Summary
Twenty years ago, the autoimmune regulator (Aire) gene was associated with autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy, and was cloned and sequenced. Its importance goes beyond its abstract link with human autoimmune disease. Aire identification opened new perspectives to better understand the molecular basis of central tolerance and self–non‐self distinction, the main properties of the immune system. Since 1997, a growing number of immunologists and molecular geneticists have made important discoveries about the function of Aire, which is essentially a pleiotropic gene. Aire is one of the functional markers in medullary thymic epithelial cells (mTECs), controlling their differentiation and expression of peripheral tissue antigens (PTAs), mTEC–thymocyte adhesion and the expression of microRNAs, among other functions. With Aire, the immunological tolerance became even more apparent from the molecular genetics point of view. Currently, mTECs represent the most unusual cells because they express almost the entire functional genome but still maintain their identity. Due to the enormous diversity of PTAs, this uncommon gene expression pattern was termed promiscuous gene expression, the interpretation of which is essentially immunological – i.e. it is related to self‐representation in the thymus. Therefore, this knowledge is strongly linked to the negative selection of autoreactive thymocytes. In this update, we focus on the most relevant results of Aire as a transcriptional and post‐transcriptional controller of PTAs in mTECs, its mechanism of action, and its influence on the negative selection of autoreactive thymocytes as the bases of the induction of central tolerance and prevention of autoimmune diseases.
Keywords: Aire, autoimmunity, immune tolerance, negative selection, thymus
Aire in the time of APECED and its first cloning in 1997
A syndrome with glandular involvement was originally described in 19291 that was later termed autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy syndrome (APECED) or autoimmune polyglandular syndrome type 1 (APS1), OMIM entry # 240300. APECED disease is a set of autoimmune manifestations with polyglandular and epithelial compromise, often accompanied by candidiasis. Originally, APECED was characterized as a recessive genetic disease of autosomal transmission with prevalence in the Finnish, Iranian Jewish and Sardinian populations.2, 3 Immunological manifestations are mainly the result of lymphocytic infiltration in the affected organs and the presence of various specific autoantibodies.3 The defect in the ability of T lymphocytes to react against specific antigens and/or defective immunoregulation could be a cause of the presence of candidiasis in these patients.3, 4
Genetic analysis showed a link between APECED and mutations on human chromosome 21, 21q22.3, a region that had not previously been associated with autoimmune diseases.5 Genetic studies allowed the identification of a new gene whose structural characteristics, such as two PHD motifs, suggested its role as a transcriptional regulator. The mutations present in this gene and associated with the presence of APECED allowed the establishment of a strong relationship between the disease and that gene, which was then denominated Autoimmune regulator (Aire) because of its possible role in the prevention of aggressive autoimmunity.3, 5, 6 In 2017, we celebrate 20 years of Aire cloning and sequencing. Of note, this gene was the first to be located outside the human MHC locus that had been associated with autoimmunity (OMIM # 240300).
However, it was not feasible to study this question easily in humans, so the role of Aire in the negative selection of autoreactive thymocytes was only demonstrated with the use of the mouse model.
Function of Aire
The human autoimmune regulator (Aire) gene is 11·9 kb long, spanning 14 exons, and encodes a 1635‐nucleotide mRNA, which in turn encodes a 545‐amino‐acid protein. In the mouse, the Aire gene is located on chromosome 10 position 39·72 cM, is 13 kb long, and encodes a 14‐exon mRNA whose encoded protein has 545 amino acid residues with 77% human–mouse identity.7 (Fig. 1).
Figure 1.
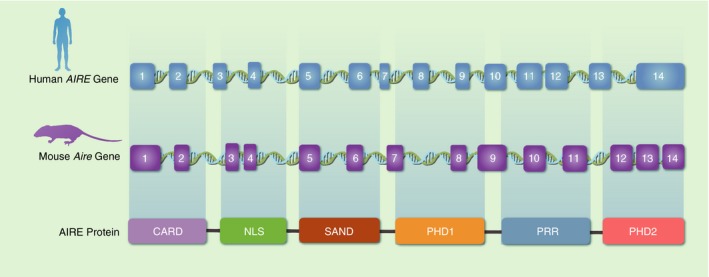
Schematic representation of the Aire gene and its respective encoded protein. Both human (chromosome 21q22.3) and mouse (chromosome 10 position 39·72 cM) Aire spans 14 exons and encodes AIRE protein with six domains.
Expression of the Aire gene is preferentially observed in the thymus, pancreas, adrenal cortex and testes;3, 8 however, it was also observed in lymph nodes, fetal liver and appendix.6 To better understand the effect of mutations in the Aire gene on its biological function and manifestations in APECED, it is important to understand its primary structure, which suggests the typical domain distribution of a protein capable of interacting with the chromatin and regulating transcriptional processes.9 The human AIRE protein has a molecular weight of 56 KDa with five subdomains: the CARD domain (caspase recruiter domain) that is important in the oligomerization of the protein; the Nuclear Location Signal that mediates its migration from the cytoplasm to the nucleus; the SAND domain (SP100, AIRE1, NucP41/P75 and DEAF1) involved in promoting protein–protein interactions; and two PHD domains, PHD‐1 and PHD‐2, that function as histone code readers, interacting and aiding in the chromatin decondensation.10, 11, 12
The inheritance of human Aire mutations is preferentially recessive and is usually accompanied by severe clinical manifestations of the disease.2 However, new dominant variants have also been described in different domains of the Aire gene, but these are usually accompanied by detectable intermediate or detectable auto‐antibodies, which would indicate nothing more than disease susceptibility.13 Consequently, each domain has a special function both in the regulation of the protein and in its transcriptional effects on the cell; additionally, depending on where the mutation occurs, Aire is affected in many ways.10, 13
In an Aire knockout (Aire−/− KO) murine model, the mice present changes in their thymic structure, deficiencies in the expression of peripheral tissue antigens (PTAs) in medullary thymic epithelial cells (mTECs), the presence of specific autoantibodies against self‐antigens, the decrease in regulatory T cells and the activation of self‐reactive T cells, promoting a general autoimmune reaction similar to that found in APECED patients. In this case, the mutation was generated in embryonic stem cells, leading to a deletion of the Aire exon 2 and resulting in a truncated protein with loss of the SAND and PHD‐1 and ‐2 domains.14, 15
Aire as a pleiotropic gene
As AIRE protein acts based on transcriptional control in mTECs and nonspecifically, several downstream genes and biological functions are subjected to its control; therefore, the Aire gene may be considered pleiotropic, and its different known functions are presented below.
Functional marker of mTEC differentiation
The expression of Aire is restricted to a few organs, among them the thymus. However, not all cells that integrate the thymic microenvironment can express this gene and its respective protein.3 Within the thymus, only mTECs can express this gene. In addition, not all mTECs express Aire at any given time, the expression of which appears to be limited to a small subpopulation of mature mTECs whose phenotype is CD80hi MHC‐IIhi.16, 17, 18, 19, 20
It has been demonstrated that the expression of Aire is an inherent property of mTECs during their differentiation programme and may occur at specific stages and/or cell states.21 Aire expression can be modulated by epigenetic factors, such as methylation,22, 23 or by molecular mechanisms, such as the induction of the nuclear factor‐κB pathway through RANKL–RANK‐mediated signalling, CD40L–CD40 or the leukotriene β‐mediated pathway, that determine the fate of the differential expression of Aire only in a subset of mTEC cells.24, 25, 26 Whether Aire might be post‐transcriptionally controlled through microRNAs (miRNAs) remains an open question.27
Populations of CD80low MHC‐IIlow mTECs, which do not express the Aire gene, not only present a significant reduction in the expression of PTAs but also greater proliferation and may also indicate a role of Aire in the control of proliferation and maintenance of the thymic structure through mechanisms of apoptosis.10, 15, 18, 19
Controlling the expression of peripheral tissue antigens
The murine genome has approximately 24 000 protein‐coding genes described so far, of which mTEChi cells express approximately 15 000 genes, which represent more than 62% of the mouse genome.18 The mTECs can express a broad number of tissue‐specific genes, a phenomenon that has not been observed in any other type of epithelial cell. This makes mTECs the most peculiar yet identified given the diversity of expressed genes and proteins. This ectopic gene expression appears to be a unique feature of the thymus and does not correspond to a defect of cell transcriptional control but rather to an inherent ability of these cells to promote promiscuous gene expression (PGE).23, 27, 28, 29, 30
Irrespective of their role in the periphery, self‐regulatory T (Treg) cells are negatively selected during the induction of central tolerance in the thymus. The thymic microenvironment is composed of different sets of antigen‐presenting cells, such as cortical TECs (cTECs), mTECs and thymic dendritic cells [(DCs) which present only PTAs that are expressed and processed by mTECs].29, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40
Aire regulates the expression of more than 3200 downstream genes in mTECs through a large transcriptional complex.10, 18, 41, 42 It is important to understand that Aire is not a conventional transcription factor because it does not bind directly to any DNA motifs of PTA genes. Rather, AIRE protein interacts and releases RNA Pol II molecules that have already started transcription but are stalled at the promoter regions of the genes – in other words, AIRE pushes them to proceed with gene transcription.43 In our view, this finding ultimately contributed to a better understanding of Aire's promiscuous action – that is, its protein favours the transcriptional activity of RNA Pol II, which is not specific and covers most, if not all, of the protein‐coding genes of a mammalian cell. Due to its vast heterogeneity, this type of expression was called PGE. It is important to note that the biological meaning of PGE is immunological – i.e., the ectopic proteins expressed in the thymus play their role as PTAs that are presented in the form of peptides and via MHC‐II to the developing thymocytes during the negative selection process.28
How Aire temporally determines the transcription of these genes is subject to extensive study. The expression of Aire‐dependent PTAs in mTECs was interpreted as stochastic and at the same time ‘ordered’. The term ‘stochastic’ was used because only a small percentage (1–3%) of mTECs express a certain PTA at any given time, although Aire promotes the expression of several PTAs in one mTEC at the same time. More than co‐expression, this seems to be a probabilistic event. The term ‘ordered’ refers to an increased likelihood of a particular set of PTA genes to be often co‐expressed in a single mTEC. Aire could determine, in an orderly way, the expression of genes that are within the same chromosomal region or forming networks between chromosomes by synchronizing their expression.10, 44, 45, 46
In fact, the AIRE protein is associated with a broad set of proteins to initiate the transcription of PTAs in mTECs that can be classified into four functional groups: proteins involved in nuclear transport, proteins that interact directly with chromatin, and proteins that can participate in both transcription and mRNA processing (Table 1). Additionally, there are proteins that, despite not interacting with AIRE, participate in the transcriptional complex associated with the expression of PTAs in mTECs.10 Many of these molecules even regulate Aire itself.47
Table 1.
AIRE partners during the promiscuous gene expression in medullary thymic epithelial cells23, 43, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109
| Protein symbol | Name | Function | AIRE domain involved |
|---|---|---|---|
| Proteins that interact concomitantly with AIRE protein | |||
| RNA Pol II | RNA Polymerase II | Starts the gene transcription after Aire‐partners‐mediated activation and phosphorylation | PHD‐2 |
| H3K4 | Unmethylated histone H3 lysine 4 | Recruits AIRE protein at the transcription start site(s) (TSS) | PHD‐1 |
| ATF7IP–MBD1 | Activating transcription factor 7‐interacting protein–methyl‐CpG‐binding domain protein 1 | Mediates AIRE protein recruitment by methylation of CpG island in repressed genes | SAND |
| DNA‐PK | DNA‐dependent protein kinase | Phosphorylates the AIRE protein, indicates where Aire starts the gene transcription by phosphorylation of H2AX histone that recognize DNA breaks | PHD‐2 |
| P‐TEFb | Positive transcription elongation factor b | Promotes that AIRE protein unleashes stalled RNA Pol II | C‐Terminus |
| HNRNPL | Heterogeneous nuclear ribonucleoprotein L | Participates in the RNA elongation mediated by pTEFb | C‐Terminus |
| BRD4 | Bromodomain‐containing 4 | Binds directly to AIRE protein and forms a complex with pTEFb and HNRNPL to induce the unleashes of stalled RNA Pol II | CARD |
| EFTUD2 | Elongation factor TU GTP‐binding domain‐containing protein 2 | Involved in the alternative splicing of RNA mediated by AIRE protein | ND |
| SNRPB | Small nuclear ribonucleoprotein‐associated proteins B | ||
| SRSF1 | Serine/arginine‐rich splicing factor 1 | ||
| SIRT1 | Deacetylase sirtuin 1 | Deacetylates lysine residues of AIRE protein leading to its activation | Lysine residues NLS (121–261) |
| HDAC1‐2 | Histone deacetylase 1 and 2 | Exports and stabilizes AIRE protein in the nucleus | |
| CBP, p300 | CREB‐binding protein | Acetylates the lysine residues of AIRE protein leading to repression of its activity | |
| HIPK2 | Homeodomain‐interacting protein kinase 2 | Phosphorylates AIRE protein suppressing its activation | PHD‐2 |
| FBXO3 | F‐box protein 3 E3 ubiquitin ligase | Ubiquitinates AIRE protein promoting and increasing its affinity to pTEFb factor | N terminus (207) |
| MARCH 8 | Membrane‐bound E3 ubiquitin ligases | Regulates the MHC‐II expression in AIRE+ medullary thymic epithelial cell (mTECs) | ND |
| PIAS1 | Protein inhibitor of activated STAT | Possible sumoylation of AIRE protein regulating its activation | CARD |
| Importins | Importins α1, α3 and α5 | Exporting of AIRE protein to the nucleus | NLS |
| TOP2A | DNA topoisomerase IIα | Assiciates with AIRE protein and promotes DNA breaks at TSS | PHD‐2 |
| Proteins that directly interact with the Aire gene (at DNA level) | |||
| ERα | Oestrogen receptor α | Induces methylation of the promoter region of Aire gene leading to decreasing of Aire expression | Promoter |
| AR | Androgen receptor | The accumulation of AR in the promoter region of Aire gene leads to its activation | Promoter |
| JMJD6 | Histone arginine demethylase | Promotes the splicing of intron 2 of Aire mRNA | Intron 2 |
| CNS1 | Conserved non‐coding sequence 1 | Conserved region of 90 nucleotides that regulate the Aire gene activation through RANK and nuclear factor‐κB signalling | Between Aire and Dnmt3l genes |
| IRF4, IRF8, TBX21 and TCF7 | Interferon regulatory factor 1 e 8, T‐box transcription factor, transcription factor 7 | Form a transcriptional complex necessary for the Aire gene expression in mTECs | Promoter |
| CTCF | CCCTC‐binding factor | Represses the Aire gene expression inhibiting the interaction of IRF4, IRF8, TBX21 and TCF7 complex with the Aire promoter | TSS and 3′ UTR |
Recently, the group of Benoist/Mathis at Harvard University in Boston, MA, have found that some AIRE partners, especially those associated with the DNA‐damage response, localize preferentially on chromatin sections known as super‐enhancers, as for example TOPO 1.48
In summary, Aire leads to the activation of PGE in mTECs when it is recruited into the transcriptional start sites (TSS) of most genes. Once joining stalled RNA Pol II and through histone modifications and deacetylation mediated by SIRT‐1, the complex allows TOP2a to generate disruptions in the regions of transcription initiation that induce epigenetic changes and relaxation of the chromatin. In turn, this allows the recruitment of the pTEFb/BRD4 complex, releasing RNA Pol II and subsequent transcription elongation.49 All these elements working in a cascade show not only how AIRE protein controls PGE but also how the Aire gene is controlled (Fig. 2, Table 1).
Figure 2.
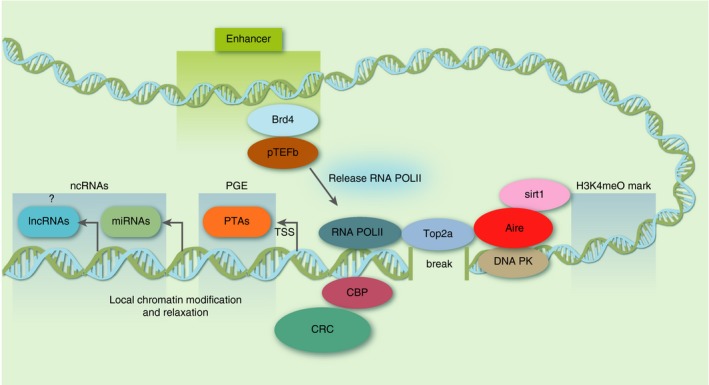
Mechanism of AIRE and its partners. AIRE protein binds to the chromatin in association with several other proteins. In this partial view, the protein complex binds to the transcription start site (TSS) of genes in medullary thymic epithelial cells (mTECs). Deacetylase sirtuin 1 (SIRT‐1) deacetylates lysine residues in AIRE leading to its activation. Then, AIRE through its PHD1 domain binds to AIRE‐dependent genes recognizing repressive epigenetic signatures that includes H3K4me0. TOP2a at the TSSs of AIRE‐dependent genes initiates breaks on DNA. DNA‐PK and other partners are activated by the DNA breaks resulting in local chromatin relaxation. The pTEFb/BRD4 complex releases stalled RNA POL II on chromatin, causing ensuing transcription elongation of AIRE‐dependent genes, including peripheral tissue antigen (PTA) ‐encoding genes and non‐PTA genes. As microRNAs are transcribed by RNA POL II, this species of non‐coding RNAs is regulated by AIRE in mTECs. The long non‐coding RNAs (lncRNAs), also transcribed by RNA POL II, could, perhaps, be regulated by AIRE? Finally, CREB‐binding protein (CBP) acetylates the lysine residues in AIRE leading to its repression. This figure is a modification of figure 2 published by Abramson J and Husebye ES. Immunol Rev 2016;271:127 (with permission of John Wiley and Sons).
Role of Aire beyond the control of PTA expression
The role of Aire in the expression of PTAs and in the prevention of the development of aggressive autoimmunity has been widely studied and debated. Nevertheless, whether Aire participates in other processes besides the negative selection of T cells remains a topic of research.10 It has been reported that Aire also participates in mechanisms of B‐cell tolerance, an event that does not occur in the thymus. Patients with APECED develop highly specific autoantibodies against a group of c.100 self‐proteins. However, some of these antibodies are neutralizing and have the function of decreasing the pathological effects of pro‐inflammatory cytokines. This could explain why APECED patients do not show symptoms of cytokine‐mediated diseases such as the interferon‐γ seen in systemic lupus erythematosus or systemic sclerosis.50 Aire also regulates the expression of genes that are not listed as autoantigens;18 many of these genes are involved, for example, in the recruitment of DCs into the thymus medulla, as in the case of the gene that encodes XCL1 ligand that binds to the XCR1 chemokine receptor.51 Aire could also influence the recruitment of cells in the thymic microenvironment through the induction of chemokine expression.
On the other hand, studies carried out independently in our group at the University of São Paulo in Ribeirão Preto, Brazil and in the Kyewski/Ucar group at the German Cancer Research Centre in Heidelberg, Germany, demonstrated the importance of Aire in the expression of miRNAs. Partial reduction of the Aire gene leads to a change in the miRNA expression of mTECs.52 Therefore, miRNAs could be important in regulating PGE in mTECs under the influence of Aire; consequently, miRNAs could be important in the organization of thymic architecture.23, 27, 53
Aire can also regulate mRNA alternative processing, possibly allowing mTECs to increase their repertoire of PTAs. Aire+ CD80hi MHC‐IIhi mTECs have an alternative splicing rate higher than that of other cell populations.18, 54, 55
Aire controlling mTEC‐thymocyte adhesion
In the thymic microenvironment, the AIRE+ and AIRE− mTECs, DCs and thymocytes continuously interact in the medulla, contributing to the central tolerance of the T cells to self‐antigens.56 The mTECs using Aire control do not only express PTAs but also molecules involved in cell–cell interaction processes and some important adhesion molecules.18 In turn, the expression of these genes is dependent on the levels of the AIRE protein, so that studies using the small interfering RNA (siRNA) technique allowed the demonstration that the reduction of Aire leads to a concomitant reduction not only in PTAs but also in the adhesion capacity of mTECs to thymocytes.57 The molecules most affected by the reduction in the Aire levels provoked by siRNA are integrins, claudins and adhesion or co‐stimulatory molecules as CD80.57 These findings were of great importance because mTEC–thymocyte adhesion is a crucial process for the negative selection of autoreactive thymocytes and induction of immunological tolerance.
Aire and apoptosis of mTECs
A controversial function of Aire is its possible role in the induction of apoptosis in mTECs.19 Due to the increase in MHC‐IIhi mTECs under Aire deficiency, a possible explanation could be that Aire promotes apoptosis in these cells. This stimulation supposes that apoptotic cells could be phagocytosed and PTAs could be presented through antigen‐presenting cells, increasing self‐representation.34 Nevertheless, this function was not yet confirmed due to conflicting observations from an inducible Cre mouse model, whose results indicated that Aire is not correlated with the mTEC lifespan.58
Promiscuous gene expression added a second controller: Fezf2
According to Sansom et al.,59 murine thymic epithelial cell populations express more than 19 000 protein‐coding genes, which confer the set of PTAs and other non‐PTA genes. Currently, no other type of cell has shown this enormous transcriptional diversity. In the mouse and this set of genes, Aire itself controls 3900 genes, which are considered Aire‐dependent, that correspond to approximately 20% of PGE. No doubt Aire is an important transcriptional controller, but other controllers should still work independently resulting in PGE. In fact, this is a long‐standing suspicion because PGE was not completely aborted under Aire annulment.60
As elegantly discussed by Klein,61 over the years, researchers have devoted their efforts to elucidating the mechanism by which Aire works; however, the regulation of Aire‐independent genes had remained elusive. Major progress occurred with the appearance in the literature of the work of Takaba et al.,62 in which the authors considered that, if there was a transcriptional regulator of Aire‐independent genes, it should be differentially expressed between cTECs and mTECs. In fact, these authors identified the forebrain‐expressed zinc finger 2 (Fezf2) that is expressed in mTECs but not in cTECs. Fezf2 had already been associated with corticospinal motor neuron differentiation and, as Fezf2‐deficient mice do not survive after weaning, the role of this gene in the immune system remained undetected.
However, when examining Fezf2‐deficient cells, a set of PTAs are down‐regulated that do not coincide with those down‐regulated in Aire‐deficient mTECs. This clearly demonstrated that Fezf2 and Aire work independently but synergistically in the control of PGE in mTECs. Much more rapidly, with the elucidation of the mode of action of Aire in chromatin, Takaba's work demonstrated that, in contrast to AIRE, FEZF2 protein directly binds to DNA near its target genes like a classical transcription factor.
Takaba's work on Fezf2, besides contributing to a better understanding of the PGE and molecular bases of central tolerance, stimulates new issues such as its role (i) in the organization of the three‐dimensional thymic structure,61 (ii) as an upstream controller of non‐PTA protein‐coding genes (chemokine or cell adhesion genes, for example), (iii) as an upstream controller of non‐coding RNAs [miRNAs and long non‐coding RNAs (lncRNAs)] and (iv) as a controller of mTEC–thymocyte adhesion, among other questions that are sure to come.
T‐cell differentiation, negative selection and central tolerance
As mentioned above, the thymus is the anatomical site where central tolerance occurs. This organ is divided into two main distinct regions, the cortex and medulla, and each provides a complex set of signals that will allow bone‐marrow‐derived progenitor cells to develop into the organ and finally become mature T cells. These cells, which were rescued from programmed cell death, acquire the ability to exit the organ and migrate to the thymus‐dependent areas of the secondary lymphoid organs.36, 63
Among the cells that compose the thymus stroma, such as DCs, macrophages, fibroblasts, B cells and others, TECs are the major component. They are located in both the cortex (cTECs) and medulla (mTECs) of the thymic lobules and have distinct phenotypes, secretomes and roles in thymocyte maturation and selection processes.64, 65 TECs produce components of the thymus microenvironment, such as insoluble extracellular matrix (ECM) molecules and soluble components like interleukins, growth factors and chemokines that mediate ECM–cell and cell–cell interactions, respectively.66
In the immune system, particularly in the thymus, cell–cell interactions are importantly mediated by T‐cell receptor (TCR)/peptide–MHC signalling, which, in combination with various other stimuli, define cell activation, proliferation and survival. In the cortex, immature double‐negative (CD4− CD8−) and double‐positive (CD4+ CD8+) thymocytes interact with cTECs, in addition to fibroblasts and macrophages that are also present.67 At this point, TCR‐β rearrangement is tested through the expression of a pre‐TCR in a process called β‐selection, and failure to generate a functional receptor results in apoptosis. Following the TCR‐α chain rearrangement, the assembly and expression of the definitive TCR‐αβ at the highly expanded double‐positive stage and TCR specificity are tested in the process known as positive selection. Here, low‐avidity interactions with self‐peptide–MHC complexes expressed by cTECs allow the survival and further differentiation of thymocytes. Next, thymocytes start losing the expression of either CD4 or CD8 and migrate from the cortex through the corticomedullary junction, reaching the thymic medulla.68
The medullary region is relatively poor in thymocytes and abundant in mTECs, which play a central role in thymocyte negative selection. Macrophages, B cells, resident and migratory dendritic cells (DCs) are also found in the medulla and can contribute to this process, as discussed below. The pathways leading to CD4 or CD8 single‐positive (CD4+ CD8− or CD4− CD8+) subsets are dependent on antigen presentation by TCR interaction with peptide–MHC class II or class I molecules, respectively, and distinct transcriptional machineries induced also by other cell surface and soluble molecules, such as Delta‐class Notch ligand Delta‐like 4 (Dll4), kit ligand and interleukin‐7.36 High‐affinity TCR interactions with MHC self‐antigens induce the deletion of thymocytes by apoptosis or skew to alternative fates such as the differentiation of Treg cells (CD4+ CD25+ FOXP3+), so contributing to the establishment of a T‐cell repertoire that can respond to non‐self‐antigens and is tolerant to self‐antigens. The particular mTEC CD80hi MHC‐IIhi subset that expresses Aire and can present PTAs is crucial for negative selection.69 These processes of positive and negative selection are the mechanisms of the generation of central tolerance that prevent aggressive autoimmunity (Fig. 3).
Figure 3.
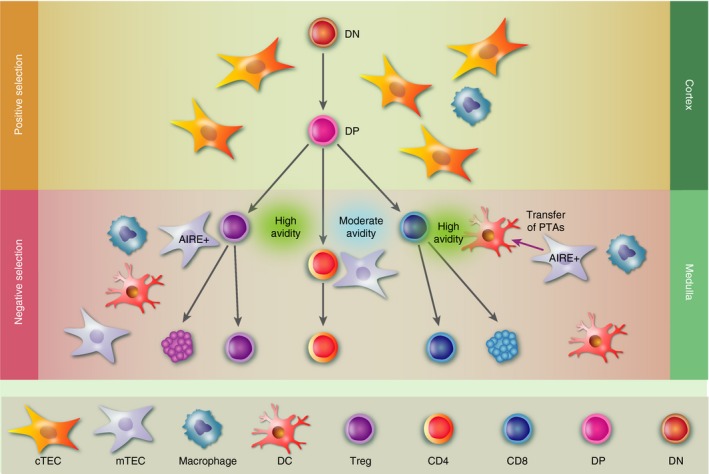
Medullary thymic epithelial cell (mTEC) –thymocyte interactions and negative selection. Negative selection occurs in the thymus in the medullary region. Double‐positive (DP; CD4+ CD8+) thymocytes from the inner cortex start losing the expression of either CD4 or CD8 and migrate from the cortex to the thymic medulla where they interact with mTECs and other bone‐marrow‐derived antigen‐presenting cells, such as dendritic cells. High‐affinity T‐cell receptor interactions with self‐antigens presented by MHC molecules induce the deletion of thymocytes by apoptosis or differentiation for regulatory T cells (CD4+ CD25+ FoxP3+ Treg cells). The particular AIRE + mTEC subtype capable of presenting peripheral tissue antigens (PTAs) is crucial for this process. AIRE + mTECs can also transfer PTAs for dendritic cells that will also play a role in negative selection. Moderate‐avidity T‐cell receptor interactions with self‐peptide and MHC class I or II complexes expressed by cortical TECs allow survival and further differentiation of thymocytes to CD4 or CD8 single‐positive subsets (CD4+ CD8− or CD4− CD8+) respectively, that will leave the thymus to the peripheral lymphoid organs.
Although the role of Aire is well defined in thymocyte negative selection, its role in Treg cell differentiation and selection remains controversial.49, 70 The generation of certain Treg clones is dependent on Aire expression. As an example, FOXP3+ MJ23tg and RT83tg Treg cells, which are found infiltrated in the tumours of mice with oncogene‐driven prostate cancer, failed to develop in the thymus and Aire −/− hosts.71 Other works have suggested that Treg cell differentiation is not dependent on Aire but on PTA–MHC presentation by other bone‐marrow‐derived antigen‐presenting cells such as DCs. Interestingly, because Aire is not expressed in these antigen‐presenting cells, it was suggested that mTEC antigens could be transferred to DCs.72, 73
Intrathymic T‐cell differentiation, including positive and negative selection processes, occur concomitantly with highly regulated migratory events, from the entrance of the progenitors until the exit of mature single‐positive cells to the periphery, a process that allows developing cells to interact with the different niches of the thymus. In this context, it was shown that Aire −/− mice show aberrant expression of migration‐related molecules, such as CCR4 and CCR7 ligands. Consequently, the Aire −/− thymus presented a delay in CD4 single‐positive cell emigration as early as 5 days after birth.74 Recent work extended these analyses in neonatal mice and showed that the Aire −/− thymus revealed significantly lower levels of CCL22 (CCR4 ligand) and CCL19 (CCR7 ligand) as well as CCL5 (ligand for CCR1, 3, 4 and 5), CCL17 (ligand for CCR8) until 6 weeks of age. Interestingly, reduced thymocyte emigration only occurred within the first 3 weeks after birth and was associated with the decreased expression of the CCR7 ligands. Conversely, an early and transient up‐regulation of sphingosine‐1‐phosphate receptor 1 expression was observed in Aire −/− thymocytes.75 Because sphingosine‐1‐phosphate receptor 1 is essential for thymocyte egress,76 one can argue that compensation mechanisms occur, allowing thymocyte emigration during this period.76 Together, these data show that the absence of Aire can also disturb peripheral tolerance by reducing the emigration of mature SP cells. Whether reduced emigration of Treg cells is involved must be investigated.
As discussed above, the absence of Aire in mTECs in vitro modulates various genes encoding proteins involved in cell adhesion and migration such as laminin, intercellular adhesion molecule 1 and vascular cell adhesion molecule 4. Functionally, Aire deficiency reduced the adhesion of thymocytes to mTECs that can modulate thymocyte migration and differentiation, contributing to abnormal tolerance or the breakdown of tolerance.57
In addition to the indirect control of adhesion, migration and egress of thymocytes, Aire can modulate DC migration and localization though the expression of XCL1. Aire −/− mice showed diminished mTEC expression of XCL1 and DC accumulation in the medulla, which could, in turn, disturb antigen transfer between mTEC and DCs.51 Interestingly, both Aire‐ and XCL1‐deficient mice presented low numbers of Treg cells. In this case, the authors hypothesized that Aire is essential for the mTEC expression of XCL1, which attracts XCR1‐expressing DCs to the medullary region and contributes to the optimal generation of Treg cells. Together, these studies show that Aire can directly regulate negative selection by controlling gene expression and, indirectly, by modulating the expression of molecules that play a role in cell–cell adhesion and cell migration.
Association between Aire gene mutations with loss of self‐tolerance in humans
More than 110 mutations along the Aire gene have been identified and associated with APECED or other autoimmune diseases in humans. Some populations present a major genetic association with a particular mutation being p.R257*, which is located within the SAND domain, the most common.9 Although APECED is a monogenic disease, it has a diverse range of clinical manifestations, often without a strict genotype–phenotype association. This suggests that not only the genetic risk, but the genetic mutation per se may be acting in immunological dysregulation and loss of self‐tolerance; environmental factors,70 or even imbalance in the thymic gene expression, might play a role in this pathogenic process.
Patients with APECED present in both children and adults with a significant decrease of CD4+ CD25+ Foxp3+ Treg cells,70 failure in thymic selection of naive T cells with defects in TCR repertoire77 or abnormal expression of cytokines.78
In addition, B cells that differentiate within the thymus express Aire 79, 80 and APECED patients present an altered B‐cell phenotype and dysregulation in B‐cell function with B‐cell reactivity to a unique set of approximately 100 self‐proteins and production of neutralizing autoantibodies against specific cytokines.81, 82
Despite the broad diversity of PTAs expressed by mTECs through PGE, patients with APECED present reactivity only against a minor fraction of self‐proteins,83 indicating that Aire may be acting beyond the selection of T cells.
Depending on the localization of mutations along the human Aire gene and the AIRE protein domain affected, the clinical manifestation can be different. For example, dominant mutations in the PHD1–2 and SAND domain affect the protein structure and correlate with the presence of organ‐specific autoimmune diseases.84 In addition, mutations in the promoter region impair the transcriptional activity of Aire 85 or mutations in the CARD/HSR domain affect the localization and export of AIRE protein from cytoplasm to the nucleus and mutations in intronic regions can alter the splicing of Aire mRNA.84, 86, 87
Summary
The current update on the molecular control of central immune tolerance that ultimately leads to negative selection in the thymus supports the participation of the Aire gene/protein and its partners. These molecular elements form a complex that finally binds to stalled RNA Pol II on the chromatin of mTECs, promoting the enzyme to proceed the transcription elongation phase of PTAs, as well as that of other non‐PTA genes. Because RNA Pol II is non‐specific and binds to the transcription start sequences of genes, this could clarify how the Aire gene controls PGE. In addition, recent evidence strongly suggests that Aire may play a role as an upstream controller of post‐transcriptional processes involving miRNA–mRNA interaction in mTECs. Many genes encoding PTAs in mTECs do not undergo Aire influence and are under the effect of the Fezf2 gene/protein that directly binds to a large set of transcription start sequences in the chromatin of these cells. With the recent association of the Fezf2 gene/protein in the control of PGE, other questions open in the light of what we have learned from Aire over the past 20 years. Are there Fezf2 DNA polymorphisms that could be associated with the loss of immune tolerance or emergence of autoimmune diseases? Are the genes associated with cell adhesion controlled by Fezf2? What are the epigenetic factors associated with the function of Fezf2? Does Fezf2 control non‐coding RNAs (miRNAs and/or lncRNAs)? Finally, what are other upstream controllers besides Aire and Fezf2 that regulate PGE in mTECs? There are two other questions still not fully resolved about the role of Aire in vivo; one of them is on the possible influence of variations of the levels of expression of Aire related to ageing thymus in the onset of aggressive autoimmunity.88 Considering the location of the Aire gene on human chromosome 21q22.3, the other question regards the effect of Aire gene dosage in children with Down syndrome.89 Finally, a new perspective was pointed out by the group of Nuno Alves at the Thymus Development and Function Laboratory (IBMC), Porto, Portugal, that communicated the participation of p53 during mTEC differentiation. Loss of p53 disrupts the integrity of the niche of mTECs while influencing the cTEC compartment.90
Disclosure
The authors declare no conflict of interest.
Acknowledgements
We thank Dr Danny Altmann, the Editor of Immunology, for the invitation to submit this update article. Our laboratories are funded by the following agencies; Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP grant # 13/17481‐1 to GAP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq grant # 306315/2013‐0 to GAP), Coordenação de Apoio ao Pessoal de Nível Superior (CAPES grant # 88881.068105/2014‐01 to GAP), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ grant # 218785/2015 to DAMC) and Fundação Oswaldo Cruz (Fiocruz) and Fondo para la Convergencia Estructural de Mercosur (FOCEM ‐ COF 03/11 to DAMC).
References
- 1. Thorpe ES. Chronic tetany and chronic mycelial stomatitis in a child aged four and one‐half years. Arch Pediatr Adolesc Med 1929; 38:328. [Google Scholar]
- 2. Ahonen P. Autoimmune polyendocrinopathy – candidosis – ectodermal dystrophy (APECED): autosomal recessive inheritance. Clin Genet 2008; 27:535–42. [DOI] [PubMed] [Google Scholar]
- 3. Björses P, Aaltonen J, Horelli‐Kuitunen N, Yaspo ML, Peltonen L. Gene defect behind APECED: a new clue to autoimmunity. Hum Mol Genet 1998; 7:1547–53. [DOI] [PubMed] [Google Scholar]
- 4. Arulanantham K, Dwyer JM, Genel M. Evidence for defective immunoregulation in the syndrome of familial candidiasis endocrinopathy. N Engl J Med 1979; 300:164–8. [DOI] [PubMed] [Google Scholar]
- 5. Aaltonen J, Björses P, Sandkuijl L, Perheentupa J, Peltonen L. An autosomal locus causing autoimmune disease: autoimmune polyglandular disease type I assigned to chromosome 21. Nat Genet 1994; 8:83–7. [DOI] [PubMed] [Google Scholar]
- 6. Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M et al Positional cloning of the APECED gene. Nat Genet 1997; 17:393–8. [DOI] [PubMed] [Google Scholar]
- 7. Blechschmidt K, Schweiger M, Wertz K, Poulson R, Christensen HM, Rosenthal A, et al The mouse Aire gene: comparative genomic sequencing, gene organization, and expression. Genome Res 1999; 9:158–66. [PMC free article] [PubMed] [Google Scholar]
- 8. Aaltonen J, Björses P. Cloning of the APECED gene provides new insight into human autoimmunity. Ann Med 1999; 31:111–6. [DOI] [PubMed] [Google Scholar]
- 9. Bruserud Ø, Oftedal BE, Wolff AB, Husebye ES. AIRE‐mutations and autoimmune disease. Curr Opin Immunol 2016; 43:8–15. [DOI] [PubMed] [Google Scholar]
- 10. Anderson MS, Su MA. AIRE expands: new roles in immune tolerance and beyond. Nat Rev Immunol 2016; 16:247–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar PG, Laloraya M, Wang C‐Y, Ruan Q‐G, Davoodi‐Semiromi A, Kao K‐J et al The autoimmune regulator (AIRE) is a DNA‐binding protein. J Biol Chem 2001; 276:41357–64. [DOI] [PubMed] [Google Scholar]
- 12. Perniola R, Musco G The biophysical and biochemical properties of the autoimmune regulator (AIRE) protein. Biochim Biophys Acta 2014; 1842:326–37. [DOI] [PubMed] [Google Scholar]
- 13. Oftedal BE, Hellesen A, Erichsen MM, Bratland E, Vardi A, Perheentupa J et al Dominant mutations in the autoimmune regulator AIRE are associated with common organ‐specific autoimmune diseases. Immunity 2015; 42:1185–96. [DOI] [PubMed] [Google Scholar]
- 14. Anderson MS. Projection of an immunological self shadow within the thymus by the Aire protein. Science 2002; 298:1395–401. [DOI] [PubMed] [Google Scholar]
- 15. Yano M, Kuroda N, Han H, Meguro‐Horike M, Nishikawa Y, Kiyonari H et al Aire controls the differentiation program of thymic epithelial cells in the medulla for the establishment of self‐tolerance. J Exp Med 2008; 205:2827–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hamazaki Y, Fujita H, Kobayashi T, Choi Y, Scott HS, Matsumoto M et al Medullary thymic epithelial cells expressing Aire represent a unique lineage derived from cells expressing claudin. Nat Immunol 2007; 8:304–11. [DOI] [PubMed] [Google Scholar]
- 17. Hubert F‐X, Kinkel SA, Webster KE, Cannon P, Crewther PE, Proeitto AI et al A specific anti‐Aire antibody reveals Aire expression is restricted to medullary thymic epithelial cells and not expressed in periphery. J Immunol 2008; 180:3824–32. [DOI] [PubMed] [Google Scholar]
- 18. St‐Pierre C, Trofimov A, Brochu S, Lemieux S, Perreault C. Differential features of AIRE‐induced and AIRE‐independent promiscuous gene expression in thymic epithelial cells. J Immunol 2015; 195:498–506. [DOI] [PubMed] [Google Scholar]
- 19. Gray D, Abramson J, Benoist C, Mathis D. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J Exp Med 2007; 204:2521–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gardner JM, Metzger TC, McMahon EJ, Au‐Yeung BB, Krawisz AK, Lu W et al Extrathymic Aire‐expressing cells are a distinct bone marrow‐derived population that induce functional inactivation of CD4+ T cells. Immunity 2013; 39:560–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kawano H, Nishijima H, Morimoto J, Hirota F, Morita R, Mouri Y et al Aire expression is inherent to most medullary thymic epithelial cells during their differentiation program. J Immunol 2015; 195:5149–58. [DOI] [PubMed] [Google Scholar]
- 22. Musco G, Peterson P. PHD finger of autoimmune regulator: an epigenetic link between the histone modifications and tissue‐specific antigen expression in thymus. Epigenetics 2008; 3:310–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ucar O, Rattay K. Promiscuous gene expression in the thymus: a matter of epigenetics, miRNA, and more? Front Immunol 2015; 6:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roberts NA, White AJ, Jenkinson WE, Turchinovich G, Nakamura K, Witers DR et al Rank signaling links the development of invariant γδ T cell progenitors and Aire+ medullary epithelium. Immunity 2012; 36:427–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Akiyama N, Takizawa N, Miyauchi M, Yanai H, Tateishi R, Shinzawa M et al Identification of embryonic precursor cells that differentiate into thymic epithelial cells expressing autoimmune regulator. J Exp Med 2016; 213:1441–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bichele R, Kisand K, Peterson P, Laan M. TNF superfamily members play distinct roles in shaping the thymic stromal microenvironment. Mol Immunol 2016; 72:92–102. [DOI] [PubMed] [Google Scholar]
- 27. Passos GA, Mendes‐da‐Cruz DA, Oliveira EH. The thymic orchestration involving Aire, miRNAs, and cell–cell interactions during the induction of central tolerance. Front Immunol 2015; 6:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol 2001; 2:1032–9. [DOI] [PubMed] [Google Scholar]
- 29. Kyewski B, Derbinski J, Gotter J, Klein L. Promiscuous gene expression and central T‐cell tolerance: more than meets the eye. Trends Immunol 2002; 23:364–71. [DOI] [PubMed] [Google Scholar]
- 30. Magalhães DAR, Silveira ELV, Junta CM, Sandrin‐Garcia P, Fachin AL, Donadi EA et al Promiscuous gene expression in the thymus: the root of central tolerance. Clin Dev Immunol 2006; 13:81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klein L, Kyewski B. Self‐antigen presentation by thymic stromal cells: a subtle division of labor. Curr Opin Immunol 2000; 12:179–86. [DOI] [PubMed] [Google Scholar]
- 32. Mizuochi T, Kasai M, Kokubo T, Kakiuchi T, Hirokawa T. Medullary but not cortical thymic epithelial cells present soluble antigens to helper T cells. J Exp Med 1992; 175:1601–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tykocinski L‐O, Sinemus A, Kyewski B. The thymus medulla slowly yields its secrets. Ann N Y Acad Sci 2008; 1143:105–22. [DOI] [PubMed] [Google Scholar]
- 34. Kyewski B, Derbinski J. Self‐representation in the thymus: an extended view. Nat Rev Immunol 2004; 4:688–98. [DOI] [PubMed] [Google Scholar]
- 35. Kyewski B, Klein L. A central role for central tolerance. Ann Rev Immunol 2006; 24:571–606. [DOI] [PubMed] [Google Scholar]
- 36. Takahama Y. Journey through the thymus: stromal guides for T‐cell development and selection. Nat Rev Immunol 2006; 6:127–35. [DOI] [PubMed] [Google Scholar]
- 37. Holländer G, Gill J, Zuklys S, Iwanami N, Liu C, Takahama Y. Cellular and molecular events during early thymus development. Immunol Rev 2006; 209:28–46. [DOI] [PubMed] [Google Scholar]
- 38. Irla M, Hugues S, Gill J, Nitta T, Hikosaka Y, Williams IR et al Autoantigen‐specific interactions with CD4+ thymocytes control mature medullary thymic epithelial cell cellularity. Immunity 2008; 29:451–63. [DOI] [PubMed] [Google Scholar]
- 39. Villasenor J, Besse W, Benoist C, Mathis D. Ectopic expression of peripheral‐tissue antigens in the thymic epithelium: probabilistic, monoallelic, misinitiated. Proc Natl Acad Sci USA 2008; 105:15854–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sousa Cardoso R, Magalhães DAR, Baião AMT, Junta CM, Macedo C, Marques MMC et al Onset of promiscuous gene expression in murine fetal thymus organ culture. Immunology 2006; 119:369–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meredith M, Zemmour D, Mathis D, Benoist C. Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat Immunol 2015; 16:942–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abramson J, Goldfarb Y. AIRE: from promiscuous molecular partnerships to promiscuous gene expression: highlights. Eur J Immunol 2016; 46:22–33. [DOI] [PubMed] [Google Scholar]
- 43. Giraud M, Yoshida H, Abramson J, Rahl PB, Young RA, Mathis D et al Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc Natl Acad Sci USA 2012; 109:535–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Derbinski J, Pinto S, Rosch S, Hexel K, Kyewski B. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc Nat Acad Sci 2008; 105:657–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brennecke P, Reyes A, Pinto S, Rattay K, Nguyen M, Küchler R et al Single‐cell transcriptome analysis reveals coordinated ectopic gene‐expression patterns in medullary thymic epithelial cells. Nat Immunol 2015; 16:933–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pinto S, Michel C, Schmidt‐Glenewinkel H, Harder N, Rohr K, Wild S et al Overlapping gene coexpression patterns in human medullary thymic epithelial cells generate self‐antigen diversity. Proc Natl Acad Sci USA 2013; 110:E3497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Herzig Y, Nevo S, Bornstein C, Brezis MR, Ben‐Hur S, Shkedy A et al Transcriptional programs that control expression of the autoimmune regulator gene Aire. Nat Immunol 2017; 18:161–72. [DOI] [PubMed] [Google Scholar]
- 48. Bansal K, Yoshida H, Benoist C, Mathis D. The transcriptional regulator Aire binds and activates super‐enhancers. Nat Immunol 2017; 18:263–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Abramson J, Husebye ES. Autoimmune regulator and self‐tolerance – molecular and clinical aspects. Immunol Rev 2016; 271:127–40. [DOI] [PubMed] [Google Scholar]
- 50. Meyer S, Woodward M, Hertel C, Vlaicu P, Haque Y, Kärner J et al AIRE‐deficient patients harbor unique high‐affinity disease‐ameliorating autoantibodies. Cell 2016; 166:582–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lei Y, Ripen AM, Ishimaru N, Ohigashi I, Nagasawa T, Jeker LT et al Aire‐dependent production of XCL1 mediates medullary accumulation of thymic dendritic cells and contributes to regulatory T cell development. J Exp Med 2011; 208:383–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Macedo C, Evangelista AF, Marques MM, Octacílio‐Silva S, Donadi EA, Sakamoto‐Hojo ET et al Autoimmune regulator (Aire) controls the expression of microRNAs in medullary thymic epithelial cells. Immunobiology 2013; 218:554–60. [DOI] [PubMed] [Google Scholar]
- 53. Oliveira EH, Macedo C, Collares CV, Freitas AC, Donate PB, Sakamoto‐Hojo ET et al Aire downregulation is associated with changes in the posttranscriptional control of peripheral tissue antigens in medullary thymic epithelial cells. Front Immunol 2016; 7:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Danan‐Gotthold M, Guyon C, Giraud M, Levanon EY, Abramson J. Extensive RNA editing and splicing increase immune self‐representation diversity in medullary thymic epithelial cells. Genome Biol 2016; 17:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Keane P, Ceredig R, Seoighe C. Promiscuous mRNA splicing under the control of AIRE in medullary thymic epithelial cells. Bioinformatics 2015; 31:986–90. [DOI] [PubMed] [Google Scholar]
- 56. Lopes N, Sergé A, Ferrier P, Irla M. Thymic crosstalk coordinates medulla organization and T‐cell tolerance induction. Front Immunol 2015; 6:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pezzi N, Assis AF, Cotrim‐Sousa LC, Lopes GS, Mosella MS, Lima DS et al Aire knockdown in medullary thymic epithelial cells affects Aire protein, deregulates cell adhesion genes and decreases thymocyte interaction. Mol Immunol 2016; 77:157–73. [DOI] [PubMed] [Google Scholar]
- 58. Nishikawa Y, Nishijima H, Matsumoto M, Morimoto J, Hirota F, Takahashi S et al Temporal lineage tracing of Aire‐expressing cells reveals a requirement for Aire in their maturation program. J Immunol 2014; 192:2585–92. [DOI] [PubMed] [Google Scholar]
- 59. Sansom SN, Shikama‐Dorn N, Zhanybekova S, Nusspaumer G, Macaulay IC, Deadman ME et al Population and single‐cell genomics reveal the Aire dependency, relief from polycomb silencing, and distribution of self‐antigen expression in thymic epithelia. Genome Res 2014; 24:1918–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Derbinski J, Gäbler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M et al Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med 2005; 202:33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Klein L. Aire gets company for immune tolerance. Cell 2015; 163:794–5. [DOI] [PubMed] [Google Scholar]
- 62. Takaba H, Morishita Y, Tomofuji Y, Danks L, Nitta T, Komatsu N et al Fezf2 orchestrates a thymic program of self‐antigen expression for immune tolerance. Cell 2015; 163:975–87. [DOI] [PubMed] [Google Scholar]
- 63. Miller JF. The golden anniversary of the thymus. Nat Rev Immunol 2011; 11:489–95. [DOI] [PubMed] [Google Scholar]
- 64. Brunk F, Michel C, Holland‐Letz T, Slynko A, Kopp‐Schneider A, Kyewski B et al Dissecting and modeling the emergent murine TEC compartment during ontogeny. Eur J Immunol 2017; 47:1153–59. [DOI] [PubMed] [Google Scholar]
- 65. Alves NL, Takahama Y, Ohigashi I, Ribeiro AR, Baik S, Anderson G et al Serial progression of cortical and medullary thymic epithelial microenvironments. Eur J Immunol 2014; 44:16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Savino W, Mendes‐da‐Cruz DA, Silva JS, Dardenne M, Cotta‐de‐Almeida V. Intrathymic T‐cell migration: a combinatorial interplay of extracellular matrix and chemokines? Trends Immunol 2002; 23:305–13. [DOI] [PubMed] [Google Scholar]
- 67. Nitta T, Suzuki H. Thymic stromal cell subsets for T cell development. Cell Mol Life Sci 2016; 73:1021–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Halkias J, Melichar HJ, Taylor KT, Ross JO, Yen B, Cooper SB et al Opposing chemokine gradients control human thymocyte migration in situ . J Clin Invest 2013; 123:2131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Holländer GA. Claudins provide a breath of fresh Aire. Nat Immunol 2007; 8:234–6. [DOI] [PubMed] [Google Scholar]
- 70. De Martino L, Capalbo D, Improda N, Lorello P, Ungaro C, Di Mase R et al Novel findings into AIRE genetics and functioning: clinical implications. Front Pediatr 2016; 4:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Malchow S, Leventhal DS, Nishi S, Fischer BI, Shen L, Paner GP et al Aire‐dependent thymic development of tumor‐associated regulatory T cells. Science 2013; 339:1219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hubert FX, Kinkel SA, Davey GM, Phipson B, Mueller SN, Liston A et al Aire regulates the transfer of antigen from mTECs to dendritic cells for induction of thymic tolerance. Blood 2011; 118:2462–72. [DOI] [PubMed] [Google Scholar]
- 73. Perry JS, Lio CW, Kau AL, Nutsch K, Yang Z, Gordon JI et al Distinct contributions of Aire and antigen‐presenting‐cell subsets to the generation of self‐tolerance in the thymus. Immunity 2014; 41:414–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Laan M, Kisand K, Kont V, Möll K, Tserel L, Scott HS et al Autoimmune regulator deficiency results in decreased expression of CCR4 and CCR7 ligands and in delayed migration of CD4+ thymocytes. J Immunol 2009; 183:7682–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jin R, Aili A, Wang Y, Wu J, Sun X, Zhang Y et al Critical role of SP thymocyte motility in regulation of thymic output in neonatal Aire–/– mice. Oncotarget 2017; 8:83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V et al Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 2004; 427:355–60. [DOI] [PubMed] [Google Scholar]
- 77. Koivula TT, Laakso SM, Niemi HJ, Kekäläinen E, Laine P, Paulin L et al Clonal analysis of regulatory T cell defect in patients with autoimmune polyendocrine syndrome type 1 suggests intrathymic impairment. Scand J Immunol 2017. (in press). [DOI] [PubMed] [Google Scholar]
- 78. Heikkilä N, Laakso SM, Mannerström H, Kekäläinen E, Saavalainen P, Jarva H et al Expanded CD4+ effector/memory T cell subset in APECED produces predominantly interferon γ . J Clin Immunol 2016; 36:555–63. [DOI] [PubMed] [Google Scholar]
- 79. Gies V, Guffroy A, Danion F, Billaud P, Keime C, Fauny JD et al B cells differentiate in human thymus and express AIRE. J Allergy Clin Immunol 2017.(in press). [DOI] [PubMed] [Google Scholar]
- 80. Yamano T, Nedjic J, Hinterberger M, Steinert M, Koser S, Pinto S et al Thymic B cells are licensed to present self‐antigens for central T cell tolerance induction. Immunity 2015; 42:1048–61. [DOI] [PubMed] [Google Scholar]
- 81. Perri V, Gianchecchi E, Scarpa R, Valenzise M, Rosado MM, Giorda E et al Altered B cell homeostasis and toll‐like receptor 9‐driven response in patients affected by autoimmune polyglandular syndrome Type 1: altered B cell phenotype and dysregulation of the B cell function in APECED patients. Immunobiology 2017; 222:372–83. [DOI] [PubMed] [Google Scholar]
- 82. Meyer S, Woodward M, Hertel C et al AIRE‐deficient patients harbor unique high‐affinity disease‐ameliorating autoantibodies. Cell 2016; 166:582–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Landegren N, Sharon D, Freyhult E, Hallgren A, Eriksson D, Edqvist PH et al Proteome‐wide survey of the autoimmune target repertoire in autoimmune polyendocrine syndrome type 1. Sci Rep 2016; 6:20104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lovewell TR, McDonagh AJ, Messenger AG, Azzouz M, Tazi‐Ahnini R. The AIRE ‐230Y polymorphism affects AIRE transcriptional activity: potential influence on AIRE function in the thymus. PLoS ONE 2015; 10:e0127476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Björses P, Pelto‐Huikko M, Kaukonen J, Aaltonen J, Peltonen L, Ulmanen I. Localization of the APECED protein in distinct nuclear structures. Hum Mol Genet 1999; 8: 259–266. [DOI] [PubMed] [Google Scholar]
- 86. Pitkänen J, Vähämurto P, Krohn K, Peterson P. Subcellular localization of the autoimmune regulator protein. Characterization of nuclear targeting and transcriptional activation domain. J Biol Chem 2001; 276:19597–602. [DOI] [PubMed] [Google Scholar]
- 87. Mora M, Hanzu FA, Pradas‐Juni M, Aranda GB, Halperin I, Puig‐Domingo M et al New splice site aceptor mutation in AIRE gene in autoimmune polyendocrine syndrome type 1. PLoS One 2014; 9:e101616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Fornari TA, Donate PB, Macedo C, Marques MM, Magalhães DA, Passos GA. Age‐related deregulation of Aire and peripheral tissue antigen genes in the thymic stroma of non‐obese diabetic (NOD) mice is associated with autoimmune type 1 diabetes mellitus (DM‐1). Mol Cell Biochem 2010; 342:21–8. [DOI] [PubMed] [Google Scholar]
- 89. Lima FA, Moreira‐Filho CA, Ramos PL, Brentani PH, Lima LA, Arrais M et al Decreased AIRE expression and global thymic hypofunction in Down syndrome. J Immunol 2011; 187:3422–30. [DOI] [PubMed] [Google Scholar]
- 90. Rodrigues PM, Ribeiro AR, Perrod C, Landry JM, Araujo L, Pereira‐Castro I et al Thymic epithelial cells require p53 to support their long‐term function in thymopoiesis in mice. Blood 2017; 130:478–88. [DOI] [PubMed] [Google Scholar]
- 91. Zumer K, Low AK, Jiang H, Saksela K, Perterlin BM. Unmodified histone H3K4 and DNA dependent protein kinase recruit autoimmune regulator to target genes. Mol Cell Biol 2012; 32:1354–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Waterfield M, Khan IS, Cortez JT, Fan U, Metzger T, Greer A et al The transcriptional regulator Aire coopts the repressive ATF7ip‐MBD1 complex for the induction of immunotolerance. Nat Immunol 2014; 15:258–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Liiv I, Rebane A, Org T, Saare M, Maslovskaja J, Kisand K et al DNA‐PK contributes to the phosphorylation of AIRE: importance in transcriptional activity. Biochim Biophys Acta 2008; 1783:74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Oven I, Brdickova N, Kohoutek J, Vaupotic T, Narat M, Peterlin BM. AIRE Recruits PTEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol Cell Biol 2007; 27:8815–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Giraud M, Jmari N, Du L, Carallis F, Nieland TJF, Perez‐Campo FM et al An RNAi screen for Aire cofactors reveals a role for Hnrnpl in polymerase release and Aire activated ectopic transcription. Proc Natl Acad Sci USA 2014; 111:1491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yoshida H, Bansal K, Schaefer U, Chapman T, Rioja I, Proekt I et al Brd4 bridges the transcriptional regulators, Aire and P‐TEFb, to promote elongation of peripheral‐tissue antigen transcripts in thymic stromal cells. Proc Natl Acad Sci USA 2015; 112:E4448 E4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Abramson J, Giraud M, Benoist C, Mathis D. Aire's partners in the molecular control of immunological tolerance. Cell 2010; 140:123–35. [DOI] [PubMed] [Google Scholar]
- 98. Chuprin A, Avin A, Goldfarb Y, Herzig Y, Levi B, Jacob A et al The deacetylase Sirt1 is an essential regulator of Aire‐mediated induction of central immunological tolerance. Nat Immunol 2015; 16:737–45. [DOI] [PubMed] [Google Scholar]
- 99. Incani F, Serra ML, Meloni A, Cossu C, Saba L, Cabras T et al AIRE acetylation and deacetylation: effect on protein stability and transactivation activity. J Biomed Sci 2014; 21:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Saare M, Rebane A, Rajashekar B, Vilo J, Peterson P. Autoimmune regulator is acetylated by transcription coactivator CBP/p300. Exp Cell Res 2012; 318:1767–78. [DOI] [PubMed] [Google Scholar]
- 101. Rattay K, Claude J, Rezavandy E, Matt S, Hofmann TG, Kyewski B et al Homeodomain‐interacting protein kinase 2, a novel autoimmune regulator interaction partner, modulates promiscuous gene expression in medullary thymic epithelial cells. J Immunol 2015; 194:921–8. [DOI] [PubMed] [Google Scholar]
- 102. Shao W, Zumer K, Fujinaga K, Peterlin BM. FBXO3 protein promotes ubiquitylation and transcriptional activity of AIRE (Autoimmune regulator) . J Biol Chem 2016; 291:17953–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Liu H, Jain R, Guan J, Vuong V, Ishido S, La Gruta NL et al Ubiquitin ligase MARCH 8 cooperates with CD83 to control surface MHC II expression in thymic epithelium and CD4 T cell selection. J Exp Med 2016; 213:1695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ilmarinen T, Kangas H, Kytömaa T, Eskelin P, Saharinen J, Seeler J‐S et al Functional interaction of AIRE with PIAS1 in transcriptional regulation. Mol Immunol 2008; 45:1847–62. [DOI] [PubMed] [Google Scholar]
- 105. Ilmarinen T, Melen K, Kangas H, Julkunen I, Ulmanen I, Eskelin P. The monopartite nuclear localization signal of autoimmune regulator mediates its nuclear import and interaction with multiple importin α molecules. FEBS J 2006; 273:315–24. [DOI] [PubMed] [Google Scholar]
- 106. Dragin N, Bismuth J, Cizeron‐Clairac G, Biferi MG, Berthault C, Serraf A et al Estrogen‐mediated downregulation of AIRE influences sexual dimorphism in autoimmune diseases. J Clin Invest 2016; 126:1525–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhu M‐L, Bakhru P, Conley B, Nelson JS, Free M, Martin A et al Sex bias in CNS autoimmune disease mediated by androgen control of autoimmune regulator. Nat Commun 2016; 7:11350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Yanagihara T, Sanematsu F, Sato T, Uruno T, Duan X, Tomino T et al Intronic regulation of Aire expression by Jmjd6 for self‐tolerance induction in the thymus. Nat Commun 2015; 6:8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Haljasorg U, Bichele R, Saare M, Guha M, Maslovskaja J, Kõnd K et al A highly conserved NF‐κB‐responsive enhancer is critical for thymic expression of Aire in mice. Eur J Immunol 2015; 45:3246–56. [DOI] [PubMed] [Google Scholar]


