Summary
Monoclonal antibody discovery and engineering is a field that has traditionally been dominated by high‐throughput screening platforms (e.g. hybridomas and surface display). In recent years the emergence of high‐throughput sequencing has made it possible to obtain large‐scale information on antibody repertoire diversity. Additionally, it has now become more routine to perform high‐throughput sequencing on antibody repertoires to also directly discover antibodies. In this review, we provide an overview of the progress in this field to date and show how high‐throughput screening and sequencing are converging to deliver powerful new workflows for monoclonal antibody discovery and engineering.
Keywords: antibodies, high‐throughput sequencing, bioinformatics, surface display
Introduction
Monoclonal antibodies are an institution in life science, as they have had a tremendous impact as clinical therapeutics. There are currently over 50 licensed monoclonal antibodies against a diverse panel of disease phenotypes (Center for Drug Evaluation and Research – List of Licensed Biological Products as of 6/2017) with another ~540 in ongoing clinical development (source: The Antibody Society). As a result, monoclonal antibodies have revolutionized the biopharmaceutical market.1, 2, 3 In addition to therapeutics, the consistency and stability of monoclonal antibodies provide superior properties for diagnostic and research reagents when compared with polyclonal sera.4
In nature, antibodies (or immunoglobulins) denote secreted effector molecules of the humoral arm of the adaptive immune response. Initially displayed as membrane‐bound receptors on the surface of B lymphocytes [B‐cell receptors (BCRs)], they constitute a highly polymorphic class of glycoproteins, thereby accounting for the myriad of pathogenic antigens that they have to recognize and fight off. The ensemble of antibodies and BCRs in an individual at any given time is often referred to as the antibody repertoire.
Antibody repertoire diversity is shaped on multiple levels, before and during antigen‐guided B‐cell differentiation and development. Initially, precursor B cells rearrange their germline variable (V), diversity (D) and joining (J) gene segments in an imprecise combinatorial process called V(D)J recombination. During this process, which occurs first at the heavy‐chain locus and then proceeds at the light‐chain locus (after allelic exclusion), (non‐)templated N/P nucleotides are inserted and/or deleted at the joining regions of the germline segments, thereby contributing additional junctional diversity to this combinatorial recombination process. Heavy and light (κ or λ) chains are then paired randomly on the repertoire level. Given the number of V, D and J segments in the human genome, combinatorial diversity can generate 1·9 × 106 combinations; when taking junctional diversity into account, a number in the order of 1011 BCRs is reached. Cells expressing self‐reactive receptors are depleted in the process of negative selection (central tolerance), and the remaining population constitutes the immature B‐cell pool of the individual. The repertoire development stages described so far take place in the bone marrow, but immature B cells migrate to the periphery where they form a population of mature naive B cells that express functional, non‐autoreactive but still antigen‐inexperienced BCRs. Following antigen engagement in peripheral secondary lymphoid organs (spleen, lymph nodes, mucosal lymphoid tissues), naive B cells become activated and form microstructures called germinal centres, where the final process of antibody diversification results in an even deeper level of repertoire specialization. This is through the process of somatic hypermutation and affinity maturation, where random mutations are introduced in the V regions of the antibody genomic loci, eventually resulting in higher‐affinity variants that are selected for improved antigen‐binding. Combined together, these mechanisms are theoretically able to generate > 1013 different antibody variants in humans.5 As will be described in the following sections, the naive as well as the highly mutated antibody repertoire have both been exploited in an effort to access and successfully harness the adaptive immune system's molecular diversity, which has been the foundation for the discovery and engineering of monoclonal antibodies.
Screening antibody repertoires
The emergence of monoclonal antibody discovery occurred nearly 40 years ago, following the invention of the hybridoma technology by George Köhler and César Milstein.6, 7 Hybridomas are comprised of the fusion between B‐cell splenocytes from antigen‐immunized mice and a myeloma tumour cell line, which results in immortalized antibody‐producing clonal cell lines that can be screened for antigen‐specific binding. Decades later, it is still common practice to discover monoclonal antibodies by immunizing mice to generate a strong humoral response, followed by hybridoma generation and screening.8, 9, 10 Therefore, characterizing the molecular diversity of antibody repertoires that occur following immunization may offer a means to augment the monoclonal antibody discovery process, while also providing valuable insight into humoral immunity.
Surface display platforms enable screening of antibody repertoires, which typically consist of recombinant libraries that are directly cloned from a pool of B cells or synthetically designed and constructed. The strength of display platforms resides in their ability to recapitulate the four key steps of the in vivo immune response in a test tube: (i) generation of genotypic diversity in variable region genes; (ii) coupling genotype with phenotype; (iii) selective pressure favouring antigen‐specific binding; (iv) expansion of selected clones and variants.11 Over the course of many years, continued development in this field has resulted in several platforms using a variety of host expression systems, which relative to one another have various advantages and disadvantages.12 For example, the format and complexity of the antibody molecule expressed [e.g. single‐chain variable fragment (scFv), full‐length IgG] is highly dependent on the host. Furthermore, the size and diversity of the library that can be physically screened is also influenced greatly by the host expression system. This is especially pertinent considering that naive libraries are not biased by any antigenic exposure and so need to be considerably larger than libraries generated from an immunized or infected individual, which are expected to be substantially enriched in specificity for a particular antigen. Finally, libraries can be generated synthetically – or semi‐synthetically – to have more control over the structure and diversity of the antibody sequence space.11, 13
The origin of surface display technologies goes back nearly 30 years ago, when George P. Smith discovered that bacteriophage (phage) coat proteins could be modified to express recombinant peptides, which could then be exploited as binding moieties for selection.14 Phage display was adapted for antibody screening and discovery shortly thereafter and has become a widely used platform ever since, most notably culminating in the first phage‐derived antibody therapeutic being approved in 2002 (Humira, adalimumab, anti‐tumour necrosis factor‐α).15 Typically, antibody fragments [scFv or fragment antigen binding (Fab)] are fused to the minor coat protein pIII (present in three to five copies on the phage surface) of the filamentous M13 phage. Phages are used to infect Escherichia coli, in which they replicate and assemble into antibody‐displaying phage particles. During the biopanning process, phage particles are subsequently selected based on binding to solid‐state antigen (i.e. plates or tubes), unbound phage is washed away and remaining antigen‐specific phages are eluted and recovered. As more cycles of selection are typically necessary to reach the desired level of enrichment,16, 17, 18 phages are often used to re‐infect bacteria so that the process can be repeated.19 The first studies with model antigens were crucial in unravelling the possibility to enrich positive clones from phage libraries,20, 21 and potential therapeutic relevance was further highlighted when high‐affinity antibodies against the HIV gp120 antigen were found in a highly diverse library (1011 range) obtained from an HIV patient.22 The high diversity that can be achieved with phage display also motivated the strategy of using a ‘single‐pot’ library; these are large, non‐immune libraries that have been generated and successfully screened to find binders for a variety of antigens.17, 23, 24, 25 A major advantage of naive libraries compared with focused immune libraries is that antibodies could potentially be discovered against antigens that do not often elicit strong immune responses (i.e. self‐antigens). Considerable efforts have also been invested in constructing synthetic libraries that aim to recapitulate the diversity present in naive repertoires. For example, a fully human library has been constructed that has an overall size of 2 × 109 members, originally designed to be adapted to multiple display hosts;26 in phage, an exemplary application allowed discovery of binders against potential disease targets.27
Antibodies in their native context are produced by eukaryotic hosts, therefore it is intuitive to expect that a eukaryotic expression system would offer certain advantages for surface display and screening. The budding yeast Saccharomyces cerevisiae has therefore become another dominant surface display system for screening recombinant antibody repertoires.28 Yeast surface display is mediated by a fusion with the Aga2p subunit of α‐agglutinin, anchored to the cell's surface through disulphide bonds with the cognate subunit Aga1p. The possibility to screen yeast display antibody libraries using fluorescence‐activated cell sorting provides valuable benefits.29, 30, 31, 32 For example, peptide expression tags (e.g. hemagglutinin, c‐Myc) are often incorporated into the antibody scaffold, making it possible to select correctly assembled proteins and normalize the antigen‐binding signal according to the surface expression level.29, 33, 34 Furthermore, protein variants with modest affinity have been enriched a hundred times with one single round of screening.30 In one of the most prominent examples by Boder et al.,35 it took only four rounds of screening to discover an scFv against fluorescein with femtomolar binding affinity.
Ribosome display,36 an alternative in vitro display platform, is able to access the highest potential diversity (1012–1013) and overcomes some of the limitations of microbial platforms.11, 37, 38, 39, 40, 41 Ribosome display relies on an intentionally ‘incomplete’ translation process where, due to the lack of a STOP codon, the polypeptide remains attached to the ribosome, and the antibody–ribosome–mRNA (ARM)42 complex can be panned against the antigen.36 The approach is entirely dependent on PCR, abolishing the need for a transformation step and the consequent loss of clones; therefore, ribosome display is particularly well suited for affinity maturation by using, for example, an error‐prone PCR.43
Sequencing antibody repertoires
Access to the genetic information encoded in antibody repertoires provides a valuable resource that can be used for both monoclonal antibody discovery and immune profiling. However, the complexity and exceptional diversity present within repertoires offers a number of unique challenges. Low‐throughput (Sanger) sequencing of antibody variable genes has been instrumental in characterizing in vivo screening platforms relying on B‐cell immortalization or cloning; indeed, when a single target is investigated, Sanger sequencing commonly reaches enough coverage and robustness to obtain accurate sequence information.44 However, no reasonable low‐throughput sequencing strategy could cover a section of the repertoire large enough to provide a comprehensive snapshot of an immune response, so limiting its utility for antibody discovery purposes.45 The rapid advancement in high‐throughput sequencing technologies (e.g. Illumina) and bioinformatics has led to an explosion of research in antibody repertoires. High‐throughput sequencing of the variable regions of B cells, conventionally referred to as immunoglobulin sequencing or Ig‐seq, not only offers the potential to unravel features of the immune response on a large scale but can also be directly implemented for monoclonal antibody discovery and engineering.45
Much of the appeal of Ig‐seq relates to pathologically and/or immunologically relevant settings; in this context, the response to influenza vaccination has been extensively evaluated.46, 47, 48 The potential use of Ig‐seq as a diagnostic tool was further demonstrated in patients with lymphocytic malignancies, where it was possible to detect highly expanded clones.49 Although Ig‐seq on a population of B cells (bulk sequencing) is advantageous, because of its cost and accessibility, thorough characterization of the repertoire would ideally require paired variable heavy‐chain (V H) and variable light‐chain (V L) sequence information, that is to say, a single‐cell level resolution. Taking advantage of a variety of different strategies, such paired sequencing methods have been established in recent years. Among the first examples, DeKosky et al.50 established a workflow where single cells deposited in a microwell plate were lysed, allowing for mRNA to be coupled to microbeads, followed by subsequent emulsion‐based PCR to produce a construct with physically linked V H and V L regions, which was then ready for high‐throughput sequencing. A further improvement of this method used a single‐cell emulsification step to bypass the initial compartmentalization in the microwells.51, 52, 53 Other single‐cell sequencing methods, such as barcode‐tagging54, 55 and primer matrices,56, 57 have also been developed and proved very valuable in interrogating the antibody repertoire (see next section).
The potential to capitalize on sequence information for the discovery of monoclonal antibodies offers one of the most promising applications for Ig‐seq. Inferring antigen specificity by combining large‐scale sequencing data and bioinformatics would offer several advantages over standard single‐cell, hybridoma, or surface display screening platforms.45 First, compared with low‐throughput single‐cell sorting and cloning, Ig‐seq delivers a more comprehensive picture of the total clonal repertoire landscape, thereby revealing somatic variants that may offer improved biophysical characteristics (binding affinity, stability) for downstream development. Second, costly laborious screening is omitted because candidate antibodies are identified in silico. Besides time and cost savings, this feature becomes even more important considering that antibody‐display selections often suffer from screening artefacts58, 59 or the propagation of false‐positive clones in selections due to a certain growth advantage.60, 61 Finally, most relevant B‐cell subsets can be comprehensively interrogated by Ig‐seq, which is not the case with hybridomas and other approaches that suffer from low immortalization efficiencies.
Antibody discovery by sequencing immune repertoires
In order to discover new monoclonal antibodies by high‐throughput sequencing and bioinformatic analysis, various immune compartments from different mammalian hosts have been investigated post‐immunization. In this regard, three distinct strategies have been proven to be successful. In a first attempt to discover monoclonal antibodies by Ig‐seq, Reddy et al.62 separately mined the V H and V L repertoires from the bone marrow plasma cell compartment of immunized mice; these plasma cells were chosen because they are responsible for the majority of functionally active circulating antibodies in the serum. Six days after the secondary immunization, the repertoire was found to be significantly polarized with a few highly expanded clones dominating the total plasma cell repertoire. Since clonal expansion (reflected by clonal frequencies in the sequencing data sets) was assumed as a sufficient metric for antigen specificity, highly expanded V H and V L genes were paired based on their relative frequencies and expressed recombinantly as scFvs, which were then verified to be predominantly antigen‐specific binders. In follow‐up studies, similar strategies were used to discover antigen‐binding clones from total splenocytes.63, 64
In addition to bulk sequencing, single‐cell sequencing that reveals native V H and V L chain pairing has also been employed for antibody discovery. For example, Wang and colleagues isolated plasmablasts from the draining lymph nodes of immunized mice and obtained V H and V L chain pairing by following the method described previously (see previous section).50, 65, 66 After sequencing paired repertoires, it was again revealed that plasma cell diversity in lymph nodes was also polarized to a great extent. After recombinant expression, the most abundant clones were antigen specific, further indicating that clonal expansion represents a viable predictor of antigen specificity. Similarly, using recently tetanus‐vaccinated human patients, antigen‐specific plasmablasts were isolated and single‐cell sequencing was performed to obtain V H and V L pairings.50 In two consecutive studies by Tan et al.,54, 55 plasmablasts from human donors were single‐cell sorted into 96‐well plates by flow cytometry and both V H and V L transcripts were tagged with a unique well‐identifier barcode at the 3′ end of the first‐strand cDNA during reverse transcription and a plate‐identifier barcode was added during subsequent library preparation. After sequencing, antibody V H and V L chain‐pairs were retrieved based on identical barcodes and candidate antibodies were chosen based on clustering frequency or similarity to the consensus sequence of a given clonal family.
Recently, single‐cell emulsion‐barcoding workflows such as InDrop67 or DropSeq68 have been developed for whole‐transcriptome RNA‐seq. A similar approach has lately been adapted for sequencing human B cells, notably from an HIV elite controller.69 In brief, single cells were encapsulated in emulsion droplets, lysed and cDNA was tagged with both unique molecular identifiers for error correction70 as well as droplet‐specific barcodes to infer chain‐pairing. In more detail, unique molecular identifiers, carrying a universal 5′ adapter sequence that was incorporated during a template‐switch reaction and droplet barcodes that were initially diluted to approximately one molecule per droplet, were PCR‐amplified. Droplet barcodes were subsequently appended to cDNA by complementary overlap extension. After final library preparation and sequencing, nearly 40 000 filtered V H : V L pairs were obtained, representing a paired recovery rate of ~11%. To identify novel potentially broadly neutralizing antibodies, V H sequences were compared against previously reported broadly neutralizing antibodies identified from the same individual, including PGT121, which was found to be very potent.71 V H sequence comparison resulted in the isolation of eight new PGT121‐like antibodies. Interestingly, separate phylogenetic tree analysis of V H and V L genes revealed a highly similar topology for the respective paired sequences. Finally, when expressed, these monoclonal antibodies showed a high level of functional activity in pseudovirus neutralization assays.
The combination of proteomics and Ig‐seq offers an alternative and more direct way to profile the functional polyclonal serum antibody response of immunized hosts.72, 73, 74 This approach starts with antigen‐based affinity purification of antibody proteins derived from serum, which reduces sample complexity by enriching for antigen specificity. Next, antibody proteins are fragmented by protease digestion and analysed by liquid chromatography coupled to tandem mass spectrometry (LC‐MS/MS). To reconstruct the original antibody sequence, whereby the complementarity determining region 3 (CDR3) peptide serves as an important sequence anchor, identified peptides are finally compared against an individual reference database created by bulk‐sequencing of the antibody V H and V L repertoires of the respective animal. The creation of such a database is necessary because publicly available databases like IMGT75 would not contain sequences present in hypervariable CDR3 or sequences that evolved post‐immunization due to somatic hypermutation. However, this approach cannot provide information on native variable V H and V L chain pairing due to proteolysis and reduction of disulphide bonds during peptide preparation. Recombinant candidate antibody selection and expression have therefore been carried out based on combinatorial chain‐pairing of most frequent clones if both V H and V L gene repertoire databases were constructed.72 If peptides were only compared against a V H repertoire database, candidate single‐domain nanobodies were directly constructed and tested74 or candidate heavy chains were paired with the total V L repertoire73 and V L‐chain shuffled libraries were screened for each candidate V H by phage display.76 Probing the human serum repertoire by a combination of LC‐MS/MS and Ig‐seq has also been successful. For example, Lavinder et al. followed two human donors after having received a tetanus toxoid booster immunization and discovered monoclonal antibodies by a dual strategy: CDR3 peptides were identified by serum proteomics and were either compared against a paired V H : V L 50 or a V H‐only database. This led to the recombinant expression of nine cognate V H : V L candidates and four V H candidates that were combinatorially paired with the respective V L plasmablast‐cDNA repertoire.77 All antibodies were confirmed to bind tetanus toxoid antigen and several were shown to be specific for the same linear epitope that is required for the toxin to enter the host cell, thereby providing a plausible explanation for the high effectiveness of the tetanus vaccine.
In another approach, Ig‐seq has been combined with methods in bioinformatics and evolution to discover antibodies from human samples. Zhu et al. used the sequence of a previously described HIV‐specific broadly neutralizing antibody (10E8) as a guide, bulk‐sequenced both V H and V L chain repertoires from the same donor and constructed phylogenetic lineage trees. Phylogenetic trees displaying comparable branch topologies relative to the 10E8 sequences enabled selective in silico chain pairing based on relative genetic distances from the 10E8 wild‐type.78 Not only did this strategy result in the isolation of 11 functional 10E8‐like antibodies with neutralizing activity but it also proved successful in a consecutive ‘cross‐donor analysis’ setting, where new variants of another known broadly neutralizing antibody were isolated in an unrelated HIV‐positive donor.79
Besides de novo discovery of monoclonal antibodies, Ig‐seq has also been used for other exciting biotechnological applications. As humanizing murine antibodies by resurfacing80, 81 or CDR‐grafting82, 83 has not always turned out to be successful, major efforts by industry have been put into making commercial mouse strains that carry the human germline immunoglobulin locus in their genome.84, 85 In this regard, Lee et al.85 used Ig‐seq as a quality‐control system to benchmark their humanized mouse strain, where they could validate that the majority of antibody transcripts were of human origin. Furthermore, sequencing data revealed a functional recombination process between human gene segments and the introduction of junctional diversity. In another compelling setting, Ig‐seq was also used to monitor the maturation trajectory of specific antibodies in the repertoire of transgenic mice after immunization with computationally designed HIV immunogens engaging germline antibody precursors.86 It could be shown that certain boosted antibodies display intermediate genetic characteristics of a highly potent class of broadly neutralizing antibodies (VRC01), thus showing how Ig‐seq could also be exploited for vaccine profiling.
Sequencing recombinant antibody repertoires
Besides directly mining natural B‐cell‐derived antibody repertoires of animals and humans, Ig‐seq can also be used to interrogate a diverse set of recombinant antibody libraries like naive,87, 88 focused,89, 90, 91 immune, semi‐synthetic92, 93 and fully synthetic libraries.94, 95, 96 These repertoires are compatible with in vitro screening platforms such as phage,87, 92, 96 yeast58 or ribosome display.97 One of the primary objectives of these sequencing efforts is to capture and elucidate characteristic library quality metrics that are indicative of the chances of successfully isolating antibodies with desired properties from a given recombinant library.
A diverse array of strategies has been reported for generating recombinant antibody libraries and their subsequent screening by surface display technologies.11, 98 However, depending on the type of recombinant repertoire being investigated, the library construction and cloning process can significantly impair the diversity and quality of the library. Quality in this sense is defined as the collection of different repertoire metrics, most importantly size, diversity and functionality, which when taken together maximize the chances of isolating a diverse panel of high‐affinity antigen‐specific clones that can be used for downstream applications.99 Depending on the desired library type and the screening platform of choice, library construction usually involves a series of molecular and cellular steps (reverse transcription, PCR, transformation), which bear the risk of introducing different forms of errors and biases and so impacting the library quality. First, as most of the commonly used reverse transcriptase variants do not have a 3′–5′ exonuclease activity, reverse transcription can introduce hotspot mutations into the resulting cDNA. Second, to achieve a broad variable‐gene coverage, multiplex‐PCR can introduce both errors and variable‐gene biases.100, 101 Third, if combinatorial libraries are constructed, assembly‐PCR can lead to frameshift mutations and/or preferential chain pairing. Fourth, even if sophisticated guidelines are being followed for the design of degenerate oligonucleotide libraries for the diversification of CDR sequence motifs,102, 103 the introduction of premature stop/cysteine codons as well as frameshift mutations cannot be circumvented because of intrinsic problems affecting oligonucleotide synthesis.104 Finally, once the recombinant antibody library is cloned, the final library size and diversity is limited by the transformation efficiency into respective host cells.
Traditionally, library quality could only be superficially assessed by Sanger‐sequencing of up to a few hundred random clones at a time. Assuming that a useful library size lies often in the range of 106 to 1011 clones, deducing library quality from such a low number of sequences provided only a slight glimpse of the total repertoire. However, with the emergence of Ig‐seq, recombinant antibody libraries could be analysed in a much more precise and comprehensive manner thereby obtaining statistically significant information on parameters such as CDR3 length distribution, variable‐gene usage, clonal frequencies and phylogenetic clonal relationships (Fig. 1). Glanville et al.87 first demonstrated how Ig‐seq could be used for the quality control of recombinant repertoires by investigating a naive scFv phage‐display library created from a pool of over 600 human donors. By sequencing both, V H and V L domain amplicons as well as rolling‐circle amplified shotgun scFv amplicons, critical library features like linker correctness, V H and V L germline gene prevalence, chain pairing, CDR3 lengths, and somatic hypermutations could successfully be evaluated. Moreover, customized capture–recapture estimates that were previously already successfully applied to the diversity assessment of the antibody repertoire in zebrafish,105 allowed the conclusion of separate diversity estimates for V H and V L, as well as for the whole phage display library, thereby also offering unprecedented insights into the human naive antibody repertoire. Building on these findings, a follow‐up study assessed the diversity of synthetic libraries that were designed by diversifying all six CDRs of a limited set of variable genes by using a positional amino acid scoring matrix deduced from human Ig‐seq data.95 Sequencing analysis of these libraries revealed that residual amino acid incorporations as well as the frequency with which they were incorporated were highly reflective of the initial in silico library design. In another example, a large‐scale rationally designed synthetic human phage display library was assessed by Ig‐seq.96 This library was created based on 36 fixed V H : V L framework pairings, which were previously found to be highly prevalent in humans and provide favourable properties for antibody expression and development. Furthermore, V H sequence diversity was based on optimized germline contributions for CDR1 and CDR2, while mimicking the natural positional frequency of amino acids in the CDR3 regions. Ig‐seq analysis revealed that the library was highly diverse, with 94% of CDR3 V H sequences observed only once. Finally, variable‐gene framework frequencies also reflected the initial library design indicating unbiased library construction.
Figure 1.
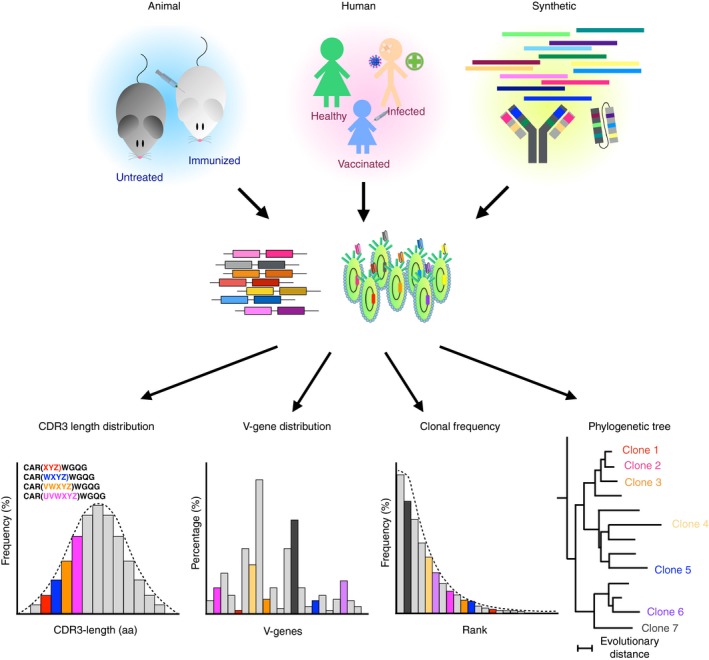
Quality control of recombinant antibody repertoires by high‐throughput sequencing. Recombinant antibody libraries can be generated from a variety of sources, such as animals (immunized or untreated), human patients (healthy, vaccinated or infected), or synthetically constructed genes. Following cloning and expression into a screening platform (e.g. phage display), these repertoires can undergo high‐throughput sequencing. Bioinformatic analysis of repertoire data delivers statistically significant information on various library parameters such as CDR3 length distribution, variable (V) gene distribution, clonal frequency and the relatedness of library members.
Combining high‐throughput screening and sequencing for monoclonal antibody discovery and engineering
In order to comprehensively access the full potential of a given natural or recombinant antibody library (usually in the range of 106–1011 clones), diversity has to be significantly reduced for high‐throughput sequencing to be able to provide a meaningful in‐depth view of potential antigen‐specific library variants. Therefore, combining Ig‐seq with a phenotype‐based sequence filter and selection pressure, such as those provided by phage or yeast display, represents a viable strategy for the selective in‐depth interrogation of antibody libraries106 (Fig. 2). Inference of antigen specificity is therefore grounded on the concept that due to the very large initial library sequence space, which becomes reduced with progressive screening, a given sequence should ideally be observed multiple times in the sequencing data sets if it has been enriched based on phenotypic binding to the antigen.
Figure 2.

Combining traditional antibody library screening with high‐throughput sequencing. The quality of a recombinant antibody library used in a screening platform (e.g. phage display) can be assessed by high‐throughput sequencing. Following biopanning and selection of this library, candidate antibodies can either be characterized in low‐throughput by traditional ELISA screening or subjected to high‐throughput sequencing. Bioinformatic analysis of high‐throughput sequencing data can reveal additional antibody sequences that were enriched during antigen‐specific selection. Antibodies discovered by traditional screening and by high‐throughput sequencing can then be cross‐referenced to find additional variants.
In an initial study by Saggy et al., the authors used B cells from antigen‐immunized mice, and conducted a side‐by‐side comparison of traditional phage‐display screening and Ig‐seq analysis. They found that both approaches resulted in the isolation of antigen‐specific antibodies exhibiting comparable affinities.63 However, out of the four clones isolated by phage display, only two corresponding V H sequences were detected in the Ig‐seq data and at a very low frequency. Likewise, the high‐frequency antigen‐specific sequences as determined by Ig‐seq were not found to be enriched by phage display. Antibody repertoire mining by Ig‐seq and phage display were therefore found to be two complementary technologies that led to the isolation of different antigen‐specific antibodies. Conversely, following a similar study design, monoclonal antibodies were discovered by either using Ig‐seq data from lymph nodes or performing yeast display on libraries generated from bone marrow and spleen from the same immunized mouse, even resulting in clones that may have originated from the same B‐cell lineage.66
In order to more fully exploit the potential of various antibody libraries, several studies reported the combined use of traditional recombinant library screening and Ig‐seq analysis.58, 61, 88, 89, 90, 91, 93, 97, 107, 108, 109 In one of the first examples by Ravn et al.107, a human synthetic scFv library was screened by phage display against an antigen (rat anti‐mouse Toll‐like receptor 4 antibody) and three rounds of panning resulted in the isolation of six specific clones. In parallel, Ig‐seq was also performed on the various selection rounds, which revealed that most of these antigen‐specific clones ranked among the top 10 most frequent variants in the final selection data set. However, some clones that were observed to be enriched by Ig‐seq were not captured by phage screening. By rescuing the six most frequently observed clones in the Ig‐seq data set (including three new clones), all antibodies were shown to be antigen‐specific. However, only clones that were previously identified by traditional ELISA screening gave a positive signal using crude supernatants or periplasmic extracts, so providing a plausible explanation as to why the other variants may have been missed by conventional screening. Likewise, when focused immune libraries from hens were explored by a combination of successive rounds of screening and high‐throughput sequencing, specific antibodies isolated by conventional panning as well as many more were identified by clustering and frequency analysis of Ig‐seq data.91 In an attempt to improve the affinity of a humanized anti‐ErbB2 monoclonal antibody, a diversified CDR library was subjected to panning and Ig‐seq, resulting in the isolation of nearly 40 mutants with improved affinity.61 Almost 50% of the highly abundant variants were missed during screening and the majority of mutants identified by conventional screening were found in the 100 most frequently observed sequences in the Ig‐seq data set. In another study, conventional screening of a recombinant single domain antibody library from immunized llamas resulted in the recovery of 13 antigen‐specific clones that showed a high degree of sequence conservation.89 To isolate a genetically more distant panel of antibodies, evaluating the clonal enrichment landscape by Ig‐seq after panning and applying phylogenetic tree analysis of the top 100 clones led to the isolation of two additional phylogenetically unrelated antigen‐specific antibodies. Furthermore, directly interrogating recombinant antibody libraries by Ig‐seq based on sequences found to be antigen‐specific by phage display screening also provided a valuable strategy to find new sequence variants that exhibited favourable biophysical characteristics.90
Concluding remarks and future perspectives
The use of high‐throughput screening and sequencing in monoclonal antibody discovery and engineering has demonstrated the capability to isolate a diverse set of antibodies against a variety of antigenic targets. Although these two approaches were initially independent of one another, continued progress has shown that they can work in synergy to gain a wealth of knowledge on both natural and recombinant antibody repertoires. Moreover, by using a two‐tier isolation strategy, both technologies can be combined to isolate antibodies with complementary properties. Furthermore, new screening platforms that combine genome editing and mammalian surface display are emerging,110 which not only enable the screening of full‐length antibodies for antigen binding but also allow for the integration of Ig‐seq to predict biophysical properties more optimal for downstream therapeutic development. Finally, new advanced approaches are emerging for identifying antigen‐specific clones based on sequence clustering algorithms,111, 112 as was recently shown for T‐cell receptors;113, 114 these methods may eventually also be employed for discovering large panels of monoclonal antibodies from Ig‐seq data sets.
Disclosures
The authors declare they have no competing interests.
References
- 1. Liu JKH. The history of monoclonal antibody development – progress, remaining challenges and future innovations. Ann Med Surg 2014; 3:113–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walsh G. Biopharmaceutical benchmarks 2014. Nat Biotechnol 2014; 32:992–1000. [DOI] [PubMed] [Google Scholar]
- 3. Weiner GJ. Building better monoclonal antibody‐based therapeutics. Nat Rev Cancer 2015; 15:361–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bradbury A, Plückthun A. Reproducibility: standardize antibodies used in research. Nature 2015; 518:27–9. [DOI] [PubMed] [Google Scholar]
- 5. Murphy K. Janeway's Immunobiology, 8th edn New York, NY, USA: Garland Science, 2012. [Google Scholar]
- 6. Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975; 256:495–7. [DOI] [PubMed] [Google Scholar]
- 7. Shulman M, Wilde CD, Köhler G. A better cell line for making hybridomas secreting specific antibodies. Nature 1978; 276:269–70. [DOI] [PubMed] [Google Scholar]
- 8. Li J, Sai T, Berger M, Chao Q, Davidson D, Deshmukh G et al Human antibodies for immunotherapy development generated via a human B cell hybridoma technology. Proc Natl Acad Sci 2006; 103:3557–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sapparapu G, Fernandez E, Kose N, Bin Cao, Fox JM, Bombardi RG et al Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature 2016; 540:443–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu Y, Lee P, Ke Y, Zhang Y, Chen J, Dai J et al Development of humanized rabbit monoclonal antibodies against vascular endothelial growth factor receptor 2 with potential antitumor effects. Biochem Biophys Res Comm 2013; 436:543–50. [DOI] [PubMed] [Google Scholar]
- 11. Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol 2005; 23:1105–16. [DOI] [PubMed] [Google Scholar]
- 12. Michnick SW, Sidhu SS. Submitting antibodies to binding arbitration. Nat Chem Biol 2008; 4:326–9. [DOI] [PubMed] [Google Scholar]
- 13. Bradbury ARM, Sidhu S, Dübel S, McCafferty J. Beyond natural antibodies: the power of in vitro display technologies. Nat Biotechnol 2011; 29:245–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 1985; 228:1315–7. [DOI] [PubMed] [Google Scholar]
- 15. Nixon AE, Sexton DJ, Ladner RC. Drugs derived from phage display: from candidate identification to clinical practice. Monoclon Antib 2013; 6:73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu Rev Immunol 1994; 12:433–55. [DOI] [PubMed] [Google Scholar]
- 17. Sheets MD, Amersdorfer P, Finnern R, Sargent P, Lindquist E, Schier R et al Efficient construction of a large nonimmune phage antibody library: the production of high‐affinity human single‐chain antibodies to protein antigens. Proc Natl Acad Sci USA 1998; 95:6157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bradbury ARM, Marks JD. Antibodies from phage antibody libraries. J Immunol Methods 2004; 290:29–49. [DOI] [PubMed] [Google Scholar]
- 19. Kretzschmar T, von Rüden T. Antibody discovery: phage display. Curr Opin Biotechnol 2002; 13:598–602. [DOI] [PubMed] [Google Scholar]
- 20. McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 1990; 348:552–4. [DOI] [PubMed] [Google Scholar]
- 21. Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By‐passing immunization. Human antibodies from V‐gene libraries displayed on phage. J Mol Biol 1991; 222:581–97. [DOI] [PubMed] [Google Scholar]
- 22. Burton DR, Barbas CF 3rd, Persson MA, Koenig S, Chanock RM, Lerner RA. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci USA 1991; 88:10134–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Griffiths AD, Malmqvist M, Marks JD, Bye JM, Embleton MJ, McCafferty J et al Human anti‐self antibodies with high specificity from phage display libraries. EMBO J 1993; 12:725–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC et al Human antibodies with sub‐nanomolar affinities isolated from a large non‐immunized phage display library. Nat Biotechnol 1996; 14:309–14. [DOI] [PubMed] [Google Scholar]
- 25. de Haard HJ, van Neer N, Reurs A, Hufton SE, Roovers RC, Henderikx P et al A large non‐immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J Biol Chem 1999; 274:18218–30. [DOI] [PubMed] [Google Scholar]
- 26. Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G et al Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol 2000; 296:57–86. [DOI] [PubMed] [Google Scholar]
- 27. Rauchenberger R, Borges E, Thomassen‐Wolf E, Rom E, Adar R, Yaniv Y et al Human combinatorial Fab library yielding specific and functional antibodies against the human fibroblast growth factor receptor 3. J Biol Chem 2003; 278:38194–205. [DOI] [PubMed] [Google Scholar]
- 28. Gai SA, Wittrup KD. Yeast surface display for protein engineering and characterization. Curr Opin Struct Biol 2007; 17:467–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol 1997; 15:553–7. [DOI] [PubMed] [Google Scholar]
- 30. VanAntwerp JJ, Wittrup KD. Fine affinity discrimination by yeast surface display and flow cytometry. Biotechnol Progress 2000; 16:31–7. [DOI] [PubMed] [Google Scholar]
- 31. Colby DW, Kellogg BA, Graff CP, Yeung YA, Swers JS, Wittrup KD. Engineering antibody affinity by yeast surface display. Methods Enzymol 2004; 388:348–58. [DOI] [PubMed] [Google Scholar]
- 32. Feldhaus MJ, Siegel RW. Yeast display of antibody fragments: a discovery and characterization platform. J Immunol Methods 2004; 290:69–80. [DOI] [PubMed] [Google Scholar]
- 33. Boder ET, Wittrup KD. Yeast surface display for directed evolution of protein expression, affinity, and stability. Methods Enzymol 2000; 328:430–44. [DOI] [PubMed] [Google Scholar]
- 34. Sheehan J, Marasco WA. Phage and yeast display. Microbiol Spectrum 2015; 3:1–17. [DOI] [PubMed] [Google Scholar]
- 35. Boder ET, Midelfort KS, Wittrup KD. Directed evolution of antibody fragments with monovalent femtomolar antigen‐binding affinity. Proc Natl Acad Sci USA 2000; 97:10701–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hanes J, Pluckthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci USA 1997; 94:4937–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Irving RA, Coia G, Roberts A, Nuttall SD, Hudson PJ. Ribosome display and affinity maturation: from antibodies to single V‐domains and steps towards cancer therapeutics. J Immunol Methods 2001; 248:31–45. [DOI] [PubMed] [Google Scholar]
- 38. Yan X, Xu Z. Ribosome‐display technology: applications for directed evolution of functional proteins. Drug Discov Today 2006; 11:911–6. [DOI] [PubMed] [Google Scholar]
- 39. Zahnd C, Amstutz P, Plückthun A. Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat Methods 2007; 4:269–79. [DOI] [PubMed] [Google Scholar]
- 40. Hanes J, Jermutus L, Weber‐Bornhauser S, Bosshard HR, Pluckthun A. Ribosome display efficiently selects and evolves high‐affinity antibodies in vitro from immune libraries. Proc Natl Acad Sci USA 1998; 95:14130–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hanes J, Schaffitzel C, Knappik A, Pluckthun A. Picomolar affinity antibodies from a fully synthetic naive library selected and evolved by ribosome display. Nat Biotechnol 2000; 18:1287–92. [DOI] [PubMed] [Google Scholar]
- 42. He M, Taussig MJ. Antibody–ribosome–mRNA (ARM) complexes as efficient selection particles for in vitro display and evolution of antibody combining sites. Nucleic Acids Res 1997; 25:5132–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schaffitzel C, Hanes J, Jermutus L, Pluckthun A. Ribosome display: an in vitro method for selection and evolution of antibodies from libraries. J Immunol Methods 1999; 231:119–35. [DOI] [PubMed] [Google Scholar]
- 44. Baum PD, Venturi V, Price DA. Wrestling with the repertoire: the promise and perils of next generation sequencing for antigen receptors. Eur J Immunol 2012; 42:2834–9. [DOI] [PubMed] [Google Scholar]
- 45. Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR. The promise and challenge of high‐throughput sequencing of the antibody repertoire. Nat Biotechnol 2014; 32:158–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Laserson U, Vigneault F, Gadala‐Maria D, Yaari G, Uduman M, Vander Heiden JA et al High‐resolution antibody dynamics of vaccine‐induced immune responses. Proc Natl Acad Sci USA 2014; 111:4928–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jiang N, He J, Weinstein JA, Penland L, Sasaki S, He XS et al Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci Transl Med 2013; 5:171ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vollmers C, Sit RV, Weinstein JA, Dekker CL, Quake SR. Genetic measurement of memory B‐cell recall using antibody repertoire sequencing. Proc Natl Acad Sci USA 2013; 110:13463–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B et al Measurement and clinical monitoring of human lymphocyte clonality by massively parallel V‐D‐J pyrosequencing. Sci Transl Med 2009; 1:12ra23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. DeKosky BJ, Ippolito GC, Deschner RP, Lavinder JJ, Wine Y, Rawlings BM et al High‐throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol 2013; 31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD et al In‐depth determination and analysis of the human paired heavy‐ and light‐chain antibody repertoire. Nat Med 2014; 21:1–8. [DOI] [PubMed] [Google Scholar]
- 52. McDaniel JR, DeKosky BJ, Tanno H, Ellington AD, Georgiou G. Ultra‐high‐throughput sequencing of the immune receptor repertoire from millions of lymphocytes. Nat Protoc 2016; 11:429–42. [DOI] [PubMed] [Google Scholar]
- 53. DeKosky BJ, Lungu OI, Park D, Johnson EL, Charab W, Chrysostomou C et al Large‐scale sequence and structural comparisons of human naive and antigen‐experienced antibody repertoires. Proc Natl Acad Sci USA 2016; 113:E2636–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tan YC, Blum LK, Kongpachith S, Ju CH, Cai X, Lindstrom TM et al High‐throughput sequencing of natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clin Immunol 2014; 151:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tan YC, Kongpachith S, Blum LK, Ju CH, Lahey LJ, Lu DR et al Barcode‐enabled sequencing of plasmablast antibody repertoires in rheumatoid arthritis. Arthritis Rheumatol 2014; 66:2706–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Busse CE, Czogiel I, Braun P, Arndt PF, Wardemann H. Single‐cell based high‐throughput sequencing of full‐length immunoglobulin heavy and light chain genes. Eur J Immunol 2013; 44:597–603. [DOI] [PubMed] [Google Scholar]
- 57. Murugan R, Imkeller K, Busse CE, Wardemann H. Direct high‐throughput amplification and sequencing of immunoglobulin genes from single human B cells. Eur J Immunol 2015; 45:2698–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ferrara F, Naranjo LA, D'Angelo S, Kiss C, Bradbury ARM. Specific binder for Lightning‐Link® biotinylated proteins from an antibody phage library. J Immunol Methods 2013; 395:83–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Christiansen A, Kringelum JV, Hansen CS, Bøgh KL, Sullivan E, Patel J et al High‐throughput sequencing enhanced phage display enables the identification of patient‐specific epitope motifs in serum. Sci Rep 2015; 5:12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. ‘t Hoen PA, Jirka SM, Ten Broeke BR, Schultes EA, Aguilera B, Pang KH et al Phage display screening without repetitious selection rounds. Anal Biochem 2012; 421:622–31. [DOI] [PubMed] [Google Scholar]
- 61. Hu D, Hu S, Wan W, Xu M, Du R, Zhao W et al Effective optimization of antibody affinity by phage display integrated with high‐throughput DNA synthesis and sequencing technologies. PLoS ONE 2015; 10:e0129125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reddy ST, Ge X, Miklos AE, Hughes RA, Kang SH, Hoi KH et al Monoclonal antibodies isolated without screening by analyzing the variable‐gene repertoire of plasma cells. Nat Biotechnol 2010; 28:965–9. [DOI] [PubMed] [Google Scholar]
- 63. Saggy I, Wine Y, Shefet‐Carasso L, Nahary L, Georgiou G, Benhar I. Antibody isolation from immunized animals: comparison of phage display and antibody discovery via V gene repertoire mining. Protein Eng Des Sel 2012; 25:539–49. [DOI] [PubMed] [Google Scholar]
- 64. Valdés‐Alemán J, Téllez‐Sosa J, Ovilla‐Muñoz M, Godoy‐Lozano E, Velázquez‐Ramírez D, Valdovinos‐Torres H et al Hybridization‐based antibody cDNA recovery for the production of recombinant antibodies identified by repertoire sequencing. Monoclon Antib 2014; 6:493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang B, Kluwe CA, Lungu OI, DeKosky BJ, Kerr SA, Johnson EL et al Facile discovery of a diverse panel of anti‐Ebola virus antibodies by immune repertoire mining. Sci Rep 2015; 5:13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang B, Lee CH, Johnson EL, Kluwe CA, Cunningham JC, Tanno H et al Discovery of high affinity anti‐ricin antibodies by B cell receptor sequencing and by yeast display of combinatorial VH:VL libraries from immunized animals. Monoclon Antib 2016; 8:1035–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V et al Droplet barcoding for single‐cell transcriptomics applied to embryonic stem cells. Cell 2015; 161:1187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M et al Highly parallel genome‐wide expression profiling of individual cells using nanoliter droplets. Cell 2015; 161:1202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Briggs AW, Goldfless SJ, Timberlake S, Belmont BJ, Clouser CR, Koppstein D et al Tumor‐infiltrating immune repertoires captured by single‐cell barcoding in emulsion. bioRxiv 2017; https://doi.org/10.1101/134841. [Google Scholar]
- 70. Shugay M, Britanova OV, Merzlyak EM, Turchaninova MA, Mamedov IZ, Tuganbaev TR et al Towards error‐free profiling of immune repertoires. Nat Methods 2014; 11:653–5. [DOI] [PubMed] [Google Scholar]
- 71. Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP et al Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 2011; 477:466–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cheung WC, Beausoleil SA, Zhang X, Sato S, Schieferl SM, Wieler JS et al A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat Biotechnol 2012; 30:447–52. [DOI] [PubMed] [Google Scholar]
- 73. Wine Y, Boutz DR, Lavinder JJ, Miklos AE, Hughes RA, Hoi KH et al Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response. Proc Natl Acad Sci USA 2013; 110:2993–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fridy PC, Li Y, Keegan S, Thompson MK, Nudelman I, Scheid JF et al A robust pipeline for rapid production of versatile nanobody repertoires. Nat Methods 2014; 11:1253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lefranc MP, Giudicelli V, Duroux P, Jabado‐Michaloud J, Folch G, Aouinti S et al IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res 2015; 43:D413–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Marks JD, Griffiths AD, Malmqvist M, Clackson TP, Bye JM, Winter G. By‐passing immunization: building high affinity human antibodies by chain shuffling. Biotechnology (N Y) 1992; 10:779–83. [DOI] [PubMed] [Google Scholar]
- 77. Lavinder JJ, Wine Y, Giesecke C, Ippolito GC, Horton AP, Lungu OI et al Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc Natl Acad Sci USA 2014; 111:2259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhu J, Ofek G, Yang Y, Zhang B, Louder MK, Lu G et al Mining the antibodyome for HIV‐1‐neutralizing antibodies with next‐generation sequencing and phylogenetic pairing of heavy/light chains. Proc Natl Acad Sci USA 2013; 110:6470–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhu J, Wu X, Zhang B, McKee K, O'Dell S, Soto C et al De novo identification of VRC01 class HIV‐1‐neutralizing antibodies by next‐generation sequencing of B‐cell transcripts. Proc Natl Acad Sci USA 2013; 110:E4088–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Padlan EA. A possible procedure for reducing the immunogenicity of antibody variable domains while preserving their ligand‐binding properties. Mol Immunol 1991; 28:489–98. [DOI] [PubMed] [Google Scholar]
- 81. Roguska MA, Pedersen JT, Keddy CA, Henry AH, Searle SJ, Lambert JM et al Humanization of murine monoclonal antibodies through variable domain resurfacing. Proc Natl Acad Sci USA 1994; 91:969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jones PT, Dear PH, Foote J, Neuberger MS, Winter G. Replacing the complementarity‐determining regions in a human antibody with those from a mouse. Nature 1986; 321:522–5. [DOI] [PubMed] [Google Scholar]
- 83. Verhoeyen M, Milstein C, Winter G. Reshaping human antibodies: grafting an antilysozyme activity. Science 1988; 239:1534–6. [DOI] [PubMed] [Google Scholar]
- 84. Murphy AJ, Macdonald LE, Stevens S, Karow M, Dore AT, Pobursky K et al Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice. Proc Natl Acad Sci USA 2014; 111:5153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lee EC, Liang Q, Ali H, Bayliss L, Beasley A, Bloomfield‐Gerdes T et al Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery. Nat Biotechnol 2014; 32:356–63. [DOI] [PubMed] [Google Scholar]
- 86. Briney B, Sok D, Jardine JG, Kulp DW, Skog P, Menis S et al Tailored immunogens direct affinity maturation toward HIV neutralizing antibodies. Cell 2016; 166:1459–1464.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR et al Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci USA 2009; 106:20216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. D'Angelo S, Kumar S, Naranjo L, Ferrara F, Kiss C, Bradbury AR. From deep sequencing to actual clones. Protein Eng Des Sel 2014; 27:301–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Miyazaki N, Kiyose N, Akazawa Y, Takashima M, Hagihara Y, Inoue N et al Isolation and characterization of antigen‐specific alpaca (Lama pacos) VHH antibodies by biopanning followed by high‐throughput sequencing. J Biochem 2015; 158:205–15. [DOI] [PubMed] [Google Scholar]
- 90. Turner KB, Naciri J, Liu JL, Anderson GP, Goldman ER, Zabetakis D. Next‐generation sequencing of a single domain antibody repertoire reveals quality of phage display selected candidates. PLoS ONE 2016; 11:e0149393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yang W, Yoon A, Lee S, Kim S, Han J, Chung J. Next‐generation sequencing enables the discovery of more diverse positive clones from a phage‐displayed antibody library. Exp Mol Med 2017; 49:e308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Venet S, Ravn U, Buatois V, Gueneau F, Calloud S, Kosco‐Vilbois M et al Transferring the characteristics of naturally occurring and biased antibody repertoires to human antibody libraries by trapping CDRH3 sequences. PLoS ONE 2012; 7:e43471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ravn U, Didelot G, Venet S, Ng KT, Gueneau F, Rousseau F et al Deep sequencing of phage display libraries to support antibody discovery. Methods 2013; 60:99–110. [DOI] [PubMed] [Google Scholar]
- 94. Ge X, Mazor Y, Hunicke‐Smith SP, Ellington AD, Georgiou G. Rapid construction and characterization of synthetic antibody libraries without DNA amplification. Biotechnol Bioeng 2010; 106:347–57. [DOI] [PubMed] [Google Scholar]
- 95. Zhai W, Glanville J, Fuhrmann M, Mei L, Ni I, Sundar PD et al Synthetic antibodies designed on natural sequence landscapes. J Mol Biol 2011; 412:55–71. [DOI] [PubMed] [Google Scholar]
- 96. Tiller T, Schuster I, Deppe D, Siegers K, Strohner R, Herrmann T et al A fully synthetic human Fab antibody library based on fixed VH/VL framework pairings with favorable biophysical properties. Monoclon Antib 2013; 5:445–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Larman HB, Xu GJ, Pavlova NN, Elledge SJ. Construction of a rationally designed antibody platform for sequencing‐assisted selection. Proc Natl Acad Sci USA 2012; 109:18523–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lerner RA. Combinatorial antibody libraries: new advances, new immunological insights. Nat Rev Immunol 2016; 16:498–508. [DOI] [PubMed] [Google Scholar]
- 99. Perelson AS, Oster GF. Theoretical studies of clonal selection: minimal antibody repertoire size and reliability of self–non‐self discrimination. J Theor Biol 1979; 81:645–70. [DOI] [PubMed] [Google Scholar]
- 100. Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM et al Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun 2013; 4:2680. [DOI] [PubMed] [Google Scholar]
- 101. Khan TA, Friedensohn S, Gorter de Vries AR, Straszewski J, Ruscheweyh HJ, Reddy ST. Accurate and predictive antibody repertoire profiling by molecular amplification fingerprinting. Sci Adv 2016; 2:e1501371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lee CV, Liang WC, Dennis MS, Eigenbrot C, Sidhu SS, Fuh G. High‐affinity human antibodies from phage‐displayed synthetic Fab libraries with a single framework scaffold. J Mol Biol 2004; 340:1073–93. [DOI] [PubMed] [Google Scholar]
- 103. Liang WC, Dennis MS, Stawicki S, Chanthery Y, Pan Q, Chen Y et al Function blocking antibodies to neuropilin‐1 generated from a designed human synthetic antibody phage library. J Mol Biol 2007; 366:815–29. [DOI] [PubMed] [Google Scholar]
- 104. Mahon CM, Lambert MA, Glanville J, Wade JM, Fennell BJ, Krebs MR et al Comprehensive interrogation of a minimalist synthetic CDR‐H3 library and its ability to generate antibodies with therapeutic potential. J Mol Biol 2013; 425:1712–30. [DOI] [PubMed] [Google Scholar]
- 105. Weinstein JA, Jiang N, White RA 3rd, Fisher DS, Quake SR. High‐throughput sequencing of the zebrafish antibody repertoire. Science 2009; 324:807–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Glanville J, D'Angelo S, Khan TA, Reddy ST, Naranjo L, Ferrara F et al Deep sequencing in library selection projects: what insight does it bring? Curr Opin Struct Biol 2015; 33:146–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ravn U, Gueneau F, Baerlocher L, Osteras M, Desmurs M, Malinge P et al By‐passing in vitro screening—next generation sequencing technologies applied to antibody display and in silico candidate selection. Nucleic Acids Res 2010; 38:e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zhang H, Torkamani A, Jones TM, Ruiz DI, Pons J, Lerner RA. Phenotype–information–phenotype cycle for deconvolution of combinatorial antibody libraries selected against complex systems. Proc Natl Acad Sci USA 2011; 108:13456–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. D'Angelo S, Glanville J, Ferrara F, Naranjo L, Gleasner CD, Shen X et al The antibody mining toolbox: an open source tool for the rapid analysis of antibody repertoires. Monoclon Antib 2014; 6:160–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Pogson M, Parola C, Kelton WJ, Heuberger P, Reddy ST. Immunogenomic engineering of a plug‐and‐(dis)play hybridoma platform. Nat Commun 2016; 7:12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yermanos A, Greiff V, Krautler NJ, Menzel U, Dounas A, Miho E et al Comparison of methods for phylogenetic B‐cell lineage inference using time‐resolved antibody repertoire simulations (AbSim). Bioinformatics 2017; doi: 10.1093/bioinformatics/btx533. [DOI] [PubMed] [Google Scholar]
- 112. Miho E, Greiff V, Roskar R, Reddy ST. The fundamental principles of antibody repertoire architecture revealed by large‐scale network analysis. bioRxiv 2017; doi: https://doi.org/10.1101/124578. [Google Scholar]
- 113. Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F et al Identifying specificity groups in the T cell receptor repertoire. Nature 2017; 547:94–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Dash P, Fiore‐Gartland AJ, Hertz T, Wang GC, Sharma S, Souquette A et al Quantifiable predictive features define epitope‐specific T cell receptor repertoires. Nature 2017; 547:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]


