SUMMARY
Mitogen-activated protein kinase cascades are important signaling modules that convert environmental stimuli into cellular responses. We show that MPK3, MPK4, and MPK6 are rapidly activated after cold treatment. The mpk3 and mpk6 mutants display increased expression of CBF genes and enhanced freezing tolerance, whereas constitutive activation of the MKK4/5-MPK3/6 cascade in plants causes reduced expression of CBF genes and hypersensitivity to freezing, suggesting that the MKK4/5-MPK3/6 cascade negatively regulates the cold response. MPK3 and MPK6 can phosphorylate ICE1, a bHLH transcription factor that regulates the expression of CBF genes, and the phosphorylation promotes the degradation of ICE1. Interestingly, the MEKK1-MKK2-MPK4 pathway constitutively suppresses MPK3 and MPK6 activities, and has a positive role in the cold response. Furthermore, the MAPKKK YDA, and two calcium/calmodulin-regulated receptor-like kinases, CRLK1 and CRLK2, negatively modulate the cold-activation of MPK3/6. Our results uncover important roles of MAPK cascades in the regulation of plant cold response.
Keywords: Freezing tolerance, Cold acclimation, ICE1, CRLKs, CBFs, MAPK cascades
In Brief
ICE1 is a central regulator of the plant cold response and its levels are tightly controlled. Zhao et al. show that cold-activated MPK3 and MPK6 phosphorylate ICE1 and promote its degradation, thus negatively regulating the cold response, whereas MPK4 positively regulates the cold response by constitutively suppressing MPK3 and MPK6 activity.
Introduction
Cold stress is an important factor that affects the geographical distribution and growth of plants. Cold temperatures can cause damages to plants, particularly when a sudden frost occurs, causing reduced crop productivity and quality (Chinnusamy et al., 2007; Thomashow, 1999). Cold acclimation is a phenomenon wherein plants increase their freezing tolerance after being exposed to low nonfreezing temperatures (Thomashow, 1999). During cold acclimation, the physiological and metabolic status of plants is altered, which is accompanied by changes in the expression of thousands of genes (Chinnusamy et al., 2007; Medina et al., 2011). The three CBF genes, which encode AP2/ERF (APETALA2/Ethylene-Responsive Factor)-type transcription factors, play critical roles in cold acclimation by regulating the expression of a subset of cold responsive (COR) genes (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Medina et al., 1999; Stockinger et al., 1997; Zhao et al., 2015). Arabidopsis plants with a knockout of all three CBF genes show a dramatic reduction in the capacity to cold acclimate and are extremely sensitive to freezing (Jia et al., 2016; Zhao and Zhu, 2016; Zhao et al., 2016).
The dominant ice1 mutant (ice1-1) was identified as a result of its reduced cold-induction of the CBF3-LUC transgene (firefly luciferase transgene driven by the CBF3 promoter) (Chinnusamy et al., 2003). In ice1-1 mutant plants, cold-induced expression of CBF genes and their downstream cold-responsive (COR) genes is reduced compared with the wild-type. As a result, the ice1-1 mutant displays hypersensitivity to freezing conditions. ICE1 encodes a MYC-like bHLH transcription factor that can bind to the MYC-recognition motif in the CBF3 promoter (Chinnusamy et al., 2003; Kim et al., 2015). ICE2 is a paralog of ICE1 in Arabidopsis, and the two proteins share 61% identity. Knock out of each gene does not cause obvious morphological phenotypes, but ice1-2 ice2-2 double mutants display a dwarf phenotype (Kim et al., 2015), suggesting that ICE1 and ICE2 are redundant.
As ICE1 is a key component in the cold response pathway, its regulation is of significant interests. HOS1 is one of the regulators that modulate the abundance of ICE1. HOS1 encodes a RING finger ubiquitin E3 ligase that interacts with ICE1 and promotes the degradation of ICE1 (Lee et al., 2001; Dong et al., 2006). In hos1 mutant plants, cold-induced degradation of ICE1 is impaired, whereas overexpression of HOS1 promotes the degradation of ICE1 under both normal and low temperatures (Dong et al., 2006). SIZ1, encoding a SUMO E3 ligase, facilitates conjugation of SUMO to ICE1. Sumoylation of ICE1 may promote ICE1 protein stability and thus leads to increased CBF gene expression and enhanced freezing tolerance (Miura et al., 2007). Most recently, OST1, which is a protein kinase in the ABA core signaling pathway, was shown to positively regulate the stability of ICE1 (Ding et al., 2015). The ost1 mutants are sensitive to freezing, whereas transgenic plants overexpressing OST1 exhibit increased freezing tolerance. OST1 enhances the stability of ICE1 by phosphorylating ICE1 at Ser278. Several studies have shown that the protein level of ICE1 is decreased under cold stress (Ding et al., 2015; Dong et al., 2006; Miura et al., 2011), but the signaling event that triggers the degradation of ICE1 remains unclear. One study showed that Ser403 of ICE1 is important for the stability of ICE1 (Miura et al., 2011). Substitution of Ser403 by alanine enhances the transactivation activity of ICE1 and blocks cold-induced degradation of ICE1. Plants overexpressing ICE1 (S403A) show increased freezing tolerance than plants overexpressing wild-type ICE1. The results imply that phosphorylation of ICE1 at Ser403 may trigger the degradation of ICE1.
MKK4/5-MPK3/6 and MEKK1-MKK1/2-MPK4 constitute two well-studied MAPK cascades that are implicated in plant responses to diverse external stimuli, including biotic and abiotic stresses (Meng and Zhang, 2013; Petersen et al., 2000; Pitzschke et al., 2014; Teige et al., 2004). Previous studies suggested that a MEKK1-MKK2-MPK4/MPK6 cascade is involved in the positive regulation of cold response and freezing tolerance (Teige et al., 2004). Low temperatures induce the kinase activity of MEKK1, which subsequently phosphorylates MKK2 (Furuya et al., 2013). Activated MKK2 phosphorylates MPK4 and MPK6 (Teige et al., 2004), which then presumably regulate downstream components to adjust the cellular status to adapt to freezing conditions. The mkk2 mutant was reported to exhibit increased sensitivity to freezing, whereas transgenic plants expressing a constitutively active form of MKK2 display increased tolerance to freezing compared with the wild-type. Furthermore, overexpression of MKK2 results in increased expression of CBF genes, even without cold treatment (Teige et al., 2004). These results indicated that a MKK2-mediated kinase cascade positively regulates cold response. However, the protein substrates of MPK4 and MPK6 in the cold response pathway have not been identified yet, and experimental evidences for the roles of MPK4 and MPK6 and their genetic interaction in freezing tolerance are still missing.
In this study, we performed a phosphoproteomics analysis to identify MAP kinases that are activated in Arabidopsis plants after low temperature treatment. We found that three MAPKs, MPK3, MPK4, and MPK6, are rapidly activated after cold treatment. We discovered that the MKK5-MPK3/MPK6 cascade negatively regulates cold response by promoting the degradation of ICE1 via phosphorylation at Ser94, Thr366, and Ser403. In contrast, the MEKK1-MKK2-MPK4 cascade positively regulates cold response, and constitutively suppresses the protein levels and kinase activities of MPK3 and MPK6. Our results thus illuminate the roles of two MAP kinase cascades in the regulation of cold responsive gene expression and freezing tolerance.
Results
MAPK cascades are activated after low temperature treatment
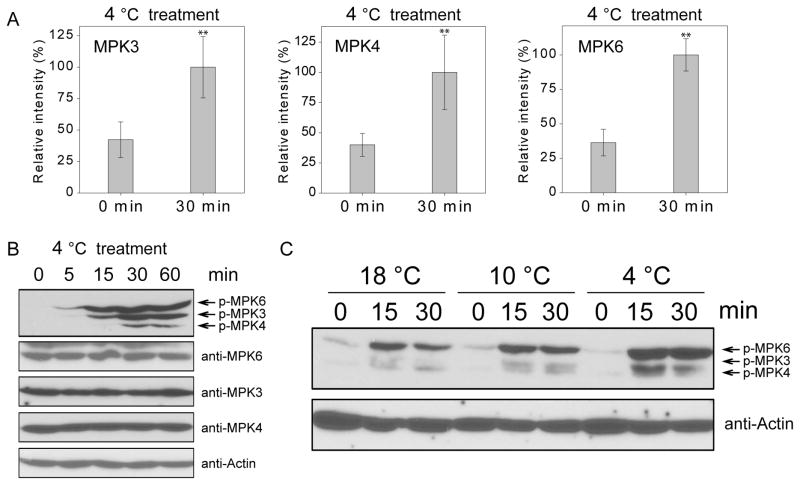
To identify the MAP kinases that are activated after low temperature treatment, we performed quantitative phosphoproteomics analysis of wild-type seedlings before and after treatment at 4°C for 30 min. The phosphoproteomics data revealed that the phosphorylation levels of two MAPKKKs, MEKK1 and YDA, were increased in response to low temperature. Of the 10 MAPKKs, only MKK2 was detected in our assay and its phosphorylation level was increased by 1.78 fold after cold treatment. The phosphorylation of 11 of the 20 MAPKs was detected (Table S1). The phosphorylation levels of MPK3, MPK4, and MPK6 showed higher increases in response to cold treatment than those of other MPKs (Table S1). Under cold treatment, the phosphorylation levels of MPK3, MPK4, and MPK6 were significantly increased (Figure 1A), whereas the phosphorylation of MPK8 was significantly decreased (p-value<0.01) (Table S1). These results are consistent with previous studies (Furuya et al., 2013; Teige et al., 2004), which showed that MEKK1, MKK2, MPK4, and MPK6 are activated after cold treatment. Here, our method detected MPK3 as another MAPK responsive to cold conditions.
Figure 1. MPK3, MPK4, and MPK6 are activated by cold treatment.
(A) Ten-day-old wild-type seedlings were treated at 4°C for 30 min or not treated (0 min). The relative phosphorylation levels of MPK3, MPK4, and MPK6 before and after cold treatment were determined by a quantitative phosphoproteomics assay. Data are means ± SD (n=3). Asterisks indicate statistically significant differences (**P < 0.01, Student’s t-test).
(B) Ten-day-old wild-type seedlings were treated at 4°C for 0, 5, 15, 30, and 60 min. Total proteins were extracted and immunoblotting assays were performed using anti-pTEpY, anti-MPK3, anti-MPK6, anti-MPK4, and anti-Actin.
(C) Ten-day-old wild-type seedlings were treated at 18°C, 10°C, and 4°C for 0, 15, and 30 min. The immunoblotting assays were performed using anti-pTEpY and anti-Actin. See also Figure S2 and Table S1.
To further corroborate that MPK3, MPK4, and MPK6 are activated by low temperature, we subjected 10-day-old seedlings to 4°C for 0, 5, 15, 30, and 60 min and performed immunoblotting using anti-pTEpY, which recognizes the phosphorylated/active form of MPK3, MPK4, and MPK6. The result supports that MPK3, MPK4, and MPK6 are all rapidly activated after low temperature treatment (Figure 1B). To exclude the possibility of elevated protein levels, we conducted immunoblotting assays using anti-MPK3, anti-MPK4, and anti-MPK6 and found that the protein levels of the three MAPKs were not affected by low temperature treatment. To further correlate the MAPK activity with temperature changes, 10-day-old seedlings were treated at 18°C, 10°C, and 4°C, respectively. With the same treatment time, the seedlings treated with lower temperature had higher phosphorylation levels of the MAPKs (Figure 1C), which suggests that the increased phosphorylation level of MAPKs is associated with the degree of temperatures.
MPK3 and MPK6 negatively regulate cold response
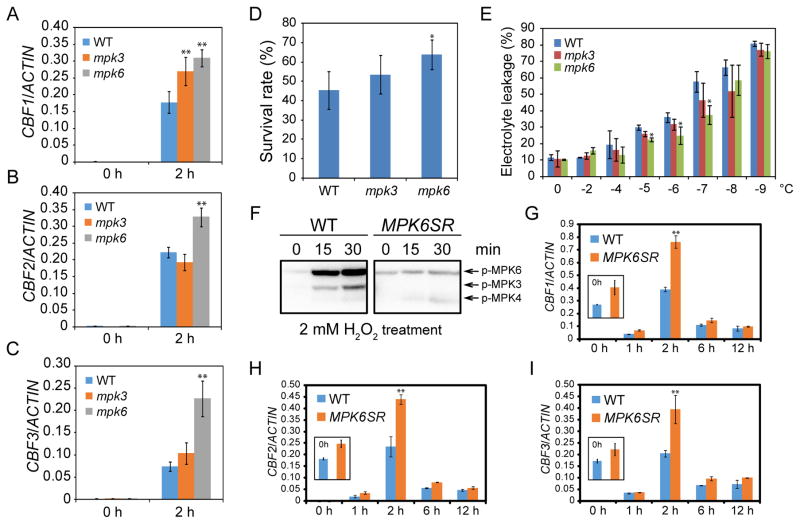
Although previous study suggested that cold-activated MPK6 may promote plant cold tolerance (Teige et al., 2004), a direct experimental evidence for how MPK6 activation may affect cold response was not presented. We evaluated the loss-of-function mutants of mpk3 and mpk6 by first examining the expression level of three CBF genes. All three CBF genes were significantly up-regulated in the mpk6 mutant, and CBF1 was also significantly upregulated in the mpk3 mutant (Figures 2A–2C), suggesting that both MPK3 and MPK6 are negative regulators of cold response. Next, we performed freezing survival assays and the result showed that mpk3 and mpk6 mutants display higher survival rates than the wild-type after freezing (Figure 2D). Electrolyte leakage assays further indicated that freezing caused more membrane damages in the wild-type than in the mpk3 and mpk6 mutants (Figure 2E). All these data support that mpk3 and mpk6 mutants have higher freezing tolerance.
Figure 2. The mpk3 and mpk6 mutants are hypersensitive to freezing.
(A–C) Ten-day-old seedlings of wild-type, mpk3, and mpk6 were treated at 4°C for 2 h. Transcript accumulation of CBF genes was assessed by qRT-PCR, and ACTIN8 was used as the internal control.
(D) Freezing survival assay. Shown are rates of survival of seedlings cold acclimated for 7 days and then treated at −7°C for 1 h.
(E) Electrolyte leakage assay was performed on wild-type, mpk3, and mpk6 plants (three well-developed leaves were used for each sample at each temperature) after cold acclimation (4°C for 7 days).
(F) The seedlings of wild-type and MPK6SR plants were pretreated with NA-PP1 (2 μM) and then treated with H2O2 for 0, 15, and 30 min. The kinase activities of MPK3, MPK4, and MPK6 were examined by using in-gel kinase assay.
(G–I) The seedlings of wild-type and MPK6SR were treated with NA-PP1 (2 μM) for 24 h and then transferred to 4°C for additional 2 h. The transcript accumulation of CBF genes was assessed by qRT-PCR, and ACTIN8 was used as the internal control.
Data are means ± SD (n=3). Asterisks indicate statistically significant differences (*P < 0.05 and **P < 0.01 by Student’s t-test). See also Figure S1.
MPK3 and MPK6 function redundantly in embryogenesis and the mpk3 mpk6 double mutant is embryo lethal. To overcome the developmental defect and to study the phenotype of the double mutant in cold response, we took advantage of a chemically inducible knockout plant, MPK6SR (genotype mpk3 mpk6 PMPK6:MPK6YG) (Xu et al., 2014), in which embryo lethality is rescued by MPK6YG (MPK6 with the Y144G amino acid substitution), the kinase activity of which can be temporally disrupted by a reversible, cell-permeable inhibitor, NA-PP1. To establish the feasibility of using MPK6SR as an inducible mpk3 mpk6 double mutant, we treated 10-day-old seedlings of MPK6SR with NA-PP1 for 24 h, followed by H2O2 treatment for 0, 15, and 30 min. The kinase activities of MPK3, MPK4, and MPK6 were examined by an in-gel kinase assay. Indeed, MPK3, MPK4, and MPK6 in wild-type plants were activated after H2O2 treatment, but in NA-PP1 pretreated MPK6SR plants, MPK4 was activated, but MPK3 and MPK6 could not be activated by H2O2 (Figure 2F). The result suggests that NA-PP1 sufficiently blocks the kinase activity of MPK6YG and that MPK6SR can be used as an inducible mpk3 mpk6 double mutant. We then subjected the NA-PP1-treated MPK6SR seedlings to low temperature and tested the expression level of CBF genes. The results showed that all three CBF genes were up-regulated in MPK6SR compared with the wild-type. Notably, even without cold treatment, the expression of CBF genes was increased in NA-PP1-pretreated MPK6SR (Figures 2G–2I), revealing a tight regulation of the CBF genes by MPK3 and MPK6.
Constitutive activation of MKK5 leads to increased sensitivity to freezing
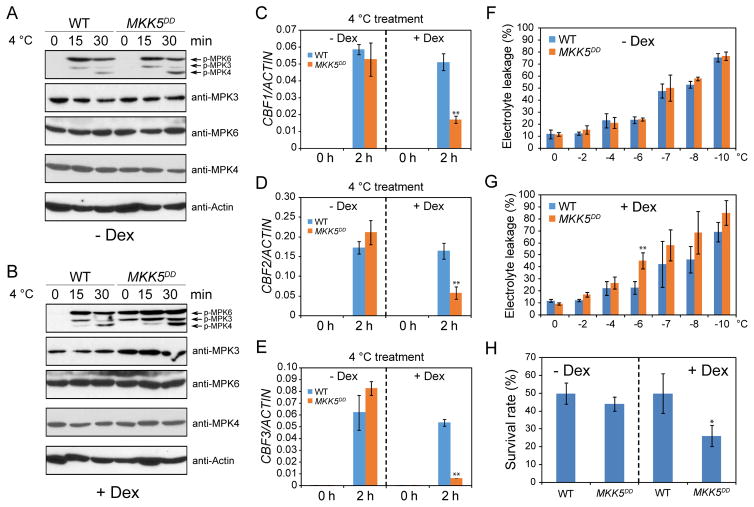
As the null mutants of mpk3 and mpk6 showed increased freezing tolerance, we wondered whether constitutive activation of MPK3 and MPK6 may lead to decreased freezing tolerance. To test this, we used MKK5DD transgenic plants, in which a constitutively active form of MKK5 (T215D/S221D) is driven by a Dexamethasone (Dex)-inducible promoter (Ren et al., 2002). Constitutive activation of MKK5 is known to cause the activation of MPK3 and MPK6 (Ren et al., 2002). Without Dex treatment, the activities and protein levels of MPK3, MPK4, and MPK6 were not changed in MKK5DD seedlings (Figure 3A). When treated with Dex, MPK3 and MPK6 were constitutively activated by the expression of MKK5DD without cold treatment, as anticipated (Figure 3B). After cold treatment, the kinase activities of MPK3 and MPK6 were obviously elevated in Dex-treated MKK5DD than in wild-type plants. In addition, we also noticed that the protein level of MPK3, but not that of MPK4 or MPK6, was increased in Dex-treated MKK5DD plants (Figure 3B).
Figure 3. Constitutive activation of the MKK5-MPK3/6 cascade leads to increased sensitivity to freezing.
(A–B) The seedlings were pre-treated with (B) or without (A) Dex for 6 h before the cold treatment. The immunoblotting assays were performed using anti-pTEpY, anti-MPK3, anti-MPK6, anti-MPK4, and anti-Actin.
(C–E) qRT-PCR analysis of CBF transcript levels. ACTIN8 was used as the internal control.
(F–G) Three-week-old plants of wild-type and MKK5DD pretreated with (G) or without (F) Dex were subjected to cold acclimation for one week and the acclimated plants were used for electrolyte leakage assay.
(H) Freezing survival assay. Shown are rates of survival of seedlings cold acclimated for 7 days and then treated at −7°C for 1 h.
Data in (C)–(H) are means ± SD (n=3). Asterisks indicate statistically significant differences (*P < 0.05 and **P < 0.01 by Student’s t-test). See also Figure S1.
We then evaluate the expression of CBF genes in the MKK5DD transgenic plants. Without Dex treatment, the wild-type and MKK5DD plants showed comparable expression levels of the three CBF genes before and after cold treatment. However, after pre-treatment of seedlings with Dex, the cold-triggered CBF expression was significantly suppressed by the expression of MKK5DD that presumably elevated the activities of MPK3 and MPK6 (Figures 3C–3E). Consistent with the gene expression data, electrolyte leakage and freezing survival assays both showed that Dex-treated MKK5DD plants exhibited higher sensitivity to freezing than the wild-type, but no significant differences were detected for the plants not treated with Dex (Figures 3F–3H). Taken together, these results strongly support that the MKK5-MPK3/6 kinase cascade negatively regulates cold response in plants.
To further establish the negative role of the MKK5-MPK3/6 cascade in cold response, we performed a protoplast assay by using the expression of CBF3-LUC as a reporter. Compared with the expression of CBF3-LUC alone, co-transfection of the transcription activator ICE1 with CBF3-LUC significantly increased the activity of luciferase in the protoplasts (Figure S1A), confirming that ICE1 is an activator of CBF3. However, co-transformation of MPK3 or MPK6 with ICE1 and CBF3-LUC significantly diminished the activity of luciferase, and the addition of MKK5DD to the protoplasts further decreased the luciferase activity (Figure S1A).
In stomatal development, YDA is the upstream MAPKKK that regulates the MKK4/5-MPK3/6 cascade (Wang et al., 2007). To assess whether YDA is also involved in cold response, wild-type and yda mutant plants were treated with 4°C for 0, 15, and 30 min, and the activities of MPK3 and MPK6 were examined by immunoblotting assay. In the yda mutant, cold-induced activation of MPK3/6 was not decreased, but instead increased (Figure S2A). In concert with the increases in MPK3/6 activities, the expression of CBF genes was significantly decreased in the yda mutant (Figures S2B–S2D). These results suggest that YDA is not the upstream MAPKKK that activates the MKK4/5-MPK3/6 cascade, but instead has a strong antagonistic role on the activities of MPK3 and MPK6 under cold conditions.
MPK4 positively regulates cold response
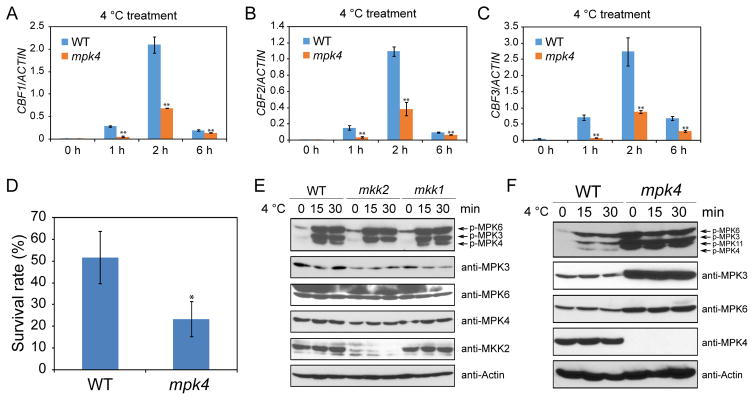
Previous studies showed that the MEKK1-MKK2-MPK4 cascade positively regulates cold response (Furuya et al., 2013; Teige et al., 2004). In this study, we found that indeed the expression levels of CBF genes were substantially down-regulated in mpk4 mutant plants (Figures 4A–4C), and the mpk4 mutant was hypersensitive to freezing (Figure 4D). Similarly, the expression of CBF genes was reduced and plant sensitivity to freezing was increased in mekk1 single and mkk1 mkk2 double mutants (Figures S3A, S3B, S3D, and S3E). MKK1 and MKK2 have redundant functions in plant development and growth (Gao et al., 2008). To assess whether MKK1 and MKK2 are also redundant in the cold response pathway, we tested the expression of CBF genes in mkk1 and mkk2 single mutants. The results showed that CBF genes were not down-regulated, but slightly up-regulated in both mkk1 and mkk2 mutants (Figure S3C). Freezing survival assays also indicated that the mkk1 and mkk2 single mutants do not have less freezing tolerance than the wild-type (Figure S3F), in contrast to a previous report that mkk2 was hypersensitive to freezing (Teige et al., 2004). We found that the cold-induced activation of MPK3 and MPK6 was not affected in either the mkk1 or mkk2 mutant (Figure 4E), suggesting that MKK1 and MKK2 may not be upstream of MPK3 and MPK6 in the cold signaling pathway. The activation of MPK4 was completely abolished in the mkk2 mutant, but was not affected in the mkk1 mutant (Figure 4E). The result shows that MKK2, but not MKK1, is necessary for cold-induced activation of MPK4. Using a MKK2-specific antibody, immunoblotting assays confirmed the absence of MKK2 protein in the mkk2 mutant (Figure 4E).
Figure 4. mpk4 mutant plants are sensitive to freezing.
(A–C) qRT-PCR analysis of CBF transcript levels in 12-day-old seedlings. ACTIN8 was used as the internal control.
(D) Freezing survival assay. Shown are rates of survival of seedlings cold acclimated for 7 days and then treated at −7°C for 1 h.
(E–F) Immunoblotting assays using anti-pTEpY, anti-MPK3, anti-MPK6, anti-MPK4, anti-MKK2, and anti-Actin.
Data in (A)–(D) are means ± SD (n=3). Asterisks indicate statistically significant differences (*P < 0.05, **P < 0.01, Student’s t-test). See also Figure S3.
Although our kinase activity data supported that MKK2 is necessary for cold-induced MPK4 activation, the mkk2 mutant did not show the anticipated phenotypes (Figures S3C and S3F), i.e. reduced CBF expression and increased sensitivity to freezing as observed in mpk4 mutants. To address this paradox, the activation of MPK3/6 was examined in the mpk4 mutant before and after cold treatment. Interestingly, the activities as well as the protein levels of MPK3 and MPK6 were highly elevated in the mpk4 mutant, even without cold treatment. The protein level of the actin control was not affected in mpk4 (Figure 4F). Similarly, the kinase activities of MPK3 and MPK6 were highly increased in mekk1 single and mkk1 mkk2 double mutants even without cold treatment (Figures S3G and S3H). Taken together, the results showed that the mekk1, mkk1 mkk2, and mpk4 mutants, in which MPK3 and MPK6 are constitutively activated, are hypersensitive to freezing, but the mkk2 mutant, in which cold-induced MPK4 activity is blocked while the activities of MPK3 and MPK6 are not affected, shows a wild-type level of freezing tolerance. These results suggest that the constitutive increase in MPK3 and MPK6 kinase activities may be the cause of the freezing-hypersensitive phenotypes of mekk1, mkk1 mkk2, and mpk4 mutants.
MPK3 and MPK6 can phosphorylate ICE1
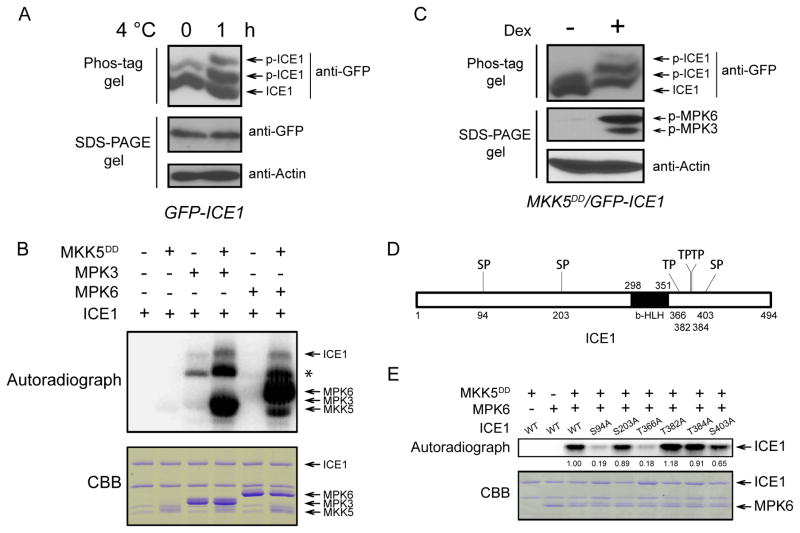
Phos-tag SDS-PAGE and phosphoproteomics assays showed that the phosphorylation of ICE1 was enhanced in vivo after cold treatment (Figures 5A and S4A) and the phosphorylation site of Ser403 was detected (Figure S4B). We tested whether ICE1 may be a substrate of MPK3 and MPK6. In the presence of MKK5DD, MPK3 and MPK6 were able to phosphorylate ICE1 (Figure 5B). To examine whether MPK4 can phosphorylate ICE1, an immunoprecipitation (IP)-kinase assay was performed. We treated wild-type seedlings with or without low temperature for 1 h and then performed IP using anti-MPK4. The immunoprecipitated MPK4 was incubated with ICE1 or the MBP control. We found that MBP but not ICE1 was phosphorylated (Figure S5). These results show that MPK3 and MPK6, but not MPK4, can phosphorylate ICE1. We crossed 35S::GFP-ICE1 transgenic plants with Dex-inducible MKK5DD plants to generate MKK5DD/35S::GFP-ICE1 plants. After Dex treatment, the MPK3 and MPK6 were activated, and we found that the ICE1 protein was phosphorylated (Figure 5C). These results suggest that the MKK5-MPK3/6 pathway is involved in cold-induced phosphorylation of ICE1 in vivo.
Figure 5. MPK3 and MPK6 can phosphorylate ICE1.
(A) Cold stress induces ICE1 phosphorylation in vivo. The GFP-ICE1 transgenic plants were treated with low temperature for 0 and 1 h. The phosphorylation of ICE1 was examined by using Phos-tag SDS-PAGE assay (top panel). The proteins of ICE1 (middle panel) and Actin (bottom panel) running in the normal SDS-PAGE gel were also examined.
(B) Phosphorylation of ICE1 by MPK3 and MPK6 in vitro. The purified proteins were mixed in the protein kinase reaction buffer for 30 min at room temperature. The proteins were separated by SDS-PAGE. The autoradiogram (upper panel) and the CBB-stained image (lower panel) of the proteins are shown. Asterisk indicates a possible ICE1 degradation product.
(C) ICE1 phosphorylation accompanies MPK3/6 activation in MKK5DD plants. The MKK5DD/35S::GFP-ICE1 plants were treated with Dex for 6 h. The total proteins were extracted and subjected to Phos-tag SDS-PAGE assay (top panel). The activity of MPK3/6 (middle panel) and the Actin protein (bottom panel) were also examined by normal SDS-PAGE gel.
(D) Diagram of the ICE1 protein with six (S/T)P motifs.
(E) Phosphorylation of wild-type and mutated form of ICE1 by MPK6 in vitro. The autoradiogram (upper panel) and the CBB-stained image (lower panel) of the proteins are shown. The band intensity was qualified using ImageJ software. See also Figures S4–S6.
By using a split luciferase complementation assay, we found that both MPK3 and MPK6 can interact with ICE1, and it appeared that ICE1 had a stronger interaction with MPK6 than with MPK3. Interestingly, the interactions of ICE1 with MPK3 and MPK6 appeared to increase after cold treatment (Figure S6).
ERK MAP kinases specifically recognize and phosphorylate serine or threonine residues followed by a proline (S/TP) (Jacobs et al., 1999). In ICE1, there are six (S/T)P motifs, including S94P, S203P, T366P, T382P, T384P, and S403P (Figure 5D). To determine which Ser or Thr of ICE1 is phosphorylated by MPK6, we mutated each of the Ser or Thr to Ala. The mutation of Ser94, Thr366, and Ser403 to Ala led to a reduction of ICE1 phosphorylation by MPK6 (Figure 5E), suggesting that these three residues are the potential sites of phosphorylation by MPK3/6.
Phosphorylation of ICE1 by MPK3/6 promotes the degradation of ICE1
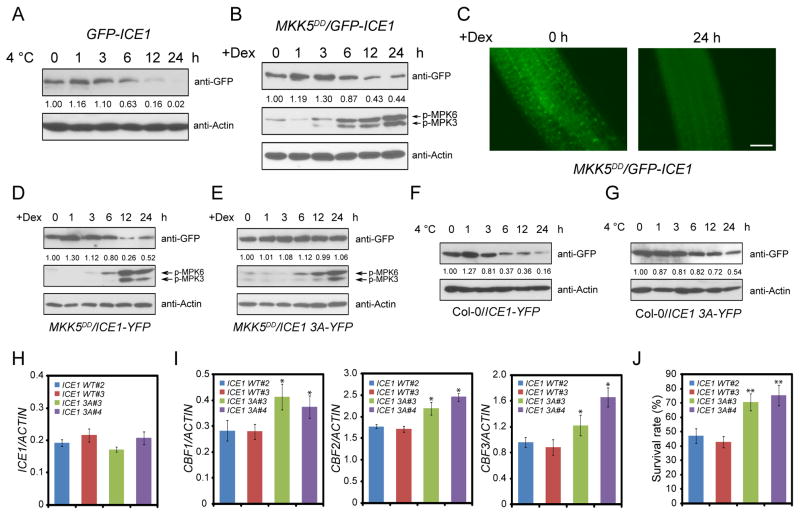
Previous studies showed that low temperature treatments trigger the degradation of ICE1 (Ding et al., 2015; Dong et al., 2006), which is verified by our immunoblotting assay (Figure 6A). We tested whether MPK6 may be involved in the regulation of ICE1 protein stability. We hypothesized that, when MPK6 is constitutively activated in plants, ICE1 would be degraded independent of cold treatment. To test this hypothesis, the MKK5DD/35S::GFP-ICE1 seedlings were treated with Dex for 0, 1, 3, 6, 12, and 24 h and immunoblotting was performed using anti-GFP and anti-pTEpY. MPK3 and MPK6 were activated to a high level at 6 h after Dex treatment. Coincidentally, ICE1 protein level was decreased after Dex treatment for 6 h (Figure 6B). The pretreatment of MKK5DD/35S::GFP-ICE1 seedlings with MG132 could slow down the degradation of ICE1 (Figure S7A), suggesting that MPK3/6-induced degradation of ICE1 is mediated by 26S proteasome pathway. Using fluorescence microscopy, we examined GFP signals in the roots of MKK5DD/35S::GFP-ICE1 before and after Dex treatment. Consistent with the immunoblotting result, the GFP signal intensity for ICE1 was decreased after Dex treatment for 24 h (Figure 6C). These results suggest that the activation of the MKK5-MPK3/6 cascade is sufficient for triggering ICE1 degradation.
Figure 6. The MKK5-MPK3/6 cascade promotes ICE1 degradation.
(A) The 35S::GFP-ICE1 transgenic plants were treated at 4°C for 0, 1, 3, 6, 12, and 24 h. The immunoblotting assays were performed using anti-GFP and anti-Actin.
(B) The MKK5DD/35S::GFP-ICE1 plants were treated with Dex for 0, 1, 3, 6, 12, and 24 h. The immunoblotting assays were performed using anti-GFP, anti-pTEpY, and anti-Actin.
(C) The seedlings of MKK5DD/35S::GFP-ICE1 were treated for 24 h with (24 h) or without (0 h) Dex. The fluorescence signal of GFP-ICE1 was detected using fluorescence microscopy. Totally 15 seedlings were examined for each treatment, and three independent experiments were performed with similar results. Shown are representative results. Bars = 50 μm.
(D–E) Native promoter-driven wild-type ICE1 and ICE1 S94A T366A S403A (ICE1 3A) were transformed to MKK5DD plants. The seedlings of MKK5DD/ICE1 pro::ICE1-YFP (D) and MKK5DD/ICE1 pro::ICE1 3A-YFP (E) transgenic plants were treated with Dex, and the immunoblotting assays were performed using anti-GFP, anti-pTEpY, and anti-Actin.
(F–G) The seedlings of Col-0/ICE1 pro::ICE1-YFP (F) and Col-0/ICE1 pro::ICE1 3A-YFP (G) were cold treated and subjected to immunoblotting assays using anti-GFP and anti-Actin. The band intensity was qualified using ImageJ software. The above experiments were repeated three times with similar results.
(H) The expression of ICE1 in Col-0/ICE1 pro::ICE1-YFP and Col-0/ICE1 pro::ICE1 3A-YFP transgenic plants was analyzed by qRT-PCR.
(I) The seedlings of Col-0/ICE1 pro::ICE1-YFP and Col-0/ICE1 pro::ICE1 3A-YFP transgenic plants were treated with low temperature for 2 h and the expression of the three CBF genes was assessed. ACTIN8 was used as the internal control.
(J) The survival rates of freezing-treated seedlings.
Data in (H)–(J) are means ± SD (n=3). Asterisks indicate statistically significant differences (*P < 0.05, **P < 0.01, Student’s t-test). See also Figure S7.
Given that the MKK5-MPK3/6 cascade negatively regulates cold response by affecting the protein stability of ICE1, we speculated that the decreased expression of CBF genes in Dex-treated MKK5DD plants may be caused by the decreased protein level of ICE1. We tested whether the defects in cold-induced expression of CBF genes in MKK5DD plants could be rescued by increased expression of ICE1. The wild-type, MKK5DD, and MKK5DD/ICE1 pro::ICE1-YFP plants were treated with Dex for 9 h and then subjected to cold treatment. The results show that the reduced expression of CBF genes in MKK5DD was indeed recovered in MKK5DD/ICE1 pro::ICE1-YFP plants (Figures S1B–S1E), supporting the genetic position of ICE1 between MKK5-MPK3/6 and CBF genes.
To investigate whether the Ser94, Thr366, and Ser403 residues of ICE1 are required for the regulation of the protein stability of ICE1, we transformed wild-type ICE1 and ICE1 3A (all three phosphor-sites mutated to Ala) driven by the native promoter of ICE1 to the MKK5DD plants to generate MKK5DD/ICE1 pro::ICE1-YFP and MKK5DD/ICE1 pro::ICE1 3A-YFP transgenic plants. The wild-type ICE1-YFP was degraded after Dex treatment in MKK5DD/ICE1 pro::ICE1-YFP seedlings (Figure 6D). However, in MKK5DD/ICE1 pro::ICE1 3A-YFP seedlings, the protein level of ICE1 3A-YFP was not decreased after Dex treatment (Figure 6E). In addition, treatment of Col-0/ICE1 pro::ICE1-YFP transgenic seedlings with low temperature led to the degradation of wild-type ICE1-YFP, whereas cold-induced degradation was delayed for ICE1 3A-YFP (Figures 6F and 6G). We also characterized the impact of the single phosphor-site mutations to protein stability of ICE1 and found that each of the three mutations could enhance the protein stability of ICE1 (Figures S7B–S7F). This result indicates that all of the three phosphorylation sites of ICE1 participate in the regulation of ICE1 stability. Consistent with the protein level of ICE1, the transgenic plants expressing ICE1 3A had higher CBF gene expression levels than the plants expressing wild-type ICE1 (Figures 6H and 6I), and also showed higher freezing tolerance (Figure 6J).
CRLK1 and CRLK2 suppress cold-induced activation of MPK3/6 and are essential for ICE1 protein accumulation
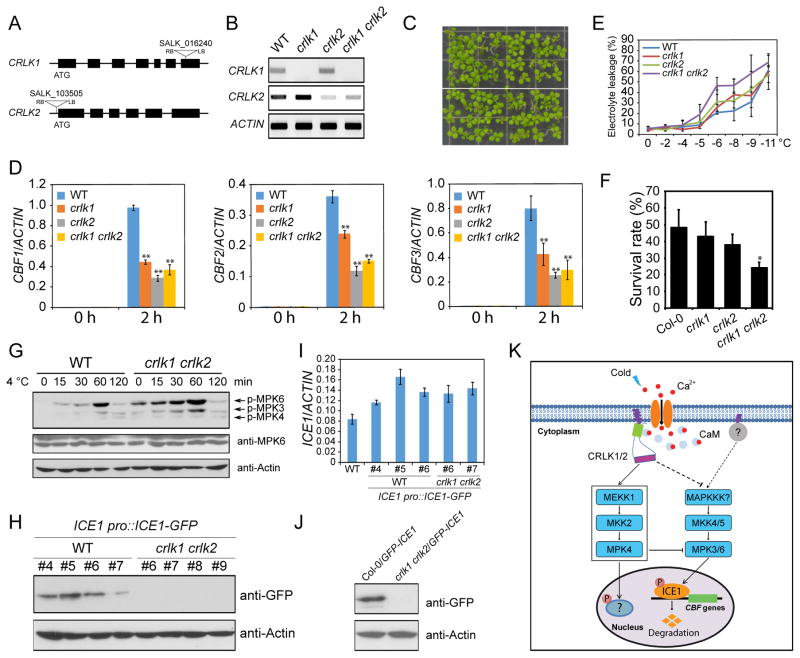
Previous studies showed that CRLK1, a calcium/calmodulin-regulated receptor-like kinase, regulates cold-responsive gene expression and freezing tolerance (Yang et al., 2010a), likely through direct interaction and phosphorylation of MEKK1 (Furuya et al., 2013; Yang et al., 2010b). CRLK2 is a paralog of CRLK1 in Arabidopsis. To further understand the role of CRLK1 and CRLK2 in cold response, we generated a crlk1 crlk2 double mutant, in which CRLK1 expression is abolished and CRLK2 expression is highly decreased (Figures 7A and 7B). Compared with wild-type plants, the crlk1 crlk2 double mutant did not show obvious morphological defects (Figure 7C). Analysis of cold-induced expression of CBFs in crlk1, crlk2 and crlk1 crlk2 mutants showed that CRLK1 and CRLK2 both contribute to the cold-induced expression of CBF genes (Figure 7D). Electrolyte leakage and freezing survival assays showed that crlk1, crlk2, and crlk1 crlk2 mutants were more sensitive to freezing than wild-type plants (Figures 7E and 7F).
Figure 7. CRLK1 and CRLK2 negatively regulate the kinase activities of MPK3 and MPK6.
(A) Diagram of CRLK1 and CRLK2 gene structures. The triangles represent the T-DNA insertion sites. LB and RB represent the left and right borders of the T-DNA.
(B) The expression of CRLK1 and CRLK2 was evaluated by semi-quantitative RT-PCR. The ACTIN gene was used as control.
(C) The phenotype of seedlings grown on MS medium for 10 days.
(D) The seedlings were treated at 4°C for 0 and 2 h. The transcript accumulation of CBF genes was assessed by qRT-PCR, and ACTIN8 was used as the internal control.
(E) Electrolyte leakage assay on plants after cold acclimation (4°C for 7 days).
(F) The survival rates of freezing-treated seedlings.
(G) The seedlings were treated at 4°C for 0, 15, 30, 60, and 120 min. The immunoblotting assays were performed using anti-pTEpY, anti-MPK6, and anti-Actin.
(H) Native promoter-driven ICE1-GFP was transformed to wild-type and crlk1 crlk2 double mutant. Four independent transgenic lines from both wild-type and crlk1 crlk2 double mutant were selected for the analysis of the protein level of ICE1. The immunoblotting assays were performed using anti-GFP and anti-Actin.
(I) Transcript level of ICE1 in the transgenic plants of WT/ICE1 pro::ICE1-GFP and crlk1 crlk2/ICE1 pro::ICE1-GFP was assessed by qRT-PCR, and ACTIN8 was used as the internal control.
(J) The Col-0/35S::GFP-ICE1 transgenic plants were crossed with crlk1 crlk2 double mutant to generate crlk1 crlk2/35S::GFP-ICE1 plants. The protein level of ICE1 in Col-0/35S::GFP-ICE1 and crlk1 crlk2/35S::GFP-ICE1 was examined using immunoblotting assay.
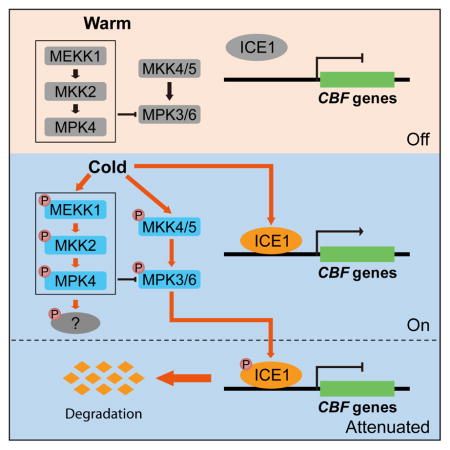
(K) A working model showing the roles of MAP kinase cascades in cold stress signaling. Both the MEKK1-MKK2-MPK4 and MKK4/5-MPK3/6 cascades are rapidly activated under cold stress. The activated MPK3 and MPK6 phosphorylate ICE1 and promote ICE1 degradation, thus attenuating CBF gene expression. The MAPKKK for the cold activation of the MKK4/5-MPK3/6 cascade is unknown. The MEKK1-MKK2-MPK4 cascade positively affects cold response, and constitutively suppresses the activity of MPK3/6. The substrates of MPK4 in the cold signaling pathway is unclear. Cold elicits a calcium signal, and two calcium/calmodulin-regulated receptor-like kinases (CRLK1 and CRLK2), and another membrane associated protein (marked as“?”), are shown as upstream positive or negative regulators of the MAPK cascades.
Data in (D), (E), (F), and (I) are means ± SD (n=3). Asterisks indicate statistically significant differences (*P < 0.05, **P < 0.01, Student’s t-test).
Immunoblotting assays showed that the activation of MPK3 and MPK6 was higher in the crlk1 crlk2 double mutant than in wild-type plants, both before and after cold treatment (Figure 7G), suggesting that CRLK1 and CRLK2 negatively affect the kinase activity of MPK3/6. Since MPK3/6 kinase activity is important for the regulation of ICE1 stability, we examined the protein level of ICE1 in crlk1 crlk2 double mutant plants. A native promoter-driven ICE1-GFP construct was transformed, and four independent transgenic lines from wild-type and crlk1 crlk2 double mutant were selected to test for the protein level of ICE1. By immunoblotting, ICE1-GFP protein was detected in the wild-type, but not in the crlk1 crlk2 double mutant background (Figure 7H). The ICE1 transcript was detected at similar levels in the wild-type and crlk1 crlk2 double mutant transgenic lines (Figure 7I). These results suggest that CRLK1 and CRLK2 are required for maintaining the protein stability of ICE1 under normal conditions. To substantiate this conclusion, we also crossed the crlk1 crlk2 double mutant with Col-0/35S::GFP-ICE1 line to generate crlk1 crlk2/35S::GFP-ICE1. Similarly, the GFP-ICE1 protein could only be detected in Col-0/35S::GFP-ICE1 plants, but not in crlk1 crlk2/35S::GFP-ICE1 plants (Figure 7J).
Discussion
How plants sense cold stress and acclimate to freezing conditions has been studied extensively in the last several decades. An important advance is the discovery of the ICE/CAMTA-CBF-COR gene expression cascade, which is critical for cold acclimation (Chinnusamy et al., 2003; Doherty et al., 2009; Jaglo-Ottosen et al., 1998; Kim et al., 2015; Stockinger et al., 1997; Zhao et al., 2016). Recently, COLD1 with a role in the generation or modulation of cold-induced calcium signal was reported as a cold sensor that is essential for chilling tolerance in rice (Ma et al., 2015). Other components involved in the positive or negative regulation of cold response include HOS1, SIZ1, OST1, MYB15, and CRLK1 (Agarwal et al., 2006; Ding et al., 2015; Dong et al., 2006; Miura et al., 2007; Yang et al., 2010a). ICE1, as a transcriptional activator of CBF genes, is mainly regulated at the posttranslational level. Several studies have shown that the protein level of ICE1 is decreased after cold treatment (Ding et al., 2015; Dong et al., 2006; Miura et al., 2011), but the signal that initiates the degradation of ICE1 is still unknown. Our work here suggests that the MKK5-MPK3/6 pathway is required for the phosphorylation of ICE1, which subsequently promotes the degradation of ICE1 under cold stress (Figure 7K).
Using a phosphoproteomics approach, we identified MAPKs that are potentially involved in cold response. Consistent with previous observations (Teige et al., 2004), MPK4 and MPK6 are activated after cold treatment. Our data showed that MPK3 is also activated in response to cold treatment. Contrary to the previous study (Teige et al., 2004), our results showed the cold-induced activation of MPK6 was not affected in the mkk2 mutant, suggesting that MKK2 is not the upstream MAP kinase kinase of MPK6 in the cold signaling pathway. In addition, although YDA is reported as an upstream component of MKK4/5-MPK3/6 in the regulatory pathway for stomatal development (Wang et al., 2007), our data showed that cold-induced activation of MPK3/6 was not decreased, but rather increased in yda mutant, suggesting that YDA may have opposite roles in the regulation of MPK3/6 kinase activity in stomatal development and cold signaling. Unlike MPK4, which negatively regulates the activity of MPK3/6 in a constitutive manner, YDA affects the activity of MPK3/6 mainly under cold stress, implicating that a YDA-mediated pathway is important for the negative regulation of MPK3/6 activity under cold conditions. It is unclear how YDA negatively regulates MPK3 and MPK6 activities in cold signaling. It is possible that YDA may somehow affect the cold-induced phosphorylation of the as yet unknown MAPKKK that is required for the activation of the MKK4/5-MPK3/6 cascade.
There is ample evidence showing that the MKK4/5-MPK3/6 module positively regulates defense response (Asai et al., 2002; Beckers et al., 2009; Meng and Zhang, 2013), whereas the MEKK1-MKK1/2-MPK4 cascade negatively regulates plant immunity (Gao et al., 2008; Petersen et al., 2000). Our study here showed that these two MAPK cascades also have opposite effects on the cold response. Many external stimuli, such as pathogens, elicitor-active epitope flg22, and cold, can all induce the activation of both MAPK cascades (Han et al., 2010; Mao et al., 2011; Roux et al., 2011; Teige et al., 2004). The mechanism underlining the antagonistic effects and simultaneous activation of these two MAPK cascades is largely unknown. Our study showed that the protein levels of MPK3 and MPK6 are increased and the kinase activities of MPK3 and MPK6 are constitutively activated in mekk1, mkk1 mkk2, and mpk4 mutants compared with wild-type plants, suggesting that the MEKK1-MKK1/2-MPK4 cascade somehow suppresses the activities and protein levels of MPK3 and MPK6, even at warm temperatures.
MKK1 and MKK2 are redundant in the regulation of plant development and flg22-mediated activation of MPK4 (Gao et al., 2008). However, our results showed that cold-induced activation of MPK4 was completely blocked in the mkk2 single mutant, but not affected in the mkk1 mutant, suggesting that MKK1 and MKK2 have different functions in cold signaling. Our phosphoproteomics data indicated that Ser65 of MKK2 was phosphorylated after cold treatment. Alignment of MKK1 and MKK2 proteins showed that Ser65 does not exist in MKK1, which may explain why MKK2, but not MKK1, is activated after cold treatment. Although the cold-induced MPK4 activation was blocked in the mkk2 mutant, mpk4 but not the mkk2 mutant is hypersensitive to freezing. The results suggest that inactivation of MPK4 alone is not the main reason leading to increased sensitivity to freezing in mekk1, mkk1 mkk2, and mpk4 mutants. While the freezing-sensitive phenotype of mpk4 mutants may be caused by the constitutive activation of MPK3 and MPK6 before cold treatment, the role of cold-activated MPK4 remains unknown.
The ICE1 protein is constitutively present in the nucleus, but the CBF genes are only up-regulated under cold stress. Thus far, how ICE1 is activated to induce the expression of CBF genes under cold stress is unknown. It is likely that posttranslational modification of ICE1 is involved in this process. We initially hypothesized that MPK3/6-mediated phosphorylation of ICE1 may be required for the activation of ICE1. However, constitutive activation of the MKK5-MPK3/6 cascade by using Dex-treated MKK5DD plants did not induce the expression of CBF genes without cold treatment, which suggests that MPK3/6-mediated phosphorylation of ICE1 is not required for the induction of the transcriptional activity of ICE1.
Instead, our results showed that MPK3/6-mediated phosphorylation promotes the degradation of ICE1 and the Ser94, Thr366, and Ser403 sites are critical for the phosphorylation-dependent degradation. Transgenic plants expressing ICE1 3A showed significant improvements in freezing tolerance, which can be accounted for by their increased expression of CBF genes and perhaps also by CBF-independent effects. The alteration of the protein level of ICE1 is correlated with the expression pattern of CBF genes. Immunoblotting assays showed that the ICE1 protein level began to decrease at 3 h after low temperature treatment, while gene expression analysis showed that the three CBF genes reached peak expression at 2–3 h after the cold stress, and then the expression decreased (Medina et al., 2011). Although the CBF regulon and other cold-activated gene regulons are critical for freezing tolerance, over-stimulation of these regulons may be detrimental to plant growth. The cold-triggered, MPK3/6-dependent phosphorylation and degradation of ICE1 is important for preventing the over-stimulation of the CBF regulon and perhaps also other cold activated gene regulons, which could be of benefit to plants to balance cold response and growth.
In summary, previous studies reported that a MEKK1-MKK2-MPK4/MPK6 pathway is activated after cold treatment (Teige et al., 2004), and CRLK1, which is stimulated by binding to calmodulin and calcium, a second messenger in plants under cold stress, physically associates with and phosphorylates MEKK1 (Furuya et al., 2013; Yang et al., 2010a, 2010b). In this study, we found that MKK2 is the upstream kinase of MPK4 but not MPK3/6 in the cold signaling pathway. The MEKK1-MKK2-MPK4 cascade constitutively suppresses the protein levels and kinase activities of MPK3 and MPK6. Cold-activated MPK3/6 phosphorylates ICE1 and promotes its degradation, which reduces the transcription of CBF genes. CRLK1 and CRLK2 positively regulate cold response, possibly by activating the MEKK1-MKK2-MPK4 pathway and by suppressing the cold-activation of the MKK4/5-MPK3/6 pathway (Figure 7K).
STAR METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-GFP | Sigma-Aldrich | Cat#11814460001 |
| Rabbit polyclonal anti-MPK3 | Sigma-Aldrich | Cat# M8318 |
| Rabbit polyclonal anti-MPK6 | Sigma-Aldrich | Cat# A7104 |
| Rabbit polyclonal anti-MPK4 | Sigma-Aldrich | Cat# A6979 |
| Rabbit polyclonal anti-pTEpY | Cell Signaling Technology | Cat#9101 |
| Mouse monoclonal anti-Actin | Agrisera | Cat#AS10 702 |
| Rabbit polyclonal anti-MKK2 | This study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MG132 | Sigma-Aldrich | Cat#C2211 |
| Phos-tag | Wako | Cat#AAL-107 |
| PerfeCTa SYBR Green Fastmix | Quanta Biosciences | Cat#95072 |
| Dexamethasone (DEX) | Sigma-Aldrich | Cat#D4902 |
| Glu-C | Sigma-Aldrich | Cat#11420399001 |
| Trypsin | Promega | Cat#V5111 |
| Leupeptin | Sigma-Aldrich | Cat#11017101001 |
| Aprotinin | Sigma-Aldrich | Cat#10236624001 |
| Antipain | Sigma-Aldrich | Cat#10791 |
| Phosphatase Inhibitor Cocktail Set II | Calbiochem | Cat#524625 |
| Protector RNase Inhibitor | Sigma-Aldrich | Cat#3335402001 |
| Critical Commercial Assays | ||
| Gateway BP Clonase II Enzyme Mix | Thermo Fisher Scientific | Cat#11789020 |
| Gateway LR Clonase II Enzyme Mix | Thermo Fisher Scientific | Cat#11791020 |
| TURBO DNA-free Kit | Ambion | Cat#AM1907 |
| M-MLV Reverse Transcriptase | Promega | Cat#M1705 |
| Protein G Agarose | Millipore | Cat#16-266 |
| Ni-NTA Agarose | QIAGEN | Cat#30210 |
| Glutathione Sepharose 4B | GE Healthcare | Cat#17-0756-01 |
| E.Z.N.A. Plant RNA Kit | Omega Bio-tek | Cat#R6827 |
| Quick Start Bradford 1x Dye Reagent | BIO-RAD | Cat#5000205 |
| QIAGEN Plasmid Midi Kit | QIAGEN | Cat#12143 |
| PrimeSTAR HS DNA Polymerase | Takara | Cat#R010A |
| Wizard SV Gel and PCR Clean-Up System | Promega | Cat#A9282 |
| E.Z.N.A. Plasmid Mini Kit I | OMEGA | Cat#D6942 |
| Experimental Models: Organisms/Strains | ||
| E. coli BL21 | N/A | |
| Agrobacterium tumefaciens (strain GV3101) | N/A | |
| Arabidopsis thaliana: WT Col-0 | N/A | |
| Arabidopsis thaliana: mpk3-1 | ABRC | SALK_151594 |
| Arabidopsis thaliana: mpk6-3 | ABRC | SALK_127507 |
| Arabidopsis thaliana: mekk1-1 | (Ichimura et al., 2006) | N/A |
| Arabidopsis thaliana: mkk1-2 | (Gao et al., 2008) | N/A |
| Arabidopsis thaliana: mkk2-1 | (Gao et al., 2008) | N/A |
| Arabidopsis thaliana: mkk1-2 mkk2-1 | (Gao et al., 2008) | N/A |
| Arabidopsis thaliana: mpk4-2 | (Xing et al., 2008) | N/A |
| Arabidopsis thaliana: MPK6SR | (Xu et al., 2014) | N/A |
| Arabidopsis thaliana: MKK5DD | (Ren et al., 2002) | N/A |
| Arabidopsis thaliana: crlk1 | (Yang et al., 2010a) | SALK_016240 |
| Arabidopsis thaliana: crlk2 | ABRC | SALK_103505 |
| Arabidopsis thaliana: crlk1 crlk2 | This study | N/A |
| Arabidopsis thaliana: yda | (Wang et al., 2007) | N/A |
| Arabidopsis thaliana: p35S::GFP-ICE1 | (Chinnusamy et al., 2003) | N/A |
| Arabidopsis thaliana: pICE1::ICE1-GFP | This study | N/A |
| Arabidopsis thaliana: pICE1::ICE1-YFP | This study | N/A |
| Arabidopsis thaliana: pICE1::ICE1 S94A-YFP | This study | N/A |
| Arabidopsis thaliana: pICE1::ICE1 T366A-YFP | This study | N/A |
| Arabidopsis thaliana: pICE1::ICE1 S403A-YFP | This study | N/A |
| Arabidopsis thaliana: pICE1::ICE1 S94A T366A S403A-YFP | This study | N/A |
| Oligonucleotides | ||
| Primers for genotyping mutants | This study; Table S2 | N/A |
| Primers for plasmid construction | This study; Table S2 | N/A |
| Primers for qRT-PCR | This study; Table S2 | N/A |
| Recombinant DNA | ||
| pET-28a HIS-MPK3 | This study | N/A |
| pET-28a HIS-MPK6 | This study | N/A |
| pET-28a HIS-MKK5DD | This study | N/A |
| pGEX-4T-1 GST-ICE1 | This study | N/A |
| pGEX-4T-1 GST-ICE1 S94A | This study | N/A |
| pGEX-4T-1 GST-ICE1 S203A | This study | N/A |
| pGEX-4T-1 GST-ICE1 T366A | This study | N/A |
| pGEX-4T-1 GST-ICE1 T382A | This study | N/A |
| pGEX-4T-1 GST-ICE1 T384A | This study | N/A |
| pGEX-4T-1 GST-ICE1 S403A | This study | N/A |
| PMDC107 pICE1::ICE1-GFP | This study | N/A |
| R4pgwb540 pICE1::ICE1-YFP | This study | N/A |
| R4pGWB540 pICE1::ICE1 S94A-YFP | This study | N/A |
| R4pGWB540 pICE1::ICE1 T366A-YFP | This study | N/A |
| R4pGWB540 pICE1::ICE1 S403A-YFP | This study | N/A |
| R4pGWB540 pICE1::ICE1 S94A T366A S403A-YFP | This study | N/A |
| pCambia MPK3-nLUC | This study | N/A |
| pCambia MPK3-cLUC | This study | N/A |
| pCambia MPK6-nLUC | This study | N/A |
| pCambia MPK6-cLUC | This study | N/A |
| pCambia ICE1-nLUC | This study | N/A |
| pCambia ICE1-cLUC | This study | N/A |
| pHBT95 ICE1 | This study | N/A |
| pHBT95 MPK3 | This study | N/A |
| pHBT95 MPK6 | This study | N/A |
| pHBT95 MPK4 | This study | N/A |
| pHBT95 MKK5DD | This study | N/A |
| Software and Algorithms | ||
| ImageJ2 | ImageJ | https://imagej.net/Welcome |
| MaxQuant | (Cox and Mann, 2008) | http://www.biochem.mpg.de/5111795/maxquant |
| SPSS | SPSS | https://www.ibm.com/us-en/marketplace/spss-statistics |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to the Lead Contact, Dr. Jian-Kang Zhu (jkzhu@sibs.ac.cn).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All Arabidopsis plants are in the Columbia background. mpk3-1 (SALK_151594), mpk6-3 (SALK_127507), mekk1-1 (SALK_052557), mkk1-2 (SALK_027645), mkk2-1 (SAIL_511_H01), mkk1-2 mkk2-1, mpk4-2 (SALK_056245), crlk1 (SALK_016240), yda (SALK_105078), 35S::GFP-ICE1, MPK6SR, and MKK5DD plants have been described previously (Beckers et al., 2009; Chinnusamy et al., 2003; Gao et al., 2008; Ichimura et al., 2006; Ren et al., 2002; Wang et al., 2007; Xing et al., 2008; Xu et al., 2014; Yang et al., 2010a). The crlk2 mutant (SALK_103505) was ordered from Arabidopsis Biological Resource Center (ABRC, Ohio State University, Columbus, OH). Transgenic plants expressing pICE1::ICE1-YFP, pICE1::ICE1 S94A-YFP, pICE1::ICE1 T366A-YFP, pICE1::ICE1 S403A-YFP, and pICE1::ICE1 S94A T366A S403A-YFP in wild-type and MKK5DD background were generated in this study. Plants were grown at 23°C with a long-day light cycle (16 h light/8 h dark). The primers used for genotyping are listed in Table S2.
METHOD DETAILS
Construction of Plasmids
To generate ICE1 construct for recombinant protein expression, the CDS sequence of ICE1 was amplified and cloned into vector pGEX-4T-1 by using BamHI and NotI restriction sites. For site-directed mutagenesis of ICE1, the primers with corresponding mutated sites were designed and explored for PCR using pGEX-4T-1-ICE1 plasmid as template. The PCR products were treated with DpnI to digest the parental double-stranded DNA. The digested PCR products were transformed to DH5α competent cells. The site mutation of ICE1 was confirmed by sequencing. For ICE1 construct used for transformation to plants, the ICE1 CDS fragment fused with ICE1 native promoter were cloned into the pENTR vector. The site mutation of ICE1 was also generated by DpnI-mediated site-directed mutagenesis. The wild-type ICE1 and site-mutated ICE1 were finally cloned into R4pGWB540 vector. The generated constructs were transformed to plants by Agrobacterium tumefaciens-mediated transformation.
Recombinant Protein Expression and in vitro Kinase Assay
To express proteins in Escherichia coli, the constructs of pGEX-4T-1-ICE1, pET-28a-MPK3, pET-28a-MPK6, and pET-28a-MKK5DD were transformed to strain BL21. A single clone of each transformant was selected and incubated in 3 ml LB liquid medium with corresponding antibiotics overnight. Transfer 3 ml solution to 300 ml LB medium and incubate for ~3 h until the OD600 reaching to 0.4–0.6. The isopropyl β-D-1-thiogalactopyranoside (0.5 μM of final concentration) was added to the culture to induce protein expression. ICE1 was purified by using glutathione agarose resin. MPK3, MPK6, and MKK5DD were purified by using Ni-NTA agarose.
For in vitro kinase assay, two steps of reactions were processed. In the first step, MKK5DD were incubated with MPK3 or MPK6 in 30 μL of reaction buffer: 25 mM Tris-HCl (pH 7.5), 12 mM MgCl2, 1 mM DTT, and 50 μM ATP. The mixture was kept at room temperature for 30 min. In the second step, 5 μL solution from reaction I was incubated with ICE1 protein in 25 μL of reaction buffer: 25 mM Tris-HCl (pH 7.5), 12 mM MgCl2, 1 mM DTT, 50 μM ATP, and 0.1 mCi [γ-32P]ATP, and allow the reaction for 30 min at room temperature. Reaction was stopped by adding SDS loading buffer. Proteins were separated by SDS-PAGE using a 10% (w/v) acrylamide gel. The phosphorylated proteins were visualized by autoradiography.
Antibody Production
To detect MKK2 protein in Arabidopsis, we produced a MKK2-specific antibody against a synthetic peptide (MEHPFLNKYDYSGINLASY) corresponding to the C terminus of MKK2. The specificity of the anti-MKK2 was tested by using mkk2 mutant.
Protein Extraction and Immunoblotting
For protein extraction, 10-day-old seedlings were collected and ground in liquid nitrogen and the proteins were extracted using the following extraction buffer: 100 mM Tris-HCl pH 7.5, 150 mM NaCl, 5% Glycerol, 1 mM DTT, 1 mM PMSF, 10 μM antipain, 10 μM aprotinin, 10 μM leupeptin, and phosphatase inhibitor cocktail set II. The total extraction was mixed well and centrifuged at 14000 rpm and 4°C for 20 min. The suspension was transferred to a new tube. The concentration of proteins was measured by using Quick Start Bradford Dye Reagent (Bio-Rad) and finally the protein concentration of each sample was adjusted to the same level. For immunoblotting, proteins were separated by SDS-PAGE (10% acrylamide gel) and transferred to supported nitrocellulose transfer membrane (Bio-Rad) by electro-transfer at 20 V for 30 min. The membrane was blocked in TBST buffer containing 5% skim milk powder and further incubated with primary antibody and secondary antibody. Finally the bands were detected using chemiluminescent HRP substrate (Millipore). For Phos-tag SDS-PAGE assay, 25 μM Phos-tag and 50 μM Mn2+ were added in SDS-PAGE gel. Antibodies used for immunoblotting were as follows: anti-GFP (1:2,000, Sigma), anti-MPK3 (1:10,000, Sigma), anti-MPK6 (1:10,000, Sigma), anti-MPK4 (1:10,000, Sigma), anti–Phospho-p44/p42 MAPK (anti-pTEpY) (1:2,000, Cell Signaling Technology), and anti-Actin (1:10,000, Agrisera).
Immunoprecipitation (IP)-kinase Assay
The IP-kinase assay was performed according to the protocol previously (Kong et al., 2012) with some modification. To purify MPK4 proteins, the 10-day-old seedlings of wild-type plants were treated with or without low temperature. The seedlings were collected and ground in liquid nitrogen. The total proteins were extracted by using extraction buffer (100 mM Tris-HCl pH 7.5, 150 mM NaCl, 5% Glycerol, 1 mM DTT, 1 mM PMSF, 10 μM antipain, 10 μM aprotinin, 10 μM leupeptin, and phosphatase inhibitor cocktail set II). For immunoprecipitation, 1 ml protein extraction was incubated with 3 μL anti-MPK4 antibody for 4 h and then the suspension was incubated with 30 μL protein G agarose beads for 2 h to capture the immunocomplex. The mixture was washed 3 times with extraction buffer and one time with kinase reaction buffer. For the kinase assay, 25 μL reaction mix, including 10 μL MPK4, ~ 1 μg ICE1 protein or 0.5 μg MBP, 1 μL ATP (40 μM), 10 μCi [γ-32P] ATP, and 2.5 μL 10× kinase reaction buffer, was incubated at room temperature for 30 min. The reaction was ended by adding SDS loading buffer and the proteins were separated by SDS-PAGE. The phosphorylated proteins were visualized by autoradiography.
Quantitative Real-time RT-PCR Analysis
To examine low temperature-induced gene expression, 10-day-old seedlings grown on MS medium were treated with 4°C. Total RNA was extracted from whole seedlings using plant RNA kit (Omega Bio-tek) according to the manufacturer’s instructions. Complementary DNA was synthesized from 2 μg total RNA using M-MLV reverse transcriptase (Promega). Quantitative real-time PCR was performed by using PerfeCTa SYBR Green Fastmix (Quanta Biosciences). Primers used for qRT-PCR are shown in Table S2.
Freezing Assay
The electrolyte leakage assay was performed according to the protocol described previously (Zhao et al., 2016). In brief, 3-week-old plants grown in soil were treated with low temperature (4°C, cold acclimation) for 7 days before freezing treatment. For MKK5DD plants, dexamethasone (30 μM) was sprayed to the surface of leaves every two days during cold acclimation. After cold acclimation, a fully developed rosette leave was excised and transferred to a small tube containing 100 μL deionized water. The three replicates were performed for each plant at each temperature point. An ice chip with similar size was added to the bottom of each tube and the tube was immediately incubated in a freezing bath (model 1187, VWRScientific) with temperature at 0°C. After placing all the tubes in the freezing bath, run the program with 1°C decrement every 30 min until reaching to −10°C. The tubes were removed from the freezing bath when the experimental temperatures were reached and placed on ice to allow gradual thawing. The leaves and solutions were then transferred to large tubes with 25 ml deionized water. The tubes were gently shaken overnight, and the conductivity of solutions was measured. The tubes were then autoclaved at 121°C for 20 min. The autoclaved solutions were shaken for additional 3 hours before the conductivity was measured again. Finally, the ratio of conductivity before and after autoclaving was calculated.
For freezing survival assay, 10-day-old seedlings (approximately 48 seedlings for each sample in each experiment) grown on the MS medium were cold-acclimated for 7 days at 4°C before being subjected to freezing condition. The acclimated plants were placed in a freezing incubator with the following program: the initial temperature was set at 4°C and the temperature was dropped 1°C every 1 h until reaching to −7°C and 1 h later returned to 4°C by the rate of 2°C every 1 h. After treatment, the seedlings were kept at 4°C for 12 h in the dark and then the seedlings were transferred to 23°C for recovery. The survival rates were counted after 5 days.
Protoplast Assay
Protoplast assay was performed as described previously (Fujii et al., 2009). In brief, leaf strips (0.5 mm) cut from the middle part of leaves of 4-weeks-old plants were submerged in 10 ml enzyme solution containing 1.5% (w/v) cellulase R-10, 0.4% (w/v) macerozyme R-10, 20 mM MES (pH 5.7), 0.4 M mannitol, 20 mM KCl, 10 mM CaCl2, 1 mM 2-mercaptoethanol, and 0.1% BSA. The leaves were infiltrated in a vaccum for 30 min and then incubated in the dark for 3 h. The protoplasts were filtered and washed by the same volume of W5 solution (2 mM MES pH 5.7 containing 154 mM NaCl, 125 mM CaCl2, and 5 mM KCl). After centrifuge at 100 g for 2 min, the protoplasts were suspended in half volume of W5 solution and kept at room temperature for 30 min. After centrifuge, the W5 solution was removed and the protoplasts were suspended in MMg solution (4 mM MES pH 5.7 containing 0.4 M mannitol and 15 mM MgCl2). For transfection, 100 μL of protoplasts (2 × 104 cells) were mixed with plasmid (5 μg) and 110 μL PEG solution (40% w/v PEG 4000 in doubly distilled water containing 0.2 M mannitol and 100 mM CaCl2) and incubated at room temperature for 20 min. After incubation, the PEG solution was removed and the protoplasts were washed by W5 olution for two times and finally the protoplasts were suspended in 1 ml W5 solution and incubated at room temperature overnight. The protoplasts were harvested by centrifuge at 100 g for 2 min, and 54 μL lysis buffer (2.5 mM Tris-phosphate pH 7.8 containing 1 mM dithiothreitol, 2 mM DACTAA, 10% (v/v) glycerol and 1% (v/v) Triton X-100) was added to lysate protoplasts. The luciferase activity was assessed by using Luciferase Assay System (Promega). To examine the activity of GUS, 2 μL protoplast lyaste was mixed with 10 μL of 4-methylumbelliferyl β-d-glucuronide (MUG) substrate mix (10 mM Tris-HCl pH 8 containing 1 mM MUG and 2 mM MgCl2) and incubated for 30 min at 37 °C. The reaction was terminated by adding 100 μl of 0.2 M Na2CO3. The fluorescence of 4-methylumbelliferone was detected by a plate reader (Wallac VICTOR2 plate reader).
Phosphoproteomics Assay
Ground Arabidopsis tissues were lysed in 6 M guanidine hydrochloride (Gnd-HCl) with EDTA-free protease inhibitor cocktail (Roche, Madison, WI) and phosphatase inhibitor cocktail (Sigma, St Louis, MO). Proteins were reduced and alkylated with 10 mM tris-(2-carboxyethyl)phosphine (TECP) and 40 mM chloroacetamide (CAA) at 95 °C for 5 min. Alkylated proteins were subject to methanol-chloroform precipitation, and precipitated protein pellets were solubilized in 8 M urea containing 50 mM triethylammonium bicarbonate (TEAB), and protein amount was quantified by BCA assay (Thermo Scientific, Rockford, IL). Protein extracts were diluted to 4 M urea and digested with Lys-C (Wako, Japan) in a 1:100 (w/w) enzyme-to-protein ratio for 3 h at 37 °C, and further diluted to a 1 M urea concentration. Trypsin (Promega, Madison, WI) was added to a final 1:100 (w/w) enzyme-to-protein ratio overnight. Digests were acidified with 10% trifluoroacetic acid (TFA) to a pH ~3, and desalted using a 100 mg of Sep-pak C18 column (Waters, Milford, MA).
Phosphopeptide enrichment was performed using polyMAC-Ti kit (Iliuk et al., 2010) (Tymora Analytical, West Lafayette, IN). Tryptic peptides (200 μg) were re-suspended in 100 μL of loading buffer (80% acetonitrile (ACN) with 1% TFA) and incubated with 25 μL of the PolyMAC-Ti reagent for 20 min. Use a magnetic rack to collect the magnetic beads to the sides of the tubes and discard the flow-through. The magnetic beads were washed with 200 μL of washing buffer 1 (80% ACN, 0.2% TFA with 25 mM glycolic acid) for 5 min and washing buffer 2 (80% ACN in water) for 30 seconds, respectively. Phosphopeptides were eluted with 200 μL of 400 mM NH4OH with 50% ACN and dried in Speed Vac.
The phosphopeptides were dissolved in 5 μL of 0.3% formic acid (FA) with 3% ACN and injected into an Easy-nLC 1000 (Thermo Fisher Scientific). Peptides were separated on a 45 cm in-house packed column (360 μm OD × 75 μm ID) containing C18 resin (2.2 μm, 100Å, Michrom Bioresources) with a 30 cm column heater (Analytical Sales and Services) set at 50 °C. The mobile phase buffer consisted of 0.1% FA in ultra-pure water (buffer A) with an eluting buffer of 0.1% FA in 80% ACN (buffer B) run over a linear 60 min gradient of 5%–30% buffer B at a flow rate of 250 nL/min. The Easy-nLC 1000 was coupled online with a Velos Pro LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific). The mass spectrometer was operated in the data-dependent mode in which a full MS scan (from m/z 350–1500 with the resolution of 30,000 at m/z 400) was followed by the five most intense ions being subjected to collision-induced dissociation (CID) fragmentation. CID fragmentation was performed and acquired in the linear ion trap (normalized collision energy (NCE) 30%, AGC 3e4, max injection time 100 ms, isolation window 3 m/z, and dynamic exclusion 60 s).
QUANTIFICATION AND STATISTICAL ANALYSIS
Phosphoproteomics Data Processing
To analyze phosphoproteomics data, the raw files were searched directly against the Arabidopsis thaliana database (TAIR10) with not redundant entries using MaxQuant software (version1.5.5.1) (Cox and Mann, 2008) with the Andromeda search engine. Initial precursor mass tolerance was set at 20 ppm, the criteria included a static carbamidomethylation of cysteines (+57.0214 Da) and variable modifications of (1) oxidation (+15.9949 Da) on methionine residues, (2) acetylation (+42.011 Da) at the N-terminus of proteins, and (3) phosphorylation (+79.996 Da) on serine, threonine or tyrosine residues. The match between runs function was enabled with 1.0 min match time window. The searches were performed with trypsin digestion and allowed a maximum of two missed cleavages on the peptides analyzed from the sequence database. The false discovery rates for proteins, peptides and phosphosites were set at 0.01. The minimum peptide length was six amino acids, and a minimum Andromeda score cut-off was set at 40 for modified peptides. A site localization probability of 0.75 was used as the cut-off for localization of phosphorylation sites.
qRT-PCR Analysis
The relative expression level of CBF genes was determined by normalizing to the levels of ACTIN gene. Student’s t-test or One-way ANOVA were used to determine the significance of difference between samples. Statistical tests were conducted by using SPSS software (https://www.ibm.com/us-en/marketplace/spss-statistics). The number of biological replicates was indicated in the Figure legends.
Freezing Assay
The number of leaves or seedlings used for electrolyte leakage assay and freezing survival assay was described in the Method. The significance of difference between samples was determined by Student’s t-test using SPSS software.
DATA AND SOFTWARE AVAILABILITY
The raw data used to compose the figures has been deposited as a Mendeley Data set: doi:10.17632/s928sc39n3.1
Supplementary Material
Figure S1 related to Figure 2 and 3.
MKK5-MPK3/6 cascade negatively regulates CBF gene expression.
Figure S2 related to Figure 1.
YDA is not required for the cold-induced activation of MPK3 and MPK6.
Figure S3 related to Figure 4.
mekk1 single mutant and mkk1 mkk2 double mutant are hypersensitive to freezing.
Figure S4 related to Figure 5.
The phosphorylation of ICE1 in vivo.
Figure S5 related to Figure 5.
MPK4 is not able to phosphorylate ICE1.
Figure S6 related to Figure 5 and 6.
MPK3 and MPK6 interact with ICE1.
Figure S7 related to Figure 6.
The phosphorylation sites of Ser94, Thr366, and Ser403 are required for the degradation of ICE1.
Table S1 related to Figure 1.
Comparison of the phosphorylation levels of the selected MAPK pathway components before and after cold treatment.
Table S2 related to STAR methods.
Primers used in this study.
Highlights.
The MKK4/5-MPK3/6 cascade negatively regulates freezing tolerance
The MEKK1-MKK2-MPK4 cascade positively regulates freezing tolerance
MPK3/6-mediated phosphorylation of ICE1 promotes ICE1 degradation
CRLK1 and CRLK2 suppress the cold-activation of MPK3/6
Acknowledgments
We thank Rebecca Stevenson for technical assistance, Prof. Shuqun Zhang for providing MPK6SR seeds, and Prof. Yuelin Zhang for providing mkk1-2 mkk2-1 seeds. This work was supported by the Strategic Priority Research Program (Grant No. XDPB0404) of the Chinese Academy of Sciences and U.S. National Institutes of Health Grant R01 GM059138 (J.-K.Z.).
Footnotes
AUTHOR CONTRIBUTIONS
C.Z., P.W., and J.-K.Z. conceived and designed the experiments; C.Z. performed most of the experiments; T.S., L.W., Z.Y., and Y.Z. constructed some plasmids; O.Z. performed split luciferase complementation assay; C.-C.H. and W.A.T. conducted phosphoproteomics assay and analyzed the phosphorylation site of ICE1; J.D. provided plasmids and assisted in data analysis; C.Z. and J.-K.Z. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal M, Hao Y, Kapoor A, Dong C-H, Fujii H, Zheng X, Zhu J-K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem. 2006;281:37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- Beckers GJM, Jaskiewicz M, Liu Y, Underwood WR, He SY, Zhang S, Conrath U. Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell. 2009;21:944–953. doi: 10.1105/tpc.108.062158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee B, Hong X, Agarwal M, Zhu JK. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu J-K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell. 2015;32:278–289. doi: 10.1016/j.devcel.2014.12.023. [DOI] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell. 2009;21:972–984. doi: 10.1105/tpc.108.063958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA. 2006;103:8281–8286. doi: 10.1073/pnas.0602874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya T, Matsuoka D, Nanmori T. Phosphorylation of Arabidopsis thaliana MEKK1 via Ca2+ signaling as a part of the cold stress response. J Plant Res. 2013;126:833–840. doi: 10.1007/s10265-013-0576-0. [DOI] [PubMed] [Google Scholar]
- Gao M, Liu J, Bi D, Zhang Z, Cheng F, Chen S, Zhang Y. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008;18:1190–1198. doi: 10.1038/cr.2008.300. [DOI] [PubMed] [Google Scholar]
- Han L, Li G-J, Yang K-Y, Mao G, Wang R, Liu Y, Zhang S. Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 2010;64:114–127. doi: 10.1111/j.1365-313X.2010.04318.x. [DOI] [PubMed] [Google Scholar]
- Ichimura K, Casais C, Peck SC, Shinozaki K, Shirasu K. MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J Biol Chem. 2006;281:36969–36976. doi: 10.1074/jbc.M605319200. [DOI] [PubMed] [Google Scholar]
- Iliuk AB, Martin VA, Alicie BM, Geahlen RL, Tao WA. In-depth analyses of kinase-dependent tyrosine phosphoproteomes based on metal ion-functionalized soluble nanopolymers. Mol Cell Proteomics. 2010;9:2162–2172. doi: 10.1074/mcp.M110.000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Jia Y, Ding Y, Shi Y, Zhang X, Gong Z, Yang S. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016;212:345–353. doi: 10.1111/nph.14088. [DOI] [PubMed] [Google Scholar]
- Kim YS, Lee M, Lee J-H, Lee H-J, Park C-M. The unified ICE–CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol Biol. 2015;89:187–201. doi: 10.1007/s11103-015-0365-3. [DOI] [PubMed] [Google Scholar]
- Kong Q, Qu N, Gao M, Zhang Z, Ding X, Yang F, Li Y, Dong OX, Chen S, Li X, et al. The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell. 2012;24:2225–2236. doi: 10.1105/tpc.112.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D, et al. COLD1 confers chilling tolerance in Rice. Cell. 2015;160:1209–1221. doi: 10.1016/j.cell.2015.01.046. [DOI] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell. 2011;23:1639–1653. doi: 10.1105/tpc.111.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J, Bargues M, Terol J, Pérez-Alonso M, Salinas J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression Is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 1999;119:463–470. doi: 10.1104/pp.119.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J, Catalá R, Salinas J. The CBFs: Three arabidopsis transcription factors to cold acclimate. Plant Sci. 2011;180:3–11. doi: 10.1016/j.plantsci.2010.06.019. [DOI] [PubMed] [Google Scholar]
- Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol. 2013;51:245–266. doi: 10.1146/annurev-phyto-082712-102314. [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ohta M, Nakazawa M, Ono M, Hasegawa PM. ICE1 Ser403 is necessary for protein stabilization and regulation of cold signaling and tolerance. Plant J. 2011;67:269–279. doi: 10.1111/j.1365-313X.2011.04589.x. [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, et al. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103:1111–1120. doi: 10.1016/s0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Datta S, Persak H. Salt stress in Arabidopsis: lipid transfer protein AZI1 and its control by mitogen-activated protein kinase MPK3. Mol. Plant. 2014;7:722–738. doi: 10.1093/mp/sst157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Yang H, Zhang S. Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J Biol Chem. 2002;277:559–565. doi: 10.1074/jbc.M109495200. [DOI] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C. The Arabidopsis leucine-rich repeat receptor–like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell. 2011;23:2440–2455. doi: 10.1105/tpc.111.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Eulgem T, D?czi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell. 2004;15:141–152. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell. 2007;19:63–73. doi: 10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Jia W, Zhang J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 2008;54:440–451. doi: 10.1111/j.1365-313X.2008.03433.x. [DOI] [PubMed] [Google Scholar]
- Xu J, Xie J, Yan C, Zou X, Ren D, Zhang S. A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J. 2014;77:222–234. doi: 10.1111/tpj.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Chaudhuri S, Yang L, Du L, Poovaiah BW. A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J Biol Chem. 2010a;285:7119–7126. doi: 10.1074/jbc.M109.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Shad Ali G, Yang L, Du L, Reddy ASN, Poovaiah BW. Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal Behav. 2010b;5:991–994. doi: 10.4161/psb.5.8.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Zhu J-K. The broad roles of CBF genes: From development to abiotic stress. Plant Signal Behav. 2016;11:e1215794. doi: 10.1080/15592324.2016.1215794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Lang Z, Zhu J-K. Cold responsive gene transcription becomes more complex. Trends Plant Sci. 2015;20:466–468. doi: 10.1016/j.tplants.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Zhang Z, Xie S, Si T, Li Y, Zhu J-K. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 2016;171:2744–2759. doi: 10.1104/pp.16.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 related to Figure 2 and 3.
MKK5-MPK3/6 cascade negatively regulates CBF gene expression.
Figure S2 related to Figure 1.
YDA is not required for the cold-induced activation of MPK3 and MPK6.
Figure S3 related to Figure 4.
mekk1 single mutant and mkk1 mkk2 double mutant are hypersensitive to freezing.
Figure S4 related to Figure 5.
The phosphorylation of ICE1 in vivo.
Figure S5 related to Figure 5.
MPK4 is not able to phosphorylate ICE1.
Figure S6 related to Figure 5 and 6.
MPK3 and MPK6 interact with ICE1.
Figure S7 related to Figure 6.
The phosphorylation sites of Ser94, Thr366, and Ser403 are required for the degradation of ICE1.
Table S1 related to Figure 1.
Comparison of the phosphorylation levels of the selected MAPK pathway components before and after cold treatment.
Table S2 related to STAR methods.
Primers used in this study.