ABSTRACT
Bacterial biofilm formation is a widespread phenomenon and a complex process requiring a set of genes facilitating the initial adhesion, maturation, and production of the extracellular polymeric matrix and subsequent dispersal of bacteria. Most studies on Escherichia coli biofilm formation have investigated nonpathogenic E. coli K-12 strains. Due to the extensive focus on laboratory strains in most studies, there is poor information regarding biofilm formation by pathogenic E. coli isolates. In this study, we genotypically and phenotypically characterized 187 human clinical E. coli isolates representing various pathotypes (e.g., uropathogenic, enteropathogenic, and enteroaggregative E. coli). We investigated the presence of biofilm-associated genes (“genotype”) and phenotypically analyzed the isolates for motility and curli and cellulose production (“phenotype”). We developed a new screening method to examine the in vitro biofilm formation ability. In summary, we found a high prevalence of biofilm-associated genes. However, we could not detect a biofilm-associated gene or specific phenotype correlating with the biofilm formation ability. In contrast, we did identify an association of increased biofilm formation with a specific E. coli pathotype. Enteroaggregative E. coli (EAEC) was found to exhibit the highest capacity for biofilm formation. Using our image-based technology for the screening of biofilm formation, we demonstrated the characteristic biofilm formation pattern of EAEC, consisting of thick bacterial aggregates. In summary, our results highlight the fact that biofilm-promoting factors shown to be critical for biofilm formation in nonpathogenic strains do not reflect their impact in clinical isolates and that the ability of biofilm formation is a defined characteristic of EAEC.
IMPORTANCE Bacterial biofilms are ubiquitous and consist of sessile bacterial cells surrounded by a self-produced extracellular polymeric matrix. They cause chronic and device-related infections due to their high resistance to antibiotics and the host immune system. In nonpathogenic Escherichia coli, cell surface components playing a pivotal role in biofilm formation are well known. In contrast, there is poor information for their role in biofilm formation of pathogenic isolates. Our study provides insights into the correlation of biofilm-associated genes or specific phenotypes with the biofilm formation ability of commensal and pathogenic E. coli. Additionally, we describe a newly developed method enabling qualitative biofilm analysis by automated image analysis, which is beneficial for high-throughput screenings. Our results help to establish a better understanding of E. coli biofilm formation.
KEYWORDS: biofilm formation, Escherichia coli, pathotypes, VideoScan
INTRODUCTION
Biofilms are microbial communities that live in a self-produced extracellular polymeric matrix consisting of exopolysaccharides (EPS), proteins, and DNA adhered to abiotic or biotic surfaces (1 – 3). Bacterial biofilms are highly problematic in clinical settings, where they can cause chronic, nosocomial, and device-related infections due to their high resistance to antibiotics and the host immune system (4, 5). It is increasingly presumed that the formation of microbial communities is associated with the pathogenicity of bacteria (6, 7) and that bacteria persisting in biofilms are the main cause for recurrent and chronic infections (8).
Escherichia coli is known as a highly versatile bacterium that can exist as a harmless commensal in the mammalian colon or as a pathogen causing significant morbidity and mortality worldwide. There are two types of pathogenic E. coli, namely, intestinal pathogenic E. coli (InPEC) and extraintestinal pathogenic E. coli (ExPEC). The ExPEC group includes uropathogenic E. coli (UPEC), which is responsible for urinary tract infections (UTI) (9) and commonly found in biofilms on the surface of urinary catheters (10). The InPEC isolates cause diarrheal infections and can be classified into eight different pathotypes based on their virulence gene repertoire and adherence pattern to epithelial HEp-2 cells. These eight pathotypes are enteropathogenic E. coli (EPEC), diffusely adherent E. coli (DAEC), enterotoxigenic E. coli (ETEC), adherent-invasive E. coli (AIEC) associated with Crohn's disease, enteroaggregative E. coli (EAEC), and Shiga toxin-producing E. coli (STEC), including enterohemorrhagic E. coli (EHEC) and enteroinvasive E. coli (EIEC) (11 – 13).
Since its first description in 1987 (14), the pathogenesis of EAEC infection remains poorly elucidated. EAEC is well known for its ability to adhere to the small and large intestinal mucosa and to form thick aggregating biofilms by surrounding itself with an extracellular matrix (15 – 17). Wakimoto et al. screened a total of 1,042 E. coli isolates from children with diarrhea and reported that EAEC isolates exhibited significantly stronger biofilm formation than non-EAEC isolates (18). In addition, Mohamed et al. determined the potential of different EAEC isolates from travelers to developing countries and confirmed that biofilm formation is a widespread phenomenon in the EAEC pathotype (19). These in vivo and in vitro observations have led to the conjecture that the genetic equipment of different pathotypes enables them to exhibit increased biofilm formation. Thus, the ability to form biofilms is a defined characteristic of certain pathotypes contributing to their pathogenesis. To date, the literature contains only one study that compares biofilm formation among a large number of different E. coli pathotypes (20), and only UPEC and EAEC are well known for their biofilm forming capabilities.
E. coli is one of the most-studied model organisms for biofilm formation analysis in vitro. Biofilm formation in E. coli requires a set of genes facilitating its initial adhesion, maturation, EPS production, and subsequent dispersal. Various studies have focused on the impact of cell surface components on the formation of bacterial surface communities of nonpathogenic E. coli K-12 strains (21 – 25). Pratt and Kolter showed that motility mediated by flagella and type 1 fimbriae contributes to biofilm formation in E. coli K-12 (21). These results were confirmed by Wood et al., who directly correlated the motility of E. coli K-12 with the biofilm-forming ability (26). However, another study demonstrated that the presence of mannoside and the lack of motility did not impair the ability of an EAEC strain to form biofilms (27). This highlights the fact that results from nonpathogenic strains do not reflect the true picture of biological processes in all E. coli isolates. Moreover, due to the extensive focus on nonpathogenic E. coli in most studies, there is scant information regarding biofilm formation by E. coli pathotypes and the impact of biofilm-associated components. Among the wide range of surface components described for E. coli, only aggregative adherence fimbriae (AAF) of EAEC were shown to be related to biofilm formation in vitro (27).
To ascertain the correlations of genotypic and phenotypic characteristics and the pathotype with biofilm formation, we investigated 187 clinical E. coli isolates. These included human intestinal commensal (fecal) E. coli (HFEC), uropathogenic E. coli (UPEC), which was described as biofilm developing, enterotoxigenic E. coli (ETEC), atypical enteropathogenic E. coli (aEPEC), enteroaggregative E. coli (EAEC), which was already described as biofilm developing, sepsis-associated E. coli (SAEC), Crohn's disease-associated E. coli (CAEC), and chicken intestinal commensal (avian fecal) E. coli (AFEC). In addition to the evaluation of the biofilm-forming capability, isolates were examined for the presence of biofilm-associated genes (genotype) and analyzed phenotypically for motility, curli expression, and cellulose production (phenotype), since E. coli biofilm formation has been associated with the expression of curli fimbriae and exopolysaccharide cellulose (28, 29).
The VideoScan technology, which is based on fully automated fluorescence microscopy, enables the analysis of multiplex assays such as microbead, droplets, and cell-based assays or assays in solution (30 – 33). We expanded this VideoScan technology during this study for high-throughput screening of biofilms.
RESULTS
Genotypic characterization of E. coli pathotypes.
Biofilm-associated genes were found in various frequencies in all eight pathotypes tested, with csgA (99.5%), bcsA (96.3%), fimH (92%), and fliC (77%) showing higher prevalences and agn43 (43.3%) and papC (33.7%) having lower prevalences in all 187 isolates. UPEC isolates carried the highest number of all detected genes per pathotype (86.9%), followed by SAEC (85.1%), while CAEC and EPEC showed the lowest numbers (71.4%).
The prevalences of agn43, fimH, fliC, and papC genes differed significantly among E. coli pathotypes. EAEC isolates exhibited a significantly higher prevalence of agn43 than EPEC (P < 0.01), ETEC (P < 0.01), CAEC (P < 0.01), and AFEC (P < 0.05) isolates. EPEC isolates showed a prevalence of agn43 that was significantly lower than isolates from the pathotypes SAEC (P < 0.01), UPEC (P < 0.05), and HFEC (P < 0.05). Furthermore, EAEC isolates were found to have a lower prevalence of fimH than AFEC, EPEC, SAEC, and UPEC isolates (P < 0.05). For fliC, a significantly lower prevalence was found in EAEC isolates than in ETEC (P < 0.05) isolates. UPEC isolates had a higher prevalence of papC than ETEC and EAEC isolates (P < 0.01). Additionally, ETEC isolates showed a lower prevalence of papC than SAEC (P < 0.01) and HFEC (P < 0.05) isolates. Detailed information about the genotypic characterizations of the examined E. coli isolates is given in Table 1.
TABLE 1.
Prevalences of biofilm-associated genes in 187 E. coli isolates
| Gene | % of isolates |
P valuea | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HFEC | UPEC | ETEC | EPEC | EAEC | SAEC | CAEC | AFEC | ||
| agn43 | 54.2 | 58.3 | 25 | 12.5b,c | 75b,d | 62.5 | 21.1 | 33.3 | 0.0001 |
| bcsA | 95.8 | 100 | 100 | 83.3 | 100 | 100 | 89.5 | 100 | 0.0165 |
| papC | 41.7 | 66.7b,e | 4.2b,f | 33.3 | 12.5 | 50 | 26.3 | 33.3 | 0.0002 |
| csgA | 95.8 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0.4470 |
| fimH | 91.7 | 100 | 87.5 | 100 | 66.7b,g | 100 | 89.5 | 100 | 0.0002 |
| fliC | 83.3 | 83.3 | 95.8 | 70.8 | 50b,h | 83.3 | 73.7 | 75 | 0.0208 |
P values for the comparison of the prevalence of a gene between all pathotypes.
Prevalence of this gene was statistically significantly different between two pathotypes, with at least a P value of <0.05.
Lower than SAEC, UPEC, and HFEC.
Higher than EPEC, ETEC, CAEC, and AFEC.
Higher than ETEC and EAEC.
Lower than SAEC and HFEC.
Lower than AFEC, EPEC, SAEC, and UPEC.
Lower than ETEC.
Curli and cellulose production in E. coli pathotypes.
After incubating at 28°C on Congo red agar plates, 52 (27.8%) isolates produced the red, dry, and rough (rdar) morphotype, showing curli and cellulose production, 61 (32.6%) isolates had the brown, dry, and rough (bdar) morphotype, showing only curli production, and 74 (39.6%) isolates produced neither curli nor cellulose and showed the smooth and white (saw) morphotype. An incubation of the bacteria at 37°C showed only 27 (14.4%) isolates produced the rdar morphotype, whereas the bdar morphotype was found in 72 (38.5%) isolates and the saw morphotype in 88 (47.1%) isolates (Fig. 1 and 2). However, no significant association between a pathotype and a specific morphotype was found. No isolate producing the pink, dry, and rough (pdar) morphotype indicative of cellulose expression without curli production was found. Detailed information about curli and cellulose production in the examined E. coli isolates is given in Table 2.
FIG 1.
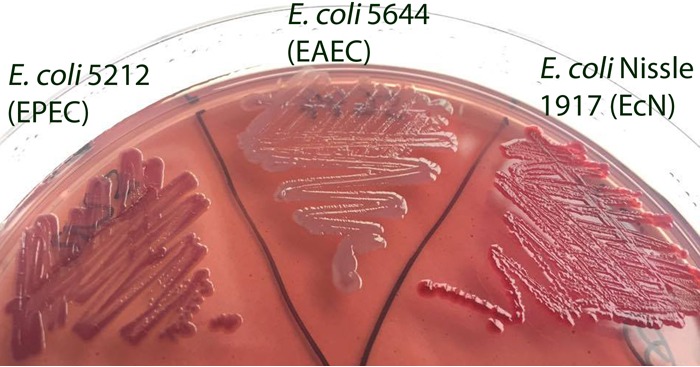
Colony morphologies and colors of different E. coli isolates on a Congo red agar plate. Bacteria were grown at 37°C for 24 h on LB agar plates without salt containing Congo red dye. E. coli Nissle 1917 developed a rough and dry colony morphology with red color based on the binding of Congo red dye (rdar morphotype). E. coli isolate 5212 (EPEC) exhibited a brown and shinier color but also a dry and rough colony morphology (bdar morphotype). The isolate E. coli 5644 showed smooth and white-colored colonies (saw morphotype).
FIG 2.

Fluorescence of different E. coli isolates on a calcofluor agar plate. Bacteria were grown at 37°C for 24 h on LB agar plates containing calcofluor. Cellulose-producing bacteria showed a fluorescent colony morphology (E. coli Nissle 1917). E. coli isolate 5644 (EAEC) exhibited no cellulose production.
TABLE 2.
Incidences of Congo red morphotype and fluorescence on calcofluor plates in 187 E. coli isolates
| Pathotype | No. (%) of isolatesa |
|||||||
|---|---|---|---|---|---|---|---|---|
| 28°C |
37°C |
|||||||
| Morphotype on CR |
Fluorescence on CF | Morphotype on CR |
Fluorescence on CF | |||||
| rdar | bdar | saw | rdar | bdar | saw | |||
| HFEC | 9 (37.5) | 5 (20.8) | 10 (41.7) | 9 (37.5) | 2 (8.3) | 7 (29.2) | 15 (62.5) | 2 (8.3) |
| UPEC | 6 (25) | 7 (29.2) | 11 (45.8) | 6 (25) | 4 (16.7) | 12 (50) | 8 (33.3) | 4 (16.7) |
| ETEC | 4 (16.7) | 11 (45.8) | 9 (37.5) | 4 (16.7) | 2 (8.3) | 11 (45.8) | 11 (45.8) | 2 (8.3) |
| EPEC | 5 (20.8) | 10 (41.7) | 9 (37.5) | 5 (20.8) | 3 (12.5) | 5 (20.8) | 16 (66.7) | 3 (12.5) |
| EAEC | 6 (25) | 5 (20.8) | 13 (54.2) | 6 (25) | 4 (16.7) | 9 (37.5) | 11 (45.8) | 4 (16.7) |
| SAEC | 9 (37.5) | 8 (33.3) | 7 (29.2) | 9 (37.5) | 3 (12.5) | 15 (62.5) | 6 (25) | 3 (12.5) |
| CAEC | 6 (31.6) | 6 (31.6) | 7 (36.8) | 6 (31.6) | 5 (26.3) | 4 (21.1) | 10 (52.6) | 5 (26.3) |
| AFEC | 7 (29.2) | 9 (37.5) | 8 (33.3) | 7 (29.2) | 4 (16.7) | 9 (37.5) | 11 (45.8) | 4 (16.7) |
Percentages based on all isolates from the pathotype. CR, Congo red; CF, calcofluor.
Motility assay.
Of the 144 (77%) E. coli isolates that were positive for fliC expression, only 114 (79%) isolates showed motility in 0.25% Luria-Bertani (LB) soft agar (Fig. 3). Twenty-five more isolates showing no amplification of fliC by PCR were also found motile on soft agar, making the total number of isolates showing motility 139 (74%). The highest number of motile isolates was found in the HFEC group (92%). The least of all motile isolates belonged to the AFEC group, with 14 motile isolates (58%). When examining the association of fliC expression and motility, we found a strong significant correlation (P < 0.01), suggesting that isolates carrying fliC were usually motile. However, no significant association was found between the different groups and motility.
FIG 3.
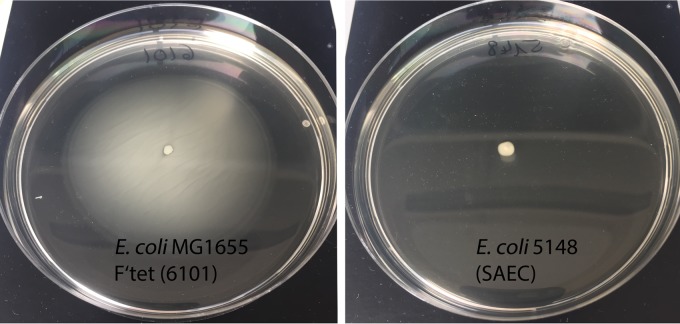
Swimming motility of E. coli on a soft agar plate. Bacteria were stabbed in the center of the agar plate and incubated at 37°C for 16 h. A positive motility was indicated by diffused growth around the point of inoculation (E. coli MG1655 F′Tet). Growth restricted along the stab point indicated a nonmotile isolate (E. coli 5148).
Biofilm formation with crystal violet assay.
In total, 187 E. coli isolates were examined for biofilm formation in LB medium using crystal violet staining. This method was applied to confirm the results achieved with VideoScan. The mean optical density at 570 nm (OD570) values were 0.100 ± 0.04 for the negative control (sterile LB medium) and 3.599 ± 0.32 for the positive control E. coli MG1655 F′Tet ΔtraD. The cutoff OD570 value for an isolate to be considered a biofilm producer (ODc) was found to be 0.208.
In total, 179 (96%) isolates were identified as biofilm forming, whereas only eight (4%) isolates were considered non-biofilm forming. OD570 readings ranged from 0.172 to 3.074. Of the biofilm-forming isolates, 77 (41%) were weak biofilm producers (OD570 of >0.208 and ≤0.416), 65 (35%) isolates were moderate biofilm producers (OD570 of >0.416 and ≤0.832), and 37 (20%) isolates were found to be strong biofilm producers (OD570 of >0.832). The majority of isolates with a strong biofilm-forming capability was found in the pathotype EAEC, with 14 (58%) isolates showing OD570 readings of >0.832. The group with the fewest isolates forming strong biofilms was the EPEC group, with only one isolate (4%).
Biofilm formation with the VideoScan technology.
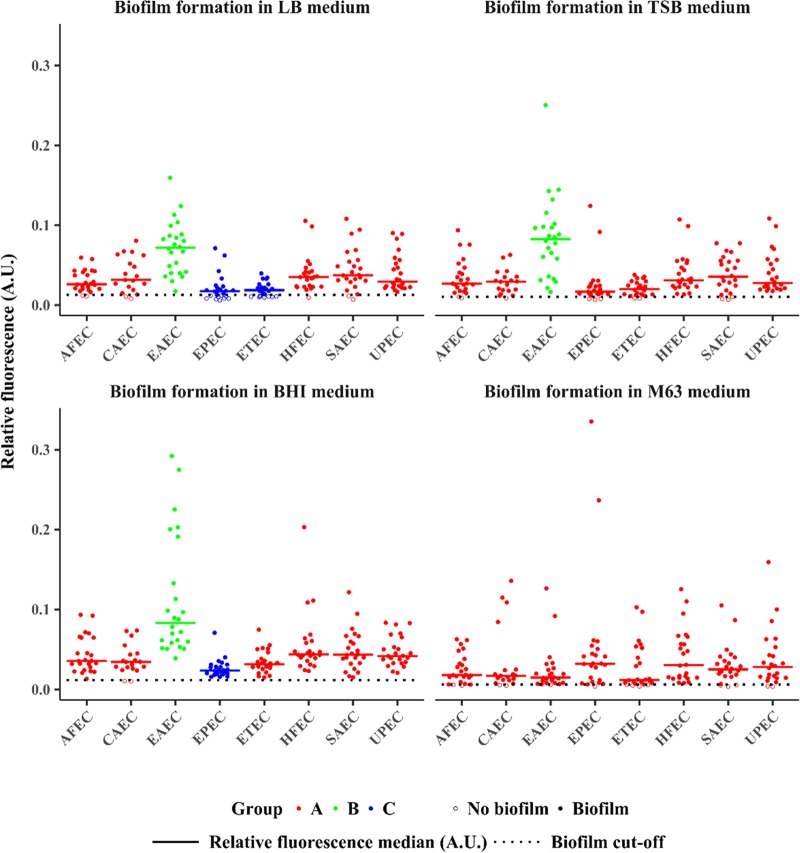
In total, 187 E. coli isolates were examined for biofilm formation in enriched LB, tryptic soy broth (TSB), and brain heart infusion (BHI) broth and in minimal medium M63 and were analyzed with the VideoScan technology. The relative fluorescence intensity (relFI) of the negative controls (medium without bacteria) was 0.0027 for LB medium, 0.0031 for TSB, 0.0055 for BHI broth, and 0.0006 for M63 medium. For the positive control E. coli MG1655 F′Tet ΔtraD, the relFI was 0.3656 in LB broth, 0.4059 in TSB, 0.3453 in BHI broth, and highest in M63 medium, at 0.7738. We defined a cutoff for each medium to distinguish between non-biofilm- and biofilm-forming isolates, as it is commonly used for crystal violet assays (34). Cutoffs were 0.0128 for LB broth, 0.0105 for TSB, 0.0115 for BHI broth, and 0.0060 for M63 medium (Fig. 4). In total, 24 isolates (13%) were considered non-biofilm forming using the VideoScan technology.
FIG 4.
Biofilm formation of E. coli isolates in four different culture media analyzed with the VideoScan technology. Dot plots showing the relative fluorescence intensities ([relFIs] AU) of SYTO 9-stained bacteria, which were examined for biofilm formation in four different media (LB, TSB, BHI, and M63). Each dot represents the median value of the relFI. The biofilm assays were performed three times with triplicates in each assay. The dotted lines mark the cutoffs for biofilm formation in each medium. Open circles indicate no biofilm and solid circles indicate biofilm formation. EAEC isolates exhibited the highest relFI in enriched media (LB, TSB, and BHI). The relFIs differed significantly between the pathotypes (0.95 confidence level). To show these differences, pathotypes were grouped. Pathotypes in group A are significantly different from pathotypes in group B. For example, in LB medium, the pathotypes AFEC, CAEC, HFEC, SAEC, and UPEC (group A) exhibit significantly higher relFIs than EPEC and ETEC (group C) and significantly lower relFIs than EAEC (group B). The groups for pathotypes were created separately for each medium.
The relFI for the examined isolates incubated in LB medium ranged from 0.0060 (EPEC isolate) to 0.1595 (EAEC isolate). In TSB medium, the lowest relFI was 0.0066 (EPEC isolate) and the highest was 0.2504 (EAEC isolate). The relFI for isolates incubated in BHI broth ranged from 0.0102 (CAEC isolate) to 0.2923 (EAEC isolate). In M63 medium, the lowest relFI was 0.0035 (SAEC isolate) and the highest was 0.3355 (EPEC isolate).
A comparison of both methods for the evaluation of biofilms in LB medium showed a significant correlation between VideoScan and crystal violet (Pearson's correlation coefficient [r] = 0.5887, P = 7.953e−19, 95% confidence interval, 0.4863 to 0.6751). The pathotype with the significantly highest capacity for biofilm formation in enriched medium was EAEC, with a mean relFI of 0.0716 in LB broth, 0.0842 in TSB, and 0.1161 in BHI medium (P < 0.01). Although EPEC isolates were found to exhibit low biofilm-forming capabilities in enriched media, this group comprised the strongest biofilm-forming isolates in minimal medium M63. The highest relFI values for all 187 tested isolates for biofilm formation were obtained using BHI medium. Figure 4 shows the distributions of relFI values for each of the E. coli pathotype isolates.
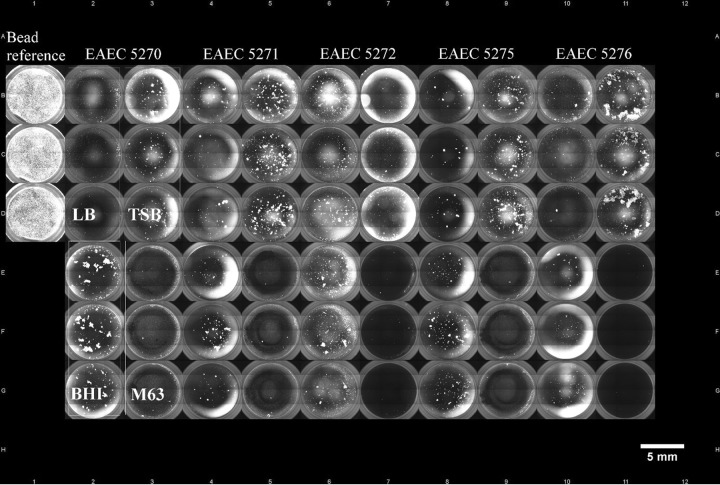
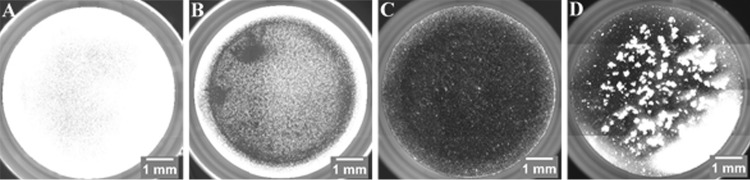
To further examine E. coli isolates for biofilm formation, overview images from each well of the 96-well plates were captured automatically. Overview images of the single wells were merged to 96-well-plate overview images for each plate (Fig. 5), which enabled better comparisons among the biofilms formed by a single isolate in different culture media and among different isolates on the plate. This qualitative analysis provided information about the general biofilm formation of a bacterial isolate, the spatial distribution, and the structure of the biofilm (e.g., flat, aggregated, etc.). Figure 6 shows overview images of single wells with E. coli isolates representing the different strengths of formed biofilms. The positive control exhibited a dense and compact biofilm, which was equally distributed within the well (Fig. 6A). Another kind of biofilm was shown by an HFEC isolate in M63 medium. The isolate built up a stronger biofilm at the edge of the well, indicated by a bright fluorescent ring, compared to at the center of the well, where bacterial cells were evenly distributed and formed a flat biofilm (Fig. 6B). In contrast to this, a UPEC isolate incubated in the same medium only formed a very thin and consistently distributed biofilm (Fig. 6C). However, no correlation was found between a pathotype and a specific kind of biofilm, except for EAEC. In addition to the significantly high biofilm-forming capability of this pathotype, EAEC isolates formed characteristic dense aggregating biofilms. In total, 21 of 24 EAEC isolates formed this typical biofilm consisting of thick bacterial aggregates (Fig. 6D).
FIG 5.
Overview image of E. coli biofilms in a 96-well plate. Merged single-well fluorescence images show a 96-well plate with SYTO 9-stained biofilms. From left to right: beads that were used as an internal reference and five EAEC isolates (5270, 5271, 5272, 5275, and 5276) are shown. Each isolate was examined for biofilm formation in four media (LB, TSB, BHI, and M63) in triplicate (box).
FIG 6.
Single-well overview images of E. coli isolates showing different strengths of biofilm formation. Fluorescence images were captured after 48 h of biofilm formation in respective media and staining with SYTO 9. (A) The positive control E. coli MG1655 F′Tet ΔtraD in LB medium exhibiting strong biofilm formation (relFI, 0.4398). (B) One HFEC isolate in M63 medium displaying an even biofilm in the center and a much more pronounced biofilm along the edge of the well (relFI, 0.1102). (C) One UPEC isolate in M63 medium forming a flat and evenly distributed biofilm in the well (relFI, 0.0369). (D) One EAEC isolate in BHI medium showing the characteristic biofilm consisting of thick bacterial aggregates (relFI, 0.2763).
DISCUSSION
We characterized 187 E. coli isolates to ascertain potential associations of biofilm-associated genes (genotype), motility, curli expression, and cellulose production (phenotype) in different E. coli pathotypes with their biofilm formation ability. To evaluate biofilm formation by a large number of commensal and pathogenic E. coli isolates, we developed and used a new screening method. Biofilm formation was analyzed by the VideoScan technology based on fully automated fluorescence microscopy to perform image-based high-throughput screening of biofilms.
A high prevalence of biofilm-associated genes was found due to the fact that the majority of the microorganisms exist in biofilms in nature. For pathogenic bacteria, biofilm formation is an important step in the host infection process and thus contributes to pathogenicity (6). Motility in E. coli is common and is considered a virulence factor in pathogenic bacteria (35 – 37). The gene encoding FliC, the main structural subunit of E. coli flagella (38, 39), was found in 144 (77%) E. coli isolates. In total, 139 (74%) isolates were motile in soft agar, of which 25 (18%) isolates showed no amplification of the fliC gene. This could be due to sequence variations in fliC among different E. coli isolates (40) and/or the expression of another or new flagellin genes (41). Although Pratt and Kolter found that motility, and not chemotaxis, is required for E. coli to form a biofilm (21), it is also possible for nonmotile strains to adhere to surfaces and develop a biofilm (42). Sheikh et al. showed that a nonmotile EAEC mutant could form biofilms on glass and plastic surfaces indistinguishable from those of the wild-type strain (27). This is in agreement with our findings, as the screening for motility revealed only 67% of EAEC isolates were motile in comparison to HFEC with 92% motile isolates; even then, EAEC isolates were found to be the strongest biofilm formers. As no correlation of motility was found with biofilm formation, our results suggest that motility is not necessarily required for biofilm formation.
Other factors playing an important role in biofilm formation are adhesins. Type I fimbriae are proteinaceous filamentous adhesins, which are commonly produced by both commensal and pathogenic E. coli isolates (43). These fimbriae have been reported to play a critical role in E. coli's ability to form biofilms on abiotic surfaces, as fimH mutants have been reported to show reduced initial attachment (21, 44, 45). The pathotype EAEC showed a prevalence of 66.7% for fimH, which was higher in all other pathotypes, suggesting different adhesive structures, e.g., aggregative adherence fimbriae, are involved in the initial adhesion of EAEC during biofilm formation (27).
Another biofilm-associated gene that was examined was papC encoding the outer membrane assembly platform, which is needed for the formation of P fimbriae. These pyelonephritis-associated pili (pap) are commonly found in UPEC organisms and they play a pivotal role in the pathogenesis of ascending urinary tract infections, such as in pyelonephritis (46, 47). By PCR, we found a high prevalence (66.7%) of papC-carrying UPEC isolates. However, the data suggested that this biofilm-associated gene cannot account for the differences observed in biofilm formation, as EAEC isolates, shown to be the strongest biofilm-forming pathotype, exhibited significantly lower prevalence for papC (P < 0.01). Additionally, no correlation for UPEC isolates between papC and biofilm formation in vitro could be found, which rather suggests a role in the in vivo colonization of uroepithelial cells (48).
One of the characteristic features that distinguishes bacteria existing in biofilms from planktonic bacteria is the extracellular matrix, with curli fimbriae being the main constituent of E. coli biofilms. CsgA, the major subunit of curli fimbriae, plays crucial roles in cell aggregation, adhesion to surfaces, and biofilm formation (28, 49, 50). Frömmel et al. screened 317 E. coli isolates for virulence-associated genes and found a prevalence of 90.9% for gene encoding CsgA (51). We have also found a very high (99.5%) prevalence in E. coli isolates, and curli have also been reported as highly conserved and present in almost all E. coli organisms (52). A second component of the matrix found in E. coli biofilms is cellulose. BcsA, acting as the catalytic subunit of the cellulose synthase, was found in 180 isolates (96.3%), as it is a widespread phenomenon in E. coli organisms (53, 54). In addition to the presence of csgA and bcsA, we verified the isolate's general ability to express curli and cellulose to ascertain a potential correlation with biofilm formation. To investigate curli and cellulose production as it is carried out in many other publications (55, 56), LB agar plates without salt were used, as low salt concentrations are known to increase curli and cellulose expression. In total, curli expression was less prevalent when incubating the isolates at higher temperatures, which is in agreement with the fact that curli expression is a temperature-regulated process (57, 58). Although curli and cellulose are known to promote biofilm formation (54, 59), we could not associate the phenotype expressing both components with biofilm formation. We observed that some of the best biofilm formers, within the group of EAEC, did not produce cellulose or curli, whereas one ETEC isolate expressed both and showed only weak biofilm formation. The fact that curli expression is repressed at 37°C, which was also confirmed in our study, showed clearly that this component is not critical for biofilm formation in the pathogenic isolates under the investigated conditions.
Finally, we investigated the occurrence of the gene agn43 or flu encoding antigen 43 (Ag43), an autoaggregation factor belonging to the group of autotransporter proteins known to promote cell-to-cell adhesion. This outer membrane protein plays a key role in biofilm formation by leading to the autoaggregation of bacterial cells and thereby facilitating the three-dimensional biofilm structure (60 – 64). Higher expression of the gene encoding Ag43 was found in biofilm-forming bacteria, and a deletion of agn43 caused a decrease in biofilm formation (24, 63). We also found high prevalences of agn43 in EAEC (75%), SAEC (62.5%), and UPEC (58.3%) isolates, which is in agreement with studies from other groups (65, 66). A strong formation of aggregates, which is promoted by Ag43 (67), was observed while screening EAEC isolates for biofilm formation in different media with our VideoScan technology. Although EAEC isolates showed the highest prevalence of agn43, no significant correlation was found between the gene and the biofilm forming ability. Therefore, we suggest that the autoaggregation factor supports biofilm formation in EAEC isolates, but it is not critical for biofilm formation on abiotic surfaces.
We have adapted the VideoScan technology and expanded it for the screening of biofilm formation. Using the crystal violet method, eight isolates (4%) were identified as non-biofilm formers, whereas 24 isolates (13%) showed no biofilm formation using the VideoScan method. Altogether, two isolates, one CAEC and one AFEC isolate, were scored as non-biofilm forming by both methods. The isolates that made up the largest part of the non-biofilm formers identified with the VideoScan technology belong to the pathotypes ETEC and EPEC. In total, 17 of 24 (71%) isolates unable to form biofilms were ETEC or EPEC isolates. In contrast, nearly all of these isolates showed weak to moderate biofilm formation using the crystal violet method. This could be explained by the fact that ETEC and EPEC isolates form biofilms at the peripheries of the wells and at the air-liquid interface, generally referred to as “pellicle” or floating biofilms (68, 69). Thus, the VideoScan method detects biofilms formed at the bottoms of the wells and distinguishes between biofilm formation on surfaces and floating biofilms, which is beneficial in testing different materials or surface coatings. In comparison to the crystal violet assay as the gold standard method for biofilm analysis, where only the strength of biofilm formation is measured, the VideoScan platform has the advantage of being an automated method, offering timesaving assay performance and creating added value, as it represents an image-based technology where the user can obtain information about the structure and spatial distribution of the formed biofilm. In summary, we found a considerable variation among the E. coli isolates in their abilities to form biofilms in vitro. As already shown in other studies (20, 70), we also found that the growth medium has a major effect on in vitro biofilm formation. The highest relFI values were obtained when using BHI medium for biofilm formation. This could be due to the fact that BHI is a highly nutritious medium containing calf brain and beef heart, and thus essential growth factors, amino acids, and vitamins enhancing bacterial growth. The most significant ability for biofilm formation was found in isolates of the pathotype EAEC (P < 0.01), and the image-based VideoScan technology demonstrated the characteristic biofilm formation pattern of EAEC, consisting of thick bacterial aggregates.
The VideoScan technology can be used as a high-throughput screening method to evaluate biofilms by automated epifluorescence microscopy. This image-based platform is cost-efficient, multiplexing (different conditions at one time), and timesaving. Furthermore, this technology can be applied to the screening of mutant libraries for biofilm formation regarding the identification of biofilm-associated genes or for testing the efficacy of antimicrobial agents. To expand the VideoScan method used for screening of biofilms, we plan to incorporate fluorescent dyes, e.g., calcofluor white, to stain different matrix components.
In conclusion, our data indicate that the presence or absence of a genotype or phenotype cannot explain the differences observed between biofilm formations of different E. coli pathotypes under the investigated conditions. These findings highlight the fact that biofilm-promoting factors shown to be critical for biofilm formation in nonpathogenic strains do not necessarily reflect their impact in clinical isolates. The biofilm of EAEC constituted a unique formation of bacterial communities, suggesting the contribution of additional, so far unknown, factors in the process of biofilm formation, supporting their survival and virulence in the host. Thus, biofilm formation of EAEC isolates could contribute to the establishment of long-term colonization and chronic infections, where the biofilm matrix protects embedded bacteria against the host immune system and antimicrobial agents.
MATERIALS AND METHODS
Bacterial isolates.
In this study, various clinical isolates from humans were investigated, including HFEC (n = 24), UPEC (n = 24), ETEC (n = 24), aEPEC (n = 24), EAEC (n = 24), SAEC (n = 24), and CAEC (n = 19) isolates, while isolates from birds (AFEC [n = 24]) were used as a control group. All isolates were epidemiologically independent, and only one E. coli isolate per sample was used. Bacterial isolates obtained from different origins (e.g., clinical laboratory or the institute's strain collection) were streaked onto CHROMagar orientation plates (Mast Diagnostica GmbH, Reinfeld, Germany) (71). One single pink colony was subcultured twice and stored in 15% glycerol at −80°C until further usage.
E. coli strain K-12 MG1655 F′Tet ΔtraD served as a positive control for biofilm formation. This strain bears a conjugative plasmid known to be a strong adhesion factor and thus mediates strong biofilm formation (72). E. coli Nissle 1917 was used as a positive control for the screening of curli fimbriae and cellulose expression (73).
Detection of biofilm-associated genes.
The presence of biofilm-associated genes was evaluated by PCR for genes csgA, agn43, papC, fliC, bcsA, and fimH. Primer sets (BioTeZ Berlin-Buch GmbH, Berlin, Germany) and sizes of PCR products are shown in Table 3. For fimH, agn43, papC, and fliC, the primer sequences were chosen from previously published studies and verified using BLAST. For bcsA and csgA, the nucleotide sequences known to be highly conserved in E. coli were taken from the GenBank sequence database (NCBI) and aligned using BLAST to verify the conservation of the gene sequences among sequenced E. coli isolates in the database.
TABLE 3.
Primers for detection of biofilm-associated genes in E. coli
| Gene | Primer |
Reference or source | Description | ||
|---|---|---|---|---|---|
| Directiona | Sequence (5′→3′) | Product size (bp) | |||
| agn43/flu | F | GGGTAAAGCTGATAATGTCG | 508 | 86 | Autoaggregation factor antigen 43 |
| R | GTTGCTGACAGTGAGTGTGC | ||||
| bcsA | F | GCTTCTCGGCGCTAATGTTG | 826 | This study | Cellulose |
| R | GAGGTATAGCCACGACGGTG | ||||
| papC | F | TGATATCACGCAGTCAGTAGC | 501 | 87 | P pili associated with pyelonephritis |
| R | CCGGCCATATTCACATAAC | ||||
| csgA | F | GCAATCGTATTCTCCGGTAG | 418 | This study | Curli fimbriae |
| R | GATGAGCGGTCGCGTTGTTA | ||||
| fimH | F | TGCAGAACGGATAAGCCGTGG | 506 | 88 | Type I fimbriae |
| R | GCAGTCACCTGCCCTCCGGTA | ||||
| fliC | F | ATGGCACAAGTCATTAATACCCAAC | 1,497 | 89 | Flagella |
| R | CTAACCCTGCAGCAGAGACA | ||||
F, forward; R, reverse.
Bacterial heat lysates were prepared by inoculating 1 ml of LB medium with a single E. coli colony and incubating at 37°C for 16 h with shaking at 180 rpm. Bacteria were centrifuged and pellets were resuspended in 300 μl sterile double-distilled water (ddH2O), followed by lysis at 99°C for 10 min and incubating 15 min on ice. After centrifugation, the DNA-containing supernatants were stored at −20°C. For each PCR reaction mixture, 1 μl of lysate was mixed with 14 μl of PCR mix in a 96-well plate using a Biomek 2000 laboratory automation workstation (Beckman Coulter GmbH, Krefeld, Germany). PCR mixtures contained the following components (final concentrations): 1× PCR buffer, 4 mM MgCl2 (Rapidozym GmbH, Berlin, Germany), 0.2 mM deoxynucleoside triphosphates (dNTPs) (Jena Bioscience GmbH, Jena, Germany), 0.4 μM each primer, and 0.5 U BioTherm Taq DNA polymerase (Rapidozym GmbH). PCR was performed in an Eppendorf cycler (Mastercycler EP gradient; Eppendorf, Hamburg, Germany) using the following program: 3 min at 95°C, 30 cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C, and a final extension for 5 min at 72°C. PCR results were analyzed using MultiNA capillary electrophoresis (MCE-202 MultiNA; Shimadzu GmbH, Duisburg, Germany). Each isolate was tested twice in independent experiments for the detection of biofilm-associated genes.
Cellulose and curli production.
In Salmonella and E. coli, the biofilm formation is associated with the expression of curli fibers and cellulose (28, 53, 74). The production of these matrix components can be examined by using Congo red dye, which binds to both cellulose and curli (55, 75). To determine the production of cellulose and curli fimbriae, E. coli isolates were streaked on LB agar plates without salt supplemented with 40 μg/ml Congo red (Carl Roth GmbH, Karlsruhe, Germany) and 20 μg/ml brilliant blue R250 (Carl Roth GmbH). After incubating for 24 h at 37°C and 48 h at 28°C, the morphotypes were determined and classified as an rdar morphotype, indicating curli and cellulose production, an bdar morphotype, indicating only curli production, and a saw morphotype, indicating neither curli nor cellulose production (55, 56).
Cellulose production was further determined by streaking the bacteria on LB agar without salt supplemented with 200 μg/ml calcofluor (fluorescence brightener 28; Sigma-Aldrich GmbH, Munich, Germany). The dye calcofluor binding to β(1-4)- and β(1-3)-linked polysaccharides is routinely used to detect cellulose-producing bacteria in agar plate assays or by fluorescence microscopy (53, 76, 77). After incubating the agar plates for 24 h at 37°C and 48 h at 28°C, the assessment of cellulose production was done by verifying the fluorescence of the bacteria under a 366-nm UV light source. Each isolate was tested for cellulose and curli production in three independent experiments. The cellulose- and curli-producing strain E. coli Nissle 1917 was used as a positive control (73).
Motility assay.
To examine motility, bacteria were inoculated in 1 ml of LB medium and incubated at 37°C for 16 h with shaking at 180 rpm. Each isolate was stabbed with sterile needle in the center of a motility agar plate (LB containing 0.25% agar) followed by an incubation at 37°C for 16 h. Positive motility was indicated by diffused growth from the point of inoculation with a diameter of growth zone of >5 mm. A negative motility test was indicated by growth along the stab point with a diameter of ≤5 mm. Each isolate was tested for motility in three independent experiments.
Biofilm formation assay.
Screening for biofilm formation was performed with a modified protocol developed by O'Toole and Kolter (78); it was carried out using standard growth media (20, 90 – 92), including LB broth, TSB, BHI broth (Sigma-Aldrich GmbH), and M63 minimal medium supplemented with 1 mM MgSO4 and 1% glucose (M63). As we screened E. coli isolates from humans, we decided to examine their biofilm-forming ability under natural ambient conditions in a human host. Isolates were grown in LB medium at 37°C for 16 h with shaking at 180 rpm (containing approximately 108 CFU/ml) and diluted 1:100 in respective media. For biofilm formation, 200 μl of a diluted culture was inoculated in a 96-well flat-bottom polystyrene microtiter plate (Greiner Bio-One GmbH, Frickenhausen, Germany). Bacteria were incubated for 48 h at 37°C under static conditions. E. coli strain MG1655 F′Tet ΔtraD served as a positive control for biofilm formation. Media without bacteria were used as negative controls. For different biofilm-staining methods, the same bacterial cultures were used as for biofilm formation and were treated equally.
Biofilm staining. (i) SYTO 9 staining.
The fluorescent nucleic acid stain SYTO 9 binds to DNA of both live and dead cells after passively diffusing through the cell membranes (79). A substantial part of the biofilm matrix is extracellular DNA (80) so that SYTO 9 can be used for the staining of total biofilm biomass. For SYTO 9 staining of biofilms, the 96-well plates were carefully washed once with isotonic saline to remove nonadherent bacteria, followed by an incubation in the dark for 10 min with isotonic saline containing 5 μM SYTO 9 green fluorescent nucleic acid stain (Thermo Fisher Scientific GmbH, Dreieich, Germany). The plates were washed again with isotonic saline and analyzed using the automated VideoScan technology. Each isolate was tested in triplicate for each of the four media in three independent experiments.
(ii) Crystal violet staining.
The basic dye crystal violet binds to negatively charged surface molecules and polysaccharides in the biofilm matrix (81). Because living and dead cells, as well as the matrix, are stained, crystal violet can be used for total biomass staining. The staining was performed as described previously by Christensen et al. (82). The 96-well plates were carefully washed once with ddH2O to remove nonadherent bacteria, stained with 0.1% crystal violet (Carl Roth GmbH) for 10 min, and washed twice with ddH2O. The extraction of surface-bound crystal violet was performed by incubating with 95% ethanol. After 10 min of incubation, the solutions were transferred to a fresh 96-well plate. Optical densities were measured at 570 nm (OD570) with a 96-well plate reader (Sunrise; Tecan GmbH, Crailsheim, Germany). Each isolate was tested in quadruplicate in LB medium in three independent experiments. The extent of biofilm formation was determined by using the classification from Stepanović et al. (34). Bacterial isolates were classified into four categories, namely, non-biofilm producer, weak, moderate, and strong, based on the means of the measured OD570 values. The cutoff value (ODc) for this classification is defined as three standard deviations above the mean OD570 of the negative control. Isolates were classified as follows: non-biofilm former, OD570 ≤ ODc; weak biofilm former, OD570 > ODc and ≤ 2× ODc; moderate biofilm former, OD570 > 2× ODc and ≤ 4× ODc; strong biofilm former, OD570 > 4× ODc.
VideoScan fluorescence imaging technology.
The VideoScan technology implemented in the Aklides system (GA Generic Assays GmbH, Dahlewitz, Germany) is a versatile platform for the analysis of fluorescent objects (30, 31, 83 – 85). The inverse motorized fluorescence microscope consists of commercial hardware components and in-house-developed software. The camera focuses automatically and captures the images, which are processed via sophisticated digital image processing methods. The VideoScan technology enables the analysis of multiplex assays such as microbead or cell-based assays (30, 31), and its utility was enhanced during the present study for the high-throughput screening of biofilms.
To evaluate the strength of the biofilm-forming capability of a bacterial isolate, the fluorescence intensity of each well of the 96-well plate was quantified by using the in-house-designed software package FastFluoScan. A filter set for SYTO 9 staining (excitation filter, 422 to 467 nm; dichroic mirror, 482 nm; emission filter, 489 to 531 nm) and an Olympus PlanApoN 1.25×/0.04 objective were used. The intensity was measured in the center of the well in a square 4 mm by 4 mm. The background fluorescence intensity of the polystyrene plate was deducted from each generated fluorescence intensity value. The fluorescence intensities of controls and SYTO 9-stained biofilms in the wells were referenced to the fluorescence intensity of internal reference microbeads (PolyAn GmbH, Berlin, Germany) containing a fluorescent dye equivalent to SYTO 9. The referenced fluorescence intensities were expressed as the median of the relFI in arbitrary units (AU). In addition, the reference beads served to compensate interarray variations. The relFI of SYTO 9-stained bacteria correlated with the amount of biofilm formation in the wells of the microtiter plates.
To further examine E. coli isolates qualitatively for biofilm formation, overview images from each well of the 96-well plates were taken automatically by focusing on the well bottoms. To obtain the overview images, the camera captured 12 smaller single images (Olympus UPlanSApo 4×/0.164 objective) per well, which were composed to one overview image showing the biofilm formed in each well (diameter of 6 mm) of the 96-well plate. Overview images of the single wells were merged to 96-well-plate overview images for each plate. The outermost columns and rows of the 96-well plate were omitted, since they were not used for the biofilm formation. Therefore, one 96-well-plate overview image contained five bacterial isolates tested for biofilm formation in four different media.
Statistical analysis.
Multiple χ2 tests were used to determine statistically significant differences for the general gene prevalences and the prevalences of genes between different pathotypes. P values for the comparison of the prevalences of a gene between all pathotypes were computed using a proportion test for multiple groups. For comparing the prevalences of a gene between different pathotypes, computed P values were adjusted using a Benjamini-Hochberg P value correction. Pearson's χ2 test with Yates' continuity correction was used to examine the correlation of fliC and motility and the correlation between VideoScan and crystal violet assay. To find significant differences in the biofilm-forming capability of a pathotype, median values for isolates belonging to different pathotypes were compared using multiple Wilcoxon tests (Benjamini-Hochberg P value correction). P values of less than 0.05 were scored as statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dennis Titze for help in performing PCRs for the detection of biofilm-associated genes. Bacterial isolates were kindly provided by Lisa Kay Nolan (Iowa State University, USA), Dirk Meissner (Labor Hameln Hildesheim, Germany), Thomas Juretzek (Carl-Thiem-Klinikum Cottbus, Germany), Alexander Swidsinski (Charité Humboldt University, Germany), and Jean-Marc Ghigo (Pasteur Institute Paris, France).
This work was supported by InnoProfile-Transfer 03IPT611A and 03IPT611X funded by the Federal Ministry of Education and Research (BMBF, Germany).
Dirk Roggenbuck has a management role in and is a shareholder of GA Generic Assays GmbH. This company is a diagnostic manufacturer. All other authors declare that they have no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01660-17.
REFERENCES
- 1.Davey ME, O'Toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64:847–867. doi: 10.1128/MMBR.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 4.Stewart PS, Costerton JW. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138. doi: 10.1016/S0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 5.Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G. 2003. The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest 112:1466–1477. doi: 10.1172/JCI200320365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsek MR, Singh PK. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol 57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 7.Parsek MR, Fuqua C. 2004. Biofilms 2003: emerging themes and challenges in studies of surface-associated microbial life. J Bacteriol 186:4427–4440. doi: 10.1128/JB.186.14.4427-4440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 9.Hooton TM, Stamm WE. 1997. Diagnosis and treatment of uncomplicated urinary tract infection. Infect Dis Clin North Am 11:551–581. doi: 10.1016/S0891-5520(05)70373-1. [DOI] [PubMed] [Google Scholar]
- 10.Nicolle LE. 2005. Catheter-related urinary tract infection. Drugs Aging 22:627–639. doi: 10.2165/00002512-200522080-00001. [DOI] [PubMed] [Google Scholar]
- 11.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 12.Clements A, Young JC, Constantinou N, Frankel G. 2012. Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes 3:71–87. doi: 10.4161/gmic.19182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogeleer P, Tremblay YDN, Mafu AA, Jacques M, Harel J. 2014. Life on the outside: role of biofilms in environmental persistence of Shiga-toxin producing Escherichia coli. Front Microbiol 5:317. doi: 10.3389/fmicb.2014.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nataro JP, Kaper JB, Robins-Browne R, Prado V, Vial P, Levine MM. 1987. Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr Infect Dis J 6:829–831. doi: 10.1097/00006454-198709000-00008. [DOI] [PubMed] [Google Scholar]
- 15.Vial PA, Robins-Browne R, Lior H, Prado V, Kaper JB, Nataro JP, Maneval D, Elsayed A, Levine MM. 1988. Characterization of enteroadherent-aggregative Escherichia coli, a putative agent of diarrheal disease. J Infect Dis 158:70–79. doi: 10.1093/infdis/158.1.70. [DOI] [PubMed] [Google Scholar]
- 16.Hicks S, Candy DC, Phillips AD. 1996. Adhesion of enteroaggregative Escherichia coli to pediatric intestinal mucosa in vitro. Infect Immun 64:4751–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nataro JP, Hicks S, Phillips AD, Vial PA, Sears CL. 1996. T84 cells in culture as a model for enteroaggregative Escherichia coli pathogenesis. Infect Immun 64:4761–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wakimoto N, Nishi J, Sheikh J, Nataro JP, Sarantuya J, Iwashita M, Manago K, Tokuda K, Yoshinaga M, Kawano Y. 2004. Quantitative biofilm assay using a microtiter plate to screen for enteroaggregative Escherichia coli. Am J Trop Med Hyg 71:687–690. [PubMed] [Google Scholar]
- 19.Mohamed JA, Huang DB, Jiang Z-D, DuPont HL, Nataro JP, Belkind-Gerson J, Okhuysen PC. 2007. Association of putative enteroaggregative Escherichia coli virulence genes and biofilm production in isolates from travelers to developing countries. J Clin Microbiol 45:121–126. doi: 10.1128/JCM.01128-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reisner A, Krogfelt KA, Klein BM, Zechner EL, Molin S. 2006. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: impact of environmental and genetic factors. J Bacteriol 188:3572–3581. doi: 10.1128/JB.188.10.3572-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 22.Danese PN, Pratt LA, Kolter R. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol 182:3593–3596. doi: 10.1128/JB.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reisner A, Haagensen JAJ, Schembri MA, Zechner EL, Molin S. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol Microbiol 48:933–946. doi: 10.1046/j.1365-2958.2003.03490.x. [DOI] [PubMed] [Google Scholar]
- 24.Schembri MA, Kjaergaard K, Klemm P. 2003. Global gene expression in Escherichia coli biofilms. Mol Microbiol 48:253–267. doi: 10.1046/j.1365-2958.2003.03432.x. [DOI] [PubMed] [Google Scholar]
- 25.Domka J, Lee J, Bansal T, Wood TK. 2007. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ Microbiol 9:332–346. doi: 10.1111/j.1462-2920.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- 26.Wood TK, González Barrios AF, Herzberg M, Lee J. 2006. Motility influences biofilm architecture in Escherichia coli. Appl Microbiol Biotechnol 72:361–367. doi: 10.1007/s00253-005-0263-8. [DOI] [PubMed] [Google Scholar]
- 27.Sheikh J, Hicks S, Dall'Agnol M, Phillips AD, Nataro JP. 2001. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol Microbiol 41:983–997. doi: 10.1046/j.1365-2958.2001.02512.x. [DOI] [PubMed] [Google Scholar]
- 28.Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol 180:2442–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Römling U. 2002. Molecular biology of cellulose production in bacteria. Res Microbiol 153:205–212. doi: 10.1016/S0923-2508(02)01316-5. [DOI] [PubMed] [Google Scholar]
- 30.Rödiger S, Schierack P, Böhm A, Nitschke J, Berger I, Frömmel U, Schmidt C, Ruhland M, Schimke I, Roggenbuck D, Lehmann W, Schröder C. 2013. A highly versatile microscope imaging technology platform for the multiplex real-time detection of biomolecules and autoimmune antibodies. Adv Biochem Eng Biotechnol 133:35–74. doi: 10.1007/10_2011_132. [DOI] [PubMed] [Google Scholar]
- 31.Frömmel U, Böhm A, Nitschke J, Weinreich J, Groß J, Rödiger S, Wex T, Ansorge H, Zinke O, Schröder C, Roggenbuck D, Schierack P. 2013. Adhesion patterns of commensal and pathogenic Escherichia coli from humans and wild animals on human and porcine epithelial cell lines. Gut Pathog 5:31. doi: 10.1186/1757-4749-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rödiger S, Burdukiewicz M, Blagodatskikh K, Jahn M, Schierack P. 2015. R as an environment for the reproducible analysis of DNA amplification experiments. R J 7:127–150. [Google Scholar]
- 33.Rödiger S, Burdukiewicz M, Schierack P. 2015. chipPCR: an R package to pre-process raw data of amplification curves. Bioinformatics 31:2900–2902. doi: 10.1093/bioinformatics/btv205. [DOI] [PubMed] [Google Scholar]
- 34.Stepanović S, Vuković D, Dakić I, Savić B, Švabić-Vlahović M. 2000. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods 40:175–179. doi: 10.1016/S0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 35.Duan Q, Zhou M, Zhu L, Zhu G. 2013. Flagella and bacterial pathogenicity. J Basic Microbiol 53:1–8. doi: 10.1002/jobm.201100335. [DOI] [PubMed] [Google Scholar]
- 36.Van Houdt R, Michiels CW. 2005. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res Microbiol 156:626–633. doi: 10.1016/j.resmic.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Josenhans C, Suerbaum S. 2002. The role of motility as a virulence factor in bacteria. Int J Med Microbiol 291:605–614. doi: 10.1078/1438-4221-00173. [DOI] [PubMed] [Google Scholar]
- 38.Macnab RM. 1996. Flagella and motility, p 123–145. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. [Google Scholar]
- 39.Macnab RM. 1992. Genetics and biogenesis of bacterial flagella. Annu Rev Genet 26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 40.Reid SD, Selander RK, Whittam TS. 1999. Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J Bacteriol 181:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ratiner YA. 1998. New flagellin-specifying genes in some Escherichia coli strains. J Bacteriol 180:979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pratt LA, Kolter R. 1999. Genetic analyses of bacterial biofilm formation. Curr Opin Microbiol 2:598–603. doi: 10.1016/S1369-5274(99)00028-4. [DOI] [PubMed] [Google Scholar]
- 43.Sauer FG, Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. 2000. Bacterial pili: molecular mechanisms of pathogenesis. Curr Opin Microbiol 3:65–72. doi: 10.1016/S1369-5274(99)00053-3. [DOI] [PubMed] [Google Scholar]
- 44.Moreira CG, Carneiro SM, Nataro JP, Trabulsi LR, Elias WP. 2003. Role of type I fimbriae in the aggregative adhesion pattern of enteroaggregative Escherichia coli. FEMS Microbiol Lett 226:79–85. doi: 10.1016/S0378-1097(03)00561-5. [DOI] [PubMed] [Google Scholar]
- 45.Orndorff PE, Devapali A, Palestrant S, Wyse A, Everett ML, Bollinger RR, Parker W. 2004. Immunoglobulin-mediated agglutination of and biofilm formation by Escherichia coli K-12 require the type 1 pilus fiber. Infect Immun 72:1929–1938. doi: 10.1128/IAI.72.4.1929-1938.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Källenius G, Svenson SB, Hultberg H, Möllby R, Helin I, Cedergren B, Winberg J. 1981. Occurrence of P-fimbriated Escherichia coli in urinary tract infections. Lancet 318:1369–1372. doi: 10.1016/S0140-6736(81)92797-5. [DOI] [PubMed] [Google Scholar]
- 47.Plos K, Connell H, Jodal U, Marklund B-I, Mårild S, Wettergren B, Svanborg C. 1995. Intestinal carriage of P fimbriated Escherichia coli and the susceptibility to urinary tract infection in young children. J Infect Dis 171:625–631. doi: 10.1093/infdis/171.3.625. [DOI] [PubMed] [Google Scholar]
- 48.Lane MC, Mobley HLT. 2007. Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int 72:19–25. doi: 10.1038/sj.ki.5002230. [DOI] [PubMed] [Google Scholar]
- 49.Olsén A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. 1993. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol Microbiol 7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 50.Uhlich GA, Cooke PH, Solomon EB. 2006. Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl Environ Microbiol 72:2564–2572. doi: 10.1128/AEM.72.4.2564-2572.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frömmel U, Lehmann W, Rödiger S, Böhm A, Nitschke J, Weinreich J, Groß J, Roggenbuck D, Zinke O, Ansorge H, Vogel S, Klemm P, Wex T, Schröder C, Wieler LH, Schierack P. 2013. Adhesion of human and animal Escherichia coli strains in association with their virulence-associated genes and phylogenetic origins. Appl Environ Microbiol 79:5814–5829. doi: 10.1128/AEM.01384-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Römling U, Bian Z, Hammar M, Sierralta WD, Normark S. 1998. Curli fibers are highly conserved between Salmonella Typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol 180:722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. 2001. The multicellular morphotypes of Salmonella Typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol 39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 54.Da Re S, Ghigo J-M. 2006. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J Bacteriol 188:3073–3087. doi: 10.1128/JB.188.8.3073-3087.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Römling U, Rohde M, Olsén A, Normark S, Reinköster J. 2000. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella Typhimurium regulates at least two independent pathways. Mol Microbiol 36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- 56.Römling U, Sierralta WD, Eriksson K, Normark S. 1998. Multicellular and aggregative behaviour of Salmonella Typhimurium strains is controlled by mutations in the agfD promoter. Mol Microbiol 28:249–264. doi: 10.1046/j.1365-2958.1998.00791.x. [DOI] [PubMed] [Google Scholar]
- 57.Olsén A, Jonsson A, Normark S. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 58.Arnqvist A, Olsén A, Pfeifer J, Russell DG, Normark S. 1992. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol Microbiol 6:2443–2452. doi: 10.1111/j.1365-2958.1992.tb01420.x. [DOI] [PubMed] [Google Scholar]
- 59.Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ Microbiol 2:450–464. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- 60.Owen P, Meehan M, de Loughry-Doherty H, Henderson I. 1996. Phase-variable outer membrane proteins in Escherichia coli. FEMS Immunol Med Microbiol 16:63–76. doi: 10.1111/j.1574-695X.1996.tb00124.x. [DOI] [PubMed] [Google Scholar]
- 61.Henderson IR, Meehan M, Owen P. 1997. Antigen 43, a phase-variable bipartite outer membrane protein, determines colony morphology and autoaggregation in Escherichia coli K-12. FEMS Microbiol Lett 149:115–120. doi: 10.1111/j.1574-6968.1997.tb10317.x. [DOI] [PubMed] [Google Scholar]
- 62.Hasman H, Chakraborty T, Klemm P. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J Bacteriol 181:4834–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kjaergaard K, Schembri MA, Hasman H, Klemm P. 2000. Antigen 43 from Escherichia coli induces inter- and intraspecies cell aggregation and changes in colony morphology of Pseudomonas fluorescens. J Bacteriol 182:4789–4796. doi: 10.1128/JB.182.17.4789-4796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schembri MA, Hjerrild L, Gjermansen M, Klemm P. 2003. Differential expression of the Escherichia coli autoaggregation factor antigen 43. J Bacteriol 185:2236–2242. doi: 10.1128/JB.185.7.2236-2242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conceição RA, Ludovico MS, Andrade CGTJ, Yano T. 2012. Human sepsis-associated Escherichia coli (SEPEC) is able to adhere to and invade kidney epithelial cells in culture. Braz J Med Biol Res 45:417–424. doi: 10.1590/S0100-879X2012007500057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lüthje P, Brauner A. 2010. Ag43 promotes persistence of uropathogenic Escherichia coli isolates in the urinary tract. J Clin Microbiol 48:2316–2317. doi: 10.1128/JCM.00611-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kjaergaard K, Schembri MA, Ramos C, Molin S, Klemm P. 2000. Antigen 43 facilitates formation of multispecies biofilms. Environ Microbiol 2:695–702. doi: 10.1046/j.1462-2920.2000.00152.x. [DOI] [PubMed] [Google Scholar]
- 68.Weiss-Muszkat M, Shakh D, Zhou Y, Pinto R, Belausov E, Chapman MR, Sela S. 2010. Biofilm formation by and multicellular behavior of Escherichia coli O55:H7, an atypical enteropathogenic strain. Appl Environ Microbiol 76:1545–1554. doi: 10.1128/AEM.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nascimento HH, Silva LEP, Souza RT, Silva NP, Scaletsky ICA. 2014. Phenotypic and genotypic characteristics associated with biofilm formation in clinical isolates of atypical enteropathogenic Escherichia coli (aEPEC) strains. BMC Microbiol 14:184. doi: 10.1186/1471-2180-14-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dewanti R, Wong AC. 1995. Influence of culture conditions on biofilm formation by Escherichia coli O157:H7. Int J Food Microbiol 26:147–164. doi: 10.1016/0168-1605(94)00103-D. [DOI] [PubMed] [Google Scholar]
- 71.Merlino J, Siarakas S, Robertson GJ, Funnell GR, Gottlieb T, Bradbury R. 1996. Evaluation of CHROMagar Orientation for differentiation and presumptive identification of Gram-negative bacilli and Enterococcus species. J Clin Microbiol 34:1788–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ghigo JM. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 73.Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol 186:5432–5441. doi: 10.1128/JB.186.16.5432-5441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Solano C, García B, Valle J, Berasain C, Ghigo J-M, Gamazo C, Lasa I. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol Microbiol 43:793–808. doi: 10.1046/j.1365-2958.2002.02802.x. [DOI] [PubMed] [Google Scholar]
- 75.Hammar M, Arnqvist A, Bian Z, Olsén A, Normark S. 1995. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol 18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x. [DOI] [PubMed] [Google Scholar]
- 76.Zogaj X, Bokranz W, Nimtz M, Römling U. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect Immun 71:4151–4158. doi: 10.1128/IAI.71.7.4151-4158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Spiers AJ, Bohannon J, Gehrig SM, Rainey PB. 2003. Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol Microbiol 50:15–27. doi: 10.1046/j.1365-2958.2003.03670.x. [DOI] [PubMed] [Google Scholar]
- 78.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 79.Boulos L, Prévost M, Barbeau B, Coallier J, Desjardins R. 1999. LIVE/DEAD BacLight: application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J Microbiol Methods 37:77–86. doi: 10.1016/S0167-7012(99)00048-2. [DOI] [PubMed] [Google Scholar]
- 80.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 81.Li X, Yan Z, Xu J. 2003. Quantitative variation of biofilms among strains in natural populations of Candida albicans. Microbiology 149:353–362. doi: 10.1099/mic.0.25932-0. [DOI] [PubMed] [Google Scholar]
- 82.Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, Melton DM, Beachey EH. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol 22:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roggenbuck D, Reinhold D, Hiemann R, Anderer U, Conrad K. 2011. Standardized detection of anti-ds DNA antibodies by indirect immunofluorescence - a new age for confirmatory tests in SLE diagnostics. Clin Chim Acta 412:2011–2012. doi: 10.1016/j.cca.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 84.Egerer K, Roggenbuck D, Hiemann R, Weyer M-G, Büttner T, Radau B, Krause R, Lehmann B, Feist E, Burmester G-R. 2010. Automated evaluation of autoantibodies on human epithelial-2 cells as an approach to standardize cell-based immunofluorescence tests. Arthritis Res Ther 12:R40. doi: 10.1186/ar2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Willitzki A, Hiemann R, Peters V, Sack U, Schierack P, Rödiger S, Anderer U, Conrad K, Bogdanos DP, Reinhold D, Roggenbuck D. 2012. New platform technology for comprehensive serological diagnostics of autoimmune diseases. Clin Dev Immunol 2012:284740. doi: 10.1155/2012/284740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ulett GC, Valle J, Beloin C, Sherlock O, Ghigo J-M, Schembri MA. 2007. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect Immun 75:3233–3244. doi: 10.1128/IAI.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Janben T, Schwarz C, Preikschat P, Voss M, Philipp HC, Wieler LH. 2001. Virulence-associated genes in avian pathogenic Escherichia coli (APEC) isolated from internal organs of poultry having died from colibacillosis. Int J Med Microbiol IJMM 291:371–378. doi: 10.1078/1438-4221-00143. [DOI] [PubMed] [Google Scholar]
- 88.Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 89.Fields PI, Blom K, Hughes HJ, Helsel LO, Feng P, Swaminathan B. 1997. Molecular characterization of the gene encoding H antigen in Escherichia coli and development of a PCR-restriction fragment length polymorphism test for identification of E. coli O157:H7 and O157:NM. J Clin Microbiol 35:1066–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nakao R, Ramstedt M, Wai SN, Uhlin BE. 2012. Enhanced biofilm formation by Escherichia coli LPS mutants defective in Hep biosynthesis. PLoS One 7:e51241. doi: 10.1371/journal.pone.0051241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Skyberg JA, Siek KE, Doetkott C, Nolan LK. 2007. Biofilm formation by avian Escherichia coli in relation to media, source and phylogeny. J Appl Microbiol 102:548–554. doi: 10.1111/j.1365-2672.2006.03076.x. [DOI] [PubMed] [Google Scholar]
- 92.Zalewska-Piątek B, Wilkanowicz S, Bruździak P, Piątek R, Kur J. 2013. Biochemical characteristic of biofilm of uropathogenic Escherichia coli Dr+ strains. Microbiol Res 168:367–378. doi: 10.1016/j.micres.2013.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.