Abstract
Background
Epidemiologists have long used case–control and related study designs to enhance variability of response and information available to estimate exposure–disease associations. Less has been done for longitudinal data.
Methods
We discuss an epidemiological study design and analysis approach for longitudinal binary response data. We seek to gain statistical efficiency by over–sampling relatively informative subjects for inclusion into the sample. In this methodological demonstration, we develop this concept by sampling repeatedly from an existing cohort study to estimate the relationship of chronic obstructive pulmonary disease to past–year smoking in a panel of baseline smokers. To account for over–sampling, we describe a sequential offsetted regressions approach for valid inferences in this setting.
Results
Targeted sampling can lead to increased statistical efficiency when combined with sequential offsetted regressions. Efficiency gains are degraded with increased prevalence of the disease response variable, with decreased association between the sampling variable and the response, and with other design and analysis parameters, providing guidance to those wishing to use these types of designs in the future.
Conclusions
These designs hold promise for efficient use of resources in longitudinal cohort studies.
Keywords: auxiliary variable dependent sampling, binary data, case-control, Generalized Estimating Equations, longitudinal data, logistic regression, marginal models, outcome-dependent sampling, outcome-related sampling
INTRODUCTION
Study designs using enhanced sampling to detect associations are ubiquitous in epidemiological research. The case–control study [1, 2, 3] is a very commonly applied design, and from its principle of enriching the sample with increased response (and possibly exposure) variability, many other efficient designs have emerged, including the nested case–control design [4], the case–cohort design [5], the case–crossover design [6, 7], and various two–phase designs [8, 9, 10].
We discuss a class of enhanced sampling study designs for longitudinal, binary response data wherein subjects are sampled prospectively with probability depending on an auxiliary variable measured at screening (or baseline) that is related to the longitudinal response. Sampled subjects are then followed over time. Even though the auxiliary sampling variable is not of interest for analyses, due to its relationship with the outcome, sampling based on it can improve efficiency and precision by increasing the event rate in the sample. A supporting aim is to describe and illustrate methods of data analysis tailored to these study designs.
To support this methodologic demonstration, we exploit an existing cohort, as subset of participant from the Lung Health Study, on the natural history of cardiovascular disease in smokers, and consider the hypothetical scenario wherein complete data are available on only a subset of needed variables measured at baseline. Then, a targeted subsample of participants from the parent cohort is drawn and referred for assessment on the entire panel of variables needed for analysis.
As an illustrative example, suppose that the aim of our study is to estimate the effect of current (past year) smoking on a time–varying chronic obstructive pulmonary disease (COPD) diagnosis. We consider the absence of COPD according to a standard clinical definition and, for illustrative purposes, severe COPD, to be defined later. Because the Lung Health Study participants tended to have mild COPD, both binary outcomes under study (COPD absence and severe COPD) are somewhat rare in the Lung Health Study cohort. This is important because, analogous to case–control studies, our designs provide the greatest benefits in precision when outcomes are rare.
In a scientifically ideal setting, individuals would be assigned to smoking or non–smoking status each year through a structured random process controlled by the investigator, as subjects were followed over time. Assuming participants adhered to their assigned smoking status, such a design would yield unambiguous estimates of the causal effect of smoking. Because this is of course not possible, we exploit available covariates to control confounding to the extent possible in an observational study. In particular, we adjust for 1–year lag of current smoking to account for time–dependent confounding between respiratory symptoms and current smoking, as well as other potential time–varying and time–invariant confounders described later in the paper.
METHODS
The Lung Health Study: Participants and Measures
The Lung Health Study [11, 12] is a 10–center randomized clinical trial on smokers with mild COPD. Lung Health Study protocols were approved by the institutional review boards at each clinical center and participants were enrolled after written informed consent was obtained. The trial was designed to test the efficacy of a smoking intervention program and the use of an inhaled bronchodilator to slow the rate of pulmonary decline over time. Data from the first five annual clinic visits in Lung Health Study are available at the National Center for Biotechnology Information database of genotypes and phenotypes ([13]; accession number phs000335.v2.p2; https://www.ncbi.nlm.nih.gov/gap). Links to instructions for downloading data are provided in the eAppendix.
Table 1 summarizes the Lung Health Study data after modest data cleaning. The original dataset contained N = 4391 subjects, and after cleaning, N = 4213 (96%) remain. Among those excluded, 78 died during follow–up. The rest were removed due to lack of available spirometry data. Whereas we recognize that bias could arise with these exclusion criteria, we felt the impact would be minimal, particularly for purposes of the present methodological investigation.
Table 1.
Demographics and features of the Lung Health Study cohort at baseline and over the course of five annual visits to study clinics: Continuous variables are summarized with the [5: 25: 50: 75: 95]th percentiles and categorical variables are summarized with proportions. ICC is the intraclass correlation coefficient for variables measured longitudinally.
| Baseline data | Longitudinal follow–up data | ||||||
|---|---|---|---|---|---|---|---|
| Year 1 | Year 2 | Year 3 | Year 4 | Year 5 | ICC | ||
| Number of participants | 4213 | ||||||
| Site 1–2–3–4–5 | 365–402–388–459–492 | ||||||
| Site 6–7–8–9–10 | 428–425–429–463–362 | ||||||
| Female | 0.37 | ||||||
| Age (years) | [37: 43: 49: 54: 58] | ||||||
| BMI (kg/m2) | [20: 23: 25: 28: 32] | ||||||
| Pack years | [17: 28: 37: 49: 76] | ||||||
| Cigarettes per day | [10: 20: 30: 40: 55] | ||||||
| Education | [10: 12: 14: 14: 20] | ||||||
| Chronic Bronchitisa | 0.04 | ||||||
| Asthmaa | 0.03 | ||||||
| COPD free | 0.21 | ||||||
| Severe COPD | 0.11 | ||||||
| Number observed | 4003 | 4022 | 3992 | 3958 | 4169 | ||
| Current smoker | 0.70 | 0.69 | 0.67 | 0.65 | 0.65 | 0.74 | |
| Dust at work | 0.28 | 0.29 | 0.27 | 0.26 | 0.25 | 0.62 | |
| COPD free | 0.23 | 0.21 | 0.19 | 0.18 | 0.16 | 0.71 | |
| Severe COPD | 0.12 | 0.14 | 0.16 | 0.18 | 0.21 | 0.79 | |
denotes that asthma and chronic bronchitis were, in fact, measured at year 1 as opposed to at the baseline screening visit. For the sake of studying the exposure and auxiliary variable stratified sampling design, we assume they were measured at baseline.
We consider a hypothetical study that seeks to estimate the effect of past–year smoking on current COPD, a longitudinal binary response realized at each of five annual clinic visits, defined for illustrative purposes in two different ways. In the Lung Health Study, spirometry was conducted during the screening visit and at all annual follow–up visits. A clinical diagnosis of COPD is realized if the ratio of post–bronchodilator forced expiratory volume in the first second of exhalation (FEV 1) to forced vital capacity (FV C) is less than 0.7; as such we define COPD absence as occurring when FEV 1/FVC ≥ 0.7. COPD is absent in approximately 21% of subjects at the screening visit and 23% of subjects at the first follow–up visit (Table 1). This is a standard definition for absence of COPD (e.g., see http://www.goldcopd.org). A second outcome, generated for methodological demonstration purposes to be rare, was severe COPD, defined at each visit as having FEV 1/FVC < 0.57. Severe COPD occurs in 11% of subjects at the screening visit and 12% of subjects at the first follow–up visit. Whereas this is not a standard definition of severe COPD, it provides a useful platform for illustrating the impact of prevalence on the efficiency of our design.
Risk factors for COPD available in the Lung Health Study database and used in analyses include (Table 1): gender, age, years of education, height, weight, and lifetime smoking status (in pack years) were collected at baseline. We use years of education as a surrogate for socioeconomic status. Through a short annual survey, dust exposure at work, asthma symptoms during the previous year, and chronic bronchitis are assessed in each wave. In annual clinic visits, current smoking is assessed via a cotinine test. Even though asthma and chronic bronchitis were measured over time, for purposes of a stratified design discussed later, we assume they are time–fixed and only use their values measured at the first follow–up visit.
Subsampling from the Cohort
Although Lung Health Study data are available on N = 4213 subjects, for methodological demonstration, we consider a hypothetical but realistic scenario wherein resources limit sampling to only n = 800 participants from the baseline sample for longitudinal follow up. Sampling may be simple random sampling of n = 800. However, with either of the two COPD endpoints, the response prevalence at any wave is fairly small, limiting information in the resulting data set.
To address this limitation, sampling may be enhanced by basing it on an auxiliary variable measured at or before baseline that is related to the COPD endpoints over time (auxiliary variable sampling). Here, we define auxiliary sampling variables to be the binary indicators of COPD absence (FEV 1/FVC ≥ 0.7) or of severe COPD (FEV 1/FVC < 0.57) at screening. In our particular implementation of auxiliary variable sampling, our goal was to sample approximately equal numbers of high– and low–risk subjects. For COPD–absence, we sampled independently from 896 high–risk subjects (i.e., COPD–absence at screening) with probability 400/896 = 0.45 and from 3317 low–risk subjects (with COPD at screening) with probability 400/3317 = 0.12. For the rarer, severe COPD outcome, we sampled high–risk participants with probability 400/445 = 0.90 and low–risk participants with probability 400/3768 = 0.11. Two key features of auxiliary variable sampling are: (i) investigator–specified sampling probabilities and (ii) oversampling of “high risk” subjects for whom the auxiliary variable equals 1. This oversampling serves to increase the observed response prevalence across waves, thereby enriching information in the sample for quantifying associations with risk factors of interest.
Sampling which also depends on a key covariate, usually rare, can additionally increase statistical efficiency for the coefficient of that covariate or its correlates. We conducted such an exposure and auxiliary variable sampling design using four strata defined by baseline COPD (either absence or severe) and baseline asthma. We sampled with probability 1.0 all subjects with asthma, and allocated the remainder of sampled subjects equally between those with and without baseline COPD absence or severe COPD, and without asthma at baseline (Table 2).
Table 2.
Exposure and auxiliary variable sampling probabilities based on chronic obstructive pulmonary disease and asthma at baseline in the Lung Health Study cohort.
| COPD–Free | ||
|---|---|---|
|
|
||
| Asthma | Yes | No |
| Yes | 17/17 = 1 | 97/97 = 1 |
| No | 343/924 = 0.37 | 343/3175 = 0.11 |
|
| ||
| Severe COPD | ||
|
|
||
| Asthma | Yes | No |
|
| ||
| Yes | 23/23 = 1 | 91/91 = 1 |
| No | 343/502 = 0.68 | 343/3597 = 0.095 |
Statistical Analysis
Regardless of sampling design, the n = 800 sampled participants will each yield up to five annual waves of data, with the dichotomous time–varying endpoint Yij being either COPD absence or severe COPD for participant i at visit j. Under a random sampling design, analysis proceeds using a population average logistic regression model across the five visits, estimated with generalized estimating equations (GEE) [14]. In this model, the set of predictors includes the time–varying current smoking indicator—the exposure of interest; time–varying adjustors included to control confounding, including 1–year lag of current smoking; and time–invariant baseline variables, also included to control confounding; complete model specification is in Table 3. Note that, because all participants are smokers at baseline, the 1–year lagged value of current smoking is 1 at wave 1 for all participants.
Table 3.
Results from Lung Health Study data analyses: full cohort (FC) analyses and auxiliary variable sampling and random sampling results averaged across 250 replicates. RS=random sampling, AVS=auxiliary variable sampling, SOR=sequential offsetted regressions, and IPW=inverse probability weighting with (known–by–design) weights 1/π(Zi)). We show logistic regression parameter estimates and standard errors (in parentheses) for the FC, and we show their average parameter and standard error estimates across 250 replicates for the RS and AVS designs. Models also included 9 dummy variables to adjust for differences across the 10 centers. For all analyses, GEE with a working independence correlation model was used.
| Covariate | FC | random sampling | AVS:SOR | AVS:IPW |
|---|---|---|---|---|
| COPD–Free analysis | ||||
| Time–varying covariates | ||||
| Current Smoker | −0.33 (0.06) | −0.34 (0.13) | −0.29 (0.11) | −0.34 (0.13) |
| Current Smoker (lag) | −0.35 (0.05) | −0.36 (0.11) | −0.37 (0.10) | −0.35 (0.11) |
| Study Year | −0.14 (0.01) | −0.15 (0.02) | −0.15 (0.02) | −0.14 (0.03) |
| Dust | −0.16 (0.06) | −0.16 (0.15) | −0.18 (0.13) | −0.15 (0.15) |
| Time–fixed covariates | ||||
| Female (vs male) | 0.28 (0.08) | 0.27 (0.18) | 0.27 (0.15) | 0.29 (0.17) |
| Baseline Age (per 10–year change) | −0.68 (0.06) | −0.68 (0.14) | −0.71 (0.12) | −0.70 (0.14) |
| Pack years (per 20–pack year change) | −0.09 (0.05) | −0.09 (0.12) | −0.07 (0.10) | −0.09 (0.12) |
| Cigarettes/day (per 10–cigs/day change) | −0.07 (0.03) | −0.07 (0.07) | −0.07 (0.06) | −0.07 (0.07) |
| Education (per 4–year change) | 0.01 (0.05) | 0.02 (0.12) | 0.03 (0.10) | 0.01 (0.12) |
| Baseline BMI (per 5 – kg/m2 change) | 0.23 (0.04) | 0.24 (0.10) | 0.19 (0.09) | 0.24 (0.10) |
| Chronic bronchitis | −0.22 (0.18) | −0.24 (0.43) | −0.43 (0.36) | −0.24 (0.41) |
| Asthma | −0.67 (0.25) | −0.74 (0.60) | −0.62 (0.50) | −0.70 (0.55) |
|
| ||||
| Severe COPD analysis | ||||
| Time–varying covariates | ||||
| Current Smoker | 0.34 (0.07) | 0.36 (0.15) | 0.35 (0.11) | 0.34 (0.15) |
| Current Smoker (lag) | 0.36 (0.06) | 0.37 (0.13) | 0.37 (0.09) | 0.37 (0.14) |
| Study Year | 0.19 (0.01) | 0.19 (0.02) | 0.18 (0.03) | 0.20 (0.03) |
| Dust | 0.04 (0.07) | 0.04 (0.17) | −0.03 (0.13) | 0.05 (0.17) |
| Time–fixed covariates | ||||
| Female (vs male) | −0.24 (0.09) | −0.26 (0.21) | −0.22 (0.16) | −0.26 (0.21) |
| Baseline Age (per 10–year change) | 0.69 (0.07) | 0.72 (0.16) | 0.69 (0.12) | 0.71 (0.16) |
| Pack years (per 20–pack year change) | 0.11 (0.05) | 0.12 (0.11) | 0.12 (0.09) | 0.12 (0.11) |
| Cigarettes/day (per 10–cigs/day change) | 0.04 (0.03) | 0.04 (0.07) | 0.01 (0.06) | 0.03 (0.07) |
| Education (per 4–year change) | −0.11 (0.06) | −0.12 (0.13) | −0.07 (0.10) | −0.11 (0.13) |
| Baseline BMI (per 5 – kg/m2 change) | −0.33 (0.06) | −0.34 (0.13) | −0.35 (0.10) | −0.35 (0.14) |
| Chronic bronchitis | 0.08 (0.18) | 0.08 (0.44) | 0.10 (0.33) | 0.11 (0.42) |
| Asthma | 0.80 (0.22) | 0.77 (0.52) | 0.68 (0.41) | 0.82 (0.49) |
Importantly, all analyses used a GEE working independence covariance structure due to the likely violation of the full covariate conditional mean assumption [15, 16], wherein the cross–sectional model of interest, namely COPD prevalence at time j as a function of baseline predictors and predictors also measured at visit j (plus lagged smoking) does not equal the full covariate conditional prevalence, namely COPD prevalence as a function of predictors measured at all visit times and taken as an ensemble. This is also referred to as a ‘no–interference’ assumption in the causal inference literature.
Under auxiliary variable sampling, some minimal notation is required to formalize the design and analysis. These study designs rely on an auxiliary variable—in this case, screening COPD absence or severe COPD—upon which sampling is based. Denote this variable Zi. Typically, Zi = 1 will indicate a high risk (for Yij = 1) stratum, and Zi = 0 a low–risk stratum. While not of direct scientific interest, if well chosen, Zi will be highly related to Yij for all j, thereby yielding enriched response prevalence (and variability), and more efficient study designs. Formally, we sample individuals with investigator–specified probability π(Zi). When the outcome is rare, π(1) will be high compared to π(0).
In addition to measuring and sampling based on Zi, in some cases we also sample based an additional time–invariant exposure (predictor) variable Vi available at baseline. Such an exposure and auxiliary variable sampling design can further improve efficiency when Vi is rare, by increasing exposure prevalence and, thereby, (co)variability between Yij and Vi. Formally, we sample from four strata (0, 0), (0, 1), (1, 0), (1, 1) with probabilities π(Zi, Vi). In most cases, π(0, 0) is low, π(1, 1) is high, and π(1, 0) and π(0, 1) depend on the relative prevalence of Zi and Vi. In our methodological demonstration, we implemented such a strategy, letting Vi indicate baseline asthma, and sampling to achieve equal numbers of subjects in the other strata defined jointly by asthma and either COPD absence or severe COPD (Table 2).
Analysis approach 1: Sequential offsetted regressions
Sequential offsetted regressions ([17]) is an analysis procedure for the designs proposed in this paper. We refer interested readers to the eAppendix and to Schildcrout and Rathouz for details [17]. Briefly, with sequential offsetted regressions, we conduct two offsetted logistic regressions to estimate parameters of scientific interest. The first, auxiliary variable regression, captures the relationship of auxiliary variable Zi to observed endpoint and predictor data (Yij,Xij). The second logistic regression is for the question of scientific interest. It is corrected for the biased auxiliary variable sampling or exposure and auxiliary variable sampling sampling design.
To understand the first logistic regression, let Si = 1 (or 0) indicate whether the ith participant in the parent cohort is (is not) sampled for longitudinal follow–up. Then, note from Bayes’ Theorem that
| (1) |
where the last factor is free of (Yij,Xij) because sampling only depends on Zi. The lefthand side is the disease odds model for Zi under the auxiliary variable sampling sample, while the first factor on the righthand side is the corresponding disease odds model in the population, i.e., the true model for Zi. It is also true that the last factor is known by design to the investigator and is equal to π(1)/π(0). Because equivalence (1) is in terms of the odds, a population logistic regression (i.e., log odds) model for the association of Zi to (Yij,Xij) will result in a sample logistic regression model of the same form, with the addition of log{π(1)/π(0)} as an offset term. Schildcrout and Rathouz [17] provide the extension to the case where sampling is also based on a stratifying covariate Vi. Sequential offsetted regressions is implemented in a Stata and R packages (available for download from http://biostat.mc.vanderbilt.edu/wiki/Main/ODSandLDA), wherein specifying log{π(1)/π(0)} = 0 yields standard GEE.
In our hypothetical auxiliary variable sampling study, we specified the auxiliary model to include the disease response Yij, and all the same predictors Xij as in the model of interest (see Table 3). Additionally, because of the potential for a weakening relationship of Yij to screening Zi with increasing j, we included the interaction between Yij and annual visit number (study year).
Once Pr(Zi = 1|Yij,Xij) is known and available, it can be combined with Pr(Si = 1|Zi) (which equals Pr(Si = 1|Zi, Yij,Xij)) to obtain Pr(Si = 1|Yij,Xij). This in turn is used to compute
for every observation in the data set.
The second logistic regression follows the same pattern as the first one, but this one is aimed at the scientific question of interest in the population. Specifically,
| (2) |
Again, in (2), because both the sample and the population model are in terms of the disease odds, a logistic regression model for disease Yij in the sample can be specified in terms of the same logistic regression model in the population, with the addition of log{B(Xij)} as an offset term. In our work, we have estimated this model using GEE with independence correlation structure, although under certain conditions, an alternative correlation structure such as exchangeable or AR(1) is allowed and could further increase statistical efficiency.
Analysis approach 2: Inverse probability weighting
Inverse probability weighting (IPW) is commonly applied when samples are intentionally (by design) or unintentionally (missing data) not representative of the source population [18, 19]. An alternative to sequential offsetted regressions, IPW is implemented by weighting each subject i (or, equivalently each observation) by the known value 1/π(Zi, Vi) to approximate the population represented by the original Lung Health Study cohort, and estimating the population model using GEE, again with an independence correlation structure. Whereas IPW is easier to implement than sequential offsetted regressions, in these designs, as we shall see, it can result in marked loss of statistical efficiency.
Comparative Analyses
Our primary analysis is of the Lung Health Study data generated under our hypothetical auxiliary variable sampling design, and analyzed using sequential offsetted regressions. As our primary aim is to demonstrate the strength and features of this methodology “on average,” we replicate the process (using auxiliary variable sampling to sample from the full cohort and using sequential offsetted regressions for analysis of sampled data) 250 times and average the results. We compare the results to those obtained from the full cohort of N = 4213, and, additionally from those obtained under a random sampling design and under the auxiliary variable sampling design analyzed with IPW. Specifically, for each replicate, each participant is either sampled with probability 800/4213 for the random sampling design, or with auxiliary variable sampling probabilities given in the Subsampling from the Cohort subsection. On average, each sample includes n = 800 participants.
Design features within auxiliary variable sampling
Whereas auxiliary variable sampling can be a powerful method for increasing statistical efficiency while controlling expenditure of research resources, there are several key features of any auxiliary variable sampling design which can impact efficiency, some of which may be under control of the investigator, and all of which should be given careful consideration. We quantify these effects, using the actual Lung Health Study data [20], to explore three data and two design and analysis features. Similar to sensitivity analysis, for each feature studied, we perturb that single feature of the original data/design/analysis, leaving the rest intact, to evaluate the impact on results.
The three data features we study are: 1) overall prevalence of the response Pr(Yij = 1), which we alter by changing the cut–point that defines COPD–free and severe COPD outcomes, 2) strength of the relationship between Zi and Yi1, i.e. OR(Yi1 | Zi), which we alter by perturbing the original Zi values, and 3) correlation among repeated measurements within each subject r(Yij, Yik), which we alter by regenerating Yi2, … Yini using a fully parametric, marginalized model [21, 22, 23] estimated from the full cohort. Additionally, we examine the impact of using a more saturated auxiliary model for Pr(Zi = 1|Yij,Xij) which includes not only Yij and all predictors in Xij but also all two–way interactions between Yij and each predictor. Finally, we consider an exposure (Vi) and auxiliary variable (Zi) sampling plan (sampling with probability π(Zi, Vi)), where Vi indicates asthma as a key predictor at baseline.
We use the inverse of sampling variance to quantify statistical efficiency. That is, the larger the sampling variance, the lower the efficiency. As such, we define relative variance (RV) as the ratio of average estimated variances,
Values greater than one are consistent with efficiency gains in auxiliary variable sampling with sequential offsetted regressions compared to random sampling. We also note that the estimators of the sampling variances are consistent for the true sampling variances [17], capturing components of variability due both to sampling the full cohort from the reference population, and to subsampling from the full cohort. To put efficiency gains of the design into context, 800 of the 4213 subjects from the Lung Health Study are randomly sampled under the random sampling design. Thus the efficiency of the full cohort analysis to the random sampling design analysis is 4123/800 = 5.26. As a final note, we conducted a distinct study wherein n = 500 (not 800) participants were sampled. The values of RV were qualitatively similar (see eAppendix).
RESULTS
In Table 3, we display logistic regression parameter and standard error estimates from full cohort analyses and average logistic regression parameter estimates and average estimated standard errors across 250 replicates from the auxiliary variable sampling and random sampling design–based analyses. As noted above, these standard error estimates capture all components of sampling variability. Focusing first on full cohort analyses for both outcomes, we can see (Table 3) that nearly all independent variables were associated with outcomes, and as expected, estimates have the opposite sign for the two outcomes. Overall, COPD is more severe in current smokers than in non–smokers, even accounting for past smoking, as measured jointly by all three of pack years at baseline, cigarette per day at baseline, and one–year lag of current smoking. The odds ratio for COPD–free and for severe COPD for smokers versus non–smokers is an estimated exp(−.33) = 0.72 and exp(0.34) = 1.41, respectively. Additionally, even allowing for variations in smoking, COPD is increasing over time: The estimated per year odds ratios of being COPD–free and for severe COPD are exp(−0.14) = 0.87 and exp(0.19) = 1.21, respectively.
Examining results from the random sampling and auxiliary variable sampling designs, we observe the following: First, for the most part, point estimates appear to be similar across the various design and analysis procedures, implying that these procedures reproduced the full cohort results quite well, on average. As expected, random sampling estimates are very similar to those from the full cohort. Auxiliary variable sampling with sequential offsetted regressions and auxiliary variable sampling with inverse probability weighting vary a bit more from the full cohort, but almost always by less than a half of a standard error. Second, random sampling and auxiliary variable sampling with inverse probability weighting produce similar average standard error estimates across replicates. Third, auxiliary variable sampling with sequential offsetted regressions produces lower average standard error estimates than the random sampling design and auxiliary variable sampling with inverse probability weighting. Finally, the increases in efficiency due to the auxiliary variable sampling with sequential offsetted regressions are more pronounced in the rarer, severe COPD outcome analysis, as compared to the COPD–free outcome analysis.
Design features within auxiliary variable sampling
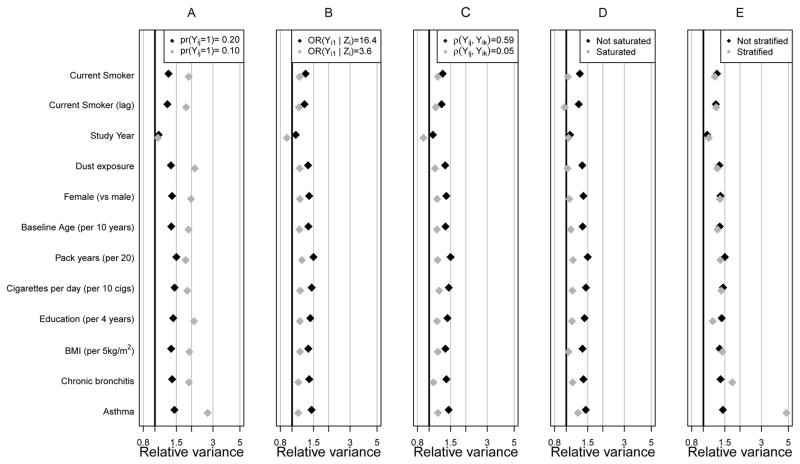
First, we focus on the COPD–free results (Figure 1). In the original Lung Health Study data analysis, denoted with black diamonds in all panels, we used a cutoff of FEV1/FVC ≥ 0.70 to define COPD–absence, resulting in Pr(Yij = 1) = 0.20, OR(Yi1 | Zi) ~ 16.4, and r(Yij, Yik) ~ 0.59. Relative variance (RV), reflecting relative efficiency, ranged from 1.09 (study year) to 1.46 (asthma) across all regression parameters, indicating 9 to 46 percent more subjects are required under a random sampling design to obtain the same precision we obtain with auxiliary variable sampling with sequential offsetted regressions. Response prevalence had a dramatic impact on RV (panel A). With cutoffs for FEV1/FVC equal to 0.70 and 0.72, the overall prevalence of COPD–absence was 0.20 and 0.10, respectively. With lower prevalence (in grey), the efficiency of the design increased dramatically. Though the RV for the study year coefficient was only 1.09, the RV exceeded 1.82 for all other estimates.
Figure 1.
Relative Variance across 250 replicates as a function of data features for the chronic obstructive pulmonary disease–free outcome: We show for a number of data features. Black diamonds denote the analyses presented in Table 3 and grey diamonds show the result of perturbing one data, analysis or design feature. Panels show the impact on relative variance of response prevalence (A), strength of the Z ~ Y relationship (B), amount of response dependence (C), richness of the auxiliary variable model for Z (D), and asthma exposure and auxiliary variable sampling (E).
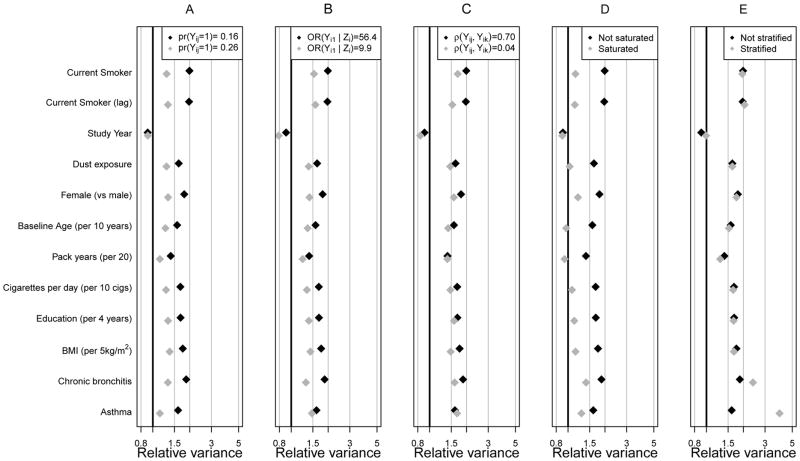
In panels B and C, we reduced the strength of the Zi ~ Yi1 relationship and the response correlation, respectively. In both cases, lower relative variances resulted, and in such circumstances, one would question the usefulness of the design. Whereas we observed similar patterns for the severe COPD model (Figure 2), RV s were higher due to lower response prevalence.
Figure 2.
Relative Variance across 250 replicates as a function of data features for the severe chronic obstructive pulmonary disease outcome: We show for a number of data features. Black diamonds denote the analyses presented in Table 3 and grey diamonds show the result of perturbing one data, analysis or design feature. Panels show the impact on relative variance of response prevalence (A), strength of the Z ~ Y relationship (B), amount of response dependence (C), richness of the auxiliary variable model for Z (D), and asthma exposure and auxiliary variable sampling (E).
To fully understand circumstances under which the designs may be useful, we examined two additional features for their impact on RV : 1) the richness of the specification of the intermediate, auxiliary variable model, Pr(Zi = 1 | Yij,Xij) used in sequential offsetted regressions analyses, and 2) the use of an exposure and auxiliary variable sampling design that creates sampling strata not only based on Zi but also on a time–fixed, baseline exposure Vi (Panels D and E of Figures 1 and 2).
In panels D of Figures 1 and 2, we show the impact that auxiliary variable model choice can have on RV. For the original analysis (labeled “not saturated”), our auxiliary model included response Yij, all predictors Xij in the model of interest, and the multiplicative interaction Yij × study yearij. For a more conservative approach, labeled “saturated”, we included the interaction between Yij and all terms in Xij. The saturated auxiliary model reduced efficiency gains for all parameters except those associated with study year. That is, by unnecessarily estimating many covariate interactions with Yij in the auxiliary model, RV s dropped substantially. Though the auxiliary model must be correct to ensure valid parameter inferences, it is reasonable to expect it to be relatively simple, because Yij should be most strongly related to Zi.
In panels E of Figures 1 and 2, we show the impact that further sampling stratification can have on efficiency. In the original analysis, two sampling strata were defined by Zi. In panel E, because asthma is very rare (Table 1), we consider a strategy to gain further efficiency on the parameter associated with presence (Vij = 1) or absence (Vij = 0) of asthma at baseline. By enriching the sample with those with asthma, we observed enormous efficiency gains even compared to the original auxiliary variable sampling design, e.g., for the asthma parameter in figure 2, compare 1.63 under auxiliary variable sampling to 4.02 under exposure and auxiliary variable sampling. We also notice that, due to its association with asthma, the exposure and auxiliary variable sampling design was far more efficient for the chronic bronchitis parameter, where the RV jumped from 1.82 under auxiliary variable sampling to 2.44 under exposure and auxiliary variable sampling. Predictors weakly associated with baseline asthma (Vi) were not strongly affected by stratifying on Vi.
DISCUSSION
In this paper, we have picked up on the classic epidemiologic notion of sampling based on disease events, and shown one family of extensions to analysis of longitudinal or clustered data. Our main aims were, first, to focus on study design, and second, to treat data analysis methods as supporting methodologies to these new and more efficient study designs. Therefore, we have not gone into depth on the technical details around estimation and inference. Finally, we focused on longitudinal data but the ideas could be easily adapted to clustered data.
For this new family of study designs, we have demonstrated a method of analysis based on population average logistic regression models which we call sequential offsetted regressions. In the context of a hypothetical study based on a real cohort, we showed that strategic sampling based on an auxiliary variable related to the binary response series, together with sequential offsetted regressions, yields increased statistical efficiency relative to random sampling, and that sequential offsetted regressions can be more statistically efficient than standard IPW. Using the auxiliary variable sampling with sequential offsetted regressions, we demonstrated sensitivity of resulting statistical efficiency to specification parameters of the design and analysis. It is worth noting that in the process of conducting sequential offsetted regressions we estimated Pr(Si = 1|Yij,Xij). Weights based on this estimate, or associated stabilized weights [24, 25, 26], could also be used in IPW–based analyses. Though standard error calculations are difficult, bootstrap–based estimators can be used.
Our objective was to describe a class of designs that extend from the case–control sampling principle to the longitudinal response data setting. It is difficult to be comprehensive and variations of these and other designs will arise according to study–specific constraints. Methodologically oriented biostatisticians will be interested in developing analysis methods for such designs.
Supplementary Material
Acknowledgments
The authors wish to thank the supported effort of the faculty and staff members of the Johns Hopkins University Bayview Genetics Research Facility, NHLBI grant HL066583 (Garcia/Barnes, PI) and NHGRI grant HG004738 (Barnes/Hansel, PI). The Lung Health Study was supported by U.S. Government contract No. N01-HR-46002 from the Division of Lung Diseases of the National Heart, Lung and Blood Institute. Data were downloaded from the NCBI database of genotypes and phenotypes (accession number phs000335.v2.p2)
Sources of funding: This project was partially funded by the NIH grants R01 HL094786 and R01 HL072966 from the National Heart Lung and Blood Institute, the Long-Range Research Initiative of the American Chemistry Council, and the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health.
Footnotes
Conflict of interest statement: The authors have no conflicts of interest. PJR is a Charter Member of a Data Safety Monitoring Board for Sunovian Pharmaceuticals, Inc., in Fort Lee, New Jersey. Sunovian is a pharmaceutical and drug development company.
Data and code availability / code: Code for conducting sequential offsetted regressions analysis is available from the first author’s website (http://biostat.mc.vanderbilt.edu/wiki/Main/ODSandLDA) and LHS are available at the National Center for Biotechnology Information database of genotypes and phenotypes (https://www.ncbi.nlm.nih.gov/gap)
References
- 1.Cornfield J. A method of estimating comparative rates from clinical data; applications to cancer of the lung, breast, and cervix. J Natl Cancer Inst. 1951 Jun;11:1269–1275. [PubMed] [Google Scholar]
- 2.Anderson JA. Separate sample logistic discrimination. Biometrika. 1972;59:19–35. [Google Scholar]
- 3.Prentice RL, Pyke R. Logistic disease incidence models and case-control studies. Biometrika. 1979;66:403–412. [Google Scholar]
- 4.Borgan O, Goldstein L, Langholz B. Methods for the analysis of sampled cohort data in the Cox proportional hazards model. ANNALS OF STATISTICS. 1995 Oct;23:1749–1778. [Google Scholar]
- 5.Prentice RL. A case-cohort design for epidemiologic cohort studies and disease prevention trials. Biometrika. 1986;73:1–11. [Google Scholar]
- 6.Maclure M. The case-crossover design - A method for studying transient effects on the risk of acute events. American Journal of Epidemiology. 1991 Jan 15;133:144–153. doi: 10.1093/oxfordjournals.aje.a115853. [DOI] [PubMed] [Google Scholar]
- 7.Navidi W. Bidirectional case-crossover designs for exposures with time trends. BIOMETRICS. 1998 Jun;54:596–605. [PubMed] [Google Scholar]
- 8.White JE. A two stage design for the study of the relationship between a rare exposure and a rare disease. American Journal of Epidemiology. 1982;115:119–128. doi: 10.1093/oxfordjournals.aje.a113266. [DOI] [PubMed] [Google Scholar]
- 9.Breslow N, Cain K. Logistic regression for two-stage case-control data. Biometrika. 1988;75(1):11–20. [Google Scholar]
- 10.Breslow N, Holubkov R. Weighted likelihood, pseudo-likelihood and maximum likelihood methods for logistic regression analysis of two-stage data. Statistics in Medicine. 1997 Jan 15;16:103–116. doi: 10.1002/(sici)1097-0258(19970115)16:1<103::aid-sim474>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 11.Connett JE, Kusek JW, Bailey WC, O’Hara P, Wu M. Design of the Lung Health Study: a randomized clinical trial of early intervention for chronic obstructive pulmonary disease. Control Clin Trials. 1993 Apr;14:3S–19S. doi: 10.1016/0197-2456(93)90021-5. [DOI] [PubMed] [Google Scholar]
- 12.Kanner RE. Early intervention in chronic obstructive pulmonary disease. A review of the Lung Health Study results. Med Clin North Am. 1996 May;80:523–547. doi: 10.1016/s0025-7125(05)70452-1. [DOI] [PubMed] [Google Scholar]
- 13.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, et al. The ncbi dbgap database of genotypes and phenotypes. Nature genetics. 2007;39(10):1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 15.Pepe MS, Anderson GL. A cautionary note on inference for marginal regression models with longitudinal data and general correlated response data. Communications in Statistics: Simulation and Computation. 1994;23:939–951. [Google Scholar]
- 16.Schildcrout JS, Heagerty PJ. Regression analysis of longitudinal binary data with time-dependent environmental covariates: bias and efficiency. Biostatistics. 2005 Oct;6:633–652. doi: 10.1093/biostatistics/kxi033. [DOI] [PubMed] [Google Scholar]
- 17.Schildcrout JS, Rathouz PJ. Longitudinal studies of binary response data following case-control and stratified case-control sampling: design and analysis. Biometrics. 2010 Jun;66:365–373. doi: 10.1111/j.1541-0420.2009.01306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robins JM, Rotnitzky A, Zhao LP. Estimation of regression coefficients when some regressors are not always observed. Journal of the American statistical Association. 1994;89(427):846–866. [Google Scholar]
- 19.Robins JM, Rotnitzky A, Zhao LP. Analysis of semiparametric regression models for repeated outcomes in the presence of missing data. Journal of the American Statistical Association. 1995;90(429):106–121. [Google Scholar]
- 20.Franklin JM, Schneeweiss S, Polinski JM, Rassen JA. Plasmode simulation for the evaluation of pharmacoepidemiologic methods in complex healthcare databases. Computational statistics & data analysis. 2014;72:219–226. doi: 10.1016/j.csda.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heagerty PJ. Marginally specified logistic-normal models for longitudinal binary data. Biometrics. 1999;55(3):688–698. doi: 10.1111/j.0006-341x.1999.00688.x. [DOI] [PubMed] [Google Scholar]
- 22.Heagerty PJ. Marginalized transition models and likelihood inference for longitudinal categorical data. Biometrics. 2002;58(2):342–351. doi: 10.1111/j.0006-341x.2002.00342.x. [DOI] [PubMed] [Google Scholar]
- 23.Schildcrout JS, Heagerty PJ. Marginalized models for moderate to long series of longitudinal binary response data. Biometrics. 2007 Jun;63:322–331. doi: 10.1111/j.1541-0420.2006.00680.x. [DOI] [PubMed] [Google Scholar]
- 24.Hernán MA, Brumback B, Robins JM. Marginal structural models to estimate the causal effect of zidovudine on the survival of HIV-positive men. Epidemiology. 2000 Sep;11:561–570. doi: 10.1097/00001648-200009000-00012. [DOI] [PubMed] [Google Scholar]
- 25.Hernán MA, Robins JM. Estimating causal effects from epidemiological data. J Epidemiol Community Health. 2006 Jul;60:578–586. doi: 10.1136/jech.2004.029496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cole SR, Hernán MA. Constructing inverse probability weights for marginal structural models. Am J Epidemiol. 2008 Sep;168:656–664. doi: 10.1093/aje/kwn164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.