TBK1 is a critical kinase required for the induction of type I IFNs and subsequent cellular antiviral responses. Yu et al. show that USP1–UAF1 deubiquitinase complex removes K48-linked polyubiquitination of TBK1, stabilizes its expression, and thus enhances antiviral responses.
Abstract
Optimal activation of TANK-binding kinase 1 (TBK1) is crucial for initiation of innate antiviral immunity and maintenance of immune homeostasis. Although several E3 ubiquitin ligases have been reported to regulate TBK1 activation by mediating its polyubiquitination, the functions of deubiquitinase on TBK1 activity remain largely unclear. Here, we identified a deubiquitinase complex, which is formed by ubiquitin specific peptidase 1 (USP1) and USP1-associated factor 1 (UAF1), as a viral infection–induced physiological enhancer of TBK1 expression. USP1–UAF1 complex enhanced TLR3/4 and RIG-I–induced IFN regulatory factor 3 (IRF3) activation and subsequent IFN-β secretion. Mechanistically, USP1 and UAF1 bound to TBK1, removed its K48-linked polyubiquitination, and then reversed the degradation process of TBK1. Furthermore, we found that ML323, a specific USP1–UAF1 inhibitor, attenuated IFN-β expression and enhanced viral replication both in vitro and in vivo. Therefore, our results outline a novel mechanism for the control of TBK1 activity and suggest USP1–UAF1 complex as a potential target for the prevention of viral diseases.
Introduction
The production of type I IFNs (IFN-α/β) is a fundamental host response to combating invading viruses (Liu et al., 2011; Wang and Fish, 2012; Hoffmann et al., 2015). Various virus structural components, including nucleic acid and surface glycoproteins, are recognized as pathogen-associated molecular patterns by pattern recognition receptors (PRRs) expressed in multiple immune cells. All PRRs use the key molecule TANK-binding kinase 1 (TBK1) to activate downstream signal transduction. TLR3/4 recruit TLR/IL-1R domain–containing adaptor protein–inducing IFN-β (TRIF) and initiate TRIF-dependent signaling (Kawai and Akira, 2010; O’Neill et al., 2013). Retinoic acid–inducible gene-I (RIG-I) recruits the signaling adaptor mitochondrial antiviral signaling (MAVS, also known as IPS-1, Cardif, or VISA; Loo and Gale, 2011; Yoneyama et al., 2015). The DNA sensor cyclic GMP-AMP synthase (cGAS) triggers a stimulator of interferon genes (STING)–dependent pathway (Cai et al., 2014). TRIF, MAVS, or STING then activates TRAF family member–associated NF-κB activator–binding kinase 1 (TBK1). Activated TBK1 then phosphorylates IFN regulatory factor 3 (IRF3) and triggers its dimerization and nuclear translocation, where it forms active transcriptional complexes that bind to IFN-stimulation response elements (ISRE) and triggers type I IFN genes transcription. Type I IFN binds to IFN α/β receptor (IFNAR) and promotes the production of numerous antiviral genes through the JAK/STAT pathway (Porritt and Hertzog, 2015). Thus, TBK1 is a critical kinase required for the induction of type I IFNs and subsequent cellular antiviral responses, and its activity must be fine-tuned during viral infection.
Ubiquitination is an important protein posttranslational modification for the control of TBK1 activity and innate antiviral immunity (Zhao, 2013; Heaton et al., 2016). Lysine 63 (K63)-linked polyubiquitination of TBK1 is required for its activation, whereas K48-linked ubiquitination of TBK1 promotes its degradation in proteasome. Previously, we reported that TRAF-interacting protein (TRIP), a virus-induced E3 ubiquitin ligase, negatively regulated innate antiviral response by promoting K48-linked polyubiquitination and proteasomal degradation of TBK1 (Zhang et al., 2012). In addition, another E3 ubiquitin ligase DTX4, can be recruited by NLR protein NLRP4 to interact with TBK1, and then promote K48-linked polyubiquitination TBK1 (Cui et al., 2012). Thus, DTX4, TRIP, and NLRP4 form a signalosome and mediate TBK1 ubiquitination (Lin et al., 2016). Although several E3 ubiquitin ligases have been reported to regulate TBK1 activation, the functions of deubiquitinating enzymes on TBK1 activity remain largely unclear. Whether any deubiquitinating enzymes exist to specifically remove K48-linked ubiquitination of TBK1, reverse the degradation process of TBK1, and thus stabilize its expression remains to be identified.
Ubiquitin specific peptidase 1 (USP1), a member of USP family, could deubiquitinate a wide range of substrates (Cohn et al., 2007; Williams et al., 2011; Ogrunc et al., 2016). The deubiquitinase activity of USP1 could be promoted by its binding partner USP1-associated factor 1 (UAF1, also called WD40 repeat containing protein 48 [WDR48] or p80; Cohn et al., 2009; Sowa et al., 2009). UAF1 and USP1 form a deubiquitinase complex, which participates in a variety of biological processes, such as regulation of DNA repair processes and tumor pathogenesis. UAF1-deficient mouse die during embryo development (Park et al., 2013), and USP1-deficient mice also exhibit a higher rate of perinatal lethality (Kim et al., 2009). However, the potential role of USP1–UAF1 complex in the immune system, especially in the innate antiviral immunity, remains unknown.
In this study, we identified USP1–UAF1 deubiquitinase complex as a physiological enhancer of TLR3/4-, RIG-I–, and cGAS-induced antiviral signaling by targeting TBK1. Mechanistically, UAF1 and USP1 bound to TBK1, removed K48-linked polyubiquitination of TBK1, and then stabilized its expression. Therefore, our results outline a novel mechanism for the control of TBK1 activity and suggest USP1–UAF1 deubiquitinase complex as a potential target for the prevention of viral diseases.
Results and discussion
Expression pattern of UAF1 and USP1 during viral infection
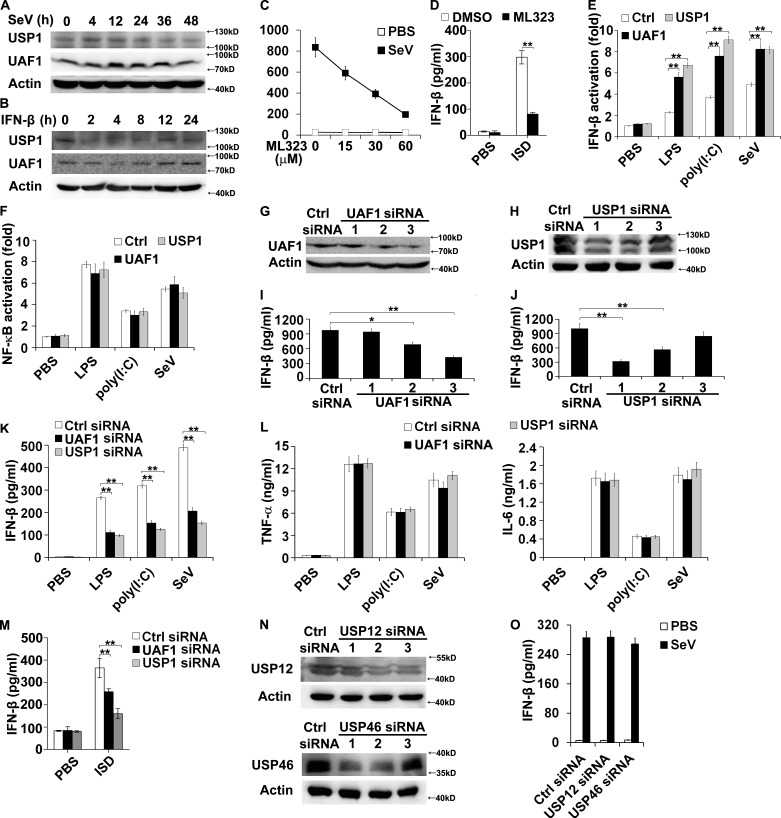
First, we examined the expression status of UAF1 and USP1 during viral infection. Sendai virus (SeV), a kind of ssRNA virus recognized by RIG-I (Loo and Gale, 2011), markedly induced UAF1 and USP1 expression in mouse primary peritoneal macrophages at the early stage of infection (Fig. 1 A). However, UAF1 and USP1 expression was decreased to normal levels at a later stage of viral infection (Fig. 1 A). Interestingly, IFN-β stimulation dramatically decreased UAF1 and USP1 expression (Fig. 1 B). Collectively, these data illustrate the dynamic changes of UAF1 and USP1 expression during viral infection, suggesting that USP1–UAF1 complex is potentially involved in the regulation of antiviral responses.
Figure 1.
USP1–UAF1 complex enhances IFN-β production. (A and B) Western blot analysis of UAF1 and USP1 expression in mouse peritoneal macrophages infected with SeV (A) or stimulated with IFN-β (B) for the indicated time periods. (C) Mouse peritoneal macrophages were treated with increasing amounts of ML323 for 4 h and then stimulated with SeV for 18 h. ELISA analyzed IFN-β production. (D) Mouse peritoneal macrophages were treated with 30 µM ML323 for 4 h and then transfected with ISD for 8 h. ELISA analyzed IFN-β production. (E and F) RAW264.7 cells were transfected with IFN-β or NF-κB reporter plasmid together with UAF1 and USP1 expression plasmids or a control plasmid and analyzed for luciferase activity after treatment with LPS for 6 h, poly(I:C) for 6 h, or SeV for 12 h. (G and H) Western blot analysis of UAF1 (G) or USP1 (H) expression in mouse peritoneal macrophages transfected with the indicated siRNA for 36 h. (I and J) ELISA analysis of IFN-β in the supernatants of peritoneal macrophages, as in G and H, stimulated with LPS. (K and L) Mouse peritoneal macrophages were transfected with control siRNA, UAF1 siRNA 3, or USP1 siRNA 1 and then stimulated with LPS for 6 h or poly(I:C) for 6 h or infected with SeV for 18 h. ELISA analysis of IFN-β, TNF-α, and IL-6 in the supernatants. (M) Mouse peritoneal macrophages were transfected with control siRNA, UAF1 siRNA 3, or USP1 siRNA 1 and then transfected with ISD for 8 h. ELISA analysis of IFN-β in the supernatants. (N) Western blot analysis of USP12 or USP46 expression in mouse peritoneal macrophages transfected with the indicated siRNA for 36 h. (O) Mouse peritoneal macrophages were transfected with control siRNA, USP12 siRNA 2, or USP46 siRNA 1 and then infected with SeV for 18 h. ELISA analysis of IFN-β in the supernatants. Data are representative of three experiments (mean ± SD of three samples in C, D, I–M, and O; mean ± SD of six samples in E and F; *, P < 0.05; **, P < 0.01; by Student’s t test). Similar results were obtained in three independent experiments in panels A, B, G, H, and N. Ctrl, control.
USP1–UAF1 complex facilitates IFN-β production
Next, we investigated the effects of USP1–UAF1 deubiquitinase complex on IFN-β production in macrophages. ML323, a selective USP1–UAF1 inhibitor (Liang et al., 2014), was used to inhibit deubiquitinase activity of USP1–UAF1 complex. ML323 treatment greatly decreased IFN-β secretion in SeV-infected macrophages in a dose-dependent manner (Fig. 1 C). ML323 treatment also markedly decreased IFN-β secretion in interferon-stimulating DNA (ISD, which can be recognized by cGAS) transfected macrophages (Fig. 1 D). In addition, UAF1 and USP1 overexpression both significantly enhanced LPS-, polyinosinic:polycytidylic acid [poly(I:C)]-, and SeV-induced IFN-β reporter gene activation (Fig. 1 E), with no effects on NF-κB activation (Fig. 1 F).
To further investigate the function of UAF1 and USP1 on IFN-β production, synthesized interfering RNAs targeting mouse UAF1 or USP1 were used to suppress endogenous UAF1 or USP1 expression. UAF1 siRNA 3 and USP1 siRNA 1, which have a higher efficiency to inhibit the expression of target proteins (Fig. 1, G and H), have a greater potential to increase LPS-induced IFN-β secretion (Fig. 1, I and J). Therefore, UAF1 siRNA 3 and USP1 siRNA 1 were used in the following experiments. UAF1 and USP1 knockdown both substantially inhibited LPS-, poly(I:C)- and SeV-induced IFN-β production in mouse peritoneal macrophages (Fig. 1 K), with no effects on TNF-α and IL-6 secretion (Fig. 1 L). In addition, UAF1 and USP1 knockdown both markedly attenuated ISD-induced IFN-β secretion in macrophages (Fig. 1 M).
UAF1 constitutes three deubiquitinating enzyme complexes, including USP1/UAF1, USP12/UAF1, and USP46/UAF1 (Cohn et al., 2009; Sowa et al., 2009). We thus examined whether USP12 or USP46 could regulate IFN-β expression. USP12 siRNA 2 and USP46 siRNA 1, which have a higher efficiency to inhibit endogenous USP12 and USP46 expression, respectively (Fig. 1 N), were used in the following experiments. USP12 and USP46 knockdown both had no effects on IFN-β secretion (Fig. 1 O). Collectively, these results indicate that USP1–UAF1 deubiquitinase complex facilitates TLR3/4-, RIG-I–, and cGAS-induced IFN-β production.
USP1–UAF1 complex facilitates IRF3 activation
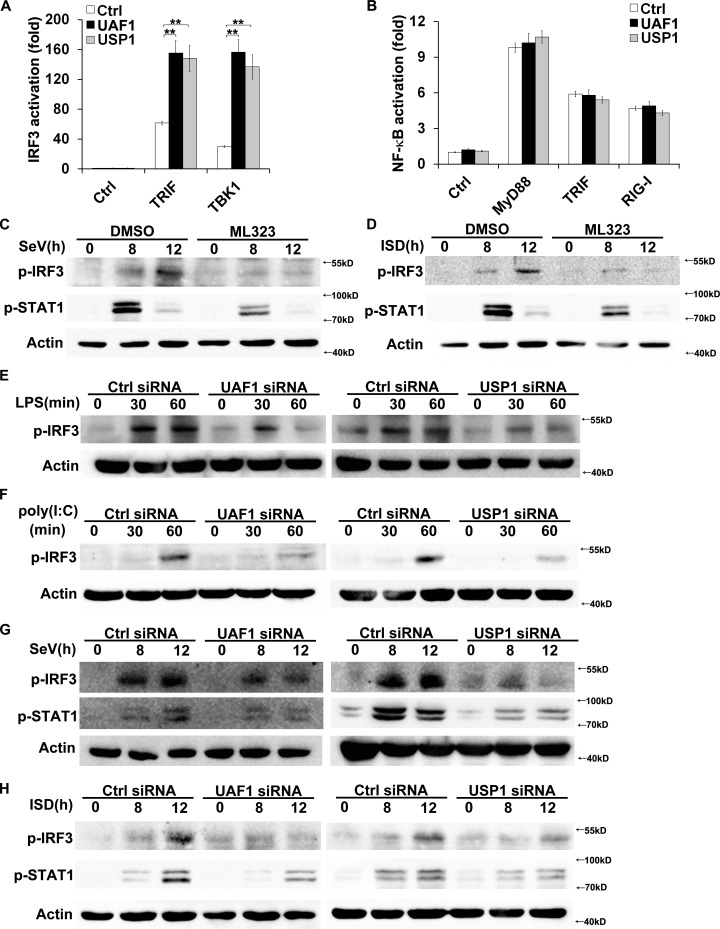
IRF3 is the key transcription factor required for IFN-β expression in TLR and RIG-I signaling (Kawai and Akira, 2010). We examined the effects of USP1–UAF1 deubiquitinase complex on IRF3 activation. UAF1 and USP1 both significantly enhanced TRIF- and TBK1-induced IRF3 reporter gene activation in HEK293T cells (Fig. 2 A). However, UAF1 and USP1 had no effects on MyD88-, TRIF-, and RIG-I–induced NF-κB activation (Fig. 2 B). In addition, ML323 treatment greatly attenuated SeV- and ISD-induced IRF3 and STAT1 phosphorylation (Fig. 2, C and D). To investigate the physiological function of USP1–UAF1 complex in IRF3 activation, mouse peritoneal macrophages were transfected with UAF1 or USP1 siRNA. UAF1 and USP1 knockdown both markedly inhibited LPS-, poly(I:C)-, SeV-, and ISD-induced phosphorylation of IRF3 (Fig. 2, E–H). Phosphorylation of STAT1 at Tyr701 after viral infection is IFN-β dependent. UAF1 and USP1 knockdown both dramatically attenuated SeV- and ISD-induced Tyr701 phosphorylation of STAT1, indicating the effect of USP1–UAF1 complex is consistent for IFN-β–mediated signaling (Fig. 2, G and H). Collectively, these data indicate that USP1–UAF1 deubiquitinase complex facilitates IRF3 activation and subsequent IFN-β signaling.
Figure 2.
USP1–UAF1 complex enhances IRF3 activation. (A and B) HEK293T cells were transfected with the indicated adaptors along with IRF3 or NF-κB reporter plasmid and UAF1 or USP1 plasmid, and luciferase activity was analyzed. (C and D) Western blot analysis of the indicated phosphorylated signaling proteins in peritoneal macrophages incubated with DMSO or 30 µM ML323 and then stimulated with SeV (C) or ISD (D). (E–H) Western blot analysis of the indicated phosphorylated signaling proteins in peritoneal macrophages transfected with control siRNA, UAF1 siRNA, or USP1 siRNA and then stimulated with LPS (E), poly(I:C) (F), SeV (G), or ISD (H). Data are representative of three experiments (mean ± SD of six samples in A and B; **, P < 0.01; by Student’s t test.). Similar results were obtained in three independent experiments in C–H. Ctrl, control.
UAF1 and USP1 interact with TBK1
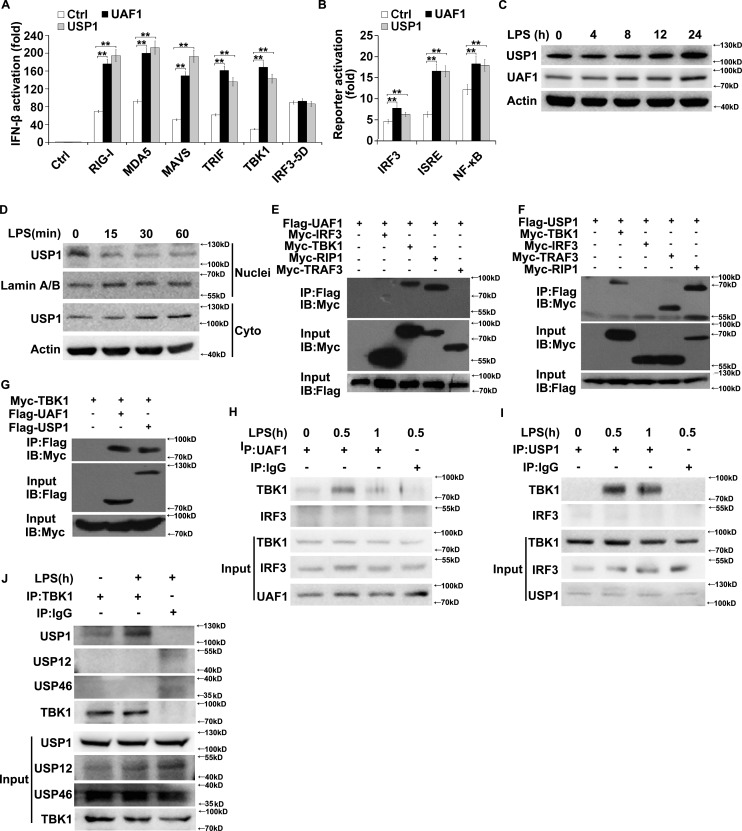
To determine the molecular targets of USP1–UAF1 deubiquitinase complex in TLR3/4- and RIG-I–induced signal transduction, the effects of UAF1 and USP1 on IFN-β promoter activation mediated by RIG-I, MDA5, MAVS, TRIF, TBK1, and IRF3 were examined in luciferase assays. UAF1 and USP1 both significantly enhanced RIG-I–, MDA5-, MAVS-, TRIF-, and TBK1-induced IFN-β promoter activation (Fig. 3 A). Wild-type IRF-3 induces marginal levels of type I IFN, whereas the IRF3 5D mutant, in which residues at positions 396, 398, 402, 404, and 405 were replaced by the phosphomimetic aspartate amino acid, induces strong activation of the IFN-β promoter (Lin et al., 1999; Zhang et al., 2012). Thus, we used IRF3 5D in our experiments. IRF3 5D–induced IFN-β promoter activation remains unchanged by UAF1 and USP1 overexpression (Fig. 3 A). These data indicate that USP1–UAF1 complex targets molecules upstream of IRF3 to inhibit IFN-β production. In addition, UAF1 and USP1 both greatly enhanced TBK1-induced IRF3, ISRE, and NF-κB activation (Fig. 3 B). Therefore, we speculate that TBK1 may be a target of USP1–UAF1 complex.
Figure 3.
UAF1 and USP1 interact with TBK1. (A) HEK293T cells were transfected with RIG-I, MDA5, MAVS, TRIF, TBK1, and IRF3 5D along with UAF1 or USP1 plasmid and IFN-β reporter plasmid, and luciferase activity was analyzed. (B) HEK293T cells were transfected with TBK1 along with UAF1 or USP1 plasmid, and IRF3, ISRE, or NF-κB reporter plasmid, and luciferase activity was analyzed. (C) Western blot analysis of UAF1 and USP1 expression in mouse peritoneal macrophages stimulated with LPS for the indicated time periods. (D) Western blot analysis of USP1 expression in nuclear or cytoplasmic fractions of mouse peritoneal macrophages stimulated with LPS for the indicated time periods. (E) Lysates from HEK293T cells transiently cotransfected with Flag-USP1 and Myc-TBK1, Myc-IRF3, Myc-TRAF3, or Myc-RIP1 expression plasmids were subjected to immunoprecipitation with anti-Flag antibody followed by Western blot analysis with anti-Myc antibody. (F) Lysates from HEK293T cells transiently cotransfected with Flag-UAF1 and Myc-IRF3, Myc-TBK1, Myc-RIP1, or Myc-TRAF3 expression plasmids were subjected to immunoprecipitation with anti-Flag antibody followed by Western blot analysis with anti-Myc antibody. (G) Lysates from HEK293T cells transiently cotransfected with Myc-TBK1 and Flag-UAF1 or Flag-USP1 expression plasmids were subjected to immunoprecipitation with anti-Flag antibody followed by Western blot analysis with anti-Myc antibody. (H–J) Lysates from mouse peritoneal macrophages stimulated with LPS were subjected to immunoprecipitation with the indicated antibody or control IgG followed by Western blot analysis with the indicated antibodies. Proteins in whole-cell lysate were used as positive control (Input). Data are representative of three experiments (mean ± SD of six samples in A and B; **, P < 0.01; by Student’s t test.). Similar results were obtained in three independent experiments in C–J.
Although USP1 expression was slightly enhanced upon LPS stimulation (Fig. 3 C), USP1 immediately translocated from the nucleus to cytoplasm after LPS stimulation in macrophages (Fig. 3 D), providing the possibility of its interaction with cytoplasmic adaptors. To further clarify the target of USP1–UAF1 complex, we examined the association between USP1–UAF1 and several adaptors. Myc-tagged TBK1, TRAF3, RIP1, and IRF3 plasmids and Flag-tagged USP1 or UAF1 plasmids were cotransfected into HEK293T cells. UAF1 and USP1 were both coprecipitated with TBK1 and RIP1 (Fig. 3, E–G). However, UAF1 and USP1 were not coprecipitated with IRF3 (Fig. 3, E and F). Next, the endogenous interaction was examined in macrophages stimulated with LPS. UAF1 and USP1 interacted with TBK1 following LPS stimulation (Fig. 3, H and I). However, no interaction was observed between USP1/UAF1 and IRF3 (Fig. 3, H and I). In addition, USP12 and USP46 were not associated with TBK1 (Fig. 3 J). Collectively, these results indicate that UAF1 and USP1 specifically bind to TBK1.
USP1–UAF1 removes K48-linked ubiquitination of TBK1 and promotes its stabilization
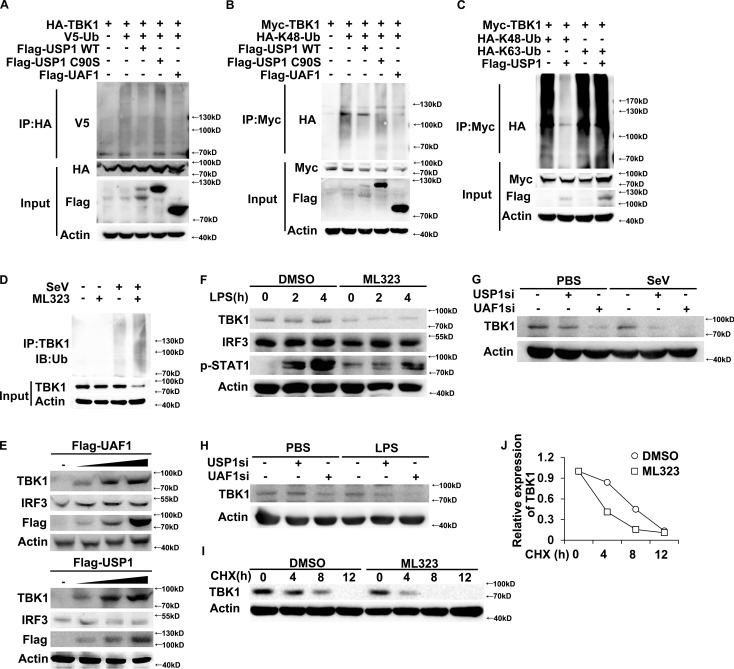
USP1–UAF1 complex possesses potent deubiquitinase activity (Cohn et al., 2007; Williams et al., 2011). We thus investigated the effects of USP1 and UAF1 on TBK1 ubiquitination. USP1 greatly inhibited TBK1 ubiquitination, whereas USP12 and USP46 had no effects on TBK1 ubiquitination (Fig. S1). Furthermore, USP1 and UAF1 both markedly inhibited total and K48-linked ubiquitination of TBK1 (Fig. 4, A and B). However, USP1 C90S mutant (a deubiquitinating enzyme activity–disrupted mutant, in which a Cys to Ser point mutation was introduced) has no effects on TBK1 ubiquitination (Fig. 4, A and B). In addition, USP1 could not remove K63-linked ubiquitination of TBK1 (Fig. 4 C). As parallel controls, USP1 and UAF1 had no effects on ubiquitination of IKK-ε and TRAF3 (Fig. S2, A and B). Under physiological conditions, the endogenous TBK1 was observed to be robustly ubiquitinated on SeV infection in macrophages (Fig. 4 D). ML323 treatment markedly increased endogenous TBK1 ubiquitination (Fig. 4 D). Collectively, these data indicate that the USP1–UAF1 complex selectively removes K48-linked ubiquitination of TBK1.
Figure 4.
USP1–UAF1 complex promotes stabilization of TBK1. (A) Western blot analysis of lysates from HEK293T cells transfected with V5-tagged ubiquitin (V5-Ub), HA-TBK1 and USP1 WT, USP1 C90S, or UAF1, followed by immunoprecipitation with anti-HA, probed with anti-V5. (B) Western blot analysis of lysates from HEK293T cells transfected with HA-tagged K48-linked ubiquitin (K48-Ub), Myc-TBK1 and USP1 WT, USP1 C90S, or UAF1, followed by immunoprecipitation with anti-Myc, probed with anti-HA. (C) Western blot analysis of lysates from HEK293T cells transfected with Myc-TBK1 and Flag-USP1, together with HA-K48-Ub or HA-K63-Ub, followed by immunoprecipitation with anti-Myc, probed with anti-HA. (D) Western blot analysis of lysates from mouse peritoneal macrophages treated with DMSO or 30 µM ML323 for 4 h and then infected with SeV for 8 h, followed by immunoprecipitation with anti-TBK1, and probed with anti-Ub. (E) Western blot analysis of extracts from HEK293T cells transfected with increasing amounts of USP1 or UAF1 expression plasmid. (F) Western blot analysis of extracts from mouse peritoneal macrophages treated with 30 µM ML323 for 4 h and then stimulated with LPS for the indicated time periods. (G and H) Western blot analysis of extracts from mouse peritoneal macrophages silenced of USP1 or UAF1 and then stimulated for various times with SeV (G) or LPS (H). (I and J) Immunoblot analysis of extracts from DMSO or 30 µM ML323 pretreated mouse peritoneal macrophages stimulated with LPS for 4 h and then treated for various times with CHX. TBK1 and IRF3 expression levels were quantitated by measuring band intensities by using ImageJ software. The values were normalized to actin (J). Similar results were obtained in three independent experiments.
K48-linked protein ubiquitination leads to the degradation of the corresponding protein by 26S proteasome (Jiang and Chen, 2011). We then investigated the effects of UAF1 and USP1 on TBK1 expression. UAF1 and USP1 overexpression both greatly enhanced TBK1 protein expression in a dose-dependent manner in HEK293T cells, with no effects on IRF3 expression (Fig. 4 E). In addition, UAF1 and USP1 overexpression both enhanced RIP1 protein expression (Fig. S3), indicating that RIP1 may be another target of USP1/UAF1, and that needs to be further investigated. Interestingly, although the association between USP1 and TRAF3 was detected (Fig. 3 F), USP1 and UAF1 overexpression had no effects on TRAF3 protein expression (Fig. S3). Furthermore, ML323 treatment dramatically decreased TBK1 expression and subsequent STAT1 phosphorylation in LPS-stimulated macrophages (Fig. 4 F). Consistently, UAF1 and USP1 knockdown also significantly decreased TBK1 protein level in both SeV- and LPS-stimulated macrophages (Fig. 4, G and H). To further investigate whether USP1–UAF1 complex could inhibit TBK1 protein degradation, a cycloheximide (CHX) chase experiment was performed. ML323 treatment greatly promoted TBK1 protein degradation (Fig. 4, I and J). All together, these data indicate that USP1–UAF1 deubiquitinase complex targets TBK1, stabilizes its expression, and thus facilitates TLR3/4- and RIG-I–induced IFN-β expression.
ML323 promotes viral replication in vitro and in vivo
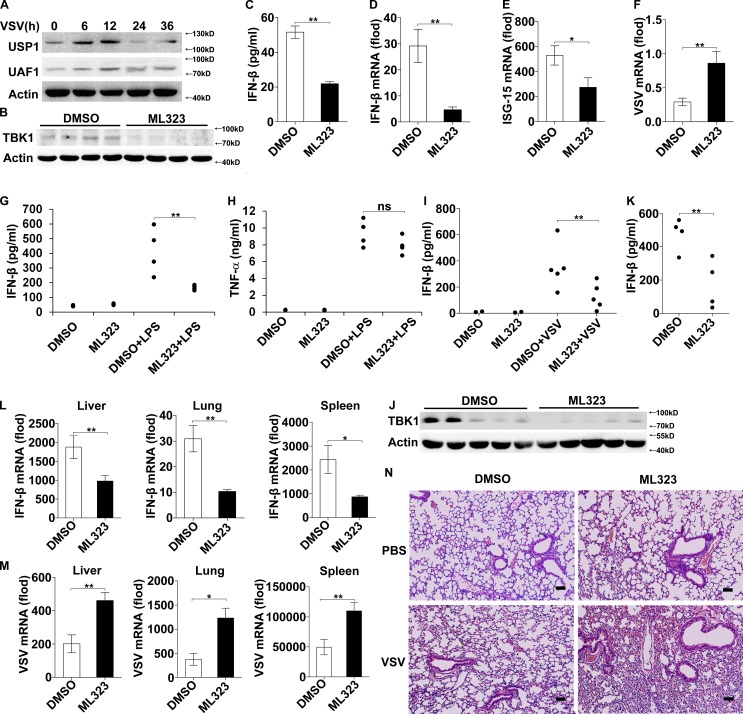
Type I IFNs are essential for the host to eliminate invading viruses (Liu et al., 2011; Wang and Fish, 2012; Hoffmann et al., 2015). We then investigated the effects of USP1–UAF1 complex on viral replication using a vesicular stomatitis virus (VSV, another kind of ssRNA virus recognized by RIG-I) infection model. UAF1 and USP1 expression was also under dynamic changes during VSV infection in macrophages. As shown in Fig. 5 A, UAF1 and USP1 expression was markedly increased at the early stage, reached a peak at 12 h, and then decreased to a normal level at 24 h after VSV infection. Next, we investigated the effects of USP1–UAF1 deubiquitinase complex on VSV replication using ML323. ML323 treatment significantly attenuated TBK1 protein expression in VSV-infected macrophages (Fig. 5 B). Accordingly, ML323 treatment inhibited subsequent IFN-β and ISG15 expression and enhanced VSV replication in macrophages (Fig. 5, C–F), indicating that ML323 could promote VSV replication in vitro.
Figure 5.
ML323 attenuates VSV-induced IFN-β and promotes viral replication in vitro and in vivo. (A) Western blot analysis of UAF1 and USP1 expression in mouse peritoneal macrophages infected with VSV for the indicated time periods. Similar results were obtained in three independent experiments. (B–F) Mouse peritoneal macrophages were treated with 30 µM ML323 for 4 h and then infected with VSV for 12 h. This experiment was repeated twice with similar results. Western blot analysis of TBK1 expression (B). ELISA analyzed IFN-β production (C). IFN-β mRNA, ISG15 mRNA, and intracellular VSV RNA replicates were measured by RT-PCR (D–F). Data are representative of three experiments (mean ± SD of three samples; **, P < 0.01; by Student’s t test). (G and H) Serum levels of IFN-β and TNF-α from C57BL/6 mice pretreated with ML323 or vehicle control as measured by ELISA 2 h after i.p. LPS injection. This experiment was repeated twice with similar results. Data shown are means ± SD (n = 4; **, P < 0.01; by Student’s t test). (I and J) C57BL/6 mice were i.p. injected with thioglycolate to elicit peritoneal macrophages. After 3 d, the mice were treated with ML323 or DMSO and then infected with VSV i.p. for 12 h. This experiment was repeated twice with similar results. IFN-β expression in lavage was examined by ELISA (I). Data shown are means ± SD (n = 5; **, P < 0.01; by Student’s t test). TBK1 expression in peritoneal exudate cells was analyzed by Western blot (J). (K–M) C57BL/6 mice were pretreated with ML323 or DMSO and then infected with VSV i.p. for 8 h. This experiment was repeated twice with similar results. Serum level of IFN-β was measured by ELISA (K). IFN-β mRNA (L) and intracellular VSV RNA replicates (M) in the liver, lung, and spleen were measured by RT-PCR. Data shown are means ± SD (n = 4; *, P < 0.05; **, P < 0.01; by Student’s t test). (N) C57BL/6 mice were pretreated with ML323 or DMSO and then infected with VSV i.p. for 18 h. H&E staining of lung tissue sections. Bars, 100 µm.
We next examined the potential functions of USP1–UAF1 complex in vivo using ML323. Mice were pretreated with ML323 before i.p. injection of LPS. ML323 treatment reduced serum level of IFN-β but did not considerably decrease the amount of TNF-α (Fig. 5, G and H), indicating that ML323 is active in vivo. We then investigated the regulatory effects of ML323 on IFN-β expression and viral replication in the context of VSV infection in vivo. ML323 treatment inhibited VSV-induced IFN-β production in peritoneal lavage (Fig. 5 I) and suppressed TBK1 protein expression in peritoneal exudate cells (Fig. 5 J). Furthermore, VSV infection induced IFN-β secretion was much less in sera of ML323-treated mice than in that of control mice (Fig. 5 K). Accordingly, IFN-β mRNA expression in the liver, lung, and spleen of ML323-treated mice was lower than that in controls (Fig. 5 L). In accordance with reduced IFN-β expression, VSV replication in the liver, lung, and spleen of ML323-treated mice was higher than that in controls (Fig. 5 M). Severe infiltration of immune cells was observed in the lungs of ML323-treated mice, compared with that of control mice after VSV infection (Fig. 5 N). Collectively, these data indicate that ML323 attenuates VSV-induced IFN-β expression and thus promotes VSV replication, both in vitro and in vivo.
Optimal productions of type I IFNs are crucial for maintaining immune homeostasis and elimination of invading viruses (Liu et al., 2011; González-Navajas et al., 2012; Wang and Fish, 2012; Hoffmann et al., 2015). Here, we identified USP1–UAF1 deubiquitinase complex as a viral infection–induced physiological enhancer of TBK1 activation and subsequent IFN-β expression. Given the vital role of TBK1 in innate antiviral immunity, the identification of a novel TBK1 physiological enhancer will be valuable for understanding the crosstalk between virus and host immune systems. In addition, as growing evidence implicates aberrant TBK1 activity in a variety of diseases (such as cancer, rheumatoid arthritis, and neurodegenerative syndromes; Shen and Hahn, 2011; Hammaker et al., 2012; Freischmidt et al., 2015), our results provide promising strategies for modulating TBK1 activation and may have therapeutic potential for the intervention of these diseases.
ML323 is a highly potent and specific inhibitor of the USP1–UAF1 deubiquitinase complex with excellent selectivity against deubiquitinases, deSUMOylase, deneddylase, and unrelated proteases (Liang et al., 2014). It has been reported that ML323 potentiates cisplatin cytotoxicity in non–small-cell lung cancer and osteosarcoma cells by inhibiting the deubiquitination of PCNA and FANCD2 (Liang et al., 2014). In the present study, we identified TBK1 as a novel substrate of USP1–UAF1 deubiquitinase complex. TBK1 is a pivotal kinase in innate antiviral immunity. Its expression and activity are crucial for IFN-β expression and initiation of antiviral immune responses (Zhao, 2013). ML323, as a selective inhibitor of USP1–UAF1, suppressed the deubiquitination of TBK1 and thus inhibited TBK1 protein degradation. Accordingly, ML323 attenuated IFN-β expression and enhanced viral replication both in vitro and in vivo. In addition, uncontrolled TBK1 activation and IFN-β production are associated with the development of multiple diseases, such as autoimmune diseases (Banchereau and Pascual, 2006; González-Navajas et al., 2012; Hammaker et al., 2012). Thus, our results indicate that ML323 may be a promising candidate for the intervention of these diseases, such as rheumatoid arthritis and systemic lupus erythematosus.
Materials and methods
Mice and cells
C57BL/6 mice were obtained from Vital River Laboratory Animal Technology Co. All animal experiments were undertaken in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, with the approval of the Scientific Investigation Board of School of Basic Medical Science, Shandong University. Mouse macrophage cell line RAW264.7, human THP-1, and human embryonic kidney (HEK293T) cells were obtained from American Type Culture Collection. Mouse primary peritoneal macrophages were prepared as described (Huai et al., 2016). The cells were cultured at 37°C under 5% CO2 in DMEM supplemented with 10% FCS (Invitrogen-Gibco), 100 U/ml penicillin, and 100 µg/ml streptomycin.
Reagents
ML323 was from Selleck Chemicals. CHX was purchased from Apexbio Company. LPS (Escherichia coli, 055:B5) and poly(I:C) were from Sigma-Aldrich. ISD was purchased from Invivogen. IFN-β was from Sino Biological Inc. The concentrations of agonists or stimuli were used as follows: 100 ng/ml LPS, 10 µg/ml poly(I:C), 10 µg/ml ISD, and 20 ng/ml IFN-β. Anti-USP1 (8033), anti-TBK1 (3054), anti-IRF3 (4302), anti–p-IRF3(Ser396; 4947), and anti–p-STAT1 (9167) were from Cell Signaling Technology. Anti-UAF1 (ab122473) and anti-V5 (ab9116) were from Abcam. Anti-Flag (F1804), anti-HA (H3663), anti-Myc (M4439), and anti–VSV-G (V4888) were from Sigma. Anti-USP12 (12608–1-AP) and anti-USP46 (13502–1-AP) were from Proteintech. Anti–Lamin A/B (BS1446) was from Bioworld Technology. Anti–β-actin (sc-81178) and protein G agarose (sc-2002) used for immunoprecipitation and horseradish peroxidase–conjugated secondary antibodies were from Santa Cruz Biotechnology. SeV was purchased from the China Center for Type Culture Collection, and the multiplicity of infection used was 1.
Plasmid constructs, sequences, and transfection
UAF1 and USP1 plasmids were constructed by PCR-based amplification of cDNA from THP-1 cells and then cloned into the pFLAG-CMV-2 eukaryotic expression vector (Sigma-Aldrich). The USP1 C90S mutation was generated by using the KOD-Plus-Mutagenesis kit (Toyobo). All constructs were confirmed by DNA sequencing. USP12 plasmid was purchased from Vigene Biosciences. USP46 plasmid was a gift from T. Gao (University of Kentucky, Lexington, KY). NF-κB reporter plasmid was purchased from Stratagene. IFN-β, ISRE, and IRF3 reporter plasmids and VSV-GFP were gifts from X. Cao (Second Military Medical University, Shanghai, China). The IRF3 reporter plasmids are composed of two plasmids, p-55UASGLuc and p-EFGAL4/IRF3 as described (Zhang et al., 2012). Expression plasmids for HA-Ub WT, HA-K48 Ub, HA-K63 Ub, RIG-I, MDA5, MAVS, TRIF, TBK1, IKK-ε, TRAF3, RIP1, IRF3, and IRF3 5D were described before (Zhang et al., 2012; Huai et al., 2016). Target sequences for transient silencing were 5′-GGUCGAGACUCCAUCAUAA-3′ (siRNA 1); 5′-GGAACAAAGACUCCAUUUA-3′ (siRNA 2) and 5′-GCCCGACCAAGUUAUUAAA-3′ (siRNA 3) for UAF1; 5′-GGCAAGUUAUGAGCUUAUA-3′ (siRNA 1), 5′-CGGCAAGGUUGAAGAACAA-3′ (siRNA 2), and 5′-GGAGAGCUCUGAAAUUUCU-3′ (siRNA 3) for USP1; 5′-CCUAAUGACAGUCUCCAAA-3′ (siRNA 1), 5′-CCCAAGAAGUUCAUCACAA-3′ (siRNA 2), and 5′-GCUUAAGAGGGUUCAGUAA-3′ (siRNA 3) for USP12; 5′-GGGAACACUCACUAACGAA-3′ (siRNA 1), 5′-GCAUUACAUCACCAUCGUA-3′ (siRNA 2), and 5′-GCUCAAGCCAUUGAGGAAU-3′ (siRNA 3) for USP46; “scrambled” control sequences were 5′-UUCUCCGAACGUGUCACGU-3′. For transient transfection of plasmids into RAW264.7 cells, jetPEI reagents were used (Polyplus-transfection). For transient transfection of plasmids into HEK293T cells, Lipofectamine 2000 reagents (Invitrogen) were used. For transient silencing, duplexes of small interfering RNA were transfected into cells with the INTERFERin Reagent (Polyplus-transfection) according to the standard protocol.
ELISA and RT-PCR
The concentration of IFN-β was measured with ELISA kits (BioLegend). The concentrations of TNF-α and IL-6 were measured with ELISA kits (Dakewe Biotech Company Ltd.). Total RNA was extracted with the RNAfast2000 RNA Extraction kit according to the manufacturer’s instructions. A LightCycler (ABI PRISM 7000) and a SYBR RT-PCR kit (Roche) were used for quantitative real-time RT-PCR analysis. Specific primers used for RT-PCR assays were 5′-ATGAGTGGTGGTTGCAGGC-3′ and 5′-TGACCTTTCAAATGCAGTAGATTCA-3′ for mIFN-β, 5′-AGAAGCAGATTGCCCAGAAG-3′ and 5′-TGCGTCAGAAAGACCTCATAGA-3′ for mISG15, 5′-TGTTACCAACTGGGACGACA’3 and 5′-CTGGGTCATCTTTTCACGGT-3′ for mβ-actin, 5′-ACGGCGTACTTCCAGATGG-3′ and 5′-CTCGGTTCAAGATCCAGGT-3′ for VSV. Data were normalized to β-actin expression in each sample.
Immunoprecipitation and Western blot analysis
For immunoprecipitation (IP), whole-cell extracts were collected 36 h after transfection and were lysed in IP buffer containing 1.0% (vol/vol) Nonidet P 40, 50 mM Tris-HCl, pH 7.4, 50 mM EDTA, 150 mM NaCl, and a protease inhibitor “cocktail” (Merck). After centrifugation for 10 min at 14,000 g, supernatants were collected and incubated with protein G Plus-Agrose Immunoprecipitation reagent (Santa Cruz) together with 1 µg monoclonal anti-Flag or 1 µg anti-Myc. After 6 h of incubation, beads were washed five times with IP buffer. Immunoprecipitates were eluted by boiling with 1% (wt/vol) SDS sample buffer. For immunoblot analysis, cells were lysed with M-PER Protein Extraction Reagent (Pierce) supplemented with a protease inhibitor mixture. Nuclear proteins were extracted by NE-PER Protein Extraction Reagent (Pierce) according to the manufacturer’s instructions. Protein concentrations in the extracts were measured with a bicinchoninic acid assay (Pierce). For Western blot analysis, immunoprecipitates or whole-cell lysates were loaded and subjected to SDS-PAGE, transferred onto nitrocellulose membranes, and then blotted as described previously (Huai et al., 2016).
Assay of luciferase activity
Luciferase activity was measured with the Dual-Luciferase Reporter Assay system according to the manufacturer’s instructions (Promega) as described previously (Zhang et al., 2012; Huai et al., 2016). Data were normalized for transfection efficiency by division of firefly luciferase activity with that of renilla luciferase.
In vivo LPS challenge
C57BL/6J mice (females, 6–8 wk old) were i.p. injected with 10 mg/kg ML323 for 6 h and then i.p. injected with 10 mg/kg LPS for 2 h. Serum levels of IFN-β and TNF-α were measured by ELISA.
Viral pathogenesis in mice
C57BL/6J mice (females, 8 wk old) were i.p. infected with VSV (5 × 107 PFU/mouse). 10 mg/kg ML323 was i.p. administered 6 h before VSV infection. Lungs from control or virus-infected mice were dissected, fixed in 10% (vol/vol) phosphate-buffered formalin, embedded into paraffin, sectioned, stained with hematoxylin and eosin (H&E) solution, and examined by light microscopy for histological changes.
Statistical analysis
All data are presented as means ± SD of three or four experiments. Statistical significance was determined with the two-tailed Student’s t test, with a p-value <0.05 considered statistically significant.
Online supplemental material
Fig. S1 shows that USP12 and USP46 have no effects on TBK1 ubiquitination. Fig. S2 shows that USP1 and UAF1 have no effects on the ubiquitination of IKK-ε and TRAF3. Fig. S3 shows that USP1 and UAF1 have no effects on the expression of IKK-ε and TRAF3.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81622030, 31570867, and 31370017), Shandong Provincial Nature Science Foundation for Distinguished Young Scholars (JQ201420), National Key Research and Developmental Program of China (2017YFC1001100), Key Research and Development Program of Shandong Province (2015GSF118159), and the Fundamental Research Funds of Shandong University (2016JC023).
The authors declare no competing financial interests.
Author contributions: Z. Yu, H. Song, M. Jia, J. Zhang, W. Wang, and Q. Li performed experiments. Z. Yu and W. Zhao designed the research and wrote the manuscript. L. Zhang provided expertise and advice. W. Zhao supervised the project.
References
- Banchereau J., and Pascual V.. 2006. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 25:383–392. 10.1016/j.immuni.2006.08.010 [DOI] [PubMed] [Google Scholar]
- Cai X., Chiu Y.H., and Chen Z.J.. 2014. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol. Cell. 54:289–296. 10.1016/j.molcel.2014.03.040 [DOI] [PubMed] [Google Scholar]
- Cohn M.A., Kowal P., Yang K., Haas W., Huang T.T., Gygi S.P., and D’Andrea A.D.. 2007. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell. 28:786–797. 10.1016/j.molcel.2007.09.031 [DOI] [PubMed] [Google Scholar]
- Cohn M.A., Kee Y., Haas W., Gygi S.P., and D’Andrea A.D.. 2009. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J. Biol. Chem. 284:5343–5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li Y., Zhu L., Liu D., Songyang Z., Wang H.Y., and Wang R.F.. 2012. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat. Immunol. 13:387–395. 10.1038/ni.2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freischmidt A., Wieland T., Richter B., Ruf W., Schaeffer V., Müller K., Marroquin N., Nordin F., Hübers A., Weydt P., et al. . 2015. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat. Neurosci. 18:631–636. 10.1038/nn.4000 [DOI] [PubMed] [Google Scholar]
- González-Navajas J.M., Lee J., David M., and Raz E.. 2012. Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 12:125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammaker D., Boyle D.L., and Firestein G.S.. 2012. Synoviocyte innate immune responses: TANK-binding kinase-1 as a potential therapeutic target in rheumatoid arthritis. Rheumatology (Oxford). 51:610–618. 10.1093/rheumatology/ker154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton S.M., Borg N.A., and Dixit V.M.. 2016. Ubiquitin in the activation and attenuation of innate antiviral immunity. J. Exp. Med. 213:1–13. 10.1084/jem.20151531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann H.H., Schneider W.M., and Rice C.M.. 2015. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 36:124–138. 10.1016/j.it.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huai W., Song H., Yu Z., Wang W., Han L., Sakamoto T., Seiki M., Zhang L., Zhang Q., and Zhao W.. 2016. Mint3 potentiates TLR3/4- and RIG-I-induced IFN-β expression and antiviral immune responses. Proc. Natl. Acad. Sci. USA. 113:11925–11930. 10.1073/pnas.1601556113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., and Chen Z.J.. 2011. The role of ubiquitylation in immune defence and pathogen evasion. Nat. Rev. Immunol. 12:35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., and Akira S.. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11:373–384. 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- Kim J.M., Parmar K., Huang M., Weinstock D.M., Ruit C.A., Kutok J.L., and D’Andrea A.D.. 2009. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev. Cell. 16:314–320. 10.1016/j.devcel.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Dexheimer T.S., Zhang P., Rosenthal A.S., Villamil M.A., You C., Zhang Q., Chen J., Ott C.A., Sun H., et al. . 2014. A selective USP1–UAF1 inhibitor links deubiquitination to DNA damage responses. Nat. Chem. Biol. 10:298–304. 10.1038/nchembio.1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Zhao Z., Yang Z., Meng Q., Tan P., Xie W., Qin Y., Wang R.F., and Cui J.. 2016. USP38 Inhibits Type I Interferon Signaling by Editing TBK1 Ubiquitination through NLRP4 Signalosome. Mol. Cell. 64:267–281. 10.1016/j.molcel.2016.08.029 [DOI] [PubMed] [Google Scholar]
- Lin R., Mamane Y., and Hiscott J.. 1999. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol. Cell. Biol. 19:2465–2474. 10.1128/MCB.19.4.2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.Y., Sanchez D.J., and Cheng G.. 2011. New developments in the induction and antiviral effectors of type I interferon. Curr. Opin. Immunol. 23:57–64. 10.1016/j.coi.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y.M., and Gale M. Jr. 2011. Immune signaling by RIG-I-like receptors. Immunity. 34:680–692. 10.1016/j.immuni.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill L.A., Golenbock D., and Bowie A.G.. 2013. The history of Toll-like receptors - redefining innate immunity. Nat. Rev. Immunol. 13:453–460. 10.1038/nri3446 [DOI] [PubMed] [Google Scholar]
- Ogrunc M., Martinez-Zamudio R.I., Sadoun P.B., Dore G., Schwerer H., Pasero P., Lemaitre J.M., Dejean A., and Bischof O.. 2016. USP1 Regulates Cellular Senescence by Controlling Genomic Integrity. Cell Reports. 15:1401–1411. 10.1016/j.celrep.2016.04.033 [DOI] [PubMed] [Google Scholar]
- Park E., Kim J.M., Primack B., Weinstock D.M., Moreau L.A., Parmar K., and D’Andrea A.D.. 2013. Inactivation of Uaf1 causes defective homologous recombination and early embryonic lethality in mice. Mol. Cell. Biol. 33:4360–4370. 10.1128/MCB.00870-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porritt R.A., and Hertzog P.J.. 2015. Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol. 36:150–160. 10.1016/j.it.2015.02.002 [DOI] [PubMed] [Google Scholar]
- Shen R.R., and Hahn W.C.. 2011. Emerging roles for the non-canonical IKKs in cancer. Oncogene. 30:631–641. 10.1038/onc.2010.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa M.E., Bennett E.J., Gygi S.P., and Harper J.W.. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell. 138:389–403. 10.1016/j.cell.2009.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.X., and Fish E.N.. 2012. The yin and yang of viruses and interferons. Trends Immunol. 33:190–197. 10.1016/j.it.2012.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.A., Maecker H.L., French D.M., Liu J., Gregg A., Silverstein L.B., Cao T.C., Carano R.A., and Dixit V.M.. 2011. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell. 146:918–930. 10.1016/j.cell.2011.07.040 [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Onomoto K., Jogi M., Akaboshi T., and Fujita T.. 2015. Viral RNA detection by RIG-I-like receptors. Curr. Opin. Immunol. 32:48–53. 10.1016/j.coi.2014.12.012 [DOI] [PubMed] [Google Scholar]
- Zhang M., Wang L., Zhao X., Zhao K., Meng H., Zhao W., and Gao C.. 2012. TRAF-interacting protein (TRIP) negatively regulates IFN-β production and antiviral response by promoting proteasomal degradation of TANK-binding kinase 1. J. Exp. Med. 209:1703–1711. 10.1084/jem.20120024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W. 2013. Negative regulation of TBK1-mediated antiviral immunity. FEBS Lett. 587:542–548. 10.1016/j.febslet.2013.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.