Abstract
Transcription factors (TFs) have emerged as essential cell autonomous mediators of subtype specific dendritogenesis; however, the downstream effectors of these TFs remain largely unknown, as are the cellular events that TFs control to direct morphological change. As dendritic morphology is largely dictated by the organization of the actin and microtubule (MT) cytoskeletons, elucidating TF-mediated cytoskeletal regulatory programs is key to understanding molecular control of diverse dendritic morphologies. Previous studies in Drosophila melanogaster have demonstrated that the conserved TFs Cut and Knot exert combinatorial control over aspects of dendritic cytoskeleton development, promoting actin and MT-based arbor morphology, respectively. To investigate transcriptional targets of Cut and/or Knot regulation, we conducted systematic neurogenomic studies, coupled with in vivo genetic screens utilizing multi-fluor cytoskeletal and membrane marker reporters. These analyses identified a host of putative Cut and/or Knot effector molecules, and a subset of these putative TF targets converge on modulating dendritic cytoskeletal architecture, which are grouped into three major phenotypic categories, based upon neuromorphometric analyses: complexity enhancer, complexity shifter, and complexity suppressor. Complexity enhancer genes normally function to promote higher order dendritic growth and branching with variable effects on MT stabilization and F-actin organization, whereas complexity shifter and complexity suppressor genes normally function in regulating proximal-distal branching distribution or in restricting higher order branching complexity, respectively, with spatially restricted impacts on the dendritic cytoskeleton. Collectively, we implicate novel genes and cellular programs by which TFs distinctly and combinatorially govern dendritogenesis via cytoskeletal modulation.
Keywords: transcription factors, dendrite, cytoskeleton, Drosophila, neurogenomics
NEURONS are highly polarized cells comprised of two structurally and functionally distinct processes, the axon, which relays signals to other neurons, and the dendrites, which receive signals from other neurons. Since dendrites are the primary site of synaptic input and signal integration, with dendritic size and the range of arborization patterns affecting connectivity, the regulation of dendritic growth and branching is extremely important for the establishment of functional neuronal networks (Lefebvre et al. 2015).
Genetic and molecular studies have demonstrated that the acquisition of class-specific dendrite morphologies is mediated by complex regulatory programs involving intrinsic factors and extrinsic cues (Jan and Jan 2010; Puram and Bonni 2013; Tavosanis 2014; Nanda et al. 2017). Many of these factors are part of, or activate, signaling pathways that eventually converge on the neuronal actin and microtubule (MT) cytoskeletons. These cytoskeletal elements form the scaffold around which cell shape is built, and the tracks along which intracellular components are transported (Rodriguez et al. 2003). Despite recent progress in dissecting the roles of transcription factor (TF) activity in regulating dendritic cytoskeletal architecture (Jinushi-Nakao et al. 2007; Ye et al. 2011; Iyer et al. 2012; Nagel et al. 2012), much remains unknown regarding the molecular mechanisms via which TFs spatio-temporally modulate cytoskeletal dynamics to direct developing and mature arbor morphologies (Santiago and Bashaw 2014). Understanding how such changes in cytoskeletal control lead to specific changes in emergent neuron shape can be facilitated by computational simulations (Samsonovich and Ascoli 2005), especially if directly and bidirectionally linked with imaging-driven, systems-level molecular investigations (Megason and Fraser 2007).
Intriguingly, two TFs, Cut (Ct) and Knot (Kn), have been shown to synergize in promoting class IV (CIV) da neuron-specific arbor morphology by each exerting distinct regulatory effects on the dendritic cytoskeleton (reviewed in Nanda et al. (2017)). Ct, a member of the evolutionarily conserved CUX family of TFs, is a homeodomain-containing molecule with functional roles in external sensory organ cell fate specification (Bodmer et al. 1987; Blochlinger et al. 1988, 1990), class-specific da neuron dendrite morphogenesis (Grueber et al. 2003a), and dendritic targeting of olfactory projection neurons (Komiyama and Luo 2007). Ct regulates dendritic diversity among da sensory neurons in an expression-level-dependent manner. Ct protein expression in da neurons is highest in class III (CIII) neurons, followed by medium and low expression levels in CIV and class II (CII) neurons, respectively, and is undetectable in class I (CI) neurons (Grueber et al. 2003a). Genetic disruption of ct leads to severe reductions in dendritic arbor complexity, particularly the formation of actin-rich structures such as short, unbranched dendrites. Conversely, ectopic misexpression of Ct in CI neurons results in supernumerary branching and the de novo formation of F-actin-rich dendritic filopodia converting typical CI dendritic morphology toward the characteristic features of CIII neurons (Grueber et al. 2003a). In mammals, Cux1/Cux2, the vertebrate homologs of Ct, also function in regulating dendritic branching, spine morphology, and synaptogenesis in the mammalian cortex revealing the Ct/Cux molecules have evolutionarily conserved roles in dendritic development and maturation (Cubelos et al. 2010; Li et al. 2010).
Similarly, the Collier/Olf1/EBF (COE) family TF Kn, which is exclusively expressed in CIV neurons, endows these neurons with an expansive and highly branched dendritic arbor by promoting MT-dependent branching and elongation. As with ct defects, loss of kn function in CIV neurons leads to significant reductions in dendritic growth and branching resulting in rudimentary arbor complexity, and conversely, ectopic misexpression of Kn in CI da neurons promotes supernumerary higher order branches coupled with excessive dendrite branch elongation (Hattori et al. 2007; Jinushi-Nakao et al. 2007; Crozatier and Vincent 2008).
The combinatorial action of Ct and Kn in specifying class-specific arbor shapes is achieved, at least in part, by differential regulatory effects on the F-actin and MT cytoskeletons (Jinushi-Nakao et al. 2007). Furthermore, Kn and Ct exert their effects on the dendritic cytoskeleton through primarily parallel pathways. Ct, acting via Rac1, promotes the formation of actin-rich dendritic filopodia, whereas Kn promotes the expression of the MT severing protein Spastin, which is thought to generate new sites for MT polymerization thereby promoting branch initiation, elongation, and arbor complexity (Jinushi-Nakao et al. 2007). Interestingly, the Krüppel-like TF Dar1, which is expressed in all da neuron subclasses, is required for Kn-mediated dendritogenesis and appears to restrict Spastin expression to achieve proper levels of this molecule in promoting dendritic growth (Ye et al. 2011). In CIV neurons, Kn suppresses Ct-induced actin-rich dendritic filopodial formation contributing to cell-type specific arborization, whereas in CIII neurons, Ct promotes the formation of these structures (Jinushi-Nakao et al. 2007). Moreover, Kn does not function in regulating Ct protein levels, however Ct controls the amplitude of Kn expression (Jinushi-Nakao et al. 2007). Despite recent advances, much remains unknown regarding the identity and function of putative targets of Ct and/or Kn, and while these molecules exert combinatorial synergistic effects on sculpting the dendritic cytoskeleton and promoting dendritic diversity, there are, as yet, no identified convergent transcriptional targets, nor do we have a complete picture of the potential cellular programs that these TFs modulate to direct cell-type-specific dendrite development.
Here, we address these knowledge gaps by specifically focusing on transcriptional programs that are directed by Ct and/or Kn via combined neurogenomic analyses, bioinformatics, genetic screens, and cytoskeletal reporter studies of putative target function in regulating dendritic architecture. We implicate a broad cross-section of molecules by which these important transcriptional regulators govern dendritic development and cytoskeletal regulation, and reveal unique and combinatorially regulated targets that contribute to dendritic diversification.
Materials and Methods
Drosophila strains
Drosophila stocks were reared at 25° on standard cornmeal-molasses-agar media. Fly strains used in this study were obtained from Bloomington (UAS-RNAi TRiP lines) and the Vienna Drosophila Research Center (UAS-RNAi GD and KK lines). Additional stocks included: GAL4221,UAS-mCD8::GFP; GAL4477,UAS-mCD8::GFP/CyO,tubP-GAL80;GAL4ppk.1.9,UAS-CD8::GFP; GAL4477;ppk-CD4::tdTomato; ppk-CD4::tdTomato (Han et al. 2011); ppk::EGFP (Grueber et al. 2003b); UAS-ct/CyO,tubP[GAL80]; GAL4221,UASmCD8::GFP; UAS-kn/CyO,tubP[GAL80];GAL4221,UASmCD8::GFP; UAS-GMA::GFP; GAL4477,UAS-mCherry::Jupiter (Trunnell et al. 2015); w, elavC155-GAL4,UASmCD8GFP,hsFLP; FRTG13,tubP-GAL80 (2R MARCM); P{ry[+t7.2]=hsFLP}1,P{w[+mC]=tubP-GAL80}LL1, w[*], P{ry[+t7.2]=neoFRT}19A; P{w[+mW.hs]=GawB}109(2)80, P{w[+mC]=UAS-mCD8::GFP.L}LL5, P{w[+mC]=SOP-FLP}73/CyO (X MARCM; Shimono et al. 2014); P{w[+m*]=GAL4}5-40, P{w[+mC]=UAS-Venus.m}1, P{w[+mC]=SOP-FLP}42; P{ry[+t7.2]=neoFRT}42D, P{w[+mC]=tubP-GAL80}LL2/CyO, y[+] (2R MARCM; Shimono et al. 2014); P{w[+m*]=GAL4}5-40, P{w[+mC]=UAS-Venus.m}1, P{w[+mC]=SOP-FLP}42; P{ry[+t7.2]=neoFRT}82B, P{w[+mC]=tubP-GAL80}LL3/TM6C, Sb[1], Tb[1] (3R MARCM; Shimono et al. 2014); Oregon-R was used as a wild-type strain. As available, a minimum of two gene-specific UAS-RNAi lines were used for mitigating off-target effects, and crosses were performed at 29°. Supplemental Material, Table S1 provides a list of all genotypes by figure and panel. Table S2 provides a list of all UAS-IR transgenes, mutant alleles, and overexpression transgenes used in this study.
Cell isolation, purification, microarray expression profiling, and qRT-PCR
The isolation and purification of da neurons was performed as previously described (Iyer et al. 2009; S. C. Iyer et al. 2013; E. P. Iyer et al. 2013) with minor modifications . Briefly, 40–50 age-matched third instar larvae expressing mCD8::GFP under the control of the class I GAL4221 driver, in the presence or absence of UAS-ct or UAS-kn, were collected and washed several times in ddH20. The larvae were then rinsed in RNAse AWAY and double-distilled water (ddH20) and finally dissected. The tissue was then dissociated to yield single cell suspensions, which were filtered using a 30-µm membrane. The filtrate was then incubated with superparamagnetic beads (Dynabeads MyOne Streptavidin T1, Invitrogen, Carlsbad, CA) coupled with biotinylated mouse anti-CD8a antibody (eBioscience) for 60 min. Finally, the da neurons plus magnetic bead complex were separated using a powerful magnetic field. The isolated neurons were washed at least five times with PBS to remove any potential nonspecific cells, and the quality and purity of isolated neurons was assessed under a stereo-fluorescent microscope equipped with phase contrast for examining the number of fluorescent (GFP-positive) vs. nonfluorescent (GFP-negative) cells. Only if the isolated cells were free of cellular debris and nonspecific (i.e., nonfluorescing) contaminants were they retained. The purified class I neuron populations (control; ectopic Ct; ectopic Kn) were then lysed in SuperAmp (Miltenyi Biotec) RNA lysis buffer followed by storage at −80°. Messenger RNA (mRNA) isolation, amplification, labeling, and microarray hybridization were conducted by Miltenyi Biotec. Two hundred fifty nanograms of complementary DNAs (cDNAs) were used as a template for Cy3 labeling followed by hybridization to Agilent whole Drosophila melanogaster genome oligo 4 × 44 K microarrays. All microarray analyses were performed in triplicate. qRT-PCR analyses were performed in quadruplicate as previously described (S. C. Iyer et al. 2013) using prevalidated Qiagen QuantiTect Primer Assays (Qiagen, Germantown, MD) for the following genes: cut (QT00501389), dmn (QT00500717), cpb (QT00927731), CCT2 (QT00918806), wdb (QT00984067), Cdlc2 (QT00927605), RhoGAP18B (QT00925113), cpa (QT00951342), ctp (QT00498183), RpL36A (QT00932155), msps (QT00975919), SkpA (QT00495845), knot (QT00502005), Ank2 (QT00960064), T-cp1 (QT00980007), and RpL7 (QT00932155). Expression data were normalized against RpL32 (QT00985677) or GAPDH2 (QT00922957) and are reported as the mean fold change in expression.
Neurogenomic analyses
Bioinformatic and statistical analyses of microarrays were performed essentially as previously described (Bhattacharya et al. 2014) with additional statistical analyses (described below). Briefly, Agilent Feature Extraction Software was used to read out and process the triplicate microarray image files. The software was used to determine feature intensities and perform background subtraction, reject outliers, and calculate statistical confidences. The raw data were quantile normalized and only those gene probes which are flagged as positive and significantly expressed above background are selected for further analysis. Microarray data, including metadata, raw data, and quantile normalized data sets have been deposited into the Gene Expression Omnibus (GEO) under accession number GSE83938. Differential expression analysis is then performed on these normalized data using three different methods: t-test, Limma (Ritchie et al. 2015), and GeneSpring GX (Agilent Technologies). While t-test is a hypothesis test, Limma uses linear models, with empirical Bayesian methods to get the differentially expressed genes. GeneSpring, on the other hand, uses ANOVA to perform these analyses. The output change in differential expression is fold change for GeneSpring and Limma, whereas for t-test it is t-value. Adjusted P-value was calculated for t-test analysis using p.adjust function in R, whereas GeneSpring and Limma provide adjusted P-value as an output. A threshold false discovery rate (FDR) corrected P-value of <0.05 (Benjamini and Hochberg 1995) and a fold change of > +1 (for overexpressed) and < −1 (for underexpressed) were considered for Limma and GeneSpring, whereas for the t-test a t-value of > +1 and value < −1 were considered. Genes discovered in >1 method were considered for further analysis. The functional annotation tool DAVID (Huang et al. 2009a,b) was used to cluster targets based on their biological functions.
Phenotypic screening and live image confocal microscopy
Virgin GAL4477,UAS-mCD8::GFP/CyO,tubP-GAL80; GAL4ppk.1.9,UAS-mCD8::GFP (CIV-GAL4) were crossed to individual, gene-specific UAS-RNAi transgenic males (Table S2) or out-crossed to wild-type Oregon-R males as control, followed by rearing at 29°. Each gene-specific UAS-RNAi strain was assigned a randomly generated numerical code and screening was conducted double-blind to the identity of the gene targeted by the UAS-RNAi construct. MARCM clonal and mutant allele phenotypic analyses were performed as previously described and compared to their respective controls (S. C. Iyer et al. 2013; Gokhale et al. 2016). Six to ten fluorescent third instar larvae were analyzed by genotype via live image confocal microscopy and representative image data were collected. For live confocal analyses, larvae were placed on a microscope slide, immersed in 1:5 (v/v) diethyl ether to halocarbon oil and covered with a 22 × 50 mm glass coverslip. Neurons expressing fluorescent protein transgenes were visualized on a Zeiss LSM 780 confocal microscope. Images were collected as z-stacks using a 20× dry objective at a step-size of 1.0–2.0 µm and 1024 × 1024 resolution.
Neurometric quantification
Maximum intensity projections of the Z-stacks were exported as a jpeg or TIFF using Zen-blue software. Once exported, images were manually curated to eliminate nonspecific auto-fluorescent spots such as the larval denticle belts using a custom designed program, Flyboys. Images were processed and skeletonized as previously described (E. P. Iyer et al. 2013). Quantitative neurometric information including total dendritic length and total dendritic branches was extracted and compiled using custom Python algorithms freely available upon request. The custom Python scripts were used to compile the output data from the Analyze Skeleton ImageJ plugin and the compiled output data were imported into Excel (Microsoft). Neurometric data were analyzed in Microsoft Excel and statistical tests were performed and plotted in GraphPad Prism 7. For Sholl analysis, we used NeuronStudio (Wearne et al. 2005) to plot the density profiles of branches as a function of distance from the cell soma and to determine the peak of maximum branch density (critical value/# of intersection) and its corresponding radius. Proportion coverage analyses were performed using an internal coverage macro plugin for ImageJ (https://github.com/JamesCSears/Internal-Coverage-Macro) (Sears and Broihier 2016). In these analyses, we employed a 10 × 10 pixel grid on the image, and quantification of coverage is defined as the proportion of grid boxes in which there is dendritic arbor against the total number of grid boxes. For reversed Strahler analysis of dendritic membrane images, we used a FIJI plugin (http://fiji.sc/Strahler_Analysis) to analyze the skeletonized images by iteratively pruning terminal branches and counting branch number in each iteration, where terminals are defined as branch order 1.
Next generation multichannel neuronal reconstructions, benchmarking, and visualizations
The two channel (GFP for F-actin and RFP for MT) image stacks (.czi file format) of da neurons from all the genetic lines were first processed in FIJI (Schindelin et al. 2012) where a third pseudochannel was created by adding the signals from the two original channels. These new files with three channels were then imported to Vaa3D (Peng et al. 2014), and the third pseudochannel was manually reconstructed into the SWC file format (Cannon et al. 1998). These initial traced SWC files and the image stacks were then reopened in Neutube (Feng et al. 2015), and additional tracing, editing, and quality checks were conducted. Remaining topological errors were programmatically repaired in batch, by building small custom scripts within the TREES toolbox (Cuntz et al. 2010) package in the MATLAB environment (MathWorks, Natick, MA). The corrected reconstruction files and the image stacks were used as input in Vaa3D plugin to create multichannel ESWC files that represent the morphology along with the intensity and volume occupied by each channel. Then the internal and external structural features were quantified using L-Measure (Scorcioni et al. 2008). Reversed Strahler order based cytoskeletal quantification of ESWC files was carried out in a new analysis program built using TREES toolbox functions (Cuntz et al. 2010). MT or F-actin quantity of a compartment is defined as (the relative signal intensity of the compartment) × (the volume of the compartment) × (the fraction of the volume occupied by the MT or F-actin signal). Total relative quantity of MT or F-actin has been quantified against path distance from the soma at 40-μm intervals (binning).
To perform benchmark tests of whether the multichannel plugin measures the intensity values correctly for arbor-wide cytoskeletal distributions, we created an artificial three-channel image stack (representing MTs, F-actin, and a third channel, which is a merge of the first two) containing a surrogate dendritic branch (cylinder). We assigned arbitrary intensity values (between 0 and 255) to all of its voxels. The voxels in the image stack are isotropic in shape and 1 μm in length in each of its three dimensions. The overall image stack dimensions are 512*512*7, and the length of the surrogate cylinder is 100 μm, with the radius of 2.5 μm. Since we assigned the voxel intensities ourselves, we knew the intensity values (between 0 and 255) of all the voxels that make up that cylinder. We then traced the artificial dendritic cylinder and created an ESWC file using our multichannel plugin. We then compared the intensity distribution (against path distance) of the generated ESWC (for both the green/F-actin and the red/MT channels) against known distributions of the artificially created surrogate cylinder (Figure S7, A–D in File S1).
To generate visual representations showing the relative distributions of F-actin and MT cytoskeletal elements in a binary fashion, as either high vs. low (see Figure S6, M and N in File S1) by genotype, we created a total of four (2*2) possible combinations (MT and F-actin high; MT high and F-actin low; MT low and F-actin high; MT and F-actin low). Here, we normalize each compartment’s intensity based on the relative volume (values ranging from 0 to 1) occupied by the particular signal and its normalized average voxel intensity (0–255, normalized to 0–1) within that compartment. For example, if a GFP/F-actin signal occupies 100% of the overall structure of a dendritic compartment (i.e., the GFP signal is present in all of the voxels that make up that dendritic compartment), and all the voxels have the maximum intensity value of 255, then the F-actin intensity value assigned to that compartment is 1*1 = 1. If the signal occupies 50% of the overall compartment, and the average intensity is 127.5 (half of maximum possible intensity), then F-actin intensity value assigned to that compartment is 0.5*0.5 = 0.25. The volume of the compartment is represented by the thickness of the branches. Each dendritic (or somatic or axonal) compartment in a neuron reconstruction is a cylinder defined by two points/two rows (start point and end point of the cylinder) and the radius value of the end point in basic SWC files (Cannon et al. 1998).
Statistics
Error bars reported in the study represent SEM. Analyses were performed using either ANOVA with correction for multiple comparisons using FDR of Benjamini, Kriega, and Yekutieli, or Student’s t-test for comparing two groups. All data sets were tested for normality (Shapiro–Wilk normality test) and homogeneity of variance (Bartlett’s test or F test) before statistical analysis. Heatmap to show sample correlation was done with gplots (Warnes et al. 2016) package of R Core Team (2014). Significance scores were: */# P < 0.05, ** P < 0.01, *** P < 0.001.
Data availability
All the new genotypes presented here are available upon request. The microarray data are available for download at GEO GSE83938. Digital reconstructions of neuronal morphology have been deposited into the NeuroMorpho.Org database (Ascoli 2006) for public distribution under the Cox and Ascoli archives.
Results
Neurogenomic dissection of Ct and Kn transcriptional effectors
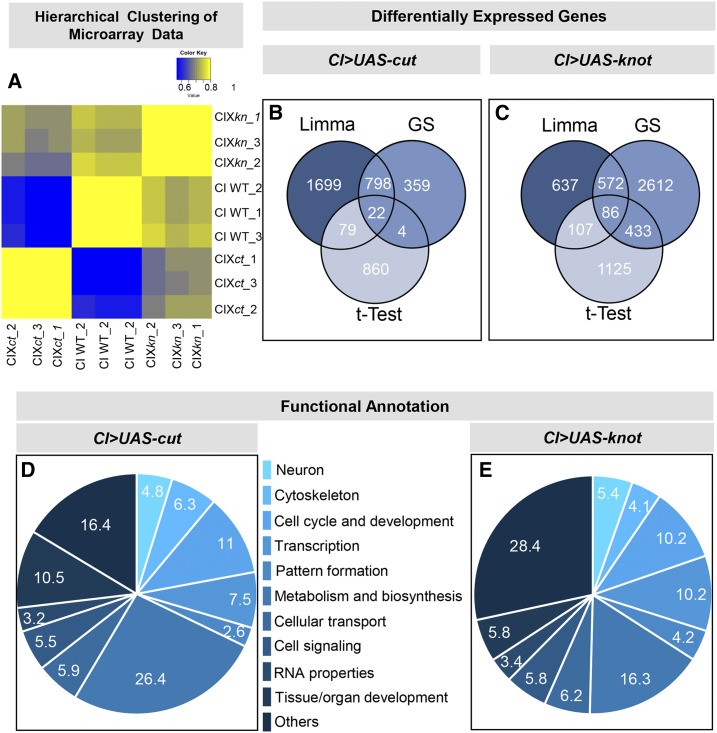
The TFs Ct and Kn have been demonstrated to regulate dendritic morphogenesis and to contribute to dendritic diversification among complex CIII and CIV da neurons (reviewed in Tavosanis (2014), Nanda et al. (2017)). Moreover, recent advances have begun to dissect the regulatory programs these TFs govern in directing dendritogenesis, including cytoskeletal regulation. While previous studies have investigated Kn transcriptional targets via DamID analyses of in vivo Kn binding sites in embryos and microarray analyses of pan-da overexpression of Kn (Hattori et al. 2013), genome-wide analyses of Ct-mediated transcriptional targets in da neurons have not been reported. To extend these previous analyses, we implemented unbiased genome-wide neurogenomic analyses of transcriptional regulatory programs mediated by Ct and/or Kn that operate in da neurons to modulate dendritic architecture. The overall neurogenomics strategy is summarized in Figure S1 in File S1. We capitalized on the observation that neither Ct nor Kn are normally expressed in CI da neurons and thus ectopic misexpression of Ct or Kn in CI neurons provides a platform for comparative analyses of Ct- or Kn-mediated gene expression relative to control CI neurons. This strategy avoids potential confounds that may arise from overexpression in all da neurons or individual classes that normally express Ct or Kn. Microarray analyses were conducted in triplicate with a high degree of correlation between replicates, and with Ct- or Kn-expressing CI neuron profiles exhibiting higher correlation levels to each other, relative to control CI neurons (Figure 1A). Differential expression analyses of microarray gene expression were performed using three different methods: (i) a two sample Hypothesis test (t-test); (ii) a one-way ANOVA followed by a Fold Change Calculation (GeneSpring); and (iii) a linear model fit approach (Linear Model for Microarray Data; Limma) (Ritchie et al. 2015). Venn diagrams reveal the number of common and unique differentially expressed target genes identified by these three distinct statistical methods for Ct-expressed genes (Figure 1B and File S2) or Kn-expressed genes (Figure 1C and File S3). Differences between the intersections of the statistical analyses are due to the nature of the statistical analysis performed and variability in stringency. Genes appearing in >1 statistical method were considered as eligible candidates for further analyses. The differentially expressed genes from the Kn microarray analyses were also cross-referenced with DamID data sets (Hattori et al. 2013) for TF binding to the putative genes’ promoter region. These analyses revealed that ∼27% of the genes identified from our Kn microarray analyses overlapped with the DamID data set shedding potential regulatory insights on putative effector genes as direct vs. indirect Kn targets (File S4). The differentially expressed genes lists were then used as an input for functional annotation using DAVID to identify enriched cellular programs or biological processes (File S2 and File S3). Gene Ontology (GO) analyses conducted via DAVID identified enrichment for genes implicated in a variety of cellular and biological processes including neuronal and cytoskeletal functions, which were of particular interest for this study given the roles of Ct and Kn in modulating dendritic architecture and the importance of cytoskeletal architecture in directing dendrite morphology (File S2 and File S3). To simplify the visualization, we clustered GO functional annotation terms for Ct and Kn under broader parent terms (Figure 1, D and E). From the lists of Ct and/or Kn differentially expressed genes that satisfied the above described statistical considerations and were linked to GO terms for neuronal and/or cytoskeletal function, we narrowed the pool of candidates for more detailed analyses to 49 putative target genes. These putative targets were selected based upon sampling genes involved in various aspects of neuronal and cytoskeletal development/function and omitting genes that had been previously characterized with respect to functions in larval dendritic development to avoid duplication (Table S2). A summary of major GO terms associated with these genes reveals their links to neuronal and cytoskeletal processes, as well as additional cellular functions for select genes that are associated with multiple GO terms (Figure S2 in File S1).
Figure 1.
Neurogenomic analyses of Ct- and Kn-mediated transcriptional programs. (A) Heatmap (Pearson’s correlation) of the triplicate microarray data (CI WT, CI X kn, and CI X ct) reveals all arrays segregate into three well-defined and distinct clusters with high interarray correlation between replicates. Ectopic expression profiles for Ct or Kn are anticorrelated to controls and show a higher degree of correlation relative to each other. (B and C) Venn diagram of the three statistical tests [Limma, GeneSpring (GS), and t-Test] used for the differential expression analyses of the microarray data. (D and E) Functional annotation analyses of Ct and Kn differentially expressed genes.
Genetic screen and phenotypic analyses of neurogenomic targets of Ct and Kn
The candidate target genes identified from neurogenomic and statistical analyses were functionally validated via an in vivo RNAi genetic screen coupled with mutant and overexpression analyses for selected genes. Given that these candidate genes were identified via ectopic expression in CI neurons, we sought to test their putative functional roles in CIV neurons that normally express both Ct and Kn. To avoid any screening bias, all transgenic RNAi lines were screened double-blind to the identity of the gene being analyzed and phenotypic analyses were performed at the third instar larval stage of development. In this study, we recorded over 1100 neuronal CIV images for knockdown phenotypes coupled with neuromorphometric analyses. The key morphological features that were analyzed included total dendritic length, total dendritic branches, number of branches as a function of distance from the soma (Sholl), field coverage (proportion covered), and branch order distribution (reversed Strahler).
Genetic screening identified putative target genes that were differentially regulated by Ct and/or Kn and that either suppressed or enhanced Ct- and/or Kn-mediated regulation of CIV da neuron dendrite morphology. The genes uncovered in our screen cover a broad range of biological functions including cytoskeletal regulation, ribosomal regulatory function, neurogenesis, MT-based transport, autophagy, dendritic development, and chaperonin activity (Figure S2 in File S1). Collectively, quantitative neuromorphometric analyses revealed that ∼70% of the genes screened had a significant phenotypic effect on CIV dendritic architecture (Figure S3 in File S1). Among these phenotypic hits, we identified groups of genes where knockdown resulted in one of three major phenotypic categories based on defects observed in dendritic arborization and denoted these as follows: (1) complexity enhancer; (2) complexity shifter; and (3) complexity suppressor. These putative downstream Ct and/or Kn target effector molecules were then selected for further analyses.
Complexity enhancer genes
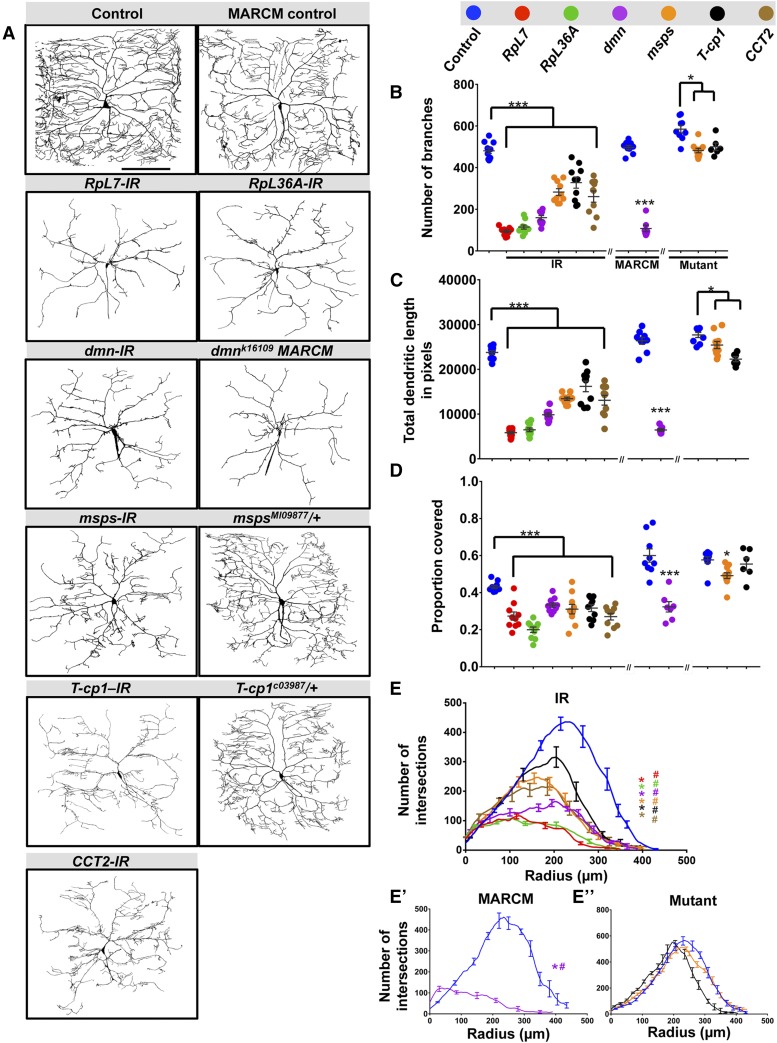
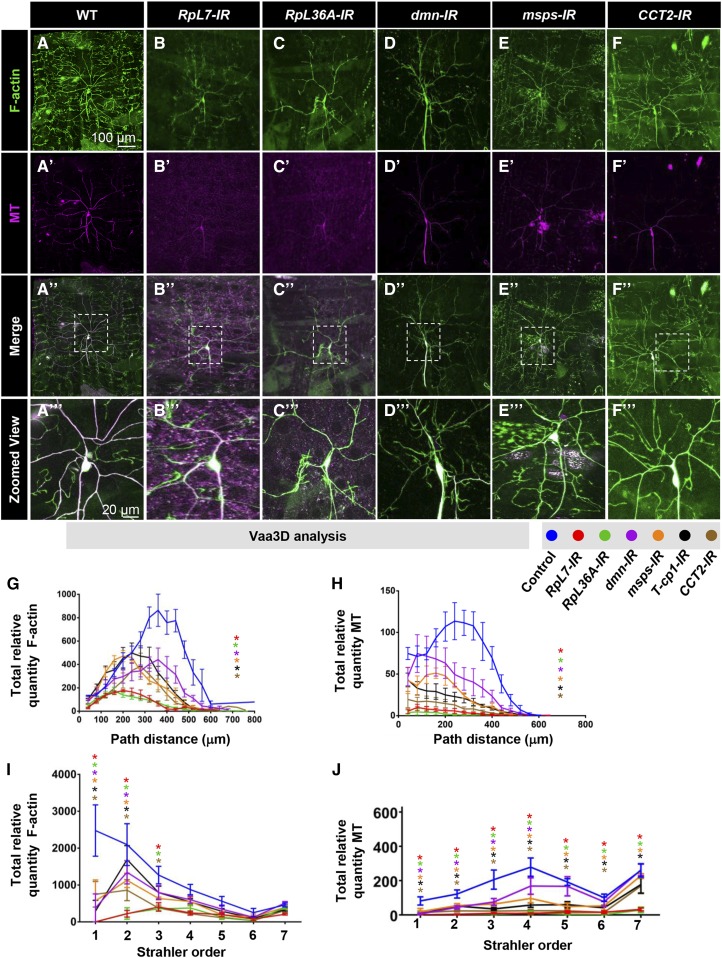
Disruption of genes that fall in this category (RpL7, RpL36A, dmn, msps, T-cp1, and CCT2) leads to a severe reduction in total dendritic complexity, which is manifested by the reductions in both total dendritic branches and concomitant reductions in total dendritic length, as well as proportion coverage (Figure 2, A–D). Morphologically, RpL7-IR, RpL36A-IR, and dmn-IR knockdowns produced similar reductions in arbor morphology, characterized by a loss of higher order branching (Figure 2A). Moreover, knockdown of these genes led to the presence of short, fine dendrites emanating from lower order branches in the region proximal to the soma, whereas higher order branching distal to the soma (termini) was suppressed relative to controls (Figure 2A and Figure S4A in File S1). For example, RpL7-IR, RpL36A-IR, and dmn-IR neurons completely lacked Strahler order 1 (terminal) dendrites as compared to controls (Figure S4A in File S1). Consistent with these findings, analyses of proportion covered likewise revealed significant reductions in field coverage (Figure 2D). While no mutant alleles are available for RpL7 or RpL36A, MARCM clonal analyses of dmn mutants revealed a phenocopy of defects observed with dmn-IR (Figure 2, A–D). Similarly, knockdown of msps, T-cp1, and CCT2 led to similar dendritic defects that likewise suppressed higher order dendritic terminal branching, overall growth, and proportion coverage, but exhibited clustered interstitial branching at an intermediate location along the proximal–distal axis relative to the soma (Figure 2, A–D and Figure S4A in File S1). Although no loss-of-function CCT2 alleles were available, we analyzed heterozygous mutant alleles for msps and T-cp1 as homozygotes were lethal prior to the third larval instar stage. Phenotypically, these heterozygous alleles were not as severe as the RNAi knockdowns (Figure 2A), although both msps and T-cp1 heterozygotes exhibited reductions in dendritic branching and total growth (Figure 2, B and C); however, only msps heterozygotes had a reduction in proportion coverage (Figure 2D).
Figure 2.
Phenotypic analyses of Ct and Kn neurogenomic targets: Complexity enhancer category. (A) Representative images of dendritic arborization in controls, gene-specific RNAi knockdowns of CIV (ddaC) neurons (IR), MARCM clones or heterozygous mutant alleles. (B and C) Quantitative analyses measuring number of branches (B) and total dendritic length (C), for RNAi knockdown (IR), MARCM, and mutant analyses. (D) Quantification of the proportion of 10 × 10 pixel squares covered by dendrite for a quadrant of the overall dendritic arbor (upper right quadrant relative to the cell body). (E–E”) Sholl analysis profiles where values are the mean (± SEM) for the number of intersections as a function of radius distance (Euclidean) from the cell body (zero), where * and # refer to the significance scores for the critical value and the corresponding radius, respectively. Statistical tests performed in: (B–E”) one-way ANOVA with FDR correction of Benjamini, Kriega, and Yekutieli. N = 6–10 neurons, and significance scores were: * P < 0.05, *** P < 0.001, # P < 0.05. Bar, 200 µm.
To quantify effects on dendritic branch distribution, Sholl analyses were used to plot the density of profiles of branches as a function of distance from the soma and compare the peak of maximum branch density (critical value) and its corresponding radius. Both parameters were dramatically reduced in all complexity enhancer gene knockdowns as compared to control (Figure 2E). RpL7-IR, RpL36A-IR, dmn-IR, and dmn MARCM clones all exhibited a strong proximal shift in the critical value and corresponding radius (Figure 2, E and E’) whereas msps-IR, T-cp1-IR, and CCT2-IR displayed a more moderate, albeit significant, proximal shift in the critical value and corresponding radius consistent with the phenotypic differences observed (Figure 2E). In contrast, the msps and T-cp1 heterozygous mutants did not reveal any significant change in measured Sholl parameters (Figure 2E’’).
As compared to the loss-of-function effects for these complexity enhancer genes, overexpression analyses of available transgenes for RpL7, RpL36A, dmn, and CCT2 revealed no obvious effects on overall dendritic growth and branching of CIV neurons (Figure S4, G and H in File S1), which may be consistent with the fact that these molecules are components of multi-protein macromolecular complexes.
To determine if these genes may also regulate axonal development, we examined CIV axonal patterning in the ventral nerve cord (VNC). Analysis of RpL7-IR and RpL36A-IR revealed variable reductions in CIV axon terminal signal intensities in the VNC; however, overall patterning of longitudinal and commissural axon tracts is relatively consistent with controls indicating that these genes may disrupt terminal axon branching/elaboration, but do not appear to disrupt axon pathfinding (Figure S5, A–C in File S1). In contrast, disruptions in dmn, msps, T-cp1, and CCT2 do not appear to have any gross defects in CIV axon patterning (Figure S5, A and D–G in File S1) suggesting the phenotypic defects are more specific to dendritic arborization. Collectively, these data indicate that RpL7, RpL36A, dmn, msps, T-cp1, and CCT2 promote the appropriate number and positions of branches along the proximal-distal axis of dendritic arbors, and are required to promote higher order branches.
Complexity shifter genes
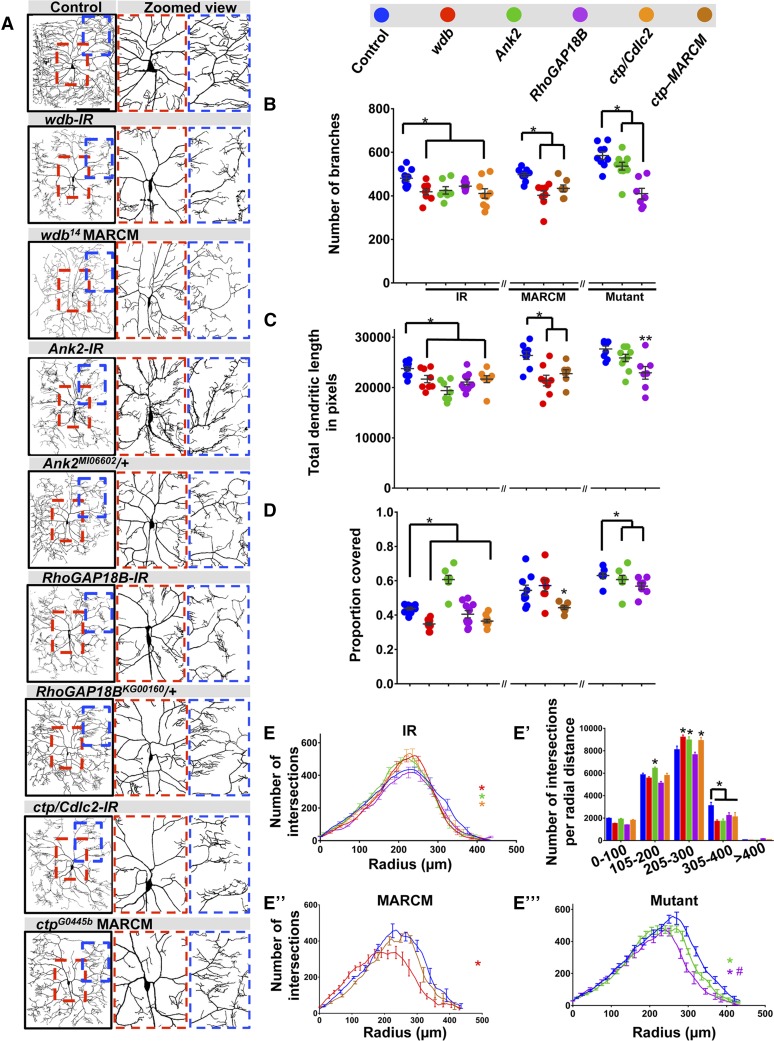
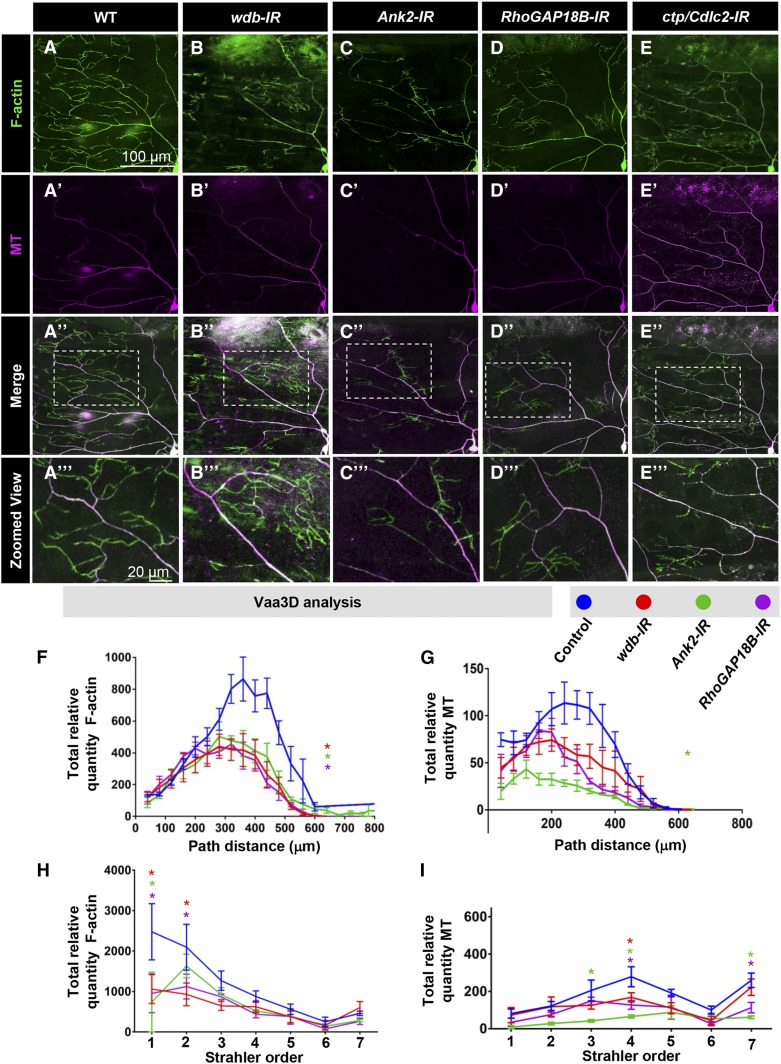
In contrast to the defects observed with the complexity enhancer gene group in which dendritic terminal branching is suppressed resulting in a loss of high order branching, we identified another set of genes (wdb, Ank2, RhoGAP18B, and ctp/Cdlc2) that when disrupted by RNAi knockdown led to a distal shift in iterative branching favoring clustered tufting of short dendrites near terminals and reduced interstitial branching proximal to the cell body (Figure 3A). Within this gene group, the one exception is Ank2 disruption, which displayed an increase in aberrant short dendritic branches emanating first and second order branches relative to the cell body (Figure 3A). Morphometric analyses revealed that RNAi knockdown of all genes in this category exhibit significant reductions in the total number of branches, which was also observed in MARCM clones (wdb, ctp) or heterozygous mutants (Ank2, RhoGAP18B) (Figure 3B). Similarly, with the exception of Ank2 heterozygotes, loss-of-function analyses for the other genes in this category revealed reductions in total dendritic length (Figure 3C). With respect to altered branching proximal to the cell body, proportion coverage analyses revealed reductions in dendritic field coverage for most loss-of-function conditions with the exception of Ank2-IR where an increase in coverage was observed consistent with the knockdown phenotype (Figure 3D). Similar to the loss-of-function effects observed with wdb and ctp, overexpression of these genes in CIV neurons resulted in reductions in dendritic branching and overall growth, with features of reduced branching near the cell body in favor of clustered short dendrites at intermediate locations between the lower order branches and the dendritic terminals (Figure S4, D–F in File S1).
Figure 3.
Phenotypic analyses of Ct and Kn neurogenomic targets: Complexity shifter category. (A) Representative images of dendritic arborization in control, gene-specific RNAi knockdowns of CIV (ddaC) neurons (IR), MARCM clones, or heterozygous mutant alleles. (B and C) Quantitative analyses measuring number of branches (B) and total dendritic length (C), for RNAi knockdown (IR), MARCM and mutant analyses. (D) Quantification of the proportion of 10 × 10 pixel squares covered by dendrite in a 400 × 400 pixel region defined around the cell body. (E, E”, and E’’’) Sholl analysis profiles where values are the mean (± SEM) for the number of intersections as a function of radius distance (Euclidean) from the cell body (zero), where * and # refer to the significance scores for the critical value and the corresponding radius, respectively. (E’) Sholl profiles for IR data plotted as histogram to reflect the total number of intersections per corresponding radial distance (Euclidean) from the soma highlighting local effects on branch distributions. Statistical tests performed in: (B–E”’) one-way ANOVA with FDR correction of Benjamini, Kriega, and Yekutieli. N = 7–10 neurons, and significance scores were: * P < 0.05, ** P < 0.01, # P < 0.05. Bar, 200 µm.
A characteristic phenotypic feature of genes in this category is the shift in the dendritic branch distribution pattern as a function of distance from the soma with RNAi-mediated knockdown. In addition to standard Sholl plots depicting changes in the critical value or corresponding radius (Figure 3, E–E’’’), we also plotted the RNAi data as the number of intersections per radial distance from the soma to enhance visualization of the local effects on branch distributions (Figure 3E’). These analyses revealed variable effects depending upon the nature of the genetic manipulation (RNAi/MARCM/heterozygous mutant) relative to their respective controls, though in all cases there were alterations in branch distribution for these genes. Analyses of wdb-IR, Ank2-IR, and ctp/Cdlc2-IR revealed an increase in the critical value of peak branch density relative to controls (Figure 3E). The increase in branch density appears to occur primarily in the 205- to 300-μm radial distance from the soma, whereas a decrease in branch density occurs in the 305- to 400-μm radial distance from the soma (Figure 3E’). In contrast, mutant analyses of wdb MARCM clones and Ank2 or RhoGAP18B heterozygous mutants revealed reductions in the critical value, and in the case of RhoGAP18B, there was also a proximal shift in the corresponding radius (Figure 3, E’’ and E’’’). In contrast to Sholl analyses, reversed Strahler analyses of the genes in this category did not identify any significant changes in branch order suggesting the altered morphology is predominantly due to effects on branch order distribution (Figure S4B in File S1).
Relative to the defects observed in dendritic arborization, analyses of CIV axon patterning for wdb-IR, RhoGAP18B-IR, and ctp/Cdlc2-IR revealed largely unaffected longitudinal and commissural axon tracts, whereas Ank2-IR appear to have some mild disruptions (Figure S5, A and H–K in File S1).
Complexity suppressor genes
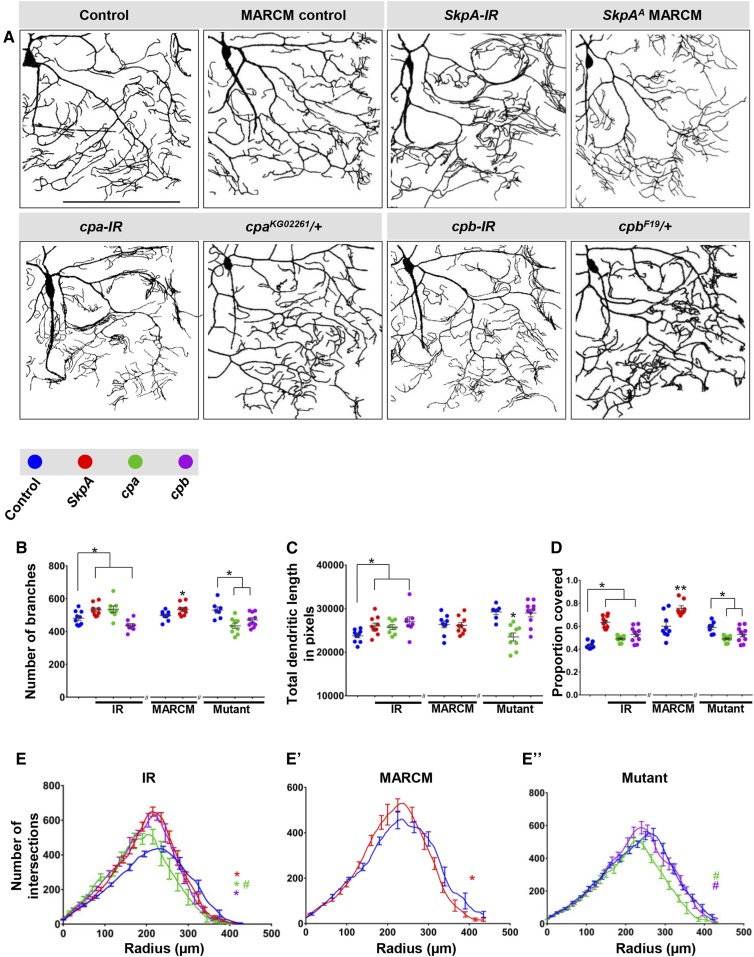
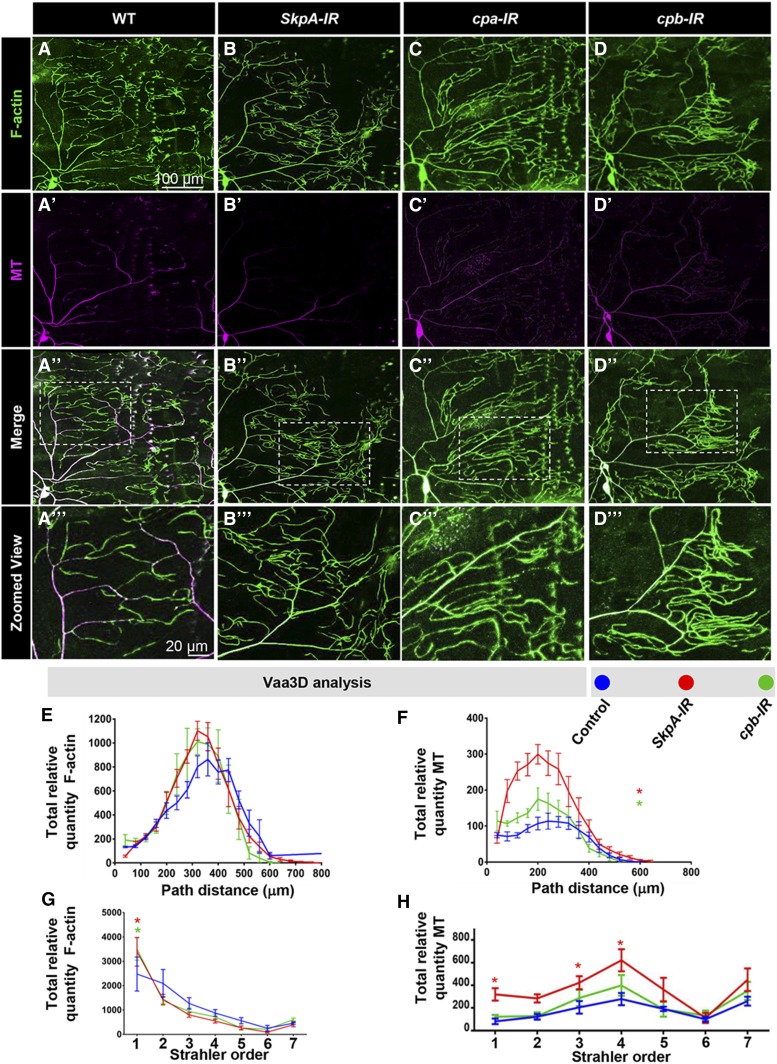
In contrast to the other two phenotypic categories, we also identified a subset of genes that when disrupted enhanced dendritic arborization (Figure 4A and Figure S4C in File S1). Knockdowns of the SCF ubiquitin ligase complex component SkpA resulted in an increase in the number of dendritic branches, total dendritic length, and proportion coverage, which was also observed, with the exception of total dendritic length, for SkpA mutant MARCM clones (Figure 4, B–D). We also identified two F-actin capping molecules, cpa and cpb, which exhibited variable effects between RNAi knockdowns and heterozygous mutant genetic backgrounds relative to their respective controls. In the case of cpa-IR, we observed increases in number of branches, total dendritic length, and proportion coverage, whereas, surprisingly, cpa heterozygotes exhibited reductions in these three categories (Figure 4, B–D). Similar to what was observed with cpa, for cpb-IR there was an increase in total dendritic length and proportion coverage; however, in cpb heterozygotes there was a reduction in the number of branches and proportion coverage (Figure 4, B–D). The differences in phenotypic effects observed for cpa and cpb disruptions may be due to variability between the genetic controls to which each is compared or perhaps the degree to which gene function is inhibited in the different genetic backgrounds (RNAi vs. heterozygous mutant). In contrast to loss-of-function effects, neither SkpA nor cpb overexpression had any significant effect on CIV dendritic growth or branching (Figure S4, G and H in File S1).
Figure 4.
Phenotypic analyses of Ct and Kn neurogenomic targets: Complexity suppressor category. (A) Representative images of dendritic arborization in controls, gene-specific RNAi knockdowns of CIV (ddaC) neurons (IR), MARCM clones, or heterozygous mutant alleles. (B and C) Quantitative analyses measuring number of branches (B) and total dendritic length (C), for RNAi knockdown (IR), MARCM, and mutant analyses. (D) Quantification of the proportion of 10 × 10 pixel squares covered by dendrite for a quadrant of the overall dendritic arbor (upper right quadrant relative to the cell body). (E–E”) Sholl analysis profiles where values are the mean (± SEM) for the number of intersections as a function of radius distance (Euclidean) from the cell body (zero), where * and # refer to the significance scores for the critical value and the corresponding radius, respectively. Statistical tests performed in: (B–E”) one-way ANOVA with FDR correction of Benjamini, Kriega, and Yekutieli. N = 7–10 neurons, and significance scores were: * P < 0.05, ** P < 0.01, # P < 0.05. Bar, 200 µm.
Sholl analyses revealed a significant increase in the peak of maximum branch density for SkpA-IR, SkpAA MARCM clones, cpa-IR, and cpb-IR compared to control (Figure 4, E and E’). With respect to cpa and cpb heterozygous mutants, there was no change in the critical value, but a significant shift in the corresponding radius where maximum branch density occurs (Figure 4E’’). Consistent with the observed phenotypic effects, reversed Strahler analyses revealed excessive higher order branching for these genes (Figure S4C in File S1).
Analyses of CIV axon projections for SkpA-IR revealed thickened longitudinal and commissural axon tracts (Figure S5L in File S1), suggesting axon terminal branching may be overelaborated, whereas cpa-IR exhibited thickened commissural axons (Figure S5M in File S1) and cpb-IR appears largely unaffected (Figure S5N in File S1).
Phenotypic and molecular validation of putative Ct and Kn effector molecules
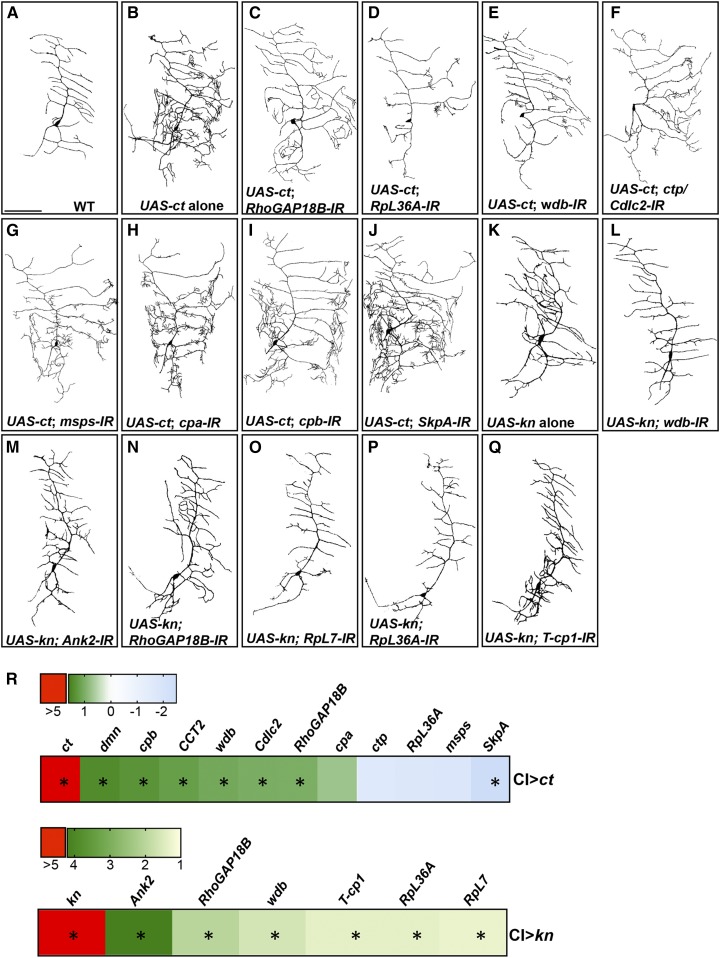
We hypothesized that if Ct or Kn functionally require these putative target genes for dendritogenesis, then knockdown of the target gene should exhibit a suppression, or possibly enhancement, of the dendritic phenotype that results from ectopic misexpression depending on the nature of the regulatory relationship (positive vs. negative) between the TF and the target gene. To test this hypothesis, CI neurons ectopically misexpressing Ct or Kn were phenotypically compared to CI neurons in which Ct or Kn were ectopically overexpressed with simultaneous expression of gene-specific RNAi for putative target genes. Consistent with this prediction, phenotypic analyses of putative Ct targets revealed that knockdown of RhoGAP18B, RpL36A, ctp/Cdlc2, wdb, and msps suppressed Ct-induced dendritic branch formation, particularly short dendritic filopodia, as well as dendritic growth as compared to control neurons misexpressing Ct alone (Figure 5, B–G and Figure S6, A and B in File S1). These findings indicate that these putative downstream effectors are required in promoting Ct-mediated dendritic branching and growth. In contrast, one putative Ct target, SkpA, exhibited an enhancement of the Ct ectopic expression phenotype leading to an overall increase in branching complexity and dendritic growth (Figure 5J and Figure S6, A and B in File S1), suggesting that Ct regulates the expression of SkpA to restrict filopodial formation and dendritic branching/growth. In the case of cpa and cpb, while we did not observe a statistically significant change in the total number of dendritic branches, we observed significant reductions in total dendritic length (Figure 5, H and I and Figure S6B in File S1). To further examine the regulatory relationships between Ct and putative target genes identified by the microarray and genetic interaction studies, we performed qRT-PCR analyses on isolated CI neurons ectopically expressing Ct vs. control CI neurons. These analyses confirmed that Ct positively regulates many of the putative target genes, but interestingly, negatively regulates SkpA (Figure 5R).
Figure 5.
Phenotypic and molecular validation of putative Ct and Kn effector molecules. Relative to wild-type controls (A), Ct ectopic misexpression in CI vpda neurons leads to a dramatic increase in dendritic branching complexity characterized by increased dendritic outgrowth, branching, and the formation of de novo dendritic filopodia (B). (C–J) CI vpda neurons overexpressing Ct with simultaneous RNAi knockdown of Ct candidate target genes. (K) Kn ectopic misexpression leads to an increase in CI vpda dendritic branching complexity and branch elongation. (L–Q) CI vpda neurons overexpressing Kn with simultaneous RNAi knockdown of Kn candidate target genes. (R) Heatmap of qRT-PCR analyses of the indicated genes in CI neurons either overexpressing Ct (top panel) or Kn (bottom panel). Numbers in the color bar legends represent mean fold change. Statistics: unpaired t-test; N = 4; and significance score was * P < 0.05. Bar, 100 µm.
Similarly, putative Kn-regulated genes that were positive hits from the CIV phenotypic analyses were examined to assess their requirement for Kn-mediated supernumerary branching and increased dendritic growth observed with Kn ectopic expression in CI neurons. Knockdown of the putative Kn target genes RpL7 and RpL36A results in a strong suppression of Kn-mediated dendritic growth and branching returning morphology to nearly wild type for CI vpda neurons (Figure 5, A, K, O, and P and Figure S6, C and D in File S1). Similarly, we observed significant suppression of total dendritic length with wdb-IR, Ank2-IR, RhoGAP18B-IR, and T-cp1-IR expression relative to Kn ectopic misexpression alone (Figure 5, K–N and Q and Figure S6D in File S1), whereas reductions in the total number of branches were observed for only RhoGAP18B-IR, RpL7-IR, and RpL36A-IR (Figure S6C in File S1). With respect to the regulatory relationship between Kn and these putative targets, qRT-PCR analyses confirmed significant upregulation for all of these genes consistent with the microarray studies (Figure 5R).
Collectively, these analyses, in combination with CIV-specific phenotypic studies, suggest a role for at least a large subset of these molecules as important downstream effectors of Ct- and/or Kn-mediated dendritic morphogenesis.
Ct and Kn effector molecules are required for dendritic cytoskeleton organization and stabilization
Ct and Kn have been demonstrated to exert their effects on dendritic morphogenesis at least in part by regulation of the arbor cytoskeleton (Jinushi-Nakao et al. 2007; Ye et al. 2011; Iyer et al. 2012; Nagel et al. 2012; Ferreira et al. 2014). To test the hypothesis that the putative Ct and/or Kn target molecules we identified in our neurogenomic-driven phenotypic analyses may regulate dendritic cytoskeletal organization, we implemented the use of multi-fluor-labeled transgenic reporters to visualize F-actin and MT organization/dynamics in combination with target molecule RNAi-mediated knockdown using live image confocal microscopy. This approach facilitates analyses of distinct subcellular organizations of F-actin and MT cytoskeletons in da neurons in both control and gene-specific disruption backgrounds (Figure 6, Figure 7, and Figure 8). Dendritic arbor cytoskeleton was visualized by using a CIV-GAL4 to drive the expression of UAS-GMA::GFP, in which the F-actin cytoskeleton is labeled by a GFP-tagged Moesin actin binding domain, and UAS-mCherry::Jupiter, in which the MT cytoskeleton is labeled by the mCherry-tagged MT associated protein Jupiter. To verify that the F-actin and MT transgene reporters do not themselves exert effects on dendrite development, we performed comparative neuromorphometric analyses of CIV neurons expressing cytoskeletal markers to CIV neurons expressing a CD8::GFP dendritic membrane marker and discovered no significant differences in arbor morphology (Figure S7, I and J in File S1).
Figure 6.
Cytoskeletal effects of Ct and Kn effector molecules in the complexity enhancer category. Representative images of CIV (ddaC) neurons labeled by class-specific GAL4 expression of UAS-GMA (F-actin) (A–F) and UAS-mCherry::Jupiter (MT) (A’–F’). (A) Wild-type (B–F) gene-specific RNAi (IR) knockdowns. Zoomed views in (A”’–F’”) represent the dashed boxes in the corresponding merge panel (A”–F”). (G–J) Quantitative analyses of multichannel next generation reconstructions of CIV neurons. (G and H) Sholl profiles of total relative F-actin quantity (G) or total relative MT quantity (H) measured as a function of path distance from the soma. Colored asterisks report significant changes in critical value. (I and J) Total relative F-actin quantity (I) or total relative MT quantity (J) by Strahler order distribution, where 7 = primary branch from cell body and 1 = terminal branches and values are the mean (± SEM). Statistical tests performed in: (G and H) one-way ANOVA with FDR correction of Benjamini, Kriega, and Yekutieli; (I and J) two-way ANOVA with FDR correction of Benjamini, Kriega, and Yekutieli. N = 3–6 neurons, and significance scores are * P < 0.05.
Figure 7.
Cytoskeletal effects of Ct and Kn effector molecules in the complexity shifter category. Representative images of CIV (ddaC) neurons labeled by class-specific GAL4 expression of UAS-GMA (F-actin) (A–E) and UAS-mCherry::Jupiter (MT) (A’–E’). (A) Wild type, (B–E) gene-specific RNAi (IR) knockdowns. Zoomed views in (A”’–E’”) represent the dashed boxes in the corresponding merge panel (A”–E”). (F–I) Quantitative analyses of multichannel next generation reconstructions of CIV neurons. (F and G) Sholl profiles of total relative F-actin quantity (F) or total relative MT quantity (G) measured as a function of path distance from the soma. Colored asterisks report significant changes in critical value. (H and I) Total relative F-actin quantity (H) or total relative MT quantity (I) by Strahler order distribution, where 7 = primary branch from cell body and 1 = terminal branches and values are the mean (± SEM). Statistical tests performed in: (F and G) one-way ANOVA with FDR correction of Benjamini, Kriega, and Yekutieli; (H and I) two-way ANOVA with FDR correction of Benjamini, Kriega, and Yekutieli. N = 3–6 neurons, and significance scores are * P < 0.05.
Figure 8.
Cytoskeletal effects of Ct and Kn effector molecules in the complexity suppressor category. Representative images of CIV (ddaC) neurons labeled by class-specific GAL4 expression of UAS-GMA (F-actin) (A–D) and UAS-mCherry::Jupiter (MT) (A’–D’). (A) Wild type, (B–D) gene-specific RNAi (IR) knockdowns. Zoomed views in (A”’–D’”) represent the dashed boxes in the corresponding merge panel (A”–D”). (E–H) Quantitative analyses of multichannel next generation reconstructions of CIV neurons. (E and F) Sholl profiles of total relative F-actin quantity (E) or total relative MT quantity (F) measured as a function of path distance from the soma. Colored asterisks report significant changes in critical value. (G and H) Total relative F-actin quantity (G) or total relative MT quantity (H) by Strahler order distribution, where 7 = primary branch from cell body and 1 = terminal branches and values are the mean (± SEM). Statistical tests performed in: (E and F) one-way ANOVA with FDR correction of Benjamini, Kriega, and Yekutieli; (G and H) two-way ANOVA with FDR correction of Benjamini, Kriega, and Yekutieli. N = 3–6 neurons, and significance scores are * P < 0.05.
While powerful methods exist to reconstruct and analyze dendritic morphology (Parekh and Ascoli 2013), quantitative characterization of dendritic developmental mechanisms remains challenging as standard descriptions of dendritic architecture do not incorporate an explicit representation of subcellular cytoskeletal compositions among distinct neuronal subtypes. To address this, we have developed novel forms of multichannel (e.g., cytoskeletal/cell membrane) digital reconstructions of dendritic morphology to enable statistical analyses of morphological changes and underlying molecular mechanisms via which global or local modulation of the cytoskeleton may affect dendritic architecture. To test whether this novel quantitative method accurately measures cytoskeletal intensity values correctly, we performed benchmark testing of the multichannel plugin on a surrogate artificial dendrite where all intensity values are known (see Materials and Methods) and then compared the intensity distributions against path distance of the multichannel reconstruction to the artificial dendrite. The results reveal a high degree of accuracy for the multichannel reconstruction output (Figure S7, A–D in File S1). In assessing quantity distributions of cytoskeletal elements, we describe changes in total F-actin or MT relative quantities as a function of 40 μm binning (see Materials and Methods) against path distance from soma (across different genotypes) rather than the average relative quantities per unit length. Therefore, the data are not normalized to length in order to focus on alterations in cytoskeletal distributions against path distance from the soma. As such, changes in quantity distribution for cytoskeletal signals are independent of total arbor length. This point is illustrated in comparisons between control and RpL7-IR CIV neurons (Figure S7, E–H in File S1). Utilizing this approach, we can qualitatively and quantitatively assess the distribution of cytoskeletal elements as a function of genotype across the dendritic arbor and discern primary defects a gene may have on cytoskeletal organization (Figure 6, Figure 7, and Figure 8 and Figure S8 in File S1).
For example, in control CIV neurons, F-actin structures extend throughout the dendritic arbor and are enriched at dendritic terminals (Figure S8, A and E in File S1), whereas MT signal intensity is more uniform across the arbor and appears correlated with branch thickness such that as the taper rate diminishes with iterative branching toward dendritic termini, so too does the signal (Figure S8, B, F, and J in File S1). Moreover, F-actin displays an uneven distribution along an arbor, as seen by presence of F-actin-rich islands along the arbor, and presence of stronger F-actin signal at branch points (Figure S8I in File S1). In contrast, RpL36A-IR expression in CIV neurons leads to a severe MT defect (Figure S8L in File S1) and exhibits a shift in F-actin distribution where the majority is accumulated proximal to the cell body relative to control distribution (Figure S8K in File S1). The composite effects of changes in cytoskeletal distributions and/or levels can also be visualized between genotypes to highlight the consequences of gene perturbation on local changes in cytoskeletal architecture (Figure S8, M and N in File S1). Therefore, we utilized this technique to assess the roles of putative Ct and/or Kn effector molecules in modulating dendritic cytoskeletal architecture.
Analyses of the complexity enhancer group revealed that knockdown of these genes variably affected the organization of actin-rich dendrite structures and had gross phenotypic defects in MT architecture/stabilization. More specifically, disruption of RpL36A (Figure 6, C–C’’’), and dmn (Figure 6, D–D’’’), leads to an abnormal accumulation of F-actin-rich branches in the proximity to soma. The predominant cytoskeletal defect in this gene group is changes in MTs, with defects in RpL7 (Figure 6, B–B’’’), RpL36A (Figure 6, C–C’’’), dmn (Figure 6, D–D’’’), msps (Figure 6, E–E’’’), and CCT2 (Figure 6, F–F’’’) severely reduced levels of MT-based dendritic cytoarchitecture relative to controls. Disruption of these genes revealed overall reductions in F-actin and MT quantities (area under the curve) when compared to control. Comparing the critical value (maximum peak of the curve) for relative quantities of F-actin or MTs revealed that all complexity enhancer genes are significantly reduced for both cytoskeletal elements (Figure 6, G and H). Next, we analyzed the distribution of the cytoskeletal components as a function of arbor branch order (Strahler order). Reductions in F-actin were observed from Strahler order 3 to Strahler order 1 (termini). Moreover, in RpL7-IR and RpL36A-IR neurons the terminal order branches were completely absent (Figure 6, I and J). However, the effects on MTs were more uniformly distributed throughout the arbor (Figure 6J). Within this category, select genes appeared to cluster together in terms of their effect on these cytoskeletal components, for instance RpL7-IR and RpL36A-IR in one cluster and the rest in the other (Figure 6, G–J). This may suggest that these genes operate in similar pathways/biological processes to modulate the dendritic cytoskeleton.
In contrast to complexity enhancer genes, those in the complexity shifter category had no obvious defects in MT cytoskeletal organization; however, consistent with an elaboration of distally tufted dendrites, wdb-IR (Figure 7, B–B’’’), Ank2-IR (Figure 7, C–C’’’), RhoGAP18B-IR (Figure 7, D–D’’’), and ctp/Cdlc2-IR (Figure 7, E–E’’’) neurons showed altered organization of F-actin-populated distal dendritic branches, suggesting a preferential role for these genes in regulating F-actin processes. Phenotypic analyses demonstrate that disruption of these genes resulted in more evident reductions (area under the curve) in F-actin (Figure 7, A–F) relative to MTs when compared to control, with an exception of Ank2-IR neurons (Figure 7, A–E and G). We observed a significant reduction in critical value for relative F-actin quantity for all of these genes (Figure 7F), as well as a modest proximal shift of the overall curves suggesting a reorganization of these cytoskeletal components. With respect to relative MT quantity, only Ank2-IR neurons exhibited a significant reduction in the critical value (Figure 7G). Strahler branch order distribution analyses revealed variable local effects on relative F-actin or MT quantity for these genes with F-actin effects restricted to terminal branch orders (Strahler order 1 and 2) and MT effects to intermediate and lowest order (primary) branch orders (Figure 7, H and I).
Finally, disruption of genes in the complexity suppressor category lead to exuberant terminal branching/elongation. We observed that SkpA-IR (Figure 8, B–B’’’), cpa-IR (Figure 8, C–C’’’), and cpb-IR (Figure 8, D–D’’’) neurons exhibited hyper-proliferation of F-actin-rich branches. Defects in these genes resulted in complex, localized effects on the dendritic cytoskeleton. SkpA-IR or cpb-IR neurons displayed a qualitative, albeit nonsignificant, increase in F-actin quantity (area under the curve) (Figure 8E) that is significantly increased at terminal branches (Figure 8G). In contrast, SkpA-IR and cpb-IR neurons had a significant increase in the critical value for relative MT quantity (Figure 8F), and SkpA-IR resulted in spatially restricted increases in MTs, particularly at higher (Strahler order 1) and intermediate (Strahler order 3, 4) order branches, when compared to control (Figure 8H), which could not have been predicted solely by qualitative analyses (Figure 8, B–B’’’).
Collectively, these studies strongly implicate Ct or Kn effectors in the complexity enhancer category in the stabilization of MTs and F-actin organization, those in the complexity shifter category in regulating F-actin organization on distally localized dendrites, and those in the complexity suppressor category in restricting F-actin-mediated terminal dendritic branching coupled with local effects on the MT cytoskeleton.
Discussion
Transcriptional control programs directing cytoskeletal-mediated dendritic architecture
Proper dendritic development is a key feature in the formation of functional neural circuitry as defects in this process have been broadly implicated in a diverse spectrum of neuropathological and neurodegenerative disease states including Alzheimer, Parkinson, Huntington diseases, schizophrenia, and various muscular dystrophies. Moreover, defects in dendritogenesis are a common neuroanatomical pathology correlated with cognitive impairments such as mental retardation (Down, Rett, and Fragile X syndromes) and autism (Fiala et al. 2002; Ramocki and Zoghbi 2008; Kulkarni and Firestein 2012). Given these observations and the critical role of the cytoskeleton in specifying and modulating dendritic shape, characterizing the molecular genetic mechanisms that govern cell-type-specific cytoskeletal architecture is pathophysiologically relevant.
TF regulation represents a critical, cell autonomous mechanism for driving cell-type-specific dendritic diversity; however, until recently, relatively little was known of the downstream effector pathways by which TFs exert control over dendritogenesis, nor do we have an understanding of the role of combinatorial TF regulation in governing neuronal development (Nanda et al. 2017). Drosophila da neurons provide a powerful neurogenetic and neurogenomic platform for probing these questions. Here, we have used this platform to uncover novel cellular and molecular mechanisms by which the TFs Ct and Kn regulate cell-type specific dendrite development, both uniquely and in a combinatorial fashion. Our neurogenomic-driven screen has identified a broad range of previously uncharacterized effector molecules that lie downstream in the Ct and/or Kn transcriptional regulatory pathways, many of which ultimately converge on the cytoskeleton to direct dendritic architecture in differential ways. Detailed phenotypic studies for a subset of these putative effector genes revealed variable defects in CIV dendritogenesis, and based upon phenotypic similarities, we classified these genes into the following groups: (1) complexity enhancer; (2) complexity shifter; and (3) complexity suppressor. Interestingly, genes that fell within these categories, and were transcriptional targets of Ct and/or Kn, shared common molecular functions and exerted similar effects on regulating the dendritic cytoskeleton. In addition, genes identified as downstream effectors of Ct and/or Kn are largely evolutionarily conserved from flies to humans, and a number of the major phenotypic hits from the screen have human orthologs that have either been directly linked to neuropathologies, though the mechanistic basis in disease etiologies is incompletely understood, or the biological process the molecule participates in (e.g., ribosomal regulatory function) has been broadly linked to human disease (Table S3).
Complexity enhancer group
The complexity enhancer group included the following cytoskeletal regulatory molecules: Dynamitin (Dmn) and Mini spindles (Msps). Other genes in this group with major regulatory roles in CIV dendritogenesis were two components of the large ribosomal subunit, RpL36A and RpL7, and two chaperonins, T-cp1 and CCT2, which are paralogous subunits that assemble to form a multi-subunit ring complex, the TCP-1 Ring Complex (TRiC) or Chaperonin Containing TCP-1 (CCT) complex. Dmn (also known as DCTN2-p50) is a component of the Dynactin complex, a large 1.2-MDa multi-subunit complex that associates with the cytoplasmic dynein complex to drive MT-based transport. Msps (also known as XMAP215) is likewise linked to regulation of the MT cytoskeleton where it has been shown to function as a processive MT polymerase adding α−β tubulin heterodimers to the plus end to promote MT polymerization (Brouhard et al. 2008). The TRiC/CCT molecules, T-cp1 and CCT2, function as molecular chaperonins catalyzing the ATP-dependent folding of ∼10% of all newly synthesized proteins, and while the spectrum of physiological substrates of the TRiC/CCT complex remain poorly defined, biochemical and genetic studies have demonstrated that this complex functions in folding of actin and tubulin cytoskeletal proteins (reviewed in Dunn et al. (2001)). Recent studies have indicated the importance of proper folding of monomeric tubulins into MT polymerization in regulating neural circuitry including neuronal morphogenesis, cellular polarization, and neurite growth and branching (Hattori et al. 2008; Okumura et al. 2015). An MT is a polymer composed of α- and β-tubulin heterodimers that are formed by a multistep process coordinated by several tubulin-folding cofactors (Lopez-Fanarraga et al. 2001). Nascent α- and β-tubulins associate with the cytosolic chaperonin complex (TRiC/CCT) suggesting that tubulin-folding cofactors may play a role in both the synthesis and degradation of tubulin heterodimers ultimately affecting the neuronal cytoskeleton (Okumura et al. 2015). Defects in the TRiC/CCT complex have been linked to both Huntington and Gaucher disease, and this complex has been shown to physically interact with polyglutamine-expanded variants of Huntingtin (Htt) where it effectively inhibits their aggregation and reduces Htt-induced neuronal toxicity (Tam et al. 2006). Moreover, improper folding of tubulin, which is dependent upon the action of the TRiC/CCT chaperonin complex, has also been implicated in hypoparathyroidism-retardation-dysmorphism and Kenny–Caffey syndromes, which is attributed to a mutation in tubulin-folding cofactors (Parvari et al. 2002).
Consistent with the known molecular functions of these genes, apart from the large ribosomal subunits, we discovered that these genes play pivotal roles in promoting dendritic arbor complexity and that the predominant cytoskeletal defect appears to be a destabilization and/or reduction in MT assembly, which results in a highly rudimentary arbor. Moreover, genes in this group also appeared to regulate the organization of F-actin-rich dendritic branching. These findings indicate that in addition to cytoskeletal regulatory molecules, like Msps and Dmn, biological processes including ribosomal regulation and chaperonin function are required for regulating the dendritic MT cytoskeleton to promote complex arborization. This regulation may occur directly via binding to MTs to promote their stabilization or assembly, or could occur as an indirect consequence via defects in ribosomal-based translation or chaperonin activity. For example, in the case of T-cp1 and CCT2, the observed defects are likely due to improper folding of tubulin monomers, which could lead to their degradation.
From a transcriptional regulation perspective, analyses of complexity enhancer genes revealed interesting regulatory patterns, both at the gene-specific level and with respect to molecules that are linked to the same cellular machinery. For example, Kn positively regulates both RpL7 and RpL36A, while in the case of the TRiC/CCT subunits, we observed distinct patterns of regulation whereby T-cp1 is positively regulated by Kn, whereas CCT2 is positively regulated by Ct in both microarray and qRT-PCR analyses. While previous studies have linked Ct to regulation of the actin cytoskeleton in da neuron dendrites (reviewed in Nanda et al. (2017)), this is an oversimplification, as here we demonstrate that Ct regulates the MT-associated regulatory molecule Dmn, revealing novel functional roles linking Ct to MT regulation.
Complexity shifter group
In contrast to the complexity enhancer group, genes in the complexity shifter group function as downstream targets of Ct and/or Kn to regulate the distribution of dendritic branching throughout the proximal–distal axis relative to the cell body. Loss-of-function analyses of the PP2A phosphatase complex regulatory subunit widerborst (wdb), the cytoskeletal regulatory molecules Ankyrin2 (Ank2) and RhoGAP18B, and the cytoplasmic dynein light chain encoding genes cut up (ctp) and Cytoplasmic dynein light chain 2 (Cdlc2) revealed largely consistent defects characterized by reduced interstitial branching proximal to the cell body in favor of a distal shift resulting in aberrant clustered terminal branches that have a tufted appearance. In the case of ctp/Cdlc2, the defects are quite interesting as they produce a phenotype that is inverted from what is observed with mutations of Dynein light intermediate chain (Dlic). In previous studies, Dlic mutations in CIV da neurons lead to hyper-proliferation of dendritic branches proximal to the cell body and leave the dendritic terminals completely stripped (Satoh et al. 2008; Zheng et al. 2008). This suggests that different classes of Dynein MT motor light chains exert distinct regulatory effects on the distribution of dendritic branches, perhaps via differential regulation of Dynein-linked cargo on MTs.
Strikingly, at the cytoskeletal level, many of these genes did not appear to have any gross defects in the MT cytoskeleton; however, terminal branching was characterized by clustered and elongated branches that were predominantly populated by F-actin, indicative of a preferential role for these molecules in modulating F-actin-mediated terminal branch structure and organization. These findings are intriguing as wdb and Ank2 have only previously been linked to functions related to the MT cytoskeleton, e.g., spindle assembly (Chen et al. 2007) and MT organization (Koch et al. 2008; Pielage et al. 2008), whereas ctp has been linked to actin filament bundle assembly (Ghosh-Roy et al. 2005). This suggests that both wdb and Ank2 may have additional cytoskeletal functions in regulating F-actin-mediated dendritic branching. In contrast, RhoGAP18B, which encodes a Rho-GTPase-activating protein, has been linked to F-actin filament organization via negative regulation of the small GTPase Rho (Kiger et al. 2003; Rothenfluh et al. 2006; Ojelade et al. 2015). When activated, Rho promotes the formation of unbranched F-actin stress fibers, promoting elongation, whereas RhoGAPs function to repress Rho-mediated F-actin organization, which could explain, at least in part, why disruption of RhoGAP18B leads to elongated, clustered F-actin-rich dendritic terminals by maintaining Rho in an activated GTP-bound state.
As with the complexity enhancer group, genes in the complexity shifter category also exhibit complex transcriptional regulation by both Ct and Kn. Ct positively regulates Cdlc2, whereas Kn positively regulates Ank2, and both wdb and RhoGAP18B are positively coregulated by Ct and Kn. Thus, in addition to previous findings linking Kn to regulation of the MT cytoskeleton (Jinushi-Nakao et al. 2007), Kn also appears to regulate the expression of genes that exert effects on actin cytoskeletal organization.
Complexity suppressor group
Finally, in the complexity suppressor gene group, knockdown of which lead to excessive dendritic growth and branching of CIV neurons, we identified three targets of Ct and/or Kn transcriptional regulation: SkpA, capping protein alpha (cpa), and capping protein beta (cpb). SkpA encodes a subunit of the Skp, Cullin, F-box (SCF)-containing ubiquitin ligase complexes and has been previously demonstrated to be functionally required for dendritic pruning in CIV neurons at the larval-to-pupal transition (Wong et al. 2013). Consistent with these previous findings, we found that SkpA function is required to restrict larval CIV dendritic complexity, which given its functional role, is likely based on ubiquitin-linked proteasomal degradation. Moreover, qRT-PCR analyses reveal SkpA is negatively regulated by Ct and studies in CI neurons identify that Ct acts via SkpA to restrict overall dendritic growth as further reduction of SkpA levels exacerbates the increased dendritic complexity observed with ectopic Ct expression alone. These results suggest that SkpA functions in restricting supernumerary dendritic elaboration and that perhaps Cut-mediated changes in dendritic arborization complexity are dependent upon regulating the levels of SkpA. At the cytoskeletal level, downregulation of SkpA leads to increases in relative F-actin and MT quantities in terminal branching relative to controls likely underlying the exuberant terminal processes observed in SkpA disrupted CI and CIV neurons. The other two genes identified in this group, cpa and cpb, have related functions in capping F-actin filaments at the barbed/plus end. Plus-end capping of F-actin by these factors restricts further F-actin polymerization and thereby limits filament growth. Consistent with these molecular functions, RNAi knockdowns of cpa or cpb leads to excessive F-actin growth, which is the characteristic cytoskeletal feature. Thus, regulated growth and branching of CIV neurons appears dependent upon the proper levels of Cpa and Cpb to cap F-actin filaments and thereby limit their growth. Both cpa and cpb were identified as Ct transcriptional targets by microarray analyses and qRT-PCR analyses confirm Ct significantly upregulates cpb expression together with a trending, albeit nonsignificant, increase in cpa levels. While knockdowns for either gene results in complexity enhancement, the CIV knockdown phenotypes are not identical, nor are the phenotypes when either Ct or Kn are ectopically expressed in CI neurons in combination with cpa-IR or cpb-IR knockdowns. These findings suggest that Ct-mediated dendritic arborization requires the action of capping factors for controlling the precise pattern of locally regulated F-actin dendritic growth and branching that ultimately contributes to cell-type-specific dendritic architectures.
Taken together, these studies set the stage for future in-depth analyses aimed at revealing mechanistic underpinnings of how these molecules exert their distinct effects on cytoskeletal-mediated dendritic architecture. More broadly, our findings demonstrate functional roles of both unique and combinatorial transcriptional regulatory programs in fine-tuning gene expression that affects cytoskeletal organization/stability and thereby contributes to both cell-type-specific dendritic architecture and the promotion of dendritic diversity.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300393/-/DC1.
Acknowledgments
We are grateful to D. P. Kiehart (Duke University), C. Q. Doe (University of Oregon/HHMI), and Y.-N. Jan for sharing the transgenic strains. We thank D. A. Patel for designing and creating custom image processing software for the Cox laboratory (Flyboys) available upon request. We thank Cox laboratory members U. Nguyen, B. D. Grantier, and D. W. Yakout and Ascoli laboratory members M. Mollajafar, S. Fantaye, and A. Penn for assistance with neuronal reconstructions. We thank H. Chen and H. Peng (Peng laboratory, Allen Institute for Brain Science) for assistance with the Vaa3D multichannel plugin. We acknowledge the Bloomington Drosophila Stock Center [National Institutes of Health (NIH) P40ODO18537] and Vienna Drosophila Resource Center (VDRC) for fly strains used in this study. This research was supported by NIH National Institute of Neurological Disorders and Stroke (NINDS) R01 NS086082 (D.N.C/G.A.A.); NIH National Institute of Mental Health (NIMH) R15 MH086928 (D.N.C.); NINDS R01 NS39600 (G.A.A.), and National Science Foundation (NSF) BRAIN EAGER DBI-1546335 (G.A.A.).
Author contributions: Conceptualization – RD, GAA, and DNC; Writing – RD and DNC; Investigation – RD, SB, AAP, JMH, SGC, and JML (experimental analyses); SB (microarray analyses and bioinformatics); EPRI (microarrays); SN and GAA (next generation multichannel reconstructions).
Footnotes
Communicating editor: L. Luo
Literature Cited
- Ascoli G. A., 2006. Mobilizing the base of neuroscience data: the case of neuronal morphologies. Nat. Rev. Neurosci. 7: 318–324. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57: 289–300. [Google Scholar]
- Bhattacharya S., Iyer E. P., Iyer S. C., Cox D. N., 2014. Cell-type specific transcriptomic profiling to dissect mechanisms of differential dendritogenesis. Genom. Data 2: 378–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blochlinger K., Bodmer R., Jack J., Jan L. Y., Jan Y. N., 1988. Primary structure and expression of a product from cut, a locus involved in specifying sensory organ identity in Drosophila. Nature 333: 629–635. [DOI] [PubMed] [Google Scholar]
- Blochlinger K., Bodmer R., Jan L. Y., Jan Y. N., 1990. Patterns of expression of cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes Dev. 4: 1322–1331. [DOI] [PubMed] [Google Scholar]
- Bodmer R., Barbel S., Sheperd S., Jack J. W., Jan L. Y., et al. , 1987. Transformation of sensory organs by mutations of the cut locus of D. melanogaster. Cell 51: 293–307. [DOI] [PubMed] [Google Scholar]
- Brouhard G. J., Stear J. H., Noetzel T. L., Al-Bassam J., Kinoshita K., et al. , 2008. XMAP215 is a processive microtubule polymerase. Cell 132: 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon R. C., Turner D. A., Pyapali G. K., Wheal H. V., 1998. An on-line archive of reconstructed hippocampal neurons. J. Neurosci. Methods 84: 49–54. [DOI] [PubMed] [Google Scholar]
- Chen F., Archambault V., Kar A., Lio P., D’Avino P. P., et al. , 2007. Multiple protein phosphatases are required for mitosis in Drosophila. Curr. Biol. 17: 293–303. [DOI] [PubMed] [Google Scholar]
- Crozatier M., Vincent A., 2008. Control of multidendritic neuron differentiation in Drosophila: the role of Collier. Dev. Biol. 315: 232–242. [DOI] [PubMed] [Google Scholar]
- Cubelos B., Sebastián-Serrano A., Beccari L., Calcagnotto M. E., Cisneros E., et al. , 2010. Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron 66: 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuntz H., Forstner F., Borst A., Häusser M., 2010. One rule to grow them all: a general theory of neuronal branching and its practical application. PLoS Comput. Biol. 6: e1000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn A. Y., Melville M. W., Frydman J., 2001. Review: cellular substrates of the eukaryotic chaperonin TRiC/CCT. J. Struct. Biol. 135: 176–184. [DOI] [PubMed] [Google Scholar]
- Feng L., Zhao T., Kim J., 2015. neuTube 1.0: a new design for efficient neuron reconstruction software based on the SWC format. eNeuro 2: ENEURO.0049–14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira T., Ou Y., Li S., Giniger E., Meyel D. J., 2014. Dendrite architecture organized by transcriptional control of the F-Actin nucleator spire. Development 141: 650–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala J. C., Spacek J., Harris K. M., 2002. Dendritic spine pathology: cause or consequence of neurological disorders? Brain Res. Brain Res. Rev. 39: 29–54. [DOI] [PubMed] [Google Scholar]
- Ghosh-Roy A., Desai B. S., Ray K., 2005. Dynein light chain 1 regulates dynamin-mediated F-actin assembly during sperm individualization in Drosophila. Mol. Biol. Cell 16: 3107–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale A., Hartwig C., Freeman A. H., Das R., Zlatic S. A., et al. , 2016. The proteome of BLOC-1 genetic defects identifies the Arp2/3 actin polymerization complex to function downstream of the schizophrenia susceptibility factor Dysbindin at the synapse. J. Neurosci. 36: 12393–12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber W. B., Jan L. Y., Jan Y. N., 2003a Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell 112: 805–818. [DOI] [PubMed] [Google Scholar]
- Grueber W. B., Ye B., Moore A. W., Jan L. Y., Jan Y. N., 2003b Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr. Biol. 13: 618–626. [DOI] [PubMed] [Google Scholar]
- Han C., Jan L. Y., Jan Y. N., 2011. Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc. Natl. Acad. Sci. 108: 9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori D., Millard S. S., Wojtowicz W. M., Zipursky S. L., 2008. Dscam-mediated cell recognition regulates neural circuit formation. Annu. Rev. Cell Dev. Biol. 24: 597–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y., Sugimura K., Uemura T., 2007. Selective expression of Knot/Collier, a transcriptional regulator of the EBF/Olf-1 family, endows the Drosophila sensory system with neuronal class-specific elaborated dendritic patterns. Genes Cells 12: 1011–1022. [DOI] [PubMed] [Google Scholar]
- Hattori Y., Usui T., Satoh D., Moriyama S., Shimono K., et al. , 2013. Sensory-neuron subtype-specific transcriptional programs controlling dendrite morphogenesis: genome-wide analysis of abrupt and Knot/Collier. Dev. Cell 27: 530–544. [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2009a Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2009b Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Iyer E. P., Iyer S. C., Sulkowski M. J., Cox D. N., 2009. Isolation and purification of Drosophila peripheral neurons by magnetic bead sorting. J. Vis. Exp. 34: e1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer E. P., Iyer S. C., Sullivan L., Wang D., Meduri R., et al. , 2013. Functional genomic analyses of two morphologically distinct classes of Drosophila sensory neurons: post-mitotic roles of transcription factors in dendritic patterning. PLoS One 8: e72434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S. C., Wang D., Iyer E. P. R., Trunnell S. A., Meduri R., et al. , 2012. The RhoGEF Trio functions in sculpting class specific dendrite morphogenesis in Drosophila sensory neurons. PLoS One 7: e33634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S. C., Ramachandran Iyer E. P., Meduri R., Rubaharan M., Kuntimaddi A., et al. , 2013. Cut, via CrebA, transcriptionally regulates the COPII secretory pathway to direct dendrite development in Drosophila. J. Cell Sci. 126: 4732–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan Y. N., Jan L. Y., 2010. Branching out: mechanisms of dendritic arborization. Nat. Rev. Neurosci. 11: 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinushi-Nakao S., Arvind R., Amikura R., Kinameri E., Liu A. W., et al. , 2007. Knot/Collier and cut control different aspects of dendrite cytoskeleton and synergize to define final arbor shape. Neuron 56: 963–978. [DOI] [PubMed] [Google Scholar]
- Kiger A., Baum B., Jones S., Jones M., Coulson A., et al. , 2003. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch I., Schwarz H., Beuchle D., Goellner B., Langegger M., et al. , 2008. Drosophila ankyrin 2 is required for synaptic stability. Neuron 58: 210–222. [DOI] [PubMed] [Google Scholar]
- Komiyama T., Luo L., 2007. Intrinsic control of precise dendritic targeting by an Ensemble of transcription factors. Curr. Biol. 17: 278–285. [DOI] [PubMed] [Google Scholar]
- Kulkarni V. A., Firestein B. L., 2012. The dendritic tree and brain disorders. Mol. Cell. Neurosci. 50: 10–20. [DOI] [PubMed] [Google Scholar]
- Lefebvre J. L., Sanes J. R., Kay J. N., 2015. Development of dendritic form and function. Annu. Rev. Cell Dev. Biol. 31: 741–777. [DOI] [PubMed] [Google Scholar]
- Li N., Zhao C. T., Wang Y., Yuan X. B., 2010. The transcription factor Cux1 regulates dendritic morphology of cortical pyramidal neurons. PLoS One 5: e10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Fanarraga M., Avila J., Guasch A., Coll M., Zabala J. C., 2001. Review: postchaperonin tubulin folding cofactors and their role in microtubule dynamics. J. Struct. Biol. 135: 219–229. [DOI] [PubMed] [Google Scholar]
- Megason S., Fraser S. E., 2007. Imaging in systems biology. Cell 130: 784–795. [DOI] [PubMed] [Google Scholar]
- Nagel J., Delandre C., Zhang Y., Forstner F., Moore A. W., et al. , 2012. Fascin controls neuronal class-specific dendrite arbor morphology. Development 139: 2999–3009. [DOI] [PubMed] [Google Scholar]
- Nanda, S., R. Das, D.N. Cox, and G.A. Ascoli, 2017 Structural plasticity in dendrites: developmental neurogenetics, morphological reconstructions, and computational modeling, pp. 1-34 in Neurobiological and Psychological Aspects of Brain Recovery, edited by L. Petrosini, Springer Press, Cham. [Google Scholar]
- Ojelade S. A., Acevedo S. F., Kalahasti G., Rodan A. R., Rothenfluh A., 2015. RhoGAP18B isoforms act on distinct Rho-family GTPases and regulate behavioral responses to alcohol via cofilin. PLoS One 10: e0137465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura M., Miura M., Chihara T., 2015. The roles of tubulin-folding cofactors in neuronal morphogenesis and disease. Neural Regen. Res. 10: 1388–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh R., Ascoli G. A., 2013. Neuronal morphology goes digital: a research hub for cellular and system neuroscience. Neuron 77: 1017–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvari R., Hershkovitz E., Grossman N., Gorodischer R., Loeys B., et al. , 2002. Mutation of TBCE causes hypoparathyroidism-retardation-dysmorphism and autosomal recessive Kenny-Caffey syndrome. Nat. Genet. 32: 448–452. [DOI] [PubMed] [Google Scholar]
- Peng H., Bria A., Zhou Z., Iannello G., Long F., 2014. Extensible visualization and analysis for multidimensional images using Vaa3D. Nat. Protoc. 9: 193–208. [DOI] [PubMed] [Google Scholar]
- Pielage J., Cheng L., Fetter R. D., Carlton P. M., Sedat J. W., et al. , 2008. A presynaptic giant ankyrin stabilizes the NMJ through regulation of presynaptic microtubules and transsynaptic cell adhesion. Neuron 58: 195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram S. V., Bonni A., 2013. Cell-intrinsic drivers of dendrite morphogenesis. Development 140: 4657–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramocki M. B., Zoghbi H. Y., 2008. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature 455: 912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2014 R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: http://www.R-project.org/. Accessed May 15, 2014.
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., et al. , 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez O. C., Schaefer A. W., Mandato C. A., Forscher P., Bement W. M., et al. , 2003. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 5: 599–609. [DOI] [PubMed] [Google Scholar]
- Rothenfluh A., Threlkeld R. J., Bainton R. J., Tsai L. T., Lasek A. W., et al. , 2006. Distinct behavioral responses to ethanol are regulated by alternate RhoGAP18B isoforms. Cell 127: 199–211. [DOI] [PubMed] [Google Scholar]
- Samsonovich A. V., Ascoli G. A., 2005. Statistical determinants of dendritic morphology in hippocampal pyramidal neurons: a hidden Markov model. Hippocampus 15: 166–183. [DOI] [PubMed] [Google Scholar]
- Santiago C., Bashaw G. J., 2014. Transcription factors and effectors that regulate neuronal morphology. Development 141: 4667–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh D., Sato D., Tsuyama T., Saito M., Ohkura H., et al. , 2008. Spatial control of branching within dendritic arbors by dynein-dependent transport of Rab5-endosomes. Nat. Cell Biol. 10: 1164–1171. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorcioni R., Polavaram S., Ascoli G. A., 2008. L-measure: a web-accessible tool for the analysis, comparison and search of digital reconstructions of neuronal morphologies. Nat. Protoc. 3: 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears J. C., Broihier H. T., 2016. FoxO regulates microtubule dynamics and polarity to promote dendrite branching in Drosophila sensory neurons. Dev. Biol. 418: 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono K., Fujishima K., Nomura T., Ohashi M., Usui T., et al. , 2014. An evolutionarily conserved protein CHORD regulates scaling of dendritic arbors with body size. Sci. Rep. 4: 4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam S., Geller R., Spiess C., Frydman J., 2006. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat. Cell Biol. 8: 1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavosanis, G., 2014 The cell biology of dendrite differentiation, pp. 23–40 in The Computing Dendrite: From Structure to Function (Springer Series in Computational Neuroscience), edited by H. Cuntz, M. W. H. Remme, and B. Torben-Nielsen. Springer, New York. [Google Scholar]
- Trunnell S. A., Bhattacharjee S., Cox D. N., 2015. Short stop Exerts Differential Effects on Class-Specific Dendritic Homeostasis Program number 486B, 56th Annual Drosophila Research Conference, Genetics Society of America, Chicago, IL. [Google Scholar]
- Warnes, G. R., B. Bolker, L. Bonebakker, R. Gentleman, W. H. A. Liaw et al., 2016 gplots: various R programming tools for plotting data. R package version 3.0.1. Available at: http://CRAN.R-project.org/package=gplots. Accessed November 1, 2016.
- Wearne S. L., Rodriguez A., Ehlenberger D. B., Rocher A. B., Hendersion S. C., et al. , 2005. New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience 136: 661–680. [DOI] [PubMed] [Google Scholar]
- Wong J. J., Li S., Lim E. K., Wang Y., Wang C., et al. , 2013. A Cullin1-based SCF E3 ubiquitin ligase targets the InR/PI3K/TOR pathway to regulate neuronal pruning. PLoS Biol. 11: e1001657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B., Kim J. H., Yang L., McLachlan I., Younger S., et al. , 2011. Differential regulation of dendritic and axonal development by the novel Kruppel-like factor Dar1. J. Neurosci. 31: 3309–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Wildonger J., Ye B., Zhang Y., Kita A., et al. , 2008. Dynein is required for polarized dendritic transport and uniform microtubule orientation in axons. Nat. Cell Biol. 10: 1172–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the new genotypes presented here are available upon request. The microarray data are available for download at GEO GSE83938. Digital reconstructions of neuronal morphology have been deposited into the NeuroMorpho.Org database (Ascoli 2006) for public distribution under the Cox and Ascoli archives.