Abstract
The role of non-coding Ribonucleic Acids (ncRNAs) in biology is currently an area of intense focus. Hematopoiesis requires rapidly changing regulatory molecules to guide appropriate differentiation and ncRNA are well suited for this. It is not surprising that virtually all aspects of hematopoiesis have roles for ncRNAs assigned to them and doubtlessly much more await characterization. Stem cell maintenance, lymphoid, myeloid and erythroid differentiation are all regulated by various ncRNAs, including microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and various transposable elements within the genome. As our understanding of the many and complex ncRNA roles continues to grow, new discoveries are challenging the existing classification schemes. In this review we briefly overview the broad categories of ncRNAs and discuss a few examples regulating normal and aberrant hematopoiesis.
Introduction
It has been proposed that less than 2% of the human genome is translated into proteins. However, somewhere between 70% [1] to over 90% [2] of the genome is transcribed, and over 60% of these transcripts are processed [1]. The RNA that does not encode conventional proteins is referred to collectively as non-coding RNA (ncRNA) and encompasses transfer RNA (tRNA), ribosomal RNA (rRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA) and small cajan body-specific RNA (scaRNA), piwi-interacting RNA (piRNA), miRNA and lncRNA amongst others (see Table 1). While relatively recent advances in deep sequencing technology has shed light on the full extent of ncRNA expression it has long been recognized ncRNA is critical in genomics. For instance, rRNA and tRNA are essential in protein synthesis [3], and messenger RNA (mRNA) splicing and nuclear organization require snRNAs and snoRNAs [4]. The recent documentation of the extensive contribution of ncRNA transcripts of all sizes and types has resulted in new and constantly evolving categories. As our understanding grows, new nomenclature will most likely develop to better categorize ncRNAs by functionality. Recently, a number of predicted ncRNAs have been documented that encode proteins, either as micropeptides hidden as short open reading frames (sORFs) within ncRNAs, or due to read-throughs of stop codon sequences [5], adding further confusion around the broad coding and non-coding classification scheme. Here we will define the current classifications of ncRNAs and briefly discuss the emerging roles they play on various aspects of hematopoiesis.
Table 1.
Classes of Non-coding RNA
| RNA | Abbreviation | Cellular Role |
|---|---|---|
| Messenger RNA | mRNA | Codes for Protein |
| Transfer RNA | tRNA | Translation |
| Ribosomal RNA | rRNA | Translation |
| Small nuclear RNA | snRNA | Splicing and other roles |
| Small nucleolar RNA | snoRNA | RNA Nucleotide modification |
| Small cajan body-specific RNA | scaRNA (Type of snoRNA) | RNA nucleotide modifications |
| Piwi interacting RNA | piRNA | Transposon Defense |
| Micro RNA | miRNA | Gene Regulation |
| Long, non-coding RNA | lncRNA | Gene Regulation and other roles |
MicroRNA (miRNAs)
miRNAs consist of 21–24 bases and are involved in regulation of post-transcriptional gene expression regulation and RNA silencing. The first example was documented in 1993 in Caenorhabditis elegans when a 21 nucleotide RNA was shown to inhibit the Lin 14 transcript [6]and was subsequently named Lin4. Since then, hundreds of miRNAs have been identified and the list continues to grow [7]. However, the name microRNA was not applied until 2001 [8].
As summarized in Figure 1, these short RNA sequences are initially generated in the nucleus from primary miRNA (pri-miRNA) transcripts. The pri-miRNAs are recognized by DiGeorge Syndrome Critical Region 8 (DGCR8) and bind the RNAse III enzyme Drosha, which in turn cleaves the RNA into precursor miRNAs (pre-miRNAs) of approximately 70 nucleotides. The pre-miRNAs are exported to the cytoplasm and subsequently processed by the RNAse III enzyme Dicer to yield the mature form [9]. Most miRNAs exert an inhibitory effect through binding a short (6–8 nucleotide sequence) in the 3′ untranslated region (3′UTR) of a target gene. Binding is facilitated by a complementary sequence (seed sequence) in the miRNA [10] and is often shared by a number of miRNAs [7], especially within the same miRNA family. Reasons for multiple miRNAs with the same seed sequence may reflect subtle differences in the inhibitory effect of each. Alternatively, because miRNA expression is very tissue specific [11] it may facilitate inhibition of the same 3′UTR sequences in different cellular contexts when a single miRNA with a particular seed sequence would not be expressed. While over 80% of miRNA-affected transcripts currently recognized share a sequence complementary to the seed sequence, seed-sequence-independent targets have also been identified [7]. MiRNAs have been associated with roles in disease, cancer and viral infection amongst others, primarily through gene regulation [12].
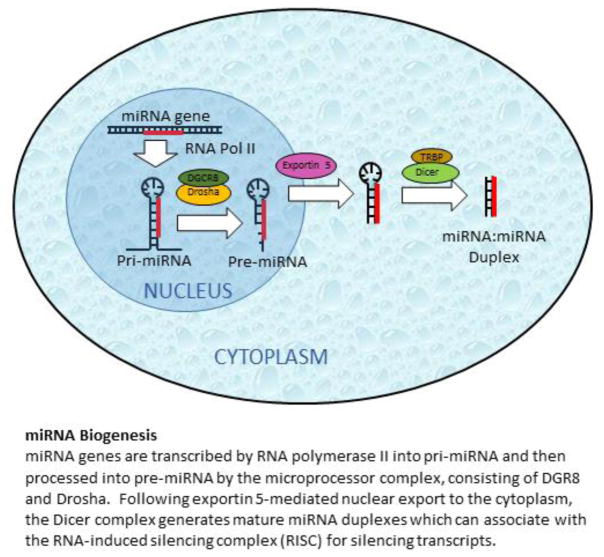
Figure 1. miRNA Biogenesis.
miRNA genes are transcribed by RNA polymerase II into pri-miRNA and then processed into pre-miRNA by the microprocessor complex, consisting of DGR8 and Drosha. Following exportin 5-mediated nuclear export to the cytoplasm, the Dicer complex generates mature miRNA duplexes which can associated with the RNA-induced silencing complex (RISC). Once associated with RISC the passenger strand is degraded leaving the guide miRNA to interact with the target mRNA.
Regulating self-renewal and differentiation of hematopoietic stem cells (HSCs) and Progenitor Cells
The first study examining miRNAs in hematopoiesis was performed in the laboratory of Harvey Lodish [13]. Since then dozens of miRNAs playing a role in both normal and malignant hematopoiesis have been identified.
Perhaps the most characterized miRNA pathways in primitive hematopoietic compartments involve the miR125 family (miR125a, miR125b1 and miR125b2), miR196b, and the miR17–92 cluster (miRNAs 17–5p, 17–3p, 18a, 19a, 20a and 92a). Four of the miR17-92 miRNAs are upregulated in HSCs compared to mature blood cells [14] with the whole cluster transcribed from a single promoter as one non-coding transcript and then processed into seven mature miRNAs [14]. This facilitates expression of the entire group simultaneously during differentiation. The importance of coordinated expression of miRNAs is illustrated by the observation that overexpression of each of the miR17-92 cluster miRNAs alone induces various blood malignancies [15]. Target genes of miR17-92 miRNAs include transcription factors involved in cell cycle regulation and differentiation [16].
MiR196b, which is highly expressed in HSCs, is encoded between the homeotic (HOX) genes HOXA9 and HOXA10 and appears to be co-regulated with HOXA9 [17]. To aid in maintaining cells in a primitive state, miR196b directly targets HOX genes upregulated during myeloid differentiation (such as HOXA7 and HOXC8) and other genes involved in differentiation [17]. During myeloid differentiation miR196b itself is downregulated by expression of the myeloid differentiation factor Growth factor independent 1 (Gfi1) [18].
The miR125 family are highly expressed in HSCs and progenitor cells compared to differentiated cells and their expression increases self-renewal, however the target genes remain uncharacterized [19]. Over-expression of miR125 provides a proliferative advantage and skews progeny towards a myeloid lineage [20].
miRNAs in Erythropoiesis and Megakaryopoiesis
Ten to 30% of genes are predicted to be targets of miRNAs [21] and many have been linked to erythropoiesis, suggesting an important role during this process. Numerous high throughput screens and functional approaches to determining the role of miRNAs in erythropoiesis have been utilized. Unfortunately, many miRNA functions characterized through high-throughput array screens have different roles when examined using more functional approaches [22]. This includes miRs15, 24, 221 and 223, which were reported as being up-regulated in multiple microarray studies, but were found to be down-regulated during erythroid maturation in non-microarray based studies [22]. The reason for such discrepancies is unclear, but reflects uncertainty regarding the exact roles of these miRNAs in the various stages of differentiation during erythropoiesis.
Other miRNAs, including miR451, have roles in promoting erythroid differentiation supported by both high-throughput and functional screens. Erythroid differentiation in K562 cells was disrupted in the absence of miR451 [23] and mice lacking miR451 exhibit defective erythropoiesis via repression of the zeta isoform of the 14-3-3 protein [24]. Further supporting a requirement for miR451 in regulating erythropoiesis, Cluster of Differentiation 34 positive (CD34+) precursor cells derived from thalassemic patients exhibit dysregulation of miR451 when differentiated towards the red blood cell lineage [25].
miRNAs play important roles during megakaryocyte differentiation from the common erythro-megakaryocyte progenitor. For example, miR181 represses lineage protein 28 homologue (Lin28) expression, which results in an increase of lethal 7 (let-7). The high levels of let-7 causes increased megakaryocytic differentiation with no appreciable role in haemin-induced erythrocytic differentiation [26]. Similarly, the miR34a transcript inhibits cellular proliferation [27] and downregulates the pro-erythropoiesis factor cellular myeloblastosis oncogene (c-Myb), to delineate these cells from erythroblasts [28]. The downregulation of miR155 is also essential for megakaryopoiesis progression. When present, miR155 represses both E26 transformation specific 1 (Ets-1) and Meis, both of which are required for megakaryocyte differentiation and inversely correlated with miR155 expression levels [29]. A central feedback loop in megakaryopoiesis involves the transcription factor RUNX1 and miR27a. High levels of runt-related transcription factor 1 (RUNX1) induce expression of miR27a which, in turn, suppresses the expression of RUNX1 [30]. Due to the developmental importance of RUNX1 in many hematopoietic lineages, its exact expression level is tightly regulated.
miRNAs in Myeloid Differentiation
Myeloid differentiation is the development of different types of mature blood cells from stem cells and several miRNAs are involved during this process. One of the most studied miRNA during granulocytic development is miR223. The expression of miR223 is significantly increased during granulocytic, but not monocytic, differentiation and induced expression of miR223 results in increased granulocytic differentiation capacity [31]. Two distinct mechanisms have been proposed for the induction of miR223. The first proposes an element within the miR223 promoter occupied by one of two transcription factors. Nuclear Factor 1A (NFI-A) and CCAAT/enhancer binding protein alpha (C/EBPα) compete for binding with NFI-A binding suppressing miR223, while C/EBPalpha binding drastically increases miR223 transcription. The second mechanism requires binding of C/EBPalpha and pu box-containing 1 (PU.1) at a more distal promoter site, with PU.1 having the strongest contribution to granulocyte differentiation [32].
Both miR21 and miR196b have also been implicated in differentiation of the granulocytic lineage, as the expression of these miRNAs inhibits granulocyte-colony stimulating factor (G-CSF) induced differentiation [18]. It has been proposed that (Gfi1) regulates miR21 and miR196b as Gfi1−/− mice display deregulated miRNAs and granulocytopoiesis [18]. Increased expression of another miRNA, miR27, has been reported during granulocytic differentiation and is required to down-modulate the granulocyte inhibitory protein acute myeloid leukemia 1 (AML1) expression [33].
In monocyte differentiation, AML1 protein levels are upregulated, partly through the suppression of the AML1-inhibiting miRNAs miR17-5a, miR20a and miR106a [34]. The master regulator of monocyte differentiation, Monocyte-colony stimulating factor receptor (M-CSFR) is partially regulated by miR424. The transcription factor, PU.1 upregulates miR-424, which targets NFI-A to inhibit its translation and subsequently the stimulation of monocyte differentiation. In turn, the decrease of NFI-A levels is important for the activation of differentiation-specific genes such as M-CSFR [35].
Some miRNAs in granulo-monocytic cells are regulated by pathogenic factors. MiR155 is induced during the monocyte inflammatory response [36] and modulates the interleukin-1 signaling pathway [37] while miR146 and miR125b are regulated in response to lipoplysaccharide (LPS) stimulation of monocytes/macrophages [38]. In granulocytes and monocytes, miR9 is induced by LPS and contributes to an inhibitory feedback loop with the transcriptional regulator of the inflammatory response nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) [39].
miRNA in Lymphoid Differentiation
MiR181a and other miR181 members are very abundant at the CD4+/CD8+ double positive (DP) stage of Thymic cell (T cell) development, comprising as much as half the miRNA molecules expressed [40, 41]. Overexpression of miR-181a results in increased DP thymocyte numbers [42] and deficiency results in a mild decrease [43]. Downstream targets of miR181a in thymocyte development include phosphatase and tensin homologue (Pten) [43] and the T cell receptor (TCR) signaling molecules Dual specificity phosphatases Dusp5 and Dusp6 and SH2-containing phosphatase 2 (Shp2) [42]. A role for miR181a is not limited to T cell development as a severe loss of Natural killer T (NKT) cells also occurs in miR181a-deficient mice [43].
The Notch signaling pathway is important for T cell development and is a target of miR150 in T cells [44], however the exact functional role is not clear. In mice, transgenic expression of miR185 targets the protein Marginal zone b1 (Mzb1) and restricts differentiation towards the DP stage, however, whether endogenous miR185 is important for T cell development remains to be determined [45].
Although the specific miRNAs responsible have not been elucidated, mice deficient in the miRNA processing proteins Drosha and Dicer, exhibited dramatic alterations in the NKT lineage. Other cell lineages were also affected to a varying extent with T regulatory (Treg) numbers reduced significantly [41, 46]. While the miRNA signatures of the different T cell lineages differ, Tregs are distinct from other T cells [46, 47]. It remains unclear if specific miRNAs are responsible for thymic Treg differentiation or if miRNA TCR regulation indirectly impacts differentiation [48].
MiR155 is a Treg-enriched miRNA that appears to contribute to Treg development by reducing Forkhead box P3 (Fox3p) expression through suppression of the Janus kinase – signal transducer and activator of transcription (JAK-STAT) signaling molecule suppressor of cytokine signaling (SOCS1) [49]. However, miR155-deficient mice display only a mild reduction in Foxp3+ cells in the thymus [49, 50]. As with other lymphoblasts, miRNAs not only play a role in Treg differentiation but are also essential for functional programming. For instance, miR146a is enriched in Tregs and is required in the control of T helper 1 (Th1) responses through specific targeting of STAT1 [49]. MiR10 is expressed in Tregs associated with Th1 responses, whereas miR182 is expressed in Th2-associated Tregs and may cross-regulate opposing networks of genes to promote the Th1- or Th2-associated phenotype [51].
The regulation of helper T cells are greatly influenced by miRNA, particularly miR17-92a cluster, miR155, miR29a and miR21. The miR17–92a cluster is required for efficient Th1 differentiation, with miR17 and miR19b having the most influence. While miR17 promotes Th1 differentiation via inhibition of transforming growth factor beta receptor 2 (TGFβRII) and cyclic-AMP response element binding protein 1 (CREB1), miR19b regulates Pten [52]. To ensure Th1 differentiation doesn’t become excessive, increased expression levels of miR155 become inhibitory to Th1 differentiation by inhibiting Interferon c Receptor a (IFNcRa) expression [53]. MiR29a inhibits expression of the transcription factors T box transcription factor (T-bet) and Eomesodermin as well as the negative regulator of the mitogen activated protein (MAP) kinase pathway, Sprouty RTK signaling antagonist 1 (Spry1) [54]. Increasing the expression of miR21 enhances gata-binding protein 3 (Gata3), and Interleukins IL-4, IL-5 and IL-13 expression by Th2 cells [55].
Numerous miRNAs promote Th17 differentiation. MiR155 is necessary for the development of disease in Th17 animal models by repressing the transcription factor Ets1 [56]. MiR17 and miR19b of the miR17–92a cluster are also required for effective Th17 differentiation and evidence suggests the relevant targets are Pten and Ikaros family zink finger 4 (IKZF4) for miR19b and miR17 respectively [57].
Understanding of the specific miRNAs involved in B cell differentiation and function is not as developed as for T cells, however it is clear that miRNA regulation is extensive. For instance, Dicer knocked out of early B cell precursors resulted in blockages in differentiation at both proB to pre-B transition [58] and mature B cells to antibody producing memory B cells [59].
MiRNAs from the miR17–92a cluster are important for early B cell development by regulating B cell survival, in part, by repressing the pro-apoptotic molecule Bim. Mice deficient in miR17–92a exhibited a greatly reduced B cell compartment, with a developmental block at the pre-B cell stage [60]. On the other hand, overexpression of miR34a has been shown to block B cell development at the pro-B to pre-B transition [61] and inhibits B lymphopoiesis through repression of the B cell oncogene Foxp1, which is required for the pro-B to pre-B transition [62].
Regulation of the latter stages of B cell development requires a number of different miRNAs, including miR150. Deficiency of miR150 results in a loss of c-Myb control and an expansion of B1 cells [63]. The differentiation of mature B cells into germinal center B cells is regulated by miR155 and B cells from miR155-deficient mice fail to express Immunoglobulin M (IgM) or class switch, resulting in impaired antibody responses [64, 65].
Piwi-interacting RNAs (piRNAs)
While microRNAs tend to span sizes 21–24 nucleotides, piRNAs are slightly larger at 26–31 nucleotides, and collectively form the largest class of small ncRNAs in animal cells [66]. They form RNA-protein complexes with the piwi class of proteins (see Figure 2 for summary). In comparison with miRNAs they lack sequence conservation and possess increased complexity. Currently, the most characterized cellular role of piRNAs is in epigentic and post-translational silencing of retrotransposons and other genetic elements [66].
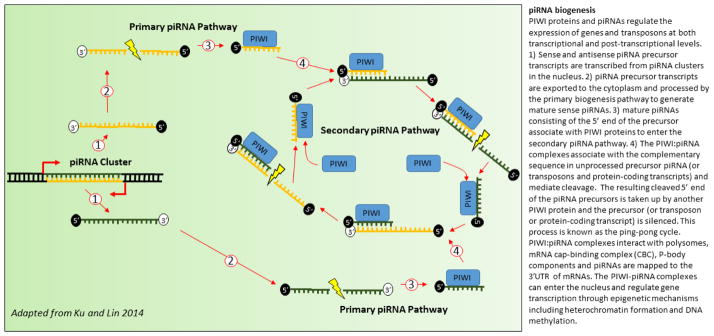
Figure 2. piRNA biogenesis (adapted from Ku and Lin 2014 [133]).
PIWI proteins and piRNAs regulate the expression of genes and transposons at both transcriptional and post-transcriptional levels. 1) Sense and antisense piRNA precursor transcripts are transcribed from piRNA clusters in the nucleus. 2) piRNA precursor transcripts are exported to the cytoplasm and processed by the primary biogenesis pathway to generate mature sense piRNAs. 3) mature piRNAs consisting of the 5′ end of the precursor then associate with PIWI proteins to enter the secondary piRNA pathway. 4) The PIWI:piRNA complexes then associate with the complementary sequence in unprocessed precursor piRNA (or transposons and protein-coding transcripts) and mediate cleavage. The resulting cleaved 5′ end of the piRNA precursors is taken up by another PIWI protein and the precursor (or transposon or protein-coding transcript) is silenced. This process is known as the ping-pong cycle. PIWI:piRNA complexes interact with polysomes, mRNA cap-binding complex (CBC), P-body components and piRNAs are mapped to the 3′UTR of mRNAs. The PIWI-piRNA complexes can enter the nucleus and regulate gene transcription through epigenetic mechanisms including heterochromatin formation and DNA methylation.
Piwi proteins and their associated RNA are involved in the maintenance of stem cell character and genome integrity, with evidence they are active in hematopoietic tissue [67]. In fact, knockdown of the piwi protein MIWI2 leads to abnormal hematopoiesis and erythroid precursors take on characteristics of more differentiated erythroid cells [67]. However knockout of this gene in mice has negligible observable loss of hematopoietic function [67]. The piwi protein PIWIL4 is over-expressed in a large proportion of AML leukemia patients and knockdown results in gross changes in histone methylation and slowed leukemic growth [66] suggesting a tightly regulated piwi pathway is essential for normal hematopoiesis. The roles of piRNAs in hematopoiesis have only just begun to be explored and it seems likely that more will be uncovered in the near future.
Long non-coding RNA (lncRNAs)
Long non-coding RNAs are defined as a large class of RNAs with sequences longer than 200 nucleotides which differentiates them from classically defined small nuclear RNAs (such as miRNAs, snoRNAs, etc). This classification is somewhat ambiguous as some classically defined small nuclear RNAs are also greater than 200 nucleotides, thus specific recognition of transcripts greater than 200 nucleotides as lncRNA has only been applied rigorously to newly recognized transcripts [68].
The number of lncRNA transcripts greatly exceeds protein-coding mRNAs with estimates between 10, 000 and 60, 000 human lncRNA genes [69–71]. This number continues to grow with deeper RNA sequencing and more sensitive and improved epigenomic technologies and computational prediction techniques [72]. These RNAs do not encode for proteins, however a number of designated lncRNAs have been determined to include short open reading frames (sORFs) that are translated into micropeptides [5, 73]. As these short peptides may have cooperative, partially overlapping, or completely separate biological functions to the role of the parental lncRNA (if indeed either has a biological role) categorizing these RNA sequences is challenging. Indeed, even mRNA includes RNA sequences that can function independently of translated protein, and include alternative splicing initiation sites and separate ORFs [5].
lncRNA classification
Despite the difficulty, there are numerous schemes currently utilized to classify lncRNAs. The most basic of these divisions is compartmentalizing lncRNAs based on their cellular roles [72]. Some lncRNAs are non-functional, which is most likely the result of transcriptional noise. The extent of these non-functional RNAs in the genome is unknown, however, current evidence suggests it is likely to be a very small percentage, while the list of functional lncRNA transcripts with authentic biological roles continues to grow exponentially [74]. LncRNAs are generally under lower selective pressure than protein coding genes but higher selective pressures than ancestral repeat sequences (considered to be under neutral selection), supporting the concept that most lncRNAs are functional. Furthermore, the promoters of IncRNAs are the region of the IncRNA gene under highest selective pressure, displaying levels of selection comparable to the promoters of protein coding genes [69, 75–78]. This suggests lncRNA and protein coding transcript regulation is equally crucial to homeostasis. Functional lncRNAs can be sub-classified as; 1) lncRNAs in which the act of transcription is sufficient for their function but the transcript itself is unnecessary and; 2) functional lncRNAs that have functional cellular roles and can act in cis- or trans- [72].
Another type of classification based on location of the transcript relative to protein-coding mRNA has also been employed [79] and to date comprises of six categories. 1) Long intergenic non-coding RNAs (lincRNAs) lies within the genomic interval between two coding genes at least 1 kilobase (kb) away from the nearest coding gene [79]. 2) Enhancer RNAs (eRNAs) displays enhancer-like activity and upregulates expression via the Mediator complex [74]. 3) Antisense lncRNAs consist of transcript overlapping with sequence within one or more exons of another transcript on the same, or opposite strand [79]. 4) Bidirectional lncRNA occurs when the sequence is located on the opposite strand from a protein coding gene whose transcription is initiated less than 1000 base pairs away [80]. 5) Intronic lncRNAs is derived from within an intron of a different coding transcript in either sense or antisense orientation. 6) Transcribed pseudogene lncRNAs result from the introduction of a premature stop codon in a duplicated copy of a functional gene, usually generated by homologous recombination [81]. As our understanding of the functions and genomic contributions of lncRNAs increases, these arbitrary definitions will undoubtedly change [82].
Regulation of lncRNAs (transcription/translation/processing)
The modes of action of lncRNAs are diverse and complex and well described in a number of reviews, including Geisler and Collier, 2013 [74]. As illustrated in Figure 3, they can act as regulators of transcription, mRNA processing, or modulators of post-transcriptional control through either translational control, regulating mRNA stability, or inhibiting miRNA activity through sponge action. At a higher level of control, lncRNAs regulate protein activity directly, act as scaffolds to facilitate higher order molecular complex assembly and localization, and act as and upon other signaling molecules [74]. Despite extensive characterization of the cellular roles of lncRNA, we are likely only in the infancy of our understanding. Considering genome-wide association studies concluded only 7% of disease or trait-associated single nucleotide polymorphisms (SNPs) occur within protein coding exons and 43% are found outside of protein coding genes altogether [83], it is likely lncRNA deregulation is an important factor in disease.
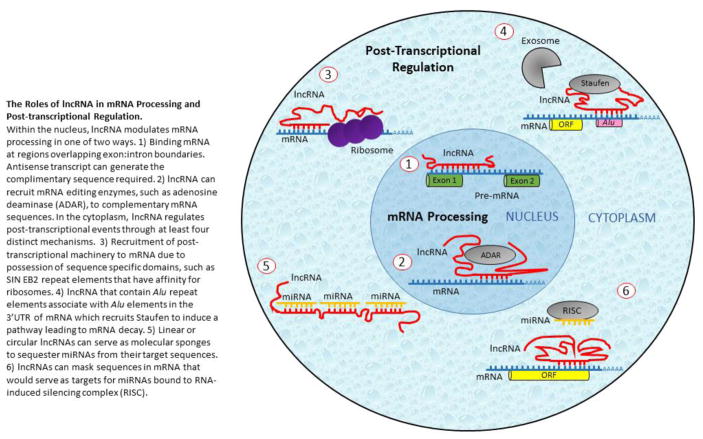
Figure 3. The Roles of lncRNA in mRNA Processing and Post-transcriptional Regulation.
Within the nucleus, lncRNA modulates mRNA processing in one of two ways. 1) Binding mRNA at regions overlapping exon:intron boundaries. Antisense transcript can generate the complimentary sequence required. 2) lncRNA can recruit mRNA editing enzymes, such as adenosine deaminase (ADAR), to complementary mRNA sequences. In the cytoplasm, lncRNA regulates post-transcriptional events through at least four distinct mechanisms. 3) Recruitment of post-transcriptional machinery to mRNA due to possession of sequence specific domains, such as SIN EB2 repeat elements that have affinity for ribosomes. 4) lncRNA that contain Alu repeat elements associate with Alu elements in the 3′UTR of mRNA which recruits Staufen to induce a pathway leading to mRNA decay. 5) Linear or circular lncRNAs can serve as molecular sponges to sequester miRNAs from their target sequences. 6) lncRNAs can mask sequences in mRNA that would serve as targets for miRNAs bound to RNA-induced silencing complex (RISC).
lncRNA in Erythroid Development
One of the most compelling studies uncovering lncRNAs in erythropoiesis documented over 100 non-annotated transcripts with erythroid-restricted expression [84]. Many of these were targets of the key erythroid transcription factors GATA, T cell acute lymphocytic leukemia protein 1 (TAL1) or Kruppel-like factor 1 (KLF1). Similarly, examination of polyadenylated lncRNAs indicated cell type specific lncRNAs between erythroblasts, megakaryocyte-erythroid precursors (MEPs) and megakaryocytes suggesting the lncRNA landscape evolves as differentiation proceeds with different lncRNAs expressed at specific differentiation steps.
The first erythropoiesis lncRNA (lincRNA-EPS) was identified in mice by RNA-sequencing as highly induced in erythroid precursors when they start synthesizing hemoglobin and other lineage-specific proteins. Knockdown of this lncRNA resulted in inhibition of differentiation and apoptosis [85]. Interestingly, in a similar study, although orthologous regions were conserved for about 90% of the erythrocyte-specific lncRNAs, only 15% of erythroid lncRNA genes expressed in mouse erythroblasts were also expressed in human erythroblasts at similar developmental and maturational stages [86]. This suggests a distinct species-specificity of lncRNAs. Even examining only lncRNA transcripts determined to be required for terminal maturation of primary murine erythroid precursors, six of the seven were not even expressed in human erythroblasts, despite orthologous genomic loci being identified [86].
lncRNA in Myeloid Development
Eosinophil Granule Ontogeny (EGO) was one of the first lncRNAs identified with a role in normal hematopoiesis, where it facilitates eosinophil differentiation of CD34+ hematopoietic progenitor through the regulation of mRNA expression of major basic protein (MBP) and eosinophil derived neurotoxin (EDN). It is transcribed antisense within an intron of the inositol triphosphate receptor type 1 (ITPR1) gene [79]. EGO is highly expressed in human bone marrow and in mature eosinophils [87].
The transcription factor PU.1 plays a critical role in monocyte lineage commitment and monocyte/macrophage maturation [88] with expression levels precisely regulated and perturbation leading to leukemias and lymphomas [89]. Both sense and antisense transcripts originate from the same promoter, with the lncRNA PU.1-AS transcribed antisense to the transcription factor PU.1. PU.1-AS was demonstrated to negatively regulate PU.1 mRNA translation through binding to PU.1 mRNA to form mRNA/AS lncRNA duplex, thus antagonizing the expression of PU.1 protein [89].
HOX antisense intergenic RNA myeloid 1 (HOTAIRM1) is specifically expressed in the myeloid lineage and is one of the best-studied lncRNAs involved in hematopoiesis. It is most highly expressed in the terminal stage of granulocytic differentiation [90], and induced during all-trans retinoid acid (ATRA)-driven granulocytic differentiation of the NB4 human acute promyelocytic leukemia cell line and normal myeloid lineage cells. Loss of HOTAIRM1 attenuates neighboring 3′ HOXA genes, including HOXA1 and HOXA4, and selectively impairs the induction of transcripts for the myeloid differentiation markers CD11b (integrin alpha M chain), CD18 (integrin beta 2 chain) and CD11c (integrin alpha X chain), while retaining the expression of CD49d (integrin alpha 4 chain) [90].
Myeloid differentiation is usually considered to be dominantly controlled by highly myeloid-specific factors, however the widespread and abundant lncRNA Nuclear Paraspeckle Assembly Transcript 1 (NEAT1) has been reported to be indispensable for normal myeloid differentiation [91]. Although not expressed in normal myelopoiesis, in an AML patient sample and an AML-derived HL-60 cell line, contain a rearrangement of the lncRNA gene Non-Structural Maintenance Of Chromosomes Element 2 Homolog (NSMCE2) that gives rise to two novel chimeric genes, PVT1-NSMCE2 and CCDC26-NSMCE2 [92]. Beyond hypothesizing an oncogenic role of plasmacytoma variant translocation 1 (PVT1) and coiled-coil domain-containing protein 26 (CCDC26) [93] a role in leukemogenesis has not been determined, however the concept of fusion transcripts of lncRNA and coding genes adds yet another layer of complexity to the lncRNA world.
lncRNA in Lymphoid Development
Analysis of human bone marrow and thymic progenitor cells spanning the earliest stages of B lymphoid and T lymphoid specification revealed over 3,000 lncRNA genes. Lymphoid commitment is characterized by lncRNA expression patterns that are highly stage specific and are more lineage specific than those of protein-coding genes [94]. Interestingly, many lncRNAs overlap shorter functional RNAs, suggesting these longer transcripts may be processed and exert their effects as smaller functional species [79].
Relative to B cells, a large number of lncRNAs have been implicated in T cell differentiation, with more than 1000 lncRNAs characterized in a genome-wide screen of mammalian human and mouse CD8+ T-cells with many of them displaying stage- or tissue-specificity [95].
Thymus-specific non-coding RNA1 (Thy-ncR1) is expressed specifically during early T cell differentiation in the thymus and a small number of human T-cell leukemia cell lines, all of which originated from immature stage III T-cells [96]. Although a mechanistic link has not been determined, and the two genes are separated by 118 kb, expression of Thy-ncR1 and the CD1 gene cluster are highly correlated. This may suggest a synergism between CD antigens and lncRNAs during T cell selection and maturation.
T cell differentiation requires the precise regulation of a group of Ca2+-regulated transcription factors called Nuclear factor of activated T-cells (NFAT) proteins. Along with the GTPase-activating protein IQ Motif Containing GTPase Activating Protein 1 (IQGAP1), the lincRNA non-coding RNA of the nuclear factor of activated T cells (NRON) forms a scaffold that protects the phosphorylated, inactive form of nuclear factor of activated T cells 1 (NFAT1) and associated inhibitory kinases. Loss of NRON results in decreased NFAT1 phosphorylation and nuclear accumulation as well as an increase in NFAT-dependent cytokines [97].
Musculoaponeurotic Fibrosarcoma Oncogene Homolog (MAF), a Th2-associated transcription factor, is required for differentiating helper T-cells toward a Th2 transcription profile. Through the exploitation of a chromosome loop, linc-MAF-4 associates closely with the MAF promoter and recruits the chromatin modifiers lysine specific demethylase 1A (LSD1) and Enhancer of Zeste Homolog 2 (EZH2) to inhibit MAF expression. The absence of linc-MAF-4 differentiation skewed differentiating helper T-cells toward a Th2 transcription profile [98].
Documented roles for lncRNAs in B cell differentiation are fewer than T cells, however the B-cell integration cluster (BIC) produces a transcript consisting of three exons spanning 13 kb and was found to be highly expressed in antigen receptor stimulated B- and T-cells as well as in macrophages and dendritic cells upon Toll-like receptor (TLR) stimulation [79]. Within this lncRNA are the processed products, miR155-5p and miR155-3p, which regulate several biological processes, including hematopoiesis, inflammation and immune responses. Biceps brachii (BIC) transcript is highly expressed Hodgkin’s lymphoma, primary mediastinal B-cell lymphoma, diffuse large B-cell lymphoma, chronic lymphocytic leukemia (CLL), AML and some solid tumors, but they are not expressed in healthy samples [99].
Analysis of lncRNA expression during human B-cell development, by array-based expression profiling of eleven distinct B-cell subsets, identified several lncRNAs within well-defined gene networks involved in specific stages of B-cell development [100], suggesting our understanding of the roles of lncRNA in this process remain in its infancy.
Pseudogenes
Pseudogenes are genes derived from another gene that show various degrees of sequence redundancy from the original “parental” gene. Due to redundancy, the pseudogene is not required for the survival of the organism and is consequently under little selective pressure, which allows multiple mutations to accumulate [101]. These mutations lead to loss of the original function, either through loss of gene expression or ability to code protein, however many pseudogenes have evolved important cellular functions different from that of the parental gene.
Pseudogenes are classified into one of two categories (processed or unprocessed), based on the mechanism facilitating duplication from the parental gene. Those pseudogenes copied from mRNA before incorporating into the genome are recognized as processed pseudogenes and lack promoters and introns [101]. Because the generation of these pseudogenes require an RNA intermediate and are retrotransposed, this group of pseudogenes are often called retrotransposons [102]. Although they usually contain poly-A tails, due to lack of promoter sequence they are considered “dead on arrival” upon insertion into the genome and do not require further mutagenesis to lose functionality [103]. Processed pseudogenes are continuously being created within a species and distinct processed pseudogene composition is evident in different human populations [103].
Non-processed pseudogenes arise from duplication of an entire gene, usually through homologous recombination, and may possess many features of coding genes including promoters, splice sites and CpG islands however disabling mutations (such as premature or frameshift mutations) or an inability to encode RNA distinguish the duplicate from the parental gene [81]. Sometimes at least part of a pseudogene is translated and over 140 human pseudogenes have been shown to be translated, however no function has been subscribed to the protein products [104].
Interestingly, a number of pseudogenes identified through DNA sequencing technologies possessing premature stop codons have been found to translate biologically important functional proteins due to translational read-through of the predicted stop codon [105]. As with other aspects of genome biology, computational analysis is predictive, and functional studies are crucial for thorough understanding.
Retrotransposons
Over half the human genome is comprised of repetitive sequences derived from pseudogenes and retrotransposons [104] (see Table 2). There are two broad categories of transposable elements (TEs) based on how they copy themselves from one location to another in the genome (summarized in Figure 4) [104]. Class 2 transposable elements are more ancient and have been referred to as DNA transposons or “jumping genes”, the discovery of which lead to the nobel prize in Physiology or Medicine for Barbara McClintock in 1983. These TEs move within the genome by a “cut-and-paste’ mechanism with no RNA intermediate.
Table 2.
Classification of repetitive elements and Contribution to the Human Genome
| Genomic Element | Sub categories | % of human genome | |
|---|---|---|---|
| Protein coding genes | N/A | Less than 2% | |
| Unprocessed Pseudogenes | N/A | 2 – 10 % | |
| Transposable Elements | Class I(DNA Transposons) | 3 % | |
| Retrotransposons | LTR-containing | 8 % | |
| LINEs | 20 % | ||
| SINEs | 13 % | ||
| sORFs | N/A | Unknown (likely <1 %) |
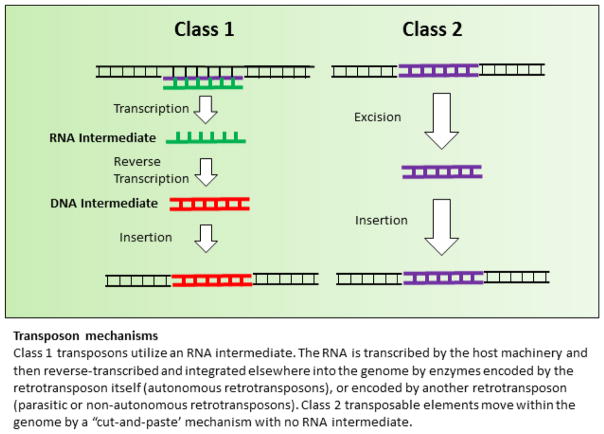
Figure 4. Transposon mechanisms.
Class 1 transposons utilize an RNA intermendiate. The RNA is transcribed by host machinery and then reverse-transcribed and integrated elsewhere into the genome by enzymes encoded by the retrotransposon itself (autonomous retrotransposons), or encoded by another retrotransposon (parasitic or non-autonomous retrotransposons). Class II transposable elements move within the genome by a “cut-and-paste’ mechanism with no RNA intermediate.
The more prevalent and polymorphic Class 1 transposable elements are commonly referred to as retrotransposons and copy themselves via an RNA intermediate. The RNA is transcribed by host machinery and then reverse-transcribed and integrated elsewhere into the genome by enzymes encoded by the retrotransposon itself (autonomous retrotransposons), or encoded by another retrotransposon (parasitic or non-autonomous retrotransposons). Autonomous retrotransposons are sub-categorized by the possession or absence of flanking long terminal repeats (LTRs). These sequences range from 300bp to 1kb, are identical in orientation and sequence on both the 5′ and 3′ ends of the proviral sequence [104], and are utilized for integrating into host DNA. LTR retrotransposons are very similar in structure to exogenous retroviruses with nonsense mutations and other sequence alterations leading to loss-of-function mutations in genes necessary for mobilization from one cell to another. Indeed, endogenous retroviruses are a family of LTR retrotransposons sharing such sequence similarity with exogenous retroviruses, the only separation being the inability to move from cell to cell [104]. In contrast, non-LTR retrotransposons utilize a mechanism referred to as target-primed reverse transcription (TPRT) to integrate into the host genome [106]. Like most LTR retrotransposons, autonomous non-LTR retrotransposons encode machinery required for reverse transcription and integration. While only a subclass of non-LTR retrotransposons, the long interspersed elements (LINEs) comprise almost 20% of the human genome with a copy number of several hundred thousand. While most have become non-functional due to nonsense mutations, around 100 full length LINEs remain functional and encodes two proteins including one (called ORF2p) possessing reverse transcriptase and enzymatic functions required for TPRT.
The largest class of non-autonomous retrotransposons in the human genome are the short interspersed elements (SINEs) of which the 300bp Alu elements comprise close to a million copies [107]. Alus hijack open reading frame-2 RNA binding protein (ORF2p) generated by LINEs to copy and integrate [108]. Although they have the potential to produce havoc through insertional mutagenesis or transcriptional perturbation, retrotransposons tend to remain inactive throughout adult life. However, aberrant activation has been associated with numerous diseases including cancer and hematological disorders. Retrotransposon activation is most active, and indeed required, early in development. Silencing of retrotransposon elements is primarily through DNA methylation, and as methylation machinery is not active during early development retrotranslocon mobility is most active at this time [109]. In appropriately differentiated and non-cancerous tissue most transposon elements are highly methylated at CpG islands [110].
Retrotransposons in regulating cell fate potential
As the zygote divides it goes from having totipotent to a pluripotent cell fate potential [111]. Cells derived from 4-cell and later blastocysts, as well as embryonic and induced pluripotent stem cells (ESCs and iPSCs respectively), have the potential to differentiate into all cell fates of the embryo (pluripotent), however they lack the ability to differentiate into extra-embryonic tissue that gives rise to the placenta and yolk sac [111]. The endogenous retrovirus family of human endogenous retrovirus-K (HERV-K), are LTR-containing retrotransposon elements silenced by DNA methylation in pluripotent and differentiated cells but highly active in totipotency and have been shown to regulate numerous genes in early development [111].
Retrotransposons in Erythropoiesis
The first de novo evidence that retrotransposition could lead to constitutional genetic defects causing human disease came from studies in hemophilia [105]. The first of these indicated 2 of 240 examined hemophilia A patients’ insertions of LINEs positioned within exon 14 of the coagulation factor VIII gene [112]. Three later reports documented Alu elements at the factor VIII locus causing a similar hemophilia A phenotype [113–115]. Hemophilia B results from a deficiency in coagulation factor IX and there have been numerous reports of retrotranlocations. Disruption at exon 5 [116] and exon 7 [117] have been attributed to LINEs, while Alu insertions disrupt exon 8 [116]and exon 5 [118, 119]. Alu insertions occur in some hemophilia C patients through deletion of the entire factor XI due to rearrangement between two flanking Alu repeat regions [120].
LINE and Alu-repeat mediated homologous recombination events also occur in thalassemia patients, resulting in loss-of-function alleles at hemoglobin loci. Hemoglobin is a major protein component of blood and carries blood to tissues as a tetramer composed of 2 α protein subunits and 2 β subunits surrounding the iron containing porphyrin ring that physically associates with oxygen. Genomic deletions in either of the duplicated α globin genes contributes to α-thalassemia while deletions in the single β globin gene is recognized as β-thalassemia. While Alu-mediated recombination dominates α-thalassemia events, LINE-mediated disruption contributes to β-thalassemias [121].
Retrotransposons in hematopoietic neoplasias
Despite the high incidence of leukemias, lymphomas and myelomas (about 10% cancer diagnosed) the role of retrotransposon elements as inherited predisposing factors or somatic mutagens is under explored [105]. Loss of DNA methylation and retrotransposon integration into clinically sensitive genes has been reported in CLL [122, 123], multiple myeloma (MM) [124].
sORFs and micropepetides
A potentially translatable sequence of in-frame sense codons beginning with a start codon and ending with a stop codon is known as on open reading frame (ORF). Translatable ORFs are the sequences of mRNA that give rise to its principle protein, often referred to as the coding DNA sequence (CDS). sORFs are distinguished from all other ORFs by size, and like longer ORFs not all are translated or even translatable [5]. The theoretical minimal size of sORFs is two codons consisting of a start and stop codon, while the upper limit remains ambiguous. Some studies describe sORFs of 200–250 codons, however the general consensus suggests an upper limit of approximately 100 [125–127].
Identification of sORFs with coding potential using conventional gene prediction software is problematic. These programs are designed to assess the coding potential of ORFs that are longer than 100 codons, based on how rich they are in features such as canonical initiation codons, termination sites, splice sites, promoter sequences, polyadenylation signals, codon usage bias, nucleotide composition and in-frame hexamer frequency [128, 129]. Consequently, many gene annotation algorithms will dismiss ORFs of smaller than 100 codons as meaningless [130, 131]. Improvements in high-throughput identification of translation start sites, sensitivity of proteomic techniques, specialized algorithms for identifying putative coding sORFs and better integration of bio-informatics, genomic and proteomic outputs have assisted our ability to predict translatable sORFs [5]. Despite improvements in identification of sORFs with coding potential, it is believed relatively few functional sequences are likely to be present within a large pool of mostly non-functional sORFs [130, 131].
Location of functional sORFs
Short and intermediate ncRNAs are considered too small (that is, <200 nucleotides in length) to support translation [132], suggesting translatable sORFs will only be found on lncRNAs. This assumption may or may not hold as ribosome profiling has detected translation initiation sites on smaller transcripts, previously thought to be non-coding [12]. Translatable sORFs have been identified within, or overlapping, CDS regions of mRNAs, the 5′ and 3′ trailer sequences, and in transcripts previously ascribed as non-coding RNAs, including lncRNAs [5]. While no role for micropeptides or other sORF products have been determined to date, our understanding of this class of molecule is just emerging and discoveries are likely just around the proverbial corner.
Concluding Remarks
Despite seemingly exponential growth in our characterization of new roles for ncRNA in hematopoiesis, our understanding is far from complete. However, it is clear these molecules are deeply embedded in the regulatory processes of hematopoiesis. From miRNAs to lncRNAs virtually all aspects of transcription, translation, localization, stabilization, structural integrity, complex formation, differentiation and evolution are intricately balanced with cell type and differentiation specific accuracy. As new roles emerge, existing classification schemes are challenged to accommodate and categorize the myriad of functions performed by these molecules encoded by regions of the genome once thought of as “junk” DNA. No doubt, as our understanding of how ncRNA modulates hematopoiesis, we will uncover new therapeutic approaches to combat hematologic afflictions.
Highlights.
A paradigm shift in our view of the role of RNA in cellular biology has occurred and the volume of new information has been overwhelming. In this review, we aim to broadly summarize recent advances and examine the trends and challenges arising in this burgeoning research area.
We examine the broad categories of non-coding RNA biology (microRNAs, lncRNAs, piRNAs as well as pseudogenes, translocons and micropeptides) with a focus on the classification and roles of the classes of non-coding RNAs, with pertinent examples relevant to hematopoiesis.
Acknowledgments
Dr. Sakamoto is funded by NIH R01DK107286 and Mark Wilkes is funded by USHHS Ruth L. Kirschstein Institutional National Research Service Award # T32 DK098132.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–8. doi: 10.1038/nature11233. Epub 2012/09/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. Epub 2012/09/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lafontaine DL. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nature structural & molecular biology. 2015;22(1):11–9. doi: 10.1038/nsmb.2939. Epub 2015/01/08. [DOI] [PubMed] [Google Scholar]
- 4.Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474(7351):395–8. doi: 10.1038/nature10165. Epub 2011/06/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nature reviews Genetics. 2014;15(3):193–204. doi: 10.1038/nrg3520. Epub 2014/02/12. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. doi: 10.1016/0092-8674(93)90529-y. Epub 1993/12/03. [DOI] [PubMed] [Google Scholar]
- 7.Lazare SS, Wojtowicz EE, Bystrykh LV, de Haan G. microRNAs in hematopoiesis. Experimental cell research. 2014;329(2):234–8. doi: 10.1016/j.yexcr.2014.08.033. Epub 2014/09/07. [DOI] [PubMed] [Google Scholar]
- 8.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science (New York, NY) 2001;294(5543):862–4. doi: 10.1126/science.1065329. Epub 2001/10/27. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107(7):823–6. doi: 10.1016/s0092-8674(01)00616-x. Epub 2002/01/10. [DOI] [PubMed] [Google Scholar]
- 10.Grosswendt S, Filipchyk A, Manzano M, Klironomos F, Schilling M, Herzog M, et al. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Molecular cell. 2014;54(6):1042–54. doi: 10.1016/j.molcel.2014.03.049. Epub 2014/05/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102(6):765–75. doi: 10.1016/s0092-8674(00)00065-9. Epub 2000/10/13. [DOI] [PubMed] [Google Scholar]
- 12.Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(37):E2424–32. doi: 10.1073/pnas.1207846109. Epub 2012/08/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science (New York, NY) 2004;303(5654):83–6. doi: 10.1126/science.1091903. Epub 2003/12/06. [DOI] [PubMed] [Google Scholar]
- 14.Jin H, Vacic V, Girke T, Lonardi S, Zhu JK. Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC molecular biology. 2008;9:6. doi: 10.1186/1471-2199-9-6. Epub 2008/01/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Vecchiarelli-Federico LM, Li YJ, Egan SE, Spaner D, Hough MR, et al. The miR-17-92 cluster expands multipotent hematopoietic progenitors whereas imbalanced expression of its individual oncogenic miRNAs promotes leukemia in mice. Blood. 2012;119(19):4486–98. doi: 10.1182/blood-2011-09-378687. Epub 2012/03/28. [DOI] [PubMed] [Google Scholar]
- 16.Xiao H, Murakami H, Suga H, Ferre-D’Amare AR. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature. 2008;454(7202):358–61. doi: 10.1038/nature07033. Epub 2008/06/13. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Huang H, Chen P, He M, Li Y, Arnovitz S, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nature communications. 2012;3:688. doi: 10.1038/ncomms1681. Epub 2012/02/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Velu CS, Baktula AM, Grimes HL. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood. 2009;113(19):4720–8. doi: 10.1182/blood-2008-11-190215. Epub 2009/03/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wojtowicz EE, Walasek MA, Broekhuis MJ, Weersing E, Ritsema M, Ausema A, et al. MicroRNA-125 family members exert a similar role in the regulation of murine hematopoiesis. Experimental hematology. 2014;42(10):909–18e1. doi: 10.1016/j.exphem.2014.06.010. Epub 2014/08/06. [DOI] [PubMed] [Google Scholar]
- 20.Chaudhuri AA, So AY, Mehta A, Minisandram A, Sinha N, Jonsson VD, et al. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28A. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(11):4233–8. doi: 10.1073/pnas.1200677109. Epub 2012/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS biology. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. Epub 2004/10/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrie CH. microRNA expression in erythropoiesis and erythroid disorders. British journal of haematology. 2010;150(2):144–51. doi: 10.1111/j.1365-2141.2009.07978.x. Epub 2009/11/17. [DOI] [PubMed] [Google Scholar]
- 23.Bruchova-Votavova H, Yoon D, Prchal JT. miR-451 enhances erythroid differentiation in K562 cells. Leukemia & lymphoma. 2010;51(4):686–93. doi: 10.3109/10428191003629362. Epub 2010/03/12. [DOI] [PubMed] [Google Scholar]
- 24.Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, et al. The miR-144/451 locus is required for erythroid homeostasis. The Journal of experimental medicine. 2010;207(7):1351–8. doi: 10.1084/jem.20100458. Epub 2010/06/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svasti S, Masaki S, Penglong T, Abe Y, Winichagoon P, Fucharoen S, et al. Expression of microRNA-451 in normal and thalassemic erythropoiesis. Annals of hematology. 2010;89(10):953–8. doi: 10.1007/s00277-010-0980-7. Epub 2010/05/13. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Zhang J, Gao L, McClellan S, Finan MA, Butler TW, et al. MiR-181 mediates cell differentiation by interrupting the Lin28 and let-7 feedback circuit. Cell death and differentiation. 2012;19(3):378–86. doi: 10.1038/cdd.2011.127. Epub 2011/10/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ichimura A, Ruike Y, Terasawa K, Shimizu K, Tsujimoto G. MicroRNA-34a inhibits cell proliferation by repressing mitogen-activated protein kinase kinase 1 during megakaryocytic differentiation of K562 cells. Molecular pharmacology. 2010;77(6):1016–24. doi: 10.1124/mol.109.063321. Epub 2010/03/20. [DOI] [PubMed] [Google Scholar]
- 28.Navarro F, Gutman D, Meire E, Caceres M, Rigoutsos I, Bentwich Z, et al. miR-34a contributes to megakaryocytic differentiation of K562 cells independently of p53. Blood. 2009;114(10):2181–92. doi: 10.1182/blood-2009-02-205062. Epub 2009/07/09. [DOI] [PubMed] [Google Scholar]
- 29.Romania P, Lulli V, Pelosi E, Biffoni M, Peschle C, Marziali G. MicroRNA 155 modulates megakaryopoiesis at progenitor and precursor level by targeting Ets-1 and Meis1 transcription factors. British journal of haematology. 2008;143(4):570–80. doi: 10.1111/j.1365-2141.2008.07382.x. Epub 2008/10/28. [DOI] [PubMed] [Google Scholar]
- 30.Ben-Ami O, Pencovich N, Lotem J, Levanon D, Groner Y. A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(1):238–43. doi: 10.1073/pnas.0811466106. Epub 2008/12/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123(5):819–31. doi: 10.1016/j.cell.2005.09.023. Epub 2005/12/06. [DOI] [PubMed] [Google Scholar]
- 32.Fukao T, Fukuda Y, Kiga K, Sharif J, Hino K, Enomoto Y, et al. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129(3):617–31. doi: 10.1016/j.cell.2007.02.048. Epub 2007/05/08. [DOI] [PubMed] [Google Scholar]
- 33.Feng J, Iwama A, Satake M, Kohu K. MicroRNA-27 enhances differentiation of myeloblasts into granulocytes by post-transcriptionally downregulating Runx1. British journal of haematology. 2009;145(3):412–23. doi: 10.1111/j.1365-2141.2009.07632.x. Epub 2009/03/21. [DOI] [PubMed] [Google Scholar]
- 34.Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nature cell biology. 2007;9(7):775–87. doi: 10.1038/ncb1613. Epub 2007/06/26. [DOI] [PubMed] [Google Scholar]
- 35.Chung SS, Hu W, Park CY. The Role of MicroRNAs in Hematopoietic Stem Cell and Leukemic Stem Cell Function. Therapeutic advances in hematology. 2011;2(5):317–34. doi: 10.1177/2040620711410772. Epub 2011/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(5):1604–9. doi: 10.1073/pnas.0610731104. Epub 2007/01/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, et al. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(8):2735–40. doi: 10.1073/pnas.0811073106. Epub 2009/02/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(33):12481–6. doi: 10.1073/pnas.0605298103. Epub 2006/08/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(13):5282–7. doi: 10.1073/pnas.0810909106. Epub 2009/03/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes & development. 2007;21(5):578–89. doi: 10.1101/gad.1522907. Epub 2007/03/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. The Journal of experimental medicine. 2008;205(9):2005–17. doi: 10.1084/jem.20081219. Epub 2008/08/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129(1):147–61. doi: 10.1016/j.cell.2007.03.008. Epub 2007/03/27. [DOI] [PubMed] [Google Scholar]
- 43.Henao-Mejia J, Williams A, Goff LA, Staron M, Licona-Limon P, Kaech SM, et al. The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity. 2013;38(5):984–97. doi: 10.1016/j.immuni.2013.02.021. Epub 2013/04/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghisi M, Corradin A, Basso K, Frasson C, Serafin V, Mukherjee S, et al. Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150. Blood. 2011;117(26):7053–62. doi: 10.1182/blood-2010-12-326629. Epub 2011/05/10. [DOI] [PubMed] [Google Scholar]
- 45.Belkaya S, Murray SE, Eitson JL, de la Morena MT, Forman JA, van Oers NS. Transgenic expression of microRNA-185 causes a developmental arrest of T cells by targeting multiple genes including Mzb1. The Journal of biological chemistry. 2013;288(42):30752–62. doi: 10.1074/jbc.M113.503532. Epub 2013/09/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, Cook T, et al. A role for Dicer in immune regulation. The Journal of experimental medicine. 2006;203(11):2519–27. doi: 10.1084/jem.20061692. Epub 2006/10/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32(6):828–39. doi: 10.1016/j.immuni.2010.05.009. Epub 2010/07/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yokota T. Guest editorial: molecular mechanisms of lymphocyte development: recent findings. International journal of hematology. 2014;100(3):218–9. doi: 10.1007/s12185-014-1645-4. Epub 2014/08/06. [DOI] [PubMed] [Google Scholar]
- 49.Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142(6):914–29. doi: 10.1016/j.cell.2010.08.012. Epub 2010/09/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. Journal of immunology (Baltimore, Md: 1950) 2009;182(5):2578–82. doi: 10.4049/jimmunol.0803162. Epub 2009/02/24. [DOI] [PubMed] [Google Scholar]
- 51.Kelada S, Sethupathy P, Okoye IS, Kistasis E, Czieso S, White SD, et al. miR-182 and miR-10a are key regulators of Treg specialisation and stability during Schistosome and Leishmania-associated inflammation. PLoS pathogens. 2013;9(6):e1003451. doi: 10.1371/journal.ppat.1003451. Epub 2013/07/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, et al. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118(20):5487–97. doi: 10.1182/blood-2011-05-355644. Epub 2011/10/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Banerjee A, Schambach F, DeJong CS, Hammond SM, Reiner SL. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. European journal of immunology. 2010;40(1):225–31. doi: 10.1002/eji.200939381. Epub 2009/10/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawant DV, Wu H, Kaplan MH, Dent AL. The Bcl6 target gene microRNA-21 promotes Th2 differentiation by a T cell intrinsic pathway. Molecular immunology. 2013;54(3–4):435–42. doi: 10.1016/j.molimm.2013.01.006. Epub 2013/02/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(44):18704–9. doi: 10.1073/pnas.0905063106. Epub 2009/10/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu RHT, Kagele DA, Runtsch MC, Bake E, Chaudhuri AA, et al. MicroRNA-155 confers encephalogenic potential to Th17 cells by promoting effector gene expression. J Immunol. 2013;190:5972–80. doi: 10.4049/jimmunol.1300351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu SQ, Jiang S, Li C, Zhang B, Li QJ. miR-17-92 cluster targets phosphatase and tensin homology and Ikaros Family Zinc Finger 4 to promote TH17-mediated inflammation. The Journal of biological chemistry. 2014;289(18):12446–56. doi: 10.1074/jbc.M114.550723. Epub 2014/03/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132(5):860–74. doi: 10.1016/j.cell.2008.02.020. Epub 2008/03/11. [DOI] [PubMed] [Google Scholar]
- 59.Xu S, Guo K, Zeng Q, Huo J, Lam KP. The RNase III enzyme Dicer is essential for germinal center B-cell formation. Blood. 2012;119(3):767–76. doi: 10.1182/blood-2011-05-355412. Epub 2011/11/26. [DOI] [PubMed] [Google Scholar]
- 60.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–86. doi: 10.1016/j.cell.2008.02.019. Epub 2008/03/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rao DS, O’Connell RM, Chaudhuri AA, Garcia-Flores Y, Geiger TL, Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33(1):48–59. doi: 10.1016/j.immuni.2010.06.013. Epub 2010/07/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu H, Wang B, Borde M, Nardone J, Maika S, Allred L, et al. Foxp1 is an essential transcriptional regulator of B cell development. Nature immunology. 2006;7(8):819–26. doi: 10.1038/ni1358. Epub 2006/07/05. [DOI] [PubMed] [Google Scholar]
- 63.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, et al. MiR-150 Controls B Cell Differentiation by Targeting the Transcription Factor c-Myb. Cell. 2016;165(4):1027. doi: 10.1016/j.cell.2016.04.056. Epub 2016/05/07. [DOI] [PubMed] [Google Scholar]
- 64.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. Regulation of the germinal center response by microRNA-155. Science (New York, NY) 2007;316(5824):604–8. doi: 10.1126/science.1141229. Epub 2007/04/28. [DOI] [PubMed] [Google Scholar]
- 65.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science (New York, NY) 2007;316(5824):608–11. doi: 10.1126/science.1139253. Epub 2007/04/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bamezai S, Rawat VP, Buske C. Concise review: The Piwi-piRNA axis: pivotal beyond transposon silencing. Stem cells (Dayton, Ohio) 2012;30(12):2603–11. doi: 10.1002/stem.1237. Epub 2012/09/22. [DOI] [PubMed] [Google Scholar]
- 67.Jacobs JE, Wagner M, Dhahbi J, Boffelli D, Martin DI. Deficiency of MIWI2 (Piwil4) induces mouse erythroleukemia cell differentiation, but has no effect on hematopoiesis in vivo. PloS one. 2013;8(12):e82573. doi: 10.1371/journal.pone.0082573. Epub 2014/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. Epub 2007/06/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22(9):1775–89. doi: 10.1101/gr.132159.111. Epub 2012/09/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22(9):1760–74. doi: 10.1101/gr.135350.111. Epub 2012/09/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nature genetics. 2015;47(3):199–208. doi: 10.1038/ng.3192. Epub 2015/01/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nature reviews Genetics. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. Epub 2015/12/17. [DOI] [PubMed] [Google Scholar]
- 73.Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160(4):595–606. doi: 10.1016/j.cell.2015.01.009. Epub 2015/02/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nature reviews Molecular cell biology. 2013;14(11):699–712. doi: 10.1038/nrm3679. Epub 2013/10/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. Epub 2011/08/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome biology. 2009;10(11):R124. doi: 10.1186/gb-2009-10-11-r124. Epub 2009/11/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. Epub 2010/10/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ponjavic J, Ponting CP. The long and the short of RNA maps. BioEssays: news and reviews in molecular, cellular and developmental biology. 2007;29(11):1077–80. doi: 10.1002/bies.20669. Epub 2007/10/16. [DOI] [PubMed] [Google Scholar]
- 79.Nobili L, Lionetti M, Neri A. Long non-coding RNAs in normal and malignant hematopoiesis. Oncotarget. 2016;7(31):50666–81. doi: 10.18632/oncotarget.9308. Epub 2016/05/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends in immunology. 2014;35(9):408–19. doi: 10.1016/j.it.2014.07.005. Epub 2014/08/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baertsch R, Diekhans M, Kent WJ, Haussler D, Brosius J. Retrocopy contributions to the evolution of the human genome. BMC genomics. 2008;9:466. doi: 10.1186/1471-2164-9-466. Epub 2008/10/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nature reviews Molecular cell biology. 2017;18(1):18–30. doi: 10.1038/nrm.2016.116. Epub 2016/11/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(23):9362–7. doi: 10.1073/pnas.0903103106. Epub 2009/05/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alvarez-Dominguez JR, Hu W, Yuan B, Shi J, Park SS, Gromatzky AA, et al. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014;123(4):570–81. doi: 10.1182/blood-2013-10-530683. Epub 2013/11/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes & development. 2011;25(24):2573–8. doi: 10.1101/gad.178780.111. Epub 2011/12/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paralkar VR, Mishra T, Luan J, Yao Y, Kossenkov AV, Anderson SM, et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood. 2014;123(12):1927–37. doi: 10.1182/blood-2013-12-544494. Epub 2014/02/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wagner LA, Christensen CJ, Dunn DM, Spangrude GJ, Georgelas A, Kelley L, et al. EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood. 2007;109(12):5191–8. doi: 10.1182/blood-2006-06-027987. Epub 2007/03/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H, et al. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nature immunology. 2003;4(10):1029–36. doi: 10.1038/ni973. Epub 2003/09/06. [DOI] [PubMed] [Google Scholar]
- 89.Ebralidze AK, Guibal FC, Steidl U, Zhang P, Lee S, Bartholdy B, et al. PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes & development. 2008;22(15):2085–92. doi: 10.1101/gad.1654808. Epub 2008/08/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113(11):2526–34. doi: 10.1182/blood-2008-06-162164. Epub 2009/01/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wei S, Wang K. Long noncoding RNAs: pivotal regulators in acute myeloid leukemia. Experimental hematology & oncology. 2015;5:30. doi: 10.1186/s40164-016-0059-9. Epub 2016/12/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chinen Y, Sakamoto N, Nagoshi H, Taki T, Maegawa S, Tatekawa S, et al. 8q24 amplified segments involve novel fusion genes between NSMCE2 and long noncoding RNAs in acute myelogenous leukemia. Journal of hematology & oncology. 2014;7:68. doi: 10.1186/s13045-014-0068-2. Epub 2014/09/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hirano T, Yoshikawa R, Harada H, Harada Y, Ishida A, Yamazaki T. Long noncoding RNA, CCDC26, controls myeloid leukemia cell growth through regulation of KIT expression. Molecular cancer. 2015;14:90. doi: 10.1186/s12943-015-0364-7. Epub 2015/05/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Casero D, Sandoval S, Seet CS, Scholes J, Zhu Y, Ha VL, et al. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nature immunology. 2015;16(12):1282–91. doi: 10.1038/ni.3299. Epub 2015/10/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. Journal of immunology (Baltimore, Md: 1950) 2009;182(12):7738–48. doi: 10.4049/jimmunol.0900603. Epub 2009/06/06. [DOI] [PubMed] [Google Scholar]
- 96.Aoki K, Harashima A, Sano M, Yokoi T, Nakamura S, Kibata M, et al. A thymus-specific noncoding RNA, Thy-ncR1, is a cytoplasmic riboregulator of MFAP4 mRNA in immature T-cell lines. BMC molecular biology. 2010;11:99. doi: 10.1186/1471-2199-11-99. Epub 2010/12/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sharma S, Findlay GM, Bandukwala HS, Oberdoerffer S, Baust B, Li Z, et al. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(28):11381–6. doi: 10.1073/pnas.1019711108. Epub 2011/06/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJ, Curti S, et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nature immunology. 2015;16(3):318–25. doi: 10.1038/ni.3093. Epub 2015/01/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Elton TS, Selemon H, Elton SM, Parinandi NL. Regulation of the MIR155 host gene in physiological and pathological processes. Gene. 2013;532(1):1–12. doi: 10.1016/j.gene.2012.12.009. Epub 2012/12/19. [DOI] [PubMed] [Google Scholar]
- 100.Petri A, Dybkaer K, Bogsted M, Thrue CA, Hagedorn PH, Schmitz A, et al. Long Noncoding RNA Expression during Human B-Cell Development. PloS one. 2015;10(9):e0138236. doi: 10.1371/journal.pone.0138236. Epub 2015/09/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vanin EF. Processed pseudogenes: characteristics and evolution. Annual review of genetics. 1985;19:253–72. doi: 10.1146/annurev.ge.19.120185.001345. Epub 1985/01/01. [DOI] [PubMed] [Google Scholar]
- 102.Dewannieux M, Heidmann T. Role of poly(A) tail length in Alu retrotransposition. Genomics. 2005;86(3):378–81. doi: 10.1016/j.ygeno.2005.05.009. Epub 2005/07/05. [DOI] [PubMed] [Google Scholar]
- 103.Graur D, Shuali Y, Li WH. Deletions in processed pseudogenes accumulate faster in rodents than in humans. Journal of molecular evolution. 1989;28(4):279–85. doi: 10.1007/BF02103423. Epub 1989/04/01. [DOI] [PubMed] [Google Scholar]
- 104.Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, et al. A draft map of the human proteome. Nature. 2014;509(7502):575–81. doi: 10.1038/nature13302. Epub 2014/05/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goodhead I, Darby AC. Taking the pseudo out of pseudogenes. Current opinion in microbiology. 2015;23:102–9. doi: 10.1016/j.mib.2014.11.012. Epub 2014/12/03. [DOI] [PubMed] [Google Scholar]
- 106.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72(4):595–605. doi: 10.1016/0092-8674(93)90078-5. Epub 1993/02/26. [DOI] [PubMed] [Google Scholar]
- 107.Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, Weichenrieder O, et al. Active Alu retrotransposons in the human genome. Genome research. 2008;18(12):1875–83. doi: 10.1101/gr.081737.108. Epub 2008/10/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. The EMBO journal. 2002;21(21):5899–910. doi: 10.1093/emboj/cdf592. Epub 2002/11/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Robbez-Masson L, Rowe HM. Retrotransposons shape species-specific embryonic stem cell gene expression. Retrovirology. 2015;12:45. doi: 10.1186/s12977-015-0173-5. Epub 2015/05/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, et al. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Molecular and cellular biology. 2002;22(2):480–91. doi: 10.1128/MCB.22.2.480-491.2002. Epub 2002/01/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Choi YJ, Lin CP, Risso D, Chen S, Kim TA, Tan MH, et al. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science (New York, NY) 2017;355(6325) doi: 10.1126/science.aag1927. Epub 2017/01/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kazazian HH, Jr, Wong CYH, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–6. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 113.Van de Water N, Williams R, Ockelford P, Browett P. A 20.7 kb deletion within the factor VIII gene associated with LINE-1 element insertion. Thrombosis and haemostasis. 1998;79(5):938–42. Epub 1998/06/03. [PubMed] [Google Scholar]
- 114.Sukarova E, Dimovski AJ, Tchacarova P, Petkov GH, Efremov GD. An Alu insert as the cause of a severe form of hemophilia A. Acta haematologica. 2001;106(3):126–9. doi: 10.1159/000046602. Epub 2001/11/20. [DOI] [PubMed] [Google Scholar]
- 115.Ganguly A, Dunbar T, Chen P, Godmilow L, Ganguly T. Exon skipping caused by an intronic insertion of a young Alu Yb9 element leads to severe hemophilia A. Human genetics. 2003;113(4):348–52. doi: 10.1007/s00439-003-0986-5. Epub 2003/07/29. [DOI] [PubMed] [Google Scholar]
- 116.Li X, Scaringe WA, Hill KA, Roberts S, Mengos A, Careri D, et al. Frequency of recent retrotransposition events in the human factor IX gene. Human mutation. 2001;17(6):511–9. doi: 10.1002/humu.1134. Epub 2001/06/01. [DOI] [PubMed] [Google Scholar]
- 117.Mukherjee S, Mukhopadhyay A, Banerjee D, Chandak GR, Ray K. Molecular pathology of haemophilia B: identification of five novel mutations including a LINE 1 insertion in Indian patients. Haemophilia: the official journal of the World Federation of Hemophilia. 2004;10(3):259–63. doi: 10.1111/j.1365-2516.2004.00895.x. Epub 2004/04/17. [DOI] [PubMed] [Google Scholar]
- 118.Vidaud D, Vidaud M, Bahnak BR, Siguret V, Gispert Sanchez S, Laurian Y, et al. Haemophilia B due to a de novo insertion of a human-specific Alu subfamily member within the coding region of the factor IX gene. European journal of human genetics: EJHG. 1993;1(1):30–6. doi: 10.1159/000472385. Epub 1993/01/01. [DOI] [PubMed] [Google Scholar]
- 119.Wulff K, Gazda H, Schroder W, Robicka-Milewska R, Herrmann FH. Identification of a novel large F9 gene mutation-an insertion of an Alu repeated DNA element in exon e of the factor 9 gene. Human mutation. 2000;15(3):299. doi: 10.1002/(SICI)1098-1004(200003)15:3<299::AID-HUMU23>3.0.CO;2-1. Epub 2000/02/19. [DOI] [PubMed] [Google Scholar]
- 120.Mitchell M, Dai L, Savidge G, Alhaq A. An Alu-mediated 31.5-kb deletion as the cause of factor XI deficiency in 2 unrelated patients. Blood. 2004;104(8):2394–6. doi: 10.1182/blood-2004-04-1318. Epub 2004/07/01. [DOI] [PubMed] [Google Scholar]
- 121.Henthorn PS, Smithies O, Mager DL. Molecular analysis of deletions in the human beta-globin gene cluster: deletion junctions and locations of breakpoints. Genomics. 1990;6(2):226–37. doi: 10.1016/0888-7543(90)90561-8. Epub 1990/02/01. [DOI] [PubMed] [Google Scholar]
- 122.Wahlfors J, Hiltunen H, Heinonen K, Hamalainen E, Alhonen L, Janne J. Genomic hypomethylation in human chronic lymphocytic leukemia. Blood. 1992;80(8):2074–80. Epub 1992/10/15. [PubMed] [Google Scholar]
- 123.Dante R, Dante-Paire J, Rigal D, Roizes G. Methylation patterns of long interspersed repeated DNA and alphoid repetitive DNA from human cell lines and tumors. Anticancer research. 1992;12(2):559–63. Epub 1992/03/01. [PubMed] [Google Scholar]
- 124.Bollati V, Fabris S, Pegoraro V, Ronchetti D, Mosca L, Deliliers GL, et al. Differential repetitive DNA methylation in multiple myeloma molecular subgroups. Carcinogenesis. 2009;30(8):1330–5. doi: 10.1093/carcin/bgp149. Epub 2009/06/18. [DOI] [PubMed] [Google Scholar]
- 125.Lease KA, Walker JC. The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant physiology. 2006;142(3):831–8. doi: 10.1104/pp.106.086041. Epub 2006/09/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hayden CA, Bosco G. Comparative genomic analysis of novel conserved peptide upstream open reading frames in Drosophila melanogaster and other dipteran species. BMC genomics. 2008;9:61. doi: 10.1186/1471-2164-9-61. Epub 2008/02/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang X, Tschaplinski TJ, Hurst GB, Jawdy S, Abraham PE, Lankford PK, et al. Discovery and annotation of small proteins using genomics, proteomics, and computational approaches. Genome research. 2011;21(4):634–41. doi: 10.1101/gr.109280.110. Epub 2011/03/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hanada K, Zhang X, Borevitz JO, Li WH, Shiu SH. A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome research. 2007;17(5):632–40. doi: 10.1101/gr.5836207. Epub 2007/03/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cheng H, Chan WS, Li Z, Wang D, Liu S, Zhou Y. Small open reading frames: current prediction techniques and future prospect. Current protein & peptide science. 2011;12(6):503–7. doi: 10.2174/138920311796957667. Epub 2011/07/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Basrai MA, Hieter P, Boeke JD. Small open reading frames: beautiful needles in the haystack. Genome research. 1997;7(8):768–71. doi: 10.1101/gr.7.8.768. Epub 1997/08/01. [DOI] [PubMed] [Google Scholar]
- 131.Claverie JM. Computational methods for the identification of genes in vertebrate genomic sequences. Human molecular genetics. 1997;6(10):1735–44. doi: 10.1093/hmg/6.10.1735. Epub 1997/01/01. [DOI] [PubMed] [Google Scholar]
- 132.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423(6937):241–54. doi: 10.1038/nature01644. Epub 2003/05/16. [DOI] [PubMed] [Google Scholar]
- 133.Ku HY, Lin H. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. National science review. 2014;1(2):205–18. doi: 10.1093/nsr/nwu014. Epub 2014/12/17. [DOI] [PMC free article] [PubMed] [Google Scholar]