FlatScope, a lensless microscope as thin as a credit card and small enough to sit on a fingertip, captures 3D fluorescence images.
Abstract
Modern biology increasingly relies on fluorescence microscopy, which is driving demand for smaller, lighter, and cheaper microscopes. However, traditional microscope architectures suffer from a fundamental trade-off: As lenses become smaller, they must either collect less light or image a smaller field of view. To break this fundamental trade-off between device size and performance, we present a new concept for three-dimensional (3D) fluorescence imaging that replaces lenses with an optimized amplitude mask placed a few hundred micrometers above the sensor and an efficient algorithm that can convert a single frame of captured sensor data into high-resolution 3D images. The result is FlatScope: perhaps the world’s tiniest and lightest microscope. FlatScope is a lensless microscope that is scarcely larger than an image sensor (roughly 0.2 g in weight and less than 1 mm thick) and yet able to produce micrometer-resolution, high–frame rate, 3D fluorescence movies covering a total volume of several cubic millimeters. The ability of FlatScope to reconstruct full 3D images from a single frame of captured sensor data allows us to image 3D volumes roughly 40,000 times faster than a laser scanning confocal microscope while providing comparable resolution. We envision that this new flat fluorescence microscopy paradigm will lead to implantable endoscopes that minimize tissue damage, arrays of imagers that cover large areas, and bendable, flexible microscopes that conform to complex topographies.
INTRODUCTION
Revolutionary advances in fabrication technologies for lenses and image sensors have resulted in the remarkable miniaturization of imaging systems and opened up a wide variety of novel applications. Examples include lightweight cameras for mobile phones and other consumer electronics, lab-on-chip technologies (1), endoscopes small enough to be swallowed or inserted arthroscopically (2), and surgically implanted fluorescence microscopes only a few centimeters thick that can be mounted onto the head of a mouse or other small animals (3). More recently, lenses themselves have been miniaturized and made extremely thin using diffractive optics (4, 5), metamaterials (6), and foveated imaging systems (7).
Notwithstanding this tremendous progress, today’s miniature cameras and microscopes remain constrained by the fundamental trade-offs of lens-based imaging systems. In particular, the maximum field of view (FOV), resolution, and light collection efficiency are all determined by the size of the lens(es). To further miniaturize microscopes while maintaining high performance, we must step outside the lens-based imaging paradigm.
Computational imaging has emerged as a powerful framework for overcoming the limitations of physical optics and realizing compact imaging systems with superior performance capabilities (8, 9). Generally speaking, computational imaging systems use algorithms that relax the constraints on the imaging hardware. For example, superresolution microscopes such as PALM and STORM use computation to overcome the physical limitations of the imager’s lenses (10, 11).
At the extreme, computation can be used to eliminate lenses altogether and break free of the traditional design constraints of physical optics. The main principle of “lensless imaging” is to design complex but invertible transfer functions between the incident light field and the sensor measurements (12–19). The acquired sensor measurements no longer constitute an image in the conventional sense, but rather data that can be coupled with an appropriate inverse algorithm to reconstruct a focused image of the scene. This redefinition of the imaging problem significantly expands the design space and enables compact yet high-performance imaging systems. For example, amplitude or phase masks placed on top of a bare conventional image sensor combined with computational image reconstruction algorithms enable inexpensive and compact photography (13–15, 20).
Lensless computational imaging has also enabled extremely lightweight and compact microscopes. For example, pioneering work has demonstrated that a bare image sensor coupled with a spacer layer and a recovery algorithm is a powerful tool for shadow imaging (1), holography (21, 22), and fluorescence (23, 24) with applications in point-of-care diagnostics and high-throughput screening. A major advantage of lensless microscopy is its ability to substantially increase the FOV. In a lens-based microscope, the FOV is inversely proportional to the magnification squared, whereas in a lens-free system the FOV is limited only by the area of the imaging sensor (25).
Despite the recent advances in ultracompact lensless microscopy, one key application area has remained infeasible: micrometer-resolution three-dimensional (3D) imaging of incoherent sources (for example, fluorescence) over volumes spanning several cubic millimeters. This application area is particularly relevant for fluorescence imaging of biological samples both in vitro and in vivo, where the illumination sources and image sensors must often be on the same side of the imaging target. The major challenge in 3D imaging of these incoherent sources is that in the absence of lenses, the incoherent point spread function lacks the high-frequency spatial information necessary for high-quality image reconstruction. Although coherent sources can exploit the high-contrast interference fringes that carry high-frequency spatial information, the absence of interference fringes when imaging incoherent sources makes direct extensions of holographic methods infeasible without additional optical elements (26).
Using a novel computational algorithm and optimized amplitude mask, we demonstrate the first 3D lensless microscopy technique that does not rely on optical coherence. The result is the world’s thinnest fluorescence microscope (less than 1 mm thick) that is capable of micrometer resolution over a volume of several cubic millimeters (Fig. 1). The major innovation that enables the “FlatScope” is an optimized 2D array of apertures (or amplitude mask) that modulates the incoherent point spread function, which makes high-frequency spatial information recoverable. In addition, the amplitude mask for FlatScope is designed to reduce the complexity of the image reconstruction algorithm such that the image can be recovered from the sensor measurements in near real time. When an arbitrary amplitude mask is placed atop an N × N pixel image sensor, the transfer function relating the unknown image to the sensor measurements is an N2 × N2 matrix containing O(N4) entries (for example, a 1-megapixel image sensor produces a matrix with ~1012 elements). The massive size of this matrix leads to two major impracticalities: First, calibrating such a microscope would require the estimation of O(N4) parameters, and second, reconstructing an image would require a matrix inversion involving roughly O(N6) computational complexity.
Fig. 1. Traditional microscope versus FlatScope.
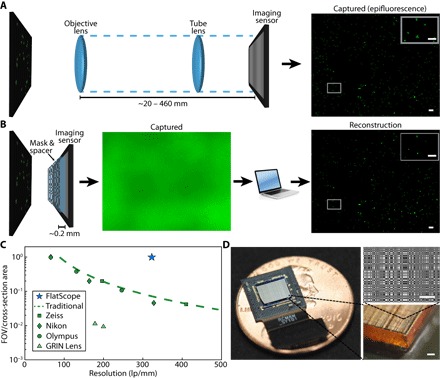
(A) Traditional microscopes capture the scene through an objective and tube lens (~20 to 460 mm), resulting in a quality image directly on the imaging sensor. (B) FlatScope captures the scene through an amplitude mask and spacer (~0.2 mm) and computationally reconstructs the image. Scale bars, 100 μm (inset, 50 μm). (C) Comparison of form factor and resolution for traditional lensed research microscopes, GRIN lens microscope, and FlatScope. FlatScope achieves high-resolution imaging while maintaining a large ratio of FOV relative to the cross-sectional area of the device (see Materials and Methods for elaboration). Microscope objectives are Olympus MPlanFL N (1.25×/2.5×/5×, NA = 0.04/0.08/0.15), Nikon Apochromat (1×/2×/4×, NA = 0.04/0.1/0.2), and Zeiss Fluar (2.5×/5×, NA = 0.12/0.25). (D) FlatScope prototype (shown without absorptive filter). Scale bars, 100 μm.
FlatScope greatly simplifies both the calibration and image reconstruction process by using a separable mask pattern composed of the outer product of two 1D functions. These separable masks have been shown to result in computational tractability (14, 19). In the special case of FlatScope with a separable mask, the local spatially varying point spread function (Fig. 2C) can be decomposed into two independent, separable terms: The first term models the effect of a hypothetical “open” mask (with no apertures) (Fig. 2, D and F), and the second term models the effect due to the coding of the mask pattern (Fig. 2, E and G). We call this superposition of two separable functions the “Texas Two-Step (T2S) model.”
Fig. 2. T2S model.
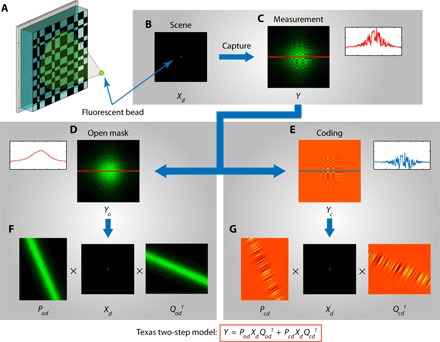
(A) Illustration of the FlatScope model using a single fluorescent bead as the scene. (B) Fluorescent point source at depth d, represented as input image Xd. (C) FlatScope measurement Y. FlatScope measurement can be decomposed as a superposition of two patterns: (D) pattern when there is no mask in place (open) and (E) pattern due to the coding of the mask. Each of the patterns is separable along x and y directions and can be written as (F and G) two separable transfer functions. The FlatScope model, which we call as the T2S, is the superposition of the two separable transfer functions.
For a 2D (planar) sample Xd at depth d, we show in section S1 that the FlatScope measurements Y satisfy
| (1) |
where Pod and Pcd operate only on the rows of Xd, and Qod and Qcd operate only on the columns of Xd (the subscripts o and c refer to “open” and “coding,” respectively). The total number of parameters in Pod, Qod, Pcd, and Qcd is O(N2) instead of O(N4). Thus, calibration of a moderate-resolution FlatScope with a 1-megapixel sensor requires the estimation of only ~4 × 106 rather than 1012 elements, and image reconstruction requires roughly 109 instead of 1018 computations. For a 3D (volumetric) sample XD, we discretize it into a superposition of planar samples Xd at D different depths d to yield the measurements
| (2) |
Separability is critical in making lensless imaging practical. As we show in section S2, the T2S model reduces the memory requirements for our prototype from 6 terabytes to 21 megabytes (a reduction of five orders of magnitude) and the reconstruction run time from several weeks for a single 2D image to 15 min for a complete 3D volume.
Reconstructing a 3D volume from a single FlatScope measurement requires that the system be calibrated, meaning that we know the separable transfer functions {Pod, Qod, Pcd, Qcd}{d = 1,2 …D}. To estimate these transfer functions for a particular FlatScope, we capture images from a set of separable calibration patterns (fig. S1). Because the calibration patterns displayed are separable, each calibration image depends only on either the row operation matrices {Pod, Pcd}{d = 1,2 …D} or the column operation matrices {Qod, Qcd}{d = 1,2 …D}; this observation massively reduces the number of images required for calibration. Using a truncated singular value decomposition, we can then estimate the columns of Pod, Qod, Pcd, and Qcd (see section S3). We perform this one-time calibration procedure for each depth plane d independently (see Materials and Methods). Given a measurement Y and the separable calibration matrices {Pod, Qod, Pcd, Qcd}{d = 1,2 …D}, we solve a regularized least-squares reconstruction algorithm to recover either a 2D depth plane Xd or an entire 3D volume XD. The gradient steps for this optimization problem are computationally tractable because of the separability of the T2S model (see section S4).
RESULTS
Experimental evaluation
To evaluate FlatScope’s performance, we constructed several prototypes (see Materials and Methods). We describe the performance of a particular prototype based on a Sony IMX219 sensor with 2×2 pixel binning (using only the green pixels in a Bayer sensor array). We used a region of 1300×1000 binned pixels to create an effective 1.3-megapixel sensor with 2.24-μm pixels. The amplitude mask is a 2D modified uniformly redundant array (MURA) (27) designed with prime number 3329, where the mask’s smallest feature size is 3 μm. We show that MURA patterns are separable in section S5.
Lateral resolution
To test the lateral resolution of the FlatScope prototype, we used test patterns composed of closely spaced double-slit resolution targets in a chrome mask with varying line spacing. We found that the lateral resolution of our prototype is less than 2 μm. In Fig. 3, we show the FlatScope results with a target containing a slit gap of approximately 1.6 μm compared to a confocal image of the same target (Fig. 3A). The FlatScope images were captured at a distance of 150 μm (Fig. 3B) and resolve the gap (Fig. 3C). A comparable microscope would require an objective with an NA ~0.16, typically found in objectives with ×4 to ×5 magnification. Note that although a 4× objective on a traditional lens-based microscope with a similar active area sensor would provide a FOV of only 0.41 mm2, FlatScope markedly increases this FOV by more than 10 times to 6.52 mm2. These results show that FlatScope can produce high-resolution images while maintaining an ultrawide FOV. In addition, our computational algorithm allows for the incorporation of image statistics, enabling us to resolve features smaller than the size of the binned pixel, the minimum aperture on the mask, and the calibration line thickness (2.24, 3, and 5 μm, respectively, for this prototype).
Fig. 3. Resolution tests with the FlatScope prototype.
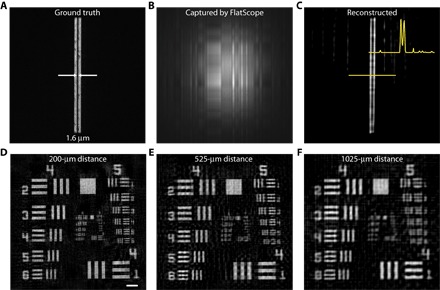
(A) Double slit with a 1.6-μm gap imaged with a 10× objective. (B) Captured FlatScope image. (C) FlatScope reconstruction of the double slit with a 1.6-μm gap. (D to F) FlatScope reconstructions of USAF resolution target at distances from the mask surface of 200 μm (D), 525 μm (E), and 1025 μm (F). Scale bar, 100 μm.
Computational depth scanning
Because depth is a free parameter in our reconstruction algorithm, FlatScope can reconstruct focused images at arbitrary distances from the prototype. To demonstrate this dynamic focusing, we captured images of a 1951 U.S. Air Force (USAF) target at distances ranging from 200 μm to ~1 mm. We reconstruct FlatScope images with no magnification, and therefore, angular resolution stays constant. As a result, we expect some lateral resolution degradation as distance increases but predict that the information captured through the mask (along with our reconstruction algorithm) will aid in maintaining high resolution through this range, especially when compared to imaging with no mask. The images reconstructed at the closest distance of 200 μm resulted in the best resolution (Fig. 3E), whereas at a distance of ~1 mm, line pairs in group 5 were still resolvable (Fig. 3G). These results confirm the capability of FlatScope to resolve images over a significant distance range while still maintaining high resolution.
When the depth of the sample is unknown, we can bring the image into focus by computationally scanning the reconstruction depth just like one would scan the focal plane of a conventional microscope. The difference with FlatScope is that the focal depth is selected after the data are captured. Thus, from a single frame of captured data, we can reconstruct a focused image without knowing the distance between the sample and the FlatScope a priori. To illustrate the computational focusing, we show, by simulation, that a resolution target has maximum contrast when the reconstruction depth matches the true distance between the target and the FlatScope (fig. S2 and movie S1).
We also compare, by simulation, the performance of imaging incoherent light sources of FlatScope versus two other lensless camera designs that share similar FOV and form factor advantages over traditional lens-based microscopes (fig. S3). Sencan et al. (28) have shown that using a bare image sensor without a mask is cost-effective for fluorescence lab-on-chip applications. To achieve high spatial resolution, sparsity constraints and close proximity to the bare sensor must be enforced so that the reconstruction (deconvolution) remains stable and tractable (29). The quality of images reconstructed from bare sensor images decays rapidly with increasing depth (fig. S3A). In contrast, the single separable model proposed by Asif et al. (14) breaks down as expected when the source distance approaches the device size, that is, in the regime where the point spread function of a point source is localized to a portion of the sensor (fig. S3C and section S6). In this regime, our new T2S model is a necessity for high-resolution reconstructions (fig. S3B).
3D volume reconstruction
In addition to the ability to reconstruct images at arbitrary distances, FlatScope is also able to reconstruct an entire 3D volume from a single image capture. To showcase this ability, we prepared a 3D sample by suspending 10-μm fluorescent beads in an agarose solution. We then captured a single image using FlatScope and reconstructed the entire 3D volume (Fig. 4, A and B). For ground truth, we captured the 3D fluorescence volume using a scanning confocal microscope (Fig. 4C). Comparing the FlatScope reconstruction to the confocal data over a depth range of >200 μm, we not only have sufficient lateral resolution to resolve the beads but also achieve an axial resolution comparable to that of the confocal microscope (Fig. 4D). Empirically, we find that the axial spread of the 10-μm beads is approximately 15 μm in the FlatScope reconstructions (Fig. 4D).
Fig. 4. 3D volume reconstruction of 10-μm fluorescent beads suspended in agarose.
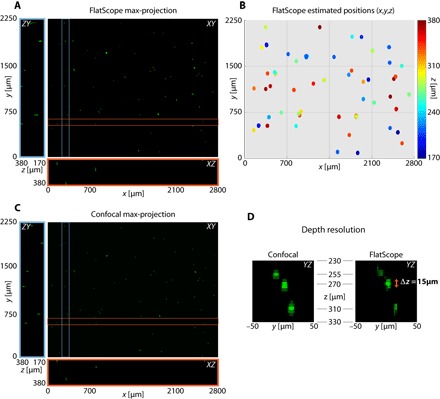
(A) FlatScope reconstruction as a maximum intensity projection along the z axis as well as a ZY slice (blue box) and an XZ slice (red box). (B) Estimated 3D positions of beads from the FlatScope reconstruction. (C) Ground truth data captured by confocal microscope (10× objective). (D) Depth profile of reconstructed beads compared to ground truth confocal images. Empirically, we can see that the axial spread of 10-μm beads is around 15 μm in FlatScope reconstruction. That is, FlatScope’s depth resolution is less than 15 μm. The three beads shown are at depths of 255, 270, and 310 μm from the top surface (filter) of the FlatScope.
Two important advantages of FlatScope’s ability to image complete 3D volumes from a single capture are data compression and speed. To obtain 3D volumes comparable to FlatScope, a confocal microscope must scan in both the lateral and axial dimensions, capturing a series of images one pixel at a time. As a result, a large amount of data must be collected. For example, to image the fluorescent beads in Fig. 4C, the confocal microscope had to overlay a total of 41 z-sections, with each z-section imaged at 4 megapixels, for a total of 164 recorded megavoxels. In contrast, FlatScope can achieve the same depth resolution with a single capture of 1.3 megapixels, which represents a 41× data compression (from depth alone). Moreover, the confocal data collection took more than 20 min, whereas FlatScope’s capture took 30 ms (a 40,000× speedup). It must be noted that whereas a confocal microscope can achieve diffraction-limited lateral resolution, the lateral resolution of FlatScope is limited to approximately 2 μm in our prototype because of the pixel size on the image sensor. The FlatScope resolution will improve as image sensor pixels scale down in size.
Despite the over 40-fold data compression ratio, we found that when we increased the density of beads by a factor of 10, we maintained better than 94% true positive rates (TPRs) in our image reconstructions. To quantify the error rates as a function of spatial sparsity of the sample, we captured images with FlatScope and reconstructed 3D volumes (fig. S4B), as well as captured ground truth images with a confocal microscope (fig. S4A) for multiple densities of 10-μm fluorescent beads in polydimethylsiloxane (PDMS). Counts of true positive (TP), false negatives (FN), and false positives (FP) were taken to determine the accuracy of the FlatScope reconstructions (fig. S4C). The TPR and false discovery rates (FDRs) were plotted, where TPR = TP/(TP + FN) and FDR = FP/(TP + FP). We observed a slight decrease in TPR as concentrations increase, but we are still able to maintain greater than 94% TPR at the highest concentration tested. FDR remains fairly constant at higher concentrations, indicating that errors in reconstruction for increasing sample size may not introduce additional false positives.
3D volumetric video
FlatScope’s radically reduced capture time enables 3D volumetric video capture at the native frame rate of the image sensor. To demonstrate this capability, we created an imaging target composed of a 3D microfluidic device with two channels separated axially by approximately 100 μm. Figure 5 shows a time lapse from a movie of a subsection of the FlatScope 3D volume reconstruction and corresponding still frames of the fluorescent beads as they travel through the two channels (full FOV, 18 frames per second captured video available as movie S2). In our experiments, FlatScope’s 3D volumetric imaging frame rate was constrained only by the native frame rate of the image sensor.
Fig. 5. 3D volumetric video reconstruction of moving 10-μm fluorescent beads.
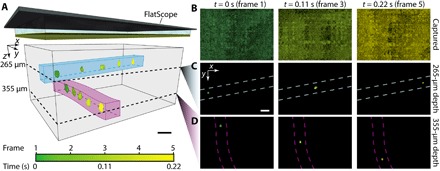
(A) Subsection of FlatScope time-lapse reconstruction of 3D volume with 10-μm beads flowing in microfluidic channels (approximate location of channels drawn to highlight bead path and depth). Scale bar, 50 μm (FlatScope prototype graphic at the top not to scale). (B) Captured images of frames 1, 3, and 5 (false-colored to match time progression). (C and D) FlatScope reconstructions of frames 1, 3, and 5 at estimated depths of 265 and 355 μm, respectively (dashed lines indicate approximate location of microfluidic channels). Reconstructed beads false-colored to match time progression. Scale bar, 50 μm.
DISCUSSION
We have demonstrated single-frame, wide-field 3D fluorescence microscopy with FlatScope over depths of hundreds of micrometers with the ability to refocus a single capture beyond 1 mm. FlatScope achieves this unprecedented performance without a lens and with all components (mask, spacer, and filter) residing in a layer less than 300 μm thick on top of a bare imaging sensor. This results in a remarkably compact form factor: Our FlatScope prototype features lateral dimensions of 8 mm × 8 mm, thickness below 600 μm, and weight of ~0.2 g. To our knowledge, no other compact lensless device is capable of 3D fluorescence imaging over hundreds of micrometers in depth. Here, we have quantified the performance of FlatScope using calibration samples, but important next steps include imaging of complex biological samples both in vitro and in vivo. One should also consider that although we have focused this work on fluorescence, the FlatScope concept may also be applied to bright-field, dark-field, and reflected-light microscopy.
The dynamic range of our FlatScope fluorescence images was degraded by the autofluorescence of the gel filter (see Materials and Methods), losing up to 64% of the sensor’s dynamic range due to autofluorescence of the filter. Using an alternative thin-film absorptive filter (30, 31), potentially in conjunction with an omnidirectional reflector (32, 33), we expect to substantially increase the dynamic range of FlatScope while reducing the prototype’s total thickness to less than 500 μm. This increase in dynamic range will enable improved imaging of biological samples with the goal of in vivo 3D fluorescence imaging. We are also planning to replace the external excitation source by integrating micro–light-emitting diodes (μLEDs) around the imaging sensor, thereby reducing the excitation light reaching the sensor and eliminating the need for additional filters. Fabrication advancements have led to μLEDs that are less than the thickness of our spacer layer, which will enable us to maintain our overall compact form factor with no significant increase in weight. Because the FOV of FlatScope is limited only by the sensor size, larger imaging sensors can easily be integrated for extreme wide-field imaging. In addition, the small footprint of FlatScope raises the exciting prospect of arraying FlatScopes on flexible substrates for conformal imaging. Overall, FlatScope opens up a new design space for 3D fluorescence microscopy, where size, weight, and performance are no longer determined by the optical properties of physical lenses.
MATERIALS AND METHODS
Fabrication of FlatScope
A 100-nm-thin film of chromium was deposited onto a 170-μm-thick fused silica glass wafer and then photolithographically patterned with Shipley S1805. The chromium was then etched, leaving the MURA pattern with a minimum feature size of 3 μm. The wafer was diced to slightly larger than the active area of the imaging sensor. The imaging sensor is Sony IMX219, which provides direct access to the surface of the bare sensor. The diced amplitude mask was aligned rotationally to the pixels of the imaging sensor under a microscope to enforce the separability under the T2S model and then epoxied to the sensor with Norland Optical Adhesive #72 using a flip-chip die bonder. To filter blue light, we used an absorptive filter (Kodak Wratten #12) cut to the size of the mask and attached using epoxy with a flip-chip die bonder in the same manner. The device was finally conformally coated with <1 μm of parylene for insulation. An overview of the fabrication process is shown in fig. S5.
Refractive index matching
The separability of the FlatScope model is based on light propagation through a homogeneous medium. A large change in refractive index across the target medium to the FlatScope interface (for example, air to glass) results in a mapping of lines in the scene to curves at the sensor plane. Because separability requires the preservation of rectangular features, the curving effect weakens the T2S model. A small change in refractive index (for example, water to glass) was observed to only minimally affect the model (fig. S6). For calibration and the experiments presented, a refractive index matching immersion oil (Cargille #50350) was used between the surface of the mask and the target.
Matching the refractive index used during calibration to the corresponding refractive index present during experiments can help further mitigate image degradation. In anticipation of experiments requiring different refractive index media, multiple calibrations can be done with a variety of refractive index media to produce a library of calibration matrices. A user would simply select the calibration matrices acquired using a refractive index medium that most closely matches the desired experiment. Because calibration is a one-time procedure, this would not affect the data capture or reconstruction time.
Calibration
To calibrate FlatScope, we used 5-μm-wide line slits fabricated in a 100-nm film of chromium on glass wafer and a LED array (green 5050 SMD) located ~10 cm below the line slit (fig. S1A). To ensure that the light passing through the calibration slit was representative of a group of mutually incoherent point sources (34), we placed a wide-angle diffuser (Luminit 80°) between the target and light source as shown in the figure. The diffuser helps to increase the angular spread of light emanating from the slit, thus mimicking an isotropic source and mitigating diffraction effects. Although FlatScope remained static, the calibration slit, diffuser, and LED array were translated with linear stages/stepper motors (Thorlabs LNR25ZFS/KST101) separately along the x and y axes (fig. S1, B and C). The horizontal and vertical slits were translated over the FOV of the FlatScope, determined by the acceptance profile of the pixels in the imaging sensor (fig. S7B). The full width at half maximum of our sensor’s pixel response profile is approximately 40°. The translation step distance of 2.5 μm was repeated at different depth planes ranging from 160 to 1025 μm (Thorlabs Z825B/TDC001), whereas a translation step distance of 1 μm was used for a single depth of 150 μm. This calibration needs to be performed only once and the calibrated matrices {Pod, Qod, Pcd, Qcd}{d = 1,2 …D} can be reused as long as the mask and sensor retain their relative positions (that is, not deformed).
Resolution measurements
Double slits were fabricated in a 100-nm film of chromium on a silica glass wafer. As with calibration, we used a LED array and wide-angle diffuser to illuminate the target. We captured ground truth images with a confocal microscope (Nikon Eclipse Ti/Nikon, CFI Plan Apo, 10× objective), measuring the width of the slits to be 4.3 μm with a gap of 1.6 μm. FlatScope images were captured at a distance of 150 μm with a 230-ms exposure; five images were averaged to increase the signal-to-noise ratio (SNR). FlatScope images of the 1951 USAF target (Thorlabs R1S1L1N) were captured with the same setup. Distances of 200, 525, and 1025 μm were captured by translating along the z axis (Thorlabs Z825B/TDC001). Exposure times were 24, 28, and 28 ms, respectively, and five images were averaged at each depth to increase the SNR.
Fluorescence measurements
A 2D sample was constructed by drop casting a 100-μl sample of 10-μm polystyrene microspheres (1.7 × 105 beads/ml, FluoSpheres yellow-green) onto a microscope slide and fixed with a standard coverslip (~170 μm thickness). A single image was captured with FlatScope 50 μm from the coverslip and using a 30-ms exposure (Fig. 1B). Epifluorescence images were captured using an Andor Zyla sCMOS camera and 4× objective (Nikon, Plan Fluor) (Fig. 1A).
The 3D sample was prepared with the fluorescent beads (3.6 × 104 beads/ml) in a 1% agarose solution. A 100-μl portion of the mixture was placed onto a well slide and fixed with a standard coverslip (~170 μm thickness). Images were captured by FlatScope identically to the 2D sample. Ground truth images of the 3D sample were captured with a depth range of 210 μm (and an area just larger than the FOV of the FlatScope prototype) using a confocal microscope (Nikon Eclipse Ti/Nikon, CFI Plan Apo, 10× objective). 3D samples with increasing density were made by spin-coating a mixture of PDMS (Sylgard, Dow Corning; 10:1 elastomer/cross-linker weight ratio) and fluorescent beads (concentrations ranging from 7.2 × 104 to 3.6 × 105 beads/ml) onto a SiO2 wafer at 500 rpm. Samples were cured at room temperature for a minimum of 24 hours. Ground truth images of the 3D PDMS samples were captured for the complete depths of the samples (average thickness of ~170 μm and an area just larger than the FOV of the FlatScope prototype) using a confocal microscope (Nikon Eclipse Ti/Nikon, CFI Plan Apo, 10× objective). The confocal imaging required scanning and stitching to match the FOV of FlatScope; z-axis measurements were captured every 5 μm.
FlatScope video of the flowing 10-μm beads (3.6 × 106 beads/ml) was captured for 640 × 480 pixels (2× 2 binned) at 18 frames per second at a distance of ~100 μm from the coverslip (~150 μm thickness) on which the microfluidic device was mounted. The resolution and capture speed were limited by the sensor. The microfluidic channels had approximate dimensions of 50 μm × 40 μm, with an axial separation of the channels of ~100 μm. The excitation light for all fluorescence images captured by FlatScope was provided by a 470-nm LED (Thorlabs M470L3), with filter (BrightLine Basic 469/35) focused on the beads at an angle of ~60°.
Microfluidic device
The microfluidic device was composed of three PDMS layers (two flow layers and one insertion layer) bonded to each other using an O2 plasma treatment. Individual flow layers were fabricated following standard soft lithography techniques. Briefly, Si wafers were spin-coated with photoresist (SU-8 2050, MicroChem), followed by a photolithography process that defined the channels. For the flow layers, ~70-μm-thick layers of PDMS (Sylgard, Dow Corning; 10:1 elastomer/cross-linker weight ratio) were spin-coated on the patterned Si wafer and cured in an oven at 90°C for 2 hours. The insertion layer (~4 mm thick) was fabricated by pouring PDMS on a blank Si wafer and cured in an oven at 90°C for 2 hours. Next, the PDMS layers were removed from the wafer and subjected to a plasma treatment (O2, 320 mtorr, 29.6 W, 30 s). The insertion layer was cut for port placement at either end of the flow layers. Layers were manually aligned and gently pressed together to promote bonding and mounted on cover glass (~150 μm thickness). A final 10-min bake at 90°C resulted in a multilayer device with a strong covalent bond between layers.
Aberration removal before reconstruction
In practice, the FlatScope prototype could suffer from the following unwanted artifacts: dead or saturated pixels on the image sensor, dust trapped beneath the spacer, and air bubbles trapped in epoxy. We refer to these artifacts, generally, as “aberrations.” Aberrations do not fit into the T2S model because they are not separable and are invariant to the scene/sample. Hence, aberrations act as strong noise in localized regions and result in erroneous reconstruction around these regions. To correct aberrations, it is not sufficient to subtract them from the measurements; we also need to fill in the appropriate regions with correct values. To achieve the correction, we observed that our captured images are fairly low-dimensional (low rank when the captured image is considered a matrix), and aberrations occur as sparse outliers. Robust principal component analysis (RPCA) (35, 36) is an effective algorithm to separate such sparse outliers from an inherently low-dimensional matrix. We used RPCA as a pre-reconstruction processing step to replace aberrations with aberration-corrected sensor values in the captured image (see fig. S8 for an experimental example).
Filter autofluorescence compensation
The Kodak Wratten filter autofluorescence induced a significant DC shift in the captured images. Given the limited dynamic range of the sensor, this led to contrast loss. We could remove the DC shift by subtracting the mean of the captured image; however, the filter did not autofluoresce uniformly, invalidating an exact DC shift assumption. Because the brightness profile due to autofluorescence was of spatially low frequency, we subtracted the lower-frequency components as obtained by a discrete cosine transform decomposition of the image. Subtracting the lower-frequency components eliminated almost all of the autofluorescence (fig. S9, A and B). We note that up to 64% of the dynamic range of the sensor was lost to autofluorescence, which limited the performance of FlatScope by reducing the signal strength. Despite this limitation, we were able to show high-quality image reconstructions (fig. S9, C and D). Shifting to a thinner and less autofluorescent filter will improve the performance of FlatScope.
Reconstruction algorithms
To be robust to various sources of noise, we formulated the reconstruction problem as a regularized least-squares minimization. The regularization was chosen on the basis of the scene. For extended scenes like the USAF resolution target, we used Tikhonov regularization. For a given depth d and calibrated matrices Pod, Qod, Pcd, and Qcd, we estimated the scene by solving a Tikhonov regularized least-squares problem
| (3) |
For sparse scenes like the double slit, we solved the Lasso problem
| (4) |
For the fluorescent samples, we solved the 3D reconstruction problem as a Lasso problem
| (5) |
Iterative techniques and rudimentary graphics processing unit (GPU) implementations were developed in Matlab to solve all the above optimization problems. Equation 3 was solved using Nesterov’s gradient method (37), whereas Eqs. 4 and 5 were solved using FISTA (fast iterative shrinkage-thresholding algorithm) (38). The gradient steps for the optimization problems are shown in section S4. As expected, the longest running time was taken by the 3D deconvolution problem, with the solution converging in under 15 min.
Calculations of resolution, FOV, and cross-section area
Resolution for lensed microscopes was considered to equal λ/2(NA), where λ is the wavelength (509 nm used for calculations) and NA is the numerical aperture of the objective. For Fig. 1C, the area of the largest commonly available format sensor for a C-Mount (4/3″ format, 22 mm diameter) was assumed with FOV = Sensor area/magnification2. The cross-section area, π(D/2)2, is the physical constraint on the beam created by the pupil diameter for each objective given by D = 2(ft/M)tan(sin−1(NA/n)), where ft is the focal length of the tube lens, M is the magnification, and n is the refractive index. For FlatScope, the cross-section area is the same as the active sensor area, because there are no additional optics to restrict the light path. The cross-section area for the GRIN lens system is assumed to be the diameter of the aspherical lens. The trend line in Fig. 1C represents the approximate maximum for traditional lensed microscope systems.
Supplementary Material
Acknowledgments
We thank A. C. Sankaranarayanan for valuable discussions and for providing feedback on this manuscript. Funding: This work was supported in part by the NSF (grants CCF-1502875, CCF-1527501, and IIS-1652633) and the Defense Advanced Research Projects Agency (grant N66001-17-C-4012). Author contributions: A.V., J.T.R., and R.G.B. developed the concept and supervised the research. J.K.A. fabricated prototypes and designed and built experimental hardware setup. V.B. developed T2S model, computational algorithms, and simulation platform. B.W.A. designed and built software for hardware interface. D.G.V. prepared samples and aided in related experimental investigations. F.Y. aided in design and fabrication of components for prototypes. J.K.A. and V.B. performed the experiments. All authors contributed to the writing of the manuscript. Competing interests: A.V., R.G.B., J.T.R., V.B., J.K.A., and B.W.A. are inventors on a patent application related to this work (application no. PCT/US2017/044448, filed on 28 July 2017). The other authors declare that they have no competing interests. Data and materials availability: Our team is committed to reproducible research. With that in mind, we will release detailed procedures and protocols that we followed in the fabrication of our prototypes. We will also release standard data sets of images and volumes (including all the data presented in the paper) along with the code for image and volume reconstructions. This will be released publicly on a website that will be updated with newer data sets as they are acquired.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/12/e1701548/DC1
section S1. Derivation of T2S model
section S2. Computational tractability
section S3. Model calibration
section S4. Gradient direction for iterative optimization
section S5. MURA: A separable pattern
section S6. FlatCam model: An approximation to the T2S model
section S7. Diffraction effects and T2S model
section S8. Light collection
section S9. Impact of sensor saturation
fig. S1. Calibration setup.
fig. S2. Digital focusing.
fig. S3. Simulation comparison of bare sensor, FlatScope, and FlatCam.
fig. S4. 3D volume reconstruction accuracy.
fig. S5. Fabrication of FlatScope.
fig. S6. Refractive index matching.
fig. S7. T2S derivation.
fig. S8. Aberration removal using RPCA.
fig. S9. Removing effects from autofluorescence.
fig. S10. T2S model error from diffraction.
fig. S11. Light collection comparison of FlatScope and microscope objectives.
movie S1. Digital focusing of simulated resolution target.
movie S2. Fluorescent beads flowing in microfluidic channels of different depths.
REFERENCES AND NOTES
- 1.Ozcan A., Demirci U., Ultra wide-field lens-free monitoring of cells on-chip. Lab Chip 8, 98–106 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Iddan G., Meron G., Glukhovsky A., Swain P., Wireless capsule endoscopy. Nature 405, 417 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Ghosh K. K., Burns L. D., Cocker E. D., Nimmerjahn A., Ziv Y., Gamal A. E., Schnitzer M. J., Miniaturized integration of a fluorescence microscope. Nat. Methods 8, 871–878 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A. Nikonorov, R. Skidanov, V. Fursov, M. Petrov, S. Bibikov, Y. Yuzifovich, Fresnel lens imaging with post-capture image processing, IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops (IEEE, 2015), pp. 33–41. [Google Scholar]
- 5.Peng Y., Fu Q., Heide F., Heidrich W., The diffractive achromat full spectrum computational imaging with diffractive optics. ACM Trans. Graph., 35, 1–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khorasaninejad M., Chen W. T., Devlin R. C., Oh J., Zhu A. Y., Capasso F., Metalenses at visible wavelengths: Diffraction-limited focusing and subwavelength resolution imaging. Science 352, 1190–1194 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Thiele S., Arzenbacher K., Gissibl T., Giessen H., Herkommer A. M., 3D-printed eagle eye: Compound microlens system for foveated imaging. Sci. Adv. 3, e1602655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D. J. Brady, Optical Imaging and Spectroscopy (Wiley, 2009). [Google Scholar]
- 9.Waller L., Tian L., Computational imaging: Machine learning for 3D microscopy. Nature 523, 416–417 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Betzig E., Patterson G. H., Sougrat R., Lindwasser O. W., Olenych S., Bonifacino J. S., Davidson M. W., Lippincott-Schwartz J., Hess H. F., Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Rust M. J., Bates M., Zhuang X., Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenbaum A., Luo W., Su T.-W., Göröcs Z., Xue L., Isikman S. O., Coskun A. F., Mudanyali O., Ozcan A., Imaging without lenses: Achievements and remaining challenges of wide-field on-chip microscopy. Nat. Methods 9, 889–895 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.P. R. Gill, D. G. Stork, Lensless ultra-miniature imagers using odd-symmetry spiral phase gratings. Computational Optical Sensing and Imaging, Arlington, VA, 23 to 27 June 2013 (Optical Society of America, 2013).
- 14.Asif M. S., Ayremlou A., Sankaranarayanan A., Veeraraghavan A., Baraniuk R. G., FlatCam: Thin, lensless cameras using coded aperture and computation. IEEE Trans. Comput. Imaging, 384–397 (2016). [Google Scholar]
- 15.G. Kim, K. Isaacson, R. Palmer, R. Menon, Lensless photography with only an image sensor. http://arxiv.org/abs/1702.06619 (2017). [DOI] [PubMed]
- 16.Boominathan V., Adams J. K., Asif M. S., Avants B. W., Robinson J. T., Baraniuk R. G., Sankaranarayanan A. C., Veeraraghavan A., Lensless imaging: A computational renaissance. IEEE Signal Process. Mag. 33, 23–35 (2016). [Google Scholar]
- 17.Horisaki R., Ogura Y., Aino M., Tanida J., Single-shot phase imaging with a coded aperture. Opt. Lett. 39, 6466–6469 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Egami R., Horisaki R., Tian L., Tanida J., Relaxation of mask design for single-shot phase imaging with a coded aperture. Appl. Opt. 55, 1830–1837 (2016). [DOI] [PubMed] [Google Scholar]
- 19.DeWeert M. J., Farm B. P., Lensless coded-aperture imaging with separable Doubly-Toeplitz masks. Opt. Eng. 54, 023102 (2015). [Google Scholar]
- 20.A. Wang, P. Gill, A. Molnar, in Proceedings of the Custom Integrated Circuits Conference (IEEE, 2009), pp. 371–374. [Google Scholar]
- 21.Ryu D., Wang Z., He K., Zheng G., Horstmeyer R., Cossairt O., Subsampled phase retrieval for temporal resolution enhancement in lensless on-chip holographic video. Biomed. Opt. Express 8, 1981–1995 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo S., Su T.-W., Tseng D. K., Erlinger A., Ozcan A., Lensfree holographic imaging for on-chip cytometry and diagnostics. Lab Chip 9, 777–787 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coskun A. F., Su T.-W., Ozcan A., Wide field-of-view lens-free fluorescent imaging on a chip. Lab Chip 10, 824–827 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ah Lee S., Ou X., Lee J. E., Yang C., Chip-scale fluorescence microscope based on a silo-filter complementary metal-oxide semiconductor image sensor. Opt. Lett. 38, 1817–1819 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenbaum A., Zhang Y., Feizi A., Chung P.-L., Luo W., Kandukuri S. R., Ozcan A., Wide-field computational imaging of pathology slides using lens-free on-chip microscopy. Sci. Transl. Med. 6, 267ra175 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Rosen J., Brooker G., Non-scanning motionless fluorescence three-dimensional holographic microscopy. Nat. Photonics 2, 190–195 (2008). [Google Scholar]
- 27.Gottesman S. R., Fenimore E. E., New family of binary arrays for coded aperture imaging. Appl. Opt. 28, 4344–4352 (1989). [DOI] [PubMed] [Google Scholar]
- 28.Sencan I., Coskun A. F., Sikora U., Ozcan A., Spectral demultiplexing in holographic and fluorescent on-chip microscopy. Sci. Rep. 4, 3760 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coskun A. F., Sencan I., Su T.-W., Ozcan A., Lensless wide-field fluorescent imaging on a chip using compressive decoding of sparse objects. Opt. Express 18, 10510–10523 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coskun A. F., Sencan I., Su T.-W., Ozcan A., Lensfree fluorescent on-chip imaging of transgenic Caenorhabditis elegans over an ultra-wide field-of-view. PLOS ONE 6, e15955 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yıldırım E., Arpali Ç., Arpali S. A., Implementation and characterization of an absorption filter for on-chip fluorescent imaging. Sens. Actuators B Chem. 242, 318–323 (2017). [Google Scholar]
- 32.Fink Y., Winn J. N., Fan S., Chen C., Michel J., Joannopoulos J. D., Thomas E. L., A dielectric omnidirectional reflector. Science 282, 1679–1682 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Richard C., Renaudin A., Aimez V., Charette P. G., An integrated hybrid interference and absorption filter for fluorescence detection in lab-on-a-chip devices. Lab Chip 9, 1371–1376 (2009). [DOI] [PubMed] [Google Scholar]
- 34.J. W. Goodman, Introduction to Fourier Optics (McGraw-Hill, 1968). [Google Scholar]
- 35.Candès E. J., Li X., Ma Y., Wright J., Robust principal component analysis? J. ACM 58, 11 (2011). [Google Scholar]
- 36.Lin Z., Ganesh A., Wright J., Wu L., Chen M., Ma Y., Fast convex optimization algorithms for exact recovery of a corrupted low-rank matrix. Comput. Adv., 1–18 (2009). [Google Scholar]
- 37.Nesterov Y., Efficiency of coordinate descent methods on huge-scale optimization problems. SIAM J. Optim. 22, 341–362 (2012). [Google Scholar]
- 38.Beck A., Teboulle M., A fast iterative shrinkage-thresholding algorithm for linear inverse problems. SIAM J. Imaging Sci. 2, 183–202 (2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/12/e1701548/DC1
section S1. Derivation of T2S model
section S2. Computational tractability
section S3. Model calibration
section S4. Gradient direction for iterative optimization
section S5. MURA: A separable pattern
section S6. FlatCam model: An approximation to the T2S model
section S7. Diffraction effects and T2S model
section S8. Light collection
section S9. Impact of sensor saturation
fig. S1. Calibration setup.
fig. S2. Digital focusing.
fig. S3. Simulation comparison of bare sensor, FlatScope, and FlatCam.
fig. S4. 3D volume reconstruction accuracy.
fig. S5. Fabrication of FlatScope.
fig. S6. Refractive index matching.
fig. S7. T2S derivation.
fig. S8. Aberration removal using RPCA.
fig. S9. Removing effects from autofluorescence.
fig. S10. T2S model error from diffraction.
fig. S11. Light collection comparison of FlatScope and microscope objectives.
movie S1. Digital focusing of simulated resolution target.
movie S2. Fluorescent beads flowing in microfluidic channels of different depths.


