Abstract
In a genetic screen for mutants showing modified splicing of an alternatively spliced GFP reporter gene in Arabidopsis thaliana, we identified mutations in genes encoding the putative U1 small nuclear ribonucleoprotein (snRNP) factors RBM25 and PRP39a. The latter has not yet been studied for its role in pre-messenger RNA (pre-mRNA) splicing in plants. Both proteins contain predicted RNA-binding domains and have been implicated in 5′ splice site selection in yeast and metazoan cells. In rbm25 mutants, splicing efficiency of GFP pre-mRNA was reduced and GFP protein levels lowered relative to wild-type plants. By contrast, prp39a mutants exhibited preferential splicing of a U2-type AT-AC intron in GFP pre-mRNA and elevated levels of GFP protein. These opposing findings indicate that impaired function of either RBM25 or PRP39a can differentially affect the same pre-mRNA substrate. Given a prior genome-wide analysis of alternative splicing in rbm25 mutants, we focused on examining the alternative splicing landscape in prp39a mutants. RNA-seq experiments performed using two independent prp39a alleles revealed hundreds of common genes undergoing changes in alternative splicing, including PRP39a itself, a second putative U1 snRNP component PRP40b, and genes encoding a number of general transcription-related proteins. The prp39a mutants displayed somewhat delayed flowering, shorter stature, and reduced seed set but no other obvious common defects under normal conditions. Mutations in PRP39b, the paralog of PRP39a, did not visibly alter GFP expression, indicating the paralogs are not functionally equivalent in this system. Our study provides new information on the contribution of PRP39a to alternative splicing and expands knowledge of plant splicing factors.
Keywords: alternative splicing, Arabidopsis thaliana, PRP39, RBM25, U1 snRNP
EUKARYOTIC messenger RNAs are produced from primary transcripts (pre-mRNAs) that undergo extensive co- and post-transcriptional processing involving 5′ cap formation, intron removal by splicing, and 3′ cleavage and polyadenylation (Moore and Proudfoot 2009). Splicing of pre-mRNAs occurs in two sequential trans-esterification steps and is executed by the spliceosome, a large, dynamic ribonucleoprotein (RNP) machine located in the nucleus. The spliceosomal cycle involves the step-wise assembly of six defined complexes: complex E, prespliceosomal complex A, precatalytic complex B, complexes B* and C, which carry out catalytic steps 1 and 2, respectively, and the postspliceosomal complex (Matera and Wang 2014). Each complex contains a distinct combination of core spliceosomal proteins and auxiliary factors, as well as one or more different small nuclear (sn) RNPs. The five snRNPs of the U2 (major) type of spliceosome feature a specific snRNA (U1, U2, U4, U5, or U6) encircled by a heteromeric ring of seven Sm/Lsm proteins together with a variable number of particle-specific proteins (Wahl et al. 2009; Will and Lührmann 2011).
The protein composition of different spliceosomal complexes has been determined through proteomic, RNA cross-linking and structural analyses carried out largely in Saccharomyces cerevisiae (budding yeast) and metazoan cells (Fabrizio et al. 2009; Wahl et al. 2009; Will and Lührmann 2011; Wahl and Lührmann 2015). Detailed proteomic and structural characterization of plant spliceosomes has lagged behind owing to the unavailability of functional spliceosomal complexes isolated from plant cells (Barta et al. 2008; Koncz et al. 2012; Reddy et al. 2012, 2013). Hence, information about the predicted protein composition of plant spliceosomes is derived primarily from comparative sequence analyses. These comparisons have revealed that the Arabidopsis thaliana (Arabidopsis) genome encodes ∼430 predicted spliceosomal proteins and splicing-related factors, most of which are conserved in budding yeast and metazoan cells (Koncz et al. 2012). Many spliceosomal and splicing-related proteins in Arabidopsis are encoded by duplicated genes, potentially allowing functional diversification of paralogous copies (Koncz et al. 2012; Reddy et al. 2012, 2013).
The spliceosome is responsible for both constitutive splicing, in which the same splice sites are always used, and alternative splicing, in which splice site usage is variable. Major modes of alternative splicing include exon skipping, intron retention, and alternative 5′ donor or 3′ acceptor splice site selection. The majority of multi-intron genes in higher organisms are subject to alternative splicing, which substantially increases transcriptome and proteome diversity (Nilsen and Graveley 2010). The regulation of alternative splicing is complex and only partly understood (Reddy et al. 2013; Staiger and Brown 2013; Ramanouskaya and Grinev 2017). Major contributors include exonic and intronic splicing enhancers and silencers that bind serine/arginine-rich (SR) proteins and heterogeneous nuclear (hn) RNPs to regulate alternative splicing in a positive and negative manner, respectively (Nilsen and Graveley 2010; Y. Wang et al. 2015). In plants, alternative splicing is important for many developmental and physiological processes, including responses to abiotic and biotic stresses and the circadian clock (Staiger and Brown 2013; Filichkin et al. 2015; Deng and Cao 2017). A number of SR proteins that regulate alternative splicing have been implicated in plant stress–response pathways (Ding et al. 2014; Filichkin et al. 2015). However, many predicted plant splicing factors have not yet been investigated for their contributions to alternative splicing and plant phenotypes.
The critical initial step in splicing and spliceosome assembly is 5′ splice site recognition, which occurs in complex E and requires base pairing between the 5′ splice site and U1 snRNA (Kaida et al. 2010; Matera and Wang 2014). In addition to U1 snRNA and seven subunits of the Sm ring structure, the U1 snRNP in budding yeast contains 10 particle-specific proteins: Prp39, Prp40, Snu71, Snu56, Snp1, Mud1, Luc7, Prp42, Nam8, and Yhc1 (Will and Lührmann 2011; Plaschka et al. 2017). Arabidopsis has predicted orthologs of most of these proteins, including PRP39 (Pre-mRNA Processing protein 39), which is encoded by two expressed paralogs, PRP39a and PRP39b (formerly termed PRP39-1 and PRP39-2, respectively: Wang et al. 2007), and RBM25 (RNA-Binding Motif protein25), termed Snu71 in budding yeast (Koncz et al. 2012). PRP39 was originally identified in a genetic screen for splicing-defective mutants in budding yeast and assigned a role in the U1 snRNP, where it facilitates stable binding of the U1 snRNA to the pre-mRNA substrate (Lockhart and Rymond 1994). RBM25/Snu71was identified as a U1 snRNP component in budding yeast (Gottschalk et al. 1998; Fortes et al. 1999) and as a protein functioning in early spliceosome formation in HeLa cells (Fortes et al. 2007). RBM25 has recently been shown to be important for abscisic-acid (ABA) responses and pre-mRNA splicing in Arabidopsis (Zhan et al. 2015; Z. Wang et al. 2015; Cheng et al. 2017). However, apart from one study that reported delayed flowering time in prp39a mutants in Arabidopsis (Wang et al. 2007), PRP39 proteins have not been investigated further for their roles in pre-mRNA splicing in plants.
To gain more information about plant splicing factors, we are conducting a forward genetic screen to identify mutants showing modified splicing of an alternatively spliced GFP reporter gene in Arabidopsis. So far this screen has yielded mutations in putative U5 snRNP protein PRP8, a large, conserved protein at the catalytic core of the spliceosome; RTF2, a novel protein that may be involved in ubiquitin-based regulation of splicing (Sasaki et al. 2015; Kanno et al. 2017); coilin, a scaffold protein for Cajal Bodies, which are the site of snRNP maturation (Kanno et al. 2016); CWC16a, a putative catalytic step1 factor; SMFa, a core snRNP protein; and SMU1, a WD40 repeat-containing protein that may be involved in recognizing spliceosomal targets for ubiquitination (Kanno et al. 2017). Here we report the identification in the screen of additional mutants that are impaired in the putative U1 snRNP factors PRP39a and RBM25, and describe the impact of prp39a mutations on genome-wide differential gene transcription and pre-mRNA splicing.
Materials and Methods
Plant material, GFP reporter gene system, and forward genetic screen
The transgenic T line containing the alternatively spliced GFP reporter gene (referred to hereafter for simplicity as “wild-type”) and all gfp-weak (gfw) and hyper-gfp (hgf) mutants derived from ethyl methane sulfonate (EMS) mutagenesis of the T line are in the Col-0 ecotype (Kanno et al. 2016, 2017). Seeds of a prp39b-1 T-DNA insertion mutant (SALK_140208) were obtained from the Nottingham Arabidopsis Stock Center. All plants were grown under long-day conditions (22–23°, 16 hr light, 8 hr dark).
Approximately 40,000 seeds of the homozygous T line were treated with EMS and mutant screening performed as described previously (Kanno et al. 2016, 2017). Based on their GFP phenotypes, putative gfw and hgf mutants are screened out in the M2 population of mutagenized seedlings, which is the first generation when a recessive mutation can be homozygous. GFP phenotypes are assessed by viewing M2 seedlings (a few days after germination) growing under sterile conditions on solid Murashige and Skoog (MS) medium using a Leica M165FC fluorescence stereomicroscope. Around 10 gfw and10 hgf complementation groups have been retrieved so far in the screen, which is still ongoing (Kanno et al. 2017). Causal mutations in gfw and hgf mutants are identified by next generation mapping (NGM) (James et al. 2013). NGM is carried out by sequencing pooled genomic DNA from at least 50 BC1F2 seedlings showing the desired GFP-weak or Hyper-GFP phenotype. BC1 F2 seedlings are obtained by backcrossing an M2 or M3 plant with the wild-type T line to produce BC1 plants, which are then allowed to self-fertilize to produce BC1F2 progeny. The mutation of interest can be determined by surveying the distribution of single nucleotide polymorphisms (SNPs) between the mutant and mapping genomes (Austin et al. 2011; James et al. 2013). SNPs are detected using CLC Genomics Workbench 6 software (QIAGEN, Valencia, CA). Here, we report the identification in the screen of multiple mutant alleles of rbm25/gfw3 and prp39a/hgf5.
Testing effect of prp39b mutation on GFP level and screening for double mutants
To test whether a homozygous mutation in PRP39b, the paralog of PRP39a, would lead to a Hyper-GFP phenotype, we crossed a wild-type plant homozygous for the T locus (T/T) with a homozygous prp39b-1 mutant (b/b). Self-fertilization of the resulting F1 plants (T/-; B/b) produced a segregating F2 population (the dash indicates hemizygosity for the transgenic T locus). F2 seeds were germinated on solid MS medium and screened ∼ 2 weeks later under a fluorescence stereomicroscope for GFP expression, which is observed with a genotype of either a T/T or T/- (hereafter written collectively as T/(T)). A subset was transferred to soil for genotyping to identify T/T; b/b plants.
To examine whether double homozygous mutant plants (a/a; b/b) were viable, we crossed a homozygous prp39a-5 mutant (T/T; a/a) to a b/b plant. Self-fertilization of the resulting F1 plants (T/-; A/a; B/b) generated a segregating F2 population. The F2 seeds were germinated on MS medium and prescreened under a fluorescence stereomicroscope for a Hyper-GFP phenotype (T/(T); a/a). Owing to a dosage effect of the GFP gene resulting from segregation of the T locus in the GFP-positive F2 population, the Hyper-GFP phenotype could be difficult to discern in some cases. Selected Hyper-GFP F2 progeny were transferred to soil for genotyping to identify T/(T); a/a; b/b plants. Primers for detecting prp39a-5 and prp39b-1 alleles are listed in Supplemental Material, Table S1.
Western blotting
Western blotting to detect GFP protein was performed as described previously using total protein extracted from 2-week-old seedlings growing on solid MS medium as described above (Fu et al. 2015; Kanno et al. 2016, 2017). GFP antibodies were purchased from Roche (Cat. No. 11 814 460 001).
Semiquantitative RT-PCR
Semiquantitative RT-PCR to detect GFP splice variants in the prp39a-3/hgf5-1 and prp39a-4/hgf5-2 mutants was performed using total RNA isolated from 2-week-old seedlings (BC1F3 generation) growing on solid MS medium as described above using a Plant Total RNA Miniprep kit (GeneMark, Taichung City, Taiwan) according to a previously published protocol (Sasaki et al. 2015). GFP and actin primers are listed in Table S1.
RNA-sequencing (RNA-seq)
Total RNA was isolated from 2-week-old seedlings of the wild-type T line and the prp39a-3/hgf5-1 and prp39a-4/hgf5-2 mutants (BC1F3 generation) growing on solid MS medium as described above. The BC1F3 generation, which has a reduced number of EMS-induced second-site mutations relative to the original M2 mutant, is produced by self-fertilization of BC1F2 plants generated as described above. Library preparation and RNA-seq were carried out (biological triplicates for each sample) as described in previous publications (Sasaki et al. 2015; Kanno et al. 2016). Whole genome resequencing on the prp39a mutants was carried out to identify any remaining EMS-induced second-site mutations that change splice sites, which were then eliminated from the analysis of alternative splicing.
RNA-seq reads were mapped to the TAIR10 genome as previously described (Kanno et al. 2016, 2017) using Bowtie2 (Langmead and Salzberg 2012) and BLAT (Kent 2002), and >98% of the reads were accepted for every replicate. RackJ (http://rackj.sourceforge.net/) was used to compute read counts for all genes, the average read depths of all exons and all introns, and read counts for all splicing junctions.
Read counts of all genes were normalized using the trimmed mean of M values method (Robinson and Oshlack 2010) and transformed into logCPM (log counts per million) using the voom method (Law et al. 2014) with parameter normalize=“none.” Adjusted reads per kilobase of transcript per million mapped values were computed based on logCPMs and used for t-tests. In this study, a gene was defined as differentially expressed if its P-value by t-test was <0.01 and its fold-change was ≥2.
Given an intron, its preference of intron retention was measured by comparing intron retention ratios in the wild-type controls to those in the mutant replicates using t-test, where the intron retention ratio was defined as the average read depth of the intron divided by the average read depth of the neighboring exons. In this approach, the underlying null hypothesis assumes that the chances for an intron to be retained are the same in the two samples; a significant P-value indicates that the chance of intron retention was altered in one of the two samples. Given an intron with P-value <0.01, it was defined as more efficiently spliced if the average ratio in the mutant replicates was smaller than a half of that in the wild-type controls; otherwise, it was defined as a case of increased intron retention if the average ratio in the mutant replicates was two times greater than that in the wild-type controls.
The preference of exon skipping events and alternative donor/acceptor events were measured using methods similar to that for intron retention events. For exon skipping events, exon skipping log-ratios in the wild-type controls were compared to those in the mutant replicates using t-tests, where the exon skipping log-ratio was defined as the log of the splice-read count supporting an exon skipping event divided by the splice-read count involving a skipped exon of the exon skipping event. For alternative donor/acceptor events, the corresponding log-ratio was defined as the log of the splice-read count supporting a splicing junction divided by the splice-read count supporting all other junctions of the same intron. Aforementioned events were further confirmed using additional t-tests carried out with log-ratios made of supporting read counts and unique read counts of individual genes. Finally, an alternative splicing event was reported if its P-values were all <0.01, and it was defined as enhanced if the average ratio of supporting read counts to unique read counts of the gene in the mutant replicate was two times greater than that in the wild-type controls; otherwise, it was defined reduced if the average ratio in the mutant replicates was less than half of that in the wild-type controls.
Data availability
Seeds of prp39a and rbm25 mutants identified in this study will be available from Arabidopsis Biological Resource Center, Ohio State University, OH. Supplemental Figures: Figure S1 shows a phenotype comparison of rbm25 mutants with wild-type T. Figure S2 presents amino acid alignments of PRP39-related proteins in selected plant species. Figure S3 presents amino acid alignments of PRP39-related proteins in model organisms. Supplemental Tables: Table S1 lists primers. Table S2 shows differentially expressed genes (DEGs) shared between two independent prp39a mutants. Table S3 lists intron retention (IR) and more efficient splicing (MES) events shared between two independent prp39a mutants. Table S4 lists exon skipping (ES) events shared between two independent prp39a mutants. Table S5 lists alternative splice site donor or acceptor (SSDA) events shared between two independent prp39a mutants. Table S6 lists the top 30 coexpressed genes for PRP39a and PRP39b. RNA sequencing data used to prepare Table S2, Table S3, Table S4 and Table S5 are available under Sequence Read Archive accession number SRP108084 [samples ‘PRP39-1-3 biological replicate’ nrs. 1-3 (prp39a-3 allele) and ‘PRP39-1-4 biological replicate’ nrs. 1-3 (prp39a-4 allele) and SRP093582 (samples ‘ST biological replicate’ nrs. 1, 2 and 4 (WT T line)].
Results
Identification of rbm25 and prp39a mutants in forward genetic screen
In the wild-type T line, three main splice variants issue from the GFP reporter gene: a long unspliced transcript, a middle-length transcript resulting from splicing a canonical GT-AG intron, and a short transcript resulting from splicing a U2-type intron with noncanonical AT-AC splice sites. Owing to a number of premature termination codons in the unspliced and GT-AG transcripts, only the AT-AC transcript can be translated into GFP protein (Figure 1). A balanced ratio of these three transcripts is associated with an intermediate level of GFP expression in the wild-type T line (Kanno et al. 2017). Our working hypothesis is that mutations in genes encoding splicing-related factors will alter the ratio of the three transcripts, resulting in either an increase or decrease in GFP mRNA levels. These changes lead, respectively, to either a Hyper-GFP (hgf) or GFP-weak (gfw) phenotype relative to the intermediate level in the wild-type T line (Kanno et al. 2016, 2017). We previously reported four hgf mutants: hgf1 (coilin; At1g13030), hgf2 (CWC16a; At1g25682), hgf3 (SMU1; At1g73720), and hgf4 (SMFa; At4g30220) (Kanno et al. 2016, 2017), and two gfw mutants: gfw1 (RTF2; At5g58020) and gfw2 (PRP8; At1g80070) (Sasaki et al. 2015; Kanno et al. 2017). Here we describe the identification of multiple alleles of a new mutant in each GFP phenotypic category.
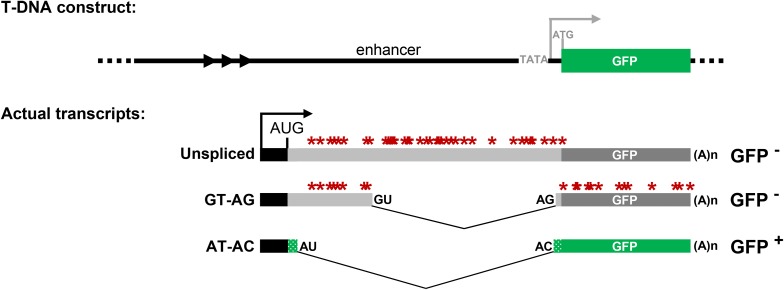
Figure 1.
Schematic drawing of T-DNA construct and alternatively spliced GFP reporter gene. Top: The original construct introduced into Arabidopsis contained a GFP reporter gene designed to be under the transcriptional control of a minimal promoter (TATA) and upstream viral enhancer element. However, neither the minimal promoter nor the downstream ATG initiation codon (gray letters) is used in the T line. Bottom: Instead, transcription of GFP pre-mRNA initiates at an upstream promoter region (black bar and arrow) to produce three alternative splicing variants that include the part of enhancer region (Kanno et al. 2008). The actual coding sequence of GFP protein (green bars), which has a unique 27-amino-acid extension (short stippled green bars), is interrupted by intronic sequences (light gray) that comprise a smaller GT-AG intron inserted within a larger U2-type intron with noncanonical AT-AC splice sites (Sasaki et al. 2015; Kanno et al. 2016). Three major GFP splice variants include a long unspliced transcript, a midlength transcript resulting from splicing of the GT-AG intron, and a shorter transcript resulting from splicing of the AT-AC intron. Only the AT-AC transcript lacks premature termination codons (PTCs, red asterisks) and can be translated into GFP protein. Arrowheads represent a short tandem repeat upstream of the promoter. The black AUG represents the main translation initiation codon. The 3′ splice sites for the GT-AG and AT-AC introns are only 3 nt apart with the noncanonical AC on the outside (Sasaki et al. 2015; Kanno et al. 2016, 2017).
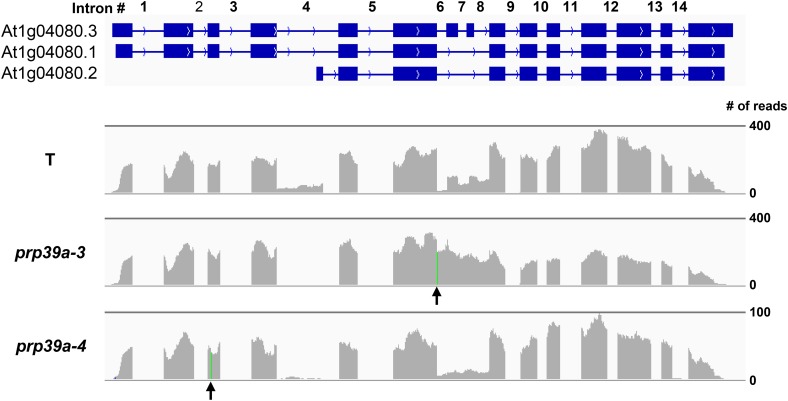
Four new hgf mutants were determined by either NGM or direct sequencing to harbor independent homozygous mutations in the gene encoding PRP39a (At1g04080). PRP39a contains seven HAT (half a tetratricopeptide) repeats, which are present in several proteins involved in RNA processing and are able to bind RNA (Hammani et al. 2012). Based on previous reports of two T-DNA insertion alleles, prp39a-1 and prp39a-2 (Wang et al. 2007), we designated our alleles prp39a-3/hgf5-1 (5′ splice site donor, sixth intron), prp39a-4/hgf5-2 (W90*), prp39a-5/hgf5-3 (N528S), and prp39a-6 (Q719*) (Figure 2, A and B). PRP39a has a paralog in Arabidopsis: PRP39b (At5g46400).
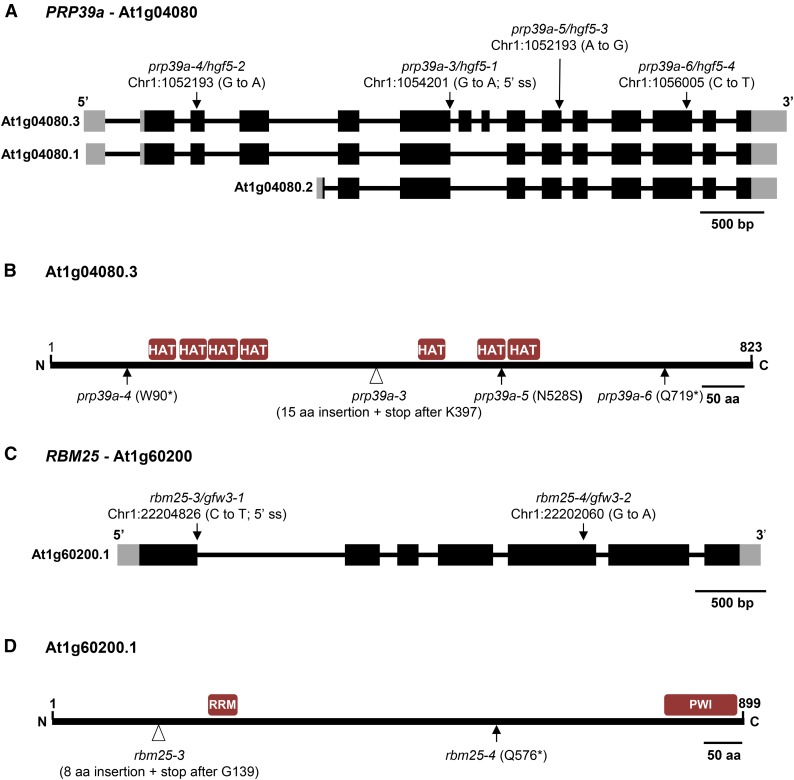
Figure 2.
PRP39a and RBM25 gene structures, positions of mutations, and protein domains. (A and B) The pre-mRNA of PRP39a (At1g04080) is alternatively spliced to produce three major splice variants (http://www.arabidopsis.org/index.jsp). One of these, At1g04080.3, encodes an 823-amino-acid protein that contains seven HAT (Half-A-Tetratricopeptide) repeats. We identified the following prp39a alleles in our screen: prp39a-3 (splice site donor, sixth intron); prp39a-4 (Trp90*); prp39a-5 (N528S); and prp39a-6 (Q719*) (Figure S2). The prp39a-3 and prp39a-4 alleles, which encode substantially truncated PRP39a proteins, are likely to be nulls. The A to G mutation in prp39a-5 is not typically induced by EMS, which produces almost exclusively to CG/AT transition mutations. It is probable that the A to G mutation, which has been recovered multiple times from different batches of M2 seeds, was present as a heterozygous mutation in a very small number of the original 40,000 seeds used for EMS mutagenesis and became homozygous in some plants during M2 seed amplification. Amino acid sequence alignments of PRP39-related proteins in selected plant species and model organisms are shown in Figure S2 and Figure S3, respectively. (C and D) RBM25 (At1g60200) is an 899-amino-acid protein containing RRM and PWI domains. We identified the following rbm25 alleles: rbm25-3 (Gln576*) and rbm25-4 (3′ splice site, first intron, resulting in an 8-amino-acid insertion followed by a stop codon after G139). Both alleles, which disrupt the full-length RBM25 protein, are likely to be nulls. Previously identified alleles resulting from point mutations are rbm25-1 (A899V) and rbm25-2 (Gln570*) (Zhan et al. 2015).
Analysis of NGM data or direct sequencing from two new gfw mutants revealed independent homozygous mutations in the gene encoding RBM25 (At1g60200), which is encoded by a single copy gene in Arabidopsis. RBM25 contains two domains capable of binding RNA: RRM (RNA Recognition Motif) and PWI (named after a highly conserved PWI tripeptide located in the N-terminal domain). In view of two previously reported mutants of RBM25 (rbm25-1 and rbm25-2; Zhan et al. 2014; Wang et al. 2015; Cheng et al. 2017), we designated our alleles rbm25-3/gfw3-1 (5′ splice site donor, first intron) and rbm25-4/gfw3-2 (Q576*) (Figure 2, C and D).
The prp39a and rbm25 mutations we identified are all recessive, as indicated by a return to the intermediate, wild-type level of GFP fluorescence in BC1 progeny produced after backcrossing the homozygous mutants to the wild-type T line (data not shown).
GFP and morphological phenotypes of rbm25 and prp39a mutants
Seedlings of the prp39a and rbm25 mutants display typical GFP-weak or Hyper-GFP phenotypes, respectively, relative to the wild-type T line (Figure 3). Western blotting using a GFP antibody confirmed that the changes in GFP fluorescence were accompanied by parallel changes in GFP protein levels, with the prp39a and rbm25 mutants exhibiting increases and decreases in GFP protein, respectively, relative to wild-type plants (Figure 4). Semiquantitative RT-PCR was performed to determine the splicing pattern of GFP pre-mRNA in the mutants. Compared to the wild-type, the prp39a mutants displayed increased levels of the short, translatable GFP mRNA resulting from splicing the AT-AC intron and reduced amounts of unspliced, untranslatable GFP pre-RNA. Conversely, the rbm25 mutants showed reduced levels of translatable transcript and increased levels of unspliced RNA (Figure 5). The GFP phenotypes of the mutants thus mirror the changes in the ratio of the three GFP splice variants.
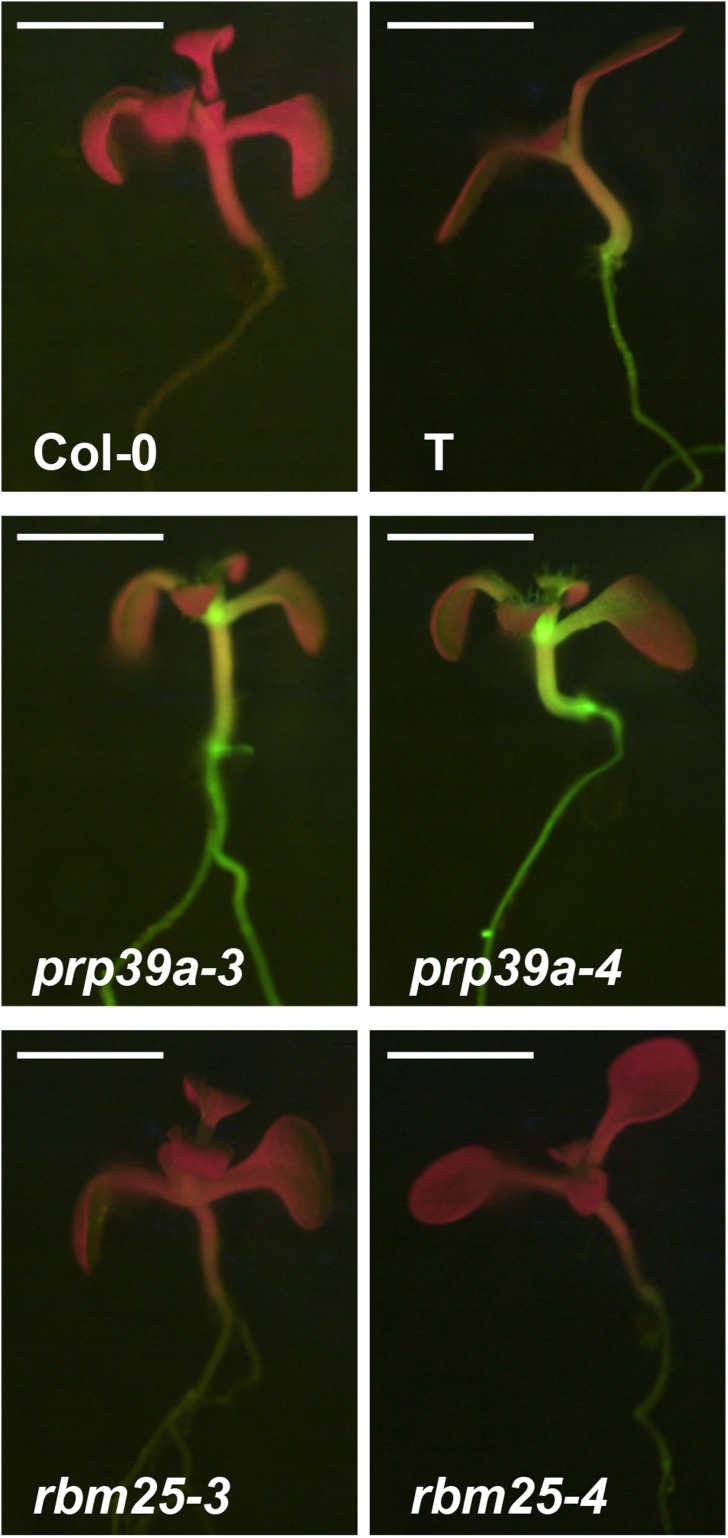
Figure 3.
GFP phenotype of prp39a and rbm25 mutants. GFP fluorescence of ∼2-week-old seedlings of nontransgenic Col-0, the wild-type T line, and selected prp39a and rbm25 mutants growing on solid MS medium as visualized under a fluorescence stereo microscope. In the prp39a mutants, GFP fluorescence is considerably increased whereas in rbm25 mutants, GFP fluorescence is decreased relative to the wild-type T line. Cotyledons (seedlings leaves) appear red owing to auto-fluorescence of chlorophyll at the excitation wavelength of GFP. The white bar indicates 2 mm.
Figure 4.
Western blots to detect levels of GFP protein in prp39a and rbm25 mutants. Total protein was extracted from ∼2-week-old seedlings of nontransgenic Col-0, the wild-type T line (GFP negative and positive controls, respectively), and selected prp39a and rbm25 mutants. Extracted proteins were separated by SDS-PAGE and blotted onto a PVDF membrane. The blots were probed with antibodies to GFP protein (top). As loading controls, the prp39a blot was washed and reprobed with an antibody to actin and a duplicate rbm25 gel was stained with Coomassie brilliant blue (CBB). The strong band is migrating at 56 kDa and is presumed to be the large subunit of ribulose bisphosphate carboxylase.
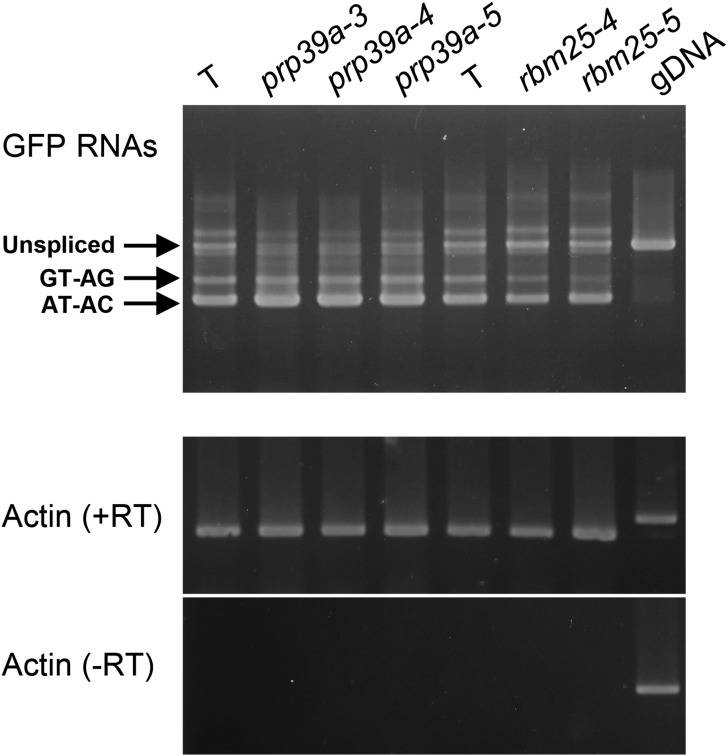
Figure 5.
RT-PCR analysis of GFP splice variants in prp39a and rbm25 mutants. Semiquantitative RT-PCR was used to investigate the accumulation of unspliced GFP transcript and two splicing variants (resulting from splicing the canonical GT-AG and noncanonical AT-AC introns, respectively) in the indicated prp39a and rbm25 mutants, and the wild-type T line. Actin is shown as a constitutively expressed control. –RT, no reverse transcriptase; gDNA (T), genomic DNA isolated from T line.
Morphological phenotypes of the mutants were assessed in BC1F3 plants, which are reduced in second-site, EMS-induced mutations relative to plants of the original M2 generation. Compared to the wild-type T line, the prp39a mutants were somewhat shorter, produced fewer seeds, and displayed a slight delay in flowering time, first starting on average 1 (prp39a-4) to 6 (prp39a-3) days later than in wild-type plants (Table 1; note that prp39a-3 showed considerable variability in flowering time). Except for reduced seed set, the rbm25-4 mutant largely resembled wild-type plants (Figure S1), similar to previous findings for rbm25-1 and rbm25-2 (Zhan et al. 2015). However, rbm25-3 plants were smaller, more spindly, somewhat yellowish, and less fecund than the wild type (Figure S1). Whether this phenotype is due to the strong rbm25-3 mutation or a closely linked second-site mutation is not known at this time because we have not yet successfully introduced a complementation construct into the rbm25-3 mutant, perhaps owing to a fertility defect.
Table 1. Phenotype comparison of prp39a mutants and wild type.
| Plant line | Germination (D.A.I.) | Transition to floweringa (days on soil) | Height (cm) | Diameter rosette leaves (cm) | Seed set (mg/plant) |
|---|---|---|---|---|---|
| prp39a-3 (n = 12) | 5 | Ave. 15.4 Range 10–20 | Ave. 32.8 Range 25–37 (n = 11) | Ave. 8.5 Range 7–10 | Ave. 90.1 Range 60–113 (n = 11) |
| prp39a-4 (n = 12) | 5 | Ave.10.3 Range 9–11 | Ave. 32.7 Range 29–36 | Ave. 4.9 Range 3.5–6 | Ave. 75.7 Range 45–115 |
| T (n = 12) | 5 | Ave. 9.5 Range 8–11 | Ave. 35.2 Range 32–38.5 | Ave. 7.3 Range 6–9 | Ave. 110.2 Range 81–149 |
To compare phenotypes of prp39a mutants compared to the wild-type T line, we grew 12 plants from each genotype on soil to maturity and seed set. The features listed above were noted for individual plants. The ranges and averages of the assessed values are indicated. D.A.I., days after imbibition; n, number of plants; Ave, average.
Most plants had 10 rosette leaves when bolting occurred.
RNA-seq analysis
The effects of rbm25 mutations on differential gene expression, alternative splicing, and ABA responses in Arabidopsis have been reported in a prior study (Zhan et al. 2015). We therefore focused our efforts on prp39a mutants, which have not previously been investigated for their genome-wide impact on alternative splicing in plants. For RNA-sequencing (RNA-seq) experiments, we isolated RNA from prp39a-3 and prp39a-4 seedlings of the BC1F3 generation. RNA-seq was carried out using biological triplicates for the two mutants and the age-matched wild-type T line.
Analysis of the RNA-seq data for GFP transcripts confirmed the semiquantitative RT-PCR data: relative to the wild-type T line, the average level of GFP mRNA resulting from splicing of the AT-AC intron increased in the prp39a mutants (from 17.9% in wild type to ∼38% in the two prp39a mutants; a 123% increase) while the average amount of unspliced, untranslatable transcript was reduced (58.6% in wild type to ∼40% in prp39a mutants; a 32% decrease). The level of untranslatable transcript resulting from splicing the GT-AG intron also decreased somewhat relative to wild type (23.5% in wild type to ∼20.5% in prp39a mutants; a 15% decrease) (Figure 6).
Figure 6.
Abundance of GFP RNA splice variants in prp39a mutants. The percentages of three major GFP RNA splice variants were ascertained from an analysis of RNA-seq data and show the average of biological triplicates. The levels of total GFP transcripts did not change significantly in prp39a mutants.
We next analyzed RNA-seq data for genome-wide DEGs and alternative splicing events, including IR, MES, ES, and alternative 5′ and 3′ splice site choice (5′_ss, 3′_ss) (Table 2). Because any change in splicing resulting from a PRP39a deficiency should be observed for >1 mutant allele, we consider here only those genes that showed statistically significant changes in both prp39a-3 and prp39a-4 mutants.
Table 2. DEGs and Alternative Splicing Events in prp39a Mutants.
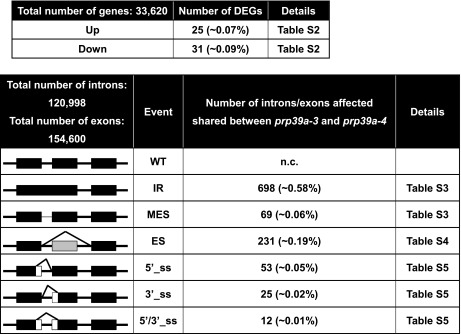 |
The major forms of alternative splicing are indicated to the left. WT, wild-type; IR, intron retention; MES, more efficient splicing; ES, exon skipping; 5′ or 3′_ss, change in 5′ splice site donor or 3′ splice site acceptor; 5′/3′_ss, change in both 5′ and 3′ splice sites. Numbers in parentheses show the percentage of total numbers of introns or exons affected.
Fifty-six shared DEGs were identified at roughly similar frequencies in “up” and “down” categories (25 up, 31 down) (Table 2). Shared IR events were relatively abundant: we detected 698 events (in 617 genes) (Table 2), indicating that >1 intron was affected in 81 multi-intron genes. Notably, a number of shared IR events were observed for general transcription factors (GTFs), including TATA binding protein 1 (TBP1) and TATA binding protein 2 (TBP2); TFIIB1, as well as Nuclear Factor Y (NF-Y)/CCAAT Binding factor (CBF) subunits A9 and B10. Two transcription elongation factors (TEFs) also appeared in the shared IR list: a TFIIS family member and SPT6L. IR events were also detected for two putative splicing-related factors: the U1 snRNP protein PRP40b and the U5 snRNP DEAD/DExH RNA helicase BRR2C (Table 3). Sixty-nine shared MES events (in 69 genes) were detected (Table 2), one of which affected intron 4 of PRP39a (Figure 7 and Table 3).
Table 3. Notable findings from analysis of alternative splicing in prp39a mutants.
| AGI | AS event | rbm25-1 | |
|---|---|---|---|
| GTFs | |||
| TBP1 | At3g13445 | IR | Zhan et al. (2015) |
| TBP2 | At1g55520 | IR | Zhan et al. (2015) |
| NFY subunit A9 | At3g20910 | IR | |
| NFY subunit B10 | At3g53340 | IR, ES | Zhan et al. (2015) |
| TFIIB1 | At2g41630 | IR | |
| TEFs | |||
| TFIIS | At5g09850 | IR | |
| SPT6L | At1g65440 | IR | |
| Splicing factors | |||
| PRP39a | At1g04080 | MES | |
| PRP40b | At3g19670 | IR, ES | |
| BRR2C | At5g61140 | IR | |
| SCL30 | At3g55460 | 5′/3′_ss |
Several transcription-related proteins and splicing factors were among the genes identified in the analysis of alternative splicing in the prp39a mutants. Promoter recognition and transcription initiation by RNAP II requires up to seven general transcription factors (GTFs), including TATA binding protein (TBP), TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH. Two of the best-studied GTFs are TBP and TFIIB, which have key roles at multiple steps of the RNAP II transcription cycle (Knutson 2013). Arabidopsis encodes two distinct TBPs (Lawit et al. 2007) and eight TFIIB-like proteins (Knutson 2013). We identified both TBP genes and TFIIB1 in our analysis of alternative splicing in prp39a mutants. NF-Y/CBF is an evolutionarily conserved trimeric transcription factor complex found in most eukaryotes (Zhao et al. 2017). All three subunits, NF-YA, NF-YB, and NF-YC, are required for binding to the CCAAT box, which is typically present 60–100 bp upstream of the transcription start site (Zhao et al. 2017). The Arabidopsis genome encodes 30 members of the NF-Y family, 10 in each subfamily (Zhao et al. 2017). In prp39a mutants, we detected changes in alternative splicing for two NF-Y/CBF protein subunits: A9 and B10. The GTFs also identified in an analysis of alternative splicing in the rbm25-1 mutant by Zhan et al. (2015) are indicated. TGIIS and SPT6L are transcription elongation factors (TEFs) that robustly copurify with RNAP II (Antosz et al. 2017). AS, alternative splicing; IR, intron retention; MEs, more efficient splicing; ES, exon skipping; 5′/3′_ss, 5′ and/or 3′ splice site change.
Figure 7.
IGV browser visualization of changes in alternative splicing of PRP39a pre-mRNA in prp39a mutants. PRP39a pre-mRNA is alternatively spliced, which generates three major annotated splice variants that differ in length and intron/exon content. The introns shown here have GT-AG splice sites (http://www.arabidopsis.org/). Changes in PRP39a pre-mRNA splicing in the prp39a mutants were observed for intron 4, which is present in its entirety only in At1g04080.1 and At1g04080.3, and in the region comprising introns 6, 7, and 8, which contains two short exons unique to At1g04080.3. Intron 4, which is the largest intron (552 nt) and relatively T-rich (∼40%), is more efficiently spliced in the two prp39a mutants. Details are shown in Table S3 (MES). According to the current annotation, intron 4 contains a transcription start site for a shorter transcript, At1g04080.2, which has at its 5′ end a unique small exon followed by a short intron that overlaps with the end of intron 4 of the other two splice variants. This short intron corresponds to the gap visible in the RNA reads at the end of intron 4 in the wild-type T line and suggests that At1g04080.2 is transcribed in 2-week-old seedlings. The Hyper-GFP phenotype observed in prp39a-4, which contains a premature termination codon (arrow, green line) preceding the transcription start site of At1g04080.2, suggests that the truncated (582-amino-acid) protein resulting from translation of this transcript is not able to functionally compensate for the loss of the full-length (823 amino acids; At1g04080.3) or nearly full-length (768 amino acids, At1g04080.1) PRP39 protein. Different profiles of RNA reads across introns 6–8 were observed between wild-type and prp39a mutants. However, these are difficult to interpret for prp39a-3, which harbors a mutation of the 5′ splice site donor of intron 6 (arrow, green line). Although the small exons 7 and 8 are only present in At1g04080.3, they are decreased in prp39a-4 relative to the wild-type T line. Whether this reduction is due to skipping of exons 7 and 8 or decreased transcription of At1g04080.3 is not known.
Two hundred thirty-one shared ES events (in 226 genes) were noted (Table 2). ES events included PRP40b and NFY/CBF, subunit B10, both of which are also on the IR list, and possibly PRP39a (Figure 7). Ninety alternative 5′-ss or 3′-ss site events were observed (Table 2). The list includes the splicing factor SCL30 (Table 3).
Tests of prp39b
To test whether a homozygous mutation in PRP39b, the paralog of PRP39a, would confer a Hyper-GFP phenotype similar to that observed with prp39a mutants, we performed the breeding scheme described in the Materials and Methods section. Fluorescence stereomicroscopy was used to assess the GFP phenotype of F2 descendants of a cross between b/b and the wild-type T line (T/T). In a population of 160 F2 seedlings, we observed only intermediate levels of GFP and no Hyper-GFP phenotypes. If T/(T); b/b plants are Hyper-GFP, the expected number would be 30 (18.75% of 160). Genotyping for the prp39b-1 mutation revealed one homozygous b/b plant among the first 10 GFP-positive F2 plants to be tested, i.e., 10%. This number approaches the expected percentage of T/(T); b/b plants in this population (18.75%), confirming that the prp39b mutation is segregating in a Mendelian manner. The collective results demonstrate that a homozygous prp39b mutation does not result in a visible Hyper-GFP phenotype.
To examine whether doubly homozygous prp39a prp39b mutants were viable, we carried out the breeding scheme described in the Materials and Methods section. Using a fluorescence stereomicroscope, we prescreened 300 F2 plants for a potentially Hyper-GFP phenotype (T/(T); a/a) and identified 54 (18%; expected percentage 25%). By genotyping 10 Hyper-GFP plants, two (20%; expected percentage 25%) were found to be doubly homozygous for prp39a and prp39b (T/(T); a/a; b/b). These findings indicate that the two mutations are segregating in a Mendelian manner and that the double homozygote is viable.
Discussion
In a forward genetic screen for mutants exhibiting changes in the splicing pattern of an alternatively spliced GFP reporter gene in Arabidopsis, we identified mutations in genes encoding two putative U1 snRNP factors: PRP39a and RBM25. Both proteins have been shown in budding yeast and mammalian cells to be components of the U1 snRNP involved in 5′ splice site selection (Lockhart and Rymond 1994; Gottschalk et al. 1998; Fortes et al. 2007; Zhou et al. 2008). PRP39a and RBM25 each contain different RNA-binding domains (HAT repeats and RRM and PWI domains, respectively), which are functionally important as indicated by the positions of the mutations retrieved in our screen. Three of the four prp39a mutations either led to truncated proteins lacking all or some of the seven HAT repeats or to a substitution of a conserved amino acid in one HAT repeat. The rbm25 mutations result in truncations removing one or both of the RNA-binding domains.
In budding yeast, both Prp39p and Rbm25/Snu71p are essential for viability (Lockhart and Rymond 1994; Gottschalk et al. 1998). By contrast, Arabidopsis mutants containing predicted null alleles of prp39a and rbm25 are viable and fertile, although seed set is reduced, particularly in the rbm25 mutants. In Arabidopsis, RBM25 is encoded by a single gene that is apparently not essential whereas PRP39 is encoded by two paralogs, PRP39a and PRP39b, which in principle could be functionally redundant. However, when introduced into the wild-type T line, a prp39b-1 mutation failed to confer a visible Hyper-GFP phenotype similar to that observed in prp39a mutants, suggesting that the two PRP39 paralogs are not functionally equivalent in this system. This finding is reinforced by the fact that we have so far recovered four independent alleles of prp39a in our screen but not yet any mutations in PRP39b. We cannot rule out, however, that PRP39a and PRP39b are able to substitute for each other in other functional capacities. The two PRP39 paralogs are ubiquitously expressed but they nevertheless show quantitative differences in their expression patterns. For example, PRP39a is more strongly expressed in the shoot apex and floral buds than PRP39b (e-FP Browser, http://www.arabidopsis.org/). The two paralogs also have differences in the identity of highly coexpressed genes, suggesting they may act in different subcomplexes or pathways (Table S6). Our ability to recover viable prp39a prp39b doubly homozygous mutants suggests that at least partial loss of function of both PRP39 paralogs is not lethal in Arabidopsis.
The flowering time of prp39a-3 and prp39-4 mutants was somewhat increased compared to the wild-type T line (on average 1–6 days, respectively, under long-day conditions). A previous study found that prp39a-1 and prp39a-2 T-DNA insertion mutants flowered on average 10 days later than wild-type plants under long-day conditions, reportedly owing to increased expression of the floral repressor FLC (Wang et al. 2007). We did not detect FLC as a differentially expressed gene or among the alternative splicing events in either the prp39a-3 or prp39a-4 mutant. The differences in delay of flowering time and changes in FLC expression in the two studies may reflect different growing conditions or the presence of second-site mutations that influence flowering time and FLC expression either independently or in conjunction with a specific prp39a allele.
From their known functions in budding yeast and mammalian cells, PRP39 and RBM25 are predicted to act at the same step of the splicing pathway, viz. during 5′ splice site recognition by the U1 snRNP. In our system, the prp39a and rbm25 mutations had opposite effects on GFP pre-mRNA splicing and consequently on GFP protein levels. The rbm25 mutants exhibited less efficient splicing of GFP pre-mRNA and higher accumulation of unspliced, untranslatable transcript, thus giving rise to a GFP-weak phenotype. Similar reduced splicing efficiency was observed in two other GFP-weak mutants retrieved in the same screen: rtf2 and prp8 (Sasaki et al. 2015; Kanno et al. 2017). By contrast, the prp39a mutants displayed preferential splicing of the AT-AC intron in GFP pre-mRNA and elevated levels of translatable GFP mRNA. The rise in GFP mRNA abundance accounts at least in part for the Hyper-GFP phenotype of prp39a mutants. The favored splicing of the AT-AC intron, which was even more pronounced in two other Hyper-GFP mutants identified in the same screen, cwc16a and smu1 (Kanno et al. 2017), is consistent with a role for these proteins in splice site choice in GFP pre-mRNA.
When reflecting on the basis of the contrasting splicing outcomes and GFP phenotypes in the rbm25 and prp39a mutants, other potential activities of U1 snRNPs can be considered. Distinct from its role in splicing, the U1 snRNP in vertebrate cells is also involved in 3′ end formation by preventing premature transcription termination and cleavage/polyadenylation (Kaida et al. 2010). Similarly, the human ortholog of RBM25 interacts with 3′ end factors and has been proposed to couple splicing to 3′-end processing (Fortes et al. 2007). Whether U1 snRNP components in plants are also involved in both splicing and 3′-end formation is not known. However, it is noteworthy that PRP39a is highly coexpressed with several cleavage and polyadenylation factors in addition to splicing proteins and other RNA-binding proteins in Arabidopsis (Table S6). Although we did not observe obvious qualitative changes in the GFP splicing pattern or in the abundance of total GFP transcripts in prp39a and rbm25 mutants, it is conceivable that PRP39a and RBM25 function to varying extents in splicing and 3′ end processing of GFP pre-mRNA, which may contribute to the opposite GFP phenotypes in the respective mutants. Alternatively, PRP39a and RBM25 may act differently in splicing in plants than in other organisms. Further work is needed to examine these possibilities.
Genome-wide analysis of splicing in prp39a mutants
Because genome-wide splicing has already been investigated in rbm25 mutants (Zhan et al. 2015), we concentrated on determining the effects of prp39a mutations on the alternative splicing profile. Among the noteworthy findings were changes in splicing-related factors including PRP39a itself, a second putative U1 snRNP component, PRP40b (Kang et al. 2009), a putative U5 RNA helicase BRR2C (Mahrez et al. 2016), and the splicing factor SCL30 (Yan et al. 2017). Cross-regulatory networks involving multiple splicing factors are well known and may help to coordinate responses of the spliceosome to various developmental and environmental signals (Barta et al. 2008). In addition, a notable number of transcription-related proteins, including GTFs, TEFs, and promoter binding proteins were detected among the alternative splicing events in prp39a mutants. These results may indicate an important role for PRP39a in proper splicing of a group of genes needed for promoter recognition as well as initiation and elongation of RNAP II transcription. A similar set of proteins involved in transcription and promoter binding was identified in a genome-wide analysis of alternative splicing in the rbm25-1 mutant (Zhan et al. 2015). The collective findings suggest that splicing of genes encoding transcription-related factors may be particularly sensitive to mutations in U1snRNP components, but more work is needed to substantiate this hypothesis.
Summary
In a forward genetic screen based on an alternatively spliced GFP reporter gene in Arabidopsis, we identified mutants impaired in putative U1 snRNP proteins PRP39a and RBM25. The contrasting effects of the respective mutations on GFP pre-mRNA splicing suggest that deficiencies of PRP39a and RBM25 can differentially affect splicing of the same RNA substrate for reasons that remain to be determined. Mutations in prp39a affect the splicing of hundreds of endogenous pre-mRNAs, including those for a number of general transcriptional proteins as well as PRP39a itself. The results pave the way for more detailed examination of the roles of PRP39a and RBM25 in the mechanism of splicing and perhaps additional aspects of pre-mRNA processing in plants.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300149/-/DC1.
Acknowledgments
We thank Shu-Jen Chou and Ai-Ping Chen at the DNA Microarray Core Laboratory of the Institute of Plant and Microbial Biology (IPMB) for preparing libraries for RNA sequencing. We are grateful to IPMB, Academia Sinica and the Taiwan Ministry for Science and Technology (grant numbers MOST 103-2311-B001-004-MY3 and MOST 105-2311-B-001-071) for financial support.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300149/-/DC1.
Communicating editor: S. Poethig
Literature Cited
- Antosz W., Pfab A., Ehrnsberger H. F., Holzinger P., Köllen K., et al. , 2017. The composition of the Arabidopsis RNA polymerase II transcript elongation complex reveals the interplay between elongation and mRNA processing factors. Plant Cell 29: 854–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin R. S., Vidaurre D., Stamatiou G., Breit R., Provart N. J., et al. , 2011. Next-generation mapping of Arabidopsis genes. Plant J. 67: 715–725. [DOI] [PubMed] [Google Scholar]
- Barta A., Kalyna M., Lorković Z. J., 2008. Plant SR proteins and their functions. Curr. Top. Microbiol. Immunol. 326: 83–102. [DOI] [PubMed] [Google Scholar]
- Cheng C., Wang Z., Yuan B., Li X., 2017. RBM25 mediates abiotic responses in plants. Front. Plant Sci. 8: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Cao X., 2017. Roles of pre-mRNA splicing and polyadenylation in plant development. Curr. Opin. Plant Biol. 35: 45–53. [DOI] [PubMed] [Google Scholar]
- Ding F., Cui P., Wang Z., Zhang S., Ali S., et al. , 2014. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genomics 15: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Dannenberg J., Dube P., Kastner B., Stark H., et al. , 2009. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell 36: 593–608. [DOI] [PubMed] [Google Scholar]
- Filichkin S., Priest H. D., Megraw M., Mockler T. C., 2015. Alternative splicing in plants: directing traffic at the crossroads of adaptation and environmental stress. Curr. Opin. Plant Biol. 24: 125–135. [DOI] [PubMed] [Google Scholar]
- Fortes P., Bilbao-Cortés D., Fornerod M., Rigaut G., Raymond W., et al. , 1999. Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev. 13: 2425–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P., Longman D., McCracken S., Ip J. Y., Poot R., et al. , 2007. Identification and characterization of RED120: a conserved PWI domain protein with links to splicing and 3′-end formation. FEBS Lett. 581: 3087–3097. [DOI] [PubMed] [Google Scholar]
- Fu J. L., Kanno T., Liang S. C., Matzke A. J., Matzke M., 2015. GFP loss-of-function mutations in Arabidopsis thaliana. G3 5: 1849–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A., Tang J., Puig O., Salgado J., Neubauer G., et al. , 1998. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4: 374–393. [PMC free article] [PubMed] [Google Scholar]
- Hammani K., Cook W. B., Barkan A., 2012. RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc. Natl. Acad. Sci. USA 109: 5651–5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James G. V., Patel V., Nordström K. J., Klasen J. R., Salomé P. A., et al. , 2013. User guide for mapping-by-sequencing in Arabidopsis. Genome Biol. 14: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D., Berg M. G., Younis I., Kasim M., Singh L. N., et al. , 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C. H., Feng Y., Vikram M., Jeong I. S., Lee J. R., et al. , 2009. Arabidopsis thaliana PRP40s are RNA polymerase II C-terminal domain-associating proteins. Arch. Biochem. Biophys. 484: 303–308. [DOI] [PubMed] [Google Scholar]
- Kanno T., Bucher E., Daxinger L., Huettel B., Böhmdorfer G., et al. , 2008. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 40: 670–675. [DOI] [PubMed] [Google Scholar]
- Kanno T., Lin W. D., Fu J. L., Wu M. T., Yang H. W., et al. , 2016. Identification of coilin mutants in a screen for enhanced expression of an alternatively spliced GFP reporter gene in Arabidopsis thaliana. Genetics 203: 1709–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Lin W. D., Fu J. L., Matzke A. J. M., Matzke M., 2017. A genetic screen implicates a CWC16/Yju2/CCDC130 protein and SMU1 in alternative splicing in Arabidopsis thaliana. RNA 23: 1068–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., 2002. BLAT – the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson B. A., 2013. Emergence and expansion of TFIIB-like factors in the plant kingdom. Gene 526: 30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Dejong F., Villacorta N., Szakonyi D., Koncz Z., 2012. The spliceosome-activating complex: molecular mechanisms underlying the function of a pleiotropic regulator. Front. Plant Sci. 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C. W., Chen Y., Shi W., Smyth G. K., 2014. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawit S. J., O’Grady K., Gurley W. B., Czarnecka-Verner E., 2007. Yeast two-hybrid map of Arabidopsis TFIID. Plant Mol. Biol. 64: 73–87. [DOI] [PubMed] [Google Scholar]
- Lockhart S. R., Rymond B. C., 1994. Commitment of yeast pre-mRNA to the splicing pathway requires a novel U1 small nuclear ribonucleoprotein polypeptide, Prp39p. Mol. Cell. Biol. 14: 3623–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrez W., Shin J., Muñoz-Viana R., Figueiredo D. D., Trejo-Arellano M. S., et al. , 2016. BRR2a affects flowering time via FLC splicing. PLoS Genet. 12: e1005924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A. G., Wang Z., 2014. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15: 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. J., Proudfoot N. J., 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700. [DOI] [PubMed] [Google Scholar]
- Nilsen T. W., Graveley B. R., 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C., Lin P. C., Nagai K., 2017. Structure of a pre-catalytic spliceosome. Nature 546: 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanouskaya T. V., Grinev V. V., 2017. The determinants of alternative RNA splicing in human cells. Mol. Genet. Genomics DOI: 10.1007/s00438-017-1350-0. [DOI] [PubMed] [Google Scholar]
- Reddy A. S., Day I. S., Göhring J., Barta A., 2012. Localization and dynamics of nuclear speckles in plants. Plant Physiol. 158: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. S., Marquez Y., Kalyna M., Barta A., 2013. Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., Oshlack A., 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Kanno T., Liang S. C., Chen P. Y., Liao W. W., et al. , 2015) An Rtf2 domain-containing protein influences pre-mRNA splicing and is essential for embryonic development in Arabidopsis thaliana. Genetics 200: 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Brown J. W., 2013. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25: 3640–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M. C., Lührmann R., 2015. SnapShot: Spliceosome Dynamics I. Cell 161: 1474–1474.e1. [DOI] [PubMed] [Google Scholar]
- Wahl M. C., Will C. L., Lührmann R., 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Wang C., Tian Q., Hou Z., Mucha M., Aukerman M., et al. , 2007. The Arabidopsis thaliana AT PRP39–1 gene, encoding a tetratricopeptide repeat protein with similarity to the yeast pre-mRNA processing protein PRP39, affects flowering time. Plant Cell Rep. 26: 1357–1366. [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu J., Huang B. O., Xu Y. M., Li J., et al. , 2015. Mechanism of alternative splicing and its regulation. Biomed. Rep. 3: 152–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Ji H., Yuan B., Wang S., Su C., et al. , 2015. ABA signalling is fine-tuned by antagonistic HAB1 variants. Nat. Commun. 6: 8138. [DOI] [PubMed] [Google Scholar]
- Will C. L., Lührmann R., 2011. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Xia X., Sun Z., Fang Y., 2017. Depletion of Arabidopsis SC35 and SC35-like serine/arginine-rich proteins affects the transcription and splicing of a subset of genes. PLoS Genet. 13: e1006663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X., Qian B., Cao F., Wu W., Yang L., et al. , 2015. An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat. Commun. 6: 8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Wu D., Kong F., Lin K., Zhang H., et al. , 2017. The Arabidopsis thaliana nuclear factor Y transcription factors. Front. Plant Sci. 7: 2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A., Ou A. C., Cho A., Benz E. J., Jr., Huang S. C., 2008. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5′ splice site selection. Mol. Cell. Biol. 28: 5924–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Seeds of prp39a and rbm25 mutants identified in this study will be available from Arabidopsis Biological Resource Center, Ohio State University, OH. Supplemental Figures: Figure S1 shows a phenotype comparison of rbm25 mutants with wild-type T. Figure S2 presents amino acid alignments of PRP39-related proteins in selected plant species. Figure S3 presents amino acid alignments of PRP39-related proteins in model organisms. Supplemental Tables: Table S1 lists primers. Table S2 shows differentially expressed genes (DEGs) shared between two independent prp39a mutants. Table S3 lists intron retention (IR) and more efficient splicing (MES) events shared between two independent prp39a mutants. Table S4 lists exon skipping (ES) events shared between two independent prp39a mutants. Table S5 lists alternative splice site donor or acceptor (SSDA) events shared between two independent prp39a mutants. Table S6 lists the top 30 coexpressed genes for PRP39a and PRP39b. RNA sequencing data used to prepare Table S2, Table S3, Table S4 and Table S5 are available under Sequence Read Archive accession number SRP108084 [samples ‘PRP39-1-3 biological replicate’ nrs. 1-3 (prp39a-3 allele) and ‘PRP39-1-4 biological replicate’ nrs. 1-3 (prp39a-4 allele) and SRP093582 (samples ‘ST biological replicate’ nrs. 1, 2 and 4 (WT T line)].