Abstract
Mosquitoes are high-impact disease vectors with the capacity to transmit pathogenic agents that cause diseases such as malaria, yellow fever, chikungunya, and dengue. Continued growth in knowledge of genetic, molecular, and physiological pathways in mosquitoes allows for the development of novel control methods and for the continued optimization of existing ones. The emergence of site-specific nucleases as genomic engineering tools promises to expedite research of crucial biological pathways in these disease vectors. The utilization of these nucleases in a more precise and efficient manner is dependent upon knowledge and manipulation of the DNA repair pathways utilized by the mosquito. While progress has been made in deciphering DNA repair pathways in some model systems, research into the nature of the hierarchy of mosquito DNA repair pathways, as well as in mechanistic differences that may exist, is needed. In this review, we will describe progress in the use of site-specific nucleases in mosquitoes, along with the hierarchy of DNA repair in the context of mosquito chromosomal organization and structure, and how this knowledge may be manipulated to achieve precise chromosomal engineering in mosquitoes.
Keywords: Mosquito, Aedes, Gene editing, TALEN, CRISPR, DNA repair
Mosquito-borne disease
Mosquitoes are important disease vectors, with none more important than the malaria vector Anopheles gambiae and the dengue vector Aedes aegypti. Malaria is believed to infect more than 200 million annually causing more than half a million deaths (2014a; 2014d), while dengue is thought to infect 50 to 100 million annually, causing mass morbidity and placing a large economic burden on developing countries (2014b;2014c). Other pathogens such as yellow fever virus, chikungunya virus, West Nile virus, eastern equine encephalitis virus, western equine encephalitis virus, and La Crosse virus are also vectored by mosquitoes (Colpitts et al. 2011; Pialoux et al. 2007; Chhabra et al. 2008) and cause substantial human morbidity and mortality.
Treatment for malaria revolves around the use of insecticide-treated bed nets (Hill et al. 2006), indoor residual insecticides (Pluess et al. 2010), and treatment of infected human hosts with anti-malarial drugs (Schlitzer 2008); for dengue, prevention is primarily through control of the vector by way of source reduction and insecticides (Kamgang et al. 2011). Techniques such as the sterile insect technique (SIT) (Alphey et al. 2010), release of insects harboring intracellular bacteria Wolbachia (Walker et al. 2011), and release of insects with dominant lethality (RIDL) (Massonnet-Bruneel et al. 2013; Fu et al. 2010; Labbé et al. 2012) are currently being investigated as additional interventions and appear to hold great promise. Also in development are techniques that aim to replace current mosquito populations with those that are pathogen resistant [reviewed in Burt (2014) and Wang and Jacobs-Lorena (2013)]. A primary technical concern of gene-drive-based control strategies is the integrity and long-term stability of any pathogen resistance transgene. Optimally, such transgenes would be placed in a chromosomal region most likely to be repaired faithfully in the case of DNA damage. However, little is known about mosquito DNA break repair pathways, or how chromosomal structure and repeat content may influence the long-term stability of such transgenes, particularly regions that might be more inclined toward mutations or transcriptional silencing.
More generally, the advent of site-specific gene editing technologies such as transcription activator-like effector nucleases (TALENs) and clustered regulatory interspaced short palindromic repeats (CRISPR)/Cas9 is poised to revolutionize the field of mosquito genetics and molecular biology and physiology (Kim and Kim 2014). These highly specific nucleases rely on host DNA repair pathways to fix the broken DNA ends, preferably in a fashion consistent with the hopes of the investigator. An unavoidable complication, however, is that each of the various end-joining and homology-based repair pathways competes with each other for access to the double-stranded DNA break (DSB). Work by others has shown that it is possible to manipulate the repair outcome in insects such as Drosophila (Beumer et al. 2008; Ciapponi et al. 2004; Yoo and McKee 2005; Wei and Rong 2007) and Bombyx mori (Ma et al. 2014). However, little is known about the hierarchy of DNA repair choice in mosquitoes, or the protein components most critical for completing each form of DSB repair. In this review, we present our current understanding of DSB repair as gleaned from model organisms such as yeast, flies, and vertebrates in the context of genes and gene families lost and duplicated in the mosquito. Additionally, we present potential strategies for manipulating the DSB response in mosquitoes with respect to protein machinery, chromosomal structure, and sequence content. An understanding of these aspects of genome engineering in mosquitoes will hopefully stimulate further investigation and generate evidence-based questions about how genomic engineering in mosquito disease vectors can be further improved.
Gene editing technologies used in mosquitoes
Homing endonucleases (HEs) were the first site-specific nucleases used to edit the mosquito genome (Windbichler et al. 2007). Known for their extreme target specificity (Stoddard 2005; Stoddard 2011), HEs are naturally occurring selfish genetic elements that can display hyper-Mendelian rates of inheritance due to their ability to be copied from a template to a target chromosome via homology-directed repair following DSB induction (Burt 2003). In the malaria mosquito, the HE I-SceI has been used to generate an artificial gene drive system that may one day be used to convert these mosquitoes into a more benign form that no longer transmits malaria parasites (Windbichler et al. 2011). The fortuitous insertion of a transposon bearing an I-SceI recognition sequence onto the Y chromosome of An. gambiae has also allowed the homology-dependent integration of additional transgenes to this location (Bernardini et al. 2014). The HE I-PpoI, which recognizes a target ribosomal DNA that is conserved amongst all eukaryotes, was used to develop transgenic strains of An. gambiae that display either male-specific sterility (Windbichler et al. 2008; Klein et al. 2012) or male-specific sex distortion (Galizi et al. 2014) phenotypes. While HEs have not been used as extensively to perform chromosomal manipulations in other mosquitoes, several HEs have been shown to recognize and cleave their target sites in a highly specific manner when these sites are present in the Ae. aegypti genome (Aryan et al. 2013a; Traver et al. 2009). Despite the fact that several hundred naturally occurring HEs have been described, the difficulty and expense in re-engineering these site-specific nucleases to recognize new and useful targets will likely restrict their application.
In contrast, customizable endonucleases such as zinc finger nucleases (ZFNs) and TALENS are modular and can be much more easily re-engineered to recognize interesting chromosomal targets (Gaj et al. 2013; Carlson et al. 2012). Both systems depend on the generation of two synthetic proteins that when heterodimerized yield an active site-specific nuclease. ZFNs use a variety of zinc finger binding domains linked together, with each binding domain coding for three specific nucleotides (Urnov et al. 2010), while TALENs use a system of linked TALE repeats, where each repeat specifies a single nucleotide (Joung and Sander 2013). Both ZFNs and TALENs couple these repeat domains to a nuclease such as FokI. ZFNs have been used successfully in Ae. aegypti (Liesch et al. 2013; Degennaro et al. 2013; McMeniman et al. 2014) and TALENs have been used in both Ae. aegypti and An. gambiae (Aryan et al. 2013b; Smidler et al. 2013). Despite these advances, the cost of synthesizing or assembling new ZFNs or TALENs is likely to prevent their widespread use in mosquito gene editing.
Thus, the most promising future for mosquito chromosomal manipulation may be with CRISPR/Cas9. The CRISPR/Cas9 system is part of the adaptive immune system within certain bacteria (Chakraborty et al. 2009; Karginov and Hannon 2010) and uses short RNA sequences to guide a DNA endonuclease (Cas9), resulting in a DSB (Sander and Joung 2014; Bassett and Liu 2014; Liu and Fan 2014; Mali et al. 2013b). The CRISPR/Cas9 system has been further adapted to use synthetic guide RNAs, further optimizing the process (Bassett et al. 2013; Ma et al. 2013; Upadhyay and Sharma 2014; Bae et al. 2014). Re-engineering new target sites is as simple as synthesizing a new small RNA molecule; this ease of use explains why Cas9-based editing has so rapidly supplanted other technologies. For mosquito gene editing, as for Drosophila, a computational search for potential cross-targeting guide RNAs before the experiment begins (Xie et al. 2014), followed by several generations of out-crossing of edited individuals (Liesch et al. 2013), should be sufficient to minimize confounding off-target effects. Other options include optimizing the concentration of sgRNAs to Cas9, and utilizing pairs of Cas9 nucleases that have been mutated only to allow nicking (or single-stranded breaks) to occur, which when used in conjunction effectively generate a DSB (Mali et al. 2013a). While there have been no published incidences of the use of CRISPR/Cas 9 in mosquitoes, unpublished data suggests that the system will in fact work in a highly efficient manner (not shown).
All nuclease-based gene editing and chromosomal manipulation tools rely intrinsically on host-mediated repair processes. In the absence of a visible marker, gene editing events are typically detected via PCR, followed by Sanger or Illumina-based sequencing, digestion of the PCR amplicon with a restriction endonuclease or mismatch-specific nuclease, or through analysis of the amplicon using high-resolution melt curve analysis (HRMA). While these systems have all proven effective to various degrees, these assays all underestimate nuclease activity due to (1) repair that correctly restores the original sequence, or (2) repair that is sufficiently deleterious as to remove one or both primer binding sites. Where the experimental goal is to produce targeted deletions, the experimenter must rely on mistakes made by the classical non-homologous end-joining (C-NHEJ) pathway, while targeted insertions rely on homology-directed repair (HDR) (Liesch et al. 2013; Bassett et al. 2014; Gratz et al. 2014). However, each of these mechanisms is capable of both highly faithful and highly deleterious repair. Thus, all assays and outcomes depend not just on the activity of the site-specific nuclease, but also on the success or failure of the target cell to repair the resultant DSB in a manner consistent with the wishes of the experimenter. Understanding how these repair pathways function, as well as how they interact and compete with each other, is thus critical to optimizing gene editing experiments. This is especially necessary in non-model organisms such as mosquitoes that are more difficult to handle at larger scales.
Double-stranded DNA break repair
Generally speaking, DSBs are repaired by C-NHEJ or HDR, but the more these mechanisms are elucidated, the more complex they appear to be. For the purposes of this review, we will cover HDR, single-strand annealing (SSA), C-NHEJ, and alternative non-homologous end joining (A-NHEJ), in reference to how they repair DSBs and how they may be manipulated to achieve the experimenters’ desired results.
Homology-directed repair is initiated when the MRN nuclease complex (Niu et al. 2010) composed of Mre11, Rad50, and Nbs1 resects the DSB (from either end). The MRN complex is aided by the endonuclease Sae2 (Lamarche et al. 2010), as well as secondary endonucleases such as Exo1, Dna2, and Sgs1 (Zhu et al. 2008). Loss of either Mre11 or Rad50 in Drosophila leads to chromosomal instability and higher cell death rates (Ciapponi et al. 2004). Interestingly, Ae. aegypti appears to have duplicated both Mre11 and Rad50 (Table 1). Once resection has been accomplished and each end of the DSB has an exposed ssDNA strand, RPA binds the ssDNA (Golub et al. 1998) and is replaced by Rad51 with the aid of mediator proteins such as BRCA2 (Klovstad et al. 2008). In yeast, this mediation is accomplished by Rad52; while Rad52 appears to be less important in some vertebrates, it appears to retain some role in humans (Liu and Heyer 2011) and is completely lost in flies and mosquitoes (Table 1). Rad51 creates a filament complex that has the ability to invade a homologous sequence of dsDNA (Yoo and McKee 2005); Rad54 works in conjunction with the Rad51 invasion filament until the appropriate homologous sequence is found (Kiianitsa et al. 2006). Once homology is detected, either δ or ε (delta or epsilon), polymerase is recruited to accurately repair the DNA lesion (Mehta and Haber 2014). Depending on the nature of the break, a Holiday junction is formed (Heyer 2004) and eventually disassembled leading to an “error-free” repair of the DSB. Rad51 and Rad54 mutants have been evaluated in Drosophila using a DNA repair assay; Rad51 was determined to be crucial for HDR, with Rad54 deemed important as well, but to a lesser extent than Rad51 (Wei and Rong 2007). In an alternative approach, RNAi-based suppression of Rad51 mRNA in Drosophila led to higher death rates in the presence of a mutagenic substance (Yoo 2006). Conversely, over-expression of Rad51 in Drosophila using a heat shock promoter also resulted in lethality, suggesting that the amount of homology-based repair must be finely controlled (Yoo and McKee 2004). HDR components have not yet been studied in mosquitoes.
Table 1.
Orthologs of DNA break repair components in mosquitoes
| Gene | NHEJ | A-NHEJ | HDR | SSA | D. melanogaster | Ae. aegypti | An. gambiae |
|---|---|---|---|---|---|---|---|
| Ku70 | X | FBgn0011774 | No gene model | AGAP002690 | |||
| Ku80 | X | FBgn0041627 | AAEL003684 | AGAP009910 | |||
| DNA-PKcs | X | Absent | AAEL008123 | AGAP003967 | |||
| Xrcc4 | X | FBgn0069301 | No ortholog identified | No ortholog identified | |||
| XLF | X | No ortholog identified | AAEL002939 | No ortholog identified | |||
| Ligase 4 | X | FBgn0030506 | AAEL0173656/AAEL017561 | AGAP000623 | |||
| Polμ/Polλ | X | Absent | Absent | Absent | |||
| Artemis | X | No ortholog identified | No ortholog identified | AGAP000597 | |||
| APLF | X | FBgn0026737 | AAEL011254 | AGAP004516 | |||
| PNKP | X | FBgn0037578 | AAEL000527 | AGAP012174 | |||
| APTX | X | FBgn0038704 | AAEL014945 | AGAP004307 | |||
| Parp1 | X | FBgn0010247 | AAEL011815 | AGAP003230 | |||
| Ligase 3 | X | FBgn0038035 | Absent | Absent | |||
| Ligase 1 | X | FBgn0262619 | AAEL017566 | AGAP009222 | |||
| Xrcc3 | X | FBgn0003480 | AAEL005399 | AGAP013180 | |||
| Xrcc1 | X | FBgn0026751 | AAEL002782 | AGAP002605 | |||
| ATM | X | FBgn0045035 | AAEL014900 | AGAP009632 | |||
| Mre11 | X | X | FBgn0020270 | AAEL010595/AAEL000034 | AGAP006797 | ||
| Rad50 | X | X | FBgn0034728 | AAEL014748/AAEL005245 | AGAP003676 | ||
| Nbs1 | X | X | FBgn0261530 | AAEL014377 | AGAP003213 | ||
| Sae2 | X | X | FBgn0029113 | AAEL010641 | AGAP008637 | ||
| Exo1 | X | FBgn0015553 | AAEL006209 | AGAP004491 | |||
| RPA | X | FBgn0010173 | AAEL012826 | AGAP001421 | |||
| Sgs1 | X | FBgn0002906 | AAEL004039 | AGAP002967 | |||
| Dna2 | X | FBgn0030170 | AAEL000201 | AGAP004685 | |||
| Rad51 | X | FBgn0003479 | AAEL006080 | AGAP013412 | |||
| Rad54 | X | FBgn0002989 | AAEL002647 | AGAP008748 | |||
| BRCA2 | X | FBgn0050169 | AAEL014774/AAEL010133 | AGAP007032 | |||
| Polδ | X | FBgn0263600 | AAEL014178 | AGAP011731 | |||
| Polσ | X | FBgn0264326 | AAEL002800 | AGAP004615 | |||
| Rad52 | X | X | Absent | Absent | Absent | ||
| Rad1 | X | FBgn0026778 | AAEL009701 | AGAP002255 | |||
| Rad10/Ercc1 | X | FBgn0028434 | AAEL008081/AAEL013693 | AGAP004029 | |||
| Msh2 | X | FBgn0015546 | AAEL014856 | AGAP010282 | |||
| Slx4 | X | FBgn0002909 | AAEL008482 | AGAP007582 | |||
| Msh3 | X | Yeast Only | Yeast Only | Yeast Only | |||
| Rad59 | X | Absent | Absent | Absent | |||
| Saw1 | X | Yeast Only | Yeast Only | Yeast Only |
NHEJ non-homologous end joining, A-NHEJ alternative NHEJ, HDR homology-directed repair, SSA single-strand annealing
Unlike traditional HDR, which relies on homologous sequences present on sister chromatids or homologous chromosomes, the SSA pathway relies on the use of homologous repeats flanking the DSB (Ivanov et al. 1996). The homologous regions can be as short as 30 bp in yeast (Sugawara et al. 2000), or as short as 18 bp in mosquitoes (Aryan et al. 2013a), and are eventually collapsed, while the genetic information between the two repeats is deleted. SSA is generally categorized as a sub-pathway of HDR because of the use of similar protein machinery in the initial resection steps. As in HDR, resecting occurs via the MRN complex and secondary exonucleases (Mehta and Haber 2014). A protein complex consisting of the scaffolding proteins Slx4 and Saw1 (Li et al. 2008a), the mismatch repair proteins Msh2 and Msh3 (Sugawara et al. 1997), and the Rad1–Rad10 endonuclease complex (Davies et al. 1995) work in conjunction to collapse the two homologous sections and remove excess nucleotides. In yeast, Rad52 then anneals the strands in conjunction with the homo-log Rad59 (Sugawara et al. 1997). Saw1 and Msh2 are not present in flies or mosquitoes, suggesting differences in both scaffolding and mismatch scanning in SSA-based repair, while the absence of Rad52 and Rad59 in flies and mosquitoes suggests that there may be alternative machinery which compensates for the annealing step (Table 1). Mice deficient in Rad1 and Rad10 orthologs showed similar phenotypes with regard to chromosomal damage and ultimately died prematurely (Tian et al. 2004; McWhir et al. 1993), while Drosophila Rad1 and Rad10 mutants are both viable and fertile but largely unstudied (Drysdale and FlyBase 2008).
The C-NHEJ pathway is distinct from HDR and SSA in both its machinery and mechanism. Upon DSB formation, Ku70 and Ku80 form a heterodimeric complex on the ends of the DSB called the Ku complex (Wang and Lees-Miller 2013). Knockout of Ku70 in Arabidopsis led to a 5- to 16-fold increase in HDR (QI et al. 2013), while knockdown of Ku70 in B. mori increased rates of HDR via a junction PCR assay (Ma et al. 2014), suggesting that Ku proteins may be a target of interest in mosquitoes. While the most current gene set (AaegL3.2) failed to identify a gene model for Ae. aegypti Ku70, a homology-based search (tblastn) using the Drosophila or Anopheles Ku70 protein sequences indicates that a likely ortholog is located on scaffold 1.240. The kinase DNA-PKcs tethers the Ku complexes and activates multiple proteins associated with the pathway (Williams et al. 2014), primarily the endonuclease Artemis, which is responsible for removing nucleotides from the DSB (MA et al. 2002, 2013). Subsequently, the polymerases μ and λ play a role in adding any additional nucleotides needed for ligation. Both polymerases μ and λ are absent in flies and mosquitoes, while Artemis appears to have been lost in both Drosophila melanogaster and Ae. aegypti (Fig. 1a and Table 1). In vertebrates, these factors are primarily associated with DNA ligation during V(D)J recombination, a process that does not occur in insects, with mutations resulting in a failure to generate proper T and B cells. Following modification of the broken DNA ends, the factors XLF and Xrcc4 form the primary scaffolding components (Mahaney et al. 2013) and recruit Lig4 to the break site, allowing for the final ligation of both ends of the DSB (Williams et al. 2014). While mutation of XLF in mice resulted in radiation sensitivity, impaired V(D)J recombination, and lower levels of lymphocytes (Li et al. 2008b), work by others suggests that Xrcc4 and Lig4 mutants are the most severe in respect to loss of NHEJ (Karanjawala et al. 2002). While in mice the absence of Lig4 results in embryonic death (Karanjawala et al. 2002), Lig4 mutants in Arabidopsis show a 3- to 4-fold increase in HDR (QI et al. 2013), and Lig4 Drosophila mutants are viable and produce fertile off-spring with rates of HDR higher than 70 %, where they had previously been less than 15 % (Beumer et al. 2008). Xrcc4 orthologs could not be identified in either Ae. aegypti or An. gambiae via homology (Blastp) or domain-based (HMMER) searches (not shown), while Lig4 appears to have been duplicated in Ae. aegypti. Other proteins such as APLF, APTX, and PNKP are also involved in NHEJ-based repair. APLF is believed to play a role as a scaffolding protein (Grundy et al. 2013) as well as an exonuclease (Li etal. 2011); APTX has been shown to remove AMP from DNA ends (Clements et al. 2004), while PNKP removes and replaces non-ligatable groups from the DSB allowing for Lig4 to complete its function (Weinfeld et al. 2011). While the end joining of this highly complex repair system is believed to be error prone in nature, recent studies suggest that it may be much more faithful than traditionally thought (Betermier et al. 2014).
Fig. 1.
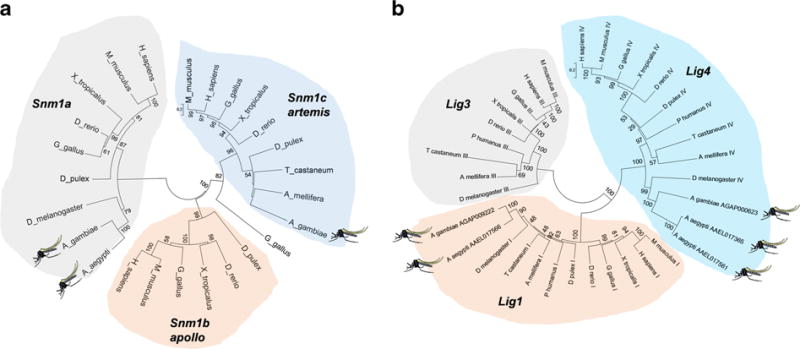
Gain and loss of NHEJ components in mosquitoes. Neighboring-joining tree produced from a clustalW alignment of Smn1-family proteins (a) or DNA ligases (b) using MEGA6 [118]. Bootstrap support (2000 replicates) is indicated on each branch if over 50 %. Abbreviations: Homo sapiens (H_sapiens), Mus musculus (M_musculus), Gallus gallus (G_gallus), Xenopus tropicalis (X_tropicalis), Danio rerio (D_rerio), Daphnia pulex (D_pulex), Pediculus humanus (P_humanus), Tribolium castaneum (T_castaneum), Apis mellifera (A_mellifera), Drosophila melanogaster (D_melanogaster), Anopheles gambiae (A_gambiae), Aedes aegypti (A_aegypti). Mosquito species are indicated with an icon
In addition to C-NHEJ, DSBs may be repaired by a Ku-independent mechanism, termed alternative non-homologous end joining (A-NHEJ). Unlike C-NHEJ, A-NHEJ is suspected to be highly error prone (Betermier et al. 2014; Deriano and Roth 2013). Like the C-NHEJ pathway, A-NHEJ ligates two broken ends of a DSB together, largely without the use of a homologous template. Unlike C-NHEJ, which initiates at the step of binding free-DNA ends, A-NHEJ appears to proceed after initiation of repair by the HDR resection machinery, with the use of the MRN complex, in conjunction with Sae2, to remove undesired nucleotides (Truong et al. 2013). Once resection has initiated, microhomology is utilized in the absence of a competing pathway to ligate the broken ends in an error-prone manner (Soni et al. 2014). The protein PARP1 competes with Ku proteins for binding of the DSB, as well as possibly recruiting other proteins to the break (Wang et al. 2006), tethering the DNA strands (Chiruvella et al. 2013) and possibly mediating translocations (Soni et al. 2014; Simsek and Jasin 2010). The primary ligase involved in this process is believed to be Lig3, which operates in conjunction with Xrcc1 (Oh et al. 2014). However, removal of Xrcc1 via mutation in hamster cells does not stop Lig3 from functioning, instead increasing its susceptibility to competition by Lig1 (Soni et al. 2014). Notably, Lig3 has been lost in mosquitoes, suggesting that Lig1 may be critical for A-NHEJ in these species (Fig. 1b and Table 1).
While repair pathways have been studied in model organisms [reviewed in Lamarche et al. (2010), Williams et al. (2014), and Chiruvella et al. (2013)], little is known about these processes in mosquitoes, which diverged from Drosophila approximately 240 Mya. While the majority of DNA break repair proteins appear to be conserved in flies and mosquitoes, there are some interesting anomalies, as noted above. While models of hierarchical repair have been proposed in model organisms (Mansour et al. 2008), similar models must be developed and tested in mosquitoes (Fig. 2). A strong grasp of how pathways are chosen will facilitate better use of endonuclease tools and more confident interpretation of experimental results. How can the various proteins involved in these pathways be manipulated to achieve the experimenter’s desired results? The suppression of end-joining repair pathways to increase repair via a homologous template has been accomplished in both flies and silkworm (Beumer et al. 2008; Ma et al. 2014), suggesting that similar results could be achieved in mosquitoes. Alternatively, disabling both HDR and C-NHEJ could potentially increase rates of targeted deletions, favoring the generation of gross repair errors such as translocations and inversions. A more complete understanding of the processes that determine the hierarchy of DNA repair pathways will allow the vector biologist to take full advantage of tools such as CRISPR/Cas9 in the most efficient manner possible.
Fig. 2.
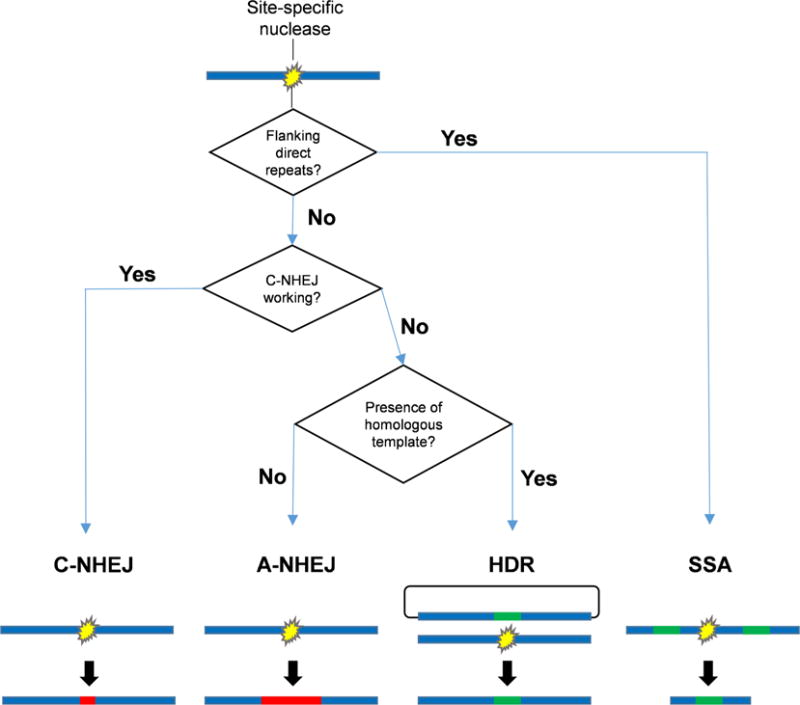
Potential hierarchy of DSB repair pathways in mosquitoes. Simplified flowchart of possible repair outcomes following DSB induction in the mosquito genome. Both known (proximity and length of repeat sequences, microhomologies, cell cycle phase, developmental stage, chromosomal organization, etc.) and unknown variables may contribute to each decision fork
Chromosomal structure and organization in mosquito genome manipulation
Manipulating the various DNA repair pathways, whether by increasing HDR with Lig4 or Ku70 knockouts (Beumer et al. 2008; Ma et al. 2014; QI et al. 2013) or increasing SSA by removing Rad51 or Rad54 (Wei and Rong 2007) may help generate favorable repair outcomes; however, other factors such as the proteins associated with DNA packaging, as well as sequence content, may also play a role in DNA repair pathway choice. Chromosomal DNA, packed into nucleosomes and wound around a histone octomer, can be generally characterized as either euchromatin or heterochromatin. Euchromatin is typically gene rich, less condensed, and transcriptionally accessible, while heterochromatin is gene poor, highly condensed, and transcriptionally repressed (Tamaru 2010). The highly condensed nature of heterochromatin has been shown to impact repair time and the recruitment of repair proteins such as Ku70 and DNA-PKcs (Lorat et al. 2012). The physical barrier presented by a more condensed heterochromatin may require the selection of a more advantageous target or the use of chemical agents known to relax chromatin structure, such as sodium butyrate or chloroquine (Murr et al. 2006). The existence of chromosome remodeling complexes such as TIP60 acetyltransferase, which acetylates histone H4 permitting the repair of DSBs [reviewed in Price and D’Andrea (2013)], may be manipulated to influence chromatin structure and DNA repair choice. Deletion of TRRAP, a component of the TIP60 remodeling complex, in murine cells leads to a 2-fold reduction in homologous recombination (Murr et al. 2006). Could over-expression of TRRAP lead to an increase in HDR? What other chromosome remodeling complexes can be modulated to impact DNA repair pathway choice?
In addition to the physical obstructions that chromatin proteins may induce, the organization of the chromosome itself also may have a role in the choice of DNA repair pathways. The An. gambiae genome is organized with pericentric and intercalary heterochromatic regions, which are gene poor, transposable element (TE) rich, and highly condensed (Sharakhova etal. 2010). The Ae. aegypti genome is much larger (~1.4 Gbp), with the typical heterochromatic versus euchromatic demarcation clouded by the presence of large regions of short interspaced repeats (Nene et al. 2007). Gene models in Ae. aegypti have expanded intronic regions (nearly four times as long as those of An. gambiae), with repetitive elements distributed throughout; such repeats and TEs make up roughly 50 % of the genome (Nene et al. 2007; Severson and Behura 2012). Because SSA-based repair requires flanking repeat sequences (Ivanov et al. 1996) and A-NHEJ appears to utilize microhomology (Truong et al. 2013), the content and organization of repetitive elements around the selected target site may influence the choice of DNA repair pathway. Ae. aegypti, which has proportionally more repetitive elements in its genome than does An. gambiae, may thus be more likely to use the SSA repair pathway for DNA break repair; we have shown previously that SSA competes favorably with NHEJ when direct repeats are present (Aryan et al. 2013a). Considering the expansion of interspersed repetitive elements in most gene models of Ae. aegypti, achieving efficient levels of HDR in this organism may be a challenge due to difficulty identifying unique sequences of sufficient length flanking a DSB site. Initial experiments have used 1–2 kb of homologous sequence on either side of the break (Liesch et al. 2013; McMeniman et al. 2014), but a thorough characterization of minimum requirements for HDR would be beneficial.
Final conclusions
Site-specific nucleases generate controlled DSBs (Carroll 2014; Kim and Kim 2014), which may be selectively used to generate deletions, insertions, inversions, and translocations (van der Weyden and Bradley 2006). While both the C-NHEJ and A-NHEJ pathways can produce deletions (Aryan et al. 2013b; Smidler et al. 2013), the contribution of each of these to perfect repair (undesired) and error-prone repair (desired) remains to be firmly established. Currently, insertion of transgenic constructs into the mosquito genome is a random process associated with the use of transposable elements (Adelman et al. 2002; Kokoza and Raikhel 2011). While site-specific integration based on HDR has been accomplished in mosquitoes (Liesch et al. 2013; McMeniman et al. 2014), efficiencies were less than 1 %. A more efficient system for insertion of a target sequence via HDR would be highly advantageous and represents a critical barrier in mosquito chromosomal engineering. Lastly, the SSA pathway may be utilized opportunistically to remove genetic information such as individual exons or entire genes when located between homologous direct repeats.
Targeted inversions have been generated in human cells via the NHEJ pathway using pairs of ZFNs targeting different sites (Lee et al. 2012); similar results were obtained in Drosophila using HEs (Egli et al. 2004). Inversions in mosquitoes, such as the 2La inversion in An. gambiae, have been suggested to confer an advantage to the organism in arid environments (White et al. 2009). Targeted inversions at various locations could be used to confirm phenotypic effects of suspected naturally occurring inversions, or the creation of novel inversion phenotypes. In Drosophila and mice, targeted inversions have been used in the creation of balancer chromosomes that are resistant to homologous recombination (Casso et al. 2000; Zheng et al. 1999). The creation of balancer chromosomes in mosquitoes would simplify screening (by fluorescent makers and homozygous lethal genes) of genetically engineered organisms and reduce rates of homologous recombination stabilizing introduced mutations. These technologies would also facilitate high-throughput methods of screening for the identification of genetic mutants and phenotypic traits. Translocations have been generated with nucleases in both Drosophila and human cells (Egli et al. 2004; Piganeau et al. 2013). Perhaps by upregulating components of the A-NHEJ pathway, which is suspected to play a role in translocations (Soni et al. 2014), targeted translocations may be generated to produce novel meiotic gene drive systems (Pearson and Wood 1980). Mosquitoes carrying a targeted translocation could be used in typical SIT fashion for population reduction through the release of males only, or could be used to drive population replacement by the continued release of both males and females. This process could also be reversible with the release of wild-type males and females.
The use of site-specific nucleases in important mosquito vector species is still an emerging field. How these tools work in different mosquito species, as well as the hierarchy of mosquito DNA repair pathways, requires further investigation. Additional inquiries into topics such as the impact of target site location on nuclease selection, the modulation of chromatin protein complexes with respect to DNA repair pathways, and the impact of DNA repair pathway choice on the desired engineering event are required to advance mosquito chromosomal engineering. A firm understanding of how DNA repair pathways can be manipulated will hasten the development of more effective vector control techniques.
Acknowledgments
The project described was supported by grants [AI085091, AI099843] from NIAID, and its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID.
Abbreviations
- A-NHEJ
Alternative non-homologous end joining
- C-NHEJ
Classical non-homologous end joining
- CRISPR
Clustered regulatory interspaced short palindromic repeats
- DSB
Double-stranded break
- HE
Homing endonuclease
- HDR
Homology-directed repair
- HRMA
High-resolution melt curve analysis
- RIDL
Release of insects with dominant lethality
- SIT
Sterile insect technique
- SSA
Single-strand annealing
- TALEN
Transcription activator-like effector nuclease
- TE
Transposable element
- ZFN
Zinc finger nuclease
Footnotes
Responsible Editors: Natalay Kouprina and Vladimir Larionov
References
- CDC and Malaria. The Centers for Disease Control and Prevention. 2014a http://www.cdc.gov/malaria/resources/pdf/fsp/cdc_malaria_program.pdf.
- Dengue and severe dengue. World Health Organization. 2014b http://www.who.int/mediacentre/factsheets/fs117/en/
- Dengue and the Aedes aegypti mosquito The. Centers for Disease Control and Prevention. 2014c http://www.cdc.gov/dengue/resources/30Jan2012/aegyptifactsheet.pdf.
- Malaria. World Health Organization. 2014d http://www.who.int/mediacentre/factsheets/fs094/en/
- Adelman ZN, Jasinskiene N, James AA. Development and applications of transgenesis in the yellow fever mosquito, Aedes aegypti. Mol Biochem Parasitol. 2002;121:1–10. doi: 10.1016/s0166-6851(02)00028-2. [DOI] [PubMed] [Google Scholar]
- Alphey L, Benedict M, Bellini R, Clark G, Dame D, Service M, Dobson S. Sterile-insect methods for control of mosquito-borne diseases: an analysis. Vector Borne Zoonotic Dis. 2010;10:295–311. doi: 10.1089/vbz.2009.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryan A, Anderson MA, Myles KM, Adelman ZN. Germline excision of transgenes in Aedes aegypti by homing endonucleases. Sci Rep. 2013a;3:1603. doi: 10.1038/srep01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryan A, Anderson MA, Myles KM, Adelman ZN. TALEN-based gene disruption in the dengue vector Aedes aegypti. PLoS One. 2013b;8:e60082. doi: 10.1371/journal.pone.0060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–5. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Liu JL. CRISPR/Cas9 and genome editing in Drosophila. J Genet Genomics. 2014;41:7–19. doi: 10.1016/j.jgg.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–8. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu JL. Mutagenesis and homologous recombination in Drosophila cell lines using CRISPR/Cas9. Biol Open. 2014;3:42–9. doi: 10.1242/bio.20137120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardini F, Galizi R, Menichelli M, Papathanos PA, Dritsou V, Marois E, Crisanti A, Windbichler N. Site-specific genetic engineering of the Anopheles gambiae Y chromosome. Proc Natl Acad Sci U S A. 2014;111:7600–5. doi: 10.1073/pnas.1404996111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betermier M, Bertrand P, Lopez BS. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 2014;10:e1004086. doi: 10.1371/journal.pgen.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer KJ, Trautman JK, Bozas A, Liu JL, Rutter J, Gall JG, Carroll D. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc Natl Acad Sci U S A. 2008;105:19821–6. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc Biol Sci. 2003;270:921–8. doi: 10.1098/rspb.2002.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A. Heritable strategies for controlling insect vectors of disease. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130432. doi: 10.1098/rstb.2013.0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson DF, Fahrenkrug SC, Hackett PB. Targeting DNA with fingers and talens. DNA Repair (Amst) 2012;1:e3. doi: 10.1038/mtna.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with targetable nucleases. Annu Rev Biochem. 2014;83:409–439. doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- Casso D, Ramirez-Weber F, Kornberg TB. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech Dev. 2000;91:451–4. doi: 10.1016/s0925-4773(00)00248-3. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Waise TM, Hassan F, Kabir Y, Smith MA, Arif M. Assessment of the evolutionary origin and possibility of CRISPR-Cas (CASS) mediated RNA interference pathway in Vibrio cholerae O395. Silicon Biol. 2009;9:245–54. [PubMed] [Google Scholar]
- Chhabra M, Mittal V, Bhattacharya D, Rana U, Lal S. Chikungunya fever: a re-emerging viral infection. Indian J Med Microbiol. 2008;26:5–12. doi: 10.4103/0255-0857.38850. [DOI] [PubMed] [Google Scholar]
- Chiruvella KK, Liang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol. 2013;5:a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciapponi L, Cenci G, Ducau J, Flores C, Johnson-Schlitz D, Gorski MM, Engels WR, Gatti M. The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr Biol. 2004;14:1360–6. doi: 10.1016/j.cub.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Clements PM, Breslin C, Deeks ED, Byrd PJ, Ju L, Bieganowski P, Brenner C, Moreira MC, Taylor AM, Caldecott KW. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair (Amst) 2004;3:1493–502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Colpitts TM, Cox J, Vanlandingham DL, Feitosa FM, Cheng G, Kurscheid S, Wang P, Krishnan MN, Higgs S, Fikrig E. Alterations in the Aedes aegypti transcriptome during infection with West Nile, dengue and yellow fever viruses. PLoS Pathog. 2011;7:e1002189. doi: 10.1371/journal.ppat.1002189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AA, Friedberg EC, Tomkinson AE, Wood RD, West SC. Role of the Rad1 and Rad10 proteins in nucleotide excision repair and recombination. J BiolChem. 1995;270:24638–41. doi: 10.1074/jbc.270.42.24638. [DOI] [PubMed] [Google Scholar]
- Degennaro M, Mcbride CS, Seeholzer L, Nakagawa T, Dennis EJ, Goldman C, Jasinskiene N, James AA, Vosshall LB. Orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature. 2013;498:487–491. doi: 10.1038/nature12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet. 2013;47:433–55. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- Drysdale R, Flybase C. FlyBase: a database for the Drosophila research community. Methods Mol Biol. 2008;420:45–59. doi: 10.1007/978-1-59745-583-1_3. [DOI] [PubMed] [Google Scholar]
- Egli D, Hafen E, Schaffner W. An efficient method to generate chromosomal rearrangements by targeted DNA double-strand breaks in Drosophila melanogaster. Genome Res. 2004;14:1382–93. doi: 10.1101/gr.2279804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G, Lees RS, Nimmo D, Aw D, Jin L, Gray P, Berendonk TU, White-Cooper H, Scaife S, Kim Phuc H, Marinotti O, Jasinskiene N, James AA, Alphey L. Female-specific flightless phenotype for mosquito control. Proc Natl A cad Sci USA. 2010;107:4550–4. doi: 10.1073/pnas.1000251107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galizi R, Doyle LA, Menichelli M, Bernardini F, Deredec A, Burt A, Stoddard BL, Windbichler N, Crisanti A. A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat Commun. 2014;5:3977. doi: 10.1038/ncomms4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub EI, Gupta RC, Haaf T, Wold MS, Radding CM. Interaction of human rad51 recombination protein with single-stranded DNA binding protein, RPA. Nucleic Acids Res. 1998;26:5388–93. doi: 10.1093/nar/26.23.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’connor-Giles KM. Highly specific and efficient CRISPR/Cas9–catalyzed homology-directed repair in Drosophila. Genetics. 2014;196:961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy GJ, Rulten SL, Zeng Z, Arribas-Bosacoma R, Iles N, Manley K, Oliver A, Caldecott KW. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 2013;32:112–25. doi: 10.1038/emboj.2012.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD. Recombination: Holliday junction resolution and crossover formation. Curr Biol. 2004;14:R56–8. doi: 10.1016/j.cub.2003.12.043. [DOI] [PubMed] [Google Scholar]
- Hill J, Lines J, Rowland M. Insecticide-treated nets. Adv Parasitol. 2006;61:77–128. doi: 10.1016/S0065-308X(05)61003-2. [DOI] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamgang B, Marcombe S, Chandre F, Nchoutpouen E, Nwane P, Etang J, Corbel V, Paupy C. Insecticide susceptibility of Aedes aegypti and Aedes albopictus in Central Africa. Parasitol Vectors. 2011;4:79. doi: 10.1186/1756-3305-4-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanjawala ZE, Adachi N, Irvine RA, Oh EK, Shibata D, Schwarz K, Hsieh CL, Lieber MR. The embryonic lethality in DNA ligase IV-deficient mice is rescued by deletion of Ku: implications for unifying the heterogeneous phenotypes of NHEJ mutants. DNA Repair (Amst) 2002;1:1017–26. doi: 10.1016/s1568-7864(02)00151-9. [DOI] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiianitsa K, Solinger JA, Heyer WD. Terminal association of Rad54 protein with the Rad51–dsDNA filament. Proc Natl Acad Sci U S A. 2006;103:9767–72. doi: 10.1073/pnas.0604240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim J-S. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Klein TA, Windbichler N, Deredec A, Burt A, Benedict MQ. Infertility resulting from transgenic I-PpoI male Anopheles gambiae in large cage trials. Pathog Glob Health. 2012;106:20–31. doi: 10.1179/2047773212Y.0000000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klovstad M, Abdu U, Schupbach T. Drosophila brca2 is required for mitotic and meiotic DNA repair and efficient activation of the meiotic recombination checkpoint. PLoS Genet. 2008;4:e31. doi: 10.1371/journal.pgen.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoza VA, Raikhel AS. Targeted gene expression in the transgenic Aedes aegypti using the binary Gal4–UAS system. Insect Biochem Mol Biol. 2011;41:637–44. doi: 10.1016/j.ibmb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé GMC, Scaife S, Morgan SA, Curtis ZH, Alphey L. Female-specific flightless (fsRIDL) phenotype for control of Aedes albopictus. PLoS Negl Trop Dis. 2012;6:e1724. doi: 10.1371/journal.pntd.0001724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarche BJ, Orazio NI, Weitzman MD. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010;584:3682–95. doi: 10.1016/j.febslet.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kweon J, Kim E, Kim S, Kim JS. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 2012;22:539–48. doi: 10.1101/gr.129635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Dong J, Pan X, Oum JH, Boeke JD, Lee SE. Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10–dependent processing of recombination intermediates. Mol Cell. 2008a;30:325–35. doi: 10.1016/j.molcel.2008.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Alt FW, Cheng HL, Brush JW, Goff PH, Murphy MM, Franco S, Zhang Y, Zha S. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in v(d)j recombination. Mol Cell. 2008b;31:631–40. doi: 10.1016/j.molcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Kanno S, Watanabe R, Ogiwara H, Kohno T, Watanabe G, Yasui A, Lieber MR. Polynucleotide kinase and aprataxin-like forkhead-associated protein (PALF) acts as both a single-stranded DNA endonuclease and a single-stranded DNA 3′ exonuclease and can participate in DNA end joining in a biochemical system. J Biol Chem. 2011;286:36368–77. doi: 10.1074/jbc.M111.287797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesch J, Bellani LL, Vosshall LB. Functional and genetic characterization of neuropeptide Y-like receptors in Aedes aegypti. PLoS Negl Trop Dis. 2013;7:e2486. doi: 10.1371/journal.pntd.0002486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Fan XD. CRISPR-Cas system: a powerful tool for genome engineering. Plant Mol Biol. 2014;85:209–18. doi: 10.1007/s11103-014-0188-7. [DOI] [PubMed] [Google Scholar]
- Liu J, Heyer WD. Who’s who in human recombination: BRCA2 and RAD52. Proc Natl Acad Sci U S A. 2011;108:441–2. doi: 10.1073/pnas.1016614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorat Y, Schanz S, Schuler N, Wennemuth G, Rube C, Rube CE. Beyond repair foci: DNA double-strand break repair in euchromatic and heterochromatic compartments analyzed by transmission electron microscopy. PLoS One. 2012;7:e38165. doi: 10.1371/journal.pone.0038165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Ye AY, Zheng W, Kong L. A guide RNA sequence design platform for the CRISPR/Cas9 system for model organism genomes. Biomed Res Int. 2013;2013:270805. doi: 10.1155/2013/270805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Chang J, Wang X, Liu Y, Zhang J, Lu W, Gao J, Shi R, Zhao P, Xia Q. CRISPR/Cas9 mediated multiplex genome editing and heritable mutagenesis of BmKu70 in Bombyx mori. Sci Rep. 2014;4:4489. doi: 10.1038/srep04489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MA Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–94. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- Mahaney BL, Hammel M, Meek K, Tainer JA, Lees-Miller SP. XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair. Biochem Cell Biol. 2013;91:31–41. doi: 10.1139/bcb-2012-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013a;31:833–8. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013b;10:957–63. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour WY, Schumacher S, Rosskopf R, Rhein T, Schmidt-Petersen F, Gatzemeier F, Haag F, Borgmann K, Willers H, Dahm-Daphi J. Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks. Nucleic Acids Res. 2008;36:4088–98. doi: 10.1093/nar/gkn347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet-Bruneel B, Corre-Catelin N, Lacroix R, Lees RS, Hoang KP, Nimmo D, Alphey L, Reiter P. Fitness of transgenic mosquito Aedes aegypti males carrying a dominant lethal genetic system. PLoS One. 2013;8:e62711. doi: 10.1371/journal.pone.0062711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcmeniman CJ, CORFAS RA, Matthews BJ, Ritchie SA, Vosshall LB. Multimodal integration of carbon dioxide and other sensory cues drives mosquito attraction to humans. Cell. 2014;156:1060–71. doi: 10.1016/j.cell.2013.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcwhir J, Selfridge J, Harrison DJ, Squires S, Melton DW. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat Genet. 1993;5:217–24. doi: 10.1038/ng1193-217. [DOI] [PubMed] [Google Scholar]
- Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–9. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CF, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O’leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JM, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Fraser-Liggett CM, Severson DW. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–23. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, Ira G, Sung P. Mechanism of the ATP-dependent DNA end-resection machinery from saccharomyces cerevisiae. Nature. 2010;467:108–11. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Harvey A, Zimbric J, Wang Y, Nguyen T, Jackson PJ, Hendrickson EA. DNA ligase III and DNA ligase IV carry out genetically distinct forms of end joining in human somatic cells. DNA Repair (Amst) 2014;21:97–110. doi: 10.1016/j.dnarep.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson AM, Wood RJ. Combining the meiotic drive gene-d and the translocation-T1 in the mosquito, Aedes-Aegypti (L) .1. Sex-ratio distortion and fertility. Genetica. 1980;51:203–210. [Google Scholar]
- Pialoux G, Gauzere BA, Jaureguiberry S, Strobel M. Chikungunya, an epidemic arbovirosis. Lancet Infect Dis. 2007;7:319–27. doi: 10.1016/S1473-3099(07)70107-X. [DOI] [PubMed] [Google Scholar]
- Piganeau M, Ghezraoui H, de Cian A, Guittat L, Tomishima M, Perrouault L, Rene O, Katibah GE, Zhang L, Holmes MC, Doyon Y, Concordet JP, Giovannangeli C, Jasin M, Brunet E. Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res. 2013;23:1182–93. doi: 10.1101/gr.147314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluess B, Tanser FC, Lengeler C, Sharp BL. Indoor residual spraying for preventing malaria. Cochrane Database Syst Rev. 2010:Cd006657. doi: 10.1002/14651858.CD006657.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price BD, D’Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152:1344–54. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QI Y, Zhang Y, Zhang F, Baller JA, Cleland SC, Ryu Y, Starker CG, Voytas DF. Increasing frequencies of site-specific mutagenesis and gene targeting in Arabidopsis by manipulating DNA repair pathways. Genome Res. 2013;23:547–54. doi: 10.1101/gr.145557.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–55. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer M. Antimalarial drugs—what is in use and what is in the pipeline. Arch Pharm (Weinheim) 2008;341:149–63. doi: 10.1002/ardp.200700184. [DOI] [PubMed] [Google Scholar]
- Severson DW, Behura SK. Mosquito genomics: progress and challenges. Annu Rev Entomol. 2012;57:143–66. doi: 10.1146/annurev-ento-120710-100651. [DOI] [PubMed] [Google Scholar]
- Sharakhova MV, George P, Brusentsova IV, Leman SC, Bailey JA, Smith CD, Sharakhov IV. Genome mapping and characterization of the Anopheles gambiae heterochromatin. BMC Genomics. 2010;11:459. doi: 10.1186/1471-2164-11-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4–ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–6. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidler AL, Terenzi O, Soichot J, Levashina EA, Marois E. Targeted mutagenesis in the malaria mosquito using TALE nucleases. PLoS One. 2013;8:e74511. doi: 10.1371/journal.pone.0074511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni A, Siemann M, Grabos M, Murmann T, Pantelias GE, Iliakis G. Requirement for Parp-1 and DNA ligases 1 or 3 but not of Xrcc1 in chromosomal translocation formation by backup end joining. Nucleic Acids Res. 2014;42:6380–92. doi: 10.1093/nar/gku298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard BL. Homing endonuclease structure and function. Q Rev Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- Stoddard BL. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Paques F, Colaiacovo M, Haber JE. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci USA. 1997;94:9214–9. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Ira G, Haber JE. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol Cell Biol. 2000;20:5300–9. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H. Confining euchromatin/heterochromatin territory: jumonji crosses the line. Genes Dev. 2010;24:1465–78. doi: 10.1101/gad.1941010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Shinkura R, Shinkura N, Alt FW. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol Cell Biol. 2004;24:1200–5. doi: 10.1128/MCB.24.3.1200-1205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver BE, Anderson MA, Adelman ZN. Homing endonucleases catalyze double-stranded DNA breaks and somatic transgene excision in Aedes aegypti. Insect Mol Biol. 2009;18:623–33. doi: 10.1111/j.1365-2583.2009.00905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong LN, Li Y, Shi LZ, Hwang PY, He J, Wang H, Razavian N, Berns MW, Wu X. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci U S A. 2013;110:7720–5. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay SK, Sharma S. SSFinder: high throughput CRISPR-Cas target sites prediction tool. Biomed Res Int. 2014;2014:742482. doi: 10.1155/2014/742482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–46. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- van der Weyden L, Bradley A. Mouse chromosome engineering for modeling human disease. Annu Rev Genomics Hum Genet. 2006;7:247–76. doi: 10.1146/annurev.genom.7.080505.115741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, Mcmeniman CJ, Leong YS, Dong Y, Axford J, Kriesner P, Lloyd AL, Ritchie SA, O/’Neill SL, Hoffmann AA. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476:450–453. doi: 10.1038/nature10355. [DOI] [PubMed] [Google Scholar]
- Wang S, Jacobs-Lorena M. Genetic approaches to interfere with malaria transmission by vector mosquitoes. Trends Biotechnol. 2013;31:185–193. doi: 10.1016/j.tibtech.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lees-Miller SP. Detection and repair of ionizing radiation-induced DNA double strand breaks: new developments in nonhomologous end joining. Int J Radiat Oncol Biol Phys. 2013;86:440–449. doi: 10.1016/j.ijrobp.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–82. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei DS, Rong YS. A genetic screen for DNA double-strand break repair mutations in Drosophila. Genetics. 2007;177:63–77. doi: 10.1534/genetics.107.077693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JN. Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem Sci. 2011;36:262–71. doi: 10.1016/j.tibs.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, Cheng C, Sangare D, Lobo NF, Collins FH, Besansky NJ. The population genomics of trans-specific inversion polymorphisms in Anopheles gambiae. Genetics. 2009;183:275–88. doi: 10.1534/genetics.109.105817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GJ, Hammel M, Radhakrishnan SK, Ramsden D, Lees-Miller SP, Tainer JA. Structural insights into NHEJ: building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Repair (Amst) 2014;17:110–20. doi: 10.1016/j.dnarep.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windbichler N, Papathanos PA, Catteruccia F, Ranson H, Burt A, Crisanti A. Homing endonuclease mediated gene targeting in Anopheles gambiae cells and embryos. Nucleic Acids Res. 2007;35:5922–33. doi: 10.1093/nar/gkm632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windbichler N, Papathanos PA, Crisanti A. Targeting the X chromosome during spermatogenesis induces Y chromosome transmission ratio distortion and early dominant embryo lethality in Anopheles gambiae. PLoS Genet. 2008;4:e1000291. doi: 10.1371/journal.pgen.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windbichler N, Menichelli M, Papathanos PA, Thyme SB, Li H, Ulge UY, Hovde BT, Baker D, Monnat RJ, Jr, Burt A, Crisanti A. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature. 2011;473:212–5. doi: 10.1038/nature09937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Shen B, Zhang C, Huang X, Zhang Y. sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One. 2014;9:e100448. doi: 10.1371/journal.pone.0100448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. Characterization of Drosophila Rad51/SpnA protein in DNA binding and embryonic development. Biochem Biophys Res Commun. 2006;348:1310–8. doi: 10.1016/j.bbrc.2006.07.211. [DOI] [PubMed] [Google Scholar]
- Yoo S, Mckee BD. Overexpression of Drosophila Rad51 protein (DmRad51) disrupts cell cycle progression and leads to apoptosis. Chromosoma. 2004;113:92–101. doi: 10.1007/s00412-004-0300-x. [DOI] [PubMed] [Google Scholar]
- Yoo S, Mckee BD. Functional analysis of the Drosophila Rad51 gene (spn-A) in repair of DNA damage and meiotic chromosome segregation. DNA Repair (Amst) 2005;4:231–42. doi: 10.1016/j.dnarep.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Zheng B, Sage M, Cai WW, Thompson DM, Tavsanli BC, Cheah YC, Bradley A. Engineering a mouse balancer chromosome. Nat Genet. 1999;22:375–8. doi: 10.1038/11949. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–94. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]


