Abstract
Fanconi anemia (FA) is an inherited cancer predisposition syndrome characterized by cellular hypersensitivity to DNA interstrand crosslinks (ICLs). To repair these lesions, the FA proteins act in a linear hierarchy: following ICL detection on chromatin, the FA core complex monoubiquitinates and recruits the central FANCI and FANCD2 proteins that subsequently coordinate ICL removal and repair of the ensuing DNA double-stranded break by homology-dependent repair (HDR). FANCD2 also functions during the replication stress response by mediating the restart of temporarily stalled replication forks thereby suppressing the firing of new replication origins. To address if FANCI is also involved in these FANCD2-dependent mechanisms, we generated isogenic FANCI-, FANCD2- and FANCI:FANCD2 double-null cells. We show that FANCI and FANCD2 are partially independent regarding their protein stability, nuclear localization and chromatin recruitment and contribute independently to cellular proliferation. Simultaneously, FANCD2—but not FANCI—plays a major role in HDR-mediated replication restart and in suppressing new origin firing. Consistent with this observation, deficiencies in HDR-mediated DNA DSB repair can be overcome by stabilizing RAD51 filament formation in cells lacking functional FANCD2. We propose that FANCI and FANCD2 have partially non-overlapping and possibly even opposing roles during the replication stress response.
INTRODUCTION
FA (Fanconi anemia) is an inherited genomic instability disorder that is characterized by bone marrow failure and a strong predisposition to cancer, predominantly leukemia and squamous cell carcinoma (1,2). A defining characteristic of FA patient cells is that they are highly sensitive to DNA ICL (interstrand crosslink)-inducing agents such as MMC (mitomycin C) and DEB (diepoxybutane). Moreover, FA cells exhibit spontaneous chromosomal aberrations that are further exacerbated upon treatment with replication inhibiting agents such as HU (hydroxyurea) or APH (aphidicolin) (1,3,4). Thus, the FA pathway constitutes an extremely important pathway for the maintenance of genome stability. Currently, 21 different FA genes have been identified and mutations in any one of them are sufficient to cause FA (5–7).
The canonical FA pathway of DNA ICL repair is thought to consist of three layers: an upstream FA core complex (8 proteins), a central protein heterodimer composed of FANCI and FANCD2 (the ID2 complex), and a growing number of downstream proteins including FANCD1/BRCA2 (breast cancer associated protein 2) and the FANCR/RAD51 (radiation sensitive 51) recombinase (5,8). Repair of the DNA ICLs occurs predominately in S-phase when they block the progression of replication forks (9,10). Following DNA ICL detection during S-phase, the FA core complex acts as an E3 ubiquitin ligase that monoubiquitinates FANCI and FANCD2, facilitating their recruitment to DNA ICLs on chromatin (11–14). Subsequently, the chromatin-bound ID2 complex coordinates downstream FA scaffolding proteins and nucleases like FANCP/SLX4 (synthetically lethal in the absence (X) of Sgs14) and FANCQ(XPF)/ERCC1 (xeroderma pigmentosumF/excision repair cross-complementing 1), respectively, along with BRCA2 and RAD51 to mediate incisions at the DNA ICL, followed by HDR (homology-dependent repair) of the newly generated DNA DSBs (double-stranded breaks) (5,8,15–20).
Recent studies from our laboratory and others discovered novel roles for FANCD2 during the replication stress response (4,21–26). Upon replication fork stalling in the presence of HU or APH, FANCD2 is recruited to the stalled forks where it performs dual roles. First, it protects the stalled replication forks from nucleolytic degradation (4,21). FANCD2 fulfills this role in concert with the upstream FA core complex and several downstream FA proteins such as BRCA2 and RAD51 (4,26). Second, FANCD2 promotes restart of the stalled replication forks while blocking the firing of new replication origins (22,24,25). Interestingly, the fork restart function of FANCD2 does not depend on the FA core complex nor on FANCD2 monoubiquitination (25). Instead, the non-ubiquitinated FANCD2 isoform binds chromatin upon replication fork stalling and cooperates with downstream FA factors such as BRCA2 and FANCJ (25) as well as non-FA DNA repair proteins such as the BLM (Bloom syndrome) helicase complex (22), CtIP (C-terminal interacting protein) (24) and FAN1 (Fanconi-associated nuclease 1) (23) to promote fork restart. Intriguingly, previous findings from our laboratory suggested that FANCD2 may fulfill some of its roles during the cellular replication stress response independently of FANCI. Using the Xenopus laevis S-phase extract system, we showed that FANCD2 dissociates from FANCI upon replication stress and is recruited to chromatin prior to FANCI (27). Moreover, FANCD2 participates in the assembly of the BLM complex independently of FANCI (22). However, if and how FANCI contributes to mechanisms of replication stress recovery is not well understood.
To dissect the roles of FANCI and FANCD2 during the replication stress response, we generated human FANCI−/−, FANCD2−/− and FANCI−/−:FANCD2−/− knockout cells in an isogenic background (the HCT116 cell line). Analysis of these cells revealed that FANCI and FANCD2 act in concert to promote DNA ICL repair, but have partially independent roles during the HU- or APH-triggered replication stress response. Our findings indicate that FANCD2 fulfills crucial roles in mediating cellular resistance to HU or APH independent of FANCI. In fact, HU- or APH-triggered cell death in FANCD2-deficient cells was FANCI-dependent, suggesting opposing roles of FANCD2 and FANCI during cell survival. Moreover, FANCD2 played a significantly more important role during the HDR-mediated, RAD51-dependent mechanisms of replication fork restart and DNA DSB repair. Our results suggest that whereas FANCD2 supports cellular survival and fork recovery in the face of APH- or HU-triggered replication stress, FANCI may in fact oppose those functions.
MATERIALS AND METHODS
Generation of FANCI-null (I−/−) and FANCD2-null (D2−/−) cells using rAAV-mediated gene targeting
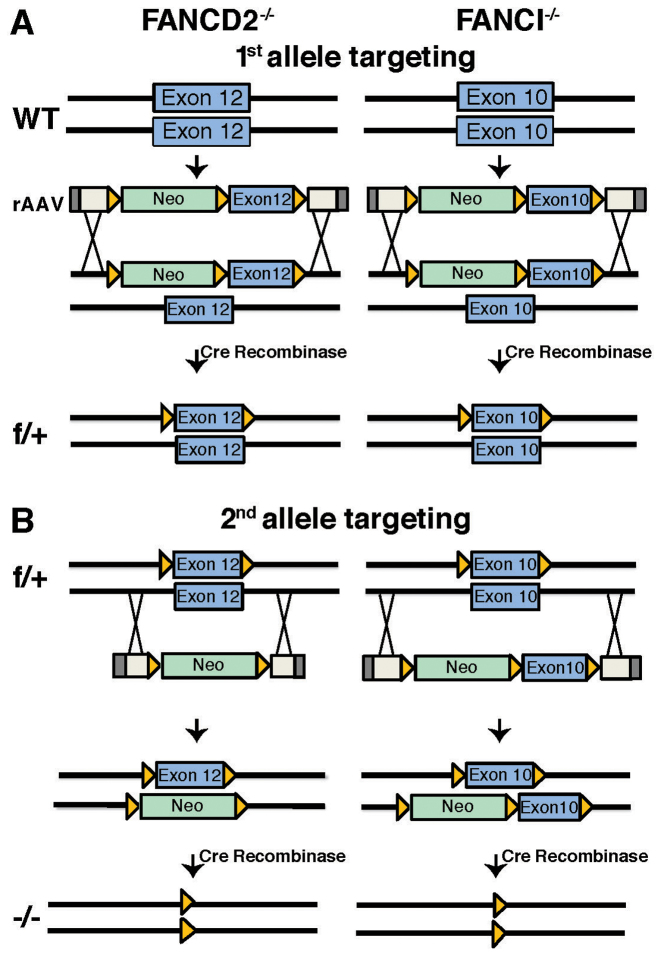
FANCI-null and FANCD2-null HCT116 cells were generated using rAAV (recombinant adeno-associated virus)-mediated gene targeting (28). Conditional and knock-out rAAV vectors targeting FANCI exon 10 and FANCD2 exon 12 were constructed using Golden Gate cloning and designed as described (28–30). We targeted FANCD2 exon 12 and FANCI exon 10 since these exons both lie within regions encoding conserved protein domains associated with heterodimer formation and putative DNA binding (31–33), and the deletion of these exons should result in frameshift mutations. The first round of targeting with a conditional vector replaced FANCI exon 10 and FANCD2 exon 12 with their respective conditional, floxed (flanked by LoxP sites) alleles along with an NEO (neomycin) selection cassette, also flanked by LoxP sites. G418-resistant clones were screened by polymerase chain reaction (PCR) to confirm correct targeting, and Cre (cyclization recombinase) transiently expressed from an adenoviral vector (hereafter AdCre) was then used to remove the NEO selection cassette as described (28–30). Retention of the FANCI floxed exon 10 and FANCD2 floxed exon 12 in the conditional allele was confirmed by PCR. The second round of FANCD2 gene targeting was performed in the FANCD2flox/+ cells with a knock-out rAAV vector in which exon 12 was replaced with an NEO selection cassette. The second round of FANCI gene targeting was performed in the FANCIflox/+ cells using the same conditional vector that had been used in the first round of targeting. G418-resistant clones were screened by PCR for correct targeting. Primer sequences for all PCR reactions are listed in Supplementary Table S1. AdCre recombinase was then used to remove both the NEO selection cassette and the conditional allele(s) and resulted in viable I−/− and D2−/− clones. Two independent I−/− clones (labeled as clones #28 and #30) and a single D2−/− clone (labeled as clone #39) were generated. These three clones and the clones described below were used interchangeably for most of the subsequent experiments.
CRISPR/Cas9 generation of FANCD2−/− and FANCI−/−:FANCD2−/− (ID2 DKO) cells
A guide RNA (gRNA) targeting FANCD2 exon 11 was designed so that Cas9 (CRISPR associated 9) cleavage would disrupt an endogenous restriction enzyme recognition site for BpuEI. The gRNA was cloned into a CRISPR (clustered regularly interspersed short palindromic repeats)/Cas9 plasmid (hSpCas9–2A-Puro/px459) as described (34). WT (wild-type) HCT116 cells were transfected with the CRISPR/Cas9 plasmid containing the gRNA targeting FANCD2 exon 11 using Lipofectamine 3000 (Life Technologies). Two days after transfection, the cells were subcloned, and individual subclones were screened for targeting by PCR amplification of exon 11 and by subsequent digestion with the restriction enzyme BpuEI (New England BioLabs, Inc.). Clones that were resistant to digestion with BpuEI were TOPO TA cloned (Life Technologies). Sequencing of the TOPO TA clones confirmed targeted bi-allelic disruption that induced frameshifts in FANCD2, generating an independent FANCD2-null cell line, designated as clone #29 (Supplementary Figure S1A).
In a highly similar fashion FANCI-null cells were also transfected with the CRISPR/Cas9 plasmid containing the gRNA targeting FANCD2 exon 11 using Lipofectamine 3000 (Life Technologies). Subsequent sequencing of the resulting TOPO TA clones confirmed targeted bi-allelic disruption that induced frameshifts in FANCD2, generating two independent ID2 DKO (double knockout) cell lines, which were arbitrarily designated as clone #1 and clone #2. Western blot analysis further confirmed that these clones were null for FANCD2 and FANCI expression. Primer sequences for all PCR reactions are listed in Supplementary Table S1.
To demonstrate unequivocally that the reagents (rAAV or CRISPR/Cas9) were irrelevant to the resulting phenotypes of the cell lines used in these experiments we also generated additional ID2 DKO clones (arbitrarily designated as clones #3 and #4). These clones were created starting with D2−/− clone #29 (CRISPR/Cas9 generated) and D2−/− clone #39 (rAAV generated) cell lines, respectively. A gRNA targeting FANCI exon 9 was designed so that Cas9 cutting would disrupt an endogenous restriction enzyme recognition site for AcuI. The gRNA was cloned into a CRISPR/Cas9 plasmid (hSpCas9(BB)-2A-GFP/ PX458; Addgene plasmid #48138). This CRISPR/Cas9 plasmid co-expresses Cas9 and GFP, and therefore GFP-expressing cells were collected two days after transfection by flow cytometry and subcloned. Subclones were screened for correct targeting by PCR amplification of FANCI exon 9 and subsequent digestion with AcuI (New England BioLabs, Inc.). Clones that were resistant to digestion with AcuI were TOPO TA cloned and ID2−/- clones #3 and #4 were sequence confirmed for biallelic frameshift-inducing mutations (Supplementary Figures S1B and C). Additionally, western blot analyses confirmed that these cell lines did not express any WT FANCI or FANCD2 protein nor was there any evidence of expression of potential truncated protein products (Supplementary Figures S1D and E). In toto, these experiments demonstrated that the cells were functional DKOs. DKO clones #3 and #4 were used exclusively in the experiments in which growth of the cell lines was assessed (Figure 4A).
Figure 4.
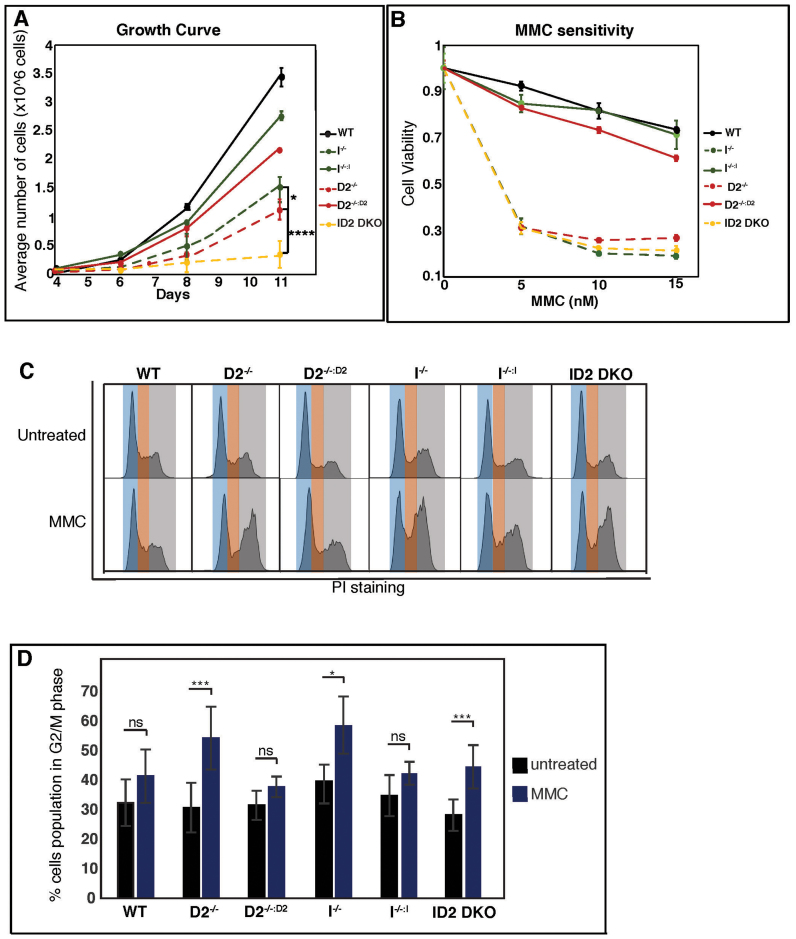
FANCD2 and FANCI contribute differently to cell proliferation and the cellular response to replication stress. (A) The absence of FANCD2 and FANCI affects cell proliferation synergistically. WT, D2−/−(clones #29 and #39), I−/− (clones #28 and #30) and ID2 DKO (clones #1 to #4) cells, as well as the complemented counterparts, were plated according to their plating efficiency and total cell counts were performed in triplicate at days 4, 6 and 8. Data points were averaged between clones of identical genetic backgrounds. Error bars represent the standard deviation. (B) FANCD2 and FANCI act in concert to promote MMC resistance. Cell viability was measured for WT, D2−/−, I−/−, ID2 DKO and complemented cell lines using CellTiter96 Aqueous staining 5 days after the addition of 0, 5, 10 or 15 nM MMC. Results were averaged from a minimum of three replicates and normalized to the average viability of the respective untreated cells. Error bars represent the standard deviation. (C) FANCD2 and FANCI act in concert to activate the MMC-triggered intra-S phase checkpoint. WT, D2−/−(clones #29 and #39), I−/− (clones #28 and #30) and ID2 DKO (clones #1 and #2) cells, as well as the complemented counterparts, were untreated or treated with 10 nM MMC for 20 h, followed by propidium iodide (PI) staining and FACS analysis. Representative histograms of the cell cycle profiles are shown for each cell line. Blue shading represents G1 phase cells, orange shading represents S phase cells and grey shading represents G2/M phase cells. (D) Graphic representation of the percentage of the indicated cell populations present in the G2/M phase of the cell cycle in the absence or presence of MMC. Data points were averaged between clones of identical genetic backgrounds. The average percentage of the G2/M cell population was determined from a minimum of three replicates. Error bars represent the standard deviation and the significance was determined by t-test.
Complementation of D2−/− and I−/− cells
For expression of hFANCD2, the human FANCD2 transcript 1 cDNA (NM_001018115) was optimized by silent mutagenesis, thereby introducing unique restriction enzyme recognition sites and removing potential prokaryotic instability motifs, cryptic splice sites, RNA instability motifs and polyA sites (Hanenberg et al., in preparation). The cDNA was purchased from GenScript (Piscataway, NJ, USA). The D2−/− cells were complemented using the optimized human FANCD2 expression cassette that was Gateway-cloned into a PiggyBac transposase vector containing a CAG (cytomegaloviral/beta-actin/beta-globin) promoter and an NEO selection cassette. G418-resistant clones were screened for FANCD2 expression by western blot analyses.
The I−/− cells were complemented by performing a third round of gene targeting to ‘knock back in’ a functional FANCI allele. This was accomplished using a CRISPR/Cas9 gRNA designed to target immediately 5′ of the LoxP scar left when exon 10 was removed to create the I−/− cells. The FANCI conditional rAAV plasmid containing the FANCI exon 10 flanked by LoxP sites and an NEO selection cassette, also flanked by LoxP sites was used as the donor template. G418-resistant clones were screened for the knock-in of FANCI exon 10, and PCR was used to confirm heterozygous knock-in in two clones. The NEO selection cassette was subsequently removed by transient treatment with AdCre recombinase, and retention of FANCI exon 10 was confirmed by PCR and DNA sequencing. Restored FANCI expression from the knock-in floxed allele was confirmed by Western blot analyses. Throughout the manuscript, the complemented cell lines are designated as (D2−/−:D2, I−/−:I) for D2- and I-null, cells respectively.
Finally, I−/− cells overexpressing FANCD2 protein were established by introducing a PiggyBac transposon containing the FANCD2 codon-optimized cDNA described above into I−/− cells (clone #28). Two FANCD2-overexpressing FANCI-null cell lines (named clones #2 and #12) were identified by Western blot analysis. These clones were designated as I−/−:D2o/e.
Cell proliferation assay
WT cells, knockout cells and complemented cells were plated in 6-well tissue culture plates according to their plating efficiency: WT cells were plated at 2500 cells/well, complemented cells (D2−/−:D2, I−/−:I) were plated at 5000 cells/well and the D2−/−, I−/− and ID2 DKO cells were plated at 6000 cells/well. Cell counts were performed in triplicate 4, 6 or 8 days after seeding.
MMC sensitivity assay
Cells were plated in triplicate in 96-well plates according to their plating efficiency. WT cells were plated at 750 cells/well, complemented cells (D2−/−:D2, I−/−:I) were plated at 1000 cells/well and the D2−/−, I−/− and ID2 DKO were plated at 1200 cells/well. On the following day, the media was removed and replaced with media containing 0, 5, 10 or 15 nM MMC. Cells were allowed to grow for 5 days and cell viability was measured with CellTiter96 Aqueous (Promega) according to the manufacturer's instructions. The viability of MMC-treated cells was normalized to the average viability of the untreated control cells for each cell line.
Cell cycle analysis
WT cells, knockout cells and complemented cells were plated in 6-well plates according to their plating efficiency. WT cells were plated at 100 000 cells/well, singly-null and complemented cells were plated at 200 000 cells/well and DKO cells were plated at 32 000 cells/well. After 24 h, the media was removed and either untreated media, media containing 100 nM APH or media containing 10 nM MMC was added to the cells. After 24 h the cells were harvested and fixed with 1% paraformaldehyde and subsequently with cold 70% ethanol. Fixed cells were washed with PBS (phosphate-buffered saline), stained (with PBS containing 40 μg/ml propidium iodide; 100 μg/ml RNase A) and RNase treated (37°C for 30 min). Samples were then filtered through a cell strainer cap (Corning) and sorted on a BD Accuri C6 instrument and analyzed using FlowJo software (V10.1).
Colony forming assay
WT cells, null cells and complemented cells were plated in 6-well plates in triplicate according to their plating efficiency. WT cells were plated at 300 cells/well, complemented cells (D2−/−:D2, I−/−:I) were plated at 400 cells/well, I−/− cells were plated at 600 cells/well and D2−/− and ID2 DKO cells were plated at 800 to 1000 cells/well. After 24 h, the media was removed and dimethyl sulfide (DMSO) media or media containing 0, 50 100 or 150 μM HU, or media containing 0, 10, 25 or 50 nM APH was added in triplicate. Cells were incubated for 12 to 14 days, washed in PBS, fixed in 10% acetic acid/10% methanol and stained with Coomassie as described (35). Colonies reaching a minimum size of 50 cells were counted and normalized to the average colony number in untreated wells.
DNA repair assays
The HDR reporter assay was performed with the DR-GFP plasmid as described (36). The DR-GFP reporter plasmid contains two mutated GFP genes in tandem, and one of the mutated GFP genes contains an I-SceI restriction enzyme recognition site. If HDR/gene conversion between the two mutated GFP genes is used to repair the DSB induced by I-SceI digestion, then GFP expression is restored.
The A-NHEJ (alternative non-homologous end joining) reporter assay was performed with the pEJ2-GFP plasmid as described (30,37). The pEJ2-GFP reporter plasmid contains a GFP expression cassette interrupted by an I-SceI restriction enzyme site and three stop codons flanked by 8 bp of microhomology. If microhomology-mediated repair (i.e. A-NHEJ) is used to repair the DSB induced by I-SceI restriction enzyme digestion, then GFP expression is restored. For both DNA repair assays, three plasmids including the reporter plasmid (DR-GFP or pEJ2-GFP), an I-SceI expression plasmid, and a mCherry expression plasmid (which was used as a transfection control) were co-transfected into the cells using Lipofectamine 3000 (Life Technologies). Seventy-two hours after transfection, the cells were fixed with 4% paraformaldehyde and sorted by FACS (fluorescence-activated cell sorting). The DNA repair efficiencies were determined as the number of dual GFP-positive and mCherry-positive cells divided by the number of mCherry-positive cells. The values were normalized to the repair efficiency observed in WT cells. For the RS-1 treatment during the HDR assay, 7.5 nM RS-1 was added to the media 2 h prior to plasmid transfection. Fresh media containing 7.5 nM RS-1 was added again 24 h after transfection.
Preparation of whole cell extracts (WCE), as well as nuclear and chromatin fractions
For WCE (whole cell extract) preparation, cells were washed in PBS, resuspended in lysis buffer (10 mM Tris pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1 mM ethylenediaminetetraacetic acid, 1 mM dithiothreitol (DTT), 0.5 mg/ml pefabloc protease inhibitor) and incubated on ice for 20 min. Cell extracts were centrifuged for 5 min at 10 000 rpm, and the supernatant was used for further analysis. Nuclear and chromatin fractions were prepared using a Subcellular Protein Fractionation Kit (Thermo Scientific).
Antibodies
The following antibodies were used for western blotting and immunofluorescence assays: anti-FANCD2 (Santa Cruz, sc-20022 and Abcam, ab2187), anti-FANCI (Bethyl Laboratories, A300–212), anti-FANCI (38), anti-Ku86 (Santa-Cruz, sc-5280), anti-CtIP (39), anti-RAD51 (Santa Cruz sc-8349), anti-GAPDH (Genetex, GTX627408), anti-tubulin (Abcam, ab7291), anti-γ-H2AX (Bethyl Laboratories, A300–081), anti-phospho-p53 (Ser15) (Santa Cruz, sc-11764-R), anti-p21 (Santa Cruz, sc-397) and anti-PCNA (Calbiochem, PC10).
Immunoblotting
Protein samples were separated on gradient gels and transferred to Immobilon P membranes (Millipore). After blocking in 5% milk, the membranes were incubated with the following primary antibodies: FANCD2 (1:1000), FANCI (1:2000), Ku86 (1:5000), GAPDH (1:5000), CtIP (1:400), PCNA (1:5000), γ-H2AX (1:10,000), phospho-p53 (1:1,000) and p21 (1:400). A horseradish peroxidase-conjugated rabbit secondary antibody (Jackson Laboratories) or a mouse secondary antibody (Biorad) were used at dilutions of 1:10 000 and 1:3000, respectively. Protein bands were visualized using an EL Plus system (Amersham).
Immunofluorescence analysis of DNA repair foci
Indirect immunofluorescence was carried out essentially as described (40). The primary antibodies used were: FANCD2 (Abcam, ab2187, 1:4000), CtIP (mouse monoclonal, 1:400) and RAD51 (Calbiochem, PC130, 1:1000). The secondary antibodies used were: Alexa Fluor 594-conjugated goat anti-rabbit (1:1000) and Alexa Fluor 488-conjugated goat anti-mouse (1:1000; Molecular Probes). For statistical analysis of nuclear foci formation, images were taken using a Leica DM LB2 microscope with a Hamamatsu Orca-ER camera. Quantification of CtIP and RAD51 foci was carried out using ImageJ.
DNA fiber assay
We used a vetted DNA fiber protocol (41). Moving replication forks were labeled with DigU (digoxigenin-dUTPs) for 25 min and then with BioU (biotin-dUTPs) for 40 min. To allow efficient incorporation of the dUTPs, a hypotonic buffer treatment (10 mM HEPES, 30 mM KCl, pH 7.4) preceded each dUTP-labeling step. To visualize labeled fibers, cells were mixed with a 10-fold excess of unlabeled cells, fixed and dropped onto slides. After cell lysis, DNA fibers were released and extended by tilting the slides. Incorporated dUTPs were visualized by immunofluorescence detection using anti-digoxigenin-Rhodamine (Roche) and streptavidin-Alexa-Fluor-488 (Invitrogen). Images were captured using a Deltavision microscope (Applied Precision) and analyzed using Deltavision softWoRx 5.5 software. All DNA fiber results shown are the means of two or three independent experiments (using 300 DNA fibers/experiment). Error bars represent the standard error of the mean (SEMS) and the significance was determined by t-test and Mann–Whitney tests. Statistical significance at P < 0.05, P < 0.01 and P < 0.001 are indicated as *, **, ***, respectively.
Molecular DNA combing assay
We carried out DNA combing assays (42) as described in the manufacturer’s protocol (Genomic Vision), with some modifications as described below. Briefly, moving replication forks were labeled with 5-chloro-20-deoxyuridine (CldU, Sigma-Aldrich) for 20 min and then with 5-iodo-20-deoxyuridine (IdU, Sigma-Aldrich) for 40 min. To achieve lysis, the cells were embedded in low-melting agarose (Bio-Rad) followed by incubation in DNA extraction buffer (0.5M MES buffer, pH 5.5, Millipore). To stretch the DNA fibers, silanized coverslips (22 × 22 mm, Genomic Vision) were dipped into the extracted DNA buffer for 13 min and then pulled out a constant speed at 300 um/s. The coverslips were then baked at 60°C for 4 h and incubated with 2.5 M HCl for denaturation. Incorporated CldU and IdU were visualized using a rat anti-BrdU antibody (dilution 1:50 ∼ 1:100 for CldU, Abcam) or a mouse anti-BrdU antibody (1:10 ∼ 1:50 for IdU, Becton Dickinson). Slides were fixed with 4% paraformaldehyde in PBS and incubated with Alexa Fluor 488-conjugated goat anti-rat antibody (dilution 1:100 ∼ 1:200, Molecular Probes/Thermo Fisher) or Alexa Fluor 568-conjugated goat anti-mouse antibody (dilution 1:100 to 1:200, Molecular Probes/Thermo Fisher) for 1 h. Finally, coverslips were mounted with ProLong Gold Antifade Reagent (Molecular Probes) and stored at −20°C. DNA fiber images were captured using LSM880 microscope (Carl Zeiss).
All DNA combing results shown are the means of two or three independent experiments (using 300 DNA fibers/experiment). Error bars represent the standard error of the mean (SEMS) and the significance was determined by a t-test. Statistical significance at P < 0.05, P < 0.01, and P < 0.001 are indicated as *, **, ***, respectively.
Statistical analysis
Error bars represent the standard deviation of the mean and P-values were determined using a Student’s t-test. Statistical significances at P < 0.05, P < 0.01 and P < 0.001 were indicated as *, ** and ***, respectively. A summary of all P-values for results shown in Figures 3–6 is provided in Supplementary Table S2.
Figure 3.
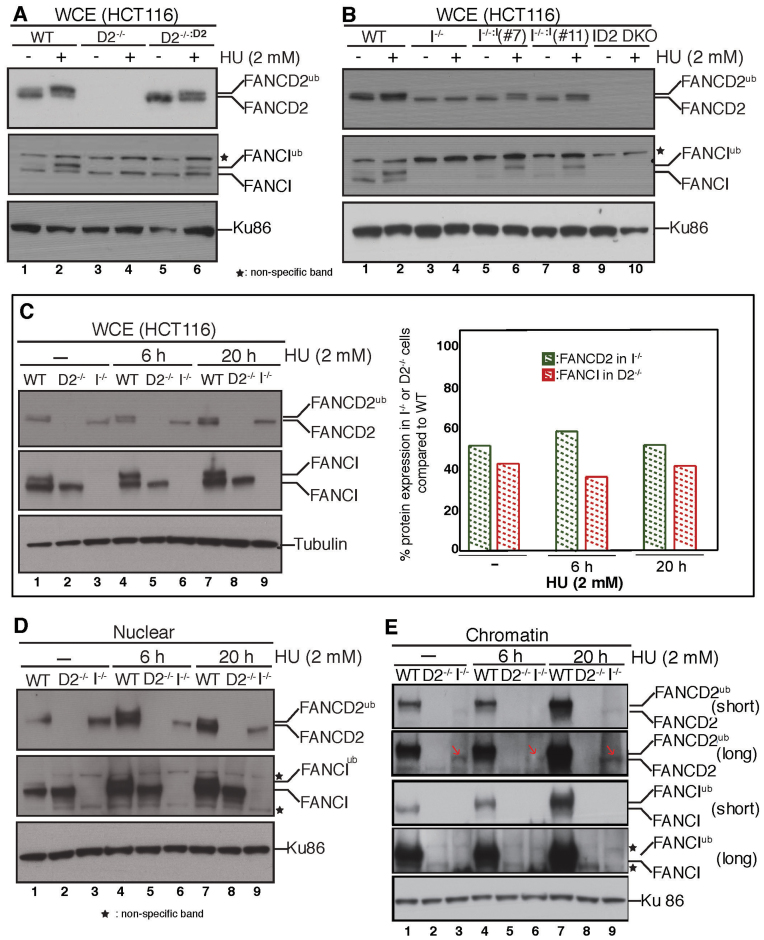
Initial characterization of the I−/−, I−/−:I, D2−/−, D2−/-:D2 and ID2 DKO cell lines. FANCD2 and FANCI are completely interdependent for their replication stress-triggered monoubiquitination. (A) WCE was prepared from WT cells (lanes 1 and 2), D2−/− cells (lanes 3 and 4) or D2−/-:D2 cells (lanes 5 and 6) that had been untreated (lanes 1, 3 and 5) or treated with 2 mM HU for 20 h (lanes 2, 4 and 6) and analyzed for the presence of FANCD2 and FANCI by western blot analysis. Ku86 was used as a loading control. (B) WCE were prepared from WT cells (lanes 1 and 2), I−/− cells (lanes 3 and 4), I−/−:I (clone 7) cells (lanes 5 and 6), I−/−:I (clone 11) cells (lanes 7 and 8) and ID2 DKO cells (lanes 9 and 10) that were untreated (lanes 1, 3, 5, 7 and 9) or treated with 2 mM HU for 20 h (lanes 2, 4, 6, 8 and 10) and analyzed for the presence of FANCD2 and FANCI by western blot analysis. Ku86 was used as a loading control. (C) FANCD2 and FANCI are partially stable in the absence of one another. Left panel: WCE were prepared from WT (lanes 1, 4 and 7), D2−/− (lanes 2, 5 and 8) and I−/− (lanes 3, 6 and 9) cells that were untreated (lanes 1–3) or treated with 2 mM HU for 6 h (lanes 4–6) or for 20 h (lanes 7–9) and analyzed for FANCD2 and FANCI expression by western blot analysis. Tubulin was used as a loading control. Right panel: Immunoblot signals shown in the left panel were analyzed by densitometry and normalized against the tubulin signals using ImageJ software. The graph shows the percentage of FANCD2 and FANCI protein levels in I−/− and D2−/- cells, respectively, compared to the WT cells. (D) Nuclear import of FANCD2 and FANCI becomes interdependent during replication stress. Nuclear fractions were isolated from WT, D2−/− and I−/− cells that were untreated (lanes 1–3) or HU-treated (lanes 4–9) for the indicated time points and analyzed for the presence of FANCD2 and FANCI. Ku86 was used as a loading control. (E) Non-ubiquitinated FANCD2 binds chromatin independently of FANCI during normal replication and following replication stress. Chromatin fractions were isolated from WT, D2−/− and I−/− cells that were untreated (lanes 1–3) or HU-treated (lanes 4–9) for the indicated time points and analyzed for the presence of FANCD2 and FANCI. Ku86 was used as a loading control.
Figure 6.
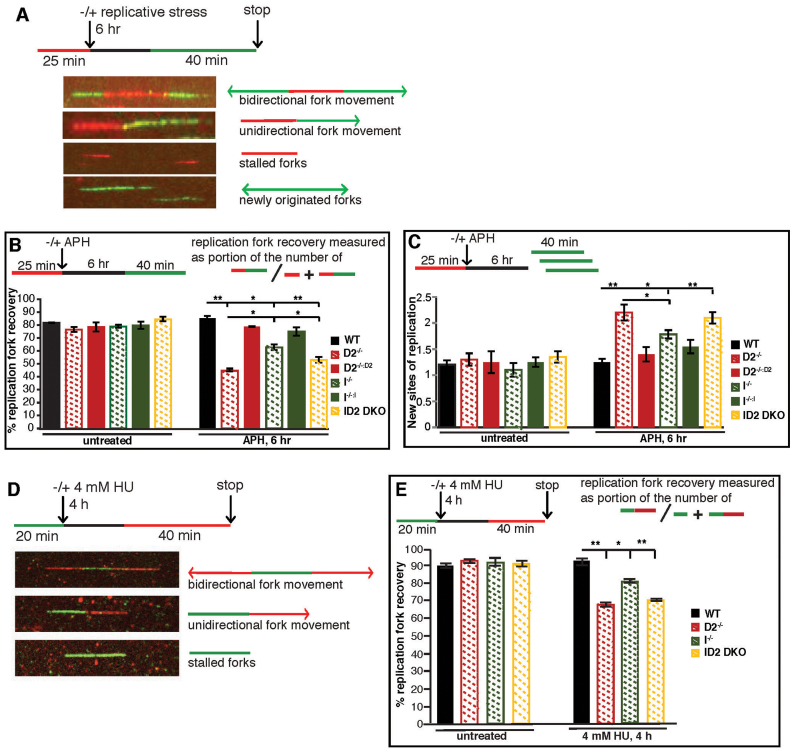
FANCD2 plays a more crucial role than FANCI to promote replication fork recovery. (A) Schematic of the replication fork restart protocol with representative images of DNA fibers. Red tracks: DigU; green tracks: BioU. (B) FANCD2 plays a more crucial role than FANCI in promoting the restart of APH-stalled replication forks. The efficiency of replication restart in WT, D2−/− (clone #39), I−/− (clone #28), and ID2 DKO (clone #1), as well as in the complemented cells was measured as the number of restarted replication forks after APH (30 μM) mediated fork stalling (DigU-BioU tracts), compared with the total number of DigU-labeled tracts (DigU plus DigU-BioU). Statistical significance at P < 0.05, P < 0.01 and P < 0.001 are indicated as *, **, ***, respectively. (C) FANCD2 plays a more crucial role than FANCI in suppressing new origin firing in the presence of stalled replication forks. WT, D2−/− (clone #39), I−/− (clone #28), and ID2 DKO (clone #1), as well as the complemented cells were analyzed for the fraction of newly fired replication origins during the 40 min post-APH recovery period. Fractions were measured as the number of green-only (BioU) tracts compared with the total number of spreading replication tracts (BioU plus DigU-BioU). Statistical significance at P < 0.05, P < 0.01 and P < 0.001 are indicated as *, **, ***, respectively. (D) Schematic of the DNA combing assay with representative images of stretched DNA fibers. Green tracks: CldU; red tracks: IdU. (E) FANCD2 plays a more crucial role than FANCI in promoting the restart of HU-stalled replication forks. The efficiency of replication restart in WT, D2−/−(clones #29 and #39), I−/− (clones #28 and #30) and ID2 DKO (clones #1 and #2) was measured as the number of restarted replication forks after HU-mediated fork stalling (CldU-IdU tracts), compared with the total number of CldU-labeled tracts (CldU plus CldU-IdU). Data points were averaged between clones of identical genetic backgrounds. Statistical significance at P < 0.05, P < 0.01 and P < 0.001 are indicated as *, **, ***, respectively.
RESULTS
Generation and complementation of human FANCI, FANCD2 and FANCI:FANCD2 DKO knockout cell lines
To knock out the FANCD2 and FANCI genes in the parental HCT116 cell line, a conditional knockout approach was used. We chose this approach since all patient-derived cells from complementation groups FA-D2 and FA-I described to date exhibit residual expression of FANCD2 (43) or FANCI (12,44,45), respectively, which raised the possibility that FANCD2 and/or FANCI might be essential in human somatic cells. The conditionally null cell lines were created using recombinant adeno-associated virus (rAAV) gene targeting techniques, as described (Figure 1) (28–30,46). Thus, the FANCD2−/− cell line (D2−/− clone #39) was generated using two rounds of rAAV gene targeting, first with the conditional vector (Figure 1A) and second with the knockout vector (Figure 1B). To unequivocally demonstrate that the technology utilized to generate the knockout cell lines was irrelevant to the cell's subsequent phenotypes we also generated (described in the ‘Materials and Methods’ section) a second FANCD2 knockout clone (D2−/− clone #29) utilizing CRISPR/Cas9 technology.
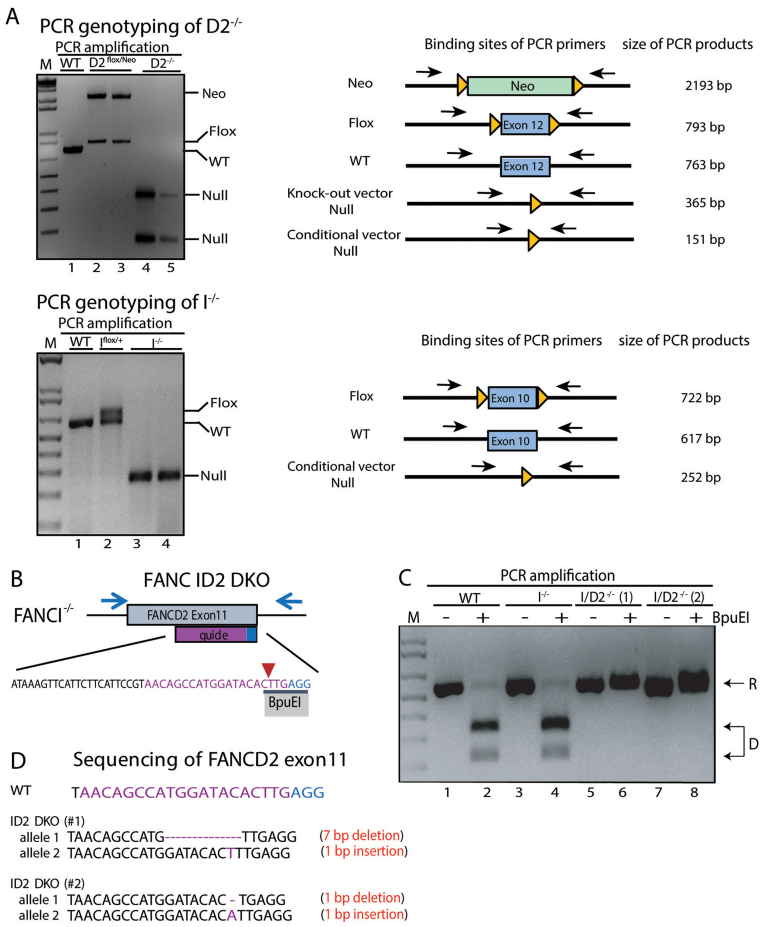
Figure 1.
rAAV-mediated generation of FANCD2−/− (D2−/−), FANCI−/− (I−/−) and FANCI−/−:FANCD2−/- (ID2 DKO) cell lines. (A) Schematic of FANCD2 (left panel) and FANCI (right panel) targeting strategies in HCT116 cells. FANCD2 exon 12 and FANCI exon 10 were targeted for deletion by rAAV gene targeting. The first allele targeting was performed using conditional rAAV vectors for FANCD2 and FANCI. The conditional vectors contain rAAV inverted terminal repeats (ITRs) (gray boxes), homology arms (white boxes), an NEO selection cassette (green boxes) and the targeted exon (blue boxes) flanked by LoxP sites (orange triangles). Targeted clones were treated with Cre recombinase to remove the NEO selection cassette. (B) The second allele targeting was performed in the FANCD2flox/+ cell line (f/+) using a FANCD2 knockout rAAV vector and in the FANCIflox/+ cell line (f/+) with the same FANCI conditional rAAV vector utilized in the first round of targeting. Targeted clones were treated with Cre recombinase to remove both the NEO selection cassette and the conditionally floxed exons, resulting in the generation of FANCD2−/− (D2−/−) and FANCI−/− (I−/−) cell lines.
Similarly, the FANCI−/− cell lines (I−/−) were generated using two rounds of targeting with a conditional rAAV vector (Figure 1A). In this fashion, two independent FANCI-null clones were obtained and designated as clone #28 and clone #30, respectively. These experiments demonstrated that our original concern about the possibility of these genes being essential was unwarranted as null cell lines corresponding to both genes were readily isolated.
Deletion of the targeted exons was confirmed by PCR amplification across the deleted exon region resulting in significantly smaller PCR products for the null alleles (Figure 2A). Since the FANCD2 exon 12 was targeted with two different rAAV vectors that were designed to generate different-sized deletions the D2−/− genotype could consequently be confirmed by the appearance of the two differently-sized PCR products (‘Null’ bands of 365 and 151 bp; Figure 2A). Confirmation of the I−/− genotype was achieved by PCR amplification across FANCI exon 10 resulting in a single PCR band representing deletion of exon 10 on both alleles (‘Null’ band of 252 bp; Figure 2A).
Figure 2.
Confirmation of D2−/−, I−/− and ID2 DKO cell lines. (A) PCR genotyping of D2−/− and I−/− cells and targeting intermediates. Left panel: analyses of DNA fragments for WT (lane 1), D2flox/Neo (lanes 2 and 3) and D2−/− (lanes 4 and 5) after PCR amplification with primers FANCD2_EX11SF and FANCD2_LoxPSR flanking the targeted exon. Right panel: analyses of DNA fragments after PCR amplification from WT (lane 1), Iflox/+ (lane 2) and I−/− (lanes 3 and 4) cells using primers FancIc_GG_LIF and FancIcond_GG_LoxR flanking the targeted exon. The PCR amplification spanning the targeted exons (exon 12 in FANCD2; exon 10 in FANCI) was used to confirm the removal of the respective exon in the D2−/− and I−/− cell lines. M: 1 kb markers. (B) Schematic of the FANCD2 targeting strategy in I−/− cells. CRISPR/Cas9-mediated gene targeting was used to functionally inactivate FANCD2 in the I−/− cell line. A guide RNA (purple sequence) was designed targeting FANCD2 exon 11 with the Cas9 cut site (red arrow) overlapping an endogenous BpuEI restriction enzyme recognition site (black bar). The PAM sequence of the sgRNA is shown in blue. Indels introduced at the Cas9 cut site should disrupt the BpuEI cleavage site. (C) Genotyping of ID2 DKO cells. PCR amplification and BpuEI restriction enzyme digestion of FANCD2 exon 11 in WT, I−/− and two ID2−/− clones (1 and 2). Analyses of DNA fragments after PCR amplification with primers FancD2_CC_ F2 and FancD2_CC_ R2 (blue arrows from panel B) from WT (lanes 1 and 2), I−/− (lanes 3 and 4) and ID2 DKO cells (lanes 5, 6, 7 and 8) that had been untreated (lanes 1, 3, 5 and 7) or treated with BpuEI (lanes 2, 4, 6 and 8). Cleavage by BpuEI produces two faster migrating fragments (D, lanes 2 and 4). Resistance (R) to BpuEI digestion is seen in lanes 6 and 8 with the two ID2 DKO clones. (D) Sequence confirmation of CRISPR/Cas9 induced bi-allelic frameshift mutations in FANCD2 in the two ID2 DKO clones #1 and #2.
To generate the FANCD2−/−:FANCI−/− doubly-null cell line (ID2 DKO), we utilized CRISPR/Cas9 gene targeting to knock out FANCD2 in the I−/− cell line (Figure 2B–D). A guide RNA was designed to target the Cas9 endonuclease to cleave within an endogenous restriction enzyme recognition site (BpuEI) in FANCD2 exon 11. Subsequently, mutagenic insertion or deletions (indels) created by end joining repair of the CRISPR/Cas9-mediated DSB in FANCD2 were detected by disruption of the BpuEI restriction enzyme site (Figure 2B). Screening of targeted clones revealed two independent clones with complete resistance to digestion with BpuEI, indicating bi-allelic disruption of FANCD2 in the I−/− cells (Figure 2C). Sequence analysis of the targeted region was used to confirm that both clones had CRISPR/Cas9-induced bi-allelic frameshift mutations in FANCD2, resulting in two independent viable ID2 DKO cell lines (Figure 2D; arbitrarily designated as clones #1 and #2, respectively). To once again demonstrate that the knockout technology used to construct the cell lines was irrelevant to their biological phenotypes, we also generated (described in the ‘Materials and Methods’ section) two additional ID2 DKO clones in a converse fashion to that described above, thus inactivating FANCI in a FANCD2-null cell line. These clones were arbitrarily designated as ID2 DKO clones #3 and #4, respectively.
Western blot analysis of WCE prepared from WT, D2−/−, I−/− and ID2 DKO cells confirmed that the genetically-null cells lacked expression of FANCD2, FANCI or both proteins, respectively, compared to the WT cells (Figure 3A and B; Supplementary Figure S1D and E). Complementation of the D2−/− cell line (D2−/−:D2) was accomplished by stable genome integration of a constitutively expressed, sequence-optimized FANCD2 cDNA via a PiggyBac transposon vector. Interestingly, we were unable to follow this complementation strategy in I−/− cells, since these cells did not maintain stable FANCI protein expression of a constitutively expressed FANCI cDNA (regardless of whether the expression vector was a transposon, retrovirus or plasmid; data not shown), similar to previous reports (45,47). Instead, complementation of the I−/− cell line was accomplished by a third round of CRISPR/Cas9-mediated knock-in of the conditional exon 10 allele, resulting in reversion of the FANCI−/− genotype to a FANCIflox/− genotype (designated as I−/−:I). Western blot analysis of the complemented cell lines showed that the D2−/−:D2 andI−/−:I cells exhibited WT-like protein expression of FANCD2 and FANCI, respectively (Figure 3Aand B). In agreement with previous reports that FANCD2 and FANCI are interdependent for their DNA damage-inducible monoubiquitination (12,27,44,48), HU treatment triggered FANCD2Ub and FANCIUb formation in WT cells, but not in I−/− or D2−/−cells, respectively (Figure 3A and B). Importantly, FANCD2Ub and FANCIUb formation was fully restored in D2−/−:D2 and I−/−:I cells (Figure 3A and B), demonstrating a functional restoration of the key monoubiquitination step in both complemented cell lines.
FANCD2 and FANCI are partially independent for their protein stability, nuclear import and chromatin recruitment and contribute separately to normal cell proliferation
Several previous studies suggested that the FANCD2 and FANCI proteins stabilize one another, while other reports did not (12,27,44,48). To address this question, we compared the expression levels of FANCD2 and FANCI in WT, I−/− or D2−/− cells that were untreated or treated with 2 mM HU. FANCD2 protein levels in WCEs were reduced by ∼50% in I−/− cells compared to WT cells regardless of the absence or presence of HU-induced DNA damage (Figure 3C). Similarly, FANCI protein levels were reduced by ∼60% in untreated or HU-treated D2−/− cells compared to WT cells (Figure 3C). Thus, approximately half of the cellular FANCD2 and FANCI protein levels remain unaffected in the absence of the respective binding partner.
FANCD2 and FANCI each contain nuclear localization signals and can bind chromatin constitutively; moreover their chromatin binding increases after replication stress induction (12,27,49–51). To determine if FANCD2 and FANCI rely on each other for their nuclear import, we analyzed nuclear extracts from WT, D2−/− or I−/− cells that were untreated or HU-treated for 6 or 24 h. In untreated conditions, D2−/− and I−/− cells contained nuclear protein levels of FANCI and FANCD2, respectively, that were comparable to those in WT cells (Figure 3D, compare lanes 1–3). In contrast, while HU treatment stimulated a strong increase in nuclear FANCD2 and FANCI protein levels in the WT cells (Figure 3D, lanes 1, 4 and 7), no such increase was observed in I−/− cells (Figure 3D, lanes 6 and 9) or D2−/− cells (Figure 3D, lanes 5 and 8). These results suggest that nuclear accumulation of FANCD2 and FANCI occurs independently in unperturbed conditions, but becomes interdependent during replication stress.
To determine the extent to which FANCD2 and FANCI chromatin recruitment occurs interdependently, we prepared chromatin factions from WT, D2−/− or I−/− cells that were either untreated or HU-treated for 6 or 24 h. In WT cells, FANCD2 and FANCI were robustly bound to chromatin and accumulated further during the course of HU treatment (Figure 3E, lanes 1, 4 and 7). In contrast, no chromatin-bound FANCI was detectable in the D2−/− cells regardless of the presence or absence of HU (Figure 3E, lanes 2, 5 and 8). Interestingly, a small amount of residual chromatin-associated FANCD2 was detected reproducibly in untreated or HU-treated I−/− cells (Figure 3E, lanes 3, 6 and 9, red arrows). Since FANCD2 cannot become monoubiquitinated in I−/− cells (Figure 3B and C), this finding demonstrates that the non-ubiquitinated FANCD2 isoform retains chromatin binding ability in the absence of FANCI. Thus, although FANCD2 and FANCI are monoubiquitinated in a strictly interdependent manner, they are partially independent of one another regarding their protein stability, nuclear accumulation and chromatin recruitment, hinting at separate cellular functions of the two factors. To further test this, we compared cellular growth between the WT or WT-like D2−/−:D2 and I−/−:I cell lines and the I−/−, D2−/− and ID2 DKO knockout cells in a cell proliferation assay. The three knockout cell lines exhibited significantly reduced cellular growth rates compared to the WT, D2−/−:D2 and I−/−:I cells, indicating that FANCD2 and FANCI both contribute to cellular proliferation (Figure 4A). Strikingly, the cellular growth rates of the three knockout cell lines were also significantly different from one another, with decreasing colony formation abilities in the following order: I−/− > D2−/− > ID2 DKO cells. These results indicate that FANCD2 and FANCI have partially non-overlapping roles to promote cellular proliferation in otherwise unperturbed conditions.
FANCD2 and FANCI act in concert during ICL repair
A hallmark of FA is the cellular hypersensitivity to ICL-inducing agents such as MMC (52), accompanied by a persistent G2/M cell cycle arrest due to a failure to arrest (and repair) cells at the intra-S-phase checkpoint (1,12,53,54). To test if FANCD2 and FANCI cooperate to promote cellular resistance to DNA ICLs, we performed a survival assay. The cells were plated with increasing concentrations of MMC and the cell viability was measured after 5 days. D2−/−, I−/− and ID2 DKO cells were highly and equally sensitive to even the lowest MMC concentration (5 nM) when compared to the WT cells; moreover the complemented D2−/−:D2 and I−/−:I cells exhibited WT-like MMC resistance, further indicating full functional complementation of these cells (Figure 4B). To analyze the cell cycle profiles of FANCD2- and/or FANCI-deficient cells in unperturbed and ICL-treated conditions, asynchronous cell populations were either untreated or treated with 10 nM MMC for 24 h, followed by staining with propidium iodide and FACS analysis. In untreated conditions, the D2−/−, I−/− and ID2 DKO cells exhibited unaltered cell cycle profiles compared to the WT cells or their complemented counterparts (Figure 4C). In contrast, MMC treatment triggered a robust and comparable G2/M arrest in D2−/−, I−/− and ID2 DKO cells compared to WT, D2−/−:D2 or I−/−:I cells (Figure 4C and D; Supplementary Figure S2). Thus, FANCD2 and FANCI act in concert to promote cellular resistance to DNA ICLs.
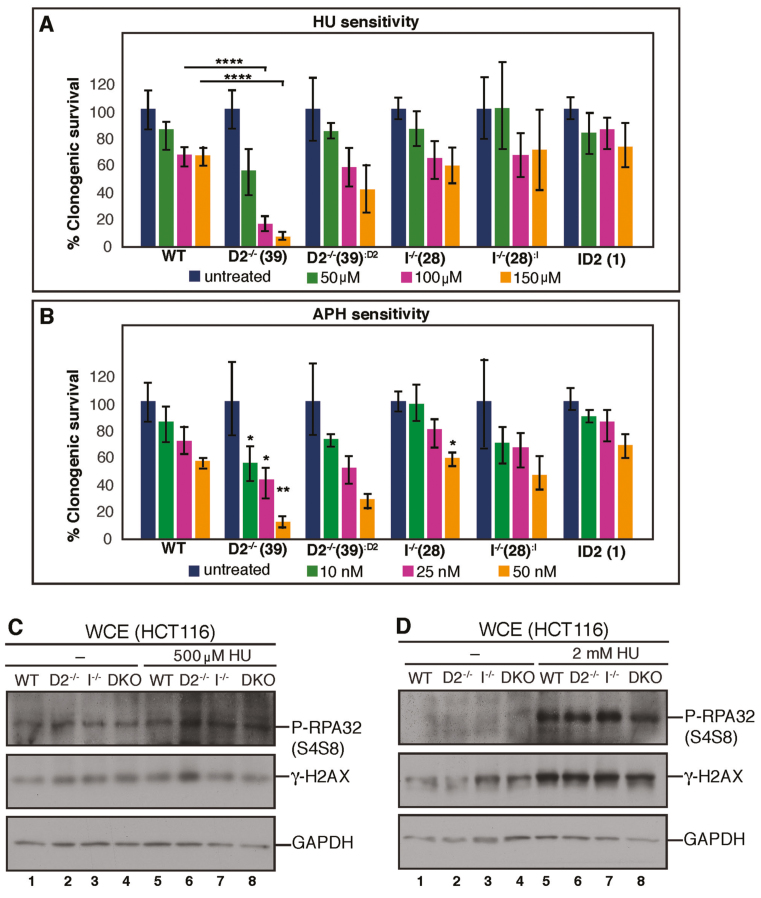
FANCD2 negatively regulates FANCI to promote cellular resistance to APH or HU
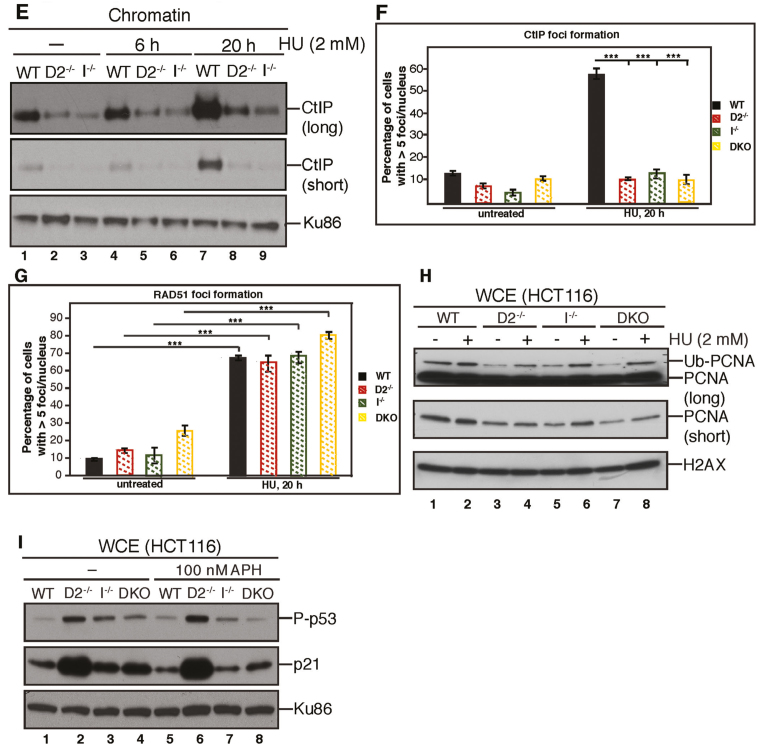
FANCD2 and FANCI accumulate on S-phase chromatin in response to the replication inhibitors APH or HU (27,49,51,55). Moreover, the Kupfer laboratory recently demonstrated that FANCD2-deficient patient cells are sensitive to HU treatment (55). To investigate if FANCD2 and FANCI both contribute to cellular APH or HU resistance, WT, D2−/−, D2−/−:D2, I−/−, I−/−:I and ID2 DKO cells were plated and either left untreated or treated with increasing doses of HU (50, 100 or 150 μM) or APH (10, 25 or 50 nM). Colony formation of each cell line was determined after 12 to 14 days. The D2−/− cells exhibited a strikingly increased, dose-dependent hypersensitivity to both HU and APH compared to WT cells (Figure 5A and B; Supplementary Figure S3A and B). In contrast, the I−/−cells did not exhibit any HU or APH hypersensitivity compared to WT cells, indicating that FANCI is dispensable for the cellular resistance to HU or APH. Strikingly, the ID2 DKO cells did not exhibit HU or APH sensitivity either, demonstrating that the depletion of FANCI restores the cellular resistance to these replication stressors in the absence of FANCD2 (Figure 5A and B; Supplementary Figure S3A and B). To further investigate this, we set out to analyze the cellular replication stress response in WT, D2−/−, I−/− or ID2 DKO cells in more detail. Since FANCD2 is predicted to protect stalled replication forks from collapsing into DNA DSBs (4,22,23), we asked if spontaneous or HU-triggered DNA DSB levels differed between D2−/− cells and the WT, I−/− or ID2 DKO cells. Cells were untreated or treated with doses of low HU (500 μM) or high HU (2 mM) for 24 h and WCEs were analyzed for the DNA DSB markers γ-H2AX and phospho-RPA2 (replication protein A; phosphorylated at serines 4 and 8). We did not observe any differences in the spontaneous or HU-triggered γ-H2AX or phospho-RPA2 levels between the four cell lines, indicating that the DNA DSB burden per se is not elevated in cells lacking FANCD2 and/or FANCI (Figure 5C and D). Next, we asked if the chromatin recruitment of HDR repair proteins during replication stress was differently affected in D2−/− cells compared to WT, I−/− or ID2 DKO cells. CtIP, a key HDR factor required for replication restart and DNA DSB repair, relies on FANCD2 for its own chromatin recruitment (24,56,57). To analyze CtIP chromatin binding and CtIP relocalization into nuclear DNA repair foci, cells were left untreated or treated with 2 mM HU for 20 h, followed by chromatin blotting or immunofluorescence analyses. In support of our previous findings, CtIP recruitment to chromatin and into DNA repair foci was severely reduced in untreated and HU-treated D2−/− cells compared to the WT cells (Figure 5E and F; Supplementary Figure S4). However, the I−/− and ID2 DKO cells exhibited CtIP recruitment defects comparable to those observed in the D2−/− cells, suggesting that CtIP recruitment to stalled or collapsed replication forks relies on both FANCD2 and FANCI. We next analyzed the HU-triggered foci recruitment of RAD51, a second key HDR protein that forms protein–DNA filaments and is required for replication fork recovery and HDR repair of DNA DSBs. As expected, WT cells showed spontaneous RAD51 foci formation (10% cells), which increased significantly following 2 mM HU treatment for 20 h (65% cells). Somewhat unexpectedly, the D2−/−, I−/− and ID2 DKO cells were fully competent for RAD51 foci formation before and after HU treatment (Figure 5G and Supplementary Figure S5), indicating that neither FANCD2 nor FANCI are crucial for the relocalization of RAD51 to HU-induced DNA repair foci. Supportive of this idea, we observed no difference in RAD51 foci formation in the untreated or HU-treated FA-D2 patient cell line, PD20, compared to its complemented counterpart, the PD20+D2 cell line (Supplementary Figure S6).
Figure 5.
FANCI promotes replication stress-induced cellular apoptosis in the absence of FANCD2. (A) FANCD2, but not FANCI, promotes cellular resistance to HU. WT, D2−/−(clone #39), I−/- (clone #28) and ID2 DKO (clone #1) cells, as well as the complemented cell lines were plated at low density and incubated with increasing doses of HU (0–150 μM) for 12–14 days to allow for single cell colony formation. Plates were fixed and stained with Coomassie, and colonies with a minimum of 50 cells were scored. Results were averaged from a minimum of three replicates and normalized to the respective untreated cells. Error bars represent the standard deviation and significance was determined by t-test. Statistical significance at P < 0.05, P < 0.01 and P < 0.001 are indicated as *, **, ***, respectively. (B) FANCD2, but not FANCI, promotes cellular resistance to APH. WT, D2−/−(clone #39), I−/− (clone #28) and ID2 DKO (clone #1) cells, as well as the complemented cell lines were plated at low density and incubated with increasing doses of APH (0–50 nM) for 12–14 days to allow for single cell colony formation. Plates were fixed and stained with Coomassie, and colonies with a minimum of 50 cells were scored. Results were averaged from a minimum of three replicates and normalized to the respective untreated cells. Error bars represent the standard deviation and significance was determined by t-test. Statistical significance at P < 0.05, P < 0.01 and P < 0.001 are indicated as *, **, ***, respectively. (C) FANCD2 and FANCI are dispensable for the prevention of spontaneous or replication stress-induced DNA DSBs. WCE were prepared from WT (lanes 1 and 5), D2−/− (lanes 2 and 6), I−/− (lanes 3 and 7) and ID2 DKO (lanes 4 and 8) cells that were untreated (lanes 1–4) or treated with 500 μM HU for 20 h (lanes 5–8) and analyzed for the induction of pRPA32 (S4/8) and γ-H2AX expression by western blot analysis. GAPDH was used as a loading control. (D) WCE was prepared from WT (lanes 1 and 5), D2−/− (lanes 2 and 6), I−/− (lanes 3 and 7) and ID2 DKO (lanes 4 and 8) cells that were untreated (lanes 1–4) or treated with 2 mM HU for 20 h (lanes 5–8) and analyzed for DNA DSB markers, pRPA32 (S4/8) and γ-H2AX by western blot analysis. GAPDH was used as a loading control. (E) FANCD2 and FANCI cooperate to recruit CtIP to chromatin in the absence or presence of replication stress. Chromatin fractions were isolated from WT, D2−/− (clone #39) and I−/− (clone #28) cells that were untreated (lanes 1–3) or treated with 2 mM HU (lanes 4–9) for the indicated time points and analyzed for the presence of CtIP by western blot analysis. Ku86 was used as a loading control. (F) FANCD2 and FANCI cooperate to promote CtIP foci formation during replication stress. WT, D2−/− (clone #39), I−/− (clone #28) and ID2 DKO (clone #1) cells were untreated or treated with 2 mM HU for 20 h and cellular nuclei were analyzed for the presence of CtIP foci. Nuclei with >5 foci were considered positive for CtIP foci formation. (G) FANCD2 and FANCI are dispensable for RAD51 foci formation during replication stress. WT, D2−/− (clone #39) and I−/− (clone #28) and ID2 DKO (clone #1) cells were untreated or treated with 2 mM HU for 20 h and cellular nuclei were analyzed for the presence of RAD51 foci. Nuclei with >5 foci were considered positive for RAD51 foci formation. (H) FANCD2 and FANCI cooperate to promote the monoubiquitination of chromatin-bound PCNA in the absence or presence of HU-triggered replication stress. Chromatin fractions were isolated from WT, D2−/− (clone #39), I−/− (clone #28) and ID2 DKO (clone #1) cells that were untreated (lanes 1, 3, 5 and 7) or treated with 2 mM HU (lanes 2, 4, 6 and 8) for 20 h and analyzed for the presence of PCNA and PCNAUb by western blot analysis. H2AX was used as a loading control. (I) FANCD2 prevents FANCI-dependent cellular apoptosis in unperturbed conditions and following replication stress. WCE were prepared from WT (lanes 1 and 5), D2−/− (lanes 2 and 6), I−/− (lanes 3 and 7) and ID2 DKO (lanes 4 and 8) cells that were untreated (lanes 1–4) or treated with 100 nM APH for 20 h (lanes 5–8) and analyzed for phospho-p53 (S15) and p21 by western blot analysis. Ku86 was used as a loading control.
In addition to its role in DNA repair, FANCD2 has also been implicated in the so-called DNA damage tolerance (DDT) pathway that allows for cellular survival in the face of persistent DNA damage and replication stress (58,59). A key activation step of this pathway is the monoubiquitination of PCNA (proliferating cell nuclear antigen), which in turn recruits DNA TLS (translesion synthesis) polymerases to promote continuous, albeit mutagenic, replicative DNA synthesis. Recent studies showed that HU-induced PCNA monoubiquitination (PCNAUb) occurs in a FANCD2-supported manner to promote cellular HU resistance (55,60). To test if PCNAUb formation is affected differently in D2−/− cells compared to WT, I−/− or ID2 DKO cells, cells were left untreated or treated with 2 mM HU for 20 h, followed by a chromatin blotting analysis. As expected, D2−/− cells exhibited a decreased spontaneous and HU-induced PCNAUb formation; however, we observed similarly reduced PCNAUb levels in the I−/− or ID2 DKO cells (Figure 5H), suggesting that HU-induced PCNA monoubiquitination relies on both FANCD2 and FANCI.
Since none of these analyses adequately explained the unique hypersensitivity of D2−/− cells to HU and APH we next assessed the status of activated p53 in all of the cell lines. WT and all three knockout cell lines were untreated or treated with APH for 20 h, followed by analysis of phospho-p53 (S15) and—as a relevant downstream p53 target—cellular p21 protein levels. Strikingly, phospho-p53 and p21 levels were highly elevated in untreated and APH-treated D2−/− cells, but not in the WT, I−/− or ID2 DKO cells (Figure 5I). These findings suggest that FANCD2 may protect cells from FANCI-mediated, replication stress-induced cell death.
FANCD2 promotes the restart of stalled replication forks independently of FANCI
FANCD2 functions to restart HU or APH-stalled replication forks, while simultaneously suppressing the firing of new replication origins (22–25). To test if FANCI shares these functions, we monitored replication events in WT, D2−/−, I−/− and ID2 DKO cells with a dual-labeling DNA fiber assay (22,41). Replication tracts were first labeled with DigU (red label) for 25 min, then left untreated or treated with 30 μM APH for 6 h to cause replication fork arrest, followed by a second labeling with BioU (green label) for 40 min (Figure 6A). Different from the efficient fork restart in WT cells, the proportion of replication forks competent for restart after APH treatment was severely reduced in D2−/− cells (45 versus 85%, P < 0.01) (Figure 6B). In parallel, the proportion of newly originated replication tracts (BioU label only) increased significantly (2-fold; P < 0.01) in D2−/− cells compared to WT cells (Figure 6C). In contrast, we observed much milder replication restart defects in the I−/− cells (63%; P < 0.05) and the ID2 DKO cells (54%; P < 0.01), accompanied by a lower number of newly fired replication origins (I−/− cells: 1.4-fold, P < 0.05; ID2 DKO cells: 1.6-fold, P < 0.01) compared to the D2−/− cells (Figure 6B and C). Importantly, the replication restart efficiencies were significantly different between the three knockout cell lines, with an increase in restart efficiency in the order: D2−/− < ID2 DKO < I−/− cells (Figure 6B). To further validate these findings in the presence of a second replication fork stalling agent, HU, we performed a technically improved DNA replication fork restart analysis using DNA combing (42). Moving replication forks were labeled with CldU (green label) for 20 min, then left untreated or treated with 4 mM HU for 4 h to cause replication fork arrest, followed by a second labeling with IdU (red label) for 40 min (Figure 6D). Comparable to what we observed following APH-mediated fork stalling, WT cells exhibited efficient fork restart following HU treatment, whereas the proportion of restarted replication forks after HU treatment was significantly reduced in D2−/− cells (68%, P < 0.01) (Figure 6E). Moreover, we observed milder replication restart defects in the I−/− cells (81%; P < 0.05) and in the ID2 DKO cells (71%; P < 0.01).
These findings indicate that FANCD2 plays a more crucial role than FANCI during replication fork restart and the inhibition of new origin firing. Moreover, the fact that FANCI depletion in D2−/− cells improved these cells’ ability to restart replication forks and suppress new origin firing, particularly after APH treatment, provocatively suggests that FANCI may have an inhibitory effect on both mechanisms in the absence of FANCD2.
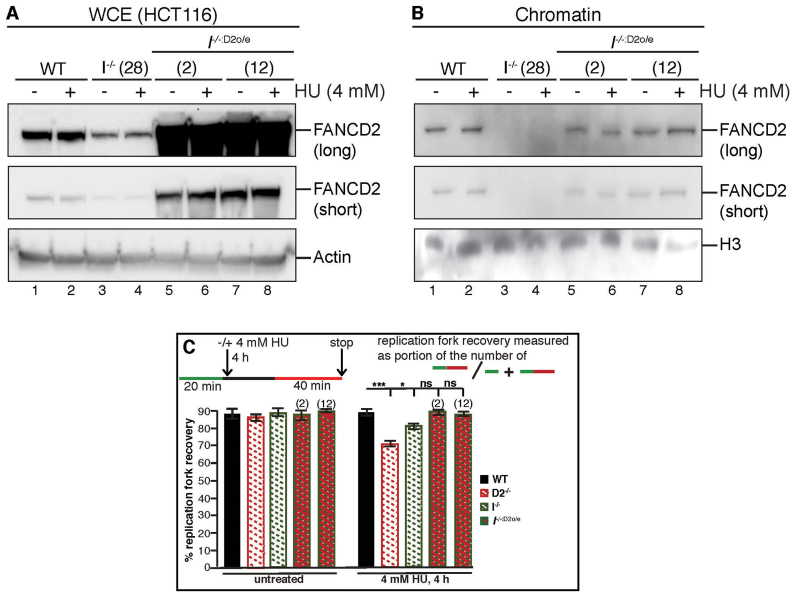
Considering that non-ubiquitinated FANCD2 retains weak chromatin binding activity in I−/− cells (see Figure 3E), we then hypothesized that the few remaining chromatin-bound FANCD2 molecules may still be functional and capable of promoting replication fork restart in the absence of FANCI. If this were indeed the case, it should be possible to completely alleviate the replication fork restart defect in I−/− cells by increasing the amount of chromatin-bound FANCD2. To test this, we overexpressed FANCD2 in I−/− cells (I−/−:D2o/e, Figure 7A), resulting in chromatin-bound, non-ubiquitinated FANCD2 levels comparable to those observed for chromatin-bound, monoubiquitinated FANCD2 in WT cells, before and after HU treatment (Figure 7B). WT, D2−/− and I−/− cells, as well as two different I−/−:D2o/e cell clones, were then analyzed for their replication fork restart efficiencies following HU treatment using the DNA combing assay. Strikingly, unlike the I−/− cells that were moderately defective for replication fork restart (81%, P < 0.05), both I−/−:D2o/e cell clones were completely proficient in restarting HU-stalled replication forks, comparable to the WT cells (Figure 7C). These results indicate that non-ubiquitinated, chromatin-bound FANCD2 is fully competent to promote replication fork restart in the absence of FANCI.
Figure 7.
Over-expressed FANCD2 fully promotes replication fork restart in the absence of FANCI. (A) Human wild-type FANCD2 can be stably overexpressed in I−/− cells. A PiggyBac transposon containing the FANCD2 codon-optimized cDNA was introduced into I−/− cells (clone #28), and two FANCD2-overexpressing I−/− cell lines (named I−/−:D2o/e, clones #2 and #12) were generated. To determine the level of FANCD2 overexpression, WCEs were prepared from WT (lanes 1 and 2), I−/− (lanes 3 and 4) and I−/−:D2o/e (lanes 5–8) cells that were untreated (lanes 1, 3, 5, 7) or treated with 4 mM HU for 4 h (lanes 2, 4, 6 and 8) and analyzed for the presence of FANCD2 by western blot analysis. Actin was used as a loading control. (B) Overexpression of FANCD2 leads to an increase in FANCD2 chromatin binding in the absence of FANCI. In parallel to the preparation of WCEs described in (A) above, chromatin fractions were isolated from these same WT (lanes 1 and 2), I−/− (lanes 3 and 4) and I−/−:D2o/e (lanes 5–8) cells that were untreated (lanes 1, 3, 5, 7) or treated with 4 mM HU for 4 h (lanes 2, 4, 6 and 8) and analyzed for the presence of FANCD2 by western blot analysis. H3 was used as a loading control. (C) FANCD2 overexpression rescues the replication fork restart defects in FANCI null cells. The efficiency of replication fork restart in WT, D2−/−, I−/− and I−/−:D2o/e cells was measured as the number of restarted replication forks after HU-mediated fork stalling (CldU-IdU tracts), compared with the total number of CldU-labeled tracts (CldU plus CldU-IdU). Statistical significance at P < 0.05, P < 0.01 and P < 0.001 are indicated as *, **, ***, respectively.
Stabilizing RAD51 nucleoprotein filaments restores HDR-mediated DNA DSB repair in cells lacking functional FANCD2
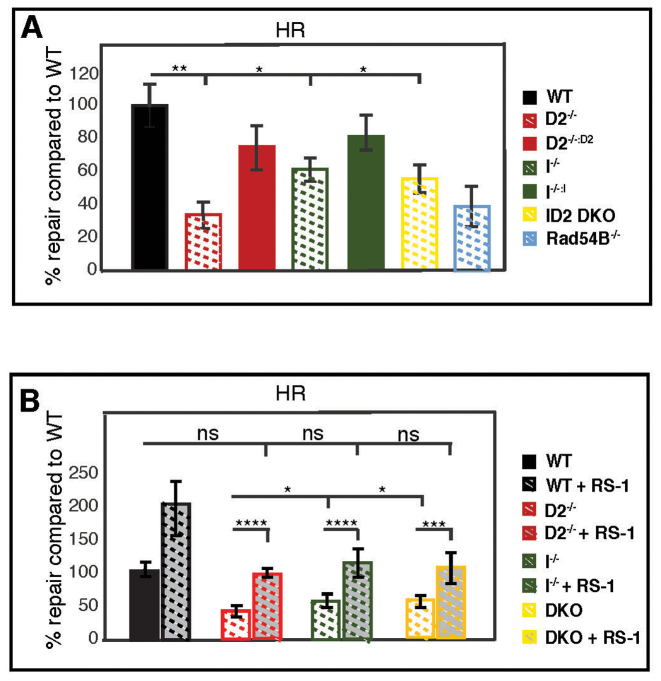
Cells can utilize HDR to repair DNA DSBs (5,61), including those created by the unhooking of a DNA ICL lesion (9,10). In addition, a growing number of HDR factors have been implicated in promoting replication fork recovery in concert with FANCD2 (4,22–26,62), suggesting that the restart machinery utilizes HDR-related mechanisms to restart DNA synthesis at a stalled fork. To investigate the roles of FANCD2 and FANCI in HR-mediated DNA DSB repair we utilized a well-established reporter plasmid DNA repair assay (36). In this assay, a plasmid containing two non-functioning GFP genes in tandem is transfected into the cells. One non-functioning GFP gene contains an I-SceI restriction enzyme recognition site and the other is truncated at both the 5′- and 3′-ends. Digestion of the plasmid with I-SceI creates a DNA DSB in the non-functional GFP gene that can subsequently use the truncated GFP gene as template to restore GFP expression by homology-directed gene conversion repair. As a control for this assay, we included the RAD54B−/−/− HCT116 cell line that has an established HDR defect (63,64). The D2−/− cells were severely deficient in HDR-mediated DNA DSB repair (34% of WT), a level that was comparable to the RAD54B−/−/− cells (Figure 8A). In contrast, the I−/− and ID2 DKO cells exhibited much milder HDR repair defects (62 and 56% of WT, respectively (Figure 8A)). These results indicate that (i) FANCD2 has a more important role than FANCI in HDR-mediated DNA DSB repair and—surprisingly that—(ii) FANCI opposes HDR-mediated DNA DSB repair in the absence of FANCD2.
Figure 8.
FANCD2 plays a crucial role to promote HDR-mediated, RAD51-dependent DNA DSB repair. (A) A GFP-HDR DNA repair assay was used to determine the HDR efficiency in WT, D2−/−(clones #29 and #39), I−/− (clones #28 and #30) and ID2 DKO (clones #1 and #2), as well as in the complemented cells. RAD54B−/−/− cells were included in the assay as a control cell line that is severely HDR deficient. In this assay, I-SceI restriction enzyme digestion creates a DSB in the HDR reporter plasmid (DR-GFP). Repair of the DSB by HDR restores GFP expression. The repair efficiency was determined by dual GFP and mCherry positive cells divided by the mCherry positive cells (transfection control). Results were averaged from a minimum of three replicates and normalized to the average repair efficiency in the WT cells. Data points were averaged between clones of identical genetic backgrounds. Statistical significance at P < 0.05, P < 0.01, P < 0.001 and P < 0.0001 are indicated as *, **, ***, **** respectively. (B) The HDR assay shown in (A) was repeated for the WT, D2−/−, I−/− and ID2 DKO cells in the absence or presence of the RAD51 stabilizer, RS-1 (7.5 mM). Results were averaged from a minimum of three replicates and normalized to the average repair efficiency in the WT cells. Statistical significance at P < 0.05, P < 0.01, P < 0.001 and P < 0.0001 are indicated as *, **, ***, **** respectively.
FANCD2 was proposed to recruit the HDR factor RAD51 to chromatin during replication stress and to stabilize RAD51 nucleoprotein filaments that are crucial to promote HDR mechanisms of replication fork recovery and DSB repair (4,65). Thus, we wanted to ask if the HDR repair deficiency observed in cells lacking functional FANCD2 can be overcome by stabilizing RAD51 filament formation. To this end, we repeated the GFP HDR assay described above in WT, D2−/−I−/− and ID2 DKO cells in the absence or presence of a RAD51 filament stabilizing agent, RS-1 (66). In the presence of RS-1, all three knockout cell lines restored their HDR repair efficiency to a level comparable to the untreated WT cells (Figure 8B). Taken together, these findings support a current model (65) in which chromatin-bound FANCD2 plays a crucial role in stabilizing RAD51 filament formation during HDR-mediated DNA repair and, consequently, replication fork recovery.
DISCUSSION
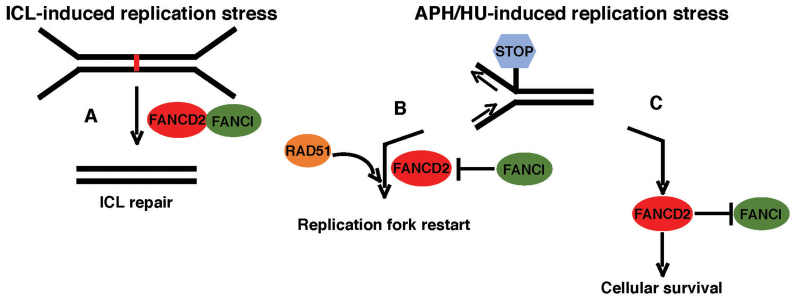
In this study, we analyzed the functions of FANCD2 and FANCI during DNA replication, using isogenic knockout cell lines. Our results suggest that FANCD2 and FANCI fulfill partially independent and at times opposing roles in the cellular replication stress response (Figure 9).
Figure 9.
Model of common and independent functions of FANCD2 and FANCI. (A) FANCD2 and FANCI act in concert during the cellular response to DNA ICLs in S-phase to promote ICL removal and cell survival. (B and C) FANCD2 and FANCI have separate roles during the cellular response to APH- or HU- stalled replication forks. (B) FANCD2—but not FANCI—is crucial to recruit RAD51 and promotes RAD51 filament formation to mediate fork recovery. (C) Simultaneously, FANCD2 promotes cell survival by inhibiting a FANCI-dependent lethal response.
A novel cell model to study common and independent roles of FANCD2 and FANCI
Our model system provides unique advantages compared to the use of FA patient cells or siRNA knockdown approaches. First, patient-derived FA-D2 and FA-I cell lines are derived from different genetic backgrounds and exhibit residual FANCD2 and FANCI protein expression (12,43,44); second, siRNA-mediated knockdown approaches cannot completely or permanently eliminate protein expression (12,24,25,44). In contrast, our system allowed for a functional dissection of the roles of FANCD2 and FANCI in identical genetic background settings and in the complete absence of any FANCD2 and/or FANCI protein expression. On a technical note, but similar to previous reports (45,47), our attempts to complement I−/− cells with a stably integrated FANCI cDNA were unsuccessful due to the rapid loss of FANCI expression, strongly suggesting that cells may not tolerate FANCI expression from a random genomic locus or, more likely, that FANCI overexpression is toxic (see also below).
Based on the fact that all known FA-D2 or FA-I patient cell lines have residual FANCD2 and FANCI (respective) expression (12,43,44), we predicted that the complete loss of one or both proteins may be lethal. However, the D2−/−, I−/− and ID2 DKO cell lines were viable, indicating that neither FANCD2 nor FANCI are essential in human somatic cells; needless to say, however, these experiments do not rule out important developmental roles for either of these genes, a likelihood that is consistent with the developmental abnormalities often observed in FA patients. While not formally essential, both proteins contribute to cellular proliferation. Importantly, the fact that ID2 DKO cells proliferated significantly slower than either of the singly-null cells demonstrates that FANCD2 and FANCI have (at least) partially non-overlapping roles during normal cellular proliferation. Consistent with this idea, in the Xenopus system the ID2 dimer dissociates upon activation in S-phase, followed by consecutive recruitment of FANCD2 and FANCI to S-phase chromatin (27). Furthermore, FANCD2 acts independently of FANCI to assemble and recruit the BLM helicase complex to replicating chromatin (22).
A separation-of-function between FANCD2 and FANCI is further supported by our finding that ∼40 to 50% of either protein is stably expressed in absence of the respective other partner, in agreement with some previous studies (12,44,48). Interestingly, nuclear accumulation of FANCD2 and FANCI occurred independently of one another in unperturbed conditions. This suggests that FANCD2 and FANCI are not interdependent for their nuclear localization per se: in unperturbed conditions, cells simply upregulate the nuclear import of residual FANCI in absence of FANCD2 and vice versa, possibly in an attempt to partially compensate for one another. During replication stress however, the reduced FANCI and FANCD2 protein levels in I−/− and D2−/− cells, respectively, become limiting and block sufficient nuclear FANCD2 and FANCI accumulation. Inconsistent with this interpretation, it has been reported that the FA-D2 patient cell line, PD20, lacks any nuclear FANCI protein compared to the complemented PD20+D2 cells (50). We suggest that this discrepancy may be because the difference in FANCI protein expression appears to be greater between the PD20 cells and the highly FANCD2-overexpressing PD20+D2 cells (12) than between our D2−/− and D2−/−:D2 cells.
Different roles for chromatin-bound and non-chromatin-bound FANCD2 and FANCI?
Importantly, the independent nuclear accumulation of FANCD2 and FANCI was not mirrored by their chromatin binding behavior, since both proteins appeared to be strongly interdependent for chromatin recruitment even in unperturbed conditions. Considering that the three knockout cell lines have significantly different growth rates, the chromatin binding ability of FANCD2 and FANCI is not indicative of a cell’s proliferation ability. This in turn suggests that FANCD2 and FANCI have functions that do not require a direct association with chromatin and supports accumulating evidence that FANCD2 and FANCI have independent roles, such as the (only FANCI-mediated) stabilization of the FA core complex on chromatin or the (only FANCD2-mediated) support of A-NHEJ DNA repair mechanisms (see also Supplementary Figure S7) (22,25,27,47,55,62,67). Our observation that non-ubiquitinated FANCD2 is present on chromatin in the absence of FANCI is in agreement with our previous findings that non-ubiquitinated FANCD2 binds chromatin in FANCI-depleted Xenopus extracts (22,27) and supports a recent study that showed that FANCD2 binds DNA prior to being monoubiquitinated (68). Moreover, we have shown that low levels of non-ubiquitinated FANCD2 accumulate on chromatin in an APH-inducible manner to promote replication fork restart, regardless of the presence or absence of a functional FA core complex (25). Finally, this current study shows that I−/− cells that have residual levels of chromatin-bound, non-ubiquitinated FANCD2 are still capable of partially supporting replication fork restart following APH or HU treatment; moreover, simply increasing the chromatin-bound FANCD2 levels via FANCD2 overexpression completely alleviated the replication restart defects in these cells. In toto, these observations argue strongly that the non-ubiquitinated FANCD2 isoform is functional in its chromatin-bound state.
Importantly, however, the residual FANCD2 chromatin binding in FANCI-deficient cells did not alleviate their ICL sensitivity, reinforcing the model of a linear FA ICL repair pathway, where the FANCI-dependent FANCD2 monoubiquitination is crucial for cellular ICL resistance (12,14,44,69) (Figure 8A). Thus, these experiments define a separation-of-function for FANCD2 monoubiquitination: it is required for ICL repair but dispensable for replication fork restart.
The striking finding that only D2−/− cells — but not I−/− or ID2 DKO cells—were sensitive to HU or APH and exhibited elevated activation of phospho-p53 and p21 expression is supported by a recent report showing HU sensitivity in FA-D2 patient cells (55). Together, these studies suggest that FANCD2 may prevent FANCI-mediated cell death following replication stress (Figure 9). Since D2−/− cells are indistinguishable from I−/− or ID2 DKO cells regarding their spontaneous or stress-induced DNA DSB accumulation, and the recruitment of HDR repair factors or the activation of the DDT pathway, FANCD2’s role in preventing APH- or HU-triggered cell death appears to be separate from its role in protecting chromosomal stability. Moreover, since I−/− and ID2 DKO cells are equally resistant to HU or APH despite harboring different residual chromatin-bound FANCD2 levels, FANCD2’s role in preventing APH/HU-induced cell death may in fact not depend on its own chromatin association. Consistent with our findings and interpretation, a role for FANCD2 in preventing apoptosis and cell death was also suggested by several groups (70–72). Moreover, a recent study showed that siRNA-mediated knockdown of FANCI, but not FANCD2, substantially reduced cellular apoptosis after UV treatment (67). Clearly, in the future, the mechanistic basis of FANCD2′s putative role in cell death induction will be a key question for the field to answer (Figure 9).
Opposing roles for FANCD2 and FANCI during HDR-mediated mechanisms of fork recovery and DNA DSB repair?
In agreement with our previous findings (22–24), FANCD2 promoted the restart of APH-stalled replication forks, a process that requires several members of the HDR repair machinery, including RAD51 (22,24,25,41,73). The fact that the HDR repair defect in D2−/− cells could be almost completely rescued by stabilizing RAD51 filaments via RS-1 supports a model in which FANCD2 functions to (i) promote replication fork restart and (ii) protect nascent DNA from nucleolytic degradation (4,22–24) by promoting RAD51 filament formation. In support of this model, a recent study showed that the FANCD2:FANCI heterodimer stabilizes RAD51 filament formation in vitro (65). It should be noted that wild-type-like RAD51 foci formation was observed in all three knockout cell lines including the D2−/− cells (Figure 5G), seemingly contradicting the idea that FANCD2 is required for RAD51 filament formation. However, findings from our laboratory and others (24,73) indicate that foci formation of HDR repair factors, such as RAD51 or CtIP, does not occur at intact stalled replication forks, but instead occurs much later once the forks have collapsed. The question, therefore, of whether RAD51 foci can form in the absence of FANCD2 remains controversial (65,74–78).
The I−/− cells exhibited milder defects in HDR-mediated replication restart and DSB repair than the D2−/− cells, initially suggesting separate roles for FANCD2 and FANCI during HDR as well. However, two additional findings suggest otherwise: first, overexpression of FANCD2 completely rescued HDR-mediated replication restart defects in I−/− cells. Second, ID2 DKO cells exhibited a less pronounced HDR defect than the D2−/− cells. Thus, FANCI’s role in HDR may be to simply stabilize FANCD2, which by itself is the actual recruiter of HDR factors. In this scenario, the residual chromatin-bound, non-ubiquitinated FANCD2 would allow I−/- cells to partially support HDR-mediated fork restart, similar to chromatin-bound FANCD2 in FA core complex-deficient cells (25). Importantly, FANCD2- and FANCI-deficient cells can compensate for their replication restart defects by the enhanced firing of new replication origins (22,41), which likely counteracts chromosomal instability in the presence of replication stress. Our finding that cells lacking FANCI modestly upregulate new origin firing during replication stress seemingly contradicts a recent study showing that FANCI is required for new origin firing during replication stress (62). However, these authors investigated the role of FANCI mostly under continuous low replication stress conditions (0.2 mM HU), whereas our study used transient, high replication stress conditions (30 μM APH for 6 h), indicating, at the very least, that FANCI is not required for new origin firing when replication forks are stalled for several hours.
Perhaps the most unexpected finding was that cells lacking both FANCD2 and FANCI exhibited intermediate HDR repair and replication fork restart defects, more severe than FANCI-deficient cells, but less severe than FANCD2-deficient cells. We suggest at least two possible explanations: (i) FANCI actively opposes FANCD2 to negatively regulate HDR or (ii) FANCD2 and FANCI work in concert during HDR, but FANCI becomes inhibitory to HDR in the absence of FANCD2 (Figure 9). FANCD2 and FANCI can form a protein complex with RAD51 and contribute to RAD51 recruitment to DNA (65). If FANCI is in fact the direct interactor with RAD51 in vivo, the nuclear (but not chromatin-bound) FANCI may actively sequester RAD51 away from the DNA in FANCD2-deficient cells, causing the particularly severe HDR phenotype observed in these cells. Importantly, in both of these scenarios it would be predicted that the overexpression of FANCI would be deleterious for HDR (and presumably, therefore, cell viability), which was indeed observed by us and others in our inability to establish FANCI-overexpressing complemented clones. Last, it may also be possible that only in the absence of both proteins is a compensatory/back-up pathway engaged that can facilitate RAD51 recruitment.
In summary, our findings suggest a model in which FANCD2 and FANCI have partially non-overlapping roles in promoting cell survival and HR-mediated fork recovery in the presence of APH or HU-induced replication stress.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the FA patients and their families for their interest in our research over the years. We are especially grateful to the Fanconi Anemia Research Fund (FARF), Inc. and the German family organizations ‘Fanconi-Anämie-Stiftung e. V.’, ‘Aktionskreis Fanconi-Anämie e.V.’ and ‘Deutsche Fanconi-Anämie-Hilfe e.V.’ for their support of our ideas and work. We would like to thank Dr Sarah Riman for technical help in generating the FANCD2-null cell line and Jihyeon Yang for technical help with cell culture and western blots. We would also like to thank Dr Anja-Katrin Bielinsky for her comments on the manuscript during its preparation.
Author Contributions: E.L.T., J.E.Y., O.D.S., E.A.H. and A.S. designed the study and wrote the manuscript. E.L.T., J.E.Y., E.A.L and Y.K. performed experiments. H.H. and C.W. generated and provided novel reagents.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [GM088351, in part, CA194871]; National Cancer Institute [CA190492]; University of Minnesota’s NCI-designated Cancer Center Brainstorm Award (in part); American Cancer Society [RSG-13–039-01-DMC, in part]; DFG [SPP 1230]; Deutsche Jose Carreras Leukämie Stiftung e. V. (to H.H.) (in part); Forschungskommission of the Medical Faculty; Strategische Forschungsverbund (to C.W.); Korean Institute for Basic Science [IBS-R022-A1–2017]. Funding for open access charge: NIH Grant [CA194871].
Conflict of interest statement. E.A.H. declares that he is a member of the scientific advisory boards of Horizon Discovery, Ltd. and Intellia Therapeutics, companies that specialize in applying gene editing technology to basic research and therapeutics. The other authors declare no conflict of interest.
REFERENCES
- 1. Kee Y., D’Andrea A.D.. Molecular pathogenesis and clinical management of Fanconi anemia. J. Clin. Invest. 2012; 122:3799–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kupfer G.M. Fanconi anemia: a signal transduction and DNA repair pathway. Yale J. Biol. Med. 2013; 86:491–497. [PMC free article] [PubMed] [Google Scholar]
- 3. Naim V., Rosselli F.. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat. Cell Biol. 2009; 11:761–768. [DOI] [PubMed] [Google Scholar]
- 4. Schlacher K., Wu H., Jasin M.. A distinct replication fork protection pathway connects fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012; 22:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ceccaldi R., Sarangi P., D’Andrea A.D.. The Fanconi anaemia pathway: new players and new functions. Nat. Rev. Mol. Cell Biol. 2016; 17:337–349. [DOI] [PubMed] [Google Scholar]
- 6. Bluteau D., Masliah-Planchon J., Clairmont C., Rousseau A., Ceccaldi R., Dubois d’Enghien C., Bluteau O., Cuccuini W., Gachet S., Peffault de Latour R. et al. . Biallelic inactivation of REV7 is associated with Fanconi anemia. J. Clin. Invest. 2016; 126:3580–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park J.Y., Virts E.L., Jankowska A., Wiek C., Othman M., Chakraborty S.C., Vance G.H., Alkuraya F.S., Hanenberg H., Andreassen P.R.. Complementation of hypersensitivity to DNA interstrand crosslinking agents demonstrates that XRCC2 is a Fanconi anaemia gene. J. Med. Genet. 2016; 53:672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kottemann M.C., Smogorzewska A.. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013; 493:356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Raschle M., Knipscheer P., Enoiu M., Angelov T., Sun J., Griffith J.D., Ellenberger T.E., Scharer O.D., Walter J.C.. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008; 134:969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Knipscheer P., Raschle M., Smogorzewska A., Enoiu M., Ho T.V., Scharer O.D., Elledge S.J., Walter J.C.. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009; 326:1698–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meetei A.R., De Winter J.P., Medhurst A.L., Wallisch M., Waisfisz Q., Van De Vrugt H.J., Oostra A.B., Yan Z., Ling C., Bishop C.E. et al. . A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 2003; 35:165–170. [DOI] [PubMed] [Google Scholar]
- 12. Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E.R. 3rd, Hurov K.E., Luo J., Ballif B.A., Gygi S.P., Hofmann K., D’Andrea A.D. et al. . Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007; 129:289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Timmers C., Taniguchi T., Hejna J., Reifsteck C., Lucas L., Bruun D., Thayer M., Cox B., Olson S., D’Andrea A.D. et al. . Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol. Cell. 2001; 7:241–248. [DOI] [PubMed] [Google Scholar]
- 14. Taniguchi T., Garcia-Higuera I., Xu B., Andreassen P., Gregory R., Kim S., Lane W., Kastan M., D’Andrea A.. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002; 109:459–472. [DOI] [PubMed] [Google Scholar]
- 15. Kim Y., Lach F.P., Desetty R., Hanenberg H., Auerbach A.D., Smogorzewska A.. Mutations of the SLX4 gene in Fanconi anemia. Nat. Genet. 2011; 43:142–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crossan G.P., van der Weyden L., Rosado I.V., Langevin F., Gaillard P.H., McIntyre R.E., Gallagher F., Kettunen M.I., Lewis D.Y., Brindle K. et al. . Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat. Genet. 2011; 43:147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stoepker C., Hain K., Schuster B., Hilhorst-Hofstee Y., Rooimans M.A., Steltenpool J., Oostra A.B., Eirich K., Korthof E.T., Nieuwint A.W. et al. . SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat. Genet. 2011; 43:138–141. [DOI] [PubMed] [Google Scholar]
- 18. Klein Douwel D., Boonen R.A., Long D.T., Szypowska A.A., Raschle M., Walter J.C., Knipscheer P.. XPF-ERCC1 Acts in Unhooking DNA Interstrand Crosslinks in Cooperation with FANCD2 and FANCP/SLX4. Mol. Cell. 2014; 54:460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamamoto K.N., Kobayashi S., Tsuda M., Kurumizaka H., Takata M., Kono K., Jiricny J., Takeda S., Hirota K.. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:6492–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang A.T., Kim T., Wagner J.E., Conti B.A., Lach F.P., Huang A.L., Molina H., Sanborn E.M., Zierhut H., Cornes B.K. et al. . A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol. Cell. 2015; 59:478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang Y., Liu Z., Wang F., Temviriyanukul P., Ma X., Tu Y., Lv L., Lin Y.F., Huang M., Zhang T. et al. . FANCD2 and REV1 cooperate in the protection of nascent DNA strands in response to replication stress. Nucleic Acids Res. 2015; 43:8325–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chaudhury I., Sareen A., Raghunandan M., Sobeck A.. FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res. 2013; 41:6444–6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chaudhury I., Stroik D.R., Sobeck A.. FANCD2-controlled chromatin access of the Fanconi-associated nuclease FAN1 is crucial for the recovery of stalled replication forks. Mol. Cell. Biol. 2014; 34:3939–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yeo J.E., Lee E.H., Hendrickson E.A., Sobeck A.. CtIP mediates replication fork recovery in a FANCD2-regulated manner. Hum. Mol. Genet. 2014; 23:3695–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raghunandan M., Chaudhury I., Kelich S.L., Hanenberg H., Sobeck A.. FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi anemia core complex. Cell Cycle. 2015; 14:342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M.. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011; 145:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sareen A., Chaudhury I., Adams N., Sobeck A.. Fanconi anemia proteins FANCD2 and FANCI exhibit different DNA damage responses during S-phase. Nucleic Acids Res. 2012; 40:8425–8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kohli M., Rago C., Lengauer C., Kinzler K.W., Vogelstein B.. Facile methods for generating human somatic cell gene knockouts using recombinant adeno-associated viruses. Nucleic Acids Res. 2004; 32:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fattah F.J., Kweon J., Wang Y., Lee E.H., Kan Y., Lichter N., Weisensel N., Hendrickson E.A.. A role for XLF in DNA repair and recombination in human somatic cells. DNA Repair (Amst). 2014; 15:39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oh S., Harvey A., Zimbric J., Wang Y., Nguyen T., Jackson P.J., Hendrickson E.A.. DNA ligase III and DNA ligase IV carry out genetically distinct forms of end joining in human somatic cells. DNA Repair (Amst). 2014; 21:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yuan F., El Hokayem J., Zhou W., Zhang Y.. FANCI protein binds to DNA and interacts with FANCD2 to recognize branched structures. J. Biol. Chem. 2009; 284:24443–24452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Joo W., Xu G., Persky N.S., Smogorzewska A., Rudge D.G., Buzovetsky O., Elledge S.J., Pavletich N.P.. Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science. 2011; 333:312–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Niraj J., Caron M.C., Drapeau K., Berube S., Guitton-Sert L., Coulombe Y., Couturier A.M., Masson J.Y.. The identification of FANCD2 DNA binding domains reveals nuclear localization sequences. Nucleic Acids Res. 2017; 45:8341–8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Latt S.A., Kaiser T.N., Lojewski A., Dougherty C., Juergens L., Brefach S., Sahar E., Gustashaw K., Schreck R.R., Powers M. et al. . Cytogenetic and flow cytometric studies of cells from patients with Fanconi's anemia. Cytogenet. Cell Genet. 1982; 33:133–138. [DOI] [PubMed] [Google Scholar]
- 36. Pierce A.J., Johnson R.D., Thompson L.H., Jasin M.. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999; 13:2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bennardo N., Cheng A., Huang N., Stark J.M.. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008; 4:e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Williams S.A., Longerich S., Sung P., Vaziri C., Kupfer G.M.. The E3 ubiquitin ligase RAD18 regulates ubiquitylation and chromatin loading of FANCD2 and FANCI. Blood. 2011; 117:5078–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu X., Baer R.. Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J. Biol. Chem. 2000; 275:18541–18549. [DOI] [PubMed] [Google Scholar]
- 40. Xu D., Guo R., Sobeck A., Bachrati C.Z., Yang J., Enomoto T., Brown G.W., Hoatlin M.E., Hickson I.D., Wang W.. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 2008; 22:2843–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Davies S.L., North P.S., Hickson I.D.. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat. Struct. Mol. Biol. 2007; 14:677–679. [DOI] [PubMed] [Google Scholar]
- 42. Bianco J.N., Poli J., Saksouk J., Bacal J., Silva M.J., Yoshida K., Lin Y.L., Tourriere H., Lengronne A., Pasero P.. Analysis of DNA replication profiles in budding yeast and mammalian cells using DNA combing. Methods. 2012; 57:149–157. [DOI] [PubMed] [Google Scholar]
- 43. Kalb R., Neveling K., Hoehn H., Schneider H., Linka Y., Batish S.D., Hunt C., Berwick M., Callen E., Surralles J. et al. . Hypomorphic mutations in the gene encoding a key Fanconi anemia protein, FANCD2, sustain a significant group of FA-D2 patients with severe phenotype. Am. J. Hum. Genet. 2007; 80:895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sims A.E., Spiteri E., Sims R.J. 3rd, Arita A.G., Lach F.P., Landers T., Wurm M., Freund M., Neveling K., Hanenberg H. et al. . FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 2007; 14:564–567. [DOI] [PubMed] [Google Scholar]
- 45. Colnaghi L., Jones M.J., Cotto-Rios X.M., Schindler D., Hanenberg H., Huang T.T.. Patient-derived C-terminal mutation of FANCI causes protein mislocalization and reveals putative EDGE motif function in DNA repair. Blood. 2011; 117:2247–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Topaloglu O., Hurley P.J., Yildirim O., Civin C.I., Bunz F.. Improved methods for the generation of human gene knockout and knockin cell lines. Nucleic Acids Res. 2005; 33:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Castella M., Jacquemont C., Thompson E.L., Yeo J.E., Cheung R.S., Huang J.W., Sobeck A., Hendrickson E.A., Taniguchi T.. FANCI regulates recruitment of the FA core complex at sites of DNA damage independently of FANCD2. PLoS Genet. 2015; 11:e1005563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ishiai M., Kitao H., Smogorzewska A., Tomida J., Kinomura A., Uchida E., Saberi A., Kinoshita E., Kinoshita-Kikuta E., Koike T. et al. . FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 2008; 15:1138–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sobeck A., Stone S., Costanzo V., de Graaf B., Reuter T., de Winter J., Wallisch M., Akkari Y., Olson S., Wang W. et al. . Fanconi anemia proteins are required to prevent accumulation of replication-associated DNA double-strand breaks. Mol. Cell. Biol. 2006; 26:425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boisvert R.A., Rego M.A., Azzinaro P.A., Mauro M., Howlett N.G.. Coordinate nuclear targeting of the FANCD2 and FANCI proteins via a FANCD2 nuclear localization signal. PLoS One. 2013; 8:e81387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Howlett N.G., Taniguchi T., Durkin S.G., D’Andrea A.D., Glover T.W.. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum. Mol. Genet. 2005; 14:693–701. [DOI] [PubMed] [Google Scholar]
- 52. Zhang F., Altorki N.K., Mestre J.R., Subbaramaiah K., Dannenberg A.J.. Curcumin inhibits cyclooxygenase-2 transcription in bile acid- and phorbol ester-treated human gastrointestinal epithelial cells. Carcinogenesis. 1999; 20:445–451. [DOI] [PubMed] [Google Scholar]
- 53. Pichierri P., Rosselli F.. The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. EMBO J. 2004; 23:1178–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kee Y., D’Andrea A.D.. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010; 24:1680–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen X., Bosques L., Sung P., Kupfer G.M.. A novel role for non-ubiquitinated FANCD2 in response to hydroxyurea-induced DNA damage. Oncogene. 2016; 35:22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Murina O., von Aesch C., Karakus U., Ferretti L.P., Bolck H.A., Hanggi K., Sartori A.A.. FANCD2 and CtIP cooperate to repair DNA interstrand crosslinks. Cell Rep. 2014; 7:1030–1038. [DOI] [PubMed] [Google Scholar]
- 57. Unno J., Itaya A., Taoka M., Sato K., Tomida J., Sakai W., Sugasawa K., Ishiai M., Ikura T., Isobe T. et al. . FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Rep. 2014; 7:1039–1047. [DOI] [PubMed] [Google Scholar]
- 58. Ghosal G., Chen J.. DNA damage tolerance: a double-edged sword guarding the genome. Transl. Cancer Res. 2013; 2:107–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Branzei D., Szakal B.. DNA damage tolerance by recombination: Molecular pathways and DNA structures. DNA Repair (Amst). 2016; 44:68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brown S., Niimi A., Lehmann A.R.. Ubiquitination and deubiquitination of PCNA in response to stalling of the replication fork. Cell Cycle. 2009; 8:689–692. [DOI] [PubMed] [Google Scholar]
- 61. Michl J., Zimmer J., Tarsounas M.. Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J. 2016; 35:909–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen Y.H., Jones M.J., Yin Y., Crist S.B., Colnaghi L., Sims R.J. 3rd, Rothenberg E., Jallepalli P.V., Huang T.T.. ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress. Mol. Cell. 2015; 58:323–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Miyagawa K., Tsuruga T., Kinomura A., Usui K., Katsura M., Tashiro S., Mishima H., Tanaka K.. A role for RAD54B in homologous recombination in human cells. EMBO J. 2002; 21:175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Oh S., Wang Y., Zimbric J., Hendrickson E.A.. Human LIGIV is synthetically lethal with the loss of Rad54B-dependent recombination and is required for certain chromosome fusion events induced by telomere dysfunction. Nucleic Acids Res. 2013; 41:1734–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sato K., Shimomuki M., Katsuki Y., Takahashi D., Kobayashi W., Ishiai M., Miyoshi H., Takata M., Kurumizaka H.. FANCI-FANCD2 stabilizes the RAD51-DNA complex by binding RAD51 and protects the 5′-DNA end. Nucleic Acids Res. 2016; 44:10758–10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jayathilaka K., Sheridan S.D., Bold T.D., Bochenska K., Logan H.L., Weichselbaum R.R., Bishop D.K., Connell P.P.. A chemical compound that stimulates the human homologous recombination protein RAD51. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:15848–15853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vinciguerra P., Godinho S.A., Parmar K., Pellman D., D’Andrea A.D.. Cytokinesis failure occurs in Fanconi anemia pathway-deficient murine and human bone marrow hematopoietic cells. J. Clin. Invest. 120:3834–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liang C.C., Li Z., Lopez-Martinez D., Nicholson W.V., Venien-Bryan C., Cohn M.A.. The FANCD2-FANCI complex is recruited to DNA interstrand crosslinks before monoubiquitination of FANCD2. Nat. Commun. 2016; 7:12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nakanishi K., Yang Y.G., Pierce A.J., Taniguchi T., Digweed M., D’Andrea A.D., Wang Z.Q., Jasin M.. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:1110–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Suzuki S., Racine R.R., Manalo N.A., Cantor S.B., Raffel G.D.. Impairment of fetal hematopoietic stem cell function in the absence of Fancd2. Exp. Hematol. 2016; 48:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xia P., Sun Y., Zheng C., Hou T., Kang M., Yang X.. p53 mediated apoptosis in osteosarcoma MG-63 cells by inhibition of FANCD2 gene expression. Int. J. Clin. Exp. Med. 2015; 8:11101–11108. [PMC free article] [PubMed] [Google Scholar]
- 72. Joksic I., Vujic D., Guc-Scekic M., Leskovac A., Petrovic S., Ojani M., Trujillo J.P., Surralles J., Zivkovic M., Stankovic A. et al. . Dysfunctional telomeres in primary cells from Fanconi anemia FANCD2 patients. Genome Integr. 2012; 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Petermann E., Orta M.L., Issaeva N., Schultz N., Helleday T.. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010; 37:492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Godthelp B.C., Artwert F., Joenje H., Zdzienicka M.Z.. Impaired DNA damage-induced nuclear Rad51 foci formation uniquely characterizes Fanconi anemia group D1. Oncogene. 2002; 21:5002–5005. [DOI] [PubMed] [Google Scholar]
- 75. Godthelp B.C., Wiegant W.W., Waisfisz Q., Medhurst A.L., Arwert F., Joenje H., Zdzienicka M.Z.. Inducibility of nuclear Rad51 foci after DNA damage distinguishes all Fanconi anemia complementation groups from D1/BRCA2. Mutat. Res. 2006; 594:39–48. [DOI] [PubMed] [Google Scholar]
- 76. Ohashi A., Zdzienicka M.Z., Chen J., Couch F.J.. Fanconi anemia complementation group D2 (FANCD2) functions independently of BRCA2- and RAD51-associated homologous recombination in response to DNA damage. J. Biol. Chem. 2005; 280:14877–14883. [DOI] [PubMed] [Google Scholar]
- 77. Digweed M., Rothe S., Demuth I., Scholz R., Schindler D., Stumm M., Grompe M., Jordan A., Sperling K.. Attenuation of the formation of DNA-repair foci containing RAD51 in Fanconi anaemia. Carcinogenesis. 2002; 23:1121–1126. [DOI] [PubMed] [Google Scholar]
- 78. Wang X., Andreassen P.R., D’Andrea A.D.. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell. Biol. 2004; 24:5850–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.