Summary
In yeast target of rapamycin complex 1 (TORC1) and Tap42-associated phosphatases regulate expression of genes involved in nitrogen limitation response and the nitrogen discrimination pathway. However, it remains unclear whether TORC1 and the phosphatases are required for sensing nitrogen conditions. Utilizing temperature sensitive mutants of tor2 and tap42, we examined the role of TORC1 and Tap42 in nuclear entry of Gln3, a key transcription factor in yeast nitrogen metabolism, in response to changes in nitrogen conditions. Our data show that TORC1 is essential for Gln3 nuclear entry upon nitrogen limitation and downshift in nitrogen quality. However, Tap42-associated phosphatases are required only under nitrogen limitation condition. In cells grown in poor nitrogen medium, the nitrogen permease reactivator kinase (Npr1) inhibits TORC1 activity and alters its association with Tap42, rendering Tap42-associated phosphatases unresponsive to nitrogen limitation. These findings demonstrate a direct role for TORC1 and Tap42-associated phosphatases in sensing nitrogen conditions and unveil an Npr1-dependent mechanism that controls TORC1 and the phosphatases in response to changes in nitrogen quality.
Keywords: TORC1, Tap42, Npr1, nitrogen, rapamycin
Graphical abstract
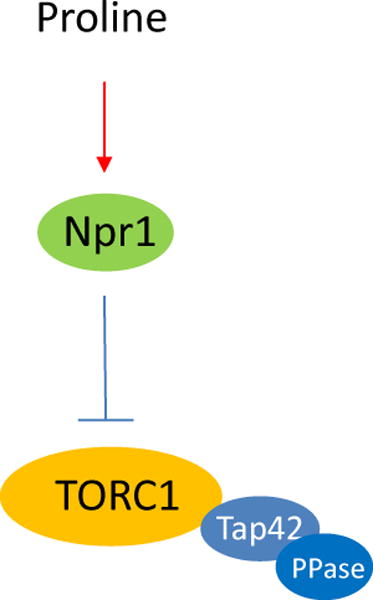
This study shows that target of rapamycin complex 1 (TORC1) and Tap42 are directly involved in sensing changes in nitrogen conditions and their activities are inhibited by the Npr1 kinase in response to downshift in nitrogen quality.
Introduction
The yeast Saccharomyces cerevisiae, like many other microorganisms, is able to utilize various nitrogen-containing compounds as nitrogen sources for growth and survival. The cells possess the ability to assess quantity as well quality of nitrogen sources and make appropriate adjustments in their transcriptional and metabolic profiles for optimal fitness (Magasanik & Kaiser, 2002, Zaman et al., 2008). For commonly used laboratory yeast strains, glutamine and ammonium are preferred nitrogen sources, whereas proline and urea are non-preferred, hence poor nitrogen sources. When grown in the presence of preferred nitrogen sources, yeast cells repress the expression of genes involved in uptake and metabolism of poor nitrogen-containing compounds. Upon nitrogen limitation or when poor nitrogen-containing compounds become the only nitrogen source, yeast cells derepress the genes required for the alternative nitrogen metabolism (Magasanik & Kaiser, 2002).
The ability for the yeast cells to adjust transcriptional profiles in response to different sources of nitrogen depends on the nitrogen discrimination pathway (NDP), which controls the expression of more than 90 NDP genes (Magasanik & Kaiser, 2002). A key transcriptional activator involved in the process is Gln3, a GATA family zinc-finger transcription factor. In cells grown in the presence of glutamine or ammonium, Gln3 is sequestered in the cytoplasm through association with Ure2. When cells encounter nitrogen limitation or being downshifted to poor nitrogen medium, Gln3 dissociates from Ure2 and translocates into the nucleus, where it activates the expression of NDP genes. Previous studies have shown that phosphorylation of Gln3 plays an important role in its association with Ure2 and nuclear translocation (Cooper, 2002). Shifting cells from good nitrogen sources to nitrogen-free condition triggers rapid dephosphorylation of Gln3 (Bertram et al., 2002, Yan et al., 2012a), which accompanies its dissociation from Ure2 and nuclear entry (Bertram et al., 2000, Cardenas et al., 1999). However, the dephosphorylation is not observed when cells are switched from preferred nitrogen to poor nitrogen medium, despite the fact that the downshift in nitrogen quality induces nuclear entry of Gln3 (Cox et al., 2004a).
A key factor involved in controlling Gln3 activity is target of rapamycin complex 1 (TORC1). TORC1 is a multi-protein complex that contains the Tor1 or Tor2 kinase as alternative catalytic subunit and three other proteins, including Kog1, Lst8 and Tco89. Kog1 serves as the regulatory subunit that defines the substrate specificity of the Tor kinases. Rapamycin, in complex with FKBP12, binds specifically to the Tor kinases within TORC1 and blocks its signaling activity (Wei & Zheng, 2011, Wullschleger et al., 2006). Treating yeast cells grown in good nitrogen with rapamycin causes Gln3 dephosphorylation and consequently its nuclear translocation and activation (Beck & Hall, 1999, Bertram et al., 2000, Cardenas et al., 1999). The drug-induced Gln3 dephosphorylation requires the Sit4 phosphatase, a unique form of type 2A protein phosphatase. Sit4 and other type 2A-like phosphatases exist in complex with Tap42, which serves as a regulatory subunit to control their activity and localization. The Tap42-phosphatase complexes are associated with TORC1 in an inactive state under normal growth condition but are released and activated in response to adverse conditions, including cell wall damage, heat, nitrogen starvation, and rapamycin treatment (Duvel & Broach, 2004, Yan et al., 2006, Yan et al., 2012a). In Tap42 deficient cells, the drug or nitrogen starvation-induced depression of NDP genes is compromised, which has led to the suggestion that signals for nitrogen conditions are transmitted through TORC1 and Tap42-associated phosphatases to regulate Gln3 and GDP gene expression (Duvel et al., 2003, Shamji et al., 2000). However, the roles of TORC1 and the phosphatases in nitrogen signaling remain unclear. Evidence exists indicating that nitrogen conditions can affect Gln3 localization independent of TORC1 and Tap42-associated phosphatases (Cox et al., 2004a, Cox et al., 2004b).
In this study, using temperature sensitive mutants of TOR2 and TAP42, we examined the roles of TORC1 and Tap42 in controlling Gln3 activation in response to changes in nitrogen conditions. Our findings show that TORC1 and Tap42 are directly involved in sensing nitrogen conditions.
Results
Tap42 is dispensable for nuclear entry of Gln3 in response to nitrogen downshift
To determine the role of Tap42-associated phosphatases in nitrogen signaling, we examined Gln3 nuclear translocation in response to changes in nitrogen conditions in Tap42 deficient cells. We selected a temperature sensitive mutant of tap42 (tap42-119) for the analysis. We have previously shown that elevated temperature is able to trigger release and activation of Tap42-associated phosphatases from TORC1 in wild type cells. When nitrogen is available, the release and activation are transient. Tap42 and the phosphatases re-associate with TORC1 in less than 2 hr and the cells regain responsiveness to other adverse conditions, including rapamycin treatment and nitrogen limitation (Yan et al., 2012a). Utilizing this feature, we examined the nuclear translocation of Gln3 in response to changes in nitrogen conditions after inactivating Tap42 by shifting the tap42 mutant cells to the nonpermissive temperature (37°C) for 2 hr. In wild type cells grown at 23°C in glutamine medium, Gln3 was mainly cytosolic and only a small fraction of cells contained nuclear localized Gln3 (Fig. 1A, left panel). A similar distribution of Gln3 was observed after the cells were shifted to 37°C for 2 hr (Fig. 1A and Supplemental Fig. 1A). When the cells were shifted to nitrogen-free medium, a large portion of cells had Gln3 accumulated in the nucleus and the growth temperature had no obvious effects on the distribution (Fig. 1A, left panel). The distribution patterns of Gln3 in tap42 mutant cells grown at the permissive temperature (23°C) were similar to those of wild type cells in either glutamine or after being shifted to nitrogen-free medium. However, after incubation of the mutant cells at 37°C for 2 hr, the nitrogen limitation-induced nuclear entry of Gln3 was largely blocked (Fig. 1A, right panel and Supplemental Fig. 1A). Consistent with the lack of nuclear accumulation of Gln3, the nitrogen limitation-induced expression of GLN1, a transcriptional target of Gln3, was impaired in the mutant cells grown at the nonpermissive temperature (Supplemental Fig. 2A).
Figure 1. Tap42 is not required for nuclear entry of Gln3 in response to nitrogen downshift.
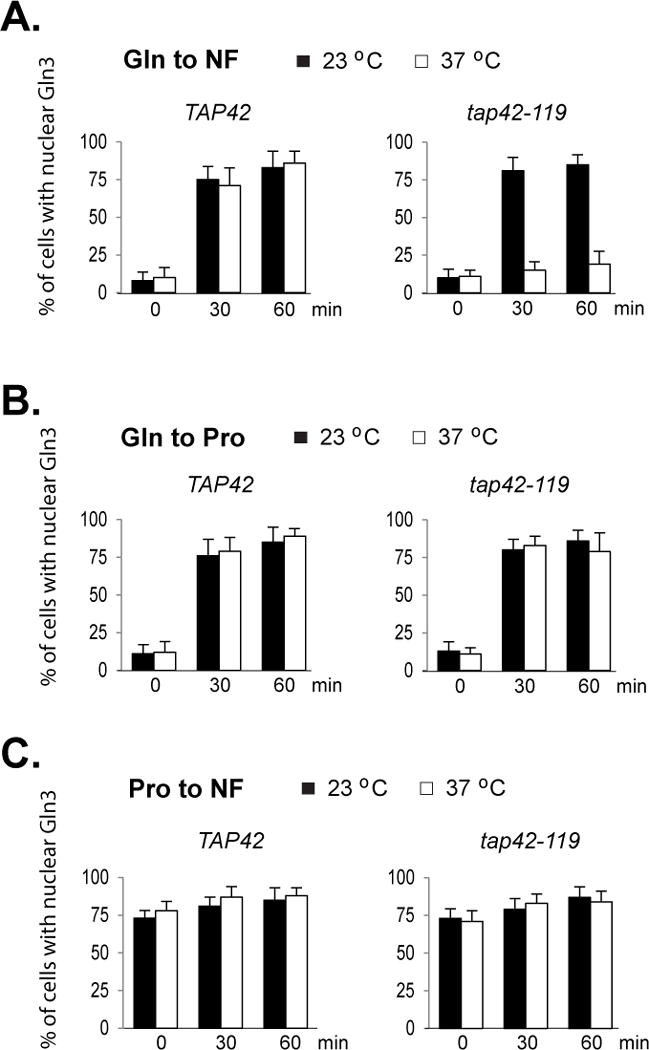
Wild type (Y339) and tap42-119 mutant (Y351) expressing GLN3-myc were grown at 23°C to early log phase in glutamine (Gln) or proline (Pro) medium. The cultures were divided and incubated for two additional hours with one half at 37°C and the other half at 23°C. A. Cells were transferred from glutamine (Gln) to nitrogen-free (NF) medium. B. Cells were transferred from glutamine (Gln) to proline (Pro) medium. C. Cells were transferred from proline to nitrogen-free medium. Cells were collected before (time 0), 30 and 60 min after the transfer. Localization of Gln3-myc in the cells was examined by confocal microscopy. The experiment was repeated three times. Percentages of cells with nuclear staining of Gln3-myc are expressed as mean ± SD.
It has been previously shown that downshift in nitrogen quality causes nuclear entry of Gln3 (Cox et al., 2002). Consistent with the finding, we found that when wild type cells grown at either 23 or 37°C were shifted from glutamine medium to proline medium, Gln3 was concentrated in the nuclei of the majority of the cells (Fig. 1B, left panel and Supplemental Fig. 1A). The downshift in nitrogen quality also induced nuclear entry of Gln3 in tap42 mutant cells grown at 23°C as well as at 37°C (Fig. 1B, right panel and Supplemental Fig. 1A). In line with the finding, we found that the nitrogen downshift was able to induce expression of GLN1 in the tap42 mutant cells grown at the nonpermissive temperature (Supplemental Fig. 2B). These observations show that inactivation of Tap42 does not block the effect of nitrogen downshift on Gln3, indicating that Tap42 is not involved in sensing changes in nitrogen quality.
When wild type and tap42 mutant cells were grown in proline medium, a large portion of the cells contained nuclear Gln3. Shifting the cells to nitrogen-free condition did not significantly affect the nuclear distribution of Gln3 (Fig. 1C).
TORC1 is required for nuclear entry of Gln3 in response to changes in nitrogen conditions
To determine the role of TORC1 in sensing changes in nitrogen conditions, we utilized a temperature sensitive mutant allele of TOR2, tor2-219. This mutant allele is temperature sensitive for growth in the both TOR1 wild type (TOR1 tor2-219) and tor1 deletion (tor1 tor2-219) backgrounds, indicating that the allele is defective for both TORC1 and TORC2 function. In the tor2-219 single mutant cells, because of the presence of wild type TOR1, TORC1 is expected to remain functional after the cells are shifted to the nonpermissive temperature. This mutant thus serves as a positive control for TORC1 activity. In addition, since the single and double mutants are both temperature sensitive for growth, the single mutant serves as an appropriate control for the double mutant, allowing us to rule out potential complications that may result from growth cessation-induced by the elevated temperature. When the tor2-219 single mutant cells were grown at 23 or 37°C in glutamine medium, only a small fraction of cells contained nuclear Gln3. Shifting the cells to nitrogen-free medium caused nuclear accumulation of Gln3 (Fig. 2A, left panel and Supplemental Fig. 1B). The distribution of Gln3 in tor1 tor2-219 double mutant cells grown in glutamine medium was similar to that of the tor2-219 single mutant cells at both 23 and 37°C (Fig. 2A, right panel). At the permissive temperature (23°C), shifting the double mutant cells from glutamine medium to nitrogen-free medium caused nuclear entry of Gln3. However, after the cells were pre-incubated at 37°C for 2 hr, which inactivated the Tor2 mutant protein, the same change in nitrogen condition failed to induce Gln3 nuclear entry (Fig. 2A, right panel and Supplemental Fig. 1B). Inactivation of the Tor2 mutant protein also blocked the nitrogen limitation-induced expression of GLN1 (Supplemental Fig. 3A). Because the tor2-219 single but not the tor1 tor2-219 double mutant contains wild type TOR1, it is expected that TORC1 remained functional in the single but not in double mutant cells when the Tor2 mutant protein was inactivated at 37°C. These findings thus indicate that a functional TORC1 is required for nitrogen limitation-induced Gln3 response.
Figure 2. TORC1 is required for nuclear entry of Gln3 in response to nitrogen limitation and downshift.
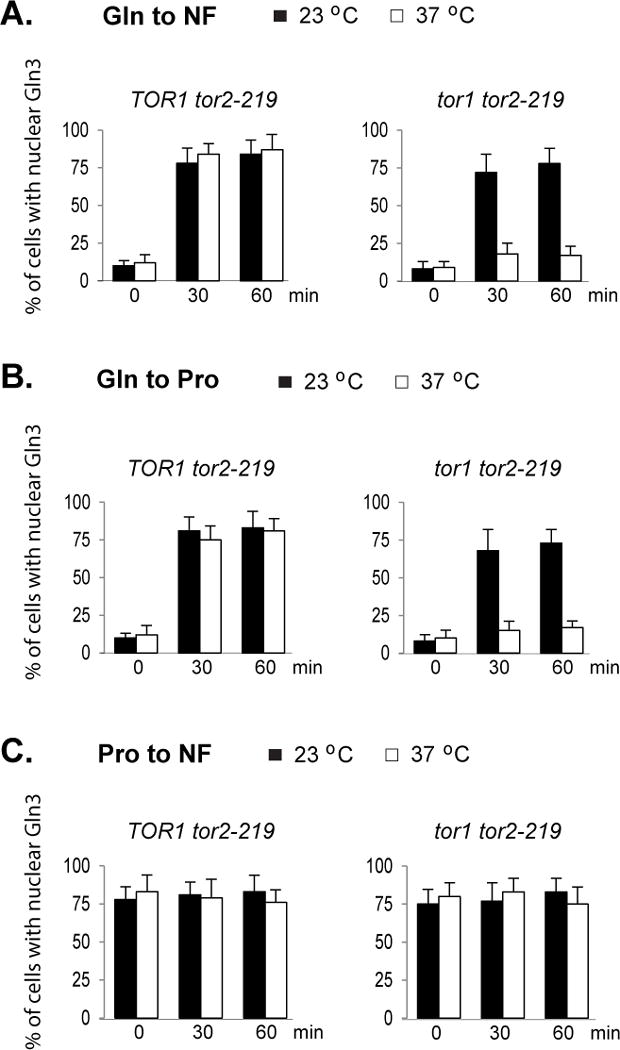
The tor2-219 (Y511) and tor1 tor2-102 mutant (Y442) cells expressing GLN3-myc were grown at 23°C to early log phase in glutamine (Gln) or proline (Pro) medium. The cultures were divided and incubated for two additional hours with one half at 37°C and the other half at 23°C. A. Cells were transferred from glutamine (Gln) to nitrogen-free (NF) medium. B. Cells were transferred from glutamine (Gln) to proline (Pro) medium. C. Cells were transferred from proline to nitrogen-free medium. Cells were collected before (time 0), 30 and 60 min after the shift. Localization of Gln3-myc in the cells was examined by confocal microscopy. The experiment was repeated three times. Percentages of cells with nuclear staining of Gln3-myc expressed as mean ± SD are shown.
Shifting tor2-219 single mutant cells (TOR1 tor2-219) grown at 23 or 37°C from glutamine medium to proline medium induced nuclear entry of Gln3 (Fig. 2B, left panel). The downshift caused similar effect on Gln3 in tor1 tor2-119 double mutant cells grown at 23°C. However, pre-incubating the double mutant cells at 37°C for 2 hr abolished the effect of the downshift on Gln3 (Fig. 2B, right panel and Supplemental Fig. 1B), suggesting that TORC1 is required for nitrogen downshift-induced nuclear entry of Gln3. Consistent with the finding, the elevated temperature also blocked nitrogen downshift-induced GLN1 expression (Supplemental Fig. 3B).
When the tor2-219 single and tor1 tor2-219 double mutant cells were grown in proline medium, a large fraction of cells contained nuclear Gln3 and shifting the cells to nitrogen-free medium did not cause significant changes in the portion of cells containing nuclear Gln3 (Fig. 2C).
Both TORC1 and Tap42 are required for rapamycin-induced nuclear localization of Gln3
When wild type cells were grown in glutamine medium at either 23 or 37°C, rapamycin treatment induced nuclear entry of Gln3 (Fig. 3A). The drug also caused nuclear entry of Gln3 when tap42 mutant cells were grown at 23°C. However, it failed to do so after the mutant cells were pre-incubated at 37°C for 2 hr (Fig. 3B and Supplemental Fig. 1C). Similarly, when tor2-219 mutant cells were grown in glutamine medium at either 23 or 37°C, rapamycin caused Gln3 to concentrate in the nucleus (Fig. 3C). When tor1 tor2-219 double mutant cells were grown at 23°C, the drug was able to trigger nuclear entry of Gln3. Pre-incubating the cells at 37°C for 2 hr abolished the effect of the drug on Gln3 (Fig. 3D and Supplemental Fig. 1C). Consistent with the effect of the drug on Gln3, the drug-induced expression of GLN1 was blocked in tap42 and tor1 tor2-219 double mutant cells grown at the nonpermissive temperature (Supplemental Fig. 4). These observations demonstrate that inactivation of Tap42 or TORC1 blocks rapamycin-induced nuclear entry of Gln3, which correlated with the impaired expression of GLN1, indicating that when cells are grown in good nitrogen medium, both Tap42 and TORC1 are required for the drug-induced effect on Gln3.
Figure 3. Both Tap42 and TORC1 are required for nuclear entry of Gln3 in response to rapamycin treatment.
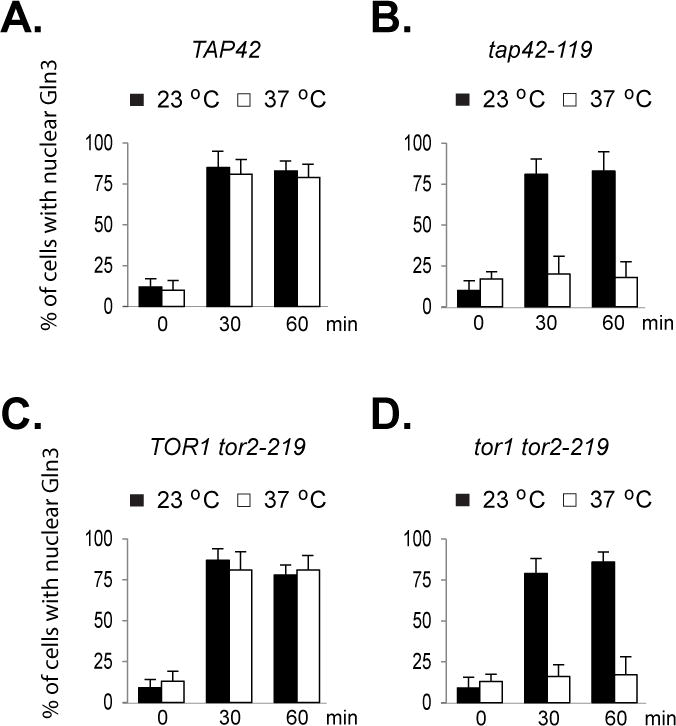
Cells expressing GLN3-myc were grown at 23°C to early log phase in glutamine (Gln) or proline (Pro) medium. The cultures were divided, with one half shifted to 37°C and the other half remained at 23°C. Upon incubation for additional 2 hr, cells were treated with rapamycin and collected before (time 0), 30 and 60 min after addition of the drug. Localization of Gln3-myc in the cells was examined by confocal microscopy. The experiment was repeated three times. Percentages of cells with nuclear staining of Gln3-myc expressed as mean ± SD are shown. A. Wild type (Y339), B. tap42-119 mutant (Y351), C. TOR1 tor2-219 (Y511), and D. tor1 tor2-219 mutant (Y442).
Collectively, the above findings demonstrate that TORC1 and Tap42 have differentiated roles in nitrogen signaling. While the former is involved in response to both nitrogen limitation and downshift, the latter is only required for nitrogen limitation-induced response.
Proline prevents rapamycin-induced the release of Tap42 from TORC1
Tap42-associated phosphatases are in complex with TORC1 under normal growth condition but are released and activated when cells are exposed to adverse conditions (Yan et al., 2012a). Previous studies have shown that Tap42-associated Sit4 phosphatase mediates Gln3 dephosphorylation in response to rapamycin treatment (Beck & Hall, 1999, Jacinto et al., 2001). Consistent with the observation, we found that when cells were shifted from glutamine to nitrogen-free medium, Tap42 was released from TORC1, as indicated by the dissociation of Tap42 from Kog1, the unique component of TORC1, in co-immunoprecipitation assays. The release was accompanied by Gln3 dephosphorylation (Fig. 4A). In contrast, the release was not observed when cells were shifted from glutamine to proline medium. Correlatively, Gln3 was not dephosphorylated (Fig. 4B). This observation is consistent with the nonessential role of Tap42 for Gln3 nuclear entry in the process (Fig.1B). When cells were shifted from proline medium to nitrogen-free medium, Tap42 remained associated with TORC1 and Gln3 dephosphorylation did not take place (Fig. 4C). Surprisingly, the presence of proline also prevented the dissociation of Tap42 from TORC1 and Gln3 dephosphorylation caused by rapamycin treatment (Fig. 4C). The effect of proline took place rapidly. After shifting cells to proline medium for 10 min, Tap42 as well as Gln3 phosphorylation became unresponsive to the drug treatment (Fig. 4D). Consistent with the response of Tap42 to proline, we found that cells were more resistant to rapamycin when grown in proline medium than in glutamine medium (Fig. 4E). These observations suggest that growing in poor nitrogen medium changes the response of cells to rapamycin treatment.
Figure 4. Proline prevents rapamycin-induced release of Tap42 from TORC1.
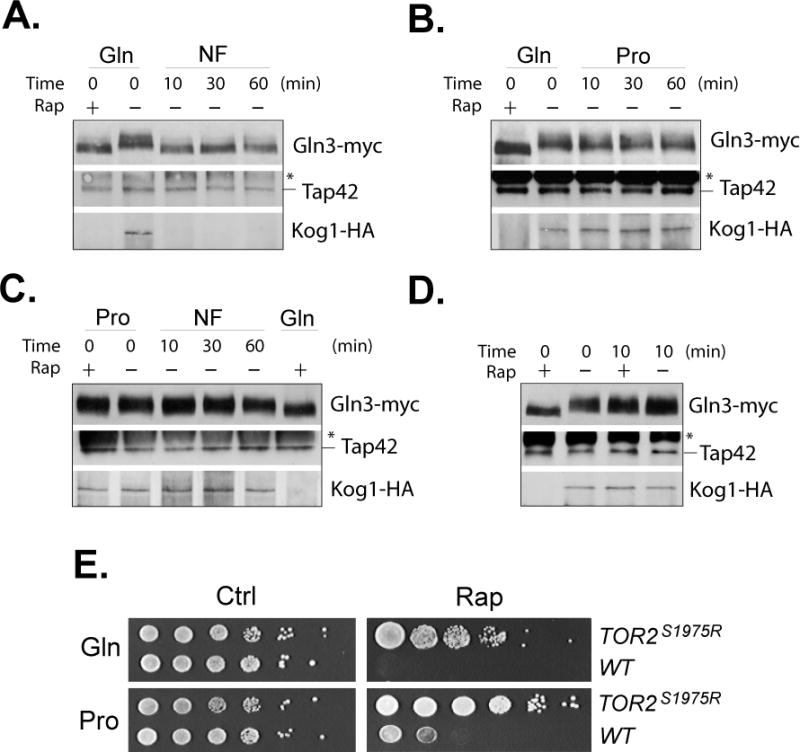
Cells expressing KOG1-HA (Y1032) and GLN3-myc grown in glutamine medium (Gln) were treated with rapamycin for 30 min (Rap +), transferred to nitrogen-free (NF) (A) or proline (Pro) medium (B) for indicated times. C. Cells grown in proline medium were treated with rapamycin for 30 min (Rap +) or transferred to nitrogen-free (NF) medium. Cells treated with rapamycin in glutamine medium were included as a control (Gln Rap+). D. Cells grown in glutamine medium were treated with rapamycin (+) or drug vehicle control (−) for 30 min or shifted to proline medium for 10 min before being treated with rapamycin or vehicle control for 30 min. Gln3-myc phosphorylation states in the cells were assayed by western blotting and the association of Tap42 with Kog1-HA by immunoprecipitation with anti-Tap42 antibody. The expression levels of Tap42 and Kog1-HA in the cells were examined by western blotting and shown in Supplemental Figure 5. E. Cultures of mid-log phase wild type (Y661) and rapamycin resistant TOR2S1975R mutant (Y032) cells were subjected to a series of 10 × fold dilution and spotted on plates containing glutamine (Gln) or proline (Pro) medium with 100 ng / ml of rapamycin (Rap) or drug vehicle control (ctrl). The plates were imaged after incubation at 30°C for 48 hr. The experiments were repeated three times and representative data are shown. * marks IgG heave chain of the Tap42 antibody used in the immunoprecipitation.
Proline does not affect stress-induced Rho1 activation
We have previously shown that release of Tap42 and its associated phosphatases from TORC1 is controlled by Rho1 GTPase, which upon activation, binds to TORC1 and causes the release (Yan et al., 2012a, Yan et al., 2012b). The lack of the Tap42 response to rapamycin can be thus explained if Rho1 fails to be activated in cells grown in proline medium. We thus examined stress-induced Rho1 activation in cells grown in proline medium using Rho1-dependent phosphorylation of Mpk1, a mitogen-activated protein kinase (MAPK) in yeast cells (Levin, 2005), as a surrogate marker. We found that in cells grown in proline medium Rho1 was activated normally in response to rapamycin treatment (Fig. 5A) and to nitrogen limitation (Fig. 5B). These findings suggest that proline does not control TORC1 through stress-induced Rho1 activation.
Figure 5. Rapamycin induces normal Mpk1 phosphorylation in proline medium.
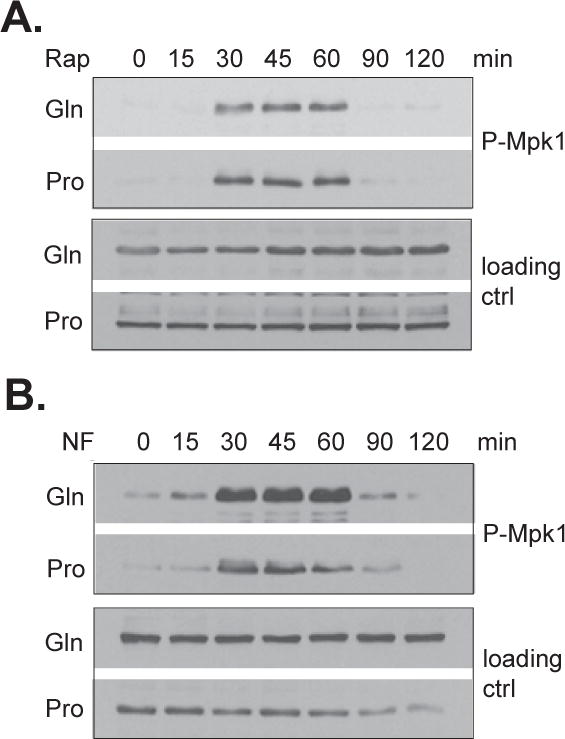
Wild type cells (Y1032) grown in glutamine (Gln) or proline (Pro) medium were treated with rapamycin (A) or transferred to nitrogen-free (NF) medium (B) for indicated times. The phosphorylation states of Mpk1 in the treated cells were examined using anti-phospho-MAPK antibody. The levels of Tpd3 were used as loading controls. The experiments were repeated three times and representative data are shown.
Npr1 prevents release of Tap42 from TORC1 in proline medium
The quick effect of proline on rapamycin-induced Tap42 release suggests that proline controls TORC1/Tap42 directly through activating a signaling event rather than indirectly through altering intracellular nitrogen conditions. To determine the underlying mechanism, we used deletion mutants and examined the role of several nutrient-related kinases, including Vps34, Gcn2, Yck1/2, and Npr1, in rapamycin-induced Tap42 release in cells grown in proline medium. This analysis identified Npr1 that prevented the release in proline medium (Fig. 6). Npr1, a serine/threonine kinase, is a key factor involved in controlling cell’s response to changes in nitrogen conditions (Crespo et al., 2004, De Craene et al., 2001, Tate et al., 2006, Vandenbol et al., 1990). We found that when cells were grown in glutamine medium, rapamycin caused dissociation of Tap42 from TORC1 in both wild type and npr1 deletion cells (Fig. 6A). However, when cells were grown in proline medium, the drug failed to dissociate Tap42 from TORC1 in wild type cells, but was still capable of doing so in npr1 deletion cells (Fig. 6B). This observation demonstrates that Npr1 prevents the rapamycin-induced release of Tap42 from TORC1 in proline medium. To further determine the role of Npr1 in regulation of TORC1/Tap42, we examined the release of Tap42 in npr1 deletion cells in response to changes in nitrogen conditions. When wild type and npr1 deletion cells grown in glutamine medium were shifted to nitrogen-free medium, Tap42 was released from TORC1 (Fig. 6C). However, when cells grown in proline medium were shifted to nitrogen-free medium, Tap42 released was blocked in wild type cells but not in npr1 deletion cells. This observation demonstrates that Npr1 prevents the release of Tap42 from TORC1 in proline medium, implying that Npr1 is able to modify TORC1 or Tap42 in response to proline.
Figure 6. Npr1 prevents the release of Tap42 from TORC1 in cells grown in proline medium.
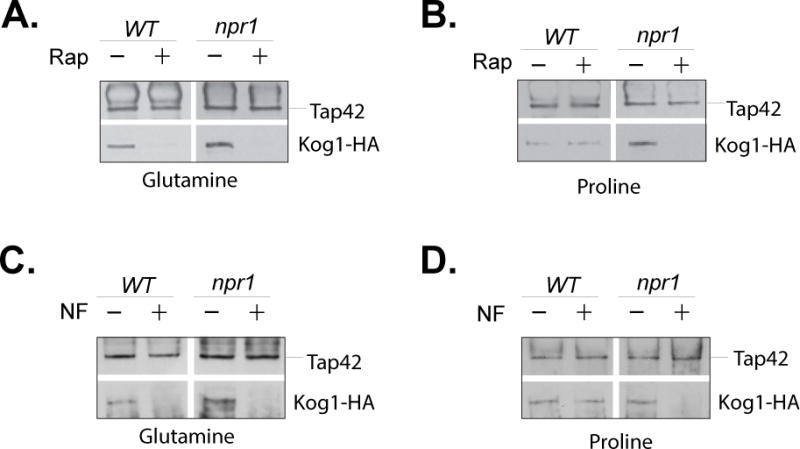
Wild type (Y1032) and npr1 deletion (Y1709) cells expressing KOG1-HA grown in glutamine (A) or proline medium (B) were treated with rapamycin (Rap +) or drug vehicle control (Rap −) for 30 min. The same pair of cells grown in glutamine (C) or proline medium (D) were transferred to nitrogen–free (NF) medium for 30 min. The association of Tap42 with Kog1-HA in the cells was examined by immunoprecipitation with anti-Tap42 antibody. The expression levels of Tap42 and Kog1-HA in the cells were examined by western blotting and shown in Supplemental Figure 6. The experiments were repeated three times and representative data are shown.
Npr1 inhibits TORC1 in response to nitrogen downshift
Npr1 kinase has been known as a downstream effector of TORC1 in nutrient signaling (MacGurn et al., 2011). However, the above findings suggest a role for the kinase upstream of TORC1 in response to changes in nitrogen conditions. To determine how Npr1 affects TORC1 activity, we assayed TORC1 signaling activity in npr1 deletion cells using TORC1-dependent phosphorylation of Sch9, a common surrogate marker for TORC1 activity in yeast cells (Urban et al., 2007). We found that when grown in glutamine medium, the npr1 deletion cells exhibited a higher TORC1 signaling activity than their wild type counterparts as indicated by the increased TORC1-dependent phosphorylation of Sch9 (Fig. 7A). Upon shifting to proline medium, the TORC1-dependent phosphorylation in wild type cells decreased over time to a level similar to that induced by rapamycin treatment (Fig. 7B). However, in npr1 deletion cells, TORC1-dependent phosphorylation of Sch9 remained unchanged after the cells were shifted to proline medium. These observations suggest that Npr1 is responsible for downregulating TORC1 activity when cells are shifted from good nitrogen to poor nitrogen sources.
Figure 7. Npr1 inhibits TORC1 activity.
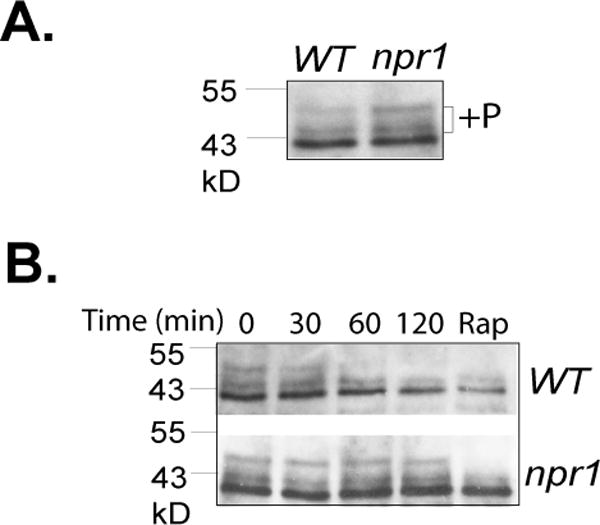
A. Wild type (Y661) and npr1 deletion (Y238) cells expressing SCH9-HA were grown in glutamine medium and TORC1-dependent phosphorylation of Sch9-HA was assayed by western blotting. B. Cells were shifted from glutamine to proline medium and aliquots of cells were collected before (time 0) and at indicated times after the shift. At the end of 2 hr incubation, the remaining cells were treated with rapamycin for 30 min (Rap). Phosphorylation of Sch9-HA in the cells was assayed by western blotting. The experiments were repeated three times and representative data are shown.
Discussion
Inactivation of Tap42 has been shown to block the effect of rapamycin on expression of genes involved in stress response as well as the nitrogen discrimination pathway (Duvel et al., 2003, Shamji et al., 2000). The role of Tap42 in the drug-induced gene expression has led to the notion that Tap42 and its associated phosphatases relay the activity of TORC1 for controlling stress response and nitrogen discrimination (Duvel & Broach, 2004). Consistent with the view, rapamycin treatment has been shown to cause release of Tap42-associated phosphatases from TORC1 and consequently, their activation (Yan et al., 2006, Yan et al., 2012a). However, our present study reveals that while Tap42 is required for the nuclear entry of Gln3 in response to nitrogen limitation, it is dispensable for the process in response to nitrogen downshift (Fig. 1 and Supplemental Fig. 1A), and hence does not play a role in the nitrogen discrimination pathway. In support of this notion, we find that Tap42 remains associated with TORC1 and that Gln3 is not dephosphorylated when yeast cells are downshifted from good nitrogen to poor nitrogen condition (Fig. 4). Our findings thus demonstrate that TORC1 and the Tap42-associated phosphatases have differentiated roles in cell’s response to changes in nitrogen conditions. While TORC1 is required for both nitrogen limitation and discrimination, the phosphatases are essential only for limitation response.
TORC1 signaling activity has been known to control expression of genes involved in stress response and the nitrogen discrimination pathway (Cardenas et al., 1999, Hardwick et al., 1999). However, its role in nitrogen signaling remains unclear. It has been suggested that TORC1 does not play a direct role in nitrogen sensing but acts in parallel to nitrogen signals to control nuclear entry of Gln3 (Zaman et al., 2008). It was found that while both rapamycin treatment and nitrogen downshift trigger Gln3 nuclear entry, the former but not the latter condition was accompanied by Gln3 dephosphorylation (Cox et al., 2004a, Tate & Cooper, 2007). The absence of Gln3 dephosphorylation indicates that Tap42-associated phosphatases are not activated during nitrogen downshift, which has led to the suggestion that TORC1 does not play a role in the process (Zaman et al., 2008). This notion is largely based on the assumption that activation of Tap42-associated phosphatases is an inevitable consequence of TORC1 inactivation, a concept deduced from analysis of the effects of rapamycin, which inhibits TORC1 and causes activation of the Tap42-associated phosphatases concurrently (Duvel et al., 2003, Shamji et al., 2000). However, in a previous study we found that activation of Tap42-associated phosphatases is indirectly regulated by TORC1 and can be uncoupled from TORC1 inactivation (Yan et al., 2012a, Yan et al., 2012b), which raises the possibility that alterations in TORC1 activity may not be always accompanied by activation of the phosphatases. Consistent with the view, the present study shows that TORC1 is required for the nuclear entry of Gln3 in response to nitrogen downshift, whereas, Tap42 is not.
How does TORC1 sense changes in nitrogen condition? Our findings indicate that Npr1 kinase is a key mediator in the process. The kinase is known for its role in controlling turnover of amino acid permeases in response to changes in nitrogen and amino acid conditions (De Craene et al., 2001, Schmidt et al., 1998, Vandenbol et al., 1990). While previous studies have shown that the kinase acts as a downstream effector of TORC1 (MacGurn et al., 2011, Schmidt et al., 1998), our findings unveil a role for the kinase upstream of TORC1 in sensing changes in nitrogen conditions. Because TORC1 exhibits a higher signaling activity in Npr1 deficient cells, it is expected that Npr1 acts as a negative regulator of TORC1. The observation that TORC1 signaling activity decreases in wild type but not npr1 deletion cells when the cells are shifted from glutamine to proline medium further suggests that the kinase is responsible for reducing TORC1 activity in response to downshift in nitrogen quality. The Npr1-mediated reduction in TORC1 is thus likely an important mechanism for the cells to accommodate poor quality of nitrogen source.
It remains unclear how Npr1 is able to regulate TORC1 activity and prevents the release of Tap42-associated phosphatases from TORC1. One possibility is that Npr1 directly phosphorylates a component of TORC1, leading to inhibition of TORC1 activity. Alternatively, Npr1, as a key regulator endocytosis of permeases, may regulate TORC1 indirectly through altering the endocytic pathway, which in turn changes TORC1 localization and/or activity. Further studies are needed to determine the molecular details underlying the Npr1-dependent regulation of TORC1. In addition, analysis of gene expression and metabolic profiles in Npr1 deficient cells will allow us to address the biological significance for preventing Tap42-associated phosphatases from activation during nitrogen downshift.
In summary, the present study identifies Npr1 as an upstream regulator of TORC1 and uncovers a novel mechanism that controls TORC1 activity in response to changes in nitrogen quality. This study paves a way for further elucidating the mechanism whereby eukaryotic cells sense and respond to changes in nitrogen conditions.
Experimental procedures
Strains, plasmids and culture media
Yeast strains used in this study are listed in Table 1 and plasmids in Table 2. Standard YP (1% yeast extract and 2% peptone) and synthetic complete (SC) dropout media were used for normal yeast cell propagation. All media contain 2% glucose as the carbon source. Plasmid pTS130 (Ycplac33-SCH9-HA3) was a gift from Michael Hall (Urban et al., 2007). Plasmid p718 contains a 13× myc epitope tagged at 3′ end of GLN3 that was generated in a previous study (Zheng & Jiang, 2005). Plasmid p948 contains a 5′ end truncated KOG1 tagged with triple HA epitope sequences at the end of the gene that has been described before (Yan et al., 2012a). The plasmid was linearized by digestion with BglII for integration of the HA-tagged KOG1 gene into yeast cells at the KOG1 locus.
Table I.
Yeast strains used in this study.
| Strains | Genotype* | Reference | Figures |
|---|---|---|---|
| Y032 | MATa ura3 leu2 his3 ade2 trp1 can1 tor2::HIS [pYcp50 (CEN URA3)-TOR2S1975R] | Lab stock | 4E |
| Y238 | MATa ura3 leu2 his3 ade2 trp1 can1 npr1::HIS3 | Lab stock | 7A, 7B |
| Y339 | MATa ura3 leu2 his3 ade2 trp1 can1 tap42::HIS3 [pRS314(CEN TRP1)-TAP42] | Lab stock | Fig. 1A, 1B, 1C, 3A |
| Y351 | MATa ura3 leu2 his3 ade2 trp1 can1 tap42::HIS3 [pRS314(CEN TRP1)-tap42-119] | (Yan et al., 2012a) | 1D, 1E, 1F, 3B |
| Y452 | MATa ura3 leu2 his3 ade2 trp1 can1 tor1::HIS3 tor2::LEU2 [pRS314(CEN TRP1)-tor2-219] | Lab stock | 2D, 2E, 2F, 3D |
| Y511 | MATα ura3 leu2 his3 ade2 trp1 can1 tor2::LEU2 [pRS314(CEN TRP1)-tor2-219] | Lab stock | 2A, 2B, 2C, 3C |
| Y661 | MATa ura3 leu2 his3 ade2 trp1 can1 | Lab stock | 4E, 7A, 7B |
| Y1032 | MATα ura3 leu2 his3 ade2 trp1 can1 kog1::KOG1-HA3 | (Yan et al., 2012a) | 4A, 4B, 4C, 4D, 5A, 5B, 6A, 6B, 6C, 6D |
| Y1709 | MATa ura3 leu2 his3 ade2 trp1 can1 npr1::HIS3 kog1::KOG1-HA3:URA3 | This study | 6A, 6B, 6C, 6D, |
All strains are derivatives of W303 background.
Table II.
Plasmids used in this study
| Plasmids | Descriptions | Reference |
|---|---|---|
| p718 | GLN3-myc13 in pRS416 (CEN URA3) | (Zheng & Jiang, 2005) |
| p948 | kog1-HA3 in pRS406 (URA3) | (Yan et al., 2012a) |
| pTS130 | SCH9-HA3 in Ycplac33 (CEN URA3) | (Urban et al., 2007) |
Antibodies and reagents
Rabbit polyclonal Tap42 antibody was described before (Jiang & Broach, 1999). Anti-Myc (9E10) (#M4439) and HA (12CA5) (#11666606001) epitope antibodies were purchased from Sigma. Anti-phospho-MAPK antibody (#9101) was obtained from Cell Signaling Technology. Rapamycin was acquired from LC laboratories (Woburn, MA) and used at a concentration of 100 ng / ml.
Nitrogen limitation and downshift treatments
Nitrogen-free medium contains 0.17% yeast nitrogen base, 2% glucose and appropriate amino acids for required auxotrophic supplements. Glutamine or proline was added to the medium at final concentration of 0.1% to make glutamine or proline medium. For nitrogen limitation and downshift treatments, yeast cells were grown in glutamine medium to early-log phase (~0.5×107cell/ml), collected by filtration, and immediately transferred to nitrogen-free or proline medium at the same density as the original culture.
Immunofluorescent microscopy
Yeast cells expressing GLN3-myc13 were grown in glutamine or proline medium at 23 or 37°C to early log phase. Cells were shifted to media with different nitrogen conditions. Aliquots of cultures were collected at various time points after the shift and fixed directly in culture medium with 3.7% formaldehyde. After incubation at room temperature for 30 min, cells were transferred to phosphate saline buffer (pH 7.4) containing 3.7% formaldehyde and incubated for additional 30 min. Fixed cells were then prepared for immunofluorescence staining as previously described (Pringle et al., 1991). Anti-myc (9E10) antibody and Alexa 488-conjugated goat anti‐mouse antibody (Molecular Probes) were used, respectively, as primary and secondary antibodies. Nuclei were stained with DAPI (4′,6‐diamidino‐2‐phenylindole dihydrochloride) prior to mounting. Fluorescent samples were visualized using Olympus FluoView™ FV1000 confocal microscope. For each condition, ~300 cells from three independent experiments were counted and analyzed for Gln3 localization. Cells were randomly selected from at least five different fields for the analysis. Data shown are percentages of cells with nuclear staining of Gln3-myc and expressed as mean ± SD. Comparison analyses between different time-points were done using Student’s t-test. A P-value < 0.05 is considered statistically significant.
Co-immunoprecipitation assay
Yeast cells expressing HA-tagged KOG1 were grown overnight at 30°C in glutamine medium to mid-log phase and transferred to either nitrogen-free or proline medium. At the indicated time points after the transfer, an aliquot of cells (5×108) was transferred to a centrifuge tube filled with ice and collected by centrifugation at 500 g for 5 min. Cells were resuspended in lysis buffer containing 50 mM Tris-Cl, pH7.4, 150 mM NaCl, 1 mM EDTA, 1mM PMSF, 1.5 × protease inhibitor cocktail (Roche Diagnostics) and lysed by vortexing with glass beads. Triton X-100 was then added to cell extracts to a final concentration of 1 % followed by incubation on ice for 15 min. Cell lysates were centrifuged at 10,000 g for 10 min to remove insoluble cell debris. Clarified cell lysates were then used for immunoprecipitation with anti-Tap42 antibody as described before (Yan et al., 2012a). The presence of Tap42 and HA-tagged Kog1 in the precipitates was examined by western blotting using anti-HA antibody.
Assay for C-terminal phosphorylation of Sch9
TORC1-dependent Sch9 phosphorylation was analyzed as described previously (Urban et al., 2007). Briefly, an aliquot of cell culture containing 2× 107 cells was directly mixed with trichloroacetic acid (final concentration 6%) followed by incubation on ice for 5 min. Cells were collected by centrifugation, washed twice with cold acetone, and dried. Cells were lysed with glass beads in 100 μl of lysis buffer containing 50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 6 M urea, 1% SDS, 1 mM PMSF, and 0.5× phosphatase inhibitor cocktail followed by incubation at 65°C for 10 min. After adjusting pH to 9.0 with 30 μl of 0.5 M CHES (pH 10.5), lysates were treated with 2.5 mM of 2-nitro-5-thiocyanatobenzoic acid overnight at room temperature. The treated lysates were boiled for 5 min after addition of 1 vol of 2 × SDS sample buffer. Sch9-HA in the lysates was detected by western blotting using anti-HA antibody.
Supplementary Material
Acknowledgments
We thank Michael Hall for providing the SCH9-HA plasmid and other laboratory members for comments and discussion during the course of the study. This study was supported in part by NSFC grants (U1601255, 81671965, 81372030) and Guangdong Proteomics Key laboratory Fund (2014B 030301044) to Yong J and NIH grants (GM068832 and CA169186) to Yu J.
Footnotes
The authors have no conflict of interest to declare for publishing the present study.
References
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, Chan TF, Zheng XF. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. The Journal of biological chemistry. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- Bertram PG, Choi JH, Carvalho J, Chan TF, Ai W, Zheng XF. Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Molecular and cellular biology. 2002;22:1246–1252. doi: 10.1128/MCB.22.4.1246-1252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes & development. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol Rev. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Kulkarni A, Tate JJ, Cooper TG. Gln3 phosphorylation and intracellular localization in nutrient limitation and starvation differ from those generated by rapamycin inhibition of Tor1/2 in Saccharomyces cerevisiae. The Journal of biological chemistry. 2004a;279:10270–10278. doi: 10.1074/jbc.M312023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Tate JJ, Cooper TG. Cytoplasmic compartmentation of Gln3 during nitrogen catabolite repression and the mechanism of its nuclear localization during carbon starvation in Saccharomyces cerevisiae. The Journal of biological chemistry. 2002;277:37559–37566. doi: 10.1074/jbc.M204879200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Tate JJ, Cooper TG. Actin cytoskeleton is required for nuclear accumulation of Gln3 in response to nitrogen limitation but not rapamycin treatment in Saccharomyces cerevisiae. The Journal of biological chemistry. 2004b;279:19294–19301. doi: 10.1074/jbc.M309240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo JL, Helliwell SB, Wiederkehr C, Demougin P, Fowler B, Primig M, Hall MN. NPR1 kinase and RSP5-BUL1/2 ubiquitin ligase control GLN3-dependent transcription in Saccharomyces cerevisiae. The Journal of biological chemistry. 2004;279:37512–37517. doi: 10.1074/jbc.M407372200. [DOI] [PubMed] [Google Scholar]
- De Craene JO, Soetens O, Andre B. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. The Journal of biological chemistry. 2001;276:43939–43948. doi: 10.1074/jbc.M102944200. [DOI] [PubMed] [Google Scholar]
- Duvel K, Broach JR. The role of phosphatases in TOR signaling in yeast. Curr Top Microbiol Immunol. 2004;279:19–38. doi: 10.1007/978-3-642-18930-2_2. [DOI] [PubMed] [Google Scholar]
- Duvel K, Santhanam A, Garrett S, Schneper L, Broach JR. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Molecular cell. 2003;11:1467–1478. doi: 10.1016/s1097-2765(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Guo B, Arndt KT, Schmelzle T, Hall MN. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Molecular cell. 2001;8:1017–1026. doi: 10.1016/s1097-2765(01)00386-0. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Broach JR. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. The EMBO journal. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn JA, Hsu PC, Smolka MB, Emr SD. TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell. 2011;147:1104–1117. doi: 10.1016/j.cell.2011.09.054. [DOI] [PubMed] [Google Scholar]
- Magasanik B, Kaiser CA. Nitrogen regulation in Saccharomyces cerevisiae. Gene. 2002;290:1–18. doi: 10.1016/s0378-1119(02)00558-9. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Adams AE, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Beck T, Koller A, Kunz J, Hall MN. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. The EMBO journal. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamji AF, Kuruvilla FG, Schreiber SL. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Current biology: CB. 2000;10:1574–1581. doi: 10.1016/s0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- Tate JJ, Cooper TG. Stress-responsive Gln3 localization in Saccharomyces cerevisiae is separable from and can overwhelm nitrogen source regulation. The Journal of biological chemistry. 2007;282:18467–18480. doi: 10.1074/jbc.M609550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JJ, Rai R, Cooper TG. Ammonia-specific regulation of Gln3 localization in Saccharomyces cerevisiae by protein kinase Npr1. The Journal of biological chemistry. 2006;281:28460–28469. doi: 10.1074/jbc.M604171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, De Virgilio C, Hall MN, Loewith R. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Molecular cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Vandenbol M, Jauniaux JC, Grenson M. The Saccharomyces cerevisiae NPR1 gene required for the activity of ammonia-sensitive amino acid permeases encodes a protein kinase homologue. Molecular & general genetics: MGG. 1990;222:393–399. doi: 10.1007/BF00633845. [DOI] [PubMed] [Google Scholar]
- Wei Y, Zheng XF. Nutritional control of cell growth via TOR signaling in budding yeast. Methods Mol Biol. 2011;759:307–319. doi: 10.1007/978-1-61779-173-4_18. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yan G, Lai Y, Jiang Y. The TOR complex 1 is a direct target of Rho1 GTPase. Molecular cell. 2012a;45:743–753. doi: 10.1016/j.molcel.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Lai Y, Jiang Y. TOR under stress: targeting TORC1 by Rho1 GTPase. Cell Cycle. 2012b;11:3384–3388. doi: 10.4161/cc.21461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Shen X, Jiang Y. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. The EMBO journal. 2006;25:3546–3555. doi: 10.1038/sj.emboj.7601239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annual review of genetics. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Jiang Y. The yeast phosphotyrosyl phosphatase activator is part of the Tap42-phosphatase complexes. Molecular biology of the cell. 2005;16:2119–2127. doi: 10.1091/mbc.E04-09-0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


