ABSTRACT
The genome of the bacterium Burkholderia thailandensis encodes three complete LuxI/LuxR-type quorum sensing (QS) systems: BtaI1/BtaR1 (QS-1), BtaI2/BtaR2 (QS-2), and BtaI3/BtaR3 (QS-3). The LuxR-type transcriptional regulators BtaR1, BtaR2, and BtaR3 modulate the expression of target genes in association with various N-acyl-l-homoserine lactones (AHLs) as signaling molecules produced by the LuxI-type synthases BtaI1, BtaI2, and BtaI3. We have systematically dissected the complex QS circuitry of B. thailandensis strain E264. Direct quantification of N-octanoyl-homoserine lactone (C8-HSL), N-3-hydroxy-decanoyl-homoserine lactone (3OHC10-HSL), and N-3-hydroxy-octanoyl-homoserine lactone (3OHC8-HSL), the primary AHLs produced by this bacterium, was performed by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) in the wild-type strain and in QS deletion mutants. This was compared to the transcription of btaI1, btaI2, and btaI3 using chromosomal mini-CTX-lux transcriptional reporters. Furthermore, the levels of expression of btaR1, btaR2, and btaR3 were monitored by quantitative reverse transcription-PCR (qRT-PCR). We observed that C8-HSL, 3OHC10-HSL, and 3OHC8-HSL are differentially produced over time during bacterial growth and correlate with the btaI1, btaI2, and btaI3 gene expression profiles, revealing a successive activation of the corresponding QS systems. Moreover, the transcription of the btaR1, btaR2, and btaR3 genes is modulated by cognate and noncognate AHLs, showing that their regulation depends on themselves and on other QS systems. We conclude that the three QS systems in B. thailandensis are interdependent, suggesting that they cooperate dynamically and function in a concerted manner in modulating the expression of QS target genes through a successive regulatory network.
KEYWORDS: Burkholderia, acyl-homoserine lactone, gene regulation, LuxR/LuxI, quorum sensing
IMPORTANCE
Quorum sensing (QS) is a widespread bacterial communication system coordinating the expression of specific genes in a cell density-dependent manner and allowing bacteria to synchronize their activities and to function as multicellular communities. QS plays a crucial role in bacterial pathogenicity by regulating the expression of a wide spectrum of virulence/survival factors and is essential to environmental adaptation. The results presented here demonstrate that the multiple QS systems coexisting in the bacterium Burkholderia thailandensis, which is considered the avirulent version of the human pathogen Burkholderia pseudomallei and thus commonly used as an alternative study model, are hierarchically and homeostatically organized. We found these QS systems to be finely integrated into a complex regulatory network, including transcriptional and posttranscriptional interactions, and further incorporating growth stages and temporal expression. These results provide a unique, comprehensive illustration of a sophisticated QS network and will contribute to a better comprehension of the regulatory mechanisms that can be involved in the expression of QS-controlled genes, in particular those associated with the establishment of host-pathogen interactions and acclimatization to the environment.
INTRODUCTION
Quorum sensing (QS) is a global regulatory mechanism of gene expression depending on bacterial density (1). Gram-negative bacteria typically possess homologues of the LuxI/LuxR system initially characterized in the bioluminescent marine bacterium Vibrio fischeri (2). The signaling molecules N-acyl-l-homoserine lactones (AHLs) produced by the LuxI-type synthases accumulate in the environment throughout bacterial growth, providing information on cell density. These AHLs activate the LuxR-type transcriptional regulators that modulate the expression of QS target genes, which usually contain a lux box sequence in their promoter region. These genes include a luxI homologue encoding a LuxI-type synthase generally located in close vicinity of a luxR homologue that codes for a LuxR-type transcriptional regulator, resulting in a typical self-inducing loop of AHLs (3).
Species belonging to the Burkholderia genus generally carry a unique AHL-based QS system referred as the CepI/CepR QS system (4). The CepI synthase is responsible for the production of N-octanoyl-homoserine lactone (C8-HSL), whereas the CepR transcriptional regulator modulates the expression of QS target genes in association with C8-HSL, including the cepI gene (4). Additionally, the cepR gene transcription can be autoregulated as well (5, 6). Multiple QS circuitries were also reported for several Burkholderia spp., such as the members of the Bptm group that consists of the nonpathogenic soil saprophyte Burkholderia thailandensis and the closely related pathogens Burkholderia pseudomallei and Burkholderia mallei responsible for melioidosis and glanders, respectively (7–9). QS was reported to be involved in the regulation of several virulence factors in B. pseudomallei and to be essential to its pathogenicity (10, 11). B. thailandensis, which is considered the avirulent version of B. pseudomallei (12), is commonly used as a surrogate model for the study of B. pseudomallei, which is considered a potential bioterrorism agent and whose manipulation is consequently restricted to biosafety level 3 (BSL3) laboratories. The members of the Bptm group contain homologous LuxI/LuxR QS systems that are involved in the biosynthesis of various AHLs (13–17). In B. thailandensis, the LuxI/LuxR QS systems are referred to as the BtaI1/BtaR1 (QS-1), BtaI2/BtaR2 (QS-2), and BtaI3/BtaR3 (QS-3) QS systems. The QS-1, QS-2, and QS-3 systems are also found in B. pseudomallei, whereas the QS-2 system is absent in B. mallei (18). These species also possess additional orphan luxR homologues, namely, btaR4 (malR) and btaR5 in B. thailandensis (7–9, 19).
The QS-1 system is composed of the btaI1 and btaR1 genes that code for the BtaI1 synthase and the BtaR1 transcriptional regulator, respectively. BtaI1 is responsible for the production of C8-HSL (13), and transcription of btaI1 is positively modulated by BtaR1 (20). The BtaI2 synthase and the BtaR2 transcriptional regulator encoded by the btaI2 and btaR2 genes, respectively, constitute the QS-2 system. BtaR2 directly activates expression of btaI2 involved in both N-3-hydroxy-decanoyl-homoserine lactone (3OHC10-HSL) and N-3-hydroxy-octanoyl-homoserine lactone (3OHC8-HSL) biosynthesis (16). The QS-3 system comprises the btaI3 gene encoding the BtaI3 synthase that also catalyzes the synthesis of 3OHC8-HSL (13), as well as the BtaR3 transcriptional regulator, the product of the btaR3 gene located next to btaI3.
The main goal of this study was to dissect the QS regulatory network of B. thailandensis E264 to reveal the interactions existing between the QS-1, QS-2, and QS-3 systems. Besides verifying previously proposed and established interactions, we uncovered several interconnections between the QS-1, QS-2, and QS-3 circuits, providing a comprehensive picture of the complex QS network in B. thailandensis E264. Ultimately, this study will contribute to a better appreciation of the QS regulatory mechanism of the expression of genes in B. thailandensis, and in particular those related to pathogenicity in B. pseudomallei.
RESULTS
The B. thailandensis QS-1, QS-2, and QS-3 systems are successively activated.
B. thailandensis E264 produces 3OHC10-HSL and to lesser extents, C8-HSL and 3OHC8-HSL (13, 16), but their levels at different stages throughout bacterial growth had never been investigated. Considering that nonsimultaneous production of AHLs in B. pseudomallei KHW was suggested (17), we hypothesized that these three AHLs are differentially produced over the growth phases of B. thailandensis E264. We thus determined the production profiles of C8-HSL, 3OHC10-HSL, and 3OHC8-HSL at various time points of the bacterial growth. Liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) was used to quantify the concentrations of these AHLs in wild-type B. thailandensis E264 cultures. We found that the amounts of 3OHC10-HSL increased rapidly through the early logarithmic growth phase (optical density at 600 nm [OD600] ≈ 3.0) and late exponential growth phase (OD600 ≈ 5.0) but decreased thereafter (Fig. 1A). Interestingly, 3OHC8-HSL concentrations kept increasing throughout bacterial growth to levels similar to the ones of 3OHC10-HSL (Fig. 1A). C8-HSL accumulated only during logarithmic growth and then remained stable in the stationary growth phase (OD600 ≈ 8.0; Fig. 1A).
FIG 1 .
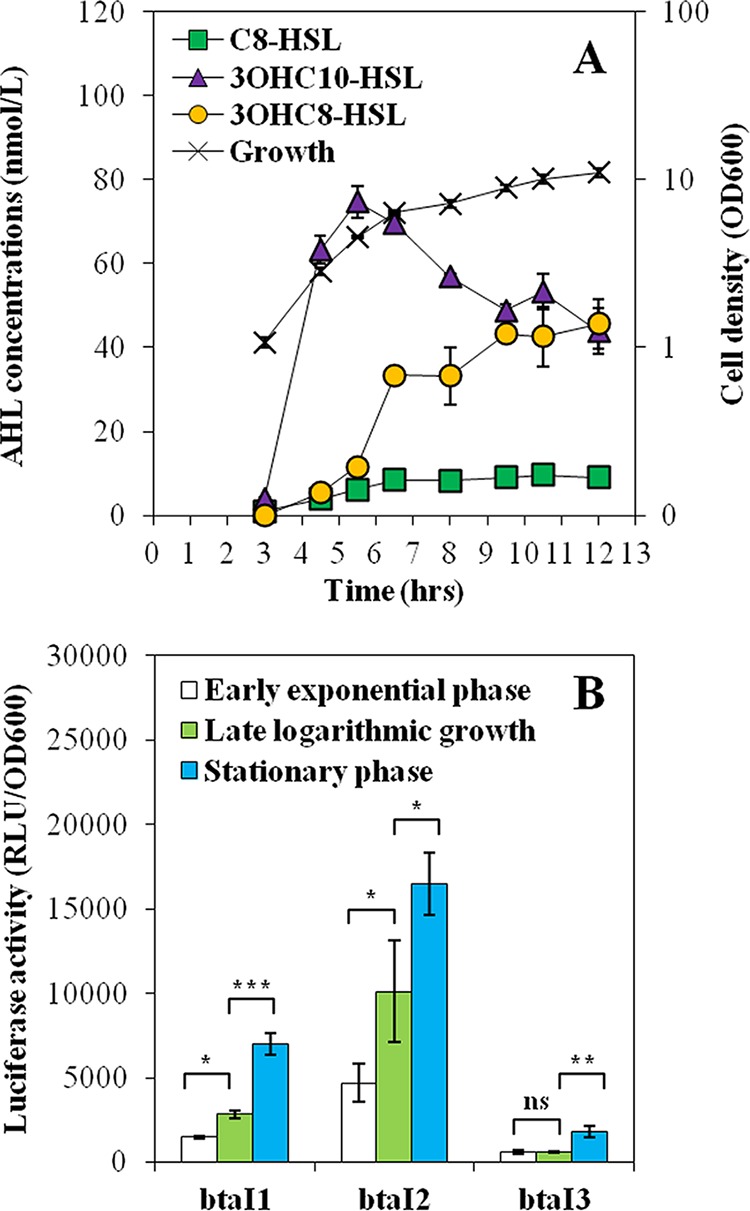
The QS-1, QS-2, and QS-3 systems are consecutively activated. (A) C8-HSL, 3OHC10-HSL, and 3OHC8-HSL concentrations were measured by LC-MS/MS throughout the different stages of bacterial growth in cultures of the wild-type E264 strain of B. thailandensis. The values are means ± standard deviations (error bars) for three replicates. (B) The luciferase activity of the chromosomal btaI1-lux, btaI2-lux, and btaI3-lux transcriptional fusions was monitored during the early exponential growth phase (OD600 ≈ 3.0), late logarithmic growth phase (OD600 ≈ 5.0), and stationary growth phase (OD600 ≈ 8.0). Luminescence is expressed in relative light units per optical density of the culture (RLU/OD600). Values that are significantly different are indicated by brackets and asterisks as follows: ***, P < 0.001; **, P < 0.01; *, P < 0.05. Values that are not significantly different (ns) are also indicated.
To gain additional insights, biosynthesis of AHLs was correlated to the expression of the btaI1, btaI2, and btaI3 genes. The activity of the chromosomal btaI1-lux, btaI2-lux, and btaI3-lux transcriptional reporters was measured during bacterial growth. In agreement with the AHL production profiles, activation of both btaI1 and btaI2 was observed from logarithmic growth (Fig. 1B), with btaI2 expression starting earlier than for btaI1 (data not shown), whereas btaI3 was not activated until stationary phase was reached (Fig. 1B). Collectively, our results point toward a successive activation of the different QS systems in B. thailandensis E264 throughout the bacterial growth phases.
The QS-1, QS-2, and QS-3 systems act in a coordinated way to finely modulate the synthesis of AHLs.
In order to verify whether the successive activation of the QS-1, QS-2, and QS-3 systems results from interactions between these QS circuits, we determined the kinetics of production of AHLs in cultures of the ΔbtaR1, ΔbtaR2, and ΔbtaR3 mutants compared to the wild-type E264 strain of B. thailandensis throughout the bacterial growth phases. We also measured expression of the AHL synthase-coding genes btaI1, btaI2, and btaI3 in the same backgrounds harboring a chromosomal btaI1-lux, btaI2-lux, or btaI3-lux transcriptional fusion.
BtaI1 produces C8-HSL, and BtaR1 is considered the main regulator of btaI1 expression (13). Therefore, we were surprised to see increased production of C8-HSL in the ΔbtaR1 mutant compared to the wild-type strain (Fig. 2A). This overproduction was principally detected after the end of the exponential phase. Nevertheless, transcription of the btaI1 gene was lower in the ΔbtaR1 mutant throughout the different stages of bacterial growth, and it was almost zero in early logarithmic growth (Fig. 2B). Because of these results, it was important to confirm that btaI1 expression is activated by BtaR1 in conjunction with C8-HSL. We monitored btaI1 expression in response to exogenous addition of C8-HSL in the wild-type B. thailandensis strain E264 and its ΔbtaR1, ΔbtaI1, and ΔbtaI1 ΔbtaI2 ΔbtaI3 mutants. The btaI1 gene exhibited comparable transcriptional profiles in the absence of BtaR1 or C8-HSL, supporting the idea that BtaR1/C8-HSL does indeed activate btaI1 transcription (see Fig. S1 in the supplemental material). Accordingly, adding exogenous C8-HSL restored btaI1 transcription in both the ΔbtaI1 and ΔbtaI1 ΔbtaI2 ΔbtaI3 mutants (Fig. S1). While expression of btaI1 was induced in the wild-type strain culture supplemented with exogenous C8-HSL, no difference was noticed for the ΔbtaR1 mutant, confirming that activation of btaI1 by this AHL involves BtaR1 (Fig. S1).
FIG 2 .
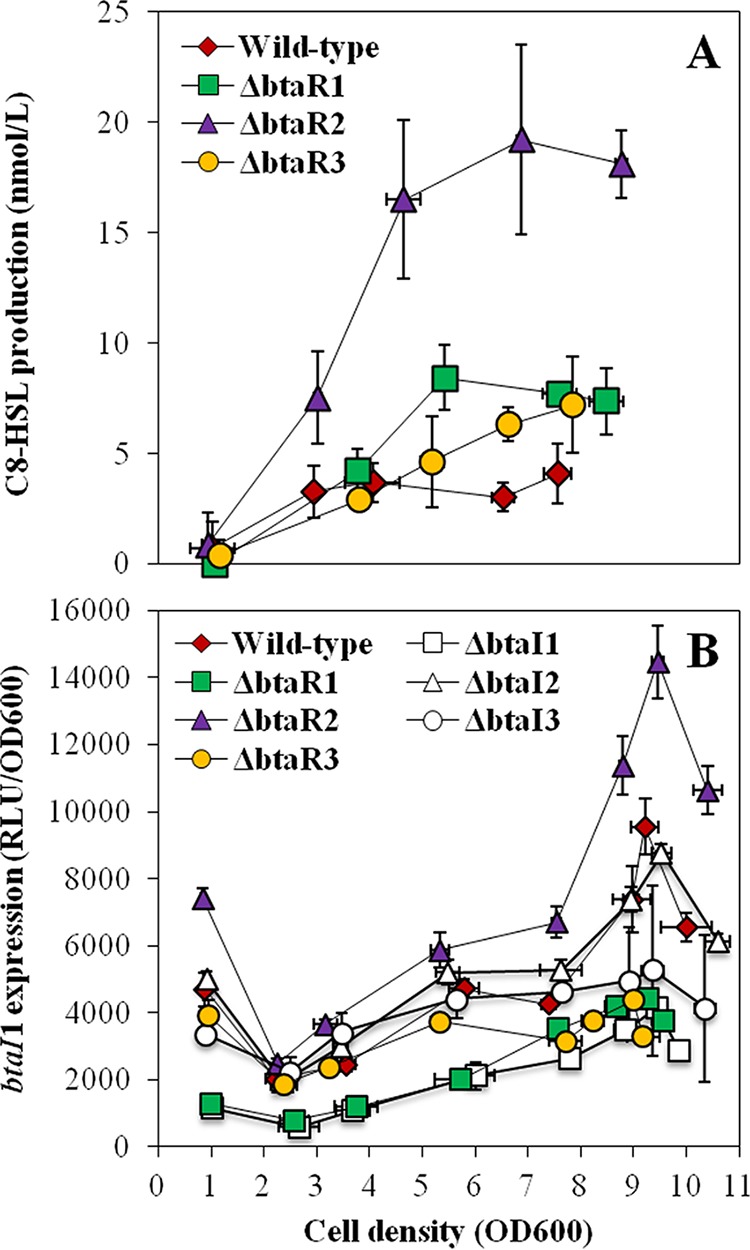
C8-HSL production and expression from the btaI1 promoter in the wild-type and QS mutant strains of B. thailandensis E264. (A) The biosynthesis of C8-HSL was quantified using LC-MS/MS at various times during growth in cultures of the wild-type strain and of the ΔbtaR1, ΔbtaR2, and ΔbtaR3 mutant strains of B. thailandensis E264. The error bars represent the standard deviations of the averages for three replicates. (B) The luciferase activity of the chromosomal btaI1-lux transcriptional fusion was monitored in cultures of the wild-type strain and of the ΔbtaR1, ΔbtaR2, ΔbtaR3, ΔbtaI1, ΔbtaI2, and ΔbtaI3 mutant strains of B. thailandensis E264. The luminescence is expressed in relative light units per optical density of the culture (RLU/OD600).
btaI1 activation requires BtaR1 and C8-HSL. The luciferase activity of the chromosomal btaI1-lux transcriptional fusion was monitored during the exponential growth phase in cultures of the B. thailandensis E264 wild-type strain and the ΔbtaR1, ΔbtaI1, and ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant strains. Cultures were supplemented with 10 µM C8-HSL. Acetonitrile only was added to the controls. The values represent the means for three replicates. The luminescence is expressed in relative light units per optical density of the culture (RLU/OD600). Download FIG S1, PDF file, 0.04 MB (43KB, pdf) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To determine whether the QS-1 system is also under BtaR2 and BtaR3 control, we investigated the effects of these transcriptional regulators on both the production of C8-HSL and expression of btaI1. Interestingly, C8-HSL concentrations were also increased in the ΔbtaR2 mutant, with a matching upregulation of btaI1 expression during logarithmic growth (Fig. 2), revealing that BtaR2 might repress the production of C8-HSL by modulating the transcription of btaI1. While C8-HSL was also overproduced in the absence of BtaR3 during stationary phase (Fig. 2A), btaI1 transcription was downregulated in the ΔbtaR3 mutant (Fig. 2B), suggesting that the negative impact of BtaR3 on C8-HSL biosynthesis is indirect and does not result from btaI1 regulation. Altogether, these data indicate that while BtaR1 constitutes the main regulator of the QS-1 system, C8-HSL biosynthesis is also directly and indirectly dependent on both BtaR2 and BtaR3, respectively.
3OHC10-HSL is produced by the BtaI2 synthase (16). While BtaR2 directly activates btaI2 expression in response to 3OHC10-HSL and 3OHC8-HSL, the latter being also produced by BtaI2 (16), the direct impact of BtaR2 on the production of these two AHLs is still untested. We observed that both 3OHC10-HSL biosynthesis and btaI2 expression were almost completely abolished in the ΔbtaR2 mutant, confirming that BtaR2 is their main regulator (Fig. 3). Despite the absence of BtaR2, we detected a slight, but consistent and highly reproducible, production of 3OHC10-HSL during stationary phase (Fig. 3A). Accordingly, transcription of btaI2 was also slightly augmented later (Fig. 3B). Thus, 3OHC10-HSL biosynthesis and btaI2 expression might not be exclusively under BtaR2 control.
FIG 3 .
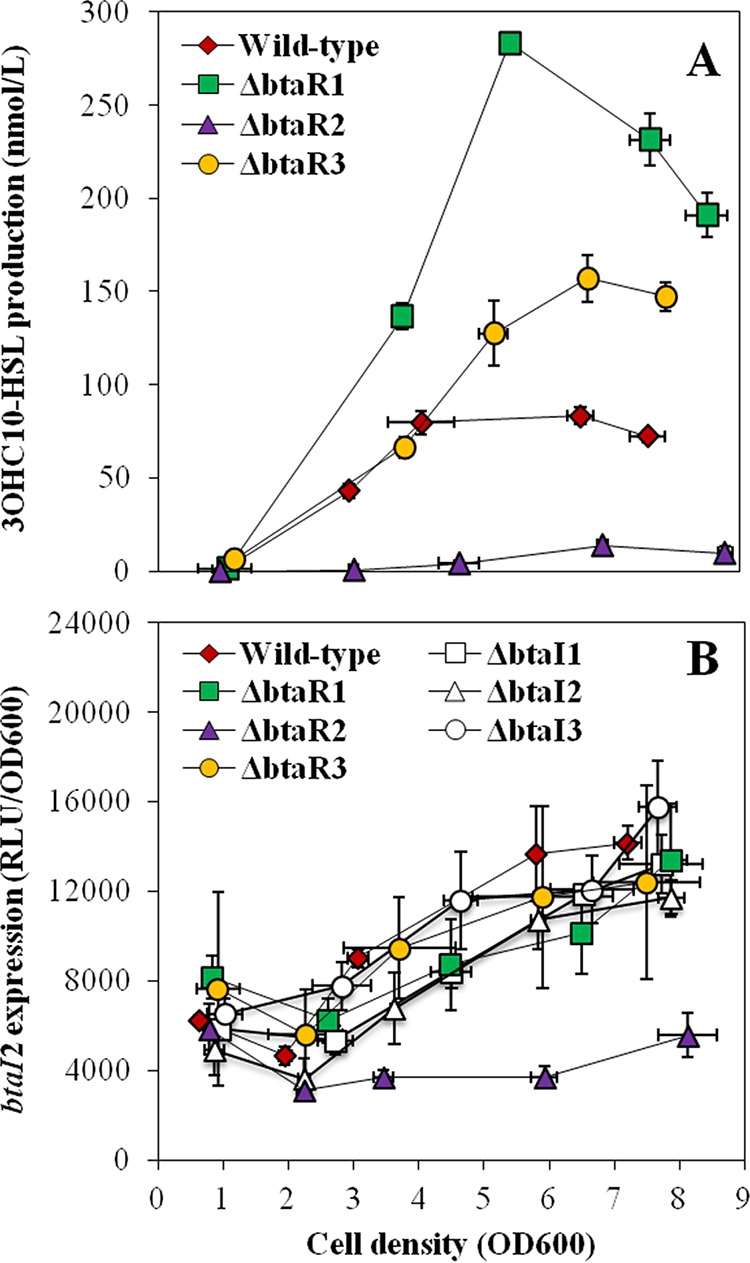
3OHC10-HSL production and expression from the btaI2 promoter in the wild-type strain and QS mutant strains of B. thailandensis E264. (A) The biosynthesis of 3OHC10-HSL was quantified using LC-MS/MS at various times during growth in cultures of the wild-type and ΔbtaR1, ΔbtaR2, and ΔbtaR3 mutant strains of B. thailandensis E264. The error bars represent the standard deviations of the averages for three replicates. (B) The luciferase activity of the chromosomal btaI2-lux transcriptional fusion was monitored in cultures of the wild-type and ΔbtaR1, ΔbtaR2, ΔbtaR3, ΔbtaI1, ΔbtaI2, and ΔbtaI3 mutant strains of B. thailandensis E264. The luminescence is expressed in relative light units per optical density of the culture (RLU/OD600).
To determine whether BtaR1 and BtaR3 also intervene in the regulation of 3OHC10-HSL production and btaI2 transcription, their effects on the QS-2 system were investigated. Interestingly, 3OHC10-HSL concentrations were strongly increased in the ΔbtaR1 mutant compared to the wild-type strain from the beginning of logarithmic growth (Fig. 3A). The levels of 3OHC10-HSL were also increased in the ΔbtaR3 mutant background, but this was observed only after the end of the exponential phase (Fig. 3A). However, in both cases, no impact on btaI2 transcription was noticed despite an increase in the amounts of 3OHC10-HSL (Fig. 3B). Collectively, these observations indicate that although BtaR1 and BtaR3 influence the biosynthesis of 3OHC10-HSL, the effects of these transcriptional regulators on the QS-2 system are indirect.
BtaI3 is mainly responsible for 3OHC8-HSL biosynthesis (13). While no discernible difference in 3OHC8-HSL concentrations was detected in cultures of the ΔbtaR3 mutant compared to cultures of the wild-type strain (Fig. 4A), the levels of btaI3 transcription were decreased (Fig. 4B). To confirm whether transcription of btaI3 is dependent on BtaR3 and on 3OHC8-HSL, btaI3 expression was measured in the wild-type strain and in the ΔbtaR3, ΔbtaI3, and ΔbtaI1 ΔbtaI2 ΔbtaI3 mutants supplemented with exogenous 3OHC8-HSL or not supplemented with 3OHC8-HSL. We found that btaI3 was similarly downregulated in the ΔbtaR3 and ΔbtaI3 mutant backgrounds, suggesting that BtaR3 activates btaI3 in response to 3OHC8-HSL (Fig. S2). Accordingly, btaI3 transcription was not affected by the addition of 3OHC8-HSL in the ΔbtaR3 mutant, but it was increased in the wild-type strain culture under the same conditions, revealing that activation of btaI3 by this AHL is linked to BtaR3 (Fig. S2). Unexpectedly, adding exogenous 3OHC8-HSL to the culture of the ΔbtaI3 mutant did not restore btaI3 transcription to wild-type levels (Fig. S2). However, we observed that expression of btaI3 was restored in the AHL-defective ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant supplemented with 3OHC8-HSL, confirming the involvement of this AHL in the activation of btaI3 (Fig. S2). Taken together, these data confirm that btaI3 is activated by BtaR3/3OHC8-HSL and suggest that expression of this gene is controlled by additional AHLs and/or alternative LuxR-type transcriptional regulators.
FIG 4 .
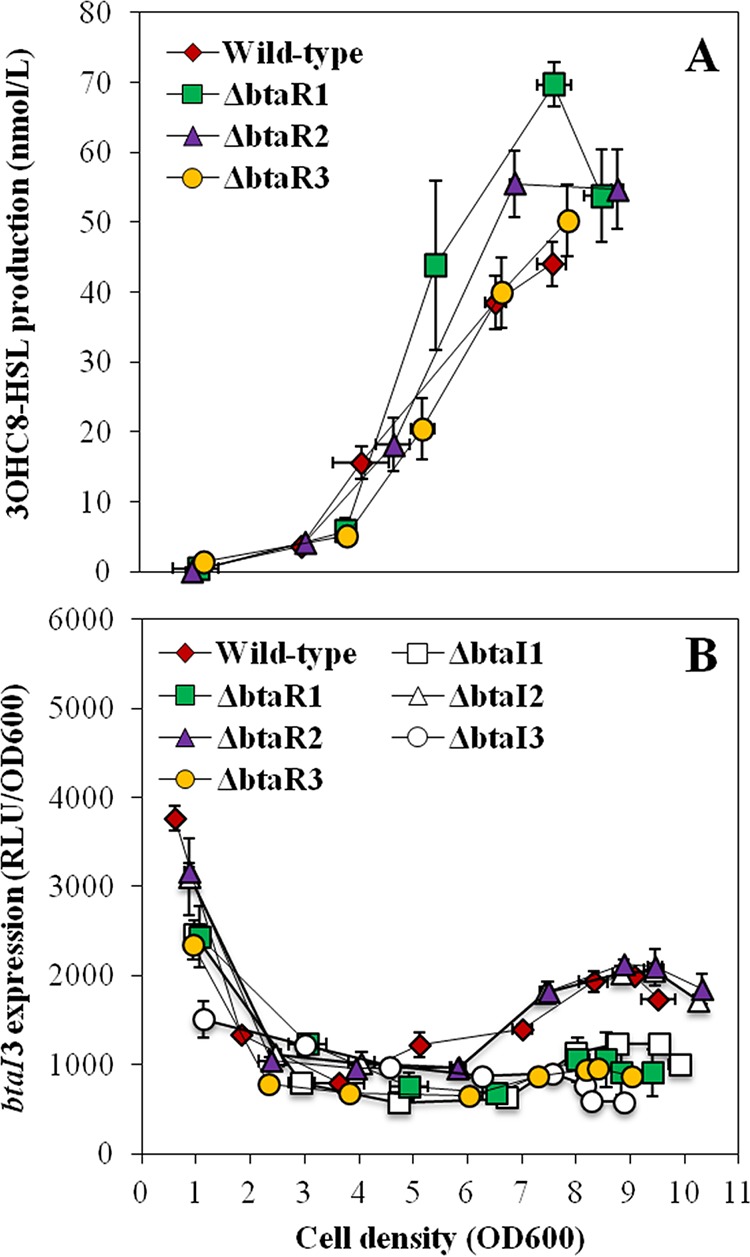
3OHC8-HSL production and expression from the btaI3 promoter in the wild-type and QS mutant strains of B. thailandensis E264. (A) The biosynthesis of 3OHC8-HSL was quantified using LC-MS/MS at various times during growth in cultures of the wild-type and ΔbtaR1, ΔbtaR2, and ΔbtaR3 mutant strains of B. thailandensis E264. The error bars represent the standard deviations of the averages for three replicates. (B) The luciferase activity of the chromosomal btaI3-lux transcriptional fusion was monitored in cultures of the wild-type and ΔbtaR1, ΔbtaR2, ΔbtaR3, ΔbtaI1, ΔbtaI2, and ΔbtaI3 mutant strains of B. thailandensis E264. The luminescence is expressed in relative light units per optical density of the culture (RLU/OD600).
btaI3 is activated by BtaR3 and 3OHC8-HSL. The luciferase activity of the chromosomal btaI3-lux transcriptional fusion was measured during stationary phase in cultures of the B. thailandensis wild-type E264 strain and ΔbtaR3, ΔbtaI3, and ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant strains. Cultures were supplemented with 10 µM 3OHC8-HSL. Acetonitrile only was added to the controls. The values represent the means for three replicates. The luminescence is expressed in relative light units per optical density of the culture (RLU/OD600). Download FIG S2, PDF file, 0.04 MB (43KB, pdf) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To confirm that the QS-3 system is not exclusively modulated by BtaR3, we investigated the influence of BtaR1 and BtaR2 on 3OHC8-HSL biosynthesis and btaI3 expression. As previously noted for C8-HSL and 3OHC10-HSL, the levels of 3OHC8-HSL were enhanced in the ΔbtaR1 mutant compared to the wild-type strain (Fig. 4A). While 3OHC10-HSL overproduction was observed during the different stages of bacterial growth (Fig. 3A), augmentation of 3OHC8-HSL concentrations occurred principally in the late exponential phase in the ΔbtaR1 mutant (Fig. 4A). Surprisingly, expression of btaI3 was lower, suggesting that the negative regulation of 3OHC8-HSL biosynthesis by BtaR1 is indirect and does not result from btaI3 modulation (Fig. 4B). Additionally, we observed an increase in 3OHC8-HSL levels in the ΔbtaR2 mutant from late logarithmic growth (Fig. 4A). Nevertheless, no obvious change in expression of btaI3 was visible, revealing that BtaR2 might not repress 3OHC8-HSL biosynthesis through regulation of btaI3 transcription as well (Fig. 4B). All in all, these findings demonstrate that the QS-1, QS-2, and QS-3 systems work collectively to regulate production of AHLs.
We also analyzed production of AHLs in the ΔbtaR4 and ΔbtaR5 mutants, and no difference with the wild-type strain production was found, revealing that neither BtaR4 nor BtaR5 was involved in the regulation of the biosynthesis of C8-HSL, 3OHC10-HSL, and 3OHC8-HSL under the conditions of our experiments (data not shown).
The btaR1, btaR2, and btaR3 genes are QS controlled.
In order to verify whether the QS modulatory cascade also involves cross-regulation between the BtaR transcriptional regulators, the levels of expression of btaR1, btaR2, and btaR3 were assessed by quantitative reverse transcription-PCR (qRT-PCR) in the wild-type B. thailandensis E264 strain and in the AHL-defective ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant during the exponential phase. Interestingly, the transcription of btaR1, btaR2, and btaR3 was significantly affected by the absence of AHLs, indicating that they are controlled by QS (Fig. 5). btaR1 transcription was increased in the ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant compared to the wild-type strain, revealing that its expression is negatively regulated by AHLs (Fig. 5A). Conversely, btaR2 and btaR3 were both downregulated in the absence of AHLs, showing that these genes are activated by QS (Fig. 5B and C). To further investigate the impact of AHLs on the expression of btaR1, btaR2, and btaR3, their transcription was measured in the ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant supplemented with exogenous C8-HSL, 3OHC10-HSL, or 3OHC8-HSL. Interestingly, the levels of expression of btaR1, btaR2, and btaR3 were restored to wild-type levels in the presence of AHLs produced by their respective cognate synthase, as well as in the presence of noncognate AHLs, suggesting that their regulation depends on themselves and on other QS systems (Fig. 5). Collectively, our results indicate that the interdependence of the QS-1, QS-2, and QS-3 systems also implicates cross-modulation between BtaR1, BtaR2, and BtaR3.
FIG 5 .
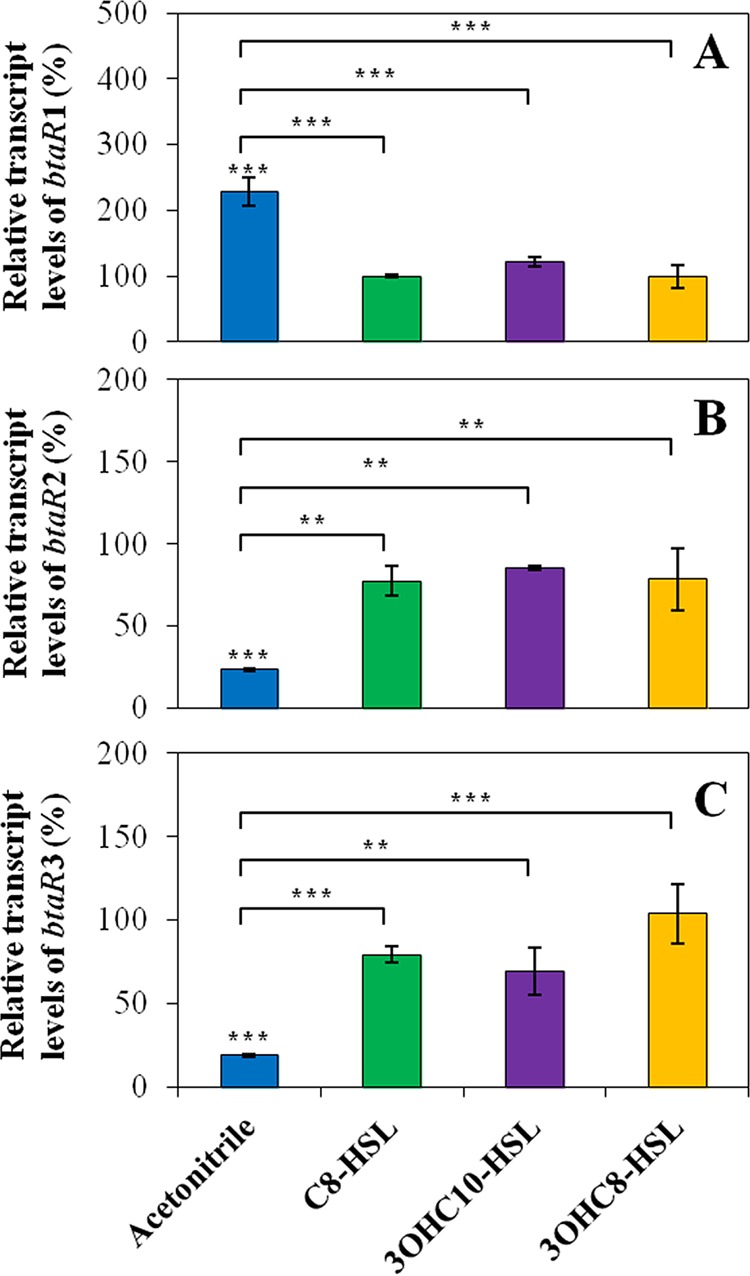
Effects of AHLs on the levels of expression of the btaR1, btaR2, and btaR3 genes. The relative transcript levels of (A) btaR1, (B) btaR2, and (C) btaR3 from the wild-type B. thailandensis E264 strain and its ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant strain were estimated by qRT-PCR. Cultures were supplemented with 10 µM C8-HSL, 3OHC10-HSL, or 3OHC8-HSL. Acetonitrile only was added to the controls. The results are presented as relative quantification of transcription of the gene compared to the wild-type strain, which was set at 100%. The error bars represent the standard deviations of the averages for three replicates. ***, P < 0.001; **, P < 0.01.
The levels of expression of btaI1, btaI2, and btaI3 are modulated by cognate and noncognate AHLs.
To further elucidate the regulatory mechanisms directing btaI1, btaI2, and btaI3 expression, the activity of the corresponding chromosomal lux transcriptional reporters was measured in the AHL-defective ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant supplemented with exogenous AHLs or not supplemented with AHLs. Since we noticed that the QS-1 and QS-2 systems were both activated in the logarithmic growth phase, whereas activation of the QS-3 system started in stationary phase (Fig. 1), experiments with btaI1-lux and btaI2-lux were done during the exponential phase, while those with btaI3-lux were performed during the stationary phase. Additionally, the impact of AHLs on the transcription of btaI1, btaI2, and btaI3 was also estimated by monitoring the activity of btaI1-lux, btaI2-lux, and btaI3-lux, respectively, in cultures of the ΔbtaI1, ΔbtaI2, and ΔbtaI3 mutants versus the wild-type B. thailandensis E264 strain throughout the bacterial growth phases.
While we demonstrated that btaI1 is positively controlled by BtaR1 and activated by BtaI1-produced C8-HSL (Fig. S1), expression of btaI1 was also enhanced in the presence of noncognate AHLs, namely, 3OHC10-HSL and 3OHC8-HSL (13), in the AHL-negative ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant background (Fig. 6A). Since we found that BtaR3 activates btaI1 as well (Fig. 2B), we tested the impact of 3OHC10-HSL and 3OHC8-HSL on btaI1 transcription in the absence of BtaR3 in order to verify whether activation of btaI1 by these AHLs could be dependent on BtaR3. No significant effect on btaI1 transcription was visible in cultures of the ΔbtaR3 mutant supplemented with either 3OHC10-HSL or 3OHC8-HSL (data not shown). This suggests that BtaR3 is necessary for activation of btaI1 by these AHLs. Collectively, these observations confirm that btaI1 is mainly activated by BtaR1/C8-HSL and might also be positively regulated by BtaR3 in conjunction with 3OHC10-HSL and 3OHC8-HSL.
FIG 6 .
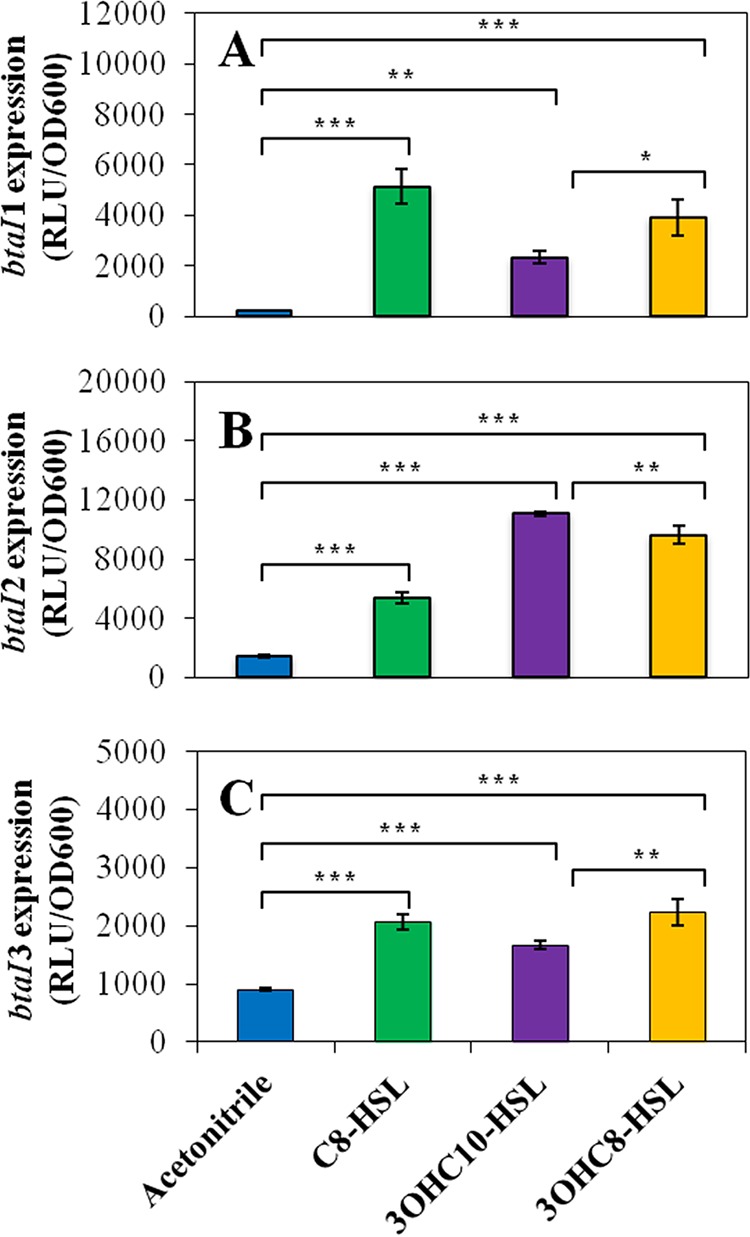
Activation of expression from the btaI1, btaI2, and btaI3 promoters by AHLs. The luciferase activity of the chromosomal (A) btaI1-lux, (B) btaI2-lux, and (C) btaI3-lux transcriptional fusions was monitored in cultures of the B. thailandensis E264 ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant strain. Cultures were supplemented with 10 µM C8-HSL, 3OHC10-HSL, or 3OHC8-HSL. Acetonitrile only was added to the controls. The error bars represent the standard deviations of the averages for three replicates. The luminescence is expressed in relative light units per optical density of the culture (RLU/OD600). ***, P < 0.001; **, P < 0.01; *, P < 0.05.
Expression of btaI2 was more strongly enhanced by 3OHC10-HSL (Fig. 6B). We also noticed a significant activation with 3OHC8-HSL (Fig. 6B). Surprisingly, activation in the presence of the noncognate C8-HSL was observed as well, revealing that expression of btaI2 is not exclusively under BtaR2 control (Fig. 6B). Additionally, we confirmed that BtaR2 directly modulates btaI2 transcription in response to 3OHC10-HSL and 3OHC8-HSL, produced by its cognate synthase BtaI2 (16), but does not function with C8-HSL (Fig. S3). Altogether, these data confirm that btaI2 is positively regulated by BtaR2 in response to both 3OHC10-HSL and 3OHC8-HSL, whereas activation by C8-HSL is independent of BtaR2.
btaI2 is directly activated by BtaR2 in response to 3OHC8-HSL or 3OHC10-HSL. The luciferase activity of the chromosomal btaI2-lux transcriptional fusion was monitored in the heterologous system, namely, E. coli DH5α also containing a BtaR2 expression vector with an arabinose-inducible promoter. Cultures were supplemented with 10 µM C8-HSL, 3OHC8-HSL, or 3OHC10-HSL. Acetonitrile only was added to the controls. The values represent the means for three replicates. The luminescence is expressed in relative light units per optical density of the culture (RLU/OD600). Download FIG S3, PDF file, 0.04 MB (42.1KB, pdf) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of btaI3 was at least doubled in cultures of the ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant when supplemented with any of the three AHLs (Fig. 6C), with 3OHC8-HSL being the most efficient AHL. Interestingly, 3OHC8-HSL had no impact in the ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant background with coaddition of C8-HSL and 3OHC10-HSL, suggesting that these AHLs might compete for btaI3 activation (Fig. S4). Similarly to 3OHC8-HSL, the expression of btaI3 was not enhanced by 3OHC10-HSL in the absence of BtaR3 (data not shown), showing that BtaR3 responds to both 3OHC8-HSL and 3OHC10-HSL to stimulate btaI3 transcription. Since all three AHLs seem able to activate expression of btaI3, we investigated whether their respective influence changes over the various growth phases. Strikingly, btaI3 was mostly activated by C8-HSL during the logarithmic growth phase, whereas activation of btaI3 by 3OHC8-HSL and 3OHC10-HSL was more prominent during the stationary phase (Fig. 7). Taken together, these results indicate that btaI3 is activated by BtaR1/C8-HSL in the exponential growth phase and is also positively regulated by BtaR3 in association with 3OHC8-HSL and 3OHC10-HSL in the stationary phase.
FIG 7 .
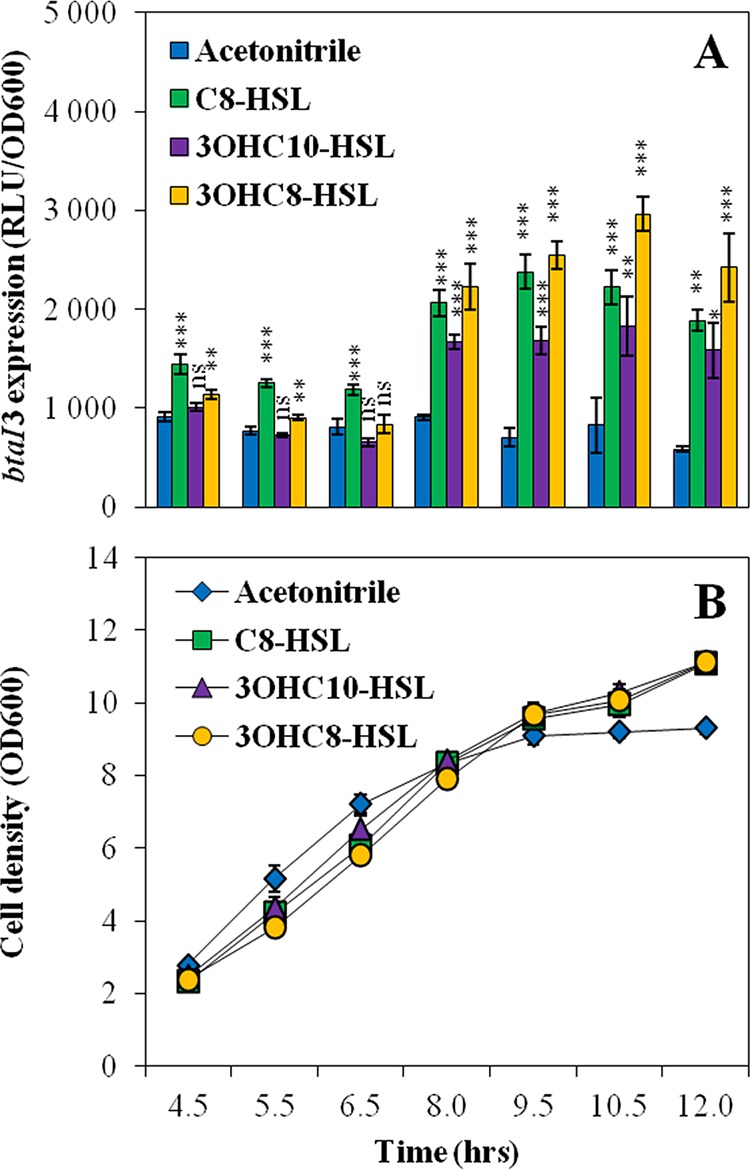
Activation of expression from the btaI3 promoter by AHLs. (A) The luciferase activity of the chromosomal btaI3-lux transcriptional fusion was monitored at (B) various times during growth in cultures of the B. thailandensis E264 ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant strain. Cultures were supplemented with 10 µM C8-HSL, 3OHC10-HSL, or 3OHC8-HSL. Acetonitrile only was added to the controls. The error bars represent the standard deviations of the averages for three replicates. The luminescence is expressed in relative light units per optical density of the culture (RLU/OD600). ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, nonsignificant.
3OHC8-HSL activation of btaI3 is dependent on C8-HSL and 3OHC10-HSL. The luciferase activity of the chromosomal btaI3-lux transcriptional fusion was measured during stationary phase in cultures of the B. thailandensis wild-type E264 strain and the ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant strain. Cultures were supplemented with 10 µM C8-HSL, 3OHC8-HSL, and 3OHC10-HSL. Acetonitrile only was added to the controls. The values represent the means for three replicates. The luminescence is expressed in relative light units per optical density of the culture (RLU/OD600). Download FIG S4, PDF file, 0.04 MB (39.7KB, pdf) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Although the QS-1, QS-2, and QS-3 systems of B. thailandensis had been previously described (13, 16, 20), a detailed picture of the interactions between the elements composing this complex QS regulatory network was missing. Since the real impact of the BtaR transcriptional regulators on the biosynthesis of their cognate AHLs and expression of adjacent btaI genes was assumed in the literature but almost never confirmed experimentally, we investigated production of AHLs in all ΔbtaR mutants and compared it with measurements of the levels of expression of btaI genes.
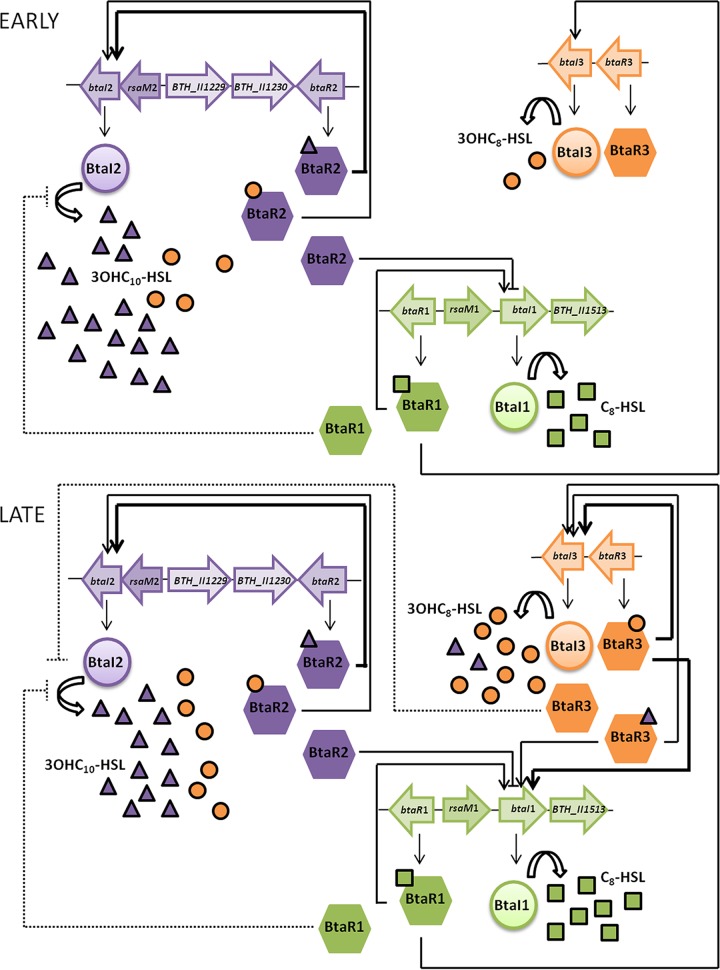
As previously described for B. pseudomallei KHW (17), we observed variations in the biosynthesis of the main AHLs as well as in the transcription of the AHL synthase-coding genes btaI1, btaI2, and btaI3 throughout the growth phases in B. thailandensis E264 (Fig. 1). These observations highlighted the timing of expression of the QS-1, QS-2, and QS-3 systems during the different stages of growth and consequently the existence of potential interactions between these QS circuits. While C8-HSL is generally considered the primary AHL produced by Burkholderia spp. (4) and is indeed predominately detected in stationary-phase cultures of B. pseudomallei K96243 and B. mallei ATCC 23344 (15, 17), we confirmed that 3OHC10-HSL is actually the most abundant AHL found in B. thailandensis E264 cultures during the different stages of growth, revealing the importance of the QS-2 system in the QS circuitry of B. thailandensis E264 (Fig. 8).
FIG 8 .
Proposed interactions between the QS-1, QS-2, and QS-3 systems.
While we confirmed that transcription of btaI2 and biosynthesis of 3OHC10-HSL are activated by BtaR2, a stronger activation by 3OHC10-HSL indicates that BtaR2 exhibits higher affinity for this AHL than for 3OHC8-HSL (Fig. 6B), which is also produced by the same synthase (16). Similarly, the bpsI2 gene that codes for the BpsI2 synthase was also shown to be substantially enhanced by 3OHC10-HSL in B. pseudomallei KHW (17). The fact remains that the levels of expression of btaI2 were similar in the wild-type E264 strain of B. thailandensis and in the non-3OHC10-HSL-producing ΔbtaI2 mutant (Fig. 3B). Considering that 3OHC8-HSL is still produced in the absence of BtaI2 (16), we must conclude that both 3OHC10-HSL and 3OHC8-HSL can induce the transcription of btaI2 (Fig. 8). Because we confirmed that BtaR2 does not function with C8-HSL (Fig. S3), an alternative LuxR-type transcriptional regulator is likely involved in its effect on btaI2 expression, highlighting an interaction between the QS-1 and QS-2 systems.
Although both BtaR1 and BtaR3 affect 3OHC10-HSL production (Fig. 3A), indicating that regulation of the biosynthesis of this AHL implies dynamic coordination between the B. thailandensis E264 QS-1, QS-2, and QS-3 circuits (Fig. 8), neither one has an effect on btaI2 expression (Fig. 3B). Nevertheless, Majerczyk et al. (20) demonstrated that btaR2 expression is stimulated by 3OHC8-HSL, and we determined that the transcription of this gene is in fact affected by the absence of all AHLs found in B. thailandensis E264 (Fig. 5B). Thus, we hypothesize that BtaR1 and BtaR3 act indirectly through btaR2 control. We also do not exclude the possibility that additional transcriptional and/or posttranscriptional regulators are involved in the modulation of the QS-2 system. Interestingly, this system contains an additional gene between btaI2 and btaR2 that is conserved in the Burkholderia genus (21). It encodes a hypothetical protein that is 37% identical to the B. cenocepacia J2315 BcRsaM (22), a homologue of the QS repressor RsaM originally identified in the plant pathogen Pseudomonas fuscovaginae (23), which we consequently renamed RsaM2 (Fig. S5). Accordingly, we observed that C8-HSL, 3OHC10-HSL, and 3OHC8-HSL concentrations were all increased in an rsaM2 mutant compared to the wild-type strain (24), indicating that RsaM2 likely intervenes in the regulation of all QS systems of B. thailandensis E264.
Genetic organization of the QS regulatory genes in B. thailandensis E264. btaI1 and btaR1 are not located next to each other and are divergently transcribed in B. thailandensis E264. The promoter region of btaI1 contains a putative lux box sequence centered 73.5 bp upstream of the btaI1 translation start site (CCCTGTAAGGGTTAACAGTT). btaI2 and btaR2 are also not located next to each other and are transcribed in the same direction on the genome of B. thailandensis E264. The promoter region of btaI2 contains a putative lux box sequence centered 65.0 bp upstream of the btaI2 translation start site (ACCTGTAGAAATCGTAGT). btaI3 and btaR3 are also transcribed in the same direction and are located next to each other in B. thailandensis E264. Download FIG S5, PDF file, 0.1 MB (125.8KB, pdf) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
As described previously for the B. pseudomallei KHW BpsI and B. mallei ATCC 23344 BmaI1 synthases (11, 15), Chandler et al. (13) demonstrated that BtaI1 is responsible for C8-HSL production. In agreement with the finding that the B. pseudomallei K96243 BpsR and B. mallei ATCC 23344 BmaR1 transcriptional regulators directly activate the BpsI- and BmaI1-encoding genes in response to C8-HSL, respectively (15, 25), Majerczyk et al. (20) reported that btaI1 transcription is positively modulated by BtaR1. We observed a strong BtaR1-dependent induction of btaI1 through C8-HSL (Fig. S1) and confirmed that the QS-1 system responds best toward its cognate AHL (Fig. 6A). While we demonstrated that BtaR1 constitutes the main regulator of btaI1 expression, we assume that BtaR1 represents the main regulator of C8-HSL biosynthesis as well. An uncoupling of AHL production and expression of the corresponding synthase was also reported in a Burkholderia RsaM-deficient strain (22, 26). BcRsaM from B. cenocepacia H111 was indeed described as an important repressor of C8-HSL biosynthesis and shown to activate the transcription of cepI and cepR encoding the LuxI-type synthase CepI and the LuxR-type transcriptional regulator CepR, respectively (22, 26). Interestingly, a gene encoding a hypothetical protein sharing 63% identity with the B. cenocepacia J2315 BcRsaM, hence called RsaM1, was also found between btaI1 and btaR1 (Fig. S5). Investigating the effect of RsaM1 on the biosynthesis of AHLs in B. thailandensis E264 showed that C8-HSL is overproduced in an rsaM1 mutant compared to the wild-type strain (24), revealing a possible link between the QS-1 system and RsaM1. Additional experiments will be necessary to fully understand the mechanisms involved in the regulation of the QS-1 system as well as the implications of the RsaM-like proteins in B. thailandensis E264.
We demonstrated that the biosynthesis of C8-HSL and transcription of btaI1 are both negatively controlled by BtaR2 (Fig. 2). Because no overexpression of the btaI1 gene was observed in the ΔbtaI2 mutant background, we assume that BtaR2 represses the QS-1 system in the absence of its ligands. This contrasts with the BtaR3-dependent regulation of btaI1 transcription in conjunction with 3OHC8-HSL, as well as with 3OHC10-HSL, albeit to a lesser extent (Fig. 8). This is also further supported by the fact that BpsR3 was reported to directly activate bpsI in response to both 3OHC8-HSL and 3OHC10-HSL, with 3OHC8-HSL eliciting the strongest response from BpsR3 (17). Considering that bmaI1 was also shown to be directly controlled by BmaR3/3OHC8-HSL (14), we suppose that BtaR3 directly activates expression of the btaI1 gene as well. However, we believe the effect of BtaR3 on the QS-1 system is more complex. While the bpsR gene encoding BpsR was reported to be positively autoregulated (11), we determined that btaR1 expression is repressed by QS (Fig. 5A). Thus, negative regulation of C8-HSL biosynthesis by BtaR3 could be linked to btaR1 modulation. Altogether, these observations further highlight the existence of interactions between the QS-1, QS-2, and QS-3 circuits and reveal that the timing of expression of the QS-1 system is dependent on both the QS-2 and QS-3 systems (Fig. 8). This might contribute to the successive activation of the B. thailandensis E264 QS circuits observed throughout bacterial growth.
Similarly to the B. pseudomallei KWH BpsI3 and B. mallei ATCC 23344 BmaI3 synthases, BtaI3 was shown to produce 3OHC8-HSL (13, 14, 17). While the B. pseudomallei KHW BpsR3 and B. mallei ATCC 23344 BmaR3 transcriptional regulators specifically respond to 3OHC8-HSL, the bpsI3 and bmaI3 genes were not reported to be activated by BpsR3 and BmaR3, respectively, in conjunction with 3OHC8-HSL (14, 17). Here, in B. thailandensis E264, we demonstrated that the transcription of btaI3 is positively controlled by BtaR3 and activated by 3OHC8-HSL (Fig. S2). However, 3OHC8-HSL-dependent activation of btaI3 seems to be conditioned by the presence of other AHLs (Fig. S4). The interaction between BtaR3 and 3OHC8-HSL, necessary to activate btaI3 expression, could be impeded by a competitive inhibition exerted by another AHL, as already proposed for B. pseudomallei KHW (17). In addition, we observed that btaI3 expression is activated by 3OHC10-HSL, albeit to a lesser extent (Fig. 6C). Indeed, the BtaR3-controlled genes identified in transcriptomic analyses were also generally affected by both 3OHC8-HSL and 3OHC10-HSL (20). This further supports the idea that BtaR3 functions with these two AHLs (Fig. 8). Considering that BpsI3 and BmaI3 were both shown to produce 3OHC10-HSL in addition to 3OHC8-HSL (14, 17), it is possible that BtaI3 intervenes in the biosynthesis of 3OHC10-HSL in B. thailandensis E264 as well.
Remarkably, positive 3OHC8-HSL- and 3OHC10-HSL-dependent regulation of btaI3 occurred in the stationary growth phase (Fig. 7), in agreement with the expression profile of this gene. Conversely, activation of btaI2 transcription by these AHLs was mainly observed during logarithmic growth. We thus hypothesize that the QS-3 system regulates the QS-2 system targets by producing 3OHC8-HSL in stationary phase, whereas production of this AHL by the QS-2 system occurs essentially during the exponential phase, implying a coordination between the QS-2 and QS-3 systems (Fig. 8). Additionally, it seems that 3OHC8-HSL is produced by BtaI2 at the expense of 3OHC10-HSL. This would explain why there is an overlap between these QS circuits when it comes to genes modulated by 3OHC8-HSL and 3OHC10-HSL (20). Importantly, while sharing common AHLs, the QS-2 and QS-3 systems are apparently not transcriptionally linked.
The BtaR1/C8-HSL-dependent control of btaI3 transcription, which starts in the exponential growth phase, is consistent with the idea that the QS-1 system is required for the expression of btaI3 (20), and might also account for the belated activation of the QS-3 circuit in comparison with the QS-1 and QS-2 systems. This again illustrates the successive expression of these QS circuits and points toward an interdependence between the QS-1 and QS-3 systems (Fig. 8). Such an interconnection has already been observed among the members of the Bptm group, as bpsI3 transcription was reported to be stimulated by the BpsI/BpsR QS system (17). Nevertheless, the precise regulatory mechanism directing the QS-3 system through BtaR1 is currently unknown. While BtaR1 seems to act by activating btaI3 transcription, we propose that the negative impact of BtaR1 on 3OHC8-HSL biosynthesis does not result from a direct interaction with the btaI3 promoter but rather could imply the effect of BtaR1 on the level of btaR3 as previously suggested (20). Additional transcriptional and/or posttranscriptional regulators might also be involved in the BtaR1-dependent modulation of the QS-3 system.
Conclusion.
The study described here provides for the first time an exhaustive portrait of the interplay between the QS-1, QS-2, and QS-3 systems in B. thailandensis E264 (Fig. 8). We observed an interdependence between the QS-1 and QS-2 systems. While we confirmed that the QS-3 system is controlled by BtaR1, we also found that BtaR3 modulates the QS-1 system, which indicates that those two systems are linked. Interestingly, such an interaction between the QS-1 and QS-3 systems seems to be conserved in the closely related species of the Bptm group (14, 17, 20). Interestingly, the QS-2 and QS-3 systems that share common AHLs seem not to be transcriptionally linked, but instead they are temporally connected by their common AHLs. We also highlighted a surprising uncoupling of AHL production and expression of the corresponding synthase in the QS-1 system, which hints that QS regulation does not always follow a classic pattern. Collectively, the results of our study suggest that there are homeostatic regulatory loops provided by the various QS systems in B. thailandensis resulting from transcriptional and posttranscriptional interactions, allowing tightly controlled coordination of the expression of genes.
Although we have found new connections and insights on the QS cascade, there are still many questions to be answered. Indeed, further work is needed to comprehend more about the mechanisms behind those links and regulation as well as the implications of recently characterized RsaM-like proteins. The temporal pattern of QS-controlled genes clearly shows that additional factors are involved (17, 20, 27).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table S1 in the supplemental material. Unless stated otherwise, all bacteria were cultured at 37°C in tryptic soy broth (TSB) (BD Difco, Mississauga, Ontario, Canada), with shaking (240 rpm) in a TC-7 roller drum (New Brunswick, Canada), or on petri dishes containing TSB solidified with 1.5% agar. When required, antibiotics were used at the following concentrations: 15 µg/ml tetracycline (Tc) and 25 µg/ml gentamicin (Gm) for Escherichia coli DH5α, while Tc was used at 200 µg/ml for Burkholderia thailandensis E264. All measurements of optical density (optical density at 600 nm [OD600]) were acquired with a Thermo Fisher Scientific NanoDrop ND-1000 spectrophotometer.
Bacterial strains used in this study. Download TABLE S1, DOCX file, 0.02 MB (22.7KB, docx) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of plasmids.
All plasmids used in this study are described in Table S2. Amplification of the promoter regions of btaI1, btaI2, and btaI3 was performed from genomic DNA from B. thailandensis E264 using the appropriate primers (Table S3). The amplified products were digested with the FastDigest restriction enzymes XhoI and BamHI (Thermo Fisher Scientific) and inserted by T4 DNA ligase (Bio Basic, Inc., Markham, ON, Canada) within the corresponding restriction sites in the mini-CTX-lux plasmid (28), generating the transcriptional reporters pSLG02, pSLG03, and pSLG04, respectively. All primers were from Alpha DNA (Montreal, Quebec, Canada).
Plasmids used in this study. Download TABLE S2, DOCX file, 0.02 MB (20KB, docx) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used for PCR. Download TABLE S3, DOCX file, 0.01 MB (11.8KB, docx) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of reporter strains.
The mini-CTX-btaI1-lux, mini-CTX-btaI2-lux, and mini-CTX-btaI3-lux transcriptional reporters were integrated into the chromosomes of B. thailandensis E264 strains through conjugation with E. coli χ7213 followed by selection with Tc. Successful chromosomal insertion of the btaI1-lux, btaI2-lux, and btaI3-lux plasmids was confirmed by PCR using the appropriate primers.
LC-MS/MS quantification of AHLs.
The concentrations of AHLs were determined from samples of B. thailandensis E264 cultures obtained at different time points during bacterial growth, by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). The samples were prepared and analyzed as described previously (29). 5,6,7,8-Tetradeutero-4-hydroxy-2-heptylquinoline (HHQ-d4) was used as an internal standard. All experiments were performed in triplicate and conducted at least twice independently.
Measurement of the activity of btaI1-lux, btaI2-lux, and btaI3-lux reporters.
The levels of expression from the promoter regions of btaI1, btaI2, or btaI3 were quantified by measuring the luminescence of B. thailandensis E264 cultures carrying the corresponding chromosomal reporters. Overnight bacterial cultures were diluted in TSB to an initial OD600 of 0.1 and incubated as described above. The luminescence was regularly determined from culture samples using a multimode microplate reader (Cytation 3; Bio-Tek Instruments, Inc., Winooski, VT, USA) and expressed in relative light units per optical density of the culture (RLU/OD600). For experiments with AHL additions, cultures were supplemented with 10 µM C8-HSL, 3OHC8-HSL, and 3OHC10-HSL (Sigma-Aldrich Co., Oakville, ON, Canada) or not supplemented with AHLs from stocks prepared in HPLC-grade acetonitrile. Acetonitrile only was added to the controls. All experiments were performed with three biological replicates and repeated at least twice.
Heterologous E. coli expression system for BtaR2 regulation of btaI2 expression.
The response of the btaI2 promoter to the BtaR2 transcriptional regulator was determined using a recombinant E. coli DH5α strain containing both the chromosomal btaI2-lux transcriptional fusion and the arabinose-inducible expression vector pJN105-btaR2. Overnight bacterial cultures of E. coli DH5α were diluted in lysogeny broth (LB) (Alpha Biosciences, Inc., Baltimore, MD) with the appropriate antibiotics and grown in triplicate at 37°C, with shaking in a TC-7 roller drum. When the cultures reached an OD600 of 0.5, they were supplemented with 10 μM C8-HSL, 3OHC8-HSL, or 3OHC10-HSL. Acetonitrile only was added to the controls. The BtaR2 expression vector was induced with 0.2% l-arabinose (wt/vol). The btaI2-lux luciferase activity was measured every 30 min during 10 h as described above. All experiments were repeated at least three times.
Quantitative reverse transcription-PCR experiments.
Total RNA from B. thailandensis E264 cultures at an OD600 of 4.0 was extracted with the PureZOL RNA isolation reagent (Bio-Rad Laboratories, Mississauga, ON, Canada) and treated twice with the TURBO DNA-Free kit (Ambion Life Technologies, Inc., Burlington, ON, Canada) according to the manufacturer’s instructions. Extractions were done on three different bacterial cultures. Quality and purity controls were confirmed by agarose gel electrophoresis and UV spectrophotometric analysis, respectively. cDNA synthesis was performed using the iScript reverse transcription supermix (Bio-Rad Laboratories), and amplification was accomplished on a Corbett Life Science Rotor-Gene 6000 thermal cycler using the SsoAdvanced universal SYBR green supermix (Bio-Rad Laboratories), according to the manufacturer’s protocol. The reference gene was ndh (30). The ndh gene displayed stable expression under the different genetic contexts tested. All primers used for cDNA amplification are presented in Table S4. Differences in gene expression between Burkholderia thailandensis E264 strains were calculated using the 2−ΔΔCT formula (31). A threshold of 0.5 was chosen as significant. All experiments were performed in triplicate and conducted at least twice independently.
Primers used for qRT-PCR. Download TABLE S4, DOCX file, 0.01 MB (11.7KB, docx) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data analysis.
Unless stated otherwise, data are reported as means ± standard deviations (SD). Statistical analyses were performed with the R software version 3.3.3 (http://www.R-project.org.) using one-way analysis of variance (ANOVA). Probability values of less than 0.05 were considered significant.
ACKNOWLEDGMENTS
We thank Everett Peter Greenberg (Department of Microbiology, University of Washington School of Medicine, Seattle, WA) for providing the B. thailandensis E264 strains. Special thanks to Sylvain Milot for his technical help.
This study was supported by Canadian Institutes of Health Research (CIHR) operating grants MOP-97888 and MOP-142466 to Eric Déziel. Eric Déziel holds the Canada Research Chair in Sociomicrobiology.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Le Guillouzer S, Groleau M-C, Déziel E. 2017. The complex quorum sensing circuitry of Burkholderia thailandensis is both hierarchically and homeostatically organized. mBio 8:e01861-17. https://doi.org/10.1128/mBio.01861-17.
REFERENCES
- 1.Fuqua WC, Winans SC, Greenberg EP. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nealson KH, Platt T, Hastings JW. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol 104:313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuqua C, Greenberg EP. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol 3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 4.Eberl L. 2006. Quorum sensing in the genus Burkholderia. Int J Med Microbiol 296:103–110. doi: 10.1016/j.ijmm.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 5.Lewenza S, Sokol PA. 2001. Regulation of ornibactin biosynthesis and N-acyl-l-homoserine lactone production by CepR in Burkholderia cepacia. J Bacteriol 183:2212–2218. doi: 10.1128/JB.183.7.2212-2218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malott RJ, Sokol PA. 2007. Expression of the bviIR and cepIR quorum-sensing systems of Burkholderia vietnamiensis. J Bacteriol 189:3006–3016. doi: 10.1128/JB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulrich RL, Deshazer D, Brueggemann EE, Hines HB, Oyston PC, Jeddeloh JA. 2004. Role of quorum sensing in the pathogenicity of Burkholderia pseudomallei. J Med Microbiol 53:1053–1064. doi: 10.1099/jmm.0.45661-0. [DOI] [PubMed] [Google Scholar]
- 8.Ulrich RL, Deshazer D, Hines HB, Jeddeloh JA. 2004. Quorum sensing: a transcriptional regulatory system involved in the pathogenicity of Burkholderia mallei. Infect Immun 72:6589–6596. doi: 10.1128/IAI.72.11.6589-6596.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulrich RL, Hines HB, Parthasarathy N, Jeddeloh JA. 2004. Mutational analysis and biochemical characterization of the Burkholderia thailandensis DW503 quorum-sensing network. J Bacteriol 186:4350–4360. doi: 10.1128/JB.186.13.4350-4360.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valade E, Thibault FM, Gauthier YP, Palencia M, Popoff MY, Vidal DR. 2004. The PmlI-PmlR quorum-sensing system in Burkholderia pseudomallei plays a key role in virulence and modulates production of the MprA protease. J Bacteriol 186:2288–2294. doi: 10.1128/JB.186.8.2288-2294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song Y, Xie C, Ong YM, Gan YH, Chua KL. 2005. The BpsIR quorum-sensing system of Burkholderia pseudomallei. J Bacteriol 187:785–790. doi: 10.1128/JB.187.2.785-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brett PJ, DeShazer D, Woods DE. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int J Syst Bacteriol 48:317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- 13.Chandler JR, Duerkop BA, Hinz A, West TE, Herman JP, Churchill ME, Skerrett SJ, Greenberg EP. 2009. Mutational analysis of Burkholderia thailandensis quorum sensing and self-aggregation. J Bacteriol 191:5901–5909. doi: 10.1128/JB.00591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duerkop BA, Herman JP, Ulrich RL, Churchill ME, Greenberg EP. 2008. The Burkholderia mallei BmaR3-BmaI3 quorum-sensing system produces and responds to N-3-hydroxy-octanoyl homoserine lactone. J Bacteriol 190:5137–5141. doi: 10.1128/JB.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duerkop BA, Ulrich RL, Greenberg EP. 2007. Octanoyl-homoserine lactone is the cognate signal for Burkholderia mallei BmaR1-BmaI1 quorum sensing. J Bacteriol 189:5034–5040. doi: 10.1128/JB.00317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duerkop BA, Varga J, Chandler JR, Peterson SB, Herman JP, Churchill ME, Parsek MR, Nierman WC, Greenberg EP. 2009. Quorum-sensing control of antibiotic synthesis in Burkholderia thailandensis. J Bacteriol 191:3909–3918. doi: 10.1128/JB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamage AM, Shui G, Wenk MR, Chua KL. 2011. N-octanoylhomoserine lactone signalling mediated by the BpsI-BpsR quorum sensing system plays a major role in biofilm formation of Burkholderia pseudomallei. Microbiology 157:1176–1186. doi: 10.1099/mic.0.046540-0. [DOI] [PubMed] [Google Scholar]
- 18.Ong C, Ooi CH, Wang D, Chong H, Ng KC, Rodrigues F, Lee MA, Tan P. 2004. Patterns of large-scale genomic variation in virulent and avirulent Burkholderia species. Genome Res 14:2295–2307. doi: 10.1101/gr.1608904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Truong TT, Seyedsayamdost M, Greenberg EP, Chandler JR. 2015. A Burkholderia thailandensis acyl-homoserine lactone-independent orphan LuxR homolog that activates production of the cytotoxin malleilactone. J Bacteriol 197:3456–3462. doi: 10.1128/JB.00425-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majerczyk C, Brittnacher M, Jacobs M, Armour CD, Radey M, Schneider E, Phattarasokul S, Bunt R, Greenberg EP. 2014. Global analysis of the Burkholderia thailandensis quorum sensing-controlled regulon. J Bacteriol 196:1412–1424. doi: 10.1128/JB.01405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choudhary KS, Hudaiberdiev S, Gelencsér Z, Gonçalves Coutinho B, Venturi V, Pongor S. 2013. The organization of the quorum sensing luxI/R family genes in Burkholderia. Int J Mol Sci 14:13727–13747. doi: 10.3390/ijms140713727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michalska K, Chhor G, Clancy S, Jedrzejczak R, Babnigg G, Winans SC, Joachimiak A. 2014. RsaM: a transcriptional regulator of Burkholderia spp. with novel fold. FEBS J 281:4293–4306. doi: 10.1111/febs.12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattiuzzo M, Bertani I, Ferluga S, Cabrio L, Bigirimana J, Guarnaccia C, Pongor S, Maraite H, Venturi V. 2011. The plant pathogen Pseudomonas fuscovaginae contains two conserved quorum sensing systems involved in virulence and negatively regulated by RsaL and the novel regulator RsaM. Environ Microbiol 13:145–162. doi: 10.1111/j.1462-2920.2010.02316.x. [DOI] [PubMed] [Google Scholar]
- 24.Le Guillouzer S, Groleau M-C, Déziel E. 2017. Two rsaM homologues encode central regulatory elements modulating quorum sensing expression in Burkholderia thailandensis. bioRxiv doi: 10.1101/192625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiratisin P, Sanmee S. 2008. Roles and interactions of Burkholderia pseudomallei BpsIR quorum-sensing system determinants. J Bacteriol 190:7291–7297. doi: 10.1128/JB.00739-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inhülsen S. 2011. Investigations on the quorum sensing circuitry in Burkholderia cenocepacia H111. PhD dissertation University of Zurich, Zurich, Switzerland. [Google Scholar]
- 27.Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becher A, Schweizer HP. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29:948–950, 952. [DOI] [PubMed] [Google Scholar]
- 29.Chapalain A, Groleau MC, Le Guillouzer S, Miomandre A, Vial L, Milot S, Déziel E. 2017. Interplay between 4-hydroxy-3-methyl-2-alkylquinoline and N-acyl-homoserine lactone signaling in a Burkholderia cepacia complex clinical strain. Front Microbiol 8:1021. doi: 10.3389/fmicb.2017.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subsin B, Chambers CE, Visser MB, Sokol PA. 2007. Identification of genes regulated by the cepIR quorum-sensing system in Burkholderia cenocepacia by high-throughput screening of a random promoter library. J Bacteriol 189:968–979. doi: 10.1128/JB.01201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
btaI1 activation requires BtaR1 and C8-HSL. The luciferase activity of the chromosomal btaI1-lux transcriptional fusion was monitored during the exponential growth phase in cultures of the B. thailandensis E264 wild-type strain and the ΔbtaR1, ΔbtaI1, and ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant strains. Cultures were supplemented with 10 µM C8-HSL. Acetonitrile only was added to the controls. The values represent the means for three replicates. The luminescence is expressed in relative light units per optical density of the culture (RLU/OD600). Download FIG S1, PDF file, 0.04 MB (43KB, pdf) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
btaI3 is activated by BtaR3 and 3OHC8-HSL. The luciferase activity of the chromosomal btaI3-lux transcriptional fusion was measured during stationary phase in cultures of the B. thailandensis wild-type E264 strain and ΔbtaR3, ΔbtaI3, and ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant strains. Cultures were supplemented with 10 µM 3OHC8-HSL. Acetonitrile only was added to the controls. The values represent the means for three replicates. The luminescence is expressed in relative light units per optical density of the culture (RLU/OD600). Download FIG S2, PDF file, 0.04 MB (43KB, pdf) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
btaI2 is directly activated by BtaR2 in response to 3OHC8-HSL or 3OHC10-HSL. The luciferase activity of the chromosomal btaI2-lux transcriptional fusion was monitored in the heterologous system, namely, E. coli DH5α also containing a BtaR2 expression vector with an arabinose-inducible promoter. Cultures were supplemented with 10 µM C8-HSL, 3OHC8-HSL, or 3OHC10-HSL. Acetonitrile only was added to the controls. The values represent the means for three replicates. The luminescence is expressed in relative light units per optical density of the culture (RLU/OD600). Download FIG S3, PDF file, 0.04 MB (42.1KB, pdf) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
3OHC8-HSL activation of btaI3 is dependent on C8-HSL and 3OHC10-HSL. The luciferase activity of the chromosomal btaI3-lux transcriptional fusion was measured during stationary phase in cultures of the B. thailandensis wild-type E264 strain and the ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant strain. Cultures were supplemented with 10 µM C8-HSL, 3OHC8-HSL, and 3OHC10-HSL. Acetonitrile only was added to the controls. The values represent the means for three replicates. The luminescence is expressed in relative light units per optical density of the culture (RLU/OD600). Download FIG S4, PDF file, 0.04 MB (39.7KB, pdf) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genetic organization of the QS regulatory genes in B. thailandensis E264. btaI1 and btaR1 are not located next to each other and are divergently transcribed in B. thailandensis E264. The promoter region of btaI1 contains a putative lux box sequence centered 73.5 bp upstream of the btaI1 translation start site (CCCTGTAAGGGTTAACAGTT). btaI2 and btaR2 are also not located next to each other and are transcribed in the same direction on the genome of B. thailandensis E264. The promoter region of btaI2 contains a putative lux box sequence centered 65.0 bp upstream of the btaI2 translation start site (ACCTGTAGAAATCGTAGT). btaI3 and btaR3 are also transcribed in the same direction and are located next to each other in B. thailandensis E264. Download FIG S5, PDF file, 0.1 MB (125.8KB, pdf) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial strains used in this study. Download TABLE S1, DOCX file, 0.02 MB (22.7KB, docx) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Download TABLE S2, DOCX file, 0.02 MB (20KB, docx) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used for PCR. Download TABLE S3, DOCX file, 0.01 MB (11.8KB, docx) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used for qRT-PCR. Download TABLE S4, DOCX file, 0.01 MB (11.7KB, docx) .
Copyright © 2017 Le Guillouzer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.