Chromosome 16 inversion, inv(16), is the signature chromosome abnormality in M4Eo subtype of acute myeloid leukemia (AML), which produces a fusion gene CBFB-MYH111. We previously generated a knockin mouse model for CBFB-MYH11 (Cbfb+/MYH11)2. Heterozygous Cbfb-MYH11 knockin mice have definitive hematopoiesis blockage and die at mid-gestation, which is similar to the phenotypes of Runx1−/− and Cbfb−/− mice3, 4, indicating that CBFβ-SMMHC, the fusion protein encoded by Cbfb-MYH11, dominantly suppresses RUNX1 and CBFβ. Chimeric and conditional Cbfb-MYH11 knockin mice develop AML when they acquire additional mutations5.
An important domain of CBFβ-SMMHC is the C-terminal region of SMMHC, which catalyzes homo-dimerization and multimerization of the fusion protein that may be functionally important6. To test the function of this region, we previously generated knockin mice that expressed a truncated CBFβ-SMMHC missing C-terminal 95 amino acids. These mice would not develop leukemia, indicating the importance of C-terminal region for leukemogenesis7.
The CBFβ-SMMHC C-terminal region contains an assembly competence domain (ACD), which is important for SMMHC multimerization8, 9, as well as a transcriptional repression domain8–10 . Specific amino acid residues in the helices D and E of ACD are important for multimerization of CBFβ-SMMHC but not for transcription repression8. To distinguish which domain is critical for the role of CBFβ-SMMHC in leukemia development, we generated the current mouse model with mutations in helices D & E of CBFβ-SMMHC to impair multimerization but leave the transcriptional repression domain intact.
Specifically, we mutated six charged residues of helices D and E to threonine, serine or alanine residues: D (NANRRKL to NSNRASL), E (QRELDEA to QAELTSA), as published previously8. We incorporated these mutations into the full length Cbfb-MYH11 knockin construct2 (Supplementary Figure S1a). Correct knockin was confirmed by southern blot hybridization (Supplementary Figure S1b) and the expression of the fusion protein CBFβ-SMMHCmDE was detectable in BM cells, at a level similar to CBFβ-SMMHC when compared to CBFβ (Supplementary Figure S1c).
Mice carrying one copy of DE mutated Cbfb-MYH11 (Cbfb+/mDE) developed normally through adulthood, while CbfbmDE/mDE embryos died in mid-gestation with central nervous system hemorrhage (Supplementary Table S1) and defective definitive hematopoiesis (Supplementary Figure S2a and b), similar to Cbfb+/MYH11 embryos2. However, Cbfb+/mDE and CbfbmDE/mDE embryos did not develop primitive hematopoiesis defect (Supplementary Figure S2c–e) as seen in Cbfb+/MYH11 and Cbfb ΔC95/ΔC95 mice7.
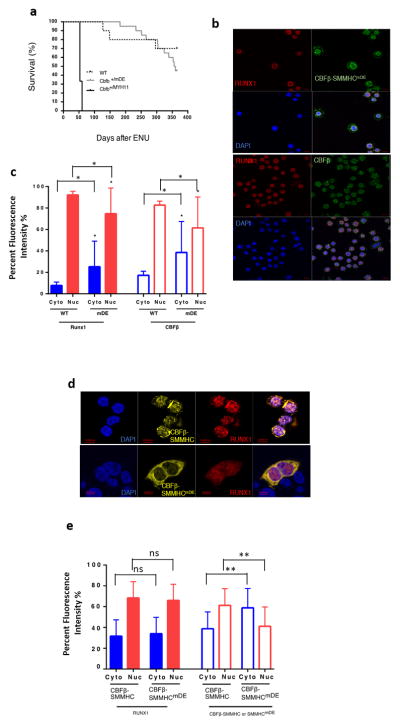
We observed Cbfb+/mDE mice for up to two years and did not see any notable abnormality. Their peripheral blood (PB) cells were largely normal except for decreased B cells and an increased Mac1+Gr-1+ myeloid cells in older mice (Supplementary Figure S3). Adult BM cells from Cbfb+/mDE mice showed no significant difference when compared to their wild type littermates in lineage differentiation, colony forming ability, cell viability, total cell count and apoptosis, except a slight increase of apoptosis in lin+ cells (Supplementary Figures S4–6). Importantly, none of the Cbfb+/mDE mice developed leukemia, even after treatment with ENU to induce additional mutations (Figure 1a).
Figure 1.
Lack of leukemia development in Cbfb+/mDE mice and decreased ability of CBFβ-SMMHCmDE protein to sequester RUNX1. (a) Survival curves of wildtype (WT) (N=10), Cbfb+/mDE (N=20), and Cbfb+/MYH11 (N=3) mice after ENU treatment. All Cbfb+/MYH11 mice died from leukemia around 2 months after ENU treatment. No leukemia development was observed in Cbfb+/mDE and WT mice. Cbfb+/mDE vs WT: p = 0.3447; Cbfb+/MYH11 vs WT or Cbfb+/mDE: p < 0.0001. (b) Immunofluorescence staining of E11.5 PB cells by anti-CBFβ (green) and anti-RUNX1 (red) antibodies. DAPI (blue) was used for nuclear staining. PB cells from a CbfbmDE/mDE mouse (upper panel) showed both cytoplasmic and nuclear stainings of CBFβ-SMMHCmDE and mainly nuclear staining of RUNX1. PB cells from a WT mouse (lower panel) showed that CBFβ is mainly co-localized in the nuclei with RUNX1. (c) and (e) Protein subcellular distributions in immunofluorescence stained E11.5 PB cells (c) and transfected 293 cells (e). Data presented as a percentage of the fluorescence intensity of each protein in each cellular fraction, cytoplasmic (Cyto) and nuclear (Nuc), compared to total fluorescence intensity of each protein in both cellular fractions. n=70 for WT cells and n=30 for CbfbmDE/mDE (mDE) cells. n=37 for cells transfected with mCherry-labeled RUNX1 and EYFP-labeled CBFβ-SMMHC; n=30 for cells transfected with mCherry-labeled RUNX1 and EYFP-labeled CBFβ-SMMHCmDE. (d) 293 cells transfected with mCherry-labeled RUNX1 (red) and EYFP-labeled CBFβ-SMMHC or CBFβ-SMMHCmDE (yellow). Upper panel: RUNX1 and CBFβ-SMMHC showed co-localization in both nucleus and cytoplasm. Lower panel: RUNX1 localized mainly to the nuclei while CBFβ-SMMHCmDE mainly stayed in the cytoplasm. Statistical significance was calculated for the differences in fluorescence intensity of RUNX1 (left panel) and CBFβ (right panel) in PB cells between wildtype and CbfbmDE/mDE embryos (c) and for the differences in fluorescence intensity of RUNX1 (left panel) and CBFβ-SMMHC/CBFβ-SMMHCmDE (right panel) in 293 cells transfected with CBFβ-SMMHC or CBFβ-SMMHCmDE (e). *: p=0.0004, **: p<0.0001, ns: not significant. Scale bars in panels b and d = 10 μM.
It was shown previously that the DE mutations decreased the ability of CBFβ-SMMHC to form multimers and enter the nuclei8. We therefore examined the subcellular localization of endogenous CBFβ-SMMHCmDE in the knockin mice. Immunofluorescence staining of PB cells from E11.5 CbfbmDE/mDE embryos showed that RUNX1 was mostly localized in the nuclei while CBFβ-SMMHCmDE was localized in both nuclei and cytoplasm (Figure 1b and c). On the other hand, RUNX1 and CBFβ were mostly co-localized in the nuclei of PB cells in WT embryos (Figure 1b and c). These observations were confirmed by western blot, since more CBFβ-SMMHCmDE than CBFβ-SMMHC was detected in the cytoplasm of bone marrow cells from adult mice (Supplementary Figure S7a). In addition, transfected 293 cells showed similar reduction of co-localization between RUNX1 and CBFβ-SMMHCmDE as well as more cytoplasmic localization of CBFβ-SMMHCmDE (Figure 1d and e) and the western blot of transfected 293 cells (Supplementary Figure 7b) confirmed the expression of transfected proteins. The reduced co-localization between RUNX1 and CBFβ-SMMHCmDE as well as more cytoplasmic localization of CBFβ-SMMHCmDE suggested that CBFβ-SMMHCmDE likely has reduced capacity to interact with RUNX1, which may be important for leukemogenesis by CBFβ-SMMHC.
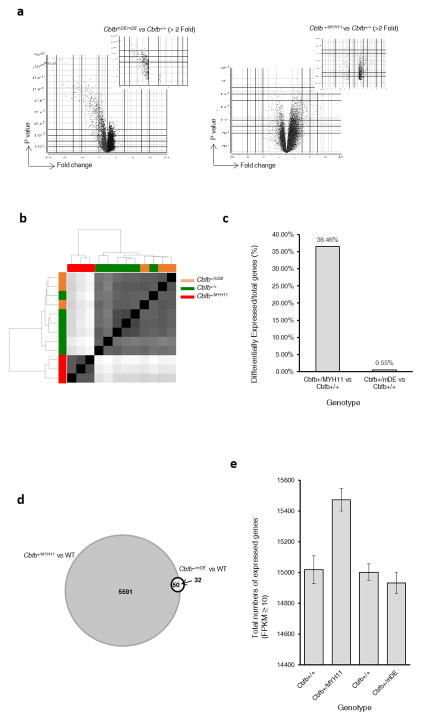
Even though CbfbmDE/mDE and Cbfb+/MYH11 embryos had a similar phenotype at midgestation, they had very different gene expression patterns, with 188 and 1725 differentially expressed genes, respectively (FDR<0.01, Fold change>2). Moreover, most differentially expressed genes in Cbfb+/MYH11 embryos were up-regulated while almost all differentially expressed genes in CbfbmDE/mDE embryos were down regulated (Figure 2a). Interestingly, canonical pathway and disease/biological functions were also affected in opposite directions in CbfbmDE/mDE and Cbfb+/MYH11embryos (Supplementary Figure S8).
Figure 2.
Gene expression changes in Cbfb+/mDE and CbfbmDE/mDE mice. (a) Volcano plots showing gene expression profile differences in PB cells between CbfbmDE/mDE and Cbfb+/+ embryos (left panel) and between Cbfb+/MYH11 and Cbfb+/+ embryos (right panel). (b) Principal component analysis of RNA-Seq data shows that the gene expression profile of Cbfb+/mDE C-KIT+ cells is more similar to C-KIT+ cells in Cbfb+/+mice. (c) Percentages of differentially expressed genes vs. all expressed genes (average of three samples). (d) Venn diagram of differentially expressed genes in Cbfb+/MYH11 and Cbfb+/mDE C-KIT+ cells (p ≤0.05, fold change ≥1.5). (e) Total numbers of expressed genes (obtained with featureCounts11; average FPKM from three samples ≥ 10) in C-KIT+ cells from mice of the indicated genotype.
We performed RNA-Seq experiments to compare the expression profile of C-KIT+ cells in the adult Cbfb+/mDE mice with C-KIT+ cells in Cbfb+/MYH11 and WT mice. As shown in Figure 2b, gene expression profile of Cbfb+/mDE cells overlapped with that of WT cells, but differed from Cbfb+/MYH11 cells. There were 5641 differentially expressed genes (FDR<0.05, Fold change ≥1.5) in Cbfb+/MYH11 cells, compared to the WT cells, accounting for 36.46% of all expressed genes. Between Cbfb+/mDE and WT cells, only 82 genes (82 of 14932, 0.55%) were differentially expressed (Figure 2c).
Among the 82 differentially expressed genes in the Cbfb+/mDE cells, 50 (60%) (Supplementary Table S2) were also differentially expressed in the Cbfb+/MYH11 cells (Figure 2d). Interestingly, some genes were differentially expressed in opposite directions, e.g., Socs2 was 2X down in Cbfb+/mDE cells, but 17.6X up in Cbfb+/MYH11cells (Supplementary Table S2).
Furthermore, Cbfb+/MYH11 cells expressed significantly higher number of genes, while Cbfb+/mDE cells expressed similar number of genes as the WT cells (Figure 2e and Supplementary Figure S9). In addition, MYH11 and MYH11mDE expression was detected in Cbfb+/MYH11 and Cbfb+/mDE cells, respectively, from the RNA-Seq data.
Overall our data demonstrated that the C-terminal multimerization domain, especially the 6 charged aa residues, is essential for leukemogenesis by CBFβ-SMMHC. The mechanism is still unclear, but likely through reduced RUNX1 sequestration, leading to fewer gene expression changes. Our findings point to an important target for developing specific therapeutic approaches for this type of leukemia.
Supplementary Material
Acknowledgments
We thank NHGRI transgenic mouse core (Elsa Escoba, Cece Rivas and Gene Elliot), Irene C. Ginty from OLM, microarray core (Abdel G. Elkahloun), microscopic core (Stephen Wincovitch), flow cytometry core (Stacy Anderson and Martha Kirby), Bioinformatics core (Niraj Trivedi) and NIH Intramural Sequencing Center for their help. The research was supported by Intramural Research Program of National Human Genome Research Institute, National Institutes of Health.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Liu P, Tarle SA, Hajra A, Claxton DF, Marlton P, Freedman M, et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261(5124):1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 2.Castilla LH, Wijmenga C, Wang Q, Stacy T, Speck NA, Eckhaus M, et al. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996 Nov 15;87(4):687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87(4):697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 4.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84(2):321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 5.Castilla LH, Garrett L, Adya N, Orlic D, Dutra A, Anderson S, et al. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nature genetics. 1999 Oct;23(2):144–146. doi: 10.1038/13776. [DOI] [PubMed] [Google Scholar]
- 6.Adya N, Stacy T, Speck NA, Liu PP. The leukemic protein core binding factor beta (CBFbeta)-smooth-muscle myosin heavy chain sequesters CBFalpha2 into cytoskeletal filaments and aggregates. Mol Cell Biol. 1998;18(12):7432–7443. doi: 10.1128/mcb.18.12.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamikubo Y, Hyde RK, Zhao L, Alemu L, Rivas C, Garrett LJ, et al. The C-terminus of CBFbeta-SMMHC is required to induce embryonic hematopoietic defects and leukemogenesis. Blood. 2013 Jan 24;121(4):638–642. doi: 10.1182/blood-2012-06-434688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, D’Costa J, Kummalue T, Civin CI, Friedman AD. Identification of a region on the outer surface of the CBFbeta-SMMHC myeloid oncoprotein assembly competence domain critical for multimerization. Oncogene. 2006 Nov 23;25(55):7289–7296. doi: 10.1038/sj.onc.1209725. [DOI] [PubMed] [Google Scholar]
- 9.Kummalue T, Lou J, Friedman AD. Multimerization via its myosin domain facilitates nuclear localization and inhibition of core binding factor (CBF) activities by the CBFbeta-smooth muscle myosin heavy chain myeloid leukemia oncoprotein. Mol Cell Biol. 2002;22(23):8278–8291. doi: 10.1128/MCB.22.23.8278-8291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lutterbach B, Hou Y, Durst KL, Hiebert SW. The inv(16) encodes an acute myeloid leukemia 1 transcriptional corepressor. Proc Natl Acad Sci U S A. 1999;96(22):12822–12827. doi: 10.1073/pnas.96.22.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014 Apr 1;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.