Abstract
BRCA2 loss-of-heterozygosity (LOH) is frequently observed in BRCA2-mutated tumors, but its biallelic loss causes embryonic lethality in mice and inhibits proliferation of normal somatic cells. Therefore, it remains unclear how loss of BRCA2 contributes to tumorigenesis. One possibility is that mutation in potential genetic interactors of BRCA2, such as TRP53, is required for cell survival/proliferation in the absence of BRCA2. In this study, using an insertional mutagenesis screen in mouse embryonic stem cells (mESC), we have identified GIPC3 (GAIP-interacting protein C-terminus 3) as a BRCA2 genetic interactor that contributes to survival of Brca2-null mESC. GIPC3 does not compensate for BRCA2 loss in the repair of double-strand breaks. Mass-spectrometric analysis resulted in the identification of G-protein signaling transducers, APPL1 and APPL2, as potential GIPC3-binding proteins. A mutant GIPC3 (His155Ala) that does not bind to APPL1/2 failed to rescue the lethality of Brca2-null mESC, suggesting that the cell viability by GIPC3 is mediated via APPL1/2. Finally, the physiological significance of GIPC3 as a genetic interactor of BRCA2 is supported by the observation that Brca2-null embryos with Gipc3 overexpression are developmentally more advanced than their control littermates. Taken together, we have uncovered a novel role for GIPC3 as a BRCA2 genetic interactor.
Keywords: BRCA2, GIPC3, genetic interactors, insertional mutagenesis, mouse ES cells, breast cancer
BREAST CANCER2 gene (BRCA2) encodes a 3418-amino-acid protein that functions as a tumor suppressor by maintaining genome integrity (Venkitaraman 2009). It is essential for homology-directed DNA repair (HDR) because of its critical role in recruitment of DNA recombinase, RAD51, to the site of DNA double-strand breaks (DSB). BRCA2 also plays an important role in protecting stalled replication forks (Sharan et al. 1997; Moynahan et al. 2001; Venkitaraman 2009; Schlacher et al. 2011). The detailed mechanism of how loss of BRCA2 results in tumorigenesis, and why it causes predominantly breast tumors remains poorly understood (Collins et al. 1995; Gudmundsson et al. 1995; Kass et al. 2016; Schneider et al. 2017). However, it is evident that BRCA2-loss is not sufficient to cause cancer. It requires the cooperation of genetic interactors that can contribute to the viability of BRCA2-deficient cells that are generated by loss of heterozygosity (LOH) in mutation carriers. Loss of Trp53 is known to delay the lethality of Brca2 null mouse embryos and conditional loss of Trp53 along with Brca2 loss in mouse mammary epithelial cells resulted in mouse mammary tumorigenesis (Ludwig et al. 1997; Jonkers et al. 2001). Importantly, TRP53 is also frequently mutated in human BRCA2-mutated breast tumors (Gretarsdottir et al. 1998). Taken together, these findings demonstrate the role of TRP53 in BRCA2-loss induced tumorigenesis. Identification of other potential BRCA2 genetic interactors will improve our understanding of BRCA2-mediated tumorigenesis.
We have undertaken a genetic screen in mouse embryonic stem cells (mESC) to identify new BRCA2 genetic interactors. The screen is based on our observation that BRCA2 loss is lethal in mESC (Sharan et al. 1997; Kuznetsov et al. 2008). The aim of the screen is to identify genes that can rescue the lethality of Brca2ko/ko mESC. We have previously reported the generation of Brca2cko/ko mESC (PL2F7) carrying a functionally null and conditional allele of Brca2. When the conditional allele is deleted in PL2F7 cells by Cre recombinase, no viable Brca2ko/ko cells are obtained (Kuznetsov et al. 2008). Using the PL2F7 cells, we have identified PARP1 and PTIP as BRCA2 genetic interactors. Knockdown of Parp1 and Ptip in PL2F7 cells resulted in viable Brca2ko/ko cells (Chaudhuri et al. 2016; Ding et al. 2016). Furthermore, Parp1 heterozygosity significantly increased the tumor incidence in K14-Cre;Brca2cko/cko mice (Ding et al. 2016).
In this study, we describe the use of a retrovirus-based insertional mutagenesis screen to identify genes that can rescue the lethality of Brca2ko/ko mESC. We transduced PL2F7 cells with Murine Stem Cell Virus expressing CRE recombinase (MSCV-Cre). CRE induces cell death due to deletion of the conditional allele of Brca2. Viral integration, on the other hand, can disrupt genes or the viral long terminal repeat can upregulate expression of flanking genes (Du et al. 2005). Cloning of the MSCV integration sites allows identification of the region containing the candidate genes. Potential genetic interactors can be identified and validated by examining their expression in the rescued Brca2ko/ko mESC and testing their ability to rescue lethality of Brca2ko/ko mESC. By using this approach, we identified GAIP interacting protein C terminus 3 (Gipc3), which encodes a cytosolic adaptor protein involved in G-protein signaling pathway (Katoh 2013), as a potential genetic interactor of Brca2. GIPC3 is a member of the GIPC (GAIP-interacting protein C-terminus) gene family, which is characterized by a single, conserved PDZ domain and GIPC homology (GH1 and GH2) domains (Katoh 2013). Loss-of-function mutations in GIPC3 have been found in families with audiogenic seizures and sensorineural hearing loss (Charizopoulou et al. 2011; Rehman et al. 2011). We have identified APPL1 and APPL2 as key GIPC3 interacting proteins that bind to its PDZ domain and are essential for GIPC3-mediated cell viability. Our findings support a role for GIPC3 as a genetic interactor of BRCA2.
Materials and Methods
Cell culture
All mESC were cultured on mitotically inactive SNL feeder cells in M15 media, which is Knockout DMEM media (Life Technologies) supplemented with 15% fetal bovine serum (Life Technologies), 0.00072% β-mercaptoethanol, 100 unit/ml penicillin, 100 μg/ml streptomycin, and 0.292 mg/ml l-glutamine at 37°, 5% CO2. PL2F7 cells were generated from AB2.2 mouse embryonic stem cell line by genetically deleting one copy of Brca2 allele, and putting two loxP sites to flox the other copy of Brca2 allele (Kuznetsov et al. 2008).
MSCV-based insertion mutagenesis in mESC
To perform insertional mutagenesis, we used MSCV to introduce Cre into the cells. This approach allowed us to perform insertional mutagenesis as well as delete the conditional allele in a single step. Stable virus-producing cells were generated by transfecting packaging cell line GP+E86 (ATCC, CRL 9642) with the plasmid. Transfected cells were selected with appropriate antibiotics (Hygromycin or G418). ES cells were transduced with the retrovirus in a 10 cm dish by coculturing PL2F7 mESC (1–2 × 106) with packaging cells (3 × 106) in the presence of 8 μg/ml polybrene (Santa Cruz Biotech) for 48 hr. Packaging cells were mitotically inactivated by mitomycin C (MMC) treatment (10 μg/ml for 2 hr). The transduced cells were washed with PBS, and then plated at lower density and selected for either in HAT medium or for antibiotic resistance. Individual colonies were picked into 96-well plates and further analyzed either by Southern hybridization as described previously (Kuznetsov et al. 2008). To identify the viral insertion site in rescued Brca2ko/ko mESC, we used the Splinkerette PCR-based method (Li et al. 1999). We extracted genomic DNA from Brca2ko/ko cells, digested with EcoRI, and ligated to the splinkerette oligos linker overnight. PCR was performed on the ligation reaction using gene-specific (Cre or Hygro) primers and splinkerette-specific primers followed by a nested PCR performed using primers recognizing the long terminal repeat of MSCV and the splinkerette linker. PCR products were loaded on 1% agarose gels. PCR bands were excised and purified using MiniElute columns (Qiagen, Valencia, CA), and sequenced directly using the Big Dye Cycle Sequencing kit (PerkinElmer, Shelton, CT) and an ABI Model 373A DNA Sequencer (Applied Biosystems, Foster City, CA).
Generation of GIPC3 stable expression mESC clones
Stable expression clones of GIPC3 were generated by murine stem cell virus (MSCV) transduction in PL2F7 cells. Empty MSCV vector (neor) was purchased from Clontech. Mouse Gipc3 gene fused with HA tag in the C-terminus was cloned into MSCV vector by EcoRI and BglII sites. To generate stable expression clones, MSCV-GIPC3 plasmid was transfected into packaging cells, GP+E86 (a mouse fibroblast cell line) by Lipofectamine 2000 (Life Technologies). After 48 hr of transfection, cells were mitotically inactivated by mitomycin C treatment (10 μg/ml, 2 hr) and mixed with SNL feeder cells at the same ratio to make mixed feeder plates. PL2F7 cells (1 × 106 cells) were plated on the mixed feeder plates with 10 μg/ml polybrene in the medium to increase transduction efficiency. After 48 hr of plating, cells were trypsinized and 10,000 cells were replated to SNL feeder and G418 selection (0.18 μg/ml) was started 24 hr after replating. G418 was replaced with M15 medium after 5 days of selection and mESC colonies were picked 3–5 days after G418 withdrawal. Stable expression clones were identified by immunoblotting using antibody against HA-tag.
Deletion of Brca2 cko allele and selection of Brca2ko/ko mESC
PGK-Cre plasmid DNA (20 μg) was electroporated into 1 × 107 mESC suspended in 0.9 ml PBS by Gene Pulser (Bio-Rad) at 230 V, 500 μF. HAT selection was started 36 hr after electroporation and lasted for 5 days, followed by selection in HT medium for 2 days and then normal M15 medium until colonies became visible. Colonies were picked into 96-well plates. For extracting genomic DNA, colonies were lysed in 50 μl mESC buffer (10 mM Tris-HCl, pH 7.4, 10 mM EDTA, 10 mM NaCl, 5 mg/ml sodium lauroyl sarcosinate, 1 mg/ml proteinase K) at 55° overnight, and DNA was precipitated by 100 μl 75 mM NaCl in absolute ethanol. Genomic DNA was rinsed by 70% ethanol and digested by EcoRV at 37° overnight for Southern blot.
Southern blot
EcoRV-digested DNA was electrophoresed on a 1% agarose gel in 1×TBE (0.1 M Tris, 0.1 M Boric acid, 2 mM EDTA, pH 8.0) and transferred to nylon membrane. DNA probe for distinguishing conditional Brca2 allele (cko, 4.8 kb) and Brca2 knockout allele (ko, 2.2 kb) was labeled by [α-32P]-dCTP by Prime-It II Random Primer Labeling Kit (Agilent Technologies) and hybridized with Hybond-N+ nylon membrane (GE Healthcare) at 65° overnight. Membrane was washed twice with SSCP buffer containing 0.1% SDS in and exposed to a phosphor image screen overnight, and subsequently developed in a Typhoon image scanner.
Western blot, immunofluorescence, and immunoprecipitation
The following antibodies were used: anti-HA tag (rat; Roche), anti-APPL1 (rabbit; Cell Signaling Technology), anti-APPL2 (rabbit; Santa Cruz), anti-ACTIN (goat; Santa Cruz), anti-RAD51 (rabbit; Calbiochem), and anti-γH2AX (mouse; Millipore). For Western blot, cells were lysed in SDS lysis buffer (2% SDS, 10% glycerol, 0.1 M dithiothreitol, and 0.2 M Tris-HCl, pH 6.8), subjected to SDS-PAGE gel electrophoresis, and subsequently transferred to nitrocellulose membrane. Blots were incubated with the indicated primary antibodies at 4° overnight, washed by PBST, and probed with corresponding horseradish peroxidase -conjugated secondary antibodies at room temperature for 2 hr and subjected to ECL (Amersham). For immunofluorescence, cells were fixed with 4% paraformaldehyde, permeabilized by 0.25% Triton X-100 in PBS, and blocked with 3% bovine serum albumin in PBS. Cells were incubated with the indicated primary antibodies at 4° overnight. After washing four times with PBST, cells were incubated with AlexaFluor 488 conjugated anti-rabbit IgG antibody and AlexaFluor 568 conjugated anti-mouse IgG antibody (Life Technologies) at room temperature for 2 hr. Nucleus was counterstained by DAPI. Images were taken on Zeiss LSM 510 confocal microscope.
For immunoprecipitation, cells were lysed in IP buffer (30 mM Tris-HCl, pH 8.0, 75 mM NaCl, 10% glycerol, 0.5% TritonX-100) containing protease inhibitors (Complete Mini protease inhibitor cocktail tablets; Roche) at 4° for 15 min, and subjected to centrifugation at 15,000 × g for 15 min at 4°. Supernatant was transferred and added 30 μl beads (Anti-Glu-Glu epitope tag affinity matrix; BioLegend) and then incubated on a rocker at 4° for 4 hr. Beads were rinse four times by IP buffer at 800 × g for 1 min each time. Proteins were dissociated from the beads by SDS lysis buffer and boiled at 95° for 10 min.
Karyotyping of mESC
Untreated mESC were arrested at metaphase by incubation with Colcemid (KaryoMax Colcemid Solution; Invitrogen, Carlsbad, CA) (10 μg/ml), 3 hr prior to harvest. Cells were collected and treated with hypotonic solution (KCl, 0.075 M) for 15 min at 37° and fixed with methanol: acetic acid 3:1. Slides were prepared and chromosomal aberrations were analyzed.
Cell viability assay
Cells were seeded at 10,000 cells per well in 96-well gelatinized plates. The drugs indicated were added 24 hr after seeding, and cell viability was measured by XTT assay after 72 hr of drug treatment. For XTT assay, cells were rinsed by PBS and incubated in DMEM medium w/o phenol red (Life Technologies), 1 mg/ml 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) and 2 mM phenazine methosulfate (PMS) at 37° for 30 min. Plates were read in iMark Microplate reader (Bio-Rad).
Mass spectrometry
The protein complex that was pulled down by immunoprecipitation was resolved on a 4–12% Bis-Tris gradient gel (Thermo Fisher Scientific), and stained with SimplyBlue SafeStain (Thermo Fisher Scientific) as per the manufacturer’s instructions. The gel bands were excised and subjected to tryptic digestion as described previously (Ory et al. 2003; Zofall et al. 2009). Digested peptide samples were extracted, desalted and resuspended in 0.1% TFA. The samples were analyzed by an Easy-nLC 1000 nanoHPLC system with a C18 Nano Trap Column, and an C18 Nano analytical column connected with a stainless steel emitter on a Nanospray Flex Ion Source, coupled online with a LTQ Velos Pro mass spectrometer (Thermo Scientific, San Jose, CA) Acquired MS/MS spectra were searched against Uniprot human protein database, using SEQUEST interfaced with BioWorks 3.3 (Thermo Scientific). The precursor ion tolerance was set at 1.4 Da, and the fragment ions tolerance was set at 0.5 Da. Methionine oxidation was set as dynamic modification and up to two miscleavages for fully tryptic peptide was allowed during the database search. The statistical cutoff threshold was set as Xcorr ≥2.0 for [M+H]1+, ≥2.5 for [M+2H]2+, and ≥3.0 for [M+3H]3+ and delta correlation (ΔCn) ≥0.1.
Generation of Gipc3 transgenic mice
Mouse Gipc3 cDNA fused with HA tag in the C-terminus was cloned into the pCCALL2 vector (a kind gift from Dr. Corrinne Lobe) by the BglII and XhoI restriction sites. pCCALL2 has the chicken β-actin promoter. This promoter and Gipc3 cDNA were separated by a floxed LacZ cassette to achieve conditional induction. The pCCALL2-Gipc3 construct was microinjected into fertilized eggs of C57/BL6 mice. Transgenic mice were determined by PCR using primers for LacZ and Gfp. Gipc3 transgenic mice were crossed with Brca2ko/+ mice (Sharan et al. 1997) to obtain Gipc3flLacZ; Brca2ko/+ mice. Mice with β-actin driven Cre were crossed with Brca2ko/+ mice to obtain β-actin Cre; Brca2ko/+ mice. We crossed Gipc3flLacZ; Brca2ko/+ mice with β-actin Cre; Brca2ko/+ to induce GIPC3 expression in the mice. All animal use is in accordance with the protocol approved by Animal Care and Use Committee of National Cancer Institute (NCI)-Frederick.
Genotyping PCR primers
Gfp:
Forward: 5′-AGCTGACCCTGAAGTTCATCTG-3′.
Reverse: 5′-GACGTTGTGGCTGTTGTAGTTG-3′.
Product size: 329 bp.
Brca2+ allele:
Forward: 5′-GCAAAAGTAGGACCAAGAGG-3′.
Reverse: 5′-TCACCTTTATGAATATAAACTG-3′.
Product size: ∼300 bp.
Brca2ko allele:
Forward: 5′-GTGAATCTTTGTCAGCAGTTCCC-3′.
Reverse: 5′-CCCACTAGCTGTATGAAAAC-3′.
Product size: ∼340 bp.
Generation of Gipc3 mutants
Mouse Gipc3 cDNA fused with pyo tag in the N-terminus and HA-tag in the C-terminus was cloned into the pcDNA3.1 (+) vector by EcoRI and XhoI sites. G115R, R123A, and H155A mutants were generated by using QuikChange Site-Directed Mutagenesis kit.
Embryo dissection and laser capture microdissection (LCM)
Timed pregnancy in mice was set up, and embryos were collected at E8.5. Embryos were dissected as per an established protocol (Nagy 2003). Dissected embryos were lysed in 50 μl mESC lysis buffer at 55° overnight. Genomic DNA was precipitated using 3 M sodium acetate (1/10 vol) and 2 vol of ethanol. Precipitated DNA was rinsed with 70% ethanol, resuspended in 1 × TE buffer and used for genotyping PCR.
For LCM, whole embryos were fixed in 10% neutral buffered formalin solution (Sigma). Embryos were embedded in Paraffin, and serially sectioned at 5 μm. The section adjacent to H&E was mounted on LCM slide (MMI CellCut Plus from Molecular Machines & Industries) for embryo collection. LCM workflow, LCM slide preparation, target dissection, and collection were carried out per previous technical book chapters (Golubeva and Rogers 2009; Golubeva et al. 2013). Collected embryos were incubated in 25 µl DNA lysis buffer (Arcturus PicoPure DNA extraction Kit; Life Technologies) for 48 hr at 55°. Genomic DNA was precipitated by adding 1 μl glycogen (Sigma), 3 M sodium acetate (1/10 vol), and 2 vol of ethanol. DNA was precipitated at −20° for at least 1.5 hr. Precipitated DNA was rinsed with 70% ethanol, resuspended in 1 × TE buffer and used for genotyping PCR.
Statistics
Statistics was performed in Prism 7 software. All error bars represent SD. P < 0.05 was considered statistically significant.
Data availability
All data necessary for the conclusions from the current study are represented in the article. Mouse strains are available on request.
Results
GIPC3 overexpression rescues BRCA2-loss induced lethality in mESC
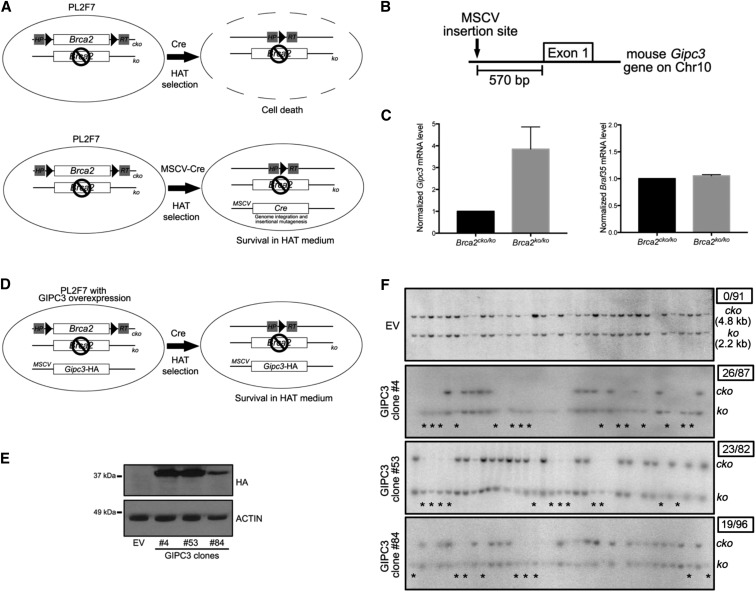
To identify genetic interactors of BRCA2, we performed a mutagenesis screen by transducing previously described PL2F7 mESC carrying a conditional (cko) and a functionally null (ko) allele of Brca2 with MSCV-Cre (Kuznetsov et al. 2008). We selected the recombinant clones for the generation of a functional HPRT minigene after Cre-mediated deletion of the conditional allele of Brca2 (Figure 1A). The HAT-resistant clones were genotyped by Southern blot to confirm the deletion of the cko allele. Cloning and sequence analysis of MSCV-Cre insertion site in viable Brca2ko/ko mESC identified Gipc3 as one of the genes flanking the insertion site on mouse chromosome 10 (Figure 1B). Quantitative PCR revealed a threefold to fourfold mRNA increase in the rescued clone, but this increase was not observed in another nearby gene Braf35 (Figure 1C). To determine whether GIPC3 overexpression can indeed rescue BRCA2-loss induced lethality, we generated PL2F7 cells stably expressing mouse GIPC3 (HA-tagged at the C-terminus). To avoid position effect, we used multiple independently generated mESC clones with GIPC3 overexpression (Figure 1, D and E). To test the rescue of Brca2-null cell lethality, we deleted the cko allele. No Brca2ko/ko clones were obtained from cells transduced with the empty vector (EV) (0/91). However, all the three GIPC3 overexpressing clones (#4, #53, and #84) resulted in several Brca2ko/ko clones (26/87, 23/82, and 19/96, respectively) (Figure 1F). These results demonstrated that GIPC3 overexpression can indeed rescue BRCA2-loss induced cell lethality.
Figure 1.
GIPC3 overexpression rescues BRCA2 loss-induced lethality in mESC (A) Schematic diagram of MSCV-Cre mediated insertional mutagenesis in PL2F7 cells. (B) Sketch of MSCV-Cre insertion site near the Gipc3 gene on mouse chromosome 10. (C) Quantitative PCR showing Gipc3 and Braf35 mRNA levels in rescued mESC (Brca2ko/ko) compared to PL2F7 cells (Brca2cko/ko). (D) Schematic diagram of mESC cell model used in the rescue experiment. (E) Western blot showing expression of GIPC3 (HA tagged) probed by HA antibody in GIPC3 stable overexpression mESC clones. Three independent clones (#4, #53, and #84) along with control clone (EV, empty vector) were examined. Actin serves as loading control. (F) Southern blot showing the rescue of Brca2ko/ko mESC lethality in GIPC3 stable overexpression clones (#4, #53, and #84). Asterisks point the rescued Brca2ko/ko clones. Numbers in the upper right boxes indicate rescued clone number over total examined clone number.
Brca2ko/ko cells rescued by GIPC3 overexpression are defective in DNA repair
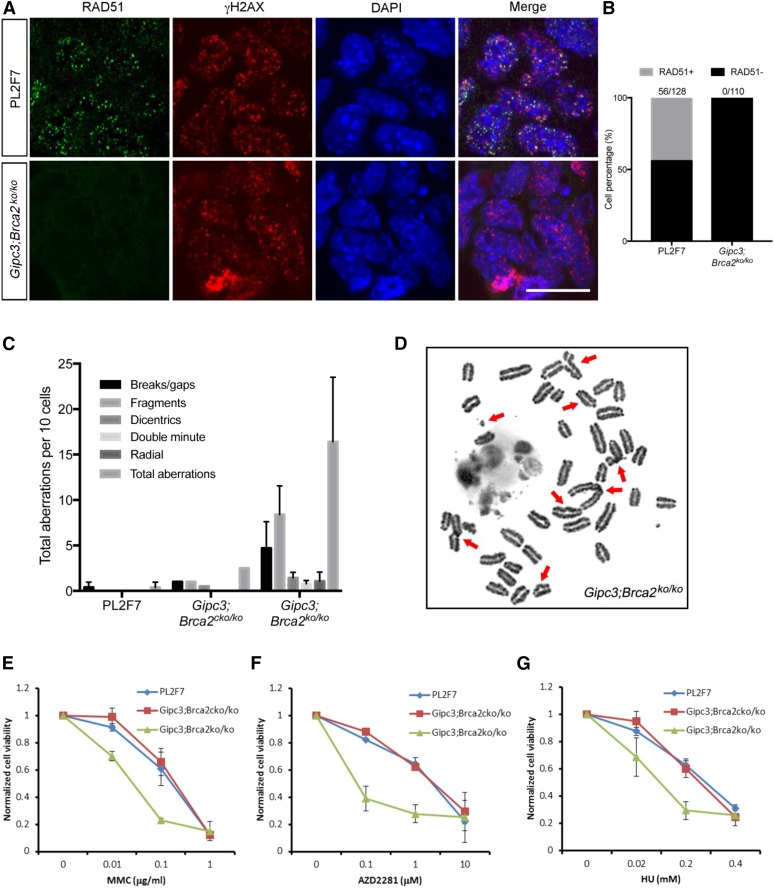
We first assessed whether GIPC3 overexpression contributed to viability of Brca2ko/ko mESC by functionally compensating for BRCA2 loss. We examined the HDR ability of rescued cells. Also, we tested their sensitivity to DNA damaging agents and assessed the genomic integrity of the cells. HDR ability was assayed by ionizing radiation-induced foci (IRIF) formation of RAD51. We exposed cells to 10 Gy of ionizing irradiation and stained with antibodies against RAD51 and DNA damage marker, γH2AX, 4 hr after irradiation. In PL2F7 cells, RAD51 IRIF were visible and colocalized with γH2AX foci. In Gipc3;Brca2ko/ko cells, although γH2AX foci were present, RAD51 IRIF was completely absent (Figure 2, A and B), indicating that GIPC3 overexpression did not restore HDR the Brca2 null cells.
Figure 2.
Brca2ko/ko mESC rescued by GIPC3 overexpression are HDR defective (A) Immunofluorescence images showing RAD51 foci formation 4 hr after 10 Gy IR in the indicated cells. γH2AX serves as DNA break marker. DAPI indicates the nucleus. Bar, 20 μm. (B) Quantification of the RAD51 foci immunofluorescence as shown in (A). Numbers on each column indicate number of cells having RAD51 foci over total number of counted cells. (C) Quantification of chromosomal aberrations in the indicated cells. (D) Representative mitotic metaphase chromosome spread image of the indicated cells as shown in (C). (E–G) XTT assay results showing the normalized cell viability of indicated cells under different drug treatment for 72 hr.
Since HDR ensures error-free repair of DSBs, we tested whether the Gipc3;Brca2ko/ko cells exhibited increased genomic instability. As expected, very few untreated PL2F7 cells and Gipc3;Brca2cko/ko cells had any chromosomal aberrations (Figure 2C). In contrast, all Gipc3;Brca2ko/ko mESC (n = 20) exhibited multiple chromosomal aberrations as shown by an increase in the number of spontaneous breaks/gaps, fragments, dicentrics and radial structure (Figure 2, C and D). These results suggest that GIPC3 overexpression did not suppress the genomic instability in the viable Brca2ko/ko mESC.
We next tested the sensitivity of Gipc3;Brca2ko/ko cells to DNA damaging agents including mitomycin C (MMC, DNA interstrand crosslinking agent), AZD2281 (olaparib, PARP inhibitor), and hydroxyurea (HU, replicative stress inducer). We treated PL2F7 cells, Gipc3;Brca2cko/ko cells, and Gipc3;Brca2ko/ko cells with different doses of MMC, AZD2281, and HU. After 72 hr of treatment, we measured cell viability by XTT assay. The sensitivity of Gipc3;Brca2cko/ko cells was comparable to that of PL2F7 cells, indicating GIPC3 overexpression per se does not affect drug sensitivity. However, Gipc3;Brca2ko/ko cells were hypersensitive to all these DNA damaging agents (Figure 2, E–G), indicating their compromised DNA repair ability, which was consistent with their defect in HDR. We conclude from these observations that GIPC3 does not compensate for loss of BRCA2 function and the rescued cells exhibit genomic instability, characteristic of BRCA2-deficient cells.
Interaction with APPL1 and APPL2 contributes to GIPC3 rescuing effect
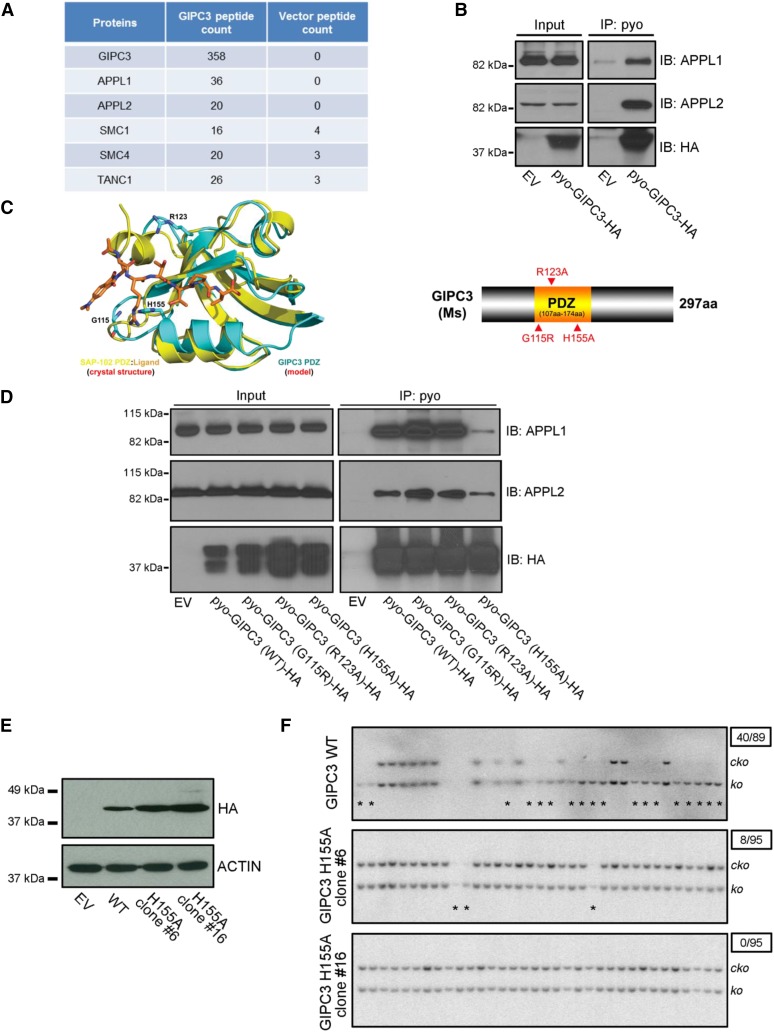
It was puzzling how Gipc3;Brca2ko/ko mESC were able to overcome the cell cycle arrest or apoptosis induced by the presence of unrepaired DSB. Since GIPC3 has a protein interacting PDZ domain, we hypothesized that it may serve as a scaffold protein in the cytoplasm. Therefore, we performed immunoprecipitation-coupled mass spectrometry (IP-MS) analysis to identify GIPC3 interacting protein(s) that can provide clues to the downstream signaling pathways that contribute to cell viability. We generated a construct to express mouse GIPC3 fused with a pyo tag (also known as Glu-Glu tag) at the N-terminus, and a HA tag at the C-terminus (pyo-GIPC3-HA). We transfected pyo-GIPC3-HA or empty vector (EV) in HEK293T cells and performed immunoprecipitation using an antibody against the pyo tag. The pulled-down protein complex was then subjected to MS analysis. The results showed that, after GIPC3 immunoprecipitation, other than GIPC3 itself, APPL1 and APPL2 peptides were the two most abundant (36 and 20 peptides, respectively) in pyo-GIPC3-HA transfected sample (Figure 3A). EV transfected samples did not show the presence of GIPC3, APPL1, or APPL2 peptides. These are multifunctional adaptor proteins that bind to membrane receptors, signaling proteins, and nuclear factors, and are involved in multiple signaling pathways (Deepa and Dong 2009). Interestingly, GIPC1, another member of the GIPC family is known to interact with APPL1 and APPL2 (Lin et al. 2006; Varsano et al. 2006). To validate the interaction between GIPC3 and APPL1/APPL2, we performed IP in HEK293T cells. As shown in Figure 3B, all the pyo-GIPC3-HA, APPL1, and APPL2 were dramatically enriched in the pyo-GIPC3-HA transfected cells compared to EV, suggesting that GIPC3 can indeed interact with APPL1 and APPL2.
Figure 3.
APPL1/2 contributes to the rescue effect of GIPC3 overexpression (A) List of top candidates of GIPC3 interaction proteins as revealed by IP-MS. (B) Western blot of immunoprecipitation in HEK293T cells showing interaction of GIPC3 with APPL1 and APPL2. GIPC3 with Pyo and HA tag (pyo-GIPC3-HA) were ectopically expressed in HEK293T cells. EV, empty vector. (C) Left, schematic illustration showing the superposition of the model of the GIPC3 PDZ domain with the crystal structure of the SAP-102 PDZ domain in complex with a fluorogenic peptide-based ligand (SAP-102:Ligand, PDB entry 3JXT). The polypeptide chains are shown as ribbon diagrams with helices as spirals, strands as arrows, and loops as tubes. The ligand and side chains are shown as stick models. Color schemes are shown and the three predicted residues are labeled. Right, schematic diagram of domain structure of mouse GIPC3 protein and point mutations in the PDZ domain that were generated. (D) Western blot of immunoprecipitation in HEK293T cells showing differential interaction of GIPC3 wildtype (WT) and mutated GIPC3 (G115R, R123A, and H155A) with APPL1 and APPL2. GIPC3 WT and mutants with Pyo and HA tag were ectopically expressed in HEK293T cells. (E) Western blot showing expression of HA tagged GIPC3 in GIPC3 WT and H155A (#6 and #16) stable expressing mESC clones. (F) Southern blot showing differential rescue efficiency of Brca2ko/ko mESC lethality in GIPC3 WT and H155A stable expressing mESC clones (#6 and #16). Asterisks point the rescued Brca2ko/ko clones. Numbers in the upper right boxes indicate rescued clone number over total examined clone number.
Does the interaction between GIPC3 with APPL1 and APPL2 contribute to the rescue of Brca2ko/ko mESC lethality? To assess this, using comparative analysis of a homologous model of the GIPC3 PDZ domain, which was built online at the Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2) (Kelley and Sternberg 2009) and the crystal structure of the SAP-102 PDZ domain in complex with a fluorogenic peptide-based ligand (Sainlos et al. 2013), we predicted Gly115, Arg123, and His155 residues in the PDZ domain of GIPC3 to be critical for protein–protein interaction (Figure 3C). We mutated each residue individually (G115R, R123A, and H155A) in the pyo-GIPC3-HA construct, and performed IP to examine whether these mutations disrupted the interaction with APPL1 and APPL2. We transfected wild-type (WT) or mutant pyo-GIPC3-HA constructs in HEK293T cells and immunoprecipitated using pyo tag antibody. The immunoprecipitation efficiency was comparable between WT and all the mutants based on Western analysis using HA tag antibody (Figure 3D). However, the H155A mutant exhibited reduced binding to APPL1 and APPL2 compared to WT (Figure 3D). In contrast, G115R, R123A had no effect on the interaction between GIPC3 and APPL1 as well as APPL2.
We generated PL2F7 cells that stably overexpressed GIPC3 H155A mutant, and examined its ability to rescue the lethality of Brca2-null cells. We used WT GIPC3-expressing cells as a positive control (Figure 3E). As shown in Figure 3F, WT GIPC3-overexpressing cells resulted in 44.9% Brca2ko/ko cells (40/89), whereas two GIPC3 H155A-mutant-overexpressing clones (#6 and #16) showed much reduced rescue efficiency, with only 8.4% (8/95) and 0% (0/95) Brca2ko/ko cells, respectively. Based on these results, we concluded that the interaction with APPL1 or APPL2 is critical for GIPC3 overexpression-mediated rescue of Brca2ko/ko mESC.
Gipc3 transgene partially rescued Brca2ko/ko mouse embryo lethality
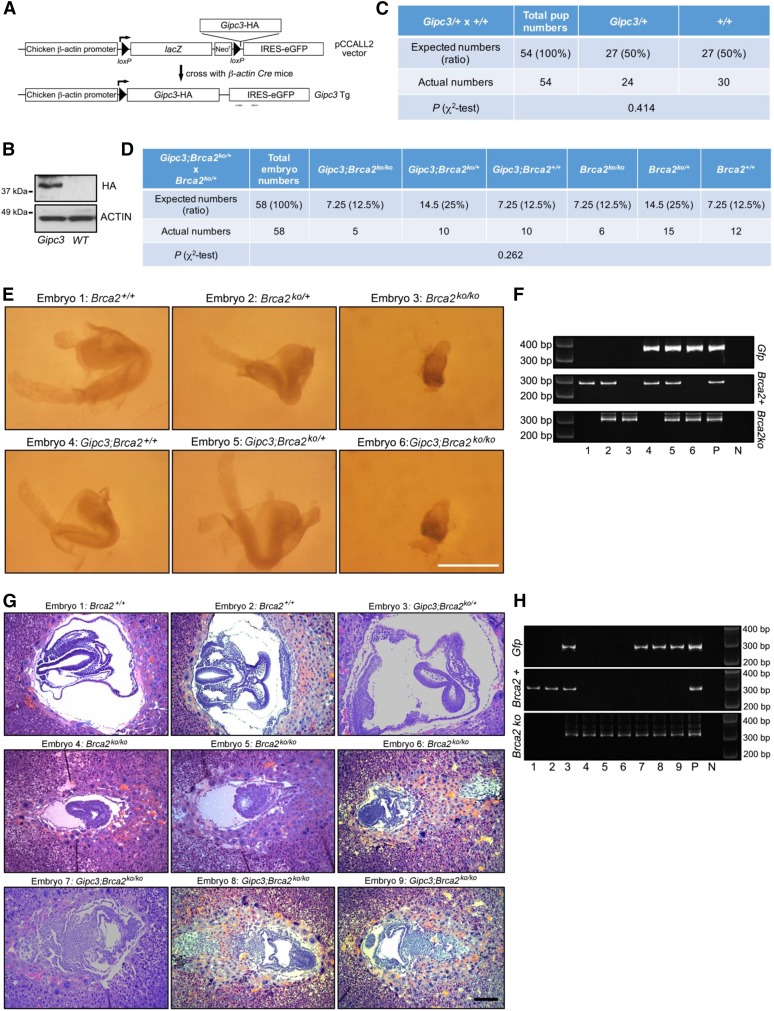
To examine the physiological significance of the genetic interaction between Gipc3 and Brca2, we generated Gipc3 transgenic mice. We cloned mouse Gipc3 cDNA with an HA tag at the C-terminus under the control of chicken β-actin promoter with a loxP-LacZ-loxP cassette (Figure 4A). We used the construct to generate transgenic mice. Transgenic lines were crossed with β-actin-promoter-driven Cre transgenic mice to delete the floxed lacZ cassette, and to activate Gipc3 transgene expression ubiquitously (Figure 4B). Gipc3 transgenic mice were phenotypically similar to their WT littermates, and were born at Mendelian ratio (Figure 4C). We then crossed Gipc3 transgenic mice with Brca2ko/+ mice, and set up mating to generate Gipc3;Brca2ko/ko. We failed to obtain viable Gipc3;Brca2ko/ko pups. To examine the phenotype of Gipc3;Brca2ko/ko embryos, we set up timed pregnancy by crossing Gipc3;Brca2ko/+ with Brca2ko/+ mice. Embryos were dissected at embryonic day 8.5 (E8.5), because Brca2ko/ko embryos are known to die around E8.5 (Ludwig et al. 1997; Sharan et al. 1997). We obtained comparable numbers of Gipc3;Brca2ko/ko and Brca2ko/ko embryos (n = 5 and 6, respectively, out of 58 embryos, Figure 4D). Their gross morphology was also indistinguishable, both being developmentally retarded compared to other embryos (Figure 4, E and F). However, when examined histologically, the Gipc3;Brca2ko/ko embryos were found to be developmentally more advanced and exhibited more complex embryonic structure compared to the Brca2ko/ko embryos (n = 3 each, Figure 4G). We confirmed the genotype of the embryos using tissues obtained by laser-captured microdissection (Figure 4H). Our results suggest that the Gipc3 transgene partially rescues the mutant phenotype of Brca2-null embryos.
Figure 4.
Gipc3 transgene partially rescued Brca2ko/ko mouse embryo lethality (A) Schematic strategy of generating Gipc3 transgenic mice. Gfp PCR primers were indicated. PCR by using Gfp primers indicates the Gipc3 transgene. (B) Western blot showing expression of Gipc3 transgene expression in whole embryo lysate harvested from E13.5 Gipc3 transgene and wildtype embryos. (C) Table showing the number of viable mice with indicated genotypes born from the indicated mating. (D) Table showing the number of E8.5 embryos with indicated genotypes obtained by the indicated mating. (E) Macroscopic images of E8.5 embryos with indicated genotypes. Bar, 1 mm. (F) Gel images showing the genotyping PCR of the embryos shown in (E). PCR by using Gfp primers indicates the Gipc3 transgene. P, Positive control; N, Negative control. (G) H&E staining of the E8.5 embryo sections with indicated genotypes. Bar, 200 μm. (H) Gel images showing the genotyping PCR of the embryos shown in (G). PCR by using Gfp primers indicates the Gipc3 transgene. P, Positive control; N, Negative control.
Discussion
Overcoming BRCA2 loss-induced cell lethality is vital for normal somatic cells to become cancerous. Loss of TRP53 is known to contribute to partial rescue of Brca2ko/ko embryos and also to promote tumorigenesis in mice (Ludwig et al. 1997; Jonkers et al. 2001). We have recently shown that protection of stalled replication forks by PARP1, PTIP, as well as MRE11 knockdown, supports viability of Brca2ko/ko mESC (Chaudhuri et al. 2016; Ding et al. 2016). Protection of stalled replication forks suppresses genomic instability as well as contributes to chemoresistance in BRCA2-deficient cells (Chaudhuri et al. 2016). In this study, we have identified GIPC3 as a novel genetic interactor of BRCA2 using a genetic screen in mESC. We have shown that overexpression of GIPC3 can rescue the lethality of Brca2-null mESC. The rescue is mediated by their downstream binding partner APPL1 and APPL2. This is based on our finding that mutant GIPC3 fails to rescue Brca2-null mESC lethality. The physiological relevance of the genetic interaction between BRCA2 and GIPC3 is demonstrated by the partial rescue of Brca2ko/ko embryos when Gipc3 is upregulated.
While the precise mechanism of cell viability by Gipc3 overexpression remains unknown, our findings show that they do not affect BRCA2-mediated HDR. APPL1/2 are known interactors of GIPC1, and our results show they mediate the rescue by GIPC3. APPL1/2 are adaptor proteins with multiple protein–protein interacting domains, and have versatile biological functions mediated via their binding partners including Rab5, OCRL, AdipoR1, DCC, FSHR, TrkA, Akt, and NuRD/MeCP1 (Deepa and Dong 2009). These proteins are associated with biological processes such as apoptosis, cell proliferation and survival, chromatin remodeling, and protein trafficking. We hypothesize that Akt-mediated prosurvival signaling is likely to be one of the contributing factors, because Akt has been shown to mediate resistance to DNA damaging chemotherapy in human tumors (Liu et al. 2014). It is possible that upregulated Akt activity due to GIPC3 overexpression overcomes the BRCA2 loss-induced DNA damage in mESC, and allows cells to survive. Future studies will be focused on dissecting the biological processes that contribute to GIPC3-mediated cell viability.
The fact that GIPC3 overexpression can rescue the lethality of Brca2-null mESC suggests that this protein can play a role when a normal somatic cell loses BRCA2 and becomes a preneoplastic cell. Cancer stem cells (CSC) are known to play an important role in tumor initiation (Beck and Blanpain 2013) and GIPC3 may be involved in CSC-driven tumor initiation. This is supported by the observation that GIPC3 is one of the genes upregulated in human breast CSC (Charafe-Jauffret et al. 2009). Although the functional significance of this finding remains unknown, it is plausible that the higher expression of GIPC3 in CSC promotes BRCA2 loss-induced tumor initiation in CSC. Future studies will be aimed at examining the role of GIPC3 in BRCA2-mediated tumorigenesis.
Acknowledgments
We thank members of our laboratory for helpful discussions and suggestions. We thank Jairaj Acharya, Kajal Biswas, Ira Daar, and Suhas Kharat for critical review of the manuscript. This research was sponsored by the Intramural Research Program, Center for Cancer Research, National Cancer Institute, United States National Institutes of Health. X.D. received a Department of Defense, Breast Cancer Research Program, postdoctoral fellowship (W81XWH-13-1-0362).
Footnotes
Communicating editor: J. Schimenti
Literature Cited
- Beck B., Blanpain C., 2013. Unravelling cancer stem cell potential. Nat. Rev. Cancer 13: 727–738. [DOI] [PubMed] [Google Scholar]
- Charafe-Jauffret E., Ginestier C., Iovino F., Wicinski J., Cervera N., et al. , 2009. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 69: 1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charizopoulou N., Lelli A., Schraders M., Ray K., Hildebrand M. S., et al. , 2011. Gipc3 mutations associated with audiogenic seizures and sensorineural hearing loss in mouse and human. Nat. Commun. 2: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri A. R., Callen E., Ding X., Gogola E., Duarte A. A., et al. , 2016. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535: 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N., McManus R., Wooster R., Mangion J., Seal S., et al. , 1995. Consistent loss of the wild type allele in breast cancers from a family linked to the BRCA2 gene on chromosome 13q12–13. Oncogene 10: 1673–1675. [PubMed] [Google Scholar]
- Deepa S. S., Dong L. Q., 2009. APPL1: role in adiponectin signaling and beyond. Am. J. Physiol. Endocrinol. Metab. 296: E22–E36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Chaudhuri A. R., Callen E., Pang Y., Biswas K., et al. , 2016. Synthetic viability by BRCA2 and PARP1/ARTD1 deficiencies. Nat. Commun. 7: 12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Jenkins N. A., Copeland N. G., 2005. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood 106: 3932–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubeva Y., Rogers K., 2009. Collection and preparation of rodent tissue samples for histopathological and molecular studies in carcinogenesis, pp. 3–60 in Methods in Molecular Biology, edited by Kozlov S. V. Humana Press, New York. [DOI] [PubMed] [Google Scholar]
- Golubeva Y., Salcedo R., Mueller C., Liotta L. A., Espina V., 2013. Laser capture microdissection for protein and NanoString RNA analysis, pp. 213–257 in Cell Imaging Techniques Methods and Protocols, edited by Taatjes D. J., Roth J. Humana Press, New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gretarsdottir S., Thorlacius S., Valgardsdottir R., Gudlaugsdottir S., Sigurdsson S., et al. , 1998. BRCA2 and p53 mutations in primary breast cancer in relation to genetic instability. Cancer Res. 58: 859–862. [PubMed] [Google Scholar]
- Gudmundsson J., Johannesdottir G., Bergthorsson J. T., Arason A., Ingvarsson S., et al. , 1995. Different tumor types from BRCA2 carriers show wild-type chromosome deletions on 13q12-q13. Cancer Res. 55: 4830–4832. [PubMed] [Google Scholar]
- Jonkers J., Meuwissen R., van der Gulden H., Peterse H., van der Valk M., et al. , 2001. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29: 418–425. [DOI] [PubMed] [Google Scholar]
- Kass E. M., Lim P. X., Helgadottir H. R., Moynahan M. E., Jasin M., 2016. Robust homology-directed repair within mouse mammary tissue is not specifically affected by Brca2 mutation. Nat. Commun. 7: 13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M., 2013. Functional proteomics, human genetics and cancer biology of GIPC family members. Exp. Mol. Med. 45: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. A., Sternberg M. J., 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4: 363–371. [DOI] [PubMed] [Google Scholar]
- Kuznetsov S. G., Liu P., Sharan S. K., 2008. Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nat. Med. 14: 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Shen H., Himmel K. L., Dupuy A. J., Largaespada D. A., et al. , 1999. Leukaemia disease genes: large-scale cloning and pathway predictions. Nat. Genet. 23: 348–353. [DOI] [PubMed] [Google Scholar]
- Lin D. C., Quevedo C., Brewer N. E., Bell A., Testa J. R., et al. , 2006. APPL1 associates with TrkA and GIPC1 and is required for nerve growth factor-mediated signal transduction. Mol. Cell. Biol. 26: 8928–8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Turner K. M., Alfred Yung W. K., Chen K., Zhang W., 2014. Role of AKT signaling in DNA repair and clinical response to cancer therapy. Neuro-oncol. 16: 1313–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig T., Chapman D. L., Papaioannou V. E., Efstratiadis A., 1997. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 11: 1226–1241. [DOI] [PubMed] [Google Scholar]
- Moynahan M. E., Pierce A. J., Jasin M., 2001. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7: 263–272. [DOI] [PubMed] [Google Scholar]
- Nagy A., 2003. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Ory S., Zhou M., Conrads T. P., Veenstra T. D., Morrison D. K., 2003. Protein phosphatase 2A positively regulates Ras signaling by dephosphorylating KSR1 and Raf-1 on critical 14–3-3 binding sites. Curr. Biol. 13: 1356–1364. [DOI] [PubMed] [Google Scholar]
- Rehman A. U., Gul K., Morell R. J., Lee K., Ahmed Z. M., et al. , 2011. Mutations of GIPC3 cause nonsyndromic hearing loss DFNB72 but not DFNB81 that also maps to chromosome 19p. Hum. Genet. 130: 759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainlos M., Iskenderian-Epps W. S., Olivier N. B., Choquet D., Imperiali B., 2013. Caged mono- and divalent ligands for light-assisted disruption of PDZ domain-mediated interactions. J. Am. Chem. Soc. 135: 4580–4583. [DOI] [PubMed] [Google Scholar]
- Schlacher K., Christ N., Siaud N., Egashira A., Wu H., et al. , 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G., Schmidt-Supprian M., Rad R., Saur D., 2017. Tissue-specific tumorigenesis: context matters. Nat. Rev. Cancer 17: 239–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan S. K., Morimatsu M., Albrecht U., Lim D. S., Regel E., et al. , 1997. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386: 804–810. [DOI] [PubMed] [Google Scholar]
- Varsano T., Dong M. Q., Niesman I., Gacula H., Lou X., et al. , 2006. GIPC is recruited by APPL to peripheral TrkA endosomes and regulates TrkA trafficking and signaling. Mol. Cell. Biol. 26: 8942–8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman A. R., 2009. Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Annu. Rev. Pathol. 4: 461–487. [DOI] [PubMed] [Google Scholar]
- Zofall M., Fischer T., Zhang K., Zhou M., Cui B., et al. , 2009. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature 461: 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data necessary for the conclusions from the current study are represented in the article. Mouse strains are available on request.