Abstract
Background:
Ambient particulate matter (PM) air pollution exposure has been associated with increases in QT interval duration (QT). However, innate susceptibility to PM-associated QT prolongation has not been characterized.
Objective:
To characterize genetic susceptibility to PM-associated QT prolongation in a multi-racial/ethnic, genome-wide association study (GWAS).
Methods:
Using repeated electrocardiograms (1986–2004), longitudinal data on in diameter (), and generalized estimating equations methods adapted for low-prevalence exposure, we estimated approximately interactions among nine Women’s Health Initiative clinical trials and Atherosclerosis Risk in Communities Study subpopulations (), then combined subpopulation-specific results in a fixed-effects, inverse variance-weighted meta-analysis.
Results:
A common variant (rs1619661; coded allele: T) significantly modified the association (). At concentrations percentile, QT increased 7 ms across the CC and TT genotypes: 397 (95% confidence interval: 396, 399) to 404 (403, 404) ms. However, QT changed minimally across rs1619661 genotypes at lower concentrations. The rs1619661 variant is on chromosome 10, 132 kilobase (kb) downstream from CXCL12, which encodes a chemokine, stromal cell-derived factor 1, that is expressed in cardiomyocytes and decreases calcium influx across the L-type channel.
Conclusions:
The findings suggest that biologically plausible genetic factors may alter susceptibility to -associated QT prolongation in populations protected by the U.S. Environmental Protection Agency’s National Ambient Air Quality Standards. Independent replication and functional characterization are necessary to validate our findings. https://doi.org/10.1289/EHP347
Introduction
Ambient particulate matter (PM) air pollution contributes substantially to cardiovascular disease morbidity and mortality (Dockery et al. 1993; Miller et al. 2007; Samet et al. 2000). Several studies have attributed part of the contribution to prolonged ventricular repolarization, a known risk factor for cardiovascular events (Dekker et al. 2004; Goldberg et al. 1991; Rautaharju et al. 2006a, b; Schouten et al. 1991), as suggested by PM-associated increases in risk of ventricular arrhythmia/sudden cardiac death (Dockery et al. 2005; Ljungman et al. 2008). Indeed, PM has been associated with increases in QT interval duration (QT) (Liao et al. 2010; Mordukhovich et al. 2016; Van Hee et al. 2011), a quantitative electrocardiographic measure of ventricular repolarization. Although QT prolongation is also related to innate variation in myocardial cation channel proteins (Arking et al. 2014) and the rate at which cation gradients across these voltage-gated channels return to their resting potential, genetic susceptibility to (i.e., modification of) PM-associated QT prolongation has not been evaluated.
The Clean Air Act nevertheless requires the U.S. Environmental Protection Agency (EPA) to create National Ambient Air Quality Standards (NAAQS) that protect populations susceptible to the adverse health effects of PM. Mindful of such regulatory obligations and their public health implications, the present study examined genome-wide variation in susceptibility to -associated QT prolongation among nine longitudinally well-characterized and racially/ethnically diverse populations of men and women living in the 48 contiguous states in the United States (U.S. ).
Methods
Study Design
The study included 22,158 participants in the Atherosclerosis Risk in Communities Study (ARIC) (ARIC Investigators 1989) and the Women’s Health Initiative (WHI) clinical trials (National Institutes of Health 1998) cohorts who were examined between 1986 and 2004. They consented to the use of DNA for genetic research, had genomic data, and had one or more high-quality, non-paced baseline or follow-up electrocardiograms (ECGs). They were not taking antiarrhythmic medication and did not have heart failure, bundle branch block (), or Wolf-Parkinson-White pattern.
ARIC participants included black and white men and women. White WHI participants were drawn from three substudies: the Genome-wide Association Research Network into Effects of Treatment (GARNET) (National Institutes of Health 2013), Modification of PM-Mediated Arrhythmogenesis in Populations (MOPMAP) (National Institutes of Health 2010), and Women's Health Initiative Memory Study (WHIMS) (Shumaker et al. 1998). Black and Hispanic WHI participants were drawn from the single nucleotide polymorphism (SNP) Health Association Resource Project (SHARe) (National Institutes of Health 2011).
Electrocardiography
At triennial participant examinations and examination sites, trained and certified technicians used standardized procedures and identical electrocardiographs (MAC PCTM; GE Marquette Electronics Inc.) to digitally record and telephonically transmit resting, 10-second, standard, simultaneous 12-lead ECGs to a central laboratory (Epidemiological Cardiology Research Center, Wake Forest School of Medicine, Winston-Salem, NC) for visual inspection, identification of technical errors/inadequate quality, and analysis using the 2001 version of the GE Marquette 12–SLTM program (GE Marquette) (ARIC Investigators 1987; WHI Study Group 1994). Analysis included reliable estimation of median QT (ms) from the orthogonal XYZ leads and median RR interval duration (RR, ms), i.e., the unit-corrected inverse of heart rate (Schroeder et al. 2004; Vaidean et al. 2005).
Genotyping, Quality Control, and Imputation
Genotypes of participants determined on the Affymetrix GeneChip SNP Array 6.0, Illumina Human Omni1-Quad v1-0 B, Affymetrix Genome-wide Human CEU I, or Human OmniExpress Exome-8v1_B Genome-wide Human platforms were subjected to platform-specific quality filters (see Table S1). In GARNET, SNP dosage was imputed using BEAGLE (Browning and Browning 2009) and 1,000 Genomes Project (1000G v3 EUR, 03/2012) reference haplotypes. In the remaining subpopulations, imputation relied on MACH (Li et al. 2010) or Minimac (Howie et al. 2012) and HapMap2 CEU reference haplotypes in ARIC, MOPMAP, and WHIMS whites; a 1:1 mix of HapMap2 CEU/YRI in ARIC and WHI SHARe blacks; and all 1000G ancestry samples in SHARe Hispanics. Coordinate ranges for all HapMap 2 (Build 36) SNPs were converted to Build 37 using liftOver (Kuhn et al. 2012).
PM Exposure Estimation
Participant addresses at the time of ECGs were accurately geocoded (Whitsel et al. 2004; Whitsel et al. 2006), and then national-scale, log-normal ordinary kriging and U.S. EPA Air Quality Systems (AQS) monitor data were used to estimate geocoded address-specific, daily mean concentrations of ambient in diameter () (Liao et al. 2006) between 1986 and 2004. Validity of the exposure-estimation method during this period was evaluated using standard cross-validation statistics: the average prediction error ( concentration at each monitor site), standardized prediction error (), root mean square standardized (), root mean square prediction error (), and mathematically calculated standard error (SE). Observed values of PE and SPE near 0, RMSS near 1, and RMS near SE have provided evidence of model validity (Liao et al. 2006; Liao et al. 2007) and justification for use of the estimates in published studies of –health outcome associations (Holliday et al. 2014; Liao et al. 2009; Shih et al. 2011; Whitsel et al. 2009; Zhang et al. 2009) in which daily concentrations were averaged over the day of and day before each ECG (). Although comparably estimated and accurate, daily mean concentrations of ambient in diameter () were not available until 1999, when EPA AQS monitoring data for became more widely available, monthly mean concentrations of ambient were spatiotemporally estimable at the same geocoded addresses between 1986 and 2004 using generalized additive mixed models, the log-transformed ratio of to predicted , and geographic information system (GIS)-based predictors. A 5- to 10-set, out-of-sample cross-validation of the estimates in which the squared Pearson correlation between excluded monthly observations and model predictions () suggested that the model performed well (Yanosky et al. 2014).
Statistical Analysis
The population was stratified by study, race/ethnicity, and sex. Within these subpopulations, modeling involved a generalized estimating-equations method implemented in R (via the boss package) (Voorman and Sitlani 2013) that was designed to detect interactions between SNPs and low-prevalence environmental exposures on a genome-wide scale using repeated measures data (Sitlani et al. 2015), in which and denote the ith participant at the jth electrocardiographic examination. Models were given by
where is QT (ms); is the intercept; is the HapMap 2 SNP dosage (0–2); is the dichotomized concentration ( an a priori threshold for abnormality, defined as the subpopulation-specific 90th percentile); is the additive interaction term , and is a vector of coefficients corresponding to , a vector of covariables comprising age (years), geographic region (in WHI) or center (in ARIC), season, calendar year, RR interval (ms), and principal components of ancestry estimated using Eigenstrat (Price et al. 2006). Fit of the fully adjusted model to dichotomized concentrations reinforced its selection over alternatives expressing QT as a linear, quadratic, linear spline, or quadratic spline function of with one to five knots, with and without restriction (Hardin and Hilbe 2012; Pan 2001). To reduce type-1 errors in detecting interactions at low-prevalence exposure, the t-reference distribution with degrees of freedom based on Satterthwaite’s approximation was used (Pan and Wall 2002; Satterthwaite 1946; Sitlani et al. 2015). SNPs with a low minor allele frequency and imputation quality also were excluded from subpopulation-specific analyses (Sitlani et al. 2015) when
where is the minor allele frequency, is the SNP imputation quality, and is the estimated number of observations with a concentration percentile. For each of the approximately 2.5 million SNPs remaining across subpopulations, subpopulation-specific interaction estimates were adjusted using genomic control, tested for homogeneity (Cochran’s Q), and combined using fixed-effects, inverse variance-weighted meta-analysis implemented in METAL (Willer et al. 2010). Two-stage, split-sample alternatives (in which subpopulations are grouped into discovery and replication populations) were considered but avoided to maximize statistical power (Skol et al. 2006).
Observed genome-wide distributions of meta-analyzed SNP-specific interaction p-value were transformed and compared with those expected under the distribution using a quantile-quantile (QQ) plot and the genomic inflation factor () (Devlin and Roeder 1999). They were also mapped by chromosomal position to produce Manhattan and regional association plots (Pruim et al. 2010).
After accounting for linkage disequilibrium (LD) among the approximately 2.5 million SNPs across racially/ethnically diverse subpopulations, a Bonferroni-corrected threshold of was used to identify genome-wide significant interactions (Barsh et al. 2012; Pe'er et al. 2008). Interaction and standard error estimates of SNPs meeting that threshold were forest plotted and used to compute predicted mean QT (ms) and 95% confidence intervals (95% CIs) by genotype and concentration, while adjusting for centered covariables. Sensitivity of results also was examined as follows: to lower thresholds for abnormality (50th–80th percentiles), longer lagged exposure averaging periods (1–4 wk), alternative exposures () (Yanosky et al. 2014), use of -antagonists, additional adjustments [temperature (); dew point (); barometric pressure (kPa); neighborhood socioeconomic status; smoker status (current, former, never); alcohol drinker status (current, former, never); total caloric intake (kcal); sedentary lifestyle] and separately, application of Bayesian meta-analytic methods allowing for ancestral population heterogeneity implemented in MANTRA (Morris 2011) (genome-wide threshold of importance: , ) (Stephens and Balding 2009).
Significant associations identified lead SNPs and variants in LD with them () for bioinformatic characterization using HaploReg V4 (Ward and Kellis 2012), the UCSC Genome BrowserTM (Kuhn et al. 2012), and the WashU Epigenome Browser (Zhou et al. 2011) with data from the Encyclopedia of DNA Elements Project (ENCODE) (Rosenbloom et al. 2010) and Roadmap Epigenomics Project (Bernstein et al. 2010). Their characterization involved searching surrounding regions of the cardiac genome (e.g., in cardiomyocytes, cardiac fibroblasts, and heart tissue) for evidence of active or altered transcription.
Results
The nine ARIC and WHI subpopulations in this study collectively represented 22,158 participants, of whom 26% were black, 7% were Hispanic, and 22% were male. On average, participants were 64.3 years old and contributed 2.9 ECGs with a mean QT of 402 ms. The two-day mean () concentration and its 90th percentile (P90) were and , i.e., below NAAQS for in place at the time of participant examinations (Table 1) (U.S. EPA 2016).
Table 1.
Characteristics of subpopulations, by study, race/ethnicity, and sex.
| Study | Race/ethnicity | Sex | Age, y | ECGs | QT, ms | a | P90 | |
|---|---|---|---|---|---|---|---|---|
| ARIC | Black | Men | 826 | 50.3 | ||||
| ARIC | Black | Women | 1,343 | 50.9 | ||||
| ARIC | White | Men | 3,976 | 49.8 | ||||
| ARIC | White | Women | 4,462 | 49.7 | ||||
| WHI GARNETb | White | Women | 1,732 | 41.5 | ||||
| WHI MOPMAPb | White | Women | 1,237 | 41.2 | ||||
| WHI SHARe | Black | Women | 3,538 | 41.8 | ||||
| WHI SHARe | Hispanic | Women | 1,562 | 43.4 | ||||
| WHI WHIMS | White | Women | 3,482 | 39.7 | ||||
| All | White (67%) | Women (78%) | 22,158 | 64.3 | 2.9 | 402 | 29.9 | 45.4 |
Note: ARIC, Atherosclerosis Risk in Communities study; ECG, electrocardiogram; GARNET, Genomics and Randomized Trials Network; MOPMAP, Modification of PM-Mediated Arrhythmogenesis in Populations; P90, 90th percentile; , particulate matter in diameter; QT, QT interval duration; SD, standard deviation; SHARe, SNP Health Association Resource; WHI, Women’s Health Initiative; WHIMS, Women's Health Initiative Memory Study.
Range, .
Controls.
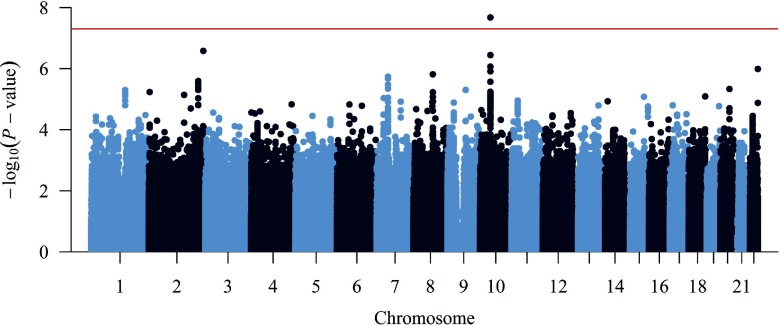
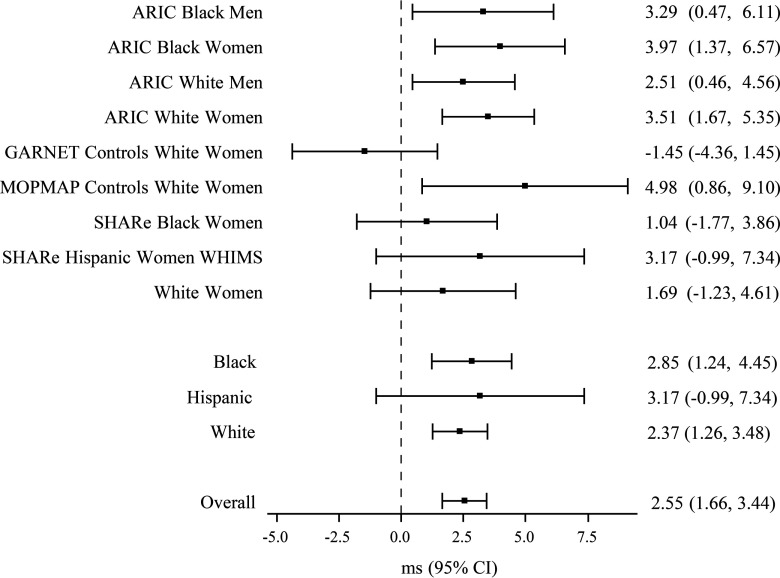
Manhattan, regional association, and QQ plots (Figures 1 and 2; see Figure S1) of interaction p-value from the trans-ethnic, fixed-effects, inverse variance-weighted meta-analysis identified one genome-wide significant association (rs1619661; ) and 22 subthreshold associations () across six independent loci (Table 2). The lead SNP, rs1619661 is on chromosome 10, approximately 132 kilobase (kb) downstream of CXCL12 (Table 2). This variant’s coded allele, T (vs. the noncoded allele, C), was common among racial/ethnic groups (T allele: 81–92%; CC genotype: 1.6%, CT: 21.8%, TT: 76.7%) and associated with QT prolongation in eight (89%) of the nine subpopulations (; Figure 3).
Figure 1.
Manhattan plot of p-value vs. chromosomal position of each SNP from the trans-ethnic, fixed-effects meta-analysis of the interactions. The red line references the genome-wide significance threshold (p-value).
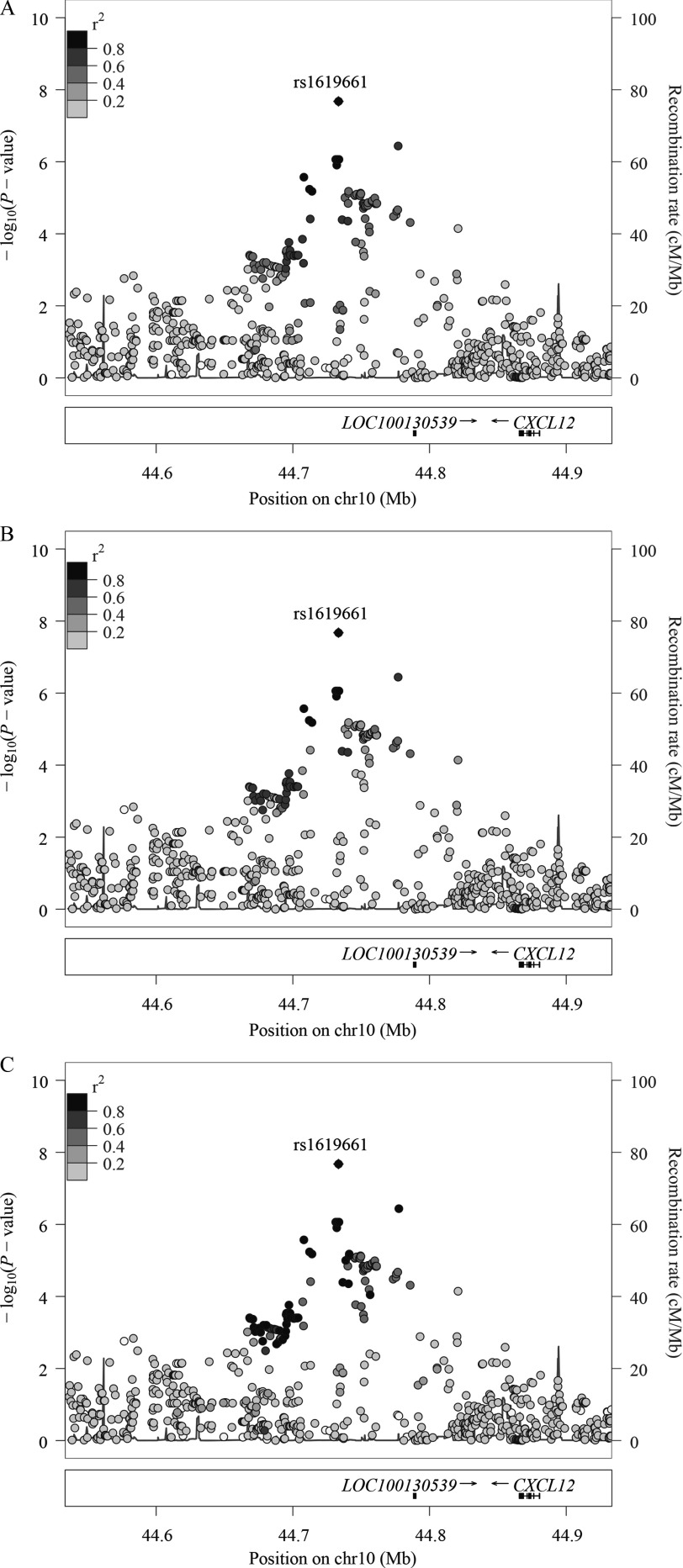
Figure 2.
Regional plots of the locus, rs1619661, identified by the trans-ethnic, fixed-effects meta-analysis of the interactions, on chromosome 10, near CXCL12. Each point represents the p-value of a SNP plotted as a function of its genomic position (build 37) and the genome-wide significance threshold (p-value). One SNP reached this threshold. The color coding of all other SNPs indicated linkage disequilibrium with this lead SNP, estimated among Africans (A), Ad-mixed Americans (B), and Europeans (C) from 1000G. Recombination rates were estimated from the 1,000 Genomes Project.
Table 2.
Findings from the trans-ethnic, fixed-effects, inverse variance-weighted meta-analysis, including sub-threshold associations ().
| Chr | Position | Lead SNP | CA /NCA | Coded allele Frequency | p-value | Interaction Estimate (SE) | Nearest Gene | SNPsa | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Black | Hispanic | White | |||||||||
| 10 | 44,733,383 | rs1619661 | T/C | 0.81 | 0.92 | 0.91 | 2.55 (0.46) | 22,158 | CXCL12 | 8 | |
| 22 | 51,065,600 | rs6151412 | G/A | 0.90 | 0.95 | 0.95 | 3.88 (0.79) | 20,921 | ARSA | 1 | |
| 8 | 83,252,586 | rs10504754 | A/G | 0.74 | 0.47 | 0.43 | 1.54 (0.32) | 22,158 | SNX16 | 1 | |
| 7 | 48,811,506 | rs13309098 | G/A | 0.88 | 0.88 | 0.93 | 2.37 (0.50) | 22,158 | ABCA13-CDC14C | 4 | |
| 2 | 213,065,465 | rs6725041 | T/C | 0.78 | 0.44 | 0.48 | 1.52 (0.32) | 22,158 | ERBB4 | 8 | |
| 20 | 39,435,700 | rs7361259 | T/C | 0.91 | 5.98 (1.39) | 2,169 | MAFB | 1 | |||
Note: CA, coded allele; CAF, coded allele frequency; Chr, chromosome; NCA, noncoded allele; SE, standard error; SNP, single nucleotide polymorphism.
Total number of significant or sub-threshold SNPs located within the same gene or in LD with the lead SNP ().
Figure 3.
Forest plot of interaction (95% confidence interval) per T allele increase in rs1619661 (genotype CT) at concentrations percentile, by study, race/ethnicity, and overall ().
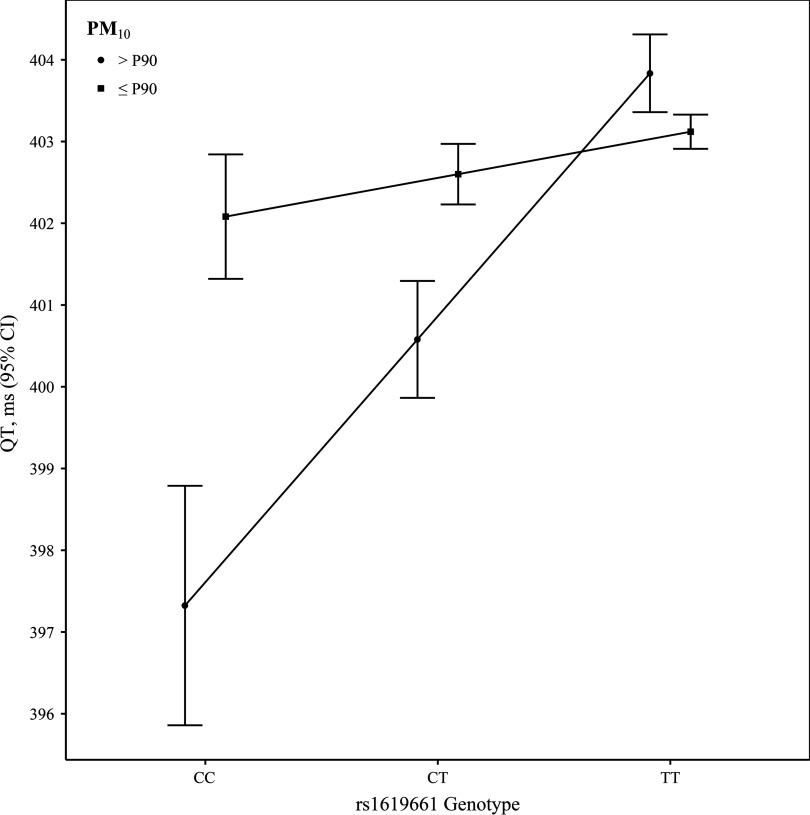
At concentrations percentile, QT increased 7 ms across the CC, CT, and TT rs1619661 genotypes: from 397 (95% CI: 396, 399) to 401 (400, 401) to 404 (403, 404) ms, but at concentrations percentile, QT only increased from 402 (401, 403) to 403 (402, 403) to 403 (403, 403) ms (Figure 4; Table S2). Associations were insensitive to additional adjustment, Bayesian meta-analysis (; ), and adoption of a threshold, the annual NAAQS for . However, they were attenuated by decreasing thresholds, increasing lagged exposure averaging periods, substituting , and restricting to -antagonist users (see Table S3, Figure S2).
Figure 4.
Predicted mean (95% confidence interval) QT (ms) per unit increase in the coded allele (T) dosage of rs1619661 at concentrations and percentile (P90), while adjusting for age, geographic region or center, season, calendar year, RR interval, and ancestry. C allele frequency range: 8–19%.
In cardiomyocytes, cardiac fibroblasts, and other (including fetal, right atrial, and left/right ventricular) heart tissue, genomic regions surrounding rs1619661 and associated SNPs included deoxyribonuclease (DNAse1) hypersensitivity areas, DNA methylation sites, enhancer/promoter histone marks, and regulatory motifs (see Figure S3 and “TITLE” in Supplemental Material).
Full results from the trans-ethnic, fixed-effects, inverse-variance meta-analysis and rs1619661 characterization using HaploReg Version 4 (Ward and Kellis 2012) and the WashU Epigenome BrowserTM (Zhou et al. 2011) are available at https://qtgwaspm.web.unc.edu/EHP/ (Gondalia 2016).
Discussion
This genome-wide association study (GWAS) of gene–environment interactions discovered a genetic variant associated with increased susceptibility of a racially and geographically diverse population of U.S. men and women to prolonged ventricular repolarization during short-duration ambient PM air pollution exposures below annual and daily thresholds established by the U.S. EPA under the Clean Air Act (U.S. EPA 2016).
Although we observed a clinically modest, 7-ms increase in QT among persons in the highest decile with two vs. zero copies of the T allele (genotype TT vs. CC, respectively), the T allele of rs1619661 tends to be so common in many U.S. populations that related but seemingly minor population-level shifts in QT may have significant public-health implications. Indeed, upper decile -associated increases in QT exceed the U.S. Food and Drug Administration (FDA) 5-ms threshold used in premarket evaluation of drug safety (U.S. FDA 2015), an increase that may also carry cardiovascular disease morbidity and mortality risk (Zhang et al. 2011).
The attendant cardiovascular risks are plausibly related to CXCL12 (Table 2)—the locus most proximate to rs1619661—which has been implicated in, for example, GWAS of coronary artery disease (Samani et al. 2007) and early-onset myocardial infarction (Kathiresan et al. 2009). CXCL12 encodes stromal cell-derived factor 1 (SDF1), an evolutionarily conserved chemokine that is expressed in cardiomyocytes (Pyo et al. 2006) and is induced by pro-inflammatory stimuli, including particulate exposures (Liberda et al. 2010). SDF1 binds to CXCR4, a seven-transmembrane, G-protein coupled receptor that is widely distributed on cardiomyocytes and neurons.
In those cell types, the ligand-receptor complex inhibits -adrenergically activated calcium influx through the L-type ion channel (Pyo et al. 2006), recently implicated in the largest GWAS of QT to date (Arking et al. 2014). Resultant shortening of the ventricular action potential (Phase 2) and its electrocardiographic manifestation, QT interval duration, was apparent in the present study among persons in the highest decile with the C allele [i.e., those individuals with the CC or CT (vs. TT) genotype]. Although contrary to PM-associated increases in QT duration observed in prior studies (Liao et al. 2010; Mordukhovich et al. 2016; Van Hee et al. 2011), this group represents only a minority of the study population. Likewise, its reversibility was reflected, albeit in this observational epidemiologic context, by the attenuation of the observed interaction among users of -adrenergic antagonists, the first-line therapy in long-QT syndromes (LaRocca et al. 2010).
The interaction also was attenuated at longer lagged exposure averaging periods in this context. This form of attenuation highlights the potential role of -adrenergic receptor-mediated blunting of sympathetic nervous system responses to chronic PM exposure. Indeed, sympathetic responses of the heart to stressors are mediated by the binding of catecholamines to cardiac -adrenergic receptor s, the density, sensitivity, and activity of which decrease with chronic stress exposure (Konarska et al. 1989; Stone 1983). Chronic stress exposures also lead to adaptive changes of neural and glial cells in the central nervous system (McEwen 2007), which controls the heart via innervation of the sinoatrial node. The attenuated interactions that we observed herein may thereby reflect physiologically desensitizing adaptations to longer-term PM exposures.
However, several limitations apply to the study of gene-environment interaction in genome-wide contexts, e.g., low power and overestimation of observed effect sizes in hypothesis-generating discovery efforts (Göring et al. 2001). To increase power, we used all nine subpopulations in the discovery effort. To further increase power, we used generalized estimating equations methods to leverage repeated measures of QT and among 22,158 participants from two well-characterized, multi-ethnic, and environmentally diverse cardiovascular disease cohorts. Furthermore, we established homogeneity and robustness of interaction estimates among the cohorts, subpopulations, and races/ethnicities in meta-analyses, which were also subjected to additional adjustment for meteorological, neighborhood-socioeconomic, and lifestyle characteristics. Finally, the trans-ethnic, fixed-effects, inverse variance-weighted meta-analysis discovered a genome-wide significant interaction in data that also provided convincing evidence of association in a Bayesian meta-analysis that allowed for racial/ethnic heterogeneity, where the interaction was found to be 1.6 million times more likely under the alternative to the null hypothesis of no association.
The 132-kb separation of rs1619661 and CXCL12 also limits the biological plausibility of their role in PM-associated QT prolongation. However, causal genes that are megabases away from GWAS-implicated lead SNPs have been identified in other settings (Musunuru et al. 2010; Smemo et al. 2014). For example, obesity-associated SNPs within the well-known FTO locus directly interact with promoter regions of IRX3 that are approximately 500 kb downstream. In fact, IRX3, which is causally linked to body mass and composition, participates in long-range interactions across a relatively large, 2-megabase region (Ragvin et al. 2010; Smemo et al. 2014). We also identified potentially active or altered transcription in regions of the cardiac genome surrounding rs1619661 and its associated SNPs with data from ENCODE. Although it is unclear whether these regions are functionally linked to CXCL12, it is plausible because of important, long-range (i.e., ) mechanisms of distal gene regulation (Sanyal et al. 2012). Nevertheless, expression assays are needed to confirm the proposed link between the rs1619661 locus and CXCL12.
Replication—a suggested gold standard for validating GWAS of main genetic effects—poses a particular challenge for gene-environment interaction studies like the one described here (Aschard et al. 2012; Aslibekyan et al. 2013; Hutter et al. 2013; Thomas et al. 2012). The extent of the challenge is related to the need for similarly powered populations with equally well-harmonized outcomes and exposures, even if they are, e.g., rare, difficult to measure, or peculiar to racial/ethnic minority populations poorly represented in large-scale GWAS to date. In the present study, a well-powered, independent replication was not feasible, given the limited availability of populations with high-quality, 12-lead ECGs; national-scale, kriged daily mean concentrations; and genome-wide SNP data. Moreover, functional validation in model organisms (Gibert et al. 2013; Stevens et al. 2015) was beyond the scope of the original project. We therefore view this discovery effort as hypothesis-generating, and given the importance of replication in protecting against type-1 error (Siontis et al. 2010), we have provided publicly accessible summary statistics (https://qtgwaspm.web.unc.edu/EHP/) to facilitate functional validation and external replication as additional data become available.
Although not reaching genome-wide significance, the subthreshold loci identified herein (Table 2) may also warrant scrutiny. ARSA (rs6151412, synonymous) and ERBB4 (rs6725041, intronic) are particularly compelling in this setting due to their functional role in transport (Brero et al. 2010; Ritzler et al. 1992). ERBB4 has additionally been associated with cardiac myopathy (García-Rivello et al. 2005), coronary artery calcification (Wojczynski et al. 2013), and cardiomyocyte proliferation (Wadugu and Kühn 2012). SNX16 (rs10504754, 498 kb upstream) has been associated with heart failure (Smith et al. 2010). MAFB (rs7361259, 118 kb upstream) has been implicated in a gene–drug interaction GWAS of rheumatoid arthritis, an inflammatory disorder associated with QT prolongation (Chauhan et al. 2015). ABCA13-CDC14C (rs13309098; 124-137 kb downstream) currently has no established link with cardiovascular disease.
Conclusions
We conclude that genetic variation may modify susceptibility to -associated QT prolongation, and pending further follow-up, cautiously postulate changes in L-type ion channel activity triggered by inflammatory responses to PM exposure as a possible mechanism. In lieu of such possibilities, previously hypothesized genetic, inflammatory, and neural mechanisms of PM-mediated arrhythmogenesis would remain largely distinct. The Clean Air Act mandates setting of NAAQS for PM that protect sensitive populations—including persons with innate factors that may increase vulnerability to PM-associated disease. Although we cannot unequivocally link genetic variation to PM-associated QT prolongation, we did discover a biologically plausible variant that may confer susceptibility, a finding that must undergo replication and functional characterization in future studies.
Supplemental Material
Acknowledgments
The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by NHLBI contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C.
The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by UL1RR025005, a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research.
The WHI program is funded by the NHLBI, U.S. Department of Health and Human Services, through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C. Funding support for WHI GARNET was provided through the NHGRI Genomics and Randomized Trials Network (GARNET) (U01 HG005152). Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GARNET Coordinating Center (U01 HG005157). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Funding support for genotyping, which was performed at the Broad Institute of MIT and Harvard, was provided by the NIH Genes, Environment and Health Initiative (GEI) (U01 HG004424). Funding for WHI SHARe genotyping was provided by NHLBI N02-HL-64278.
This work was supported by National Institute of Environmental Health Sciences grant R01-ES017794 (E.A.W.) and a National Research Service Award (R.G.) provided by a National Heart, Lung, and Blood Institute training grant 5-T32-HL007055.
The study was reviewed and approved by the University of North Carolina at Chapel Hill Biomedical IRB (Study 03-0809).
References
- ARIC Investigators. 1987. Atherosclerosis Risk in Communities (ARIC) Operations Manual 5. Electrocardiography. Version 1.0. Chapel Hill, NC: ARIC Coordinating Center, School of Public Health, University of North Carolina. [Google Scholar]
- ARIC Investigators, 1989. The atherosclerosis risk in communities (ARIC) study: Design and objectives. The ARIC investigators. Am J Epidemiol 129:687–702, 10.1093/oxfordjournals.aje.a115184 [DOI] [PubMed] [Google Scholar]
- Arking DE, Pulit SL, Crotti L, van der Harst P, Munroe PB, Koopmann TT, et al. 2014. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet 46:826–836, PMID: 24952745, 10.1038/ng.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschard H, Lutz S, Maus B, Duell EJ, Fingerlin TE, Chatterjee N, et al. 2012. Challenges and opportunities in genome-wide environmental interaction (GWEI) studies. Hum Genet 131:1591–1613, PMID: 22760307, 10.1007/s00439-012-1192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslibekyan S, Claas SA, Arnett DK. 2013. To replicate or not to replicate: The case of pharmacogenetic studies establishing validity of pharmacogenomic findings: From replication to triangulation. Circ Cardiovasc Genet 6:409–412, 10.1161/CIRCGENETICS.112.000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsh GS, Copenhaver GP, Gibson G, Williams SM. 2012. Guidelines for genome-wide association studies. PLoS Genet 8:e1002812, PMID: 22792080, 10.1371/journal.pgen.1002812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. 2010. The NIH roadmap epigenomics mapping consortium. Nat Biotechnol 28:1045–1048, PMID: 20944595, 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brero A, Ramella R, Fitou A, Dati C, Alloatti G, Gallo MP, et al. . 2010. Neuregulin-1beta1 rapidly modulates nitric oxide synthesis and calcium handling in rat cardiomyocytes. Cardiovasc Res 88:443–452, PMID: 20634213, 10.1093/cvr/cvq238. [DOI] [PubMed] [Google Scholar]
- Browning BL, Browning SR, Ramella R, Fitou A, Dati C, Alloatti G, Gallo MP, et al. 2009. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84:210–223, PMID: 19200528, 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan K, Ackerman MJ, Crowson CS, Matteson EL, Gabriel SE. 2015. Population-based study of QT interval prolongation in patients with rheumatoid arthritis. Clin Exp Rheumatol 33:84–89, PMID: 25572282. [PMC free article] [PubMed] [Google Scholar]
- Dekker JM, Crow RS, Hannan PJ, Schouten EG, Folsom AR. 2004. Heart rate-corrected QT interval prolongation predicts risk of coronary heart disease in black and white middle-aged men and women: The ARIC study. J Am Coll Cardiol 43:565–571, PMID: 14975464, 10.1016/j.jacc.2003.09.040. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K. 1999. Genomic control for association studies. Biometrics 55:997–1004, 10.1111/j.0006-341X.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Dockery DW, Pope CA 3rd, Xu X, Spengler JD, Ware JH, Fay ME, et al. 1993. An association between air pollution and mortality in six U.S. cities. N Engl J Med 329:1753–1759, PMID: 8179653, 10.1056/NEJM199312093292401. [DOI] [PubMed] [Google Scholar]
- Dockery DW, Luttmann-Gibson H, Rich DQ, Link MS, Mittleman MA, Gold DR, et al. 2005. Association of air pollution with increased incidence of ventricular tachyarrhythmias recorded by implanted cardioverter defibrillators. Environ Health Perspect 113:670–674, PMID: 15929887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA (U.S. Food and Drug Administration). 2015. Guidance for industry: E14 clinical evaluation of QT/QTC interval prolongation and proarrhythmic potential for non-antiarrhythmic drugs. Center for Drug Evaluation and Research. Center for Biologics Evaluation and Research. [Google Scholar]
- García-Rivello H, Taranda J, Said M, Cabeza-Meckert P, Vila-Petroff M, Scaglione J, et al. 2005. Dilated cardiomyopathy in erb-b4-deficient ventricular muscle. Am J Physiol Heart Circ Physiol 289:H1153–H1160. [DOI] [PubMed] [Google Scholar]
- Gibert Y, Trengove M, Ward A. 2013. Zebrafish as a genetic model in pre-clinical drug testing and screening. Curr Med Chem 20:2458–2466, PMID: 23521675. [DOI] [PubMed] [Google Scholar]
- Goldberg RJ, Bengtson J, Chen ZY, Anderson KM, Locati E, Levy D. 1991. Duration of the QT interval and total and cardiovascular mortality in healthy persons (the Framingham Heart Study experience). Am J Cardiol 67:55–58, PMID: 1986505. [DOI] [PubMed] [Google Scholar]
- Gondalia R. 2016. Online material: Genome-wide association study of susceptibility to particulate matter-associated QT prolongation. https://qtgwaspm.web.unc.edu/EHP [accessed 6 March 2016]. [DOI] [PMC free article] [PubMed]
- Göring HH, Terwilliger JD, Blangero J. 2001. Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet 69:1357–1369, PMID: 11593451, 10.1086/324471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin JW, Hilbe JM. 2012. Generalized Estimating Equations, second edition: Chapman & Hall/CRC. [Google Scholar]
- Holliday KM, Avery CL, Poole C, McGraw K, Williams R, Liao D, et al. 2014. Estimating personal exposures from ambient air pollution measures: Using meta-analysis to assess measurement error. Epidemiology (Cambridge, Mass.) 25:35–43, 10.1097/EDE.0000000000000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. 2012. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44:955–959, PMID: 22820512, 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter CM, Mechanic LE, Chatterjee N, Kraft P, Gillanders EM. 2013. Gene-environment interactions in cancer epidemiology: A national cancer institute think tank report. Genet Epidemiol 37:643–657, PMID: 24123198, 10.1002/gepi.21756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, et al. 2009. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet 41:334–341, PMID: 19198609, 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M, Stewart RE, McCarty R. 1989. Habituation of sympathetic-adrenal medullary responses following exposure to chronic intermittent stress. Physiol Behav 45:255–261, PMID: 2756012. [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Haussler D, Kent WJ. 2012. The UCSC genome browser and associated tools. Briefings in Bioinformatics:bbs038. [DOI] [PMC free article] [PubMed]
- LaRocca TJ, Schwarzkopf M, Altman P, Zhang S, Gupta A, Gomes I, et al. 2010. β2-adrenergic receptor signaling in the cardiac myocyte is modulated by interactions with CXCR4. J Cardiovasc Pharmacol 56:548–559, PMID: 20729750, 10.1097/FJC.0b013e3181f713fe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. 2010. Mach: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 34:816–834, PMID: 21058334, 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D, Peuquet DJ, Duan Y, Whitsel EA, Dou J, Smith RL, et al. 2006. GIS approaches for the estimation of residential-level ambient PM concentrations. Environ Health Perspect 114:1374–1380, PMID: 16966091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D, Peuquet DJ, Lin H-M, Duan Y. 2007. National kriging exposure estimation: Liao et al. respond. Environ Health Perspect 115:A338, 10.1289/ehp.10205R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D, Shaffer ML, Rodriguez-Colon S, He F, Li X, Wolbrette DL, et al. 2010. Acute adverse effects of fine particulate air pollution on ventricular repolarization. Environ Health Perspect 118:1010–1015, PMID: 20363686, 10.1289/ehp.0901648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D, Whitsel EA, Duan Y, Lin H-M, Quibrera PM, Smith R, et al. 2009. Ambient particulate air pollution and ectopy–the environmental epidemiology of arrhythmogenesis in Women's Health Initiative Study, 1999–2004. J Toxicol Environ Health A 72:30–38, 10.1080/15287390802445483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberda EN, Cuevas AK, Gillespie PA, Grunig G, Qu Q, Chen LC. 2010. Exposure to inhaled nickel nanoparticles causes a reduction in number and function of bone marrow endothelial progenitor cells. Inhal Toxicol 22 Supp 2: 95–99, PMID: 20936915, 10.3109/08958378.2010.515269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungman PL, Berglind N, Holmgren C, Gadler F, Edvardsson N, Pershagen G, et al. 2008. Rapid effects of air pollution on ventricular arrhythmias. Eur Heart J 29:2894–2901, PMID: 19004842, 10.1093/eurheartj/ehn463. [DOI] [PubMed] [Google Scholar]
- McEwen BS. 2007. Physiology and neurobiology of stress and adaptation: Central role of the brain. Physiol Rev 87:873–904, PMID: 17615391, 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- Miller KA, Siscovick DS, Sheppard L, Shepherd K, Sullivan JH, Anderson GL, et al. 2007. Long-term exposure to air pollution and incidence of cardiovascular events in women. N Engl J Med 356:447–458, PMID: 17267905, 10.1056/NEJMoa054409. [DOI] [PubMed] [Google Scholar]
- Mordukhovich I, Kloog I, Coull B, Koutrakis P, Vokonas P, Schwartz J. 2016. Association between particulate air pollution and QT interval duration in an elderly cohort. Epidemiology (Cambridge, Mass.) 27:284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP. 2011. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol 35:809–822, PMID: 22125221, 10.1002/gepi.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru K, Strong A, Frank-Kamenetsky M, Lee LE, Ahfeldt T, Sachs KV, et al. 2010. From noncoding variant to phenotype via sort1 at the 1p13 cholesterol locus. Nature 466:714–719, PMID: 20686566, 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health. 1998. Design of the Women's Health Initiative clinical trial and observational study. The Women's Health Initiative Study Group. Control Clin Trial 19:61–109. [DOI] [PubMed] [Google Scholar]
- National Institutes of Health. 2010. Modification of PM-mediated arrhythmogenesis in populations. http://projectreporter.nih.gov/project_info_description.cfm?aid=7984809&icde=19283008 [accessed 6 March 2016].
- National Institutes of Health. National Heart, Lung, and Blood Institute (NHLBI). 2011. SNP Health Association Resource Project. https://www.nhlbi.nih.gov/resources/geneticsgenomics/programs/share.htm [accessed 6 March 2015].
- National Institutes of Health. National Human Genome Research Institute (NHGRI). 2013. Genomics and randomized trials network (GARNET) http://www.genome.gov/27541119 [accessed 6 March 2016]. [Google Scholar]
- Pan W. 2001. Akaike’s information criterion in generalized estimating equations. Biometrics 57:120–125, PMID: 11252586. [DOI] [PubMed] [Google Scholar]
- Pan W, Wall MM. 2002. Small‐sample adjustments in using the sandwich variance estimator in generalized estimating equations. Stat Med 21:1429–1441, PMID: 12185894, 10.1002/sim.1142. [DOI] [PubMed] [Google Scholar]
- Pe'er I, Yelensky R, Altshuler D, Daly MJ. 2008. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol 32:381–385, PMID: 18348202, 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909, PMID: 16862161, 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. 2010. Locuszoom: Regional visualization of genome-wide association scan results. Bioinformatics 26:2336–2337, PMID: 20634204, 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo RT, Sui J, Dhume A, Palomeque J, Blaxall BC, Diaz G, et al. 2006. CXCR4 modulates contractility in adult cardiac myocytes. J Mol Cell Cardiol 41:834–844, PMID: 17010372, 10.1016/j.yjmcc.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragvin A, Moro E, Fredman D, Navratilova P, Drivenes ∅, Engström PG, et al. 2010. Long-range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3. Proc Natl Acad Sci USA 107:775–780, PMID: 20080751, 10.1073/pnas.0911591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautaharju PM, Kooperberg C, Larson JC, LaCroix A. 2006a. Electrocardiographic predictors of incident congestive heart failure and all-cause mortality in postmenopausal women: The Women's Health Initiative. Circulation 113:481–489. [DOI] [PubMed] [Google Scholar]
- Rautaharju PM, Kooperberg C, Larson JC, LaCroix A. 2006b. Electrocardiographic abnormalities that predict coronary heart disease events and mortality in postmenopausal women: The Women's Health Initiative. Circulation 113:473–480. [DOI] [PubMed] [Google Scholar]
- Ritzler J, Sawhney R, van Kessei AG, Grzeschik K-H, Schinzel A, Berchtold M. 1992. The genes for the highly homologous Ca 2+-binding proteins oncomodulin and parvalbumin are not linked in the human genome. Genomics 12:567–572, 10.1016/0888-7543(92)90449-3. [DOI] [PubMed] [Google Scholar]
- Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, et al. 2010. Encode whole-genome data in the UCSC genome browser. Nucleic Acids Res 38:D620–D625, PMID: 19920125, 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al. 2007. Genomewide association analysis of coronary artery disease. N Engl J Med 357:443–453, PMID: 17634449, 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samet JM, Dominici F, Curriero FC, Coursac I, Zeger SL. 2000. Fine particulate air pollution and mortality in 20 U.S. cities, 1987–1994. N Engl J Med 343:1742–1749, PMID: 11114312, 10.1056/NEJM200012143432401. [DOI] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. 2012. The long-range interaction landscape of gene promoters. Nature 489:109–113, PMID: 22955621, 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite FE. 1946. An approximate distribution of estimates of variance components. Biometrics Bull: 110–114, 10.2307/3002019. [DOI] [PubMed] [Google Scholar]
- Schouten EG, Dekker JM, Meppelink P, Kok FJ, Vandenbroucke JP, Pool J. 1991. QT interval prolongation predicts cardiovascular mortality in an apparently healthy population. Circulation 84:1516–1523, 10.1161/01.CIR.84.4.1516. [DOI] [PubMed] [Google Scholar]
- Schroeder EB, Whitsel EA, Evans GW, Prineas RJ, Chambless LE, Heiss G. 2004. Repeatability of heart rate variability measures. J Electrocardiol 37:163–172, PMID: 15286929. [DOI] [PubMed] [Google Scholar]
- Shih RA, Griffin BA, Salkowski N, Jewell A, Eibner C, Bird CE, et al. 2011. Ambient particulate matter air pollution and venous thromboembolism in the women's health initiative hormone therapy trials. Environ Health Perspect 119:326–331, PMID: 21036692, 10.1289/ehp.1002256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker SA, Reboussin BA, Espeland MA, Rapp SR, McBee WL, Dailey M, et al. 1998. The Women's Health Initiative Memory Study (WHIMS): A trial of the effect of estrogen therapy in preventing and slowing the progression of dementia. Control Clin Trials 19:604–621, PMID: 9875839, https://www.ncbi.nlm.nih.gov/pubmed/9875839. [DOI] [PubMed] [Google Scholar]
- Siontis KC, Patsopoulos NA, Ioannidis JP. 2010. Replication of past candidate loci for common diseases and phenotypes in 100 genome-wide association studies. Eur J Hum Genet 18:832–837, PMID: 20234392, 10.1038/ejhg.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitlani CM, Rice KM, Lumley T, McKnight B, Cupples LA, Avery CL, et al. 2015. Generalized estimating equations for genome-wide association studies using longitudinal phenotype data. Stat Med 34:118–130, PMID: 25297442, 10.1002/sim.6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. 2006. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 38:209–213, PMID: 16415888, 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim K-H, Gamazon FR, Sakabe NJ, Gomez-Marin C, et al. 2014. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507:371–375, PMID: 24646999, 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NL, Felix JF, Morrison AC, Demissie S, Glazer NL, Loehr LR, et al. 2010. Association of genome-wide variation with the risk of incident heart failure in adults of European and African ancestry a prospective meta-analysis from the cohorts for heart and aging research in genomic epidemiology (charge) consortium. Circ Cardiovasc Genet 3(3):256–266, PMID: 20445134, 10.1161/CIRCGENETICS.109.895763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Balding DJ. 2009. Bayesian statistical methods for genetic association studies. Nat Rev Genet 10:681–690, PMID: 19763151, 10.1038/nrg2615. [DOI] [PubMed] [Google Scholar]
- Stevens J, Padilla S, Tennant A, Conolly R, DeMarini D, Gilmour I. et al. 2015. A simplified and rapid screening assay using zebrafish to assess cardiac effects of air pollution-derived particulate matter In: Society of Toxicology. San Diego, CA. [Google Scholar]
- Stone EA. 1983. Adaptation to stress and brain noradrenergic receptors. Neurosci Biobehav Rev 7:503–509, PMID: 6322067. [DOI] [PubMed] [Google Scholar]
- Thomas DC, Lewinger JP, Murcray CE, Gauderman WJ. 2012. Invited commentary: GE-whiz! Ratcheting gene-environment studies up to the whole genome and the whole exposome. Am J Epidemiol 175:203–207, PMID: 22199029, 10.1093/aje/kwr365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA (U.S. Environmental Protection Agency). 2016. Particulate matter (PM) standards - Table of Historical PM NAAQS. http://epa.gov/ttn/naaqs/standards/pm/s_pm_history.html> [accessed March 6 2016].
- Vaidean GD, Schroeder EB, Whitsel EA, Prineas RJ, Chambless LE, Perhac JS, et al. 2005. Short-term repeatability of electrocardiographic spatial T-wave axis and QT interval. J Electrocardiol 38:139–147, PMID: 15892024, 10.1016/j.jelectrocard.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Van Hee VC, Szpiro AA, Prineas R, Neyer J, Watson K, Siscovick D, et al. 2011. Association of long-term air pollution with ventricular conduction and repolarization abnormalities. Epidemiology (Cambridge) 22:773–780, PMID: 21918454, 10.1097/EDE.0b013e31823061a9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorman A, Sitlani CM. 2013. Boss: Boosted one-step statistics: Fast and accurate approximations for GLM, GEE and mixed models for use in GWAS. Part R package version 2.1. [Google Scholar]
- Wadugu B, Kühn B. 2012. The role of neuregulin/ErbB2/ErbB4 signaling in the heart with special focus on effects on cardiomyocyte proliferation. Am J Physiol Heart Circ Physiol 302:H2139–H2147, PMID: 22427524, 10.1152/ajpheart.00063.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M. 2012. Haploreg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40:D930–D934, PMID: 22064851, 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsel EA, Rose KM, Wood JL, Henley AC, Liao D, Heiss G. 2004. Accuracy and repeatability of commercial geocoding. Am J Epidemiol 160:1023–1029, PMID: 15522859, 10.1093/aje/kwh310. [DOI] [PubMed] [Google Scholar]
- Whitsel EA, Quibrera PM, Smith RL, Catellier DJ, Liao D, Henley AC, et al. 2006. Accuracy of commercial geocoding: Assessment and implications. Epidemiol Perspect Innov 3:8, PMID: 16857050, 10.1186/1742-5573-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsel EA, Quibrera PM, Christ SL, Liao D, Prineas RJ, Anderson GL, et al. 2009. Heart rate variability, ambient particulate matter air pollution, and glucose homeostasis: The environmental epidemiology of arrhythmogenesis in the Women's Health Initiative. Am J Epidemiol 169:693–703, PMID: 19208727, 10.1093/aje/kwn400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHI Study Group. 1994. WHI Manuals. Volume 2. Procedures. Section 13. ECG Procedures Seattle, WA: WHI Clinical Coordinating Center, Fred Hutchinson Cancer Research Center. [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. 2010. Metal: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26:2190–2191, 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojczynski MK, Li M, Bielak LF, Kerr KF, Reiner AP, Wong ND, et al. 2013. Genetics of coronary artery calcification among African Americans, a meta-analysis. BMC Med Genet 14:75, PMID: 23870195, 10.1186/1471-2350-14-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanosky JD, Paciorek CJ, Laden F, Hart JE, Puett RC, Liao D, et al. 2014. Spatio-temporal modeling of particulate air pollution in the conterminous United States using geographic and meteorological predictors. Environ Health 13:63, PMID: 25097007, 10.1186/1476-069X-13-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Post WS, Blasco-Colmenares E, Dalal D, Tomaselli GF, Guallar E. 2011. Electrocardiographic QT interval and mortality: A meta-analysis. Epidemiology 22:660–670, PMID: 21709561, 10.1097/EDE.0b013e318225768b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z-M, Whitsel EA, Quibrera PM, Smith RL, Liao D, Anderson GL, et al. 2009. Ambient fine particulate matter exposure and myocardial ischemia in the environmental epidemiology of arrhythmogenesis in the Women's Health Initiative (EEAWHI) Study. Environ Health Perspect 117:751–756, PMID: 19479017, 10.1289/ehp.0800046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Maricque B, Xie M, Li D, Sundaram V, Martin EA, et al. 2011. The human epigenome browser at Washington University. Nat Methods 8:989–990, PMID: 22127213, 10.1038/nmeth.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.