Singh et al. examined microRNA expression and physiological requirements in type 2 innate lymphoid cells (ILC2s). The miR-17∼92 cluster promotes ILC2 growth, cytokine expression, and function in allergic inflammation.
Abstract
MicroRNAs (miRNAs) exert powerful effects on immunity through coordinate regulation of multiple target genes in a wide variety of cells. Type 2 innate lymphoid cells (ILC2s) are tissue sentinel mediators of allergic inflammation. We established the physiological requirements for miRNAs in ILC2 homeostasis and immune function and compared the global miRNA repertoire of resting and activated ILC2s and T helper type 2 (TH2) cells. After exposure to the natural allergen papain, mice selectively lacking the miR-17∼92 cluster in ILC2s displayed reduced lung inflammation. Moreover, miR-17∼92–deficient ILC2s exhibited defective growth and cytokine expression in response to IL-33 and thymic stromal lymphopoietin in vitro. The miR-17∼92 cluster member miR-19a promoted IL-13 and IL-5 production and inhibited expression of several targets, including SOCS1 and A20, signaling inhibitors that limit IL-13 and IL-5 production. These findings establish miRNAs as important regulators of ILC2 biology, reveal overlapping but nonidentical miRNA-regulated gene expression networks in ILC2s and TH2 cells, and reinforce the therapeutic potential of targeting miR-19 to alleviate pathogenic allergic responses.
Introduction
Type 2 innate lymphoid cells (ILC2s) are a potent source of the type 2 signature cytokines IL-13 and IL-5 that drive allergic inflammation (Scanlon and McKenzie, 2012; Diefenbach et al., 2014; Klose and Artis, 2016). ILC2s are present in sputum and bronchoalveolar lavage (BAL) in human asthma (Christianson et al., 2015; Gordon et al., 2016; Smith et al., 2016). They respond to the epithelial cell–derived cytokines IL-33, IL-25, and thymic stromal lymphopoietin (TSLP) and may play a role in the initiation and/or maintenance of chronic airway inflammation. The mechanisms underlying ILC2 development, homeostasis, and function have been the subject of intense investigation. The signaling modules and transcriptional programs that control ILC2 biology bear striking resemblance to those operative in tissue-homing TH2 cells that mediate similar functions in adaptive immunity (Van Dyken et al., 2016). However, very little is known about the posttranscriptional regulation of ILC2 gene expression and immune function.
MicroRNAs (miRNAs) are ubiquitous posttranscriptional repressors of gene expression that play critical roles in lymphocyte homeostasis and allergic responses (Deshpande et al., 2015; Pua and Ansel, 2015). Each miRNA regulates multiple mRNA targets, and individual mRNAs may be regulated by multiple miRNAs. In this way, miRNA-directed regulatory networks powerfully influence biological processes despite the modest quantitative effect produced by each miRNA-target interaction. ILC2s express miR-155, and miR-155–deficient mice exhibit defective ILC2 and TH2 cell expansion and allergic airway (AA) inflammation (Okoye et al., 2014; Johansson et al., 2017). However, the expression and function of miRNAs in ILC2s remains largely unexplored.
The miR-17∼92 cluster of miRNAs is a particularly potent and pleiotropic regulator of B and T cell development, proliferation, survival, activation, differentiation, and cytokine production (Mogilyansky and Rigoutsos, 2013). The cluster is transcribed as a single primary miRNA transcript that is processed by the Drosha/Dgcr8 microprocessor complex and Dicer to produce six distinct mature miRNAs. These miRNAs can be classified by their target-determining seed sequences into four families: the miR-17 family (miR-17 and miR-20a), the miR-18 family (miR-18a), the miR-19 family (miR-19a and miR-19b-1), and the miR-92 family (miR-92a-1). miR-19a is uniquely up-regulated in airway-infiltrating T cells from asthmatic subjects, and it promotes TH2 cell cytokine production at least in part through direct targeting of the signaling inhibitors Pten, Socs1, and Tnfaip3 (which encodes A20; Simpson et al., 2014).
In the current study, we profiled and compared the miRNA transcriptomes of lung ILC2s and TH2 cells. miRNA expression was absolutely required to maintain ILC2 homeostasis in vivo, and the miR-17∼92 cluster was partially responsible for this function. In addition, miR-17∼92 promoted IL-13 and IL-5 production in response to IL-33 and ILC2-driven type 2 inflammation in vivo. We established RNA transfection of ILC2s and showed that miR-19a or siRNAs against its targets, Socs1 or Tnfaip3, can rescue the defective cytokine production of miR-17∼92–deficient ILC2s. We conclude that miRNAs are crucial regulators of ILC2 biology that mediate similar but nonidentical programs of posttranscriptional gene regulation in innate and adaptive lymphocytes.
Results
ILC2 miRNAome shares features with TH2 cells
Small RNA sequencing was performed to define the ILC2 miRNAome. RNA was obtained from CD45+Lin–Thy1.2+CD4–ST2+CD25+ ILC2s sorted from dissociated mouse lung tissue with or without three consecutive days of oropharyngeal IL-33 treatment. IL-33 instillation in the lungs expands and activates ILC2s (Bartemes et al., 2012; Kamijo et al., 2013), and we consistently recovered approximately twice as many ILC2s after IL-33 treatment. ILC2s were also analyzed after in vitro culture for 10 d in IL-33, TSLP, and IL-7. For comparison, RNA was collected from CD4+CD25–CD62LhiCD44lo naive T cells from spleen and lymph nodes; CD4+4get+ TH2 cells from BAL after the induction of AA inflammation in 4get mice containing a bicistronic mRNA linking a readily identifiable reporter, enhanced GFP (EGFP), to IL-4 gene expression (Mohrs et al., 2001); and in vitro differentiated TH2 cells. Among all samples, 46–95% of sequence reads that mapped to the genome aligned to mature miRNAs. Contaminating rRNA, tRNA, and mRNA fragments comprised, on average, just 9%, 4%, and 5% of mapped reads, respectively (Table S1).
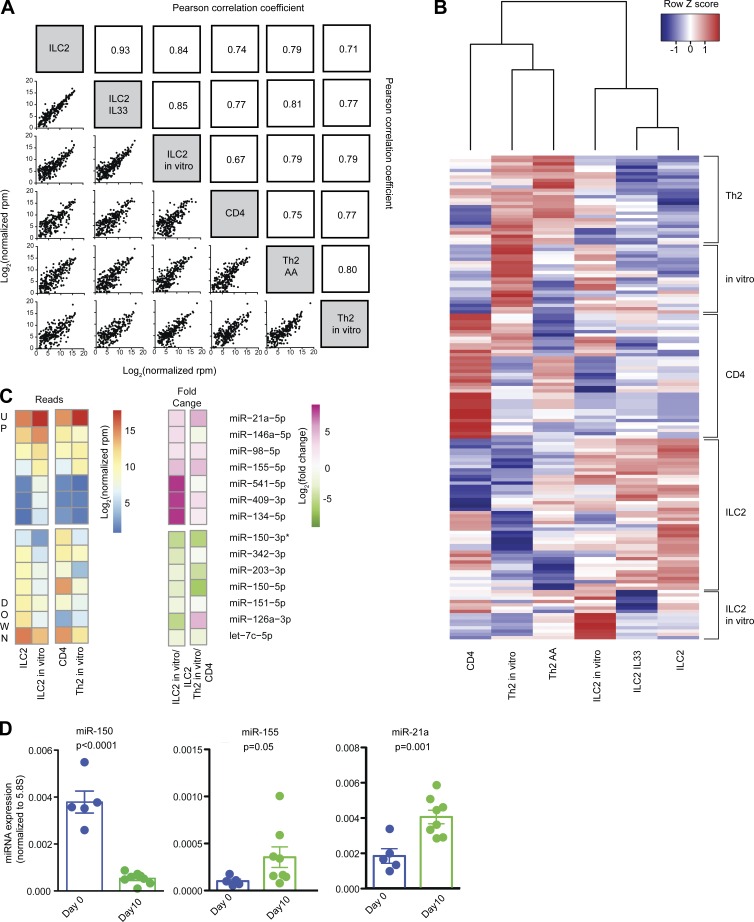
Overall, there was a strong positive correlation between the global miRNA expression profiles of all ILC2 and T cell samples (Fig. 1 A). However, pairwise comparisons revealed differences between sample groups (Table S2). Unsupervised hierarchical clustering of 147 miRNAs with significant differential expression (adjusted P < 0.01) in any pair of sample groups clustered the data by cell type (Fig. 1 B). Blocks of miRNAs with a shared signature were highlighted by this analysis, the largest being an “ILC2” block, a naive “CD4” block, and an “in vitro” block common to cultured ILC2 and T cells (Table S3). The expression profiles of ILC2s and ILC2s harvested after IL-33 administration in vivo were nearly identical. However, 14 miRNAs were significantly differentially expressed in ILC2s after 10 d of in vitro culture in IL-33 (Fig. 1 C and Table S2).
Figure 1.
Small RNA sequencing expression analysis of miRNAs in ILC2s and T cells. (A) Pearson correlation coefficients for global miRNA expression profiles among all samples. Sample groups include ILC2s isolated from naive mouse lung (ILC2), ILC2s isolated from mouse lung after IL-33 treatment (ILC2 IL33), ILC2 cultured in vitro for 10 d (ILC2 in vitro), naive CD4 T cells (CD4), TH2 cells isolated from BAL of mice with AAs (TH2 AA), and TH2 cells polarized in vitro (TH2 in vitro). (B) Unsupervised hierarchical clustering of all DESeq2 differentially expressed miRNAs. A list of individual miRNAs in each subgroup is included in Table S2. (C) Heat map of log2 normalized read counts and fold changes of miRNAs significantly (adjusted P < 0.01) up- or down-regulated in ILC2s after culture in vitro with IL-33, TSLP, and IL-7. Values for naive CD4 T cells and T cells after 5 d of TH2 polarizing culture are also provided for comparison. (D) Quantitative PCR confirmation of RNA sequencing data for the indicated miRNAs. Bars represent mean ± SEM. P-values were calculated with Student’s t test.
The expression of this group of 14 miRNAs was also investigated in naive CD4+ T cells and in vitro–cultured TH2 cells, revealing both shared and unique expression changes after in vitro activation of ILC2s and their adaptive lymphoid counterparts. miR-21a, miR-98, and miR-155 were similarly up-regulated, and miR-203, miR-151, and let-7c were similarly down-regulated, in both ILC2s and TH2 cells (Fig. 1 C). miR-150 was also down-regulated in both cell types, though the effect size was larger in TH2 cells. Quantitative PCR analyses confirmed that miR-150 was down-regulated, and miR-155 and miR-21 up-regulated, in ILC2s cultured for 10 d in IL-33 (Fig. 1 D). miR-146, a feedback inhibitor of NF-κB signaling that can be up-regulated in activated T cells under some conditions (Curtale et al., 2010; Rusca et al., 2012; Yang et al., 2012), was also up-regulated in ILC2s cultured in IL-33, but not in T cells cultured in TH2-polarizing conditions (Fig. 1 C; Monticelli et al., 2005).
A few miRNAs exhibited markedly different expression and regulation patterns in ILC2s and T cells. miR-126a expression was down-regulated from a mean of 8,000 reads per million (rpm) to 200 rpm after ILC2 culture in IL-33, but was up-regulated from 10 rpm in naive CD4+ T cells to 100 rpm during in vitro TH2 cell differentiation (Fig. 1 C and Table S4). In addition, three members of the large Mirc7 miRNA cluster on mouse chromosome 12, miR-541, miR-409, and miR-134, had very low baseline expression in ILC2s and naive CD4+ T cells but were up-regulated more than 100-fold in cultured ILC2s. These miRNAs remained poorly expressed in cultured TH2 cells, but they were expressed in 4get+ TH2 cells freshly isolated from AA samples at ∼30-fold higher abundance than in naive CD4+ T cells. The remaining members of the Mirc7 cluster were all expressed in relatively low amounts, typically <10 rpm. Full aligned miRNA sequence data are compiled in Table S4.
Severe reduction of lung ILC2s in miRNA-deficient mice
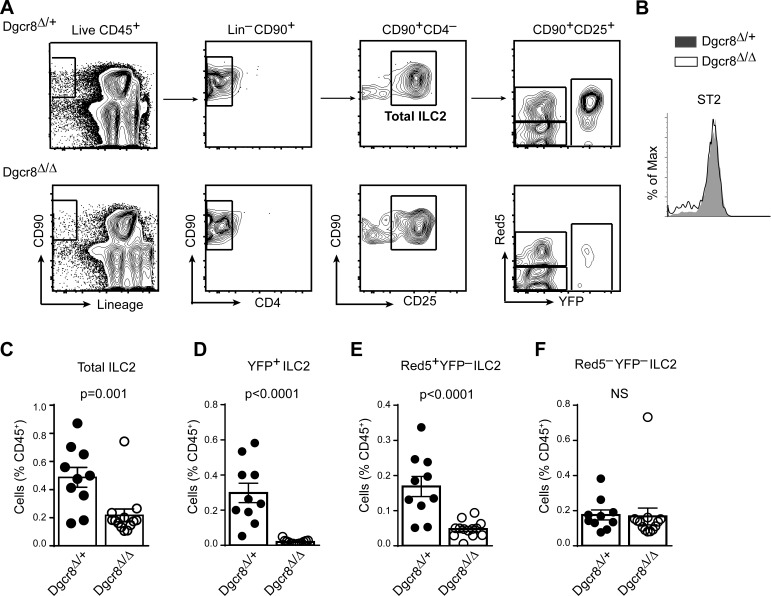
To investigate the overall requirements for miRNAs in ILC2 biology, we generated mice that lack the essential canonical miRNA biogenesis factor Dgcr8 in fluorescently marked ILC2s (hereafter termed Dgcr8Δ/Δ). Conditional Dgcr8-deficient mice (Rao et al., 2009) were intercrossed with Red5 mice (Nussbaum et al., 2013) in which Il5 is replaced with sequences encoding Cre recombinase and the fluorescent protein tdTomato, and with R26-YFP transgenic mice (Luche et al., 2007) to introduce a durable reporter of Cre expression. Total ILC2s in the lungs of Dgcr8+/− (Dgcr8-sufficient controls) and Dgcr8Δ/Δ (Dgcr8-deficient) mice were analyzed as live CD45+ CD90+lin−CD4−CD25+ (Fig. 2 A). Lung ILC2s identified in this way also expressed the IL-33 receptor, ST2 (Fig. 2 B). Global loss of canonical miRNAs in these cells severely impaired ILC2 homeostasis. Flow cytometric analyses showed a marked reduction of total lung ILC2s in Dgcr8Δ/Δ mice compared with Dgcr8-sufficient controls (Fig. 2 C). A very severe defect was revealed by restricting analysis to YFP+ ILC2s, indicating that Dgcr8 deficiency is incompatible with long-term ILC2 survival (Fig. 2 D). Further analysis of the impact of miRNA deficiency on ILC2 function in these animals was precluded by the near absence of YFP+ Dgcr8Δ/Δ ILC2s (Fig. 2 E). Even ILC2s that expressed the Red5 tdTomato but had not yet established R26-YFP reporter expression were affected (Fig. 2 F). In contrast, there was no difference in the frequency of YFP–Red5– ILC2s that lack evidence of current or prior Cre expression, indicating that ILC2s that retain miRNA expression did not expand to fill the niche left vacant by Dgcr8-deficient ILC2s. Collectively, these results demonstrate that Dgcr8 deletion leads to ILC2 depletion, underscoring the importance of miRNA biogenesis for the maintenance of normal ILC2s in the lung.
Figure 2.
Severe reduction of lung ILC2s in Red5 Dgcr8Δ/Δ mice. (A) Contour plots depict the gating strategy for lung ILC2s in Dgcr8Δ/+ and Dgcr8Δ/Δ mice. Total ILC2s are identified as live CD45+CD90+lin–CD4–CD25+ cells. Among total ILC2s, tdTomato (Red5) fluorescence indicates Cre expression, and YFP expressed from the R26-YFP reporter indicates Cre activity. (B) Flow cytometric analysis of ST2 median fluorescence intensity (MFI) in the total ILC2s of Dgcr8Δ/+ and Dgcr8Δ/Δ lung. (C–F) Flow cytometric quantification of total ILC2s (C), YFP+ ILC2s (D), Red5+ YFP–ILC2s (E), and Red5– YFP–ILC2s (F) in the lung of Dgcr8Δ/+ and Dgcr8Δ/Δ mice. Bar graphs show ILC2 frequency as percentage of live CD45+ cells (mean ± SEM). Each data point represents an individual mouse. Data were pooled from two independent experiments each with five to seven mice per group. P-values were calculated with Student’s t test.
The miR-17∼92 cluster regulates lung ILC2 homeostasis
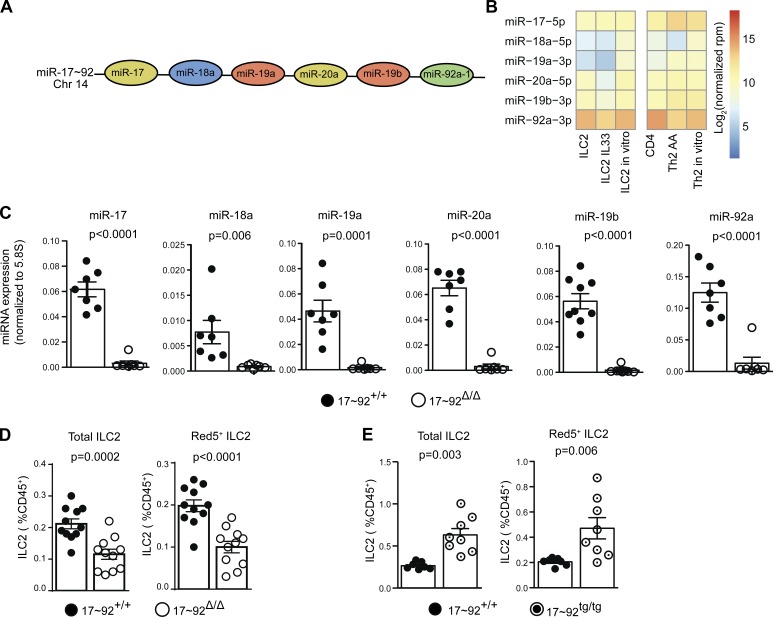
Because of the severity of the defect caused by Dgcr8 deficiency, further investigation of miRNA regulation of ILC2 immune function requires examination of specific miRNAs expressed in ILC2s. The miR-17∼92 cluster of miRNAs exert powerful effects on lymphocyte homeostasis, differentiation, and effector function, including TH2 cell cytokine production (Mogilyansky and Rigoutsos, 2013; Simpson et al., 2014). All six mature miRNA members of the miR-17∼92 cluster (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1; Fig. 3 A) were expressed in freshly isolated and IL-33–stimulated ILC2s at abundances similar to those expressed in naive CD4+ T cells and TH2 cells (Fig. 3 B). To determine whether the miR-17∼92 cluster regulates ILC2 homeostasis and function, we intercrossed mice in which the entire miR-17∼92 cluster is flanked by loxP sites with Red5 mice (hereafter termed 17∼92Δ/Δ). In addition, Cre-activatable R26–miR-17∼92 transgenic mice were intercrossed with Red5 mice to generate animals with fluorescently marked 17∼92tg/tg ILC2s overexpressing the miR-17∼92 cluster. miR-17∼92 cluster miRNA expression in Red5+ 17∼92Δ/Δ ILC2s was below the limit of detection or very low compared with miR-17∼92–sufficient control Red5+ 17∼92+/+ ILC2s (Fig. 3 C).
Figure 3.
miR-17∼92 cluster expression and requirement in lung ILC2s. (A) Schematic of the miR-17∼92 cluster miRNAs and their chromosomal positions. Colors indicate miRNAs families. (B) Heat map of log2 normalized read counts of miR-17∼92 cluster miRNAs in the indicated cell types (see Fig. 1 legend). (C) Quantitative PCR quantification of the indicated miRNAs in 10-d-cultured 17∼92Δ/Δ and 17∼92+/+ ILC2s. Symbols indicate individual cultures seeded with cells from a pool of two to three mice each. Data were pooled from three independent experiments. (D and E) Flow cytometric quantification of total lung ILC2 and Red5+ ILC2 frequency (see Fig. 2 A legend) in 17∼92Δ/Δ (D) and 17∼92tg/tg (E) mice compared with matched 17∼92+/+ controls. Each ILC2 culture was started with 10,000 (B) or 3,500 (C) ILC2s sorted from individual mouse or pools of two to six mice and cultured with IL-33, TSLP, and IL-7 for 10 d. Each data point represents the ILC2 frequency expressed as percentage of live CD45+ cells in an individual mouse. Data were pooled from two independent experiments. Bars represent mean ± SEM. P-values were calculated with Student’s t test.
The miR-17∼92 cluster regulated the maintenance of lung ILC2s. The frequency of total and Red5+ ILC2s was reduced by almost 50% in 17∼92Δ/Δ mouse lungs compared with miR-17∼92–sufficient Red5+ 17∼92+/+ control mice (Fig. 3 D). Conversely, 17∼92tg/tg mice bore more than twice the normal number of total and Red5+ ILC2s (Fig. 3 E). Thus, miR-17∼92 cluster miRNAs account for a significant proportion of the miRNA requirements for lung ILC2 homeostasis, although other miRNAs must also play major roles in this process.
The miR-17∼92 cluster promotes ILC2 function in AA inflammation
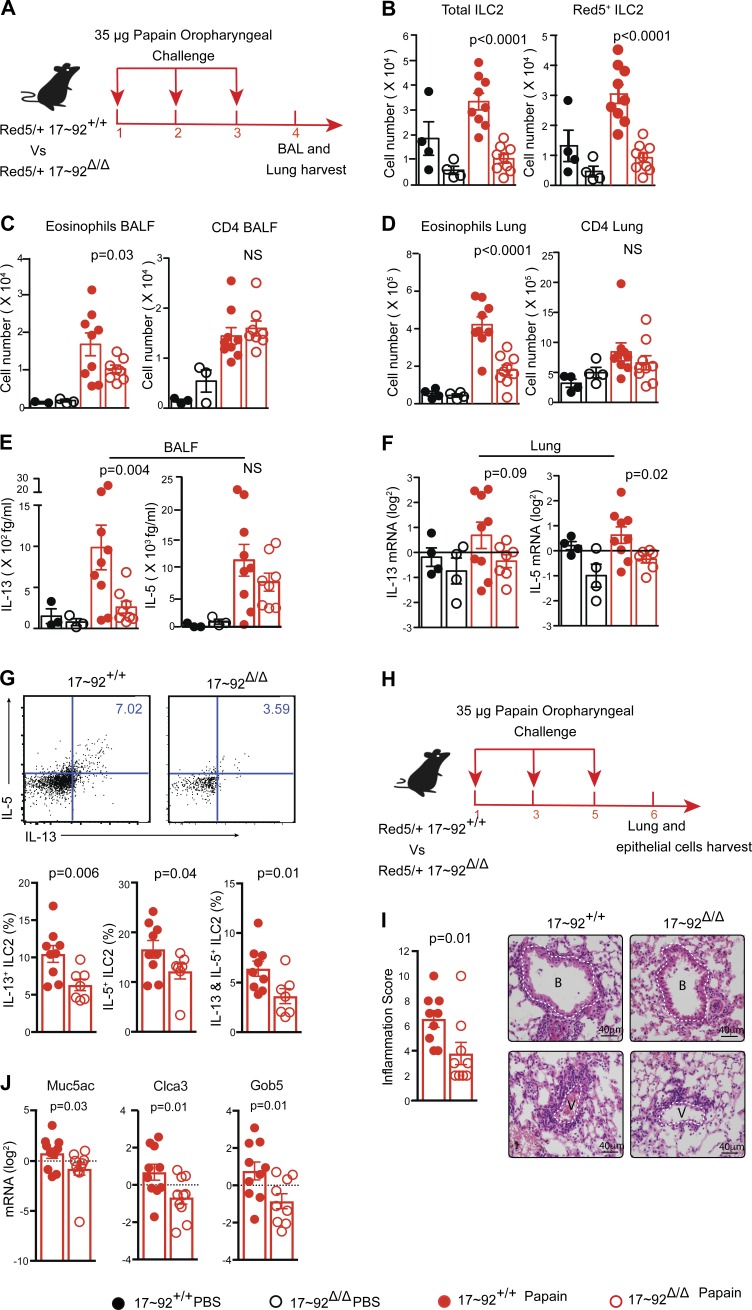
Tissue-resident ILC2 cells expand and secrete IL-5 and IL-13 in response to agents that incite type 2 inflammation. We used a mouse model of AA inflammation to test the generation and function of ILC2s in vivo and investigate the requirements for miR-17∼92 in an ILC2-mediated type 2 response. Mouse airways were challenged with the allergen papain on three consecutive days, and lung tissue samples were harvested on day 4 (Fig. 4 A). ILC2s expanded in response to papain in both 17∼92+/+ and 17∼92Δ/Δ mice, although the total number of ILC2s present in the lung was significantly lower in 17∼92Δ/Δ mice (Fig. 4 B). To characterize and quantify airway inflammation, we analyzed BAL and digested lung tissue cells by flow cytometry. Airway and lung eosinophil infiltration was reduced in 17∼92Δ/Δ mice, whereas CD4+ T cell numbers were similar in both groups of mice (Fig. 4, C and D). Consistent with the observed type 2 inflammation, IL-13 was significantly induced by papain in the BAL fluid of 17∼92+/+, but not 17∼92Δ/Δ mice (Fig. 4 E). IL-5 protein was induced in the BAL of both groups of mice, but the Il5 mRNA was more abundant in 17∼92+/+ total lung cells (Fig. 4 F). Il13 mRNA exhibited a similar trend but did not reach statistical significance. Papain-treated 17∼92Δ/Δ mice exhibited significantly reduced frequencies of IL-5+, IL-13+, and IL-5/IL-13 double-positive lung ILCs compared with 17∼92+/+ control mice (Fig. 4 G). Thus, miR-17∼92 deficiency in ILC2s decreased type 2 cytokine expression and eosinophilia, the hallmarks of allergic lung inflammation.
Figure 4.
miR-17∼92 cluster enhances ILC2 function in AA inflammation. (A) 4-d experimental model of papain-induced airway inflammation. Red5 heterozygous 17∼92Δ/Δ and 17∼92+/+ mice received 35 µg papain in 40 µl PBS or PBS alone oropharyngeally on three consecutive days, and lung tissue samples were harvested on day 4. (B) Flow cytometric enumeration of total ILC2s and Red5+ ILC2s in the lung. (C and D) Flow cytometric quantification of the number of eosinophils (Siglec-F+CD11c–) and CD4+ T cells recovered in BAL (C) and digested lung tissue (D). (E) Cytometric bead array (CBA) measurement of IL-13 and IL-5 in BAL fluid. (F) Quantitative PCR (qPCR) quantification of Il13 and Il5 mRNA in total lung cells, normalized to Gapdh mRNA. (G) Representative flow cytometric analysis of intracellular cytokine staining (top). Numbers in quadrants indicate percentage of cytokine-producing live, Lin−CD45+CD90+CD4− ILCs. Bar graphs (bottom) show frequencies of lung ILCs expressing IL-5, IL-13, or both IL-5 and IL-13. (H) 6-d experimental model of papain-induced airway inflammation. (I) Inflammation score from hematoxylin and eosin staining of lung sections (see Materials and methods). Note the thicker and more circumferential inflammatory infiltrates cuffing a representative 17∼92+/+ versus 17∼92Δ/Δ mice bronchiole (B, top) and vessel (V, bottom). The thickness of the bronchiole or vessel wall without inflammation is highlighted by a dashed white line for reference. Bars, 40 µm. (J) qPCR quantification of IL-13–induced repair genes in epithelial brushing of papain-treated mice. Data are representative of three independent experiments each with 5–10 mice in each group (A–F). Bars represent mean ± SEM. P-values were calculated using one-way ANOVA with Sidak’s multiple comparison between papain-treated 17∼92+/+ and 17∼92Δ/Δ mice (B–F) or with Student’s t test (G–J).
To observe lung pathology, mice were challenged with papain on days 1, 3, and 5, and lung samples were harvested for histology 6 d after the initiation of papain treatment (Fig. 4 H). A persistent reduction in total inflammation was observed in 17∼92Δ/Δ mice (Fig. 4 I). Only sparse focal epithelial cell mucus metaplasia was observed by periodic acid–Schiff staining in tissue sections of either 17∼92+/+ or 17∼92Δ/Δ mice, precluding quantitative analysis (not depicted). However, IL-13–responsive genes that promote mucus production and tissue repair, including Muc5ac, Clca3, and Gob5 (Kuperman et al., 2002; Thai et al., 2005), were significantly lower in the epithelial brushings of papain-challenged 17∼92Δ/Δ mice (Fig. 4 J). Collectively, these data show that the miR-17∼92 cluster promotes the in vivo function of ILC2s in the induction of AA inflammation. Because a minor fraction of ILC2s remained Red5– in papain-challenged mice (Fig. 4 B), these results may underestimate the physiological effect of miR-17∼92 deficiency in ILC2s.
The miR-17∼92 cluster promotes ILC2 growth and regulates cytokine production
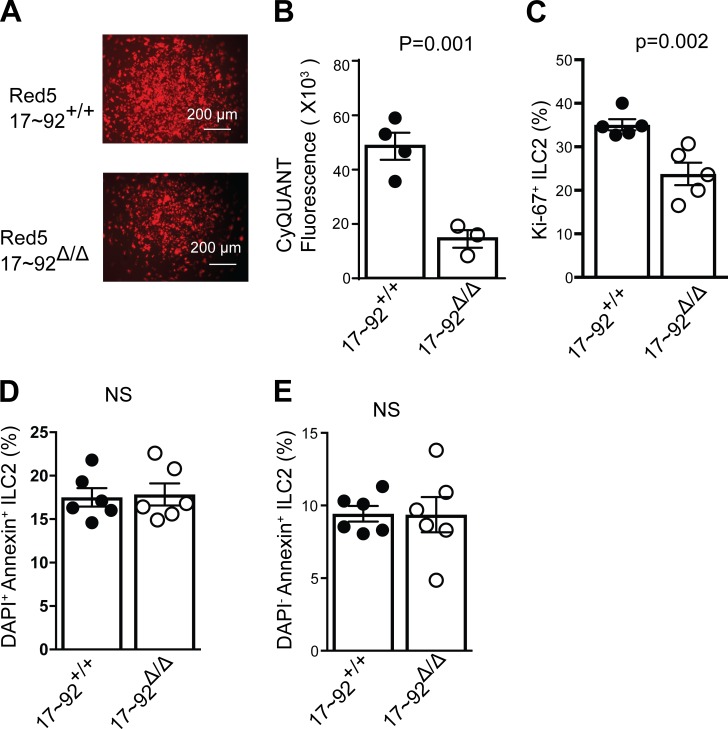
To understand the mechanisms by which the miR-17∼92 cluster promotes ILC2-mediated type 2 immune responses, equal numbers of lung ILC2s from 17∼92+/+ or 17∼92Δ/Δ mice were purified and cultured in IL-33, TSLP, and IL-7. 17∼92Δ/Δ ILC2s expressed ST2, TSLPR, and IL7R normally (not depicted). These stimuli induced ILC2 proliferation in vitro, but fewer fluorescent Red5+ cells were visible in 17∼92Δ/Δ cultures within 4 d (Fig. 5 A). We used CyQUANT assays to accurately measure ILC2s outgrowth in vitro, revealing a 60% reduction in the 17∼92Δ/Δ ILC2s present at day 4 compared with control 17∼92+/+ cultures (Fig. 5 B). Similar results were observed throughout the 10-d culture period. To test whether the miR-17∼92 cluster affected ILC2 proliferation, we examined Ki67 expression at day 3. A significantly higher percentage of 17∼92+/+ ILC2s were Ki67+ compared with 17∼92Δ/Δ ILC2s (Fig. 5 C). Annexin V staining indicated no difference in the rate of apoptosis (Fig. 5, D and E). These data indicate that miR-17∼92 is required for normal cytokine-driven ILC2 proliferation in vitro.
Figure 5.
The miR-17∼92 cluster promotes ILC2s proliferation. (A) Microscopic view of tdTomato fluorescence in Red5+ ILC2s. Bars, 200 µm. (B) CyQUANT NF cell proliferation assay fluorescent quantification of 17∼92Δ/Δ and 17∼92+/+ ILC2s after culture for 4 d. (C) Quantification of Ki67+ 17∼92Δ/Δ and 17∼92+/+ ILC2s frequencies after culture for 3 d. (D and E) Apoptosis in 17∼92Δ/Δ and 17∼92+/+ ILC2s was assessed using Annexin V and DAPI staining on day 4 of culture. Bar graphs show the percentage of dead apoptotic ILC2s (DAPI+ Annexin+; D) and live apoptotic ILC2s (DAPI− Annexin−; E). NS, not significant. Data in A, B, D, and E are representative of at least 20, 4, and 2 independent experiments, respectively. Bars represent mean ± SEM. P-values were calculated with Student’s t test (B–E).
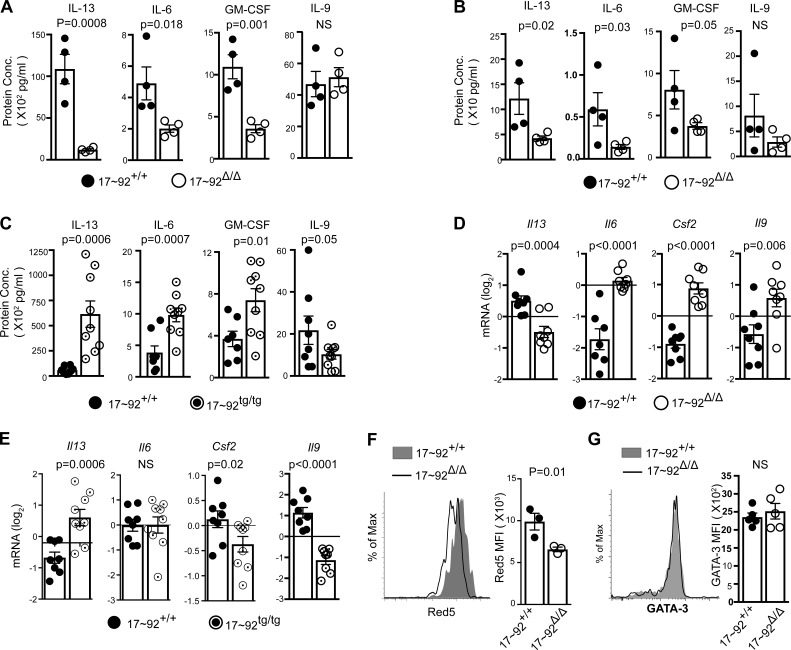
ILC2 cytokine production was also independently affected by miR-17∼92 deficiency. 17∼92Δ/Δ ILC2s produced less IL-13, IL-6, and GM-CSF over 10 d of culture (Fig. 6 A). Because there were fewer ILC2s present in 17∼92Δ/Δ cultures at day 4, we also measured early cytokine secretion at day 2 and observed a similar result (Fig. 6 B). Conversely, 17∼92tg/tg ILC2s produced more IL-13, IL-6, and GM-CSF than did their 17∼92+/+ counterparts at 10 d of culture (Fig. 6 C). IL-2, IL-4, and IL-10 were not detected in these culture conditions. For further confirmation and to determine whether cytokine expression differences could be observed on a per-cell basis independent of cell number, we also measured cytokine transcripts. Indeed, 17∼92Δ/Δ ILC2s exhibited reduced Il13 mRNA expression (Fig. 6 D), and 17∼92tg/tg ILC2s had increased Il13 expression compared with control 17∼92+/+ ILC2s (Fig. 6 E). However, this defect was specific to Il13, as 17∼92Δ/Δ ILC2s expressed higher levels of Il6, Csf2 (which encodes GM-CSF), and Il9 (Fig. 6 D). Consistent with these data, 17∼92tg/tg ILC2s had decreased expression of Csf2 and Il9 compared with control 17∼92+/+ ILC2s (Fig. 6 E). The enhanced expression of Il6, Csf2, and Il9 in 17∼92Δ/Δ ILC2s indicates that they were fully capable of cytokine expression in response to IL-33, TSLP, and IL-7 in vitro, yet they display a specific defect in the production of type 2 cytokines. 17∼92Δ/Δ ILC2s also showed a per-cell reduction in IL-5 expression as indicated by reduced Red5-encoded tdTomato fluorescence intensity (Fig. 6 F). Consistent with previous findings in TH2 cells, miR-17∼92 did not affect GATA-3 expression in ILC2s (Fig. 6 G). Collectively, these data demonstrate important roles for the miR-17∼92 cluster in promoting ILC2 proliferative capacity and regulating cytokine production.
Figure 6.
The miR-17∼92 cluster regulates cytokines production from ILC2s. (A and B) IL-13, IL-6, GM-CSF, and IL-9 measured by cytometric bead array (CBA) in supernatants from 10- and 2-d cultured 17∼92Δ/Δ and 17∼92+/+ ILC2s, respectively. (C) IL-13, IL-6, GM-CSF, and IL-9 measured by CBA in supernatants from 10-d cultures of 17∼92+/+ and 17∼92tg/tg ILC2s. (D and E) Quantitative PCR quantification of Il13, Il6, Csf3, and Il9 in 10-d cultured ILC2s, normalized to Gapdh. (F) Red5 tdTomato median fluorescence intensity (MFI) in 4-d-cultured ILC2s, assessed by flow cytometry as an indirect measure of Il5 expression. (G) Flow cytometry analysis of GATA-3 expression in 17∼92Δ/Δ and 17∼92+/+ ILC2s at day 3 of culture, presented as MFI of GATA-3 expression in live singlet. Each culture was started with 3,500 (A–F) ILC2s, sorted from an individual mouse or pools of two to three mice, and cultured with IL-33, TSLP, and IL-7. Data in A and B are representative of at least 10 independent experiments. Data in D are pooled from three independent experiments. Data in F are representative of two independent experiments. Bars represent mean ± SEM. P-values were calculated with Student’s t test (A–G).
miR-19a promotes IL-13 and IL-5 production in ILC2s
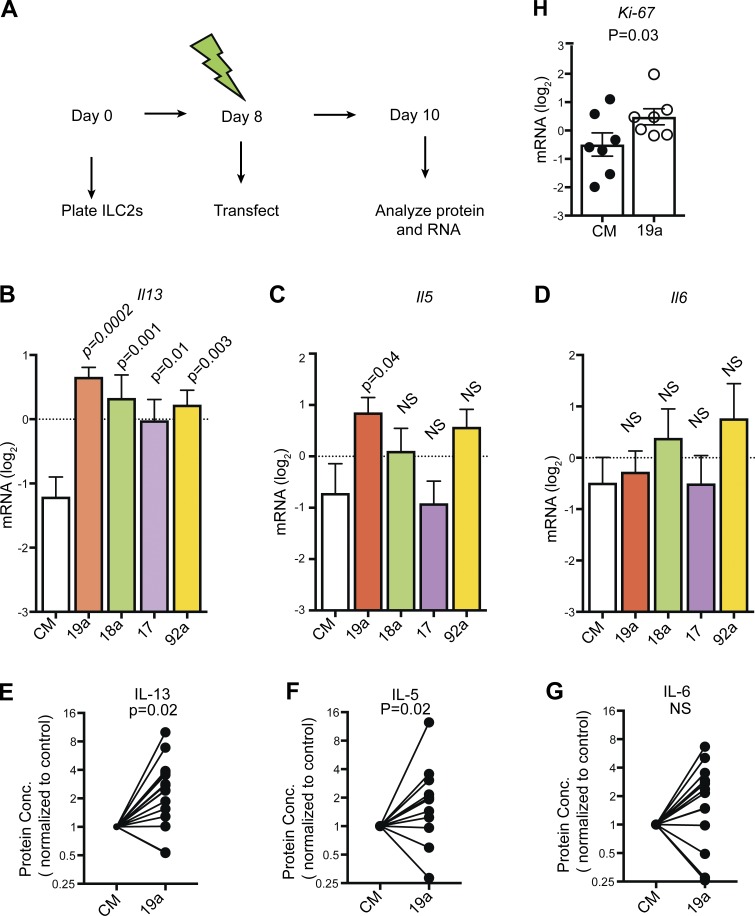
The miR-17∼92 cluster promotes type 2 cytokine expression in TH2 cells, mainly as a result of the function of the miR-19 family (Simpson et al., 2014). We established a protocol for next-generation transfection of ILC2s with small RNAs and used it to restore miR-17∼92 family members’ activity in 17∼92Δ/Δ ILC2s by transfecting them with miR-19a, miR-18a, miR-17, and miR-92a mimics (Fig. 7 A). All four miRNA mimics increased Il13 mRNA expression in 17∼92Δ/Δ ILC2s compared with control mimics, although miR-19a transfection induced Il13 expression the most (Fig. 7 B). miR-19a transfection also significantly increased Il5 mRNA expression (Fig. 7 C), but it did not affect Il6 (Fig. 7 D). 17∼92Δ/Δ ILC2s transfected with miR-19a mimic secreted significantly more IL-13 (Fig. 7 E) and IL-5 (Fig. 7 F), but not IL-6 protein (Fig. 7 G), than cells transfected with control mimic. Importantly, miR-19a rescued defective IL-13 and IL-5 production in the absence of other endogenous miR-17∼92 family miRNAs, even though it was delivered after 8 d of culture. 17∼92Δ/Δ ILC2s transfected with miR-19a also exhibited significantly higher expression of Ki67 mRNA than control cells transfected with control mimic, indicating that miR-19a affects ILC2 proliferation as well (Fig. 7 H). We conclude that miR-19a is an important member of the miR-17∼92 cluster that promotes proliferation and enhances IL-13 and IL-5 production by ILC2s in a cell-intrinsic manner.
Figure 7.
Cell-intrinsic defect in IL-13 and IL-5 production in 17∼92Δ/Δ ILC2s rescued by miR-19a. (A) 10-d experimental model of next-generation transfection of ILC2s. Each culture was started with 10,000–12,000 17∼92Δ/Δ ILC2s sorted from the pool of three to six mice and cultured with IL-33, TSLP, and IL-7. Cells were transfected on day 8 with control mimic (CM) or mimics of miR-19a, miR-18a, miR-17, and miR-92a and harvested on day 10. (B–D) Quantitative PCR (qPCR) quantification of Gapdh-normalized Il13, Il5, and Il6 mRNA from 17∼92Δ/Δ ILC2s transfected with CM or miR-19a, miR-18a, miR-17, and miR-92a mimic. (E–G) IL-13, IL-5, and IL-6 measured by cytometric bead array (CBA) in supernatants from 17∼92Δ/Δ ILC2s transfected with miR-19a or control miRNA mimic. Data are pooled from five independent experiments and are normalized to matched control mimic transfection in each biological replicate. Bars represent mean ± SEM. (H) qPCR quantification of Gapdh-normalized Ki-67 mRNA expression from 17∼92Δ/Δ ILC2s transfected with miR-19a or control miRNA mimic. Bars represent mean ± SEM. Data in B–D and H are pooled from two independent experiments. P-values were calculated with one-way ANOVA with Dunnett’s multiple comparison post hoc test (comparing each column to CM; B–D), paired Student’s t test on unnormalized data (E–G), or Student’s t test (H).
miR-19a targets genes that limit IL-13 and IL-5 production in ILC2s
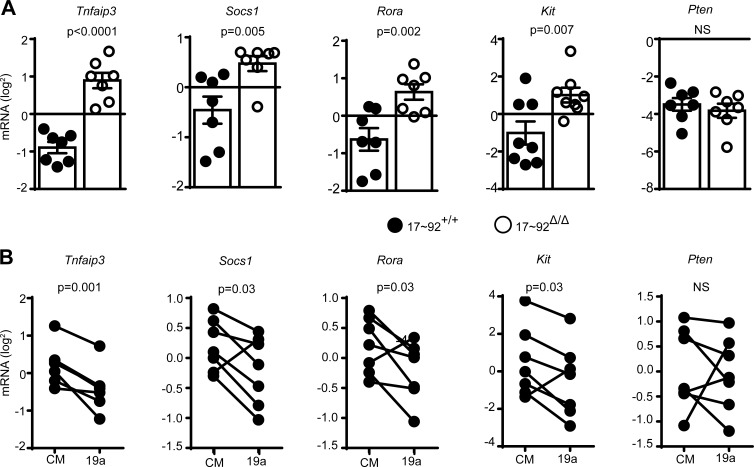
To investigate how miR-19 regulates IL-13 production in ILC2s, a candidate gene approach was taken. We tested several miR-19 target genes that had been previously confirmed in other cell types (Hsu et al., 2011). The mRNAs encoding four of these candidates (Tnfaip3, Socs1, Rora, and Kit) were more abundant in cultured 17∼92Δ/Δ ILC2s than in control 17∼92+/+ ILC2s (Fig. 8 A), consistent with targeting by one or more miR-17∼92 cluster miRNAs in ILC2s. miR-19 was sufficient to mediate repression of Tnfaip3, Socs1, Rora, and Kit in 17∼92Δ/Δ ILC2s, as transfection with miR-19a mimic significantly reduced the expression of their corresponding mRNAs compared with cells transfected with control mimic (Fig. 8 B). Surprisingly, the well-known miR-19 target Pten was affected by neither miR-17∼92 deficiency nor miR-19a mimic transfection in ILC2s (Fig. 8, A and B).
Figure 8.
miR-19 regulates target gene expression in ILC2s. (A) Quantitative PCR (qPCR) quantification of the indicated candidate miR-19a target mRNAs in 10-d-cultured 17∼92+/+ and 17∼92Δ/Δ ILC2s. (B) qPCR quantification of mRNAs in 17∼92Δ/Δ ILC2s transfected with control mimic (CM) or miR-19a mimic (19a). Data are normalized to Gapdh mRNA. Each culture was started with 3,500 (A) or 10,000 (B) ILCs, sorted from pools of two to six mice. Data are pooled from three independent experiments (A and B). Bars represent mean ± SEM. P-values were calculated with Student’s t test (A) or paired Student’s t test (B).
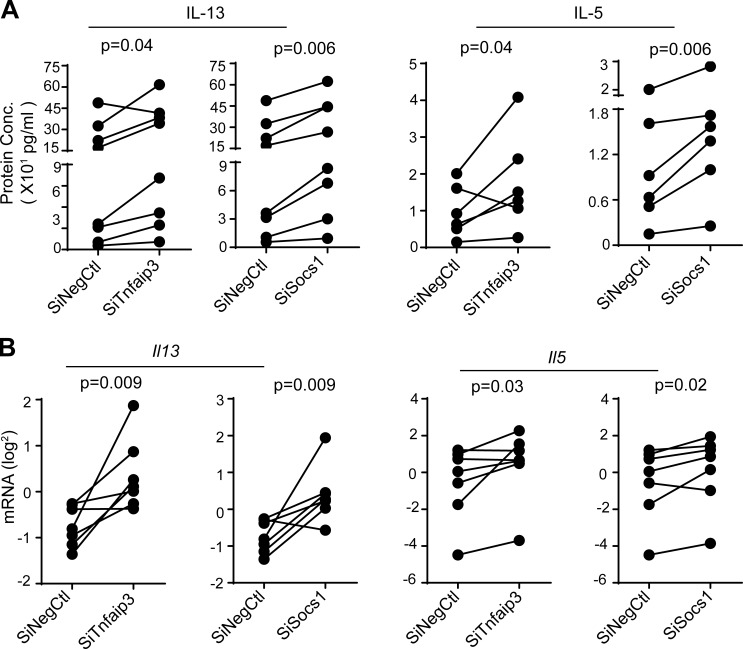
The functional impact of miR-19 targets on ILC2 cytokine expression was tested by transfecting 17∼92Δ/Δ ILC2s with siRNAs against Tnfaip3 and Socs1. Compared with control siRNA, Tnfaip3 and Socs1 siRNAs increased both IL-13 and IL-5 cytokine production and mRNA expression in ILC2s (Fig. 9, A and B). Thus, these two miR-19 targets specifically regulated IL-13 and IL-5 production in ILC2s. Collectively, these data indicate that miR-19 promotes type 2 cytokine production and regulates overlapping but nonidentical sets of functionally relevant target genes in ILC2 and TH2 cells.
Figure 9.
miR-19 targets Tnfaip3 and Socs1 negatively regulate IL-13 and IL-5 expression in ILC2s. (A) IL-13 and IL-5 measured by cytometric bead array (CBA) in supernatants from 17∼92Δ/Δ ILC2s transfected with siRNAs targeting Tnfaip3 or Socs1 or negative control siRNA (siNegCtl). (B) Quantitative PCR quantification of Il13 and Il5 mRNA, normalized to Gapdh mRNA. Each culture was started with 10,000 ILC2s sorted from pools of five to seven mice. Data are pooled from three independent experiments. P-values were calculated with paired Student’s t test.
Discussion
Extensive study of ILC2s in recent years has changed our perception of TH2 cells as the central cytokine-secreting drivers of allergic inflammation. ILC2s have emerged as a potent early source of type 2 cytokines, important contributors to airway disease models, and a potential link between innate and adaptive phases of type 2 responses (Scanlon and McKenzie, 2012; Martinez-Gonzalez et al., 2015). Several key transcription factors and cytokine signals required for ILC2 development and maturation have been identified, yet knowledge of the gene expression networks that govern ILC2 biology remains underdeveloped (Walker and McKenzie, 2013; von Moltke and Locksley, 2014). In the current study, we demonstrate that miRNAs are absolutely critical regulators of ILC2 homeostasis, and the miRNAs of the miR-17∼92 cluster in particular are required for normal ILC2 survival, cytokine production, and allergic responses. One of these, miR-19, promoted IL-13 production and targeted inhibitors of the NF-κB and STAT pathways that mediate innate cytokine signals upstream of Il13 expression in ILC2s. These findings illustrate how the network logic of miRNA function allows them to regulate common biological outcomes through overlapping but distinct target gene repertoires in different cellular contexts, such as ILC2s and TH2 cells.
The expression and importance of miRNAs in T, B, and natural killer cells has been well established (Fehniger et al., 2010; Baumjohann and Ansel, 2013; Beaulieu et al., 2013; de Yébenes et al., 2013) but miRNAs were heretofore largely unexplored in other lymphocytes. Global disruption of miRNA biogenesis by inactivating Dgcr8 in Il5-expressing cells provoked extensive loss of ILC2s, revealing a critical role for miRNAs in ILC2 homeostasis. In addition, we identified the miRNA repertoire of resting and cytokine-activated murine ILC2s, providing critical information and a framework for formulating questions about miRNA participation in the regulatory circuits that control ILC2 biology. Consistent with their similar gene expression and cytokine production profiles (Van Dyken et al., 2016), ILC2s and TH2 cells exhibited broadly shared miRNA repertoires. It is tempting to speculate that miRNAs expressed in both ILC2s and TH2 cells might regulate common pathways that determine their similar immune effector phenotypes. Indeed, our analysis of miR-19 demonstrates that overlapping but nonidentical miRNA:target networks govern common functional behaviors in ILC2s and TH2 cells.
In addition, ILC2s actively reset their miRNA repertoire when stimulated with TSLP and IL-33, similar to the behavior of T cells activated by antigen stimulation (Bronevetsky et al., 2013). This finding suggests that the posttranscriptional down-regulation of mature miRNAs and concurrent changes in primary miRNA transcription that rapidly establish a new pattern of miRNA expression in activated T cells may operate similarly downstream of cytokine signals in ILC2s. We observed this dynamic cytokine-induced change in miRNA expression in ILC2s stimulated in vitro, but very similar profiles of miRNA expression in ILC2s harvested from lungs with and without IL-33 exposure in vivo. Either IL-33 is insufficient to drive miRNA expression changes in the context of the tissue microenvironment, or miRNA expression recovers to homeostatic levels rapidly after the IL-33 source is exhausted.
A few miRNAs exhibited markedly different expression patterns in ILC2s and TH2 cells. For example, miR-126 was abundant in ILC2s and strongly down-regulated upon cytokine activation, but scarcely expressed in naive T cells and moderately up-regulated in differentiated TH2 cells in vitro and in vivo. Because miR-126a antagonists inhibit the response to house dust mite allergen in a mouse model of asthma (Mattes et al., 2009), these findings raise the possibility that miR-126a may be functionally important for ILC2 sentinel function in the initiation of type 2 immune responses. It is also possible that a subset of tissue-resident TH2 cells may further modulate their expression of miR-126 and other miRNAs to more closely resemble the pattern observed in ILC2s.
The miR-17∼92 cluster has many established functions in lymphocytes. It promotes the proliferation and survival of T cells and regulates the differentiation and function of several effector T cell subsets, including TH2 cells (Xiao et al., 2008; Jiang et al., 2011; Steiner et al., 2011; Baumjohann et al., 2013; de Kouchkovsky et al., 2013; Kang et al., 2013; Simpson et al., 2014; Han et al., 2015). All six miR-17∼92 cluster miRNAs were expressed similarly in ILC2s and TH2 cells, and ILC2s lacking the cluster also exhibited defective induction of allergic inflammation in vivo and in proliferation and type 2 cytokine production in response to TSLP, IL-7, and IL-33 in vitro. IL-33 is a potent activator of lung ILC2s (Bartemes et al., 2012; Kamijo et al., 2013), and the genes encoding IL-33 and its receptor ST2 are major susceptibility loci for human asthma (Moffatt et al., 2010). ST2 surface expression was normal in resting and activated 17∼92Δ/Δ ILC2s. Reduced IL7R expression was observed in miR-17∼92–deficient common lymphoid progenitors and thymocytes (Regelin et al., 2015). However, we observed no difference in IL7R or TSLPR expression in cultured ILC2s. Thus, modulation of signaling or gene expression downstream of these cytokine receptors must be responsible for the altered responses of 17∼92Δ/Δ ILC2s.
In miR-17∼92–deficient TH2 cells, miR-19a and miR-19b restored IL-13 production, but other miRNAs of the cluster did not (Simpson et al., 2014). miR-19a also restored IL-13 production in 17∼92Δ/Δ ILC2s. We identified two important pieces of the miR-19 target network in ILC2s: Tnfaip3 and Socs1. Both of these targets encode inhibitors of cytokine signaling pathways, indicating that miR-19 supports the production of IL-13 and IL-5 by simultaneously amplifying signals downstream of IL-33 and IL-7/TSLP. Tnfaip3 encodes A20, a powerful negative regulator of the NF-κB pathway previously identified as an important target of miR-19 in macrophages and TH2 cells (Gantier et al., 2012; Simpson et al., 2014). SOCS1 inhibits the JAK-STAT pathway downstream of IL-7 and other related cytokines, and also limits type 2 cytokine production in TH2 cells (Simpson et al., 2014). Our analysis in ILC2s revealed a limiting role for both of these miR-19 target genes, as depleting each individually with siRNAs was sufficient to markedly increase IL-13 and IL-5 production. These findings suggest that both A20 and SOCS1 are important miR-19 targets that regulate the ILC2 effector function. Genome-wide approaches to determine the full miR-19 target network in ILC2s would probably reveal additional targets involved in ILC2 biology.
The tumor suppressor Pten is another important miR-19 target that has been implicated in miRNA control of many biological processes (Baumjohann and Ansel, 2013). However, Pten expression was unaltered by miR-17∼92 deficiency or miR-19a mimic transfection into ILC2s. Genetic rescue of the Pten overexpression observed in miR-17∼92–deficient TH2 cells partially restored IL-13 production (Simpson et al., 2014), but apparently this mode of regulation is not operational in ILC2s. In contrast, two miR-19 targets that are not highly expressed in TH2 cells were regulated by miR-19 in ILC2s: Kit and Rora. c-Kit is a receptor tyrosine kinase often used to identify mouse and human ILC2 cells. Rora is a direct target of multiple miR-17∼92 cluster miRNAs, and aberrant expression of the transcription factor that it encodes, RORα, disrupts the fidelity of lineage-specific gene expression in miR-17∼92–deficient follicular helper T cells (Baumjohann et al., 2013). RORα is critical for ILC2 development, but little is known about its contribution to functional aspects of ILC2 biology (Wong et al., 2012). Polymorphisms in the human RORA locus are associated with asthma susceptibility (Moffatt et al., 2010). Future studies may uncover additional functionally relevant miR-17∼92 targets in ILC2s.
In addition to augmenting IL-13 and IL-5 production, miR-17∼92 surprisingly reduced ILC2 expression of Il9, Il6, and Csf2 (GM-CSF). These latter effects were not rescued by transfection with mimics corresponding to each miRNA family in the miR-17∼92 cluster, although transgenic overexpression of the entire cluster reduced Il9 and Csf3 expression. These findings verify that 17∼92Δ/Δ ILC2s are capable of efficient cytokine production and highlight the specificity of miR-19 regulation of IL-13 and IL-5. In addition, they suggest that multiple miR-17∼92 miRNAs act in concert to coordinate regulatory networks that control ILC2 cytokine production and expansion in response to IL-33.
In conclusion, this study provides concrete evidence that miRNAs regulate diverse aspects of ILC2 biology. Using mouse genetic tools and a novel protocol for ILC2 transfection with siRNAs and miRNA mimics, we dissected the mechanism by which the miR-17∼92 cluster regulates type 2 cytokine production. miR-19a regulates IL-13 production through overlapping but nonidentical downstream target networks in ILC2s and TH2 cells, suggesting that miR-19a itself might be an attractive therapeutic target (Lu and Rothenberg, 2013). Alternatively, targeting the same signaling pathways modulated by miR-19 could be an effective intervention for asthma and allergic diseases (Edwards et al., 2009). Our findings open new avenues for understanding the gene expression programs and signaling pathways that underlie the onset of allergic responses by innate and adaptive lymphocytes.
Materials and methods
Mice
Mice with loxP-flanked alleles encoding miR-17∼92 (Mirc1fl/fl, 008458; The Jackson Laboratory) and Rosa26-Mir-17∼92-transgenic mice Gt(ROSA)26Sortm3(CAG-MIR17-92,-EGFP)RSKY, 008517; The Jackson Laboratory) were crossed to Il5tm1.1(cre)LKy (Red5) mice (Nussbaum et al., 2013) to delete the floxed alleles specifically in IL-5–producing cells. Dgcr8tm1.1Blel (Dgcr8fl/fl); Gt(ROSA)26Sortm3(CAG-EYFP)Hze (Rosa-YFP; Rao et al., 2009) were provided by R. Blelloch (University of California, San Francisco) and crossed with Red5 mice to generate Red5 Dgcr8fl/fl ROSA-YFP mice (here Dgcr8Δ/Δ). Il4tm1LKy (4get) mice have been described previously (Mohrs et al., 2001). Male and female mice used were 5–9 wks of age on the C57BL/6 background (≥10 backcross generations). All mice were housed and bred in the specific pathogen–free barrier facility at the University of California, San Francisco. The Institutional Animal Care and Use Committee at the University of California, San Francisco, approved all animal experiments.
Lung digestion
For flow cytometric analysis or sorting of ILC2s, lungs were perfused with PBS via the right ventricle until it blanched. Lung lobes were excised and processed with a gentleMACS automated tissue dissociator in C tubes (Miltenyi Biotec), running program “m_lung_01,” followed by shaking incubation for 35 min at 37°C, 175 rpm, in RPMI medium containing 50 µg/ml Liberase TM and 25 µg/ml DNAs1 (Roche). After incubation, a second dissociation was performed with program “m_lung_02.” After tissue dissociation, cell suspensions were filtered through a 70-μm nylon mesh, washed, and treated with BD Pharm Lyse lysing solution (BD Biosciences) to lyse red blood cells before final suspension in PBS with 3% FBS.
Flow cytometry and sorting
Monoclonal, murine-specific antibodies from BioLegend included anti-B220 (RA3-6B2), anti–Gr-1 (RB6-8C5), anti-F4/80 (BM8), anti-CD3 (145-2C11), anti-CD49b (DX5), anti-NK1.1 (PK136), anti-CD4 (RM4-5), anti-CD11c (N418), anti-CD4 (GK 1.5), anti-CD8a (53–6.7), and anti–IL-5 (TRFK5). Antibodies from eBioscience included anti-CD25 (PC61.5), anti-CD90.2 (53–2.1), anti-CD4 (GK1.5), anti-CD4 (RM4.5), anti-CD11b (M1/70), and anti–Ly-6G (RB6-8C5). Antibodies from BD Biosciences included anti-CD11C (HL3), Streptavidin, anti-CD45 (30-F11), anti–Siglec-F (E50-2440), anti-Ki67 (SolAI5), anti–IL-13 (eBio13A), and anti-GATA3 (TWAJ). Antibodies from TONBO Biosciences included anti-CD11b (M1/70). Nonspecific antibody binding was blocked with anti-CD16/CD32 plus 2% normal mouse/rat serum. Dead cells were excluded with Fixable Viability Dye eFluor780 (eBioscience). Sample data were acquired with a LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo software. Cells were sorted using a FACS Aria II (Beckman Coulter).
For measurement of intracellular cytokine expression, cells were isolated ex vivo and stimulated in complete high-glucose DMEM supplemented with 10% heat-inactivated FBS, penicillin/streptomycin/l-glutamine, 2-mercaptoethanol with 50 ng/ml phorbol 12-myristate 13-acetate (Sigma-Aldrich), 500 ng/ml ionomycin (Sigma-Aldrich), and 10 µg/ml brefeldin A (Sigma-Aldrich) for 4 h. Cells were stained with the viability dye eFluor780 and fixed in 4% PFA for 8 min at room temperature. Cytokine staining was done in permeabilization buffer containing 0.5% saponin (Invitrogen). For analysis of transcription factor expression, cultured cells were fixed and permeabilized according to the manufacturer’s instructions (Foxp3/Transcription Factor Staining Buffer Set; eBioscience).
In vitro cultures and transfection
Sorted ILC2s were cultured (37°C, 10% CO2) at 3,000–10,000 cells per well in 200 µl high-glucose DMEM supplemented with 10% heat-inactivated FBS, penicillin/streptomycin/l-glutamine, 2-mercaptoethanol, recombinant mouse cytokines, IL-33, TSLP, and IL-7 at 10 ng/ml (R&D Systems). At harvest, cell-free culture supernatants were collected, and cells were analyzed by flow cytometry or collected in TRIzol LS (Thermo Fisher Scientific) for RNA extraction. On day 8 of culture, ILC2s were transfected with miRNA mimics and siRNAs (Dharmacon). miRIDIAN miRNA mimics were used at 500 nM per transfection. siGENOME SmartPools and ON-TARGETplus Individual siRNAs were used at 500 nM. Cells were transfected using the Neon Transfection system (Invitrogen) as previously described (Steiner et al., 2011). Cells and culture supernatants were harvested on day 10 for further analysis.
CyQUANT NF cell proliferation assay
CyQUANT NF Cell Proliferation Assay kit (Thermo Fisher Scientific) was used to measure proliferation of 4-d-cultured ILC2s, which were analyzed on a FLUOstar Optima plate reader (BMG Labtech).
Cytokine protein analysis
For ILC2 culture supernatants and BAL, IL-13, IL-5, IL-9, IL-6, and GM-CSF were quantified by cytometric bead array using a LSRII flow cytometer and FCAP Array analysis software (BD Biosciences).
RNA isolation and quantitative RT-PCR
ILC2s, CD4+ T cells, and TH2 cells were sorted directly into TRIzol LS (Thermo Fisher Scientific). Cultured ILC2s, TH2 cells, or gentleMACS-homogenized lung cells were also frozen in TRIzol LS (Thermo Fisher Scientific), and RNA was isolated using the miRNeasy Micro kit (Qiagen). RNA was reverse-transcribed with the SuperScript III kit (Invitrogen). Primers used for SYBR Green PCRs are as follows: Gapdh: 5′-GTGTTCCTACCCCCAATGTGT-3′, 5′-ATTGTCATACCAGGAAATGAGCTT-3′; Il9: 5′-ATGTTGGTGACATACATCCTTGC-3′, 5′-TGACGGTGGATCATCCTTCAG-3′; Il5: 5′-CTCTGTTGACAAGCAATGAGACG-3′, 5′-TCTTCAGTATGTCTAGCCCCTG-3′; Il13: 5′-CCTGGCTCTTGCTTGCCTT-3′, 5′-GGTCTTGTGTGATGTTGCTCA-3′; Csf2: 5′-GTCTCTAACGAGTTCTCCTTC A-3′, 5′-CCTTGAGTTTGGTGAAATTGCC-3′; Il6: 5′-AGCCAGAGTCCTTCAGAGA-3′, 5′-TCCTTAGCCACTCCTTCTGT-3′; Rora: 5′-CTTCTCGGTGGTTCTTCTGT-3′, 5′-TTCTGCATTCGTACTGATGTCA-3′; Socs-1: 5′-ATTCCACTCCTACCTCTCCAT-3′, 5′-CAGAAAAATGAAGCCAGAGACC-3′; Tnfaip3: 5′-AGAACCAGAGATTCCATGAAGC-3′, 5′-CTGTGTAGTTCGAGGCATGT-3′; Kit: 5′-CCATAGGACCAGACATCACTT-3′, 5′-CTAGCCAGAGACATCAGGAATG-3′; Pten: 5′ TAATATACATAGCGCCTCTGACTG-3′, 5′-CGGACTGGTGTAATGATTTGTG-3′; Muc5ac: 5′-CAGGACTCTCTGAAATCGTACCA-3′, 5′-AAGGCTCGTACCACAGGGA-3′; Clca3: 5′-CTGTCTTCCTCTTGATCCTCCA-3′, 5′-CGTGGTCTATGGCGATGACG-3′; and Gob5: 5′-CATCGCCATAGACCACGACG-3′, 5′-TTCCAGCTCTCGGGAATCAAA-3′.
In vivo allergic inflammation model
Mice were anesthetized using isoflurane inhalation and challenged with 35 µg papain (Calbiochem) in 40 µl PBS administered to the lungs by oropharyngeal aspiration daily for three consecutive days. Samples were harvested on day 4. BAL was performed by three consecutive washes of 1 ml complete medium. The number of cells in BAL and lung samples was quantified by FACS analysis with AccuCount Particles (Spherotech). For lung pathology, mice were challenged with 35 µg papain in 40 µl PBS on days 1, 3, and 5, and samples were harvested on day 6. A blinded pathologist scored inflammation in lungs on day 6 using routine hematoxylin and eosin–stained paraffin sections. Both the right and left lungs were examined. Perivascular, peribronchiolar, and interstitial/alveolar inflammation was scored for percentage involvement (0% = 0, 1–25% = 1, 26–50% = 2, 51–75% = 3, and 76–100% = 4). Perivascular and peribronchiolar inflammation was also scored for the mean thickness of the inflammatory cuff (0 cells = 0, 1–3 cells = 1, and ≥4 cells = 2). The values were summed for each sample to determine the inflammation score. One mouse was excluded because of histological evidence of acute bronchopneumonia, and two mice were excluded for a paucity of bronchioles available for evaluation.
Small RNA sequencing
5–50 ng total RNA from sorted and/or cultured ILC2 and T cell samples were used to build small RNA libraries as previously described (Williams et al., 2013). Libraries were sequenced on a HiSeq2500 (Illumina). Data were demultiplexed and adapters trimmed using the FastX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Sequence alignment and small RNA read counts were obtained using the exceRpt small RNA-seq Pipeline (v3; https://www.genboree.org/epigenomeatlas/genboreeWorkbench.rhtml). Comparative expression analysis was performed with DESeq2 (Love et al., 2014). Raw RNA sequencing data were deposited in the GEO database (GSE96934). Three or four biological replicates were collected for each cell type and condition. ILC2 cells were sorted from the lungs of control mice (ILC2) or the lungs of mice killed 1 d after treatment for three consecutive days with 500 ng IL-33 (R&D Systems) instilled by oropharyngeal aspiration (ILC2 IL33). Sorted ILC2s from control mice were also cultured in vitro for 10 d with 10 ng/ml IL-33, TSLP, and IL-7 from R&D Systems (ILC2 in vitro). Naive CD4+ T cells were sorted from pooled spleen and lymph nodes of control mice (CD4). TH2 cells were generated in vitro under polarizing conditions including 500 U/ml IL-4 supernatant and anti-IFNγ (TH2 in vitro) as described (Pua et al., 2016). An in vivo population of TH2 cells was generated by the induction of AA responses to OVA as previously described (Pua et al., 2016), with slight modification: 4get mice were sensitized with three weekly i.p. injections of OVA-alum and challenged with OVA in the lung for three consecutive days by oropharyngeal aspiration, and CD4+GFP+ TH2 cells were then sorted from BAL fluid 1 d after the final challenge (TH2 AA).
For ILC2, ILC2 IL33, CD4, and TH2 AA, 8,000-1,000,000 cells were sorted on either FACSAria II or FACSAria IIIu (BD Biosciences) directly into TRIzol LS (Thermo Fisher Scientific). For sorting, the following gating strategies were used: ILC2, ILC2 IL33, and ILC2 in vitro: CD45+Thy1+lin–CD4–CD25+ST2+; CD4: CD4+CD62LhiCD44loCD25–CD69–; and TH2 AA: CD45+CD11c−CD11b−CD4+CD3+4get+. For ILC2 in vitro, 10,000 sorted cells were seeded into wells, and the entire cell population was lysed in TRIzol LS after 10 d of culture. For TH2 in vitro, 3,000,000 cells were collected from TH2-polarizing cultures at day 5 and lysed with TRIzol LS. Total RNA was isolated using a miRNeasy Micro kit (Qiagen), and RNA was quantified using an RNA Pico kit on the Bioanalyzer (Agilent).
Statistical analysis
Excel (Microsoft) and Prism (Graph Pad Software) were used for data analysis. Unless otherwise specified, p-values were calculated using unpaired Student’s t tests. For comparisons of multiple groups, ANOVA was used with appropriate post hoc testing.
Online supplemental material
Table S1 summarizes mapped reads for small RNA sequencing of ILC2s and T cells. Table S2 details differential expression analyses for all pairwise comparisons of ILC2, ILC2 IL33, ILC2 in vitro, CD4, TH2 AA, and TH2 small RNA sequencing miRNA expression data. Table S3 lists miRNAs in groups of hierarchically clustered rows from differential expression analysis of small RNA sequencing data in ILC2 and T cell samples. Table S4 provides DESeq2 normalized miRNA expression for ILC2 and T cell samples. Tables S1–S4 are provided as zipped Excel files.
Supplementary Material
Acknowledgments
All sequencing was performed at the Center for Advanced Technology at the University of California, San Francisco, and the Vincent J. Coates Genomic Sequencing Laboratory at the University of California, Berkeley. We thank Vinh Nguyen for his assistance in ILC2 sorts.
This work was supported by US National Institutes of Health grants (R01HL109102, P01HL107202, and U19CA179512); a Leukemia and Lymphoma Society Scholar award (K.M. Ansel); the National Multiple Sclerosis Society; the University of California, San Francisco Program for Breakthrough Biomedical Research, funded in part by the Sandler Foundation; and the Emmy Noether Program (BA 5132/1-1) of the Deutsche Forschungsgemeinschaft (D. Baumjohann).
The authors declare no competing financial interests.
Author contributions: P.B. Singh designed, performed, and analyzed most of the experiments; H.H. Pua analyzed RNAseq and histology data; and H.C. Happ helped to perform in vivo experiments. C. Schneider, J. von Moltke, R.M. Locksley, and D. Baumjohann helped design and interpret some experiments. D. Baumjohann and K.M. Ansel initiated the project. K.M. Ansel helped design, analyze and interpret all experiments. P.B. Singh and K.M. Ansel wrote the manuscript. All authors reviewed and approved the manuscript.
Footnotes
Abbreviations used:
- AA
- allergic airway
- BAL
- bronchoalveolar lavage
- ILC2
- type 2 innate lymphoid cell
- miRNA
- microRNA
- rpm
- reads per million
- TSLP
- thymic stromal lymphopoietin
References
- Bartemes K.R., Iijima K., Kobayashi T., Kephart G.M., McKenzie A.N., and Kita H.. 2012. IL-33-responsive Lineage−CD25+CD44hi lymphoid cells mediate innate type-2 immunity and allergic inflammation in the lungs. J. Immunol. 188:1503–1513. 10.4049/jimmunol.1102832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D., and Ansel K.M.. 2013. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat. Rev. Immunol. 13:666–678. 10.1038/nri3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D., Kageyama R., Clingan J.M., Morar M.M., Patel S., de Kouchkovsky D., Bannard O., Bluestone J.A., Matloubian M., Ansel K.M., and Jeker L.T.. 2013. The microRNA cluster miR-17∼92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat. Immunol. 14:840–848. 10.1038/ni.2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu A.M., Bezman N.A., Lee J.E., Matloubian M., Sun J.C., and Lanier L.L.. 2013. MicroRNA function in NK-cell biology. Immunol. Rev. 253:40–52. 10.1111/imr.12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronevetsky Y., Villarino A.V., Eisley C.J., Barbeau R., Barczak A.J., Heinz G.A., Kremmer E., Heissmeyer V., McManus M.T., Erle D.J., et al. . 2013. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J. Exp. Med. 210:417–432. 10.1084/jem.20111717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson C.A., Goplen N.P., Zafar I., Irvin C., Good J.T. Jr., Rollins D.R., Gorentla B., Liu W., Gorska M.M., Chu H., et al. . 2015. Persistence of asthma requires multiple feedback circuits involving type 2 innate lymphoid cells and IL-33. J. Allergy Clin. Immunol. 136:59–68.e14. 10.1016/j.jaci.2014.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtale G., Citarella F., Carissimi C., Goldoni M., Carucci N., Fulci V., Franceschini D., Meloni F., Barnaba V., and Macino G.. 2010. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood. 115:265–273. 10.1182/blood-2009-06-225987 [DOI] [PubMed] [Google Scholar]
- de Kouchkovsky D., Esensten J.H., Rosenthal W.L., Morar M.M., Bluestone J.A., and Jeker L.T.. 2013. microRNA-17-92 regulates IL-10 production by regulatory T cells and control of experimental autoimmune encephalomyelitis. J. Immunol. 191:1594–1605. 10.4049/jimmunol.1203567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande D.A., Dileepan M., Walseth T.F., Subramanian S., and Kannan M.S.. 2015. MicroRNA regulation of airway inflammation and airway smooth muscle function: Relevance to asthma. Drug Dev. Res. 76:286–295. 10.1002/ddr.21267 [DOI] [PubMed] [Google Scholar]
- de Yébenes V.G., Bartolomé-Izquierdo N., and Ramiro A.R.. 2013. Regulation of B-cell development and function by microRNAs. Immunol. Rev. 253:25–39. 10.1111/imr.12046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach A., Colonna M., and Koyasu S.. 2014. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 41:354–365. 10.1016/j.immuni.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M.R., Bartlett N.W., Clarke D., Birrell M., Belvisi M., and Johnston S.L.. 2009. Targeting the NF-kappaB pathway in asthma and chronic obstructive pulmonary disease. Pharmacol. Ther. 121:1–13. 10.1016/j.pharmthera.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehniger T.A., Wylie T., Germino E., Leong J.W., Magrini V.J., Koul S., Keppel C.R., Schneider S.E., Koboldt D.C., Sullivan R.P., et al. . 2010. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res. 20:1590–1604. 10.1101/gr.107995.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantier M.P., Stunden H.J., McCoy C.E., Behlke M.A., Wang D., Kaparakis-Liaskos M., Sarvestani S.T., Yang Y.H., Xu D., Corr S.C., et al. . 2012. A miR-19 regulon that controls NF-κB signaling. Nucleic Acids Res. 40:8048–8058. 10.1093/nar/gks521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon E.D., Simpson L.J., Rios C.L., Ringel L., Lachowicz-Scroggins M.E., Peters M.C., Wesolowska-Anderson A., Gonzalez J.R., MacLeod H.J., Christian L.S., et al. . 2016. Alternative splicing of interleukin-33 and type 2 inflammation in asthma. Proc. Natl. Acad. Sci. USA. 113:8765–8770. 10.1073/pnas.1601914113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.-C., Vidigal J.A., Mu P., Yao E., Singh I., González A.J., Concepcion C.P., Bonetti C., Ogrodowski P., Carver B., et al. . 2015. An allelic series of miR-17 ∼ 92-mutant mice uncovers functional specialization and cooperation among members of a microRNA polycistron. Nat. Genet. 47:766–775. 10.1038/ng.3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S.-D., Lin F.-M., Wu W.-Y., Liang C., Huang W.-C., Chan W.-L., Tsai W.T., Chen G.Z., Lee C.J., Chiu C.M., et al. . 2011. miRTarBase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 39(suppl_1):D163–D169. 10.1093/nar/gkq1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Li C., Olive V., Lykken E., Feng F., Sevilla J., Wan Y., He L., and Li Q.J.. 2011. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 118:5487–5497. 10.1182/blood-2011-05-355644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson K., Malmhäll C., Ramos-Ramírez P., and Rådinger M.. 2017. MicroRNA-155 is a critical regulator of type 2 innate lymphoid cells and IL-33 signaling in experimental models of allergic airway inflammation. J. Allergy Clin. Immunol. 139:1007–1016.e9. 10.1016/j.jaci.2016.06.035 [DOI] [PubMed] [Google Scholar]
- Kamijo S., Takeda H., Tokura T., Suzuki M., Inui K., Hara M., Matsuda H., Matsuda A., Oboki K., Ohno T., et al. . 2013. IL-33-mediated innate response and adaptive immune cells contribute to maximum responses of protease allergen-induced allergic airway inflammation. J. Immunol. 190:4489–4499. 10.4049/jimmunol.1201212 [DOI] [PubMed] [Google Scholar]
- Kang S.G., Liu W.-H., Lu P., Jin H.Y., Lim H.W., Shepherd J., Fremgen D., Verdin E., Oldstone M.B., Qi H., et al. . 2013. MicroRNAs of the miR-17∼92 family are critical regulators of T(FH) differentiation. Nat. Immunol. 14:849–857. 10.1038/ni.2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C.S.N., and Artis D.. 2016. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol. 17:765–774. 10.1038/ni.3489 [DOI] [PubMed] [Google Scholar]
- Kuperman D.A., Huang X., Koth L.L., Chang G.H., Dolganov G.M., Zhu Z., Elias J.A., Sheppard D., and Erle D.J.. 2002. Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat. Med. 8:885–889. 10.1038/nm734 [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T.X., and Rothenberg M.E.. 2013. Diagnostic, functional, and therapeutic roles of microRNA in allergic diseases. J. Allergy Clin. Immunol. 132:3–13, quiz :14. 10.1016/j.jaci.2013.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche H., Weber O., Nageswara Rao T., Blum C., and Fehling H.J.. 2007. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur. J. Immunol. 37:43–53. 10.1002/eji.200636745 [DOI] [PubMed] [Google Scholar]
- Martinez-Gonzalez I., Steer C.A., and Takei F.. 2015. Lung ILC2s link innate and adaptive responses in allergic inflammation. Trends Immunol. 36:189–195. 10.1016/j.it.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Mattes J., Collison A., Plank M., Phipps S., and Foster P.S.. 2009. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc. Natl. Acad. Sci. USA. 106:18704–18709. 10.1073/pnas.0905063106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt M.F., Gut I.G., Demenais F., Strachan D.P., Bouzigon E., Heath S., von Mutius E., Farrall M., Lathrop M., and Cookson W.O.C.M.. GABRIEL Consortium . 2010. A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med. 363:1211–1221. 10.1056/NEJMoa0906312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilyansky E., and Rigoutsos I.. 2013. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 20:1603–1614. 10.1038/cdd.2013.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrs M., Shinkai K., Mohrs K., and Locksley R.M.. 2001. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity. 15:303–311. 10.1016/S1074-7613(01)00186-8 [DOI] [PubMed] [Google Scholar]
- Monticelli S., Ansel K.M., Xiao C., Socci N.D., Krichevsky A.M., Thai T.H., Rajewsky N., Marks D.S., Sander C., Rajewsky K., et al. . 2005. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 6:R71 10.1186/gb-2005-6-8-r71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum J.C., Van Dyken S.J., von Moltke J., Cheng L.E., Mohapatra A., Molofsky A.B., Thornton E.E., Krummel M.F., Chawla A., Liang H.E., and Locksley R.M.. 2013. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 502:245–248. 10.1038/nature12526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye I.S., Czieso S., Ktistaki E., Roderick K., Coomes S.M., Pelly V.S., Kannan Y., Perez-Lloret J., Zhao J.L., Baltimore D., et al. . 2014. Transcriptomics identified a critical role for Th2 cell-intrinsic miR-155 in mediating allergy and antihelminth immunity. Proc. Natl. Acad. Sci. USA. 111:E3081–E3090. 10.1073/pnas.1406322111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pua H.H., and Ansel K.M.. 2015. MicroRNA regulation of allergic inflammation and asthma. Curr. Opin. Immunol. 36:101–108. 10.1016/j.coi.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pua H.H., Steiner D.F., Patel S., Gonzalez J.R., Ortiz-Carpena J.F., Kageyama R., Chiou N.T., Gallman A., de Kouchkovsky D., Jeker L.T., et al. . 2016. MicroRNAs 24 and 27 suppress allergic inflammation and target a network of regulators of T helper 2 cell-associated cytokine production. Immunity. 44:821–832. 10.1016/j.immuni.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P.K., Toyama Y., Chiang H.R., Gupta S., Bauer M., Medvid R., Reinhardt F., Liao R., Krieger M., Jaenisch R., et al. . 2009. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ. Res. 105:585–594. 10.1161/CIRCRESAHA.109.200451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regelin M., Blume J., Pommerencke J., Vakilzadeh R., Witzlau K., Łyszkiewicz M., Ziętara N., Saran N., Schambach A., and Krueger A.. 2015. Responsiveness of developing T cells to IL-7 signals is sustained by miR-17:d. J. Immunol. 195:4832–4840. 10.4049/jimmunol.1402248 [DOI] [PubMed] [Google Scholar]
- Rusca N., Dehò L., Montagner S., Zielinski C.E., Sica A., Sallusto F., and Monticelli S.. 2012. MiR-146a and NF-κB1 regulate mast cell survival and T lymphocyte differentiation. Mol. Cell. Biol. 32:4432–4444. 10.1128/MCB.00824-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon S.T., and McKenzie A.N.J.. 2012. Type 2 innate lymphoid cells: New players in asthma and allergy. Curr. Opin. Immunol. 24:707–712. 10.1016/j.coi.2012.08.009 [DOI] [PubMed] [Google Scholar]
- Simpson L.J., Patel S., Bhakta N.R., Choy D.F., Brightbill H.D., Ren X., Wang Y., Pua H.H., Baumjohann D., Montoya M.M., et al. . 2014. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat. Immunol. 15:1162–1170. 10.1038/ni.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.G., Chen R., Kjarsgaard M., Huang C., Oliveria J.-P., O’Byrne P.M., Gauvreau G.M., Boulet L.P., Lemiere C., Martin J., et al. . 2016. Increased numbers of activated group 2 innate lymphoid cells in the airways of patients with severe asthma and persistent airway eosinophilia. J. Allergy Clin. Immunol. 137:75–86.e8. 10.1016/j.jaci.2015.05.037 [DOI] [PubMed] [Google Scholar]
- Steiner D.F., Thomas M.F., Hu J.K., Yang Z., Babiarz J.E., Allen C.D.C., Matloubian M., Blelloch R., and Ansel K.M.. 2011. MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity. 35:169–181. 10.1016/j.immuni.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai P., Chen Y., Dolganov G., and Wu R.. 2005. Differential regulation of MUC5AC/Muc5ac and hCLCA-1/mGob-5 expression in airway epithelium. Am. J. Respir. Cell Mol. Biol. 33:523–530. 10.1165/rcmb.2004-0220RC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken S.J., Nussbaum J.C., Lee J., Molofsky A.B., Liang H.-E., Pollack J.L., Gate R.E., Haliburton G.E., Ye C.J., Marson A., et al. . 2016. A tissue checkpoint regulates type 2 immunity. Nat. Immunol. 17:1381–1387. 10.1038/ni.3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J., and Locksley R.M.. 2014. I-L-C-2 it: type 2 immunity and group 2 innate lymphoid cells in homeostasis. Curr. Opin. Immunol. 31:58–65. 10.1016/j.coi.2014.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.A., and McKenzie A.N.J.. 2013. Development and function of group 2 innate lymphoid cells. Curr. Opin. Immunol. 25:148–155. 10.1016/j.coi.2013.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams Z., Ben-Dov I.Z., Elias R., Mihailovic A., Brown M., Rosenwaks Z., and Tuschl T.. 2013. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc. Natl. Acad. Sci. USA. 110:4255–4260. 10.1073/pnas.1214046110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.H., Walker J.A., Jolin H.E., Drynan L.F., Hams E., Camelo A., Barlow J.L., Neill D.R., Panova V., Koch U., et al. . 2012. Transcription factor RORα is critical for nuocyte development. Nat. Immunol. 13:229–236. 10.1038/ni.2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Srinivasan L., Calado D.P., Patterson H.C., Zhang B., Wang J., Henderson J.M., Kutok J.L., and Rajewsky K.. 2008. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 9:405–414. 10.1038/ni1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Boldin M.P., Yu Y., Liu C.S., Ea C.-K., Ramakrishnan P., Taganov K.D., Zhao J.L., and Baltimore D.. 2012. miR-146a controls the resolution of T cell responses in mice. J. Exp. Med. 209:1655–1670. 10.1084/jem.20112218 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.