Abstract
Rationale: Nontuberculous mycobacteria (NTM) are ubiquitous environmental microorganisms. Infection is thought to result primarily from exposure to soil and/or water sources. NTM disease prevalence varies greatly by geographic region, but the geospatial factors influencing this variation remain unclear.
Objectives: To identify sociodemographic and environmental ecological risk factors associated with NTM infection and disease in Colorado.
Methods: We conducted an ecological study, combining data from patients with a diagnosis of NTM disease from National Jewish Health’s electronic medical record database and ZIP code–level sociodemographic and environmental exposure data obtained from the U.S. Geological Survey, the U.S. Department of Agriculture, and the U.S. Census Bureau. We used spatial scan methods to identify high-risk clusters of NTM disease in Colorado. Ecological risk factors for disease were assessed using Bayesian generalized linear models assuming Poisson-distributed discrete responses (case counts by ZIP code) with the log link function.
Results: We identified two statistically significant high-risk clusters of disease. The primary cluster included ZIP codes in urban regions of Denver and Aurora, as well as regions south of Denver, on the east side of the Continental Divide. The secondary cluster was located on the west side of the Continental Divide in rural and mountainous regions. After adjustment for sociodemographic, drive time, and soil variables, we identified three watershed areas with relative risks of 12.2, 4.6, and 4.2 for slowly growing NTM infections compared with the mean disease risk for all watersheds in Colorado. This study population carries with it inherent limitations that may introduce bias. The lack of complete capture of NTM cases in Colorado may be related to factors such as disease severity, education and income levels, and insurance status.
Conclusions: Our findings provide evidence that water derived from particular watersheds may be an important source of NTM exposure in Colorado. The watershed with the greatest risk of NTM disease contains the Dillon Reservoir. This reservoir is also the main water supply for major cities located in the two watersheds with the second and third highest disease risk in the state, suggesting an important possible source of infection.
Keywords: nontuberculous mycobacteria, watershed epidemiology, spatial, Bayesian, Poisson
Nontuberculous mycobacteria (NTM) disease has become increasingly prevalent across North America, especially among older adults (1–4). NTM are ubiquitous environmental organisms (5). Beyond previously described immunosuppressive conditions, host genetic factors, and environmental factors important to the epidemiology of NTM disease (6–8), infection is thought to result primarily from exposure to soil and/or water sources (9). Although disease prevalence varies greatly by geographic region (10, 11), the geospatial factors influencing this variation are poorly understood. Researchers in previous studies have identified geographic regions with increased disease rates (10, 11), as well as regions with high numbers of mycobacteria isolated from environmental samples, as compared with other surveyed regions (12, 13). Few studies have involved systematic assessments of the influence of environmental factors on NTM disease prevalence. Statistical analysis can be used to identify regions of high risk, enabling subsequent study of associated factors. Researchers in two previous studies employed spatial analytic techniques to identify high-risk regions and identify factors associated with disease distribution (14, 15). Their findings warrant further investigation into the spatial patterns and environmental predictors of NTM disease.
Our study uses the concept of “watershed epidemiology,” whereby connections between ecosystems and human health are explored over broad spatial scales (16). A watershed is “the area of land where all of the water under it or draining off of it goes to the same place and includes both surface and ground water” (17, p. 1). Consequently, two individuals living far from one another within the same watershed may have more similar exposures than two individuals living near one another but in different watersheds. The use of watersheds can place cases into a construct with defined natural boundaries and environmental relevance (17). From a population-level perspective, data related to geophysical, pollutant sources, and physicochemical properties of water, to name a few, can be applied across watersheds to estimate associations between ecosystems and human health (16). To systematically explore the role of water exposure on NTM infection risk in Colorado, we used watersheds as our main variable of interest while controlling for other known risk factors.
With over 150 NTM species identified (18), NTM represent a genetically diverse group of environmental organisms (19, 20). Previous literature indicates that the genetic diversity of some NTM species differs by geographic region, implying that environmental sources and routes of transmission may differ by species (21). Mycobacteria are defined by growth rate as either slowly growing or rapidly growing. These groups are phylogenetically distinct (22, 23), and it is logical to explore whether different environmental sources are involved. Our data enabled examination of slowly growing and rapidly growing NTM species from patient isolates, both together and separately.
A better understanding of the regional ecology and environmental sources of NTM is crucial because patients undergo lengthy and complex treatment regimens and are often reinfected despite initial cure. Regional factors may provide insight into the characterization of optimal growth environments, which ultimately influence disease prevalence in different geographic settings (24). Using an ecological design with data from patients with NTM disease resident in the State of Colorado who were treated at National Jewish Health (NJH), a leading respiratory hospital in Denver, we aimed to identify geospatial patterns of NTM disease and to identify sociodemographic and environmental variables associated with NTM infection risk in Colorado.
Methods
Patient data were obtained from the NJH electronic medical record database. Our study population included patients with a diagnosis of NTM treated at NJH who were resident in Colorado during the study period that spanned from February 2008 through June 2015. Patients with NTM disease diagnosed with cystic fibrosis were excluded from the study to ensure that we had a homogeneous patient population. This study was approved by the NJH Institutional Review Board (HS-2901).
Data Collection
NTM species categories
NTM species from patient isolates were grouped by (1) all NTM species, (2) slowly growing NTM species, and (3) rapidly growing NTM species. Individual species in categories 2 and 3 are listed in Table E1 in the online supplement. Outcome variables in the regression models were case counts per ZIP code for each category.
Sociodemographic and environmental exposure data
U.S. Census Bureau
U.S. ZIP Code Tabulation Areas (ZCTAs) and state base layers, ZCTA land areas, and ZCTA-level sociodemographic data were obtained from the U.S. Census Bureau American FactFinder online portal (25, 26).
U.S. Department of Agriculture
Soil data (pH; salinity; cation exchange capacity; calcium carbonate; permeability; bulk density; water infiltration rates; and soil texture, e.g., clay, sand, silt) were obtained from the State Soil Geographic database (27).
U.S. Geological Survey
Geochemical soil properties were obtained from the U.S. Geological Survey National Geochemical Survey database (mrdata.usgs.gov/geochem/) (28). Data on the U.S. population served by ground and surface water was obtained from the U.S. Geological Survey National Water Use Information Program (29). Watershed boundaries were obtained from the U.S. Geological Survey Watershed Boundary Dataset (30).
Watershed delineation
The U.S. Geological Survey partitions the U.S. land surface into a hierarchical system of watershed polygons. We used the hydrologic unit code level 8 watersheds (30) as an independent variable, which includes 92 watersheds throughout Colorado.
Statistical Analysis
Our initial analyses, using SaTScan software, were focused on identifying clusters of ZIP codes that had unusually large NTM risk relative to ZIP codes outside the cluster. Our second analyses, involving Bayesian regression, were focused on explaining why the clusters may have greater risk.
SaTScan
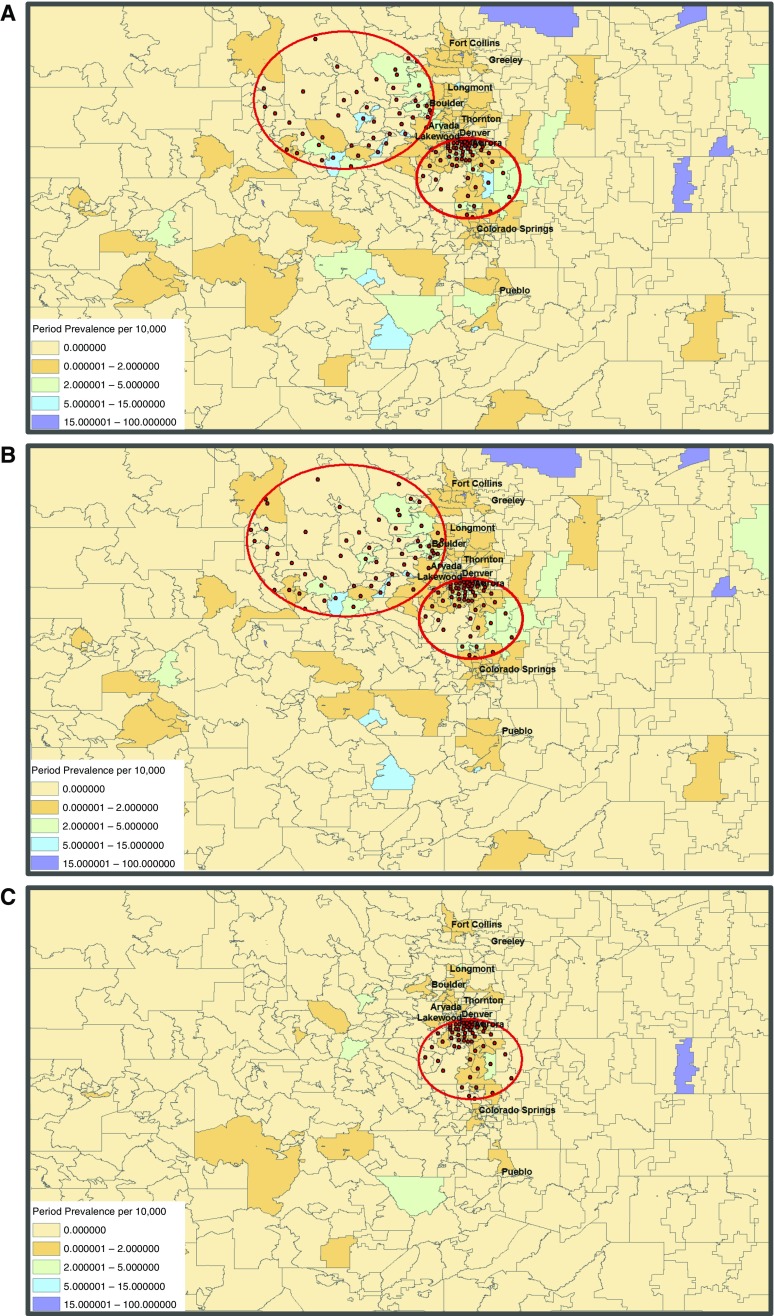
Statistical analyses were performed using spatial scan methods implemented in SaTScan (version 9.4.1; Kulldorff M. and Information Management Services, Inc., Rockville, MD). All ZIP codes with a population of at least 1 were used to represent the at-risk population. Given the ubiquitous nature of NTM, the maximum at-risk population was set to include up to 50% of the population (the maximum geographical cluster size allowable in the SaTScan software) across all ZIP codes. We sought to identify high-risk clusters of NTM disease. A high-risk cluster is defined as a collection of ZIP codes where the total number of observed cases is greater than what would be expected under the null hypothesis of constant risk across all regions. We estimated the relative risk (RR) for each cluster. This is the ratio of the risk of NTM for of all ZIP codes in the cluster to the risk of NTM in all ZIP codes outside the cluster. A discrete Poisson distribution was selected to model the case counts using the “purely spatial” analysis option. In Figures 1A–1C, red dots indicate centroid centers of ZIP codes that were included in the cluster.
Figure 1.
Period prevalence per 10,000 population and significant high-risk clusters of nontuberculous mycobacteria (NTM) disease in Colorado. Gray lines delineate ZIP code boundaries throughout the state. Red circles indicate significant clusters. Red dots indicate centroid center of each ZIP code included in the significant cluster. (A) All NTM species. (B) Slowly growing NTM species. (C) Rapidly growing NTM species.
Bayesian Poisson regression analyses
Analysis of data was performed using the R (version 3.2.2) statistical software packages “spdep” (31), “arm” (32), “ggmap” (33), and “geoRglm” (34). Our model included the log of the population for each ZIP code as an offset term to account for the differing population in each region. We ran Bayesian generalized linear models assuming overdispersed, Poisson-distributed discrete responses (case counts by ZIP code) with the log link function (32).
In all regression models, ZIP code was the unit of analysis. Exposure variables (all soil, sociodemographic covariates, and population proportion served by ground water) were averaged by ZIP code in ArcGIS 10.2 (Esri, Redlands, CA) and then entered as explanatory variables in our regression models. Drive time between ZIP code centroids and NJH were calculated using the ggmap package (33). We categorized ZIP codes on the basis of whether they were within a 30-minute drive to NJH. For ZIP codes that intersected multiple watersheds, we calculated the fraction of the ZIP code land area overlapping each watershed. In our regression models, the RR of an NTM diagnosis in a given watershed was estimated by comparing the risk of each watershed relative to the weighted mean risk of disease of all watersheds throughout Colorado. Sociodemographic and drive-time variables were included to control for potential confounding induced by using a hospital-based population. Any soil variable that changed watershed estimates by more than 10% or that was statistically significant in an analysis of deviance test was retained in the final model. We tested whether residual spatial autocorrelation remained by using a populated-adjusted version of Moran’s I test (35). If residual spatial autocorrelation remained, we replaced our Bayesian generalized linear model with a Bayesian Poisson kriging model (34).
Some variables, such as soil pH, soil bulk density, soil permeability, geochemical soil properties, age, sex, race, income, education, and population density, were selected to replicate and confirm prior findings reported in the literature (14, 15). Other soil- and water-related variables, including soil texture (clay, sand, or silt), proportion of the population served by ground water versus surface water, and watershed boundaries, were selected to explore hypotheses related to the exposure of soil or water or a combination of the two.
Results
Our dataset included 479 patients with NTM disease with Colorado ZIP codes who sought treatment at NJH, a respiratory hospital in Denver, from February 2008 through June 2015. The characteristics of our study population are summarized in Table 1.
Table 1.
Descriptive statistics of the Colorado state resident nontuberculous mycobacteria patient population treated at National Jewish Health
| Characteristic | All NTM Species (n = 479) | Slowly Growing NTM Species (n = 399) | Rapidly Growing NTM Species (n = 115) | Slowly + Rapidly Growing NTM Species (n = 35) |
|---|---|---|---|---|
| Age*, yr, mean ± SD | 70.14 ± 12.27 | 70.41 ± 11.93 | 69.87 ± 12.14 | 72.42 ± 9.51 |
| Female sex, n (%) | 351 (73.28) | 304 (76.2) | 74 (64.4) | 27 (77.1) |
| Married, n (%) | 289 (60.33) | 244 (61.2) | 69 (60.0) | 24 (68.6) |
| White race*, n (%)a | 304 (84.44) | 262 (85.6) | 69 (83.1) | 27 (93.1) |
Definition of abbreviation: NTM = nontuberculous mycobacteria.
Missing data (48 patients did not report age; 119 patients did not report race).
SaTScan
SaTScan identified two significant (P < 0.05) clusters for the all NTM species group and the slowly growing NTM group (Figures 1A and 1B), as well as one significant cluster for the rapidly growing group (Figure 1C). The RR of having an NTM infection diagnosis with any NTM species was 2.4 for ZIP codes within the primary cluster (P = 0.00001; radius, 42 km), including regions in Denver, Aurora, and south of Denver, compared with all ZIP codes outside this cluster. The risk of having NTM disease for ZIP codes inside the cluster was almost 2.5 times greater than the risk of having NTM disease in ZIP codes outside the cluster. The second highest statistically significant cluster had an RR of 2.2 (P = 0.032; radius, 72 km). When we examined the spatial scan statistics for slowly growing NTM species, we observed similarly significant clusters as those in the all NTM species group. This is intuitive because slowly growing NTM infections comprised the majority of the all NTM species group. The RRs of the primary and secondary clusters for the slowly growing group were 2.3 (P = 0.00001; radius, 41.9 km) and 2.2 (P = 0.011; radius, 79.5 km), respectively. For the rapidly growing group, we observed one significant cluster (at approximately the same location as the small cluster of the all NTM species and slowly growing NTM groups) with an RR of 3.1 (P = 0.00004; radius, 41.8 km).
Bayesian Poisson Regression Analyses
Having identified spatial clusters where the risk of NTM appeared to be greater than what could be expected under a hypothesis of constant NTM risk in all regions, we then sought to identify factors that could explain the higher risk in these clusters. To this end, a Bayesian Poisson regression model was constructed to model disease risk as a function of various environmental and sociodemographic variables.
All NTM species and slowly growing NTM species
Older age was a significant risk factor for all NTM species and slowly growing NTM; higher education was significant only for infections caused by slowly growing NTM species. Low soil pH was a significant risk factor for infections caused by slowly growing NTM species (Table 2). For all NTM species, the inclusion of soil texture (clay, sand, or silt) significantly improved the model fit. For each 1% increase in sand at the expense of silt, each 1% increase in clay at the expense of silt, and each 1% increase in sand at the expense of clay, the RRs were 0.15, 0.02, and 8.1, respectively. These findings implicate increasing silt concentrations as a risk factor for disease and increasing clay concentrations as protective against disease. In analyses using patient infections caused by slowly growing NTM, for every 1 SD increase of manganese soil concentration, the RR was 0.72, and for every 1 SD increase of phosphorus soil concentration, the RR was 1.3.
Table 2.
Multivariable Bayesian Poisson regression models of ecological risk factors significantly associated with all nontuberculous mycobacteria species, slowly growing nontuberculous mycobacteria species, and rapidly growing nontuberculous mycobacteria species
| All NTM Species | Relative Risk 95% CI (P Value) | Slowly Growing NTM Species | Relative Risk 95% CI (P Value) | Rapidly Growing NTM Species | Relative Risk 95% CI (P Value) |
|---|---|---|---|---|---|
| Watershed | Significant* | Watershed | Significant* | Watershed | NS |
| Education: bachelor’s degree (%) | 1.72 | Education: bachelor’s degree (%) | 2.66 | Education: bachelor’s degree (%) | 1.05 |
| 0.92–3.24 | 1.39–5.07 | 0.25–4.49 | |||
| (0.09) | (0.0031) | (0.95) | |||
| Age: ≥65 yr (%) | 1.09 | Age: ≥65 yr (%) | 1.12 | Age: ≥65 yr (%) | 1.06 |
| 1.05–1.13 | 1.07–1.16 | 0.99–1.14 | |||
| (1.4 × 10−6) | (3.8 × 10−8) | (0.11) | |||
| Median income (per $1,000) | 1.004 | Median income (per $1,000) | 1.01 | Median income (per $1,000) | 1.01 |
| 0.996–1.02 | 0.999–1.02 | 0.99–1.03 | |||
| (0.27) | (0.093) | (0.53) | |||
| Population (per 1,000)/square mile | 1.14 | Population (per 1,000)/square mile | 1.12 | Population (per 1,000)/square mile | 1.68 |
| 0.92–1.40 | 0.88–1.42 | 1.15–2.46 | |||
| (0.23) | (0.37) | (0.0078) | |||
| Uninsured (%) | 0.98 | Uninsured (%) | 0.99 | Uninsured (%) | 1.01 |
| 0.96–1.008 | 0.96–1.02 | 0.95–1.06 | |||
| (0.17) | (0.58) | (0.84) | |||
| Female sex (%) | 0.99 | Female sex (%) | 0.97 | Female sex (%) | 1.03 |
| 0.94–1.06 | 0.92–1.04 | 0.89–1.19 | |||
| (0.93) | (0.43) | (0.66) | |||
| Drive time >30 min from NJH | 1.11 | Drive time >30 min from NJH | 0.99 | Drive time >30 min from NJH | 1.78 |
| 0.78–1.56 | 0.68–1.46 | 0.86–3.67 | |||
| (0.57) | (0.99) | (0.12) | |||
| Soil pH (1 unit) | 0.75 | Soil pH (1 unit) | 0.69 | Population (per 1,000) served by ground water | 1.02 |
| 0.57–1.008 | 0.51–0.95 | 0.99–1.03 | |||
| (0.057) | (0.022) | (0.05) | |||
| Soil bulk density (1 unit) | 0.99 | Soil bulk density (1 unit) | 0.99 | Soil manganese† (1 SD) | 1.23 |
| 0.992–1.006 | 0.992–1.007 | 1.01–1.49 | |||
| (0.82) | (0.93) | (0.038) | |||
| Soil permeability (1 unit) | 1.00 | Soil permeability (1 unit) | 1.00 | Soil phosphorus‡ (1 SD) | 0.99 |
| 0.99–1.001 | 0.99–1.001 | 0.62–1.27 | |||
| (0.79) | (0.69) | (0.50) | |||
| Soil calcium carbonate (1 unit) | 1.07 | Soil calcium carbonate (1 unit) | 1.06 | ||
| 0.98–1.17 | 0.96–1.16 | ||||
| (0.12) | (0.23) | ||||
| Soil texture | 8.1 | Soil manganese† (1 SD) | 0.72 | ||
| Sand–clay§ (%) | 1.29–50.5 | 0.56–0.94 | |||
| (0.025) | (0.0136) | ||||
| Sand–silt§ (%) | 0.15 | Soil phosphorus‡ (1 SD) | 1.25 | ||
| 0.026–0.90 | 1.01–1.55 | ||||
| (0.037) | (0.0396) | ||||
| Clay–silt§ (%) | 0.02 | ||||
| 0.0009–0.36 | |||||
| (0.0086) |
Definition of abbreviations: CI = confidence interval; NJH = National Jewish Health; NS = not significant; NTM = nontuberculous mycobacteria.
Boldface estimates are statistically significant. Watershed refers to all 92 watershed boundaries as defined by the hydrologic unit code level 8 watersheds.
Significant watershed effects are detailed in Figures 2B and 3B.
One SD in manganese soil concentration is equivalent to 466.5 ppm.
One SD in phosphorus soil concentration is equivalent to 0.052 percent by weight.
Holding the third soil variable constant.
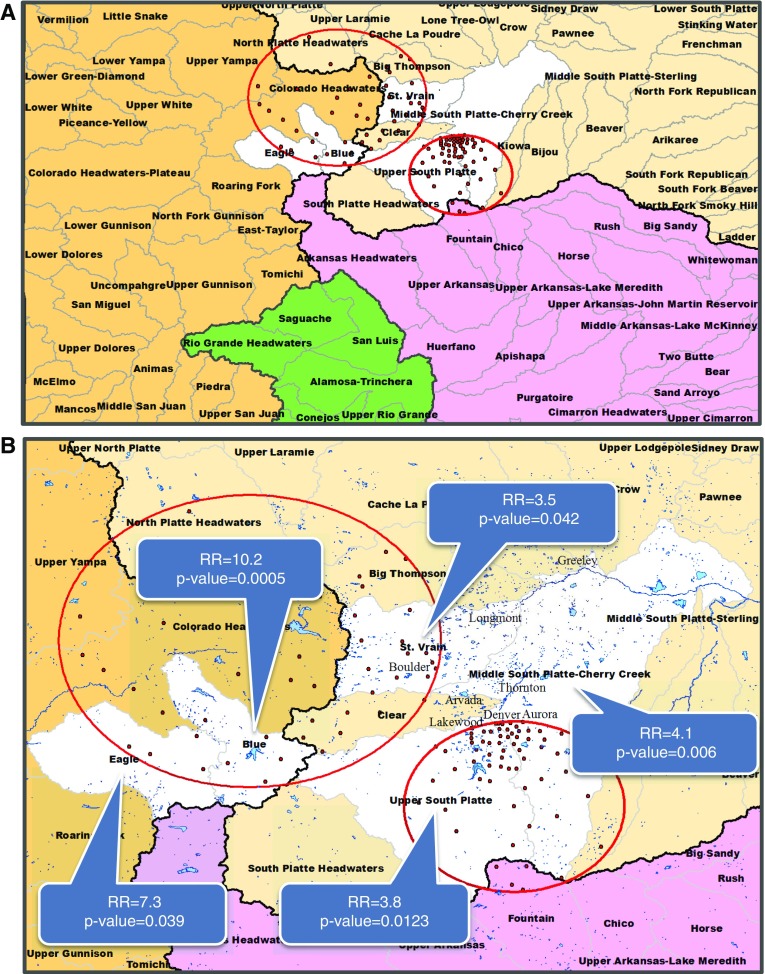
Multiple watersheds were significantly associated with disease. For all NTM species, the Blue watershed had an RR of 10.2 compared with the mean risk of all 92 watersheds, whereas the Upper South Platte (USP) and the Middle South Platte Cherry Creek (MSPCC) watersheds had RRs of 3.8 and 4.1, respectively. The Eagle and St. Vrain watersheds yielded statistically significant risk increases similar in magnitude to those of the Blue, USP, and MSPCC watersheds, though with higher P values (Figures 2A and 2B). After controlling for multiple testing using a Bonferroni correction (3 models), we observed that the Eagle and St. Vrain watersheds did not retain statistical significance.
Figure 2.
Significant watersheds for all nontuberculous mycobacteria (NTM) species. The four major watersheds in Colorado are represented by four colors: Missouri (beige), Upper Colorado (brown), Arkansas-White-Red (purple), and Rio Grande (green). Black lines represent watershed boundaries of the four major watersheds. Black line separating the Upper Colorado watershed (brown) from the remaining three watersheds represents the Continental Divide. Statistically significant watersheds are shown in white. (A) Four major watersheds are divided into 92 watersheds (boundaries delineated by gray lines) in Colorado. Watershed names are printed in boldface type. (B) Watersheds narrowed into region of interest. Watershed names are printed in boldface type. Major city names are in printed in gray. Red circles indicate significant clusters of disease caused by all NTM infections. Red dots indicate centroid center of each ZIP code included in the significant cluster. Blue areas indicate lakes, reservoirs, and rivers. RR = risk ratio.
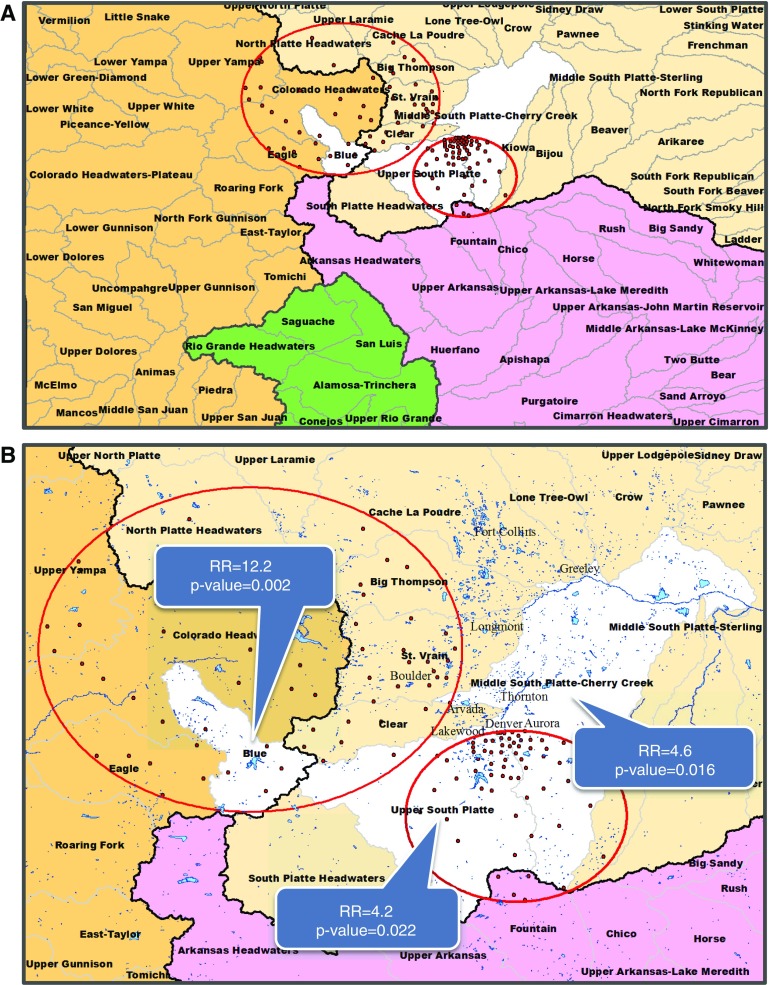
For infections caused by slowly growing NTM species, the RR of the Blue watershed was 12.2 compared with the mean risk of all watersheds. Consistent with the all NTM species group, it was the most significant watershed in the state, followed by the USP and MSPCC watersheds that had RRs of 4.2 and 4.6, respectively, compared with the mean risk of all 92 watersheds (Figures 3A and 3B). After controlling for multiple testing (three models), we found that the USP watershed did not exceed the threshold for significance.
Figure 3.
Significant watersheds for slowly growing nontuberculous mycobacteria (NTM) species. The four major watersheds in Colorado are represented by four colors: Missouri (beige), Upper Colorado (brown), Arkansas-White-Red (purple), and Rio Grande (green). Black lines represent watershed boundaries of the four major watersheds. Black line separating the Upper Colorado watershed (brown) from the remaining three watersheds represents the Continental Divide. Statistically significant watersheds are shown in white. (A) Four major watersheds are divided into 92 watersheds (boundaries delineated by gray lines) in Colorado. (B) Watersheds narrowed into region of interest. Watershed names are printed in boldface type. Major city names are printed in gray. Red circles indicate significant clusters of disease caused by slowly growing NTM infections. Red dots indicate centroid center of each ZIP code included in the significant cluster. Blue areas indicate lakes, reservoirs, and rivers. RR = risk ratio.
Rapidly growing NTM species
For rapidly growing species, every 1-SD increase in manganese soil concentration was associated with an RR of 1.2. For a 1,000-person increase in the average ZIP code–level population per square mile, the RR was 1.7 while all other variables were held constant.
Moran’s I test and Bayesian kriging model
After modeling the infections caused by all NTM species and slowly growing NTM species using the previously described environmental and sociodemographic variables, Moran’s I tests of residual spatial autocorrelation yielded nonsignificant P values, so the null hypothesis of no residual spatial autocorrelation could not be rejected. In other words, we could not detect evidence of additional clustering of NTM infections after accounting for the previously discussed risk factors, which suggests that the covariates in our model explain most of the variation in the inhomogeneous risk pattern of NTM cases in our study area. However, the model using patient infections caused by rapidly growing NTM species was significant. Consequently, we ran a Bayesian kriging model to control for the residual spatial autocorrelation present in these data. This model did not demonstrate statistical significance for any risk variable. Thus, no conclusions could be drawn regarding covariates related to infections caused by rapidly growing NTM species.
Discussion
NTM are opportunistic human pathogens that normally inhabit drinking water, natural water, and soils (36). However, epidemiological researchers have only begun to systematically assess the relative importance of these different habitats in the risk of infection and disease (14, 15). By using watersheds as a proxy for general water exposure, a statistically significant watershed effect could theoretically imply that exposure to water is a source of NTM infection in Colorado.
We found significant spatial variability in the prevalence of NTM disease using NJH patient data spanning from February 2008 through June 2015. Our SaTScan results revealed two statistically significant high-risk clusters. The primary cluster contained ZIP codes located in urban regions such as Denver, Aurora, and locations south of Denver. The risk of having NTM disease for people in ZIP codes inside the primary cluster was almost 2.5 times greater than the risk of people having NTM disease in ZIP codes outside the cluster. The secondary cluster included ZIP codes located within mountainous, rural, and more sparsely populated regions. The disease risk for individuals in ZIP codes located within the secondary cluster was over two times that of people in ZIP codes located outside the cluster.
SaTScan results can be used to assess possible disease transmission factors. Specifically, one can model the relationship between exposure variables and disease risk to see if the variables can adequately explain the pattern of disease distribution observed in the study area. The decision to use watersheds as an exposure variable was determined by having observed significant disease clusters on both sides of the Continental Divide, with the knowledge that water is pumped from west to east across the Continental Divide.
Our regression results indicated that soil acidity was significantly associated with increased disease risk, specifically for infections caused by the slowly growing NTM species. This finding is consistent with previous literature indicating that NTM preferentially grows in acidic soil (36, 37). Our findings also indicated that soil with low manganese concentrations was positively associated with infections caused by slowly growing infections. Adjemian and colleagues (14) reported a protective odds ratio for pulmonary NTM disease with increasing manganese soil concentrations. Our results for infections caused by the slowly growing species are consistent with their findings. Although their data could not separate infections caused by slowly growing from rapidly growing NTM species, the majority of their cases likely comprised infections caused by the slowly growing NTM species.
We also explored the role of soil texture (sand, silt, or clay) as a predictor of disease risk. Clay was significantly protective against disease for all NTM species, whereas silt was a significant risk factor. Clay and silt soils differ in their particle size and associated physical properties. The coarser-textured silt soils would be expected to be more permeable than clay soils and to have higher infiltration rates. These data suggest that perhaps NTM growth conditions are enhanced in regions where the interaction between soil and water can occur. The association of silt with infections caused by slowly growing species was only marginally significant. Because this variable did not influence the model for rapidly growing infections, the lower number of infections caused by slowly growing species (and therefore lower sample size) compared with all NTM species may in part explain the marginal significance.
Sociodemographic variables, as a combined set of variables, consistently demonstrated statistical significance in all models. Higher education and older age were significantly associated with disease risk at the ZIP code level, consistent with previous literature (14, 15, 38). Whereas education may simply be a proxy for access to medical care, older age may in fact represent a biological correlate of disease, where older age is a predisposing factor to illness.
Watershed Effect
For infections caused by slowly growing NTM species, the Blue watershed had an RR of 12.2 compared with the mean risk of all 92 watersheds in Colorado. In the USP and MSPCC watersheds, we observed RRs over 3 for all NTM species and over 4 for infections caused by slowly growing NTM species (Figures 2A and 2B).
Over 80% of all NTM species of patient isolates were slowly growing NTM species. Therefore, with a more homogeneous phenotype, the watershed estimates for infections caused by slowly growing species were more pronounced than what we observed for all NTM species. The Blue watershed consistently demonstrated the greatest increased risk of NTM disease in Colorado. After adjusting for sociodemographic, drive time, and soil variables, we observed that the Blue watershed effect increased further. The USP and MSPCC watersheds demonstrated the second and third highest significance levels in the state. Curiously, the Blue watershed contains rural and more sparely populated regions. Furthermore, it is mountainous and at higher elevation, with more precipitation and cooler temperatures. In contrast, the USP and MSPCC watersheds include more urban and densely populated regions. They are at lower elevation, and as a result these regions are dryer, with hotter temperatures and less precipitation.
The Dillon Reservoir (the main water supply for Denver) is located in the Blue watershed, and it is connected by a system of water tunnels to the USP and MSPCC watersheds, where the cities of Denver and Aurora are located (39, 40). Dillon Reservoir water is pumped across the Continental Divide through the Roberts Tunnel and enters the South Platte River. Water then gets diverted to purification facilities along the South Platte River before being delivered to the Denver area (Figure E1). Whereas the city of Aurora does not receive water directly from the Dillon Reservoir, Aurora water is mixed with Dillon Reservoir water in the South Platte River, effectively leading Aurora to receive the same water as Denver. Both the USP and MSPCC watersheds receive South Platte River water, which in large part originates from the Dillon Reservoir.
The Eagle and St. Vrain watersheds were also associated with increased disease risk for all NTM species. The St. Vrain watershed is connected to the Denver water supply via the Moffat Tunnel, where water is pumped from the west side of the Continental Divide at Winter Park into South Boulder Creek (Figure E2). The Eagle watershed association could be explained by easy travel and accessibility to the Blue watershed. A more heterogeneous phenotype (mixed infections caused by slowly growing and rapidly growing NTM species) makes the results from the all NTM species group less reliable than our findings for the slowly growing NTM species.
The association of the Blue, USP, and MSPCC watersheds with increased disease risk suggests that water pumped from the west side of the Continental Divide, possibly originating in the Dillon Reservoir, may be transporting NTM organisms into Denver and Aurora drinking-water treatment facilities. Denver Water and Aurora Water use chlorine, chlorine dioxide, and chloramines as microbe disinfectants. The literature confirms that slowly growing NTM species, specifically Mycobacterium avium and Mycobacterium intracellulare (over 90% of our slowly growing NTM population), have been recovered from drinking water distribution systems (41–44) and are highly resistant to chlorine, chloramine, and chlorine dioxide (42, 45–47). Taylor and coworkers (46) reported the disinfectant concentration and time to 99.9% inactivation of water-grown M. avium strains using chlorine dioxide and chlorine to be 100-fold greater and 580–2,300 times greater, respectively, than that of Escherichia coli. The use of these disinfectants, which kill other bacterial competitors, allows for NTM persistence in drinking-water distribution systems by creating an unoccupied niche where NTM are free to use any nutrients for their growth (36).
Because NTM are commonly found in water, especially in drinking-water distribution systems (24, 47–53) and tap water (54, 55), municipal water supplies may be viewed as a potential source of infection. Colorado represents a unique geographic and hydrologic system where, to a large extent, we can separate the potential effect of source water from that of municipal water by using watersheds as a unit of exposure. In contrast to most of the United States, where water is infrequently pumped across watersheds, the Front Range cities (Fort Collins, Boulder, Denver, Aurora, Colorado Springs, and Pueblo) receive a proportion of their water supply from different locations and different watersheds along the western slope of the Continental Divide. In other words, cities’ municipal water systems collect water from multiple watersheds, some of which are far from city limits.
The various municipal systems along the Front Range are similar with respect to types of disinfectants used, as well as the time period for which each of these systems was developed (J. Oropoza Fort Collins, email communication, April 2017. M. Wind, Boulder, email communication, April 2017, P. Benskin, Aurora, email communication, April 2017. Colorado springs utility website, https://www.csu.org/CSUDocuments/waterqualityreport2016.pdf. https://www.csu.org/CSUDocuments/watertour.pdf Denver Water utility website, https://www.denverwater.org/sites/default/files/2017-05/treated-water-quality-summary-report-2015_0.pdf. Pueblo utility website, https://www.pueblowater.org/images/pdfs/CCR2017.pdf). Despite these similarities, we did not observe disease clusters in other Front Range cities, except Denver and Aurora.
The Blue watershed contains a water source that is common to all watersheds that have demonstrated a statistically significant increased risk of disease in Colorado (Blue, USP, and MSPCC watersheds). Ultimately, if the belief is that individuals are exposed to NTM because these organisms exist in the water pipes, this study suggests that an environmental source is initiating the presence of NTM in the pipes, resulting in subsequent exposure.
Limitations
NTM is not a reportable condition in most U.S. states (56), and, as such, it is difficult to obtain a true population-based sample. Our referral hospital-based study population carries with it inherent limitations that may introduce bias. Because we do not have complete capture of cases, we underestimate the true risk of disease in Colorado. Patients with NTM disease who were resident in Colorado and were not referred to NJH for treatment may differ in factors related to disease severity, education and income levels, and insurance status. In an attempt to control for these differential referral biases across ZIP codes, we adjusted for sociodemographic variables in our models. Additionally, referral patterns between healthcare institutions are unknown because they depend strongly on medical specialties and the clinician social networks (C. Daley, oral communication, March 2017). Therefore, the NTM patient population at NJH may not be representative of the general NTM patient population in Colorado. These limitations should be acknowledged when interpreting our findings.
Because NJH is a national referral hospital for NTM treatment, our study population likely reflects the majority of severe and hard-to-treat cases that arise in Colorado, though we would not a priori expect the distribution of these cases to differ by watershed. Additionally, we did not use other NJH patients treated for a different disease as an offset in our Poisson model, because the at-risk population for NTM may be quite different from the at-risk population for another disease. We also included a drive-time variable to control for potential oversampling of patients residing in the Denver region. Last, we lacked information on duration of residence in ZIP codes. As a result, we could not exclude patients who moved to Colorado after diagnosis. This number of patients is small (G. Huitt, email communication, April 2017) and likely did not qualitatively affect our results.
Conclusions
Ecological risk factors for environmentally transmitted infections often receive little attention, despite the fact that they can be major predictors for disease (57). We identified two high-risk clusters of disease and high-risk watersheds located within the same geographic regions. These clusters and watersheds were identified via two separate analytic approaches.
Our regression findings implicate watersheds. That being said, we cannot infer on the basis of our data whether the Dillon Reservoir is a source of infection, whether rivers feeding into the reservoir carry an abundance of these organisms, or whether a source of infection exists at all. However, our data suggest that some unknown set of environmental conditions may be promoting NTM growth on the west side of the Continental Divide. Exploring watershed effects is the first step in determining whether exposure to water is a predictor of NTM disease. Further research is required to identify NTM in the water supply and to elucidate the specific water characteristics that may promote NTM growth, persistence, and subsequent infection.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Laura Medalie (U.S. Geological Survey) for assistance with ArcGIS software, Dr. David Wolock (U.S. Geological Survey) for providing soil data from the U.S. Department of Agriculture, and Madelyn Percy (University of North Carolina at Chapel Hill) for consultation on soil properties.
Footnotes
Supported by National Library of Medicine institutional training grant 5T15LM009451 from the National Institutes of Health (NIH) (E.M.L.), the Boettcher Foundation Webb Waring Award (J.R., M.S.), and NIH grant R01 CA157528 and National Science Foundation grant 1463642 (J.F.).
Author Contributions: E.M.L.: participated in the conception of the study; E.M.L. and J.R.: participated in the collection of the data; E.M.L., D.K., J.L.C., and J.F.: participated in the design of the study; E.M.L., J.L.C., and J.F.: participated in the analysis and interpretation; and E.M.L., J.L.C., J.F., D.K., J.R., and M.S.: drafted the manuscript for important intellectual content.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Marras TK, Daley CL. Epidemiology of human pulmonary infection with nontuberculous mycobacteria. Clin Chest Med. 2002;23:553–567. doi: 10.1016/s0272-5231(02)00019-9. [DOI] [PubMed] [Google Scholar]
- 2.Prevots DR, Shaw PA, Strickland D, Jackson LA, Raebel MA, Blosky MA, Montes de Oca R, Shea YR, Seitz AE, Holland SM, et al. Nontuberculous mycobacterial lung disease prevalence at four integrated health care delivery systems. Am J Respir Crit Care Med. 2010;182:970–976. doi: 10.1164/rccm.201002-0310OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winthrop KL, McNelley E, Kendall B, Marshall-Olson A, Morris C, Cassidy M, Saulson A, Hedberg K. Pulmonary nontuberculous mycobacterial disease prevalence and clinical features: an emerging public health disease. Am J Respir Crit Care Med. 2010;182:977–982. doi: 10.1164/rccm.201003-0503OC. [DOI] [PubMed] [Google Scholar]
- 4.Kendall BA, Winthrop KL. Update on the epidemiology of pulmonary nontuberculous mycobacterial infections. Semin Respir Crit Care Med. 2013;34:87–94. doi: 10.1055/s-0033-1333567. [DOI] [PubMed] [Google Scholar]
- 5.Hoefsloot W, van Ingen J, Andrejak C, Angeby K, Bauriaud R, Bemer P, Beylis N, Boeree MJ, Cacho J, Chihota V, et al. Nontuberculous Mycobacteria Network European Trials Group. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: an NTM-NET collaborative study. Eur Respir J. 2013;42:1604–1613. doi: 10.1183/09031936.00149212. [DOI] [PubMed] [Google Scholar]
- 6.Dirac MA, Horan KL, Doody DR, Meschke JS, Park DR, Jackson LA, Weiss NS, Winthrop KL, Cangelosi GA. Environment or host? A case–control study of risk factors for Mycobacterium avium complex lung disease. Am J Respir Crit Care Med. 2012;186:684–691. doi: 10.1164/rccm.201205-0825OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szymanski EP, Leung JM, Fowler CJ, Haney C, Hsu AP, Chen F, Duggal P, Oler AJ, McCormack R, Podack E, et al. Pulmonary nontuberculous mycobacterial infection: a multisystem, multigenic disease. Am J Respir Crit Care Med. 2015;192:618–628. doi: 10.1164/rccm.201502-0387OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prevots DR, Marras TK. Epidemiology of human pulmonary infection with nontuberculous mycobacteria: a review. Clin Chest Med. 2015;36:13–34. doi: 10.1016/j.ccm.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falkinham JO., III Surrounded by mycobacteria: nontuberculous mycobacteria in the human environment. J Appl Microbiol. 2009;107:356–367. doi: 10.1111/j.1365-2672.2009.04161.x. [DOI] [PubMed] [Google Scholar]
- 10.Billinger ME, Olivier KN, Viboud C, de Oca RM, Steiner C, Holland SM, Prevots DR. Nontuberculous mycobacteria-associated lung disease in hospitalized persons, United States, 1998–2005. Emerg Infect Dis. 2009;15:1562–1569. doi: 10.3201/eid1510.090196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adjemian J, Olivier KN, Seitz AE, Holland SM, Prevots DR. Prevalence of nontuberculous mycobacterial lung disease in U.S. Medicare beneficiaries. Am J Respir Crit Care Med. 2012;185:881–886. doi: 10.1164/rccm.201111-2016OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falkinham JO, III, Parker BC, Gruft H. Epidemiology of infection by nontuberculous mycobacteria. I. Geographic distribution in the eastern United States. Am Rev Respir Dis. 1980;121:931–937. doi: 10.1164/arrd.1980.121.6.931. [DOI] [PubMed] [Google Scholar]
- 13.Brooks RW, George KL, Parker BC, Falkinham JO, III, Gruff H. Recovery and survival of nontuberculous mycobacteria under various growth and decontamination conditions. Can J Microbiol. 1984;30:1112–1117. doi: 10.1139/m84-174. [DOI] [PubMed] [Google Scholar]
- 14.Adjemian J, Olivier KN, Seitz AE, Falkinham JO, III, Holland SM, Prevots DR. Spatial clusters of nontuberculous mycobacterial lung disease in the United States. Am J Respir Crit Care Med. 2012;186:553–558. doi: 10.1164/rccm.201205-0913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou MP, Clements AC, Thomson RM. A spatial epidemiological analysis of nontuberculous mycobacterial infections in Queensland, Australia. BMC Infect Dis. 2014;14:279. doi: 10.1186/1471-2334-14-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordan SJ, Benson WH. Sustainable watersheds: integrating ecosystem services and public health. Environ Health Insights. 2015;9(Suppl 2):1–7. doi: 10.4137/EHI.S19586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolok AS, Beseler CL, Chen XH, Shea PJ. The watershed as a conceptual framework for the study of environmental and human health. Environ Health Insights. 2009;3:1–10. doi: 10.4137/EHI.S1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tortoli E. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin Microbiol Rev. 2003;16:319–354. doi: 10.1128/CMR.16.2.319-354.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sapriel G, Konjek J, Orgeur M, Bouri L, Frézal L, Roux AL, Dumas E, Brosch R, Bouchier C, Brisse S, et al. Genome-wide mosaicism within Mycobacterium abscessus: evolutionary and epidemiological implications. BMC Genomics. 2016;17:118. doi: 10.1186/s12864-016-2448-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rindi L, Garzelli C. Genetic diversity and phylogeny of Mycobacterium avium. Infect Genet Evol. 2014;21:375–383. doi: 10.1016/j.meegid.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Ichikawa K, van Ingen J, Koh WJ, Wagner D, Salfinger M, Inagaki T, Uchiya K, Nakagawa T, Ogawa K, Yamada K, et al. Genetic diversity of clinical Mycobacterium avium subsp. hominissuis and Mycobacterium intracellulare isolates causing pulmonary diseases recovered from different geographical regions. Infect Genet Evol. 2015;36:250–255. doi: 10.1016/j.meegid.2015.09.029. [DOI] [PubMed] [Google Scholar]
- 22.Dai J, Chen Y, Lauzardo M. Web-accessible database of hsp65 sequences from Mycobacterium reference strains. J Clin Microbiol. 2011;49:2296–2303. doi: 10.1128/JCM.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bochner BR. Global phenotypic characterization of bacteria. FEMS Microbiol Rev. 2009;33:191–205. doi: 10.1111/j.1574-6976.2008.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Reyn CF, Waddell RD, Eaton T, Arbeit RD, Maslow JN, Barber TW, Brindle RJ, Gilks CF, Lumio J, Lähdevirta J, et al. Isolation of Mycobacterium avium complex from water in the United States, Finland, Zaire, and Kenya. J Clin Microbiol. 1993;31:3227–3230. doi: 10.1128/jcm.31.12.3227-3230.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.U.S. Census Bureau. Census 2010 summary file 1. September 2012 [accessed 2015 July 7]. Available from: https://factfinder.census.gov/faces/nav/jsf/pages/searchresults.xhtml?refresh=t.
- 26.U.S. Census Bureau. American Community Survey 5-year dataset. 2013. [accessed 2015 Jul 7]. Available from: https://www.census.gov/programs-surveys/acs/technical-documentation/table-and-geography-changes/2015/5-year.html.
- 27.U.S. Department of Agriculture, Natural Resources Conservation Service, National Soil Survey Center. State Soil Geographic (STATSGO) database. 1991. [accessed 2015 Jun 15]. Available from: https://water.usgs.gov/GIS/metadata/usgswrd/XML/ds866_ssurgo_variables.xml.
- 28.U.S. Geological Survey. National Geochemical Survey database. 2012. [accessed 2016 Feb 8]. Available from: https://mrdata.usgs.gov/geochem/doc/averages/countydata.htm.
- 29.U.S. Geological Survey. National Water Use Information Program. [Accessed 2015 Jun 12]. Available from: http://waterdata.usgs.gov/nwis/wu.
- 30.U.S. Department of Agriculture, Natural Resources Conservation Service (USDA-NRCS) The Watershed Boundary Dataset (WBD) [Accessed 2015 Jun 12]. Available from: https://www.nrcs.usda.gov/wps/portal/nrcs/main/national/water/watersheds/
- 31.Bivand R, Piras G. Comparing implementations of estimation methods for spatial econometrics. J Stat Softw. 2015;63(18):1–36. [Google Scholar]
- 32.Gelman A, Hill J. Data analysis using regression and multilevel/hierarchical models. Cambridge, UK: Cambridge University Press; 2006. [Google Scholar]
- 33.Kahle D, Wickham H. ggmap: spatial visualization with ggplot2. R J. 2013;5:144–161. [Google Scholar]
- 34.Christensen OF, Ribeiro P., Jr geoRglm: a package for generalized linear spatial models. R News. 2002;2:26–28. [Google Scholar]
- 35.Waller LA, Gotway CA. Applied spatial statistics for public health data. Hoboken, NJ: Wiley Interscience; 2004. [Google Scholar]
- 36.Falkinham JO., III Environmental sources of nontuberculous mycobacteria. Clin Chest Med. 2015;36:35–41. doi: 10.1016/j.ccm.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 37.Portaels F, Pattyn SR. Growth of mycobacteria in relation to the pH of the medium. Ann Microbiol (Paris) 1982;133:213–221. [PubMed] [Google Scholar]
- 38.Prevots DR, Adjemian J, Fernandez AG, Knowles MR, Olivier KN. Environmental risks for nontuberculous mycobacteria: individual exposures and climatic factors in the cystic fibrosis population. Ann Am Thorac Soc. 2014;11:1032–1038. doi: 10.1513/AnnalsATS.201404-184OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denver Water. [Accessed 2016 Jun 1]. Available from: http://www.denverwater.org/SupplyPlanning/WaterSupply/
- 40.Denver Water. Water watch report. [Accessed 2016 Jun 1]. Available from: https://www.denverwater.org/sites/default/files/water-watch-report.pdf.
- 41.Covert TC, Rodgers MR, Reyes AL, Stelma GN., Jr Occurrence of nontuberculous mycobacteria in environmental samples. Appl Environ Microbiol. 1999;65:2492–2496. doi: 10.1128/aem.65.6.2492-2496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carson LA, Bland LA, Cusick LB, Favero MS, Bolan GA, Reingold AL, Good RC. Prevalence of nontuberculous mycobacteria in water supplies of hemodialysis centers. Appl Environ Microbiol. 1988;54:3122–3125. doi: 10.1128/aem.54.12.3122-3125.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pelletier PA, du Moulin GC, Stottmeier KD. Mycobacteria in public water supplies: comparative resistance to chlorine. Microbiol Sci. 1988;5:147–148. [PubMed] [Google Scholar]
- 44.Fischeder R, Schulze-Röbbecke R, Weber A. Occurrence of mycobacteria in drinking water samples. Zentralbl Hyg Umweltmed. 1991;192:154–158. [PubMed] [Google Scholar]
- 45.Falkinham JO., III Factors influencing the chlorine susceptibility of Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum. Appl Environ Microbiol. 2003;69:5685–5689. doi: 10.1128/AEM.69.9.5685-5689.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor RH, Falkinham JO, III, Norton CD, LeChevallier MW. Chlorine, chloramine, chlorine dioxide, and ozone susceptibility of Mycobacterium avium. Appl Environ Microbiol. 2000;66:1702–1705. doi: 10.1128/aem.66.4.1702-1705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.King DN, Donohue MJ, Vesper SJ, Villegas EN, Ware MW, Vogel ME, Furlong EF, Kolpin DW, Glassmeyer ST, Pfaller S. Microbial pathogens in source and treated waters from drinking water treatment plants in the United States and implications for human health. Sci Total Environ. 2016;562:987–995. doi: 10.1016/j.scitotenv.2016.03.214. [DOI] [PubMed] [Google Scholar]
- 48.Lecuona M, Abreu R, Rodríguez-Álvarez C, Castro B, Campos S, Hernández-Porto M, Mendoza P, Arias A. First isolation of Mycobacterium canariasense from municipal water supplies in Tenerife, Canary Islands, Spain. Int J Hyg Environ Health. 2016;219:48–52. doi: 10.1016/j.ijheh.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 49.Amha YM, Anwar MZ, Kumaraswamy R, Henschel A, Ahmad F. Mycobacteria in municipal wastewater treatment and reuse: microbial diversity for screening the occurrence of clinically and environmentally relevant species in arid regions. Environ Sci Technol. 2017;51:3048–3056. doi: 10.1021/acs.est.6b05580. [DOI] [PubMed] [Google Scholar]
- 50.Briancesco R, Alaimo C, Bonanni E, Delle Site A, Di Gianfilippo F, Grassano L, Moscatelli R, Ottaviano C, Paradiso R, Quintiliani S, et al. An Italian investigation on non-tuberculous mycobacteria in an urban water supply. Ann Ig. 2014;26:264–271. doi: 10.7416/ai.2014.1984. [DOI] [PubMed] [Google Scholar]
- 51.Thomson RM, Carter R, Tolson C, Coulter C, Huygens F, Hargreaves M. Factors associated with the isolation of nontuberculous mycobacteria (NTM) from a large municipal water system in Brisbane, Australia. BMC Microbiol. 2013;13:89. doi: 10.1186/1471-2180-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sartori FG, Leandro LF, Montanari LB, de Souza MG, Pires RH, Sato DN, Leite CQ, de Andrade Prince K, Martins CH. Isolation and identification of environmental mycobacteria in the waters of a hemodialysis center. Curr Microbiol. 2013;67:107–111. doi: 10.1007/s00284-013-0341-6. [DOI] [PubMed] [Google Scholar]
- 53.Le Dantec C, Duguet JP, Montiel A, Dumoutier N, Dubrou S, Vincent V. Occurrence of mycobacteria in water treatment lines and in water distribution systems. Appl Environ Microbiol. 2002;68:5318–5325. doi: 10.1128/AEM.68.11.5318-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gangadharam PR, Lockhart JA, Awe RJ, Jenkins DE. Letter: Mycobacterial contamination through tap water. Am Rev Respir Dis. 1976;113:894. doi: 10.1164/arrd.1976.113.6.894. [DOI] [PubMed] [Google Scholar]
- 55.Chang CT, Wang LY, Liao CY, Huang SP. Identification of nontuberculous mycobacteria existing in tap water by PCR-restriction fragment length polymorphism. Appl Environ Microbiol. 2002;68:3159–3161. doi: 10.1128/AEM.68.6.3159-3161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Totten SE, Marola JL, Salfinger M. Review of state reporting requirements for tuberculosis and nontuberculous mycobacteria: a case for more uniformity; Presented at the 2015 American Society for Microbiology Meeting. May 31, 2015, New Orleans, LA. [Google Scholar]
- 57.Diez Roux AV. The study of group-level factors in epidemiology: rethinking variables, study designs, and analytical approaches. Epidemiol Rev. 2004;26:104–111. doi: 10.1093/epirev/mxh006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.