Abstract
Most cells divide symmetrically into two approximately identical cells. There are many examples, however, of asymmetric cell division that can generate sibling cell size differences. Whereas physical asymmetric division mechanisms and cell fate consequences have been investigated, the specific problem caused by asymmetric division at the transcription level has not yet been addressed. In symmetrically dividing cells the nascent transcription rate increases in parallel to cell volume to compensate it by keeping the actual mRNA synthesis rate constant. This cannot apply to the yeast Saccharomyces cerevisiae, where this mechanism would provoke a never-ending increasing mRNA synthesis rate in smaller daughter cells. We show here that, contrarily to other eukaryotes with symmetric division, budding yeast keeps the nascent transcription rates of its RNA polymerases constant and increases mRNA stability. This control on RNA pol II-dependent transcription rate is obtained by controlling the cellular concentration of this enzyme.
INTRODUCTION
During exponential growth, total cell mass and volume increase exponentially. This increase should be compensated by an equivalent increase in the number of molecules (RNA or proteins) to maintain ribostasis and proteostasis (1–3). At the single cell level, the simplest consequence of ribostasis for mRNA levels would be the mRNA concentration ([mRNA]) remaining constant with cell volume. However, compensatory differences in mRNA synthesis and degradation rates [mRNA] between cells of different sizes could exist. Recently, a study on mRNA levels and transcription rates in different sized cells was done in mammalian fibroblasts (4). These authors found that those cells adapted RNA polymerase II (RNA pol II) nascent transcription rates (nTRII) to cell volume in order to keep [mRNA] ribostasis at the single cell level. A previous study conducted with exponential growing populations of Schizosaccharomyces pombe yeast cell size mutants (5) obtained a similar result: nTRII increases linearly with the population average cell volume. In both studies, the measured parameter was nTR, which conceptually differs from the actual mRNA synthesis rate (SRII) (see (6) for a detailed discussion). nTRII evaluates the number of transcribed mRNA molecules, but the chemical equilibrium between mRNA synthesis and degradation uses [mRNA] instead of number of molecules per cell. Accordingly, the SRII is the right parameter to be considered when dealing with equilibrium and ribostasis:
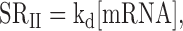 |
where kd is the mRNA first-order degradation constant that allows the mRNA half-life (HL) to be calculated. SRII can be approximated by dividing nTRII per cell volume. Thus, a reinterpretation of both studies (4,5) can conclude that SRII is kept constant because nTRII scales in parallel with cell volume. A corollary of those results is that as SRII and [mRNA] are independent of cell volume, kd, and therefore the HL, are also invariable (Figure 1A).
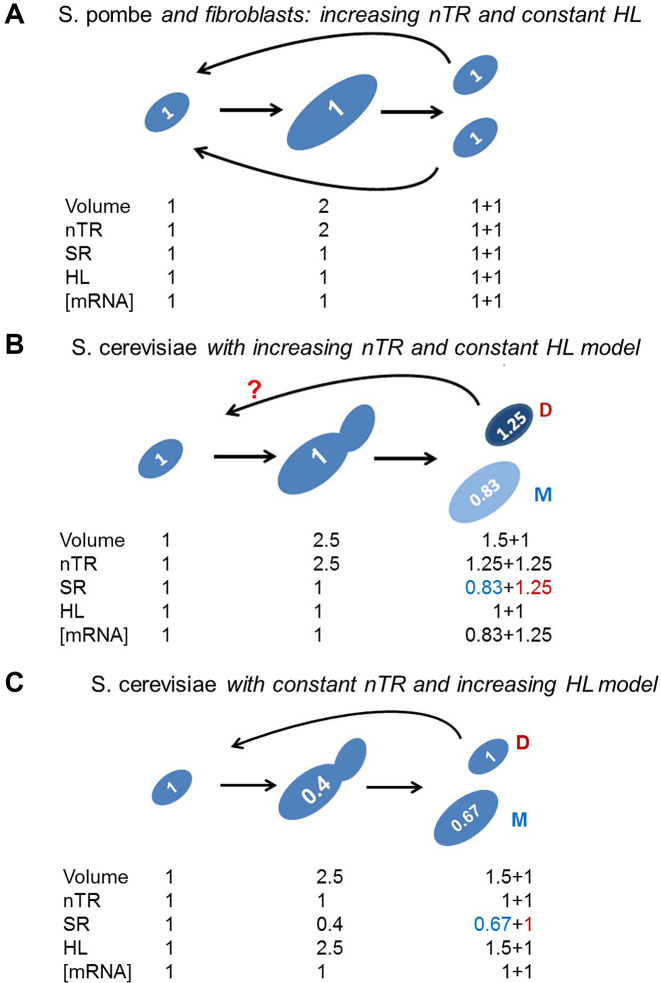
Figure 1.
Asymmetrical division in S. cerevisiae provokes a conceptual problem in transcription rate control along successive cell generations. (A) In symmetrical cell division increased cell size is paralleled by nTR in such a way that SR is kept constant, and both identical daughter cells have approximately the same volume, nTR and SR, as their previous generation. This has been observed in S. pombe and human fibroblasts (4,5), where no change in [mRNA] and, therefore in the mRNA half-life (HL), has been detected. (B) With asymmetrical division that produces a large mother (M) and a small daughter (D) cells, a similar model for nTR control would produce daughter cells with higher SRs than the previous generation, which would render this model unsatisfactory to explain actual behavior in S. cerevisiae. (C) We propose a model in which nTR remains constant with volume, which would provoke a lower SR inversely to increased volume. If [mRNA] ribostasis is strictly conserved, an increase in HL will appear. Numbers inside cells represent SR values. Note that genome replication occurs in between second and third growth stages in the three models shown. In the case of model C nTR should be duplicated at the end of the replication (2) to be then divided (1+1) between daughter and mother cells.
These two biological systems divide by cell fission to (stochastically) produce similar volume cells (7). However, other cellular systems divide asymmetrically and produce two sibling cells of different sizes. The appearance, after each division, of two cells with different cell volumes imposes a new scenario for transcription rate control. Asymmetric cell division (ACD) is a mechanism that generates cell diversity in single cells or in multicellular organisms, while maintaining self-renewing stem cell populations (8,9). Different experimental systems have been widely studied to discover the molecular mechanisms that partition RNA and proteins between sibling cells, which cause changes in cell behavior and fate (10). Not always do sibling cells have different volumes, but it is a frequent outcome of ACD (8), e.g. the development of Drosophila brain depends on neuroblasts, a type of stem cell that produces markedly smaller daughter cells (11). However, the consequences of different sized sibling cells on general gene expression have not yet been studied.
The yeast Saccharomyces cerevisiae is a particularly good example of ACD that involves marked differences in size. Cell volume control in this yeast differs to S. pombe given that it buds produce a smaller daughter cell that is phenotypically different from the larger mother cell (12). ACD conditions many budding yeast life circumstances. For instance, mother cells experience aging and die after a number of generations (13,14), a phenomenon that does not happen in symmetrically dividing cells such as S. pombe (15).
No detailed study has been done on the influence of cell volume on mRNA turnover in S. cerevisiae to date, despite old studies having addressed the evolution of mRNA turnover in the cell cycle (16–18), a process during which cell volume changes. This yeast is distantly related (330 to 420 million years from its common ancestor) to S. pombe (19). In fact many genes, cell cycle parameters and transcription regulation are quite different between these two yeast species (7,20–22).
Cell size in microorganisms is influenced by different parameters, including ploidy (5,23) and growth rates (24) which, in turn, depend on culture conditions (25–27). The volume of individual cells also changes during their cell cycle (28). Changes in cell volume in all these instances represent different physiological situations and can, therefore, be affected by additional parameters, such as the fermentative/respiratory quotient. Thus the selection of a particular experimental strategy to investigate the dependence of S. cerevisiae mRNA turnover with cell volume may be obscured by indirect effects.
In this study we used different experimental strategies and re-visited previously published studies to conduct a comprehensive study about changes in mRNA turnover with cell volume in an asymmetric dividing cell (S. cerevisiae) and to extract robust conclusions from various results. We conclude that in budding yeast mRNA turnover decreases with cell volume in both global synthesis and degradation rates, and in such a way that mRNA ribostasis is basically maintained similarly to that found in other eukaryotes. However, we found that S. cerevisiae keeps all its RNA polymerases nTR constant in spite of volume changes. With nTRII this is achieved by controlling the expression of RNA pol II itself. We postulate new regulatory models for budding yeast that differ from that found in other cellular systems with symmetrical cell division. This suggests that the quantitative constraints imposed by ACD have influenced the evolution of different regulatory mechanisms for cells to possess symmetric or asymmetric division.
MATERIALS AND METHODS
Yeast strains, media and growth conditions
The S. cerevisiae strains used herein are listed in Supplementary Figure S4C. Yeast cells were grown in liquid YPD (2% glucose, 2% peptone, 1% yeast extract). Experimental assays were performed with cells grown for at least seven generations until OD600 0.5 at 28°C.
Standard procedures were followed for synchronization at START and flow cytometry (29,30).
Cell volume and other cellular determinations
The median values of cell volumes were calculated by a Coulter-Counter Z series device (Beckman Coulter, USA). Absolute values in femtoliters and relative values are shown in Supplementary Figure S4C.
We obtained the growth rate (GR) by growing 50 ml of yeast cultures in 250-ml flasks with shaking (190 rpm) at 28°C. Aliquots were taken every 30 min in the exponential phase and their OD600 (from 0.05 to 0.7) were measured. The GR (in h−1) in the exponential phase was calculated from growth curves.
RNA extraction and poly(A) RNA measurements
To determine RNA amount, cells were grown in rich media until the exponential phase total RNA was extracted by phenol:chloroform extraction as described in (31) in three biological triplicates and was quantified by OD260 estimation in a Nanodrop device (Thermo-Fisher). Serial dilutions of total RNA were then spotted on a nylon membrane (Nytran SPC, GE Healthcare) and hybridized with a specific oligo d(T)40 probe, terminally labeled with Polynucleotide Kinase (Roche) and γ-32P-ATP. Membranes were exposed to an Imaging plate (BAS-MP, Fujifilm) and scanned by a Fujifilm FLA3000 Phosphorimager. The signal intensity of the spots was quantified with the Array Vision software. These data were used for poly(A) cell concentration calculations, as described in (32). Alternatively, the poly(A) quantification in individual cells was done essentially as described (doi.org/10.1101/044735), but an oligo d(T)30V labeled with Cy3 was used. These samples were analyzed in an LSR Fortessa cytometer (Becton Dikinson). Poly(A) amount was taken as estimator of total mRNA and then divided by cell volume to obtain total mRNA concentration ([mRNA]).
Global mRNA half-life determination by thiolutin shutoff
To determine the global mRNA half-life, we used a similar dot-blot strategy to that described in the previous section, except for the fact that RNA samples were collected at 0, 5, 12 and 25, min after transcriptional shut-off, following the addition of thiolutin to 5 μg/ml. All the samples were flash-frozen in liquid nitrogen and RNA extraction was then performed. The detailed protocol is described in (32).
Determination of nascent transcription rates and synthesis rates by different methods
Genomic Run-On (GRO) analyses were done as in (33) with the modifications described (34). All the data sets were obtained from the exponentially growing yeast populations grown at 28°C in YPD. The nascent transcription rate values for the individual genes were measured as ratios with regard to a wild-type haploid (BY4741) strain. Individual values were summed to obtain the global RNA pol II transcription rate estimates (nTRII).
Some data were obtained from other published datasets (35,36) (see below) by a different technique (cDTA, (35)), which calculates mature mRNA transcription rates. These rates, however, are not corrected by cell volume and are, thus, chemically equivalent to nTRII. nTR and SR represent different aspects of the same phenomenon (see (6) for a further explanation). In this paper, for the sake of simplicity, we used the acronym SR to refer to the molecular process of RNA synthesis by any RNA polymerase and to differentiate between RNA polymerases using the subindex (e.g. SRII, SRI). The SR can be inferred from experimental nTR data (GRO) by assuming that a stable percentage of nascent mRNA molecules reaches the cytoplasm and from cDTA data by dividing them by relative cell volumes, as described (37). The meta-analysis of these data sets is described below.
For the total SR (SRT: RNA pol I + II + III), the total radioactivity incorporated during a run-on experiment (nTRT) was determined by TCA precipitation on a glass-fiber filter and then divided by cell volume. For these experiments, the S. cerevisiae cells grown under the same conditions as above were collected in 3-ml aliquots by centrifugation at 4000 rpm for 2 min. For the control sample (blank), the cell pellet was resuspended in 5 ml of distilled water and was recollected. Then the pellet was resuspended in 1 ml of distilled water and transferred to an eppendorf tube. For the experimental samples the cell pellet was resuspended in 5 ml of 0.5% Sarkosyl and was recollected. Then the pellet was resuspended in 1 ml of Sarkosyl and transferred to an eppendorf tube. The cells in eppendorf tubes were centrifuged at 6000 rpm for 1 min. The supernatant that contained Sarkosyl (samples) or water (blank) was removed and the pellet was resuspended in 7.2 μl of distilled water. The run-on pulse was performed by adding, per sample, 9.87 μl of the transcription mix (7.5 μl of 2.5× Transcription buffer: 50 mM Tris-HCl pH 7.7, 500 mM KCl, 80 mM MgCl2, 1 μl of 10 mM ATP, CTP and GTP, 0.375 μl of 0.1 M DTT, 0.66 μl of 3 μM UTP and 0.34 μl of 3 μM [α-33P] UTP (Perkin Elmer, 3000 Ci /mmol, 10 μCi/μl to a final volume of 18 μl). To allow transcription elongation, the mix was incubated by agitation (650 rpm) for 5 min at 30 C. The pulse was stopped by adding 82 μl of cold distilled water to the mix and being stored on ice. To measure the total amount of radioactivity present in the mix (‘Total’), 10 μl of the reaction were directly spotted onto paper and dried in an aerated heater at 65°C. To measure the percentage of radioactivity that had been incorporated into the nascent RNA chain, another 10-μl volume of the mix was spotted onto glass fiber paper and dried in the aerated heater, followed by nucleic acid precipitation performed in technical duplicates. For nucleic acid precipitation, glass fiber paper was soaked in 4 ml of 10% (v/v) of trichloroacetic acid (TCA) and incubated at 4°C for 20 min. TCA was removed by decanting and 4 ml of cold TCA (10% v/v) was added again, followed by incubation at 4°C for 10 min. TCA was removed and glass fiber paper was washed with 3 ml of cold 70% (v/v) EtOH, followed by washing with 3 ml of cold 96% (v/v) EtOH. Glass fiber paper was dried in a heater at 65°C. Once dried, 5 ml of scintillation liquid was added to each vial for radioactive counting. For each individual sample, the percentage of incorporation was calculated as: (precipitated/total) x 100] – blank, where ‘blank’ was calculated as [(precipitated/total) × 100] in the control sample.
RT-qPCR analysis of the RPB1 mRNA levels in strains with different volumes
The quantification of the expression of Rpb1, the largest subunit of RNA polymerase II, was measured by RT-qPCR and normalized against the ACT1 mRNA levels. Specific primers were designed to this aim: Rpb1-F: 5′-CCAGAAGTGGTCACACCATATAA-3′ and Rpb1-R: 5′-GGTCTCCGCTATCACGAATG-3′. Reverse transcription of mRNA was carried out using an oligo d(T)15VN with Thermo-Scientific Maxima Reverse Transcriptase (Thermo Scientific). cDNA was labeled with SYBR Premix Ex Taq (Tli RNase H Plus) from Takara and the Cq values were obtained from the CFX96 Touch™ Real-Time PCR Detection System (BioRad).
Western blot analysis for RNA polymerase II quantification
Western blots were performed as described previously (32) using three different antibodies for RNA pol II detection: anti-POLR2C antibody (Abcam) against Rpb3 subunit, anti –Rpb1 N-terminal (γ-80, Santa Cruz Biotechnology) and anti-phospho-S2 (Abcam). The first two detect all RNA pol II molecules and the third one detects elongating Ser2-phosphorylated molecules. Total protein was quantified in the same samples by the Bradford method (38).
Meta-analysis of the experimental data sets
To evaluate the correlation between cell volume and mRNA turnover in asynchronous exponential growth phase cells, a set of 38 yeast mutants was used (35). In that study, the global SRII, [mRNA] and the average half-life (HL, inverse of kd) were determined according to the cDTA protocol. They were all represented as being relative to their wild type. Those data were processed as previously described (37) and plotted against the cell volumes obtained from various sources (23,39). The observed Pearson's correlation coefficient, r, and the P-value of the statistically significant deviation from the null hypothesis of no correlation (r = 0) were calculated. With the R package BayesVarSel (40), a Bayesian analysis of the inclusion probabilities among GR, SRII and volume confirmed the SRII dependence on cell volume (see Supplementary Figure S3).
To evaluate the correlation between cell volume and mRNA turnover during individual cell growth throughout the cell cycle, data from the study of (36) were used. In that study, SRII, RA and HL (kd) were also determined according to the cDTA protocol.
Enrichment analyses for gene categories in the set of polyploid and cell size mutant strains
The gene expression values obtained from the GRO experiments on the strains listed in Supplementary Figure S4C were done in triplicate and the acquired raw data were normalized using the ArrayStat statistics software (Imaging Research Inc.) by the median absolute deviation (MAD) approach. Volume-gene expression covariation was quantified using a modified version of Pearson's correlation (see (41) for details) between both values. A gene-to-gene analysis of the differential behavior, as regards to the global average, was performed by the Significance Analysis of Microarrays (SAM) method (41) with a false discovery rate of q<0.05. A gene set enrichment analysis was applied to previously detected gene sets with different behaviors. A unilateral Fisher's exact test was applied where the gene sets to be compared were the Gene Ontology (GO) groups. Analyses were run with R packages (40). The whole R code used in this paper is found as Supplementary Material.
RESULTS
Global mRNA synthesis and degradation rates lower with the individual cell volume during the cell cycle
As a first approach to study the influence of cell volume on gene expression in S. cerevisiae, we analyzed global mRNA synthesis and degradation according to cell volume across the cell cycle. We envisioned that the direct transposition of the established model for symmetrically dividing cells to the budding yeast would be unviable (compare Figure 1A and B). By assuming that both copies of the genome should have identical nTR, and that the cell volume was ∼50% higher in mother cells (28), SRs would differ after cytokinesis. In fact if nTR increased in parallel to cell volume during the cell cycle, as in symmetrically dividing cells, the daughter cell would have an increased SR as regards the original value at the beginning of cycle. This would provoke a never-ending increase in the SR of newborn daughter cells, unless the budding yeast had a mechanism to compensate (in advance) changes in SR (Figure 1B). Moreover, if HL remained constant, which seems to happen in cells that undergo symmetric division (4,5), [mRNA] would also increase in daughter cells and mRNA ribostasis would be compromised. Alternatively, we reasoned that S. cerevisiae should have a type of control of its transcription rate as regards cell volume that differs from the linear increase in nTR seen in S. pombe (5) and human cells (4). We hypothesized an alternative model where nTR would remain constant and SR would decrease with volume. If there was a compensatory change in SR and HL, [mRNA] ribostasis would be preserved (Figure 1C).
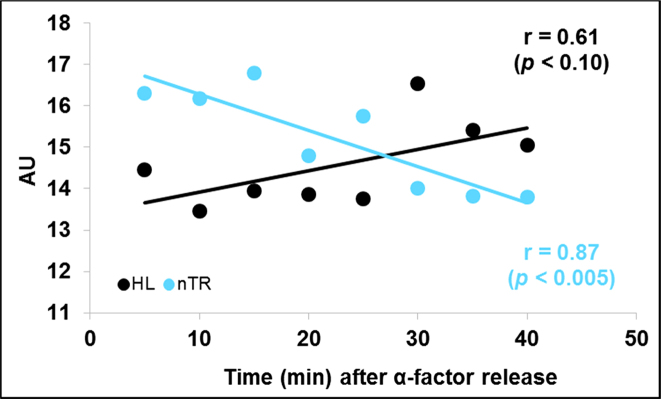
To test this hypothesis we first ran a meta-analysis of the published RNA pol II data from (36). The exhaustive analysis done in that paper contained many data points during three cell cycles using S. cerevisiae cells synchronized with α-factor. We analyzed only the data from 0 to 40 min, which corresponded to the G1 phase of the first cell cycle before budding. Our meta-analysis results indicated no increase in nTRII, but a reduced mRNA turnover (decrease in nTRII and increase in HL) associated with cell cycle progression in S. cerevisiae (Figure 2). As we do not have experimental cell volumes for (36) data points, it was impossible to determine actual SRII variation. However, as HL is independent on concentration and, therefore, on cell volumes, there was an actual increase in global mRNA stability with cell volume, which agreed with our predicted model.
Figure 2.
Meta-analysis of the published data for mRNA turnover dependence on cell volume in synchronized cells. Meta-analysis of the results from Eser et al. (36). Although the original authors use the cDTA method, which measures newborn mature mRNAs, and they described their data as synthesis rates (SRII) given that they do not take into account cell volumes their transcription data should be considered formally equivalent to nascent transcription rates (nTRII). The cDTA study also provided values for mRNA degradation rates that can be converted into mRNA half-lives (HL). We show here that nTRII lowers and the HL increases with time after α-factor release. There is no cell volume measurement in this experiment, although it is conceivable that it increases with time.
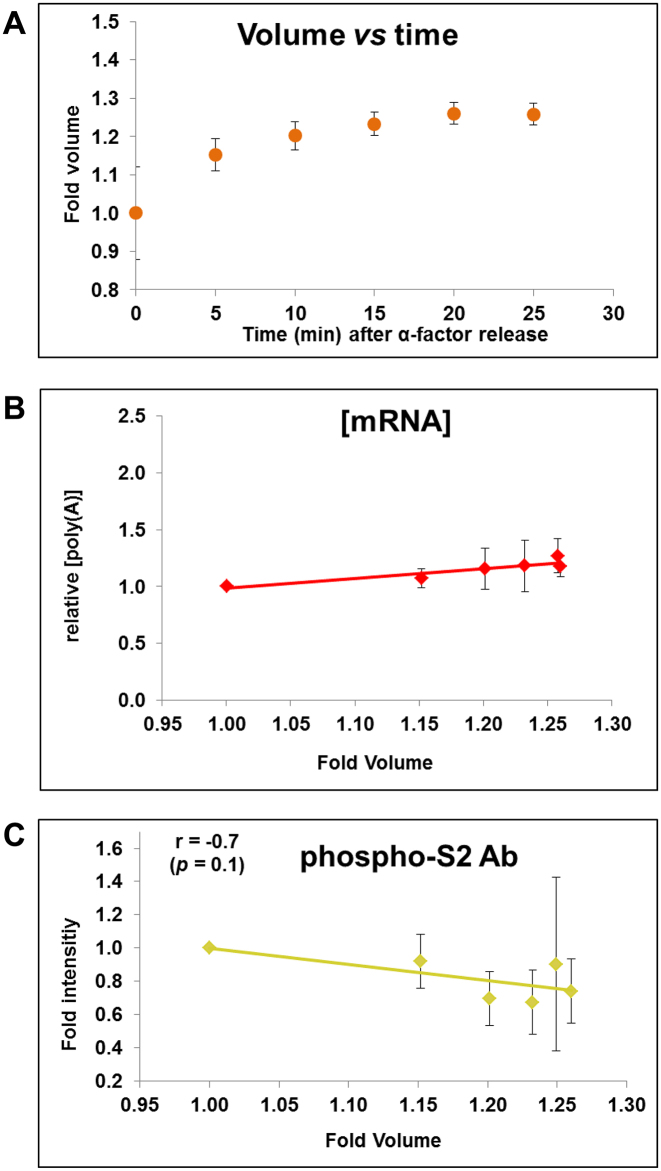
In order to complete the investigation of ribostasis during the cell cycle we performed a cell synchronization experiment with α-factor and release, in which we measured the average cell volume by Coulter Counter, total RNA by phenol extraction, and mRNA by dot-blot and flow cytometry. We focused on the first 25 min after α-factor release which, in our hands, corresponded to the G1 period (Supplementary Figure S1) before budding (28,36). Figure 3A shows how cell volume increases by ∼25% during a similar time course to that depicted in the experiment of (36). In our experiment, total [RNA] remained constant (Supplementary Figure S2) and [mRNA] has a slight increase (Figure 3B) as regards cell volume during the cell cycle. Thus it seems that mRNA ribostasis was maintained by a compensatory change in SRII and mRNA stability during the cell volume changes throughout the cell cycle.
Figure 3.
Ribostasis and proteostasis analysis in synchronized cells. We performed a similar experiment to that of Eser et al. (36) (Figure 2) by making a α-factor synchronization of yeast cells (see text for details) and we measured cell volume (A) and [mRNA] (B) at different times after α-factor release. [mRNA] was calculated as the poly(A) amount per cell (divided by cell volume) by an assay based on fluorescent oligo-d(T) hybridization and cytometry quantification. Equal protein amounts were used in Western blot analyses for RNA pol II quantification with the phospho-S2 antibody, which measures elongating RNA pol II (C). Four independent biological replicates were used to calculate the average and standard deviation (SD) values. Total [RNA] and [protein] from the same samples are seen in Supplementary Figure S2A and B. A representative Western blot is shown in Supplementary Figure S2C. The levels of the total and elongating RNA pol II were normalized against the internal glucose 6-phosphate dehydrogenase (G-6-PDH) control.
Protein concentration remains constant, but active RNA pol II decreases during G1 phase
In order to know the reason for the drop in SRII with cell volume, we analyzed the active RNA pol II concentration in the same samples by western blot using an antibody that quantifies the molecules engaged in elongation (anti Ser2P). Figure 3C depicts that whereas the total protein concentration is constant (Supplementary Figure S2B), elongating RNA pol II decreases in concentration by up to 25% in parallel to the increased cell volume through G1 after the alpha factor release. This result suggests that S. cerevisiae decreases SRII by keeping the number of elongating RNA pol II molecules onto chromatin (constant nTRII) constant in spite of an increasing cell volume.
Global mRNA synthesis rate (SRII) lowers and mRNA stability increases with the average cell volume in asynchronous cultures
Cell volumes vary in a yeast cell according to several circumstances. In the first part of this study we saw that variations in cell volume during growth during the G1 period did not behave as they do in other eukaryotes. Given that the experimental setup using cell synchronized cultures can impose some constraints or biases to studying the regulation mRNA synthesis rate with cell volume, we decided to do studies that used different yeast strains with variable cell volumes caused either by different genotypes or different ploidies. In actively growing yeast, as cell cultures are composed of cells in different cell cycle stages and of various replicative ages, the values obtained in this study represent the average (median) cell volumes for the whole population. Previous experiments done in the yeast S. pombe have shown that the total mRNA amount per yeast cell can increase with cell volume to maintain [mRNA], which occurs through an increase in the global SRII (5), but with no changes in mRNA stabilities. In S. cerevisiae it has been previously shown that global [mRNA] remains within certain limits (±50%) in different physiological situations (32,35,37). Interestingly enough, this mRNA ribostasis is maintained by a coordinated parallel change in its synthesis (SRII) and degradation rates (inverse of HL) (37,42).
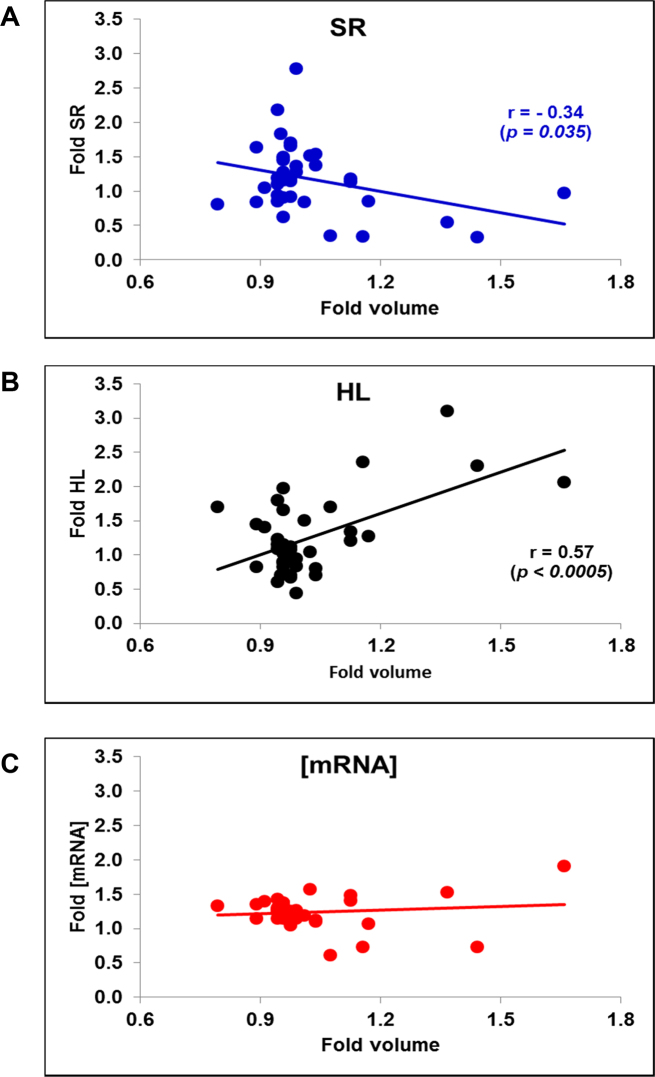
We first made a meta-analysis of the data published by (35), where 38 yeast mutants of different cell volumes were analyzed for [mRNA], SRII and HL by the cDTA method. To obtain SRII values from the original nTRII ones, we used cell volumes of this strain list, which were taken from (23,39). We found a decreasing tendency of SRII and an increasing one in HL with increased volume (Figure 4A and B). [mRNA], however, displayed a flat tendency (Figure 4C). Thus it seems that S. cerevisiae tends to keep ribostasis in spite of a decreasing mRNA turnover.
Figure 4.
Meta-analysis of published data on different cell size mutant strains. We used the data for the average total mRNA synthesis rate (SR), [mRNA] and mRNA stability (HL) from 38 mutant strains described in Sun et al. (35). The original nTR and RA data were corrected by cell volume of exponentially growing cell cultures of the same mutants taken from references (23,39). Pearson's correlation coefficient (r) and the associated P-value for the non-flat plots are shown. (A) SR; (B) HL, (C) [mRNA].
The general tendency to decrease SRII and to increase mRNAs HL with cell volume seen in the meta-analysis of the mutants was clear, but there is a potential problem. This data set showed an inverse correlation between GR and cell volume (37). We previously described the direct dependence of SRII with GR (37). Thus the inverse correlation between cell volume and SRII can be an indirect result of this. In order to test this possibility, we used a Bayesian model. The results of this statistical analysis confirmed that there was a true dependence of SRII on cell volume, apart from its dependence on GR (Supplementary Figure S3).
On this general tendency, the particular physiology of each mutant provokes noise and some particular mutants especially behave discrepantly to the general rule (e.g. some the outliers in Figure 4A). To solve this problem and to experimentally confirm the previous meta-analysis, we used a series of polyploid strains with an identical genotype constructed by the D. Pellman laboratory (43) and two haploid mutants (whi5 and cln3) known to have very different cell volumes, but with similar growth rates to their wild type (BY4741, see Supplementary Figure S4). We measured the average cell volume and DNA content (see Supplementary Figure S4A–C). We also measured vacuole size in each strain to rule out that total cell volumes were not good evaluators of cytoplasm volumes. By vacuole-specific staining, we determined vacuole and cytoplasmic volumes, and showed a linear 1:1 correlation between total cell volumes and cytoplasmic volumes (Supplementary Figure S5).
In this study we used a different method to quantify mRNA concentrations and synthesis rates. Our Genomic Run-On method (GRO (33)) allows the quantification of global nTRII which, by knowing cell volume, can be used as a proxy of SRII (6). Figure 5A shows a significant decrease in global SRII (the sum of all the synthesis rates for all the genes) with cell volume, whereas the global [mRNA] plot shows no significant slope. The calculated global mRNA stability (HL = [mRNA]/SRII) increased (i.e. kd decreased) with cell volume.
Figure 5.
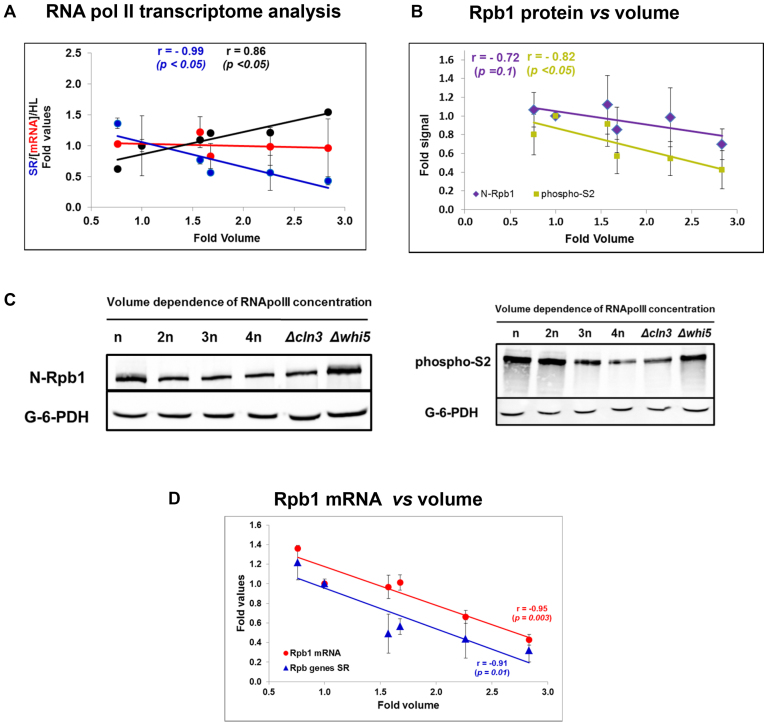
Examination of mRNA turnover in polyploid and cell size mutant strains. (A) Global transcriptional study. Genomic Run-On (GRO) in the wild type haploid BY4741 and isogenic polyploid strains was performed to obtain a nascent transcription rate (nTRII). nTRII was divided by the median cell volume as a proxy of SRII. [mRNA] was calculated as described in Figure 3 legend. Essentially the same result was obtained by hybridization with radioactive oligo d(T) in a dot-blot protocol (not shown). The whole cell population mRNA half-life (HL) was determined by dividing [mRNA] by SRII. (B) RNA pol II protein regulation. Identical protein amounts were used to perform Western blot analyses for RNA pol II quantification by the antiSer2-phosphorylated antibody that measures elongating RNA pol II, and the Anti-N-terminal-Rpb1 antibody that recognizes all the RNA pol II molecules. The shown quantification corresponds to the averages and standard deviations (SD) of four independent biological replicates. Total protein concentration does not vary with cell volume (see Supplementary Figure S8). The levels of total and elongating RNA pol II were normalized against the internal G-6-PDH control. Examples of representative Western blots are shown in panel (C). The same study on aliquots with the same samples and with Anti-Rbp3 antibody is shown in Supplementary Figure S8. (C) The RT-qPCR analysis of the Rpb1 mRNA levels in the same cells normalized against ACT1 mRNA. (D) Regulation of the RNA pol II subunits that encode mRNAs. The qRT-PCR analysis of RPB1 mRNA (red dots) was done on the samples of the same yeast strains set. Data represent the average and SD of three biological repeats. The average SR and SEM of the three genes RPB1, RPB2 and RPB3, which encode the three largest RNA pol II subunits in the same strains, is also shown (blue triangles). These individual SR gene data correspond to the GRO data used for the total SRII used in panel (A).
We found that the SRII of most genes followed the general tendency within the statistical limits, which is consistent with previous studies into budding yeast (43,44) and S. pombe (5). Only some gene functional categories showed a particular divergent tendency. For instance, the genes related with transposons and mitochondria and respiration lower SRII more slowly with volume as regards the average population, and those related with plasma membrane lowered more quickly (Supplementary Figure S6). The three RNA pol II largest subunits genes (RBP1, RPB2 & RPB3) also displayed behavior that resembled the general tendency (see below).
Finally, in order to verify the increase in the global mRNA stabilities calculated from SRII and [mRNA], we decided to use an independent approach to evaluate the mRNA stabilities in the set of polyploid strains. For this purpose, we used a method based on oligo dT hybridization of the identical total RNA amounts in a dot-blot (see Materials and Methods). In this way we obtained a decay curve that represented the average stability of the global poly(A) mRNA population. Supplementary Figure S7 displays a clear increase in the global mRNA stability with cell volume. This result ratified the previous result (Figure 5A), in which the average global stability of mRNAs was mathematically inferred, and confirmed that mRNA turnover decreased with average cell volume in exponentially growing yeast cell cultures.
RNA pol II concentration lowers with cell size
In our previous experiment done with synchronized cells, we showed that the number of active elongating RNA pol II molecules onto chromatin remained constant despite the increase in cell volume (Figure 3C), which led SRII to decrease. We hypothesized that this situation would also happen in experiments with exponential unsynchronized cultures, and would cause SRII to lower (as seen in Figure 5A).
We checked this hypothesis by a Western blot with three different antibodies against total (anti-Rpb3 and anti-N-terminal Rpb1) and elongating RNA pol II (anti Ser2P). Figure 5B and C and Supplementary S8 indicate that all three antibodies led to a 3-fold change in the RNA pol II concentration (when comparing the whi5 mutant and the 4n polyploid), which was almost antiparallel to the cell volume increase (3.2-fold, see Supplementary Figure S4). This result confirms that S. cerevisiae specifically controls RNA pol II levels as regards cell volume, but differently to human cells, which keep the RNA pol II concentration constant in parallel to the total protein concentration (4).
We wondered if RNA pol II concentration regulation in response to cell volume took place at the mRNA level. In order to answer this question, we performed an RT-qPCR analysis of the RPB1 (which encodes the largest subunit of RNA pol II) mRNA levels. We found that the level of this mRNA lowered with cell volume as regards both total RNA (mostly rRNA-tRNA, not shown) and ACT1 control (Figure 5D). Since the SRII of this gene and the other two large RNA pol II subunits (measured in the GRO experiments described in Figure 5A) behaved identically (Figure 5D), we deduced that RPB1 expression decreased with cell volume given its drop in SRII. As this decrease was identical to that of the global SRII drop (see Figure 5A), we concluded that, in fact, the actual reason for the lower RPB1 (and possibly for the other specific RNA pol II subunits) expression was that it did not undergo the general stabilization observed for the global transcriptome.
The different behavior of the other two RNA polymerases
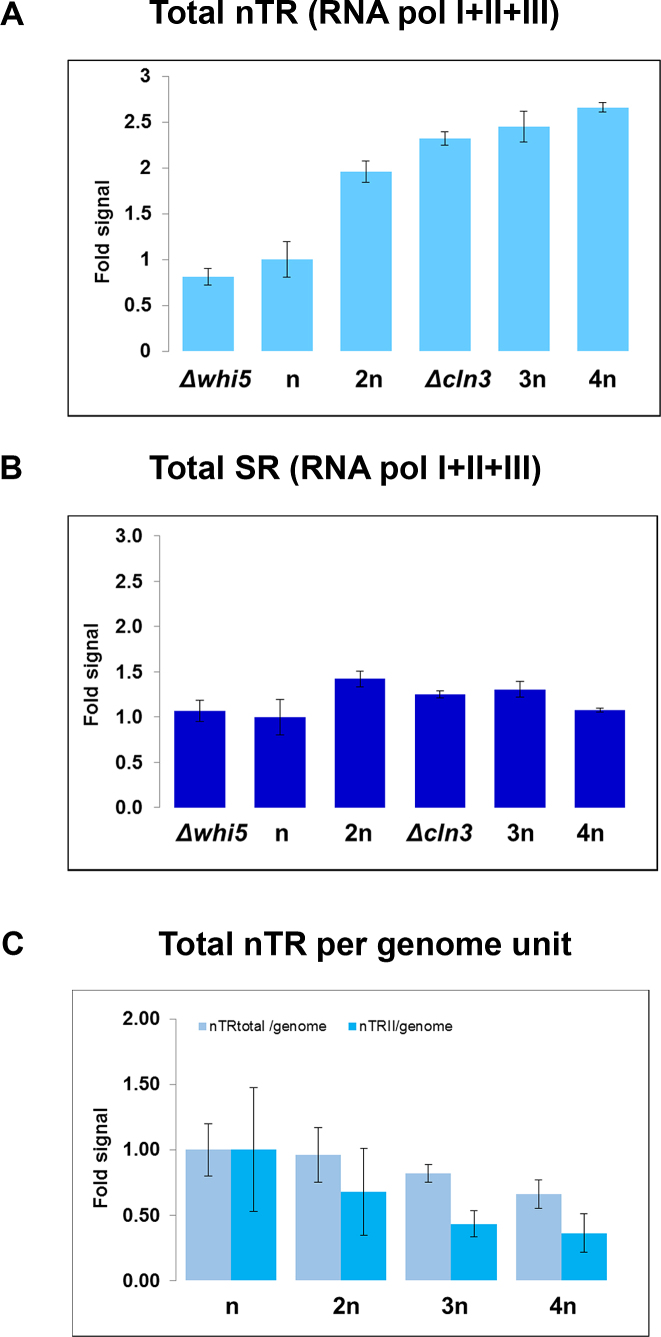
Interestingly, the evaluation of total nTR (nTRT), the sum of all three RNA polymerases, showed an increasing profile with volume (Figure 6A), which caused an approximate constant SRT (Figure 6B). Although our method did not distinguish SRII from the SR of the other two RNA polymerases as SRI + III was ∼75% of the total SR (45), we concluded that the behavior of RNA pol II lowering SRII with cell volume was specific of this polymerase, and that RNA pol I+III, which transcribe ncRNAs, kept SRI + III constant with cell volume in the population average cell volume experiment.
Figure 6.
Total transcription in the polyploid and cell size mutant strains. (A) Changes in the overall nTRT (RNA pol I+ II + II) in relation to the wild-type strain (BY4741) for the different yeast strains, calculated by a run-on protocol as described in the Methods section. (B) nTRT was then divided by the relative cell volume to obtain SRT. (C) The results from both nTRT (panel A) and nTRII (Figure 5A blue dots) are represented here as per genome copy by dividing the ploidy of each strain (Supplementary Figure S4C). All the data have been relativized against the values of wild strain BY4741, and correspond to the average and SD of three experiments.
As we used cells with different ploidies (n–4n), we wondered how the genome copy number would influence transcription rates. When dealing with genome copy changes, we distinguished between nTR/cell from a nTR/genome copy, which was more convenient because it would reflect the actual density of transcribing RNA polymerases onto their templates (6). When we represented the nTR/genome of the series of yeast strains from our study we saw that whereas nTRT/genome activity slightly decreased (25% from n to 4n, see Figure 6C), the nTRII/genome decreased almost proportionally to ploidy (70% from n to 4n, Figure 6C). In fact the 25% decrease in nTRT, which included 25% of RNA pol II activity approximately, can be explained almost entirely by the drop in the RNA pol II component. Thus we concluded that there was no dose compensation in RNA pol I nTR in S. cerevisiae. On the contrary, RNA pol II showed pronounced dose compensation by splitting nTRII among the actual genome copies. Given the increase in cell volume with ploidy (Supplementary Figure S4C), this dose compensation led the actual SRII to decrease from haploid to tetraploid (Figure 5A).
Model for the S. cerevisiae control of mRNA turnover
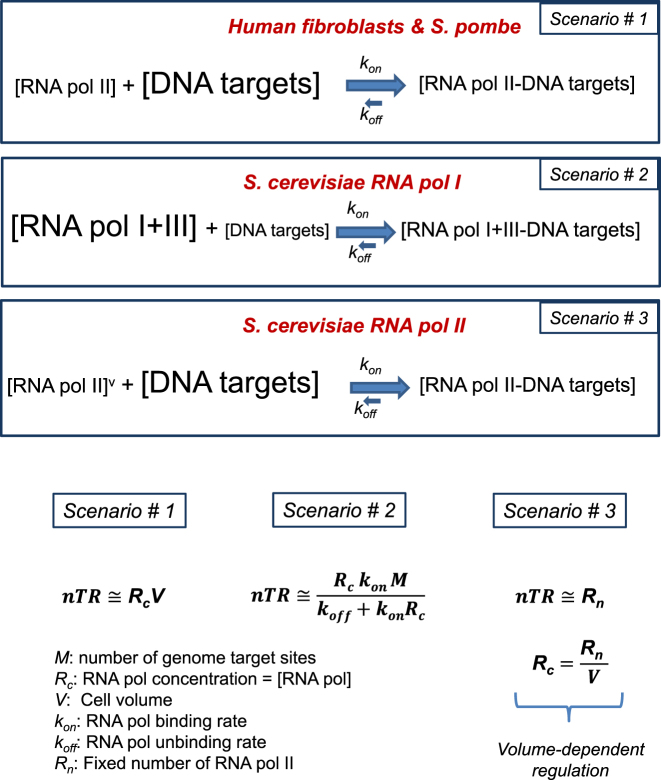
After considering all the previous results and arguments, we propose a general model for cell control over RNA transcription (Figure 7). This model is an extension of the model proposed in (4) for fibroblasts and S. pombe, and of that discussed in (46). Eukaryotic cells with a symmetrical division have a limiting amount of RNA pol compared to its targets. By assuming a very high association constant for RNA pol binding and a limiting concentration of this enzyme, the nTR becomes dependent on cell volume (scenario #1). This situation has been shown for RNA pol II in mammalian cells (4). However, if DNA targets are limiting in relation to RNA pol, which is the case of budding yeast RNA pol I, nTR becomes independent on cell volume (scenario #2). Finally if the limiting RNA pol concentration lowers with cell volume, nTR also becomes constant and independent on cell size (scenario #3). This scenario matches the experimental results that we found for RNA pol II-dependent genes expression in S. cerevisiae.
Figure 7.
Model for transcription rate control during the cell cycle in eukaryotes with symmetric and asymmetric cell division. In symmetrically dividing cells, such as human fibroblasts and S. pombe, RNA pol II is much less concentrated than their DNA targets. As there is a strong bias of the equilibrium toward the bound state (kon >> koff), all the RNA pol II molecules are bound to chromatin and nTR is strictly dependent on the RNA pol II number of molecules (Scenario #1). In asymmetrically dividing cells, such as S. cerevisiae, nTR does not depend on the RNA pol number of molecules, either because there is a vast excess of them over their targets (e.g. for RNA pol I+III) in which it becomes dependent on the number of DNA targets number (Scenario #2), or because the RNA pol concentration is regulated in such a way that it varies with cell volume (represented as an v superscript in the figure). This scenario (#3) corresponds to RNA pol II. In it nTR is constant and proportional to the actual number of active RNA pol II molecules. Note that scenarios #1 and #3 allow the regulation of the nascent transcription rate by controlling the concentration of RNA pol II, whereas scenario #2 does not allow regulation at the RNA polymerase level because it is in vast excess.
DISCUSSION
Synthesis and degradation rates (mRNA turnover) determine concentrations of mRNAs. We found in the yeast S. cerevisiae that the global mRNA turnover rate lowers with cell volume, while the global mRNA concentration remains approximately constant. In our study we found not only a decreases in global synthesis rates with cell volume when we used data from two different techniques (GRO and cDTA), but also increases in mRNA stability when we used data from three different ones (GRO, cDTA ad transcriptional shutoff). We also found a similar behavior in the asynchronous cell cultures of different well-known cell-size mutant strains, culture conditions or ploidy, and in synchronized cells throughout the G1 phase. In all these cell volume change cases, we observed an inverse change in mRNA synthesis and degradation rates, while the global mRNA concentration remained constant. Thus the conclusion that we obtained is very robust. It is noteworthy that our results refer to the global mRNA population, but do not apply to all individual mRNAs. Some examples of differential expression and asymmetric distribution of mRNA species have been described in mother and daughter cells in budding yeast (47).
Some past papers have addressed the behavior of total RNA and mRNA synthesis rates during the cell cycle in budding yeast (16,18). Although they have not specifically studied mRNA turnover dependence on cell volume, it is possible to re-visit those results by comparing cell cycle changes for measured nTR, SR and cell volumes. In (18) found that nTRI remained constant throughout the G1 phase in spite of an increasing volume, indicating a drop in SRI). Their study also found that nTRII was constant (SRII lowered) with cell volume increases, what well matches our results. Our current results revealed that, similarly to that observed in other eukaryotes, total [RNA], [mRNA] and [protein] remained approximately constant in relation to cell volume in budding yeast. It seems that proteostasis remained by keeping the protein translation (and degradation) rates constant for all cells (16). This could correlate with total [RNA] ribostasis given that total RNA is composed mainly of translation-related ncRNA (rRNA + tRNA), as observed by (18). Additionally, we found that budding yeast preserves [mRNA] ribostasis by a parallel change in its synthesis and degradation rates. To do so, S. cerevisiae keeps total nTRII constant and provokes a change in SRII inversely to the change in cell volume. This result contrasts with previously published results in S. pombe and mammalian cells, in which nTRII increased as regards cell volume by keeping SRII constant. Thus in those organisms, and unlike budding yeast, [mRNA] homeostasis was preserved because no change in SR and HL took place.
We propose that the special behavior of RNA pol II in budding yeast is related to ACD (Figure 1). Unlike other eukaryotes, S. cerevisiae generates two cells with a large difference in volume: mother and daughter differ by almost 2-fold in cell volume (e.g. see (28)). Thus the number of free-moving molecules (RNAs, proteins) should be proportionally segregated to maintain the concentration in both cells after division. As the genome, however, should be split into two identical copies, and not only at the DNA sequence level, but also at the structural and functional levels, many nuclear proteins, like RNA polymerases, should segregate equally between mother and daughter cells as no significant differences in volume have been detected between mother and daughter nuclei in late anaphases (30). A similar weak nuclear volume scaling with cell size has been observed in fibroblasts (4). An increase in any kind of SR in daughter cell is unviable because it would seriously hinder the required cycling nature of SR, unless a resetting mechanism would compensate (in advance) the predicted SRII excess of daughter cells (see Figure 1B).
In both S. pombe and fibroblasts, other authors have found the conservation of RNA pol II concentration with cell volume (4,5), which falls in line with the observed general proteostasis. Moreover, measurements of chromatin-bound RNA pol II in S. pombe (5) and the in situ labeling of transcription at single cell level in primary fibroblasts (4) have demonstrated a parallel increase in nTRII with cell volume, where SRII remained constant. In contrast, our results showed that the global and elongating RNA pol II concentration lowered in S. cerevisiae in cell cycle experiments and when different cell size strains were compared (Figures 3B and 6A), which parallels the drop in SRII.
This change in SRII with cell size would provoke alterations in [mRNA] ribostasis if not compensated by changes in mRNA stability. The changes we observed in mRNA stability (Figures 2, 4B and Supplementary Figure S6) can be explained by the well-known cross-talk between transcription and mRNA degradation, which we (48) and others (49,50) have demonstrated. Thus we hypothesize that the control for nTRII based on the transcriptional regulation of RNA pol II according to cell volume determines the actual global SRII and, afterward, ‘automatic’ cross-talk mechanisms compensate global mRNA stability to keep mRNA ribostasis. The dependence of mRNA turnover with cell size in budding yeast does not, thus, need a different biological reason from its asymmetric division. Interestingly, the regulation of RNA pol II with volume, which makes this differential phenomenon possible, and which we describe in budding yeast, takes place by uncoupling the SR of their encoding genes from their mRNA degradation (42). As a result, RNA pol II subunits’ mRNA concentrations lower with cell volume (Figure 5D). So, as expected, the key regulatory element of this phenomenon escapes the general regulation of gene expression with cell volume.
What is the relationship between SRII and genome content? The dependence of the transcriptional activity of this RNA polymerase with the gene copy number has been studied by other authors from different perspectives. In principle it would appear that, by keeping all other variables constant, nTRII should be proportional to the gene copy number, provided that transcriptional machinery is in excess as regards the whole set of templates (as with RNA pol I+III; see below). This is the usual case for single gene duplications, and perhaps for larger duplications like aneuploidies. This topic is controversial (51–53). Another perspective is the case of genome duplication in the S phase. General behavior involves an increase in nTRII in parallel to genome replication as a passive consequence of gene duplication (18). However, when carefully looking at particular genome regions, it has been found that freshly replicated genes do not increase nTRII in relation to unreplicated ones because of the inhibitory effect of newly synthesized histone H3 acetylated in lysine K56 (54). This can also be the case for genome replication in fibroblasts, as observed (4). The genome multiplicity effect on polyploid strains has, however, not been investigated to date at the transcriptional level. Our results indicate that the amount of active RNA pol II is limiting as total activity is maintained by splitting it between genome copies (Figure 6C).
Although we focused our work mainly on RNA pol II, the problem caused by asymmetric division should be the same as for the other two nuclear RNA pol in budding yeast (I and III). We have no data on the cell cycle, but authors from (18) showed that throughout the cell cycle, except for the S phase where it steeply doubles, nTRI is kept constant. This result is consistent with an excess amount of these two RNA polymerases, which would transcribe at a rate dependent on their template (that is limiting). In fact, it has been recently shown that budding yeast RNA pol I is stored in an inactive dimeric form when it is not bound to its targets (55). This indicates a potential regulatory mechanism for the RNA pol I/rDNA balance, as we proposed in scenario #2 (Figure 7). Doubling in the S phase (18) fits this model well. In our experiments with polyploid and cell size mutants, we found an increasing nTRI+III (constant SR I+III) when we compared asynchronous cultures (Figure 6A and B). Polyploid cells would increase nTRI+III by increasing the available templates to these RNA polymerases. A similar result has been found (5) in S. pombe using mutants that duplicated its genome (5). The mechanism that budding yeast haploid mutants cln3 and whi5 use to change their nTR remains to be discovered, but we suggest that it is related with the plasticity shown by rDNA repeats in the number (56) and the proportion of active copies (57).
To summarize, we propose that budding yeast uses two different strategies to deal with the problem created by the different sizes of daughter and mother cells (Figure 7): in one scenario (#2) there is a much higher concentration of RNA polymerases than their targets, which should be saturated with them. nTR is constant and independent on cell volume. This scenario explains RNA pol I behavior. Any increase in their targets (during replication, or in polyploids) involves a proportional increase in nTRI. We anticipate that this scenario would be the general one for eukaryotes as it seems to also apply for S. pombe and for RNA pol I in S. pombe (5). For RNA pol II in S. cerevisiae, however, the constant nTRII during G1 throughout the cell cycle, and when comparing cells with different sizes, fits a different scenario (#3). In this scenario the splitting of RNA pol II among the genome copies seen in polyploids would satisfy a model with a constant, but limiting, amount of RNA pol II. We propose the existence of a mechanism that senses cell volume irrespectively of the target number and regulates the level of the mRNAs that encode RNA pol II subunits by uncoupling their stability from the general cross-talk mechanism that keeps global mRNA ribostasis.
Given that scenarios #1 and 3 should be a compulsory consequence when a cell divides into two unequal sized ones (see Figure 1), the problem posed herein may affect other kinds of cells with ACD. In nature there are more examples of asymmetric division that involve differences in cell size. Asymmetric cell division is a mechanism used by prokaryotes and eukaryotes alike to generate cellular diversity. Stem cells particularly rely on ACD to self-renew while simultaneously generating a differentiating sibling (8,9). This is frequently linked to the size differences between the two sibling cells, as in the neuroblast system in Drosophila (11). Polyploid giant cancer cells (PGCCs) is yet another example for which budding yeast has been used as a model (58). In Caenorhabditis elegans embryos, the zygote bisects into two cells of unequal sizes, a larger anterior cell and smaller posterior cell (10). ACD also plays a fundamental key in plant development (10). Our S. cerevisiae results predict that other organisms with ACD may follow a similar regulation of [mRNA] ribostasis as the problem of RNA synthesis rate differences seems a basic feature of those dividing cells that produce different sized siblings.
AVAILABILITY
The Gene Expression Omnibus (GEO) accession number for new experimental genomic data is GSE96803.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr D. Pellman for his generous gift of polyploid strains; Dr A. Tresch for sharing cDTA data; Dr R. Gaviria, (S.C.S.I.E., UVEG) for her statistical analyses; Dr D. Gresham for providing us with a detailed poly(A) determination protocol; Dr F. Estruch for his generous gift of the fluorescently labelled oligo d(T) and N-terminal Rpb1 antibody and Drs J.C. Igual and J. Moreno for their useful comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish MiNECO and European Union funds (FEDER) [BFU2013-48643-C3-3-P and BFU2016-77728-C3-3-P to J.E.P.-O. and BFU2013–48643-C3-1-P and BFU2016-77728-C3-1-P to S.C.]; Regional Valencian Government [PROMETEO II 2015/006 to J.E.P.-O.]; Junta de Andalucía and European Union funds (FEDER) [P12-BIO-1938 MO to S.C]; National Institutes of Health [1R01GM126557-01 to A.S.]. V.B. is the recipient of an FPI fellowship from MiNECO]. Funding for open access charge: [BFU2016-77728-C3-3-P].
Conflict of interest statement. None declared.
REFERENCES
- 1. Powers E.T., Balch W.E.. Diversity in the origins of proteostasis networks [mdash] a driver for protein function in evolution. Nat. Rev Mol. Cell. Biol. 2013; 14:237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walters R.W., Parker R.. Coupling of Ribostasis and Proteostasis: Hsp70 Proteins in mRNA Metabolism. Trends Biochem. Sci. 2015; 40:552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bustamante C.J., Kaiser C.M., Maillard R.A., Goldman D.H., Wilson C.A.M.. Mechanisms of cellular proteostasis: insights from single-molecule approaches. Annu. Rev. Biophys. 2014; 43:119–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Padovan-Merhar O., Nair G.P., Biaesch A.G., Mayer A., Scarfone S., Foley S.W., Wu A.R., Churchman L.S., Singh A., Raj A.. Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms. Mol. Cell. 2015; 58:339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhurinsky J., Leonhard K., Watt S., Marguerat S., Bähler J., Nurse P.. A coordinated global control over cellular transcription. Curr. Biol. 2010; 20:2010–2015. [DOI] [PubMed] [Google Scholar]
- 6. Pérez-Ortín J.E., Medina D.A., Chávez S., Moreno J.. What do you mean by transcription rate?. BioEssays. 2013; 35:1056–1062. [DOI] [PubMed] [Google Scholar]
- 7. Turner J.J., Ewald J.C., Skotheim J.M.. Cell size control in yeast. Curr. Biol. 2012; 22:R350–R359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roubinet C., Cabernard C.. Control of asymmetric cell division. Curr. Opin. Cell Biol. 2014; 31:84–91. [DOI] [PubMed] [Google Scholar]
- 9. Chen C., Fingerhut J.M., Yamashita Y.M.. The ins(ide) and outs(ide) of asymmetric stem cell division. Curr. Opin. Cell Biol. 2016; 43:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pillitteri L.J., Guo X., Dong J.. Asymmetric cell division in plants: mechanisms of symmetry breaking and cell fate determination. Cell. Mol. Life Sci. 2016; 73:4213–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Homem C.C.F., Knoblich J.A.. Drosophila neuroblasts: a model for stem cell biology. Development. 2012; 139:4297–4310. [DOI] [PubMed] [Google Scholar]
- 12. Henderson K.A., Gottschling D.E.. A mother's sacrifice: what is she keeping for herself. Curr. Opin. Cell Biol. 2008; 20:723–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jazwinski S.M. Longevity, genes, and aging. Science. 1996; 273:54–59. [DOI] [PubMed] [Google Scholar]
- 14. Tissenbaum H.A., Guarente L.. Model organisms as a guide to mammalian aging. Dev. Cell. 2002; 2:9–19. [DOI] [PubMed] [Google Scholar]
- 15. Spivey E.C., Jones S.K. Jr, Rybarski J.R., Saifuddin F.A., Finkelstein I.J.. An aging-independent replicative lifespan in a symmetrically dividing eukaryote. Elife. 2017; 6:e20340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elliott S.G., McLaughlin C.S.. Regulation of RNA synthesis in yeast III. Mol. Gen. Genet. MGG. 1979; 169:237–243. [DOI] [PubMed] [Google Scholar]
- 17. Elliott S.G. The yeast cell cycle: coordination of growth and division rates. Prog. Nucleic Acid Res. Mol. Biol. 1983; 28:143–176. [DOI] [PubMed] [Google Scholar]
- 18. Fraser R.S.S., Carter B.L.A.. Synthesis of polyadenylated messenger RNA during the cell cycle of Saccharomyces cerevisiae. J. Mol. Biol. 1976; 104:223–242. [DOI] [PubMed] [Google Scholar]
- 19. Sipiczki M. Where does fission yeast sit on the tree of life. Genome Biol. 2000; 1, doi:10.1186/gb-2000-1-2-reviews1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nurse P., Thuriaux P., Nasmyth K.. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. MGG. 1976; 146:167–178. [DOI] [PubMed] [Google Scholar]
- 21. Dujon B. Yeast evolutionary genomics. Nat. Rev. Genet. 2010; 11:512–524. [DOI] [PubMed] [Google Scholar]
- 22. Booth G.T., Wang I.X., Cheung V.G., Lis J.T.. Divergence of a conserved elongation factor and transcription regulation in budding and fission yeast. Genome Res. 2016; 26:799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jorgensen P., Nishikawa J.L., Breitkreutz B.-J., Tyers M.. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002; 297:395–400. [DOI] [PubMed] [Google Scholar]
- 24. Aldea M., Jenkins K., Csikász-Nagy A.. Growth rate as a direct regulator of the start network to set cell size. Front. Cell Dev. Biol. 2017; 5:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tyson C.B., Lord P.G.. Dependency of size of Saccharomyces cerevisiae cells on growth rate. J. Bacteriol. 1979; 138:92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klis F.M., de Koster C.G., Brul S.. Cell Wall-Related Bionumbers and Bioestimates of Saccharomyces cerevisiae and Candida albicans. Eukaryot. Cell. 2014; 13:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Slavov N., Budnik B.A., Schwab D., Airoldi E.M., van Oudenaarden A.. Constant growth rate can be supported by decreasing energy flux and increasing aerobic glycolysis. Cell Rep. 2014; 7:705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ferrezuelo F., Colomina N., Palmisano A., Garí E., Gallego C., Csikász-Nagy A., Aldea M.. The critical size is set at a single-cell level by growth rate to attain homeostasis and adaptation. Nat. Commun. 2012; 3:1012. [DOI] [PubMed] [Google Scholar]
- 29. Rose M., Winston F., Hieter P.. Methods in Yeast Genetics — A Laboratory Course Manual. 1990; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 30. Winey M., Goetsch L., Baum P., Byers B.. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol. 1991; 114:745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garrido-Godino A.I., García-López M.C., García-Martínez J., Pelechano V., Medina D.A., Pérez-Ortín J.E., Navarro F.. Rpb1 foot mutations demonstrate a major role of Rpb4 in mRNA stability during stress situations in yeast. Biochim. Biophys. Acta - Gene Regul. Mech. 2016; 1859:731–743. [DOI] [PubMed] [Google Scholar]
- 32. Benet M., Miguel A., Carrasco F., Li T., Planells J., Alepuz P., Tordera V., Pérez-Ortín J.E.. Modulation of protein synthesis and degradation maintains proteostasis during yeast growth at different temperatures. Biochim. Biophys. Acta - Gene Regul. Mech. 2017; 1860:794–802. [DOI] [PubMed] [Google Scholar]
- 33. García-Martínez J., Aranda A., Pérez-Ortín J.E.. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell. 2004; 15:303–313. [DOI] [PubMed] [Google Scholar]
- 34. García-Martínez J., Pelechano V., Pérez-Ortín J.E.. Becskei A. Genomic-Wide Methods to Evaluate Transcription Rates in Yeast BT - Yeast Genetic Networks: Methods and Protocols. 2011; Totowa: Humana Press; 25–44. [DOI] [PubMed] [Google Scholar]
- 35. Sun M., Schwalb B., Pirkl N., Maier K.C., Schenk A., Failmezger H., Tresch A., Cramer P.. Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol. Cell. 2013; 52:52–62. [DOI] [PubMed] [Google Scholar]
- 36. Eser P., Demel C., Maier K.C., Schwalb B., Pirkl N., Martin D.E., Cramer P., Tresch A.. Periodic mRNA synthesis and degradation co‐operate during cell cycle gene expression. Mol. Syst. Biol. 2014; 10:717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. García-Martínez J., Delgado-Ramos L., Ayala G., Pelechano V., Medina D.A., Carrasco F., González R., Andrés-León E., Steinmetz L., Warringer J. et al. . The cellular growth rate controls overall mRNA turnover, and modulates either transcription or degradation rates of particular gene regulons. Nucleic Acids Res. 2016; 44:3643–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976; 72:248–254. [DOI] [PubMed] [Google Scholar]
- 39. Truong S.K., McCormick R.F., Polymenis M.. Genetic determinants of cell size at birth and their impact on cell cycle progression in Saccharomyces cerevisiae. G3 Genes|Genomes|Genetics. 2013; 3:1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. R Development Core Team R: A Language and Environment for Statistical Computing. 2015.
- 41. Tusher V.G., Tibshirani R., Chu G.. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. García-Martínez J., Troulé K., Chávez S., Pérez-Ortín J.E.. Growth rate controls mRNA turnover in steady and non-steady states. RNA Biol. 2016; 13:1175–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Storchova Z., Breneman A., Cande J., Dunn J., Burbank K., O’Toole E., Pellman D.. Genome-wide genetic analysis of polyploidy in yeast. Nature. 2006; 443:541–547. [DOI] [PubMed] [Google Scholar]
- 44. de Godoy L.M.F., Olsen J. V, Cox J., Nielsen M.L., Hubner N.C., Fröhlich F., Walther T.C., Mann M.. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008; 455:1251–1254. [DOI] [PubMed] [Google Scholar]
- 45. Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999; 24:437–440. [DOI] [PubMed] [Google Scholar]
- 46. Schmoller K.M., Skotheim J.M.. The biosynthetic basis of cell size control. Trends Cell Biol. 2015; 25:793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Darzacq X., Powrie E., Gu W., Singer R.H., Zenklusen D.. RNA asymmetric distribution and daughter / mother differentiation in yeast. Curr. Opin. Microbiol. 2003; 6:614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Haimovich G., Medina D.A., Causse S.Z., Garber M., Millán-Zambrano G., Barkai O., Chávez S., Pérez-Ortín J.E., Darzacq X., Choder M.. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013; 153:1000–1011. [DOI] [PubMed] [Google Scholar]
- 49. Collart M.A. The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip. Rev. RNA. 2016; 7:438–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Collart M.A., Reese J.C.. Gene expression as a circular process. RNA Biol. 2014; 11:320–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hose J., Yong C.M., Sardi M., Wang Z., Newton M.A., Gasch A.P.. Dosage compensation can buffer copy-number variation in wild yeast. Elife. 2015; 4:e05462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Torres E.M., Springer M., Amon A.. No current evidence for widespread dosage compensation in S. cerevisiae. Elife. 2016; 5:e10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gasch A.P., Hose J., Newton M.A., Sardi M., Yong M., Wang Z.. Further support for aneuploidy tolerance in wild yeast and effects of dosage compensation on gene copy-number evolution. Elife. 2016; 5:e14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Voichek Y., Bar-Ziv R., Barkai N.. Expression homeostasis during DNA replication. Science. 2016; 351:1087–1090. [DOI] [PubMed] [Google Scholar]
- 55. Torreira E., Louro J.A., Pazos I., González-Polo N., Gil-Carton D., Duran A.G., Tosi S., Gallego O., Calvo O., Fernández-Tornero C.. The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription. Elife. 2017; 6:e20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kobayashi T., Sasaki M.. Ribosomal DNA stability is supported by many ‘buffer genes’—introduction to the Yeast rDNA Stability Database. FEMS Yeast Res. 2017; 17:fox001. [DOI] [PubMed] [Google Scholar]
- 57. Dammann R., Lucchini R., Koller T., Sogo J.M.. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993; 21:2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang S., Mercado-Uribe I., Xing Z., Sun B., Kuang J., Liu J.. Generation of cancer stem-like cells through the formation of polyploid giant cancer cells. Oncogene. 2014; 33:116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.