In natural forests, tree leaf surfaces host diverse bacterial communities whose structure and composition are primarily driven by host species identity. Tree leaf bacterial diversity has also been shown to influence tree community productivity, a key function of terrestrial ecosystems. However, most urban microbiome studies have focused on the built environment, improving our understanding of indoor microbial communities but leaving much to be understood, especially in the nonbuilt microbiome. Here, we provide the first multiple-species comparison of tree phyllosphere bacterial structures and diversity along a gradient of urban intensity. We demonstrate that urban trees possess characteristic bacterial communities that differ from those seen with trees in nonurban environments, with microbial community structure on trees influenced by host species identity but also by the gradient of urban intensity and by the degree of isolation from other trees. Our results suggest that feedback between human activity and plant microbiomes could shape urban microbiomes.
KEYWORDS: biodiversity, bioindicators, microbial communities, microbial ecology, phyllosphere-inhabiting microbes, plant-microbe interactions, urban gradient, urban microbiome
ABSTRACT
Tree leaf-associated microbiota have been studied in natural ecosystems but less so in urban settings, where anthropogenic pressures on trees could impact microbial communities and modify their interaction with their hosts. Additionally, trees act as vectors spreading bacterial cells in the air in urban environments due to the density of microbial cells on aerial plant surfaces. Characterizing tree leaf bacterial communities along an urban gradient is thus key to understand the impact of anthropogenic pressures on urban tree-bacterium interactions and on the overall urban microbiome. In this study, we aimed (i) to characterize phyllosphere bacterial communities of seven tree species in urban environments and (ii) to describe the changes in tree phyllosphere bacterial community structure and diversity along a gradient of increasing urban intensity and at two degrees of tree isolation. Our results indicate that, as anthropogenic pressures increase, urban leaf bacterial communities show a reduction in the abundance of the dominant class in the natural plant microbiome, the Alphaproteobacteria. Our work in the urban environment here reveals that the structures of leaf bacterial communities differ along the gradient of urban intensity. The diversity of phyllosphere microbial communities increases at higher urban intensity, also displaying a greater number and variety of associated indicator taxa than the low and medium urban gradient sites. In conclusion, we find that urban environments influence tree bacterial community composition, and our results suggest that feedback between human activity and plant microbiomes could shape urban microbiomes.
IMPORTANCE In natural forests, tree leaf surfaces host diverse bacterial communities whose structure and composition are primarily driven by host species identity. Tree leaf bacterial diversity has also been shown to influence tree community productivity, a key function of terrestrial ecosystems. However, most urban microbiome studies have focused on the built environment, improving our understanding of indoor microbial communities but leaving much to be understood, especially in the nonbuilt microbiome. Here, we provide the first multiple-species comparison of tree phyllosphere bacterial structures and diversity along a gradient of urban intensity. We demonstrate that urban trees possess characteristic bacterial communities that differ from those seen with trees in nonurban environments, with microbial community structure on trees influenced by host species identity but also by the gradient of urban intensity and by the degree of isolation from other trees. Our results suggest that feedback between human activity and plant microbiomes could shape urban microbiomes.
INTRODUCTION
While the human population in urban centers is estimated to increase by 2 to 4 billion this century (1), the focus of public health research is expanding from the benefits of urban plant communities (air quality, physical activity, social cohesion, and stress reduction) (2) to include the potential roles of the urban microbiota. The positive influence of urban vegetation on human physical health has been demonstrated many times (3, 4), but it could also play an unexpected role in the microbial communities that they support and in their contribution to urban biodiversity. Studies using high-throughput sequencing techniques are rapidly improving our understanding of the urban microbiome, defined as the ensemble of microbial organisms residing or transiting in the urban environment (5). Land use type (e.g., forest, rural, urban) has been shown to impact airborne microbial communities (6, 7), and recent work has demonstrated that the local vegetation influences the airborne bacterial community composition and abundance in urban settings (8) and natural settings (9). Most urban microbiome research has been done on the built environment (indoor space of human-built structure) (but see references 8, 10, and 11), improving our understanding of urban microbial communities but leaving much to be defined, especially in the nonbuilt environmental microbiome. In addition, the surrounding plant community has been suggested to influence the microbial community of key buildings frequented by the human population (i.e., hospitals, schools, and homes) (12–14). Therefore, characterizing the assembly and dynamics of the urban plant microbiome is crucial to ascertain the influence of higher anthropogenic pressures on these microbial communities and to describe their potential contribution to the urban microbiome.
The phyllosphere, defined as the leaf surfaces of plants, is estimated to have a surface area of 4 × 108 km2 across the globe (15) and thus represents a major potential source of local microbial organisms (16, 17). In addition to its contribution to the urban microbiome, the canopy of urban trees provides a variety of services such as reducing local temperature, limiting water runoff, and increasing air quality (18). Recent research on the phyllosphere has found host species identity to be the key driver of leaf microbial community structure in both tropical ecosystems (19–23) and temperate ecosystems (24–28). However, to our knowledge few studies have described the changes in plant-associated microbiota from the natural environment to urban environments (but see reference 29 for bacterial communities on Hedera sp. and reference 30 for fungal communities on Quercus macrocarpa), leaving much to be learned on how the plant microbiome changes with increasing anthropogenic pressures. In this report, we focus on tree phyllosphere bacterial communities along a gradient of urban intensity to quantify the similarities and differences in community structure in the urban plant microbiome.
In the urban ecology literature, many different study-specific methodologies have been employed to estimate the intensity of human influence in an area, reducing the potential to compare results between experiments. In an attempt to standardize our work, we use the well-described index by Sanderson et al. (31) which has also been employed previously in a similar plant community urban gradient study (32). This index allows the evaluation of the anthropogenic impact on terrestrial ecosystems through multiple parameters, including population density, built-up areas, road access, landscape use, and electric power infrastructure, using nine public data sets (31). This index focuses on direct measures of human infrastructures and population, which are suggested to have the greatest and most direct effect on terrestrial ecosystems.
The urban environment differs strikingly from the natural forest environment mainly through an increase in the biotic and abiotic stresses caused directly and indirectly by anthropogenic activities. The increase in anthropogenic pressures in urban areas reduces tree fitness and longevity (33) and could modify many important functional traits (32). Numerous studies have shown that anthropogenic activities increase levels of leaf macronutrients (nitrogen, potassium, sulfur), micronutrients (boron, manganese, selenium), and trace elements (cadmium, lead, zinc) for urban trees (30, 34, 35). Higher temperatures in the urban environment influence vegetation phenology (36–38) and are intensified by city growth and global warming (39). The urban heat island phenomenon (40) results from the increase in the levels of nonpenetrating surfaces (41) and the decrease in the levels of vegetation cover (42) in cities. Thermal accumulation could drive enzymatic processes, affecting microbial communities directly, and could also increase the presence of insect ectotherms (43), which are known disease vectors (44). This increase in insect pest abundance in urban areas (45, 46) could also be intensified by changes in host plant quality and natural enemy efficiency (47). In addition to these stresses, urban trees frequently suffer from limited access to water and nutrients (48, 49), root development limitation (see reference 50 for a review), and photosynthetic biomass loss and tree lesions (51). These stresses have been shown to affect plant survival (52–54) and to induce numerous physiological responses, a phenomenon that could cause profound changes in urban tree leaf microbial communities. Therefore, urban biotic and abiotic conditions could provoke changes in the tree phyllosphere microbial community, potentially impacting host fitness and modifying the local pool of urban microbial organisms. However, little is known about the identity and dynamics of the urban tree phyllosphere.
To improve our understanding of the urban tree microbiome, we aimed (i) to characterize the bacterial communities present in tree phyllosphere bacterial communities of the urban environment and (ii) to describe the changes in tree phyllosphere bacterial community structure and diversity along a gradient of increasing urban intensity and at two degrees of tree isolation. Our hypotheses are (i) that the increasing urban intensity influences the abundance of the main taxonomical groups of bacteria usually described in the natural temperate forest phyllosphere literature and (ii) that the amount of stresses on trees imposed by higher anthropogenic pressures in urban agglomerations (nutrient enrichment, heat increase, physical stress, etc.) impacts phyllosphere bacterial community structure and reduces its diversity from low urban intensity to higher urban intensity settings. While previous urban microbiome studies have focused on air and built environment microorganisms (but see references 8, 10, and 11), our report provides key information on the urban plant-associated microbiota at different levels of urban intensity and offers new explanatory paths to better understand the gap between natural and urban environment microbiomes.
RESULTS
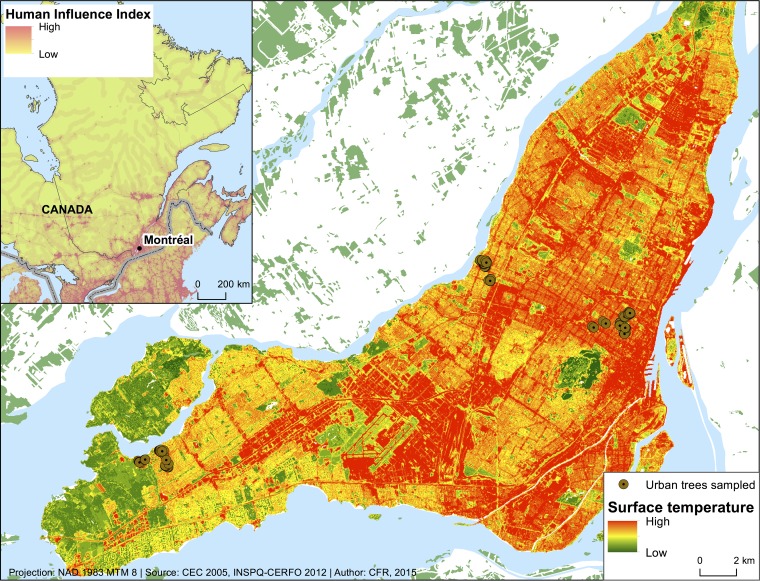
To characterize the tree phyllosphere microbiome found in urban settings, we sampled seven tree species (Acer platanoides, Acer rubrum, Acer saccharum, Celtis occidentalis, Fraxinus americana, Fraxinus pennsylvanica, and Picea glauca) at three sites in Montreal, Canada, along a gradient of increasing anthropogenic pressures. This urban intensity gradient was based on a composite index of human influence (IHI [31]) as employed by Nock et al. (32) where plots are classified in three categories: low, medium, and high anthropogenic pressures (Fig. 1). At each site, six individuals per tree species were randomly selected from the public district database to be sampled: three in parks (with tree neighbors and close plant community) and three in streets (no close tree neighbors and no close plant community) (see Table S1 in the supplemental material).
FIG 1 .
Locations of trees sampled along an urban gradient (three intensities: low, medium, and high) on the Island of Montreal, Canada. Temperature is used as a layover in this figure as an example of anthropogenic pressures along the gradient of intensity. The map was created using data from references 90, 91, and 92.
Description of the three study sites sampled during summer of 2014. Download TABLE S1, DOCX file, 0.02 MB (17KB, docx) .
Copyright © 2017 Laforest-Lapointe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Our data set contained 8,671 operational taxonomic units (OTUs) with 3,634 to 32,060 sequences per sample for a total of 1,755,757 quality sequences from 126 samples. We rarefied the samples to 3,000 sequences each, with 18 samples excluded due to insufficient sequence reads as a result of extraction or sequencing errors.
Our results show that leaf bacterial community composition varies among individual trees, among tree species, and along the urban gradient (Fig. 2). Along the urban gradient, phyllosphere bacterial communities were dominated by Alphaproteobacteria, averaging 40.8%, 39.4%, and 31.9% of sequences in communities from settings of low, medium, and high urban intensity, respectively. The five most abundant bacterial classes in the urban phyllosphere were Alphaproteobacteria, Gammaproteobacteria, Betaproteobacteria, Cytophagia, and Actinobacteria. Only the members of the class Alphaproteobacteria displayed a significant change in relative abundance along the urban gradient (post hoc Tukey’s tests on analysis of variance [ANOVA]), with differences of 3.4% and 2.0% comparing the high-intensity sites to the low-intensity and medium-intensity sites, respectively (F = 6.7, P < 0.005) (Fig. 3).
FIG 2 .
Relative abundance (%) of sequences from bacterial classes in the phyllosphere microbiome. Data represent the class community composition of (a) Acer platanoides; (b) Acer rubrum; (c) Acer saccharum; (d) Fraxinus americana; (e) Fraxinus pennsylvanica; (f) Picea glauca; and (g) Celtis occidentalis. Med, medium.
FIG 3 .
Relative abundances of the five most abundant classes along the urban gradient. Letters indicate the groups identified through a post hoc test of Tukey multiple comparisons of means at a 95% family-wise confidence level.
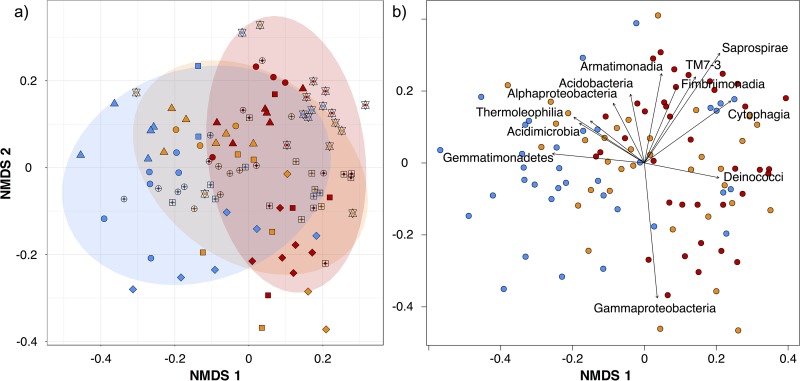
We quantified the relative influences of multiple drivers on leaf bacterial community taxonomic and phylogenetic structure by conducting a permutational multivariate analysis of variance (PERMANOVA [55]) on Bray-Curtis dissimilarities and weighted Unifrac (56) distances among samples (Table 1). We compared the relative influences of multiple drivers of leaf bacterial community composition along the gradient of urban intensity: host species identity (Acer platanoides, Acer rubrum, Acer saccharum, Celtis occidentalis, Fraxinus americana, Fraxinus pennsylvanica, and Picea glauca), tree isolation (park or street), and urban intensity (low, medium, and high). Because the two models yielded highly similar results in terms of significant variables and explanatory power, here we present only the results of analyses based on Bray-Curtis dissimilarities. We illustrated bacterial community taxonomic structure patterns by performing a nonmetric multidimensional scaling (NMDS) ordination of Bray-Curtis dissimilarities (Fig. 4). The strongest driver of urban phyllosphere community structure was host species identity (R2 = 19.4%, P = 0.001; PERMANOVA on Bray-Curtis distances; Table 1). Urban intensity (R2 = 6.1%, P = 0.001) and tree isolation (R2 = 1.8%, P = 0.001) were both weaker but significant drivers of leaf community structure. All second-level interactions were significant, the strongest being the interaction between urban intensity and host species identity (R2 = 12.1%, P = 0.001), and the third-level interaction was also significant (R2 = 8.0%, P = 0.017). At the class taxonomic level, we found multiple significant correlations among the relative abundances of various bacterial groups with the two axes of the NMDS (Fig. 4b), including Gemmatimonadetes, Deinococci, Acidomicrobia, and Thermoleophilia with NMDS axis 1 and Gammaproteobacteria, Acidobacteria, Alphaproteobacteria, Armatimonadia, Saprospirae, TM7-3, Fimbriimonadia, and Cytophagia with NMDS axis 2. Because of the significance of the second- and third-level interactions, we quantified how each of the host tree species bacterial community structure was explained by the urban gradient and tree isolation by building individual host species models (Table 2). Urban intensity had the strongest explanatory power for the leaf bacterial communities of A. rubrum (R2 = 32.1%, P = 0.003), followed by C. occidentalis (R2 = 25.4%, P < 0.001), F. americana (R2 = 24.3%, P < 0.001), and F. pennsylvanica (R2 = 19.1%, P = 0.007). The effect of urban intensity was not significant for A. saccharum (R2 = 23.8.1%, P = 0.102) and was only marginally significant for P. glauca (R2 = 14.7%, P = 0.078). Since host species identity was the main driver of leaf bacterial community structure across the urban gradient, we built a model accounting for phylogenetic relatedness between host tree species to identify the explanatory power of different phylogenetic levels. The angiosperm/gymnosperm level explained 4.5% of the total variation in phyllosphere bacterial community structure, whereas the genus and species levels explained, respectively, 7.2% and 8.3% of the variation.
TABLE 1 .
Bacterial community structure explained by host species identity, urban intensity, and tree isolation (street or park) and their interactionsa
| Variableb | Bray-Curtis dissimilarity |
Weighted Unifrac distance |
||||
|---|---|---|---|---|---|---|
| F value | R2 | Pr(>F) | F value | R2 | Pr(>F) | |
| 1st level | ||||||
| Host species identity | 4.97 | 19.38 | 0.001 | 5.03 | 19.73 | 0.001 |
| Urban intensity | 4.66 | 6.05 | 0.001 | 5.07 | 6.63 | 0.001 |
| Tree isolation | 2.78 | 1.81 | 0.001 | 2.92 | 1.91 | 0.009 |
| 2nd level | ||||||
| Urban intensity* | ||||||
| Tree isolation | 1.33 | 1.73 | 0.056 | NS | NS | NS |
| Host species* | ||||||
| Tree isolation | 1.74 | 6.77 | 0.001 | 1.77 | 6.96 | 0.004 |
| Urban intensity* | ||||||
| Host species | 1.55 | 12.10 | 0.001 | 1.49 | 11.71 | 0.005 |
| 3rd level | ||||||
| Urban intensity* | ||||||
| Host species* | 1.23 | 7.98 | 0.017 | NS | NS | NS |
| Tree isolation | ||||||
Data represent results of PERMANOVA analysis of Bray-Curtis dissimilarities and weighted Unifrac distances. The models explained, respectively, 56% and 47% of the variation in bacterial taxonomical and phylogenetic community structure. Pr(>F), P value; NS, not significant.
An asterisk represents interaction between variables.
FIG 4 .
Nonmetric multidimensional scaling (NMDS) ordination of variation in bacterial community structure of tree phyllosphere along a gradient of urban intensity. Data represent ordination based on Bray-Curtis distances among 108 samples. Samples (points) are colored based on the urban gradient (blue for low intensity, orange for medium intensity, and red for high intensity). In panel a, shapes represent host species identity (squares for Acer platanoides; circles for Acer rubrum; triangles for Acer saccharum; diamonds for Celtis occidentalis; sun crosses for Fraxinus Americana; squares with cross for Fraxinus pennsylvanica; stars for Picea glauca); ellipses indicate 1 standard deviation confidence intervals around samples from urban gradient intensity. In panel b, arrows represent the significant (P < 0.001) correlations between NMDS axes versus the relative abundances of bacterial classes in communities.
TABLE 2 .
Bacterial community structure of individual host species explained by host urban intensity and tree isolation (street or park) and their interactiona
| Host species | Urban intensity |
Tree isolation |
Urban intensity × tree isolation |
Total R2 (%) | |||
|---|---|---|---|---|---|---|---|
| R2 (%) | P | R2 (%) | P | R2 (%) | P | ||
| A. platanoides | NS | 16.0% | 0.030 | NS | 16 | ||
| A. rubrum | 32.1 | 0.001 | NS | 10.3 | 0.034 | 42.4 | |
| A. saccharum | 24.2 | 0.001 | 9.2 | 0.012 | NS | 33.4 | |
| C. occidentalis | 25.4 | 0.001 | 13.3 | 0.003 | NS | 38.7 | |
| F. americana | 24.3 | 0.001 | NS | NS | 24.3 | ||
| F. pennsylvanica | 19.1 | 0.009 | NS | NS | 19.1 | ||
| P. glauca | NS | 11.3 | 0.010 | NS | 11.3 | ||
Data represent results of PERMANOVA analysis of Bray-Curtis dissimilarities.
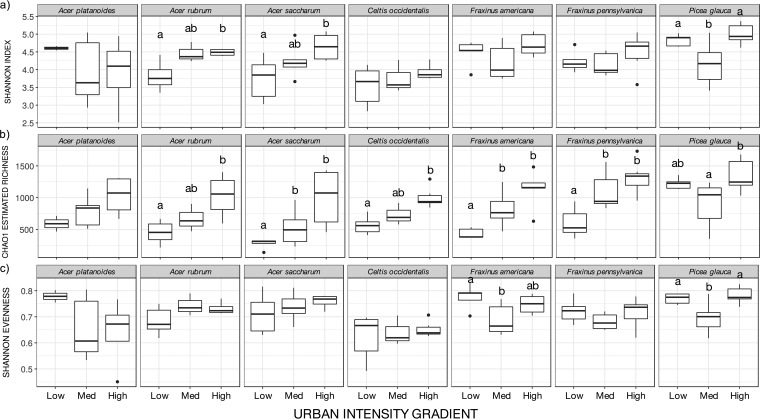
To further describe the changes in leaf bacterial community structure and diversity, we built a linear model to measure the relative influences of host species identity, urban intensity, and tree isolation on tree leaf bacterial alpha-diversity (Shannon diversity index). We compared the strengths of the variables in the best model by an ANOVA, which included urban intensity (F = 7.067, P = 0.001) and host species (F = 5.901, P = <0.001) as well as their interaction (F = 1.961, P = 0.037). Comparing the leaf bacterial alpha-diversity levels across sites, the highest level of urban intensity exhibited greater leaf bacterial alpha-diversity (mean = 4.5; Shannon index) than the low urban intensity (mean = 4.1, P = 0.004, post hoc Tukey’s test on ANOVA) and the medium urban intensity (mean = 4.1, P = 0.005, post hoc Tukey’s test on ANOVA). Since the interaction between the urban intensity and host species identity was significant, we computed per-host species alpha-diversity indices (Chao 1 and Shannon diversity), as well as community evenness (Shannon evenness) (Fig. 5). All species but A. platanoides showed an increase in estimated Chao 1 richness along the urban gradient, whereas only A. rubrum and A. saccharum demonstrated an increase in the Shannon diversity index value with increased urban intensity. Community evenness fluctuated across sites only for F. americana and P. glauca, which displayed a loss in community evenness at the medium level of the urban gradient.
FIG 5 .
Community diversity indices of phyllosphere bacterial communities for different host species along the urban gradient. (a) Shannon diversity. (b) Chao 1 estimated richness. (c) Shannon evenness.
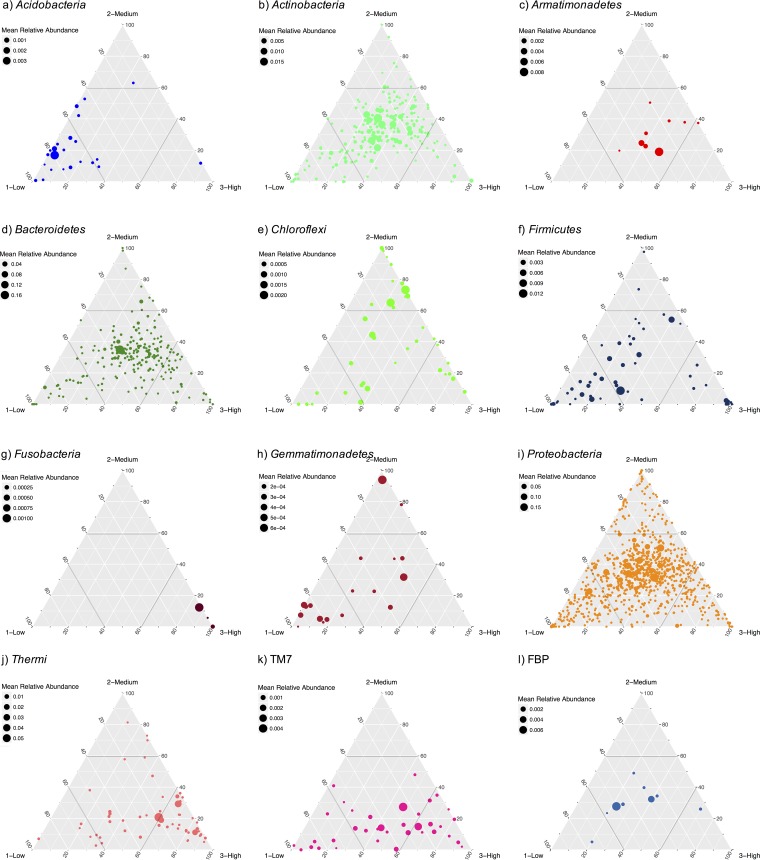
In order to identify significant indicator species in the leaf microbiome of each of the sites of the urban gradient, we estimated the indicator values for each OTU (57). Following the methodology presented by Wilhelm et al. (58), OTUs with an indicator value higher than 0.3 and a P value of <0.05 were classified as indicator species (Table S2). We found 21 indicators associated with the low level of the urban gradient (including OTUs from the orders Rhizobiales, Sphingomonadales, Rhodospirillales, and Burkholderiales), 9 associated with the medium level (including OTUs from the orders Actinomycetales, Rhodospirillales, and Sphingomonadales), and 37 indicators associated with the high level (including OTUs from the orders Cytophagales, Deinococcales, Rhodospirillales, and Actinomycetales). In addition, we built a three-axis ternary plot (Fig. 6) for the 12 most abundant phyla to visualize the relative proportions of OTUs at the three sites along the urban gradient. Each of OTUs was fitted in an x-y-z coordinate system, with each corner of the triangle representing a site along the urban gradient. The ternary plot showed that many OTUs were present in similar proportions at the three sites of the urban gradient but that some were relatively more abundant at a specific site (Fig. 6). In agreement with the indicator analysis, a greater number of OTUs showed an association with either the low level or high level of the urban gradient than with the medium level.
FIG 6 .
Ternary plots of per-site mean relative abundances of OTUs. Panels represent the 12 most abundant phyla. Each point represents an OTU, and its position indicates the proportion of its relative abundance at the different sites along the urban gradient. Points closer to the ternary plot corners indicate that a greater proportion of the total relative abundance of this OTU was found in this particular environment. Point colors indicate the phylum of the OTU. The lines inside the ternary plot indicate the 60% level of relative abundance of each of the sites. Only the 275 OTUs with a per-site mean relative abundance of more than 0.1% are shown.
Significant associations between bacterial OTUs and sites along the urban gradient detected by indicator species analysis (57). Panels represent (a) low (Pierrefonds), (b) medium (Ahuntsic), and (c) high (Mont-Royal) anthropogenic pressures. Download TABLE S2, DOCX file, 0.2 MB (210.1KB, docx) .
Copyright © 2017 Laforest-Lapointe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
In this report, we describe the tree phyllosphere bacterial communities at three sites along a gradient of urban intensity (based on a planet-wide index of human influence; 31) and two levels of tree isolation among seven different tree species. In agreement with previous studies of the natural tree phyllosphere, our results show that the composition of tree leaf bacterial communities of the urban environment is determined mainly by the identity of the host species. However, we confirmed our first hypothesis, i.e., that urban intensity and, to a lesser extent, tree isolation also influence the identity of bioindicator taxa, community composition, and diversity. While the results presented in this work are new and informative for future urban plant microbiome studies, we acknowledge that our study did not describe leaf bacterial load, a factor that could reveal density-dependent effects on population structure in the phyllosphere. We suggest that future studies could address these issues using measurements of microbial load through quantitative PCR (qPCR) performed on extracted DNA (i.e., 16S), cell counting, and culture-based approaches (59). Nonetheless, in the context of the urban microbiome, this work provides an unprecedented description of urban plant-associated microbiomes, providing key information for future studies of the impact of urban conditions on leaf microbial communities as well as on the overall urban microbiome. In addition, studying the urban plant microbiome increases our knowledge of the potential sources of the air and built-environment microbiome, which offers great insights for the eventual management of the urban microbiome. Our results could also impact the agriculture and the food industry, where research effort is now being put into designing biological control agents as a means to replace common pest-management interventions that have a strong negative impact on human and environment health (i.e., pesticides). Recent studies showcase successful biocontrol examples with both bacteria (60) and fungi (61). However, our work shows that the plant leaf microbiome is a function of local environmental conditions; therefore, future studies could benefit from considering this factor while designing leaf microflora biocontrol interventions.
In previous studies of tree phyllosphere microbial communities in natural environments, host species identity had often been found to be the strongest determinant of community structure (20, 24, 26). Here, our results also show that host species identity is a stronger driver of leaf bacterial community structure than urban intensity or tree isolation (Table 1; Fig. 4). Although host tree species identity is the main determinant of leaf bacterial community structure, this driver explained 20% of the variation. Therefore, we cannot assume that controlling the urban forest composition would allow us to manage the entire local urban microbiome. The remaining 80% of the variation could be linked to other local (random dispersal, site history, control from other microorganisms, animal and insect vectors), regional (air contamination), and national phenomena such as large-scale atmospheric air movements (62).
In accordance with previous descriptions of the tree phyllosphere (20, 26–28, 63), the urban phyllosphere bacterial communities are dominated by the Alphaproteobacteria (Fig. 2). However, our results demonstrate that the abiotic and biotic changes induced by the urban environment reduced the relative abundance of this bacterial taxonomic group (Alphaproteobacteria; Fig. 3) and enriched or depleted the relative abundances of many specific OTUs (Fig. 6). Similar effects on the phyllosphere of ivy (Hedera sp.) have been found by Smets et al. (29), who showed shifts in leaf bacterial communities between nonurban and urban sites in relation to atmospheric contamination. Jumpponen and Jones (30) also showed that tree phyllosphere fungal communities differed from nonurban to urban environments, in parallel with a general enrichment of foliar macronutrients in urban trees. In addition to changes in air quality and leaf composition, the differences in tree leaf bacterial community composition that we observed could also have been driven by the increased stress level experienced by urban trees or by changes in some key leaf traits induced by the urban environment.
Considering that the urban gradient index used in this work is based on data representing a combination of parameters such as population density, built-up areas, road access, landscape use, and electric power infrastructure from nine public data sets (31), future studies of urban leaf microbiome dynamics should aim to disentangle the relative influences of these parameters. Anthropogenic stresses on urban trees dramatically reduce the life span of trees to a few years (33, 64, 65). Understanding how tree-microbe interactions potentially accentuate or reduce these stresses is key to helping to improve the management of the urban forest, especially in the face of a changing climate, intensified world trade, and increased pest and disease abundance in urban areas. Urban trees often suffer from limited access to water, which has strong consequences for growth by affecting many processes linked to photosynthesis such as respiration, protein synthesis, and secondary carbohydrate metabolism (66, 67) and therefore also for leaf gas exchanges (68). Another factor with consequences for the leaf microhabitat is heavy metal deposition on leaves driven by air contamination. If water availability influences the potential interactions between the host and microbes at the leaf surface, the deposition of heavy metals (i.e., Cd, Pb, Zn, and Ni) on the leaf in urban environments (69) can also influence the growth capacity of phyllosphere microbes following their metabolic pathways. This hypothesis also concurs with our results demonstrating a greater number of indicators (OTUs) associated with the highest level of urban intensity. Future studies could test the hypothesis that increased anthropogenic stresses select for specific OTUs with higher resistance to stress. Therefore, human population density, local temperature, access to water, air contamination, physical procedures performed on trees, and leaf urban microconditions are all potential drivers of the changes in tree leaf microbiome.
Several studies have suggested that urban areas retain only a limited quantity of biodiversity (70–72), whereas other empirical studies have suggested that these areas could support diverse assemblies of organisms (73, 74). Our results show that, regarding the tree leaf bacterial communities, diversity increases with urban intensity, which allows us to reject our second hypothesis. Although this report cannot provide a mechanistic explanation for this augmentation, we hypothesize that the increased leaf bacterial diversity at high urban intensity could be linked to the local human population density and the heavy metal deposition. The local human population contributes to the location-specific pool of bacterial colonizers, which act as a source for tree leaf microbiomes (75). In parallel, the local air contamination could modify drastically phyllosphere microhabitat growth conditions and toxicity, therefore impacting the microbe-microbe interactions in the phyllosphere.
Increasing levels of anthropogenic pressures, including land use changes, biogeochemical changes, global warming, and exotic species invasion, cause an augmentation of tree stress and a corresponding diminution of longevity and productivity (52–54). Here, we show that the degree of tree isolation, as well as its interactions with host species identity or with urban intensity or with both variables, participates in driving tree phyllosphere community structure (Table 1). In 1983, Baldwin and Schultz (76) and Rhoades (77) revealed that conspecific trees had the capacity to influence the gene expression of neighboring trees through the emission of volatile organic compounds (VOC). These studies revealed that plants possess key mechanisms to “communicate” with each other when attacked by pathogens or pests, suggesting that trees can potentially benefit from proximity to other trees. However, little is known about the influence of the tree leaf microbiome on tree VOC emissions. Urban trees are submitted to multiple anthropogenic stresses of different lengths and intensities, leading to photosynthetic biomass loss and tree lesions (51) which could impact retroactively their interactions with leaf microbiota. In combination with previous work on a tree diversity experiment (28), our results highlight the potential role of the plant neighborhood in driving its leaf microbial community structure. The significant interactions of host tree species with urban intensity or with the degree of isolation or with both factors suggest that each tree species leaf microbiome was influenced differently by these environmental factors (Table 1). Further longitudinal experiments are needed to follow the temporal changes in tree-associated microbiota transplanted from natural to urban environments that occur as the host tree adapts to its new abiotic and biotic conditions. Future studies of the tree microbiome should also aim at describing how the change in the phyllosphere microbiome from natural to urban environments affects tree host fitness. Because of the size and longevity of trees, alternative greenhouse experiments with model organisms such as Arabidopsis thaliana could offer a great opportunity to disentangle the influences of the urban environment (i.e., temperature, air contamination, access to water, and physical lesions) and leaf microbial communities. Future experiments developing a “germfree” or “microbiome-controlled” plant growth system in greenhouses will allow researchers to separate the relative influences of the environment and the host microbiome.
Our results support previous findings showing that rural and urban microbial communities differ in composition (6, 7, 78). The proportion of green spaces and species diversity have been suggested as potential drivers of these natural environment-urban environment differences in community composition (8), but our work shows that the plant-associated microbiota per se is different from what is usually found in the natural environment as described in previous work in the natural temperate forest (26, 27). Urban abiotic and biotic conditions linked directly and indirectly to human actions are potential drivers of the changes in leaf microbial community structure. Therefore, future studies comparing the relative influences of the increased stress, the sources of microbial input, and the host capacity to select their microbiota in urban settings on the plant-associated microbiome are required to identify clearly the causes of this shift in the urban plant microbiome. In particular, future greenhouse experiments characterizing the influence of physical lesion, nutrient deficiency, water limitation, and air contamination on the plant microbiome and plant fitness will provide key information to support the development of the management of the urban microbiome. Such studies have the potential of identifying the most effective interventions to manage the urban plant microbiome (i.e., increasing plant diversity, increasing plant cover, reducing heat islands, reducing air contamination, introducing specific plant species). Although this work focused on bacterial communities, leaf-inhabiting fungi, yeasts, bacteriophages, and small eukaryotes could be impacted by the higher intensity of anthropogenic pressures but could also interact with tree leaf bacterial communities. Therefore, upcoming studies of the urban plant microbiome focusing on these organisms will definitely provide crucial information to the field, especially in considering the tree phyllosphere as a vector for the airborne microbiome. In addition, including an estimation of the leaf microbial load would allow future work to directly assess the quantitative effect of increased anthropogenic pressures on the phyllosphere microbiome. Finally, our work here also highlights the importance of future studies aimed at understanding the impact of this microbial enrichment in tree leaf microbial communities on host tree health through pathogen infections and pest attacks.
MATERIALS AND METHODS
Study sites.
The three study sites are located along an urban intensity gradient on the Island of Montreal, Canada, characterized by a cold and humid continental climate with temperate summer, and were classified as follows: Pierrefonds (45°27′26″N; 73°53′14″W) represented low urban intensity; Ahuntsic (45°33′22″N; 73°39′49″W) represented medium intensity; and Mont-Royal (45°31′32″N; 73°34′00″W) represented high intensity (Fig. 1). We assessed the urban intensity of the locations of the sampled trees based on a composite index of human influence (IHI) as described by Nock et al. (32). This index incorporates information on human infrastructures and presence, movements, landscape use, and electric infrastructure (31) to estimate the footprint of human influence. The IHI of the trees sampled ranged from 38 to 60 in function with respect to site identity and tree isolation.
Bacterial community collection.
All samples were acquired on 31 July 2014. For each randomly chosen mature tree (at least 20 cm in diameter), we clipped 50 to 100 g of shade leaves at midcanopy height (1 to 2 m above the bottom of the tree’s canopy) into sterile roll bags with surface-sterilized shears. For bacterial community collection and amplification, we used the protocols described by Kembel et al. (20). We collected microbial communities from the leaf surface by agitating the samples in a diluted Redford buffer solution and then resuspended cells in 500 μl of PowerSoil bead solution (MoBio, Carlsbad, CA). We extracted DNA from isolated cells using a PowerSoil kit according to the manufacturer’s instructions and stored the DNA at −80°C.
DNA library preparation and sequencing.
Samples were amplified using a one-step PCR step and normalized with primers designed to attach a 12-bp barcode and Illumina adaptor sequence to the fragments during PCR (79). For all samples, we used chloroplast-excluding primers targeting the V5-V6 region (799F and 1115R [24]) of the 16S rRNA gene. These primers contained a heterogeneity spacer along with the Illumina linker sequence (forward [799F], 5′-CAAGCAGAAGACGGCATACGAGATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTxxxxxxxxxxxx-HS-AACMGGATTAGATACCCKG-3′; reverse [1115R], 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTxxxxxxxxxxxx-HS-AGGGTTGCGCTCGTTG-3′) where x represents barcode nucleotides and HS represents a 0-to-7-bp heterogeneity spacer. Each sample was submitted to a single PCR procedure using a 25-µl mixture containing 5 µl 5× HF buffer (Thermo Scientific), 0.5 µl deoxynucleoside triphosphates (dNTPs) (10 µM), 0.5 µl forward primer (10 µM), 0.5 µl reverse primer (10 µM), 0.75 µl dimethyl sulfoxide (DMSO), 0.25 µl Phusion HotStart II polymerase (Thermo Scientific), 1 µl DNA, and 16.5 µl molecular-grade water. The reaction was performed using 30 s of initial denaturation at 98°C followed by 35 cycles of 15 s at 98°C, 30 s at 60°C, and 30 s at 72°C, with a final 10-min elongation at 72°C. The resulting products of natural forest samples were cleaned using a MoBio UltraClean PCR cleanup kit. We isolated an ~445-bp fragment by electrophoresis in a 2% agarose gel and recovered DNA with a MoBio GelSpin kit. We prepared multiplexed 16S libraries by mixing equimolar concentrations of DNA and sequenced the DNA library using Illumina MiSeq 250-bp paired-end sequencing at Genome Quebec. The urban samples were processed with an Invitrogen Sequalprep PCR cleanup and normalization kit (Frederick, MD) and were then pooled with equal concentrations and sequenced on an Illumina MiSeq platform at the University of Montreal. To ensure the quality of our sequencing, we included our negative and positive controls in the sequencing runs.
We processed the raw sequence data with PEAR (80) and QIIME (81) pipelines to merge paired-end sequences to a single sequence of a length of approximately 350 bp, to eliminate low-quality sequences (mean quality score of <30 or with any series of five bases with a quality score of <30), and to demultiplex sequences into samples. We eliminated chimeric sequences using the Uclust and Usearch algorithms (82). We then binned the remaining sequences into operational taxonomic units (OTUs) at a 97% sequence similarity cutoff level using the Uclust algorithm (82). We determined the taxonomic identity of each OTU using the BLAST algorithm and the Greengenes database (83) as implemented in QIIME (81).
Statistical analyses.
To exclude any spurious OTUs that might have been created by PCR or sequencing errors, we filtered the OTUs to remove those that were represented by fewer than 5 sequences. Rarefactions and analyses were repeated 100 times and showed no qualitative differences across iterations. Therefore, we present the result of a single random iteration. We performed analyses with the ape (84), picante (85), phyloseq (86), ggtern (87), and vegan (88) packages in R (89).
Data availability.
The data sets supporting the conclusions of this article are available in the Urban Tree phyllosphere Microbiome project repository on Figshare at https://figshare.com/s/07342fc588afdf8d1499 for the R code; https://figshare.com/s/16b8c237fe9b46d3da9b for the metadata; and, finally, https://figshare.com/s/af205b286c6f88f4b1f1 for the 16S RNA sequences.
ACKNOWLEDGMENTS
We declare that each experiment complies with the current laws of the country in which the experiment was performed. We declare that we have no competing interest.
Financial support was obtained from grants contributed by the Natural Sciences and Engineering Research Council of Canada (NSERC), by the Fonds de Recherche du Québec Nature et Technologies (FRQNT), and by the Canada Research Chairs Program.
The original version of the manuscript appears in the thesis of I.L.-L. hosted on the Université du Québec à Montréal website.
We thank T. Dawson for support in the laboratory and R. Fréchon for support in the field.
I.L.-L., C.M., and S.W.K. designed the study; I.L.-L. collected data and performed DNA extraction and amplification; I.L.-L. analyzed the data; I.L.-L., C.M., and S.W.K. wrote the manuscript.
REFERENCES
- 1.United Nations Department of Economic and Social Affairs, Population Division 2015. World urbanization prospects, 2015 revision: highlights. United Nations, New York, NY: https://esa.un.org/unpd/wpp/publications/files/key_findings_wpp_2015.pdf. Accessed 15 August 2016. [Google Scholar]
- 2.Hartig T, Mitchell R, De Vries S, Frumkin H. 2014. Nature and health. Annu Rev Publ Health 35:207–228. doi: 10.1146/annurev-publhealth-032013-182443. [DOI] [PubMed] [Google Scholar]
- 3.Maas J, Verheij RA, Groenewegen PP, de Vries S, Spreeuwenberg P. 2006. Green space, urbanity, and health: how strong is the relation? J Epidemiol Commun Health 60:587–592. doi: 10.1136/jech.2005.043125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richardson EA, Mitchell R. 2010. Gender differences in relationships between urban green space and health in the United Kingdom. Soc Sci Med 71:568–575. doi: 10.1016/j.socscimed.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 5.King GM. 2014. Urban microbiomes and urban ecology: how do microbes in the built environment affect human sustainability in cities? J Microbiol 52:721–728. doi: 10.1007/s12275-014-4364-x. [DOI] [PubMed] [Google Scholar]
- 6.Burrows SM, Elbert W, Lawrence MG, Pöschl U. 2009. Bacteria in the global atmosphere–part 1: review and synthesis of literature data for different ecosystems. Atmos Chem Phys 9:9263–9280. doi: 10.5194/acp-9-9263-2009. [DOI] [Google Scholar]
- 7.Bowers RM, McLetchie S, Knight R, Fierer N. 2011. Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J 5:601–612. doi: 10.1038/ismej.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mhuireach G, Johnson BR, Altrichter AE, Ladau J, Meadow JF, Pollard KS, Green JL. 2016. Urban greenness influences airborne bacterial community composition. Sci Total Environ 571:680–687. doi: 10.1016/j.scitotenv.2016.07.037. [DOI] [PubMed] [Google Scholar]
- 9.Lymperopoulou DS, Adams RI, Lindow SE. 2016. Contribution of vegetation to the microbial composition of nearby outdoor air. Appl Environ Microbiol 82:3822–3833. doi: 10.1128/AEM.00610-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Afshinnekoo E, Meydan C, Chowdhury S, Jaroudi D, Boyer C, Bernstein N, Maritz JM, Reeves D, Gandara J, Chhangawala S, Ahsanuddin S, Simmons A, Nessel T, Sundaresh B, Pereira E, Jorgensen E, Kolokotronis SO, Kirchberger N, Garcia I, Gandara D, Dhanraj S, Nawrin T, Saletore Y, Alexander N, Vijay P, Hénaff EM, Zumbo P, Walsh M, O’Mullan GD, Tighe S, Dudley JT, Dunaif A, Ennis S, O’Halloran E, Magalhaes TR, Boone B, Jones AL, Muth TR, Paolantonio KS, Alter E. 2015. Geospatial resolution of human and bacterial diversity with city-scale metagenomics. Cell Syst 1:72–87. doi: 10.1016/j.cels.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tischer C, Weikl F, Probst AJ, Standl M, Heinrich J, Pritsch K. 2016. Urban dust microbiome: impact on later atopy and wheezing. Environ Health Perspect 124:1919–1923. doi: 10.1289/EHP158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kembel SW, Jones E, Kline J, Northcutt D, Stenson J, Womack AM, Bohannan BJ, Brown GZ, Green JL. 2012. Architectural design influences the diversity and structure of the built environment microbiome. ISME J 6:1469–1479. doi: 10.1038/ismej.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meadow JF, Altrichter AE, Kembel SW, Kline J, Mhuireach G, Moriyama M, Northcutt D, O’Connor TK, Womack AM, Brown GZ, Green JL, Bohannan BJ. 2014. Indoor airborne bacterial communities are influenced by ventilation, occupancy, and outdoor air source. Indoor Air 24:41–48. doi: 10.1111/ina.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meadow JF, Altrichter AE, Kembel SW, Moriyama M, O’Connor TK, Womack AM, Brown GZ, Green JL, Bohannan BJ. 2014. Bacterial communities on classroom surfaces vary with human contact. Microbiome 2:7. doi: 10.1186/2049-2618-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris CE, Kinkel LL. 2002. Fifty years of phyllosphere microbiology: significant contributions to research in related fields, p 353–363. In Lindow SE, Poinar E, Elliot V (ed). Phyllosphere microbiology. APS Publishing, Minneapolis, MN. [Google Scholar]
- 16.Whipps JM, Hand P, Pink D, Bending GD. 2008. Phyllosphere microbiology with special reference to diversity and plant genotype. J Appl Microbiol 105:1744–1755. doi: 10.1111/j.1365-2672.2008.03906.x. [DOI] [PubMed] [Google Scholar]
- 17.Lighthart B, Shaffer BT, Frisch AS, Paterno D. 2009. Atmospheric culturable bacteria associated with meteorological conditions at a summer-time site in the mid-Willamette Valley, Oregon. Aerobiologia 25:285–295. doi: 10.1007/s10453-009-9133-7. [DOI] [Google Scholar]
- 18.Pataki DE, Carreiro MM, Cherrier J, Grulke NE, Jennings V, Pincetl S, Pouyat RV, Whitlow TH, Zipperer WC. 2011. Coupling biogeochemical cycles in urban environments: ecosystem services, green solutions, and misconceptions. Front Ecol Environ 9:27–36. doi: 10.1890/090220. [DOI] [Google Scholar]
- 19.Kim M, Singh D, Lai-Hoe A, Go R, Rahim RA, Ainuddin AN, Chun J, Adams JM. 2012. Distinctive phyllosphere bacterial communities in tropical trees. Microb Ecol 63:674–681. doi: 10.1007/s00248-011-9953-1. [DOI] [PubMed] [Google Scholar]
- 20.Kembel SW, O’Connor TK, Arnold HK, Hubbell SP, Wright SJ, Green JL. 2014. Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc Natl Acad Sci U S A 111:13715–13720. doi: 10.1073/pnas.1216057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kembel SW, Mueller RC. 2014. Plant traits and taxonomy drive host associations in tropical phyllosphere fungal communities. Botany 92:303–311. doi: 10.1139/cjb-2013-0194. [DOI] [Google Scholar]
- 22.Lambais MR, Crowley DE, Cury JC, Büll RC, Rodrigues RR. 2006. Bacterial diversity in tree canopies of the Atlantic forest. Science 312:1917–1917. doi: 10.1126/science.1124696. [DOI] [PubMed] [Google Scholar]
- 23.Lambais MR, Lucheta AR, Crowley DE. 2014. Bacterial community assemblages associated with the phyllosphere, dermosphere, and rhizosphere of tree species of the Atlantic forest are host taxon dependent. Microb Ecol 68:567–574. doi: 10.1007/s00248-014-0433-2. [DOI] [PubMed] [Google Scholar]
- 24.Redford AJ, Bowers RM, Knight R, Linhart Y, Fierer N. 2010. The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ Microbiol 12:2885–2893. doi: 10.1111/j.1462-2920.2010.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redford AJ, Fierer N. 2009. Bacterial succession on the leaf surface: a novel system for studying successional dynamics. Microb Ecol 58:189–198. doi: 10.1007/s00248-009-9495-y. [DOI] [PubMed] [Google Scholar]
- 26.Laforest-Lapointe I, Messier C, Kembel SW. 2016. Host species identity, site and time drive temperate tree phyllosphere bacterial community structure. Microbiome 4:27. doi: 10.1186/s40168-016-0174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laforest-Lapointe I, Messier C, Kembel SW. 2016. Tree phyllosphere bacterial communities: exploring the magnitude of intra- and inter-individual variation among host species. PeerJ 4:e2367. doi: 10.7717/peerj.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laforest-Lapointe I, Paquette A, Messier C, Kembel SW. 2017. Leaf bacterial diversity mediates plant diversity and ecosystem function relationships. Nature 546:145–147. doi: 10.1038/nature22399. [DOI] [PubMed] [Google Scholar]
- 29.Smets W, Wuyts K, Oerlemans E, Wuyts S, Denys S, Samson R, Lebeer S. 2016. Impact of urban land use on the bacterial phyllosphere of ivy (Hedera sp.). Atmos Environ 147:376–383. doi: 10.1016/j.atmosenv.2016.10.017. [DOI] [Google Scholar]
- 30.Jumpponen A, Jones KL. 2010. Seasonally dynamic fungal communities in the Quercus macrocarpa phyllosphere differ between urban and nonurban environments. New Phytol 186:496–513. doi: 10.1111/j.1469-8137.2010.03197.x. [DOI] [PubMed] [Google Scholar]
- 31.Sanderson EW, Jaiteh M, Levy MA, Redford KH, Wannebo AV, Woolmer G. 2002. The human footprint and the last of the wild: the human footprint is a global map of human influence on the land surface, which suggests that human beings are stewards of nature, whether we like it or not. BioScience 52:891–904. doi: 10.1641/0006-3568(2002)052[0891:THFATL]2.0.CO;2. [DOI] [Google Scholar]
- 32.Nock CA, Paquette A, Follett M, Nowak DJ, Messier C. 2013. Effects of urbanization on tree species functional diversity in eastern North America. Ecosystems 16:1487–1497. doi: 10.1007/s10021-013-9697-5. [DOI] [Google Scholar]
- 33.Nowak DJ, McBride JR. 1991. Comparison of Monterey pine stress in urban and natural forests. J Environ Manag 32:383–395. doi: 10.1016/S0301-4797(05)80074-X. [DOI] [Google Scholar]
- 34.Pouyat RV, McDonnell MJ. 1991. Heavy metal accumulations in forest soils along an urban-rural gradient in southeastern New York, USA. Water Air Soil Pollut 57–58:797–807. doi: 10.1007/BF00282943. [DOI] [Google Scholar]
- 35.Kaye JP, Groffman PM, Grimm NB, Baker LA, Pouyat RV. 2006. A distinct urban biogeochemistry? Trends Ecol Evol 21:192–199. doi: 10.1016/j.tree.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 36.Roetzer T, Wittenzeller M, Haeckel H, Nekovar J. 2000. Phenology in central Europe—differences and trends of spring phenophases in urban and rural areas. Int J Biometeorol 44:60–66. doi: 10.1007/s004840000062. [DOI] [PubMed] [Google Scholar]
- 37.White MA, Nemani RR, Thornton PE, Running SW. 2002. Satellite evidence of phenological differences between urbanized and rural areas of the eastern United States deciduous broadleaf forest. Ecosystems 5:260–273. doi: 10.1007/s10021-001-0070-8. [DOI] [Google Scholar]
- 38.Zhang XY, Friedl MA, Schaaf CB, Strahler AH, Schneider A. 2004. The footprint of urban climates on vegetation phenology. Geophys Res Lett 31:L12209. doi: 10.1029/2004GL020137. [DOI] [Google Scholar]
- 39.Kalnay E, Cai M. 2003. Impact of urbanization and land-use change on climate. Nature 423:528–531. doi: 10.1038/nature01675. [DOI] [PubMed] [Google Scholar]
- 40.Oke TR. 1973. City size and urban heat island. Atmos Environ 7:769–779. doi: 10.1016/0004-6981(73)90140-6. [DOI] [Google Scholar]
- 41.Hart MA, Sailor DJ. 2009. Quantifying the influence of land-use and surface characteristics on spatial variability in the urban heat island. Theor Appl Climatol 95:397–406. doi: 10.1007/s00704-008-0017-5. [DOI] [Google Scholar]
- 42.Jenerette GD, Harlan SL, Stefanov WL, Martin CA. 2011. Ecosystem services and urban heat riskscape moderation: water, green spaces, and social inequality in Phoenix, USA. Ecol Appl 21:2637–2651. doi: 10.1890/10-1493.1. [DOI] [PubMed] [Google Scholar]
- 43.Briere J, Pracros P, Le Roux A, Pierre J. 1999. A novel rate model of temperature-dependent development for arthropods. Environ Entomol 28:22–29. doi: 10.1093/ee/28.1.22. [DOI] [Google Scholar]
- 44.Lounibos LP. 2002. Invasions by insect vectors of human disease. Annu Rev Entomol 47:233–266. doi: 10.1146/annurev.ento.47.091201.145206. [DOI] [PubMed] [Google Scholar]
- 45.Bennett AB, Gratton C. 2012. Local and landscape scale variables impact parasitoid assemblages across an urbanization gradient. Landscape Urban Plan 104:26–33. doi: 10.1016/j.landurbplan.2011.09.007. [DOI] [Google Scholar]
- 46.San Martin Y Gomez G, Van Dyck H. 2012. Ecotypic differentiation between urban and rural populations of the grasshopper Chorthippus brunneus relative to climate and habitat fragmentation. Oecologia 169:125–133. doi: 10.1007/s00442-011-2189-4. [DOI] [PubMed] [Google Scholar]
- 47.Raupp MJ, Shrewsbury PM, Herms DA. 2010. Ecology of herbivorous arthropods in urban landscapes. Annu Rev Entomol 55:19–38. doi: 10.1146/annurev-ento-112408-085351. [DOI] [PubMed] [Google Scholar]
- 48.Wiersum LK, Harmanny K. 1983. Changes in the water-permeability of roots of some trees during drought stress and recovery, as related to problems of growth in urban environment. Plant Soil 75:443–448. doi: 10.1007/BF02369978. [DOI] [Google Scholar]
- 49.Flückiger W, Braun S. 1999. Stress factors of urban trees and their relevance for vigour and predisposition for parasite attacks. In International Symposium on Urban Tree Health, p 325–334. doi: 10.17660/ActaHortic.1999.496.40. [DOI] [Google Scholar]
- 50.Day SD, Wiseman PE, Dickinson SB, Harris JR. 2010. Tree root ecology in the urban environment and implications for a sustainable rhizosphere. Arboric Urban Forestry 36:193–205. [Google Scholar]
- 51.Sieghardt M, Mursch-Radlgruber E, Paoletti E, Couenberg E, Dimitrakopoulus A, Rego F, Hatzistathis A, Randrup TB. 2005. The abiotic urban environment: impact of urban growing conditions on urban vegetation, p 281–323. In Konijnendijk C, Nilsson K, Randrup T, Schipperijn J (ed), Urban forests and trees: a reference book. Springer Verlag, Berlin, Germany. doi: 10.1007/3-540-27684-X_12. [DOI] [Google Scholar]
- 52.Mittler R. 2006. Abiotic stress, the field environment and stress combination. Trends Plant Sci 11:15–19. doi: 10.1016/j.tplants.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 53.Niinemets U. 2010. Mild versus severe stress and BVOCs: thresholds, priming and consequences. Trends Plant Sci 15:145–153. doi: 10.1016/j.tplants.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 54.Niinemets Ü. 2010. Responses of forest trees to single and multiple environmental stresses from seedlings to mature plants: past stress history, stress interactions, tolerance and acclimation. Forest Ecol Manag 260:1623–1639. doi: 10.1016/j.foreco.2010.07.054. [DOI] [Google Scholar]
- 55.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Aust Ecol 26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 56.Lozupone C, Hamady M, Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dufrêne M, Legendre P. 1997. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr 67:345–366. doi: 10.1890/0012-9615(1997)067[0345:SAAIST]2.0.CO;2. [DOI] [Google Scholar]
- 58.Wilhelm L, Besemer K, Fragner L, Peter H, Weckwerth W, Battin TJ. 2015. Altitudinal patterns of diversity and functional traits of metabolically active microorganisms in stream biofilms. ISME J 9:2454–2464. doi: 10.1038/ismej.2015.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emerson JB, Adams RI, Román CMB, Brooks B, Coil DA, Dahlhausen K, Ganz HH, Hartmann EM, Hsu T, Justice NB, Paulino-Lima IG, Luongo JC, Lymperopoulou DS, Gomez-Silvan C, Rothschild-Mancinelli B, Balk M, Huttenhower C, Nocker A, Vaishampayan P, Rothschild LJ. 2017. Schrödinger’s microbes: tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 5:86. doi: 10.1186/s40168-017-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang LY, Xie YS, Cui YY, Xu J, He W, Chen HG, Guo JH. 2015. Conjunctively screening of biocontrol agents (BCAs) against fusarium root rot and fusarium head blight caused by Fusarium graminearum. Microbiol Res 177:34–42. doi: 10.1016/j.micres.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 61.Hue AG, Voldeng HD, Savard ME, Fedak G, Tian X, Hsiang T. 2009. Biological control of fusarium head blight of wheat with Clonostachys rosea strain ACM941. Can J Plant Pathol 31:169–179. doi: 10.1080/07060660909507590. [DOI] [Google Scholar]
- 62.Barberán A, Henley J, Fierer N, Casamayor EO. 2014. Structure, inter-annual recurrence, and global-scale connectivity of airborne microbial communities. Sci Total Environ 487:187–195. doi: 10.1016/j.scitotenv.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 63.Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, von Mering C, Vorholt JA. 2009. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci U S A 106:16428–16433. doi: 10.1073/pnas.0905240106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berrang P, Karnosky DF, Stanton BJ. 1985. Environmental factors affecting tree health in New York City. J Arboric 11:185–189. [Google Scholar]
- 65.Gilbertson P, Bradshaw AD. 1985. Tree survival in cities: the extent and nature of the problem. J Arboric 9:131–142. doi: 10.1080/03071375.1985.9746706. [DOI] [Google Scholar]
- 66.Cregg BM. 1995. Plant moisture stress of green ash trees in contrasting urban sites. J Arboric 21:271–276. [Google Scholar]
- 67.Kramer PJ. 1987. The role of water stress in tree growth. J Arboric 13:33–38. [Google Scholar]
- 68.Cregg BM, Dix ME. 2001. Tree moisture stress and insect damage in urban areas in relation to heat island effects. J Arboric 27:8–17. [Google Scholar]
- 69.Baycu G, Tolunay D, Özden H, Günebakan S. 2006. Ecophysiological and seasonal variations in Cd, Pb, Zn, and Ni concentrations in the leaves of urban deciduous trees in Istanbul. Environ Pollut 143:545–554. doi: 10.1016/j.envpol.2005.10.050. [DOI] [PubMed] [Google Scholar]
- 70.Blair RB. 1999. Birds and butterflies along an urban gradient: surrogate taxa for assessing biodiversity? Ecol Appl 9:164–170. doi: 10.1890/1051-0761(1999)009[0164:BABAAU]2.0.CO;2. [DOI] [Google Scholar]
- 71.Cincotta RP, Engelman R. 2000. Nature's place: human population and the future of biological diversity, p 79 Population Action International, Washington, DC. [Google Scholar]
- 72.McKinney ML. 2002. Urbanization, biodiversity, and conservation: the impacts of urbanization on native species are poorly studied, but educating a highly urbanized human population about these impacts can greatly improve species conservation in all ecosystems. Bioscience 52:883–890. doi: 10.1641/0006-3568(2002)052[0883:UBAC]2.0.CO;2. [DOI] [Google Scholar]
- 73.Kühn I, Brandl R, Klotz S. 2004. The flora of German cities is naturally species rich. Evol Ecol Res 6:749–764. [Google Scholar]
- 74.Wania A, Kühn I, Klotz S. 2006. Plant richness patterns in agricultural and urban landscapes in Central Germany—spatial gradients of species richness. Landsc Urban Plan 75:97–110. doi: 10.1016/j.landurbplan.2004.12.006. [DOI] [Google Scholar]
- 75.Knief C, Ramette A, Frances L, Alonso-Blanco C, Vorholt JA. 2010. Site and plant species are important determinants of the Methylobacterium community composition in the plant phyllosphere. ISME 4:719–728. doi: 10.1038/ismej.2010.9. [DOI] [PubMed] [Google Scholar]
- 76.Pakarinen J, Hyvärinen A, Salkinoja-Salonen M, Laitinen S, Nevalainen A, Mäkelä MJ, Haahtela T, Von Hertzen L. 2008. Predominance of Gram-positive bacteria in house dust in the low‐allergy risk Russian Karelia. Environ Microbiol 10:3317–3325. doi: 10.1111/j.1462-2920.2008.01723.x. [DOI] [PubMed] [Google Scholar]
- 77.Baldwin IT, Schultz JC. 1983. Rapid changes in tree leaf chemistry induced by damage: evidence for communication between plants. Science 221:277–279. doi: 10.1126/science.221.4607.277. [DOI] [PubMed] [Google Scholar]
- 78.Rhoades DF. 1983. Responses of alder and willow to attack by tent caterpillars and webworms: evidence for pheromonal sensitivity of willows, p 55–68. In Hedin PA (ed), Plant resistance to insects. American Chemical Society, Washington, DC. doi: 10.1021/bk-1983-0208.ch004. [DOI] [Google Scholar]
- 79.Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J. 2014. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 83.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 85.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 86.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hamilton N. 2016. ggtern: an Extension to “ggplot2”, for the creation of ternary diagrams. R package version 2.1.5 https://CRAN.R-project.org/package=ggtern. [Google Scholar]
- 88.Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ; Suggests MASS . 2007. The vegan package. Community Ecology Package 10, p 631–637. [Google Scholar]
- 89.R Development Core Team 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 90.WCS/CIESIN 2005. Last of the wild data version 2 2005 (LWP-2): global human footprint dataset (geographic). Wildlife Conservation (WCS) and Center for International Earth Science Information Network (CIESIN), New York, NY. [Google Scholar]
- 91.Government of Quebec 2015. ILOTS de chaleur/fraicheur urbains et température de surface. Government of Quebec, Quebec, Canada: http://www.donneesquebec.ca/recherche/fr/dataset/ilots-de-chaleur-fraicheur-urbains-et-temperature-de-surface. [Google Scholar]
- 92.Boulfroy E, Khaldoune J, Grenon F, Fournier R, Talbot B. 2013. Conservation des îlots de fraîcheur urbains—description de la méthode suivie pour identifier et localizer les îlots de fraîcheur et de chaleur (méthode en 9 niveaux). Report 2012-11c. CERFO et Université de Sherbrooke, Quebec, Canada. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of the three study sites sampled during summer of 2014. Download TABLE S1, DOCX file, 0.02 MB (17KB, docx) .
Copyright © 2017 Laforest-Lapointe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Significant associations between bacterial OTUs and sites along the urban gradient detected by indicator species analysis (57). Panels represent (a) low (Pierrefonds), (b) medium (Ahuntsic), and (c) high (Mont-Royal) anthropogenic pressures. Download TABLE S2, DOCX file, 0.2 MB (210.1KB, docx) .
Copyright © 2017 Laforest-Lapointe et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The data sets supporting the conclusions of this article are available in the Urban Tree phyllosphere Microbiome project repository on Figshare at https://figshare.com/s/07342fc588afdf8d1499 for the R code; https://figshare.com/s/16b8c237fe9b46d3da9b for the metadata; and, finally, https://figshare.com/s/af205b286c6f88f4b1f1 for the 16S RNA sequences.