Abstract
The eukaryotic kinetochore is a sophisticated multi-protein machine that segregates chromosomes during cell division. To ensure accurate chromosome segregation, it performs three major functions using disparate molecular mechanisms. It operates a mechanosensitive signaling cascade known as the Spindle Assembly Checkpoint (SAC) to detect and signal the lack of attachment to spindle microtubules, and delay anaphase onset in response. After attaching to spindle microtubules, the kinetochore generates the force necessary to move chromosomes. Finally, if the two sister kinetochores on a chromosome are both attached to microtubules emanating from the same spindle pole, they activate another mechanosensitive mechanism to correct the monopolar attachments. All three functions maintain genome stability during cell division. The outlines of the biochemical activities responsible for these functions are now available. How the kinetochore integrates the underlying molecular mechanisms is still being elucidated. In this review, we will discuss how the nanoscale protein organization in the kinetochore, which we refer to as kinetochore ‘architecture’, organizes its biochemical activities to facilitate the realization and integration of emergent mechanisms underlying its three major functions. For this discussion, we will use the relatively simple budding yeast kinetochore as a model, and extrapolate insights gained from this model to elucidate functional roles of the architecture of the much more complex human kinetochore.
Introduction
Multi-protein assemblies and machines assume tremendously diverse composition and organization to perform complex cell biological functions. An excellent example of a protein assembly is the endocytic coat, which is a transient, continuously evolving assemblage of many interacting proteins [1]. At the other extreme, is the nuclear pore. The core scaffold of the nuclear pore is a long-lived structure containing precisely organized copies of many proteins [2]. In both cases, the protein architecture, defined as the nanoscale spatial organization of component proteins within the protein assembly or machine, decides how component proteins cooperate with one another to realize their functions. Reductionist methods have been extremely successful in defining structure-function relationships for individual proteins. However, to fully understand multi-protein machines, integrative approaches that define how individual components give rise to emergent functions, and establish ‘architecture-function’ relationships, are also necessary. The eukaryotic kinetochore presents an excellent case to study architecture-function relationships.
Much is now known about the structures, biochemical activities, and the biophysics of the component proteins of the kinetochore that execute its three major functions (Figure 1A, ref. [3]). However, this knowledge does not fully reveal the underlying molecular mechanisms, explain how the kinetochore integrates these mechanisms into one framework, or predict the possibility of cross-talk among its functions [4]. For this, the spatial organization of the biochemical activities must be considered. Complicating this analysis, however, is the fact that most eukaryotic kinetochores bind multiple microtubules dynamically. For example, the human kinetochore simultaneously interacts with the plus-ends of ~ 20 microtubules that exist as a mixed population of both polymerizing and depolymerizing microtubules. Furthermore, the ~ 200 nm diameter disk-shaped human kinetochore is densely populated with a large and diverse set of proteins, most of which are in multi-copy. In this context, the kinetochore found in the budding yeast Saccharomyces cerevisiae is a particularly suitable model, because it stably binds to the plus-end of one microtubule in metaphase [5, 6]. It thus represents the basic functional unit of the eukaryotic kinetochore – one kinetochore-microtubule attachment. Important aspects of the architecture of the yeast kinetochore-microtubule attachment in metaphase have been quantified. Models of kinetochore architecture created from these and structural data provide the starting point needed to study architecture-function relationships [7, 8].
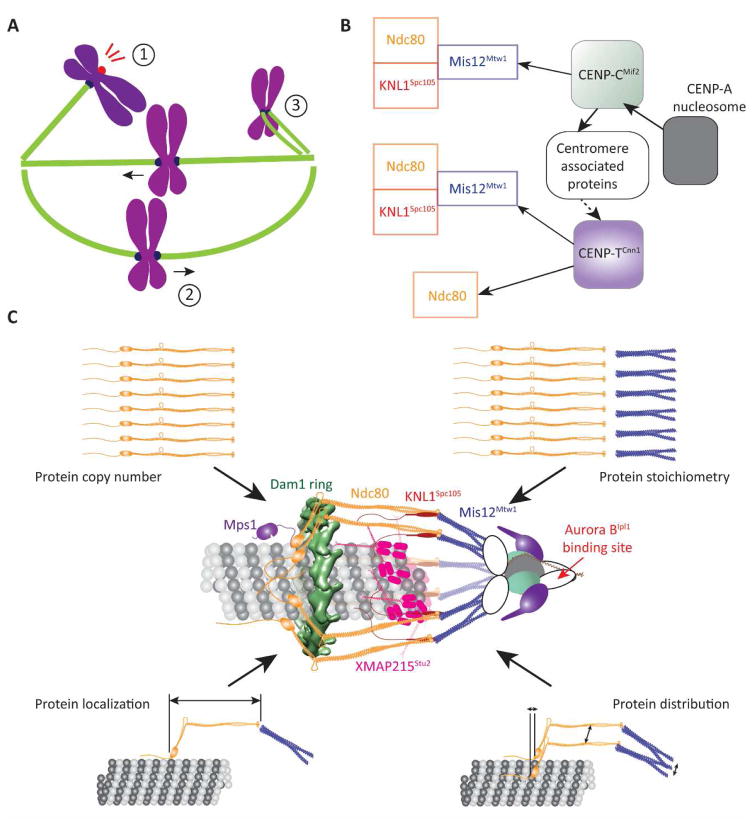
Figure 1. The function and protein architecture of the kinetochore.
A Cartoon of a mitotic spindle displaying the three main kinetochore functions: (1) Activation of the Spindle Assembly Checkpoint, (2) generation of bidirectional chromosome movement that is coupled to microtubule polymerization and depolymerization, and (3) correction of monopolar attachment of sister kinetochores. B (left to right = microtubule plus-end to centromere) The conserved, dual pathways (solid arrows – direct interaction, dashed arrow – indirect interaction) that assemble the KMN network, which forms the interface of the kinetochore with the microtubule plus-end. C Reconstruction of the protein architecture of the budding yeast kinetochore using fluorescence microscopy measurements and protein structures [7, 8, 14, 15, 32, 33, 48–50, 73]. Centromere-associated proteins are represented by white, oblong shapes.
The core protein machinery of the yeast kinetochore is conserved. Therefore, the architecture-function relationships derived from budding yeast will provide insight into the operation of the highly complex human kinetochore. Indeed, a recent study proposed an elegant conceptualization of the human kinetochore as the two-dimensional convolution of multiple yeast kinetochore-like subunits over a disk-shaped surface [9]. Nevertheless, the human kinetochore is built for entirely different performance specifications: it must coordinate the activities of its multiple microtubule binding sites to move the chromosome over longer distances (~ 5 μm versus < 0.5 μm in budding yeast, ref. [10]), against much larger opposing forces (>100 pN versus ~ 7 pN in yeast, refs. [11, 12]). In this review, we will use the budding yeast kinetochore as a starting point for the discussion of architecture-function relationships. We will then highlight how these relationships may fit into the complex architecture of the human kinetochore, and the areas in which the two kinetochores likely diverge.
The composition, assembly pathways, and biochemical activities of the kinetochore
We begin the discussion by briefly describing the essential biochemical activities that execute the three major functions of the kinetochore (please see the recent review by Musacchio and Desai [3] for a comprehensive discussion of the molecular biology of the kinetochore). We will refer to each protein by the name for the human ortholog followed by a super-scripted name of the corresponding budding yeast protein, if it is different. From the functional perspective, the protein composition and assembly of the kinetochore can be simplified as follows (Figure 1B). The kinetochore interacts with the microtubule and with SAC signaling proteins via a network of three protein complexes: KNL1Spc105, Mis12Mtw1, and Ndc80, collectively referred to as the KMN network [13–15]. This interface is assembled by two parallel pathways initiated by the proteins CENP-CMif2 and CENP-TCnn1 [16–20]. CENP-CMif2 and CENP-TCnn1 are assembled on a well-defined territory on each chromosome, known as the centromere, through their interactions with the centromere-specific histone H3 variant CENP-ACse4 [9, 19, 21]. Although the CENP-CMif2 and CENP-TCnn1 pathways are conserved, their contribution to kinetochore function is species-specific: both pathways are required for the function of the human kinetochore, whereas only the CENP-CMif2 pathway is required in budding yeast [22].
Remarkably, the three mechanisms of end-on microtubule attachment, SAC signaling, and error correction ultimately focus on just two proteins that directly interface the kinetochore with the microtubule and the SAC signaling machinery: Ndc80 and KNL1Spc105. To activate the SAC, the Calponin-Homology (CH) domains of Ndc80, which are globular domains located at the microtubule-binding end of the complex, bind the SAC activator, Mps1 kinase [8, 23–25]. Mps1 phosphorylates conserved motifs within KNL1Spc105 to enable these motifs to recruit a number of SAC signaling proteins, and form the Mitotic Checkpoint Complex [26–30]. KNL1Spc105 also recruits phosphatases that antagonize Mps1 to facilitate SAC silencing [26]. In addition to the crucial role of recruiting Mps1 for SAC signaling, the CH-domains of Ndc80 also function as the primary binding site that establishes end-on microtubule attachment [31, 32]. To maintain end-on attachments and to generate force, Ndc80 recruits several accessory microtubule-associated proteins (e.g. Dam1 complex in fungi, Ska complex, Astrin/SKAP, etc. in metazoa [33–35]). Finally, the kinetochore destabilizes monopolar attachments by directing the Aurora BIpl1 kinase toward the microtubule-binding domains of Ndc80 and other proteins, thereby weakening their affinity for the microtubule [36–42]. This description of the biochemical activities of kinetochore proteins does not fully explain the underlying molecular mechanisms; knowledge of kinetochore architecture is required to elucidate how these activities cooperate. Furthermore, the functional roles of any reorganization of the kinetochore induced by microtubule attachment or dynamic changes within the architecture during kinetochore movement must also be studied.
The protein architecture in the end-on kinetochore-microtubule attachment
The end-on morphology of the kinetochore-microtubule attachment is highly conserved in all eukaryotes that have been studied to date [6]. Kinetochore researchers recognized early on that this morphology plays an integral role in its functional mechanisms, and proposed generalized models centered on the end-on morphology to explain the functional mechanisms [43–45]. To test the implementation of these model mechanisms, however, it is necessary to first define the biochemical properties and structures of kinetochore components, and then their organization within the end-on kinetochore-microtubule attachment. The latter part has proven to be a significant challenge. The kinetochore is a network of several protein components, most of which are present in multiple copies. Many of these components contain inherently flexible domains and linkages [3]. Additionally, the microtubule plus-end likely re-organizes this protein network in a functionally significant manner [46, 47]. These issues pose a major obstacle for structural biological approaches in defining its architecture. Resolving the positions of individual molecules in the densely packed kinetochore is also beyond the capabilities of super-resolution microscopy. Therefore, alternative approaches are necessary to determine the architecture.
One such approach is to re-construct kinetochore architecture by answering simpler questions pertaining to its key features (Figure 1C). How many molecules of each protein component does one kinetochore incorporate, and how variable is this number? What is the average position of each component, and are these positions variable? What is the axial and circumferential distribution of protein molecules about their average positions? Quantitative answers to these questions obtained from diverse fluorescence microscopy methods and combined with the known structures of kinetochore proteins established a detailed model of the architecture of the KMN network in the budding yeast kinetochore-microtubule attachment [7, 48–50]. Although this architecture invokes certain assumptions, specifically the circular symmetry of kinetochore proteins around the microtubule diameter and the relative positions of the CH-domains and the Dam1 ring, it has enabled powerful predictions regarding the emergent mechanisms of kinetochore function (discussed in the sections below).
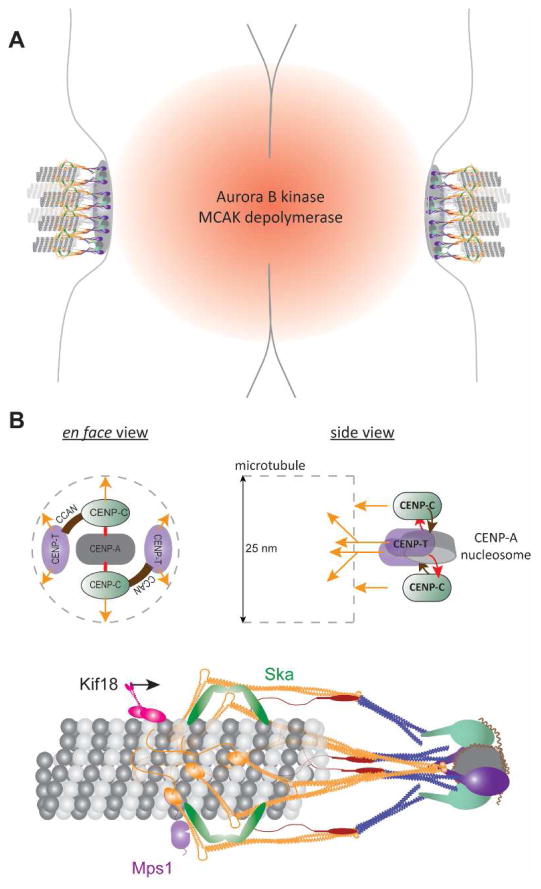
Much work is still needed to synthesize a similar understanding of the architecture of the human kinetochore. The average copy numbers of KMN network molecules per kinetochore, and their organization along the axis of the microtubule in metaphase is known [51, 52]. However, their distribution about the average positions and over the disk-shaped surface of the centromere is unknown, a problem that is significantly complicated by the fact that human kinetochores contain multiple microtubule binding sites (Figure 2A). Identification of the CENP-ACse4 nucleosome as the minimal foundation for assembling the KMN network will simplify this problem to some extent [9, 18]. This finding is useful for proposing a model for the ‘local’ kinetochore architecture, defined as the organization of kinetochore proteins in one kinetochore-microtubule attachment. The bilateral symmetry the CENP-ACse4 nucleosome will impose an orientation and spacing on the two CENP-CMif2 molecules that it recruits (Figure 2B, top). This patterning of CENP-CMif2 will then direct the spatial organization of other centromeric proteins, including CENP-TCnn1. Thus, the spatial organization of centromeric proteins will ultimately dictate the patterning of KMN network molecules, and hence the architecture of the interface of the human kinetochore with the microtubule plus-end (Fig. 2B, bottom). Beyond the local architecture of KMN molecules within one attachment lies the broader architecture of the kinetochore: the distribution of many such attachments across the disk-shaped surface of the centromere. This broader architecture will influence the ability of the kinetochore to interact simultaneously with many microtubule plus-ends. Defining both the local and broader architecture of the human kinetochore remains a major challenge for the field.
Figure 2. Protein architecture of the human kinetochore.
A Cartoon of the organization of sister kinetochores on a human chromosome. The inter-centromeric localization of Aurora BIpl1 and MCAK is highlighted. B Top: Schematic displays a hypothetical spatial manifestation of the biochemical pathways of kinetochore assembly. Orange arrows indicate pathways of Ndc80 recruitment; grey dashed lines represent the microtubule. Note that CENP-TCnn1 recruits two Ndc80 molecules. Bottom: Cartoon of a hypothetical local architecture of the kinetochore-microtubule attachment in humans.
Kinetochore architecture encodes a mechanism for sensing end-on attachment
In most eukaryotes, the kinetochore is unattached at the beginning of mitosis. To avoid chromosome missegregation, it delays cell division by activating the SAC. Typically, the kinetochore first binds laterally to the microtubule lattice, and then converts this interaction into a stable end-on attachment. SAC inactivation occurs once end-on attachments form [23, 53]. Recent work reveals two mechanisms that the kinetochore can use to detect end-on attachment and silence the SAC in response [8, 23, 24]. Both mechanisms rely on the dual role of the CH-domains of Ndc80 as the Mps1 binding site and as the interface for end-on attachment. The first mechanism, studied in human cells, proposes that Mps1 and the microtubule plus-end compete for binding to the CH-domains of Ndc80 [23, 24]. Consequently, end-on attachments displace Mps1 from the kinetochore so that it can no longer phosphorylate KNL1Spc105. The second mechanism, which comes from studies in budding yeast, suggests an integral role for kinetochore architecture in implementing the attachment-mediated SAC silencing [8].
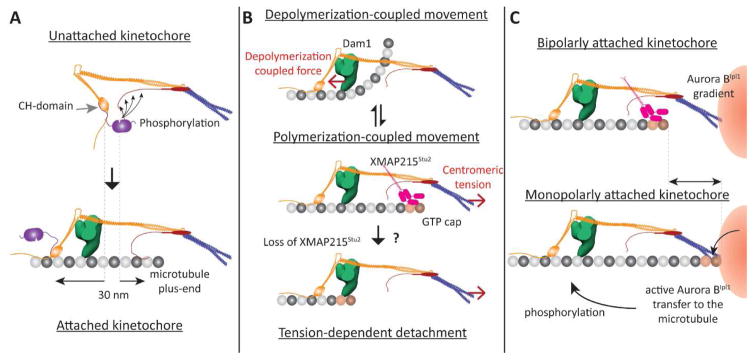
The signaling state of the yeast kinetochore is determined by a single change in its architecture that is elicited by end-on attachment. In the unattached, SAC active kinetochore, the CH-domains of Ndc80 are located within 10 nm of the phosphodomain of KNL1Spc105 (Fig. 3A, top). Therefore, Mps1 bound to the CH-domains robustly phosphorylates KNL1Spc105 and the SAC proteins that it recruits, initiating the SAC. In the attached, SAC inactive kinetochore, the CH-domains and KNL1Spc105 phosphodomain are ~ 30 nm apart [50]. This prevents Mps1 from phosphorylating KNL1Spc105, thereby disrupting SAC signaling (Fig. 3A, bottom). If the 30 nm gap is experimentally abridged, the yeast kinetochore becomes unable to sense end-on attachment, and the SAC becomes constitutively active [8]. Thus, the yeast kinetochore relies on the separation between Mps1 and its target to detect end-on attachments.
Figure 3. Proposed architecture-function relationships for the yeast kinetochore (1-D representations of the kinetochore shown).
A Role of kinetochore architecture in SAC inactivation: Separation of the CH-domains of Ndc80 and the phosphodomain of KNL1Spc105 by end-on attachment (highlighted by dashed lines) disrupts the phosphorylation of KNL1Spc105 by the Mps1 kinase (magenta) bound to the CH-domain. B Proposed roles of the architecture of microtubule-binding proteins in generating bidirectional movement. Top: When the plus-end is depolymerizing, the Dam1 ring (green) mechanically opposes the curling of tubulin protofilaments, and experiences a pushing force (red arrow). Middle panel: We propose that XMAP215Stu2 localizes to the kinetochore by recognizing the GTP-tubulin cap on the polymerizing plus-end. Its microtubule-destabilizing activity reverts the plus-end back to the depolymerizing state. Bottom panel: In the absence of XMAP215 activity, centromeric tension can slide the kinetochore off the growing plus-end. C Potential roles for kinetochore architecture in correcting monopolar attachment: The position of the plus-end may be significantly different in kinetochores with bipolar and monopolar attachment (highlighted by the arrow, top and bottom panels respectively). The proximity of the lattice to the centromere may facilitate the transport of hyper-activated Aurora BIpl1 kinase to its targets – microtubule-binding kinetochore proteins.
Despite recent progress, two significant questions about the mechanism of SAC silencing remain unresolved. The first question is whether the silencing mechanism exclusively relies on the presence of end-on attachment, or whether it also requires force generation by such this attachment. Recent observations suggest that force generation is not necessary for SAC silencing [54, 55]. This conclusion is consistent with the biochemical competition mechanism for SAC silencing, which does not require any force. It is also consistent with the architecture-based model of SAC silencing: the displacement of two protein domains is unlikely to require a large force [8, 56]. Future biophysical analyses of SAC silencing by the kinetochore will unequivocally establish whether SAC silencing requires significant force generation by end-on attachment. The second question relates to the assumption of the binary, switch-like activation and inactivation of the SAC in the two models. This description is superficially valid for the yeast kinetochore, which can exist in only one of two states: attached or unattached. The binary state description does not apply to the human kinetochore, because it attaches dynamically to ~ 20 microtubule plus-ends that turn over completely in about 4 minutes [57]. Furthermore, study of metaphase kinetochores in Potaroo Kidney (PtK1) cells suggest that the kinetochore possesses ~ 15% excess microtubule-binding capacity that is unused even in metaphase [58]. To distinguish between partial attachment from a complete lack of attachments, the human kinetochore may use either additional regulation or more complex mechanisms to silence the SAC. For example, to inactivate the SAC, the human kinetochore may use either a temporal threshold defined by a minimum time period that the kinetochore must spend in the attached state, or a number threshold defined by the minimum number of microtubules bound by the kinetochore. Very little is known about the existence or nature of such mechanisms.
The role of kinetochore architecture in driving persistent, bidirectional chromosome movement
As suggested by its name, the major function of the kinetochore is to drive chromosome movement. It produces the force necessary for generating movement by harnessing microtubule polymerization dynamics. It is reasonable to expect that the architecture of the microtubule-binding kinetochore proteins is tailored to suit the changing form and position of tubulin dimers at the plus-end. The kinetochore also recruits motor proteins and microtubule-associated proteins (MAPs) as accessory factors for attachment and force generation. These proteins are expected to occupy positions dictated by their interactions with kinetochore proteins and the microtubule. The relatively simple and well-defined microtubule-binding machinery of the yeast kinetochore and its persistent interaction with one microtubule plus-end in metaphase provides the ideal opportunity to study the significance of its architecture to force generation.
Ndc80 is the linchpin of end-on kinetochore-microtubule attachment (Figure 3B). It uses three microtubule-binding domains: a positively-charged disordered N-terminal tail of the Ndc80 subunit and the CH-domains of the Ndc80 and Nuf2 subunits [59]. The disordered tail binds to the negatively-charged tubulin tails. This binding assists in the initial contact between the kinetochore and the microtubule lattice [60]. The CH-domains bind in the groove between tubulin monomers along a straight tubulin protofilament in the microtubule lattice, but they cannot do so if this groove is distorted, as in a curling, depolymerizing protofilament [32, 61]. This property of the CH-domains is essential for forming end-on kinetochore-microtubule attachments [13, 62]. Ndc80 structure appears to be tailored for both lateral and end-on attachment: it contains a flexible hinge in its front section to enable the CH-domains access to the binding groove between tubulin monomers by making a 40° angle to the microtubule axis [63]. The conformation of Ndc80 in metaphase yeast kinetochores suggests that they can assume the preferred orientation for the CH-domains to bind to the lattice (Figure 3B, top, [7]).
Ndc80 positions the kinetochore at the plus-end, but it cannot hold on to a dynamic plus-end against high opposing forces [64]. In budding yeast, the Dam1 complex, which is recruited by Ndc80, is essential for force generation [38, 65]. Dam1 molecules likely assemble in the form of an oligomeric ring encircling the microtubule [7, 33, 66]. The Dam1 ring mechanically opposes the outward curling of tubulin protofilaments during depolymerization, and thus, experiences a poleward force [67, 68]. However, to generate force in this manner, the Dam1 ring must be positioned at the edge of the microtubule lattice where it can encounter curling protofilaments [69]. Therefore, it is not surprising that Dam1 localizes in close proximity to the CH-domains of Ndc80, which likely bind to the microtubule lattice near the plus-end (Figure 3B, top [7, 50]). It is reasonable to expect that the Dam1 ring is positioned on the centromeric side of the CH-domains so that it can transmit the force generated to the centromere via Ndc80 [7]. The mechanical opposition to microtubule depolymerization offered by the Dam1 complex also institutes a crucial regulatory mechanism known as ‘tension-dependent rescue’ of the depolymerizing plus-end [11, 38, 70]. As the opposing pull of sister kinetochores on the centromere increases, the Dam1 ring inhibits the conformational change that the tubulin dimers undergo to depolymerize. This force-dependent inhibition promotes the transition from microtubule depolymerization to polymerization. Consequently, the kinetochore switches its direction of movement, and relieves centromeric tension in the process. Tension-dependent rescue is necessary for persistent attachment of sister kinetochores to spindle microtubules.
The high combined affinity of the Ndc80-Dam1 ring assembly likely makes its diffusion along the microtubule lattice very slow [67]. If diffusion is much slower than the rate of microtubule polymerization, then biased-diffusion may not be able keep pace with the polymerizing microtubule tip. The higher affinity of the Dam1 complex for GTP-tubulin, which is present only at the growing microtubule tip, over GDP-tubulin present in the lattice enables Dam1 monomers and small oligomers to track growing microtubule tips [38, 71]. However, whether Dam1 rings can also do this is unclear. Indeed, yeast kinetochore particles have not been observed to track polymerizing microtubule plus-ends in vitro, unless they are experimentally assisted by the imposition of an external force directed toward the plus-end [11]. Centromeric strain will drag the kinetochore toward the plus-end in vivo, but this process depends on the magnitude of the strain, and it is independent of the growth of the plus-end (Figure 3B, middle). This means that the movement induced by high centromeric tension could potentially slide the kinetochore off the polymerizing microtubule tip, and additional mechanisms may be necessary to mitigate this possibility. In this context, the involvement of XMAP215Stu2 in yeast kinetochore motility is noteworthy, but also perplexing. Although XMAP215Stu2 is a well-known tubulin polymerase, it destabilizes microtubules during mitosis in yeast cells [72, 73]. Consistent with this finding, XMAP215Stu2 localizes in a region within the yeast kinetochore where the polymerizing plus-end is expected to be located (Figure 3B, middle) [7]. On the other hand, in vitro findings suggest that XMAP215Stu2 stabilizes attachment of yeast kinetochores to the microtubule tip in a tension-dependent manner [74]. We suggest a simple model to reconcile these observations (Figure 3B, middle & bottom panels). We propose that XMAP215Stu2 recognizes polymerizing microtubule plus-ends within yeast kinetochores and promotes their transition to depolymerization. This prevents the plus-ends from outpacing the kinetochore, and minimizes the possibility of the kinetochore sliding off a growing microtubule tip under high centromeric tension.
The conserved mechanics and biochemistry underlying microtubule plus-end dynamics suggest that the basic mechanisms of harnessing force and persistent bidirectional movement used by the budding yeast kinetochore will be conserved in human kinetochores. This is reflected in the similar organization of microtubule-binding activities in the yeast and human kinetochore [50, 52]. Ndc80 is similarly organized and required for end-on attachment in both kinetochores [50, 52, 75]. However, the human kinetochore attaches to ~ 20 microtubule plus-ends on average, and these attachments are dynamic: they turn over with a half-life of ~ 4 minutes (in RPE-1 cells [76] compared to ~ 25–30 minutes duration of mitosis [10]). The human kinetochore also employs a much larger array of accessory motors and MAPs to elaborate on the basic mechanisms of force generation. The motors include MCAK, Kif18A, Dynein, and CENP-E [77–80]. The MAPs include the Ska complex, Astrin/SKAP, CLASP, EB1, and XMAP215Stu2 [34, 35, 81–84]. The mechanisms of kinetochore recruitment and organization of some of these accessory proteins are similar to the corresponding mechanisms in yeast [52, 77–80, 85]. For example, the recruitment and location of the Ska complex in the human kinetochore appears similar to that of Dam1 in the yeast kinetochore [85]. The collective activities of these proteins in microtubule-binding and in modulating plus-end polymerization dynamics establish and maintain end-on kinetochore-microtubule attachment, generate force, and coordinate the activities of sister kinetochores to drive persistent, bidirectional chromosome movement.
One puzzling aspect of the large array of accessory motors and MAPs employed by the human kinetochore is the apparent redundancy in their activities and kinetochore-specific functions. For example, Ska and XMAP215 are involved in microtubule attachment and force generation; EB1, CLASP, and Astrin/SKAP promote plus-end polymerization [34, 74, 84, 86, 87]. This redundancy may be necessary in part to achieve robust kinetochore functionality. However, it is also possible that the unique kinetochore position of some of these proteins ascribes unique functions to these proteins. For example, MCAK and Kif18A can both destabilize the plus-end [77, 88]. However, MCAK improves the coordinated movement of sister kinetochores, whereas Kif18A promotes the mutually antagonistic activity of the sisters and reduces their coordinated movements [77, 78]. In addition to differences in the biochemical activities of these motors, how they encounter the microtubule plus-end in the kinetochore may give rise to the differences in their function. MCAK localizes at the centromere, whereas Kif18A walks along the microtubule to reach the plus-ends (Figure 2A–B, [89]). Due to its centromeric localization, MCAK may selectively destabilize only those plus-ends that polymerize to extend more than usual towards the centromere. In contrast, Kif18A-mediated plus-end destabilization may be dependent on microtubule length/age [78, 90]. Thus, differences in function may arise from differences in protein position. Therefore, to fully understand how the human kinetochore brings about bidirectional chromosome movement, the biochemical activities as well as the nanoscale architecture of its microtubule-binding machinery must be studied. Additionally, any temporal coordination of these activities and dynamics of protein architecture are also likely to play key roles in kinetochore motility [91]. A major challenge in the field is to formulate a comprehensive model that explains how the human kinetochore synthesizes the activities of its microtubule-binding proteins to control plus-end dynamics, and generate persistent, bidirectional chromosome movement.
Potential roles of kinetochore architecture in correcting monopolar attachments
Although bipolar attachment of sister kinetochores to microtubules emanating from opposite spindle poles is strongly favored, two types of erroneous attachments occur frequently: merotelic and monopolar [57, 92, 93]. The first type of erroneous attachments is known as merotelic attachment, wherein a single kinetochore attaches to microtubules emanating from both poles. There is no kinetochore-based mechanism for the resolution of merotelic attachments; these attachments are destabilized, and bipolar attachments stabilized, by a finely tuned regulation of the dynamicity of spindle microtubules [57, 94]. The second type of erroneous attachments are known as syntelic or monopolar attachments, wherein both sister kinetochores in a pair are attached to the same pole. These attachments are destabilized by a dedicated, kinetochore-based error correction process. Two elements of this process are clear. First, it directs Aurora BIpl1 kinase activity toward microtubule-binding kinetochore proteins to weaken their microtubule-binding affinity [36–42, 95]. Second, the activity of the error correction process is strongly correlated with a sustained lack of centromeric tension, which is a characteristic unique to sister kinetochores with monopolar attachments [36]. Interestingly, sister kinetochores with bipolar attachments also experience periodic and transient loss of centromeric tension, but they do not activate the error correction process. How the kinetochore senses a prolonged absence of centromeric tension, and activates Aurora BIpl1 in response, is unclear. Potential mechanisms that can explain this error correction process have been discussed in excellent reviews (e.g. see [92, 96]). Therefore, we will only describe the prevalent model, and focus on the potential role of kinetochore architecture in the error correction process.
The prevalent model postulates that centromeric tension in the human kinetochore separates Aurora BIpl1 from its phosphorylation targets in a manner that is superficially similar to the mechanism of attachment-dependent SAC signaling [36, 97]. Aurora BIpl1 dynamically localizes to chromatin situated in between the sister centromeres (Figure 2A). Here, the high local concentration of Aurora BIpl1 stimulates its auto phosphorylation and hence hyper-activation. Super-imposition of a phosphatase activity on this Aurora BIpl1 hyper-activation region creates a steep gradient in Aurora BIpl1 kinase activity (Figure 3C) [97]. Sister kinetochores with monopolar attachment fall within the region of Aurora BIpl1 hyperactivity, whereas kinetochores with bipolar attachment deform the centromere and emerge out of it. Consequently, microtubule-binding proteins in these kinetochores become dephosphorylated, and microtubule attachment is stabilized. Although this elegant mechanism is strongly supported by data from vertebrate kinetochores, it does not explain a key observation from budding yeast. Budding yeast kinetochores correct monopolar attachments even when Aurora BIpl1 is unable to localize to the centromere [98]. Therefore, additional mechanisms in the error correction process remain to be discovered in both yeast and humans.
Kinetochore architecture is clearly important in the prevalent model of the error correction process, because it determines the positions of Aurora BIpl1 targets relative to the centromere-localized Aurora BIpl1. However, the architecture of kinetochores with monopolar and bipolar attachments is likely to be similar given that they both possess end-on attachment. A key difference in kinetochore architecture under the two scenarios may be that the plus-end is in close proximity to the centromere only in kinetochores with monopolar attachment (Figure 3C). One possibility is that the error correction mechanism uses the proximity between centromere-localized Aurora BIpl1 and the microtubule plus-end to locally concentrate, and then effectively transport active Aurora BIpl1 along the microtubule to microtubule-bound kinetochore proteins [98]. However, the role of kinetochore architecture, if any, in mechanisms that direct Aurora BIpl1 activity selectively to kinetochores with monopolar attachments remains poorly understood.
Concluding remarks
The nanoscale architecture of the kinetochore can provide insight into how it integrates three truly disparate mechanisms in one molecular framework. Comparison of the yeast and human kinetochores also reveals how the basic integration of the three mechanisms may be enhanced to meet species-specific functional requirements. In the integrative model of the eukaryotic kinetochore, Ndc80 emerges as the focal point of all three mechanisms. It acts as the terminal of a switch that controls the SAC, the conformational sensor that positions the kinetochore at the plus-end, an organizer of microtubule-binding activities in the kinetochore, and a major target of phosphoregulation by Aurora BIpl1. Therefore, the spatial patterning of Ndc80 in the kinetochore and its architecture relative to the microtubule plus-end will play key roles in shaping the emergent mechanisms underlying all three kinetochore functions. Future studies of the kinetochore that explicitly test the role of kinetochore architecture in its functional and regulatory mechanisms will lead us to a comprehensive understanding of one of the most fascinating multi-protein machines in cell biology.
Acknowledgments
This work was supported by R01 GM105948 to APJ. The authors thank Iain Cheeseman & Jennifer Deluca for helpful discussions and a critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Picco A, Mund M, Ries J, Nedelec F, Kaksonen M. Visualizing the functional architecture of the endocytic machinery. Elife. 2015:4. doi: 10.7554/eLife.04535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 3.Musacchio A, Desai A. A Molecular View of Kinetochore Assembly and Function. Biology. 2017:6. doi: 10.3390/biology6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nezi L, Musacchio A. Sister chromatid tension and the spindle assembly checkpoint. Current Opinion in Cell Biology. 2009;21:785–795. doi: 10.1016/j.ceb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McIntosh JR, O’Toole E, Zhudenkov K, Morphew M, Schwartz C, Ataullakhanov FI, Grishchuk EL. Conserved and divergent features of kinetochores and spindle microtubule ends from five species. J Cell Biol. 2013;200:459–474. doi: 10.1083/jcb.201209154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aravamudhan P, Felzer-Kim I, Gurunathan K, Joglekar AP. Assembling the protein architecture of the budding yeast kinetochore-microtubule attachment using FRET. Curr Biol. 2014;24:1437–1446. doi: 10.1016/j.cub.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aravamudhan P, Goldfarb AA, Joglekar AP. The kinetochore encodes a mechanical switch to disrupt spindle assembly checkpoint signalling. Nat Cell Biol. 2015;17:868–879. doi: 10.1038/ncb3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weir JR, Faesen AC, Klare K, Petrovic A, Basilico F, Fischbock J, Pentakota S, Keller J, Pesenti ME, Pan D, et al. Insights from biochemical reconstitution into the architecture of human kinetochores. Nature. 2016;537:249–253. doi: 10.1038/nature19333. [DOI] [PubMed] [Google Scholar]
- 10.Magidson V, O’Connell Christopher B, Lon arek J, Paul R, Mogilner A, Khodjakov A. The Spatial Arrangement of Chromosomes during Prometaphase Facilitates Spindle Assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–579. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicklas RB. Measurements of the force produced by the mitotic spindle in anaphase. J Cell Biol. 1983;97:542–548. doi: 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 14.Petrovic A, Keller J, Liu Y, Overlack K, John J, Dimitrova YN, Jenni S, van Gerwen S, Stege P, Wohlgemuth S, et al. Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores. Cell. 2016;167:1028–1040. e1015. doi: 10.1016/j.cell.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dimitrova YN, Jenni S, Valverde R, Khin Y, Harrison SC. Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly. Cell. 2016;167:1014–1027. e1012. doi: 10.1016/j.cell.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 2012;148:487–501. doi: 10.1016/j.cell.2011.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim S, Yu H. Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J Cell Biol. 2015;208:181–196. doi: 10.1083/jcb.201407074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huis In ‘t Veld PJ, Jeganathan S, Petrovic A, Singh P, John J, Krenn V, Weissmann F, Bange T, Musacchio A. Molecular basis of outer kinetochore assembly on CENP-T. Elife. 2016:5. doi: 10.7554/eLife.21007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gascoigne Karen E, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman Iain M. Induced Ectopic Kinetochore Assembly Bypasses the Requirement for CENP-A Nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rago F, Gascoigne KE, Cheeseman IM. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr Biol. 2015;25:671–677. doi: 10.1016/j.cub.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klare K, Weir JR, Basilico F, Zimniak T, Massimiliano L, Ludwigs N, Herzog F, Musacchio A. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J Cell Biol. 2015;210:11–22. doi: 10.1083/jcb.201412028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schleiffer A, Maier M, Litos G, Lampert F, Hornung P, Mechtler K, Westermann S. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat Cell Biol. 2012;14:604–613. doi: 10.1038/ncb2493. [DOI] [PubMed] [Google Scholar]
- 23.Hiruma Y, Sacristan C, Pachis ST, Adamopoulos A, Kuijt T, Ubbink M, von Castelmur E, Perrakis A, Kops GJ. CELL DIVISION CYCLE. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science. 2015;348:1264–1267. doi: 10.1126/science.aaa4055. [DOI] [PubMed] [Google Scholar]
- 24.Ji Z, Gao H, Yu H. CELL DIVISION CYCLE. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science. 2015;348:1260–1264. doi: 10.1126/science.aaa4029. [DOI] [PubMed] [Google Scholar]
- 25.Kemmler S, Stach M, Knapp M, Ortiz J, Pfannstiel J, Ruppert T, Lechner J. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009;28:1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.London N, Ceto S, Ranish JA, Biggins S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr Biol. 2012;22:900–906. doi: 10.1016/j.cub.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shepperd LA, Meadows JC, Sochaj AM, Lancaster TC, Zou J, Buttrick GJ, Rappsilber J, Hardwick KG, Millar JB. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr Biol. 2012;22:891–899. doi: 10.1016/j.cub.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.London N, Biggins S. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev. 2014;28:140–152. doi: 10.1101/gad.233700.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faesen AC, Thanasoula M, Maffini S, Breit C, Muller F, van Gerwen S, Bange T, Musacchio A. Basis of catalytic assembly of the mitotic checkpoint complex. Nature. 2017 doi: 10.1038/nature21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji Z, Gao H, Jia L, Li B, Yu H. A sequential multi-target Mps1 phosphorylation cascade promotes spindle checkpoint signaling. Elife. 2017:6. doi: 10.7554/eLife.22513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 32.Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 34.Welburn JPI, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, Iii, Cheeseman IM. The Human Kinetochore Ska1 Complex Facilitates Microtubule Depolymerization-Coupled Motility. Developmental Cell. 2009;16:374–385. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manning AL, Bakhoum SF, Maffini S, Correia-Melo C, Maiato H, Compton DA. CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. EMBO J. 2010;29:3531–3543. doi: 10.1038/emboj.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt JC, Kiyomitsu T, Hori T, Backer CB, Fukagawa T, Cheeseman IM. Aurora B kinase controls the targeting of the Astrin-SKAP complex to bioriented kinetochores. J Cell Biol. 2010;191:269–280. doi: 10.1083/jcb.201006129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welburn JPI, Vleugel M, Liu D, Yates JR, Iii, Lampson MA, Fukagawa T, Cheeseman IM. Aurora B Phosphorylates Spatially Distinct Targets to Differentially Regulate the Kinetochore-Microtubule Interface. Molecular Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan YW, Jeyaprakash AA, Nigg EA, Santamaria A. Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. J Cell Biol. 2012;196:563–571. doi: 10.1083/jcb.201109001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shang C, Hazbun TR, Cheeseman IM, Aranda J, Fields S, Drubin DG, Barnes G. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol Biol Cell. 2003;14:3342–3355. doi: 10.1091/mbc.E02-11-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- 43.Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. Proc Natl Acad Sci USA. 1985;82:4404–4408. doi: 10.1073/pnas.82.13.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McIntosh JR. Structural and mechanical control of mitotic progression. Cold Spring Harb Symp Quant Biol. 1991;56:613–619. doi: 10.1101/sqb.1991.056.01.070. [DOI] [PubMed] [Google Scholar]
- 45.Inoue S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magidson V, He J, Ault JG, O’Connell CB, Yang N, Tikhonenko I, McEwen BF, Sui H, Khodjakov A. Unattached kinetochores rather than intrakinetochore tension arrest mitosis in taxol-treated cells. J Cell Biol. 2016;212:307–319. doi: 10.1083/jcb.201412139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aravamudhan P, Felzer-Kim I, Joglekar AP. The budding yeast point centromere associates with two Cse4 molecules during mitosis. Curr Biol. 2013;23:770–774. doi: 10.1016/j.cub.2013.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki A, Badger BL, Salmon ED. A quantitative description of Ndc80 complex linkage to human kinetochores. Nat Commun. 2015;6:8161. doi: 10.1038/ncomms9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wan X, O’Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krefman NI, Drubin DG, Barnes G. Control of the spindle checkpoint by lateral kinetochore attachment and limited Mad1 recruitment. Mol Biol Cell. 2015 doi: 10.1091/mbc.E15-05-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Etemad B, Kuijt TE, Kops GJ. Kinetochore-microtubule attachment is sufficient to satisfy the human spindle assembly checkpoint. Nat Commun. 2015;6:8987. doi: 10.1038/ncomms9987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tauchman EC, Boehm FJ, DeLuca JG. Stable kinetochore-microtubule attachment is sufficient to silence the spindle assembly checkpoint in human cells. Nat Commun. 2015;6:10036. doi: 10.1038/ncomms10036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joglekar AP. A Cell Biological Perspective on Past, Present and Future Investigations of the Spindle Assembly Checkpoint. Biology. 2016:5. doi: 10.3390/biology5040044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Godek KM, Kabeche L, Compton DA. Regulation of kinetochore-microtubule attachments through homeostatic control during mitosis. Nat Rev Mol Cell Biol. 2015;16:57–64. doi: 10.1038/nrm3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McEwen BF, Heagle AB, Cassels GO, Buttle KF, Rieder CL. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J Cell Biol. 1997;137:1567–1580. doi: 10.1083/jcb.137.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci USA. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka TU. Kinetochore-microtubule interactions: steps towards bi-orientation. EMBO J. 2010;29:4070–4082. doi: 10.1038/emboj.2010.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 63.Wang HW, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–875. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol. 2010;189:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Umbreit NT, Miller MP, Tien JF, Ortola JC, Gui L, Lee KK, Biggins S, Asbury CL, Davis TN. Kinetochores require oligomerization of Dam1 complex to maintain microtubule attachments against tension and promote biorientation. Nat Commun. 2014;5:4951. doi: 10.1038/ncomms5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grishchuk EL, Spiridonov IS, Volkov VA, Efremov A, Westermann S, Drubin D, Barnes G, Ataullakhanov FI, McIntosh JR. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc Natl Acad Sci USA. 2008;105:6918–6923. doi: 10.1073/pnas.0801811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- 69.Efremov A, Grishchuk EL, McIntosh JR, Ataullakhanov FI. In search of an optimal ring to couple microtubule depolymerization to processive chromosome motions. Proc Natl Acad Sci USA. 2007;104:19017–19022. doi: 10.1073/pnas.0709524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Franck AD, Powers AF, Gestaut DR, Gonen T, Davis TN, Asbury CL. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–837. doi: 10.1038/ncb1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lampert F, Mieck C, Alushin GM, Nogales E, Westermann S. Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. J Cell Biol. 2013;200:21–30. doi: 10.1083/jcb.201210091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kosco KA, Pearson CG, Maddox PS, Wang PJ, Adams IR, Salmon ED, Bloom K, Huffaker TC. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol Biol Cell. 2001;12:2870–2880. doi: 10.1091/mbc.12.9.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ayaz P, Ye X, Huddleston P, Brautigam CA, Rice LM. A TOG:alphabeta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science. 2012;337:857–860. doi: 10.1126/science.1221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miller MP, Asbury CL, Biggins S. A TOG Protein Confers Tension Sensitivity to Kinetochore-Microtubule Attachments. Cell. 2016;165:1428–1439. doi: 10.1016/j.cell.2016.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kabeche L, Compton DA. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature. 2013;502:110–113. doi: 10.1038/nature12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wordeman L, Wagenbach M, von Dassow G. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J Cell Biol. 2007;179:869–879. doi: 10.1083/jcb.200707120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Varma D, Monzo P, Stehman SA, Vallee RB. Direct role of dynein motor in stable kinetochore-microtubule attachment, orientation, and alignment. J Cell Biol. 2008;182:1045–1054. doi: 10.1083/jcb.200710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vitre B, Gudimchuk N, Borda R, Kim Y, Heuser JE, Cleveland DW, Grishchuk EL. Kinetochore-microtubule attachment throughout mitosis potentiated by the elongated stalk of the kinetochore kinesin CENP-E. Mol Biol Cell. 2014;25:2272–2281. doi: 10.1091/mbc.E14-01-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pereira AL, Pereira AJ, Maia AR, Drabek K, Sayas CL, Hergert PJ, Lince-Faria M, Matos I, Duque C, Stepanova T, et al. Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol Biol Cell. 2006;17:4526–4542. doi: 10.1091/mbc.E06-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cassimeris L, Becker B, Carney B. TOGp regulates microtubule assembly and density during mitosis and contributes to chromosome directional instability. Cell motility and the cytoskeleton. 2009;66:535–545. doi: 10.1002/cm.20359. [DOI] [PubMed] [Google Scholar]
- 83.Tirnauer JS, Canman JC, Salmon ED, Mitchison TJ. EB1 targets to kinetochores with attached, polymerizing microtubules. Mol Biol Cell. 2002;13:4308–4316. doi: 10.1091/mbc.E02-04-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Draviam VM, Shapiro I, Aldridge B, Sorger PK. Misorientation and reduced stretching of aligned sister kinetochores promote chromosome missegregation in EB1- or APC-depleted cells. Embo J. 2006;25:2814–2827. doi: 10.1038/sj.emboj.7601168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang G, Kelstrup CD, Hu XW, Kaas Hansen MJ, Singleton MR, Olsen JV, Nilsson J. The Ndc80 internal loop is required for recruitment of the Ska complex to establish end-on microtubule attachment to kinetochores. J Cell Sci. 2012;125:3243–3253. doi: 10.1242/jcs.104208. [DOI] [PubMed] [Google Scholar]
- 86.Friese A, Faesen AC, Huis in ‘t Veld PJ, Fischbock J, Prumbaum D, Petrovic A, Raunser S, Herzog F, Musacchio A. Molecular requirements for the inter-subunit interaction and kinetochore recruitment of SKAP and Astrin. Nat Commun. 2016;7:11407. doi: 10.1038/ncomms11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 89.Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow JR. Aurora B regulates MCAK at the mitotic centromere. Developmental Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 90.Varga V, Leduc C, Bormuth V, Diez S, Howard J. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 2009;138:1174–1183. doi: 10.1016/j.cell.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 91.Dumont S, Salmon ED, Mitchison TJ. Deformations within moving kinetochores reveal different sites of active and passive force generation. Science. 2012;337:355–358. doi: 10.1126/science.1221886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lampson MA, Grishchuk EL. Mechanisms to Avoid and Correct Erroneous Kinetochore-Microtubule Attachments. Biology. 2017:6. doi: 10.3390/biology6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gregan J, Polakova S, Zhang L, Tolic-Norrelykke IM, Cimini D. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol. 2011;21:374–381. doi: 10.1016/j.tcb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cimini D, Cameron LA, Salmon ED. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr Biol. 2004;14:2149–2155. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 95.Zaytsev AV, Sundin LJ, DeLuca KF, Grishchuk EL, DeLuca JG. Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions. J Cell Biol. 2014;206:45–59. doi: 10.1083/jcb.201312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sarangapani KK, Asbury CL. Catch and release: how do kinetochores hook the right microtubules during mitosis? Trends Genet. 2014;30:150–159. doi: 10.1016/j.tig.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zaytsev AV, Segura-Pena D, Godzi M, Calderon A, Ballister ER, Stamatov R, Mayo AM, Peterson L, Black BE, Ataullakhanov FI, et al. Bistability of a coupled Aurora B kinase-phosphatase system in cell division. Elife. 2016;5:e10644. doi: 10.7554/eLife.10644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Campbell CS, Desai A. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature. 2013;497:118–121. doi: 10.1038/nature12057. [DOI] [PMC free article] [PubMed] [Google Scholar]