ABSTRACT
Clostridium perfringens is a gastrointestinal pathogen capable of causing disease in a variety of hosts. Necrotic enteritis in chickens is caused by C. perfringens strains that produce the pore-forming toxin NetB, the major virulence factor for this disease. Like many other C. perfringens toxins and antibiotic resistance genes, NetB is encoded on a conjugative plasmid. Conjugative transfer of the netB-containing plasmid pJIR3535 has been demonstrated in vitro with a netB-null mutant. This study has investigated the effect of plasmid transfer on disease pathogenesis, with two genetically distinct transconjugants constructed under in vitro conditions, within the intestinal tract of chickens. This study also demonstrates that plasmid transfer can occur naturally in the host gut environment without the need for antibiotic selective pressure to be applied. The demonstration of plasmid transfer within the chicken host may have implications for the progression and pathogenesis of C. perfringens-mediated disease. Such horizontal gene transfer events are likely to be common in the clostridia and may be a key factor in strain evolution, both within animals and in the wider environment.
IMPORTANCE Clostridium perfringens is a major gastrointestinal pathogen of poultry. C. perfringens strains that express the NetB pore-forming toxin, which is encoded on a conjugative plasmid, cause necrotic enteritis. This study demonstrated that the conjugative transfer of the netB-containing plasmid to two different nonpathogenic strains converted them into disease-causing strains with disease-causing capability similar to that of the donor strain. Plasmid transfer of netB and antibiotic resistance was also demonstrated to occur within the gastrointestinal tract of chickens, with approximately 14% of the isolates recovered comprising three distinct, in vivo-derived, transconjugant types. The demonstration of in vivo plasmid transfer indicates the potential importance of strain plasticity and the contribution of plasmids to strain virulence.
KEYWORDS: Clostridium perfringens, conjugation, in vivo plasmid transfer, necrotic enteritis, pathogenicity, virulence
INTRODUCTION
Horizontal gene transfer (HGT) is a driver of the diversification and evolution of bacterial species. HGT involves the transfer of DNA between strains or species, providing new and varied functionality to pathogens or other bacterial species (1, 2). In particular, HGT is an alarming issue in both human health and livestock production due to the transfer of antibiotic resistance genes to and between pathogens (3). This has resulted in intense monitoring of antibiotic-resistant pathogenic bacteria and bans or restrictions on the use of antibiotics and antimicrobial growth promoters in livestock production.
Three independent gene transfer mechanisms, conjugation, transduction, and transformation, can result in HGT (1). Conjugation, where plasmids mediate their own transfer from donor to recipient cells through direct contact, has been repeatedly demonstrated to be an efficient mechanism of gene transfer within a wide range of bacterial species, including the diverse plasmid-carrying species Clostridium perfringens (4 – 6). HGT of antibiotic resistance genes has been demonstrated both in vitro and in vivo in various animal models; for example, studies have demonstrated in vivo DNA transfer in the chicken gastrointestinal tract. HGT has been established between native chicken Lactobacillus species (7) and an antibiotic resistance plasmid in Klebsiella sp., and Escherichia coli has been shown to transfer to other Escherichia coli strains both with and without antibiotic selection (8, 9). Tetracycline resistance genes have also been transferred between Campylobacter jejuni strains in vivo (10).
C. perfringens plasmids have been a major focus of studies for many years due to their prevalence, diversity, and carriage of key virulence genes (11 – 13). Plasmids within C. perfringens contain an array of toxins, antibiotic resistance genes, bacteriocins, metabolism genes, and regulatory elements (5, 11). The varied pathogenic potential of C. perfringens is due, at least in part, to the production of specific toxins (14, 15). Many of these toxins are encoded by plasmids, including three of four toxins used in the C. perfringens typing scheme (11). Most of the large clostridial plasmids have a cluster of genes, covering ∼40 kb, which encode the conjugal transfer machinery. This region is commonly referred to as tcp (16, 17). The tcp region has been extensively characterized (11, 16 – 19). Toxin genes carried on plasmids that have a tcp locus include cpb, etx, iapA or iapB, cpb2, tpeL, netB, and cpe (11, 12, 20 – 22). Some plasmids encode more than one toxin or a combination of toxins and other factors, such as those for antibiotic resistance (11, 12, 21).
Conjugation of tcp-encoding plasmids has been demonstrated in vitro with the tetracycline resistance-encoding plasmids pCW3 and pJIR3537 (5, 20), a cpe-containing plasmid pMRS4969 (4), and type D epsilon toxin-encoding plasmids CN1020 and CN3718 (6). More recently, derivatives of the netB plasmid pJIR3535 and the cpb2 plasmid pJIR3844, from a necrotic enteritis-causing strain, EHE-NE18, were demonstrated to be conjugative (5). Although conjugation of these tcp plasmids has been demonstrated reliably and repeatedly in vitro, very little is known about how these plasmids and their host strains behave in their natural habitat of the chicken gastrointestinal tract.
NetB-producing strains of C. perfringens cause necrotic enteritis in chickens, an enteric disease which is a significant health, welfare, and economic issue in the commercial poultry industry (23). NetB is a 33-kDa secreted beta-barrel-pore-forming toxin that causes lesions or necrotic foci in the chicken gastrointestinal tract, leading to a reduced feed-to-conversion ratio and, in severe cases, death (24 – 26). NetB is encoded on conjugative plasmids, such as pJIR3535 and pNetB-Ne10 (5, 27). Conjugative transfer of this plasmid has been demonstrated in vitro with a netB-null mutant; however, in vivo transfer of this plasmid and the effect of this transfer on disease pathogenesis have not been previously examined (5).
In several recent studies, NetB-encoding strains have been shown to originate from several distinct genetic backgrounds, with varied genomic content carried on both plasmids and the chromosome (28 – 31). In particular, the presence of the toxin plasmids carrying netB, cpb2, and tpeL, as well as bacitracin and tetracycline resistance genes, are variably present in chicken strains with both similar and diverse chromosomes (26, 28, 32, 33). This variability in plasmid content and chromosomal content between strains has led to the hypothesis that plasmid transfer of these virulence and antibiotic resistance plasmids occurs naturally in the environment, contributing to the genomic diversity of strains and potentially contributing to virulence through the emergence of new and varied strains of necrotic enteritis-causing C. perfringens. This hypothesis is tested in the experiments reported here.
RESULTS
Conjugation and confirmation of in vitro-derived transconjugants.
Two genetically distinct recipients were used in this study, BER-NE33 and PBD1. The two strains are nonpathogenic chicken isolates with two distinct chromosomes and do not carry a netB-carrying plasmid. BER-NE33 was isolated in 2010 from a chicken suffering from necrotic enteritis and contains a single plasmid, an atypical tetracycline resistance plasmid of ∼70 kb. Although isolated from a diseased bird, the strain has been shown to be incapable of causing disease (26), presumably because it does not carry the essential virulence gene netB. PBD1, in contrast, was isolated in 2008 from a healthy chicken with no signs of disease, and it does not carry any plasmids. Nalidixic acid-resistant mutants of each strain were derived to allow easy differentiation of donor strains and recipient strains. A rifampin-resistant spontaneous mutant of EHE-NE18, in which the NetB-encoding plasmid had been marked with an erythromycin resistance-encoding expression cassette, was used as the plasmid donor strain (see Fig. S1 in the supplemental material).
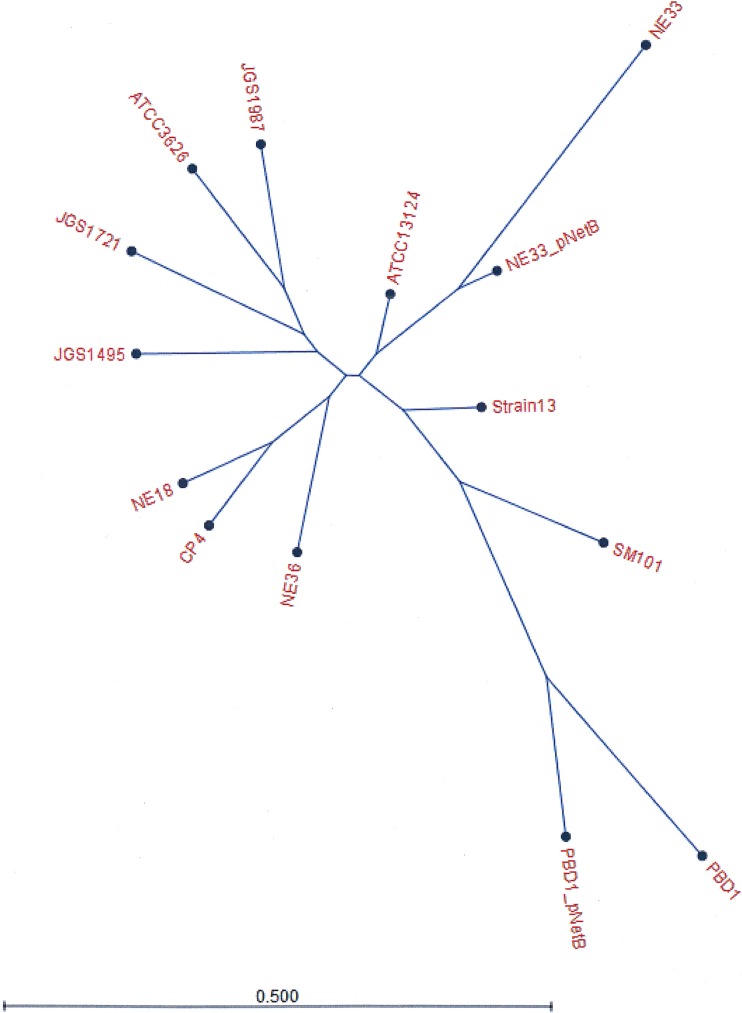
Matings were performed using the solid-plate mating technique. The conjugative transfer efficiency was expressed as the number of transconjugants per donor cell, as shown in Table S1. Transconjugants were isolated by selecting and screening for colonies that were resistant to nalidixic acid and erythromycin but sensitive to rifampin. Transconjugants were confirmed by whole-genome sequencing (Fig. 1) and pulsed-field gel electrophoresis (PFGE) analysis to examine the movement of plasmids and chromosomal elements. The PFGE profiles of transconjugants were identical to the recipient strains, apart from a new plasmid band, showing that only plasmids were transferred and there were no major chromosomal rearrangements (Fig. S2 and S3). Various combinations of plasmids were observed in the transconjugants (Fig. S2 and S3). For further investigation into disease pathogenesis, tetracycline-sensitive transconjugants were chosen and sequenced. Genome sequencing determined that only the erythromycin resistance-carrying pNetB and pBeta2 plasmids were present in the transconjugants, and no chromosomal regions were affected by the mating procedure. Transconjugants were shown by Western blotting to produce NetB in the culture supernatants of the cultures used in the subsequent necrotic enteritis disease induction experiments (Fig. S4).
FIG 1.
Phylogenetic tree showing evolutionary relationships of total gene content (gene presence and absence) between strains PBD1 and NE33 before and after the mating procedure. Other strains were included in the comparison to root the tree.
In vitro plasmid transfer converts two nonpathogenic C. perfringens strains into virulent necrotic enteritis-causing strains.
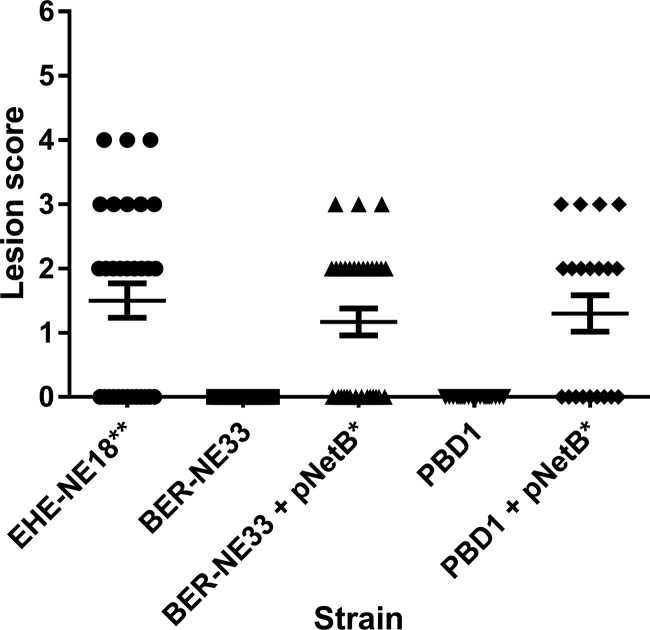
The virulence of donor, recipient, and transconjugant strains was tested in the necrotic enteritis disease model. As shown in Fig. 2, birds challenged with the nalidixic acid-resistant recipients PBD1 and BER-NE33 had no detectable disease. However, the birds challenged with the pNetB-containing transconjugants had significant amounts of disease to levels like those seen in the group challenged with the erythromycin-marked virulent EHE-NE18 strain, with an average lesion score of ∼1.5 observed in all three groups. Statistically, there are no significant differences between the donor strains EHE-NE18 and the two transconjugants in terms of disease causing capability in the necrotic enteritis disease induction model. However, statistical significance was observed between EHE-NE18 and both recipient strains PBD1 and NE33 (P < 0.01), which was also observed between each of the transconjugants and their corresponding recipient strains (P < 0.05; Fig. 2). Clearly, the conjugation of pNetB can change two different nonpathogenic strains into virulent strains.
FIG 2.
Virulence of C. perfringens strains in the necrotic enteritis disease induction model. The lesion scores of individual 24-day-old broiler chickens challenged with different C. perfringens strains are shown. The horizontal bars represent the average lesion score for each group, and the error bars represent the standard error for each group. Intestinal lesions in the small intestine (duodenum to ileum) were scored as previously reported: 0, no gross lesions; 1, thin or febrile walls; 2, focal necrosis or ulceration (one to five foci); 3, focal necrosis or ulceration (six to 15 foci); 4, focal necrosis or ulceration (16 or more foci); 5, patches of necrosis 2 to 3 cm long; and 6, diffuse necrosis typical of field cases. The results presented are the pooled data from three independent trials, n = 30. The strains tested are as follows: EHE-NE18, BER-NE33, BER-NE33/pNetB, PBD1, and PBD1/pNetB. A one-way ANOVA Kruskal-Wallis test with a Dunn's posttest showed a statistical difference between the recipient strains NE33 and PBD1 to the donor strain EHE-NE18 (***, P < 0.01), a statistical difference between each pNetB transconjugant and the original recipient strain (**, P < 0.05), and no statistical difference between EHE-NE18 and either of the transconjugants.
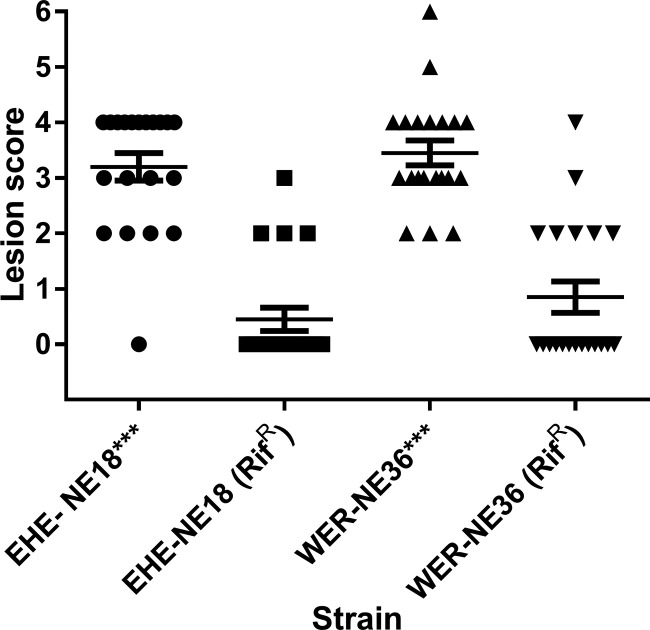
Rifampin-resistant mutant strains of pathogenic C. perfringens are significantly attenuated in virulence.
Rifampin-resistant spontaneous mutants of two pathogenic C. perfringens strains, EHE-NE18 and WER-NE36, were selected by passaging on increasing concentrations of rifampin on tryptose sulfite cycloserine (TSC) solid medium. The rifampin-resistant strains were required for mating experiments to allow easy monitoring and recovery of the marked strains from the in vivo infection experiments. The presence of netB was monitored and shown to be unaffected during this process. The two rifampin-marked strains and their wild-type isogenic parent strains were examined in the necrotic enteritis disease model to investigate the effect on virulence of the mutations to give rise to rifampin resistance (Fig. 3). In two independent trials, reduced virulence was observed with the resistant derivatives. Pooled data from the two trials showed that lesions were observed in 20 out of 20 chickens challenged with the wild-type WER-NE36 and 19 out of 20 chickens challenged with wild-type EHE-NE18. However, lesions were observed in only 7 out of 20 chickens in the rifampin-resistant WER-NE36 mutant strain-challenged groups and 4 out of 20 chickens in rifampin-resistant EHE-NE18 mutant strain-challenged groups. A statistical significance at a P value of <0.01 was observed between the rifampin-resistant mutants and their corresponding wild types.
FIG 3.
The effect of rifampin resistance marking on the virulence of C. perfringens strains in the necrotic enteritis disease induction model. The lesion scores of individual 24-day-old broiler chickens challenge with different C. perfringens strains are shown. The horizontal bars represent the average lesion score for each group, and the error bars represent the standard error for each group. Intestinal lesions in the small intestine (duodenum to ileum) were scored as previously reported: 0, no gross lesions; 1, thin or febrile walls; 2, focal necrosis or ulceration (one to five foci); 3, focal necrosis or ulceration (six to 15 foci); 4, focal necrosis or ulceration (16 or more foci); 5, patches of necrosis 2 to 3 cm long; and 6, diffuse necrosis typical of field cases. The results are from two separate trials. The strains tested are as follows: WER-NE36, EHE-NE18, and rifampin-resistant strains WER-NE36 and EHE-NE18, n = 20. A one-way ANOVA Kruskal-Wallis test with a Dunn's posttest showed a statistical difference between the rifampin-marked strains of WER-NE36 and EHE-NE18 from their wild-type forms, with P values of 0.001 for both strains examined.
In vivo plasmid transfer in the chicken gastrointestinal tract.
In vivo plasmid transfer was investigated by inoculating birds with donor and recipient strains and then screening digesta and swabs from lesions for transconjugants. No selective pressure, such as antibiotic treatment, was used. To maximize the chances of identifying transconjugants, a rifampin-resistant mutant of WER-NE36 carrying the erythromycin resistance gene-marked pNetB plasmid was used as the donor, and a nalidixic acid-resistant BER-NE33 strain was used as the recipient. WER-NE36 does not contain a tetracycline resistance conjugative plasmid, but BER-NE33 does. With this combination of strains, it was possible to detect plasmid transfer in both directions, specifically, the pTet plasmid from BER-NE33 to WER-NE36 and the erythromycin resistance gene-marked pNetB plasmid from WER-NE36 to BER-NE33. As one strain is nonpathogenic and the other is partially attenuated, an increase in disease in cochallenge birds compared to infection with each individual strain should indicate the presence of transconjugants which have acquired properties enabling them to be more virulent.
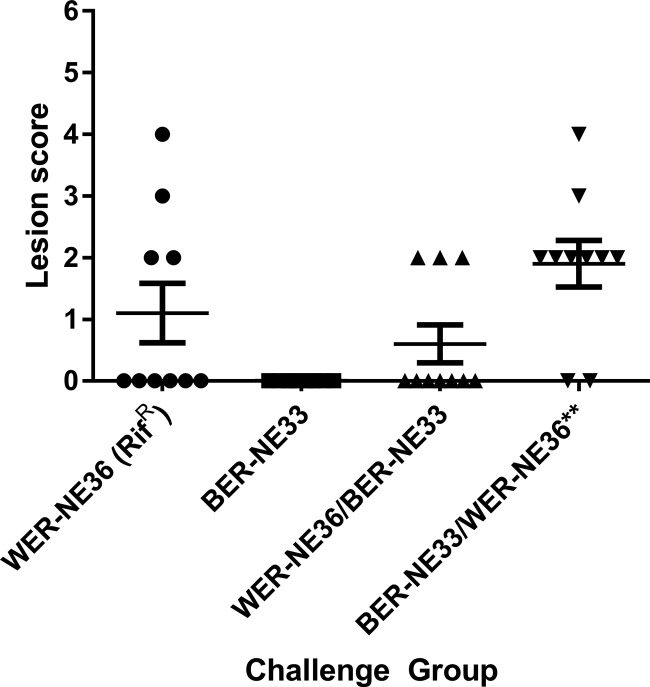
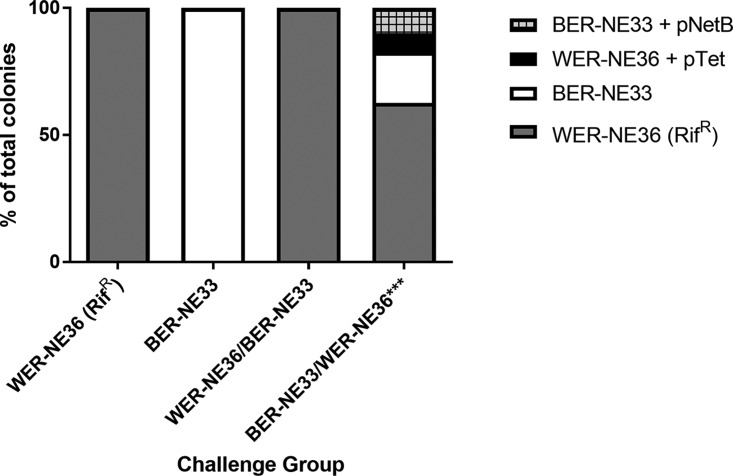
The infection-based challenge was performed over 2 days, with one strain used to inoculate birds in the morning and the second strain in the evening, such that coinfection was achieved within a 12-h period. After 2 days of challenge, chickens were examined for lesions (Fig. 4). In the group challenged with the rifampin-resistant WER-NE36 strain, four of the 10 challenged birds had visible lesions. The group challenged with the nalidixic acid-resistant BER-NE33 strain had no lesions. In the first cochallenged group, with WER-NE36 inoculated in the morning and BER-NE33 in the evening, three of 10 chickens had lesions. Only the rifampin-resistant WER-NE36 strain was reisolated from the birds in this group. In contrast, in the cochallenge group with BER-NE33 inoculated in the morning and WER-NE36 in the evening, eight of 10 chickens had necrotic enteritis lesions. A combination of WER-NE36, BER-NE33, and a mix of transconjugants was isolated from the gastrointestinal tracts of these chickens. Transconjugants were only isolated from birds with lesions. Screening of the antibiotic resistance patterns of the reisolated C. perfringens colonies identified that 63% were WER-NE36 (rifampin resistant [Rifr] erythromycin resistant [Ermr] donor), 20% were BER-NE33 (nalidixic acid resistant [Nalr] tetracycline resistant [Tetr] recipient), 7% were WER-NE36 transconjugants (Rifr Tetr), and 10% were BER-NE33 transconjugants (Ermr Nalr) (Fig. 5). Transconjugants were confirmed by PFGE and PCR (Fig. S5).
FIG 4.
Virulence of C. perfringens strains in the necrotic enteritis disease induction model with cochallenge. The lesion scores of individual 24-day-old broiler chickens challenge with different C. perfringens strains are shown. The horizontal bars represent the average lesion score for each group, and the error bars represent the standard error for each group. Intestinal lesions in the small intestine (duodenum to ileum) were scored as previously reported: 0, no gross lesions; 1, thin or febrile walls; 2, focal necrosis or ulceration (one to five foci); 3, focal necrosis or ulceration (six to 15 foci); 4, focal necrosis or ulceration (16 or more foci); 5, patches of necrosis 2 to 3 cm long; and 6, diffuse necrosis typical of field cases. The results are from a single trial. The groups tested were as follows: WER-NE36 (RifR), BER-NE33, and groups sequentially challenged, first with WER-NE36 (RifR) followed by BER-NE33 and then BER-NE33 followed by WER-NE36 (RifR) (n = 10). A one-way ANOVA Kruskal-Wallis test with a Dunn's posttest showed a statistical difference between the recipient strains BER-NE33 and cochallenge group BER-NE33/WER-NE36 (***, P < 0.01), and no statistical significance was observed between rifampin-marked WER-NE36 and BER-NE33 or to the WER-NE36/BER-NE33 cochallenge group. Transconjugants were isolated only from the BER-NE33/WER-NE36 cochallenge group.
FIG 5.
The distribution of isolated colonies from the in vivo cochallenge model. The antibiotic resistance profile indicates the nature of the colonies as the % of total colonies isolated. In the group challenged with BER-NE33 in the morning and WER-NE36 in the afternoon, 63% of the colonies were the rifampin-marked WER-NE36 donor, 20% of the colonies were BER-NE33, 10% of the colonies isolated were shown to be BER-NE33 transconjugants, and 7% were shown to be WER-NE36 transconjugants.
Cochallenge with PBD1 and EHE-NE18 was also performed. However, within these challenge groups, no PBD1 was recovered from any of the chickens. Only the donor EHE-NE18 was reisolated. This indicates displacement of the recipient strain, PBD1, from the gastrointestinal tract before conjugation could occur.
Plasmid transfer confers a selective advantage.
In almost all cases, the transconjugants from both the in vitro and the in vivo experiments contained two plasmids at a minimum, pNetB-ErmR and the beta2-encoding plasmid pJIR3844. In fact, we were unable to find a single colony that did not contain the beta2-encoding plasmid. The in vitro mating was performed at a 1:10 donor-to-recipient ratio, which was done because the donor strains tended to kill or inhibit the growth of the recipient strains. Once a transconjugant was obtained, the transconjugants of PBD1 and BER-NE33 were no longer inhibited by the donor strains and could, in turn, inhibit other strains, while the wild-type strains were not inhibitory (Table 1), suggesting that the transconjugants had inherited the inhibitory factor and were also immune to it. Inhibition was also examined using supernatants of six strains, BER-N33, PBD1, EHE-NE18, WER-NE36, and the two transconjugants, BER-NE33/pNetB-ErmR and PBD1/pNetB-ErmR, against lawn cultures of the same set of strains. It was observed that the pathogenic strains EHE-NE18 and WER-NE36 could inhibit the growth of the nonpathogenic strains BER-NE33 and PBD1, and, in turn, the nonpathogenic strains were unable to inhibit the growth of pathogenic strains. However, the two transconjugants BER-NE33/pNetB-ErmR and PBD1/pNetB-ErmR could resist inhibition of the native pathogenic strains and gained the ability to inhibit the growth of their wild-type strains and the pathogenic strains. This ability was removed when supernatants were boiled or subject to trypsin activity, leading us to believe a protein encoded by a plasmid is responsible for the inhibitory effects observed. This suggests that plasmid transfer confers a selective advantage in strains containing these plasmids, since they can outcompete strains within the same environment by inhibiting their growth.
TABLE 1.
Inhibitory activities of in vitro-constructed transconjugants compared to the wild-type strains
| Lawn | Inhibitory activity by test straina |
|||||
|---|---|---|---|---|---|---|
| BER-NE33 | BER-NE33/pNetB | PBD1 | PBD1/pNetB | EHE-NE18 | WER-NE36 | |
| BER-NE33 | − | ++ | − | ++ | + | ++ |
| PBD1 | − | ++ | − | ++ | + | ++ |
| NE18 | − | + | − | ++ | − | ++ |
| NE36 | − | − | − | − | − | − |
| PBD1/pNetb | − | + | − | − | + | ++ |
| NE33/pNetb | − | − | − | + | + | ++ |
−, no inhibition; +, <5 mm inhibition; ++, >5 mm inhibition.
DISCUSSION
In this study, we have shown that transfer of the NetB-encoding plasmid can convert nonpathogenic strains into disease-causing strains. Using two different strains, PBD1 and BER-NE33, as recipients, transconjugants of the two strains were shown to cause similar levels of disease. This was somewhat surprising, as the chromosomal contents of these two strains are very different (Fig. 2). Genome sequencing of the two transconjugants used as challenge strains, in comparison to their native state lacking pNetB, showed that only the plasmids pNetB-ErmR and the beta2-encoding plasmid pJIR3844 were transferred to the recipient strains and that no chromosomal elements were transferred during the mating procedure. This raises a critical issue for C. perfringens diseases, as there is potential for any C. perfringens strain to become a disease-causing strain when an appropriate toxin-encoding plasmid is acquired via lateral gene transfer (LGT), regardless of the chromosomal background. Providing plasmids are maintained and compatible through their partitioning systems (34), the genomic potential of a strain could be rapidly expanded or diversified by LGT, resulting in the adaptation of C. perfringens strains to new and varied host environments.
Conjugation, resulting in the transfer and persistence of antibiotic resistance genes and virulence determinants, is a significant issue in both hospital settings and livestock industries (2). This issue is of particular concern in commercial animal production systems, resulting in the banning of the use of antibiotics as in-feed growth promoters in the European Union and increasing regulatory restrictions on their use in other countries, such as the United States (3, 35). Although HGT is a significant issue, there have been relatively few studies to determine its occurrence using in vivo systems. Some studies have investigated the transfer of antibiotic resistance genes (9, 36 – 39) both across species and within them, but little has been reported on the transfer of key virulence factors, such as toxins. The possibility that plasmid transfer can occur inside animals has been considered since the 1960s and has been shown in a variety of models, mostly under antibiotic selective pressure (8, 10, 40, 41). This study demonstrated that both the virulence and antibiotic resistance-encoding plasmids of necrotic enteritis-associated C. perfringens are conjugative within the chicken gastrointestinal tract, without selection.
C. perfringens transconjugant strains with different genetic backgrounds can cause disease in chickens when they carry the netB-containing virulence plasmid. The question therefore arises as to why the genetic background of nonpathogenic strains is more diverse than natural necrotic enteritis-causing strains (31). The chicken gastrointestinal tract has been shown, through several different studies, to contain a diverse indigenous C. perfringens population, with many unique and varied genotypes observed using multilocus sequence typing (MLST) and PFGE (30, 42, 43), with lower diversity in necrotic enteritis outbreaks (29). The lack of varied pathogenic genotypes in commercial flocks may be because the native necrotic enteritis-causing strains are more adapted to the chicken gastrointestinal tract, and although transfer of plasmids and virulence to other strains can occur, over many generations, the original strain may outcompete the transconjugants due to other genetic factors carried on the chromosome that provide a selective advantage, such as the bacteriocin perfrin gene (cpp) (44).
An interesting observation from this study is that growth of the recipient strains PBD1 and BER-NE33 was inhibited by the donor strains WER-NE36 and EHE-NE18, which provided some initial difficulty in obtaining transconjugants. During the isolation of potential transconjugants, zones of inhibition were observed surrounding the transconjugants. Upon further investigation, the transconjugants were determined to inhibit other strains, including the original donor and recipient strains, and to resist inhibition by the same strains. We do not believe the inhibitory effect is due to the previously described bacteriocin gene cpp (44), as the transconjugants do not carry the gene. These data suggest that acquisition of the virulence plasmids pNetB-ErmR and pJIR3844 confers a selective advantage over non-plasmid-carrying strains in an environment where multiple strains are present. Previous studies have shown that pathogenic C. perfringens strains can displace commensal and nonpathogenic C. perfringens from the gastrointestinal tract of chickens (45, 46). The displacement of indigenous C. perfringens strains has been thought of as a very important factor in niche competition in the gastrointestinal tract (43 – 46). However, in this study, we demonstrated that conjugative transfer of plasmids can provide resistance to inhibition to recipient strains. Therefore, it appears that if conjugation occurs before inhibition, the emergence of new and varied strains is possible.
This study did not examine long term cochallenge with multiple strains or the ecological interactions of the strains; as such, there may be genomic differences between strains that provide an advantage over different strains containing the same plasmids. The isolation of netB-negative strains from chickens suffering necrotic enteritis is also commonplace (28, 47). This could be explained if the strain causing the outbreak is not as well adapted to the chicken host. If unfit transconjugants or strains are the cause of a necrotic enteritis outbreak, there may be potential stability issues with that particular strain background and the virulence plasmids or a compatibility issue with other existing plasmids (28, 34). This could result in mild levels of disease and the loss of virulence plasmids.
Although rifampin resistance is commonly used to selectively mark C. perfringens strains (4, 16, 48), we additionally demonstrated that rifampin-resistant mutants were dramatically attenuated in the necrotic enteritis disease model. This was also true for pNetB transconjugants created with a rifampin-marked chromosome (data not shown). Rifampin resistance and attenuation have been demonstrated in other bacteria, such as Flavobacterium psychrophilum (49). Rifampin marking of strains could therefore also influence other C. perfringens virulence studies performed in the gastrointestinal tract of other animals, such as sheep and cows.
HGT has been demonstrated to occur in a wide range of bacteria that inhabit the gastrointestinal tract (GIT) (50). HGT is a major driver for strain evolution and contributes to the pangenome of a species (51, 52). As most of the major C. perfringens toxins, enzymes, and antibiotic resistance genes are a part of the accessory genome (11, 12), it is unlikely that they originated from a common ancestor and therefore have likely been acquired over time through mechanisms of HGT, including conjugation.
C. perfringens is ubiquitously found in the environment (e.g., soil) and is part of the normal microbiota in humans and a diverse range of animals (53). Whether strains colonize particular hosts or can transiently colonize a range of host species, the movement of toxin and antibiotic resistance plasmids between strains in the environment becomes an enhanced risk for human and animal health (2). If horizontal gene transfer enables any strain background to become a new or antigenically distinct pathogenic strain or act as a reservoir for new and varied virulence factors, it becomes a very important factor for C. perfringens epidemiology, genetics, and pathogenesis.
In conclusion, conjugative plasmid transfer between C. perfringens strains may have a significant role in necrotic enteritis (NE) virulence and pathogenesis. In vivo and in vitro plasmid transfer can convert nonpathogenic C. perfringens strains into NE-causing strains with the transfer of virulence plasmids to nonpathogenic strains, and antibiotic resistance plasmids can be acquired by pathogenic strains.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All C. perfringens strains used in this study are listed in Table 2. These strains were grown in tryptone-peptone-glucose (TPG) medium, tryptose sulfite cycloserine (TSC), or heart infusion (HI) broth. For conjugation experiments, HI agar (Oxoid) or TSC (Oxoid) was used. When required, solid media and broths were supplemented with antibiotics at the following concentrations: thiamphenicol (Tm), 10 μg/ml; tetracycline (Tc), 10 μg/ml; rifampin (Rif), 40 μg/ml, nalidixic acid (Nal), 40 μg/ml; and erythromycin (Erm), 50 μg/ml. Agar cultures were incubated overnight at 37°C anaerobically (10% CO2, 10% H2, and 80% N2).
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| EHE-NE18 | Wild type | 25 |
| EHE-NE18 (Rifr) | EHE-NE18 Rifr Tetr | This study |
| EHE-NE18 pNetB (Ermr) | EHE-NE18/pNetB-ErmR Rifr Tetr | This study |
| WER-NE36 | Wild type | 59 |
| WER-NE36 (Rifr) | WER-NE36 Rifr | This study |
| WER-NE36 pNetB (Ermr) | WER-NE36/pNetB-ErmR Rifr | This study |
| BER-NE33 | Wild type | 26 |
| BER-NE33 (Nalr) | BER-NE33 Nalr Tetr | This study |
| PBD1 | Wild type | 60 |
| PBD1 (Nalr) | PBD1 Nalr | This study |
| BER-NE33 | BER-NE33/pNetB-ErmR Nalr Tetr | Transconjugant |
| BER-NE33 | BER-NE33/pNetB-ErmR Nalr | Transconjugant |
| PBD1 | PBD1/pNetB-ErmR Nalr Tetr | Transconjugant |
| PBD1 | PBD1/pNetB-ErmR Nalr | Transconjugant |
| BER-NE33 | BER-NE33/pNetB-ErmR Nalr Tetr | Transconjugant |
| WER-NE36 | WER-NE36/pNetB-ErmR Rifr Tetr | Transconjugant |
| Plasmid | ||
| pNetB-ErmR | pJIR3536 (ermR_TargetTron insertion) between netI and ORF2a | This study |
ORF2, open reading frame 2.
Construction of antibiotic-resistant strains.
In order to insert an erythromycin resistance gene into the netB-carrying plasmid and thus allow selection, mutagenesis was performed as described previously for the virR gene, using a derivative of the pJIR3566 TargeTron vector. Insertion was directed into the sense strand between the genes netI and ORF-2, with a target sequence of 45 bp as follows: AAAAATTTAAATAAAGCTTAATCATGAAGA-intron-GAAGTATAGTAATTC. To retarget the group II intron, primer-mediated mutations by PCR were carried out with the IBS, ESB2, EBS1d, and EBS universal primers, in accordance with the instructions from the TargeTron gene knockdown system (Sigma-Aldrich). The 250-bp gel-extracted retargeting PCR product was digested with HindIII and BsrGI and ligated into similarly digested pJIR3566 DNA. The ligation mixture was used to transform E. coli DH5α into an erythromycin-resistant strain, and plasmid DNAs from a selection of the resultant transformants were isolated and sequenced. A vector that contained the desired altered nucleotides was chosen for subsequent mutagenesis studies. Introduction of the TargeTron plasmid, subsequent screening, and mutant confirmation were performed as previously reported (5).
Spontaneous rifampin- and nalidixic acid-resistant strains were selected by plating 100 μl of overnight TPG broths of the appropriate strains onto TSC agar plates supplemented with antibiotics at the concentrations mentioned previously. Strains with single or multiple antibiotic resistances were then confirmed by plating onto various combinations of antibiotics to ensure that the correct resistance profiles were obtained.
Western blot experiments.
A single colony from a freshly streaked and overnight incubated TSC agar plate was inoculated into tryptone-peptone-glucose (TPG) broth and incubated overnight at 37°C. Fresh TPG broth was inoculated at a 1:10 dilution and incubated until an optical density at 600 nm (OD600) of 1.0. Cultures were centrifuged at 13,000 rpm to pellet the cells. Supernatants were removed, and an equal volume of 20% trichloroacetic acid (TCA) was added to the culture supernatants and incubated on ice for 10 min. The supernatant-TCA mixture was centrifuged at 13,000 rpm to precipitate the proteins. The pellets were then washed in 100% cold acetone (4°C); this was repeated until residual color from the media was removed, leaving a white fluffy precipitate. The precipitates were then resuspended in phosphate-buffered saline agar (PBSA) and stored at 4°C.
TCA-precipitated samples were run on duplicate NuPAGE Bis-Tris (SDS-PAGE) (Thermo Fisher Scientific) protein gels for 1 h at 200 V. One gel was stained for 1 h with SimplyBlue stain and destained for 2 h in water. The second gel was transferred to a nitrocellulose membrane using the iBlot protocol (Thermo Fisher Scientific). The transferred membrane was washed twice for 10 min in Tris-buffered saline (TBS) and blocked overnight in 5% skim milk in TBS at 4°C. The blocking agent was removed, and the membrane was washed 3 times in TBS with 5% Tween for 10 min each cycle. The membrane was added to a mixture containing the primary antibody polyclonal anti-rabbit netB IgG in 1% skim milk in TBS at a 1:10,000 dilution and incubated at room temperature for 1 h. The primary antibody was removed, and the membrane was washed 3 times in TBS with 5% Tween for 10 min each cycle. The membrane was added to a mixture containing the secondary antibody horseradish peroxidase (HRP)-conjugate goat anti-rabbit IgG (Thermo Fisher Scientific) in 1% skim milk in TBS at a 1:10,000 dilution and incubated at room temperature for 1 h. The secondary antibody was removed, and the membrane was washed 3 times in TBS with 5% Tween for 10 min each cycle. The membrane was then dried using a sterile paper towel. The membrane was placed into a black sunlight-blocking container, and Pierce ECL+ substrate (Thermo Fisher Scientific) was added, as per the manufacturer's specifications, and incubated at room temperature for 5 min. Detection was done using photographic paper.
Conjugation experiments.
All in vitro matings were carried out as described previously (5). The transconjugants were selected on TSC agar supplemented with the required antibiotics.
Animal trials. (i) Ethics statement.
All animal experiments were assessed, approved, and monitored by the CSIRO Australian Animal Health Laboratory Animal Ethics Committee (approval numbers AEC1517 and AEC1697) and in accordance with national (Australian Code for the Care and Use of Animals for Scientific Purposes, 2013) and state (Victorian Prevention of Cruelty to Animals Act 1986 and part 4 of the Prevention of Cruelty to Animals Regulations 2008) legislation.
(ii) Necrotic enteritis induction model to test in vitro-derived transconjugants.
The inoculum was prepared according to the method of Keyburn et al. (54). Briefly, C. perfringens strains were streaked onto C. perfringens-specific agar (TSC; Oxoid). After incubation under anaerobic conditions at 37°C for 24 h, colonies were transferred to 10 ml of fluid thioglycolate (FTG; Difco) and incubated anaerobically at 37°C for 18 h; 1 ml was then used to inoculate 20 ml of cooked meat medium (CMM; Oxoid) and again incubated anaerobically at 37°C for 18 h. This was diluted to 1:20 in FTG medium and incubated under the same conditions. Commercial 1-day-old Ross 308 broiler chickens were fed an antibiotic-free chicken starter diet containing 20% protein for the 19 days prechallenge. The day before the challenge period, the feed was changed to a high-protein fish meal, and this feed was maintained during the challenge period. Ten birds per treatment group were challenged with C. perfringens on days 19 to 20 in-feed with a 1:1 mixture of culture and feed (vol/wt), which was performed twice daily, morning and night. Water was available ad libitum. Birds were euthanized by carbon dioxide asphyxiation on day 23; the birds were dissected, and the small intestine was cut along its length to reveal the luminal surface. Intestinal lesions were scored on a scale of 0 to 6 using a previously described scheme (55). Intestinal swabs from the small intestine and necrotic enteritis lesions were collected from the birds. A one-way analysis of variance (ANOVA) Kruskal-Wallis test with a Dunn's posttest was used to determine statistical significance between groups.
(iii) Cochallenge model.
The culture preparation was performed as described above. Birds were challenged with C. perfringens on days 21 and 22 in-feed with a 1:1 mixture of culture and feed (vol/wt), which was performed twice daily, morning and night, with alternating challenge strains. For example, in one challenge group, strain WER-NE36 was fed to chickens in the morning and strain BER-NE33 was fed to the chickens at night, while in another group, the opposite was performed. Water was available ad libitum. Birds were euthanized on day 23, and intestinal lesions were scored as described above.
(iv) Detection of transconjugants in chicken gut digesta/mucosa.
Swabs were taken from lesions or general gastrointestinal contents from chickens during necropsy, after disease was scored. Swabs were plated on TSC agar (Oxoid). Single colonies were then taken and patched onto TSC plates with different combinations of antibiotics, at concentrations described previously, to determine the antibiotic resistance profile of each colony.
PFGE analysis.
C. perfringens strains were grown overnight in TPG broth, and the bacterial pellets were suspended in 1% PFGE-certified agarose gel. Agarose plugs were incubated for 1 h with gentle shaking at 37°C in lysis buffer (0.5 M EDTA [pH 8.0], 0.5% N-lauryl-Sarkosyl, 0.25% lysozyme) and subsequently incubated in 2% proteinase buffer overnight at 55°C. For each isolate, a portion of a plug was equilibrated in 200 μl of restriction buffer at room temperature for 20 min and then digested with 20 U of SmaI at 25°C overnight. Electrophoresis was performed in a 1% PFGE-certified agarose gel and separated with the CHEF-DR-III PFGE system (Bio-Rad Laboratories) in 0.5× Tris-borate-EDTA (TBE) buffer at 14°C at 300 V for 20 h with a ramped pulse time of 4 to 38 s at an angle of 120°. The gels were stained with GelRed and visualized by UV light. Midrange and low-range PFG markers (New England BioLabs) were used as reference size standards on the gels.
Inhibition assay.
Bacteria were grown overnight in heart infusion (HI) broth (Oxoid). Lawns of bacteria were prepared by diluting the overnight culture in phosphate-buffered saline (PBS) to a density of McFarland 1, and 100 μl of the suspension was spread on HI agar plates. Drops of 20 μl from actively growing cultures (HI broth) were spotted onto the lawns, or single colonies were transferred onto the lawns. Antimicrobial activity was indicated by the absence of a bacterial lawn around the colony.
DNA extraction, genome sequencing, and bioinformatic analysis.
Strains were cultured from −80°C glycerol stocks onto fresh TSC plates (Oxoid). Single colonies were used to seed overnight cultures in tryptone-peptone-glucose (TPG) broth. The overnight broth was used to seed fresh TPG broth and incubated for 8 h. Cultures were then centrifuged for 30 min, and the supernatant was discarded. The pellets were subject to a phenol-chloroform DNA extraction. Genomes were sequenced using Illumina MiSeq methods. Illumina MiSeq read data were quality trimmed using EA-utils version 1.1.2 fastq-mcf program (55); sequences with an average quality score lower than 15 in a 5-bp sliding window were trimmed. Reads with lengths less than 50 bp or more than 70% low complexity were discarded. MiSeq reads were assembled using IDBA-UD version 1.1.1 with default settings (56). Plasmid contigs were scaffolded against the completed plasmid sequences that have been previously published (5). Genomes were annotated with Prokka (57), and the phylogenetic tree and strain comparisons were produced using Roary (58) against the previously published genomes obtained from the National Center for Biotechnology Information database.
Supplementary Material
ACKNOWLEDGMENTS
This research was conducted within the Poultry Cooperative Research Centre (CRC 2.1.5), established and supported under the Australian Government's Cooperative Research Centres Program. We thank the staff of the CSIRO Werribee Animal Facility for assistance with chicken trials.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01814-17.
REFERENCES
- 1.Nakamura Y, Itoh T, Matsuda H, Gojobori T. 2004. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet 36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- 2.Schjørring Krogfelt KA. 2011. Assessment of bacterial antibiotic resistance transfer in the gut. Int J Microbiol 2011:312956. doi: 10.1155/2011/312956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castanon JIR. 2007. History of the use of antibiotic as growth promoters in European poultry feeds. Poult Sci 86:2466–2471. doi: 10.3382/ps.2007-00249. [DOI] [PubMed] [Google Scholar]
- 4.Brynestad S, Sarker MR, McClane BA, Granum PE, Rood JI. 2001. Enterotoxin plasmid from Clostridium perfringens is conjugative. Infect Immun 69:3483–3487. doi: 10.1128/IAI.69.5.3483-3487.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bannam TL, Yan X-X, Harrison PF, Seemann T, Keyburn AL, Stubenrauch C, Weeramantri LH, Cheung JK, McClane BA, Boyce JD, Moore RJ, Rood JI. 2011. Necrotic enteritis-derived Clostridium perfringens strain with three closely related independently conjugative toxin and antibiotic resistance plasmids. mBio 2(5):e00190-. doi: 10.1128/mBio.00190-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes ML, Poon R, Adams V, Sayeed S, Saputo J, Uzal FA, McClane BA, Rood JI. 2007. Epsilon-toxin plasmids of Clostridium perfringens type D are conjugative. J Bacteriol 189:7531–7538. doi: 10.1128/JB.00767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vieira de Souza F, Roque R, Silva Moreira JL, Resende de Souza M, Nicoli JR, Neumann E, Cantini Nunes Á. 2012. Transfer of antibiotic resistance determinants between lactobacilli isolates from the gastrointestinal tract of chicken. Benef Microbes 3:137–144. doi: 10.3920/BM2011.0058. [DOI] [PubMed] [Google Scholar]
- 8.Guillot JF, Boucaud JL. 1988. In vivo plasmid transfer between strains of Escherichia coli in the chicken digestive system. Pathol Biol (Paris) 36:655–659. (In French.) [PubMed] [Google Scholar]
- 9.Bidet P, Burghoffer B, Gautier V, Brahimi N, Mariani-Kurkdjian P, El-Ghoneimi A, Bingen E, Arlet G. 2005. In vivo transfer of plasmid-encoded ACC-1 AmpC from Klebsiella pneumoniae to Escherichia coli in an infant and selection of impermeability to imipenem in K. pneumoniae. Antimicrob Agents Chemother 49:3562–3565. doi: 10.1128/AAC.49.8.3562-3565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avrain L, Vernozy-Rozand C, Kempf I. 2004. Evidence for natural horizontal transfer of tetO gene between Campylobacter jejuni strains in chickens. J Appl Microbiol 97:134–140. doi: 10.1111/j.1365-2672.2004.02306.x. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Adams V, Bannam TL, Miyamoto K, Garcia JP, Uzal FA, Rood JI, McClane BA. 2013. Toxin plasmids of Clostridium perfringens. Microbiol Mol Biol Rev 77:208–233. doi: 10.1128/MMBR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freedman JC, Theoret JR, Wisniewski JA, Uzal FA, Rood JI, McClane BA. 2015. Clostridium perfringens type A–E toxin plasmids. Res Microbiol 166:264–279. doi: 10.1016/j.resmic.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams V, Li J, Wisniewski JA, Uzal FA, Moore RJ, McClane BA, Rood JI. 2014. Virulence plasmids of spore-forming bacteria. Microbiol Spectr 2:1–24. doi: 10.1128/microbiolspec.PLAS-0024-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popoff MR. 2014. Clostridial pore-forming toxins: powerful virulence factors. Anaerobe 30:220–238. doi: 10.1016/j.anaerobe.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 15.McDonel JL. 1986. Toxins of Clostridium perfringens types A, B, C, D and E, p 477–517. In Dorner F, Drews J (ed), Pharmacology of bacterial toxins. Pergamon Press, New York, NY. [Google Scholar]
- 16.Wisniewski JA, Teng WL, Bannam TL, Rood JI. 2015. Two novel membrane proteins, TcpD and TcpE, are essential for conjugative transfer of pCW3 in Clostridium perfringens. J Bacteriol 197:774–781. doi: 10.1128/JB.02466-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wisniewski JA, Traore DA, Bannam TL, Lyras D, Whisstock JC, Rood JI. 2015. TcpM, a novel relaxase that mediates transfer of large conjugative plasmids from Clostridium perfringens. Mol Microbiol 99:884–896. doi: 10.1111/mmi.13270. [DOI] [PubMed] [Google Scholar]
- 18.Wisniewski JA, Rood JI. 2017. The Tcp conjugation system of Clostridium perfringens. Plasmid 91:28–36. doi: 10.1016/j.plasmid.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Bantwal R, Bannam TL, Porter CJ, Quinsey NS, Lyras D, Adams V, Rood JI. 2012. The peptidoglycan hydrolase TcpG is required for efficient conjugative transfer of pCW3 in Clostridium perfringens. Plasmid 67:139–147. doi: 10.1016/j.plasmid.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 20.Bannam TL, Teng WL, Bulach D, Lyras D, Rood JI. 2006. Functional identification of conjugation and replication regions of the tetracycline resistance plasmid pCW3 from Clostridium perfringens. J Bacteriol 188:4942–4951. doi: 10.1128/JB.00298-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han X, Du X-D, Southey L, Bulach DM, Seemann T, Yan X- X, Bannam TL, Rood JI. 2015. Functional analysis of a bacitracin resistance determinant located on ICECp1, a novel Tn916-like element from a conjugative plasmid in Clostridium perfringens. Antimicrob Agents Chemother 59:6855–6865. doi: 10.1128/AAC.01643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi S, Wada A, Shibasaki S, Annaka M, Higuchi H, Adachi K, Mori N, Ishikawa T, Masuda Y, Watanabe H, Yamamoto N, Yamaoka S, Inamatsu T. 2009. Spread of a large plasmid carrying the cpe gene and the tcp locus amongst Clostridium perfringens isolates from nosocomial outbreaks and sporadic cases of gastroenteritis in a geriatric hospital. Epidemiol Infect 137:108–113. doi: 10.1017/S0950268808000794. [DOI] [PubMed] [Google Scholar]
- 23.Wade B, Keyburn AL. 2015. The true cost of necrotic enteritis. World Poult 31:16–17. [Google Scholar]
- 24.Keyburn AL, Bannam TL, Moore RJ, Rood JI. 2010. NetB, a pore-forming toxin from necrotic enteritis strains of Clostridium perfringens. Toxins 2:1913–1927. doi: 10.3390/toxins2071913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keyburn AL, Boyce JD, Vaz P, Bannam TL, Ford ME, Parker D, Di Rubbo A, Rood JI, Moore RJ. 2008. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog 4:e26. doi: 10.1371/journal.ppat.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keyburn AL, Yan X-X, Bannam TL, Van Immerseel F, Rood JI, Moore RJ. 2010. Association between avian necrotic enteritis and Clostridium perfringens strains expressing NetB toxin. Vet Res 41:21. doi: 10.1051/vetres/2009069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allaart JG, de Bruijn ND, van Asten AJAM, Fabri THF, Gröne A. 2012. NetB-producing and beta2-producing Clostridium perfringens associated with subclinical necrotic enteritis in laying hens in the Netherlands. Avian Pathol 41:541–546. doi: 10.1080/03079457.2012.729809. [DOI] [PubMed] [Google Scholar]
- 28.Lepp D, Gong J, Songer JG, Boerlin P, Parreira VR, Prescott JF. 2013. Identification of accessory genome regions in poultry Clostridium perfringens isolates carrying the netB plasmid. J Bacteriol 195:1152–1166. doi: 10.1128/JB.01032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chalmers G, Bruce HL, Hunter DB, Parreira VR, Kulkarni RR, Jiang Y-F, Prescott JF, Boerlin P. 2008. Multilocus sequence typing analysis of Clostridium perfringens isolates from necrotic enteritis outbreaks in broiler chicken populations. J Clin Microbiol 46:3957–3964. doi: 10.1128/JCM.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chalmers G, Martin SW, Hunter DB, Prescott JF, Weber LJ, Boerlin P. 2008. Genetic diversity of Clostridium perfringens isolated from healthy broiler chickens at a commercial farm. Vet Microbiol 127:116–127. doi: 10.1016/j.vetmic.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Lacey JA, Johanesen PA, Lyras D, Moore RJ. 2016. Genomic diversity of necrotic enteritis-associated strains of Clostridium perfringens: a review. Avian Pathol 45:302–307. doi: 10.1080/03079457.2016.1153799. [DOI] [PubMed] [Google Scholar]
- 32.Lepp D, Roxas B, Parreira VR, Marri PR, Rosey EL, Gong J, Songer JG, Vedantam G, Prescott JF. 2010. Identification of novel pathogenicity loci in Clostridium perfringens strains that cause avian necrotic enteritis. PLoS One 5:e10795. doi: 10.1371/journal.pone.0010795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coursodon CF, Glock RD, Moore KL, Cooper KK, Songer JG. 2012. TpeL-producing strains of Clostridium perfringens type A are highly virulent for broiler chicks. Anaerobe 18:117–121. doi: 10.1016/j.anaerobe.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Adams V, Watts TD, Bulach DM, Lyras D, Rood JI. 2015. Plasmid partitioning systems of conjugative plasmids from Clostridium perfringens. Plasmid 80:90–96. doi: 10.1016/j.plasmid.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Casewell M, Friis C, Marco E, McMullin P, Phillips I. 2003. The European ban on growth-promoting antibiotics and emerging consequences for human and animal health. J Antimicrob Chemother 52:159–161. doi: 10.1093/jac/dkg313. [DOI] [PubMed] [Google Scholar]
- 36.Boguslawska J, Zycka-Krzesinska J, Wilcks A, Bardowski J. 2009. Intra- and interspecies conjugal transfer of Tn916-like elements from Lactococcus lactis in vitro and in vivo. Appl Environ Microbiol 75:6352–6360. doi: 10.1128/AEM.00470-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bratoeva MP, John JF. 1994. In vivo R-plasmid transfer in a patient with a mixed infection of shigella dysentery. Epidemiol Infect 112:247–252. doi: 10.1017/S0950268800057654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson TJ, Singer RS, Isaacson RE, Danzeisen JL, Lang K, Kobluk K, Rivet B, Borewicz K, Frye JG, Englen M, Anderson J, Davies PR. 2015. In vivo transmission of an IncA/C plasmid in Escherichia coli depends on tetracycline concentration, and acquisition of the plasmid results in a variable cost of fitness. Appl Environ Microbiol 81:3561–3570. doi: 10.1128/AEM.04193-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morelli L, Sarra PG, Bottazzi V. 1988. In vivo transfer of pAM beta 1 from Lactobacillus reuteri to Enterococcus faecalis. J Appl Bacteriol 65:371–375. doi: 10.1111/j.1365-2672.1988.tb01905.x. [DOI] [PubMed] [Google Scholar]
- 40.Balis E, Vatopoulos AC, Kanelopoulou M, Mainas E, Hatzoudis G, Kontogianni V, Malamou-Lada H, Kitsou-Kiriakopoulou S, Kalapothaki V. 1996. Indications of in vivo transfer of an epidemic R plasmid from Salmonella enteritidis to Escherichia coli of the normal human gut flora. J Clin Microbiol 34:977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Syvanen M. 2012. Evolutionary implications of horizontal gene transfer. Annu Rev Genet 46:341–358. doi: 10.1146/annurev-genet-110711-155529. [DOI] [PubMed] [Google Scholar]
- 42.Nauerby B, Pedersen K, Madsen M. 2003. Analysis by pulsed-field gel electrophoresis of the genetic diversity among Clostridium perfringens isolates from chickens. Vet Microbiol 94:257–266. doi: 10.1016/S0378-1135(03)00118-4. [DOI] [PubMed] [Google Scholar]
- 43.Hibberd MC, Neumann AP, Rehberger TG, Siragusa GR. 2011. Multilocus sequence typing subtypes of poultry Clostridium perfringens isolates demonstrate disease niche partitioning. J Clin Microbiol 49:1556–1567. doi: 10.1128/JCM.01884-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timbermont L, De Smet L, Van Nieuwerburgh F, Parreira VR, Van Driessche G, Haesebrouck F, Ducatelle R, Prescott J, Deforce D, Devreese B, Van Immerseel F. 2014. Perfrin, a novel bacteriocin associated with netB positive Clostridium perfringens strains from broilers with necrotic enteritis. Vet Res 45:40. doi: 10.1186/1297-9716-45-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barbara AJ, Trinh HT, Glock RD, Glenn Songer J. 2008. Necrotic enteritis-producing strains of Clostridium perfringens displace non-necrotic enteritis strains from the gut of chicks. Vet Microbiol 126:377–382. doi: 10.1016/j.vetmic.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 46.Timbermont L, Lanckriet A, Pasmans F, Haesebrouck F, Ducatelle R, Van Immerseel F. 2009. Intra-species growth-inhibition by Clostridium perfringens is a possible virulence trait in necrotic enteritis in broilers. Vet Microbiol 137:388–391. doi: 10.1016/j.vetmic.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 47.Bailey MA, Macklin KS, Krehling JT. 2014. Low prevalence of netB and tpeL in historical Clostridium perfringens isolates from broiler farms in Alabama. Avian Dis 59:46–51. doi: 10.1637/10866-051914-Reg. [DOI] [PubMed] [Google Scholar]
- 48.Pedersen K, Bjerrum L, Nauerby B, Madsen M. 2003. Experimental infections with rifampicin-resistant Clostridium perfringens strains in broiler chickens using isolator facilities. Avian Pathol 32:403–411. doi: 10.1080/0307945031000121158. [DOI] [PubMed] [Google Scholar]
- 49.Gliniewicz K, Wildung M, Orfe LH, Wiens GD, Cain KD, Lahmers KK, Snekvik KR, Call DR. 2015. Potential mechanisms of attenuation for rifampicin-passaged strains of Flavobacterium psychrophilum. BMC Microbiol 15:179. doi: 10.1186/s12866-015-0518-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huddleston JR. 2014. Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect Drug Resist 7:167–176. doi: 10.2147/IDR.S48820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 52.Aminov RI. 2011. Horizontal gene exchange in environmental microbiota. Front Microbiol 2:158. doi: 10.3389/fmicb.2011.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park JY, Kim S, Oh JY, Kim HR, Jang I, Lee HS, Kwon YK. 2015. Characterization of Clostridium perfringens isolates obtained from 2010 to 2012 from chickens with necrotic enteritis in Korea. Poult Sci 94:1158–1164. doi: 10.3382/ps/pev037. [DOI] [PubMed] [Google Scholar]
- 54.Keyburn AL, Sheedy SA, Ford ME, Williamson MM, Awad MM, Rood JI, Moore RJ. 2006. Alpha-toxin of Clostridium perfringens is not an essential virulence factor in necrotic enteritis in chickens. Infect Immun 74:6496–6500. doi: 10.1128/IAI.00806-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aronesty E. 2011. ea-utils: command-line tools for processing biological sequencing data. https://github.com/ExpressionAnalysis/ea-utils.
- 56.Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 57.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 58.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stanley D, Keyburn AL, Denman SE, Moore RJ. 2012. Changes in the caecal microflora of chickens following Clostridium perfringens challenge to induce necrotic enteritis. Vet Microbiol 159:155–162. [DOI] [PubMed] [Google Scholar]
- 60.Wade B, Keyburn AL, Seemann T, Rood JI, Moore RJ. 2015. Binding of Clostridium perfringens to collagen correlates with the ability to cause necrotic enteritis in chickens. Vet Microbiol 180:299–303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.