Abstract
Desiccation tolerance is commonly regarded as one of the key features for the colonization of terrestrial habitats by green algae and the evolution of land plants. Extensive studies, focused mostly on physiology, have been carried out assessing the desiccation tolerance and resilience of the streptophytic genera Klebsormidium and Zygnema. Here we present transcriptomic analyses of Zygnema circumcarinatum exposed to desiccation stress. Cultures of Z. circumcarinatum, grown in liquid medium or on agar plates, were desiccated at ~86% relative air humidity until Y(II) ceased. In general, the response to dehydration was much more pronounced in Z. circumcarinatum, cultivated in liquid medium for one month, compared to filaments grown on agar plates for seven and twelve months. Cultivation on solid medium enables the alga to acclimate to dehydration much better and an increase in desiccation tolerance was clearly correlated to increased culture age. Moreover, gene expression analysis revealed that photosynthesis was strongly repressed upon desiccation treatment in the liquid culture while only minor effects were detected in filaments cultivated on agar plates for seven months. Otherwise, both samples showed an induction of stress protection mechanisms such as ROS scavenging (Early light-induced proteins, glutathione metabolism) and DNA repair as well as the expression of chaperones and aquaporins. Additionally, Z. circumcarinatum, cultivated in liquid medium, upregulated sucrose synthesizing enzymes and strongly induced membrane modifications in response to desiccation stress. These results corroborate the previously described hardening and associated desiccation tolerance in Zygnema in response to seasonal fluctuations in water availability.
Keywords: Desiccation Tolerance, Gene Expression, Streptophytic Algae, Transcriptomics, Zygnema
Introduction
The colonization of terrestrial habitats by plants is accompanied by a number of abiotic stress factors such as high irradiance and dehydration (Borstlap 2002, Holzinger & Pichrtová 2016). Tolerating water stress is crucial for survival of land plants as well as terrestrial algae and has been addressed extensively (Borstlap 2002, Dinankar & Bartels 2013, Holzinger et al. 2014, Oliver 2007). Most land plants are able to actively regulate their water status (homoiohydry) but lost the ability to tolerate desiccation. However, the so-called resurrection plants, such as Craterostigma plantagineum, possess desiccation tolerance (poikilohydry; Norwood et al. 2003) and are not able to actively regulate their water content (Holzinger & Karsten 2013). Hence, C. plantagineum and other resurrection plants were used as models for desiccation stress tolerance (Bartels & Salamini 2001, Dinankar & Bartels 2013). Additionally, other higher plants (Basu et al. 2016, Ma et al. 2015, Oliver et al. 2011) and mosses were studied to unravel the mechanisms of desiccation response and resilience (Gao et al. 2015, Shinde et al. 2012).
Over the past years, substantial information about the impact of desiccation stress on streptophytic green algae, including Zygnema spp. and Klebsormidium spp., became available (for summary see Holzinger & Karsten 2013, Karsten & Holzinger 2014, Holzinger & Pichrtová 2016). Klebsormidium generally occupies moist terrestrial habitats (Lokhorst 1996) while Zygnema occurs in hydro-terrestrial environments, meaning in or in close vicinity to freshwater bodies or streams (Davey 1991, Hawes 1990). Upon decreasing air humidity, water is rapidly lost but these poikilohydric organisms have the ability to tolerate dehydration in the vegetative state to a certain degree. However, in Zygnema desiccation tolerance is strongly dependent on the physiological state of the cell. Stress tolerance increases during maturation of the algae (Pichrtová et al. 2014, Herburger et al. 2015) which is associated with the transition from vegetative cells to pre-akinetes and akinetes (McLean & Pessoney 1971). The maturation process is accompanied by changes in the fatty acid composition which have recently been studied by Pichrtová et al. (2016a).
To shed light on the molecular mechanisms of desiccation response and tolerance in streptophyte green algae, transcriptomic profiling was performed for Klebsormidium crenulatum revealing reaction patterns similar to land plants when exposed to water stress (Holzinger et al. 2014). Dehydration in plants and algae is linked to a number of defense mechanisms, e.g. protection of the photosynthetic apparatus by the expression of early light-induced proteins (ELIPs), synthesis of low-molecular-weight osmolytes to maintain turgor pressure, induction of ROS scavenging and increase of the chaperone transcript pool (late embryogenesis abundant (LEA) and heat-shock proteins (Hsps)) (Fernández-Marín et al. 2016, Wang et al. 2004). The above listed defense systems were also induced in the basal streptophyte alga K. crenulatum upon harsh desiccation over silica gel as demonstrated by Holzinger et al. (2014).
In contrast to Klebsormidiophyceae, which are located closer to the basis of the Streptophyta (Becker & Marin 2009), Zygnematophyceae are the sister lineage of the land plants (Wickett et al. 2014). This has been proven through several phylogenetic analyses but is also confirmed by the fact that zygnematophycean algae possess a modified plastid, the 'embryoplast', which played a key role in the development of the land plants (de Vries et al. 2016, Ruhfeld et al. 2014, Wodniok et al. 2011, Zhong et al. 2013). Thus, the investigation of their water stress tolerance on a molecular level is particularly interesting from an evolutionary point of view. While the dehydration-induced changes in physiology of Zygnema circumcarinatum and K. crenulatum are similar, with a drastic reduction of Y(II), the kinetics of water loss are different (Herburger & Holzinger 2015, Lajos et al. 2016). Herburger & Holzinger (2015) reported an elevated callose content in the cell walls of K. crenulatum, which enables shrinkage of the whole cell, compared to Z. circumcarinatum which forms rigid cellulosic secondary walls. In terms of water loss, Z. circumcarinatum reduces the protoplast volume more rapidly compared to Klebsormidium at a relative air humidity (RH) lower than 85%, however, this observation was reversed for higher RH (Lajos et al. 2016). To gain deeper insights into the mechanisms of desiccation tolerance in Z. circumcarinatum and study the differences to K. crenulatum, transcriptomic analyses were performed on algal cultures, either grown in liquid medium or on agar plates and, subsequently, subjected to dehydration. Furthermore, differently matured cultures (one, seven and twelve months) were used to investigate the influence of culture age, which is associated with pre-akinete formation in cultures older than seven months. In contrast to Klebsormidium, the genome of Zygnema has not been published yet. Thus, transcriptomic data also provide a valuable information resource and give insights into a plethora of molecular mechanisms.
Results
Physiological response to desiccation
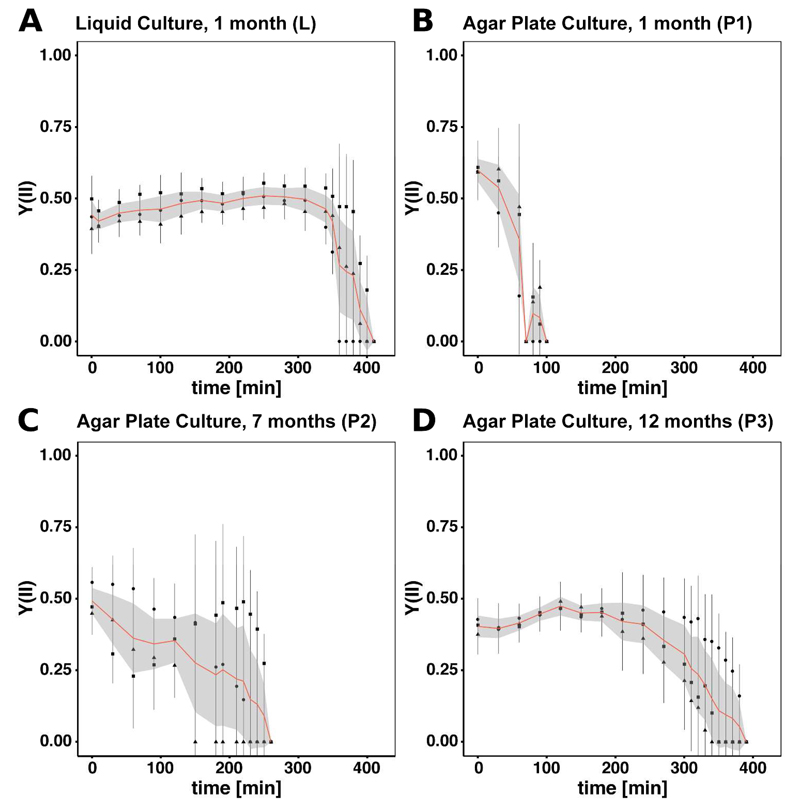
All samples were desiccated over KCl and the Y(II) was monitored. An experimental overview is given in Figure 1. For all samples dehydration stress was applied until Y(II) dropped to zero. Hence, the physiological state of all filaments is comparable. The biomass of the liquid culture (L) and the 12 months old agar plates (P3) maintained photosynthesis the longest until approximately 390 min and 360 min, respectively (Figure 2A/D). Y(II) of the filaments, cultivated for one month on solid medium (P1), drops first at about 90 min (Figure 2B) while Y(II) of the 7 months old culture (P2) reaches zero at approximately 220 min (Figure 2C). Table 1 displays the desiccation time and observed water loss for all samples. When comparing the time required for the cells cultivated on agar plates to reduce Y(II) to zero, increased age is clearly correlated (r = 0.98) with a prolonged activity. All three cultures grown on agar (P1, P2, P3) are differing significantly in desiccation time (p < 0.001). No significant difference has been observed between P3 and L, however, P2 and P1 differ significantly from L. Concerning water loss, no significant differences were detected between L, P2 and P3. Sample P1 differs significantly in water content reduction from L (p < 0.001), P2 (p < 0.01) and P3 (p < 0.01).
Figure 1.
Experimental Setup. Four different culture conditions were chosen: one month old culture grown in liquid medium (L), one month old culture grown on solid medium (P1), seven months old culture grown on solid medium (P2), 12 months old culture grown on solid medium (P3). Samples were taken as controls and for desiccation treatment and RNA was extracted subsequently.
Figure 2.
Y(II) measured over the course of desiccation for all samples. Each triplicate was measured three times at different positions of the filter. The bar indicates standard deviation per replicate while square, triangle and circle indicate the mean value. The mean of all replicates is displayed in red giving the lower and upper Gaussian confidence limit in light grey. A) One month old culture grown in liquid medium (L). B) One month old culture grown on solid medium (P1). C) Seven months old culture grown on solid medium (P2). D) 12 months old culture grown on solid medium (P3).
Table 1.
Physiological results of desiccation stress experiment. Desiccation time means the time that elapsed from the start of the desiccation treatment until Y(II) dropped to zero.
| Sample ID | Cultivation | Desiccation Time [min] | Water loss [%] |
|---|---|---|---|
| L | liquid medium, 1 mo | 390 ± 26.5 | 93.7 ± 2 |
| P1 | solid medium, 1 mo | 90 ± 17.3 | 81 ± 3.4* |
| P2 | solid medium, 7 mo | 223 ± 40.4 | 92.8 ± 3.2 |
| P3 | solid medium, 12 mo | 360 ± 26.5 | 89 ± 0.01 |
n=3 if not indicated otherwise, *n=2
Sequencing outcome and reference library
The triplicates for P3 were pooled for sequencing due to low RNA quantities. Hence, this group was excluded from analysis of differential gene expression. Furthermore, one replicate of P2D exhibited a low extraction yield and, thus, was not sequenced. The sequencing results for all libraries are summarized in supplementary Table S1. For the reference, which was pooled from all samples, 13,241 Mbp were obtained, and for all samples a total of 85,721 Mbp was sequenced.
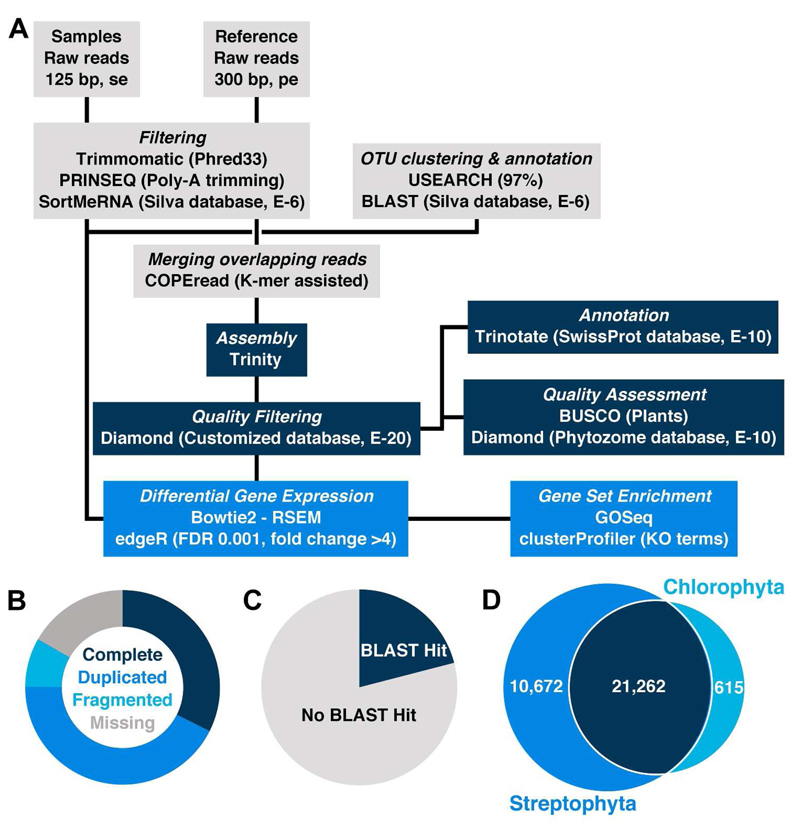
Figure 3A gives an overview of the bioinformatic pipeline used for raw read processing, assembly and further analysis. The assembly of the quality filtered and trimmed reference reads yielded a total of 135,572 contigs with an N50 of 950 bp, and a smallest and largest contig size of 224 bp and 24,724 bp, respectively. To assess the completeness of the established reference transcriptome, Benchmarking Universal Single-Copy Orthologs (BUSCO) were used. As displayed in Figure 3B, we found 76% to be complete, 8% to be fragmented while only 16% were missing. Compared to a variety of other transcriptomes, these values can be regarded as excellent (Simão et al. 2015). Annotating the assembly against the Swiss-Prot database resulted in 28,427 (21%) hits with an e-value smaller than E-10 (Figure 3C). Moreover, all contigs were tested for homology to amino acid sequences, retrieved from complete streptophyte and chlorophyte genomes, resulting overall in higher annotation rates for streptophytic than for chlorophytic sequences (Figure 3D, Figure S2 in the supporting information). For all examined e-values ranging from E-3 to E-20, all assembled contigs shared most sequences with Physcomitrella patens and Klebsormidium flaccidum (Figure S2). The homology comparison of the assembly to all streptophytic and all chlorophytic sequences showed that our assembly shared 21,262 with both groups while 10,672 and 615 sequences were exclusively aligned to streptophytic and chlorophytic proteins, respectively (Figure 3D). In order to evaluate the coverage of metabolic networks, the assigned KO numbers (5.9% of all contigs) were mapped onto the KEGG metabolic pathways map (ko01100, Supplemental Figure S3). In general, we observed a very good coverage with the most important pathways (e.g. carbohydrate metabolism, amino acid metabolism, fatty acid metabolism, nucleotide metabolism, respiration) being complete. The annotation rate for GO terms was 28% of all contigs.
Figure 3.
A) Bioinformatic Pipeline. Raw Reads of all samples (controls and treated) as well as the reference (pool of all samples) were filtered using Trimmomatic, PRINSEQ and SortMeRNA. The remaining paired-end reads of the reference were merged, if possible, using COPEread and subsequently assembled with Trinity. The filtered rRNA reads of the samples were clustered and annotated using USEARCH and BLAST. The assembly was further quality filtered against a customized database containg sequences originating from Physcomitrella, Klebsormidium and Naegleria. After quality assessment with BUSCO and Diamond the contigs were annotated with Trinotate. The sample reads were mapped onto the contigs using Bowtie2, read counts were calculated with RSEM and differential gene expression was performed with edgeR. Finally, GOSeq and clusterProfiler were used for gene set enrichment analyses. More details on the procedure can be found in the material and methods section. B) The donut chart displays the results of the BUSCO analysis which was carried out to assess the completeness of the assembly. The categories complete, duplicated, fragmented and missing are represented by 310, 408, 78 identified and 160 not identified orthologs, respectively. C) In total, 28,427 contigs of the assembly could be annotated at E-10 while 107,145 could not. D) Venn-diagram depicting the number of contigs mapping to sequences of selected Streptophyta (blue), Chlorophyta (turquoise) or both (dark blue) at E-10.
rRNA analysis
The Z. circumcarinatum culture used in this study was only recently established (Herburger et al. 2015). It is unialgal but neither axenic nor characterized with respect to possible contamination by heterotrophic eukaryotes. The rRNA reads, that were filtered out during quality control, were used to investigate the presence of putative contaminations in the algal culture (Supplemental Figure S4). The largest fraction of all rRNA reads could be annotated as Zygnema sp. However, a large number of different bacterial rRNA species as well as a few eukaryotic rRNAs, with most of them mapping to the genus Naegleria sp., was detected. We also observed a large number of putative rRNAs which could not be annotated at all (up to 20% for P3C). Overall, rRNA reads related to Zygnema sp. or with no significant similarity to any organism in BLAST analyses constituted about 90% of the rRNA except for the P1C and P1D. For these samples a large contamination with bacterial (up to 40%) and eukaryotic (Naegleria sp., up to 17%, among others) sequences was observed. Hence, P1C and P1D were excluded from analyses if not indicated otherwise.
Expression analysis
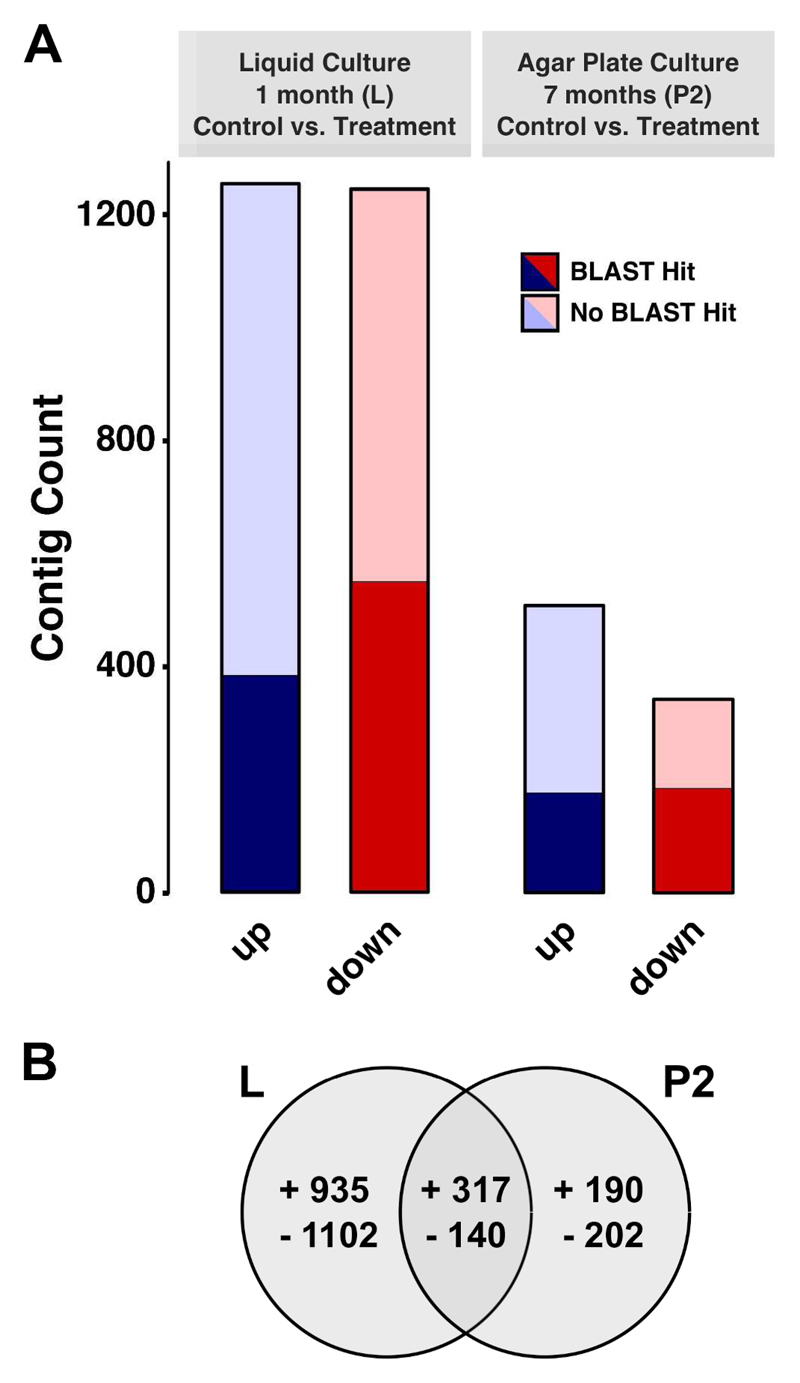
Differential expression analysis was carried out for control versus desiccated samples of the liquid culture (LC to LD) and the seven months old solid culture (P2C to P2D). The analysis was performed without P1C and P1D as well as P3C and P3D due to high levels of contamination and low RNA extraction yields, respectively. A total of 2,886 (2.1%) transcripts exhibited differential expression (FDR less or equal to 0.001) upon desiccation treatment compared to the corresponding control in at least one of the two group comparisons (L, P2). For the liquid culture (L), we observed the strongest reaction with 2,494 contigs regulated while only 849 were differentially expressed in group P2 (Figure 4A, supplementary Table S5). Between 30% and 54% of the upregulated and downregulated contigs in those groups were successfully annotated. The largest part of the annotated sequences showed similarities to proteins of Viridiplantae. Regarding the overlap of genes responsive to desiccation in both groups, a total of 457 contigs were regulated; 317 were induced while 150 were repressed (Figure 4B). A total of 2,037 and 392 transcripts exhibited differential expression solely in group L and P2, respectively. To assess the correlation of the replicates of each sample, a principal component analysis was performed and sample correlations were visualized as a heatmap (Supplementary Figure S6). We found that the individual replicates cluster closer according to the sample affiliation than randomly when looking at the first two principal components (Figure S6A). The same is true for the Pearson correlation matrix, where clustering of the associated replicates as well as the controls and treated samples was observed (Figure S6B).
Figure 4.
A) The total of up- (dark, light blue) and downregulated (dark, light red) contigs of Z. circumcarinatum under desiccation in group L and P2. Only contigs with a FDR of less than or equal to 0.001 were considered. All contigs were annotated against the Swiss-Prot database using BLASTx with an e-value of less than or equal to E-10. B) Overview of total up- and downregulated contigs of Z. circumcarinatum upon desiccation in group L and P2. Only contigs with a FDR of less than or equal to 0.001 were considered. The Venn diagram displays the number of regulated genes shared between both groups.
Gene set enrichment analyses
In order to identify desiccation related metabolic pathways, a KEGG pathway enrichment analysis based on KO annotations was performed for the up- and downregulated transcripts in both analysed groups. A total of 16,910 contigs could be annotated with KO terms. Significantly enriched pathways were found for the liquid culture dehydration treatment (L). Among upregulated transcripts, we found the “starch and sucrose metabolism” to be enriched while “photosynthesis” and “glyoxylate and dicarboxylate metabolism” were enriched in the downregulated contigs (Table 2).
Table 2.
Enriched KEGG pathways.
| Group | Regulation | KEGG ID | Pathway |
|---|---|---|---|
| L | up | ko00500 | Starch and sucrose metabolism |
| down | ko00195 | Photosynthesis | |
| ko00630 | Glyoxylate and dicarboxylate metabolism |
The concept of GO categorization enables the comparison of homologous genes in different organisms (Ashburner et al. 2000). One or multiple GO terms, belonging to one of the three root categories, are assigned to each protein, similar parent categories are grouped and, subsequently, tested for enrichment in one sample compared to another (Ashburner et al. 2000, Young et al. 2010).
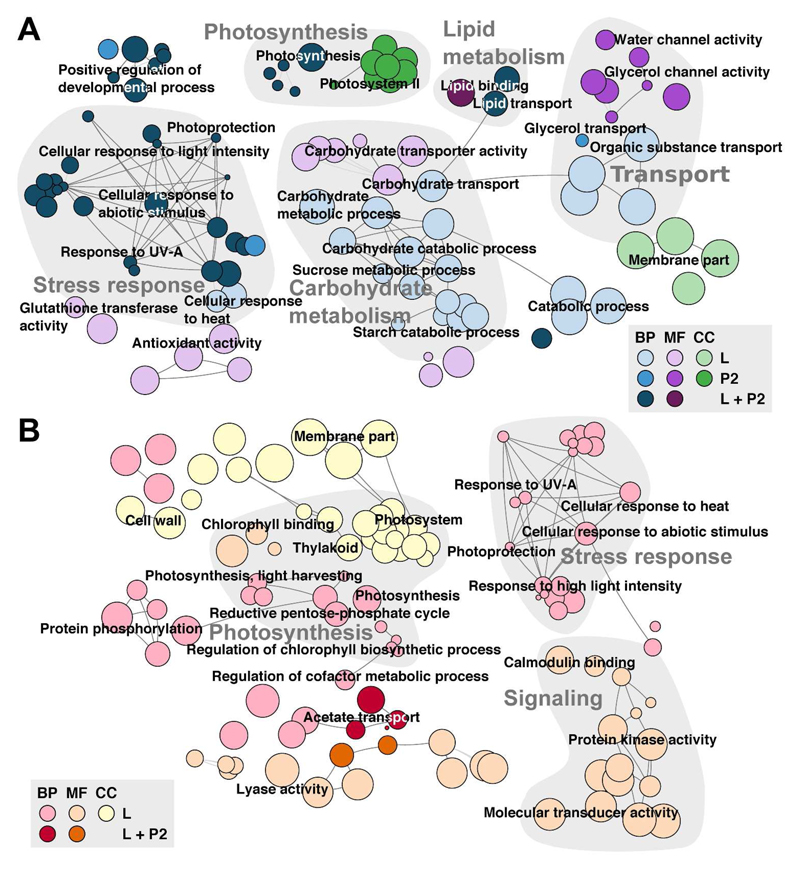
For example, the large subunit of RuBisCo was assigned the “biological process” photosynthesis, the “molecular function” ribulose-bisphosphate carboxylase activity and the “cellular component” chloroplast. In our study, GO terms could be assigned to 26,879 sequences forming the basis for a GO enrichment analysis. Enriched root categories for each group and directed regulation are summarized in Table 3 featuring more enriched GO terms for group L, with 80 and 107 up- and downregulated, respectively, than for group P2 with 55 and 6 terms up- and downregulated, respectively. Most enriched GO terms belonged to the root category “biological process” while “molecular function” and “cellular component” were represented to a lesser extent. In Figure 5, a network of enriched non-redundant GO terms in up- and downregulated gene sets is displayed with major categories highlighted in light grey. Both “photosynthesis” and “stress response” were enriched in up- and downregulated contigs while “carbohydrate metabolism”, “lipid metabolism” and “transport” appeared upregulated and “signaling” downregulated. Lists of all enriched GO terms are included in the supplemental Table S7.
Table 3.
Outcome of GO enrichment analysis displaying solely root category distribution (CC = “Cellular component”, MF = “Molecular function”, BP = “Biological process”). Detailed information is included in supplemental Table S9.
| Group | Regulation | CC | MF | BP |
|---|---|---|---|---|
| L | up | 4 | 16 | 60 |
| down | 26 | 31 | 50 | |
| P2 | up | 8 | 8 | 39 |
| down | 0 | 2 | 4 |
Figure 5.
GO Network displaying all enriched categories in both groups L and P2 as well as A) up- and B) downregulation. The root categories are “Biological Process” (BP; blue or red), “Molecular Function” (MF; violet or orange) and “Cellular Component” (CC; green or yellow). Edges depict shared terms. Highlighted in grey are selected groups such as photosynthesis, lipid metabolism, transport, carbohydrate metabolism, stress response and signaling.
Individual analysis of desiccation responsive genes
Based on gene set enrichment analyses, we studied individual differentially expressed transcripts responsive to the applied desiccation treatment (Supplemental Table S5). Our main focus lies on photosynthesis, carbohydrate and lipid metabolism, transporter proteins and signaling as well as stress protection. Selected genes, exhibiting differential expression, and the detected fold changes (all given in log2 hereinafter) are displayed in Table 4.
Table 4.
Selection of contigs showing differential expression in response to desiccation stress (The complete list can be found in Table S5). Contigs are divided into the following groups: Photosynthesis and photorespiration, carbohydrate metabolism, lipid metabolism, transporter proteins, signaling, stress protection. Selected contigs are displayed with ID, annotation, e-value and fold change (log2 transformed) for group L and P2.
| Contig ID | Annotation | E-value | L | P2 |
|---|---|---|---|---|
| Photosynthesis and photorespiration | ||||
| TR14384|c0_g2_i1 | Photosystem I subunit II | 2.18E-85 | -3.2 | - |
| TR18990|c0_g9_i1 | Photosystem I subunit IV | 5.94E-22 | -2.7 | -4.7 |
| TR1369|c1_g1_i1 | Photosystem I subunit III | 8.25E-76 | -2.5 | - |
| TR59163|c0_g5_i2 | Photosystem I subunit V | 1.36E-28 | -3.0 | - |
| TR21504|c0_g2_i1 | Photosystem I subunit VI | 8.11E-40 | -2.3 | - |
| TR16905|c0_g2_i1 | Photosystem I subunit X | 1.42E-34 | -2.3 | - |
| TR33976|c0_g1_i1 | Photosystem II oxygen-evolving enhancer protein 1 | 3.57E-135 | -3.0 | - |
| TR48275|c1_g1_i1 | Photosystem II oxygen-evolving enhancer protein 3 | 3.9E-49 | -3.4 | - |
| TR20185|c2_g1_i1 | Photosystem II 22kDa protein (PsbS) | 1.08E-83 | -4.6 | -4.1 |
| TR24382|c0_g1_i1 | Photosystem II protein (PsbY) | 4.85E-13 | -3.4 | - |
| TR71565|c0_g1_i1 | Photosystem II protein (Psb27) | 2.46E-36 | -2.5 | - |
| TR64030|c0_g1_i1 | Light-harvesting chlorophyll-protein complex I subunit A4 | 4.67E-116 | -2.9 | - |
| TR37377|c0_g1_i2 | Photosystem I light harvesting complex protein 5 | 2E-93 | -3.0 | - |
| TR62754|c5_g48_i1 | Photosystem II light harvesting complex protein 2.2 | 5.38E-82 | -6.2 | - |
| TR12320|c6_g1_i1 | Light-harvesting chlorophyll B-binding protein 3 | 1.83E-96 | -3.5 | - |
| TR37376|c0_g1_i2 | Light harvesting complex photosystem II | 5.2E-121 | -3.8 | -3.9 |
| TR25593|c4_g1_i1 | Light harvesting complex of photosystem II 5 | 1.57E-117 | -2.7 | - |
| TR1329|c0_g2_i1 | Light harvesting complex photosystem II subunit 6 | 3.79E-92 | -2.8 | - |
| TR75181|c0_g1_i1 | ATPase delta chain | 6.28E-52 | -2.3 | - |
| TR3752|c0_g5_i1 | ATPase subunit b’ | 2E-37 | -2.7 | -4.3 |
| TR4441|c0_g1_i2 | Plastocyanin | -3.75E-37 | -2.5 | - |
| TR31328|c0_g1_i1 | Chlorophyllide a oxygenase | 0 | -2.1 | - |
| TR68443|c0_g1_i3 | Magnesium chelatase subunit | 0 | -4.5 | -4.4 |
| TR8034|c11_g29_i1 | Early light-induced protein, chloroplastic (ELI) | 1.83E-15 | - | 12.2 |
| TR58021|c0_g5_i1 | Early light-induced protein 1, chloroplastic (ELIP1) | 1.43E-21 | 3.5 | 5.2 |
| TR4192|c1_g15_i1 | High molecular mass early light-inducible protein, chloroplastic (HV58) | 1.86E-21 | 5.2 | 4.3 |
| TR4440|c0_g2_i1 | Low molecular mass early light-inducible protein, chloroplastic (HV60) | 4.1E-16 | -3.4 | - |
| TR73556|c0_g9_i2 | (S)-2-Hydroxy-acid oxidase | 0 | -2.5 | -3.5 |
| TR29652|c0_g1_i1 | Serine-glyoxylate transaminase | 0 | -3.4 | -3.6 |
| TR68913|c1_g1_i2 | Glycine dehydrogenase | 0 | -2.6 | -4.2 |
| TR54933|c0_g2_i1 | Glutamate-glyoxylate aminotransferase | 0 | -3.0 | - |
| TR48225|c1_g1_i1 | Glycerate dehydrogenase | 0 | -3.5 | - |
| Carbohydrate metabolism | ||||
| TR23256|c0_g1_i1 | Glycogen phosphorylase | 0 | 8.3 | - |
| TR75230|c1_g1_i1 | alpha-Amylase | 1.95E-175 | 4.3 | - |
| TR70181|c1_g1_i2 | beta-Amylase | 0 | 2.7 | - |
| TR24697|c0_g3_i4 | Isoamylase | 6.66E-40 | 2.6 | - |
| TR25586|c0_g1_i1 | 4-alpha-Glucanotransferase | 1.47E-45 | 2.2 | - |
| TR45454|c0_g2_i1 | Sucrose-phosphatase | 6.91E-108 | 3.6 | - |
| TR61067|c0_g1_i2 | Sucrose synthase | 0 | 3.0 | - |
| Lipid metabolism | ||||
| TR39622|c0_g1_i1 | Lysophospholipid acyltransferase | 4.12E-110 | 2.0 | - |
| TR53082|c0_g1_i1 | Diacylglycerol kinase | 2.68E-160 | 2.0 | - |
| TR16611|c0_g1_i2 | alpha-Galactosidase | 1.91E-154 | 2.3 | 3.5 |
| TR42973|c1_g1_i1 | Sulfoquinovosyltransferase | 4.73E-12 | 3.7 | - |
| TR31318|c0_g1_i1 | Phospholipase D1/2 | 0 | 3.7 | - |
| TR28615|c2_g10_i1 | Phosphoethanolamine N-methyltransferase | 5.8E-102 | 2.9 | 3.2 |
| TR41908|c1_g1_i4 | Phosphatidylserine synthase 2 | 1.62E-177 | - | 3.8 |
| TR13652|c0_g1_i13 | 2-Acylglycerol O-acyltransferase 1 | 2.54E-89 | - | 3.5 |
| Transporter proteins | ||||
| TR43432|c0_g2_i2 | Probable aquaporin TIP1-2 | 8.15E-17 | 13.3 | 13.5 |
| TR43432|c0_g3_i1 | Aquaporin TIP2-1 | 2.25E-32 | 2.9 | 2.7 |
| TR61568|c0_g2_i1 | Aquaporin TIP2-3 | 2.22E-33 | 4.1 | - |
| TR34049|c0_g1_i2 | Plastidic glucose transporter 2 | 2.02E-138 | 4.2 | 3.6 |
| TR23238|c0_g1_i2 | Sucrose transport protein 3 | 2.09E-147 | 2.7 | - |
| TR31|c0_g1_i1 | Glucose-6-phosphate/phosphate translocator 1 | 1.22E-164 | 2.1 | - |
| TR40733|c1_g1_i1 | sugar transport protein 13 | 8.24E-177 | 5.8 | 5.5 |
| TR41946|c0_g1_i8 | sugar-transport protein ERD6-like 16 | 3.33E-71 | 9.0 | - |
| Signaling | ||||
| TR52105|c7_g5_i3 | Leucine-rich repeat receptor-like serine/threonine-protein kinase BAM2 | 2.56E-18 | 3.4 | - |
| TR58701|c1_g2_i3 | Leucine-rich repeat receptor-like serine/threonine-protein kinase FLS2 | 7.92E-65 | -5.4 | -6.7 |
| TR34848|c0_g2_i1 | Calcium-dependent protein kinase 17 | 1.25E-81 | 3.4 | - |
| TR6882|c0_g1_i1 | Calcium-dependent protein kinase 20 | 5.18E-76 | -6.1 | - |
| Stress protection | ||||
| TR10757|c0_g3_i1 | Chaperone protein ClpB1 | 7.43E-16 | 3.4 | - |
| TR35960|c0_g2_i2 | Proteasome assembly chaperone 2 | 4.89E-28 | 11.2 | - |
| TR41947|c0_g1_i3 | Chaperone protein DnaJ | 1.06E-10 | 3.9 | - |
| TR75210|c0_g1_i2 | Molecular chaperone Hsp31 | 1.57E-10 | 2.5 | 4.2 |
| TR39621|c0_g1_i2 | Glutathione S-transferase | 4.85E-53 | 2.9 | 5.0 |
| TR14048|c0_g1_i1 | Peroxisomal catalase | 0 | 3.7 | 6.7 |
| TR58823|c0_g2_i1 | Peroxiredoxin | 6.92E-54 | 2.3 | 3.4 |
| TR57779|c0_g1_i1 | Peptide methionine sulfoxide reductase | 4.35E-79 | 2.5 | 3.0 |
| TR35953|c0_g2_i4 | (Chloroquine-resistance transporter)-like transporter 3 | 7.29E-81 | 9.0 | - |
| TR50557|c0_g2_i12 | Nijmegen breakage syndrome 1 protein | 3.14E-13 | 9.5 | 9.7 |
| TR35997|c1_g1_i8 | DNA-damage-repair/toleration protein | 1.42E-42 | 3.0 | - |
| TR49464|c0_g1_i1 | Late embryogenesis abundant protein 4 (LEA4; AT3G53040) | 8.1E-19 | 5.1 | 3.6 |
| TR39628|c0_g2_i1 | Late embryogenesis abundant protein 4 (LEA4; AT2G18340) | 4.6E-24 | 5.0 | - |
| TR69744|c2_g23_i1 | Late embryogenesis abundant protein 4 (LEA4; AT4G36600) | 1.1E-14 | 5.3 | 3.4 |
| TR60896|c0_g1_i1 | Late embryogenesis abundant protein 5 (LEA5; AT2G40170) | 6.5E-27 | - | 9.8 |
Photosynthesis and photorespiration
Transcriptomic analysis of photosynthetic processes revealed a strong downregulation of components of both PSs in group L. A repression of transcripts encoding parts of PS I and PS II with fold changes of -2.3 to -3.4 was observed. The strongest downregulated transcript was the PS II 22kDa protein (PsbS) with a fold change of -4.6. In group P2, only PS I subunit IV and PS II 22kDa protein exhibited fold changes of -4.7 and -4.1, respectively. Furthermore, several contigs coding for light-harvesting complexes as well as some proton transporting ATPase subunits and plastocyanin, which is part of the electron transport chain, were downregulated in group L. Detected fold changes lay in the range of -2.3 to -3.8 while the PS II light harvesting complex protein 2.2 displayed a rather strong repression of -6.2. In addition, the putative chlorophyllide a oxygenase and the magnesium chelatase subunit, which are both part of the chlorophyll metabolism, were repressed 2.1- and 4.5-fold, respectively. In contrast, group P2 exhibited a weaker downregulation. Upon desiccation, the light harvesting complex PS II, ATPase subunit b’ and magnesium chelatase subunit were repressed 3.9-, 4.3- and 4.4-fold, respectively. Rather striking is the plethora of ELIPs that showed differential expression during dehydration. Desiccation of the liquid culture caused the transcription level of ELIP1 and the chloroplastic high molecular mass ELIP to increase while the chloroplastic low molecular mass ELIP was repressed. The solid culture of Z. circumcarinatum (P2) showed an upregulation of the chloroplastic ELIP (ELI), ELIP1 and the chloroplastic high molecular mass ELIP. The chloroplastic high molecular mass ELIP exhibits the strongest induction with a fold change of 12.2.
Desiccation also caused a repression of enzymes involved in photorespiration in both comparisons. The transcript pools of the (S)-2-hydroxy-acid oxidase, serine-glyoxylate transaminase and glycine dehydrogenase showed a decline for L and P2 while group L also exhibits a downregulation of glutamate-glyoxylate aminotransferase and glycerate dehydrogenase. The detected fold changes ranged from -2.5 to -4.2.
Carbohydrate metabolism
Starch degradation and sucrose formation were induced during dehydration in group L while group P2 shows only minor effects. A variety of starch consuming enzymes were upregulated such as glycogen phosphorylase, alpha- and beta-amylase, isoamylase, 4-alpha-glucanotransferase as well as the sucrose synthesizing enzymes sucrose-phosphatase and sucrose-synthase. The strongest induction was observed for glycogen phosphorylase with an 8.3-fold upregulation while the others exhibit fold changes between 2.2 and 4.3.
Lipid metabolism
Investigating both the glycerolipid and glycerophospholipid metabolism, an enhanced gene expression of certain enzymes was found. Group L showed an increased transcript level for the lysophospholipid acyltransferase, diacylglycerol kinase, alpha-galactosidase, sulfoquinovosyltransferase, phosphoethanolamine N-methyltransferase and phospholipase D1/2. The response of group P2 was less pronounced with an upregulation of alpha-galactosidase, phosphoethanolamine N-methyltransferase, phosphatidylserine synthase 2 and 2-acylglycerol O-acyltransferase 1. Transcript levels were increased between 2- and 3.8-fold.
Transporter and signaling
The upregulation of the contig TR43432|c0_g2_i2, which was annotated to be an aquaporin (AQP) of type TIP (tonoplast intrinsic protein), was most pronounced considering all differentially expressed transcripts. Additionally, TIP2-1 in both groups L and P2 was enhanced and TIP2-3 only in group L. Moreover, the expression of various putative sugar transporters (plastidic glucose transporter 2, sucrose transport protein 3, glucose-6-phosphate/phosphate translocator 1, sugar transport protein 13, sugar-transport protein ERD6-like 16) was strongly induced in the liquid sample and to a lesser extent also in group P2. The sugar-transport protein ERD6-like 16 exhibited a fold-change of 9 while the induction of other sugar transport proteins lay in the range of 2.1- to 5.8- fold.
A complex regulation of signaling pathways in group L was identified. Mainly transcripts, which were showing similarities to the family of serine/threonine-protein kinases but also other kinases and transcription factors, were differentially expressed. Leucine-rich repeat receptor and receptor-like serine/threonine-protein kinases and calcium-dependent protein kinases (CPK), such as the leucine-rich repeat receptor-like serine/ threonine-protein kinases BAM2 and FLS2 as well as the calcium-dependent protein kinases 17 and 20, were the most prominent transcripts. Overall, more signaling related contigs were repressed than upregulated.
Stress response
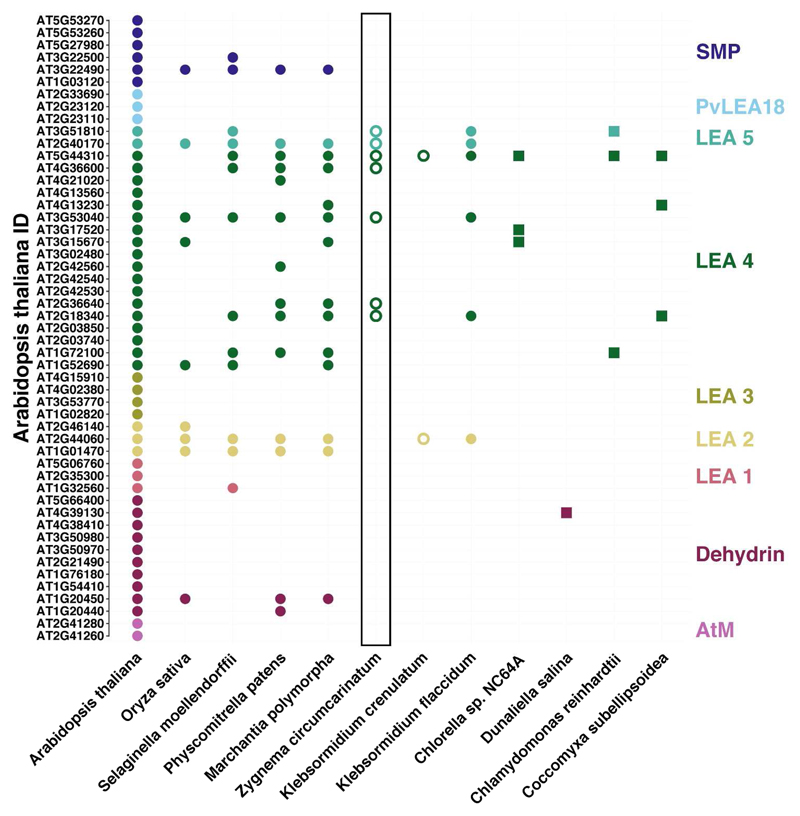
An upregulation of other stress protection associated genes was discovered. Chaperone and Hsp encoding genes, such as the chaperone protein ClpB1, proteasome assembly chaperone 2, the chaperone protein DnaJ and the molecular chaperone Hsp31, appeared to be highly transcribed during desiccation, primarily in group L, while genes involved in ROS scavenging, such as the glutathione-S-transferase, peroxisomal catalase, peroxiredoxin, peptide methionine sulfoxide reductase, and (chloroquine-resistance transporter)-like transporter 3, were induced in both groups. For the desiccated liquid culture, the proteasome assembly chaperone 2 exhibits the largest change of 11.2-fold. Transcripts similar to the Nijmegen breakage syndrome 1 protein and DNA-damage-repair/toleration protein, both involved DNA repair, also showed an enhanced expression. Astonishingly, the Nijmegen breakage syndrome 1 protein was upregulated 9.5- and 9.7-fold in group L and P2, respectively. Furthermore, a large number of LEA proteins showed high induction, consisting only of LEA4 proteins in group L while group P2 showed the upregulation of both LEA4 and LEA5 proteins. The LEA5 gene exhibited an 9.8-fold increase in expression. The BLAST analysis of several Chlorophyta and Streptophyta (Figure 6) reveals LEA group distribution among Viridiplantae species. We observed that all studied streptophyte genomes contained at least LEA proteins from group LEA2, LEA4 and LEA5 while chlorophytes mostly only possessed LEA2 proteins with some exceptions (in our example, D. salina).
Figure 6.
LEA proteins found in A. thaliana and described by Hundermark & Hincha (2008). Annotations were retrieved from Phytozome v11.0 for A. thaliana, C. reinhardtii, C. subellipsoidea, D. salina, M. polymorpha, P. patens and S. moellendorffii. The same is true for the transcripts of O. sativa which were annotated using diamond BLASTx with E-9. The transcripts of Chlorella sp. NC64A, K. crenulatum and K. flaccidum were downloaded from the JGI Genome Portal, Holzinger et al. (2014) and the Klebsormidium flaccidum genome project, respectively, and processed accordingly. Points indicate streptophytes while squares stand for chlorophytes. Solid and hollow symbols represent sequences derived from genomes or only transcriptomes, respectively.
Overall, photosynthesis appears to be repressed in group L while several ELIPs are upregulated in both groups. Desiccation stress also causes an upregulation of transcripts involved in the lipid metabolism and transport in L and P2. Furthermore, group L exhibits an induction of the carbohydrate metabolism. Both groups increase the transcript pool of ROS scavenging and chaperone proteins.
Discussion
Photosynthesis and photorespiration
Dehydration has an extremely negative influence on photosynthesis as water is crucial for structural integrity and functionality of the algal cell and also acts as an electron donor in the electron transport chain (Fernández-Marín et al. 2016). Herburger et al. (2015) observed a complete loss of photosynthetic activity in Z. circumcarinatum during prolonged desiccation. However, older cultures appeared to be more tolerant towards dehydration as photosynthesis was maintained longer compared to younger cells (Herburger et al. 2015). These findings are in agreement with our observations for Z. circumcarinatum cultivated on agar (Figure 2B-D). Culture P1 abandoned photosynthetic activity first, then the effective quantum yield of P2 dropped while P3 resisted the longest. According to Herburger et al. (2015), this increased tolerance is likely caused by the formation of pre-akinetes which can be regarded as a stress tolerant resting stage (Pichrtová et al. 2016b). Typical features of pre-akinetes are hardened cell walls, accumulation of starch and lipid bodies in the cytoplasm as well as reduced growth and physiological activity (Herburger et al. 2015, Pichrtová et al. 2016b). Compared to the agar cultures, filaments grown in liquid medium appeared to maintain photosynthesis as long as P3. However, this effect is clearly linked to the clinging water which could not completely removed by blotting. During the desiccation treatment, the excess water had to evaporate before the algal biomass could be desiccated effectively.
The dehydration of Z. circumcarinatum, cultivated in liquid medium for one month, led to a strong repression of transcripts encoding components of PS I and II as well as light-harvesting proteins indicating photoinhibition. In contrast, K. crenulatum (Holzinger et al. 2014) showed an upregulation of genes related to photosynthesis in response to desiccation. Holzinger et al. (2014) suggest that this mechanism serves as a preparation to rapidly resume photosynthesis upon rehydration. Similar results were obtained for Trebouxia gelatinosa (Carniel et al. 2016). However, Carniel et al. (2016) compared a couple of different transcriptomic studies on desiccation of Viriplantae detecting a repression of photosynthetic transcripts in Syntrichya ruralis (desiccation tolerant moss), C. plantagineum (resurrection plant), Haberlea rhodopensis (resurrection plant) and Xerophyta humilis (resurrection plant). Moreover, the resurrection plant Myrothamnus flabellifolia also exhibited a downregulation of photosynthesis genes when desiccated (Ma et al. 2015). Surprisingly, mainly genes, encoding parts of PS I and II, the electron transport chain and the ATP synthase, were repressed in Myrothamnus flabellifolia (Ma et al. 2015). This is also true for Z. circumcarinatum, as demonstrated in the present study. Ma et al. (2015) argue that thereby excitation energy and, thus, ROS production likely are reduced. Moreover, the level of transcripts, involved in chlorophyll biosynthesis, decreased. Similar findings were obtained for desiccated Vitis vinifera (grapevine) leaves (Salman et al. 2016) and salt stressed Oryza sativa (rice) seedlings (Turan & Tripathy 2015). The observed impairment of the chlorophyll biosynthesis is probably occurring to avoid ROS formation (Farrant et al. 2003). As an additional protection, the expression of ELIPs was enhanced in group L and P2. ELIPs are photoprotectants and belong to the chlorophyll a/b-binding (CAB) superfamily, respond to abiotic stress, mainly to high light and UVR, and are located in the thylakoid membrane (Hayami et al. 2015, Hutin et al. 2003, Norén et al. 2003). Similar reactions were observed for other green algae, for example, Chlamydomonas reinhardtii and Dunaliella bardawil, both of which increased the ELIP transcript pool in response to high light stress which is also common for various higher plants such as Arabidopsis (Lers et al. 1991, Teramoto et al. 2004). Less studies have been dedicated to the relationship of cold stress and ELIP expression in algae. Król et al. (1997) reported an induction of ELIPs in Dunaliella salina when exposed to low temperatures. Spirogyra varians (Zygnematales) also exhibits an accumulation of ELIP-like transcripts when cultivated at 4°C (Han & Kim 2013). Nevertheless, the accumulation of ELIPs is also commonly associated with desiccation stress (Dinakar & Bartels 2013, Ma et al. 2015, Zeng et al. 2002). These proteins protect the thylakoid membranes against photooxidative damage by scavenging free chlorophyll molecules and act as sinks for excitation energy (Heddad et al. 2012, Zeng et al. 2002). Paradoxically, some contigs of Zygnema, exhibiting similarities to ELIPs, are negatively regulated. However, Holzinger et al. (2014) also found a complex regulation of ELIP-related transcripts for K. crenulatum hinting at a multigenetic family with several ELIPs being responsive to desiccation stress (Hutin et al. 2003, Marraccini et al. 2012, Zeng et al. 2002).
A decrease in photorespiratory transcripts upon desiccation was detected of both groups of Z. circumcarinatum suggesting a repression of photorespiration. In contrast, photorespiration can function as a protection of the photosynthetic apparatus against photoinhibition (Wingler et al. 1999). Especially, during drought stress when carbon dioxide fixation and, thus, the consumption of electrons is reduced (Wingler et al. 1999). However, protection of the PSs by photorespiration is not essential during desiccation (Wingler et al. 2000). Furthermore, dehydration is reported to reduce photorespiratory activity in plants which may be caused by the decreased photosynthetic activity (Levitt 1980).
Carbohydrate metabolism
A common protective mechanism against water stress is the accumulation of low-molecular-weight osmolytes which can be sugars, polyols and proteins (Dinankar & Bartels 2013, Fernández-Marín et al. 2016, Hinsha et al. 1996, Holzinger & Pichrtová 2016, Ma et al. 2015). By increasing the amount of osmoprotectants in the cell, a negative osmotic potential is achieved, membranes are stabilized and protein protection is enhanced (Bisson & Kirst 1995). Nagao et al. (2008) found that K. flaccidum accumulates the osmolyte sucrose during cold acclimation which contributes to a higher freezing tolerance. Moreover, the alga Chlorella vulgaris exhibits an increase in sucrose and raffinose content in response to cold shock treatment (Salerno & Pontis 1989). However, sucrose is also typically formed upon desiccation stress in plants and algae (Cruz de Carvalho et al. 2014, Dinankar & Bartels 2013, Holzinger & Pichrtová 2016, Ramanjulu & Bartels 2002, Sadowsky et al. 2016). Sadowsky et al. (2016) reported increased sucrose levels in an Antarctic Trebouxia strain to counteract desiccation. Our data indicate a metabolic shift towards sucrose as starch degrading as well as sucrose biosynthetic enzymes were upregulated in dehydrated filaments. The KEGG enrichment analysis also clearly indicated an enhancement of the "starch and sucrose metabolism". A similar strategy is pursued by K. crenulatum inducing transcripts encoding sucrose synthase and the sucrose phosphate synthase (Holzinger et al. 2014). The authors suggest that raffinose family oligosaccharides also function as osmoprotectants because several enzymes, belonging to the galactinol/raffinose metabolism, exhibited a higher expression in desiccated cells (Holzinger et al. 2014). Z. circumcarinatum did not show this expression pattern in response to water stress. However, the sucrose phosphate synthase of Z. cirumcarinatum contains a conserved phosphorylation site (results not shown) which is typically found in angiosperms and known to become modified upon osmotic stress (Winter & Huber 2000). Hence, the sucrose metabolism of Z. circumcarinatum is most likely not only regulated by transcription but also posttranslational modifications.
Callose is an important polysaccharide found to be involved in response to different abiotic stress factors, e.g. in drought stress in plants, such as Gossypium hirsutum L. (cotton; McNairn 1972), but also algae such as K. crenulatum (Herburger & Holzinger 2015). Complementary, the desiccation transcriptome of Klebsormidium revealed an upregulation of the callose synthase complex confirming the significance of this carbohydrate during dehydration events (Holzinger et al. 2014). Albeit the occurrence of this enzyme in the Zygnema transcriptome no differential expression was detected. These results are in agreement with Herburger & Holzinger (2015) who reported a stable callose content throughout desiccation for 2.5 h. As the callose synthase is located in the plasma membrane and the protoplast is retracting from the cell wall upon dehydration callose incorporation is prevented (Herburger & Holzinger 2015).
Lipid metabolism and membranes
The fact, that low temperatures, dehydration etc. initially target biomembranes, highlights the importance of membrane modification upon water stress to preserve integrity and fluidity (Dinankar & Bartels 2013, Holzinger et al. 2014, Perlikowski et al. 2016, Valledor et al. 2013). For example, decreased temperatures cause membrane modifications in the green alga C. reinhardtii (Valledor et al. 2013, Wang et al. 2016). Gasulla et al. (2013) observed similar tendencies in C. plantagineum induced by desiccation treatment. Monogalactosyldiacylglycerol was removed from the thylakoid membranes and either transformed to digalactosyldiacylglycerol or hydrolyzed to form diacylglycerol (Gasulla et al. 2013). In contrast, our results indicate the conversion from digalactosyldiacylglycerol to monogalactosyldiacylglycerol to be amplified as the putative alpha-galactosidase is upregulated in group L. However, parts of the glycero- and glycerophospholipid metabolism are enhanced during desiccation suggesting other membrane modifications. Similarly, the lichen phycobiont Asterochloris erici exhibited elevated levels of phosphatidic acid upon desiccation indicating that phospholipase D is involved in stress protection mechanisms (Gasulla et al. 2016). Z. circumcarinatum induced phospholipase D1/2 during dehydration stress confirming that phospholipase D1/2 is part of the stress response.
Transporter proteins and signaling
Major intrinsic proteins (MIPs), or AQPs, establish channels for passive transportation of small uncharged substances, such as water or glycerol, across the membrane (Anderberg et al. 2011 & 2012, Barkla et al. 1999). AQPs in embryophytes comprise seven groups: GlpF-like intrinsic proteins (GIPs), hybrid intrinsic proteins (HIPs), X intrinsic proteins (XIPs), small basic intrinsic proteins (SIPs), nodulin-26 like intrinsic proteins (NIPs), plasma membrane intrinsic proteins (PIPs) and TIPs (Danielson et al. 2008). Anderberg et al. (2011) studied different chlorophytes and found AQPs of these groups only in Trebouxiophyceae (PIP, GIP). Z. circumcarinatum, a charophyte alga, is more closely related to land plants and expresses TIPs, NIPs and SIPs while only TIPs appeared to play an important role in desiccation. Both groups, L and P2, showed a strong induction of TIPs during water stress which was also observed for other AQPs in the plants Arabidopsis thaliana and C. plantagineum (Dinankar & Bartels 2012, Ramanjulu & Bartels 2002) as well as the alga T. gelatinosa (Carniel et al. 2016). Carniel et al. (2016) argue that AQPs are protection against damage during rehydration by increasing the permeability of biomembranes to water.
As mentioned above, the formation of sugars and other osmolytes is increased during water stress, however, these molecules also need to be distributed within the cell (Jarzyniak & Jarsiński 2014).Thus, desiccation tolerance is dependent on sugar transportation within the cell. Liu et al. (2016) reported an increased drought tolerance in A. thaliana associated with the expression of the hexose facilitator AtSWEET4. Similar results, indicating desiccation induced expression of sugar transporters, were obtained analyzing Caragana korshinskii (leguminous shrub; Li et al. 2016) and Saccharum spp. (sugar cane; Zhang et al. 2016).
The signaling in plants during desiccation generally involves several hormones such as abscisic acid, cytokinin and ethylene (Campo et al. 2014, Holzinger & Becker 2015, Van de Poel et al. 2016, Zhou et al. 2014) as well as calcium-dependent and serine/threonine-protein kinases (Campo et al. 2014, Ramanjulu & Bartels 2002). Especially abscisic acid plays a major role in the desiccation stress response linking host and plastid signaling which is considered a key step in the land plant evolution (de Vries et al. 2016). In contrast to K. crenulatum (Holzinger & Becker 2015), the up-regulation of hormone specific signaling transcripts was not evident in Zygnema. However, we found an intricate expression pattern of threonine/serine- and calcium-dependent protein kinases in response to dehydration in Z. circumcarinatum. Other studies obtained desiccation induced signaling networks with similar complexity, e.g. for K. crenulatum and V. vinifera (Holzinger et al. 2014, Salman et al. 2016).
Stress protection
The formation of ROS, occurring during different abiotic stress conditions, is extremely harmful making ROS scavenging crucial for cell survival (e.g. Dong et al. 2016, Cruz de Carvalho 2008, Cruz de Carvalho et al. 2012, Han et al. 2011, Heinrich et al. 2016, Kranner et al. 2005). For example, high light stress triggers the expression of ROS scavenging enzymes in the green alga C. reinhardtii and the marine diatom Thalasiosira pseudonana (Dong et al. 2016, Ericksson et al. 2015). Similarly, the dinoflagellate Symbiodinium involves a number of ROS defense proteins and antioxidants when exposed to heat stress (Gierz et al. 2017). The same holds true for cold stress which induces the accumulation of antioxidants in S. varians to counteract ROS formation (Han et al. 2011). Our data suggests ROS generation upon desiccation as Z. circumcarinatum expresses a multitude of enzymes related to ROS protection: the glutathione-S-transferase conjugates glutathione (GSH) to hydrophobic molecules (Rezaei et al. 2013), the peroxisomal catalase acts directly on ROS (Gorrini et al. 2013), peroxiredoxin catalyzes the reduction of peroxides (Dayer et al. 2008), the peptide methionine sulfoxide reductase reduces methionine sulfoxide back to methionine (Weissbach et al. 2002) and (chloroquine-resistance transporter)-like transporter 3 transports GSH (Noctor et al. 2011). Interestingly, the induction of the glutathione-S-transferase, peroxisomal catalase, peroxiredoxin and peptide methionine sulfoxide reductase was higher in group P2 than L suggesting a more pronounced ROS stress response of filaments grown on agar plates than filaments from a liquid cultivation. An upregulation of peroxiredoxin and the peptide methionine sulfoxide reductase was also detected in T. pseudonana exposed to high light stress (Dong et al. 2016). Hence, both enzymes are certainly involved in a general ROS coping mechanism. Moreover, group L strongly increases the transcript pool of the (chloroquine-resistance transporter)-like transporter 3. These transporter proteins play a major role in GSH homeostasis in A. thaliana as they connect the plastid and the cytosolic thiol pool (Maughan et al. 2010). Thus, (chloroquine-resistance transporter)-like transporters are essential to counteract ROS stress (Maughan et al. 2010). Furthermore, DNA damage, which is also linked to ROS formation (Cruz de Carvalho 2008, Heinrich et al. 2015), was addressed by a strong upregulation of repair enzymes in desiccation stressed Z. circumcarinatum. For example, the Nijmegen breakage syndrome 1 protein was highly induced with fold changes of 9.5 and 9.7 in group L and P2, respectively, suggesting a higher risk of DNA damages associated with desiccation (Akutsu et al. 2007, Cruz de Carvalho 2008).
Abiotic stress generally leads to aggregation and conformational changes in protein which is lethal to the cell (Wang et al. 2004). In response, plants and algae express chaperones and Hsps which are assisting the refolding of proteins and protect them from aggregation (Al-Whaibi 2011, Mitra et al. 2013, Schulz-Raffelt et al. 2007, Van de Poel 2016, Wang et al. 2004). For example, C. reinhardtii raises the transcription level of Hsp90A when heat shocked at 40 °C (Schulz-Raffelt et al. 2007). Kobayashi et al. (2014) found an increased expression of small Hsps in the red alga Cyanidioschyzon merolae and C. reinhardtii in response to heat stress. Heinrich et al. (2012a) and Dong et al. (2016) reported that the brown macroalga Saccharina latissima and the diatom T. pseudonana, respectively, induce various chaperones and Hsps when treated with high light intensities. Similarily, cold stress triggers the expression of two chaperones in the Antarctic diatom Chaetoceros neogracile (Park et al. 2008). The same holds true for desiccation which causes an increased expression of certain chaperones and Hsps in Selaginella lepidophylla (lycophyte, Carniel et al. 2016), Physcomitrella patens (moss, Wang et al. 2009), Pyropia orbicularis (red alga, López-Cristoffanini et al. 2015) and Asterochloris erici (green alga, Gasulla et al. 2013). Z. circumcarinatum showed an induction of chaperones and Hsps as well, which is probably a preparation for protein refolding upon rehydration (Carniel et al. 2016). Clp proteins are chaperones capable of refolding protein complexes and are induced in response to various stress factors (Lee et al. 2006). In O. sativa, a number of Clp proteins showed an increased transcription during drought stress (Hu et al. 2009) while the cytosolic chaperone ClpB1 of A. thaliana is involved in chloroplast development and acclimation to increased temperatures (Lee et al. 2006). However, Z. circumcarinatum probably raised the transcription level of the chaperone ClpB1 in response to desiccation stress to enable remodeling of aggregated and misfolded proteins. Another important group of stress induced proteins are the co-chaperones DnaJ (Wang et al. 2014). Wang et al. (2014) reported an increase in drought tolerance in transgenic tobacco resulting from an overexpression of the chloroplast-targeted chaperone DnaJ. Furthermore, an induction of DnaJ in Saccharum in response to desiccation was demonstrated by de Andrade et al. (2015). The applied desiccation treatment caused Z. circumcarinatum to induce the co-chaperone as part of its stress response. Interestingly, DnaJ expression is also triggered by high light exposure in S. latissimi and T. pseudonana (Dong et al. 2016, Heinrich et al. 2012a). Furthermore, our data shows an upregulation of the transcription factor Hsp31 upon water stress which is in agreement with the findings in O. sativa by Wang et al. (2011). Another important component of the protein quality control are proteasomes which selectively eliminate dysfunctional proteins (Hanssum et al. 2014). In response to changing conditions and environmental stress, the demand of proteasomes increases immensely forcing the cell to increase the proteasome pool (Hanssum et al. 2014). The assembly process is promoted by proteasome assembly chaperones such as proteasome assembly chaperone 2 (Le Tallec et al. 2007). The desiccated filaments of Z. circumcarinatum will most likely accumulate a number of misfolded proteins which need to be degraded. To be able to assemble proteasomes, the alga requires adequate chaperones. Additionally, a number of putative LEA proteins are upregulated. LEA proteins are proposed as water stress specific chaperones (Goyal et al. 2005, Hatanaka et al. 2014, Shinde et al. 2012) which can be grouped depending on their sequence motifs/patterns (Hundertmark & Hincha 2008). The drought induced expression of LEA proteins has been already reported for several species, e.g. M. flabellifolia (Ma et al. 2015), P. patens (Shinde et al. 2012) and K. crenulatum (Holzinger et al. 2014), confirming our findings. An interesting aspect is the distribution of these chaperones across Viridiplantae which reveals evolutionary relationships of the different subfamilies. Based on the classification by Hundertmark & Hincha (2008), the Z. circumcarinatum transcriptome covered sequences belonging to LEA4 and LEA5 (Figure 6). LEA4 is likely the group, which emerged first, as it shows the biggest diversity of all and is present in all investigated algae and plants except D. salina. LEA5 was probably also emerging early because it was found in all streptophyta and C. reinhardtii. All analyzed streptophyta genomes featured LEA2 proteins but this group is missing in Z. circumcarinatum. The absence of LEA2 is likely linked to the missing induction in response to any condition tested. Finally, the LEA protein group SMP probably evolved during the development of mosses from streptophytic green algae as all embryophyta possess these proteins.
Conclusion
In this study, the molecular response of the conjugating green alga Z. circumcarinatum to desiccation stress was investigated using differential gene expression analysis. To assess the effect of hardening, the impact of dehydration on a young liquid culture and a seven months old agar culture of Zygnema was analyzed. Our results found a 3-fold stronger transcriptional response of filaments grown in liquid medium compared to older filaments cultivated on agar. These findings are a clear indication of a pre-acclimation to low water availability of the algal culture grown on agar for seven months. In agreement with earlier observations, photosynthesis related genes are highly repressed in group L while the response of group P2 is much less pronounced. Furthermore, water withdrawal causes membrane modifications and the expression of several transporters such as aquaporins and carbohydrate transporter proteins. Desiccation also induces the accumulation of sucrose, a common osmolyte, to counteract the rapid loss of water. Finally, a number of stress related molecules are produced, e.g. ELIPs, chaperones such as LEA proteins, proteins involved in ROS scavenging and DNA repair proteins. Overall, we conclude that culture age and conditions highly influence the physiological state of the algal filaments and the acclimation to water stress. However, it is difficult to mimic natural conditions in a laboratory environment as natural habitats are influenced by stochastic parameters such as weather and soil quality. Thus, future experiments shall include the transcriptomic analysis of field samples collected in different seasons with different water availability.
Material and Methods
Algal strain and cultivation
Zygnema sp. (SAG 2419), previously isolated from a sandy shore near the river Saalach in Salzburg, Austria, was used for the experiments. The alga was either cultivated for one month in liquid Waris-H medium (McFadden & Melkonian 1986) or on 1.5% agar plates, containing BBM medium, as previously described by Herburger et al. (2015). The algal strain clusters in phylogenetic analysis close to Zygnema circumcarinatum (Herburger and Holzinger 2015), thus, we use this species name in the present study. However, an unambiguous morphological determination of the species was not possible as zygospores were not detected.
Desiccation experiment
Four different cultures of Z. circumcarinatum were prepared: a one month old liquid culture (L) and one month (P1), seven months (P2) and twelve months old cultures (P3) grown on agar plates (solid medium; Figure 1). The liquid culture of Z. circumcarinatum was harvested by centrifugation (300 xg) and blotted onto cellulose membrane filters (pore size 0.45 mm, Sigma Aldrich, St. Louis, MO, USA) while the biomass, grown on agar plates, was transferred directly to the filters without any blotting. The triplicates of all samples were placed into separate desiccation chambers, which were previously described (Karsten et al. 2014), and desiccated over a saturated KCl solution at a RH of approximately 86% according to Pichrtová et al. (2014). Y(II) was monitored during the desiccation event, using a portable PAM (Model 2500, Heinz Walz, Effeltrich, Germany), and samples were taken when Y(II) reached zero. The water loss of the biomass was determined prior to and after incubation in the desiccation chamber. The reduction of the water content was determined according to the following formula:
Reduction [%] = 100 – desiccated biomass/fresh biomass x 100
Both for Y(II) and water loss, differences between each group were assessed in R by a one-way ANOVA (p < 0.01) followed by Tukey’s post-hoc test (HSD, p < 0.01). Correlation coefficients were calculated according to Pearson (1895).
RNA isolation and sequencing
Extraction of RNA was performed as previously described by Holzinger et al. (2014) with some modifications. Six filters from each cultivation (L, P1, P2, P3), containing either control samples (C, n = 3) or samples, subjected to desiccation (D, n = 3), were treated with 450 µl of Life Guard Soil Preservation solution (MO BIO Laboratories, Carlsbad, CA USA) and frozen in liquid nitrogen. Subsequently, 2% CTAB buffer (in 100 mM Tris, 50 mM EDTA, pH=8) was added and the samples were ground in a mortar as previously described by Heinrich et al. (2012b). Furthermore, 20 µl DTT were added to the mixture followed by a centrifugation step. The supernatant was mixed with ethanol and, finally, RNA was extracted using the peqGold Plant RNA Kit (peqlab/VWR International, Erlangen, Germany) according to the manufacturer’s instructions. RNA samples were further purified as described by Rippin et al. (2016). After DNase I treatment (Thermo Fisher Scientific, Waltham, MA, USA), the RNeasy MinElute Cleanup kit (Qiagen, Hilden, Germany) was utilized for concentration and clean-up. After assessing RNA quantity using the NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) the RNA content of one replicate of P2D and all replicates of P3C and P3D, was insufficient for sequencing. Thus, the P2D replicate was excluded and the triplicates of each of the samples P3C and P3D were pooled in equal amounts. Finally, a reference was pooled from all samples in equal amounts.
After RNA quality control with the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA), two expression libraries for the reference and one for each of the sample replicates and the two triplicate pools were prepared by Eurofins Genomics (Ebersberg, Germany). Furthermore, mRNA was enriched using oligo-(dT) beads followed by fragmentation, random-primed cDNA synthesis and Illumina compatible adaptor ligation. The two normalized reference libraries were sequenced on one lane of an Illumina MiSeq, 300 bp paired end mode, and all sample libraries together on three different lanes of an Illumina HiSeq 2500, 125 bp single mode, using the MiSeq Control Software 2.5.0.5 or HiSeq Control Software 2.2.38, RTA 1.18.54 or RT 1.18.61 and bcl2fastq-1.8.4 (Illumina, San Diego, CA, USA).
Bioinformatic analyses
Raw reads of the reference were first quality trimmed and filtered using Trimmomatic 0.35 (Bolger et al. 2014) and PRINSEQ lite 0.20.4 (Schmieder & Edwards 2011). Subsequently, rRNA sequence reads were separated by SortMeRNA 2.1 (Kopylova et al. 2012) employing the SILVA SSU NR Ref 119 and LSU Ref 119. Before assembly, COPEread (Liu et al. 2012) was utilized to stitch overlapping reads together. The assembly of the reference was done with Trinity 2.0.6 (Grabherr et al. 2011) and the quality was assessed with scripts from the Trinity package and BUSCO plants 1.1b (Simão et al. 2015). An additional quality filtering step was carried out annotating all contigs with Diamond 0.8.24 (Buchfink et al. 2015) against a custom-made database which contained the protein sequences of Physcomitrella patens (Phytozome database version 12), Klebsormidium flaccidum (Hori et al. 2014) and Naegleria gruberi (Fritz-Laylin et al. 2010). Contigs scoring an e-value higher than E-20 against N. gruberi were removed. Control and treatment reads were subjected to trimming with Trimmomatic 0.35, rRNA filtering with SortMeRNA 2.1 and were subsequently mapped to the reference assembly using Bowtie2 2.2.9 (Langmead & Salzberg 2012) estimating the abundance with RSEM (Li & Dewey 2011). For differential gene expression analysis, the R package edgeR (Robinson et al. 2010) was employed, analyzing LC, LD, P2C and P2D and proceeding only with differentially expressed genes possessing an FDR (Benjamini & Hochberg 1995) smaller than 0.001 and a fold change of at least 4. Contig annotation was performed with the Trinotate pipeline 3.0.0 (http://trinotate.github.io/), including TransDecoder 2.1 (http://transdecoder.github.io/), NCBI BLAST+ 2.3.0 (Altschul et al. 1990), HMMER 3.1b (Finn et al. 2011), SignalP 4.1 (Petersen et al. 2011), TMHMM 2.0c (Krogh et al. 2001), RNAmmer 1.2 (Lagesen et al. 2007) as well as the databases Swiss-Prot and PFAM 3.1b2. The e-value cutoff was set to E-10. Two gene set enrichment analyses were carried out in R, GO term enrichment with GoSeq 1.26.0 (Young et al. 2010) and KEGG pathway enrichment with clusterProfiler 3.2.11 (Yu et al. 2012), setting the FDR in both cases to 0.001. Diamond 0.8.24 was used for annotations against the Phytozome database version 12 and the Chlorella sp. NC64A genome (Blanc et al. 2010), the Klebsormidium crenulatum transcriptome (Holzinger et al. 2014) and the Klebsormidium flaccidum genome. Filtered rRNA reads were clustered into OTUs utilizing USEARCH 5.2.2 (Edgar 2013) and annotated against the Silva SSU database 123.1 by feeding them into the QIIME 1.9.1 (Caporaso et al. 2010) script assign_taxonomy.py (e-value cutoff E-6). All raw reads and assembled contigs were submitted to the SRA database and will become available upon publication of the study.
Supplementary Material
Acknowledgements
The authors kindly acknowledge the help of Karin Komsic-Buchmann, Cologne, Germany, and Beatrix Jungwirth, Innsbruck, Austria.
Funding
This study was supported by Austrian Science Fund (FWF) [P24242-B16, I1951-B16 to A.H.].
Abbreviations
- ANOVA
Analysis of variance
- BBM
Bold’s basal medium
- BLAST
Basic local alignment search tool
- CTAB
Cetyl trimethylammonium bromide
- DTT
Dithiothreitol
- ERD
Early-response-to-dehydration protein
- FDR
False discovery rate
- GO
Gene ontology
- HSD
Honestly significant difference
- KEGG
Kyoto encyclopedia of genes and genomes
- KO
KEGG orthology
- OTU
Operational taxonomic unit
- PAM
Pulse-amplitude modulated fluorometer
- ROS
Reactive oxygen species
- RuBisCo
Ribulose-1,5-bisphosphat-carboxylase/-oxygenase
- UVR
Ultraviolet radiation
- Y(II)
Effective quantum yield of photosystem II
Footnotes
Disclosure
The authors declare that they do not have a conflict of interest.
Subject Areas:
Environmental and Stress Responses
Regulation of Gene Expression
References
- Akutsu N, Iijima K, Hinata T, Tauchi H. Characterization of the plant homolog of Nijmegen breakage syndrome 1: Involvement in DNA repair and recombination. Biochem Biophys Res Commun. 2007;353:394–398. doi: 10.1016/j.bbrc.2006.12.030. [DOI] [PubMed] [Google Scholar]
- Al-Whaibi MH. Plant heat-shock proteins: A mini review. J King Saud Univ - Sci. 2011;23:139–150. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Molucular Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderberg HI, Danielson JÅ, Johanson U. Algal MIPs, high diversity and conserved motifs. BMC Evol Biol. 2011;11:110. doi: 10.1186/1471-2148-11-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderberg HI, Kjellbom P, Johanson U. Annotation of Selaginella moellendorffii major intrinsic proteins and the evolution of the protein family in terrestrial plants. Front Plant Sci. 2012;3:33. doi: 10.3389/fpls.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkla BJ, Vera-Estrella R, Pantoja O, Kirch HH, Bohnert HJ. Aquaporin localization - How valid are the TIP and PIP labels? Trends Plant Sci. 1999;4:86–88. doi: 10.1016/s1360-1385(99)01388-6. [DOI] [PubMed] [Google Scholar]
- Bartels D, Salamini F. Desiccation tolerance in the resurrection plant Craterostigma plantagineum. A contribution to the study of drought tolerance at the molecular level. Plant Physiol. 2001;127:1346–1353. [PMC free article] [PubMed] [Google Scholar]
- Basu S, Ramegowda V, Kumar A, Pereira A. Plant adaptation to drought stress. F1000Research. 2016:5. doi: 10.12688/f1000research.7678.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B, Marin B. Streptophyte algae and the origin of embryophytes. Ann Bot. 2009;103:999–1004. doi: 10.1093/aob/mcp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Bisson MA, Kirst GO. Osmotic acclimation and turgor pressure regulation in algae. Naturwissenschaften. 1995;82:461–471. [Google Scholar]
- Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell. 2010;22:2943–2955. doi: 10.1105/tpc.110.076406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borstlap AC. Early diversification of plant aquaporins. Trends Plant Sci. 2002;7:529–530. doi: 10.1016/s1360-1385(02)02365-8. [DOI] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Campo S, Baldrich P, Messeguer J, Lalanne E, Coca M, San Segundo B. Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol. 2014;165:688–704. doi: 10.1104/pp.113.230268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carniel FC, Gerdol M, Montagner A, Banchi E, De Moro G, Manfrin C, et al. New features of desiccation tolerance in the lichen photobiont Trebouxia gelatinosa are revealed by a transcriptomic approach. Plant Mol Biol. 2016;91:319–339. doi: 10.1007/s11103-016-0468-5. [DOI] [PubMed] [Google Scholar]
- Cruz de Carvalho MH. Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant Signal Behav. 2008;3:156–165. doi: 10.4161/psb.3.3.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz de Carvalho R, Bernardes A, Soares R, Almeida AM, Coelho AV, Marques J, et al. Differential proteomics of dehydration and rehydration in bryophytes: Evidence towards a common desiccation tolerance mechanism. Plant, Cell Environ. 2014;37:1499–1515. doi: 10.1111/pce.12266. [DOI] [PubMed] [Google Scholar]
- Cruz de Carvalho R, Catalá M, Marques da Silva J, Branquinho C, Barreno E. The impact of dehydration rate on the production and cellular location of reactive oxygen species in an aquatic moss. Ann Bot. 2012;110:1007–1016. doi: 10.1093/aob/mcs180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson JAH, Johanson U. Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol. 2008;8:45. doi: 10.1186/1471-2229-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MC. The seasonal periodicity of algae on Antarctic fellfield soils. Holarct Ecol. 1991;14:112–120. [Google Scholar]
- Dayer R, Fischer BB, Eggen RIL, Lemaire SD. The peroxiredoxin and glutathione peroxidase families in Chlamydomonas reinhardtii. Genetics. 2008;179:41–57. doi: 10.1534/genetics.107.086041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Andrade JCF, Terto J, Silva JV, Almeida C. Expression profiles of sugarcane under drought conditions: Variation in gene regulation. Genet Mol Biol. 2015;38:465–469. doi: 10.1590/S1415-475738420140288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Stanton A, Archibald JM, Gould SB. Streptophyte terrestrialization in light of plastid evolution. Trends Plant Sci. 2016;21:467–476. doi: 10.1016/j.tplants.2016.01.021. [DOI] [PubMed] [Google Scholar]
- Dinakar C, Bartels D. Desiccation tolerance in resurrection plants: New insights from transcriptome, proteome and metabolome analysis. Front Plant Sci. 2013;4:482. doi: 10.3389/fpls.2013.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Dong Y, Cui L, Balamurugan S, Gao J, Lu S, et al. High light stress triggers distinct proteomic responses in the marine diatom Thalassiosira pseudonana. BMC Genomics. 2016;17:994. doi: 10.1186/s12864-016-3335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–1000. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Erickson E, Wakao S, Niyogi KK. Light stress and photoprotection in Chlamydomonas reinhardtii. Plant J. 2015;82:449–465. doi: 10.1111/tpj.12825. [DOI] [PubMed] [Google Scholar]
- Farrant JM, Willigen C Vander, Loffell DA, Bartsch S, Whittaker A. An investigation into the role of light during desiccation of three angiosperm resurrection plants. Plant, Cell Environ. 2003;26:1275–1286. [Google Scholar]
- Fernández-Marín B, Holzinger A, García-plazaola JI. Handbook of Photosynthesis. 2016. Photosynthetic strategies of desiccation-tolerant organisms; pp. 663–681. [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39:29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Gao B, Zhang D, Li X, Yang H, Zhang Y, Wood AJ. De novo transcriptome characterization and gene expression profiling of the desiccation tolerant moss Bryum argenteum following rehydration. BMC Genomics. 2015;16:416. doi: 10.1186/s12864-015-1633-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasulla F, Barreno E, Parages ML, Camara J, Jiménez C, Dörmann P, et al. The role of phospholipase D and MAPK signaling cascades in the adaption of lichen microalgae to desiccation: Changes in membrane lipids and phosphoproteome. Plant Cell Physiol. 2016;57:1908–1920. doi: 10.1093/pcp/pcw111. [DOI] [PubMed] [Google Scholar]
- Gasulla F, Dorp K, Dombrink I, Za U, Gisch N, Do P, et al. The role of lipid metabolism in the acquisition of desiccation tolerance in Craterostigma plantagineum: A comparative approach. Plant J. 2013;75:726–741. doi: 10.1111/tpj.12241. [DOI] [PubMed] [Google Scholar]
- Gasulla F, Jain R, Barreno EVA, Guéra A, Balbuena TS, Thelen JAYJ, et al. The response of Asterochloris erici (Ahmadjian) Skaloud et Peksa to desiccation: A proteomic approach. Plant, Cell Environ. 2013;36:1363–1378. doi: 10.1111/pce.12065. [DOI] [PubMed] [Google Scholar]
- Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- Goyal K, Walton LJ, Tunnacliffe A. LEA proteins prevent protein aggregation due to water stress. Biochem J. 2005;388:151–157. doi: 10.1042/BJ20041931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson Da, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierz SL, Forêt S, Leggat W. Transcriptomic analysis of thermally stressed Symbiodinium reveals differential expression of stress and metabolism genes. Front Plant Sci. 2017;8:271. doi: 10.3389/fpls.2017.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JW, Kim GH. An ELIP-like gene in the freshwater green alga, Spirogyra varians (Zygnematales), is regulated by cold stress and CO2 influx. J Appl Phycol. 2013;25:1297–1307. [Google Scholar]
- Han JW, Yoon M, Kupper FD, Klochkova TA, Oh J-S, Rho J-R, et al. Accumulation of galloyl derivatives in a green alga, Spirogyra varians, in response to cold stress. J Appl Phycol. 2012;24:1279–1286. [Google Scholar]
- Hanssum A, Zhong Z, Rousseau A, Krzyzosiak A, Sigurdardottir A. An inducible chaperone adapts proteasome assembly to stress. Mol Cell. 2014;55:566–577. doi: 10.1016/j.molcel.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka R, Furuki T, Shimizu T, Takezawa D, Kikawada T, Sakurai M, et al. Biochemical and structural characterization of an endoplasmic reticulum-localized late embryogenesis abundant (LEA) protein from the liverwort Marchantia polymorpha. Biochem Biophys Res Commun. 2014;454:588–593. doi: 10.1016/j.bbrc.2014.10.130. [DOI] [PubMed] [Google Scholar]
- Hawes I. Effects of freezing and thawing on a species of Zygnema (Chlorophyta) from the Antarctic. Phycologia. 1990;29:326–331. [Google Scholar]
- Hayami N, Sakai Y, Kimura M, Saito T, Tokizawa M, Iuchi S, et al. The responses of Arabidopsis early light-induced protein2 to ultraviolet B, high light, and cold stress are regulated by a transcriptional regulatory unit composed of two elements. Plant Physiol. 2015;169:840–855. doi: 10.1104/pp.15.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddad M, Engelken J, Adamska I. Photosynthesis: Plastid Biology, Energy Conversion and Carbon Assimilation, Advances in Photosynthesis and Respiration. 2012. Light stress proteins in viruses, cyanobacteria and photosynthetic eukaryota; pp. 299–317. [Google Scholar]
- Heinrich S, Frickenhaus S, Glöckner G, Valentin K. A comprehensive cDNA library of light- and temperature-stressed Saccharina latissima (Phaeophyceae) Eur J Phycol. 2012b;47:83–94. [Google Scholar]
- Heinrich S, Valentin K, Frickenhaus S, John U, Wiencke C. Transcriptomic analysis of acclimation to temperature and light stress in Saccharina latissima (Phaeophyceae) PLoS One. 2012a;7:e44342. doi: 10.1371/journal.pone.0044342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich S, Valentin K, Frickenhaus S, Wiencke C. Temperature and light Interactively modulate gene expression in Saccharina latissima (Phaeophyceae) J Phycol. 2015;51:93–108. doi: 10.1111/jpy.12255. [DOI] [PubMed] [Google Scholar]
- Heinrich S, Valentin K, Frickenhaus S, Wiencke C. Origin matters - Comparative transcriptomics in Saccharina latissima (Phaeophyceae) J Exp Mar Bio Ecol. 2016;476:22–30. [Google Scholar]
- Herburger K, Lewis LA, Holzinger A. Photosynthetic efficiency, desiccation tolerance and ultrastructure in two phylogenetically distinct strains of alpine Zygnema sp. (Zygnematophyceae, Streptophyta): Role of pre-akinete formation. Protoplasma. 2015;252:571–589. doi: 10.1007/s00709-014-0703-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herburger K, Holzinger A. Localization and quantification of callose in the streptophyte green algae Zygnema and Klebsormidium: Correlation with desiccation tolerance. Plant Cell Physiol. 2015;56:2259–2270. doi: 10.1093/pcp/pcv139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hincha DK, Sonnewald U, Willmitzer L, Schmitt JM. The role of sugar accumulation in leaf frost hardiness - Investigations with transgenic tobacco expressing a bacterial pyrophosphatase or a yeast invertase gene. J Plant Physiol. 1996;147:604–610. [Google Scholar]
- Holzinger A, Becker B. Desiccation tolerance in the streptophyte green alga Klebsormidium: The role of phytohormones. Commun Integr Biol. 2015;8:e1059978. doi: 10.1080/19420889.2015.1059978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger A, Pichrtova M. Abiotic stress tolerance of charophyte green algae: New challenges for omics techniques. Front Plant Sci. 2016;7 doi: 10.3389/fpls.2016.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger A, Kaplan F, Blaas K, Zechmann B, Komsic-Buchmann K, Becker B. Transcriptomics of desiccation tolerance in the streptophyte green alga Klebsormidium reveal a land plant-like defense reaction. PLoS One. 2014;9:e110630. doi: 10.1371/journal.pone.0110630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger A, Karsten U. Desiccation stress and tolerance in green algae: Consequences for ultrastructure, physiological, and molecular mechanisms. Front Plant Sci. 2013;4:327. doi: 10.3389/fpls.2013.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun. 2014;5 doi: 10.1038/ncomms4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Hu G, Han B. Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci. 2009;176:583–590. doi: 10.1016/j.plantsci.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Hundertmark M, Hincha DK. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9:118. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin C, Nussaume L, Moise N, Moya I, Kloppstech K, Havaux M. Early light-induced proteins protect Arabidopsis from photooxidative stress. PNAS. 2003;100:4921–4926. doi: 10.1073/pnas.0736939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarzyniak KM, Jasinski M. Membrane transporters and drought resistance - A complex issue. Front Plant Sci. 2014;5:687. doi: 10.3389/fpls.2014.00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten U, Herburger K, Holzinger A. Dehydration, temperature, and light tolerance in members of the aeroterrestrial green algal genus Interfilum (Streptophyta) from biogeographically different temperate soils. J Phycol. 2014;50:804–816. doi: 10.1111/jpy.12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten U, Holzinger A. Green algae in alpine biological soil crust communities: Acclimation strategies against ultraviolet radiation and dehydration. Biodivers Conserv. 2014;23:1845–1858. doi: 10.1007/s10531-014-0653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Harada N, Nishimura Y, Saito T, Nakamura M, Fujiwara T, et al. Algae sense exact temperatures: Small heat shock proteins are expressed at the survival threshold temperature in Cyanidioschyzon merolae and Chlamydomonas reinhardtii. Genome Biol Evol. 2014;6:2731–2740. doi: 10.1093/gbe/evu216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova E, Noé L, Touzet H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- Kranner I, Stabentheiner E, Kranner I, Cram WJ, Zorn M, Wornik S, et al. Antioxidants and photoprotection in a lichen as compared with its isolated symbiotic partners. PNAS. 2005;102:3141–3146. doi: 10.1073/pnas.0407716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer E. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Król M, Maxwell DP, Huner NPA. Exposure of Dunaliella salina to low temperature mimics the high light-induced accumulation of carotenoids and the carotenoid binding protein (Cbr) Plant Cell Physiol. 1997;38:213–216. [Google Scholar]
- Lagesen K, Hallin P, Rødland EA, Stærfeldt HH, Rognes T, Ussery DW. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajos K, Mayr S, Buchner O, Blaas K, Holzinger A. A new microscopic method to analyse desiccation-induced volume changes in aeroterrestrial green algae. J Microsc. 2016;263:192–199. doi: 10.1111/jmi.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tallec B, Barrault M-B, Courbeyrette R, Guérois R, Marsolier-Kergoat M-C, Peyroche A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol Cell. 2007;27:660–674. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Lee U, Rioflorido I, Hong S, Larkindale J, Waters ER, Vierling E. The Arabidopsis ClpB/Hsp100 family of proteins: Chaperones for stress and chloroplast development. Plant J. 2006;49:115–127. doi: 10.1111/j.1365-313X.2006.02940.x. [DOI] [PubMed] [Google Scholar]
- Lers A, Levy H, Zamir A. Co-regulation of a gene homologous to early light-induced genes in higher plants and ß-carotene biosynthesis in the alga Dunaliella bardawil. J Biol Chem. 1991;266:13698–13705. [PubMed] [Google Scholar]
- Levitt J. Responses of plants to environmental stresses: water, radiation, salt, and other stresses, Physiological ecology. Academic Press; 1980. [Google Scholar]
- Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Fan C, Li Y, Zhang J, Sun J, Chen Y, et al. Effects of drought and salt-stresses on gene expression in Caragana korshinskii seedlings revealed by RNA-seq. BMC Genomics. 2016;17:200. doi: 10.1186/s12864-016-2562-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Yuan J, Yiu SM, Li Z, Xie Y, Chen Y, et al. COPE: An accurate k-mer-based pair-end reads connection tool to facilitate genome assembly. Bioinformatics. 2012;28:2870–2874. doi: 10.1093/bioinformatics/bts563. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Y, Yang C, Tian Z, Li J. AtSWEET4, a hexose facilitator, mediates sugar transport to axial sinks and affects plant development. Sci Rep. 2016;6 doi: 10.1038/srep24563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokhorst GM. Comparative taxonomic studies on the genus Klebsormidium (Charophyceae) in Europe, Cryptogamic studies. John Wiley & Sons Incorporated; 1996. [Google Scholar]
- Ma C, Wang H, Macnish AJ, Estrada-Melo AC, Lin J, Chang Y, et al. Transcriptomic analysis reveals numerous diverse protein kinases and transcription factors involved in desiccation tolerance in the resurrection plant Myrothamnus flabellifolia. Hortic Res. 2015;2:15034. doi: 10.1038/hortres.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraccini P, Vinecky F, Alves GSC, Ramos HJO, Elbelt S, Vieira NG, et al. Differentially expressed genes and proteins upon drought acclimation in tolerant and sensitive genotypes of Coffea canephora. J Exp Bot. 2012;63:4191–4212. doi: 10.1093/jxb/ers103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan SC, Pasternak M, Cairns N, Kiddle G, Brach T, Jarvis R, et al. Plant homologs of the Plasmodium falciparum chloroquine-resistance transporter, Pf CRT, are required for glutathione homeostasis and stress responses. PNAS. 2010;107:2331–2336. doi: 10.1073/pnas.0913689107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI, Melkonian M. Use of Hepes buffer for microalgal culture media and fixation for electron microscopy. Phycologia. 1986;25:551–557. [Google Scholar]
- McLean RJ, Pessoney GF. Contributions in Phycology. 1971. Formation and resistance of akinetes of Zygnema; pp. 145–52. [Google Scholar]
- McNairn RB. Phloem translocation and heat-induced callose formation in field-grown Gossypium hirsutum L. Plant Physiol. 1972;50:366–370. doi: 10.1104/pp.50.3.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra J, Xu G, Wang B, Li M, Deng X. Understanding desiccation tolerance using the resurrection plant Boea hygrometrica as a model system. Front Plant Sci. 2013;4:446. doi: 10.3389/fpls.2013.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao M, Matsui K, Uemura M. Klebsormidium flaccidum, a charophycean green alga, exhibits cold acclimation that is closely associated with compatible solute accumulation and ultrastructural changes. Plant, Cell Environ. 2008;31:872–885. doi: 10.1111/j.1365-3040.2008.01804.x. [DOI] [PubMed] [Google Scholar]
- Noctor G, Queval G, Mhamdi A, Chaouch S, Foyer CH. Glutathione. The Arabidopsis Book. 2011:1–32. doi: 10.1199/tab.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norén H, Svensson P, Stegmark R, Funk C, Adamska I, Andersson B. Expression of the early light-induced protein but not the PsbS protein is influenced by low temperature and depends on the developmental stage of the plant in field-grown pea cultivars. Plant, Cell Environ. 2003;26:245–253. [Google Scholar]
- Norwood M, Toldi O, Richter A, Scott P. Investigation into the ability of roots of the poikilohydric plant Craterostigma plantagineum to survive dehydration stress. J Exp Bot. 2003;54:2313–2321. doi: 10.1093/jxb/erg255. [DOI] [PubMed] [Google Scholar]
- Oliver MJ. Plant Desiccation Tolerance. Blackwell Publishing Ltd; 2007. Lessons on dehydration tolerance from desiccation-tolerant plants; pp. 11–50. [Google Scholar]
- Oliver MJ, Jain R, Balbuena TS, Agrawal G, Gasulla F, Thelen JJ. Proteome analysis of leaves of the desiccation-tolerant grass, Sporobolus stapfianus, in response to dehydration. Phytochemistry. 2011;72:1273–1284. doi: 10.1016/j.phytochem.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Park SK, Jin ES, Lee MY. Expression and antioxidant enzymes in Chaetoceros neogracile, an Antarctic alga. Cryo letters. 2007;29:351–361. [PubMed] [Google Scholar]
- Pearson K. Note on regression and inheritance in the case of two parents. Proc R Soc London. 1895;58:240–242. [Google Scholar]
- Perlikowski D, Kierszniowska S, Sawikowska A, Krajewski P, Rapacz M, Eckhardt A, et al. Remodeling of leaf cellular glycerolipid composition under drought and re-hydration conditions in grasses from the Lolium-Festuca complex. Front Plant Sci. 2016;7 doi: 10.3389/fpls.2016.01027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Pichrtová M, Arc E, Stöggl W, Kranner I, Hájek T, Hackl H, et al. Formation of lipid bodies and changes in fatty acid composition upon pre-akinete formation in Arctic and Antarctic Zygnema (Zygnematophyceae, Streptophyta) strains. FEMS Microbiol Ecol. 2016a;92:fiw096. doi: 10.1093/femsec/fiw096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichrtova M, Hájek T, Elster J. Annual development of mat-forming conjugating green algae Zygnema spp. in hydro-terrestrial habitats in the Arctic. Polar Biol. 2016b;39:1653–1662. [Google Scholar]
- Pichrtová M, Kulichová J, Holzinger A. Nitrogen limitation and slow drying induce desiccation tolerance in conjugating green algae (Zygnematophyceae, Streptophyta) from polar habitats. PLoS One. 2014;9:e113137. doi: 10.1371/journal.pone.0113137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanjulu S, Bartels D. Drought-and desiccation-induced modulation of gene expression in plants. Plant, Cell Environ. 2002;25:141–151. doi: 10.1046/j.0016-8025.2001.00764.x. [DOI] [PubMed] [Google Scholar]
- Rezaei MK, Shobbar ZS, Shahbazi M, Abedini R, Zare S. Glutathione S-transferase (GST) family in barley: Identification of members, enzyme activity, and gene expression pattern. J Plant Physiol. 2013;170:1277–1284. doi: 10.1016/j.jplph.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Rippin M, Komsic-Buchmann K, Becker B. RNA isolation from biological soil crusts: Methodological aspects. Arch Hydrobiol Suppl Algol Stud. 2016;151:21–37. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG. From algae to angiosperms - Inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol Biol. 2014;14:23. doi: 10.1186/1471-2148-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salerno GL, Pontis HG. Raffinose synthesis in Chlorella vulgaris cultures after a cold shock. Plant Physiol. 1989;89:648–651. doi: 10.1104/pp.89.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowsky A, Mettler-Altmann T, Ott S. Metabolic response to desiccation stress in strains of green algal photobionts (Trebouxia) from two Antarctic lichens of southern habitats. Phycologia. 2016;55:703–714. [Google Scholar]
- Salman MH, Zhang C, Pervaiz T, Zheng T, Zhang CB, Lide C, et al. Gene regulation mechanism in drought-responsive grapevine leaves as revealed by transcriptomic analysis. bioRxiv. 2016:65136. [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz-Raffelt M, Lodha M, Schroda M. Heat shock factor 1 is a key regulator of the stress response in Chlamydomonas. Plant J. 2007;52:286–295. doi: 10.1111/j.1365-313X.2007.03228.x. [DOI] [PubMed] [Google Scholar]
- Shinde S, Nurul Islam M, Ng CKY. Dehydration stress-induced oscillations in LEA protein transcripts involves abscisic acid in the moss, Physcomitrella patens. New Phytol. 2012;195:321–328. doi: 10.1111/j.1469-8137.2012.04193.x. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Teramoto H, Itoh T, Ono T. High-intensity-light-dependent and transient expression of new genes encoding distant relatives of light-harvesting chlorophyll-a/b proteins in Chlamydomonas reinhardtii. Plant Cell Physiol. 2004;45:1221–1232. doi: 10.1093/pcp/pch157. [DOI] [PubMed] [Google Scholar]
- Turan S, Tripathy BC. Salt-stress induced modulation of chlorophyll biosynthesis during de-etiolation of rice seedlings. Physiol Plant. 2015;152:477–491. doi: 10.1111/ppl.12250. [DOI] [PubMed] [Google Scholar]
- Valledor L, Furuhashi T, Hanak A, Weckwerth W. Systemic cold stress adaptation of Chlamydomonas reinhardtii. Mol Cell Proteomics. 2013;12:2032–2047. doi: 10.1074/mcp.M112.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Poel B, Cooper ED, Straeten D, Van Der, Chang C, Delwiche CF. Transcriptome profiling of the green alga Spirogyra pratensis (Charophyta) suggests an ancestral role for ethylene in cell wall metabolism, photosynthesis, and abiotic stress responses. Plant Physiol. 2016;172:533–545. doi: 10.1104/pp.16.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Pan Y, Zhao X, Zhu L, Fu B, Li Z. Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice. BMC Genomics. 2011;12:149. doi: 10.1186/1471-2164-12-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Cai G, Kong F, Deng Y, Ma N, Meng Q. Overexpression of tomato chloroplast-targeted DnaJ protein enhances tolerance to drought stress and resistance to Pseudomonas solanacearum in transgenic tobacco. Plant Physiol Biochem. 2014;82:95–104. doi: 10.1016/j.plaphy.2014.05.011. [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Wang XQ, Yang PF, Liu Z, Liu WZ, Hu Y, Chen H, et al. Exploring the mechanism of Physcomitrella patens dessication tolerance through a proteomic strategy. Plant Physiol. 2009;149:1739–1750. doi: 10.1104/pp.108.131714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-B, Liu M-F, Zhang X-F, Zhang A-J, Wang B, Zheng Z, et al. Composition and regulation of thylakoid membrane of Antarctic ice microalgae Chlamydomonas sp. ICE-L in response to low-temperature environment stress. J Mar Biol Assoc United Kingdom. 2016:1–9. [Google Scholar]
- Weissbach H, Etienne F, Hoshi T, Heinemann SH, Lowther WT, Matthews B, et al. Peptide methionine sulfoxide reductase: Structure, mechanism of action, and biological function. Arch Biochem Biophys. 2002;397:172–178. doi: 10.1006/abbi.2001.2664. [DOI] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci. 2014;111:E4859–E4868. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Quick WP, Bungard RA, Bailey KJ, Lea PJ, Leegood RC. The role of photorespiration during drought stress: An analysis utilizing barley mutants with reduced activities of photorespiratory enzymes. Plant, Cell Environ. 1999;22:361–373. [Google Scholar]
- Wingler A, Lea PJ, Quick WP, Leegood RC. Photorespiration: Metabolic pathways and their role in stress protection. Philos Trans R Soc B Biol Sci. 2000;355:1517–1529. doi: 10.1098/rstb.2000.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter H, Huber SC. Regulation of sucrose metabolism in higher plants: Localization and regulation of activity of key enzymes. Crit Rev Biochem Mol Biol. 2000;35:253–289. doi: 10.1080/10409230008984165. [DOI] [PubMed] [Google Scholar]
- Wodniok S, Brinkmann H, Glöckner G, Heidel AJ, Philippe H, Melkonian M, et al. Origin of land plants: Do conjugating green algae hold the key? BMC Evol Biol. 2011;11:104. doi: 10.1186/1471-2148-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Chen X, Wood AJ. Two early light-inducible protein (ELIP) cDNAs from the resurrection plant Tortula ruralis are differentially expressed in response to desiccation, rehydration, salinity, and high light. J Exp Bot. 2002;53:1197–1205. doi: 10.1093/jexbot/53.371.1197. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Hu W, Zhu F, Wang L, Yu Q, Ming R, et al. Structure, phylogeny, allelic haplotypes and expression of sucrose transporter gene families in Saccharum. BMC Genomics. 2016;17:88. doi: 10.1186/s12864-016-2419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Xi Z, Goremykin VV, Fong R, Mclenachan PA, Novis PM, et al. Streptophyte algae and the origin of land plants revisited using heterogeneous models with three new algal chloroplast genomes. Mol Biol Evol. 2013;31:177–183. doi: 10.1093/molbev/mst200. [DOI] [PubMed] [Google Scholar]
- Zhou L, Xu H, Mischke S, Meinhardt LW, Zhang D, Zhu X, et al. Exogenous abscisic acid significantly affects proteome in tea plant (Camellia sinensis) exposed to drought stress. Hortic Res. 2014;1:14029. doi: 10.1038/hortres.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.