Extreme suppression of lateral floret development in deficiens barley is the result of a single amino acid substitution in the homeodomain-leucine zipper class I transcription factor VRS1.
Abstract
Increasing grain yield is an endless challenge for cereal crop breeding. In barley (Hordeum vulgare), grain number is controlled mainly by Six-rowed spike 1 (Vrs1), which encodes a homeodomain leucine zipper class I transcription factor. However, little is known about the genetic basis of grain size. Here, we show that extreme suppression of lateral florets contributes to enlarged grains in deficiens barley. Through a combination of fine-mapping and resequencing of deficiens mutants, we have identified that a single amino acid substitution at a putative phosphorylation site in VRS1 is responsible for the deficiens phenotype. deficiens mutant alleles confer an increase in grain size, a reduction in plant height, and a significant increase in thousand grain weight in contemporary cultivated germplasm. Haplotype analysis revealed that barley carrying the deficiens allele (Vrs1.t1) originated from two-rowed types carrying the Vrs1.b2 allele, predominantly found in germplasm from northern Africa. In situ hybridization of histone H4, a marker for cell cycle or proliferation, showed weaker expression in the lateral spikelets compared with central spikelets in deficiens. Transcriptome analysis revealed that a number of histone superfamily genes were up-regulated in the deficiens mutant, suggesting that enhanced cell proliferation in the central spikelet may contribute to larger grains. Our data suggest that grain yield can be improved by suppressing the development of specific organs that are not positively involved in sink/source relationships.
The domestication of cereal crops such as rice (Oryza sativa), maize (Zea mays), and barley (Hordeum vulgare) began about 10,000 years ago (Doebley et al., 2006). Throughout domestication, plant forms have changed to produce larger edible parts, reduced seed dormancy, and loss of shattering (Doebley et al., 2006). Together with these changes, some unnecessary organs such as long barbed awns in rice (Hua et al., 2015) and high tillering in maize (Studer et al., 2011) have been lost, resulting in higher grain yields. Grain yield is affected by inflorescence architecture in cereal plants such as rice, maize, barley, and wheat (Triticum aestivum; Komatsuda et al., 2007; Shomura et al., 2008; Wang et al., 2012; Bommert et al., 2013; Koppolu et al., 2013; Poursarebani et al., 2015). In rice, a model grass plant, several major quantitative trait loci related to grain size, such as GW2, qSW5, GS5, OsSPL16, and An-1, have been identified (Song et al., 2007; Shomura et al., 2008; Li et al., 2011; Wang et al., 2012; Luo et al., 2013). However, little is known about the genetic basis of grain size in Triticeae species, including economically important crops such as wheat, barley, and rye (Secale cereale), except for TaGW2-A1, which is a rice GW2 ortholog in wheat (Simmonds et al., 2016), and HvDEP1 in barley, an ortholog of rice Dense and Erect Panicle 1 (Wendt et al., 2016).
As a model organism for plant development in the Triticeae, barley has been useful due to its simple diploid genome and the availability of a large number of different morphological mutants (Druka et al., 2011). In the genus Hordeum, including cultivated barley, the inflorescence is called a spike and is composed of three unifloreted spikelets at each rachis node (Bothmer et al., 1995). When all three spikelets are fertile and produce grains, the spike is called six-rowed, whereas only central spikelets are fertile in a two-rowed spike. Barley row type is one of the most important traits affecting yield, especially for grain number; therefore, advancing its molecular understanding is key to yield improvement. Row type is controlled by at least five loci: Six-rowed spike 1 (Vrs1 [syn = HvHox1]), Vrs2, Vrs3, Vrs4, and Intermedium spike-c (Int-c [syn = Vrs5]), and all five genes have been identified (Komatsuda et al., 2007; Ramsay et al., 2011; Koppolu et al., 2013; Bull et al., 2017; van Esse et al., 2017; Youssef et al., 2017). In four of these (Vrs1, Vrs2, Vrs4, and Vrs5), wild-type alleles encode transcription factors: homeodomain-leucine zipper class I (HD-Zip I), SHORT INTERNODES, RAMOSA2, and TEOSINTE BRANCHED1, respectively; in Vrs3, the wild-type allele putatively encodes a histone H3K9 demethylase. Loss of function at any of these five genes results in varying levels of fertile lateral spikelet development. The selection of naturally occurring six-rowed alleles of Vrs1 and Int-c during the domestication of barley positively affected production (Komatsuda et al., 2007; Ramsay et al., 2011).
HD-Zip I genes like Vrs1 have evolved through a series of gene duplications and divergence through subfunctionalization or neofunctionalization (Ariel et al., 2007; Sakuma et al., 2013). HvHox2, which is a paralog of Vrs1 (HvHox1), is highly conserved among cereals and expressed broadly (Sakuma et al., 2010). So far, a loss-of-function mutant of HvHox2 has not been reported; thus, the gene has been considered as essential for plant development. In contrast, Vrs1 plays more specific roles in the control of lateral spikelet fertility. Vrs1 is expressed in the lateral florets, especially in the pistil. At least 58 loss-of-function mutants have been described; therefore, it seems to be a driving force of genetic variation in barley spike development (Komatsuda et al., 2007; Sakuma et al., 2013).
Deficiens barley plants have a two-rowed spike with extremely rudimentary lateral spikelets/florets compared with canonical two-rowed cultivars (Fig. 1A). The deficiens phenotype is a naturally occurring variant and potentially an allele of Vrs1 (Habgood and Chambi, 1984). Deficiens barley is endemic in Ethiopia, where the unique spike of labile, which displays a variable number of fertile spikelets at each rachis node, also is localized (Woodward, 1949; Youssef et al., 2014). Old studies report that deficiens barley affects the grain size of central spikelets (Powers, 1936; Habgood and Chambi, 1984). Currently, deficiens types dominate U.K. winter barley grain production, and hence the area grown, and are increasing in spring barley (Supplemental Fig. S1). Although breeders recognize the agricultural importance of a perceived yield advantage, the molecular genetic basis of deficiens is not understood. This study sought to identify the deficiens locus through positional cloning and to elucidate the molecular mechanism underlying the extreme suppression of the floral organs of lateral spikelets in barley.
Figure 1.
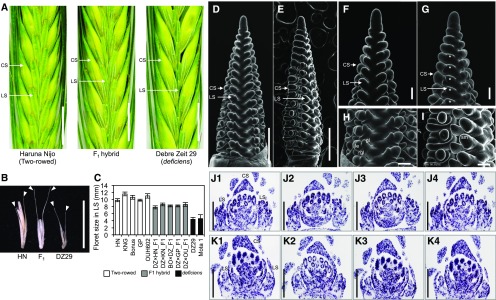
The inflorescence of deficiens barley. A, Morphology of the inflorescence (from left to right, canonical two-rowed spike of cv Haruna Nijo [HN]; F1 plant between Debre Zeit 29 [DZ29] and HN; and deficiens landrace DZ29). Central spikelets of DZ29 are larger than those of HN, and lateral spikelets of DZ29 are smaller than those of HN. B, Dissected lateral spikelets. Arrowheads indicate glumes. C, Variation of floret size in the lateral spikelet. GP, Golden Promise; KNG, Kanto Nakate Gold. D to I, Scanning electron microscopy images of two-rowed cultivar Bonus (D, F, and H) and DZ29 (E, G, and I) at the awn primordium stage. Asterisks indicate smaller lateral spikelet meristems in DZ29 compared with Bonus. J and K, In situ mRNA hybridization of histone H4 shows weaker expression in the lateral spikelet of DZ29 (K) than that of Bonus (J) but stronger expression in the central spikelet (all the organs) of DZ29 than that of Bonus. Four serial sections are shown for each cultivar. CS, Central spikelet; gl, glume; le, lemma; LS, lateral spikelet; sm, spikelet meristem. Bars = 1 cm in A and B, 500 µm in D, E, J, and K, and 100 µm in F to I.
RESULTS
Phenotype of deficiens Barley
Deficiens barley (DZ29) shows extremely rudimentary lateral floral organs, although glume development in its lateral spikelets was comparable to that of canonical two-rowed barley (Fig. 1, A and B). To better understand the deficiens phenotype, the size of florets in the lateral spikelets was measured at the anthesis stage in field conditions. Deficiens barley produced 4- to 5-mm-long florets in its lateral spikelets, significantly less than the 10- to 12-mm lengths of canonical two-rowed varieties. F1 plants from five different parental crosses showed an intermediate phenotype (7–9 mm) compared with parental lines (Fig. 1C), showing a low degree of dominance (0.26, 0.17, 0.23, 0.41, and 0.27 for DZ × HN, DZ × KN, BO × DZ, DZ × GP, and DZ × OU, respectively). To investigate the effect of deficiens on the development of lateral spikelets (especially florets) in more detail, scanning electron microscopy images were compared (Fig. 1, D and E). Barley spike development progresses from the basal to the apical part gradually (Kirby and Appleyard, 1981; Youssef et al., 2017). At the apical part of the spike, when triple spikelet primordia had differentiated, DZ29 showed a smaller lateral spikelet meristem compared with the canonical two-rowed type (Fig. 1, F and G). In the canonical type, the lateral spikelet meristems have differentiated into glume and lemma primordia at the basal part, whereas those of DZ29 remained as undifferentiated spikelet meristems (Fig. 1, H and I) with suppressed floral differentiation (Fig. 1, D–I).
To explore the cellular activity of the spikelet meristems, RNA in situ hybridization was conducted using histone H4 antisense probe as a marker for active cell division. The results showed that the transcripts of histone H4 were detectable in both DZ29 and canonical two-rowed barley (Fig. 1, J and K; Supplemental Fig. S2). The related abundance of the transcript in the lateral spikelets compared with that in the central spikelets is less in DZ29 (Fig. 1K) compared with canonical two-rowed barley (Fig. 1J), although the in situ hybridization is not quantitative (Supplemental Fig. S2). Interestingly, stronger signals were observed in the central spikelet of DZ29, suggesting enhanced cell proliferation. These phenotypic data suggest that deficiens causes an enduring suppression of lateral spikelet primordia from an early stage of spike development onward.
Map-Based Identification of deficiens
To map the deficiens gene, five filial generation 2 (F2) segregating populations were produced, and the genotype of each F2 individual was inferred by F3 progeny analysis to show that the genotype segregated as a monogenic factor (Supplemental Fig. S3; Supplemental Table S1). Depending upon the cross, the gene was localized to within a 1- to 2.6-centimorgan (cM) interval on chromosome 2H (Supplemental Fig. S4). No recombinants with the HvHox1_DraIII marker, defined by a single-nucleotide polymorphism (SNP) located on the promoter region of HvHox1 (–359 bp from the transcription start site), were observed. The marker order in the region was conserved across all five mapping populations, indicating no chromosomal rearrangements (Supplemental Fig. S4).
Larger scale F2 populations of 2,096 and 2,264 individuals for DZ29 × OUH602 and DZ29 × KNG, respectively, refined the deficiens locus to a 0.02-cM region flanked by the markers e40m36-1110S_AccIII and BC12348_HhaI (Fig. 2A; Supplemental Table S2). The interval is composed of 458,564 bp in Morex BAC sequences (accession no. EF067844; Komatsuda et al., 2007), and only one predicted gene, Vrs1 (HvHox1), encoding an HD-Zip I transcription factor, was found in the interval. A sequence comparison of the Vrs1 locus (1,580 bp upstream, 1,538-bp coding region, and 1,183 bp downstream) among mapping parents revealed a base pair change that encoded a single amino acid substitution (S184G) at the Ser-rich motif of the C-terminal region (CTR) specific to DZ29 (renamed the Vrs1.t1 allele; Fig. 2B; Supplemental Fig. S5). Resequencing Vrs1 in 48 northwest (NW) European winter barley cultivars (24 canonical two-rowed and 24 deficiens cultivars) revealed that all 24 accessions of the deficiens type shared the Vrs1.t1 allele (Supplemental Table S3). Furthermore, according to the NCBI database, all sequenced accessions of deficiens barley have an identical amino acid sequence to Vrs1.t1 allele (Supplemental Table S3). This suggests that the amino acid change in this region may be responsible for the deficiens phenotype.
Figure 2.
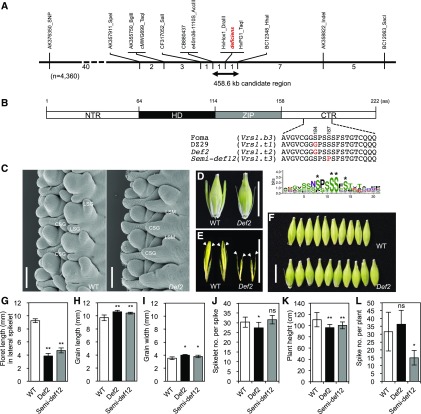
Fine-mapping of deficiens and analysis of induced mutants. A, Genetic map of the deficiens region. The numbers beneath the line indicate the number of recombinants recovered. One recombination corresponds to a genetic distance of 0.01 cM. B, Protein sequence of VRS1. Independent single amino acid (aa) substitutions were identified in DZ29 and induced mutants (S184G and S187P) at the CTR. The Ser-rich motif is highly conserved among plant HD-Zip I proteins (n = 87). Asterisks indicate putative phosphorylation sites. NTR, N-terminal region; HD, homeodomain; ZIP, Leu zipper. C, Scanning electron microscopy images of cv Foma (wild type [WT]) and the Def2 mutant at the awn primordium stage. In Def2, the lateral spikelet meristem is highly suppressed. CSG, Central spikelet glume; LSG, lateral spikelet glume; LSM, lateral spikelet meristem. D, Triple spikelet morphology. E, Dissected lateral spikelets. Arrowheads indicate glumes. F, The Def2 mutant shows bigger grain size. G to L, Phenotypic comparison between cv Foma (WT) and deficiens mutants. G, Floret length in the lateral spikelet. H, Grain length. I, Grain width. J, Central spikelet number per spike. K, Plant height. L, Spike number per plant. Data are given as means ± sd (n = 20). Asterisks indicate that means are significantly different at the 1% (**) and 5% (*) probability levels; ns, not significantly different. Bars = 100 µm in C and 1 cm in D to F.
To test this hypothesis, seven Deficiens mutants and 35 Semi-deficiens mutants derived from canonical two-rowed cultivars (Foma, Bonus, Kristina, and Lina) were phenotyped and resequenced (Supplemental Table S4). Two of the 42 induced mutants, Deficiens 2 (Def2; named the Vrs1.t2 allele) and Semi-deficiens 12 (Semi-def12; named the Vrs1.t3 allele), derived from cv Foma, showed a clear deficiens spike phenotype with rudimentary lateral spikelets (Fig. 2, C–E and G; Supplemental Fig. S6). These two mutants had independent single amino acid substitutions, S184G and S187P, respectively, at the CTR (Fig. 2B; Supplemental Table S4), suggesting that a single amino acid substitution of Ser in the Ser-rich motif was responsible for the deficiens phenotype (Supplemental Fig. S7). The remaining 40 lines did not show a deficiens phenotype or a sequence variant, suggesting that either the character has been lost from the gene bank stocks or the phenotype was environmentally controlled and not genetic (Supplemental Table S4). Scanning electron microscopy images of Def2 showed that lateral spikelet meristems seem to arise from the central spikelet glume (Fig. 2C; Supplemental Fig. S8). Interestingly, the Def2 mutant shares an identical SNP (A→G at position +775 relative to the start codon) with deficiens cultivars, including DZ29. To date, Vrs1 has been resequenced in more than 400 lines, but neither the nucleotide sequences nor the haplotypes of Def2 or Semi-def12 mutants were found in any databases, suggesting that this change is novel, originating from a mutation in cv Foma. Interestingly, the Ser-rich motif around positions 184 and 187 was highly conserved among plant HD-Zip I transcription factors (n = 87; 22 species in Embryophyta; Fig. 2B; Supplemental Table S5; Supplemental Data S1). Since Ser is known as a potential phosphorylation site, we examined the possibility of the phosphorylation of Ser using the program NetPhos 3.1. The results showed that the Ser residues at the CTR in canonical two-rowed genotypes are predicted targets of phosphorylation, whereas deficiens alleles showed lower potential than the threshold (Supplemental Fig. S9), suggesting an altered phosphorylation status in deficiens lines.
To understand the phenotypic effect of the Deficiens mutations, several agronomic traits were compared between cv Foma (wild type) and its derived Def2 and Semi-def12 mutants. Both deficiens mutants showed about 10% increases in grain length and width compared with the wild type (Fig. 2, F, H, and I). Moreover, both deficiens mutants showed reduced plant height compared with the wild type (Fig. 2K). The significant difference of spikelet number per spike was observed only in Def2, and the significant difference of spike number per plant was observed only in Semi-def12.
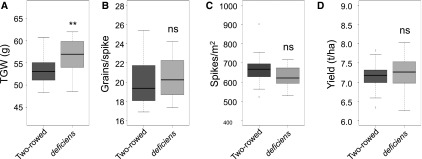
The phenotypic effect of deficiens (Vrs1.t1) was then assessed in the collection of 48 contemporary NW European winter barley cultivars (24 canonical two-rowed and 24 deficiens lines) referred to previously for yield and yield-related traits (Fig. 3). The deficiens lines have a significantly higher thousand grain weight (56.5 g) than the canonical two-rowed lines (53.5 g; t [23], P = 0.002). In this set of germplasm, the number of spikes per m2 was slightly, but not significantly, lower for the deficiens lines (629.8) compared with the two-rowed lines (665.3; t [23], P = 0.051), and deficiens does not significantly influence yield (deficiens = 7.2 t ha−1, two-rowed = 7.1 t ha−1) or grains per spike (deficiens = 20.5, two-rowed = 20.1).
Figure 3.
Mean effect of the deficiens allele for grain yield. A, Thousand grain weight (TGW). B, Grains per ear. C, Spikes per m2. D, Yield (t ha−1). Data were obtained from five site/season field trials of 24 two-rowed and 24 deficiens NW European cultivars grown under a winter feed barley regime (200 kg total nitrogen ha−1). Asterisks indicate that means are significantly different at the 1% (**) probability levels; ns, not significantly different.
Expression Pattern of Vrs1 in the Def2 Mutant
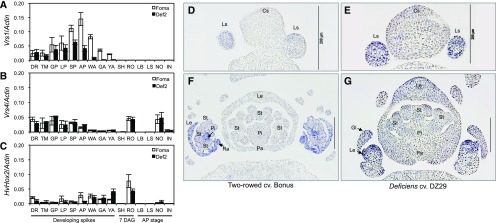
Quantifying mRNA levels at different developmental stages revealed no significant differences in Vrs1 transcript abundance at the beginning of spike development (from the double-ridge to the lemma primordium stage) between the wild type and the Def2 mutant. This indicates that Vrs1 is normally regulated at this stage (Fig. 4A). After the lemma primordium stage, however, the Vrs1 transcript level was decreased significantly in both Def2 (Fig. 4A) and DZ29 (Supplemental Fig. S10). To determine whether the reduced Vrs1 transcript level was affected by the upstream regulator Vrs4, we examined Vrs4 expression levels but found no significant difference between the wild type and Def2 (Fig. 4B). The expression pattern of HvHox2, which is a paralog of Vrs1, also was not significantly different (Fig. 4C).
Figure 4.
Transcript patterns of Vrs1, Vrs4, and HvHox2. A to C, qRT-PCR comparison of Vrs1 (A), Vrs4 (B), and HvHox2 (C) in selected tissues between cv Foma and the Def2 mutant. DR, Double-ridge stage; TM, triple-mound stage; GP, glume primordium stage; LP, lemma primordium stage; SP, stamen primordium stage; AP, awn primordium stage; WA, white anther stage; GA, green anther stage; YA, yellow anther stage; SH, shoots; RO, roots; LB, leaf blade; LS, leaf sheath; NO, nodes; IN, internodes. D to G, In situ mRNA hybridization with Vrs1 antisense probe. Vrs1 is expressed predominantly in the lateral spikelet meristem at the glume primordium stage of Bonus (Vrs1.b3; D) and DZ29 (Vrs1.t1; E). At the white anther stage, Vrs1 is localized mainly in lemma, pistil, and rachilla in Bonus (F), whereas Vrs1 is expressed in floret meristem and lemma primordia in DZ29 (G). Cs, Central spikelet; Gl, glume; Le, lemma; Ls, lateral spikelet; Pi, pistil; Ra, rachilla; St, stamen. Bars = 200 µm.
The hypothesis that a different tissue localization of the Vrs1 transcript also could be responsible for the deficiens phenotype guided us to test this possibility by RNA in situ hybridization using Vrs1-specific probes (Sakuma et al., 2013). Vrs1 mRNA was clearly localized to the lateral spikelet meristem in both deficiens and canonical two-rowed barley at the glume primordium stage and the white anther stage, respectively (Fig. 4, D–G). These quantitative real-time (qRT)-PCR and in situ hybridization data suggest that the amino acid alterations of VRS1, but not the mRNA differences, were responsible for the deficiens phenotype.
Genome-Wide Transcriptional Regulation by Def2
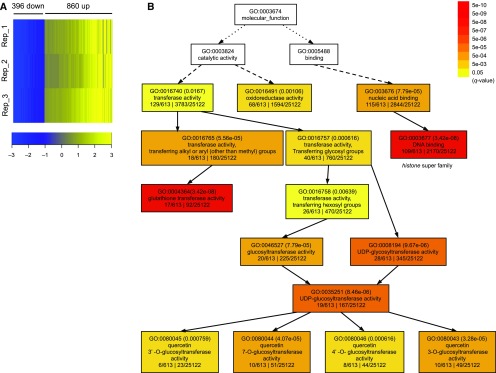
To identify the transcriptional changes underlying the morphological differences during immature spike development, RNA-sequencing (RNA-seq) was conducted using developing spikes at the awn primordium stage of both the wild type and the Def2 mutant. Based on scanning electron microscopy analysis at this developmental stage, the meristem of the lateral spikelet remains undifferentiated in Def2 (Fig. 2C). We reasoned that such a clear morphological difference should enable the detection of differentially expressed genes (DEGs). RNA-seq analysis revealed 1,256 DEGs (q < 0.05, P < 0.05, and log2 fold change > 1 or < −1) that comprised 860 (68.5%) up-regulated and 396 (31.5%) down-regulated genes in the Def2 mutant relative to the wild type (Fig. 5A; Supplemental Table S6). Gene Ontology (GO) enrichment analysis of the DEGs revealed that genes up-regulated in Def2 (Supplemental Data S3) were highly enriched for molecular functions related to DNA binding (GO:0003677; P = 5.80e–11) and glutathione transferase activity (GO:0004364; P = 1.20e–10; Fig. 5B; Supplemental Table S7). The DNA-binding class includes 78 histone superfamily genes, indicating the promotion of cell proliferation in Def2; this might be related to the fact that grain size was increased in deficiens alleles (Fig. 2). In contrast, genes down-regulated in Def2 were not generally enriched for any GO terms (Supplemental Data S4), although we did detect the down-regulation of cytokinin oxidase/dehydrogenase (CKX) genes, which are involved in cytokinin degradation (HORVU3Hr1G105360 and HORVU3Hr1G075920). Reduced expression of CKX genes generally causes cytokinin accumulation and more meristem activity (Ashikari et al., 2005; Han et al., 2014). The reduction of Vrs1 (HORVU2Hr1G092290) expression in Def2 also was detected in RNA-seq analysis, which validates the findings of the qRT-PCR experiment.
Figure 5.
Transcriptional regulation by the Deficiens mutation. A, Heat map of DEGs in the Def2 mutant (Vrs1.t2 allele) compared with cv Foma. B, Hierarchical tree graph of GO enrichment for the up-regulated DEG. Numbers in parentheses indicate q values. Solid, dashed, and dotted lines represent two, one, and zero enriched terms at both ends connected by the line, respectively.
The Origin of deficiens Barley
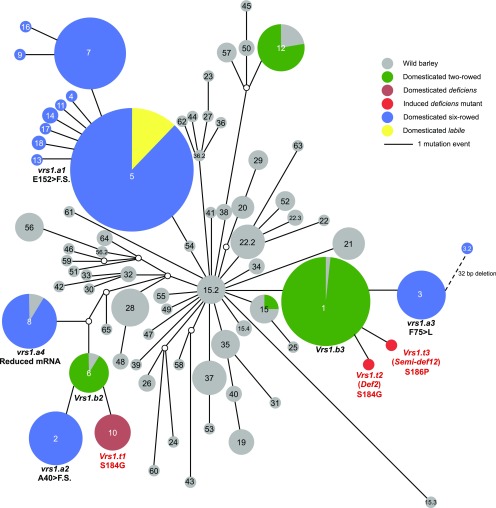
The deficiens landraces are distributed predominantly in Ethiopia, and a monophyletic origin of deficiens is hypothesized. To infer the origin of the deficiens allele (Vrs1.t1), we reanalyzed the sequence of the Vrs1 region (2,062 bp length) from 321 domesticated barley and 136 wild barley accessions (Saisho et al., 2009). Haplotype network analysis revealed that the Vrs1.t1 allele (haplotype 10) was derived from the Vrs1.b2 allele (haplotype 6) through a single nucleotide substitution in exon 3, which resulted in the S184G amino acid substitution, implying that Vrs1.b2 was the immediate ancestral allele of Vrs1.t1 (Fig. 6). Landraces carrying haplotype 6 are distributed predominantly in North Africa (Supplemental Table S8). Interestingly, the vrs1.a2 allele (haplotype 2), a six-rowed spike, also was derived from the same Vrs1.b2 allele through a single nucleotide insertion in exon 2, which results in a frame shift of the amino acid sequence (Fig. 6, A40>F.S.). Landraces carrying the vrs1.a2 allele are distributed throughout North Africa, southern Europe, and North America. Both Vrs1.t1 (contributing grain size) and vrs1.a2 (contributing grain number) alleles originated from North Africa. While Vrs1.t1 was endemic to Ethiopia, vrs1.a2 expanded to North Africa and southern Europe. We consider it remarkable that the opposite yield components, grain size and grain number, are controlled by different alleles derived from the same Vrs1 gene and that ancient peoples selected both mutations independently.
Figure 6.
Haplotype network analysis of Vrs1. The median-joining network was constructed from the haplotypes based on the resequencing of 321 domesticated and 136 wild barley accessions. Circle sizes correspond to the frequency of individual haplotypes. Lines represent genetic distances between haplotypes. Open circles indicate inferred intermediate haplotypes.
Considering contemporary NW European winter cultivars, we identified three SNPs in the coding sequence of Vrs1 leading to amino acid substitutions, D8G, D26E, S184G, and one SNP in the second intron of this gene (Supplemental Table S3). These four SNPs are in complete linkage disequilibrium, and the two haplotypes segregate based on the canonical two-rowed/deficiens phenotype of each cultivar. All of the deficiens lines contain the Vrs1.t1 allele, shared with accessions originating from Ethiopia (Supplemental Table S3).
DISCUSSION
A Single Amino Acid Substitution in VRS1 Is Responsible for deficiens Barley
Deficiens was previously considered one of multiple alleles at the Vrs1 locus (Woodward, 1947), but its causal mutation had not been discovered. In this study, detailed phenotypic observation confirmed that deficiens alleles extremely suppress floret development in the lateral spikelets. Our genetic analysis strongly suggests that a point mutation leading to a single amino acid substitution at the conserved Ser-rich motif in the CTR (motif 4) of VRS1 is responsible for the developmental fate of the lateral spikelet. This Ser-rich motif is highly conserved among plant HD-Zip I transcription factors, but its biological function(s) remained elusive. Next to Thr and Tyr, Ser is an amino acid residue commonly phosphorylated by several kinases in eukaryotes. Large-scale phosphorylation mapping analysis revealed 85% of Ser residues in Arabidopsis (Arabidopsis thaliana) to be phospho-Ser (Sugiyama et al., 2008). Phosphorylation influences the stability and activity of transcription factors (Hunter and Karin, 1992; Fujiwara et al., 2008). When phosphorylated, CONSTANS (CO), a flowering promoter encoding a B-box zinc finger, influences the rate of turnover of the CO protein via the activity of the CONSTITUTIVE PHOTOMORPHOGENIC1 ubiquitin ligase (Sarid-Krebs et al., 2015). Phosphorylated CO is the preferred substrate for degradation. In this study, almost all Ser residues in the Ser-rich motif (motif 4) were predicted as phospho-Ser in the canonical two-rowed allele of Vrs1 but not in its deficiens alleles, because of a single amino acid substitution in motif 4. From our results, we hypothesize that hypophosphorylated VRS1 in deficiens remains functional for longer during a critical stage in plant development, resulting in stronger suppression of floret development in the lateral spikelets. The expression analysis revealed that the deficiens phenotype is not due to an overexpression of Vrs1 but most likely acts at the protein level (Fig. 4). Reduced Vrs1 transcript levels from the stamen primordium stage onward are probably due to the much smaller size of the lateral spikelets compared with the wild type, noting that Vrs1 mRNA is localized predominantly in the lateral spikelets (Fig. 4, D–G).
Since Vrs1 evolved through gene duplication and neofunctionalization, the mutation frequency is notably higher than in its paralog HvHox2 (Sakuma et al., 2010, 2013). Six of the 58 vrs1 mutants identified so far have a mutation at the CTR (motifs 4 and 10 in this study), which is considered as a transcriptional activation domain (Komatsuda et al., 2007; Arce et al., 2011). This region is in the vicinity of the phosphorylation region, and the mutation sites were C194→S (Int-d.11), Q196→stop (Int-d.36), Q197→stop (Int-d.40), and A203→frame shift (hex-v.27, hex-v.30, and hex-v.31). Previous yeast two-hybrid assays showed that the CTR (motifs 4 and 10 in this study) of VRS1 functions as a transcriptional activator (Sakuma et al., 2013). Therefore, the six vrs1 mutants mentioned above could each be considered to represent a loss of transactivation activity as a transcription factor. By identifying its role in the suppression of floret primordia, this study, to our knowledge, is the first to uncover a biological function of the Ser-rich motif in HD-Zip I transcription factors.
Higher Yield Potential of deficiens Barley
Deficiens is now the major spike type of winter barleys grown in NW Europe and is increasing in the European spring barley crop (Supplemental Fig. S1). Therefore, we hypothesize that deficiens spike types have replaced canonical two-rowed types due to a higher grain yield potential associated with the altered spike architecture. Our results strongly support this hypothesis, since two independent mutant accessions showed enlarged grains, which is positively related to higher grain yield. It is hard to think of why deficiens resulted in reduced height; a possible explanation is a change in resource partitioning or additional mutations at other loci that reduce height. A previous study showed that the sterile lateral spikelets of canonical two-rowed barleys do not contribute to photosynthesis, suggesting that there is potential conflict between the central and lateral spikelets for assimilate supply (Habgood and Chambi, 1984). The notion that vrs1 loss-of-function mutants produce more but smaller grains compared with the wild type supports this model. Therefore, improvement of grain size could be achieved by suppressing the development of specific organs (in this study, lateral spikelets) that do not contribute to yield potential.
Our RNA-seq analysis revealed that a number of histone genes were up-regulated in the Def2 mutant, suggesting active cell proliferation in the central spikelet, which, in turn, may result in bigger grain size. This hypothesis is partially supported by the in situ hybridization analysis with histone H4 probe showing higher signals in the central spikelet and lower signals in the lateral spikelet of deficiens barley (Fig. 1, J and K). Since lateral florets were strongly suppressed at the awn primordium stage, the up-regulation of histone genes would be a central spikelet event and an indirect effect through the suppression of lateral floret development. In rice, higher expression of GW6a enhances grain weight and yield by increasing acetylation levels of histone H4 and the up-regulation of cell cycle genes (Song et al., 2015). Interestingly, the rare allele elevating GW6a expression has escaped human selection during rice domestication and modern breeding. Enlarged grain size and, thus, thousand grain weight in deficiens barley, as observed in both mutant germplasm and NW European cultivars, could be analogous to the GW6a event in rice, albeit at an independent locus. However, the precise mechanism by which up-regulating cell cycle genes prefertilization leads to increases in grain size remains to be established.
Haplotype network analysis revealed that both Vrs1.t1 and vrs1.a2 alleles originated from a point mutation of the Vrs1.b2 allele. Based on geographical data, it stands to reason that mutation of Vrs1.b2 to Vrs1.t1 occurred in Ethiopia and was selected there, while vrs1.a2 was selected in northwest Africa and then distributed to southern Europe. Remarkably, two different alleles with opposing phenotypic effects, enlarged grain size and increased grain number, were selected independently during barley domestication, presumably due to different selective forces for these two competing components of yield. Vrs1.t1 can be considered a rare allele, and its utilization has increased in NW European winter and spring barley only recently.
In summary, we identified that the deficiens spike architecture of cultivated barley is the result of a single amino acid substitution in a conserved motif of the major row-type protein VRS1. The substitution leads to a repression of the development of the lateral florets on the barley spike. Our phenotypic analysis suggested that deficiens alleles (Vrs1.t1, Vrs1.t2, and Vrs1.t3) ultimately contribute to increasing the size of grain in the central spikelets. Haplotype analysis indicates that the natural deficiens Vrs1.t1 allele originated from a point mutation in the two-rowed allele Vrs1.b2, an event that probably occurred in Ethiopia. Unlocking the precise molecular cause of the deficiens spike morphology will enable direct selection for higher yielding two-rowed barley varieties and enhance our biological understanding of the molecular mechanisms underpinning the elaboration of barley inflorescence development.
MATERIALS AND METHODS
Plant Materials
The deficiens barley (Hordeum vulgare var deficiens) DZ29 (SV043) and Mota 1 (SV039) together with the wild barley line OUH602 (H. vulgare ssp. spontaneum) were obtained from the Institute of Plant Science and Resources at Okayama University. The canonical two-rowed cultivars HN, KNG, BO, Foma, and GP were obtained from the National Institute of Agrobiological Sciences (NIAS) in Tsukuba, Japan. The Def and Semi-def mutants listed in Supplemental Table S4 were obtained from the Nordic gene bank NordGen. A total of 24 deficiens and 24 canonical two-rowed cultivars grown under a feed nitrogen regime were analyzed here. They were first placed on the U.K. National List between 1992 and 2012 and between 1992 and 2007, respectively, representing, as far as possible, a balanced data set and focusing on contemporary, relevant germplasm. The field trials were grown over five site/season combinations in Scotland, England, and Germany for harvest years 2010 and 2011 with known available soil mineral nitrogen content. Each trial was supplemented with three fertilizer regimes in a split-plot design with partial replication: no nitrogen application, a malting nitrogen application, and a feed nitrogen application. Plots were sown at a constant density and kept free from disease with a standard fungicide regime. Grab samples were taken from each plot at physiological maturity and used to estimate spikes per m2 and grains per spike. The plots were harvested, grain was dried to a constant moisture content, weighed, and cleaned, and a subsample was used to estimate thousand grain weight using a Marvin digital seed analyzer (GTA Sensorik). The weighed grain from each plot together with the plot area was used to estimate grain yield in t ha−1. Not all lines were in trial for 2 years, so best linear unbiased predictors (BLUPs) were calculated for each genotype and trait with the nitrogen treatment as a fixed effect and all others, including the interaction of line with fertilizer, as random effects. BLUPs were generated using the REML directive in Genstat 14 (http://www.vsni.co.uk/software/genstat/). All lines were genotyped previously with the barley iSelect 9K SNP platform (Comadran et al., 2012).
Scanning Electron Microscopy Analysis
Immature spikes were fixed in 4% formaldehyde in phosphate buffer. After dehydration and critical point drying, samples were examined in a Hitachi S4100 or a KEYENCE VHX-D510 scanning electron microscope at 5-kV acceleration voltage.
Basic Mapping
Five F2 populations were developed by crossing DZ29 with five different two-rowed accessions (KNG, HN, BO, GP, and OUH602). F2 plants were grown until full maturity under field condition at NIAS. To construct a basic genetic map, 78 to 96 plants were used from each of the five F2 populations. The deficiens genotype of F2 plants was determined by progeny testing 24 to 30 F3 plants from each F2 individual in the field of the Central Agricultural Experiment Station in Hokkaido, Japan, which enabled classification into the three genotypic classes expected for the segregation of a single Mendelian gene. The length of the lateral florets from the center of at least three spikes per plant was measured after spike emergence was complete. F2 individuals were genotyped using selected polymorphic DNA markers from the insertion-deletion, SNP, cleaved-amplified polymorphic sequence, and derived cleaved-amplified polymorphic sequence markers described by Sakuma et al. (2010). The primers are listed in Supplemental Table S9, and the resulting genotypic data were used to construct linkage maps using MAPMAKER/EXP version 3.0 (Lander et al., 1987).
Fine-Mapping
The F2 segregants of DZ29 × KNG and DZ29 × OUH602 were genotyped with two DNA markers, AK376350_SNP and BC12063_SNP3, that we found to flank the deficiens locus. F3 progeny in which a recombination had occurred between the two flanking markers were genotyped with de novo DNA markers to further delimit the deficiens locus (Supplemental Table S9).
Resequencing Vrs1 in NW European Winter Barley Cultivars
Genomic DNA was extracted using bench-grown 2-week-old leaf tissue with a QIAamp 96 DNA kit on a QIAcube HT automated DNA extraction platform (Qiagen). DNA quality and quantity were assessed using a NanoDrop 2000 (Thermo Scientific). PCR amplifications and Sanger sequencing were carried out as described previously (Houston et al., 2012). DNA sequence trimming and alignments were performed using Geneious (vR10; Biomatters). Details of the primers used are provided in Supplemental Table S9.
Peptide Motif Analysis and Functional Prediction
To obtain VRS1 homologs from several plant species, a BLASTP search (query: BAF43315) was made against Phytozome version 12.0 (https://phytozome.jgi.doe.gov/pz/portal.html) and the IPK Barley BLAST Server (http://webblast.ipk-gatersleben.de/barley_ibsc/). Peptide sequences with a threshold of E < 1e–10 were retrieved, and 87 peptide sequences (Supplemental Data S1), including VRS1 and HvHOX2 (BAI49294), were used for motif analysis. The MEME database was used for discovering peptide motifs (Bailey et al., 2006). The prediction of potential phosphorylation amino acid sites was performed using NetPhos 3.1 (Blom et al., 1999, 2004).
In Situ RNA Hybridization Analysis
Immature spikes at the glume primordium stage and the white anther stage (Kirby and Appleyard, 1981) from field-grown plants were freshly sampled and fixed in 4% paraformaldehyde and 0.25% glutaraldehyde in 0.05 m sodium phosphate buffer, pH 7.2. RNA probes of Vrs1 and histone H4 were developed as described by Sakuma et al. (2013). Primers are listed in Supplemental Table S9. In situ hybridization was conducted as described by Komatsuda et al. (2007).
RNA Extraction and qRT-PCR
Immature spikes were developmentally staged from double ridge stage to yellow anther stage using a stereoscopic microscope (Kirby and Appleyard, 1981). Total RNA was extracted from immature spikes, seedling shoots, and roots 7 d after germination and from leaf blades, leaf sheaths, nodes, and internodes at the awn primordium stage using TRIzol (Invitrogen). RNA was quantified using a NanoDrop 2000 (Thermo Fisher Scientific). To remove genomic DNA contamination, RNA was treated with RNase-free DNase (Takara Bio). First-strand cDNA was synthesized with SuperScript III (Invitrogen), and first-strand cDNA derived from 20 ng of RNA was used as a PCR template. Transcript levels of each gene were measured by qRT-PCR using the ABI Prism 7900HT sequence detection system (Applied Biosystems) and the THUNDERBIRD SYBR qPCR Mix Kit (Toyobo) according to the manufacturers’ protocols. Primers used for qRT-PCR are listed in Supplemental Table S9. qRT-PCR analysis was performed at least twice for each sample with biological replicates of at least three independent RNA extractions per sample. The barley Actin gene (accession no. DN182500) was used to normalize the RNA level for each sample.
RNA-Seq
Total RNA was extracted from immature spikes at the awn primordium stage of cv Foma and a Def2 mutant (NGB115174). Total RNA was measured using an Agilent 2100 Bioanalyzer (Agilent Technologies) and used for the construction of sequencing libraries. Strand-specific RNA libraries were prepared using the TruSeq Stranded mRNA Sample Prep Kit (Illumina) following the instructions in the TruSeq Stranded mRNA Sample Preparation Guide Rev.E (Illumina). High-throughput sequencing was conducted using a HiSeq 2500 (Illumina; Supplemental Table S10). Sequences were aligned to barley pseudomolecules (Mascher et al., 2017; 160404_barley_pseudomolecules_masked.fasta) with TopHAT2 (Kim et al., 2013). Gene expression was estimated as read counts for each gene locus by featureCounts (Liao et al., 2013) using the gene annotation file Hv_IBSC_PGSB_r1_HighConf.gtf. Expression levels were normalized by the TMM method, and P values were calculated by an exact negative binomial test along with the gene-specific variations estimated by the empirical Bayes method in edgeR (Robinson et al., 2010). The Benjamini-Hochberg method was applied on the P values to calculate q values and to control the false discovery rate. DEGs were defined as q < 0.05, P < 0.05, and log2 fold change > 1 or < −1.
GO Term Enrichment Analysis
GO terms of barley genes were assigned using Arabidopsis GO terms as follows. Barley protein sequences (160517_Hv_IBSC_PGSB_r1_proteins_HighConf_REPR_annotation.fasta) were compared with the peptide sequence in Arabidopsis (Arabidopsis thaliana [TAIR10]; Berardini et al., 2015) using BLAST software (blastp) with a threshold of E < 1e–20 (Altschul et al., 1990). GO terms with the most similar sequence in Arabidopsis were used as the GO terms of corresponding barley genes. A total of 25,122 barley genes were assigned with more than one GO term (Supplemental Data S2). GO term enrichment analysis of up-regulated genes (Supplemental Data S3) and down-regulated genes (Supplemental Data S4) were performed separately using agriGO (Du et al., 2010) with Fisher’s exact test and the Benjamini, Hochberg, and Yekutieli method to control false discovery rate.
Haplotype Analysis
A haplotype analysis was performed based on resequencing data of the Vrs1 locus (2,062 bp) from 321 domesticated barley accessions and 136 wild barley accessions (Supplemental Table S8; Saisho et al., 2009). Sequence alignments were performed with ClustalW using MEGA7 software (Kumar et al., 2016). A median-joining network (Bandelt et al., 1999) was constructed using the software programs DNA Alignment version 1.3.3.2 and Network version 5.0.0.1 (Fluxus Technology) with default parameters (epsilon = 0; frequency > 1 criterion = inactive; The ratio of transversion:transition = 1:1; Criterion = Connection cost; External rooting = inactive; MJ square option = inactive).
Accession Numbers
The Vrs1 sequence data have been deposited in the DDBJ nucleotide core database under accession numbers LC216328 to LC216332, except for the NW European cultivars listed in Supplemental Table S3, which have been deposited in NCBI under accession numbers MF776900 to MF776946. The RNA-seq data have been submitted to the DDBJ Sequence Read Archive under accession number DRA005563.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Summary of the tons of certified grain production each year in the United Kingdom.
Supplemental Figure S2. Additional replicates of the RNA in situ hybridization of histone H4.
Supplemental Figure S3. Frequency distributions of floret length in the lateral spikelet.
Supplemental Figure S4. Comparative basic genetic maps of deficiens on chromosome 2H.
Supplemental Figure S5. Alignment of deduced amino acid sequences of VRS1.
Supplemental Figure S6. Spike phenotype of deficiens mutants.
Supplemental Figure S7. Representation of motifs in VRS1 homologs.
Supplemental Figure S8. Scanning electron microscopy views of barley spikes.
Supplemental Figure S9. Predicted phosphorylation sites of Ser in VRS1 protein.
Supplemental Figure S10. Expression pattern of Vrs1 and HvHox2 in DZ29.
Supplemental Table S1. Segregation analysis of deficiens in five F2 mapping populations.
Supplemental Table S2. Mapping population used for fine-mapping of deficiens.
Supplemental Table S3. Vrs1 sequence information of deficiens barley.
Supplemental Table S4. List of Def and Semi-def mutants with description of mutational events in Vrs1.
Supplemental Table S5. Motif analysis of HD-Zip I transcription factor in plants (n = 87).
Supplemental Table S6. Differentially regulated genes in Def2 mutants compared with the wild type.
Supplemental Table S7. GO enrichment analysis for up-regulated DEGs in the Def2 mutant.
Supplemental Table S8. Haplotype information of 321 domesticated and 136 wild barleys.
Supplemental Table S9. Primer information used in this study.
Supplemental Table S10. Summary of the RNA-seq experiment.
Supplemental Data S1. Peptide sequences used for motif analysis.
Supplemental Data S2. GO terms of 25,122 barley genes.
Supplemental Data S3. GO terms of up-regulated genes.
Supplemental Data S4. GO terms of down-regulated genes.
Acknowledgments
We thank Daisuke Saisho (Okayama University), Naoki Sentoku (NIAS), and Hiroyuki Tsuji (Yokohama City University) for help and advice. We are grateful to Harumi Koyama and Mari Sakuma (NIAS), Corinna Trautewig (IPK), and Malcolm Macaulay (James Hutton Institute) for excellent technical assistance. We thank Peter Werner, Guillaume Barol-Baron, and David Harrap of KWS UK Ltd. for allowing us to use some of their data from the trials. We also acknowledge the assistance of Hazel Bull, Richard Keith, and Chris Warden of the James Hutton Institute.
Footnotes
This research was funded by the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation grant no. TRS1002 to T.K. and S.S.); a Grant-in-Aid from the Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellow for Research Abroad (to S.S.); a Grant-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science (JSPS) (no. 16K18635 to S.S.). The field trials work was conducted under a work package of the EU-FP7, “Improving nutrient use efficiency in major European food, feed and biofuel crops to reduce negative environmental impacts of crop production” (NUE Crops) EU-FP7 222-645 (2009-2014), led by Carlo Leifert of the University of Newcastle.
Articles can be viewed without a subscription.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Arce AL, Raineri J, Capella M, Cabello JV, Chan RL (2011) Uncharacterized conserved motifs outside the HD-Zip domain in HD-Zip subfamily I transcription factors: a potential source of functional diversity. BMC Plant Biol 11: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel FD, Manavella PA, Dezar CA, Chan RL (2007) The true story of the HD-Zip family. Trends Plant Sci 12: 419–426 [DOI] [PubMed] [Google Scholar]
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34: W369–W373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37–48 [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E (2015) The Arabidopsis Information Resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53: 474–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294: 1351–1362 [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4: 1633–1649 [DOI] [PubMed] [Google Scholar]
- Bommert P, Nagasawa NS, Jackson D (2013) Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat Genet 45: 334–337 [DOI] [PubMed] [Google Scholar]
- Bothmer R, Jacobsen N, Baden C, Jorgensen R, Linde-Laursen I (1995) An Ecogeographical Study of the Genus Hordeum, Ed 2 IBPGR, Rome [Google Scholar]
- Bull H, Casao MC, Zwirek M, Flavell AJ, Thomas WTB, Guo W, Zhang R, Rapazote-Flores P, Kyriakidis S, Russell J, et al. (2017) Barley SIX-ROWED SPIKE3 encodes a putative Jumonji C-type H3K9me2/me3 demethylase that represses lateral spikelet fertility. Nat Commun 8: 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comadran J, Kilian B, Russell J, Ramsay L, Stein N, Ganal M, Shaw P, Bayer M, Thomas W, Marshall D, et al. (2012) Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat Genet 44: 1388–1392 [DOI] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127: 1309–1321 [DOI] [PubMed] [Google Scholar]
- Druka A, Franckowiak J, Lundqvist U, Bonar N, Alexander J, Houston K, Radovic S, Shahinnia F, Vendramin V, Morgante M, et al. (2011) Genetic dissection of barley morphology and development. Plant Physiol 155: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han L, Suh SS, Salomé PA, McClung CR, Somers DE (2008) Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem 283: 23073–23083 [DOI] [PubMed] [Google Scholar]
- Habgood RM, Chambi JY (1984) Effects of the deficiens allele on ear development and yield in barley. Ann Appl Biol 105: 159–166 [Google Scholar]
- Han Y, Zhang C, Yang H, Jiao Y (2014) Cytokinin pathway mediates APETALA1 function in the establishment of determinate floral meristems in Arabidopsis. Proc Natl Acad Sci USA 111: 6840–6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston K, Druka A, Bonar N, Macaulay M, Lundqvist U, Franckowiak J, Morgante M, Stein N, Waugh R (2012) Analysis of the barley bract suppression gene Trd1. Theor Appl Genet 125: 33–45 [DOI] [PubMed] [Google Scholar]
- Hua L, Wang DR, Tan L, Fu Y, Liu F, Xiao L, Zhu Z, Fu Q, Sun X, Gu P, et al. (2015) LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell 27: 1875–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T, Karin M (1992) The regulation of transcription by phosphorylation. Cell 70: 375–387 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby EJM, Appleyard M (1981) Cereal Development Guide, Ed 1 Cereal Unit, National Agricultural Centre, Kenilworth, UK [Google Scholar]
- Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, Perovic D, Stein N, Graner A, Wicker T, Tagiri A, et al. (2007) Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA 104: 1424–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppolu R, Anwar N, Sakuma S, Tagiri A, Lundqvist U, Pourkheirandish M, Rutten T, Seiler C, Himmelbach A, Ariyadasa R, et al. (2013) Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc Natl Acad Sci USA 110: 13198–13203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181 [DOI] [PubMed] [Google Scholar]
- Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J, et al. (2011) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet 43: 1266–1269 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2013) The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Liu H, Zhou T, Gu B, Huang X, Shangguan Y, Zhu J, Li Y, Zhao Y, Wang Y, et al. (2013) An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25: 3360–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, et al. (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544: 427–433 [DOI] [PubMed] [Google Scholar]
- Poursarebani N, Seidensticker T, Koppolu R, Trautewig C, Gawroński P, Bini F, Govind G, Rutten T, Sakuma S, Tagiri A, et al. (2015) The genetic basis of composite spike form in barley and ‘Miracle-Wheat’. Genetics 201: 155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers L. (1936) The nature of the interaction of genes affecting four quantitative characters in a cross between Hordeum deficiens and Hordeum vulgare. Genetics 21: 398–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay L, Comadran J, Druka A, Marshall DF, Thomas WT, Macaulay M, MacKenzie K, Simpson C, Fuller J, Bonar N, et al. (2011) INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat Genet 43: 169–172 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saisho D, Pourkheirandish M, Kanamori H, Matsumoto T, Komatsuda T (2009) Allelic variation of row type gene Vrs1 in barley and implication of the functional divergence. Breed Sci 59: 621–628 [Google Scholar]
- Sakuma S, Pourkheirandish M, Hensel G, Kumlehn J, Stein N, Tagiri A, Yamaji N, Ma JF, Sassa H, Koba T, et al. (2013) Divergence of expression pattern contributed to neofunctionalization of duplicated HD-Zip I transcription factor in barley. New Phytol 197: 939–948 [DOI] [PubMed] [Google Scholar]
- Sakuma S, Pourkheirandish M, Matsumoto T, Koba T, Komatsuda T (2010) Duplication of a well-conserved homeodomain-leucine zipper transcription factor gene in barley generates a copy with more specific functions. Funct Integr Genomics 10: 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarid-Krebs L, Panigrahi KC, Fornara F, Takahashi Y, Hayama R, Jang S, Tilmes V, Valverde F, Coupland G (2015) Phosphorylation of CONSTANS and its COP1-dependent degradation during photoperiodic flowering of Arabidopsis. Plant J 84: 451–463 [DOI] [PubMed] [Google Scholar]
- Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M (2008) Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet 40: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Simmonds J, Scott P, Brinton J, Mestre TC, Bush M, Del Blanco A, Dubcovsky J, Uauy C (2016) A splice acceptor site mutation in TaGW2-A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Theor Appl Genet 129: 1099–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39: 623–630 [DOI] [PubMed] [Google Scholar]
- Song XJ, Kuroha T, Ayano M, Furuta T, Nagai K, Komeda N, Segami S, Miura K, Ogawa D, Kamura T, et al. (2015) Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc Natl Acad Sci USA 112: 76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J (2011) Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet 43: 1160–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y (2008) Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol 4: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse GW, Walla A, Finke A, Koornneef M, Pecinka A, von Korff M (2017) Six-Rowed Spike3 (VRS3) is a histone demethylase that controls lateral spikelet development in barley. Plant Physiol 174: 2397–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, et al. (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44: 950–954 [DOI] [PubMed] [Google Scholar]
- Wendt T, Holme I, Dockter C, Preuß A, Thomas W, Druka A, Waugh R, Hansson M, Braumann I (2016) HvDep1 is a positive regulator of culm elongation and grain size in barley and impacts yield in an environment-dependent manner. PLoS ONE 11: e0168924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward RW. (1947) The Ih, I, i allels in Hordeum deficiens genotypes of barley. J Am Soc Agron 39: 474–482 [Google Scholar]
- Woodward RW. (1949) The inheritance of fertility in the lateral florets of the four barley groups. Agron J 41: 317–322 [Google Scholar]
- Youssef HM, Eggert K, Koppolu R, Alqudah AM, Poursarebani N, Fazeli A, Sakuma S, Tagiri A, Rutten T, Govind G, et al. (2017) VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat Genet 49: 157–161 [DOI] [PubMed] [Google Scholar]
- Youssef HM, Koppolu R, Rutten T, Korzun V, Schweizer P, Schnurbusch T (2014) Genetic mapping of the labile (lab) gene: a recessive locus causing irregular spikelet fertility in labile-barley (Hordeum vulgare convar. labile). Theor Appl Genet 127: 1123–1131 [DOI] [PubMed] [Google Scholar]