Abstract
The bacterial signal recognition particle (SRP) is part of the machinery that targets ribosomes synthesizing membrane proteins to membrane-embedded translocons co-translationally. Recognition of nascent membrane proteins occurs by virtue of a hydrophobic signal-anchor sequence (SAS) contained in the nascent chain, usually at the N terminus. Here we use fluorescence-based stopped-flow to monitor SRP-ribosome interactions with actively translating ribosomes while an SRP substrate is synthesized and emerges from the peptide exit tunnel. The kinetic analysis reveals that, at cellular concentrations of ribosomes and SRP, SRP rapidly binds to translating ribosomes prior to the emergence of an SAS and forms an initial complex that rapidly rearranges to a more stable engaged complex. When the growing peptide reaches a length of ∼50 amino acids and the SAS is partially exposed, SRP undergoes another conformational change which further stabilizes the complex and initiates targeting of the translating ribosome to the translocon. These results provide a reconciled view on the timing of high-affinity targeting complex formation, while emphasizing the existence of preceding SRP recruitment steps under conditions of ongoing translation.
INTRODUCTION
The universally conserved signal recognition particle (SRP) pathway directs nascent proteins for co-translational membrane translocation or insertion. In Escherichia coli, SRP consists of one protein (Ffh) and a 114 nucleotide-long RNA (4.5S RNA) (1,2). SRP binds to ribosomes synthesizing membrane proteins and recognizes a hydrophobic signal anchor sequence (SAS) that is usually located at or near the N terminus of the nascent chain. Targeting is accomplished with the assistance of the SRP receptor (FtsY in bacteria) which interacts with the translocon at the membrane. In bacteria, the translocon is a ternary protein complex consisting of SecY, SecE and SecG. FtsY is activated for binding to SRP in complex with the translocon (3) and membrane phospholipids (4,5). Thus, the assembly of the quaternary transfer complex is restricted to the translocon at the membrane. The structure of the quaternary complex shows that the translocon is positioned in the vicinity of the peptide exit by interactions with FtsY (6). Following transfer of the SAS to the SecYEG translocon, SRP and FtsY hydrolyze GTP, which leads to disassembly of the targeting complex allowing SRP and FtsY to enter another round of targeting (for reviews see (7,8)). The translating ribosome is then anchored to the translocon such that the peptide exit tunnel of the ribosome forms a continuous conduit with the translocon pore (9,10). Nascent peptides containing hydrophobic signal sequences pass through this conduit and leave the central pore of the translocon through the open lateral gate to move into the phospholipid bilayer. Opening of the lateral gate is promoted on binding of ribosomes and SAS insertion into the translocon (11). The entire process, from targeting to membrane insertion, occurs co-translationally. Ribosomes synthesizing SRP-dependent secretory proteins are targeted to the translocon co-translationally in the same way, except that the SRP-specific signal sequence is cleaved off after membrane insertion.
Recognition of the nascent SAS and targeting of the translating ribosome to the membrane is a multistep process. In the first step of targeting in bacteria, SRP is recruited to ribosomes bearing SRP-specific nascent peptide chains comprising an SAS. The analysis of SRP interaction with stalled ribosome-nascent-chain complexes (RNCs) revealed that SRP binds to ribosomes with reasonably high affinity (Kd = 50–100 nM) even in the absence of a nascent chain (12–14). The affinity for SRP increases nearly 100-fold (Kd = 1 nM) when the ribosome carries a nascent chain just filling the peptide exit tunnel (35 amino acids) (12). Interestingly, this high affinity was observed regardless of whether or not the nascent chain comprised an SRP-specific SAS. When the nascent chain is long enough to be exposed outside the peptide exit tunnel, the high-affinity interaction is maintained only for SRP substrates, i.e. nascent peptides comprising a signal or signal-anchor sequence (12–14). Exposure of a non-SRP substrate nascent chain reduces SRP binding affinity substantially, thus leading to complex disassembly. In eukaryotes, SRP appears to bind strongly to short-chain RNCs when an SAS resides within the peptide exit tunnel, and further stabilization occurs after emergence of the signal sequence outside the ribosome (15,16). The kinetic analysis of SRP binding to stalled RNCs in the bacterial system identified three phases of SRP binding differentiated mainly by their dissociation rates (13). According to that analysis, SRP rapidly scans all ribosomes and becomes kinetically stabilized when the peptide exit tunnel is partially filled with nascent peptides of any sequence. This ‘standby mode’ would allow SRP to await the emergence of the nascent chain before either binding an SAS and progressing to a stabilized targeting complex or rapidly dissociating from non-substrate RNCs.
Recent single-molecule fluorescence and ribosome-profiling studies (17,18) revealed no evidence of SRP interaction with translating RNCs until after the nascent chain emerged from the peptide exit tunnel, thus challenging the existence of the scanning and standby modes of SRP. While it is difficult to argue against the existence of transient recruitment steps on the basis of negative data alone, there is a fundamental difference between our previous work and the more recent studies, which used stalled or actively translating RNCs, respectively. Here, we address this controversy by a detailed quantitative analysis of the kinetics and thermodynamics of SRP interaction with actively translating ribosomes. We employ real-time fluorescence and fluorescence resonance energy transfer (FRET) to monitor SRP binding to ribosomes translating in a purified in-vitro translation system from Escherichia coli. The main findings are that SRP rapidly scans ribosomes very early at the onset of translation, forming a ’standby’ complex that, upon (partial) emergence of an SRP-specific SAS from the ribosome, rearranges to a high-affinity targeting complex. Subsequently, the targeting complex can interact with the SRP receptor at the translocon to form a quaternary complex in which the nascent peptide is released from SRP and inserted into the translocon to initiate membrane insertion.
MATERIALS AND METHODS
Materials
If not stated otherwise, experiments were carried out in buffer A (25 mM HEPES, pH 7.5, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, 10% glycerol) at 37°C. A few experiments were carried out in HiFi buffer containing polyamines (20 mM Tris, pH 7.5, 70 mM NH4Cl, 30 mM KCl, 3.5 mM MgCl2, 8 mM putrescine, 0.5 mM spermidine), as indicated. 4.5S RNA and Lep mRNAs were prepared by in-vitro transcription as described previously (19). Ffh from E. coli with a Bodipy label at position 430 and ribosomes with an MDCC label at position 21 on protein L23 were prepared according to published methods (13). Briefly, the E. coli Ffh variant bearing a single cysteine at position 430 was recombinantly expressed, purified, and labeled with Bodipy-FL-maleimide (Thermo Fisher Scientific). Ribosomes incorporating an L23 variant with cysteine at position 21 were isolated from E. coli and the individual subunits purified prior to labeling of 50S subunits with MDCC-maleimide (Invitrogen). Following activation in buffer A containing 20 mM MgCl2 at 37°C for 30 min, 70S ribosomes were reconstituted from the subunits, flash-frozen in liquid nitrogen, and stored at –80°C. Initiation factors and elongation factors from E. coli were expressed as recombinant proteins and purified while total aminoacyl tRNA was prepared from E. coli total tRNA (Roche) according to protocols published elsewhere (20–22). Pyruvate kinase (from rabbit muscle) was purchased from Roche, and proteinase K (from Tritirachium album) from Sigma.
Preparation of ribosome complexes
Initiation complexes were prepared in buffer A by incubating MDCC-labeled ribosomes (1 μM) with initiation factors (IF1, IF2 and IF3; 1.5 μM each), mRNA (5–10 μM), fMet-tRNAfMet (2.5 μM), and GTP (1 mM) for 1 h at 37°C. Initiation efficiency was optimized to >95% by titration of each mRNA and confirmed by nitrocellulose filtration. SRP was formed by incubating 4.5S RNA (30 μM), Bpy-Ffh (30 μM), and GTP (1 mM) in buffer A at 25°C for 5 min. EF-Tu was incubated with GTP, phosphoenol pyruvate (PEP), pyruvate kinase, and EF-Ts in buffer A for 15 min at 37°C to convert all EF-Tu to the GTP-bound form prior to translation.
Co-translational SRP recruitment
Recruitment of SRP to translating ribosomes was monitored at 37°C in a stopped-flow apparatus (Applied Photophysics) by exciting the donor fluorophore MDCC at 410 nm and recording fluorescence emission after passing through a KV 530 long-pass filter (Schott). Translation elongation was induced by rapid mixing of MDCC-labeled initiation complex (25 μM, final concentration after mixing) with a solution containing EF-Tu (15 μM), EF-G (2 μM), EF-Ts (0.1 μM), total aminoacyl-tRNA, and Bpy-labeled SRP. Leu-tRNALeu in the total aminoacyl-tRNA was 14C-labeled and present at 1.35 μM in translation reactions. This provided a 3-fold excess of Leu-tRNALeu over leucine codons in the longest mRNA, Lep75 mRNA, which encodes nine leucines. All solutions contained GTP (1 mM), PEP (3 mM), and pyruvate kinase (0.1 μg/ml). Between seven and nine replicates of each experiment were carried out for 500 s.
Global fitting analysis
Global fitting analysis was carried out using KinTek Global Kinetic Explorer Professional Version 4.0.15023 (23). Replicate traces were averaged prior to global fitting. To accommodate the different starting fluorescence of each average trace (which increased linearly with Bpy-SRP concentration), a different fluorescence offset was provided for each fluorescence signal. These offsets were initially fitted, but constrained for the final fit. The kinetic constants and errors reported are the best-fit values and standard errors in the fit, respectively.
Translation time courses
Translation of Lep75 mRNA was performed at 37°C using components and concentrations equivalent to those used in the stopped-flow experiments, except that Bpy-labeled Met-tRNAfMet was used to prepare initiation complexes and SRP was omitted. Aliquots were removed at various times during translation and quenched by addition of 2% NH3. Peptides carrying an N-terminal Bpy label were released hydrolytically by incubation at 37°C for 30 min and separated on Tris-Tricine SDS-PAGE as previously described (24). Gels were imaged using an FLA-9000 fluorescence scanner (Fujifilm) and 488 nm laser excitation. Bands corresponding to peptides with 50 or more amino acids were identified relative to a Lep50 standard, and quantified with ImageJ (free download from NIH). Translation efficiencies were calculated as percent of the intensity at 500 s and averages and standard deviations were computed for each time point based on three or four independent experiments.
Proteinase K digestion
Translation of Lep75 mRNA was performed at 37°C with the same concentrations and conditions as in the stopped-flow experiments, except that SRP was omitted and unlabeled 70S ribosomes were used for initiation complex formation. At various time points, aliquots containing 2.5 pmol of ribosomes were removed, mixed with PK (1.4 mg/ml in 5 mM CaCl2, final concentrations) that had been pre-incubated at 37°C. Digestion was carried out for 10 s prior to quenching in 0.5 M KOH. Subsequently, peptides were released from tRNA by incubating at 37°C for 30 min and precipitated by adding equal volumes of 10% TCA. After standing on ice for at least 30 min, precipitated peptides were filtered over 0.45 μm nitrocellulose filters and rinsed with 10 ml of cold 5% TCA. Filters were washed with cold 30% isopropanol prior to dissolving in scintillation cocktail, and radioactive f[3H]Met was quantified by scintillation counting.
RESULTS
Two phases of co-translational SRP binding to translating ribosomes
In order to monitor SRP binding to translating ribosomes, we employed a FRET system with the donor fluorophore (MDCC) attached to ribosomal protein L23 and the acceptor (Bodipy FL, Bpy) positioned in the M domain of SRP protein Ffh. This FRET pair was used previously to monitor interactions between SRP and ribosomes (13). These labels do not impair SRP-ribosome interactions as equilibrium titrations yielded Kd values around 100 nM comparable with other labels at different positions (12–14). Synthesis of leader peptidase (Lep), an inner-membrane protein with an N-terminal SAS (residues 4–22) that is recognized by SRP, was carried out on donor-labeled ribosomes in a highly efficient in-vitro translation system from E. coli (24,25). To initiate translation, we rapidly mixed donor-labeled 70S initiation complexes containing Lep mRNA of different lengths with a solution containing elongation factors, total aminoacyl-tRNA, GTP and acceptor-labeled SRP in a stopped-flow apparatus. We followed the interaction of SRP with ribosomes in real time by monitoring FRET-based acceptor fluorescence changes. Control measurements were performed with donor-only and acceptor-only complexes.
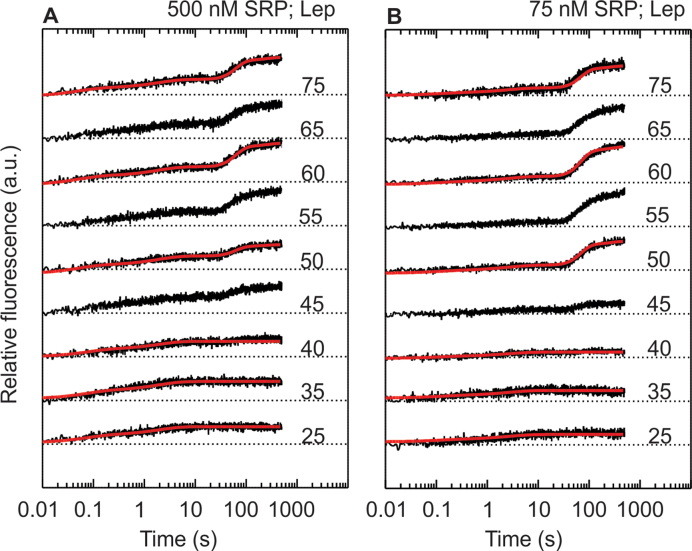
We started by translating an mRNA coding for the first 75 amino acids of Lep in the presence of Bpy-labeled SRP at high concentration (500 nM; Figure 1A, top panel). The resulting fluorescence time course reveals a rapid fluorescence increase during the first 10 s and an additional increase starting at about 40 s. Preliminary analysis of the time courses by exponential fitting reveals that the rapid phase comprises two kinetic steps. With longer nascent peptides, an additional slower fluorescence increase is observed starting at ∼40 s. The donor-only control reveals that the rapid fluorescence increase is predominantly due to a change in donor fluorescence, indicating a change in the environment of the label. The signal increase starting at 40 s is due to FRET, indicating a rearrangement of SRP on the ribosome (Supplementary Figure S1). To determine the length of the nascent chain that is required to induce these signal changes, the experiments were carried out with Lep mRNAs of different lengths (from 25 to 75 codons) (Figure 1A). The rapid, biphasic increase of the signal is observed independent of mRNA length, suggesting that it represents binding of SRP to translating ribosomes carrying short nascent peptides before the SAS emerges from the polypeptide exit tunnel. The rapid fluorescence increase is independent of the presence of an SAS in the growing peptide, as RNCs synthesizing HemK, which lacks an SAS, induce the same effect (Supplementary Figure S2). Starting at Lep mRNA lengths of 45–50 codons, the slow FRET increase becomes apparent (at 40 s).This slow FRET increase is observed only when the nascent peptide contains an SAS.
Figure 1.
Rapid and slow phases of SRP recruitment to translating ribosomes. Translation of the first 25–75 amino acids of Lep was carried out in a stopped-flow apparatus in the presence of 500 nM (A) or 75 nM (B) SRP (final concentrations) while the fluorescence of Bpy-labeled SRP was monitored. Translation was initiated by rapidly mixing MDCC-labeled initiation complexes (25 nM) with translation mix and SRP at 37°C (Methods). Fluorescence traces are offset for clarity, and dotted lines indicate the respective initial fluorescence. Red lines are obtained from global fitting of the associated data sets to the model in Figure 3.
The time preceding the FRET increase starting at 40 s likely reflects the minimum time required to synthesize a peptide chain of 45–50 amino acids. To test this directly, time courses of Lep75 mRNA translation were performed and translation products analyzed by SDS-PAGE. Products were visualized by the fluorescence of Bpy at the N terminus of the nascent chain which was introduced using Bpy-labeled Met-tRNAfMet for initiation (24) (Supplementary Figure S3). These experiments show that nascent chains of 50 amino acids and longer appear after 40 s of translation. Thus, the initial rapid recruitment of SRP to the ribosome is followed by a conformational rearrangement, the rate of which is limited by translation. The rearrangement is reflected in a FRET increase and correlates with the appearance of (part of) the Lep-SAS outside the peptide exit tunnel.
We also performed co-translational SRP recruitment experiments at lower concentration of Bpy-labeled SRP (75 nM). The resulting fluorescence traces (Figure 1B) are qualitatively similar to those observed at high SRP concentration, although the relative fluorescence change during the rapid binding phase is smaller. This suggests that SRP rapidly binds to ribosomes carrying short nascent chains with relatively low affinity (Kd > 75 nM). In contrast, the final fluorescence levels are similar at both concentrations of SRP for Lep-RNCs with 50 amino acids or more (Figure 1) indicating that after the nascent chain reaches a length of 50 amino acids, SRP binding is already saturated at 75 nM. This suggests an increase in affinity (Kd < 75 nM) when the nascent chain reaches a length of 50 amino acids.
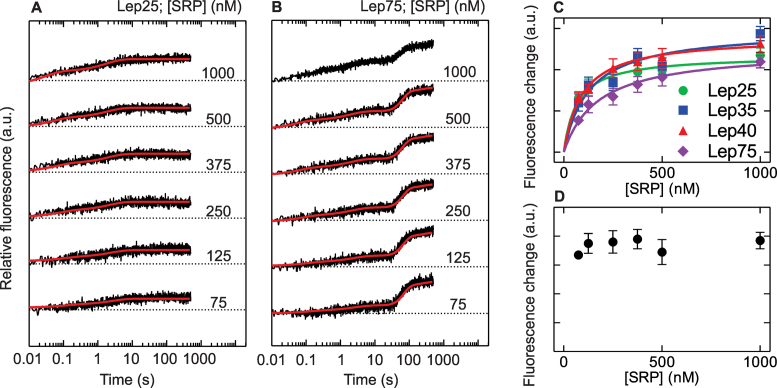
To obtain better estimates for the affinities of SRP–RNC interactions, we systematically varied the SRP concentration during co-translational recruitment to various Lep-RNCs (Figure 2). Increasing the concentration of SRP increases the relative amplitude of the rapid phase for RNCs with different lengths of the nascent peptide (up to 40 amino acids) until saturation is reached (Figure 2C), supporting that the rapid fluorescence increase is due to SRP binding. Hyperbolic fits provided Kd values ranging from 100 to 200 nM for initial SRP binding to those RNCs. A similar affinity is obtained from the analysis of the rapid phase of SRP binding to Lep75-RNCs by exponential fitting of the traces truncated at 30 s (Figure 2C). The total amplitude does not change with SRP concentration (Figure 2D), consistent with the high affinity of SRP for long-chain Lep-RNCs.
Figure 2.
Concentration dependence of SRP binding to translating ribosomes. (A, B) Translation of Lep25 mRNA (A), or Lep75 mRNA (B) was carried out in the stopped-flow apparatus in the presence of Bpy-labeled SRP at various concentrations (75 nM–1 μM, final concentrations), and FRET between MDCC-labeled ribosomes (25 nM final concentration) and Bpy-labeled SRP was monitored. Fluorescence traces are offset for clarity and dotted lines indicate the respective starting fluorescence. Red lines are obtained from global fitting of the associated data sets to the model in Figure 3. (C) Combined amplitudes of the biphasic fluorescence change during the rapid phase of binding were obtained by double-exponential fitting of time courses up to 10 s, disregarding the delay and the slow fluorescence increase seen with Lep75-RNC. Hyperbolic fitting yielded Kd values for the initial binding complex of 100–200 nM. (D) Concentration-independent total fluorescence change during cotranslational SRP binding to Lep75 RNCs. Error margins in C and D represent standard errors of the double-exponential fits.
Quantitative kinetic analysis of SRP–RNC complex formation by global fitting
To determine rates of SRP binding and subsequent rearrangements we adopted global fitting using a comprehensive kinetic model. We started by globally fitting traces that showed only the rapid fluorescence change (Lep 25, Lep35 and Lep40), at all SRP concentrations tested (18 traces total; Supplementary Figure S4). In order to adequately fit this data set, SRP binding had to be modeled as a reversible two-step process with initial binding followed by a conformational change; a one-step model did not produce satisfactory fits (Supplementary Figure S4). Since SRP interacts with all Lep-RNCs similarly, nascent-chain synthesis was not included in the preliminary model. The kinetic parameters obtained from this fitting (Table 1) indicate that, during the rapid phase, SRP binds to translating ribosomes with a reasonably high affinity (90 nM), consistent with the data in Figure 2C.
Table 1. Kinetic parameters for co-translational SRP recruitment.
| Parameter | Lep ≤ 50 | All Lep-RNCs |
|---|---|---|
| k +1 (μM−1s−1) | 21 ± 3 | 28 ± 4 |
| k -1 (s−1) | 4 ± 1 | 6 ± 2 |
| k +2 (s−1) | 0.4 ± 0.1 | 0.34 ± 0.06 |
| k -2,short (s−1) | 0.20 ± 0.05 | 0.29 ± 0.03 |
| k -2,long (s−1) | - | 0.024 ± 0.002 |
| K d Lep < 50 (nM) | 90 ± 40 | 180 ± 70 |
| K d Lep > 50 (nM) | - | 15 ± 6 |
Parameters were obtained by global fitting of the time courses depicted in Figures 1 and 2 and Supplementary Figures S3, S5 and S7. Global fitting was performed using either a two-step binding model including only short-chain RNCs (Lep≤50; 18 traces), or the model defined in Figure 3 for all Lep-RNCs (24 traces).
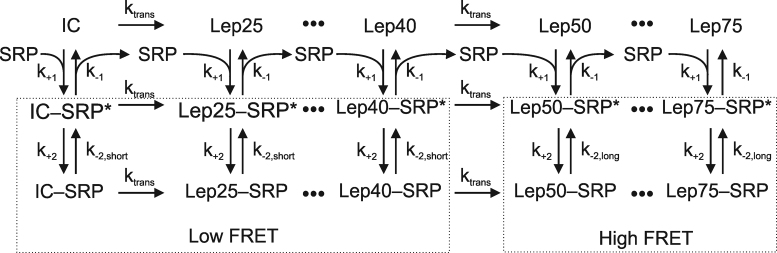
In order to fit both rapid and slow phases of the time courses it was necessary to incorporate nascent-chain synthesis into the global fitting model. The full kinetic model consists of a series of reversible two-step binding reactions connected by irreversible steps of nascent-chain elongation (Figure 3). For the short nascent chains, SRP binding is modeled by a two-step process as described above. To account for the increase in affinity of the SRP–RNC complex upon growth of the nascent chain from 40 to 50 amino acids, we have introduced an additional parameter, k-2,long. This decision is based upon an earlier study on the interaction of SRP with stalled RNCs (13). That study showed that the high affinity of SRP binding to the Lep50-RNC, compared to non-translating ribosomes, results from a lower reverse rate of a conformational rearrangement.Adequate precision of the fit is obtained by including 23 SRP-binding experiments performed with various lengths of mRNA and various SRP concentrations. For simplicity, only six intermediates are modeled between initiation complexes (ICs) and Lep75-RNCs. Furthermore, the rate of translation characterized by ktrans, the average number of amino acids incorporated into peptide per second, is assumed to be the same for all RNCs regardless of nascent chain length. Because the rate of translation in the E. coli system is not affected by SRP binding (26,27), the model assumes that ribosomes with bound SRP translated at the same rate as ribosomes without SRP.
Figure 3.
Kinetic model for SRP binding to ribosomes translating Lep mRNAs of various lengths. Translation is modeled (left to right) as a conversion of initiation complex (IC) into Lep75-RNC in a series of irreversible steps with six intermediates. A constant average rate of translation (in aa/s) is assumed for all constructs. The reversible formation of an initial (RNC–SRP*) complex is identical for all nascent chain lengths, and is followed by conformational rearrangement to RNC–SRP. A switch from low to high SRP affinity is modeled upon transition from Lep40 to Lep50 by a reduced rate of the reverse conformational rearrangement (k-2) (see text). The results of the global fit (red lines) are shown along with the respective time courses in Figures 1 and 2 and in Supplementary Figures S2, S3, S5 and S7.
To provide direct information about the rate of translation, and thus improve the fit of ktrans, the average of four independent translation time courses is also included in global fitting (Supplementary Figure S3B). Because the affinity of SRP for binding to RNCs carrying long nascent chains is very high, we used additional experimental data to constrain the parameter k-2,long. Thus, we included in the global fit a time course of dissociation of the MDCC–Lep75–RNC–Bpy–SRP complex that is induced by rapid mixing with excess unlabeled SRP and monitored by the FRET decrease over time (Supplementary Figure S5). The fits, shown as red lines along with the corresponding traces, describe the observed traces with high accuracy, despite the complexity of the signal changes. The kinetic parameters obtained from global fitting (Table 1) reveal that SRP rapidly binds to RNCs with nascent chains of any length, producing intermediate complexes (RNC–SRP*) that rearrange to more stable complexes (RNC–SRP). After incorporation of 50 amino acids, the RNC–SRP affinity is increased substantially, as k-2 decreases 10-fold, from 0.29 s−1 (k-2, short) to 0.024 s−1 (k-2 long), yielding an overall Kd = 15 ± 6 nM for these complexes.
SAS emergence from the peptide exit tunnel triggers SRP rearrangement
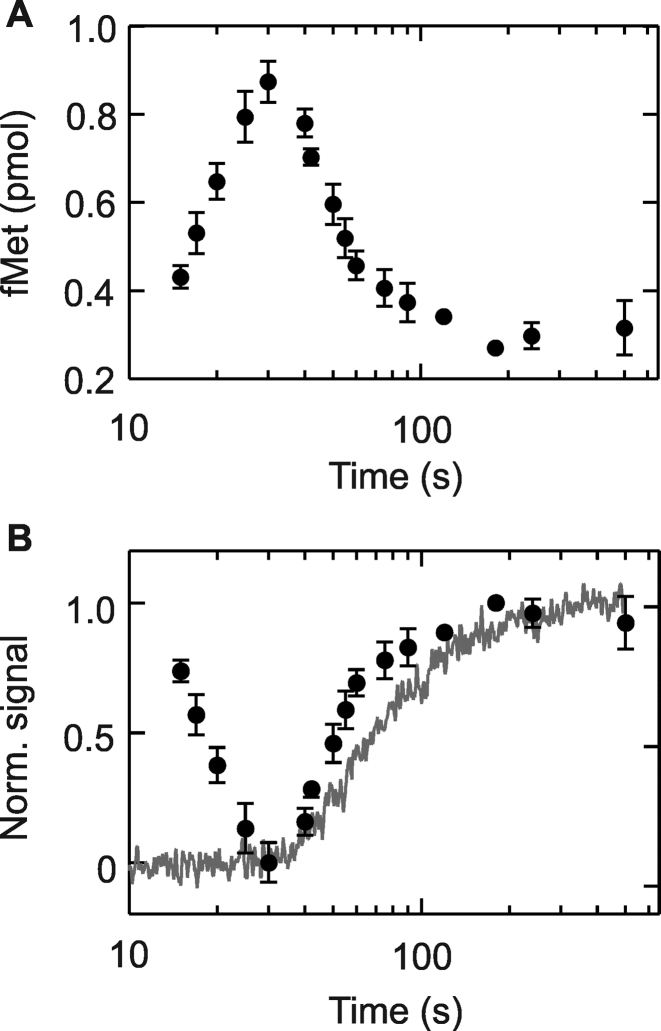
Previous crosslinking data suggested that a Lep nascent chain of about 40 amino acids can reach SRP bound near the exit site as well as the SecY translocon (28). In contrast, here we observe that 50 amino acids are required for stabilizing SRP on the RNC. This suggests that at least part of the Lep SAS has to emerge from the peptide exit tunnel of the ribosome to stabilize SRP binding. In order to directly test when the N terminus of the nascent Lep peptide emerges from the ribosome in our in-vitro experiments, we probed the proteinase sensitivity of the N-terminal fMet of Lep in a co-translational assay. In this assay, proteinase K (PK) is added to the translation mixture at different time points and allowed to act for 10 s, after which the nascent chain is released hydrolytically, precipitated with TCA and counted to determine the amount of nascent chain still carrying radioactive f[3H]Met (Figure 4A). The amount of f[3H]Met precipitated initially increases, presumably reflecting the increasing efficiency of TCA precipitation of longer peptides. However, at 30 s the f[3H]Met recovery starts to decrease, indicating that the N terminus of the nascent chain becomes accessible for PK due to exposure outside the peptide exit tunnel. When PK sensitivity is directly compared with co-translational SRP recruitment monitored by FRET it is clear that the nascent chain becomes accessible to PK prior to rearrangement of the SRP–RNC complex (Figure 4B).
Figure 4.
Delay between emergence of the nascent chain and SRP rearrangement. (A) Nascent-chain emergence from the peptide exit tunnel monitored by PK cleavage of the N terminus. After initiating translation of Lep75 mRNA with 3H-labeled fMet at the N terminus, aliquots were taken and PK digestion was performed for 10 s and quenched by adding KOH. After quenching, the Lep peptide was released by alkaline hydrolysis, precipitated with TCA, collected on nitrocellulose filters, and quantified by counting 3H (Materials and Methods). (B) Overlay of normalized PK data from panel A and SRP rearrangement monitored by FRET. The data from A are inverted and normalized (closed circles) and are plotted along with the normalized fluorescence change from co-translational recruitment of SRP to the RNC monitored by FRET (gray trace). Error margins represent standard deviations (n ≥ 3).
The synthesis of Lep50 coincides with the rearrangement of the SRP–RNC complex and is also delayed relative to the onset of PK accessibility (Supplementary Figure S2). Exponential fitting of the traces in Figure 4B provides transit times, τ, for the different processes. Based on the transit time of Lep50 synthesis (τ = 72 ± 4 s), the average translation rate is about 0.7 amino acids/s. Transit times for the other processes reveal that the nascent chain becomes exposed to PK at a length of 36 amino acids (τ = 53 ± 4 s) while the rearrangement of the SRP-RNC complex takes place later, at 46 amino acids (τ = 67 ± 2 s). Thus, the rearrangement takes place when at least 10 amino acids have emerged from the peptide exit tunnel of the ribosome, seven of which belong to the Lep SAS, which in total comprises 19 amino acids. This result indicates that SRP can bind to a partially exposed hydrophobic signal-anchor sequence.
Comparison of present and previous data
The present observation of SRP binding to translating ribosomes carrying short nascent chains is at variance with results obtained by single-molecule FRET measurements employing other fluorescence labels at different positions (17). In those experiments, which were performed at 25°C in Tris-based HiFi buffer containing polyamines, SAS-independent binding of SRP to translating ribosomes carrying short nascent peptide chains was not observed. Rather, the SRP–RNC complex was apparently only visible after emergence of the SAS, prompting the suggestion that emergence of the nascent chain causes an increase in the rate of association. One possible reason for missing SAS-independent SRP recruitment may be the use of different fluorescence labels and/or positions. Another potential explanation for the discrepancy is provided by the observation that polyamines suppress the fluorescence change related to SAS-independent SRP binding (Supplementary Figure S6). Fitting those data by two different approaches suggests that the rate of the SAS-independent phase changes with the SRP concentration in a hyperbolic manner, consistent with a bimolecular binding event followed by a rearrangement. In contrast, the rate of the SAS-dependent slow step is intrinsically independent of SRP concentration. The global rate of SAS-dependent recruitment has a residual concentration dependence owing to the contribution of the first SAS-independent phase (Supplementary Figure S6), rather than an increased bimolecular association rate constant (17). Thus, even in buffer containing polyamines, the initial SAS-independent recruitment of SRP is followed by an engagement step after the SAS becomes exposed.
DISCUSSION
Previous equilibrium and pre-steady-state kinetic analyses identified an early, SAS-independent recruitment of SRP to non-translating ribosomes or stalled RNCs carrying short nascent chains. Here, we show that early recruitment occurs with actively translating ribosomes as well. The formation of the early complex, which is of moderate affinity, is independent of the presence or exposure of an SAS. It proceeds as a rapid two-step reaction, i.e., binding followed by a rearrangement. When a nascent peptide comprising an N-terminal SAS reaches a minimum length of 46 amino acids, of which at least 10 are exposed, SRP binding to the SAS induces a rearrangement of the complex accompanied by a ten-fold increase in affinity (Kd = 15 nM at 37°C). The affinity increase is due to a decrease of the dissociation rate constant (from k-2short to k-2long;Table 1), similar to what was observed when stalled RNCs were compared with non-translating ribosomes (7,13). The two phases for SRP binding observed here correspond to steps 2 and 3 identified in the previous study performed with stalled Lep50-RNCs (13). A binding step equivalent to the bimolecular step 1 of the previous study is not observed in the present work. Presumably the higher temperature used here accelerates binding such that the present time resolution is insufficient to resolve binding as a separate step.
The transition to a high-affinity complex in the actively translating system occurs with longer nascent-chain lengths compared to what was observed previously with stalled RNCs. With stalled RNCs 32–35 amino acids of Lep (or even nascent chains of 35 amino acids lacking a signal sequence) were sufficient to reach the maximum SRP-binding affinity (12). The present data indicate that at conditions of ongoing translation the stabilization of the RNC–SRP complex occurs after translation of about 50 codons. This is consistent with recent single-molecule fluorescence and ribosome profiling studies which suggest that RNCs are most likely bound by SRP when the N terminus of the transmembrane domain is located 40–55 amino acids away from the peptidyl transferase center of the ribosome (17,18). On the other hand, the N terminus of the nascent peptide becomes accessible to protease at a length of just 36 amino acids, indicating that at that peptide length the N terminus starts to emerge from the exit tunnel. Ten more amino acids are required to elicit high-affinity SRP binding to the ribosome, presumably by exposing part of the Lep SAS. This implies that SRP can either interact with a partial SAS or that SRP interacts with an entire SAS that is partially contained within the exit tunnel. The latter scenario is supported by recent evidence obtained by cryo-EM and crosslinking suggesting that the M domain of SRP protein Ffh can insert into the peptide exit tunnel and interact with ribosomal elements and the nascent peptide (10,29).
The different lengths of nascent chains required to form high-affinity stalled or actively translating SRP–RNC complexes may, as suggested by others (17), result from different SRP-binding properties of stalled and translating RNCs. However, the SRP binding site at the peptide exit is located far away from the regions of the ribosome where factors bind/dissociate or subunit/intersubunit motions occur and is not, therefore, likely to change conformation or experience competition during normal translation. Furthermore, the nascent chain, which presumably undergoes backbone rearrangements on the microsecond to sub-microsecond timescale (30), should have ample time to attain its preferred conformation within the peptide exit tunnel of actively translating ribosomes and stalled RNCs alike. On the other hand, the conditions in which SRP binding to translating or non-translating ribosomes was measured have significant differences that should be noted. The co-translational experiments are carried out at 37°C in the presence of translation factors at micromolar concentrations and GTP. Experiments with stalled RNCs, in contrast, were performed at lower temperature, 25°C, with purified complexes without translation factors and in the presence of non-hydrolyzable GDPNP (12,13). These differences in reaction conditions likely result in the different lengths of the nascent chain required to stabilize SRP on stalled or actively translating RNCs. It has been shown previously that early SRP recruitment to stalled RNCs involves signaling from inside the exit tunnel to the SRP binding site via protein L23, although mutations of L23 that interfered with signaling did not cause a phenotype under normal growth conditions (12). This may indicate that the signaling mechanism is more efficient at 25°C than at 37°C. Despite the differences in conditions and measured Kd values, we note that the overall mechanism of initial SRP binding and subsequent conformational change is independent of conditions and pertains to both stalled RNCs and translating ribosomes.
Comparison of the rates for SRP association measured in this study to rates measured on non-translating ribosomes and stalled RNCs suggests that translation itself does not influence the kinetics of SRP interaction with the ribosome. Thus, we expect the kinetic parameters reported here, which are measured at a translation rate of 0.7 aa/s, are applicable in E. coli, where protein synthesis is faster (∼10 aa/s). In order to discuss targeting by SRP in the context of the cell, we have computed transit times based on our global fitting results (Figure 5) and cellular concentrations of factors. We find that rapid binding and dissociation of SRP (τon = 1 ms, assuming a ribosome concentration of 30 μM; τoff = 0.2 s) allows SRP to scan multiple ribosomes in <1 s. The second kinetic step is a conformational change with a transit time of 3 s that is still independent of SAS emergence from the ribosome. We refer to this complex as the ‘engaged state’. Notably, structural models of free and ribosome-bound SRP reveal a major rearrangement of the SRP NG domain allowing it to engage protein L23 on the ribosome (10,31). Since our FRET donor is attached to protein L23, it is conceivable that the rapid rearrangement observed in the present study corresponds to the engagement of the NG domain of SRP protein Ffh with L23. Although we do not know the structure of the initial binding complex, the distance between donor and acceptor may be too large to allow for significant FRET to take place–one possible reason why there is no FRET observed during initial binding.
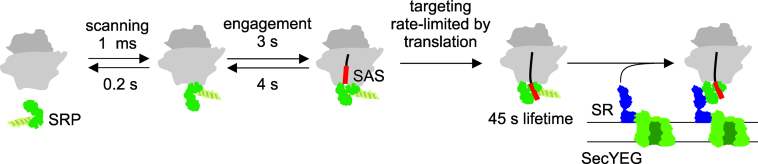
Figure 5.
Model for co-translational SRP recruitment to RNCs. Binding of SRP to translating RNCs is rapid at cellular ribosome concentrations (30 μM), and rapid dissociation gives rise to a scanning mode where SRP can sample multiple ribosomes in less than 1 s. A conformational change requiring a transit time of 3 s yields an engaged complex with substantially longer lifetime. After the nascent chain reaches a length of 50 amino acids, the complex is kinetically stabilized an additional 10-fold. The lifetime of this complex is 45 s, sufficiently long to support targeting to the SecYEG–SRP receptor (SR) complex at the membrane. Transit times were calculated as inverse net rate constants (τ = 1/knet) and net rate constants were computed from rate constants obtained via global fitting as follows: k+1,net = k+1·k+2/(k+2+k-1); k+2,net = k+2; k-1,net = k-1; k-2,net = k-2·k-1/(k-1+k+2).
The engaged SRP–RNC complex has a lifetime of 4 s, which may explain why the complex is not isolated in selective ribosome profiling studies in E. coli (18) or yeast (32). In the cell, the formation of an early SRP–RNC complex provides a ‘standby phase’ for SRP which persists until after the nascent peptide has emerged from the peptide exit tunnel. Under in-vivo conditions the ribosome would have time to synthesize a nascent chain of 30 amino acids prior to reaching the engaged complex and the nascent chain would still lie within the peptide exit tunnel. During the 4 s lifetime of the engaged complex, the ribosome could extend the nascent chain to 70 amino acids, exposing about 30 amino acids outside the ribosome. These 30 amino acids (10,29)define a search space for SRP to identify an SAS. Once SRP has recognized an SAS, or part of an SAS, the kinetic stability of the SRP–RNC complex is increased about ten-fold (Table 1). The long lifetime of the rearranged RNC–SRP complex (45 s) allows for efficient recruitment of the SecYEG–FtsY complex to form the quaternary targeting complex in which the nascent peptide is transferred from SRP to the translocon. Taken together, the present results are consistent with SRP scanning translating ribosomes until the recognition of an SAS induces a rearrangement that promotes the formation of the targeting complex.
Early SRP recruitment has also been demonstrated for eukaryotic SRP in that stalled yeast RNCs carrying a signal sequence within the peptide exit tunnel bind SRP more strongly than RNCs without signal sequence (15). Similarly, experiments with stalled mammalian RNCs revealed an early recruitment of SRP before a hydrophobic targeting sequence was exposed, forming an engaged RNC–SRP complex (33). Scanning could ensure early recognition of hydrophobic targeting sequences by SRP on translating ribosomes. It should be mentioned, though, that ribosome profiling in vivo did not reveal these early RNC–SRP interactions in yeast (32) or E. coli (18). This may be attributed to the low stability of these early complexes (complex lifetime is ∼4 s) or to an interference by factors other than SRP that may interact with RNCs in vivo.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Anna Pfeifer and Franziska Hummel for expert technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation (DFG) in the framework of the Collaborative Research Center (SFB) [1190]. Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bernstein H.D., Zopf D., Freymann D.M., Walter P.. Functional substitution of the signal recognition particle 54-kDa subunit by its Escherichia coli homolog. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:5229–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ribes V., Römisch K., Giner A., Dobberstein B., Tollervey D.. E. coli 4.5S RNA is part of a ribonucleoprotein particle that has properties related to signal recognition particle. Cell. 1990; 63:591–800. [DOI] [PubMed] [Google Scholar]
- 3. Draycheva A., Bornemann T., Ryazanov S., Lakomek N.-A., Wintermeyer W.. The bacterial SRP receptor, FtsY, is activated on binding to the translocon. Mol. Microbiol. 2016; 102:152–167. [DOI] [PubMed] [Google Scholar]
- 4. Stjepanovic G., Kapp K., Bange G., Graf C., Parlitz R., Wild K., Mayer M.P., Sinning I.. Lipids trigger a conformational switch that regulates signal recognition particle (SRP)-mediated protein targeting. J. Biol. Chem. 2011; 286:23489–23497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lam V.Q., Akopian D., Rome M., Henningsen D., Shan S.O.. Lipid activation of the signal recognition particle receptor provides spatial coordination of protein targeting. J. Cell Biol. 2010; 190:623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jomaa A., Fu Y.H., Boehringer D., Leibundgut M., Shan S.O., Ban N.. Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome. Nat. Commun. 2017; 8:15470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saraogi I., Akopian D., Shan S.O.. Regulation of cargo recognition, commitment, and unloading drives cotranslational protein targeting. J. Cell Biol. 2014; 205:693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kudva R., Denks K., Kuhn P., Vogt A., Muller M., Koch H.G.. Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res. Microbiol. 2013; 164:505–534. [DOI] [PubMed] [Google Scholar]
- 9. Frauenfeld J., Gumbart J., Sluis E.O., Funes S., Gartmann M., Beatrix B., Mielke T., Berninghausen O., Becker T., Schulten K. et al. . Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 2011; 18:614–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jomaa A., Boehringer D., Leibundgut M., Ban N.. Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat. Commun. 2016; 7:10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ge Y., Draycheva A., Bornemann T., Rodnina M.V., Wintermeyer W.. Lateral opening of the bacterial translocon on ribosome binding and signal peptide insertion. Nat. Commun. 2014; 5:5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bornemann T., Joeckel J., Rodnina M.V., Wintermeyer W.. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat. Struct. Mol. Biol. 2008; 15:494–499. [DOI] [PubMed] [Google Scholar]
- 13. Holtkamp W., Lee S., Bornemann T., Senyushkina T., Rodnina M.V., Wintermeyer W.. Dynamic switch of the signal recognition particle from scanning to targeting. Nat. Struct. Mol. Biol. 2012; 19:1332–1337. [DOI] [PubMed] [Google Scholar]
- 14. Zhang X., Rashid R., Wang K., Shan S.O.. Sequential checkpoints govern substrate selection during cotranslational protein targeting. Science. 2010; 328:757–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berndt U., Oellerer S., Zhang Y., Johnson A.E., Rospert S.. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:1398–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flanagan J.J., Chen J.-C., Miao Y., Shao Y., Lin J., Bock P.E., Johnson A.E.. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomorlar affinity that does not diminish as the nascent chain lengthens. J. Biochem. 2003; 278:18628–18637. [DOI] [PubMed] [Google Scholar]
- 17. Noriega T.R., Chen J., Walter P., Puglisi J.. Real-time observation of signal recognition particle binding to actively translating ribosomes. eLife. 2014; 3:e04418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schibich D., Gloge F., Pöhner I., Björkholm P., Wade R.C., vonHeijne G., Bukau B., Kramer G.. Global profiling of SRP interaction with nascent polypeptides. Nature. 2016; 536:219–223. [DOI] [PubMed] [Google Scholar]
- 19. Bornemann T., Holtkamp W., Wintermeyer W.. Interplay between trigger factor and other protein biogenesis factors on the ribosome. Nat. Commun. 2014; 5:4180. [DOI] [PubMed] [Google Scholar]
- 20. Milon P., Konevega A.L., Peske F., Fabbretti A., Gualerzi C.O., Rodnina M.V.. Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria. Method Enzymol. 2007; 430:1–30. [DOI] [PubMed] [Google Scholar]
- 21. Cunha C.E., Belardinelli R., Peske F., Holtkamp W., Wintermeyer W., Rodnina M.V.. Dual use of GTP hydrolysis by elongation factor G on the ribosome. Translation. 2013; 1:e24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wieden H.-J., Gromadski K., Rodnin D., Rodnina M.V.. Mechanism of elongation factor (EF)-Ts-catalyzed nucleotide exchange in EF-Tu. J. Biol. Chem. 2002; 277:6032–6036. [DOI] [PubMed] [Google Scholar]
- 23. Johnson K., Simpson Z.B., Blom T.. Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 2009; 387:20–29. [DOI] [PubMed] [Google Scholar]
- 24. Holtkamp W., Kokic G., Jäger M., Mittelstaet J., Komar A.A., Rodnina M.V.. Cotranslational protein folding on the ribosome monitored in real time. Science. 2015; 350:1104–1107. [DOI] [PubMed] [Google Scholar]
- 25. Rudorf S., Tommen M., Rodnina M.V., Lipowsky R.. Deducing the kinetics of protein synthesis in vivo fromt he transition rates measured in vitro. PLOS Comp. Biol. 2014; 10:e1003909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gu S.Q., Peske F., Wieden H.J., Rodnina M.V., Wintermeyer W.. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA. 2003; 9:566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Raine A., Ullers R., Pavlov M., Luirink J., Wikberg J.E.S., Ehrenberg M.. Targeting and insertino of heterologous membrane proteins in E. coli. Biochimie. 2003; 85:659–668. [DOI] [PubMed] [Google Scholar]
- 28. Houben E.N., Zarivach R., Oudega B., Luirink J.. Early encounters of a nascent membrane protein: specificity and timing of contacts inside and outside the ribosome. J. Cell Biol. 2005; 170:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Denks K., Sliwinski N., Erichsen V., Borodkina B., Origi A., Koch H.-G.. The signal recognition particle contacts uL23 and scans substrate translation inside the ribosomal tunnel. Nat. Microbiol. 2017; 2:16265. [DOI] [PubMed] [Google Scholar]
- 30. Callender R.H., Dyer R.B., Gilmanshin R., Woodruff W.H.. Fast events in protein folding: the time evolution of primary processes. Annu. Rev. Phys. Chem. 1998; 49:173–202. [DOI] [PubMed] [Google Scholar]
- 31. Janda C.Y., Li J., Oubridge C., Hernandez H., Robinson C.V., Nagai K.. Recognition of a signal peptide by the signal recognition particle. Nature. 2010; 465:507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chartron J.W., Hunt K.C.L., Frydman J.. Cotranslational signal-independent SRP preloading during membrane targeting. Nature. 2016; 536:224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Voorhees R.M., Hegde R.S.. Structurs of the scanning and engaged states of the mammalian SRP-ribosome complex. eLife. 2015; 4, doi:10.7554/eLife.07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.