ABSTRACT
Fluctuating environments and individual physiological diversity force bacteria to constantly adapt and optimize the uptake of substrates. We focus here on two very similar two-component systems (TCSs) of Escherichia coli belonging to the LytS/LytTR family: BtsS/BtsR (formerly YehU/YehT) and YpdA/YpdB. Both TCSs respond to extracellular pyruvate, albeit with different affinities, typically during postexponential growth, and each system regulates expression of a single transporter gene, yjiY and yhjX, respectively. To obtain insights into the biological significance of these TCSs, we analyzed the activation of the target promoters at the single-cell level. We found unimodal cell-to-cell variability; however, the degree of variance was strongly influenced by the available nutrients and differed between the two TCSs. We hypothesized that activation of either of the TCSs helps individual cells to replenish carbon resources. To test this hypothesis, we compared wild-type cells with the btsSR ypdAB mutant under two metabolically modulated conditions: protein overproduction and persister formation. Although all wild-type cells were able to overproduce green fluorescent protein (GFP), about half of the btsSR ypdAB population was unable to overexpress GFP. Moreover, the percentage of persister cells, which tolerate antibiotic stress, was significantly lower in the wild-type cells than in the btsSR ypdAB population. Hence, we suggest that the BtsS/BtsR and YpdA/YpdB network contributes to a balancing of the physiological state of all cells within a population.
IMPORTANCE Histidine kinase/response regulator (HK/RR) systems enable bacteria to respond to environmental and physiological fluctuations. Escherichia coli and other members of the Enterobacteriaceae possess two similar LytS/LytTR-type HK/RRs, BtsS/BtsR (formerly YehU/YehT) and YpdA/YpdB, which form a functional network. Both systems are activated in response to external pyruvate, typically when cells face overflow metabolism during post-exponential growth. Single-cell analysis of the activation of their respective target genes yjiY and yhjX revealed cell-to-cell variability, and the range of variation was strongly influenced by externally available nutrients. Based on the phenotypic characterization of a btsSR ypdAB mutant compared to the parental strain, we suggest that this TCS network supports an optimization of the physiological state of the individuals within the population.
KEYWORDS: histidine kinase, nutrient limitation, overflow metabolism, persister cells
INTRODUCTION
Typical two-component systems (TCSs) consist of a membrane-bound histidine kinase (HK), which perceives a stimulus, and a cytoplasmic response regulator (RR), which triggers an appropriate response (1, 2). Escherichia coli contains 30 TCSs in all. Members of the LytS/LytTR family make up one prominent class of TCSs, representatives of which are found in many microorganisms. Examples include AgrC/AgrA from Staphylococcus aureus, which is involved in the transition from the persistent, avirulent state to the virulent phenotype (3), while FsrC/FsrA from Enterococcus faecalis is responsible for the production of virulence-related proteases (4), and VirS/VirR from Clostridium perfringens induces the synthesis of exotoxins and collagenase (5, 6). In our laboratory, we are studying the only two known members of the LytS/LytTR family in E. coli: BtsS/BtsR (previously YehU/YehT) and YpdA/YpdB (7–10). These two TCSs not only share the same domain structure, they also display over 30% identity at the amino acid sequence level (9). BtsS/BtsR activation leads to the expression of yjiY, YpdA/YpdB activation results in yhjX expression (Fig. 1). Both target genes code for transporters, which belong to different transporter families: YjiY is a member of the CstA family, and YhjX has been assigned to the oxalate/formate antiporter (OFA) family (7, 8, 11). In addition, the cyclic AMP (cAMP) receptor protein (CRP) complex (CRP-cAMP) upregulates yjiY at the transcriptional level (7), whereas the carbon storage regulator A (CsrA) upregulates yhjX and downregulates yjiY at the posttranscriptional level.
FIG 1.
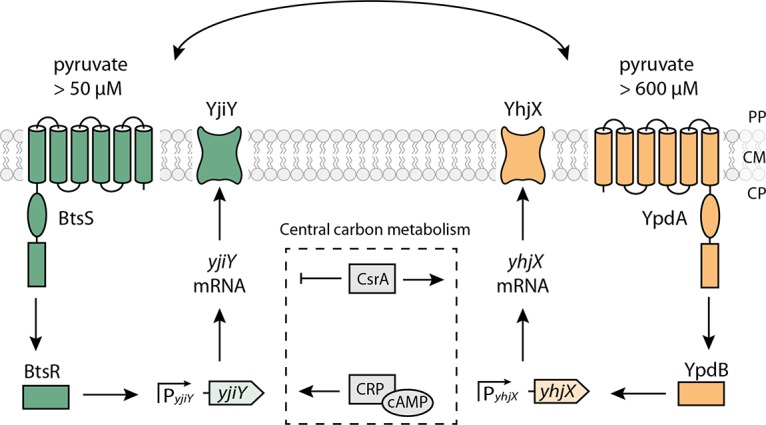
Model of the nutrient-sensing BtsS/BtsR and YpdA/YpdB network in E. coli. The scheme summarizes the signal transduction cascades triggered by the BtsS/BtsR and YpdA/YpdB systems and the influence of other regulatory elements. Activating (→) and inhibitory (⊦) effects are indicated. PP, periplasm; CM, cytoplasmic membrane; CP, cytoplasm. See the text for details.
In previous studies we found functional interconnectivity of the two TCSs (9). Deletion of either component of the TCS or its target gene influences the level of expression of the target gene regulated by the other TCS and vice versa (9). In addition, in vivo protein-protein interaction assays suggested that the two systems form a single, large signaling unit (Fig. 1). Moreover, when E. coli was grown in tryptone-based (LB) medium, both systems are activated at the onset of the post-exponential growth phase (9). A more refined study revealed that the BtsS/BtsR system is activated in the presence of extracellular pyruvate (at a threshold concentration of 50 μM) under nutrient-depleted conditions (10). Biochemical studies confirmed that BtsS is a high-affinity pyruvate receptor (Kd = 58.6 μM) (10). Recently, the corresponding YjiY transporter was characterized as a high-affinity pyruvate/H+ symporter (12). The YpdA/YpdB system also responds to extracellular pyruvate, albeit at a higher threshold concentration of 600 μM (8).
The biological significance of the BtsS/BtsR and YpdA/YpdB network is still unclear. To explore this issue, we determined the activation states of the two systems at the single-cell level in E. coli populations. Using separate fluorescence reporter strains for each system, we found a correlation between the available nutrient resources and the degree of heterogeneity in the transcriptional responses of the target gene promoters in individual cells. Based on this finding and further phenotypic analyses, we suggest that the BtsS/BtsR and YdpA/YpdB systems play a role in optimization of the physiological status of the individual cells within the population.
RESULTS
Heterogeneous activation of PyhjX-gfp and PyjiY-gfp.
For the BtsS/BtsR and YpdA/YpdB systems, population-based studies have shown that the promoters of their respective target genes, yjiY and yhjX, are activated in cells which face nutrient limitation and sense the presence of external pyruvate (9, 10). Since both systems are linked to form a network, we analyzed the activation of these two promoters at the single-cell level. We constructed fluorescent reporter strains by fusing the promoter regions of yhjX and yjiY to gfp and introduced each fusion separately into the genome of E. coli MG1655 via single homologous recombination at the native locus. Using this strategy, the regulatory inputs to the native promoters of yjiY and yhjX were maintained (9), as the promoter fused to gfp is inserted upstream of the original one (13). The fluorescence intensity of green fluorescent protein (GFP) was used to quantify the activity of the two promoters, thus allowing us to study the transcriptional activation of yjiY and yhjX in single cells. The growth rates in LB medium of strains containing a chromosomal copy of either promoter fusion (from now on referred to as PyhjX-gfp and PyjiY-gfp) were similar to that of the MG1655 wild-type strain (see Fig. S1 in the supplemental material).
From population studies it is known that in cells grown in LB medium, which is rich in amino acids and leads to the overflow of pyruvate, both promoters are activated at the onset of the post-exponential growth phase (9). Hence, as expected, at the single-cell level neither PyhjX-gfp nor PyjiY-gfp showed any activity during the exponential growth phase (Fig. 2A and C, before activation) in LB medium. However, when cells reached the end of the exponential growth phase, we observed activation of the yhjX promoter, as indicated by a shift of the distribution of fluorescence intensities to higher levels (Fig. 2A and B, after activation) in the majority of the population, albeit with a high degree of cell-to-cell variability as seen in the width of the Gaussian distribution (noise value [standard deviation divided by the mean] = 0.27). Less than 4% of the population was found to be nonfluorescent and therefore did not respond (the threshold of activation is marked by the dashed line in Fig. 2B). To differentiate these cells from dead cells, we stained cells with propidium iodide and found that dead cells made up only 0.4% of the population (data not shown).
FIG 2.
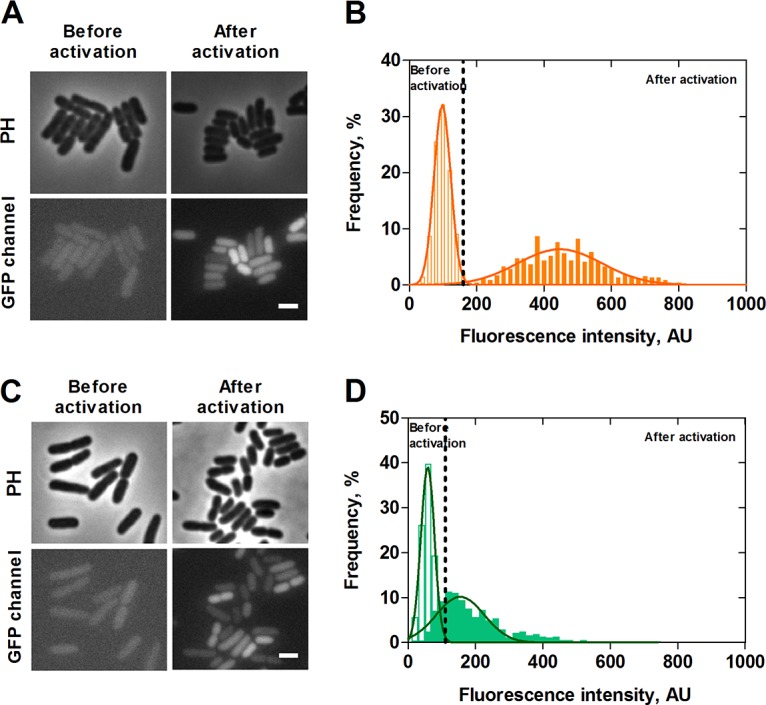
Single-cell analysis of PyhjX and PyjiY activation during growth in LB medium. E. coli cells expressing gfp under the control of the yhjX or yjiY promoter, respectively, were grown in LB medium, and fluorescence micrographs were taken before (exponential growth phase) and after activation (post-exponential growth phase) of the two TCSs. Representative fluorescence and phase-contrast images of PyhjX-gfp and PyjiY-gfp reporter strains are shown in panels A and C, respectively. The corresponding distributions of the fluorescence intensity of the PyhjX-gfp and PyjiY-gfp reporter strains are depicted in panels B and D. Unfilled bars refer to values prior to activation, and filled bars refer to values observed after activation. Dashed lines represent the threshold of activation for each of the reporter strains. A total of 200 cells were analyzed in each experiment, and frequency refers to the percentage of cells with the indicated intensity (see Materials and Methods for details). The continuous curves represent Gaussian fits based on the histograms of the fluorescence intensity. PH, phase contrast; AU, arbitrary units. Scale bar, 2 μm. Experiments were performed independently three times.
Cells of the PyjiY-gfp strain cultivated in LB medium also showed heterogeneous activation upon entry into the post-exponential growth phase. These strains exhibited an even higher noise value of 0.52 and a higher percentage of nonresponding cells (9%) (Fig. 2D) (the percentage of dead cells was determined to be 0.6%).
To determine the basal noise level of a promoter in cells at this growth phase, we performed a control experiment, in which gfp expression is controlled by a synthetic vegetative promoter (pXGSF). Cells harboring the vector pXGSF activate this promoter at the post-exponential growth phase (R. Hengge, unpublished data). In this experiment the promoter was activated in all the cells, and the variability was lower (i.e., 0.13) than that observed for either PyjiY-gfp or PyhjX-gfp. Taken together, these results indicate a heterogeneous, almost unimodal pattern of transcriptional activation for each of the two target genes of the BtsS/BtsR and YpdA/YpdB systems at the end of the exponential phase, when cells are grown in LB medium.
The degree of heterogeneity of PyhjX-gfp activation depends on the external pyruvate concentration.
Although the exact nature of the primary stimulus for the YpdA/YpdB system remains elusive, we know from previous studies that PyhjX is activated in cells which are exposed to extracellular pyruvate concentrations greater than 0.6 mM (9). Aiming to further explore the single-cell behavior of this promoter activity, we analyzed the pyruvate dependence of the activation of YpdA/YpdB by determining the fluorescence intensities of PyhjX-gfp reporter cells cultivated in M9 minimal medium supplemented with increasing concentrations of pyruvate (succinate was added to keep the total carbon concentration constant at 20 mM) (Fig. 3). As expected, a pyruvate concentration below the threshold (0.3 mM) failed to activate the YpdA/YpdB system in single cells. At pyruvate concentrations above the threshold, all cells in the population homogenously activate the yhjX promoter. The presence of 0.6 mM pyruvate in the medium generated a low, but detectable PyhjX-gfp signal in the cells and the presence of 1 mM pyruvate shifted the expression level toward higher values. Interestingly, the response was markedly less heterogeneous (noise value of 0.18) in cells grown under these conditions than in the cells grown in LB medium (noise value of 0.27). Further increases in the external pyruvate concentration (2 and 10 mM) boosted the signal intensities, while the variability further decreased (to 0.09 and 0.07, respectively) (Fig. 3). These results reveal a correlation between external pyruvate availability and PyhjX-gfp activation.
FIG 3.
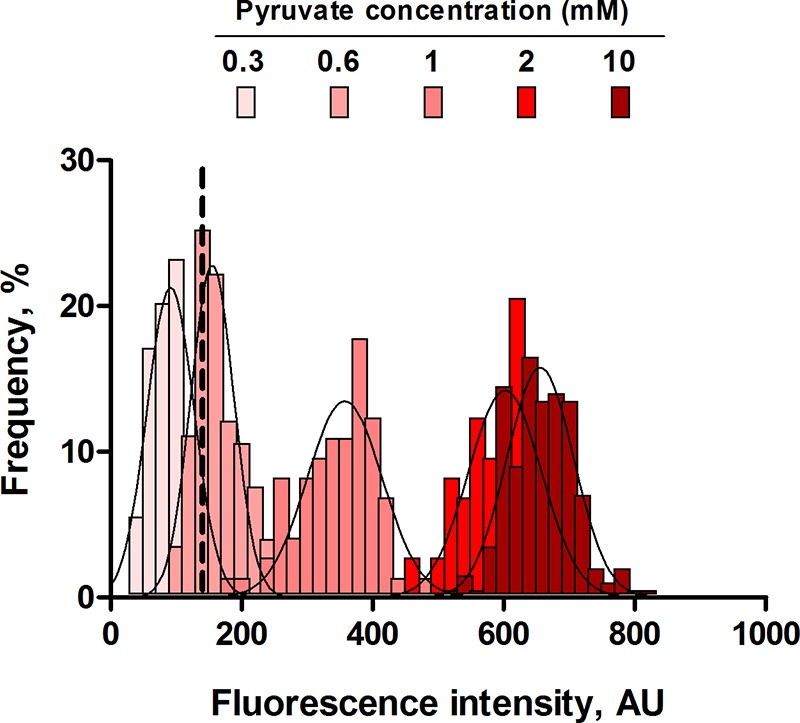
Effects of different external pyruvate concentrations on PyhjX-gfp activation at the single-cell level. E. coli cells expressing gfp under the control of the PyhjX promoter were grown in M9 minimal medium containing increasing concentrations of pyruvate (supplemented with succinate; final carbon concentration, 20 mM) and analyzed by fluorescence microscopy. A total of 200 cells was analyzed in each experiment at the time point of maximal expression, and frequency refers to the percentage of cells with the indicated intensity (see Materials and Methods). Histograms of the fluorescence intensities of cells were fitted using a Gaussian distribution (solid line). The dashed line represents the threshold of activation for the reporter strain. AU, arbitrary units. Experiments were performed independently three times.
The degree of heterogeneity of PyjiY-gfp activation is influenced both by the external pyruvate level and the metabolic state of the cells.
As previously described, growth of cells in M9 minimal medium with pyruvate as sole carbon source (20 mM) is not sufficient to activate the PyjiY promoter, because both extracellular pyruvate and nutrient limitation are needed to trigger BtsS/BtsR activation (10). Therefore, our reporter strain had first to be exposed to nutrient limitation (growth in 0.1× LB medium for 1 h) before pyruvate was added. Under these conditions, no activation of PyjiY-gfp was detected (data not shown), in accordance with our previous studies (10). Pyruvate was then added to the cell culture at a final concentration of 20 mM, and cells were analyzed by fluorescence microscopy at various time points. Cells responded within 70 min and exhibited a higher average gfp intensity than cells grown in LB medium, which confirmed the strong response of the BtsS/BtsR system to pyruvate after exposure of cells to nutrient limitation (Fig. 4A). Remarkably, in this case activation of PyjiY-gfp remained heterogeneous (noise value of 0.27) in spite of the abundance of pyruvate. Subsequently, we tested five different pyruvate concentrations to assess the pyruvate concentration dependence of BtsS/BtsR activation (Fig. 4B). Below the threshold of 50 μM (0.01 mM) to which BtsS/BtsR responds, there was no detectable PyjiY-gfp signal. As expected, only a few cells produced a weak GFP signal in an environment containing 0.05 mM pyruvate. Starting at a concentration of 0.1 mM pyruvate, PyjiY activation was found in all cells, but with a high degree of cell-to-cell variability (noise value of 0.27). At higher pyruvate concentrations, the signal intensity increased, but the noise values were unchanged. The broad Gaussian distribution found at 1 mM pyruvate resembled the profile found for cells at 20 mM pyruvate (Fig. 4A). A t test was performed on the mean values of the two distributions, and the P value was determined to be 0.88. This value revealed that there is no significant difference between the cellular responses at 1 and 20 mM pyruvate. These results confirmed at the single-cell level that BtsS/BtsR-mediated activation of PyjiY-gfp is not only dependent on the pyruvate concentration but is also influenced by internal nutrient limitation.
FIG 4.
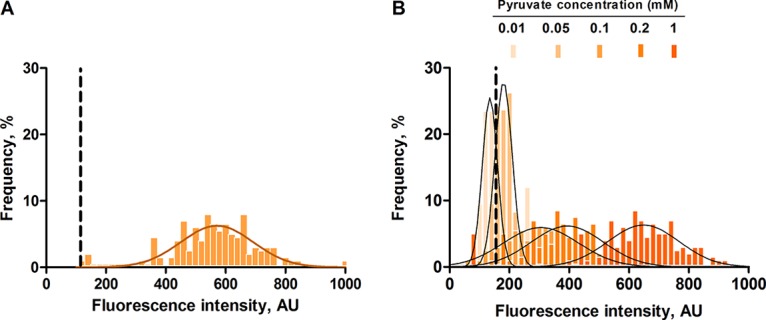
Effects of different external pyruvate concentrations on PyjiY-gfp activation at the single-cell level. E. coli cells expressing gfp under the control of the PyjiY promoter were grown in a nutrient-poor environment (0.1× LB medium) for 1 h. The medium was then supplemented with 20 mM pyruvate (A) or with increasing pyruvate concentrations (B), and the cells were subsequently analyzed by fluorescence microscopy. A total of 200 cells were analyzed for each experiment, and frequency is represented as a percentage of the cells (refer to Materials and Methods for a detailed explanation). Histograms of the fluorescence intensities of cells were fitted using a Gaussian distribution (solid line). Dashed lines represent the threshold of activation for the reporter strain. AU, arbitrary units. Experiments were performed three independent times. For further details, see the legends to Fig. 2 and 3.
Cellular physiology in the post-exponential growth phase.
We have shown thus far that transcriptional activation of both target genes of the BtsS/BtsR and YpdA/YpdB network occurs heterogeneously. Furthermore, their activation is influenced by the availability of external pyruvate, albeit with different thresholds.
Since the two systems are activated in the post-exponential growth phase in LB medium, we decided to explore the impact of the BstS/BtsR and YpdA/YpdB systems on the overall physiological state of E. coli during this growth phase. In order to do so, we investigated individual cells of both E. coli MG1655 (the wild-type [WT] strain) compared to a strain lacking both systems: MG1655 ΔbtsSR ΔypdAB (abbreviated as the btsSR ypdAB mutant).
Fast-growing cells express high levels of 16S RNA from the rrnB P1 promoter (14). Cells with an inactive rrnB P1 promoter are likely to be dormant, antibiotic-tolerant persisters (14, 15). Recently, the strength of rrnB P1 promoter activation was shown to correlate with intracellular ATP levels (16). We therefore fused the ribosomal rrnB P1 promoter to gfp as previously described (14) and integrated this construct into the genomes of the two strains as a marker for their physiological states.
As expected, E. coli MG1655 WT rrnB P1-gfp showed a Gaussian distribution of GFP signal intensities, with a mean fluorescence value of 510 arbitrary units (AU) and noise level of 0.16 (Fig. 5). In contrast, the btsSR ypdAB rrnB P1-gfp mutant had a lower overall rrnB P1-gfp activity (average fluorescence intensity of 398 AU), which indicates a lower rate of ribosome synthesis within the population. Most strikingly, a bistable distribution of the signal was observed. These results suggest that, in the absence of both systems, the population differentiates into two subpopulations, one with a normal and another with a reduced ribosome synthesis rate.
FIG 5.
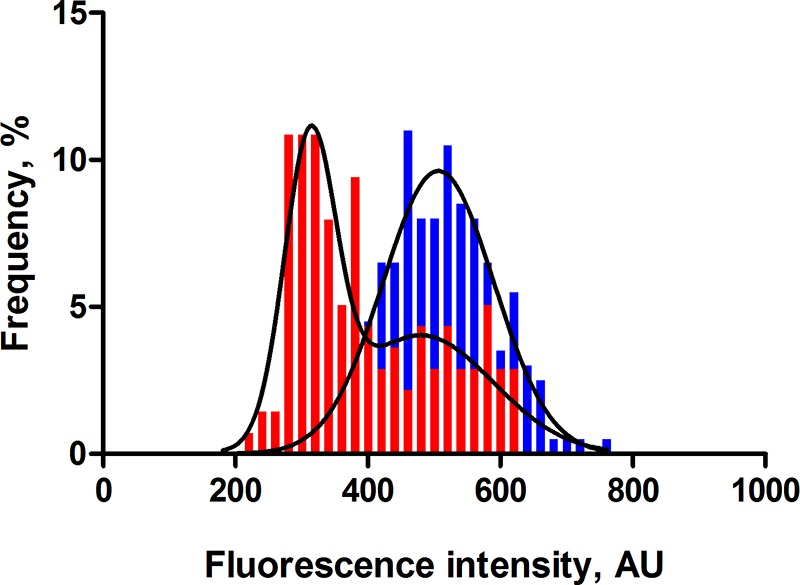
In the absence of the BtsSR/YpdAB network, rrnB P1 promoter activity is low and bistable. Wild-type E. coli MG1655 (blue) or mutant btsSR ypdAB (red) cells harboring a chromosomally encoded rrnB P1-gfp fusion were grown in LB medium and examined by fluorescence microscopy. For further details, see the legends to Fig. 2 and 3. A total of 200 cells were analyzed for each experiment at the post-exponential growth phase, and frequency is represented as a percentage of the cells (refer to Materials and Methods for detailed explanation). Histograms of the fluorescence intensities of cells were fitted using a Gaussian distribution (solid line). AU, arbitrary units. Experiments were performed three independent times.
The BtsSR/YpdAB network promotes protein overproduction.
To test the idea that BtsS/BtsR and YpdA/YpdB systems together act to optimize the physiological state of cells within the population, we set out to metabolically challenge the btsSR ypdAB mutant and compare its response to that of the parental E. coli MG1655 WT strain. Interestingly, E. coli C41 (DE3), also known as the Walker strain, has been optimized for maximal overproduction of membrane and globular proteins (17). Subsequently, the genome of this strain was sequenced and, among other mutations, a point mutation in btsS was found that led to constitutive expression of yjiY (18). Based on these data, we hypothesized that the BtsS/BtsR and YpdA/YpdB systems might help cells to cope with the metabolic burden of protein overproduction.
In order to test this hypothesis, both strains (WT and the btsSR ypdAB mutant) were transformed with the overexpression vector pBAD24-gfp, which carries gfp under the control of an arabinose-inducible promoter. Before induction with arabinose, fluorescence microscopy of single WT and btsSR ypdAB mutant cells showed no apparent GFP signals, and flow cytometry confirmed that the maximal fluorescence intensity of green cells (∼750 AU) was low (Fig. 6A and C), indicating little or no GFP expression. One hour after induction with 0.2% (wt/vol) arabinose, cells of the WT population were producing GFP, which was clearly detected as an increase in the maximum fluorescence observed by flow cytometry (to ∼1,100 AU). This result was corroborated by the detection of labeled single cells with fluorescence microscopy (Fig. 6B). In contrast, flow cytometric analysis of the mutant under inducing conditions detected two peaks: one at 1,100 AU, as in the WT, and a second at 750 AU. The accompanying micrographs revealed the presence of fluorescent and nonfluorescent cells (Fig. 6D). Therefore, the low-intensity peak in the flow cytometer represents cells that are unable to produce GFP in large amounts. The C41 (DE3) strain was also tested and was found to be capable of a homogeneously high protein overproduction, as expected (data not shown).
FIG 6.
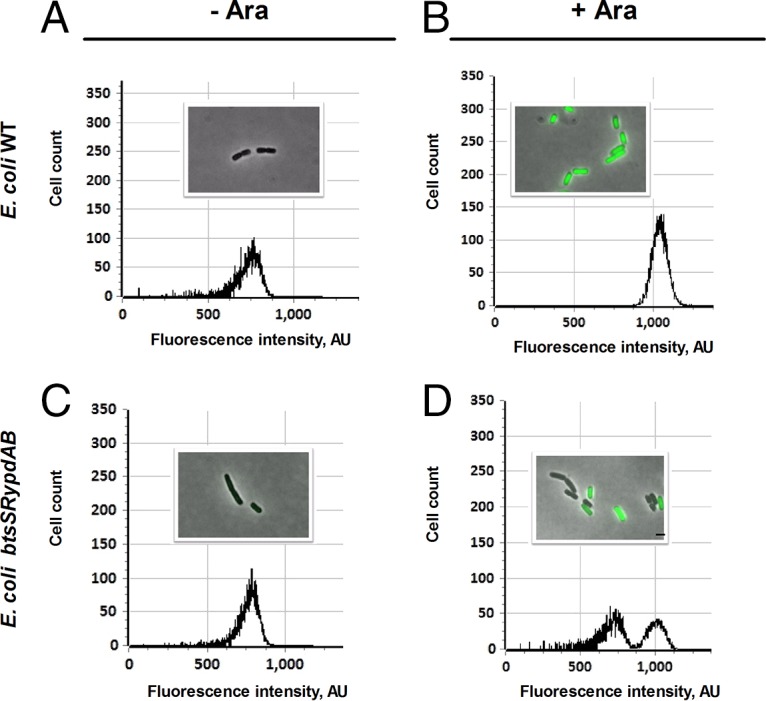
The BtsSR/YpdAB network promotes homogeneous protein overproduction in all cells. Wild-type (WT) or btsSR ypdAB mutant cells harboring the overproduction vector pBAD24-gfp were grown in LB medium. Samples were taken before and after the addition of the inducer arabinose (Ara) (0.2% [wt/vol]). The cells were analyzed by fluorescence microscopy and flow cytometry. Distributions of fluorescent cell counts and representative views of WT cells before and after addition of arabinose are shown in panels A and B, while the corresponding data for the btsSR ypdAB mutant are depicted in panels C and D. About 2,000 events were recorded for each plot. Cell counts represent the numbers of cells, and fluorescence intensity is expressed in arbitrary units (AU). Scale bar, 2 μm. Experiments were performed independently three times.
We also tested the overproduction of (i) GFP under the control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible lac promoter (pCOLA-Plac-gfp), (ii) the periplasmic protein DppA fused to the Tat translocation sequence (19) and under the control of the arabinose promoter (pBAD24-RR-gfp-dppA), and (iii) the membrane protein LysP fused to a different fluorophore and under the control of an arabinose-inducible promoter (pBAD33-lysP-mcherry) (see Table S1 in the supplemental material). The results obtained for the IPTG-inducible GFP reporter were similar to those for the arabinose-controlled system. The btsSR ypdAB mutant was hardly able to overproduce the periplasmic DppA or the membrane protein LysP. In summary, the BtsS/BtsR and YpdA/YpdB sensing network helps E. coli to cope with the additional metabolic burden imposed by protein overproduction.
The BtsSR/YpdAB network limits the proportion of persister cells in WT populations.
We hypothesized that the heterogeneous distribution of the capacity for protein overproduction among the btsSR ypdAB mutant population might be related to the presence of a subpopulation of cells that are unable to sense nutrient limitation and consequently fail to activate transporters to acquire needed resources. Persister cells survive exposure to antibiotics owing to their altered metabolic activity and low growth rate, but they can subsequently resume growth to form an antibiotic-sensitive population (20). We were interested to know whether the BtsS/BtsR and YpdA/YpdB network has an influence on persister cell formation. To address this question, we performed population-based studies by exposing growing WT or btsSR ypdAB mutant cells to ampicillin and determining the number of CFU. Only cells able to recover from the stress will form CFU. We subdivided a growing culture and exposed cells to ampicillin before and after the natural activation of the signaling systems, namely, at an optical density at 600 nm (OD600) of 0.4 (exponential growth phase) and 1.2 (post-exponential growth phase) (9). After treatment with ampicillin a biphasic time-dependent killing curve was observed, which is typical for persister formation (Fig. 7) (20). When cells were exposed to ampicillin prior to activation of the signaling systems, the two strains exhibited almost identical patterns of response, characterized by a steep initial decrease in CFU, followed by a slower killing rate, revealing persister cells (Fig. 7A). Ampicillin treatment of cells after activation of the signaling systems resulted in a considerably higher level of persister cells in the mutant (2.15%) than the WT (0.14%) population (Fig. 7B).
FIG 7.
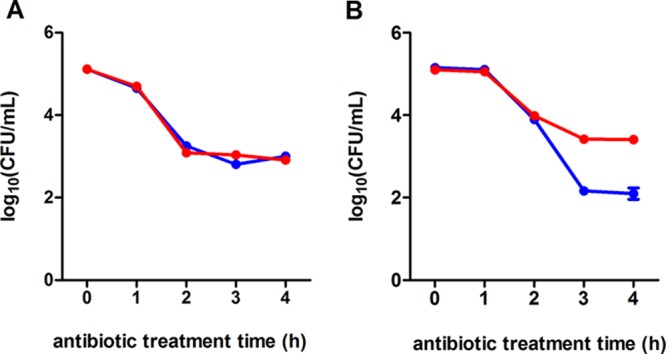
The BtsSR/YpdAB network reduces the proportion of persister cells in populations. Either WT (blue lines) or mutant btsSR ypdAB (red lines) cells were grown in LB medium. Before (exponential growth phase) (A) and after (B) activation (post-exponential growth phase) of the systems, the cells were exposed to ampicillin (200 μg/ml). Samples were taken and analyzed for CFU. Three independent experiments were performed, and error bars indicate the standard deviations of the means.
In parallel, the minimum time taken to kill 99% of the population (MDK99) (21) was determined for both strains after exposure to ofloxacin. The value for the WT was determined to be 0.49 h, and for the btsSR ypdAB mutant it was 1.98 h, which is compatible with the higher fraction of persisters in the mutant population (see Fig. S2 in the supplemental material).
These results reveal a novel role for the BtsS/BtsR and YpdA/YpdB signaling network in reducing the percentage of persister cells in a growing population. They are also in accordance with the idea of a contribution of both systems to help individual cells to replenish nutrient resources.
DISCUSSION
BtsS/BtsR (formerly YehU/YehT) is one of the most widespread TCSs in bacteria and is found in many human and plant pathogens. Although most gammaproteobacteria contain this system, some, including Escherichia, Citrobacter, and Serratia, have a second homologous system, YpdA/YpdB (22). Both belong to the LytS/LytTR family. Previous systematic studies failed to identify a function for these TCSs (23, 24). We have now identified the HK BtsS as a high-affinity pyruvate receptor (Kd = 58.6 μM) and YpdA/YpdB as a system that responds to higher levels (>0.6 mM) of the same compound (8, 10). The target genes regulated by the two systems code for the high-affinity pyruvate/H+ symporter YjiY (recently renamed BtsT [12]) and a transporter of unknown function, YhjX. However, the biological significance of the BtsS/BtsR and YpdA/YpdB systems has remained unclear.
Therefore, we first investigated the activation of the target genes of each system at the single-cell level using promoter fusions. We found that in clonal populations the chromosomally integrated copies of either PyjiY-gfp or PyhjX-gfp were heterogeneously activated when grown in LB medium, which is rich in amino acids (Fig. 2), and that induction of PyjiY-gfp was slightly more variable than that of PyhjX-gfp. In both cases, a predominantly unimodal Gaussian distribution of activation levels was observed, and only a very small percentage of cells remained in the OFF state. This pattern of activation differs markedly from the “all-or-nothing,” switch-like gene expression described for the lac or ara promoter (25, 26). However, the heterogeneous, but unimodal activation of yhjX and yjiY can, in principle, be explained by the multiple factors known to affect their expression: (i) binding of the respective transcriptional activators BtsR and YdpB (27), (ii) the influence of the cAMP/CRP protein (PyjiY), (iii) fine-tuning by the carbon starvation regulator CsrA (Fig. 1), and (iv) variations in the physiological state between cells (see below).
YpdA/YpdB-mediated activation of PyhjX was found to be dependent on the concentration of pyruvate in the medium and became homogenous when cells were grown in minimal medium containing pyruvate (20 mM) as the sole carbon source (Fig. 3). In contrast, under all tested conditions BtsS/BtsR-mediated activation of PyjiY was characterized by high cell-to-cell variability, which was virtually unaffected by the amount of pyruvate in the medium (Fig. 4). It is important to note that the BtsS/BtsR systems, whose target gene codes for a high-affinity pyruvate transporter, is only activated by external pyruvate when cells concurrently face nutrient limitation (Fig. 4) (10). The high degree of heterogeneity might reflect variations in the nutritional state of individual cells and differing needs for the high-affinity pyruvate transporter YjiY. Therefore, the BtsS/BtsR system responds only when cells are in need of a high-affinity uptake transporter to scavenge traces of available nutrients, e.g., pyruvate.
It has been proposed that cellular metabolism is both inherently stochastic and a generic source of phenotypic heterogeneity (28). In this general context, the results of our single-cell studies can be accommodated by the following model for the role of the two LytS/LytTR-type systems in E. coli. Under certain conditions, e.g., during growth in LB medium, cells excrete pyruvate due to overflow metabolism. Subsequently, other nutrients are depleted, and cells sense the availability of pyruvate. Depending on the external pyruvate concentration and their particular nutritional needs, individual E. coli cells activate either the high-affinity BtsS/BtsR and/or the low-affinity YpdA/YpdB system upon entry into the post-exponential growth phase. The interplay between transporters with different affinities for the same substrate has already been described, and this seems to be a successful strategy under nutrient limitation (29).
By using a reporter for the rate of ribosome synthesis, we found that only populations of reporter cells harboring the nutrient-sensing network exhibited unimodal activation of the rrnB P1 promoter, whereas the btsSR ypdAB mutant was characterized by a bimodal expression pattern (Fig. 5). The heterogeneous activation of either PyjiY or PyhjX in individual WT cells allows uptake of nutrients, e.g., pyruvate, according to the individual requirement of the cells. This results in a unimodal distribution of the activation level of the rrnB P1 promoter characteristic of growing cells. It should be noted that previous physiological studies revealed that E. coli has more than one pyruvate transporter (30), although only YjiY has thus far been characterized as high-affinity pyruvate transporter (12). Therefore, we assume that individuals within the population of the btsSR ypdAB mutant can cope with the lack of the sensing/transport of pyruvate and have normal ribosome synthesis. In addition, we imposed a metabolic burden by forcing cells to overproduce particular proteins. This is a natural scenario, since many pathogens have to produce virulence factors, exoenzymes, siderophores, etc., in large amounts. Although all WT cells managed to cope with this burden, about 50% of the mutant cells failed to overproduce the test protein, GFP, a pattern which we also observed for the activation of the rrnB P1 promoter (Fig. 6). It should be noted that the evolved E. coli C41(DE3) strain, which has been optimized for protein overproduction, has a point mutation in btsS that leads to stimulus-independent expression of yjiY (18). In light with the results presented here, the constitutive expression of the high-affinity pyruvate transporter YjiY in strain C41 guarantees a sufficient uptake of pyruvate in all cells independent from external or internal factors. Finally, a population-based persister assay revealed that btsSR ypdAB populations contain a higher percentage of antibiotic-tolerant persister cells (dormant cells) than do WT populations (Fig. 7).
Taking these results into account, the model described above can be further extended. Sensing of external pyruvate by the BtsS/BtsR and YpdA/YpdB systems and the tight regulation of expression of the two transporters YjiY and YhjX depending on the needs of the individual cell ensures an optimization of the physiological state within the whole population to withstand upcoming metabolic stress. These findings are important not only in light of the host colonization of pathogenic species and their persistence but also for metabolic engineering.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. coli strains, including their genotypes, and the plasmids used in this study are listed in Table 1. Mutants were constructed using an E. coli Quick-and-Easy gene deletion kit (Gene Bridges) and a BAC modification kit (Gene Bridges), as previously reported (31). Both kits rely on the Red/ET recombination technique (31). The oligonucleotide sequences are available on request.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or descriptiona | Source or reference |
|---|---|---|
| E. coli strains | ||
| MG1655 | F− λ− ilvG rfb50 rph-1 | 35 |
| ST18 | S17lpir ΔhemA | 36 |
| DH5α | fhuA2 lacΔU169 phoA glnV44 ϕ80′ lacZΔM15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | 37 |
| MG 35 | MG1655 ΔbtsSR ΔypdAB | This study |
| MG 2 | MG1655 ΔyehUT = ΔbtsSR | 7 |
| MG 20 | MG1655 ΔypdAB | 8 |
| MG1655 PyhjX-gfp | Integration of PyhjX-gfp at the native locus in E. coli MG1655 | This study |
| MG1655 PyjiY-gfp | Integration of PyjiY-gfp at the native locus in E. coli MG1655 | This study |
| MG1655 PrrnB P1-gfp | Integration of PrrnB P1-gfp at the native locus in E. coli MG1655 | This study |
| MG 35 PrrnB P1-gfp | Integration of PrrnB P1-gfp at the native locus in E. coli MG 35 | This study |
| Plasmids | ||
| pRed/ET | λ-RED recombinase in pBAD24; Ampr | Gene Bridges |
| pCP20 | FLP-recombinase, λcI 857+, λpR Repts; Ampr Cmr | 38 |
| pNPTS138-R6KT | mobRP4+ ori-R6K sacB, suicide plasmid; Kanr | 13 |
| pNPTS138-R6KT-PyhjX-gfp | 300 bp of PyhjX fused to gfp and cloned into EcoRI/PspOMI sites of pNPTS138-R6KT; Kanr | This study |
| pNPTS138-R6KT-PyjiY-gfp | 300 bp of PyjiY fused to gfp and cloned into EcoRI/PspOMI sites of pNPTS138-R6KT; Kanr | This study |
| pNPTS138-R6KT-PrrnB P1-gfp | 300 bp of PrrnB P1 fused to gfp and cloned into EcoRI/PspOMI sites of pNPTS138-R6KT; Kanr | This study |
| pXGSF | gfp under the control of a vegetative synthetic promoter | G. Klauck and R. Hengge, unpublished data |
| pBAD24 | Arabinose-inducible PBAD promoter, pBR322 ori; Ampr | 39 |
| pBAD24-gfp | gfp cloned in the EcoRI and NcoI sites of pBAD24 | 26 |
| pBAD24-RR-gfp | gfp-mut2 cloned in the NheI and HindIII sites of pBAD24 (p8754 derivative) | 19 |
| pBAD24-RR-gfpmut-dppA | dppA cloned in the HindIII site of pBAD24-RR-gfpmut2 | This study |
| pCOLA Duet-1 | Expression vector, ColA ori; Kanr | Merck |
| pCOLA-Plac-gfp | gfp under the control of the IPTG-inducible lac promoter cloned in the BamHI and HindIII sites of pCOLA-Duet-1 | This study |
| pBAD33-lysP | lysP in pBAD33; Cmr | 40 |
| pBAD33-lysP-mcherry | mcherry cloned in the XbaI and SalI sites of pBAD33-lysP; Cmr | This study |
Cmr, chloramphenicol resistance; Kanr, kanamycin resistance; Ampr, ampicillin resistance.
E. coli MG1655 strains (Table 1) were grown overnight in lysogeny broth (LB; 10 g/liter NaCl, 10 g/liter tryptone, 5 g/liter yeast extract). After inoculation, bacteria were routinely grown in LB medium under agitation (200 rpm) at 37°C. For solid medium, 1.5% (wt/vol) agar was added. Where appropriate, media were supplemented with antibiotics (kanamycin sulfate, 50 μg/ml; ampicillin sodium salt, 100 μg/ml). For the “low-nutrient environment” experiments, cells from an overnight culture in LB were inoculated into 0.1× diluted LB at a starting OD600 of 0.05 and grown for 1 h. Pyruvate was then added to the cultures to a final concentration of 0.01, 0.05, 0.1, 0.2, 1, or 20 mM.
E. coli MG1655 strains were also grown overnight in M9 minimal medium with 0.5% (wt/vol) glucose as sole carbon source. Bacteria were then inoculated into M9 minimal medium supplemented with increasing concentrations of pyruvate (0.3, 0.6, 1, 2, and 10 mM), and the total carbon source concentration was adjusted to 20 mM using succinate. The conjugation strain E. coli ST18 was grown in the presence of 50 μg/ml 5-aminolevulinic acid.
Construction of fluorescence reporters.
Molecular manipulations were carried out according to standard protocols (32). Plasmid DNA and genomic DNA were isolated using a HiYield plasmid minikit (Sued-Laborbedarf) and a DNeasy blood and tissue kit (Qiagen), respectively. DNA fragments were purified from agarose gels using a HiYield PCR cleanup and gel extraction kit (Sued-Laborbedarf). Q5 DNA polymerase (New England BioLabs) was used according to the supplier's instructions. Restriction enzymes and other DNA-modifying enzymes were also purchased from New England BioLabs and used according to the manufacturer's directions. Replicative plasmids were transferred into E. coli strains using competent cells prepared as described previously (33).
For construction of the promoter-gfp fusions, 300-bp segments of the region immediately upstream of the coding sequence were amplified using oligonucleotide pairs containing EcoRI/PspOMI restriction sites. The resulting promoter fragments were ligated into the γ-origin-dependent vector pNPTS138-R6KT-gfp after restriction with EcoRI/PspOMI. Chromosomal insertions of promoter-gfp into the designated E. coli strains were achieved by integrating the resultant suicide vectors pNPTS138-R6KT-PyhjX-gfp and pNPTS138-R6KT-PyjiY-gfp via RecA-mediated single homologous recombination as described previously (13). The donor strain E. coli ST18, containing the required plasmids, was cultivated together with the recipient E. coli MG1655 strain in LB medium, supplemented with additives as described, to an OD600 of about 0.8. Recombination-positive clones were selected on kanamycin plates, and correct chromosomal integration was checked by PCR and sequencing. To prevent duplication instability, the reporter strains were always cultivated in the presence of kanamycin.
Single-cell fluorescence microscopy and analysis.
To measure promoter activity in individual cells of the reporter strains, cells were cultivated as described above in a rotary shaker. Samples were taken (10 μl) and analyzed on an agarose pad (0.5% [wt/vol] agarose in phosphate-buffered saline [PBS; pH 7.4]), which was placed on a microscope slide and covered with a coverslip.
Images were taken on a Leica microscope (DMI 6000B) equipped with a Leica DFC 365 Fx camera (Andor, 12 bit). An excitation wavelength of 460 nm and a 512-nm emission filter with a 75-nm bandwidth were used for visualization of GFP fluorescence, and an excitation wavelength of 546 nm and a 605-nm emission filter with the same bandwidth were used for visualization of red fluorescence. At least 200 cells per condition were analyzed. The digital images were analyzed using Fiji (34), and statistical analysis was performed using Prism version 5.03 for Windows (GraphPad Software, La Jolla, CA). The background fluorescence was subtracted from each field of view.
The noise was calculated by dividing the standard deviation by the mean. The higher the noise value the more heterogeneous the distribution. The percentage of dark cells was determined from the number of cells whose fluorescence levels overlapped with the negative control (before activation) and the total number of cells quantified. The frequency distributions depict the fraction of values which lie within the range of values that define the bin. The bin range was kept constant at 20 AU. Propidium iodide (Invitrogen, Eugene, OR) was added to the cell cultures at a final concentration of 5 μM to stain dead cells (red fluorescence).
Overproduction experiments.
Overnight cultures of E. coli MG1655 transformed with the plasmid pBAD24-gfp were diluted 100-fold in 20 ml of fresh LB medium supplemented with 100 μg/ml of ampicillin sodium salt and incubated aerobically at 37°C until an OD600 was reached 0.6 (early exponential phase). The cells were induced with l-arabinose 0.2% (wt/vol) for 1 h. Before and after induction, 100-μl samples were taken, diluted 1:1,000 in PBS, and analyzed in a BD Accuri C6 flow cytometer equipped with a solid-state laser (488-nm emission; 20 mW). The green fluorescence emission from GFP was collected by the FL1 filter (BP 533/30 filter). Forward-angle light scatter (FSC) and side-angle light scatter (SSC) were collected in the FSC detector and SSC filter (BP 488/10 filter), respectively. The detection threshold was adjusted for FSC to eliminate noise, and the gate was set on the FSC-SSC dot plot to exclude debris. The sheath flow rate was 14 μl/min, and no more than 100 events/s were acquired. For each sample run, a maximum of 2,000 events were collected. Analysis of data was carried out using Cytospec software (http://www.cyto.purdue.edu/Purdue_software).
Persister cell assay.
To determine the number of persister cells, the number of CFU per ml was measured after exposure of the culture to 200 μg/ml ampicillin. Overnight cultures were diluted 100-fold in 20 ml of fresh LB medium and incubated aerobically at 37°C until the OD600 reached 0.4 or 1.2. Aliquots were then transferred to a new 100-ml flask (final OD600 = 1), and the antibiotic was added. Every hour during antibiotic treatment, samples were taken, serially diluted in PBS, plated on LB agar, and incubated at 37°C for 16 h. CFU were counted as a measure of surviving persister cells. Persisters were calculated as the surviving fraction by dividing the number of CFU per milliliter in the culture after incubation with the antibiotic by the number of CFU per milliliter in the culture before addition of the antibiotic. Each experiment was repeated on three different days.
For calculation of the minimum duration of killing (MDK99), the procedure described above was performed using ofloxacin (at a final concentration of 5 μg/ml) as the antibiotic. The MDK99 value corresponds to the time (in hours) needed to kill 99% of the initial population.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nicola Lorenz and Tobias Bauer for strain construction and Lena Stelzer for excellent technical assistance. We thank Regine Hengge and Gisela Klauck for providing plasmids.
This study was financially supported by the Deutsche Forschungsgemeinschaft grants SPP1617 and Exc114/2 and projects JU270/13-2 (K.J.) and KO 4537/1-2 (D.K.).
The funders had no role in study, design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00536-17.
REFERENCES
- 1.Mascher T, Helmann JD, Unden G. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev 70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 3.Sidote DJ, Barbieri CM, Wu T, Stock AM. 2008. Structure of the Staphylococcus aureus AgrA LytTR domain bound to DNA reveals a beta fold with an unusual mode of binding. Structure 16:727–735. doi: 10.1016/j.str.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin X, Singh KV, Weinstock GM, Murray BE. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect Immun 68:2579–2586. doi: 10.1128/IAI.68.5.2579-2586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimizu T, Shima K, Yoshino K, Yonezawa K, Shimizu T, Hayashi H. 2002. Proteome and transcriptome analysis of the virulence genes regulated by the VirR/VirS system in Clostridium perfringens. J Bacteriol 184:2587–2594. doi: 10.1128/JB.184.10.2587-2594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rood JI. 1998. Virulence genes of Clostridium perfringens. Annu Rev Microbiol 52:333–360. doi: 10.1146/annurev.micro.52.1.333. [DOI] [PubMed] [Google Scholar]
- 7.Kraxenberger T, Fried L, Behr S, Jung K. 2012. First insights into the unexplored two-component system YehU/YehT in Escherichia coli. J Bacteriol 194:4272–4284. doi: 10.1128/JB.00409-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fried L, Behr S, Jung K. 2013. Identification of a target gene and activating stimulus for the YpdA/YpdB histidine kinase/response regulator system in Escherichia coli. J Bacteriol 195:807–815. doi: 10.1128/JB.02051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behr S, Fried L, Jung K. 2014. Identification of a novel nutrient-sensing histidine kinase/response regulator network in Escherichia coli. J Bacteriol 196:2023–2029. doi: 10.1128/JB.01554-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behr S, Kristoficova I, Witting M, Breland EJ, Eberly AR, Sachs C, Schmitt-Kopplin P, Hadjifrangiskou M, Jung K. 2017. Identification of a high-affinity pyruvate receptor in Escherichia coli. Sci Rep 7:1388. doi: 10.1038/s41598-017-01410-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pao SS, Paulsen IT, Saier MH. 1998. Major facilitator superfamily. Microbiol Mol Biol Rev 62:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kristoficova I, Vilhena C, Behr S, Jung K. 2017. BtsT: a novel and specific pyruvate/H+ symporter in Escherichia coli. J Bacteriol doi: 10.1128/JB.00599-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fried L, Lassak J, Jung K. 2012. A comprehensive toolbox for the rapid construction of lacZ fusion reporters. J Microbiol Methods 91:537–543. doi: 10.1016/j.mimet.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 14.Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. 2006. Persisters: a distinct physiological state of Escherichia coli. BMC Microbiol 6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartlett MS, Gourse RL. 1994. Growth rate-dependent control of the rrnB P1 core promoter in Escherichia coli. J Bacteriol 176:5560–5564. doi: 10.1128/jb.176.17.5560-5564.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shan Y, Gandt AB, Rowe SE, Deisinger JP, Conlon BP, Lewis K. 2017. ATP-dependent persister formation in Escherichia coli. mBio 8:1–14. doi: 10.3391/mbi.2017.8.1.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miroux B, Walker JE. 1996. Overproduction of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol 260:289–298. [DOI] [PubMed] [Google Scholar]
- 18.Schlegel S, Genevaux P, de Gier JW. 2015. De-convoluting the genetic adaptations of Escherichia coli C41(DE3) in real time reveals how alleviating protein production stress improves yields. Cell Rep 10:1758–1766. doi: 10.1016/j.celrep.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 19.Santini CL, Bernadac A, Zhang M, Chanal A, Ize B, Blanco C, Wu LF. 2001. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J Biol Chem 276:8159–8164. doi: 10.1074/jbc.C000833200. [DOI] [PubMed] [Google Scholar]
- 20.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 21.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance, and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 22.Behr S, Brameyer S, Witting M, Schmitt-Kopplin P, Jung K. 2017. Comparative analysis of LytS/LytTR-type histidine kinase/response regulator systems in γ-proteobacteria. PLoS One 12:e0182993. doi: 10.1371/journal.pone.0182993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol 46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhou L, Lei X-H, Bochner BR, Wanner BL. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J Bacteriol 185:4956–4972. doi: 10.1128/JB.185.16.4956-4972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozbudak EM, Thattai M, Lim HN, Shraiman BI, Van Oudenaarden A. 2004. Multistability in the lactose utilization network of Escherichia coli. Nature 427:737–740. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]
- 26.Megerle JA, Fritz G, Gerland U, Jung K, Rädler JO. 2008. Timing and dynamics of single cell gene expression in the arabinose utilization system. Biophys J 95:2103–2115. doi: 10.1529/biophysj.107.127191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behr S, Heermann R, Jung K. 2016. Insights into the DNA-binding mechanism of a LytTR-type transcription regulator. Biosci Rep 36:e00326. doi: 10.1042/BSR20160069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiviet DJ, Nghe P, Walker N, Boulineau S, Sunderlikova V, Tans SJ. 2014. Stochasticity of metabolism and growth at the single-cell level. Nature 514:376–379. doi: 10.1038/nature13582. [DOI] [PubMed] [Google Scholar]
- 29.Levy S, Kafri M, Carmi M, Barkai N. 2011. The competitive advantage of a dual-transporter system. Science 334:1408–1412. doi: 10.1126/science.1207154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreth J, Lengeler JW, Jahreis K. 2013. Characterization of pyruvate uptake in Escherichia coli K-12. PLoS One 8:6–12. doi: 10.1371/journal.pone.0067125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heermann R, Zeppenfeld T, Jung K. 2008. Simple generation of site-directed point mutations in the Escherichia coli chromosome using Red(R)/ET(R) Recombination. Microb Cell Fact 7:14. doi: 10.1186/1475-2859-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 33.Inoue H, Nojima H, Okayama H. 1990. High-efficiency transformation of Escherichia coli with plasmids. Gene 96:23–28. doi: 10.1016/0378-1119(90)90336-P. [DOI] [PubMed] [Google Scholar]
- 34.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 36.Thoma S, Schobert M. 2009. An improved Escherichia coli donor strain for diparental mating. FEMS Microbiol Lett 294:127–132. doi: 10.1111/j.1574-6968.2009.01556.x. [DOI] [PubMed] [Google Scholar]
- 37.Taylor RG, Walker DC, McInnes RR. 1993. E. coli host strains significantly affect the quality of small-scale plasmid DNA preparations used for sequencing. Nucleic Acids Res 21:1677–1678. doi: 10.1093/nar/21.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- 39.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tetsch L, Koller C, Haneburger I, Jung K. 2008. The membrane-integrated transcriptional activator CadC of Escherichia coli senses lysine indirectly via the interaction with the lysine permease LysP. Mol Microbiol 67:570–583. doi: 10.1111/j.1365-2958.2007.06070.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


