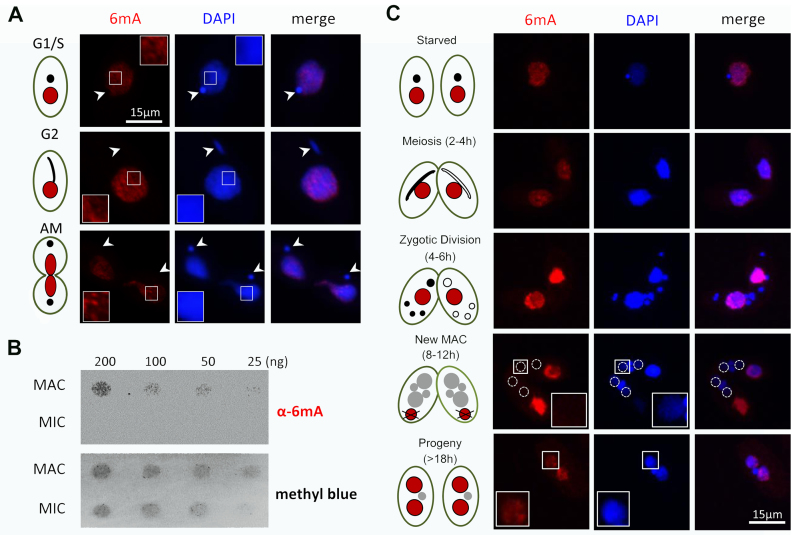
Early studies have shown that substantial amounts of 6mA in Tetrahymena can be detected by enzymatic digestion (16) and chromatography (14,41). To confirm these findings, we performed immunofluorescence staining with 6mA-specific antibodies. In vegetative cells, strong 6mA signals in an unevenly distributed pattern were detected in the transcriptionally active MAC (Figure 1A). In contrast, no signal was detected in the transcriptionally silent MIC (Figure 1A, white arrowheads). To exclude the possibility that the detected signals were due to contaminating RNA with 6mA, we pre-treated samples with different concentrations of RNase A, but the signal intensity remained unaffected (Supplementary Figure S2). Differential distribution of 6mA in MAC and MIC was also confirmed by dot blot analysis, in which 6mA signals were only detectable in genomic DNA from purified MAC, but not from MIC (Figure 1B). Importantly, 6mA levels in MAC stayed at similar levels in G1/S, G2 and amitotic phases of cell division (Figure 1A and Supplementary Figure S3), suggesting that maintenance methylation occurs quickly and efficiently after DNA replication, in a timeframe substantially shorter than the 3-h Tetrahymena cell cycle (http://www.lifesci.ucsb.edu/∼genome/Tetrahymena/genetics.htm).
Figure 1.
Cellular distribution of 6mA in Tetrahymena. (A) 6mA occurs in vegetative cells with no obvious cell-cycle difference, shown by immunofluorescence staining with the modification-specific antibody. Note the absence of 6mA signal in MIC (arrowheads). Bar = 15 μm. (B) Differential distribution of 6mA in MAC and MIC, revealed by dot blot analysis using a specific anti-6mA antibody. Methylene blue hydrate staining was performed to determine the amount of loaded DNA. (C) 6mA occurs de novo in the newly developed MAC during conjugation. Cell stages are distinguished by nuclear events during conjugation. In the early phase of conjugation (starved, meiosis and zygotic), 6mA only exists in the parental MAC. Along with degradation of the old MAC, 6mA signal gradually builds up in the new MAC (dotted line circles). Bar = 15 μm.
We further analyzed the dynamic distribution of 6mA during Tetrahymena conjugation (Figure 1C). In early conjugating cells, 6mA was detected in the parental MAC, but not in the meiotic MIC with active transcription of non-coding RNA (42). 6mA signals in the parental MAC increased dramatically during formation of gametes and zygotes but remained absent in MIC-derived gametes and zygotes (Figure 1C). Zygotic products give rise to both new MIC and new MAC (43). 6mA is eventually established in the latter by de novo deposition (Figure 1C) (19,20). Intriguingly, we found that 6mA was not detected in the early developing new MAC (Figure 1C), even though transcription and transcription-associated epigenetic marks—including H2A.Z and histone hyper-acetylation (Supplementary Figure S4)—are already present in abundance (44,45). We only detected 6mA signals in the new MAC several hours later at the last stage of conjugation, in which there were two new MACs and one new MIC, with the old MAC having been reabsorbed (Figure 1C). Our results are in agreement with a previous report that de novo methylation of anlagen DNA occurs several hours after pair separation (19). The uncoupling of 6mA and transcription in the meiotic MIC and the early developing new MAC indicates that 6mA is not required for the transcription of all genes. The delay in its buildup in the new MAC suggests that de novo deposition of 6mA in late conjugating cells is a slow process, which may be distinct from the fast maintenance methylation in vegetatively growing cells.
6mA is preferentially associated with the 5′ end of the gene body and AT motif (5′-AT-3′)
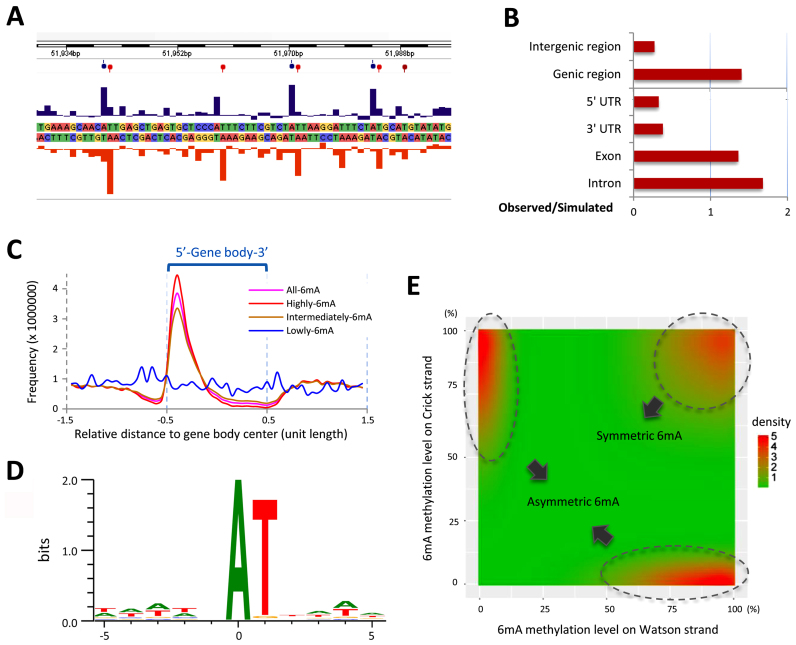
To directly interrogate the precise positions of 6mA, we performed single-molecule real-time sequencing (SMRT sequencing), using genomic DNA from vegetatively growing SB210 cells—the reference strain for the macronuclear genome sequencing project (25). SMRT sequencing generated 784 599 reads, corresponding to 71 × average coverage of the Tetrahymena MAC genome (Supplementary Table S1A). It revealed the typical kinetic signature at the modified sites (Figure 2A). 6mA was detected on 525 464 adenines, corresponding to 0.66% of the total adenines in the Tetrahymena genome. This value is comparable to the previous report (0.65–0.80%) (14). Among these identified sites, 283 759 6mA with high confidence (coverage > 10 ×, Qv > 20) were selected for subsequent analysis (Supplementary Table S1B). The methylation ratio was therefore reduced from 0.66 to 0.3%. The ‘discrepancy’ between the previous result and ours may stem from different ways of data processing: previous studies counted all detected methylated sites, while we only selected methylated sites with high confidence. It may therefore reflect variations of methylation levels/sites in a large population of cells. It has been estimated that ∼3% of substantially methylated 6mA sites in Tetrahymena reside in the sequence of GATC, therefore sensitive to DpnI digestion (20,46). This estimate is consistent with our results (number of methylated GATC/number of methylated adenines = 10690/283759 = 3.8%) (Supplementary Table S1C).
Figure 2.
Genomic distribution of 6mA in Tetrahymena. (A) Representative interpulse duration (IPD) ratios of SMRT sequencing data of vegetative Tetrahymena cells (SB210). Columns indicate the IPD peaks and balloons indicate the detected 6mA signals on both the sense (blue) and antisense (red) strands based on the IPD ratio. (B) Comparison of observed versus simulated distributions of 6mA in the different regions of the Tetrahymena genome indicates that 6mA is enriched in genic regions, including exons and introns. See Supplementary Table S1D for details. (C) 6mA sites with different degrees of methylation have different distributions in the gene body (bin = 5% of gene body length). The gene body is scaled to unit length and is extended on each side by one unit length. Distribution of all 6mA is in pink. Distributions of 6mA sites with high/intermediate/low methylation levels are in red, brown and blue, respectively. (D) SMRT sequencing identified 5′-AT-3′ as the most representative motif. (E) Distribution of methylation levels of 6mA modifications on Watson–Crick strands indicated that asymmetric 6mA (top left and bottom right) were not an insufficiently methylated status of symmetric 6mA (top right). Plotted by ggplot2 in R, bandwidth = c (0.25,0.25).
6mA was preferentially localized at the gene body rather than intergenic regions (Figure 2B and Supplementary Table S1D). This contrasts with the distribution in Caenorhabditis elegans, in which 6mA is evenly distributed in all genomic features (6). This also differs from Chlamydomonas reinhardtii in which 6mA forms bimodal peaks around TSS (5). Composite analysis of 6mA distribution in 13 177 well-annotated long genes (>1 kb) revealed that Tetrahymena 6mA did not accumulate upstream of TSS. Instead, 6mA accumulates downstream of TSS, toward the 5′ end of the gene body (Figures 2C and 4F).
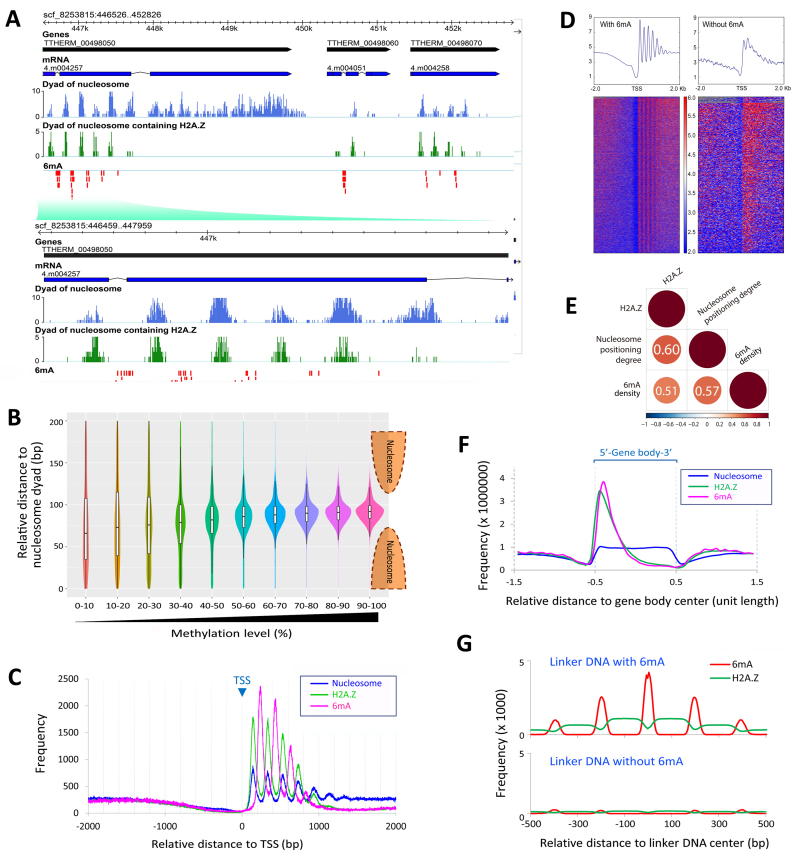
Figure 4.
6mA is preferentially associated with the linker DNA between well-positioned nucleosomes and/or nucleosomes containing the histone variant H2A.Z. (A) GBrowse snapshot of the genomic data. The tracks from top to bottom are gene models (black), mRNA transcripts (blue), dyads of nucleosomes (light blue), dyads of nucleosomes containing H2A.Z (green) and 6mA modifications (red), respectively. Note that 6mA signals are located in inter-nucleosomal regions, toward the 5′ end of gene bodies. (B) The methylation level of 6mA is correlated with the distance to the nucleosome dyad. Note that 6mA with high methylation levels are excluded from the nucleosomal DNA. The violin plots showed the density of 6mA at different relative distances to the nucleosome dyad in different methylation level groups (filled with different colors). The box plots in the violin plots indicate the median and the interquartile range in each group. (C) Distribution profiles of nucleosome (blue), H2A.Z (green) and 6mA (pink) around TSS. (D) Nucleosomes exhibit a more consistent phase relative to TSS in genes marked with 6mA (left) than genes without 6mA (right). A total of 15 841 TSS of well-modeled genes are used, including 13 485 TSS of genes marked with 6mA and 2356 TSS of genes without 6mA. (E) Correlation matrix of H2A.Z, nucleosome positioning degree, and density of highly-methylated 6mA. Correlation coefficients and correlation color dots are shown (p < 0.01). Negative correlations are in blue and positive ones in red, as the color bar shown below. (F) Distribution profiles of nucleosome (blue), H2A.Z (green) and 6mA (pink) on the gene body showing that they are all enriched toward the 5′ end of gene bodies (bin = 5% of H2A.Z peak length). Genes are scaled to unit length and one unit length is extended to each side. (G) Relative distribution of 6mA (red) and H2A.Z (green). Note that 6mA is enriched in the well-defined linker DNA regions (45–55 bp) flanked by H2A.Z-containing nucleosomes. Bin = 5 bp, normalized to 1 million tags.
Methylation levels varied almost continuously from 0 to 100% at different 6mA positions (Supplementary Figure S5A and Figure 3A), which is expected for the polyploid MAC (∼45 copies) and is consistent with previous reports (20,41,47). We found that 6mA sites with low methylation levels (<20%, at 0.1% of total 6mA sites) (Supplementary Figure S5B and Table S1B) did not accumulate towards the 5′ end of the gene body (blue in Figure 2C). In contrast, 6mA sites with intermediate methylation levels (20–80%, at 35.1% of total 6mA sites) (Supplementary Figure S5B and Table S1B) were enriched towards the 5′ end of the gene body, while 6mA sites with high methylation levels (80–100%, at 64.8% of total 6mA sites) (Supplementary Figure S5B and Table S1B) were even more so enriched (brown and red, respectively in Figure 2C). These findings strongly suggest that 6mA is targeted to specific genomic regions in the context of transcription.
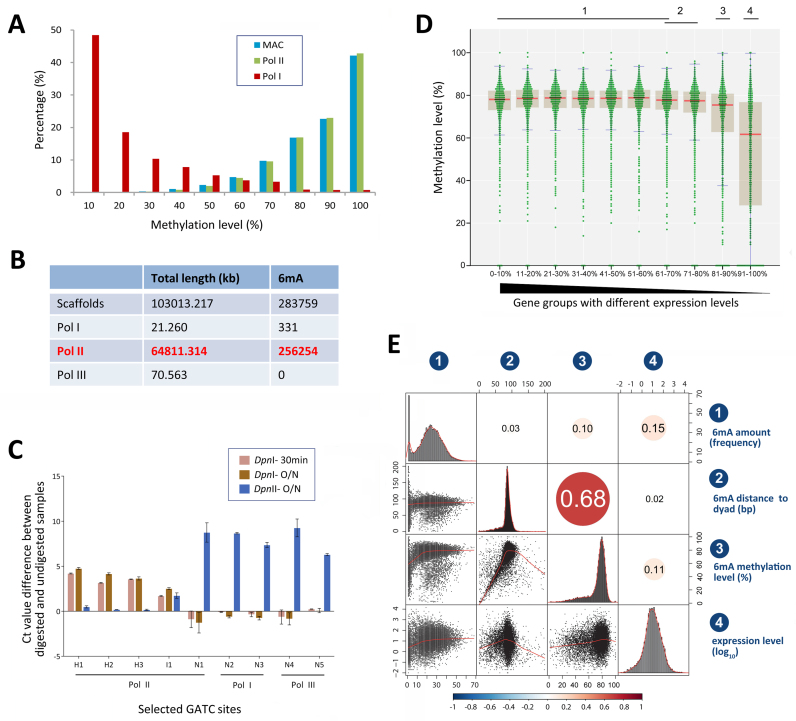
Figure 3.
6mA is preferentially associated with RNA Polymerase II-transcribed genes. (A) Comparison of 6mA methylation levels on bulk genomic DNA (MAC, blue), Polymerase II transcribed gene regions (Pol II, green) and rDNA (Pol I, red). (B) No 6mA methylation is localized on RNA Polymerase III transcribed genes. Pol III genes are listed in Supplementary Table S2. (C) qPCR validation of nine selected GATC sites located on genes transcribed by different RNA polymerases (Pol I, II and III). Genomic DNA of SB210 was digested at 37°C with DpnI for 30 min or overnight or with DpnII overnight. qPCR was performed with primers flanking GATC sites or with internal control primers (Supplementary Table S3). Y-axis represents methylation level that is reflected by normalized Ct value difference between digested and undigested samples. H1-H3: highly methylated sites. I1: intermediately methylated sites. N1-N5: unmethylated sites. (D) Methylation levels of 6mA distributed in the 1 kb region downstream the TSS of genes with different expression levels. Genes are ranked from high to low by their expression levels and divided into 10 quantiles (Q1-Q10). Each 6mA is shown as a green dot. Median of methylation level in each group of genes is marked with a red line. Inter-quartile ranges (IQR) are plotted as brown boxes. Confidence intervals are marked with blue lines. Ten groups of genes with different expression levels are divided into four homogeneous subsets according to ANOVA analysis (significance of each subset compared with others are 0.040, 0.016, 1.000 and 1.000, respectively, under condition of α = 0.01; see Supplementary Table S4). (E) Correlation matrix of different attributes of genes: (i) 6mA amount in 1 kb region downstream TSS, (ii) relative distance of 6mA to nucleosome dyad in this region, (iii) methylation level of 6mA in this region and (iv) gene expression levels (log10). Correlation coefficients and correlation color dots were shown in the higher triangle of the correlation matrix (P < 0.01). Negative correlations are in blue and positive ones in red, as the color bar shown below. The histograms of the attributes are shown on the diagonal. Bivariate scatterplots among attributes, with fitted lines, are plotted in the lower triangle of the correlation matrix.
Most 6mA (96%) occurred at the sequence of 5′-AT-3′ (16), with no additional surrounding consensus sequence (Figure 2D), while others (4.0%) occurred in non-AT motifs (Supplementary Figure S5B-C and Table S1E). This is consistent with an early report based on nearest-neighbor analysis (16). For 6mA sites with high methylation levels, T was almost the exclusive 3′ neighbor (99.4% of AT motif, 0.6% non-AT motif; Supplementary Figure S5B-C and Table S1E). For 6mA sites with intermediate and low methylation levels, non-AT motifs were dramatically and progressively increased, at 9.8 and 82.7%, respectively (Supplementary Figure S5B-C and Table S1E). Based on these results, we conclude that 6mA DNA methylase(s) preferentially targets the AT motif.
The palindromic AT motif for 6mA sites in Tetrahymena is reminiscent of the palindromic CG motif for 5mC sites in other eukaryotic systems (48–50), with implications in the maintenance of DNA methylation as epigenetic marks. We found that 54.2% of the AT motif sites with 6mA were methylated on both strands of the DNA molecule (symmetric methylation), while 45.8% were methylated asymmetrically on only one strand (Supplementary Figure S5B and Table S1E). Of the asymmetric AT motif sites, 60.5% were highly methylated (Supplementary Figure S5B and Table S1E). Interestingly, there is no correlation between strand specificity of 6mA methylation and mRNA transcription; in other words, the methylated adenine in the asymmetric AT motif is not preferentially located in either the coding or non-coding strand (Supplementary Figure S5D). Most importantly, asymmetric 6mA is not an insufficiently methylated status of symmetric 6mA, as the enriched signals of asymmetric and symmetric 6mA are well separated in their distributions, without gradual transitions between them (Figure 2E). The presence of a large population of asymmetrically methylated AT motif sites distinguishes 6mA maintenance from that of 5mC, which is exclusively based on specific targeting of hemi-methylated CG motif by DNMT1 to restore the symmetry of 5mC methylation (51).
6mA is preferentially associated with Pol II-transcribed genes, but only weakly correlated with transcription levels
The enrichment of 6mA in the gene body prompted us to investigate whether it was preferentially associated with genes transcribed by one of the three eukaryotic RNA polymerases (Pol I, II and III) (52). We first examined 6mA distribution on the rDNA mini-chromosome, which is transcribed by Pol I (53). Our SMRT sequencing data achieved ∼6,669 × coverage of rDNA, due to its high copy number in MAC (54). Surprisingly, 6mA sites identified in rDNA did not display the typical kinetic signature (a distinct peak at the modified base position) as 6mA sites on Pol II-transcribed genes (Figure 2A), being instead mostly embedded in clustered peaks (Supplementary Figure S6A). Additionally, the average interpulse duration (IPD) ratio of 6mA in rDNA was significantly lower than that in Pol II-transcribed genes (1.431 versus 4.016, P = 9.8207E-195) (Supplementary Figure S6B), which largely prevented high confidence calling. Moreover, methylation levels of 6mA sites in rDNA were much lower on average, with only 1.2% being highly methylated (Figure 3A and Supplementary Table S1C). In contrast to 6mA in Pol II-transcribed genes, residing mostly in AT motif sites, 6mA in rDNA showed no preference for thymine (T) at the +1 position (Supplementary Figure S6C). In fact, no rDNA 6mA was found at the 5′-GATC-3′ motif (Supplementary Table S1C), which is in strong contrast to previous analysis by DpnI digestion (41). We conclude that 6mA in Pol I-transcribed rDNA is at very low levels, much of which may be attributed to either false positives or background methylation events comparable to 6mA sites with low methylation levels in Pol II-transcribed genes (Figure 3A).
We also examined Pol III-transcribed genes. In total, 849 Pol III genes are identified in the Tetrahymena macronuclear genome, including 672 tRNAs, 172 5S rRNAs, 4 U6 snRNAs and 1 snoRNA (Supplementary Table S2) (25). Most Pol III-transcribed genes are very short (average 74.7 bp), the longest being 253 bp (Supplementary Table S2). No 6mA methylation was detected on these genes (Figure 3B). This may be attributed to the fact that Pol III-transcribed genes were mostly associated with the nucleosomal DNA rather than the linker DNA (Supplementary Figure S6D).
We employed restriction-enzyme digestion to validate the SMRT sequencing results (Supplementary Table S3) (5). DpnI cuts methylated GATC sites with a strong preference for symmetric rather than asymmetric methylation, while DpnII is blocked by any 6mA (55,56). We compared DpnI-digested, DpnII-digested and undigested genomic DNA, and determined the methylation status of several GATC sites by qPCR with flanking primers (Figure 3C). One unmethylated and four methylated (three at high levels, one intermediate) GATC sites were selected for Pol II-transcribed genes. No GATC sites contained 6mA detectable by SMRT sequencing on Pol I- or Pol III-transcribed genes. We therefore randomly chose two sites for each category. This assay showed that the three sites in Pol II-transcribed genes with high methylation levels detected by SMRT sequencing were all sensitive to DpnI digestion, but not DpnII (Figure 3C). There were only small differences between 30 min and overnight digestion by DpnI (Figure 3C), supporting that most MAC alleles of these sites were symmetrically rather than asymmetrically methylated. In contrast, the sites with no methylation detected by SMRT sequencing were sensitive to DpnII but not DpnI digestion (Figure 3C). The site with intermediate methylation levels detected by SMRT sequencing showed partial sensitivity to both DpnI and DpnII digestion (Figure 3C). This result provides an independent verification of the SMRT sequencing results, and further supports our conclusion that 6mA is preferentially associated with Pol II-transcribed genes.
The association with Pol II-transcribed genes prompted us to further investigate whether 6mA is correlated with transcription levels. We compared 6mA methylation levels in 10 gene quantiles ranked from high to low by their expression levels (Figure 3D). 6mA methylation levels remained at essentially the same levels in quantiles 1–6 (average methylation levels 75.2%), covering a wide range of expression levels (14352.94 ≥ RPKM ≥ 6.42). 6mA methylation levels dropped only slightly (though statistically significantly, Supplementary Table S4) for quantiles 7–9 (average methylation levels 70.4%; 6.42 ≥ RPKM ≥ 0.81), and more precipitously for the last quantile of least expressed genes (Supplementary Table S4) (average methylation levels 50.6%; 0.81 ≥ RPKM ≥ 0.01). Further analysis showed that 6mA methylation levels were only weakly correlated with expression levels of the associated genes (Pearson correlation coefficient = 0.11) (Figure 3E). Similarly, the total amount of 6mA in a gene body was also weakly correlated with its expression level (Pearson correlation coefficient = 0.15) (Figure 3E). Taken together, our results support the assertion that 6mA is less likely to be associated with silent genes, though it is not an unambiguous epigenetic mark for active genes.
6mA is preferentially associated with the linker DNA between well-positioned nucleosomes and/or nucleosomes containing histone variant H2A.Z
Early studies with enzymatic digestion revealed that some 6mA sites were preferentially localized in the linker DNA regions (17,18). We have recently mapped nucleosome distributions in Tetrahymena MAC by Illumina sequencing of DNA fragments generated by limited micrococcal nuclease digestion (MNase-Seq) (22,34). Here, our genome-wide analysis of 6mA in Tetrahymena MAC confirms its preferential association with the linker DNA as a general phenomenon. GBrowse views clearly showed that 6mA sites were accumulated in between nucleosome dyads (Figure 4A). Phasogram analysis and spectrum analysis of 6mA distribution revealed a very strong periodicity of ∼200 bp (Supplementary Figure S7A), the same as that of nucleosome arrays (34,57). Interestingly, a weak but discernible 3bp periodicity was revealed (Supplementary Figure S7A inset and 7B). The 3bp periodicity is characteristic of protein-coding sequences, perhaps arising from the selective pressure for proteins codons (58,59). Prominent 3 bp periodicity in 6mA distribution is consistent with its enrichment in protein-coding sequences (Figure 2B), probably underlain by the triplet protein codon in the gene body (60). Composite analysis of nucleosome distribution around 6mA sites confirmed the inverse relationship between 6mA and nucleosome distributions, with the two distribution curves oscillating in the opposite phase (Supplementary Figure S7C). Indeed, we found that 6mA sites, especially those with high methylation levels, were strongly excluded from the nucleosomal DNA (Figure 4B), and there was a strong positive correlation between methylation levels and distances to the nucleosome dyad (Pearson correlation coefficient = 0.68) (Figure 3E). The relationship between 6mA and nucleosome distributions in Tetrahymena is similar to that of 6mA in the green algae Chlamydomonas (5) and DNMT5-dependent 5mC in some algae and fungi (61), indicative of mechanistic coupling between the DNA methylation and nucleosome positioning.
Our previous work has demonstrated that nucleosome distributions in Tetrahymena are affected by both cis-determinants and trans-determinants (34). Trans-determinants, in particular transcription-associated trans-determinants, mainly contribute to the formation of stereotypical nucleosome arrays in the gene body in the MAC. These nucleosomes are usually well positioned (with high degrees of nucleosome positioning), due to ATP-dependent chromatin remodeling activities (62,63). Composite analysis of 6mA sites aligned to TSS revealed distinct peaks of 6mA downstream of TSS, in between the regularly spaced stereotypical nucleosome arrays in the gene body (Figure 4C). We observed that the linker DNA region between the +1 and +2 nucleosomes contained the highest level of 6mA. 6mA levels gradually diminished, as the stereotypical nucleosome arrays decayed and degrees of nucleosome positioning decreased further into the gene body (Figure 4C). By contrast, almost no 6mA was detected upstream of the +1 nucleosome (Figure 4C), despite a prominent nucleosome-free region at the promoter and large separations between TSS and the +1 nucleosome (average 138 bp) (34). We also observed that the stereotypical nucleosome arrays were prominent in the gene body with 6mA sites, as reflected by strong oscillations of nucleosome levels, with large peak-to-trough separations (Figure 4D). Gene bodies without 6mA showed a much noisier nucleosome distribution pattern, with small peak-to-trough separations (Figure 4D). Compared to genes with 6mA, genes without 6mA have significantly lower nucleosome positioning degree (p < 0.01), significantly shorter gene length (p < 0.01), but showed no significant difference in gene expression levels (p > 0.05) (Supplementary Figure S8). These observations revealed that 6mA was associated with well-positioned nucleosomes (Supplementary Figure S7D). This was further confirmed by correlation analysis, showing a strong positive correlation (Pearson correlation coefficient = 0.57) between 6mA density and degrees of positioning of adjacent nucleosomes (Figure 4E). Our results strongly suggest 6mA methylation is connected with transcription-associated trans-determinants that promote nucleosome positioning.
The enrichment of 6mA in the 5′ region of the gene body and its association with well-positioned nucleosomes are reminiscent of the behavior of the conserved histone variant H2A.Z (45,64). Prompted by this, we performed ChIP-Seq to map the genome-wide distribution of H2A.Z in Tetrahymena MAC. We observed striking associations between 6mA and H2A.Z. GBrowse views clearly showed that 6mA sites were accumulated between H2A.Z-containing nucleosomes (Figure 4A and Supplementary Figure S7D). Composite analysis confirmed H2A.Z was enriched in the 5′ region of the gene body in a pattern similar to that of 6mA (Figure 4F). We also performed composite analysis of all linker DNA regions: those with 6mA were flanked with high levels of H2A.Z, while those without were flanked with minimal levels of H2A.Z (Figure 4G). Additionally, composite analysis of H2A.Z aligned to TSS revealed the stereotypical nucleosome arrays in the gene body, with an oscillation pattern even stronger than that of all nucleosomes (Figure 4C), supporting the association between H2A.Z-containing and well-positioned nucleosomes. Indeed, we found strong positive correlations in pair-wise comparison of 6mA, H2A.Z and degrees of nucleosome positioning (Figure 4E).
DISCUSSION
Tetrahymena thermophila was among the first eukaryotes in which substantial amount of 6mA was found in DNA (14,16,18,41,46). Using 6mA-specific antibodies, we have systematically examined 6mA distribution in the Tetrahymena life cycle, confirming its exclusive association with the transcriptionally active MAC. We also provide, by SMRT sequencing, the first comprehensive, base pair-resolution map of 6mA sites in the MAC. Detailed analysis of the genomic distribution of 6mA, together with that of all nucleosomes and H2A.Z-containing nucleosomes, reveal many characteristics of this new epigenetic mark. Specifically, 6mA is preferentially associated with: (i) the AT motif, (ii) the linker DNA region, (iii) the 5′ end of the gene body, (iv) Pol II-transcribed genes, (v) well-positioned nucleosomes and (vi) H2A.Z-containing nucleosomes. These results indicate the presence of 6mA-specific methyltransferase(s) in Tetrahymena, and provide important clues for the molecular mechanism(s) by which this epigenetic mark is established and maintained.
6mA sites are determined by both DNA sequence and chromatin environment
We have shown that 6mA occurs preferentially in the 5′-AT-3′ motif. The preference is even stronger for highly methylated 6mA sites. This is indicative of the specificity of Tetrahymena 6mA-specific methyltransferase(s). However, in the AT-rich Tetrahymena genome (overall AT > 75%) (25,54), only a very small portion of adenine and the AT-motif are methylated ((17) and this study). Indeed, it has been reported that 6mA methylation is lost when a DNA fragment containing a methylated site is moved to a different genomic locus (65,66), suggesting that the genomic context and chromatin environment are crucial for 6mA occurrence.
We show that 6mA is enriched in the linker DNA, supporting early reports in which only a few sites were analyzed (17,20). This pattern is similar to the pattern of DNMT5-dependent CG methylation in diverse algae (61). It is also similar to that of 6mA in the green algae Chlamydomonas (5). More strikingly, these linker DNA regions with 6mA in Tetrahymena are usually flanked by well-positioned nucleosomes and/or H2A.Z-containing nucleosomes. Also of note, incorporation and removal of nucleosomal H2A.Z are catalyzed by a pair of highly conserved ATP-dependent chromatin remodelers, the SWR (in reference to Swr1 protein, a Swi2/Snf2-related adenosine triphosphatase) complex and the INO80 complex, respectively (67). Therefore, these results strongly suggest that 6mA methylation is connected to transcription-associated trans-determinants for nucleosome distribution, in particular H2A.Z and its cognate ATP-dependent chromatin remodeling complexes.
Our results also raise an important question concerning the cause and effect of 6mA deposition and nucleosome positioning. On the one hand, 6mA deposition, especially symmetric methylation of the AT motif, is likely to be excluded by steric hindrance (16,68,69) if the associated DNA is assembled into a nucleosome. Well-positioned nucleosomes may therefore force 6mA to be deposited preferentially in the linker DNA. On the other hand, 6mA, like other DNA modifications, may directly affect DNA structure, altering the intrinsic affinity of a DNA sequence for nucleosome assembly and thus shifting the nucleosome distribution. Indeed, it has been shown that periodically distributed 5mC clusters contribute directly to nucleosome positioning (61). We also favor the scenario that 6mA is one of the trans-determinants for nucleosome distribution, contributing significantly to nucleosome positioning.
6mA is associated with Pol II-transcribed genes
6mA has been associated with transcription in various ways. In the green algae Chlamydomonas, 6mA marks the TSS of actively transcribed genes (5). In early-diverging fungi, 6mA are associated with transcriptionally active genes (11). In C. elegans, a positive reciprocal regulation has been discovered among regulators controlling 6mA and the transcription activation marker H3K4me2 (6). In Drosophila, some 6mA peaks are enriched in the gene bodies of transposons, and deletion of the 6mA demethylase leads to increased expression of these transposons (10). During mouse embryonic stem cell differentiation, 6mA is linked to gene silencing of young LINE-1 transposons (12). It has long been speculated that 6mA in Tetrahymena is connected with transcription, as it is only present in the transcriptionally active MAC (14). However, the level and pattern of 6mA remain constant in the MAC of vegetative and starved cells (14,20,41,70) whereas the transcription level upon starvation was dramatically reduced (71), thus challenging the correlation of 6mA with transcription. The results of the present study do not support that 6mA is required for transcription in Tetrahymena. Firstly, 6mA levels are only weakly correlated with transcription levels in the MAC: there were a number of highly expressed genes with low 6mA levels, and many minimally expressed genes with high 6mA levels. Secondly, 6mA is absent in the meiotic MIC and the early developing MAC, both of which feature strong and global transcription activities. Previous studies have shown that the meiotic MIC generates abundant non-coding RNA (42), while the early developing MAC generates both mRNA and non-coding RNA (72,73). Nevertheless, 6mA may be an integral part of the chromatin environment affecting Pol II-transcribed genes and its transcriptional effect may be mediated by nucleosome distribution, as discussed above. Alternatively, 6mA may be recognized by its cognate reader(s), which in turn interacts with the transcriptional and co-transcriptional machineries.
A large number of 6mA sites in Tetrahymena are only intermediately methylated. This can lead to two different 6mA states, methylated or unmethylated, in the 45 copies of the same allele in the Tetrahymena MAC. This heterogeneity may fine-tune gene expression among the 45 copies. If the 6mA states are maintained after DNA replication (see below), the assortment of these epi-alleles—with the same DNA sequence but different epigenetic states—into different progeny can increase population-level phenotypic plasticity.
6mA is a potential epigenetic mark likely maintained by a methyltransferase(s)
In the Tetrahymena MAC, 6mA accumulates at robust levels and shows a highly specific pattern of distribution. This indicates that it is deposited by a specific methyltransferase(s), rather than random uncatalyzed methylation. 6mA distribution pattern in Tetrahymena is most similar to that in the green algae Chlamydomonas (5), and to a lesser degree, to that in early-diverging fungi (11). Indeed, 6mA levels in these organisms are orders of magnitude higher than in the several animals studied so far (7,8,12). Furthermore, 6mA in these organisms is more directly and positively linked to transcription. Based on these observations, we propose evolutionary conservation of 6mA, its methyltransferases, and relevant functions in these unicellular eukaryotes. 6mA in animals, even if enzymatically catalyzed, may have diverged in the molecular machinery involved and relevant functions.
To qualify as an epigenetic mark, 6mA needs to be maintained after DNA replication. Indeed, 6mA pattern is likely to be very stable, at least at the population level, as indicated by the consistency between our mapping result and earlier reports (14,18,20,41,70). Furthermore, the presence of a large number of 6mA sites with high methylation levels during the vegetative life cycle indicate that 6mA deposition occurs very quickly after DNA replication. It should also be noted that 6mA methylation also occurs on non-replicating DNA in starved cells, which may be attributed to methylation turnover (74). We therefore conclude that the specific methyltransferase(s) in Tetrahymena is capable of effectively maintaining the distinct 6mA pattern.
How is the 6mA pattern transmitted to the next generation? First, we believe we can rule out the mechanism based on preferential methylation of hemi-methylated sites, as is the case for DNA cytosine methylation maintenance by DNMT1 (75). This is due to the presence in Tetrahymena of a large number of AT sites asymmetrically methylated only on one strand. The proximity to the nucleosome dyad is comparable between symmetric and asymmetric AT motif sites (Supplementary Figure S7E). Therefore, it remains possible that there are two distinct methyltransferases responsible for symmetrically and asymmetrically methylated AT sites, respectively. We have not ruled out the possibility that the methylated AT motif, in particular its asymmetrically methylated state, may be specifically recognized by its cognate reader, which in turn recruits 6mA-specific methyltransferase to propagate the mark locally (but not necessarily to generate the symmetrically methylated AT motif). We favor the hypothesis that the distinct 6mA pattern in the gene body is maintained by a methyltransferase(s) recruited to the promoter region of Pol II-transcribed genes with other chromatin regulators, including various ATP-dependent chromatin remodelers (76,77). The methyltransferase may even access the linker DNA in coordination with ATP-dependent chromatin remodelers. In this way, 6mA may be maintained just like transcription-associated histone modifications, such as H3K4 methylation (78). It should be noted that several potential 6mA methyltransferases and demethylases in Tetrahymena have been predicted (5,79,80), but we have not been able to validate any of these candidates by functional analyses. Identification of 6mA-specific methyltransferase(s) in Tetrahymena, a model organism ideal for biochemical and genetic studies, will allow us to further explore the molecular mechanism and function of this rediscovered epigenetic mark in eukaryotes.
DATA AVAILABILITY
The latest SB210 MAC genome can be found at the TGD (http://ciliate.org). GEO accession number for publicly available Tetrahymena datasets: GSE96521.
Supplementary Material
ACKNOWLEDGEMENTS
Our thanks are due to Dr. Weibo Song (Ocean University of China, China) for his kind help in preparing the draft and illustrations. We also thank Dr. Alan Warren (Natural History Museum, UK) for English editing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Science Foundation of China [31522051 to S.G.]; Qingdao National Laboratory for Marine Science and Technology (Aoshan Talents Program) [2015ASTP to S.G.]; Fundamental Research Funds for the Central Universities [201562029 to S.G.]; National Institutes of Health [R01 GM087343 to Y.L.]; National Science Foundation [MCB 1411565 to Y.L.]. Funding for open access charge: Natural Science Foundation of China [31522051] (to S.G.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Zacharias W. Jost JP, Saluz HP. Methylation of cytosine influences the DNA structure. DNA Methylation. 1993; 64:Basel: Birkhäuser Verlag; 27–38. [DOI] [PubMed] [Google Scholar]
- 2. Reisenauer A., Kahng L.S., McCollum S., Shapiro L.. Bacterial DNA methylation: a cell cycle regulator?. J. Bacteriol. 1999; 181:5135–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bird A. The essentials of DNA methylation. Cell. 1992; 70:5–8. [DOI] [PubMed] [Google Scholar]
- 4. Hattman S., Kenny C., Berger L., Pratt K.. Comparative study of DNA methylation in three unicellular eucaryotes. J. Bacteriol. 1978; 135:1156–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fu Y., Luo G., Chen K., Deng X., Yu M., Han D., Hao Z., Liu J., Lu X., Doré L.C.. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell. 2015; 161:879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Greer E.L., Blanco M.A., Gu L., Sendinc E., Liu J., Aristizábal-Corrales D., Hsu C.-H., Aravind L., He C., Shi Y.. DNA methylation on N6-adenine in C. elegans. Cell. 2015; 161:868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koziol M.J., Bradshaw C.R., Allen G.E., Costa A.S., Frezza C., Gurdon J.B.. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat. Struct. Mol. Biol. 2016; 23:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu J., Zhu Y., Luo G., Wang X., Yue Y., Wang X., Zong X., Chen K., Yin H., Fu Y.. Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat. Commun. 2016; 7:13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ratel D., Ravanat J.L., Berger F., Wion D.. N6-methyladenine: the other methylated base of DNA. Bioessays. 2006; 28:309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang G., Huang H., Liu D., Cheng Y., Liu X., Zhang W., Yin R., Zhang D., Zhang P., Liu J.. N6-methyladenine DNA modification in Drosophila. Cell. 2015; 161:893–906. [DOI] [PubMed] [Google Scholar]
- 11. Mondo S.J., Dannebaum R.O., Kuo R.C., Louie K.B., Bewick A.J., LaButti K., Haridas S., Kuo A., Salamov A., Ahrendt S.R.. Widespread adenine N6-methylation of active genes in fungi. Nat. Genet. 2017; 49:964–968. [DOI] [PubMed] [Google Scholar]
- 12. Wu T.P., Wang T., Seetin M.G., Lai Y., Zhu S., Lin K., Liu Y., Byrum S.D., Mackintosh S.G., Zhong M.. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature. 2016; 532:329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cummings D.J., Tait A., Goddard J.M.. Methylated bases in DNA from Paramecium aurelia. Biochim. Biophys. Acta. 1974; 374:1–11. [DOI] [PubMed] [Google Scholar]
- 14. Gorovsky M.A., Hattman S., Pleger G.L.. [6N] Methyl adenine in the nuclear DNA of a eucaryote, Tetrahymena pyriformis. J. Cell Biol. 1973; 56:697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rae P.M., Spear B.B.. Macronuclear DNA of the hypotrichous ciliate Oxytricha fallax. Proc. Natl. Acad. Sci. U.S.A. 1978; 75:4992–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bromberg S., Pratt K., Hattman S.. Sequence specificity of DNA adenine methylase in the protozoan Tetrahymena thermophila. J. Bacteriol. 1982; 150:993–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karrer K.M., VanNuland T.A.. Methylation of adenine in the nuclear DNA of Tetrahymena is internucleosomal and independent of histone H1. Nucleic Acids Res. 2002; 30:1364–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harrison G.S., Findly R.C., Karrer K.. Site-specific methylation of adenine in the nuclear genome of a eucaryote, Tetrahymena thermophila. Mol. Cell. Biol. 1986; 6:2364–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harrison G.S., Karrer K.M.. DNA synthesis, methylation and degradation during conjugation in Tetrahymena thermophila. Nucleic Acids Res. 1985; 13:73–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pratt K., Hattman S.. Deoxyribonucleic acid methylation and chromatin organization in Tetrahymena thermophila. Mol. Cell. Biol. 1981; 1:600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sweet M.T., Allis C.D.. Spector DL, Goldman RD, Leinwand LA. Culture and biochemical analysis of cells. Cells: a Laboratory Manual. 1998; 1:NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 22. Chen X., Gao S., Liu Y., Wang Y., Wang Y., Song W.. Enzymatic and chemical mapping of nucleosome distribution in purified micro-and macronuclei of the ciliated model organism, Tetrahymena thermophila. Sci. China Life Sci. 2016; 59:909–919. [DOI] [PubMed] [Google Scholar]
- 23. Cuddapah S., Barski A., Cui K., Schones D.E., Wang Z., Wei G., Zhao K.. Native chromatin preparation and Illumina/Solexa library construction. Cold Spring Harb. Protoc. 2009; 2009, doi:10.1101/pdb.prot5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eisen J.A., Coyne R.S., Wu M., Wu D., Thiagarajan M., Wortman J.R., Ren Q., Amedeo P., Jones K.M., Tallon L.J.. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006; 4:e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stover N.A., Punia R.S., Bowen M.S., Dolins S.B., Clark T.G.. Tetrahymena Genome Database Wiki: a community-maintained model organism database. Database. 2012; 2012:bas007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zang C., Schones D.E., Zeng C., Cui K., Zhao K., Peng W.. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009; 25:1952–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A.. Circos: an information aesthetic for comparative genomics. Genome Res. 2009; 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stein L.D. Using GBrowse 2.0 to visualize and share next-generation sequence data. Brief. Bioinform. 2013; 14:162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Valouev A., Johnson S.M., Boyd S.D., Smith C.L., Fire A.Z., Sidow A.. Determinants of nucleosome organization in primary human cells. Nature. 2011; 474:516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dieci G., Conti A., Pagano A., Carnevali D.. Identification of RNA polymerase III-transcribed genes in eukaryotic genomes. Biochim. Biophys. Acta. 2013; 1829:296–305. [DOI] [PubMed] [Google Scholar]
- 33. Griffiths-Jones S. Annotating noncoding RNA genes. Annu. Rev. Genomics Hum. Genet. 2007; 8:279–298. [DOI] [PubMed] [Google Scholar]
- 34. Xiong J., Gao S., Dui W., Yang W., Chen X., Taverna S.D., Pearlman R.E., Ashlock W., Miao W., Liu Y.. Dissecting relative contributions of cis-and trans-determinants to nucleosome distribution by comparing Tetrahymena macronuclear and micronuclear chromatin. Nucleic Acids Res. 2016; 44:10091–10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xiong J., Lu X., Zhou Z., Chang Y., Yuan D., Tian M., Zhou Z., Wang L., Fu C., Orias E.. Transcriptome analysis of the model protozoan, Tetrahymena thermophila, using deep RNA sequencing. PLoS One. 2012; 7:e30630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T.. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014; 42:W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Norusis M. SPSS 16.0 Statistical Procedures Companion. 2008; Upper Saddle River: Prentice Hall Press. [Google Scholar]
- 38. Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M.. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007; 35:W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao S., Xiong J., Zhang C., Berquist B.R., Yang R., Zhao M., Molascon A.J., Kwiatkowski S.Y., Yuan D., Qin Z.. Impaired replication elongation in Tetrahymena mutants deficient in histone H3 Lys 27 monomethylation. Genes Dev. 2013; 27:1662–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao X., Wang Y., Wang Y., Liu Y., Gao S.. Histone methyltransferase TXR1 is required for both H3 and H3. 3 lysine 27 methylation in the well-known ciliated protist Tetrahymena thermophila. Sci. China Life Sci. 2017; 60:264–270. [DOI] [PubMed] [Google Scholar]
- 41. Blackburn E.H., Pan W.C., Johnson C.C.. Methylation of ribosomal RNA genes in the macronucleus of Tetrahymena thermophila. Nucleic Acids Res. 1983; 11:5131–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chalker D.L., Yao M.-C.. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 2001; 15:1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nanney D.L. Nucleo-cytoplasmic interaction during conjugation in Tetrahymena. Biol. Bull. 1953; 105:133–148. [Google Scholar]
- 44. Pfeffer U., Ferrari N., Tosetti F., Vidali G.. Histone acetylation in conjugating Tetrahymena thermophila. J. Cell Biol. 1989; 109:1007–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stargell L.A., Bowen J., Dadd C.A., Dedon P.C., Davis M., Cook R.G., Allis C.D., Gorovsky M.A.. Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila. Genes Dev. 1993; 7:2641–2651. [DOI] [PubMed] [Google Scholar]
- 46. Capowski E.E., Wells J.M., Karrer K.M.. Maintenance of methylation patterns in Tetrahymena thermophila. Gene. 1988; 74:101–104. [DOI] [PubMed] [Google Scholar]
- 47. White T., McLaren N., Allen S.. Methylation site within a facultatively persistent sequence in the macronucleus of Tetrahymena thermophila. Mol. Cell. Biol. 1986; 6:4742–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramsahoye B.H., Biniszkiewicz D., Lyko F., Clark V., Bird A.P., Jaenisch R.. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smith Z.D., Meissner A.. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013; 14:204–220. [DOI] [PubMed] [Google Scholar]
- 50. Ziller M.J., Müller F., Liao J., Zhang Y., Gu H., Bock C., Boyle P., Epstein C.B., Bernstein B.E., Lengauer T.. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011; 7:e1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Song J., Rechkoblit O., Bestor T.H., Patel D.J.. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science. 2011; 331:1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Archambault J., Friesen J.D.. Genetics of eukaryotic RNA polymerases I, II, and III. Microbiol. Rev. 1993; 57:703–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reeder R.H. McKnight S, Yamamoto KR. Regulation of transcription by RNA polymerase I. Transcriptional Regulation. 1992; NY: Cold Spring Harbor Laboratorypress; 315–348. [Google Scholar]
- 54. Gorovsky M.A. Genome organization and reorganization in Tetrahymena. Annu. Rev. Genet. 1980; 14:203–239. [DOI] [PubMed] [Google Scholar]
- 55. Luo G., Wang F., Weng X., Chen K., Hao Z., Yu M., Deng X., Liu J., He C.. Characterization of eukaryotic DNA N6-methyladenine by a highly sensitive restriction enzyme-assisted sequencing. Nat. Commun. 2016; 7:11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vovis G.F., Lacks S.. Complementary action of restriction enzymes endo R· DpnI and endo R· DpnII on bacteriophage f1 DNA. J. Mol. Biol. 1977; 115:525–538. [DOI] [PubMed] [Google Scholar]
- 57. Beh L.Y., Müller M.M., Muir T.W., Kaplan N., Landweber L.F.. DNA-guided establishment of nucleosome patterns within coding regions of a eukaryotic genome. Genome Res. 2015; 25:1727–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gutierrez G., Oliver J., Marin A.. On the origin of the periodicity of three in protein coding DNA sequences. J. Theor. Biol. 1994; 167:413–414. [DOI] [PubMed] [Google Scholar]
- 59. Tsonis A.A., Elsner J.B., Tsonis P.A.. Periodicity in DNA coding sequences: implications in gene evolution. J. Theor. Biol. 1991; 151:323–331. [DOI] [PubMed] [Google Scholar]
- 60. Crick F.H. The origin of the genetic code. J. Mol. Biol. 1968; 38:367–379. [DOI] [PubMed] [Google Scholar]
- 61. Huff J.T., Zilberman D.. Dnmt1-independent CG methylation contributes to nucleosome positioning in diverse eukaryotes. Cell. 2014; 156:1286–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Havas K., Whitehouse I., Owen-Hughes T.. ATP-dependent chromatin remodeling activities. Cell. Mol. Life Sci. 2001; 58:673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jaskelioff M., Gavin I.M., Peterson C.L., Logie C.. SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol. Cell. Biol. 2000; 20:3058–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Guillemette B., Bataille A.R., Gévry N., Adam M., Blanchette M., Robert F., Gaudreau L.. Variant histone H2A. Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005; 3:e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Karrer K.M., VanNuland T.A.. Position effect takes precedence over target sequence in determination of adenine methylation patterns in the nuclear genome of a eukaryote, Tetrahymena thermophila. Nucleic Acids Res. 1998; 26:4566–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Van Nuland T.A., Capowski E.E., Karrer K.M.. Position effect for adenine methylation in the macronuclear DNA of Tetrahymena. Gene. 1995; 157:235–237. [DOI] [PubMed] [Google Scholar]
- 67. Billon P., Côté J.. Precise deposition of histone H2A. Z in chromatin for genome expression and maintenance. Biochim. Biophys. Acta. 2012; 1819:290–302. [DOI] [PubMed] [Google Scholar]
- 68. Rando O.J., Ahmad K.. Rules and regulation in the primary structure of chromatin. Curr. Opin. Cell Biol. 2007; 19:250–256. [DOI] [PubMed] [Google Scholar]
- 69. Rube H.T., Song J.S.. Quantifying the role of steric constraints in nucleosome positioning. Nucleic Acids Res. 2014; 42:2147–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Karrer K.M., Stein-Gavens S.. Constancy of adenine methylation in Tetrahymena macronuclear DNA. J. Protozool. 1990; 37:409–414. [DOI] [PubMed] [Google Scholar]
- 71. Bannon G.A., Calzone F.J., Bowen J.K., Allis C.D., Gorovsky M.A.. Multiple, independently regulated, polyadenylated messages for histone H3 and H4 in Tetrahymena. Nucleic Acids Res. 1983; 11:3903–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Noto T., Kataoka K., Suhren J.H., Hayashi A., Woolcock K.J., Gorovsky M.A., Mochizuki K.. Small-RNA-mediated genome-wide trans-recognition network in Tetrahymena DNA elimination. Mol. Cell. 2015; 59:229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Weiske-Benner A., Eckert W.A.. Differentiation of nuclear structure during the sexual cycle in Tetrahymena thermophila: I. Development and transcriptional activity of macronuclear anlagen. Differentiation. 1985; 28:225–236. [Google Scholar]
- 74. Harrison G.S., Karrer K.M.. Methylation of replicating and nonreplicating DNA in the ciliate Tetrahymena thermophila. Mol. Cell. Biol. 1989; 9:828–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Razin A., Szyf M.. DNA methylation patterns formation and function. Biochim. Biophys. Acta. 1984; 782:331–342. [DOI] [PubMed] [Google Scholar]
- 76. Kornberg R.D. Eukaryotic transcriptional control. Trends Biochem. Sci. 1999; 24:M46–M49. [PubMed] [Google Scholar]
- 77. Yen K., Vinayachandran V., Pugh B.F.. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near+ 1 nucleosomes. Cell. 2013; 154:1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Okitsu C.Y., Hsieh J.C.F., Hsieh C.-L.. Transcriptional activity affects the H3K4me3 level and distribution in the coding region. Mol. Cell. Biol. 2010; 30:2933–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Iyer L.M., Abhiman S., Aravind L.. Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci. 2011; 101:25–104. [DOI] [PubMed] [Google Scholar]
- 80. Iyer L.M., Zhang D., Aravind L.. Adenine methylation in eukaryotes: Apprehending the complex evolutionary history and functional potential of an epigenetic modification. Bioessays. 2016; 38:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The latest SB210 MAC genome can be found at the TGD (http://ciliate.org). GEO accession number for publicly available Tetrahymena datasets: GSE96521.