Abstract
We have developed a novel approach for creating membrane-spanning protein-based pores. The construction principle is based on using well-defined, circular DNA nanostructures to arrange a precise number of pore-forming protein toxin monomers. We can thereby obtain, for the first time, protein pores with specifically set diameters. We demonstrate this principle by constructing artificial alpha-hemolysin (αHL) pores. The DNA/αHL hybrid nanopores composed of twelve, twenty or twenty-six monomers show stable insertions into lipid bilayers during electrical recordings, along with steady, pore size-dependent current levels. Our approach successfully advances the applicability of nanopores, in particular towards label-free studies of single molecules in large nanoscaled biological structures.
INTRODUCTION
In recent years, nanopores have become established as versatile tools for a wide range of nanotechnological applications (1–7). In particular, the use of nanopores as ultra-sensitive sensor elements for rapid and label-free analysis of single molecules has garnered considerable interest. However, an important challenge for such nanopore-based sensing that remains is to control the geometrical and chemical properties of the sensor channels. This is the very challenge we have addressed in our work.
The preparation of nanometre-scaled pores can be realized by perforating either (i) a semifluid membrane via membrane-spanning pores, or (ii) a thin solid-state film (e.g. silica, silicon nitrate or graphene) via, for instance, a focused ion or electron beam (8–11). Although solid-state nanopores provide a higher mechanical and chemical stability, and are less sensitive to buffer conditions, the most commonly utilized platforms for biological applications are lipid membrane-inserted protein pores (12). This is largely due to the high reproducibility of the pore structure, as well as the atomically precise positioning of chemical groups through genetic engineering, both of which cannot be achieved so far using solid-state nanopore fabrication methods.
Several natural proteins, with either α-helical- or β-strand-containing transmembrane domains, have been studied for their suitability as sensors in stochastical sensing and sequencing. The identified eligible pores are typically assemblies of identical monomers, which gives the channel a characteristic size and shape. The channel width of these pores is so far constrained to 1–4 nm, which notably limits their application (4,11–17).
Ideally, the pore should be selected according to the size of its central constriction, i.e. wide enough to allow passage of the analyte and yet sufficiently narrow to enable well-resolved analyte-specific current signatures. It has been shown that the same type of nanopore can be utilized for differently sized analytes by, e.g. refining the channel diameter via interior deposition of annular organic molecules (18), or electrophoretic separation of pores with different monomer numbers (19). However, apart from these pioneering studies, the repertoire of available shapes and dimensions for biological pores remains rather restricted. Another concern for protein pores is the limited control over the number and precision of surface functionalities that can be obtained within a homo-oligomeric arrangement.
In order to expand the assortment of fluid membrane-spanning pores, several research groups have focused on synthesis of membrane-spanning channels using peptides, synthetic organic components, or DNA (20–32). Amongst these molecules, DNA is considered the most promising in terms of compensating for the drawbacks of protein pores. That is because the DNA duplex formation is highly programmable via sequence-specific base pairing, which provides full control over the geometry and position of chemical modifications within self-assembled DNA constructs (33–36).
Prior art of DNA nanopores follows the principle of parallel arrangement of crosslinked DNA duplexes in a honeycomb- or square-like pattern. Larger pore constructs are mainly assembled using the scaffolded DNA origami technique (35) whereas smaller constructs are based on the binding of short DNA oligonucleotides. In order to facilitate the membrane insertion of the DNA nanostructures, they are functionalized at well-defined positions with lipophilic molecules (27,32). So far, different DNA constructs with channel diameters of 0.8, 2, 4.2 or 6 nm have been demonstrated (24–27,31).
Despite the enormous potential of DNA for the de novo design of pores, several unresolved issues limit their applications in lipid bilayer-based nanopore sensing. These include undesirable ion permeability, uncontrolled gating due to the DNA-duplex flexibility, poor insertion efficiency especially in planar lipid bilayers, and ill-defined functionality (27–32,37). Additionally, the spatial resolution of chemical modification of DNA nanostructures is inferior to that of proteins, due to the helical nature of the DNA duplex (34).
Here we show that the drawbacks of both protein- and DNA-based nanopores can be largely mitigated by merging them into novel, functional hybrid pores. We demonstrate this principle by templating alpha-hemolysin (αHL) monomers on well-defined DNA nanostructures into artificial pores.
The DNA nanostructures in our study are based on a single-stranded circular DNA molecule whose dimensions match those of the targeted pore. Various binding sites along the DNA template allow for the attachment of multiple α-hemolysin monomers. Using atomic force and electron microscopy, along with electrophoretic mobility assays, we verified the well-defined dimensional scalability of the DNA/protein hybrid nanopores. Electrical recordings in planar lipid bilayers corroborated the stepwise insertion of these constructs via stable and size-dependent current levels. Further, we developed a geometrical model to describe the formation of the transmembrane domain of DNA-templated α-hemolysin pores.
MATERIALS AND METHODS
αHL monomer engineering and synthesis
Introduction of the K237C mutation into the αHL gene and protein expression was done as previously described (38). The vector also encoded a D8H6 tag at the C-terminus that was used for purification by Ni2+-NTA chromatography. The αHL proteins were eluted using a buffer (50 mM Tris, 0.5 M NaCl, 250 mM imidazole, 0.1% v/v Triton X-100, pH 8.0) that was supplemented with 1 mM tris(2-carboxyethyl)phosphine (TCEP), pH 8.0) in order to prevent the formation of disulfide bonds between the proteins. To verify the expression of K237C-αHL mutants, eluates in Laemmli buffer were run at 200 V for 40 min in an ‘Any kD’ precast protein gel in TGS (25 mM Tris–HCl, 192 mM glycine, 0.1% w/v SDS) running buffer. The concentration of the eluates was determined photometrically using a NanoDrop Spectrophotometer (Thermo Scientific).
Preparation of ssDNA-modified αHL monomers
650 μM amine-modified oligonucleotide (the ssDNA tail’) was treated with 12 mM N-ε-maleimidocaproyl-oxysulfosuccinimide ester (Sulfo-EMCS) in a 50 μl final volume (0.1 M phosphate buffer (pH 7.85)) for 45 min at 35°C on a shaker. During the preceding process, monomeric K237C-αHL mutants were separated from αHL heptamers by centrifugation through a 100-kDa MWCO filter device (Amicon, Merck Millipore, USA) for 30 min at 4°C (0.2 M phosphate buffer (pH 6.6)). The reducing agent TCEP was then removed from the eluate containing the 30-kDa αHL monomers using a 10-kDa MWCO filter unit (Amicon, Merck Millipore, USA) per the manufacturer's instructions. To exchange the buffer, the centrifugation was repeated twice with the addition of 200 μl 0.2 M phosphate buffer (pH 6.6). At the same time, excess of Sulfo-EMCS was removed by two-time MicroSpin G-25 column (IllustraGE healthcare Life Science, UK) filtration. The purified Sulfo-EMCS modified oligonucleotide and monomeric K237C-αHL mutants were mixed at a ratio of 10:1 on a shaker for 1.5 h at 30°C, then for 20 min at 25°C. Finally, the proteins were separated from unbound DNA molecules, and the buffer exchanged to a 1× TEM buffer (5 mM Tris, 1 mM EDTA, 12.5 mM Mg(CH3COO)2, pH 8.05), by filtration using a 10-kDa MWCO filter device (Amicon, Merck Millipore, USA). To verify the oligonucleotide-protein conjugation, the sample was mixied at a 1:1 ratio with Laemmli buffer and run for 40 min at 200 V in an ‘Any kD’ precast protein gel in TGS running buffer.
Preparation of circular DNA structures
Circular DNA structures with 12, 20 or 26 binding domains were prepared similarly. The procedure involved the stepwise ligation of shorter DNA oligonucleotides to a closed circular single-stranded DNA, and the subsequent addition of oligonucleotides that hold the complementary sequence to both: one helical turn of the ssDNA circle at its 5′ end and the ssDNA tail’ element. The respective oligonucleotide sequence for the ligation of the single-stranded DNA circle (Outer Circle, Cap and CapComp) and for its transition into its double-stranded form (Inner Circle) are given in Supplementary Table S1.
First, 2.75 μM of each Outer Circle strand was mixed with a 3-fold molar excess of each Cap-L strand in 1 × Quick Ligation Reaction Buffer, incubated for 20 min at room temperature on a shaker before 1 μl of Quick T4 DNA Ligase was added. The mixture was kept on a shaker for 2 h at 25°C. In order to remove the Cap-L strands, 33 μM of the Cap-L complementary strands CapComp were added. The mixture was heated to 80°C for 5 min and then slowly cooled down to room temperature. The reaction mixture was electrophoretically separated on a nondenaturing polyacrylamide gel (120 V, 70 minutes) in 0.5 × Tris–acetate–EDTA (TAE) buffer. SYBR Gold Nucleic Acid Gel Stain was used to stain the gel. The band representing the correctly ligated Outer Circle oligonucleotides was excised and homogenized in 300 μl diffusion buffer (0.5 M NH4C2H3O2, 10 mM Mg(C2H3O2)2). After incubation on a shaker at room temperature overnight, the gel debris was removed by filtration for 30 min at 10 000 g in a Proteus Clarification Mini Spin Column (Generon) and the DNA recovered by ethanol precipitation. The recovered linear single-stranded DNA molecule was circularized by adding 3-fold molar excess of the corresponding Cap-C strand in 1 × Quick Ligation Reaction Buffer. 1 μl Quick T4 DNA Ligase was added after 20 min and the reaction kept at room temperature for 2 h. The Cap-C complementary CapComp oligonucleotide was added, heated to 80°C for 5 min, followed by slowly cooling down of the solution. The ssDNA circle was purified by polyacrylamide gel electrophoresis (PAGE) as previously described. After separation of the gel debris, the corresponding Inner Circle strands were added at a ratio of 1:5 per binding side along the circular DNA scaffold strand in a 1 × TEM buffer. The reaction was incubated on a shaker for 3 h at room temperature and, then, the excess of Inner Circle strands was removed by filtration through a 30-kDa MWCO filter device (Amicon, Merck Millipore, USA) using 1 × TEM buffer.
The DNA nanostructures were analyzed by native gel electrophoresis in a 0.5 × TAE buffer at 120 V for 70 min. Unless otherwise stated, a 10% nondenaturing polyacrylamide gel was used. For verification of complete double-stranded DNA nanostructures, the gel and buffer was supplemented with 6 mM MgCl2.
Conjugation of the circular DNA structure and αHL monomers
The DNA-tail’-modified K237C-αHL mutants were passed through a 100-kDa MWCO filter device, and then mixed with the DNA circles in 1 × TEM buffer at a DNA tail to DNA tail’ ratio of 1:5. The mixture was placed on a shaker at room temperature for 15 min, and then incubated at 4°C overnight. Excess αHL monomers were removed by filtration of the mixture using a 100-kDa MWCO filter device (Amicon, Merck Millipore).
Liposome preparation
1 mg/ml DPhPC solution was dried down from chloroform/methanol (9:1 v/v) under a nitrogen stream in a round glass vial, and kept in a desiccator for 3–4 h. The lipid film was hydrated with 1 × TEM buffer to give a 1 mg/ml total lipid solution. The resulting dispersion was vortexed and then extruded 21 times through two 100 nm polycarbonate filters using a mini-extruder apparatus (Avanti Polar Lipids) to produce large unilamellar DPhPC liposomes.
Transmission electron microscopy (TEM)
DNA/αHL constructs were mixed with DPhPC liposomes at a lipid to protein ratio of 1:1, and the mixture was shaken at room temperature for 15 min and further incubated overnight at 4°C. 5 μl droplet of the solution was pipetted onto a freshly glow-discharged copper/formvar/carbon grid (400 mesh, 3.05 mm diameter; Plano GmbH). After 5 min, the droplet was wicked away and the grid gently washed with double distilled water. Then, 5 μl of 2% (w/v) uranyl acetate was applied to the grid for 5 min and wicked away. Images of the DNA/αHL constructs inserted into the liposomes were acquired on a FEI Talos F200X TEM equipped with a Ceta 16M camera operated at an accelerating voltage of 200 kV. Analysis of the images was done using the Fiji image processing software (39).
Atomic force microscopy (AFM)
The samples were prepared for AFM images on a freshly cleaved mica surface that was pre-treated with 5 mM MgCl2 for 2 min before rinsing with distilled water and drying under a nitrogen stream. 5 μl of the DNA nanostructure sample was applied and the excess solution wicked away after 2 min. The sample was then washed twice with 70 μl distilled water and dried under a nitrogen stream. Finally, the assembled circular DNA structures were imaged on an Agilent 5500 AFM with a Mac Mode III module (Keysight Technologies). AFM images were taken with NSC 15 AFM tips (Mikromasch) in tapping mode in air, and the images were analyzed with WSxM 5.0 software (Nanotec, Inc.) (40). All images were processed prior to analysis by applying simple flattening with offset as subtract type.
Single-channel electrical recording
Electrical recordings were performed on a PLB formed across a 125-μm aperture in a 25-μm-thick Teflon film (Goodfellow) separating two Delrin chambers, at 21.0 ± 2.0°C. The aperture was first treated with 1 μl hexadecane in pentane (1:100, v/v) on each side. After the pentane had evaporated, 0.5 mL buffer solution (25 mM Tris, 50 μM EDTA, pH 7.99, supplemented with 0.1 M KCl unless stated otherwise) was added to each compartment such that the aperture was below the buffer level. A drop of DPhPC in pentane (10 mg/ml) was added on the buffer surface of each chamber. A lipid bilayer was formed across the Teflon aperture by lowering and raising the buffer level of the aperture in each compartment. Ag/AgCl electrodes were used for application of a bias potential (given as Vtrans – Vcis) and electrical current measurements. The electrodes were connected to the headstage of a patch–clamp amplifier (Axopatch 200B, Molecular Devices) operating in voltage-clamp mode. Current signals were low-pass-filtered (80 dB/decade) with a corner frequency of 2 kHz, and sampled at 10 kHz with a Digidata 1440A digitizer (Axon Instruments). After the bilayer was stable for 10 min, 0.5–1 μl of the purified DNA/αHL hybrid structure (typically 10 ng/μl) was added to the cis chamber. A potential of +100 mV (unless otherwise stated) was applied to assist the bilayer insertion and opening of the construct. Data were collected from freshly prepared DNA/protein hybrid structures: three independent assemblies for the [21+]12-DNA/αHL12, [21−]20-DNA/αHL20 and [11−]20-DNA/αHL20 constructs; five independent assembly reactions for the [11+]20-DNA/αHL20 and [21+]20-DNA/αHL20 constructs; four independent assemblies for [21+]26-DNA/αHL26 constructs. In order to block the ion current flow through the pore, 0.5 μl of a 15.6 nM λ-DNA solution was added to either the cis- or trans-side of an icosameric αHL pore that was stably inserted for at least 15 minutes. Data analysis was performed with the pClamp 9.2 software suite (Molecular Devices).
Statistical analysis
Unless otherwise indicated, all presented data is mean ± S.D.
β-barrel modelling
Spatial modelling of the αHL barrel (see Supplementary Section 7) was done using a self-written C++ program. The 3D coordinates of the amino-acid atoms that form the β-barrel (residues 111–126; 132–147) of chain G of the crystal structure (PDB entry: 7ahl) were used as basic variables and read in and projected onto a 2D plane. The tilt was corrected to match the barrel axis and align the two β-strands parallel to one other. This resulting β-strand unit was duplicated according to the number of required units. The relative positions of the atoms within each β-strand unit were fixed, whereas the positions of the atoms of adjacent β-hairpins were offset against each other by 2n amino acids (with 2n ≤ total number of amino acids in the β-strand). Finally, the coordinates were projected onto a 3D tubular structure and exported as a PDB-file. In addition, a log-file was generated summarizing the diameter and height of the modeled β-barrel as obtained from the coordinates. The C++ program can be provided upon request.
RESULTS AND DISCUSSION
Modular construction principles for DNA/αHL hybrid structures
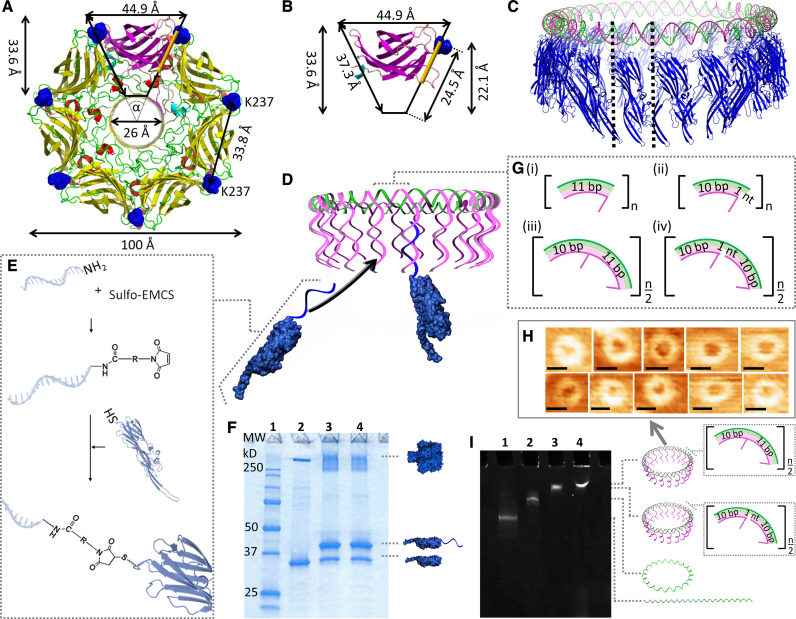
The well-characterized, mushroom-shaped αHL occurs naturally as a homoheptamer (41), and is one of the most commonly studied proteins in nanopore sensing (11,42–46). αHL’s 293-amino acid (aa) long protomer can be regarded in plan view as a trapezoid (Figure 1A and B). The residue K237 at the top of the protomer is separated by 33.8 Å from the same residue in the adjacent protomer. Since this length is very similar to the 34 Å helical pitch of B-DNA (47), we designed a circular DNA template that has a single strand of DNA protruding from every helical turn (Figure 1C and D). The sequence of this strand was optimized to be free of any hairpin structure. Therefore, the strand provides an accessible attachment point for 37.8 kDa K237C-αHL mutants that were chemically cross-linked at C237 to a complementary amide-modified oligonucleotide (Figure 1E and F; Supplementary Figure S1). The three nucleotides of the protruding strand nearest to the circular DNA remained unpaired to provide sufficient flexibility for the monomer arrangement without risking entanglement. To facilitate the circular shape of the DNA nanostructure, its design was based on sequences with a reported intrinsic curvature, i.e. five base pairs (bp)-long A-tracts in each helical turn (48–49).
Figure 1.
Design of the DNA/αHL20 hybrid pore. Top view of (A), the overall structure of the wild-type αHL pore, and (B), the β-sandwich domain of a single protomer. The residue K237 is highlighted in blue. The protein pore can be simplified as a ring of 7 triangles with an angle, α, of ∼51.43°. The positions for possible monomer-monomer interactions within the cap domain in the region are highlighted in orange. (C) Comparison of the dimensions of a 220-bp long DNA ring (green, magenta) and the circular arrangement of 20 αHL monomers (blue). (D) Illustration of the assembly of the DNA/αHL hybrid structure. The DNA ring is composed of a closed single strand (the ‘scaffold strand’, green) that includes segments complementary to the 5′ end of the DNA protruding strands. K237C-αHL mutants that are modified with an oligonucleotide which is complementary to the 3′ end of these DNA strands can be arranged along the resulting DNA structure. (E) Schematic representation of the nucleic acid modification of a single αHL cys-mutant through a chemical crosslinker. (F) SDS-polyacrylamide gel showing unheated samples of unmodified (lane 2) and ssDNA-modified (lanes 3 and 4) K237C-αHL mutants. Lane 1 contains a protein standard; lanes 3 and 4 represent two different reactions under the same conditions. (G) Design of the repetitive sequence segments of the DNA template based on either 11 bp per one helical turn (i,ii), or 21 bp per two helical turns (iii, iv). (H) AFM images of DNA structures based on 21 bp per helical turn. Scale bars = 27 nm. (I) MgCl2-supplemented native PAGE analysis of 210-bp-DNA template assemblies. Lane 1: linear ssDNA strand; lane 2: circularized ssDNA strand; lane 3: dsDNA [21+]20-circle; lane 4: dsDNA [21−]20-circle.
Concept for the circular DNA structure assembly
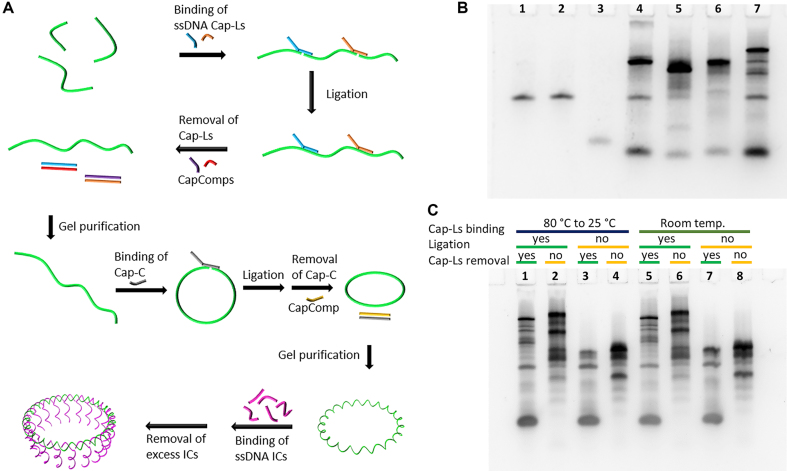
In order to prepare well-defined annular DNA nanostructures (Supplementary Figure S4), we used a design that is based on a circular single-stranded (ss) scaffold (called Outer Circle) with a diameter that matches the targeted DNA/αHL hybrid pore. This Outer Circle comprises appropriate segments in the form of one or two helical turns. The segments guide the arrangement of several shorter DNA oligonucleotides (called Inner Circle) that, in turn, hold each of the complementary sequences to the oligonucleotide-modified αHL monomer. As schematically illustrated in Figure 2A, the preparation of the Outer Circle involves a two-step ligation. The first ligation chemically links shorter oligonucleotides into a long linear single-stranded DNA molecule. This step is necessary due to the oligonucleotide-length limitations of chemical oligonucleotide synthesis. The second ligation joins both ends of the linear DNA molecule into a circle. In detail, the sequences for the Outer Circles for the first ligation are connected through Cap-L oligonucleotides that bind specifically to the two sequence domains at the 5′ and 3′ end of two Outer Circle oligonucleotides. A third sequence domain at the 5′ end of the Cap-L strand remains unbound. After ligation, the Cap-L strands are removed through strand displacement by the addition of their fully complementary oligonucleotide called CapComp. The resulting linear ssDNA strand is PAGE purified. For instance, for a linear twenty segment-long Outer Circle strand, the upper band of the gel was cut out (lane 7 of the PAGE assay in Figure 2B). The connection of the Outer Circle oligonucleotides through Cap sequences can be performed at room temperature. Annealing by heating to 80°C and slowly cooling down to 25°C resulted in a similar yield of the linearized ssDNA template (Figure 2C). The purified Outer Circle strand is closed into a ssDNA circle by adding a Cap-C oligonucleotide with a sequence that is complementary to both ends of the linear ssDNA molecule. Finally, the Cap-C is removed through strand displacement. After PAGE purification, the circular ssDNA template is converted into its double-stranded (ds) form by the addition of the corresponding Inner Circle oligonucleotides (Figure 2A; Supplementary Figure S4B).
Figure 2.
Preparation of the circular DNA template with 20 segment elements. (A) Schematical representation of the two-step ligation of several Outer Circle oligonucleotides (highlighted in green) into a single-stranded DNA circle. The addition of the Inner Circle oligonucleotides (ICs) results in the completion of the dsDNA nanostructure. (B) Native PAGE analysis of the ligation of the Outer Circle oligonucleotides. Lane 1: sequence OC01; lane 2: sequence OC02; lane 3: sequence OC03; lane 4: ligation of OC01 and OC02; lane 5: ligation of OC02 and OC03; lane 6: ligation of OC01 and OC03; lane 7: ligation of OC01, OC02 and OC03. (C) Native PAGE analysis of ligated and non-ligated Outer Circle oligonucleotides under different conditions for the binding of the Cap oligonucleotides. Annealing of the Cap sequences by heating-cooling treatment (lanes 1–4) or at room temperature (lanes 5–8). Resulting products were ligated for samples in lanes 1, 2, 5, 6. Caps were removed for lanes 1, 3, 5, 7.
Formation of a homo-oligomeric αHL icosamer
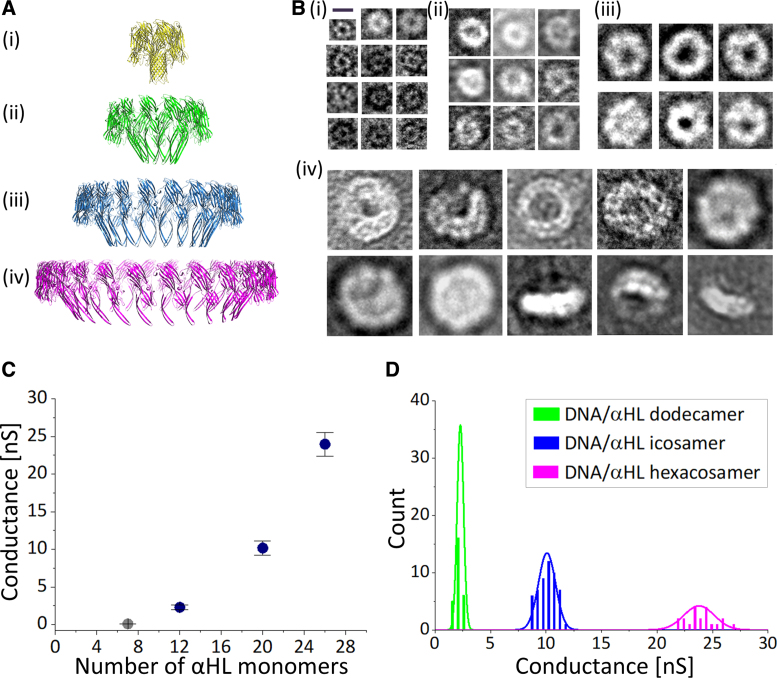
First we focused on the arrangement of twenty αHL monomers. Since one helical turn in B-DNA consists of 10.5 bp, we examined four different sequence designs for their suitability to align the protruding strands in the same direction. These were either repetitive segments of 11 bp per one helical turn, or 21 bp per two helical turns, with or without an unpaired nucleotide (nt) to compensate for possible tension (Figure 1G). In other words, the 21-bp long segments comprise two protruding strands each, while the 11bp-long segments have one protruding strand each. Hence, the total number of segments (n) for a DNA structure based on 21-bp segments is half of that for one based on 11-bp segments to yield a DNA template with the same number of protruding strands. We denoted the DNA structures as [bi]j, where b represents the number of bp per segment, i the presence (+) or absence (–) of the unpaired nt, and j the number of protruding strands. The stepwise assembled DNA structures (Figure 1I; Supplementary Figure S2B) were of an annular shape with—taking into account the AFM-tip size—a diameter comparable to the theoretical value of ∼23 nm (Figure 1H; Supplementary Figures S2A and 3). In contrast, adding DNA-modified αHL monomers resulted in better-defined circular structures with a 2-fold increase in height compared to the DNA structures alone (Supplementary Figure S7). Furthermore, transmission electron microscopy (TEM) images of the constructs in the presence of liposomes showed ring-shaped particles that were predominantly bound to lipid bilayer membranes. In the case of [11+]20- and [21+]20-DNA structures, the diameters were 19.81 ± 1.27 nm (n = 13; Supplementary Figure S5) and 19.54 ± 1.45 nm (n = 96; Figure 3B(iii)), respectively. K237C-αHL mutants in the absence of the DNA template had a diameter typical for αHL heptamers of 10.11 ± 1.08 nm (n = 121; Figure 3B(i)).
Figure 3.
Different stoichiometries of the DNA/αHL hybrid pores investigated in this study. (A) Comparison of the dimensions of a (i) heptameric, (ii) dodecameric, (iii) icosameric and (iv) hexacosameric αHL pore based on a monomer width of 33.8 Å. (B) Example TEM micrographs of liposomes-adhered DNA-modified K237C-αHL mutants in (i) the absence of a DNA structure, or assembled along a circular DNA with (ii) 12, (iii) 20, and (iv) 26, sequence segments. Scale bar in (i) is 10 nm and applies to all images. (C) Conductance dependence on pore stoichiometry of DNA-modified K237C-αHL mutants. (D) Conductance histogram for the different DNA/αHL hybrid pores.
Single-channel characterization of αHL icosamer in planar lipid bilayers (PLBs)
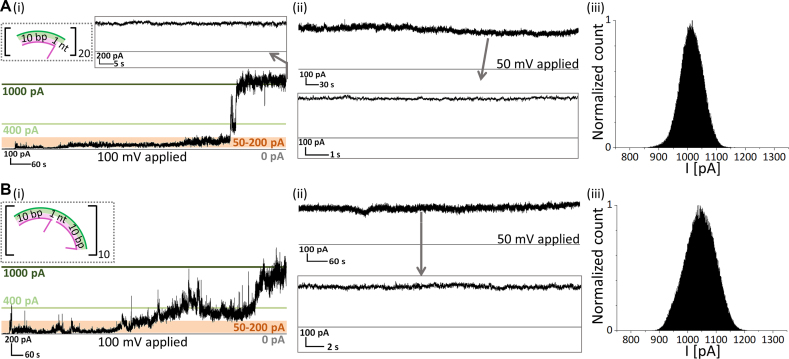
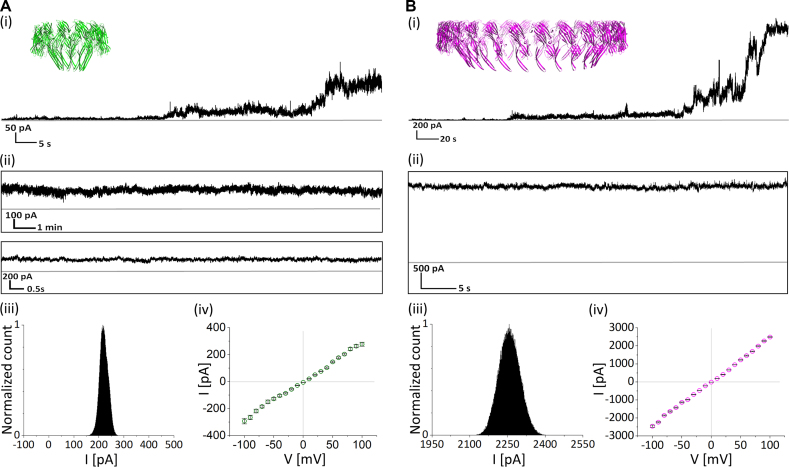
We investigated the functionality of the four different icosameric hybrid constructs using single-channel recordings in 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC)-based planar lipid bilayers (PLBs). The purified structures were added to the cis-side of the setup in a buffer solution containing 0.1 M KCl. While applying positive voltage, the unpaired-nt containing icosameric constructs inserted into the lipid bilayer with a steady increase in the transmembrane current, starting with conductance fluctuations between 0.5 and 2 nS, followed by a first sharp increase to a conductance level of ∼4 nS and a subsequent second jump to ∼10 nS (Figure 4A(i) and B(i); Supplementary Figure S10). Eventually, the accompanying fluctuations decreased to ∼0.2 nS resulting in an open pore that was stable for at least two hours (Figure 4A(ii) and B(ii)). We define the four found states as pre-pore, intermediate pore, pre-open pore and open pore, respectively. The open pore was not subject to any closing step and was also unaffected by the salt concentration (Supplementary Figure S12).
Figure 4.
Electrical recordings of DNA-templated αHL icosamers. (A, i), Stepwise increase in ionic current during the insertion of one [11+]20-based hybrid pore. (ii) Single channel recording after insertion at 50 mV. (iii) All-point histogram of a 1-min-long current trace of an open pore at 100 mV. (B) Insertion trace (i) and single channel recordings at 50 mV (ii) and all-point histogram of a 1-min-long current trace at 100 mV (iii) of an [21+]j-based icosamer.
The open pore state of [11+]20- and [21+]20-DNA/αHL hybrid structures was characterized by conductances of 9.98 ± 1.36 nS (n = 43) and 10.16 ± 0.94 nS (n = 53), respectively, as well as an ohmic I–V response (Figure 5C, blue circles; Supplementary Figure S11). The latter indicates that the DNA template does not influence the conductance for positive and negative potentials, although it may contribute to increased noise (Supplementary Table S4; Figure 4A(iii) and B(iii)).
Figure 5.
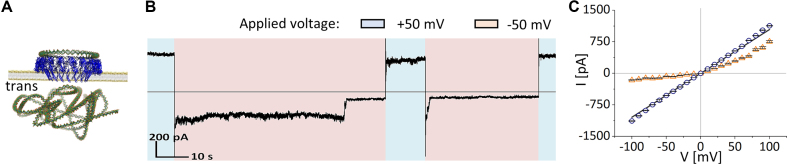
Blockage of icosameric αHL pore. (A) λ-DNA molecules were added to the trans-side after a purified DNA/αHL hybrid structure was incorporated into a DPhPC bilayer. (B) Electrical recording of a [21+]20-icosameric hybrid pore that is stochastically blocked by a λ-DNA molecule at applied negative potential (–50 mV; shaded in red) and opened at applied positive potential (50 mV; shaded in blue). (C) Current-voltage dependence of an unblocked pore (blue circles) versus a blocked pore (orange triangles).
In contrast, the insertion of nt-free hybrid structures involved only the pre-pore and intermediate pore states. In detail, single channel recordings showed that addition of [11−]20-DNA/(αHL)20 hybrid structures resulted in a rupture of the lipid bilayer upon opening of the pore (Supplementary Figure S8A). Structures with a [21−]20-DNA template were stable for several minutes as an intermediate pore (G = 4.48 ± 0.52 nS (n = 13 experiments)) (Supplementary Figure S8B). The rare opening of these intermediate pores also led to a subsequent collapse of the lipid bilayer at 9.98 ± 0.84 nS (n = 3). For DNA templates based on 11 bp/turn, a continued 17.14°-displacement of each ssDNA tail results in a hybrid construct where the αHL protomers are pointing in different directions along the 360° rotational axis of the DNA circle (Figure 6A). The most likely explanation is that the [11−]20-DNA/(αHL)20 hybrid structures wrap around the lipids, which induces a rupture of the lipid bilayer. αHL protomers along DNA templates based on 21 bp/2 turns alternate between the direction of the first αHL protomer (0°) and an offset of 17.14° (Figure 6C). These findings emphasize that nt-additions to the DNA segment design are needed to compensate for rotational deviations and ensure functionality of the hybrid construct. This is regardless of whether there are 10 or 11 bp between the ssDNA tails (Figure 6B and D).
Figure 6.
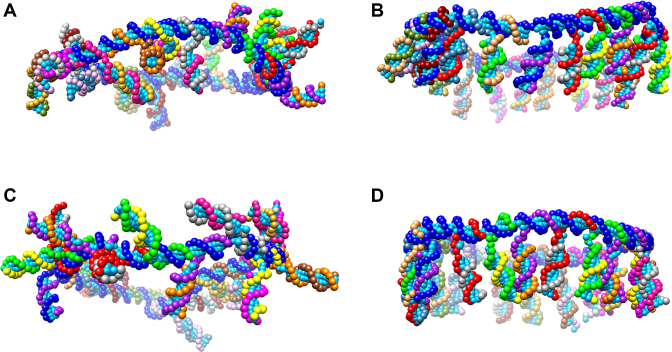
Coarse-grained DNA model of (A) [11−]20, (B) [11+]20, (C) [21−]20 and (D) [21+]20, DNA templates. Self-assembly of the oligonucleotides was simulated using the OxDNA2 software (50).
Verification of the pore formation through blockage
We further verified the pore formation by blocking the icosameric pores after stable insertion with lambda DNA (λ-DNA). λ-DNA has a radius of gyration of ∼530 nm, i.e. 27-fold larger than the outer diameter of the icosamer and, therefore, will block the pore upon its attempt to pass through. We observed complete blockage for [11+]20- and [21+]20-DNA/αHL hybrid structures when λ-DNA was added to the cis- or trans-side of the setup (Figure 5; Supplementary Figures S13–S15). The blockage occurred in a stepwise manner, reflecting the increasing compactness of the λ-DNA molecule as it attempted to pass through the pore. Reversing the direction of the applied potential resulted in the release of the λ-DNA molecule from the pore in a stepwise manner similar to the blockage. The process of closing and opening of the open pore could be repeated multiple times, illustrating the high stability of the channel once the pore is completely inserted into the bilayer.
Functional DNA/αHL hybrid structures of different diameters
Next we examined the feasibility of our approach to template different numbers of αHL monomers by assembling dodecameric and hexacosameric pores (Figure 3A) on appropriate [21+]j-based DNA structures. As observed for the icosamer, these constructs appeared as annular structures on TEM images (Figure 3B(ii), (iv); Supplementary Figure S6). The measured diameters for the dodecamer and hexacosamer were 13.70 ± 1.50 nm (n = 164) and 28.42 ± 1.88 nm (n = 371), respectively, which are similar to the theoretical values of 13.64 and 29.55 nm, respectively. The last three TEM micrographs in Figure 3B(iv) show the side view of the hexacosamer found outside the liposomes. These structures have a height of ∼10 nm and feature a graded circular shape that resembles the heptameric αHL mushroom-shape.
Single-channel recordings in PLBs revealed that the dodecamer and hexacosamer exhibited similar transitions upon pore formation, stability and I-V characteristic response as observed for the icosamer under the same conditions (Figure 7). An open-pore conductance of 2.28 ± 0.29 nS (n = 27 experiments) and 23.96 ± 1.57 nS (n = 21 experiments) was found for the dodacemeric and hexacosameric constructs, respectively (Figure 3C and D).
Figure 7.
Electrical recordings of DNA-templated αHL dodecamer and hexacosamer. Typical ionic current recorded for (A) a DNA/αHL dodecamer, and (B) a DNA/αHL hexacosamer, showing their (i) insertion, (ii) stability, (iii) all-point histogram of a 1-min-long current trace of an open pore at 100 mV, and (iv) current-voltage dependence.
Theoretical validation of the pore diameter and conductance
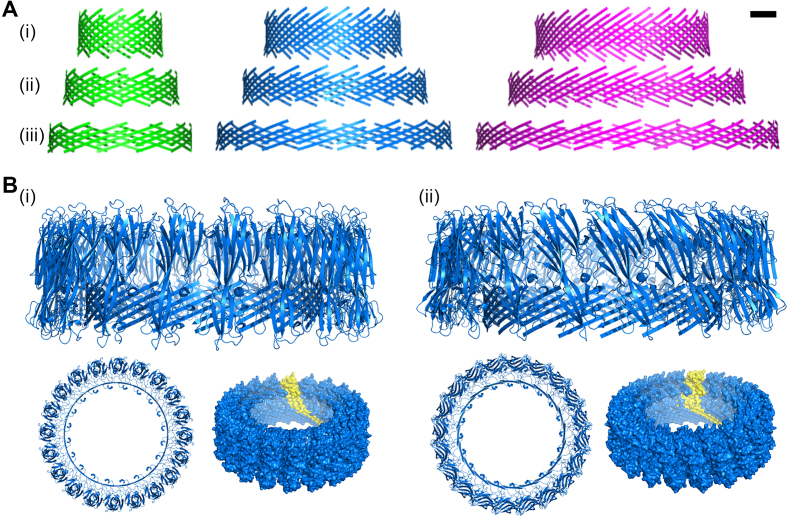
Theoretically, the extension of αHL pores with monomers involves the angle between adjacent protomers broadening from 128.57° (αHL heptamer) towards 180° (Supplementary Figure S24a). This would enlarge the β-hairpin distance to at most 31.6 Å. Hypothetically, this gap can be compensated for by the horizontal alignment of the 65-Å long β-strands (41). To test for possible β-barrel formations, we developed a geometrical model that is based on equal offsets of an even number of amino acids (aa's) between adjacent β-hairpins (Supplementary Section 7). As can be seen from the generated PDB-models presented in Figure 8A (as well as the respective values in Supplementary Table S5), an offset of 4 aa's for the dodecamer, 6 aa's for the icosamer, and 8 aa's for the hexacosamer, resulted in (i) pores with outer diameters similar to the experimentally measured ones, and (ii) β-barrel diameters of 6.53, 14.95 and 25.02 nm, respectively. Furthermore, the calculated conductances for the heptameric (0.18 nS), dodecameric (2.24 nS), icosameric (10.28 nS), and hexacosameric (22.05 nS) pores fit the experimental findings. Although the experimental data implies a quadratic relationship between the conductance and the number of monomers (Figure 3C), a comparison with the theoretical values indicates that this dependence instead scales stepwise. That is due to the naturally occurring offset adjustment following the increasing β-hairpin distance (Supplementary Figure S23). Our model indicates that the upper distance limit can be overcome by a 10-aa offset. This is accompanied by a reduction of β-barrel height to ∼1.43 nm, which is less than the DPhPC bilayer thickness (2.55 nm) (51). In general, this is the case for pores that are based on an offset of 6−14 residues (Figure 8A and B). However, our observation in PLB recordings indicates stable insertions for the hexacosamer with a β-barrel length of 1.75 nm.
Figure 8.
Predicted structure of the DNA/αHL pore. (A) Modeled structure that represents the β-sheet forming residues of the transmembrane domain of a dodecameric (green), icosameric (blue) and hexacosameric (magenta) hybrid pore with an offset of (i) 4, (ii) 6, and (iii) 8 residues, respectively. Scale bar = 2 nm. (B) Possible structure formation of 20 αHL monomers with (i) an adjusted orientation of the protein cap domain to match the β-hairpin tilt for a 6-aa offset, and (ii) same alignment of the cap domain as for the natural occurring heptameric pore. Shown are the side view (top) and top view (bottom left) of the cartoon representation, and the surface representation with one monomer highlighted in yellow (bottom right).
Comparison of wild-type and DNA-templated αHL pore characteristics
The wild-type αHL pore is typically characterized during single-channel recordings by a one-step insertion and a stable open pore conductance of about 0.09 nS. Similarly, the formation of the ssDNA-modified K237C αHL heptameric pore involves only one step (Supplementary Figure S9A(ii)). The stable conductance of 0.07 nS is reduced, and accompanied by a higher noise, compared to the wild-type αHL pore (Supplementary Table S4). Arranging the ssDNA-modified K237C αHL monomers along a DNA template likewise results in highly stable open pores, but compared to the wild-type αHL pore, the traces of the investigated DNA/αHL hybrid pores are characterized by a stepwise insertion and a higher noise. This is likely due to the presence of a flexible, highly negatively charged DNA template, and the fact that the αHL monomeric structure is optimized for heptameric oligomerization. For instance, possible missing linkages between the αHL cap domains (see Supplementary Section 8) might result in the destabilization of the hybrid pore, which is accompanied by a fluctuation of the electrical current. We anticipate that engineering the αHL cap regions to increase the interactions between the protomer cap domains would lower the noise. Furthermore, disconnection of the αHL pore from the DNA nanostructure after its complete insertion could potentially reduce the level of noise as well. We plan to investigate both options in follow-up studies.
The formation of a membrane-spanning αHL pore requires the initial oligomerization of the monomers (Supplementary Section 9). Wild-type αHL monomers typically self-assemble along a lipid membrane. Single-channel recordings of ssDNA-modified K237C αHL monomers indicate that the duration of the oligomerization depends on the monomer concentration (Supplementary Figure S9A). In other words, the higher the number of available monomers along a membrane, the higher the likelihood that the monomers come into contact. For the DNA/αHL hybrid pores, in contrast, the monomers are already in close proximity. This in turn implies that the templated DNA pore will have a higher incorporation yield than the wild-type pore, especially for lower αHL concentrations. This reasoning is supported by our finding that, at the same protein concentration used to investigate the DNA/αHL hybrid pores, hardly any pore formation was observed in the absence of a DNA template (Supplementary Figure S9A(i)).
CONCLUSIONS AND PROSPECTS
In summary, we have developed a new approach for assembling functional nanostructures that are composed of both natural polypeptides and artificial DNA domains. To our knowledge, this is the first demonstration that more than seven αHL monomers can form a functional pore. The porous αHL/DNA structures offer greater versatility in terms of a potential heteromeric composition along with a high precision for single point modifications. This will enable single molecule experiments of precisely located, multiple reaction partners within the pore. We envision utilizing these pores in a variety of contexts, such as size-dependent sorting of self-assembled nanostructures, and controlled release of therapeutics in drug delivery systems. Furthermore, our approach will facilitate the oligomerization of different pore-forming proteins and their mutants into heteromeric constructs with specific numbers of monomers. This will provide detailed biophysical insight into the mechanism of membrane-insertion and oligomerization of pore-forming proteins, and allow for the construction of various novel hybrid pores with defined properties. In addition, the design principles for DNA nanostructures explicitly defined in this work can be applied to arrange diverse monomers into functional proteins, which can potentially be used to construct artificial proteins. Finally, this approach will likely lead to innovative applications of biological nanopores, such as protein sequencing, and single molecule analysis of structures larger than ∼4 nm (52), e.g. the in vitro and in vivo study of amyloid formation and assemblies.
Supplementary Material
ACKNOWLEDGEMENTS
The AFM and TEM imaging was carried out using the Core Technology Platform resources at NYU Abu Dhabi. A.H.-K. acknowledges Hagan Bayley and his group (University of Oxford) for discussion and collaboration during 2013–2015. In particular, A.H.-K. thanks Ofer I. Wilner for assistance with protein expression. The authors also thank Justin Blau and Claude Desplan (New York University) for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
New York University Abu Dhabi Research Enhancement Fund award [RE082]; Al Jalila Foundation Research Center seed grant [AJF201624]. Funding for open access charge: Al Jalila Foundation Research Center seed grant [AJF201624].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bell N.A.W., Keyser U.F.. Digitally encoded DNA nanostructures for multiplexed, single-molecule protein sensing with nanopores. Nat. Nanotechnol. 2016; 11:645–651. [DOI] [PubMed] [Google Scholar]
- 2. Quick J., Loman N.J., Duraffour S., Simpson J.T., Severi E., Cowley L., Bore J.A., Koundouno R., Dudas G., Mikhail A. et al. . Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016; 530:228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Litvinchuk S., Tanaka H., Miyatake T., Pasini D., Tanaka T., Bollot G., Mareda J., Matile S.. Synthetic pores with reactive signal amplifiers as artificial tongues. Nat. Mater. 2007; 6:576–580. [DOI] [PubMed] [Google Scholar]
- 4. Manrao E.A., Derrington I.M., Laszlo A.H., Langford K.W., Hopper M.K., Gillgren N., Pavlenok M., Niederweis M., Gundlach J.H.. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 2012; 30:349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banghart M., Borges K., Isacoff E., Trauner D., Kramer R.H.. Light-activated ion channels for remote control of neuronal firing. Nat. Neurosci. 2004; 7:1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rodriguez-Larrea D., Bayley H.. Multistep protein unfolding during nanopore translocation. Nat. Nanotechnol. 2013; 8:288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kong J., Bell N.A.W., Keyser U.F.. Quantifying nanomolar protein concentrations using designed DNA carriers and solid-state nanopores. Nano Lett. 2016; 16:3557–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dekker C. Solid-state nanopores. Nat. Nanotechnol. 2007; 2:209–215. [DOI] [PubMed] [Google Scholar]
- 9. Li J., Stein D., McMullan C., Branton D., Aziz M.J., Golovchenko J.A.. Ion-beam sculpting at nanometre length scales. Nature. 2011; 412:166–169. [DOI] [PubMed] [Google Scholar]
- 10. Bayley H., Cremer P.S.. Stochastic sensors inspired by biology. Nature. 2001; 413:226–230. [DOI] [PubMed] [Google Scholar]
- 11. Kasianowicz J.J., Brandin E., Branton D., Deamer D.W.. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:13770–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Majd S., Yusko E.C., Billeh Y.N., Macrae M.X., Yang J., Mayer M.. Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Curr. Opin. Biotechnol. 2010; 21:439–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fahie M.A., Yang B., Mullis M., Holden M.A., Chen M.. Selective detection of protein homologues in serum using an OmpG nanopore. Anal. Chem. 2015; 87:11143–11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pastoriza-Gallego M., Rabah L., Gibrat G., Thiebot B., Gisou van der Goot F., Auvray L., Betton J.-M., Pelta J.. Dynamics of unfolded protein transport through an aerolysin pore. J. Am. Chem. Soc. 2011; 133:2923–2931. [DOI] [PubMed] [Google Scholar]
- 15. Wang S., Haque F., Rychahou P.G., Evers B.M., Guo P.. Engineered nanopore of Phi29 DNA-packaging motor for real-time detection of single colon cancer specific antibody in serum. ACS Nano. 2013; 7:9814–9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van Meervelt V., Soskine M., Maglia G.. Detection of two isomeric binding configurations in a protein–aptamer complex with a biological nanopore. ACS Nano. 2014; 8:12826–12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carter J.-M., Hussain S.. Robust long-read native DNA sequencing using the ONT CsgG Nanopore system. Wellcome Open Res. 2017; 2:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gu L.-Q., Cheley S., Bayley H.. Capture of a single molecule in a nanocavity. Science. 2001; 291:636–640. [DOI] [PubMed] [Google Scholar]
- 19. Soskine M., Biesemans A., De Maeyer M., Maglia G.. Tuning the size and properties of ClyA nanopores assisted by directed evolution. J. Am. Chem. Soc. 2013; 135:13456–13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Montenegro J., Ghadiri M.R., Granja J.R.. Ion channel models based on self-assembling cyclic peptide nanotubes. Acc. Chem. Res. 2013; 46:2955–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thomson A.R., Wood C.W., Burton A.J., Bartlett G.J., Sessions R.B., Brady R.L., Woolfson D.N.. Computational design of water-soluble α-helical barrels. Science. 2014; 346:485–488. [DOI] [PubMed] [Google Scholar]
- 22. Sakai N., Mareda J., Matile S.. Artificial β-Barrels. Acc. Chem. Res. 2008; 41:1354–1365. [DOI] [PubMed] [Google Scholar]
- 23. Geng J., Kim K., Zhang J., Escalada A., Tunuguntla R., Comolli L.R., Allen F.I., Shnyrova A.V., Cho K.R., Munoz D. et al. . Stochastic transport through carbon nanotubes in lipid bilayers and live cell membranes. Nature. 2014; 514:612–615. [DOI] [PubMed] [Google Scholar]
- 24. Göpfrich K., Zettl T., Meijering A.E.C., Hernández-Ainsa S., Kocabey S., Liedl T., Keyser U.F.. DNA-tile structures induce ionic currents through lipid membranes. Nano Lett. 2015; 15:3134–3138. [DOI] [PubMed] [Google Scholar]
- 25. Göpfrich K., Li C.-Y., Ricci M., Bhamidimarri S.P., Yoo J., Gyenes B., Ohmann A., Winterhalter M., Aksimentiev A., Keyser U.F.. Large-conductance transmembrane porin made from DNA origami. ACS Nano. 2016; 10:8207–8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krishnan S., Ziegler D., Arnaut V., Martin T.G., Kapsner K., Henneberg K., Bausch A.R., Dietz H., Simmel F.C.. Molecular transport through large-diameter DNA nanopores. Nat. Commun. 2016; 7:12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Langecker M., Arnaut V., Martin T.G., List J., Renner S., Mayer M., Dietz H., Simmel F.C.. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science. 2012; 338:932–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Plesa C., Ananth A.N., Linko V., Gülcher C., Katan A.J., Dietz H., Dekker C.. Ionic permeability and mechanical properties of DNA origami nanoplates on solid-state nanopores. ACS Nano. 2014; 8:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li C.-Y., Hemmig E.A., Kong J., Yoo J., Hernández-Ainsa S., Keyser U.F., Aksimentiev A.. Ionic conductivity, structural deformation, and programmable anisotropy of DNA origami in electric field. ACS Nano. 2015; 9:1420–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seifert A., Göpfrich K., Burns J.R., Fertig N., Keyser U.F., Howorka S.. Bilayer-spanning DNA nanopores with voltage-switching between open and closed state. ACS Nano. 2014; 9:1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burns J.R., Stulz E., Howorka S.. Self-assembled DNA nanopores that span lipid bilayers. Nano Lett. 2013; 13:2351–2356. [DOI] [PubMed] [Google Scholar]
- 32. Burns J.R., Göpfrich K., Wood J.W., Thacker V.V., Stulz E., Keyser U.F., Howorka S.. Lipid-bilayer-spanning DNA nanopores with a bifunctional porphyrin anchor. Angew. Chem. Int. Ed. 2013; 52:12069–12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang F., Jiang S., Wu S., Li Y., Mao C., Liu Y., Yan H.. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat. Nanotechnol. 2015; 10:779–784. [DOI] [PubMed] [Google Scholar]
- 34. Rothemund P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature. 2006; 440:297–302. [DOI] [PubMed] [Google Scholar]
- 35. Douglas S.M., Dietz H., Liedl T., Högberg B., Graf F., Shih W.M.. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009; 459:414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saccà B., Meyer R., Erkelenz M., Kiko K., Arndt A., Schroeder H., Rabe K.S., Niemeyer C.M.. Orthogonal protein decoration of DNA origami. Angew. Chem. Int. Ed. 2010; 49:9378–9383. [DOI] [PubMed] [Google Scholar]
- 37. Yoo J., Aksimentiev A.. Molecular dynamics of membrane-spanning DNA channels: conductance mechanism, electro-osmotic transport, and mechanical gating. J. Phys. Chem. Lett. 2015; 6:4680–4687. [DOI] [PubMed] [Google Scholar]
- 38. Mantri S., Sapra T., Cheley S., Sharp T.H., Bayley H.. An engineered dimeric protein pore that spans adjacent lipid bilayers. Nat. Commun. 2013; 4:1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. . Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Horcas I., Fernandez R., Gomez-Rodriguez J.M., Colchero J., Gomez-Herrero J., Baro A.M.. WSxM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007; 78:013705. [DOI] [PubMed] [Google Scholar]
- 41. Song L.Z., Hobaugh M.R., Shustak C., Cheley S., Bayley H., Gouaux J.E.. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996; 274:1859–1866. [DOI] [PubMed] [Google Scholar]
- 42. Braha O., Gu L.-Q., Zhou L., Lu X., Cheley S., Bayley H.. Simultaneous stochastic sensing of divalent metal ions. Nat. Biotechnol. 2000; 18:1005–1007. [DOI] [PubMed] [Google Scholar]
- 43. Clarke J., Wu H.-C., Jayasinghe L., Patel A., Reid A., Bayley H.. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 2009; 4:265–270. [DOI] [PubMed] [Google Scholar]
- 44. Gu L.-Q., Braha O., Conlan S., Cheley S., Bayley H.. Stochastic sensing of organic analytes by a pore-forming protein containing a molecular adapter. Nature. 1999; 398:686–690. [DOI] [PubMed] [Google Scholar]
- 45. Harrington L., Alexander L.T., Knapp S., Bayley H.. Pim kinase inhibitors evaluated with a single-molecule engineered nanopore sensor. Angew. Chem. Int. Ed. 2015; 54:8154–8159. [DOI] [PubMed] [Google Scholar]
- 46. Rosen C.B., Rodriguez-Larrea D., Bayley H.. Single-molecule site-specific detection of protein phosphorylation. Nat. Biotechnol. 2014; 5:807–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Watson J.D., Crick F.H.. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953; 171:737–738. [DOI] [PubMed] [Google Scholar]
- 48. Dlakic M., Park K., Griffith J.D., Harvey S.C., Harrington R.E.. The organic crystallizing agent 2-methyl-2,4-pentanediol reduces DNA curvature by means of structural changes in A-tracts. J. Biol. Chem. 1996; 271:17911–17919. [DOI] [PubMed] [Google Scholar]
- 49. Han W., Dlakic M., Zhu Y.J., Lindsay S.M., Harrington R.E.. Strained DNA is kinked by low concentrations of Zn2+. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:10565–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Šulc P., Romano F., Ouldridge T.E., Rovigatti L., Doye J.P.K., Louis A.A.. Sequence-dependent thermodynamics of a coarse-grained DNA model. J. Chem. Phys. 2012; 137:135101. [DOI] [PubMed] [Google Scholar]
- 51. Eddy M.T., Ong T.C., Clark L., Teijido O., van der Wel P.A.C., Garces R., Wagner G., Rostovtseva T., Griffin R.G.. Lipid Dynamics and Protein-Lipid Interactions in 2D Crystals Formed with the b-barrel Integral Membrane Protein VDAC1. J. Am. Chem. Soc. 2012; 134:6375–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Soskine M., Biesemans A., Maglia G.. Single-molecule analyte recognition with ClyA nanopores equipped with internal protein adaptors. J. Am. Chem. Soc. 2015; 137:5793–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.