Abstract
Arabidopsis thaliana INNER NO OUTER (INO) is a YABBY protein that is essential for the initiation and development of the outer integument of ovules. Other YABBY proteins have been shown to be involved in both negative and positive regulation of expression of putative target genes. YABBY proteins have also been shown to interact with the corepressor LEUNIG (LUG) in several systems. In support of a repressive role for INO, we confirm that INO interacts with LUG and also find that INO directly interacts with SEUSS (SEU), a known corepressive partner of LUG. Further, we find that INO can directly interact with ADA2b/PROPORZ1 (PRZ1), a transcriptional coactivator that is known to interact with the histone acetyltransferase GENERAL CONTROL NONREPRESSIBLE PROTEIN 5 (GCN5, also known as HAG1). Mutations in LUG, SEU, and ADA2b/PRZ1 all lead to pleiotropic effects including a deficiency in the extension of the outer integument. Additive and synergistic effects of ada2b/prz1 and lug mutations on outer integument formation indicate that these two genes function independently to promote outer integument growth. The ino mutation is epistatic to both lug and ada2b/prz1 in the outer integument, and all three proteins are present in the nuclei of a common set of outer integument cells. This is consistent with a model where INO utilizes these coregulator proteins to activate and repress separate sets of target genes. Other Arabidopsis YABBY proteins were shown to also form complexes with ADA2b/PRZ1, and have been previously shown to interact with SEU and LUG. Thus, interaction with these corepressors and coactivator may represent a general mechanism to explain the positive and negative activities of YABBY proteins in transcriptional regulation. The LUG, SEU, and ADA2b/PRZ1 proteins would also separately be recruited to targets of other transcription factors, consistent with their roles as general coregulators, explaining the pleiotropic effects not associated with YABBY function.
Keywords: ovule, integument, polarity, YABBY gene, ADA2b
OVULES are the plant reproductive organs that develop into seeds. In Arabidopsis, ovule primordia form from the regions near the carpel margins. Each primordium initiates two lateral sheathing organs, the integuments, which arise independently from the central region (chalaza) along the proximal–distal axis. The inner integument grows as a radially symmetrical structure to envelop the terminal nucellus. The outer integument grows asymmetrically, initiating and growing more extensively on the gynobasal side of the ovule, extending to envelop both the inner integument and nucellus, and resulting in an amphitropous (recurved) shape at anthesis (Robinson-Beers et al. 1992; Schneitz et al. 1995). The integuments enable the formation of the female gametophyte (Skinner et al. 2001) and form the seed coat after fertilization.
Several genes that regulate integument growth in Arabidopsis have been identified [reviewed in Skinner et al. (2004) and Colombo et al. (2008)]. INNER NO OUTER (INO) is one such gene that encodes a YABBY protein, and loss of INO function results in the absence of the outer integument and disruption of gametophyte development in the ovule (Baker et al. 1997; Schneitz et al. 1997; Villanueva et al. 1999). INO is the only YABBY gene expressed in the ovule, and expression is directly associated with outer integument growth being only in the abaxial layer on the side of the outer integument with maximal growth (Bowman and Smyth 1999; Sawa et al. 1999; Siegfried et al. 1999; Villanueva et al. 1999). Misexpression of INO on the opposite side of the ovule, as is observed in superman mutants, results in growth on both sides of the outer integument forming a more radially symmetrical structure (Gaiser et al. 1995; Meister et al. 2002). This indicates that, in the context of the chalaza, INO is sufficient for outer integument initiation and growth. Conservation of the specific expression pattern and function of INO orthologs in the outer integument has been observed in all other angiosperm species where it has been examined (Villanueva et al. 1999; Yamada et al. 2003, 2011; McAbee et al. 2005; Lora et al. 2011; Skinner et al. 2016).
The YABBY (YAB) gene family in Arabidopsis includes reproductively-expressed CRABS CLAW (CRC) and INO, and also FILAMENTOUS FLOWER (FIL), YAB2, YAB3, and YAB5, which exhibit both vegetative and reproductive expression (Villanueva et al. 1999; Bowman 2000). YABBY proteins regulate organ polarity and laminar expansion and include two conserved motifs: an amino-terminal region including a Cys2Cys2 zinc finger, and a carboxyl-terminal YABBY region with similarity to the DNA-binding domains of high mobility group (HMG) transcription factors (Bowman and Smyth 1999; Sawa et al. 1999; Siegfried et al. 1999; Villanueva et al. 1999; Kanaya et al. 2001). While sequence-specific DNA binding of YABBY proteins was not found in initial direct binding assays (Kanaya et al. 2002), evidence of their association with DNA has been found by chromatin immunoprecipitation (ChIP) (Shamimuzzaman and Vodkin 2013) and protein binding to DNA microarrays (Franco-Zorrilla et al. 2014). The mechanisms by which INO and other YABBY proteins regulate transcription to affect development are not well understood. FIL was shown to both activate and repress putative target genes, and to activate gene expression in yeast, and evidence for similar functions was found for YAB3 (Bonaccorso et al. 2012). A repressive role for YABBY proteins is also supported by the observation that YABBY proteins in Antirrhinum majus (including the An. majus ortholog of INO) can interact with the corepressor protein STYLOSA, an ortholog of the Arabidopsis LEUNIG (LUG) protein (Navarro et al. 2004; Sridhar et al. 2004). Similar interactions between YABBY proteins and LUG were also found in Arabidopsis, as were interactions with the related LEUNIG_HOMOLOG (LUH) protein (Stahle et al. 2009). YABBY proteins were also found to interact with SEUSS (SEU), a corepressor known to also partner with LUG (Franks et al. 2002; Navarro et al. 2004; Sridhar et al. 2004), and similar interactions were seen with the related SEUSS LIKE 1 (SLK1), SLK2, and SLK3 proteins (Stahle et al. 2009). Neither LUG nor SEU contain DNA-binding motifs; rather, these proteins are hypothesized to repress target gene expression through specific targeting by DNA-binding transcription factors (Conner and Liu 2000; Franks et al. 2002; Sridhar et al. 2004), and through interaction with histone deacetylase and components of the mediator complex (Gonzalez et al. 2007). Genetic analyses show that LUG, SEU, and SLK genes affect multiple aspects of plant development, including contributing to the growth of the outer integument (Roe et al. 1997; Franks et al. 2002; Bao et al. 2010).
Herein we confirm that, as for other YABBY proteins, INO interacts with both LUG and SEU. In addition to interaction with these corepressors, we find a direct interaction between INO and other YABBY proteins and a transcriptional coactivator protein, ADA2b [also known as PROPORZ1 (PRZ1)]. ADA2 proteins, originally characterized in yeast and also examined in Arabidopsis, form multi-protein complexes and directly interact with the histone acetyltransferase (HAT) GENERAL CONTROL NONREPRESSIBLE PROTEIN 5 (GCN5) (Grant et al. 1997; Mao et al. 2006). As seen for LUG and SEU, we find that ADA2b also supports outer integument extension. Through genetic analysis, we find that ADA2b and LUG function independently to promote outer integument growth, consistent with a bifunctional nature for INO as both a repressor and activator of target genes. Together, these results show that interaction with these corepressors and coactivator may represent a general mechanism to explain the positive and negative activities of YABBY proteins in transcriptional regulation.
Materials and Methods
Vector construction
PCR was used to add BamHI and XbaI sites 11 bp upstream and 160 bp downstream of the INO coding region (Villanueva et al. 1999), respectively, and the resulting fragment was cloned into pLITMUS28 (New England Biolabs, Beverly, MA) at these same sites, resulting in pRJM264. The GAL4BD-INO fusion was constructed by transferring the INO cDNA from pRJM264 to pGBKT7 (Clontech) using BamHI and XbaI, creating pTLG176. An N-terminal truncation of INO was created by PCR using the primers INOYabBam5′ and INOYabPst3′ with pRJM264 as a template, and the resulting product was cloned into pLIMUS28 to create pTLG134, and then transferred to pGBKT7 using BamHI/PstI to create pTLG178. A C-terminal truncation of INO was created by PCR using the primers INO-5′A2 and INOZnPst3′ using pRJM264 as a template, and the resulting product was cloned to pLITMUS28 to create pTLG133, and then transferred to pGBKT7 using BamHI/PstI to create pTLG177.
The GAL4BD-FIL fusion was constructed by adding EcoRI and BamHI sites to the FIL cDNA from Bowman and Smyth (1999) and inserting it at these sites into pGBKT7, creating pKLP90. GAL4BD-YAB2 and YAB5 fusions were constructed by transferring PCR-amplified cDNAs using primers NcoYAB2cDNA5′ and YABb2cDNA3′2, and NcoYAB5cDNA5′ and YAB5cDNA3′, with cDNA clones of these two proteins, and inserting the coding regions into pGBKT7 using NcoI/EcoRI, to create pKLP94 and pKLP89, respectively. GAL4BD-YAB3 and CRC fusions were constructed by transferring PCR-amplified cDNAs using primers YAB3cDNAf and YAB3cDNAr, and CRCcDNAf and CRCcDNAr, with cDNA clones of YAB3 and CRC, and inserting the sequences into pGBKT7 using EcoRI/PstI to create pMKS131 and pMKS130, respectively.
A full-length cDNA clone of ADA2a was cloned into pCR4-TOPO (Invitrogen, Carlsbad, CA) by PCR amplification using the primers ADA2aNcoIf and ADA2aXhoIr and the plasmid U09217 from the Arabidopsis Biological Resource Center at Ohio State (GenBank ID AY040045) as a template to create pELG03. GAL4AD-ADA2a was created by transferring the ADA2a cDNA in pELG03 to pGADT7 using NcoI/XhoI to create pKLP71.
A full-length cDNA clone of ADA2b was cloned into the pGADT7 yeast two-hybrid vector by PCR amplification using the primers 5′ADA2b and 3′ADA2b using the clone U13318 from ABRC (GenBank ID AY143911) as a template, and then cloned to pGAD424 (Clontech) using the restriction enzymes BamHI/SalI to create pTLG179. TGAL4AD-ADA2b was created by transferring the ADA2b cDNA in pTLG179 NcoI/SalI to pGADT7 NcoI/XhoI to create pKLP131.
A full-length cDNA clone of LUG (Conner and Liu 2000) was used as a template for PCR with the primers LUGf and LUG2r, the resulting product was cloned to pJET1.2 (Fermentas) to create pMKS144, and then cloned into pENTR4 (Invitrogen) using NcoI/XhoI to create pMKS146. The insert in pMKS146 was then transferred to pDEST-GADT7 (Rossignol et al. 2007) using LR clonase (Invitrogen) to create pMKS227, a GAL4AD-LUG fusion.
A full-length cDNA clone of SEU, pCRII-HFFL7 (Sridhar et al. 2004), was transferred to pLITMUS28 using HindIII/PstI to create pMKS197, and subsequently transferred to pENTR4 using NcoI/XhoI to create pMKS198. The insert in pMKS198 was then transferred to pDEST-GADT7 using LR clonase to create pMKS203, a GAL4AD-SEU fusion.
Yeast two-hybrid assays
Yeast transformation, mating, and cDNA library screening were performed as described with a previously described Arabidopsis pistil cDNA library (Kelley et al. 2012). For the initial screen, pTLG176 (BD-INO) was transformed into Y187 (Clontech) and then mated with the cDNA library. Mating mixtures were screened on yeast media lacking leucine, tryptophan, adenine, and histidine. Plasmids were recovered from yeast and sequenced. Pairwise interaction tests were performed by directly transforming plasmids into AH109 (Clontech), and colonies were restreaked on selective media at least twice for verification. Quantitative ortho-nitrophenyl-β-D-galactopyranoside assays to measure β-galactosidase activity as an indication of protein–protein interactions in yeast were performed according to the manufacturer’s instructions (Yeast Protocols Handbook, Clontech)
Plant material
Arabidopsis plants were grown under long-day conditions as previously described (McAbee et al. 2005). The alleles used in this study were ada2a (SALK_150349), ada2b-1 (Vlachonasios et al. 2003), ino-1 (Villanueva et al. 1999), lug-1 (Liu and Meyerowitz 1995), and seu-1 (Franks et al. 2002). All plants were genotyped using PCR-based markers to confirm the presence of each mutation and the primers used are described in Supplemental Material, Table S1. The P-PRZ1::PRZ1:GFP (P-ADA2b::ADA2b:GFP) transgenic line (Anzola et al. 2010) was a kind gift from Christian Luschnig [University of Natural Resources and Applied Life Sciences (BOKU), Vienna, Austria].
Microscopy
Scanning electron microscopy and ovule clearings were performed as described (Kelley et al. 2009). Differential interference contrast light microscopy was performed as described (Meister et al. 2002). Confocal images were taken with a Zeiss LSM 710 system using 488 nm laser excitation and recording 493–598 nm emission wavelengths (Zeiss [Carl Zeiss], Thornwood, NY). Images were edited in Adobe Photoshop CS5.
Data availability
Plant materials are available from the cited sources from which they were obtained. Plasmids are available from the corresponding author on request.
Results
Identifying INO-interacting proteins
We performed a yeast two-hybrid screen to identify putative INO-interacting proteins, using the full-length INO protein fused to the GAL4 DNA-binding domain (BD-INO) as bait and a library of Arabidopsis pistil cDNA fused to the coding region of the GAL4 activation domain to produce prey. Several positive clones were identified, including a clone encoding the N-terminal region of the ADA2a transcriptional coactivator protein (Stockinger et al. 2001) and one encoding the C-terminal 85% of the LUG transcription corepressor protein (Conner and Liu 2000). Because these were both consistent with the hypothesized action of INO as a transcription factor, they were selected for further analysis.
INO interacts with ADA2a and ADA2b in yeast
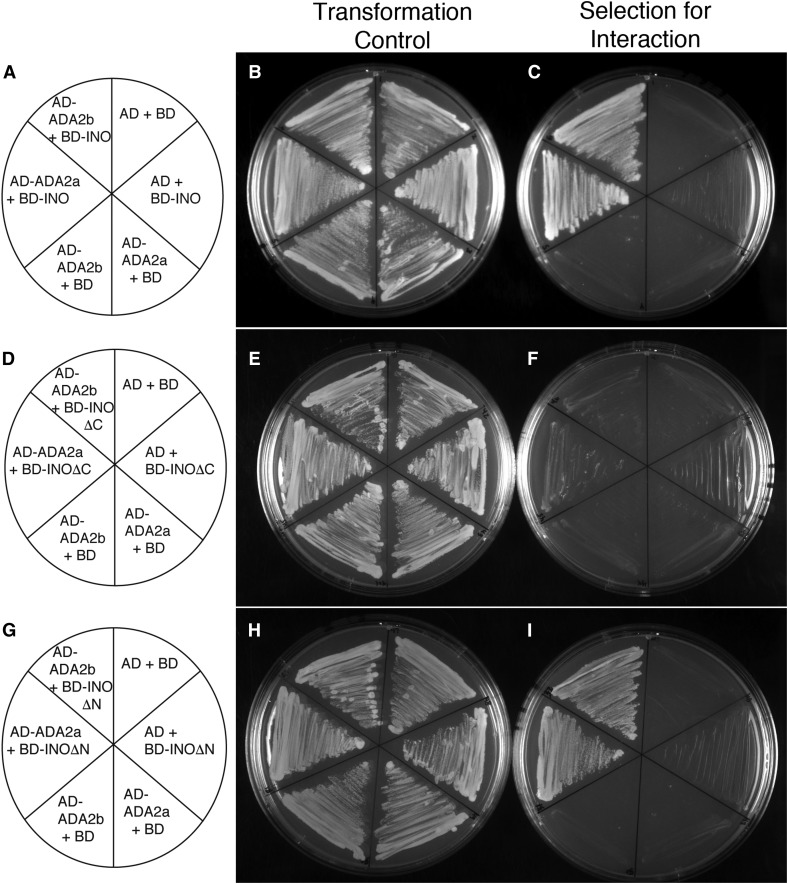
We produced fusions of full-length ADA2a, and the related paralog ADA2b (PRZ1), and showed that both of these proteins interact with INO in yeast (Figure 1). We examined the interaction of the ADA2 proteins with two conserved domains of the INO protein: the N-terminal region including the zinc finger motif (INOΔC) and the C-terminal region including the YABBY domain (INOΔN) (Bowman and Smyth 1999; Villanueva et al. 1999). Both ADA2a and ADA2b proteins showed interaction with the C-terminal region of INO that includes the YABBY domain in a plating assay (Figure 1I). This interaction was confirmed for ADA2b with enzyme activity assays (Figure S1 in File S1). Both ADA2a and ADA2b proteins also displayed a statistically significant interaction with the N-terminal region of INO (INOΔC) in the enzyme activity assay (Figure S1 in File S1), but this interaction was less apparent in the plating assay (Figure 1F). Thus, the C-terminal region of INO may be the primary determinant of interaction with these coactivators, but there may also be some contribution from the N-terminal region.
Figure 1.
Yeast two-hybrid assays detect interactions between ADA2a and ADA2b and INO. (A, D, and G) Maps showing which proteins were produced in yeast strains streaked on the adjacent plates. “AD” and “BD” indicate the activation and DNA-binding domains of yeast GAL4, respectively. (B, E, and H) Streaks of indicated yeast strains on media lacking leucine and tryptophan, selecting only for the presence of the two expression plasmids in each strain. (C, F, and I) Streaks of indicated yeast strains on media lacking leucine, tryptophan, adenine, and histidine selected for interaction of the indicated proteins. The results show that none of the tested constructs significantly auto-activated, but that both AD-ADA2a and AD-ADA2b productively interact with both BD-INO and BD-INOΔN. A possible weak interaction of the ADA2 fusions with BD-INOΔC is also visible.
The interaction of ADA2 proteins with the conserved YABBY region of INO suggested that this interaction may be a general feature of YABBY proteins, not an exclusive interaction with INO. Yeast two-hybrid assays with the other Arabidopsis YABBY proteins revealed interactions of both ADA2a and ADA2b with FIL, YAB2, YAB3, and YAB5, but no interactions were observed with CRC (Figure S2 in File S1 and Table 1). As previously reported (Bonaccorso et al. 2012), the BD-FIL fusion autoactivates reporter gene expression in yeast; however, we found that the growth of yeast (as a measure of protein interaction to activate selectable gene expression) is strongly enhanced when an ADA2 protein is also present (Figure S2 in File S1 and Table 1). Thus, our yeast two-hybrid data indicate that the ADA2 coactivator proteins can interact with all Arabidopsis YABBY proteins excepting CRC. The absence of observed phenotypic effects of loss-of-function mutations in ADA2a [Vlachonasios et al. (2003) and see below] led us to focus our further efforts on ADA2b.
Table 1. Yeast two-hybrid analysis of ADA2 interactions with YABBY proteins.
| AD | |||
|---|---|---|---|
| BD | Empty | ADA2A | ADA2B |
| Empty | — | — | — |
| FIL | + | +++ | +++ |
| YAB2 | — | + | ++ |
| YAB3 | — | +++ | +++ |
| YAB5 | — | + | ++ |
| CRC | — | — | — |
| INO | — | +++ | +++ |
AD, activation domain; BD, DNA-binding domain.
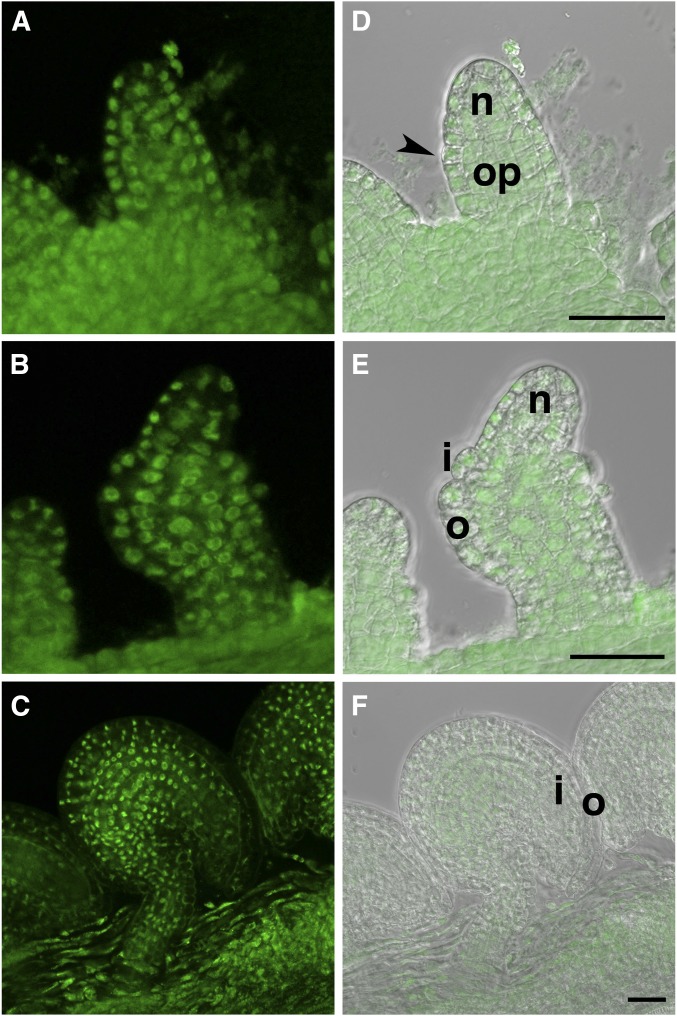
For the interaction of ADA2b and INO to be biologically relevant, the two proteins must be present in the same cells and cellular compartment in the plant. We evaluated the expression pattern and cellular localization of ADA2b in ovules by examining the ovules of a line containing a reporter gene comprising the entire genomic copy of ADA2b with an in-frame fusion to a GFP coding region (Anzola et al. 2010). We found the transgene to be expressed in all cells of the ovule throughout development, with the fusion protein product being confined to the nucleus (Figure 2). INO is expressed in cells of the outer layer of the outer integument during the development of this organ, where it is also confined to the nucleus (Meister et al. 2002). Thus, the two proteins are present in the same cellular compartment in cells of the outer integument.
Figure 2.
Accumulation of ADA2b-GFP (PRZ-GFP) fusion in ovule cells. The expression pattern of a P-PRZ1::PRZ1:GFP (P-ADA2b::ADA2b:GFP) fusion in a genomic fragment that complemented the prz1 mutant (Sieberer et al. 2003; Anzola et al. 2010) was evaluated by laser confocal microscopy. Left panels are GFP fluorescence and the right panels are the fluorescent images overlaid on differential interference contrast images. Images are of ovules at stages: (A and D) 2-I; (B and E) 2-IV; and (C and F) 4-V. Expression is in all cells at all stages of ovule development and the fusion protein accumulates in the nucleus. Arrowhead, site of outer integument initiation; i, inner integument; n, nucellus; o, outer integument; and op, ovule primordium. Bar, 25 µm.
Loss of ADA2b function results in defective ovule development
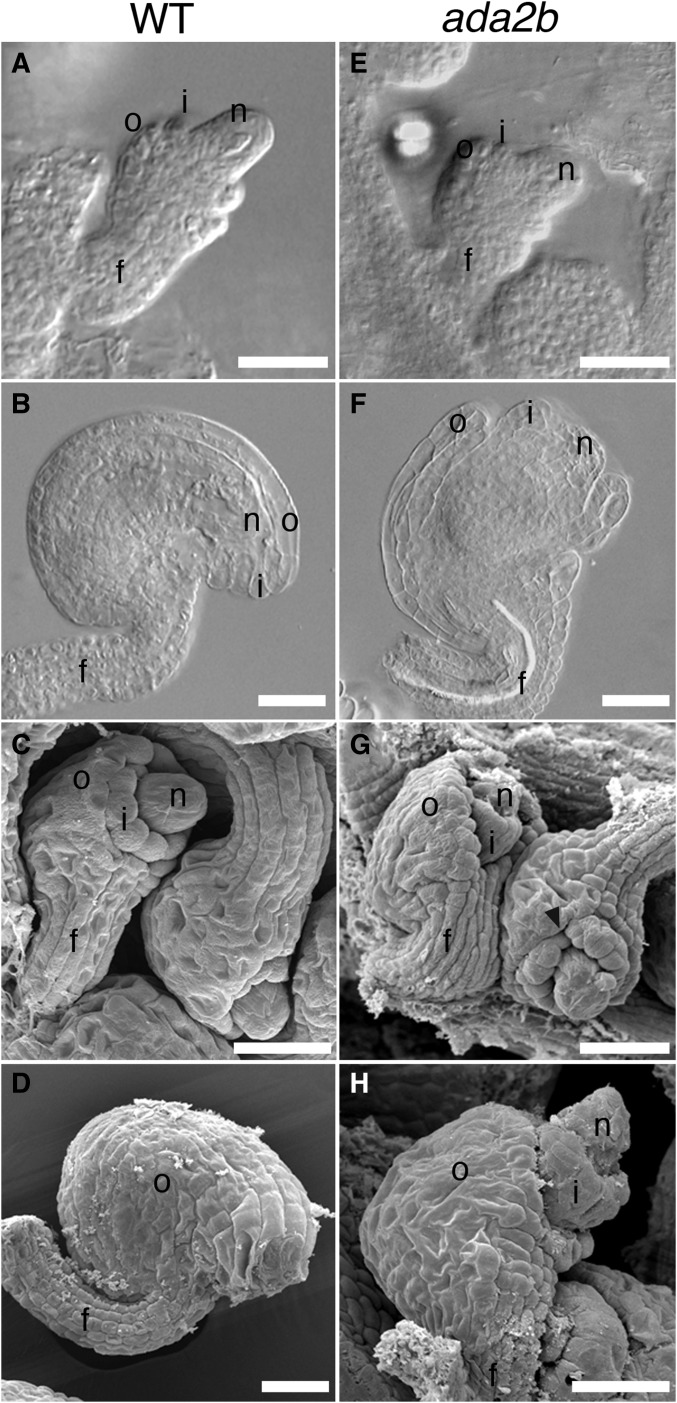
Previously, the ADA2b loss-of-function allele, ada2b-1, was shown to have pleiotropic effects on plant growth and development, while loss of ADA2a did not have visible effects (Vlachonasios et al. 2003; Hark et al. 2009). Consistent with these previous reports, we found that ada2a mutants did not exhibit observable defects in vegetative or reproductive parts, including ovules (data not shown). In the ada2b-1 mutant, in addition to the previously described vegetative defects, we observed defects in ovule development. Initiation and extension of the integuments from the chalaza through stage 2-V [stages according to Schneitz et al. (1995)] were unaffected and appeared similar to wild-type (Figure 3, A and E). However, at later stages of ovule development, both the outer and inner integuments exhibited growth defects. Growth of the outer integument was variable, but always reduced compared to wild-type, resulting in an exposed inner integument and nucellus (Figure 3, F–H). The growth of the outer integument was asymmetric, as in wild-type, resulting in a recurved shape of the ovule at anthesis (Figure 3, F and H). Growth of the inner integument was also reduced, and the nucellus was exposed beyond the inner integument at anthesis (Figure 3, F–H). We observed that inconsistent growth in the margin of the inner integument produced gaps in this structure, and saw that the funiculus was increased in diameter in some ovules (Figure 3G).
Figure 3.
ada2b mutation affects ovule development. Differential interference contrast (A, B, E, and F) and scanning electron (C, D, G, and H) micrographs of wild-type (A–D) and ada2b (E–H) ovules. At stage 2-IV, ovules of ada2b (E) are similar to those of wild-type at this same stage (A). At stage 3-III, discontinuous growth of the inner integument results in fissures in this structure (arrowhead) in ada2b (G), while this structure is continuous in wild-type at this same stage (C). At the completion of prefertilization ovule development at stage 3-V, the integuments of wild-type ovules have covered the inner integument (B and D), while at this same stage the inner integument and nucellus are both exposed in ada2b due to decreased extension of the outer integument (F and H). f, funiculus; i, inner integument; n, nucellus; and o, outer integument. Bar, 25 µm.
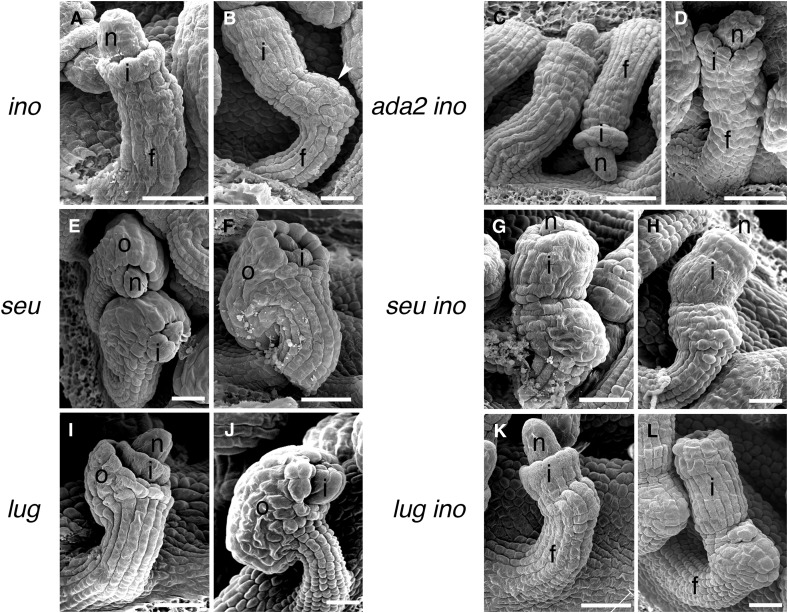
The ino-1 mutation eliminates the outer integument but the remainder of the ovule, including the inner integument, appears to continue to develop normally (Baker et al. 1997; Villanueva et al. 1999) (Figure 4, A and B). Consistent with a combination of the effects of the two mutations, the ino-1 ada2b-1 double mutant lacked an outer integument and exhibited reduced inner integument growth (Figure 4, C and D). Gaps in the margin of the inner integument similar to those seen in ada2b-1 were also present (Figure 4D). The double mutant ovules also lacked the outgrowth present on the gynoapical side (toward the apex of the gynoecium) of the chalaza that is present on ino-1 mutant ovules (cf. Figure 4, B and D). The double mutant ovules varied in the visibility of the groove delineating the base of the inner integument with many lacking this feature, and cells in the funiculus became progressively less organized relative to wild-type (Figure 4, C and D). Thus, ino-1 was epistatic to ada2b-1 in promotion of outer integument growth, but ada2b-1 had additional effects on ovule development in the inner integument, chalazal region, and funiculus, structures where INO is not expressed and does not play a role.
Figure 4.
Interactions of ada2b, lug, and seu mutations with ino in ovule development. Scanning electron micrographs of ovules of the indicated single and double mutants are shown. As previously reported (Villanueva et al. 1999), ino-1 ovules initiate only an inner integument (A) and lack outer integuments at maturity (B). The inner integument and a thickening of the chalazal region (arrowhead) are exposed by the absence of the outer integument. In the combination of ada2b with ino (C and D), the chalazal thickening is absent, the boundary at the insertion of the inner integument is often indistinct, and fissures are visible in the inner integument (D). Both seu and lug mutants initiate both integuments (E and I), but the outer integuments of both mutants exhibit reduced growth and fail to cover the inner integument at maturity (F and J). The inner integument of seu mutants also shows reduced growth and fails to cover the nucellus at maturity (F), and this is more clearly visible when the outer integument is eliminated in the seu ino-1 double mutant (G and H). The inner integument appears unaffected in lug and in the lug ino-1 double mutant (K and L). f, funiculus; i, inner integument (primordium); n, nucellus; and o, outer integument. Bar, 25 µm.
INO interacts with INO, LUG, and SEU in yeast
It was previously reported that some YABBY proteins were able to form homodimers and heterodimers (Stahle et al. 2009). Evaluation of INO protein in the yeast two-hybrid assay showed formation of an INO homodimer in yeast (Figure S3 in File S1 and Table 2). The potential interaction of INO with other YABBY proteins was not investigated because INO is the only YABBY gene expressed in the ovule (Bowman and Smyth 1999; Siegfried et al. 1999; Villanueva et al. 1999). Assays with truncated forms of INO showed that the C-terminal region (INOΔN), including the conserved YABBY domain but not the zinc finger region, was sufficient to promote the self-interaction of the INO protein in yeast (Figure S3 in File S1 and Table 2).
Table 2. Yeast two-hybrid analysis of INO interactions with INO, LUG, and SEU.
| AD | ||||
|---|---|---|---|---|
| BD | Empty | INO | LUG | SEU |
| Empty | — | — | — | — |
| INO | — | +++ | +++ | +++ |
| INOΔC | — | +++ | +++ | +++ |
| INOΔN | — | — | — | + |
AD, activation domain; BD, DNA-binding domain.
The yeast two-hybrid screen with BD-INO also identified a positive interaction with a truncated LUG protein. We found that INO also interacted with the full-length LUG protein (Figure S3 in File S1 and Table 2), as previously seen for FIL/YAB2/3 vegetatively expressed Arabidopsis YABBY proteins (Stahle et al. 2009). Interaction of these YABBY proteins with the LUG partner corepressor SEU has also been demonstrated (Stahle et al. 2009), and we found that INO also interacted with SEU in the yeast two-hybrid assay (Figure S3 in File S1 and Table 2). The regions of INO necessary for the interaction with LUG and SEU were tested, and the data indicated that the N-terminal region of INO (INOΔC), including the zinc finger region, was sufficient to promote the interaction with the LUG and SEU proteins (Figure S3 in File S1 and Table 2). There was also evidence of some interaction of SEU with the C-terminal region of INO (INOΔN) (Figure S3 in File S1 and Table 2).
Roles of LUG and SEU in ovule development
Both LUG and SEU are expressed in ovules, including the cells known to express INO (Conner and Liu 2000; Bao et al. 2010). Previous reports have also shown that lug and seu mutants exhibit ovule integument defects characterized by a reduction in outer integument growth relative to the inner integument (Roe et al. 1997; Franks et al. 2002). Here, we observed similar phenotypes of an exposed portion of the inner integument at anthesis, but note that the initiation and growth of the outer integument appeared similar to wild-type at early stages of ovule development for both mutants (Figure 4).
As in the ino-1 single mutant (Figure 4, A and B), in lug-1 ino-1 and seu-1 ino-1 double mutants the outer integument was not initiated (Figure 4, G, H, K, and L). Thus, the ino-1 phenotype in the outer integument was epistatic to both the lug-1 and seu-1 mutant phenotypes. The absence of the outer integument in the ino-1 lug-1 and ino-1 seu-1 mutants also allowed a more complete examination of the growth and development of the inner integument. lug-1 ino-1 did not differ from the ino-1 single mutant and the inner integument covered the nucellus at anthesis (Figure 4L). In contrast, in seu-1 ino-1, the extent of inner integument growth was reduced, resulting in ovules with an exposed nucellus at anthesis (Figure 4, G and H). Unlike the ada2b-1 mutant, which showed both reduced inner integument growth and also gaps or divisions in the overall structure of the integument, the continuous structure of the inner integument was maintained in seu-1 mutants. In contrast to the ada2b-1 ino-1 mutant, mutant combinations with seu-1 and lug-1 did not alter the funiculus or affect the gynoapical outgrowth on the chalaza and maintained the distinct groove at the base of the inner integument.
Genetic interaction of lug and ada2b
While effects of the ada2b, lug, and seu mutants included reduced outer integument growth, the growth of this structure was always greater than observed in the strong ino-1 allele or even the weaker ino-4 allele (Villanueva et al. 1999; Figure 3 and Figure 4). This indicates that some INO function was present to promote partial growth of the outer integument in both coactivator and corepressor mutant lines. Thus, we combined the two classes of coregulator mutations, producing lug-1 ada2b-1 double mutants, and found that this produced greater defects in outer integument growth than either single mutant, as well as other effects on ovule development (Figure 5). The outer integument of the lug-1 ada2b-1 mutant initiated on the correct gynobasal side of the ovule and grew asymmetrically around the ovule, but the growth of the outer integument was deficient at earlier stages in ovule development than in either single mutant (Figure 5). While some ovules simply had reduced outer integuments (Figure 5A), the directional growth of the outer integument was more commonly seriously disrupted (Figure 5, B and C). For 52 of 94 ovules in 28 examined carpels, the outer integument formed a large group of cells at the chalazal region that lacked directional growth or grew in the opposite direction toward the funiculus (Figure 5, B and C). Furthermore, in some ovules, disruption of integument development was even more extreme, with multiple aberrant structures emerging from the chalaza (Figure 5C). This is a novel effect not observed in either single mutant. In all cases, there was less growth of the outer integument in the lug-1 ada2b-1 double mutant than in either of the two single mutants. The additive and synergistic aspects of the double mutant indicate that LUG and ADA2b act independently to promote outer integument growth. As a consequence of the reduced and aberrant outer integument growth, the overall shape of the mature ovule was also compromised (Figure 5, B and C).
Figure 5.
Synergistic interactions of ada2b and lug mutations on ovule and carpel development. (A–C) ovules of lug ada2b double mutants at stage 12. While occasional ovules have a relatively normal but reduced outer integument (A), the majority of ovules show a synergistic effect on the outer integument, where it forms an amorphous collar of tissue (B and C). The inner integument may also be affected (C). Ovules of the lug ada2b ino triple mutant at stage 12 (D–G) most closely resemble those of the ino ada2b double mutant, with an absent outer integument and often indistinct base of the inner integument (D). Growth to stage 13 shows aberrant expansion of the funiculus, which can protrude up around the inner integument (G) (arrowhead). Some very aberrant ovules can have two nucelli (G) (*), indicating probable fusion of two ovule primordia. Carpels of ada2b mutants (I) resemble those of wild-type (H), but lug mutants have aberrant carpels (J) with horn-like protrusions (arrowheads) interrupting the stigma. Such horns are absent in lug ada2b double mutants (K) and carpel tissue is reduced such that often only a single valve is present in the gynoecium. i, inner integument (primordium); n, nucellus; o, outer integument; and v, valve. Bar, 20 µm in (A–C), 25 µm in (D–G), and 100 µm in (H–K).
The addition of the ino-1 mutation to the combination of lug-1 and ada2b-1 largely eliminated the production of cells in the region normally occupied by the outer integument (Figure 5, D–G). The resulting ovules often resemble ino-1 ada2b-1 ovules (compare Figure 4, C and D with Figure 5, D–F). Some ovules were more aberrant, showing multiple nucelli on structures that resembled fused adjacent ovules (Figure 5G), possibly indicating defects in ovule spacing in the gynoecium.
Pleiotropic mutant phenotypes of lug-1 in leaf and floral specification and morphology, and ada2b-1 in cell elongation/proliferation and leaf/flower morphology, have been previously reported (Liu and Meyerowitz 1995; Liu et al. 2000; Sieberer et al. 2003; Vlachonasios et al. 2003; Hark et al. 2009). These phenotypic effects were also observed in the lug-1 ada2b-1 double mutant, including the carpelloid sepals characteristic of the lug-1 mutant (Liu and Meyerowitz 1995) and the reduced stature and altered leaf morphology characteristic of the ada2b-1 mutant. However, in addition to the novel ovule phenotype, this double mutant also exhibited novel defects in the structure of the gynoecium. The lug-1 mutant is characterized by unfused carpels with a horn-like protrusion at the tip of the carpel valve (Liu and Meyerowitz 1995) (Figure 5J), and the ada2b-1 mutant did not show any significant carpel alterations (Figure 5I). In contrast, the lug-1 ada2b-1 double mutant frequently developed a gynoecium with decreased ovary tissue, often forming only a single ovary (Figure 5K). This phenotype was variable in the extent of fusion of the gynoecium along the medial domain, the presence of horn-like protrusions from the carpel valve regions, and also the number of carpels.
Discussion
Mutations in ADA2b, SEU, and LUG all produce similar decreases in growth of the outer integument, where extension is variable but always less than that of wild-type. These observations, combined with the observation of expression of all three genes in the outer integument, demonstrate roles for the protein products of these genes in outer integument growth and extension. As a known coactivator (Vlachonasios et al. 2003) and two known interacting corepressors (Franks et al. 2002; Navarro et al. 2004; Sridhar et al. 2004), these proteins would perform their functions through regulation of transcription, but would require a partner protein with DNA-binding activity to target specific genes. The observation that these coregulators can interact with INO suggests INO as such a partner for the regulation of outer integument growth. Notably, INO is a master regulator of outer integument formation, being required for both the initiation and subsequent growth of this structure (Villanueva et al. 1999). The observed interaction of INO with SEU and LUG is further supported by prior observation of interaction of these corepressors with INO orthologs, other YABBY proteins in other species (Navarro et al. 2004), and with other YABBY proteins in Arabidopsis (Stahle et al. 2009). Our observation that ADA2b (PRZ1) can interact with a range of other YABBY proteins further supports the interaction. Indeed, evidence suggests that vegetative YABBY proteins have both negative and positive effects on transcription (Bonaccorso et al. 2012), and our results would provide a mechanistic explanation for this.
Our observation that a severe ino loss-of-function mutation was epistatic to seu, lug, and ada2b mutations in the outer integument is what is expected if INO is a tether enabling the gene-specific function of these coregulators in the outer integument. The effects of mutations in these coregulators on other plant processes outside of the outer integument would involve DNA-binding proteins other than INO and so are not altered by ino mutations. The repressive effects of the corepressors and the activating effect of the ADA2b coactivator would be expected to be directed at different sets of genes in outer integument growth. Eliminating INO as a vector of both coactivators and corepressors would be expected to have more extreme effects than mutations in either single class of coregulator. This is indeed what we observed, with complete loss of the outer integument in the strong ino mutants, and only partial loss of outer integument growth observed for either loss of coactivator or corepressor function. This model is further supported by the observation that effects of combining corepressor and coactivator mutations (in the ada2b lug mutant) are both additive and synergistic. Mutants with defects in genes in both classes would be expected to have a more severe effect on growth of the outer integument than either single coregulator mutation, and this is what was observed. However, some outer integument growth persisted in the ada2b lug double mutant, but this was eliminated by the addition of an ino mutation. This indicated participation of INO in the residual outer integument growth. We note that a paralog of LUG, LUH, has been shown to have activity that is partially redundant with the activity of LUG (Sitaraman et al. 2008), and this could account for the residual outer integument growth activated by INO in the ada2b lug double mutant.
This model for INO as a tether binding the coregulators to DNA relies on INO being a DNA-binding transcription factor or a bridge to such a factor. INO is nuclear-localized (Meister et al. 2002), as would be expected for a transcription factor. Initial attempts to directly identify sequences specifically bound by a YABBY protein (FIL) found only weak, nonspecific DNA-binding activity (Kanaya et al. 2002). However, more recently, Franco-Zorrilla et al. (2014) used DNA microarrays to detect binding of FIL and YAB3 to specific DNA sequences. Also, using ChIP, Shamimuzzaman and Vodkin (2013) were able to detect binding of soybean FIL/YAB3 orthologs to specific sites in the genome and were able to find evidence for a consensus binding motif that was different from the motif identified in the microarray study. The latter study supports YABBY proteins as DNA-binding proteins in vivo, where there might be additional factors (including other necessary proteins) facilitating DNA binding that are absent in in vitro and heterologous assays. If INO is a DNA-binding protein then our observation of an ability of INO to interact with itself via the YABBY domains could indicate that it acts as a homodimer. Prior work has shown both homo- and heterodimerization between YABBY proteins (Kanaya et al. 2001; Stahle et al. 2009). However, because INO is the only YABBY gene that has been shown to be expressed in ovules (Bowman and Smyth 1999; Villanueva et al. 1999), only the homodimer is a possibility for INO protein in vivo.
Evaluation of the regions of INO binding ADA2 and LUG/SEU indicates that the two classes of proteins have differential affinity for different INO regions. While some binding was detected for each class to both the N- and C-terminal regions of INO, binding of the coactivator was strongest to the C-terminal region of INO, while binding of corepressors was preferential to the N-terminal region. These results indicate that, like some other transcription factors (Kurokawa et al. 1995), INO could potentially simultaneously interact with both corepressors and coactivators.
Both ADA2b and the paralogous ADA2a were found to interact with INO. The absence of phenotypic effects of the ada2a mutation, even in combination with an ada2b background, led us to not study ada2a mutants further. Evidence for ADA2b as a coactivator that facilitates recruitment of the HAT GCN5 to DNA-binding factors is strong (Vlachonasios et al. 2003; Hark et al. 2009; Kornet and Scheres 2009; Anzola et al. 2010). This type of mechanism has been demonstrated for the activation of cold response genes via transcription factor CBF1 recruitment of ADA2b and GCN5 (Stockinger et al. 2001; Mao et al. 2006). The HAT activity of GCN5 is recruited by other transcription factors to affect a wide variety of additional processes in plants, including maintenance of the stem cell niche (Kornet and Scheres 2009), floral meristem activity (Bertrand et al. 2003), light responses (Benhamed et al. 2006), and auxin responses (Anzola et al. 2010; Weiste and Droge-Laser 2014). To these, we would add a likely involvement in ovule development through interaction with INO, and possibly participation in polarity determination via interaction with other members of the YABBY gene family. Notably, tubular or filamentous growths were observed in the inflorescence of the gcn5 mutant, and this resembles effects of fil mutants (Cohen et al. 2009). This is consistent with GCN5/ADA2b also functioning in the lateral organ YABBY pathway.
Several lines of evidence support the function of SEU and LUG acting as a transcriptional corepressor complex (Sridhar et al. 2004; Stahle et al. 2009; Grigorova et al. 2011). While we studied these genes individually, mutant combinations of lug and seu did not lend themselves to the study of mutant ovules because they cause such severe effects on plant and flower growth that formation of ovule-like structures is rare (Franks et al. 2002). This is likely due to participation in multiple associations with a variety of transcription factors during gynoecium development (Pfluger and Zambryski 2004; Sridhar et al. 2006), likely including other YABBY proteins (Stahle et al. 2009). The effects of lug and seu mutations are likely not indications of the complete loss-of-function of the corepressor activity because both have at least partially redundant paralogous proteins: LUH (Sridhar et al. 2004) and SLK1/2/3 (Bao et al. 2010). Thus, the mutations examined in this work may not show the full extent of participation of these classes of corepressors in ovule development. Recently, evidence that LUH might also participate in transcriptional activation as well as repression has been reported (Lee et al. 2015), and LUG/LUH associations could therefore also partly account for gene activation by YABBY proteins.
The challenge of determining the actual biochemical role of proteins genetically identified as being important in development is daunting. It is facilitated by the incremental assembly of multiple pieces of independently acquired evidence, as undertaken in the current work. The importance of YABBY proteins in the development and polarity of lateral organs and ovule integuments is well established (Bowman and Smyth 1999; Villanueva et al. 1999; Sarojam et al. 2010). The specific patterns of expression of these genes are consistent with their direct participation in the development of these structures (Bowman and Smyth 1999; Villanueva et al. 1999). Their nuclear localization and similarities to known DNA-binding proteins suggested that they might be DNA-binding transcription factors. ChIP and DNA-binding assays now provide further support for this (Shamimuzzaman and Vodkin 2013; Franco-Zorrilla et al. 2014). Prior results indicating interactions with SEU and LUG (and both paralogous proteins and orthologs in other species) provided a first indication of a possible mechanism of action, but did not explain the apparent positive and negative action on gene expression. Our results showing interaction with both coactivator and corepressor complexes now provide a possible mechanism for these conflicting activities. Additional interactions of the YABBY proteins with other proteins would be necessary to provide the discrimination required to differentially associate the activating and repressive complexes with their specific downstream target genes. Such factors might be expected to recognize both a component of the repressor or activator complex as well as target genes using their regulatory sequences. At least one possible candidate for this may have already been described. The NOZZLE/SPOROCYTELESS (NZZ/SPL) protein has been shown to associate with INO (Sieber et al. 2004) and has more recently been shown to repress expression of specific genes through association with the TOPLESS (TPL) corepressor (Wei et al. 2015). An association with INO could form a multifunctional repressive complex combining the LUG/SEU and TPL activities to target specific genes during integument development. Alternative associations could specifically target INO bound to ADA2b to a different set of genes for activation.
Further insight could be provided by the identification of direct targets of INO regulation and determination of the subset of genes regulated by INO that are also regulated by ADA2b or by LUG and SEU. Interestingly, the DAMAGED DNA-BINDING PROTEIN 1B (DDB1B) gene [encoding part of the CULLIN4 (CUL4) E3 ligase that acts in photomorphogenesis (Schroeder et al. 2002; Bernhardt et al. 2006)] is upregulated 19-fold in flowers of lug mutants (Gonzalez et al. 2007), and is also upregulated 20-fold in ino mutant pistils (Skinner and Gasser 2009). Thus, this gene could be a target of an INO/LUG/SEU complex. The ETTIN (ETT) gene is upregulated twofold in both lug flowers and ino pistils (Gonzalez et al. 2007; Skinner and Gasser 2009). ETT expression must be repressed in the outer integument for normal development of this structure (Kelley et al. 2012) and so ETT is another potential target for a repressive INO/LUG/SEU complex. Additional potential targets for repression and for activation by INO remain to be discovered.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300140/-/DC1.
Acknowledgments
We thank Kristina Passerini and Eliana Goldstein for technical assistance and Christian Luschnig for plant material. This work was supported by the National Science Foundation (grant IOS-1354014).
Footnotes
Communicating editor: S. Poethig
Literature Cited
- Anzola J. M., Sieberer T., Ortbauer M., Butt H., Korbei B., et al. , 2010. Putative Arabidopsis transcriptional adaptor protein (PROPORZ1) is required to modulate histone acetylation in response to auxin. Proc. Natl. Acad. Sci. USA 107: 10308–10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. C., Robinson-Beers K., Villanueva J. M., Gaiser J. C., Gasser C. S., 1997. Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics 145: 1109–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao F., Azhakanandam S., Franks R. G., 2010. SEUSS and SEUSS-LIKE transcriptional adaptors regulate floral and embryonic development in Arabidopsis. Plant Physiol. 152: 821–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M., Bertrand C., Servet C., Zhou D. X., 2006. Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18: 2893–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt A., Lechner E., Hano P., Schade V., Dieterle M., et al. , 2006. CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J. 47: 591–603. [DOI] [PubMed] [Google Scholar]
- Bertrand C., Bergounioux C., Domenichini S., Delarue M., Zhou D. X., 2003. Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J. Biol. Chem. 278: 28246–28251. [DOI] [PubMed] [Google Scholar]
- Bonaccorso O., Lee J. E., Puah L., Scutt C. P., Golz J. F., 2012. FILAMENTOUS FLOWER controls lateral organ development by acting as both an activator and a repressor. BMC Plant Biol. 12: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. L., 2000. The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 3: 17–22. [DOI] [PubMed] [Google Scholar]
- Bowman J. L., Smyth D. R., 1999. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126: 2387–2396. [DOI] [PubMed] [Google Scholar]
- Cohen R., Schocken J., Kaldis A., Vlachonasios K. E., Hark A. T., et al. , 2009. The histone acetyltransferase GCN5 affects the inflorescence meristem and stamen development in Arabidopsis. Planta 230: 1207–1221. [DOI] [PubMed] [Google Scholar]
- Colombo L., Battaglia R., Kater M. M., 2008. Arabidopsis ovule development and its evolutionary conservation. Trends Plant Sci. 13: 444–450. [DOI] [PubMed] [Google Scholar]
- Conner J., Liu Z., 2000. LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc. Natl. Acad. Sci. USA 97: 12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., Lopez-Vidriero I., Carrasco J. L., Godoy M., Vera P., et al. , 2014. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 111: 2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks R. G., Wang C., Levin J. Z., Liu Z., 2002. SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129: 253–263. [DOI] [PubMed] [Google Scholar]
- Gaiser J. C., Robinson-Beers K., Gasser C. S., 1995. The Arabidopsis SUPERMAN gene mediates asymmetric growth of the outer integument of ovules. Plant Cell 7: 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D., Bowen A. J., Carroll T. S., Conlan R. S., 2007. The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol. Cell. Biol. 27: 5306–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. A., Duggan L., Cote J., Roberts S. M., Brownell J. E., et al. , 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11: 1640–1650. [DOI] [PubMed] [Google Scholar]
- Grigorova B., Mara C., Hollender C., Sijacic P., Chen X. M., et al. , 2011. LEUNIG and SEUSS co-repressors regulate miR172 expression in Arabidopsis flowers. Development 138: 2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark A. T., Vlachonasios K. E., Pavangadkar K. A., Rao S., Gordon H., et al. , 2009. Two Arabidopsis orthologs of the transcriptional coactivator ADA2 have distinct biological functions. Biochim. Biophys. Acta 1789: 117–124. [DOI] [PubMed] [Google Scholar]
- Kanaya E., Watanabe K., Nakajima N., Okada K., Shimura Y., 2001. Zinc release from the CH2C6 zinc finger domain of FILAMENTOUS FLOWER protein from Arabidopsis thaliana induces self-assembly. J. Biol. Chem. 276: 7383–7390. [DOI] [PubMed] [Google Scholar]
- Kanaya E., Nakajima N., Okada K., 2002. Non-sequence-specific DNA binding by the FILAMENTOUS FLOWER protein from Arabidopsis thaliana is reduced by EDTA. J. Biol. Chem. 277: 11957–11964. [DOI] [PubMed] [Google Scholar]
- Kelley D. R., Skinner D. J., Gasser C. S., 2009. Roles of polarity determinants in ovule development. Plant J. 57: 1054–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D. R., Arreola A., Gallagher T. L., Gasser C. S., 2012. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 139: 1105–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornet N., Scheres B., 2009. Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 21: 1070–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa R., Soderstrom M., Horlein A., Halachmi S., Brown M., et al. , 1995. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature 377: 451–454. [DOI] [PubMed] [Google Scholar]
- Lee N., Park J., Kim K., Choi G., 2015. The transcriptional coregulator LEUNIG_HOMOLOG inhibits light-dependent seed germination in Arabidopsis. Plant Cell 27: 2301–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Franks R. G., Klink V. P., 2000. Regulation of gynoecium marginal tissue formation by LEUNIG and AINTEGUMENTA. Plant Cell 12: 1879–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. C., Meyerowitz E. M., 1995. LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121: 975–991. [DOI] [PubMed] [Google Scholar]
- Lora J., Hormaza J. I., Herrero M., Gasser C. S., 2011. Seedless fruits and the disruption of a conserved genetic pathway in angiosperm ovule development. Proc. Natl. Acad. Sci. USA 108: 5461–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Pavangadkar K. A., Thomashow M. F., Triezenberg S. J., 2006. Physical and functional interactions of Arabidopsis ADA2 transcriptional coactivator proteins with the acetyltransferase GCN5 and with the cold-induced transcription factor CBF1. Biochim. Biophys. Acta 1759: 69–79. [DOI] [PubMed] [Google Scholar]
- McAbee J. M., Kuzoff R. K., Gasser C. S., 2005. Mechanisms of derived unitegmy among Impatiens species. Plant Cell 17: 1674–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister R. J., Kotow L. M., Gasser C. S., 2002. SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development 129: 4281–4289. [DOI] [PubMed] [Google Scholar]
- Navarro C., Efremova N., Golz J. F., Rubiera R., Kuckenberg M., et al. , 2004. Molecular and genetic interactions between STYLOSA and GRAMINIFOLIA in the control of Antirrhinum vegetative and reproductive development. Development 131: 3649–3659. [DOI] [PubMed] [Google Scholar]
- Pfluger J., Zambryski P., 2004. The role of SEUSS in auxin response and floral organ patterning. Development 131: 4697–4707. [DOI] [PubMed] [Google Scholar]
- Robinson-Beers K., Pruitt R. E., Gasser C. S., 1992. Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell 4: 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe J. L., Nemhauser J. L., Zambryski P. C., 1997. TOUSLED participates in apical tissue formation during gynoecium development in Arabidopsis. Plant Cell 9: 335–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol P., Collier S., Bush M., Shaw P., Doonan J. H., 2007. Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J. Cell Sci. 120: 3678–3687. [DOI] [PubMed] [Google Scholar]
- Sarojam R., Sappl P. G., Goldshmidt A., Efroni I., Floyd S. K., et al. , 2010. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 22: 2113–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S., Watanabe K., Goto K., Kanaya E., Morita E. H., et al. , 1999. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13: 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz K., Hulskamp M., Pruitt R. E., 1995. Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. Plant J. 7: 731–749. [Google Scholar]
- Schneitz K., Hulskamp M., Kopczak S., Pruitt R., 1997. Dissection of sexual organ ontogenesis: a genetic analysis of ovule development in Arabidopsis thaliana. Development 124: 1367–1376. [DOI] [PubMed] [Google Scholar]
- Schroeder D. F., Gahrtz M., Maxwell B. B., Cook R. K., Kan J. M., et al. , 2002. De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr. Biol. 12: 1462–1472. [DOI] [PubMed] [Google Scholar]
- Shamimuzzaman M., Vodkin L., 2013. Genome-wide identification of binding sites for NAC and YABBY transcription factors and co-regulated genes during soybean seedling development by ChIP-Seq and RNA-Seq. BMC Genomics 14: 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P., Petrascheck M., Barberis A., Schneitz K., 2004. Organ polarity in Arabidopsis. NOZZLE physically interacts with members of the YABBY family. Plant Physiol. 135: 2172–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer T., Hauser M. T., Seifert G. J., Luschnig C., 2003. PROPORZ1, a putative Arabidopsis transcriptional adaptor protein, mediates auxin and cytokinin signals in the control of cell proliferation. Curr. Biol. 13: 837–842. [DOI] [PubMed] [Google Scholar]
- Siegfried K. R., Eshed Y., Baum S. F., Otsuga D., Drews D. N., et al. , 1999. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 128: 4117–4128. [DOI] [PubMed] [Google Scholar]
- Sitaraman J., Bui M., Liu Z., 2008. LEUNIG_HOMOLOG and LEUNIG perform partially redundant functions during Arabidopsis embryo and floral development. Plant Physiol. 147: 672–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner D. J., Gasser C. S., 2009. Expression-based discovery of candidate ovule development regulators through transcriptional profiling of ovule mutants. BMC Plant Biol. 9: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner D. J., Baker S. C., Meister R. J., Broadhvest J., Schneitz K., et al. , 2001. The Arabidopsis HUELLENLOS gene, which is essential for normal ovule development, encodes a mitochondrial ribosomal protein. Plant Cell 13: 2719–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner D. J., Hill T. A., Gasser C. S., 2004. Regulation of ovule development. Plant Cell 16: S32–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner D. J., Brown R. H., Kuzoff R. K., Gasser C. S., 2016. Conservation of the role of INNER NO OUTER in development of unitegmic ovules of the Solanaceae despite a divergence in protein function. BMC Plant Biol. 16: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar V. V., Surendrarao A., Gonzalez D., Conlan R. S., Liu Z., 2004. Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 101: 11494–11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar V. V., Surendrarao A., Liu Z. C., 2006. APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133: 3159–3166. [DOI] [PubMed] [Google Scholar]
- Stahle M. I., Kuehlich J., Staron L., von Arnim A. G., Golz J. F., 2009. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 21: 3105–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger E. J., Mao Y., Regier M. K., Triezenberg S. J., Thomashow M. F., 2001. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29: 1524–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J. M., Broadhvest J., Hauser B. A., Meister R. J., Schneitz K., et al. , 1999. INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev. 13: 3160–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachonasios K. E., Thomashow M. F., Triezenberg S. J., 2003. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15: 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B., Zhang J., Pang C., Yu H., Guo D., et al. , 2015. The molecular mechanism of SPOROCYTELESS/NOZZLE in controlling Arabidopsis ovule development. Cell Res. 25: 121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiste C., Droge-Laser W., 2014. The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat. Commun. 5: 3883. [DOI] [PubMed] [Google Scholar]
- Yamada T., Ito M., Kato M., 2003. Expression pattern of INNER NO OUTER homologue in Nymphaea (water lily family, Nymphaeaceae). Dev. Genes Evol. 213: 510–513. [DOI] [PubMed] [Google Scholar]
- Yamada T., Yokota S., Hirayama Y., Imaichi R., Kato M., et al. , 2011. Ancestral expression patterns and evolutionary diversification of YABBY genes in angiosperms. Plant J. 67: 26–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plant materials are available from the cited sources from which they were obtained. Plasmids are available from the corresponding author on request.