Abstract
Fungal pathogens like Candida albicans exhibit several survival mechanisms to evade attack by antifungals and colonize host tissues. Rta3, a member of the Rta1-like family of lipid-translocating exporters has a 7-transmembrane domain (7TMD) topology, similar to the G-protein-coupled receptors (GPCR) and is unique to the fungal kingdom. Our findings point towards a role for the plasma membrane localized Rta3 in providing tolerance to miltefosine, an analog of alkylphosphocholine, by maintaining mitochondrial energetics. Concurrent with miltefosine susceptibility, the rta3Δ/Δ strain displays increased inward translocation (flip) of fluorophore-labelled phosphatidylcholine (PC) across the plasma membrane attributed to enhanced PC-specific flippase activity. We also assign a novel role to Rta3 in the Bcr1-regulated pathway for in vivo biofilm development. Transcriptome analysis reveals that Rta3 regulates expression of Bcr1 target genes involved in cell surface properties, adhesion, and hyphal growth. We show that rta3Δ/Δ mutant is biofilm-defective in a rat venous catheter model of infection and that BCR1 overexpression rescues this defect, indicating that Bcr1 functions downstream of Rta3 to mediate biofilm formation in C. albicans. The identification of this novel Rta3-dependent regulatory network that governs biofilm formation and PC asymmetry across the plasma membrane will provide important insights into C. albicans pathogenesis.
Introduction
In yeast, ATP-binding cassette (ABC) drug transporters are not only considered determinants of multidrug resistance but also play crucial roles in controlling lipid levels. In C. albicans, a fungal pathogen that infects immunocompromised individuals, upregulation of genes encoding the Tac1-regulated ABC drug transporters (CDR1 and CDR2) is a predominant cause for the development of antifungal resistance (Coste et al., 2004, Prasad et al., 2015). As a consequence, drug-resistant isolates display enhanced efflux of azole antifungals leading to reduced inhibition of their target enzyme, lanosterol 14α-demethylase of the ergosterol biosynthesis pathway. Presence of gain-of-function mutations in Tac1 renders it hyperactive, resulting in the simultaneous induction of CDR1 and CDR2 along with RTA3, IFU5 and HSP12 in azole-resistant isolates (Coste et al., 2004, Liu et al., 2007). The significance of coordinate regulation has been inferred from studies with Saccharomyces cerevisiae, wherein Pdr5, the major ABC drug efflux pump and genes involved in sphingolipid biosynthesis are induced co-ordinately via the transcription factors Pdr1/Pdr3 (Devaux et al., 2002). These studies argue for the role of the Pdr (pleiotropic drug resistance) pathway in co-ordinately controlling lipid levels on the plasma membrane and multidrug resistance in the budding yeast (Kolaczkowski et al., 2004, Panwar et al., 2006, Johnson et al., 2010, Khakhina et al., 2015).
Studies on the noteworthiness of Tac1 co-regulated genes have not been established. Thus, amongst the Tac1 co-regulated genes, RTA3 (named for Resistance to 7-aminocholesterol) caught our attention in particular as it was annotated as a putative lipid translocase in Candida Genome Database (CGD). Lipid translocators can direct inward- or outward- transbilayer movement of phospholipids and are referred to as flippases and floppases, respectively (Fadeel et al., 2009, Panatala et al., 2015). The balanced action of these proteins is crucial for generating plasma membrane asymmetry such that the aminophospholipids are sequestered in the cytoplasmic leaflet of the membrane, whereas choline lipids are enriched in the outer leaflet (Devaux et al., 2008). Any perturbation in membrane asymmetry serves as a signal for activating multiple cellular events (Nichols, 2002). Other than the identification of Cdr1 and Cdr2 as floppases, molecular entities that can function as flippases or signals that regulate the activity of these lipid translocators to generate membrane asymmetry remain unidentified in C. albicans (Smriti et al., 2002).
The problem of drug resistance is not limited to free living planktonic forms of Candida, but also extends to surface attached communities such as biofilms (Jabra-Rizk et al., 2004, Nobile et al., 2015). C. albicans biofilms formed on implanted medical devices (abiotic surfaces) such as catheters serve as a source of infectious cells that can cause deep seated and blood stream infections and are associated with high levels of antifungal resistance (Jabra-Rizk et al., 2004, Nobile et al., 2015). Drug transporters are not only upregulated during planktonic growth but also remain up regulated during biofilm development enabling C. albicans to persist in the presence of antifungals (Ramage et al., 2002).
A typical biofilm architecture comprises of layers of yeast and hyphal cells interlaced with each other and stabilized by adhesive interactions between these cell types (Nobile et al., 2015). The transcriptional network that controls biofilm formation is comprised of six transcription factors (Efg1, Tec1, Bcr1, Ndt80, Brg1 and Rob1), wherein all of these except for Bcr1 regulate hyphal morphogenesis (Schweizer et al., 2000, Ramage et al., 2006, Nobile et al., 2012). The downstream targets of these transcription factors comprise of genes involved in adhesion, hyphal morphogenesis, matrix production and drug resistance. Bcr1 regulates the expression of adhesins, including the agglutinin-like Als proteins Als1 and Als3, and the hyphal cell wall protein Hwp1 that promote adherence during biofilm formation (Nobile et al., 2005, Nobile et al., 2006a, Nobile et al., 2006b, Nobile et al., 2008). Bcr1 thus serves as a positive regulator for cell-cell and cell-substrate adhesion (Nobile et al., 2008, Finkel et al., 2012). In addition to the identification of transcription factors, the role of Ras- and Tor1-mediated signalling pathways in regulating cell-cell adhesion during biofilm formation via Bcr1, Efg1, Nrg1, Tec1 and Tup1 has also been established (Bastidas et al., 2009, Inglis et al., 2013). In aggregate, these studies point towards the complexity of the gene regulatory programs that control biofilm formation in C. albicans.
The predicted topology of Rta3 revealed the presence of 7-transmembrane domains (TMD), similar to the family of 7-TM receptor proteins often associated with the G-protein-coupled receptors (GPCR). The homologs of RTA3 are referred to as the Rta1-family of proteins or the lipid translocating exporters (LTEs) in S. cerevisiae (Manente et al., 2009). In addition to RTA3, C. albicans has three additional genes, orf19.6224, RTA2 and RTA4, coding for the Rta1-family of proteins. Noteworthy is that these proteins lack an overall sequence conservation with the classical GPCRs. The role of Rta2 in modulating azole tolerance and in ER stress resistance has been established, while the others remain uncharacterized (Jia et al., 2008, Thomas et al., 2015). These proteins are unique to the fungal kingdom and can be considered as potential therapeutic targets.
Herein, through the genetic analysis of rta3Δ/Δ cells, we have characterized distinct cellular processes orchestrated by Rta3, a 7-TM receptor protein. First, we show that rta3Δ/Δ cells display increased susceptibility to miltefosine, an alkylphosphocholine drug. Second, by using short chain fluorescent labelled phosphatidylcholine reporter; nitrobenz-2-oxa-1, 3-diazol-4-yl (NBD)-phosphatidylcholine (PC), we forge a link between Rta3 and maintainance of the asymmetric distribution of phosphatidylcholine (PC) across the plasma membrane. Third, we provide evidence to show that Rta3 regulates biofilm formation in vivo by functioning upstream of Bcr1, the transcription factor that is central to this cellular process. Fourth, we demonstrate that Rta3 is also required for maintenance of mitochondrial membrane energetics and that altered mitochondrial membrane potential (MMP) affects Bcr1 expression.
Results
Tac1-regulated Rta3p is a plasma membrane localised protein required for tolerance to miltefosine
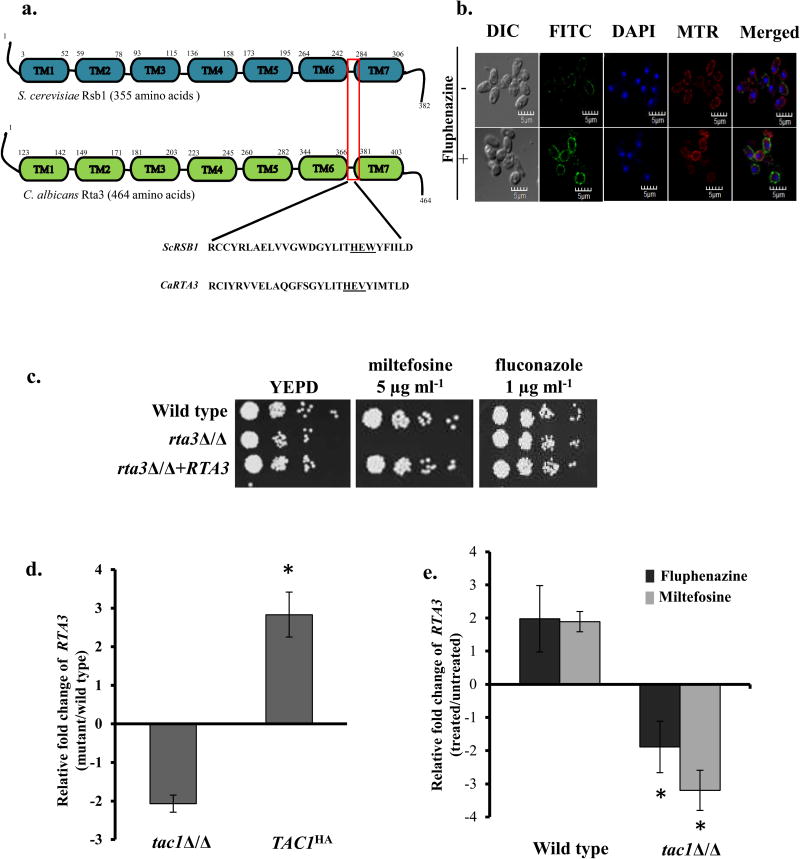
Given that Rta3 is an ortholog of Rsb1, a 7- transmembrane receptor protein of S. cerevisiae, we first analyzed its amino acid sequence for the existence of transmembrane segments. The TMHMM program predicts the presence of seven transmembrane domains in CaRta3, similar to ScRsb1. Next, we aligned the amino acid sequences of ScRsb1 and the Rta family members (Rta1 (orf19.6224), Rta2, Rta3 and Rta4) of C. albicans using Clustal Omega (Figure S1). The amino acid sequence of C. albicans Rta3 displayed 37.8% similarity and 24.5% identity with that of ScRsb1 as calculated from pairwise sequence alignment (Clustal Omega). In addition to the presence of 7-TMDs in all the Rta proteins, the alignment also revealed conservation of the Rta1 family signature sequence as identified in a previous analysis (Manente et al., 2009). The signature sequence consists of a His-Glu-Tyr/Trp motif (indicated in grey box) (Figure S1), part of a 28 amino acid stretch present in the extracellular loop connecting TMD6 and TMD7 (Figure S1). Figure 1a represents the location of 7TMDs and the conserved signature sequence in CaRta3 and ScRsb1. This analysis suggests that the defining features of the Rta1-like family of proteins in S. cerevisiae also remain conserved in C. albicans.
FIGURE 1. Rta3p is localised to the plasma membrane and is required for miltefosine tolerance.
(a) Schematic representation of the conserved transmembrane domain structure of C. albicans Rta3 and S. cerevisiae Rsb1. The red box indicates the 28 amino acid signature sequence of the Rta1-like family of proteins in the region spanning TMS6 and TMS7. Underlined amino acids represent conserved His-Glu-Tyr/Trp motif within the ScRSb1 sequence. The Tyr is replaced with a Val in CaRTA3. Abbreviations: TM, transmembrane. (b) Indirect immunofluorescence of Rta3-Myc in the wild type strain grown in the absence and presence of 20 µg ml−1 of fluphenazine. Cells were grown in YEPD till OD600 1 and treated for 30 min with 10 µg ml−1 fluphenazine before visualization. DIC represents phase images and FITC represents Rta3-Myc staining. Co-staining with DAPI and MitoTracker™ Red (MTR) is also shown. Approximately 200 cells were visualized using confocal microscopy (c) Fivefold serial dilutions of cell suspensions were spotted onto YEPD plates supplemented with miltefosine and fluconazole and incubated at 30 °C for 48 h. (d) qPCR based expression analysis of RTA3 in tac1Δ/Δ and TAC1HA cells. Fold change (mutant/wild type) is calculated by 2−ΔΔCT, and normalised to ACT1 (endogenous control), with the wild type strain as a calibrator. Values are mean ± S.D and are derived from three independent experiments. Fold change in TAC1HA is statistically significant compared to tac1Δ/Δ, *P <0.01, Student’s t-test. (e) qPCR based expression analysis of RTA3 in wild type and tac1Δ/Δ strain treated with fluphenazine (10 µg ml−1, 30 min) and miltefosine (5 µg ml−1, 120 min) The drugs were added to the secondary culture at an OD600 of 1. Values are mean ± S.D and are derived from three independent experiments. Fold change in drug treated tac1Δ/Δ is statistically significant compared to drug treated wild type, *P <0.01, Student’s t-test.
ScRsb1 was identified as a transporter of the sphingoid bases such as phytosphingosine (PHS) and dihydrosphingosine (DHS) (Kihara et al., 2002, Kihara et al., 2004). Further studies demonstrated that (i) ScRSb1 is localized on the plasma membrane (ii) its deletion leads to increased susceptibility to PHS and (iii) it affects susceptibility to PHS by regulating endocytosis of tryptophan transporter (Tat2) instead of functioning as transporter of PHS (Johnson et al., 2010). These observations are in accord with the characteristics of the 7-transmembrane receptor proteins that mediate various cellular processes in response to extracellular stimuli and are not known to function as transporters or translocators (Katritch et al., 2013). The presence of 7TMDs is also considered the hallmark of the GPCR family of proteins that upon activation by extracellular stimuli can relay signals to the heterotrimeric G-proteins (Katritch et al., 2013). Strikingly, ScRsb1 was shown to regulate Tat2 activity through an arrestin-mediated mechanism, independent of the involvment of G-proteins (Johnson et al., 2010).
Considering that Rta3 contains 7-transmembrane domains, we first sought to determine the cellular localization of Rta3. In order to assess this, we used indirect immunofluorescence to visualize Rta3-Myc in wild type cells in the absence and presence of fluphenazine; a well documented inducer of Rta3 expression (Coste et al., 2004). Rta3-Myc was primarily localized on the plasma membrane as indicated by the staining largely on the periphery of the cell both in the presence and absence of fluphenazine (Figure 1b). Compatible with its role as an inducer of Rta3 expression fluphenazine increased the fluorescence intensity of plasma membrane localized Rta3-Myc by 3-fold (measured by Olympus FV110A SW 1.7 viewer software) (Figure 1b). This data establishes the plasma membrane localization of Rta3, which is in congruence with its predicted 7-transmembrane topology, similar to ScRsb1. Taken together, we speculate that Rta3 may function as a regulatory protein, similar to ScRsb1, instead of as a transporter.
To determine the function of Rta3 in C. albicans, rta3Δ/Δ and RTA3 reconstituted strains were constructed using the SAT1 flipper strategy (Reuss et al., 2004) and confirmed by Southern blot analysis (Figure S2). Taking into consideration that Rta3 influences fluconazole susceptibility in an azole-resistant clinical isolate of C. albicans (Whaley et al., 2016), we first examined the effects of deletion of RTA3 in a laboratory wild type C. albicans strain (SC5314) on susceptibility to fluconazole. The rta3Δ/Δ cells displayed wild type susceptibility to fluconazole, suggesting that RTA3 does not contribute to azole tolerance in SC5314 (Figure 1c). The mutant was tested for susceptibility to alkylphosphocholine analog; miltefosine (hexadecylphosphocholine), as absence of ScRSB1 in S. cerevisiae renders cells susceptible to this drug (Johnson et al., 2010). The alkylphosphocholine class of drugs show anti-cancer activity and have the potential to be used in the treatment of protozoal and fungal diseases (Croft et al, 1996, Konstantinov et al, 1998, Widmer et al, 2006, Tong et al, 2007). Interestingly, rta3Δ/Δ cells displayed increased susceptibility to miltefosine (Figure 1c). Reintroduction of an RTA3 allele restored the growth defect on miltefosine to wild type levels.
Fluphenazine triggered up regulation of RTA3 via the transcription factor Tac1, has been demonstrated through a genome wide transcriptome study (Coste et al, 2004). We show that while the expression of RTA3 was 2-fold down regulated in tac1Δ /Δ cells, there was a significant (3-fold) up regulation of RTA3 in the TAC1HA (hyperactive Tac1) strain background (Figure 1d). This suggests that expression of RTA3 is dependent on a functional Tac1. Additionally, we demonstrate an increase in RTA3 expression by 2-fold in wild type cells treated with fluphenazine and miltefosine (Figure 1e). The drug-induced up regulation of RTA3 expression was abrogated in tac1Δ /Δ cells (Figure 1e). This data suggests that RTA3 is a target of the transcription factor, Tac1 and that in addition to fluphenazine, miltefosine also up regulates the expression of RTA3 via Tac1. Coupled together, we interpret these data to implicate Rta3 as a Tac1-regulated plasma membrane localized protein that modulates tolerance to miltefosine.
Intracellular accumulation of M-C6-NBD-PC is enhanced in the absence of Rta3
Resistance to miltefosine is associated with defects in internalization of its fluorescent structural analog NBD-labelled phosphatidylcholine (PC) in S. cerevisiae (Hanson et al., 2002). NBD-PC shares the glycerophosphocholine structure with miltefosine and has a similar degree of hydrophobicity. NBD-labelled lipids are used as reporters to study transport and intracellular trafficking of phospholipids in mammalian as well as in yeast cells (Haldar et al., 2013). NBD-labelled-PC and -phosphatidylethanolamine (NBD-PE) are internalized predominantly by inward-directed transbilayer transport in S. cerevisiae, referred to as flip with endocytosis as a minor contributor (Grant et al, 2001). Subsequent to its flip across the plasma membrane, the short acyl chain in the sn-2 position of NBD-PC/PE permits their passive diffusion into the cytosol followed by spontaneous redistribution to the intra-organellar membranes and to the vacuolar lumen. Eventually while majority of NBD-PC is degraded into water-soluble products after it is trafficked to the vacuole, NBD-PE remains intact and labels the mitochondria and nuclear envelope/ER (Kean et al., 1993). Additionally, experiments performed at low temperature (2 °C) where endocytosis and flop (but not flip) are blocked, demonstrate that vacuolar localization of NBD-PC is endocytosis independent and is largely dependent on the flip movement. The identity of the putative transporter(s) responsible for NBD-PC, NBD-PS and NBD-PE flip is not known. At ambient temperature, the net intracellular accumulation of these analogs, represents the sum of their inwardly (flip) and outwardly (flop) directed transbilayer translocation across the plasma membrane. A favoured flip movement in NBD-phospholipid labelled cells would lead to an increase in the net internalization of these lipids which can be confirmed by microscopy and flow cytometry.
In this study, we first labelled the wild type strains of S. cerevisiae (BY4741) and C. albicans (SC5314) with NBD-PC at 30 °C for 45 min after which cells were washed with SDC (0.67% of yeast nitrogen base (Difco), 2% glucose and 0.02% complete amino acid supplement) incubated for an additional 30 minutes and observed under microscopy. We observed significant differences in the labelling pattern between the two wild type strains. NBD-PC predominantly labels the vacuole in S. cerevisiae concurrent with earlier studies (Hanson et al., 2002), whereas it is excluded from the vacuole and enriched in the mitochondria in Candida (Fig. S3a).
In order to analyze time-dependent distribution of NBD-PC, we labelled C. albicans wild type cells with NBD-PC at 30 °C up to 15 min, followed by washing with ice cold SC-azide before imaging by fluorescence microscopy. We observed enrichment of NBD-PC in the mitochondria at all time points (0 min onwards) while the mean fluorescence intensity (MFI) gradually increased with time (Fig. S3b). Next, we performed a similar experiment at 2 °C (where flop is blocked) to investigate if there was change in the localization pattern of NDB-PC at low temperature. For this, the time-dependent internalization of NBD-PC in wild type cells was monitored at 2 °C, followed by a chase at 30 °C for 30 min prior to microscopy. Even under these experimental conditions, NBD-PC is excluded from the vacuole and labels the mitochondria, with a concommitant increase in the MFI with time (Fig. S4). These data suggests that the intracellular sorting of NBD-PC differs between C. albicans and S. cerevisiae.
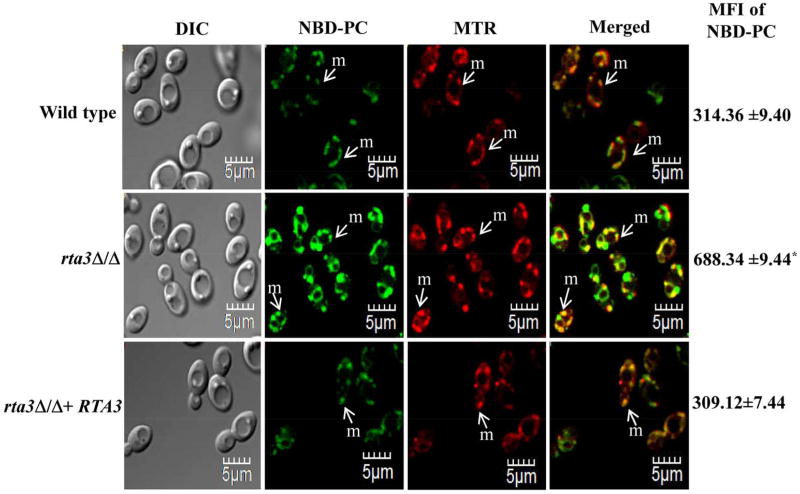
Thereafter, in order to correlate the increased miltefosine susceptibility of rta3Δ/Δ cells to the internalization of NBD-PC, we compared the dynamics of NBD-PC internalization between the mutant and the wild type at 30 °C, using the aforementioned protocol. Confocal images show that although there was no change in localisation of NBD-PC in wild type and the rta3Δ/Δ cells at 30 °C, we observed a 2-fold increase in the mean intracellular fluorescence intensity (MFI) of NBD-PC in the mutant compared to the wild type (Figure 2). We also included NBD-PE in our assay to assess whether the absence of RTA3 affects the transbilayer transport of an aminophospholipid. NBD-PE was also found to be enriched largely in the mitochondria in wild type C. albicans (Figure S5b), similar to NBD-PC. In the mutant, only a moderate increase in the intracellular NBD-PE accumulation was observed (Figure S5b). The reconstituted strain showed wild type levels of NBD-PC/PE accumulation. As high MFI in rta3Δ/Δ cells is reflective of enhanced intracellular NBD-PC accumulation, we hypothesize that this could be due to an increase in the inwardly directed transbilayer movement (flip) of NBD-PC across the plasma membrane.
FIGURE 2. rta3Δ/Δ displays increased internalization of NBD-PC.
Strains were grown to an OD600 of 1, labelled with 5 µM NBD-labelled PC and co-stained with 20 nM MitoTracker™ Red (MTR) for 45 min at 30 °C. Cells were washed, kept in fresh SDC media for 30 min at 30 °C, washed with ice cold SC-azide three times and visualized by confocal microscopy. Values on the right indicate mean fluorescence intensity (MFI) of internalized NBD-PC in all strains measured by flow cytometry (*P < 0.01, Student’s t-test). Data represent the mean ± S.D of three independent experiments. Approximately 200 cells were visualized using confocal microscopy. m denotes mitochondria.
Enhanced internalization of NBD-PC is attributed to increased PC-specific flippase activity
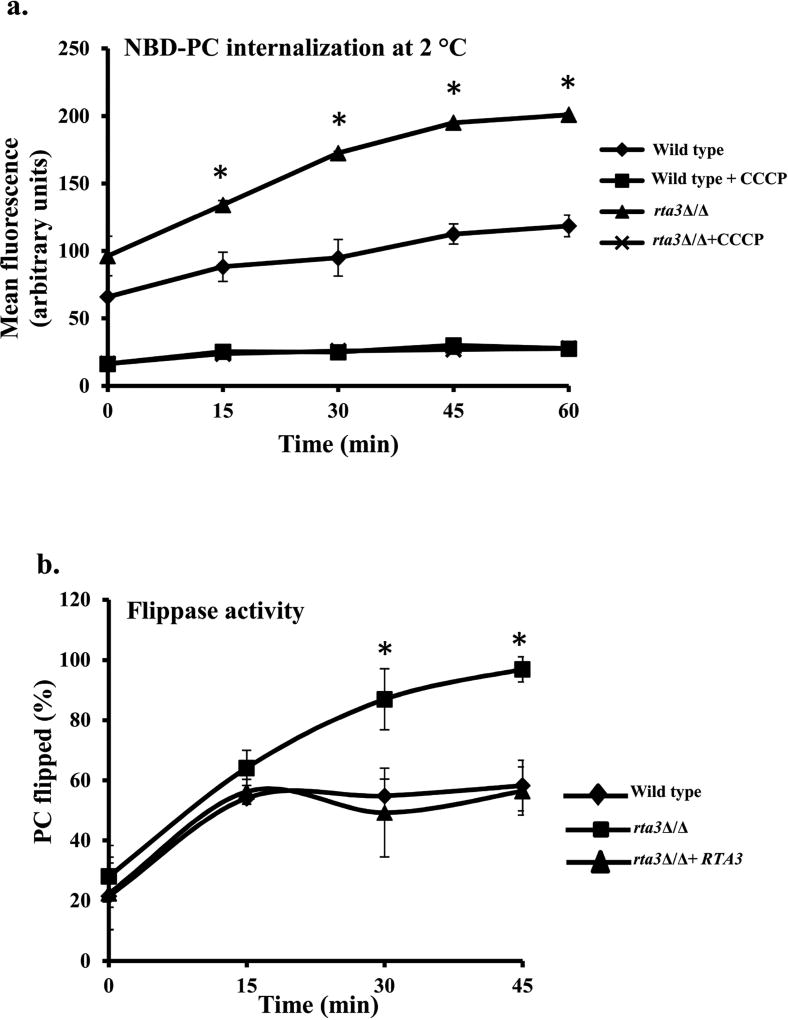
At ambient temperature, as there is ample rate of flop, it is not possible to determine if the increase in net accumulation of NBD-PC is a result of flop, flip or a sum of both. Measuring intracellular accumulation at low temperature (2 °C) provides a mean to investigate the average rate of flip activity, independent of flop; flop is almost zero at this temperature for a large population of live cells as demonstrated in S. cerevisiae (Riezman, 1985, Grant et al, 2001, Hanson et al, 2002). Therefore, in order to test our hypothesis we measured the net accumulation of NBD-PC at 2 °C in the rta3Δ/Δ cells. Prior to subjecting the cells to flow cytometry, confocal imaging of cells labelled with NBD-PC at 2 °C was done to confirm that the fluorescence originated exclusively from internalized NBD-PC. Figure S5a is a representative image demonstrating that both in the wild type and the rta3Δ/Δ mutant, NBD-PC is largely distributed to the mitochondria at 2 °C, similar to the labelling pattern obtained at 30 °C. Therefore, flow cytometric measurement of cell-associated fluorescence was interpreted to reflect net intracellular accumulation of NBD-PC. Thereafter, by measuring MFI at different time points (0–60 min), we find that the cell-associated internalized NBD-PC fluorescence gradually increases with time at 2 °C in both the wild type and rta3Δ/Δ mutant (Figure 3a). The net accumulation of NBD-PC was 1.5 fold at 0 and 15 min and ≥1.7 at 35, 45, and 60 min time points in the mutant, implying that its flip was faster compared to the wild type (Figure 3a).
FIGURE 3. Enhanced NBD-PC accumulation in rta3Δ/Δ is due to increased flippase activity.
(a) Cells untreated or treated with 50 µM CCCP were labelled at 2 °C with 2 µM NBD-PC. At the indicated time points, aliquots of cells were removed, washed with cold SC-azide and mean fluorescence intensity (MFI) of internalized NBD-PC was measured by flow cytometry. Values are the mean ±S.D. of four independent experiments. *P < 0.01, Student’s t-test, MFI of rta3Δ/Δ (triangular marker) is statistically significant compared to wild type (diamond marker). (b) Flippase activity was measured in indicated strains by using 2 µM NBD-PC and the impermeant quencher sodium dithionite (25 mM). At indicated times, MFI of NBD-PC in the presence or absence of dithionite was measured by flow cytometry. Percentage of internalized NBD-PC (or PC flipped in %) was represented as the normalized FD/Ftotal ratio. For 30 min and 45 min time points, % PC flipped in rta3Δ/Δ were found to be statistically significant compared to wild type (*P < 0.01, Student’s t-test).
Furthermore, in S. cerevisiae low temperature flip of NBD-phospholipids is dependent on the proton electrochemical gradient across the plasma membrane that is maintained by Pma1 (Hanson et al, 2001, Stevens et al., 2007). Hence, if this gradient is disturbed by using CCCP, a protonophore, Pma1 is unable to maintain the proton gradient due to depletion of ATP, thereby inhibiting flip movement. As a control, NBD-PC accumulation was measured in the presence of CCCP to confirm that enhanced internalization is solely attributed to flip movement. As shown in Figure 3a, the flip movement of NBD-PC was inhibited by CCCP in both wild type and rta3Δ/Δ. Moreover, in contrast to NBD-PC, the net accumulation of NBD-PE between the mutant and the wild type remained unchanged at 2 °C (data not shown). This suggested that the marginal increase in intracellular MFI of NBD-PE at 30 °C as mentioned above and shown in Figure S5b was not due to increased flip. This set of data suggests that the difference in the MFI (mean fluorescence intensity) of internalized NBD-PC between the wild type and the rta3Δ/Δ cells can solely be attributed to increased flip (inwardly directed) in the mutant.
In order to assess whether the enhanced flip of PC in the rta3Δ/Δ cells was due to an increase in the flippase activity that transports PC across the plasma membrane, we measured the NBD-PC fluorescence by flow cytometry in the presence or absence of sodium dithionite (a impermeant reducing agent and quencher of NBD), a well known method in S. cerevisiae (McIntyre et al, 1991, Popescu et al, 2010). The total lipid upload is dependent on the adsorption of NBD-PC in the exoplasmic leaflet of the plasma membrane and the flippase activity that transports it across the bilayers. Dithionite binds and exclusively reduces the fluorescent lipid present in the outer leaflet, producing non-fluorescent derivatives. This method provides information about the proportion of the fluorescent lipid incorporated in the outer leaflet and that which has migrated to locations within the cell. Therefore, the faster the movement of the fluorescent lipid into the inner leaflet (increased flip), the lesser lipid would be quenched by dithionite, and the intracellular MFI would continue to remain high with time. Flippase activity, therefore, can be assessed by measuring the intracellular MFI of NBD-PC in the presence (FD) or absence (FTotal) of dithionite as measured by flow cytometry as described in Materials and Methods. In line with this, Figure 3b shows that the percent flippase activity was similar in rta3Δ/Δ cells compared with wild type cells at T0 (28±10 vs 21.44±11), while there was significant difference at T45 (96.89%± 4.2 vs 58.23± 8.3). The flippase activity in the reconstituted strain remained similar to the wild type. The flippase activity for NBD-PE in the mutant was equivalent to that in the wild type, reaffirming that the absence of RTA3 does not affect the distribution of NBD-PE, consistent with internalization assays at ambient temperature and 2 °C (Figure S3c). Coupled together, our results show that loss of RTA3 triggers an enhanced PC-specific flippase activity.
Transcriptional profiling of the rta3Δ/Δ cells reveals differential regulation of biofilm associated genes
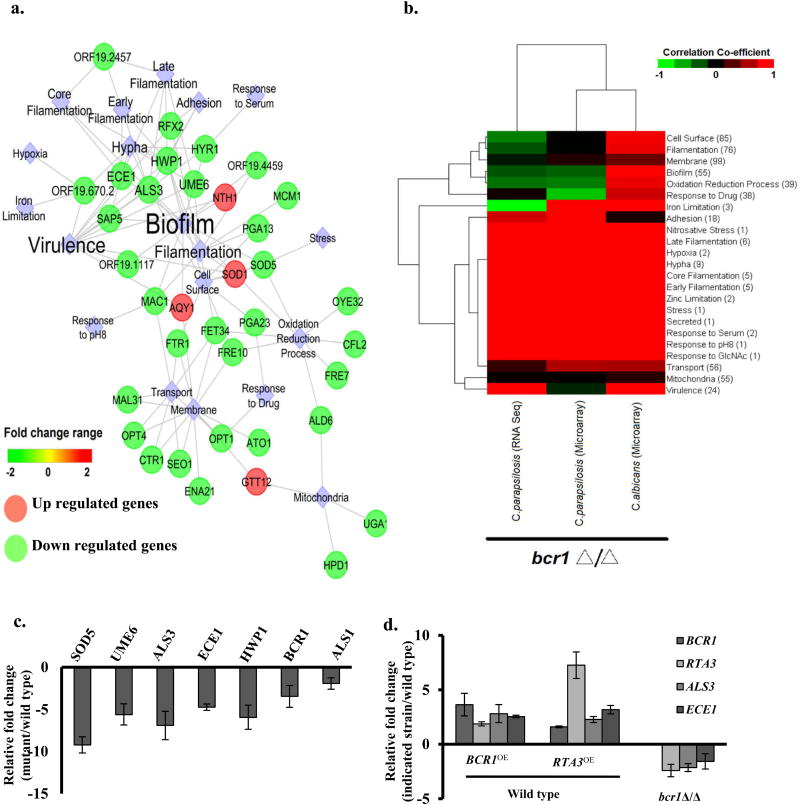
In order to acquire an insight into the global consequences of deleting RTA3, we compared mRNA profiles of wild type and rta3Δ/Δ cells. After filtering, the entire data set resulted in a total of 115 statistically significant differentially regulated ORFs (P-value<0.05, >1.5-fold up- or down regulated). Interestingly, we noticed that 67 out of 115 genes were down regulated and were enriched for the GO processes related to cell adhesion, biofilm formation, filamentation, virulence, iron assimilation, oxidation-reduction and mitochondrial function. The up regulated genes were significantly enriched for the GO function ‘repressed in spider biofilms’ and were mostly uncharacterized ORFs with no known function (Table S5). GO enrichment analysis show that genes related to biofilm formation topped the list of up regulated genes, followed by genes involved in membrane structure, hyphal formation and transport.
An overview of the dataset comprising of the 115 statistically significant differentially regulated ORFs, reveal a core network consisting of pathways or genes that are associated with specific biological processes (Figure 4a). Analysis of this network point to a few striking findings pertaining to biofilm-associated genes. Firstly, a cluster of genes that include biofilm-associated genes such as adhesin genes (ALS3 and HWP1), GPI-anchored cell wall protein genes (ECE1 and PGA13), genes involved in hyphal growth (UME6 and SOD5) and copper transport (CTR1 and FRE7) were severely altered (down regulated) in the rta3Δ/Δ cells (Table 1, Figure 4a). Secondly, a majority of genes in the network are targets of the transcription factor Bcr1, one of the best characterized regulators of biofilm formation, indicating that RTA3 is required for full expression of a set of Bcr1-target genes. (Nobile et al, 2006a, Finkel et al, 2011, Fanning et al, 2012). Third, we also observed down regulation of a subset of genes associated with ‘mitochondria and oxidation-reduction process’ (Figure 4a). These subsets of genes are connected via ALD6, predicted to encode a putative aldehyde dehydrogenase that is annotated as rat catheter/spider biofilm induced gene. ALD6 is involved in alcohol production during fermentation and is up regulated in the hypoxic microenvironment of mature biofilms (Fox et al, 2015). Alcohol production may be involved in providing energy and/or may serve as signalling molecules for cell-cell communication (Mukherjee et al, 2006, Rane et al, 2012, Chauhan et al, 2013). Furthermore, the mitochondria and membrane/transport networks are connected via glutathione-transferase putative gene (GTT12) and the oligopeptide transporter (OPT1). Gtt12 contributes to defence against hydrogen peroxide induced oxidative stress during stationary phase arrest in C. albicans (Michan et al, 2009), while up regulation of OPT genes has been associated with early biofilm stages (Yeater et al, 2007). Genes involved in iron acquisition such as FRE10, FET34, FTR1 also connect multiple cellular processes in the network. We surmise that cellular processes requisite for adaptation to metabolic changes within a biofilm are affected upon absence of RTA3. These observations suggest that the absence of RTA3 results in transcriptional remodelling of processes associated largely with biofilm formation in the mutant. We also noted that a majority of the genes in the core network are also differentially regulated in bcr1Δ/Δ cells as inferred from the data obtained from Nobile et al, 2012 (Nobile et al, 2012). This prompted us to perform a comparative analysis of the transcriptional profiles of C. albicans rta3Δ/Δ and bcr1Δ/Δ (Cabcr 1 Δ/Δ). Considering that Bcr1 is also a major regulator of biofilm formation in C. parapsilosis, we also included a transcriptional profile of a C. parapsilosis bcr1Δ /Δ (Cpbcr1Δ /Δ) strain. We first extracted a list of 23 differentially expressed biological functions/pathway and genes associated with these pathways from the transcription profile of rta3Δ/Δ cells. Next we subjected this list and the gene list obtained from the transcription profiling data of the bcr1Δ /Δ cells as described by Holland et al and Ding et al (Ding et al, 2011, Holland et al, 2014); GSE33490 and GSE57451 to correlation covariance estimation. Based on the unsupervised hierarchical clustering (Figure 4b) we observed that firstly, the Carta3Δ/Δ mutant displayed positive correlation exclusively to Cabcr1Δ/Δ (and not to Cpbcr1Δ/Δ) for genes associated with biofilm formation (cell surface, oxidation-reduction processes and iron limitation). Positive correlation between the rta3Δ/Δ and Cpbcr1Δ/Δ included other processes known to be essential to biofilm formation such as hypoxia, filamentation (early/core/late), nitrosative stress and zinc limitation. Secondly, genes differentially regulated exclusively in rta3Δ/Δ cells (and not in either Cabcr 1 Δ/Δ or Cpbcr1Δ/Δ) are associated largely with mitochondrial functions and biological functions like membrane and transport, suggesting that these processes are likely to be preferentially modulated by RTA3 (Figure 4b). All told, the network visualization and comparative analysis suggests that the transcriptional program elicited by the absence of RTA3 reflects the role of this gene as a central component that may govern biofilm formation.
FIGURE 4. Transcriptional profiling of rta3Δ/Δ mutant.
(a) Visualization of the coexpression network in rta3Δ/Δ representing the structured rta3Δ/Δ distinct modules of differentially expressed genes involved in specific biological processes. Clustering was based on over-representation analysis using Cytoscape v8.0. Circles are genes while diamonds represent cellular processes. (b) Gene expression profile of rta3Δ/Δ cells was compared to the profile of the bcr1Δ/Δ cells from the indicated strains. Red indicates higher correlation, black indicates poor correlation, and green indicates negative correlation. The numbers within the bracket for each biological function/pathway indicate the number of genes differentially regulated in rta3Δ/Δ cells and involved in that particular function/pathway. (c) qPCR based expression analysis of selected genes in rta3Δ/Δ. (d) qPCR based expression analysis of selected genes in the indicated strains. Fold change (mutant/wild type) is calculated by 2−ΔΔCT and normalized to ACT1 (endogenous control) with the wild type strain as a calibrator. Values are mean ± S.D and are derived from three independent experiments.
Table 1.
Functional categories of C. albicans genes whose transcript levels in the rta3Δ/Δ strain are 1.5-fold up- and down- regulated (P≤ 0.05)
| Category and System Name |
Gene | Function | Fold Change |
|---|---|---|---|
| Adhesion | |||
| orf19.1816 | ALS3a,b | Cell wall adhesin; epithelial adhesion | −7.27 |
| orf19.1756 | GPD1b | Glycerol-3-phosphate dehydrogenase biosynthesis | 1.84 |
| orf19.691 | GPD2b | Surface protein similar to glycerol 3-P dehydrogenase | 1.68 |
| orf19.5804 | HYU1 | Putative hydantoin utilization protein A | 1.93 |
| Cell Surface | |||
| orf19.6420 | PGA13b | GPI-anchored cell wall protein involved in cell wall synthesis | −4.35 |
| orf19.3740 | PGA23 | Putative GPI-anchored protein of unknown function | −1.69 |
| General stress | |||
| orf19.2849 | AQY1b | Aquaporin water channel; osmotic shock resistance | 1.68 |
| orf19.359 | GTT12 | glutathione transferase activity | 1.72 |
| orf19.5068 | IRE1b | Putative protein kinase; role in cell wall regulation | 2.53 |
| Hyphal formation and Virulence | |||
| orf19.3374 | ECE1b,b | Hypha-specific protein | −22.23 |
| orf19.1321 | HWP1a,b | Hyphal cell wall proteina | −19.98 |
| orf19.4975 | HYR1b | GPI-anchored hyphal cell wall protein | −1.64 |
| orf19.5585 | SAP5b | Secreted aspartyl proteinase | −2.16 |
| orf19.2770.1 | SOD1b | GPI-anchored cell wall protein involved in cell wall synthesis | −4.35 |
| orf19.1822 | UME6a,b | Zn(II)2Cys6 transcription factor | −4.06 |
| Iron Assimilation | |||
| orf19.4215 | FET34b | Multicopper ferroxidase | −1.78 |
| orf19.6139 | FRE7b | Copper-regulated cupric reductase | −2.89 |
| orf19.1415 | FRE10b | Major cell-surface ferric reductase under low-iron conditions | −1.80 |
| orf19.7219 | FTR1 | High-affinity iron permease | −2.17 |
| Membrane Structure | |||
| orf19.6169 | ATO1b | Putative fungal-specific transmembrane protein | −8.44 |
| orf19.700 | SEO1b | Protein with similarity to permeases | −2.10 |
| Metabolic Enzymes | |||
| orf19.5565 | HPD1b | 3-hydroxypropionate dehydrogenase; involved in degradation of toxic propionyl-CoA | −9.45 |
| Mitochondrial Function and Oxidation-Reduction Processes | |||
| orf19.5806 | ALD5b | NAD-aldehyde dehydrogenase | −2.00 |
| orf19.742 | ALD6b | Putative aldehyde dehydrogenase | −2.43 |
| orf19.2060 | SOD5a,b | Cu and Zn-containing superoxide dismutase; protects against oxidative stress | −6.58 |
| Transport | |||
| orf19.3646 | CTR1b | Copper transporter | −2.90 |
| orf19.6993 | GAP2 b | General amino acid permease; | −2.08 |
| orf19.3981 | MAL31 b | Putative high-affinity maltose transporter | −1.90 |
| orf19.2602 | OPT1b | Oligopeptide transporter | −1.78 |
| orf19.5673 | OPT7b | Putative oligopeptide transporter | −2.21 |
Genes that were validated by qPCR
Genes that are annotated as flow model/RPMI/Spider/rat catheter biofilm induced or repressed in CGD
Rta3 and Bcr1 are mutually regulated
We validated the microarray data by performing qPCR analysis and observed good correlation in transcript levels between microarray and qPCR data for adhesin genes ECE1, ALS3, ALS1 and HWP1 that are Bcr1 targets as well as for SOD5 and UME6 (Figure 4c). Furthermore, the expression of BCR1 was 3-fold down regulated in rta3Δ/Δ cells, consistent with the down regulation of its target adhesin genes (ALS3 and ECE1) (Figure 4c). Interestingly, the expression of RTA3 was also down regulated (2-fold) in the absence of Bcr1 (Figure 4d), also inferred from the transcriptional profiling of bcr1Δ/Δ cells in a previous study (Nobile et al, 2012).
To determine whether forced overexpression of RTA3 and BCR1 affects the expression of each other, we created strains in which the endogenous promoter of both the genes were replaced by the constitutively active TDH3 promoter in wild type cells (WT-RTA3OE and WT-BCR1OE). Overexpression of RTA3 in the wild type cells resulted in 1.5-fold increased expression of BCR1 and its target genes (Figure 4d). On the other hand, overexpression of BCR1 in the wild type cells not only led to an increased expression of its target genes but also resulted in an increased expression of RTA3 by 2-fold (Figure 4d). We infer that the transcriptional repression and overexpression of RTA3 and BCR1 is dependent on each other, suggesting that RTA3 and BCR1 are mutually regulated.
Rta3 functions upstream of Bcr1 for biofilm formation in vivo
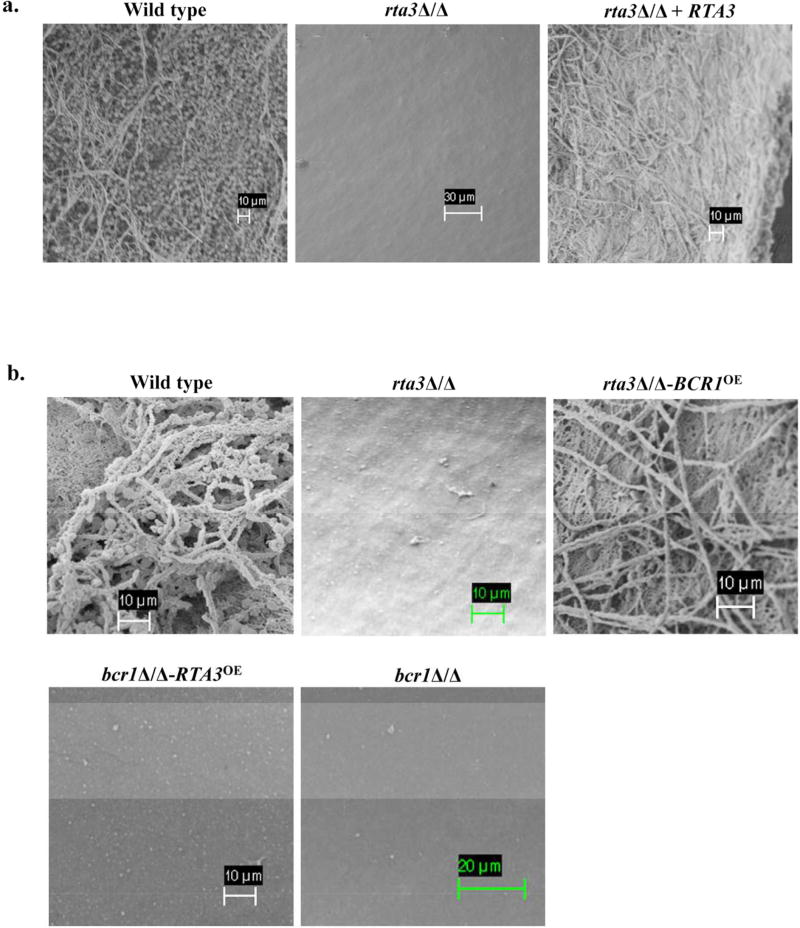
In lieu of the transcriptional profiling data, we proposed that RTA3 may function in biofilm development by promoting adherence via BCR1 and its target adhesin genes. Therefore, we subjected the rta3Δ/Δ mutant to two standard in vitro biofilm assays, an optical density assay and a silicone square assay (Lohse et al, 2017). We observed no significant biofilm defects in the rta3Δ/Δ strain versus the wild type in these two vitro biofilm assays (Figure S6a and S6b). We also note that the absence of RTA3 does not affect hyphae formation under hyphae inducing conditions (spider medium or in the presence of serum (data not shown)). For the in vivo biofilm formation assay in a rat venous catheter model (Andes et al, 2004), implanted catheters were inoculated with the wild type, rta3Δ/Δ cells, or the reconstituted strain and biofilm formation was visualized after 24 h by scanning electron microscopy (SEM). While both the wild type and the reconstituted strain produced mature biofilms, the catheter surface of the rta3Δ/Δ mutant was devoid of biofilm material (Figure 5). Taken together, this data suggests that Rta3 is required for biofilm formation in vivo, but not in vitro.
FIGURE 5. Rta3 regulates biofilm formation in vivo via Bcr1.
(a) Scanning electron micrograph of biofilm grown on a rat central venous catheter. Central venous catheters inoculated with the indicated strains were introduced into rats and incubated for 24 hours. Catheters were then removed and visualized by scanning electron microscopy. (b) Scanning electron micrograph of biofilm grown on a rat central venous catheter for overexpression study.
We hypothesized that if the down regulated expression of BCR1 is the cause of the biofilm defect in the rta3Δ/Δ cells, then increasing the expression of BCR1 in the rta3Δ/Δ mutant background should rescue the biofilm defect. Next, we created a TDH3-BCR1 overexpression strain in rta3Δ/Δ cells (rta3Δ/Δ-BCR1OE) and show increased expression levels of BCR1 and its target genes ALS3 and ECE1 (Figure S6c). Scanning electron micrograph of catheter inoculated with rta3Δ/Δ-BCR1OE shows partial restoration of biofilm formation (Figure 5b). We infer that (i) BCR1 may be mediating RTA 3-dependent biofilm formation in vivo by promoting the expression of the adhesin genes and (ii) partial restoration of the biofilm defect in our mutant maybe due to the absence of RTA3-specific targets that may be requisite for functioning in concert with Bcr1-dependent target genes, to restore biofilm formation to wild type levels.
Next, to determine the genetic relationship between RTA3 and BCR1 in governing biofilm we introduced the TDH3-RTA3 construct into the bcr1Δ /Δ cells (bcr1Δ/Δ-RTA3OE). In vitro adhesion assay on this strain show that the increased expression of RTA3 did not reverse the adhesion defect of bcr1Δ /Δ cells (Figure S4a). As expected, increased expression of RTA3 was not able to rescue the biofilm defect of the bcr1Δ /Δ cells in vivo (Figure 5b) and also did not promote the expression of the adhesin genes (Figure S6c). This suggests that RTA3 requires a functional Bcr1 for expression of adhesin genes and consequently biofilm formation (Figure 5b). Taken together, our data suggests that Rta3 regulates biofilm formation by functioning upstream of Bcr1.
Bcr1 expression and Rta3 are linked via mitochondrial membrane energetics
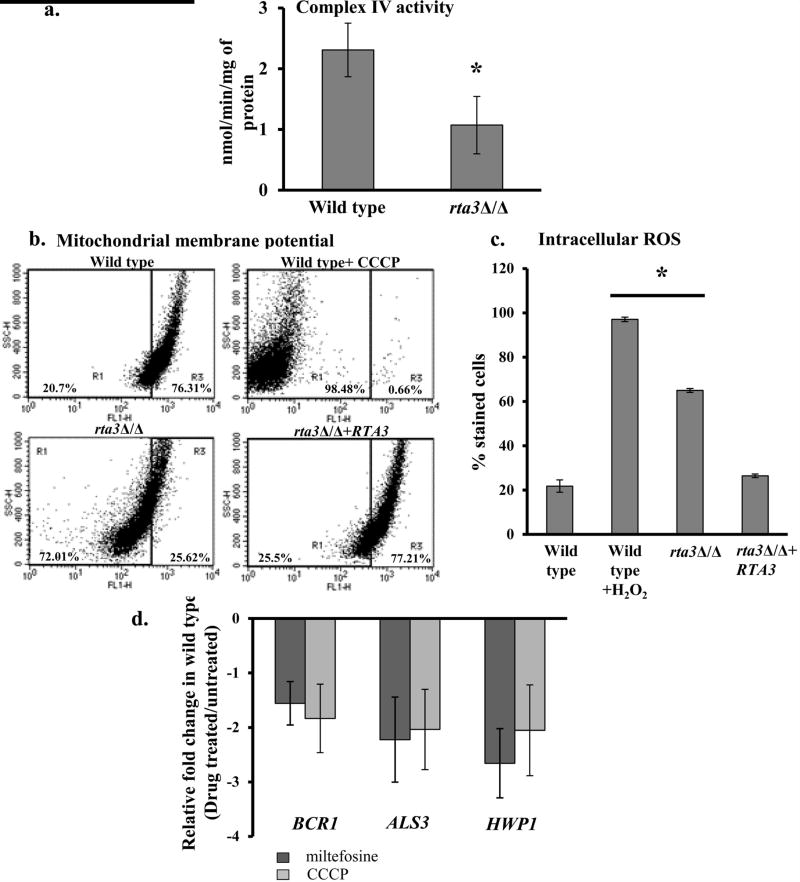
In S. cerevisiae, miltefosine enters the cell and penetrates the inner mitochondrial membrane wherein it inhibits cytochrome c oxidase (COX) and perturbs the membrane potential to cause cell death (Zuo et al, 2011). Based on this, in order to assess the basis of miltefosine susceptibility in rta3Δ/Δ cells, we analyzed the complex IV activity in a purified mitochondrial preparation from the wild type and the mutant. The rta3Δ/Δ cells displayed a 50% reduction in the activity of complex IV versus wild type (Figure 6a). Further to measure the mitochondrial membrane potential (MMP), we used a mitochondria-specific voltage-dependent dye, DiOC6(3). This dye aggregates to fluorescence green in healthy mitochondria. However, in cells with depolarized mitochondria, the dye no longer accumulates and instead is distributed throughout the cell resulting in a decrease in green fluorescence (Vayssiere et al, 1994). As shown in Figure 6b, deletion of RTA3 resulted in a significant decrease (72%) in the DiOC6(3) fluorescent signal, in accord with a wild type sample treated with 50 µM CCCP (98%) used as a positive control. The decrease in the DiOC6(3) fluorescent signal reflects depolarization of the mitochondrial membrane. Therefore, the results suggest that deletion of RTA3 could induce the breakdown of MMP. As impaired mitochondria is associated with increase in ROS levels, we hypothesized that depolarized mitochondria in the rta3Δ/Δ cells will lead to high ROS in the mutant. Cellular ROS measured by DFCDA showed 3-fold increase in the mutant, in line with a decrease in the MMP (Figure 6c). Taken together, this set of data suggests that the absence of RTA3 sensitizes the mutant to miltefosine by lowering the mitochondrial membrane potential with a commensurate increase in ROS levels.
FIGURE 6. RTA3 and BCR1 expression are linked via mitochondrial energetics.
(a) The decrease in absorbance at 550 nm resulting from the oxidation of reduced cytochrome c was measured with purified mitochondrial preparations of wild type and rta3Δ/Δ cells. Specific activity of Complex IV was calculated as mentioned in Materials and Methods. *P< 0.05, Students t-test. (b) Flow cytometric analysis of cells grown until early log phase and stained with DiOC6(3), for mitochondrial membrane potential. The mutant displays a greater proportion of cells with depolarized mitochondria (left quadrant), similar to wild type cells treated with 50 µM CCCP used as a positive control. (c) Strains were grown until early log phase and incubated with 10 µM DFCDA for 30 min for intracellular ROS measurement. Wild-type cells were treated with 10 mM H2O2 for 30 min prior to staining as a positive control. *P< 0.005, Students t-test. (d) qPCR analysis of BCR1 and indicated target genes in wild type cells grown to an OD600 of 1 in the absence and presence of CCCP (50 µM; 30 min) and miltefosine (5 µg ml−1; 60 min). Fold change (treated/untreated) is calculated by 2−ΔΔCT, and normalised to ACT1 (endogenous control). Values are mean ± S.D. and are derived from three independent RNA preparations.
Considering that MMP is a key indicator of cell health and injury and given that deletion of RTA3 caused mitochondrial depolarization (Figure 6b), we asked the question if depolarized mitochondria could be the signal that forms the basis of the phenotypes obtained in rta3Δ/Δ cells? In order to address this, we mimicked the mitochondrial depolarization status of the rta3Δ/Δ cells by treating the wild type cells with compounds that can induce mitochondrial depolarization. For this, we treated the wild type cells with CCCP (50 µM; 30 min), an uncoupler of the electron transport chain and analyzed the cells for DiOC6(3) fluorescence by flow cytometry. We also included miltefosine in this experiment as this compound intercalates into the mitochondrial membrane to disrupt the MMP in S. cerevisiae (Zuo et al, 2011). CCCP and miltefosine exposure resulted in a change in the DiOC6(3) fluorescence versus the untreated wild type cells, suggesting that these compounds were able to induce the depolarization of mitochondria (Figure S7). We note that >99% cells stained negative with propidium iodide implying that drug treatment under the conditions mentioned did not result in cell death (data not shown). Next, we determined the effect of depolarized mitochondria (in drug treated cells) on the transbilayer transport of NBD-PC and the expression of BCR1 and its target genes. While there was no effect of perturbed MMP on the flip of NBD-PC (data not shown), we observed 2- fold down regulation of BCR1 and its target genes in drug treated cells (Figure 6d), suggesting that perturbation in mitochondrial membrane energetics may be the cause for the decreased expression of BCR1 in rta3Δ/Δ cells. We conclude that pharmacological or genetic perturbation (by deleting RTA3) of mitochondrial membrane energetics results in the down regulation of BCR1 expression, in turn causing a biofilm defect in the rta3Δ/Δ cells.
Discussion
The defining feature of Rta3 (named for Resistance to 7-aminocholesterol) is the presence of 7-TMDs with an extracellular amino terminus and an intracellular carboxyl terminal (Figure 7), a hallmark of the G-protein coupled receptors (GPCRs). In agreement with its 7TMD topology, we show that C. albicans Rta3 is localized at the plasma membrane (Figure 1b), similar to Rsb1 of S. cerevisiae and thus is most likely to function as a regulatory protein rather than a transporter or translocator. Moreover, Rta3 and proteins of this family represent targets for new antifungals owing to their exclusivity in the fungal kingdom. RTA3 is a downstream target of the transcription factor Tac1 (Coste et al., 2004) (Figure 1d) and is coregulated with CDR1 and CDR2, two well-documented drug efflux pumps in C. albicans. Except for a single study where Rta3 was implicated to affect susceptibility to fluconazole in an azole-resistant clinical isolate (Whaley et al., 2016), its functional relevance remains unexplored in C. albicans. Our findings indicate that absence of Rta3 affects a spectrum of biological processes highlighting the role of this 7-TM receptor protein in multiple regulatory pathways.
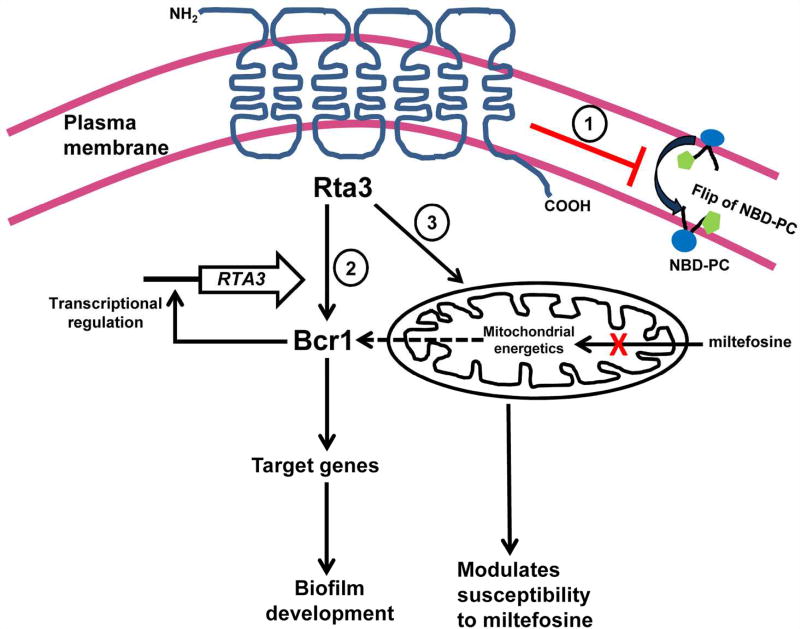
FIGURE 7. A model for the distinct roles of RTA3 in regulating plasma membrane asymmetry of phosphatidylcholine (PC), biofilm formation and miltefosine susceptibility in C. albicans.
Rta3, a member of the 7-transmembrane receptor superfamily, is localized in the plasma membrane and influences three distinct cellular processes in C. albicans. 1- Rta3 negatively regulates (red blunt arrow) flip of fluorophore tagged phosphotidylcholine (NBD-PC), ensuring that plasma membrane asymmetry of PC is maintained in C. albicans. 2- Rta3 functions upstream of Bcr1, a transcription factor which activates its target adhesin genes to promote biofilm formation. The mutual transcriptional regulation of Rta3 and Bcr1 ensures a rapid response to conditions that favour biofilm formation. 3- Rta3 also has a role in the maintenance of mitochondrial energetics and tolerance to miltefosine. Miltefosine is an alkylphosphocholine analogue and acts on cells by lowering the mitochondrial membrane potential. Our results indicate that Bcr1 expression and Rta3 may be linked via mitochondrial membrane energetics as indicated by the dotted arrow.
Loss of Rta3 not only affects tolerance to miltefosine, an alkylphosphocholine analog but also alters mitochondrial membrane energetics. Miltefosine-induced apoptosis-like cell death in the budding yeast is due to its direct interaction with COX9, a subunit of the complex IV of the electron transport chain in S. cerevisiae (Zuo et al, 2011). This results in the disassembly of complex IV, causing disruption of the electron transport chain, which in turn affects the mitochondrial membrane potential (MMP). Consistent with this, we propose that altered mitochondrial parameters in rta3Δ/Δ cells may sensitize it to a lower dose of miltefosine than is required for the wild type cells (Figure 1c and 6a–c). Although a reduced flop or export of miltefosine in rta3Δ/Δ as a cause for the increased susceptiblity to miltefosine cannot be ruled out. Additionally, Tac1-regulated RTA3 is essential for curbing miltefosine induced stress as transcript levels of RTA3 are elevated in wild type cells treated with miltefosine (Figure 1e). The causal relationship of RTA3 deletion and an altered MMP is an interesting area that requires further enquiry. Although the potential of miltefosine to treat invasive fungal infections is well documented (van Blitterswijk et al, 2008), its target in C. albicans remains unidentified. Recent studies demonstrate the ability of miltefosine to kill azole-resistant clinical isolates and inhibit biofilm formation by C. albicans (Vila et al, 2015, Vila et al, 2016). In this context, identification of Rta3 as one of the determinant of miltefosine tolerance may lead to the identification of its target in C. albicans.
The asymmetric arrangement of phospholipids on the plasma membrane is not a static situation but is maintained by a balance between the inward (flip) and the outward (flop) translocation of phospholipids across the membrane. The outer leaflet of the plasma membrane is enriched in PC and sphingolipids. The rapid translocation of phospholipids across the plasma membrane can be monitored by using fluorescent labelled short chain analogs of phospholipids, such as NBD labelled phospholipids (Haldar et al, 2013). The behaviour of NBD labelled phospholipids may differ from that of endogenous phospholipids. Nevertheless, studies in red cells and platelets show that reporter phospholipids reflect the behaviour of their endogenous counterparts (Stuart et al, 1995) Thus, these reporter lipids provide dependable means of monitoring transport activity of lipid translocators. Our data demonstrates that loss of Rta3 perturbs the dynamic equilibrium of PC by increasing its inwardly-directed (flip) movement across the plasma membrane. This results in the enrichment of PC on the inner leaflet of the plasma membrane and consequently its transfer to intracellular membranes by passive diffusion. This conclusion is based in part on the demonstration that NBD-PC internalization was high in rta3Δ/Δ cells at 30 °C (ambient) and also at 2 °C, where flop is inhibited (Figure 2 and 3a). Additionally, the dynamics of NBD-PC uptake in rta3Δ/Δ cells indicate an increase in the apparent PC-specific flippase activity because the exogenous addition of dithionite (quencher) maintained the accelerated internalization of NBD-PC (Figure 3b). We thus propose that the increased flip of PC may be attributed to the role of Rta3 as a negative regulator of PC-specific flippase activity (Figure 7). In conclusion, given the 7 TMD topology of Rta3, this study shows that functional Rta3, instead of functioning as a lipid translocator itself, may have a role in maintaining the asymmetric distribution of PC across the plasma membrane, possibly by modulating regulatory pathway(s) that signal unidentified PC-specific flippase(s) or floppase(s) in C. albicans.
The down regulation of genes that promote adherence as inferred from the transcriptional profiling data formed the basis for assaying the rta3Δ/Δ cells for biofilm formation (Figure 4a and c). The proposal that Rta3 is a positive regulator of biofilm formation and functions upstream to Bcr1 stems from the following observations. First, the catheter inoculated with the rta3Δ/Δ cells was essentially devoid of any material, similar to bcr1Δ /Δ, suggesting that there is a defect in early events of biofilm formation in vivo (Figure 5a). Second, the overexpression of BCR1 in rta3Δ/Δ cells partially rescues the biofilm defect of the mutant (Figure 5b). Third, the biofilm defect phenotype of the bcr1Δ /Δ cells was not rescued by the increased expression of RTA3 (Figure 5b). Our results therefore argue that Rta3 is pivotal for biofilm formation and that Bcr1 is one of the key downstream effector molecule of the Rta3-dependent regulatory pathway that contributes to biofilm formation (Figure 7).
Despite contributing to biofilm formation in vivo, failure of the rta3Δ/Δ cells to show adhesion defects in vitro suggests that the role of Rta3 in contributing to biofilm formation is dependent on a cue that is host-specific. In situations where this ‘cue’ is not present (such as in vitro conditions), Rta3 may not be essential for initiating biofilm formation. Many adherence regulators, such as ZFU2, CRZ2 and ZCF28, upon deletion fail to display an adhesion defect in vitro but display biofilm formation defects only in vivo (Finkel et al., 2012), similar to rta3Δ/Δ cells. These differences can be attributed to the fact that biofilm formation in vivo involves dynamic interactions with host factors. Hence, the molecular components that control biofilm formation in vivo may not necessarily affect this process to the same extent in vitro.
The results of our transcript analysis are indicative of a positive transcriptional feedback loop between Rta3 and downstream Bcr1 transcription factor (Figure 4c and d). Mutual transcriptional regulation similar of Rta3 and Bcr1 has also been observed in Rlm1-Slt2 MAPK and Ace2-Cek1 signalling pathways (Garcia et al., 2016, van Wijlick et al., 2016). These signalling pathways are modulated by feedback loops where the downstream components of the pathway regulate upstream elements of the same pathway. Positive feedback regulation amplifies the signal while negative feedback regulation helps in attenuation of the response. It is possible that in unique niches of the mammalian host, expression of Rta3 and BCR1 may be interdependent. The mutual regulation of Bcr1 and Rta3 may regulate the magnitude and duration of the signalling event entailing response to host specific cues and thus biofilm formation. In addition to mutual regulation, our study also shows a mitochondrial membrane energetics dependent expression of BCR1 (Figure 6d). We show that BCR1 is down regulated, similar to rta3Δ/Δ, in wild type cells treated with compounds that induce the breakdown of MMP (Figure S5). Based on these data, it is evident that Rta3 governs biofilm formation largely by regulating Bcr1.
Whether the distinct roles of Rta3 in regulating plasma membrane PC asymmetry and biofilm formation are linked remains to be investigated. Although we do not provide experimental evidence that can link enhanced flipping of PC to an adhesion defect and consequently biofilm formation, a study with Hela cells shows the existence of such a possibility. Enhanced flipping of PC mediated by a flippase ATP101A has been shown to be associated with changes in cell shape leading to delay in cell adhesion and cell spreading onto the extracellular matrix in Hela cells (Naito et al, 2015). Compatible with this, it is likely that enhanced flipping of PC in rta3Δ/Δ cells may be the basis for the observed adherence defect in vivo, leading to a defect in biofilm formation. How increase in the PC levels in the inner leaflet of the plasma membrane affect adhesion can be explained as follows. Firstly, as loss of phospholipid asymmetry signals multiple cellular events (Nichols, 2002) it is possible that the increased level of PC on the inner leaflet triggers a signalling event that converges to Bcr1 to down regulate its expression. Secondly, it is known that PS and phosphatidylinositiol 4,5-bisphosphate (PIP2), predominantly localized in the inner leaflet of the plasma membrane are vital for remodelling the actin-cytoskeleton, thus affecting the processes of adhesion and cell spreading (Yin et al, 2003, Naito et al, 2015). Thus, enhanced flipping of PC may perturb the PS and PIP2 equilibrium on the cytoplasmic leaflet, in turn inhibiting adhesion.
Tac1 regulated drug transporters (Cdr1 and Cdr2) are responsible for asymmetric distribution of phosphoglycerides across the plasma membrane (Smriti et al, 2002), in addition to contributing to the development of azole-resistance in C. albicans. Considering the role of Rta3 in maintaining the asymmetric distribution of PC across the plasma membrane, the physiological role of Tac1 coregulated genes under unstressed conditions may be to maintain phospholipid asymmetry in the plasma membrane. Furthermore, the function of RTA3 in conferring tolerance to another broad spectrum antifungal, miltefosine, highlights the physiological relevance of Tac1 regulated genes in defending Candida cells against xenobiotics present in the environment. The fact that Rta3 is unique to the fungal kingdom opens the possibility of interfering with the ability of C. albicans to thrive as a biofilm on indwelling devices in the human host. We also propose that the principal purpose of co-regulation of Rta3 with drug transporters in C. albicans may be to enable this pathogen to efficiently switch to the highly drug-resistant biofilm mode of growth in the human host, upon perceiving a host-specific stimuli. As a consequence, this pathogenic fungus is empowered to resist the onslaught of antifungals in both planktonic as well as the biofilm modes of growth in the host environment. In view of the fact that biofilm-specific drugs do not exist for C. albicans, identification of Rta3 as a novel component that primarily regulates Bcr1-dependent adhesion may lead to new strategies to prevent biofilm formation in this human fungal pathogen.
Experimental procedures
Strains, chemicals and growth conditions
The C. albicans strains used in this study are listed in Table S1. The strains were maintained as frozen stocks and propagated at 30 °C on the following media: YEPD (1% yeast extract, 2% peptone, 2% glucose, 2.5% agar) liquid media and agar plates. SDC media consists of 0.67% of yeast nitrogen base (Difco), 2% glucose and 0.02% complete amino acid supplement. SC medium is SDC lacking glucose but containing 2% sorbitol. YEPD plates containing 200 µg ml−1 nourseothricin (Werner Bioreagants) were used to select for deletion mutants. The following supplements, fluconazole (Sigma), miltefosine (Sigma), sodium dithionite (Sigma), cytochrome c (Sigma), sodium azide (Sigma), β-mercaptoethanol (Sigma), Zymolase-20T (MP biomedicals), Poly-L-lysine (Sigma), MitoTracker™ Red FM, DAPI (Molecular Probes/Invitrogen), carbonyl cyanide m-chlorophenyl hydrazone (CCCP); 2,7 dichlorofluorescein diacetate (DFCDA), Dihexyloxacarbocyanine iodide (DiOC6(3)) (Sigma) and NBD-PE and NBD-PC (Avanti Polar Lipids (Alabaster, AL), were added to the media/buffer at concentrations described.
Strain construction
(i) Deletion cassette construction- The first and second allele of the RTA3 gene was disrupted using the SAT1 flipper in the plasmid pSFS2B. Two different disruption cassettes were constructed for the two alleles of RTA3 in the disruption vector pSFS2B. For the RTA3 disruption construct, a 1000 bp 5′ upstream noncoding region (NCR) of RTA3 (5′ RTA3NCR) was amplified from SC5314 genomic DNA with primers RTA3P1 and RTA3P2 (Table S3), which introduced KpnI and XhoI restriction sites and cloned into 5′ end of the SAT1-FLP cassette in pSFS2B using the same enzymes. A 500 bp region of 3′ RTA3NCR was amplified with primers RTA3P3 and RTA3P4, which introduced NotI and SacII sites and was cloned in the 3′ end of the SAT1-FLP cassette (which already contains 5′ RTA3NCR). The plasmid thus constructed, containing the disruption cassette for deletion of the first allele of RTA3 is referred to pAS1 (Table S2). For deleting the second allele, 3’ end of pAS1 was replaced by 500 bp of the RTA3 ORF (amplified by primers RTA3P5 and RTA3P6) with NotI and SacII restriction sites, generating pAS2 (Table S2). The RTA3 reconstitution construct was made by amplifying a 2.3 kb fragment containing the RTA3 ORF (1.3 kb) and the 5′ NCR (1.0 kb) by using primers RTA3P1 and RTA3P7 that introduced KpnI and Xho1 sites The fragment was ligated into the KpnI-Xho1 digested pAS1 which already contained the 3′RTA3NCR. This procedure resulted in the RTA3 reconstitution plasmid (pAS3).
The wild-type strain, SC5314, was electroporated with the first round disruption cassette and deletion mutants were selected on 200 µg/ml nourseothricin (AS10). To obtain nourseothricin-sensitive derivatives of transformants, strains were grown in YPM (1% yeast extract, 2% peptone, 2% maltose) and plated on 25 µg/ml nourseothricin. These nourseothricin-sensitive heterozygous mutants (AS11) were then used for the second round of transformation generating the homozygous null mutant strain AS12. The CaSAT1 construct was flipped out from AS12, resulting in AS13. For reconstituted strain 2.3 kb fragment from pAS3 digested with KpnI and Xho1 was transformed in AS13 to yield, AS14. Proper integration at each step was confirmed by Southern hybridization. C-myc tagging of RTA3 gene- The C-terminal Myc-tagging plasmid, pADH34, containing a 13X Myc epitope tag preceding the SAT1-flipper cassette was used for epitope tagging as previously described (Nobile et al. 2009). The PCR amplicon obtained with primers RTA3mycFnostop and RTA3mycRUTR constitutes a 13X myc epitope tag, SAT1 flipper cassette, 65 bp region homologous to RTA3 ORF minus its stop codon on the 5’ end of the myc tag and a 65 bp region homologous to RTA3 UTR downstream of the stop codon on the 3’end of the SAT flipper cassette. This PCR product was transformed into SC5314 (wild type) to obtain strains AS15. Correct integration of the C-terminal 13X myc epitope tag and SAT1 flipper was verified by colony PCR using detection primers Rta3upstreamcheckF, AHO300, Rta3downstreamR and AHO301. The primer pairs Rta3upstreamcheckF and AHO300, and Rta3downstreamR and AHO302 were used in colony PCR to confirm the flipping out of the SAT1-flipper cassette. To obtain AS16 CaSAT was flipped out from AS15. The 13Xmyc epitope tag and the region of homology to the 3’end of RTA3 used for integration of the SAT1-flipper cassette was confirmed by sequencing the PCR product generated using primers Rta3upstreamcheckF and AHO283. Overexpression strain construction- The TDH3-RTA3 and TDH3-BCR1 overexpression C. albicans strains (Table S1) were constructed using plasmid pCJN542 (Nobile et al., 2008) (Primers listed in Table S3). These primers amplify the Ashbya gossypii TEF1 promoter, the C. albicans NAT1 ORF, the A. gossypii TEF1 terminator and the C. albicans TDH3 promoter with 100 bp of hanging homology to promoter region of RTA3 and BCR1. The transformation into C. albicans strains was done as described earlier and nourseothricin+ transformants were screened using detection primers listed in Table S3.
Complex IV activity assay
The protocol used was as described earlier by Spinazzi et al (Spinazzi et al., 2012). The complex IV assay was performed at 550 nm and the change in absorbance, resulting from the oxidation of reduced cytochrome c was measured. To reduce cytochrome c, 0.45 mM oxidized cytochrome c was dissolved in 20 mM potassium phosphate buffer (pH 7.0) and a few grains of sodium dithionite were added. Reduced cytochrome c brought to a final concentration of 20 µM in potassium phosphate buffer was checked for effective cytochrome c reduction by calculating the ratio of the absorbance values at 550 nm versus 565 nm. Cytochrome c oxidase activity was measured spectrophotometrically by adding 100 µl of 100 mM potassium phosphate buffer, 100 µl of reduced cytochrome c and the volume was adjusted with H2O in a cuvette. The reaction was started by adding 20 µg of mitochondrial protein (mitochondria isolation and purification was performed as described in li et al (Li et al., 2011)) and absorbance of the sample was measured at 550 nm for 2 min. The specificity of cytochrome c reduction was verified by inhibiting cytochrome c oxidase activity with 50 µl of 40 mM NaN3. Specific activity of complex IV was calculated by using the Beer-Lambert law equation.
Measurement of mitochondrial membrane potential
For this experiment cells were grown to early-log phase in YEPD at 30 °C, and labelled with 2 nM of DiOC6(3) for 30 minutes and analyzed by flow cytometry. As a positive control, cells were treated with 50 µM of CCCP for 40 min. In another experiment, wild type cells were treated with 50 µM of CCCP for 40 min and with 5 µg ml−1 of miltefosine for 60 min, followed by staining with 2 nM of DiOC6(3) for 30 min.
Immunofluorescence staining
Briefly, C. albicans strain carrying the Rta3-Myc was grown at 30 °C, and induced with 10 µg ml−1 fluphenazine for 30 min, in YEPD to OD600=0.5 (Total OD=10 for the experiment). Cell pellets of untreated and treated cultures were washed with 1X PBS, followed by fixing at room temperature in 4.5% formaldehyde for 1 h. The protocol followed was as described previously by Inglis et al (Inglis et al., 2002). The cells were resuspended in SP buffer (1.2 M sorbitol, 0.1 M potassium phosphate) with 1 to 2 µl of β-mercaptoethanol and 60 µl of Zymolase-20T (1mg ml−1) (MP biomedicals) and incubated at 37 °C with gentle shaking (300 rpm) for 30 min. 40 µl of cells was transferred to a polylysine-coated coverslip, and cell wall digestion was continued for 10 min at room temperature, while the cells settled onto the slide wells. To flatten cells, slides were subsequently submerged in methanol (at −20 °C) for 5 min, followed by acetone (-20 °C) for 30 seconds.
Cells were blocked at room temperature for 2 h in phosphate-buffered saline (PBS) with 1% bovine serum albumin (BSA) and 0.1% Tween 20. 20 µl anti Myc primary antibody (9B11 clone, Mouse, Cell Signaling Technology) was hybridized to cells at a 1:300 dilution (diluted in PBS with 1% BSA) for 1 h at room temperature. 20 µl of Cy2 conjugated donkey anti-mouse secondary antibodies (Jackson ImmunoResearch 715–225–151) at a 1:200 dilution was hybridized to cells in the dark for 1 h. Excess of antibodies were removed by washing the coverslip twice with 1X PBS and aspirated before the slides were treated with Fluormount-G (Southern Biotechnology Associates, Inc.) Digital images were captured by confocal using an 100X oil objective lens.
Microarray analysis
For transcriptional profiling, wild-type and mutant cells (three biological replicates) were grown overnight in 10 ml of YEPD at 30 °C and 200 rpm, subcultured to a starting OD600 of 0.3 in 10 ml of YEPD and grown until an OD600 of 1.5 at 30 °C and 200 rpm. RNA was extracted from three biological replicates of both strains using an RNeasy minikit (Qiagen). The cDNA samples were hybridized on to a custom C. albicans 8 ×15K microarray designed by Genotypic Technology Pvt., Ltd., Bangalore, India. All microarray data is available from GEO accession number GSE77774. Differentially expressed transcripts between mutant and wild type were identified by applying fold-change threshold of absolute fold-change ≥ 1.5 and a statistically significant t-test P-value threshold adjusted for false discovery rate of less than 0.05 derived using student t-test and visualized using Volcano Plot. Unsupervised hierarchical clustering of differentially expressed genes was done using Euclidian algorithm with centroid linkage rule to identify gene clusters. Gene ontology (GO) analysis was carried out using the GO Term Finder at the CGD (Candida Genome Database; http://www.candidagenome.org/cgi-bin/GO/goTerm Finder). For generating the core network, significantly enriched biological categories / gene ontology / pathways in the mutant, compared to wild type were subjected to network identification using BridgeIsland Software (Bionivid Technology Pvt Ltd, Bangalore, India), resulting in identification of key nodes and edges. Comparison of the transcription profile of rta3Δ/Δ cells with the profile of bcr1Δ /Δ cells was performed by accessing the profiles of the latter from the GEO database with accession numbers GSE33490 and GSE57451. The biological functions/pathways which we found to be altered in rta3Δ/Δ cells along with the list of genes involved therein were subjected to correlation covariance analysis with the three transcription profiles of bcr1Δ /Δ cells. The correlation coefficient values thus obtained were subjected to unsupervised hierarchical clustering using Pearson Uncentered algorithm with average linkage rule using Cluster 3.0 software.
Internalization of phospholipids into yeast cells
(i) For labelling experiments strains were grown to early-log phase in SDC at 30 °C, and labelled with 5 µM NBD-PC (DMSO solubilized), and 20 nM MitoTracker™ Red at OD600 1 for 45 minutes as described in Hanson et al, 2002 . Cells were washed three times with SDC and kept at 30 °C for additional 30 min. The cells were finally washed with ice-cold SC-azide three times and kept on ice until further analysis by microscopy or flow cytometry. (ii) For low temperature internalization assays, cells grown to OD600 of 1 were chilled in an ice bath for 10 min. Cells were treated 50 µM CCCP (resuspended in DMSO) for 30 min at 30 °C, prior to labelling by 5 µM NBD-PC. Untreated and CCCP treated cultures were returned to the ice bath and aliquots of labelled cells were removed at the indicated times and washed three times with SC-azide prior to microscopy and flow cytometric analysis. (ii) Flippase activity: Flow cytometric analysis of the flippase activity on wild type, rta3Δ/Δ and rta3Δ/Δ+RTA3 cells was done by using NBD-PC and sodium dithionite, an impermeant reducing agent used to quench the NBD-PC exposed on the cell surface (Popescu et al., 2010). Early log phase cells of wild type and rta3Δ/Δ were labelled with 2 µM NBD-PC. At the indicated times, an aliquot of cells were transferred to tubes with and without 25 mM sodium dithionite in SDC and washed with cold SC-azide. Residual MFI of NBD-PC in the presence (FD) or absence (FTotal) of dithionite was measured by flow cytometry. Percentage of internalized NBD-PC was represented as the normalized FD/Ftotal ratio. Triton X-100 (0.1%) was added to the dithionite group to allow quenching of internalized NBD-PC in control experiments. Residual FD/Ftotal ratio in permeabilized samples was less than 5%. Data are mean ± SEM of more than or equal to 10000 gated events.
Flow cytometry
Flow cytometry was performed using a FACSCalibur flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, CA) equipped with an argon laser emitting at 488 nm. Cells were grown to early-log phase (OD600 1) in YEPD at 30 °C, labelled with desired flourophore and analyzed. Fluorescence was measured on the FL1 fluorescence channel equipped with a 530-nm band-pass filter for NBD-PC, DiOC6(3) and DFCDA staining. A total of 10,000 events were counted. The data was analyzed using CellQuest V software.
In vivo C. albicans venous catheter biofilm model
A jugular vein rat central venous catheter infection model was used for in vivo biofilm studies, as previously described (Andes et al., 2004). After 24 h of C. albicans infection, catheters were removed from the rat. The distal 2 cm of catheter material was removed and subjected to scanning electron microscopy (SEM) for assaying biofilm growth.
Statistical analysis
Data were plotted and analysed using the GraphPad Software. Intergroup comparisons were made using the Student’s t-test. P-value of ≤ 0.05 was considered significant.
Ethics Statement
All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Wisconsin according to the guidelines of the Animal Welfare Act, The Institute of Laboratory Animal Resources Guide for the Care and Use of Laboratory Animals and Public Health Service Policy under protocol MV1947.
Other Procedures
Quantitative real time PCR (primers used for qPCR analysis are listed in Table S4), serial dilution spotting assay, southern hybridization, reactive oxygen species measurement and confocal microscopy were carried out as described previously (Thomas et al., 2013, Thomas et al., 2015).
Supplementary Material
Acknowledgments
We thank Joachim Morschhauser, Aaron Mitchell and Suzanne Noble for providing us with the strains and plasmids used in this study. Steven Claypool and Megha Gulati are acknowledged for assisting on crucial experiments. We are thankful to the Advanced Instrumentation Research Facility (AIRF), Jawaharlal Nehru University, for and technical assistance from Ashok Sahu in performing the confocal microscopy. Genotypic Technology Pvt., Ltd., and Bionivid Technology, Bangalore, India, are acknowledged for performing the microarray experiments and analysis, respectively. A very special thanks to Wylie Nichols for setting the stage for this study, with his useful insights. We appreciate Shyamal Goswami for providing valuable suggestions during the preparation of the manuscript.
FUNDING
This study was supported by grants from the Department of Biotechnology (BT/PR10362/BRB/10/596/2007 and BT/PR10324/MED/29/808/2013) and Department of Science and Technology (SR/FT/L-26/2006) to SLP. Additional funding from umbrella grants (DST-PURSE, Capacity Build-up and UGC-Resource Networking) provided to Jawaharlal Nehru University are acknowledged. AS acknowledges Senior Research Fellowship from ICMR, SS acknowledges UGC-Junior Research Fellowship, ET acknowledges UGC sponsored Dr D. S. Kothari Post-Doctoral Fellowship Scheme, SR acknowledges CSIR-Junior Research Fellowship, FH and DA acknowledge UGC-NON-NET Fellowship, SR acknowledges Senior Research Fellowship from DBT. PB and CJN were supported by the National Institutes of Health (NIH) grant R00AI100896 to CJN.
References
- Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infection and immunity. 2004;72:6023–6031. doi: 10.1128/IAI.72.10.6023-6031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas RJ, Heitman J, Cardenas ME. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog. 2009;5:e1000294. doi: 10.1371/journal.ppat.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan NM, Shinde RB, Karuppayil SM. Effect of alcohols on filamentation, growth, viability and biofilm development in Candida albicans. Brazilian journal of microbiology : [publication of the Brazilian Society for Microbiology] 2013;44:1315–1320. doi: 10.1590/S1517-83822014005000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryotic cell. 2004;3:1639–1652. doi: 10.1128/EC.3.6.1639-1652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft SL, Snowdon D, Yardley V. The activities of four anticancer alkyllysophospholipids against Leishmania donovani, Trypanosoma cruzi and Trypanosoma brucei. J Antimicrob Chemother. 1996;38:1041–1047. doi: 10.1093/jac/38.6.1041. [DOI] [PubMed] [Google Scholar]
- Devaux F, Carvajal E, Moye-Rowley S, Jacq C. Genome-wide studies on the nuclear PDR3-controlled response to mitochondrial dysfunction in yeast. FEBS letters. 2002;515:25–28. doi: 10.1016/s0014-5793(02)02387-6. [DOI] [PubMed] [Google Scholar]
- Devaux PF, Herrmann A, Ohlwein N, Kozlov MM. How lipid flippases can modulate membrane structure. Biochimica et biophysica acta. 2008;1778:1591–1600. doi: 10.1016/j.bbamem.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Ding C, Vidanes GM, Maguire SL, Guida A, Synnott JM, Andes DR, Butler G. Conserved and divergent roles of Bcr1 and CFEM proteins in Candida parapsilosis and Candida albicans. PloS one. 2011;6:1. doi: 10.1371/journal.pone.0028151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadeel B, Xue D. The ins and outs of phospholipid asymmetry in the plasma membrane: roles in health and disease. Critical reviews in biochemistry and molecular biology. 2009;44:264–277. doi: 10.1080/10409230903193307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning S, Xu W, Solis N, Woolford CA, Filler SG, Mitchell AP. Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryotic cell. 2012;11:896–904. doi: 10.1128/EC.00103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nature reviews. Microbiology. 2011;9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, et al. Portrait of Candida albicans adherence regulators. PLoS Pathog. 2012;8:e1002525. doi: 10.1371/journal.ppat.1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox EP, Bui CK, Nett JE, Hartooni N, Mui MC, Andes DR, et al. An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol Microbiol. 2015;96:1226–1239. doi: 10.1111/mmi.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia R, Sanz AB, Rodriguez-Pena JM, Nombela C, Arroyo J. Rlm1 mediates positive autoregulatory transcriptional feedback that is essential for Slt2-dependent gene expression. Journal of cell science. 2016;129:1649–1660. doi: 10.1242/jcs.180190. [DOI] [PubMed] [Google Scholar]
- Grant AM, Hanson PK, Malone L, Nichols JW. NBD-labeled phosphatidylcholine and phosphatidylethanolamine are internalized by transbilayer transport across the yeast plasma membrane. Traffic (Copenhagen, Denmark) 2001;2:37–50. doi: 10.1034/j.1600-0854.2001.020106.x. [DOI] [PubMed] [Google Scholar]
- Haldar S, Chattopadhyay A. Application of NBD-Labeled Lipids in Membrane and Cell Biology. In: Mély Y, Duportail G, editors. Fluorescent Methods to Study Biological Membranes. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013. pp. 37–50. [Google Scholar]
- Hanson PK, Grant AM, Nichols JW. NBD-labeled phosphatidylcholine enters the yeast vacuole via the pre-vacuolar compartment. Journal of cell science. 2002;115:2725–2733. doi: 10.1242/jcs.115.13.2725. [DOI] [PubMed] [Google Scholar]
- Hanson PK, Nichols JW. Energy-dependent flip of fluorescence-labeled phospholipids is regulated by nutrient starvation and transcription factors, PDR1 and PDR3. The Journal of biological chemistry. 2001;276:9861–9867. doi: 10.1074/jbc.M009065200. [DOI] [PubMed] [Google Scholar]
- Holland LM, Schroder MS, Turner SA, Taff H, Andes D, Grozer Z, et al. Comparative phenotypic analysis of the major fungal pathogens Candida parapsilosis and Candida albicans. PLoS Pathog. 2014:10. doi: 10.1371/journal.ppat.1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis DO, Johnson AD. Ash1 protein, an asymmetrically localized transcriptional regulator, controls filamentous growth and virulence of Candida albicans. Molecular and cellular biology. 2002;22:8669–8680. doi: 10.1128/MCB.22.24.8669-8680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis DO, Sherlock G. Ras signaling gets fine-tuned: regulation of multiple pathogenic traits of Candida albicans. Eukaryotic cell. 2013;12:1316–1325. doi: 10.1128/EC.00094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabra-Rizk MA, Falkler WA, Meiller TF. Fungal biofilms and drug resistance. Emerging infectious diseases. 2004;10:14–19. doi: 10.3201/eid1001.030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia XM, Ma ZP, Jia Y, Gao PH, Zhang JD, Wang Y, et al. RTA2, a novel gene involved in azole resistance in Candida albicans. Biochemical and biophysical research communications. 2008;373:631–636. doi: 10.1016/j.bbrc.2008.06.093. [DOI] [PubMed] [Google Scholar]
- Johnson SS, Hanson PK, Manoharlal R, Brice SE, Cowart LA, Moye-Rowley WS. Regulation of yeast nutrient permease endocytosis by ATP-binding cassette transporters and a seven-transmembrane protein, RSB1. The Journal of biological chemistry. 2010;285:35792–35802. doi: 10.1074/jbc.M110.162883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annual review of pharmacology and toxicology. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean LS, Fuller RS, Nichols JW. Retrograde lipid traffic in yeast: identification of two distinct pathways for internalization of fluorescent-labeled phosphatidylcholine from the plasma membrane. The Journal of cell biology. 1993;123:1403–1419. doi: 10.1083/jcb.123.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhina S, Johnson SS, Manoharlal R, Russo SB, Blugeon C, Lemoine S, et al. Control of Plasma Membrane Permeability by ABC Transporters. Eukaryotic cell. 2015;14:442–453. doi: 10.1128/EC.00021-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Igarashi Y. Identification and characterization of a Saccharomyces cerevisiae gene, RSB1, involved in sphingoid long-chain base release. The Journal of biological chemistry. 2002;277:30048–30054. doi: 10.1074/jbc.M203385200. [DOI] [PubMed] [Google Scholar]
- Kihara A, Igarashi Y. Cross talk between sphingolipids and glycerophospholipids in the establishment of plasma membrane asymmetry. Molecular biology of the cell. 2004;15:4949–4959. doi: 10.1091/mbc.E04-06-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski M, Kolaczkowska A, Gaigg B, Schneiter R, Moye-Rowley WS. Differential regulation of ceramide synthase components LAC1 and LAG1 in Saccharomyces cerevisiae. Eukaryotic cell. 2004;3:880–892. doi: 10.1128/EC.3.4.880-892.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinov SM, Eibl H, Berger MR. Alkylphosphocholines induce apoptosis in HL-60 and U-937 leukemic cells. Cancer chemotherapy and pharmacology. 1998;41:210–216. doi: 10.1007/s002800050730. [DOI] [PubMed] [Google Scholar]
- Li D, Chen H, Florentino A, Alex D, Sikorski P, Fonzi WA, Calderone R. Enzymatic dysfunction of mitochondrial complex I of the Candida albicans goa1 mutant is associated with increased reactive oxidants and cell death. Eukaryotic cell. 2011;10:672–682. doi: 10.1128/EC.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TT, Znaidi S, Barker KS, Xu L, Homayouni R, Saidane S, et al. Genome-wide expression and location analyses of the Candida albicans Tac1p regulon. Eukaryotic cell. 2007;6:2122–2138. doi: 10.1128/EC.00327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Gulati M, Valle Arevalo A, Fishburn A, Johnson AD, Nobile CJ. Assessment and Optimizations of Candida albicans In Vitro Biofilm Assays. Antimicrobial agents and chemotherapy. 2017:61. doi: 10.1128/AAC.02749-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manente M, Ghislain M. The lipid-translocating exporter family and membrane phospholipid homeostasis in yeast. FEMS Yeast Res. 2009;9:673–687. doi: 10.1111/j.1567-1364.2009.00513.x. [DOI] [PubMed] [Google Scholar]
- McIntyre JC, Sleight RG. Fluorescence assay for phospholipid membrane asymmetry. Biochemistry. 1991;30:11819–11827. doi: 10.1021/bi00115a012. [DOI] [PubMed] [Google Scholar]
- Michan C, Pueyo C. Growth phase-dependent variations in transcript profiles for thioredoxin- and glutathione-dependent redox systems followed by budding and hyphal Candida albicans cultures. FEMS Yeast Res. 2009;9:1078–1090. doi: 10.1111/j.1567-1364.2009.00558.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee PK, Mohamed S, Chandra J, Kuhn D, Liu S, Antar OS, et al. Alcohol dehydrogenase restricts the ability of the pathogen Candida albicans to form a biofilm on catheter surfaces through an ethanol-based mechanism. Infection and immunity. 2006;74:3804–3816. doi: 10.1128/IAI.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Takatsu H, Miyano R, Takada N, Nakayama K, Shin HW. Phospholipid Flippase ATP10A Translocates Phosphatidylcholine and Is Involved in Plasma Membrane Dynamics. The Journal of biological chemistry. 2015;290:15004–15017. doi: 10.1074/jbc.M115.655191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols JW. Internalization and trafficking of fluorescent-labeled phospholipids in yeast. Seminars in cell & developmental biology. 2002;13:179–184. doi: 10.1016/s1084-9521(02)00046-0. [DOI] [PubMed] [Google Scholar]
- Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, et al. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006a:2. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Johnson AD. Candida albicans Biofilms and Human Disease. Annual review of microbiology. 2015;69:71–92. doi: 10.1146/annurev-micro-091014-104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Current biology : CB. 2005;15:1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- Nobile CJ, Nett JE, Andes DR, Mitchell AP. Function of Candida albicans adhesion Hwp1 in biofilm formation. Eukaryotic cell. 2006b;5:1604–1610. doi: 10.1128/EC.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, Mitchell AP. Complementary adhesin function in C. albicans biofilm formation. Current biology : CB. 2008;18:1017–1024. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panatala R, Hennrich H, Holthuis JC. Inner workings and biological impact of phospholipids flippases. Journal of cell science. 2015;128:2021–2032. doi: 10.1242/jcs.102715. [DOI] [PubMed] [Google Scholar]
- Panwar SL, Moye-Rowley WS. Long chain base tolerance in Saccharomyces cerevisiae is induced by retrograde signals from the mitochondria. The Journal of biological chemistry. 2006;281:6376–6384. doi: 10.1074/jbc.M512115200. [DOI] [PubMed] [Google Scholar]
- Popescu NI, Lupu C, Lupu F. Extracellular protein disulfide isomerase regulates coagulation on endothelial cells through modulation of phosphatidylserine exposure. Blood. 2010;116:993–1001. doi: 10.1182/blood-2009-10-249607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Banerjee A, Khandelwal NK, Dhamgaye S. The ABCs of Candida albicans Multidrug Transporter Cdr1. Eukaryotic cell. 2015;14:1154–1164. doi: 10.1128/EC.00137-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, Bachmann S, Patterson TF, Wickes BL, Lopez-Ribot JL. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J Antimicrob Chemother. 2002;49:973–980. doi: 10.1093/jac/dkf049. [DOI] [PubMed] [Google Scholar]
- Ramage G, Martinez JP, Lopez-Ribot JL. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res. 2006;6:979–986. doi: 10.1111/j.1567-1364.2006.00117.x. [DOI] [PubMed] [Google Scholar]
- Rane HS, Bernardo SM, Walraven CJ, Lee SA. In vitro analyses of ethanol activity against Candida albicans biofilms. Antimicrobial agents and chemotherapy. 2012;56:4487–4489. doi: 10.1128/AAC.00263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Riezman H. Endocytosis in yeast: several of the yeast secretory mutants are defective in endocytosis. Cell. 1985;40:1001–1009. doi: 10.1016/0092-8674(85)90360-5. [DOI] [PubMed] [Google Scholar]
- Schweizer A, Rupp S, Taylor BN, Rollinghoff M, Schroppel K. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol Microbiol. 2000;38:435–445. doi: 10.1046/j.1365-2958.2000.02132.x. [DOI] [PubMed] [Google Scholar]
- Smriti, SmritiKrishnamurthy S, Dixit BL, Gupta CM, Milewski S, Prasad R. ABC transporters Cdr1p, Cdr2p and Cdr3p of a human pathogen Candida albicans are general phospholipid translocators. Yeast (Chichester, England) 2002;19:303–318. doi: 10.1002/yea.818. [DOI] [PubMed] [Google Scholar]
- Spinazzi M, Casarin A, Pertegato V, Salviati L, Angelini C. Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nature protocols. 2012;7:1235–1246. doi: 10.1038/nprot.2012.058. [DOI] [PubMed] [Google Scholar]
- Stevens HC, Nichols JW. The proton electrochemical gradient across the plasma membrane of yeast is necessary for phospholipid flip. The Journal of biological chemistry. 2007;282:17563–17567. doi: 10.1074/jbc.M700454200. [DOI] [PubMed] [Google Scholar]
- Stuart MC, Bevers EM, Comfurius P, Zwaal RF, Reutelingsperger CP, Frederik PM. Ultrastructural detection of surface exposed phosphatidylserine on activated blood platelets. Thrombosis and haemostasis. 1995;74:1145–1151. [PubMed] [Google Scholar]
- Thomas E, Roman E, Claypool S, Manzoor N, Pla J, Panwar SL. Mitochondria influence CDR1 efflux pump activity, Hog1-mediated oxidative stress pathway, iron homeostasis, and ergosterol levels in Candida albicans. Antimicrobial agents and chemotherapy. 2013;57:5580–5599. doi: 10.1128/AAC.00889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas E, Sircaik S, Roman E, Brunel JM, Johri AK, Pla J, Panwar SL. The activity of RTA2, a downstream effector of the calcineurin pathway, is required during tunicamycin-induced ER stress response in Candida albicans. FEMS Yeast Res. 2015;15:29. doi: 10.1093/femsyr/fov095. [DOI] [PubMed] [Google Scholar]
- Tong L, Phan TK, Robinson KL, Babusis D, Strab R, Bhoopathy S, et al. Effects of human immunodeficiency virus protease inhibitors on the intestinal absorption of tenofovir disoproxil fumarate in vitro. Antimicrobial agents and chemotherapy. 2007;51:3498–3504. doi: 10.1128/AAC.00671-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk WJ, Verheij M. Anticancer alkylphospholipids: mechanisms of action, cellular sensitivity and resistance, and clinical prospects. Current pharmaceutical design. 2008;14:2061–2074. doi: 10.2174/138161208785294636. [DOI] [PubMed] [Google Scholar]
- van Wijlick L, Swidergall M, Brandt P, Ernst JF. Candida albicans responds to glycostructure damage by Ace2-mediated feedback regulation of Cek1 signaling. Mol Microbiol. 2016 doi: 10.1111/mmi.13494. [DOI] [PubMed] [Google Scholar]
- Vayssiere JL, Petit PX, Risler Y, Mignotte B. Commitment to apoptosis is associated with changes in mitochondrial biogenesis and activity in cell lines conditionally immortalized with simian virus 40. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:11752–11756. doi: 10.1073/pnas.91.24.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila T, Ishida K, Seabra SH, Rozental S. Miltefosine inhibits Candida albicans and non-albicans Candida spp. biofilms and impairs the dispersion of infectious cells. International journal of antimicrobial agents. 2016;48:512–520. doi: 10.1016/j.ijantimicag.2016.07.022. [DOI] [PubMed] [Google Scholar]
- Vila TV, Chaturvedi AK, Rozental S, Lopez-Ribot JL. In Vitro Activity of Miltefosine against Candida albicans under Planktonic and Biofilm Growth Conditions and In Vivo Efficacy in a Murine Model of Oral Candidiasis. Antimicrobial agents and chemotherapy. 2015;59:7611–7620. doi: 10.1128/AAC.01890-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whaley SG, Tsao S, Weber S, Zhang Q, Barker KS, Raymond M, Rogers PD. The RTA3 Gene, Encoding a Putative Lipid Translocase, Influences the Susceptibility of Candida albicans to Fluconazole. Antimicrobial agents and chemotherapy. 2016;60:6060–6066. doi: 10.1128/AAC.00732-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer F, Wright LC, Obando D, Handke R, Ganendren R, Ellis DH, Sorrell TC. Hexadecylphosphocholine (miltefosine) has broad-spectrum fungicidal activity and is efficacious in a mouse model of cryptococcosis. Antimicrobial agents and chemotherapy. 2006;50:414–421. doi: 10.1128/AAC.50.2.414-421.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeater KM, Chandra J, Cheng G, Mukherjee PK, Zhao X, Rodriguez-Zas SL, et al. Temporal analysis of Candida albicans gene expression during biofilm development. Microbiology (Reading, England) 2007;153:2373–2385. doi: 10.1099/mic.0.2007/006163-0. [DOI] [PubMed] [Google Scholar]
- Yin HL, Janmey PA. Phosphoinositide regulation of the actin cytoskeleton. Annual review of physiology. 2003;65:761–789. doi: 10.1146/annurev.physiol.65.092101.142517. [DOI] [PubMed] [Google Scholar]
- Zuo X, Djordjevic JT, Bijosono Oei J, Desmarini D, Schibeci SD, Jolliffe KA, Sorrell TC. Miltefosine induces apoptosis-like cell death in yeast via Cox9p in cytochrome c oxidase. Molecular pharmacology. 2011;80:476–485. doi: 10.1124/mol.111.072322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.