SUMMARY
Gene expression can be post-transcriptionally regulated via dynamic and reversible RNA modifications. N1-methyladenosine (m1A) is a recently identified mRNA modification; however, little is known about its precise location and biogenesis. Here, we develop a base-resolution m1A profiling method, based on m1A-induced misincorporation during reverse transcription, and report distinct classes of m1A methylome in the human transcriptome. m1A in 5′-UTR, particularly those at the mRNA cap, associate with increased translation efficiency. A different, small subset of m1A exhibit a GUUCRA tRNA-like motif, are evenly distributed in the transcriptome and are dependent on the methyltransferase TRMT6/61A. Additionally, we show that m1A is prevalent in the mitochondrial-encoded transcripts. Manipulation of m1A level via TRMT61B, a mitochondria-localizing m1A methyltransferase, demonstrates that m1A in mitochondrial mRNA interferes with translation. Collectively, our approaches reveal distinct classes of m1A methylome and provide a resource for functional studies of m1A-mediated epitranscriptomic regulation.
eTOC blurb
Li et al. developed a single-nucleotide-resolution technology, named “m1A-MAP”, to profile m1A in the human transcriptome, and revealed distinct classes of m1A methylome in nuclear- and mitochondrial-encoded transcripts.
INTRODUCTION
More than 100 different types of post-transcriptional modifications have been identified so far (Machnicka et al., 2013). Recent breakthroughs in sequencing technologies have greatly advanced our understanding to the location, regulation, and function of RNA modifications in the transcriptome (Frye et al., 2016; Fu et al., 2014; Helm and Motorin, 2017; Li et al., 2016b), leading to the emerging field of epitranscriptomics (He, 2010; Saletore et al., 2012). One such example is N1-methyladenosine (m1A), a prevalent modification in non-coding RNA (ncRNA) and messenger RNA (mRNA) (Anderson and Droogmans, 2005; Dominissini et al., 2016; Li et al., 2016a; Roundtree et al., 2017). m1A was first documented more than 50 years ago (Dunn, 1961); later it was found to be a primordial RNA modification across the three major phylogenetic domains (Machnicka et al., 2013). In human cells, m1A is found at position 9 and 58 of human mitochondrial and cytoplasmic tRNAs, catalyzed by TRMT10C, TRMT61B and TRMT6/61A, respectively (Chujo and Suzuki, 2012; Ozanick et al., 2005; Vilardo et al., 2012); it is also present at position 1322 of 28S rRNA, catalyzed by NML (Waku et al., 2016). Its unique physicochemical property has also endowed m1A with pivotal roles in maintaining the proper structure and function of these ncRNAs (Roundtree et al., 2017). m1A in tRNA has also been systematically evaluated by microarray and sequencing (Cozen et al., 2015; Saikia et al., 2010; Zheng et al., 2015); more recently, m1A58 is shown to be reversible by ALKBH1, demonstrating an example of reversible tRNA modification in translation regulation (Liu et al., 2016). In addition to ncRNAs, m1A is also found to be a dynamic modification in mammalian mRNA, with strong enrichment in the 5′-UTR (Dominissini et al., 2016; Li et al., 2016a).
Despite such rapid progress, a high-resolution profile of the mammalian m1A methylome is still lacking, significantly limiting our understanding and functional characterization of this newly discovered mRNA modification. Previous m1A profiling technologies have a resolution of about tens of nucleotides to several hundred nucleotides, primarily determined by the size of RNA fragments in these experiments (Dominissini et al., 2016; Li et al., 2016a). In addition, the methyltransferase(s) and functional consequence of mRNA m1A modification is poorly understood. Hence, except for a handful positions in rRNA and tRNA, little is known about the precise location, regulation and function of m1A in the human transcriptome.
Here, we report a base-resolution method to profile m1A in the human transcriptome. Our method is based on m1A-induced misincorporation during reverse transcription and reveals distinct classes of m1A methylome: a major group of m1A sites that are enriched in 5′-UTR, a small subset of GUUCRA(“R” denotes a purine)-tRNA like m1A sites with relatively even distribution in the transcriptome, and prevalent m1A modification in the CDS of 10/13 mitochondrial(mt)-encoded transcripts. m1A sites in the 5′-UTR, particularly those located at the first and second nucleotide of mRNA transcripts (or “cap+1” and “cap+2” position), are associated with increased translation efficiency. In contrast, m1A in the CDS of mt-mRNA inhibits translation. Collectively, our approaches reveal distinct classes of base-resolution m1A methylome in the nuclear- and mitochondrial-encoded transcripts, and provide an in-depth resource towards elucidating the functions of m1A methylation in mRNA.
DESIGN
Because m1A can cause both truncation and misincorporation during cDNA synthesis (Hauenschild et al., 2015; Zubradt et al., 2017), we first established the truncation and mutation profiles of different reserve transcriptases (RTases). We systematically compared the performance of several commercially available RTases (including AMV, SuperScript II, SuperScript III and TGIRT) under different conditions (Figure S1). We found that m1A can precisely induce misincorporation at the site of modification, while m1A-induced truncation is less accurate and can occur to the neighboring nucleotides. In addition, the truncation profile could be complicated by RNA secondary structures and the fragmentation process needed for library preparation. We concluded that the mutation profile contains a higher signal/noise ratio and is more precise in detecting the exact position of m1A. Among the RTases we tested, TGIRT demonstrated excellent read-through efficiency and relatively high mutation frequency at the site of m1A (Figure S1B), consistent with the recent DMS-MaPseq and DM-tRNA-seq results (Zheng et al., 2015; Zubradt et al., 2017). Moreover, we employed a ligation-based strand-specific library preparation protocol (Van Nostrand et al., 2016), which ensures that the m1A-induced mutation is within the sequenced fragment (see Method Details).
Because we only observed ~40–50% mutation rate at m1A1322 in 28S rRNA (Figure S2A), which is known to be of high modification level, we further examined the quantitative capability of TGIRT. We chemically synthesized two model RNA sequences with site-specific m1A modification. For m1A sites with ~97–98% modification level (measured by quantitative mass spectrometry) (Figure S2B), we consistently observed ~66–75% misincorporation (Figure S2C); the mismatch rate dropped non-linearly when we gradually lowered the modification level. Even with ~50% m1A modification, a mismatch rate of only ~9–10% was observed (Figure S2C). These findings suggest that the observed mutation rate is an underestimation of the actual modification level. While the TGIRT-based procedure can still detect m1A sites of high modification level, sequencing RNA directly with TGIRT may not be able to capture the m1A sites with averaging modification level in the transcriptome (~20% as previously measured by microarray) (Dominissini et al., 2016). To improve the sensitivity for transcriptome-wide m1A detection, we decided to couple the TGIRT-based procedure with an antibody mediated pre-enrichment step and an additional in vitro demethylation step (Figure 1A). We first showed that in vitro demethylation reaction mediated by the demethylase AlkB is more efficient than the Dimroth reaction, demonstrating ~98% and ~80% efficiency (Figure S2D), respectively. In addition, the extended treatment of RNA in alkaline condition during the Dimroth reaction leads to excessive RNA degradation (Figure S2E), potentially causing loss of RNA molecules. By integrating the enrichment and demethylation steps, we successfully maximized the dynamic range of m1A-induced mutational signature for m1A1322 in 28S rRNA (~47%, ~95% and ~0.9% in the Input, (−) and (+) demethylase samples, respectively), allowing sensitive and confident m1A detection (Figure 1B). We termed our approach misincorporation-assisted profiling of m1A, or m1A-MAP.
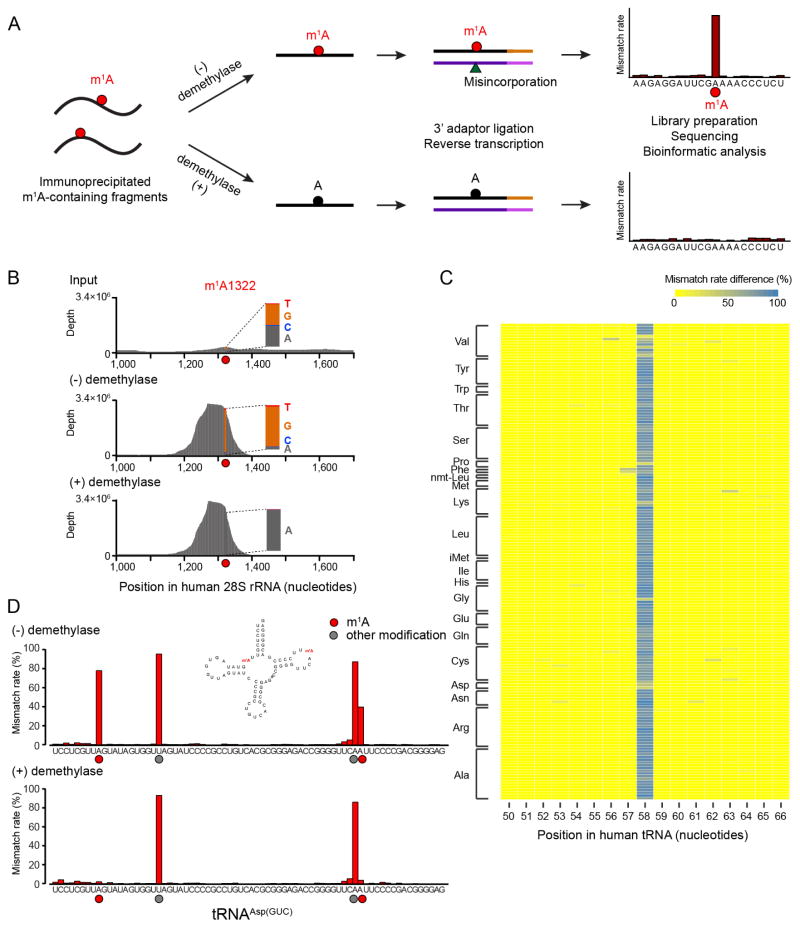
Figure 1. m1A-MAP utilizes m1A-induced misincorporation to detect m1A sites at single-nucleotide resolution.
(A) Scheme of m1A-MAP. We optimized the conditions of RT so as to allow efficient misincorporation in cDNA synthesis. The use of an m1A antibody pre-enriches the m1A-containing RNA fragments, thereby maximizing the misincorporation signal; and the use of demethylase treatment improves the confidence of detection. An m1A modification is called depending on the difference and fold change of mismatch rate between the (−) and (+) demethylase samples (see Method Details).
(B) m1A-MAP maximizes the misincorporation signal for m1A1322 on 28S rRNA.
(C) m1A-MAP detects m1A58 for the cytosolic tRNAs. Shown here is the difference of mismatch rate between the (−) and (+) demethylase samples.
(D) m1A-MAP detects an m1A site at position 9 in the cytosolic human tRNAAsp(GUC). The mismatch rate for m1A58 is also reduced after demethylase treatment, while two other modifications (at position 20 and 57) are not sensitive to demethylation, representing other types of RNA modifications.
See also Figure S1 and S2.
RESULTS
m1A-MAP detects m1A in tRNA
We first applied m1A-MAP to tRNA. In mammals, m1A can occur at position 9, 14 and 58 of tRNA (Anderson and Droogmans, 2005). m1A14 has been reported only in tRNAPhe and is considered to be very rare (Machnicka et al., 2013); we did not observe any m1A modification at position 14 for cytosolic tRNAs in HEK293T cells (Table S1). m1A58 is conserved across the three domains of life; previous tRNA microarray and sequencing data has reported hypomodified tRNAs at this position (Cozen et al., 2015; Saikia et al., 2010; Zheng et al., 2015). Our results confirmed that m1A58 is globally present in the cytosolic tRNAs (Figure 1C and S2F). A recent study reported m1A at position 9 in cytosolic tRNAAsp(GUC) (Clark et al., 2016); this site is also detected by m1A-MAP (Figure 1D). Collectively, these observations suggest that m1A-MAP is highly sensitive in detecting m1A at single-base resolution.
Single-nucleotide resolution m1A methylome in the transcriptome
We then sought to detect transcriptome-wide m1A methylome at single-base resolution. We defined two parameters to evaluate the m1A-MAP data: difference of mismatch rate and fold change of mismatch rate (see Method Details). To minimize the effect of mismatch rate variation during m1A identification, we rigorously tested our threshold and identified 740 m1A sites in the 293T transcriptome (Table S1–3). To evaluate potential false positives caused by m1A-independent mismatch, we performed a “reverse calculation” in which we artificially took the mismatch rate in (−) demethylase as the background and (+) demethylase as the signal, and retrieved only 17 such sites (see Method Details). Moreover, we also systematically evaluated the mutation pattern of the identified m1A sites in the transcriptome. Using m1A sites in tRNA as positive controls, we found that m1A-induced mutation is more strongly influenced by its 5′-nucleotide than the 3′-nucleotide; importantly, a similar sequence-dependent feature is also observed for m1A sites in mRNA (Figure 2A and S3A). Therefore, we conclude that our strict threshold allowed us to confidently detect transcriptome-wide m1A sites at single-nucleotide resolution.
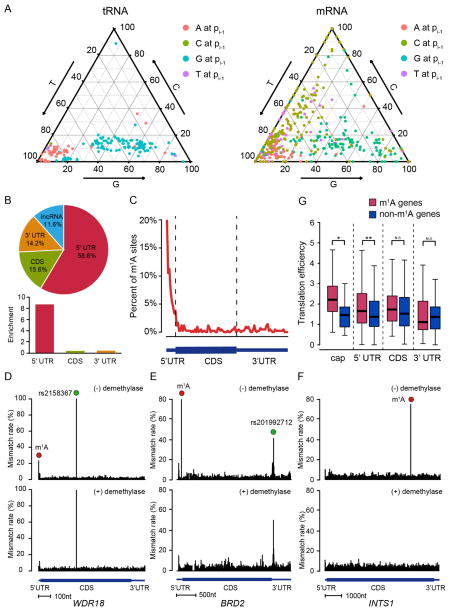
Figure 2. Single-nucleotide resolution m1A methylome in the human transcriptome.
(A) Mutation pattern of m1A sites in mRNA resembles that in tRNA. Shown here is the sequence-dependent mutation profile of m1A sites with regard to the immediate 5′ nucleotide.
(B) The pie chart shows the percentage of m1A sites in each non-overlapping segment.
(C) Distribution of m1A sites across mRNA segments. Each segment was normalized according to its average length in Refseq annotation.
(D–F) Representative views of a typical m1A site at the cap+1 position (the first transcribed nucleotide of mRNA) of WDR18 (D), the 5′-UTR of BRD2 (E), and the CDS of INTS1 mRNA (F). The demethylation-insensitive signals in (D) and (E), which are indicated as green dots, are known SNPs. The scale bars are indicated at the bottom of each panel.
(G) m1A in the cap (24 sites at the cap+1 and 3 sites at the cap+2 position) and 5′-UTR positively correlates with increased translation efficiency. Transcripts with comparable expression level but without m1A sites were chosen as the negative control for each category.
See also Figure S3.
Out of the 740 m1A modifications in the transcriptome (Figure S3B), 473 sites are located in mRNA and lncRNA molecules (Figure S3C and Table S2). Majority of these sites are within the 5′-UTR (Figure 2B and 2C), consistent with the previous finding (Dominissini et al., 2016; Li et al., 2016a). Our single-base profile also reveals multiple features of the m1A methylome: for instance, we found 24 m1A methylation sites that are present exactly at the first nucleotide of the 5′ end of the transcripts (Figure 2D and Table S2). Out of the 24 cap+1 m1A methylation, 22 are found in mRNA while the other 2 are located in lncRNA (Table S2). We further validated these cap+1 m1A sites using an antibody-independent, locus-specific approach (Figure S3D). We also found 3 additional transcripts with m1A methylation at the second nucleotide. For m1A located in CDS, while we did not observe a tendency for codon position, we did notice a mild preference for codon types, with Arg(CGA) being the most frequently modified by m1A (Figure S3E). No m1A is detected for AUG start codons. Representative examples of m1A sites identified from different mRNA regions are shown (Figure 2D–F and S3F). Two additional sites of high mutation, which are insensitive to the demethylase treatment, also appeared in the WDR18 and BRD2 examples (Figure 2D and 2E). By referring to the SNP database, we found that these two positions belong to annotated SNP sites, demonstrating the robustness of our approach in distinguishing true m1A sites from false signals (SNP, other modifications and etc.).
Because m1A is enriched in the 5′-UTR, we examined whether m1A could be involved in translation regulation. We performed ribosome profiling and compared the translation efficiency for transcripts with or without m1A. We found that m1A within the 5′-UTR positively correlates with the translation efficiency of mRNA (Figure 2G), in agreement with a previous report (Dominissini et al., 2016). This positive correlation is even stronger for m1A at the cap+1 position, but is not observed for m1A located in CDS nor 3′-UTR. This observation hints that m1A within different regions of mRNA may have different biological functions.
A subset of m1A sites demonstrate a GUUCRA consensus motif
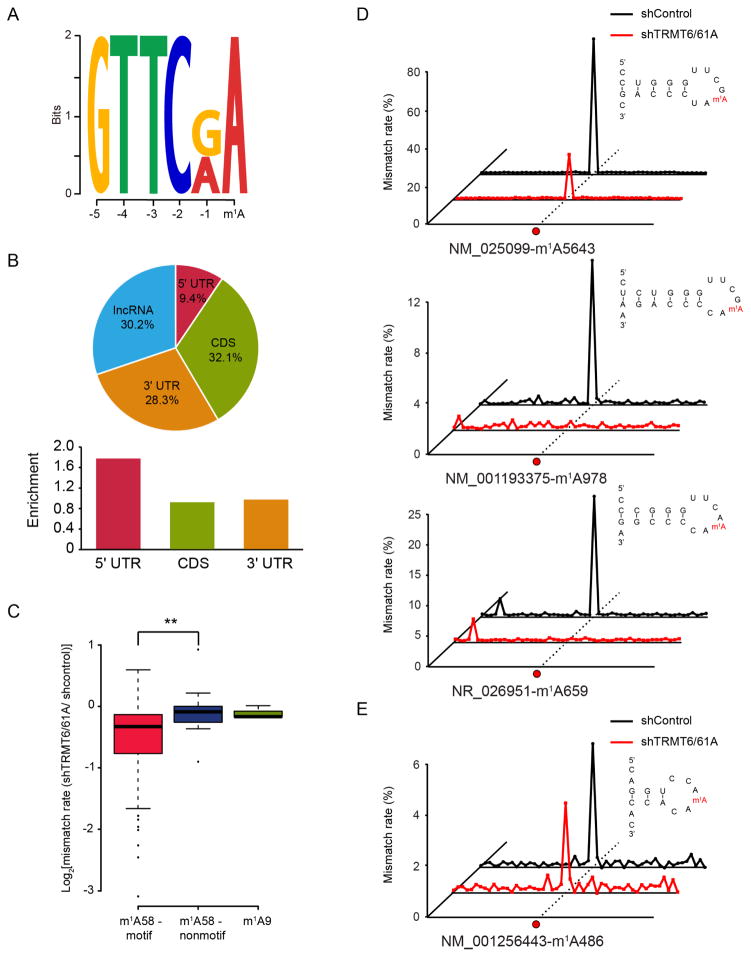
An unbiased motif detection using DREME revealed that a subset of m1A (53 sites) are found within a strong GUUCRA sequence (Figure 3A). Interestingly, these sites demonstrate a very different distribution pattern: instead of being enriched in the 5′-UTR, these sites are evenly distributed in the transcriptome (Figure 3B). Because this motif is reminiscent of the m1A-containing TψC loop in tRNA, we hypothesized that the tRNA methyltransferase complex TRMT6/61A could be responsible for these mRNA m1A sites. We first performed direct m1A sequencing (without antibody enrichment) to RNA population below 200 nt to examine the substrate specificity of TRMT6/61A. We found that the m1A58 sites within the GUUCRA motif experienced a global decrease of mutation rate in the TRMT6/61A knock-down sample, which was not observed for m1A58 sites that do not confine to the motif (Figure 3C, S4A and S4B). This result suggests that TRMT6/61A-mediated m1A methylation is highly sequence-specific, consistent with evidence from crystal structures (Finer-Moore et al., 2015). We then analyzed the secondary structure for the 53 mRNA m1A sites, and found highly conserved structural features compared to the T-loop of tRNA (Figure S4C and S4D). We also picked 3 m1A sites (in CDS, 3′-UTR and lncRNA, respectively) and examined their response after TRMT6/61A knock-down using a locus-specific approach (Figure 3D). This approach is based on targeted amplification using specific primers (see Method Details), hence enabling us to interrogate these sites with high sequencing depth. The results unambiguously demonstrated a decrease in the mismatch rate for these 3 sites after TRMT6/61A knock-down (Figure 3D). As a comparison, a non-motif m1A site located in a different structural context demonstrated an unaltered modification status (Figure 3E). Taken together, these observations suggest that in addition to tRNA, TRMT6/61A is also responsible for a subset of m1A sites in mRNA.
Figure 3. The tRNA methyltransferase complex TRMT6/61A catalyzes a subset of m1A methylation in mRNA.
(A) Motif analysis revealed a GUUCRA tRNA-like consensus for a group of m1A in mRNA, E-value=4.8e-009.
(B) Pie chart showing the percentage of m1A sites in each non-overlapping segment. Comparing to the non-motif m1A sites in mRNA, these sites are evenly distributed in the transcriptome.
(C) TRMT6/61A specifically targets m1A within the consensus sequence, while doesn’t work on m1A in non-motif sequence of the T-loop nor m1A at the 9th position of tRNA, p-value <0.005.
(D) Representative views of three mRNA targets of TRMT6/61A. The predicted RNA secondary structures are also shown, revealing a conserved stem-loop structure that harbors the GUUCRA motif.
(E) An example of non-motif mRNA m1A site, showing similar mismatch rates in the control and TRMT6/61A knock-down samples.
See also Figure S4.
Distinct m1A methylome in the mitochondrial transcriptome
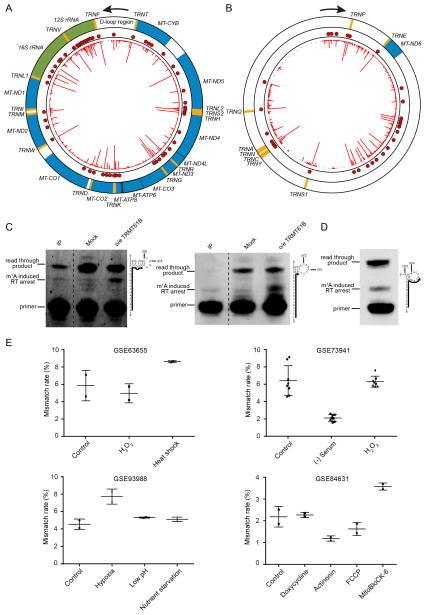
In addition to the nuclear-encoded transcripts, we also detected prevalent m1A modification in the mitochondrial (mt) transcriptome. mt-tRNAs are known to contain m1A at position 9 and 58 (Suzuki et al., 2011), catalyzed by TRMT10C and TRMT61B (Chujo and Suzuki, 2012; Vilardo et al., 2012), respectively. m1A-MAP showed that all the 14 mt-tRNAs bearing an adenosine residue at position 9 are m1A modified; for position 58, m1A was detected for the 3 known and 2 additional mt-tRNA molecules (Figure 4A, 4B and Table S3). For mt-rRNA, the only known m1A site is at position 947 of 16S rRNA (Bar-Yaacov et al., 2016). Interestingly, we additionally detected 7 and 10 m1A sites in 16S and 12S mt-rRNA, respectively (Figure 4 and Table S3). This is very different from cytosolic rRNA, where only one m1A site in 28S rRNA (m1A1322) was found. Unlike m1A947 in 16S mt-rRNA and m1A1322 in 28S rRNA (both are highly modified), the newly detected m1A sites are relatively low in modification level (Table S3). We further validated the presence of m1A in 12S mt-rRNA by both quantitative MS and a primer extension assay (Figure 4C, S5A and S5B).
Figure 4. Distinct m1A methylome in the mitochondrial transcriptome.
(A) m1A methylome of the heavy strand. Orange, green and blue colors represent tRNA, rRNA and mRNA, respectively. The red line in the inner circle represents the difference of mismatch rate of individual nucleotide while each red dot represents an identified m1A site.
(B) m1A methylome of the light strand.
(C) Detection of two novel m1A sites at position 378 and 575 in 12S mt-rRNA using a primer extension assay. In the immunoprecipitated samples (denoted as “IP”), the m1A modification at the two sites can be unambiguously detected. In the mock control samples (without immunoprecipitation), the bands corresponding to m1A378 and m1A575 induced reverse transcription (RT) arrest were weak, indicating a relatively low stoichiometry of these sites. Upon TRMT61B overexpression, strong bands were observed, suggesting that these two sites can be targeted by TRMT61B. The primers are shown as solid lines next to the RNA sequence.
(D) Detection of m1A1374 in MT-ND5 mt-mRNA using the primer extension assay. A strong band that corresponds to m1A1374 induced RT arrest can be found in the wild-type sample without immunoprecipitation, indicating a high modification level of this site. The primer is shown as solid lines next to the RNA sequence.
(E) m1A1374 in MT-ND5 is dynamically regulated by different stress conditions. Mismatch signal of m1A1374 could be consistently detected in multiple publicly available RNA-seq data and is regulated upon environmental stress.
See also Figure S5.
In human mitochondria, mRNAs are transcribed from the heavy and light strands as polycistronic units (Falkenberg et al., 2007; Mercer et al., 2011). The processed mt-mRNAs lack a cap at the 5′ end and contain no or short untranslated regions (Richter-Dennerlein et al., 2015; Rorbach and Minczuk, 2012; Temperley et al., 2010). We identified 22 m1A sites from 10/13 mitochondrial genes, in which 21 are residing in CDS and 1 is located in the 3′-UTR (Figure 4A and 4B). This is distinct to the m1A methylome in the nuclear-encoded transcripts, where m1A is enriched in the 5′-UTR. In addition, no preference for codon types was observed, yet m1A appears to be more likely present at the third position of a codon in the CDS of mt-mRNA (Figure S5C). By analyzing the published CLASH results in which microRNA and their direct mRNA targets are captured (Helwak et al., 2013), we found two m1A sites that are located within the experimentally verified targets of microRNA (Figure S5D). In fact, these m1A sites reside exactly within mRNA sequences that form base-pairing with the seed regions of microRNAs. More m1A sites in mt-mRNA were found within the predicted mt-mRNA targets of the microRNA seed regions (Figure S5D). Lastly, we also identified 25 m1A sites within the intergenic spacers. 24/25 m1A sites are in the light strand (Figure 4B); some of these m1A sites could be within the 3′-UTR of MT-ND6, for which there is no current consensus of its 3′ end (Slomovic et al., 2005).
We further validated m1A1374 in the MT-ND5 mRNA, and observed a clear band corresponding to m1A1374-induced RT arrest in the Input sample (without immunoprecipitation) using the primer extension assay (Figure 4D). Because the modification level of this site appears to be the highest among the m1A sites in mt-mRNAs (Table S3), we also surveyed publicly available RNA-seq data for the presence of m1A at this position (Kondo et al., 2017; Li et al., 2016a; Li et al., 2015; Quiros et al., 2017). Although the RT conditions differ greatly among the datasets, we were able to consistently find a mutation rate of ~2–9% for this position (Figure 4E). Interestingly, the modification level of m1A1374 appears to be dynamically regulated by different stress conditions (Figure 4E). For instance, its modification level is upregulated under heat shock and hypoxia conditions, while is down-regulated upon serum starvation and actinonin treatment, a drug that alters mitochondrial proteostasis. Hence, similar to the nuclear genome-encoded mRNA m1A methylome (Dominissini et al., 2016; Li et al., 2016a), m1A in mt-mRNA is also dynamically regulated.
m1A in mt-mRNA interferes with mitochondrial translation
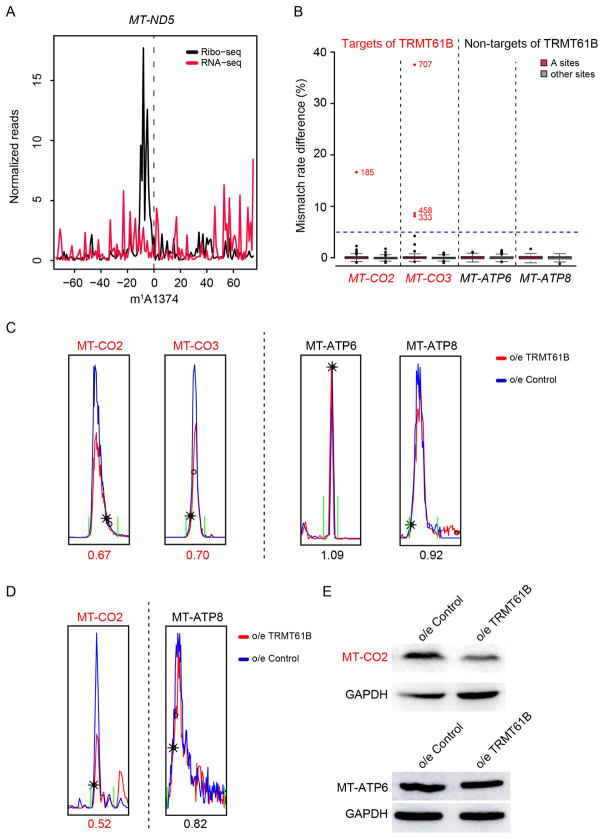
We next sought to examine the biological consequence of m1A in the mt-mRNAs. Translation requires accurate base pairing between mRNA codons and the cognate tRNAs; however, m1A is known to block the canonical A:U base pairing. These facts prompted us to hypothesize that m1A in mt-mRNA, which are primarily located in CDS, could interfere with translation in mitochondria. We first integrated published mitochondria ribosome profiling data with m1A-MAP identified m1A methylome in mitochondria (Rooijers et al., 2013). We found a strong signal of mitochondrial ribosome stalling at the m1A site on MT-ND5 (Figure 5A and S5E), whose modification level is the highest among all m1A sites in mt-mRNA. Due to the difficulty of an in vitro mitochondrial translation system (Smits et al., 2010), we sought to enzymatically manipulate the modification level of the mt-mRNAs. Two enzymes are known to introduce m1A in human mitochondria: TRMT10C generates m1A as well as m1G at position 9 in mitochondrial tRNAs, while TRMT61B is responsible for m1A at position 58 in some mitochondrial tRNAs and position 947 in 16S mt-rRNA (Bar-Yaacov et al., 2016; Chujo and Suzuki, 2012; Vilardo et al., 2012). Because TRMT10C is a subunit of the mitochondrial RNase P complex and is not specific for adenosine, we focused on TRMT61B. We utilized sequencing and a qPCR-based assay to quantitatively evaluate the modification status of the m1A sites in mt-mRNAs (see Method Details). While TRMT61B knock-down mildly reduced the m1A level (Figure S5F and S5G), TRMT61B overexpression could greatly increase the m1A modification level (Figure S5F and S5H). More specifically, we found that 5 (MT-CO1, MT-CO2, MT-CO3, MT-CYB, MT-ND4L) out of the 10 m1A-modified mt-mRNAs were targeted by TRMT61B (Figure 5B and Table S4). This observation suggests that in addition to mt-tRNA and mt-rRNA, TRMT61B could also modify mt-mRNA. Because of the high efficiency of TRMT61B overexpression in increasing the m1A level, we used mass spectrometry to quantitatively measure the mitochondrial protein level upon TRMT61B overexpression (Figure S5I). Indeed, TRMT61B overexpression led to a reduced protein level for MT-CO2 and MT-CO3 (Figure 5C and S5J), which are components of Complex IV; importantly, for MT-ATP6 and MT-ATP8 that are not targeted by TRMT61B, their protein levels remain unaffected. We also used metabolic labelling to further demonstrate that TRMT61B overexpression-induced translation repression is specific to mt-mRNA targets of TRMT61B, but not to non-targets of TRMT61B (Figure 5D). We further confirmed this observation using Western blot for MT-CO2 and MT-ATP6, a target and non-target of TRMT61B, respectively (Figure 5E). Of note, upon TRMT61B knock-down, the protein productions for several non-targets of TRMT61B were reduced (Figure S5K), presumably due to the decreased modification level in tRNA and rRNA m1A sites (which are originally highly modified and beneficial for translation) (Bar-Yaacov et al., 2016; Chujo and Suzuki, 2012). Collectively, these results suggest that m1A in mt-mRNA interferes with translation in mitochondria.
Figure 5. m1A in the mitochondrial mRNA interferes with translation.
(A) Mitochondrial ribosome stalling at the m1A site of the MT-ND5 mRNA. The density of the 5′ end of footprints was calculated for each position surrounding the m1A site. Ribosome profiling and RNA-seq data was taken from a published study (see Method Details).
(B) TRMT61B overexpression greatly increased the m1A modification level for several sites in MT-CO2 and MT-CO3, while the modification status of MT-ATP6 and MT-ATP8 remained unchanged. The red dots represent the targets of TRMT61B in MT-CO2 and MT-CO3.
(C) Extracted ion chromatograms of MT-CO2 and MT-CO3 (two targets of TRMT61B) showed decreased protein level upon TRMT61B overexpression, while those of MT-ATP8 and MT-ATP6 (non-targets of TRMT61B) remained unchanged.
(D) Extracted ion chromatograms of nascent MT-CO2 and MT-ATP8 upon TRMT61B overexpression. The nascent protein level of MT-CO2 decreased while that of MT-ATP8 remained unaffected.
(E) Western blot of MT-CO2 and MT-ATP6 upon TRMT61B overexpression.
See also Figure S5.
DISCUSSION
In this study, we developed a single-nucleotide resolution method for transcriptome-wide identification of m1A in human cells. m1A-MAP utilizes the m1A-induced misincorporation in cDNA synthesis to achieve base-resolution m1A detection. This enabled us to identify m1A modification not only at the mRNA cap, but also within a GUUCRA tRNA-like sequence motif. In principle, such misincorporation-dependent strategy could be applied to estimate the modification status of RNA sites of interest. However, our results in both rRNA and model RNA sequences strongly suggest that such estimation should be done with caution: TGIRT underestimates m1A modification level and m1A-induced mismatch decreases in a non-linear fashion as the modification level decreases. Additionally, the sequence context of RNA has also been reported to affect the mutation rate (Hauenschild et al., 2015; Zubradt et al., 2017). Hence, while direct sequencing (without enrichment) could still detect m1A sites with high modification level, m1A sites with averaging modification level or those located within a non-optimal context for mismatch induction could be missed. Therefore, coupling the pre-enrichment step to the mutational signature is necessary to improve the detection sensitivity. In addition, to achieve high confidence, we employed an in vitro demethylation step, which enabled us to distinguish true m1A sites from false signals (SNP, other modifications, and etc.) in the transcriptome. The combined use of mutational signature, the pre-enrichment step and the in vitro demethylation step enabled m1A-MAP to achieve high sensitivity and confidence. Of note, AlkB also demethylates several other methylations in RNA (e.g, m3C and m1G) (Cozen et al., 2015; Zheng et al., 2015), hence with the help of specific antibodies for these methylations, our strategy could in principle be applied to these modifications as well.
Our study revealed that two known m1A modification machinery, TRMT6/61A and TRMT61B, could work on mRNA as well. The hetero complex TRMT6/61A recognizes the sequence and structure of the tRNA T-loop and installs an m1A at the 58 position (Finer-Moore et al., 2015). Consistent with this knowledge, we found that the TRMT6/61A-dependent mRNA m1A sites are also confined to a hairpin structure mostly frequently with a 7nt loop, reminiscent of the tRNA T-loop. In contrast, we did not find an obvious sequence context for the m1A in mt-mRNA. In fact, TRMT61B appears to be a promiscuous enzyme that also modifies mt-tRNA and mt-rRNA; the substrate specificity and the underlying mechanism of TRMT61B in the human mitochondria remains to be determined. In addition, the fact that both TRMT6/61A and TRMT61B are known tRNA modification enzymes also reminds us of the modification machinery for other mRNA modifications. For instance, eukaryotic ψ synthases PUS1, PUS7 and TRUB1 can work on both tRNA and mRNA (Carlile et al., 2014; Li et al., 2015; Lovejoy et al., 2014; Safra et al., 2017; Schwartz et al., 2014a); yet the modification complex for m6A, consisting at least METTL3, METTL14, WTAP and KIAA1429, appears to be dedicated to mRNA (Bokar et al., 1997; Liu et al., 2014; Ping et al., 2014; Schwartz et al., 2014b). In the case of m1A, the enzyme(s) responsible for the majority of the modification sites in mRNA remains to be identified. It would be interesting to see if such machinery is specific for mRNA or promiscuous for multiple RNA substrates.
Our base-resolution m1A profiles reveal distinct m1A methylome in the human transcriptome. m1A is enriched in the 5′-UTR; and only 5′-UTR m1A sites, but not those in CDS nor 3′-UTR, are correlated with higher translation efficiency. Different from the m1A sites in the nuclear-encoded transcripts, m1A in mt-mRNA are primarily located in CDS and inhibit translation. Interestingly, m1A in mt-mRNA can be dynamically regulated, as shown in the case of m1A in MT-ND5. Of note, m1A517 in MT-ND2 overlaps with a known site of non-synonymous mutation that is associated with oral cancer (https://www.mitomap.org/MITOMAP). In addition, we also identified a notable group of m1A methylation adjacent to the mRNA cap; because the first two nucleotides of the 5′ end of mRNA are known to contain ribose methylation, future experiments will be needed to examine whether they are m1A or m1Am modifications. A related but different modification, m6Am, is also known to be present at this position. Yet, m6Am does not induce misincorporation during reverse transcription, nor become sensitive to demethylation by AlkB, as we showed in Figure S3D. Hence, m1A-MAP is capable of specific detection of methylation at the N1—but not the N6—position of adenosine. Whether the positive association of cap and 5′-UTR m1A with translation is due to improved mRNA stability (similar to m6Am) (Mauer et al., 2017; Schwartz et al., 2014b) or other mechanisms remains to be further studied.
Our results also revealed prevalent m1A methylation in mt-mRNA; this is due to the utilization of a stranded library preparation strategy in this study as compared to the previous methods (Dominissini et al., 2016; Li et al., 2016a). Similar stranded library preparation strategy has also enabled the identification of pseudouridylation in mt-mRNA (Antonicka et al., 2017). We also showed that such methylation interferes with translation. By manipulating the m1A level via TRMT61B, we monitored the corresponding changes of the mitochondrial protein level by both quantitative mass spectrometry and Western blot. TRMT61B modifies both mt-tRNA and mt-rRNA (Bar-Yaacov et al., 2016; Chujo and Suzuki, 2012); and these m1A sites are thought to be beneficial for their functions in translation. Because m1A sites in mt-mRNA have an opposing effect in translation, changes of protein synthesis level after TRMT61B knock-down could be a mixed result of contrary m1A sites in these different components (rRNA, tRNA and mRNA) of translation. Conversely, because16S rRNA and mt-tRNALeu(UUR) are already of high m1A level, we envisioned that TRMT61B overexpression should lead to a greater increase of m1A level in mRNA than in these ncRNAs. In this simplified scenario, we indeed observed greatly increased modification level for several mt-mRNAs upon TRMT61B overexpression and detected reduced protein level for such mt-mRNA transcripts. Importantly, protein levels for non-targets of TRMT61B, which are either devoid of m1A or contain TRMT61B-independent m1A sites, remain unaffected upon TRMT61B overexpression. Hence, TRMT61B overexpression-mediated translation repression is caused by mt-mRNA m1A and is specific to its mt-mRNA targets. For the mt-mRNAs that contain TRMT61B-independent m1A sites, additional methyltransferase(s), for instance TRMT10C that is also localized in the mitochondrial, may be involved. In terms of the mechanism of m1A-mediated translational suppression, multiple possibilities need to be considered. For instance, due to the presence of a base-pairing interfering modification in the CDS, m1A could serve as a road block for the mitochondrial ribosome. In fact, recent in vitro translation experiments on synthetic RNA sequences have shown that m1A in CDS represses translation; the effect is stronger when m1A is at codon position 1 and 2, while m1A at the third position also mitigates translation (You et al., 2017). Thus, not only the stoichiometry but also the exact position of m1A in mt-mRNA could influence protein synthesis. In addition, we showed that the presence of m1A in mt-mRNA could interfere with base pairing to microRNA, which is thought to enhance translation in mitochondria (Zhang et al., 2014). While both the speculated mechanisms point to a suppressive role of m1A in mitochondria translation, alternative hypothesis and mechanism should also be tested in future experiments. Nevertheless, our discovery that m1A in mt-mRNA interferes with translation improves our understanding of translation regulation in human mitochondria.
In summary, our study demonstrated distinct classes of m1A methylome in the nuclear- and mitochondrial-encoded transcripts. Our single-nucleotide resolution m1A technology allowed the comprehensive profiling of m1A in the human transcriptome, providing a reference and resource for future investigations to elucidate the biological functions and mechanisms of this epitranscriptomic mark.
Limitations
Our current procedure of m1A-MAP favors sensitivity and accuracy over the quantitative ability, due to the involvement of a pre-enrichment step using an m1A antibody. For the m1A sites located in abundant RNAs (rRNA, tRNA and a limited subset of mRNA), their modification levels could be estimated by comparing the mismatch rates in the Input sample with a stoichiometry curve (similar to Figure S2C). However, for the majority of mRNA molecules in the transcriptome, only limited sequencing reads could be obtained by transcriptome-wide sequencing experiments. Thus, stoichiometry for m1A sites in less abundant mRNA molecules may not be reliably estimated by their mismatch rates in the Input sample. To address this limitation, the modification level of m1A-MAP identified m1A sites can be estimated via additional locus-specific sequencing to the Input samples, which would allow sufficient sequencing depth while remain cost-effective. The exact stoichiometry of these sites can then be determined by fitting the mismatch rates in the locus-specific sequencing into the stoichiometry curve of synthetic spike-in RNAs that contain the exact sequence of the RNA molecules of interest.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests may be directed to, and will be fulfilled by the corresponding author Chengqi Yi (chengqi.yi@pku.edu.cn).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HEK293T was maintained at 37 °C in DMEM medium (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin (Gibco).
METHOD DETAILS
Cell Culture and Antibodies
HEK293T (ATCC, CRL-11268) was used in this study and maintained in DMEM medium (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin (Gibco). Monoclonal mouse anti-m1A antibody was purchased from MBL (D345-3). Polyclonal rabbit anti-TRMT6 antibody was purchased from Santa Cruz Biotechnology (sc-271752). MT-CO2 antibody (55070-1-AP) and MT-ATP6 antibody (55313-1-AP) was purchased from Proteintech Group. Monoclonal mouse anti-β-Actin antibody was purchased from CWBiotech (CW0096). Monoclonal mouse anti-GAPDH antibody was purchased from CWBiotech (CW0100M). The secondary antibodies used were anti-mouse-IgG-HRP (CW0102; CWBiotech) and anti-rabbit-IgG-HRP (CW0103; CWBiotech).
RNA isolation
Total RNA was isolated from cells using TRIzol, according to the manufacturer’s instructions (Invitrogen). An additional DNase I treatment step was performed to avoid DNA contamination. For polyA+ RNA isolation, small RNA was firstly depleted using MEGAclear™ Transcription Clean-Up Kit (Ambion), followed by two successive rounds of polyA+ selection using oligo(dT)25 Dynabeads (Invitrogen). For small RNA isolation, RNA smaller than 200 nt was recovered from the flow-through fraction in the small RNA depletion step by ethanol precipitation.
shRNA knock-down of TRMT6/61A
The oligoes targeting TRMT6 and TRMT61A were annealed and cloned into the pLKO vector according to the TRC shRNA library protocol (http://www.broadinstitute.org/rnai/public/), respectively. The oligo sequences were listed below: TRMT6-FWD: CCGGGGGAAAGTTCTGAGTATTTATCTCGAGATAAATACTCAGAACTTTCCCTTTTTG; TRMT6-RVS: AATTCAAAAAGGGAAAGTTCTGAGTATTTATCTCGAGATAAATACTCAGAACTTTCCC; TRMT61A-FWD: CCGGGAGGCCAGAGGCACCTTATATCTCGAGATATAAGGTGCCTCTGGCCTCTTTTTG; TRMT61A-RVS: AATTCAAAAAGAGGCCAGAGGCACCTTATATCTCGAGATATAAGGTGCCTCTGGCCTC. A scrambled shRNA was used as the mock control. Lentiviruses were packaged by co-transfecting HEK293T cells with pLKO-TRMT6, pLKO-TRMT61A, pCMV-dR8.91 and VSV-G plasmids, following the instructions from Broad Institute. The supernatant from transfected cells was harvested after 2 days and used to infect HEK293T cells followed by puromycin selection for 5 days. Knock-down efficiency was verified by Western blot and qPCR. qPCR primers were listed as follows: TRMT6-qFWD: CTGTCTTTGCTGGACTTTGTGGC; TRMT6-qRVS: AGACAGCCTGAGGTTGATGACC; TRMT61A-qFWD: TCCTCTACTCCACAGACATCGC; TRMT61A-qRVS: CAATGGTGCGGATGATGGCGTG.
Quantification of m1A and m6A level by LC-MS/MS
200 ng isolated RNA or 100 ng model RNA oligo was digested into nucleosides by 0.5 U nuclease P1 (Sigma, N8630) in 20 μL buffer containing 10 mM ammonium acetate, pH 5.3 at 42 °C for 6 h, followed by the addition of 2.5 μL 0.5 M MES buffer, pH 6.5 and 0.5 U alkaline phosphatase (Sigma, P4252). The mixture was incubated at 37 °C for another 6 h and diluted to 50 μL. 5 μL of the solution was injected into LC-MS/MS. The nucleosides were separated by ultra-performance liquid chromatography with a C18 column, and then detected by triple-quadrupole mass spectrometer (AB SCIEX QTRAP 5500) in the positive ion multiple reaction-monitoring (MRM) mode. The mass transitions of m/z 282.0 to 150.1 (m1A), m/z 282.0 to 150.1 (m6A), m/z 268.0 to 136.0 (A) were monitored and recorded. Concentrations of nucleosides in RNA samples were deduced by fitting the signal intensities into the stand curves.
Stoichiometry curve construction using model oligoes
Two pairs of synthetic RNA oligoes containing either m1A or A were used to examine the capacity of m1A in causing mis-incorporation during revers transcription in a quantitative manner. The oligo sequences were listed as follows: m1A-1: CGCGGCUCGm1AGCCCGCGUGCGGGCCUCUUUCAGGCCGCU; A-1: CGCGGCUCGAGCCCGCGUGCGGGCCUCUUUCAGGCCGCU; m1A-2: CGGCGGCCCGGGACCGm1AGACCCGGCCCCGGCUCCCC; A-2: CGGCGGCCCGGGACCGAGACCCGGCCCCGGCUCCCC. The m6A contamination in the m1A oligoes, which was introduced during the oligo purification process, was measured using quantitative LC-MS/MS. The m1A-oligo was mixed with the A-oligo to ratios: 100%, 75%, 50%, 25%, 12.5%, 6.25% and 0%, respectively. The mixed m1A/A oligoes were subjected to library construction using specific RT primers as listed below:
RT-m1A/A-1: ACACGACGCTCTTCCGATCTagcggcctgaaagaggc;
RT-m1A/A-2: ACACGACGCTCTTCCGATCTggggagccggggcc.
Cloning, Expression and Purification of AlkB
A truncated AlkB with deletion of the N-terminal 11 amino acids was cloned into pET30a (Novagen) and transformed to E. coli BL21(DE3) followed by culturing in LB medium at 37 °C until the OD600 reached 0.6–0.8, and further incubating at 30 °C for additional 4 h with the addition of 1 mM IPTG. Proteins were purified using Ni-NTA chromatography (GE Healthcare) and gel-filtration chromatography (Superdex 200, GE Healthcare) followed by Mono-Q anion exchange chromatography (GE Healthcare). Such purification procedure effectively avoided RNA contamination from E coli. (the expression host).
In vitro Demethylation treatment
We employed two different strategies for the removal of methyl-group at the N1 position of adenosine.
In vitro demethylation mediated by the purified AlkB protein: 10 μg full length polyA+ RNA was fragmented at 94 °C for 5 min using magnesium RNA fragmentation buffer (NEB) and fragmented polyA+ RNA was desalted and concentrated by ethanol precipitation. 10 μg (~0.2 nmol) fragmented polyA+ RNA was denatured at 65 °C for 5 min, and then subjected to demethylation reaction in a 500 μL demethylation mixture containing 0.4 nmol purified AlkB, 50 mM MES, pH 6.5, 283 μM of (NH4)2Fe(SO4)2·6H2O, 300 μM 2-ketoglutarate, 2 mM L-ascorbic acid, 1 U/μL SUPERaseIn RNase Inhibitor (Invitrogen). The demethylation reaction was incubated at 37 °C for 2 h and quenched by adding 5 mM EDTA. The demethylated RNA was then purified by phenol chloroform extraction.
In vitro demethylation mediated by the Dimroth rearrangement: 10 μg full length polyA+ RNA was incubated in alkaline buffer (0.1 M Na2CO3/NaHCO3, 5mM EDTA, pH 10.2) at 65 °C for 3 h, and then the treated RNA was purified by ethanol precipitation.
m1A-MAP
40 μg polyA+ RNA was fragmented to ~150 nt using magnesium RNA fragmentation buffer (NEB). m1A-containing RNA fragments were enriched through m1A immunoprecipitation as previous described (Li et al., 2016a). 10 ng (~0.2 pmol) of the immunoprecipitated m1A-containing RNA fragments were subjected to the AlkB demethylation treatment. Demethylation reaction was performed in a 20 μL demethylation mixture containing 0.4 pmol purified AlkB, 50 mM MES, pH 6.5, 283 μM of (NH4)2Fe(SO4)2·6H2O, 300 μM 2-ketoglutarate, 2 mM L-ascorbic acid and 0.4 U/μL RNase inhibitor, following with an incubation at 37 °C for 2 h. The reaction was quenched by the addition of 5 mM EDTA, and the demethylated RNA was purified by phenol chloroform extraction followed with ethanol precipitation.
Fragmented polyA+ RNA (as “Input”), immunoprecipitated RNA [as “(−) demethylase”] and demethylated immunoprecipitated RNA [as “(+) demethylase”] were subjected to library construction. The library construction was performed according to the eCLIP library construction protocol with several modifications (Van Nostrand et al., 2016). For dephosphorylation of 3′ ends, RNA samples were treated by PNK (NEB) with an incubation at 37 °C for 1 h, and followed by a heat-inactivation of PNK at 65 °C for 20 min. The dephosphorylated RNA was purified by ethanol precipitation and then subjected to 3′ RNA linker ligation using T4 RNA ligase2, truncated KQ (NEB) at 25 °C for 2 h. The 3′ RNA linker sequence is: 5′rAPP-AGATCGGAAGAGCGTCGTG-3SpC3. The excess RNA adaptor was digested by adding 1 μL 5′ Deadenylase (NEB) into the ligation mix followed by an incubation at 30 °C for 1 h, and then adding 1 μL RecJf (NEB), incubating at 37 °C for another 1 h. The enzymes were then heat-inactivated at 70 °C for 20 min, and the RNA was purified by ethanol precipitation. The RNA pellet was dissolved in 10 μL H2O and then 1 μL 2 μM RT primer (ACACGACGCTCTTCCGATCT) was added. RNA-primer mix was denatured at 80 °C for 2 min and then chilled on ice immediately. RT reaction buffer (50 mM Tri–HCl pH 8.3, 75 mM KCl, and 3 mM MgCl2, final), dNTPs (1 mM, final), DTT(5 mM, final), RNase Inhibitor (1 U/μL, final) and 1 μL TGIRT (InGex) were added into the denatured RNA-primer mix and reverse transcription was carried out at 57 °C for 2 h. Excess RT primer was digested by the addition of 1 μL Exonuclease I (NEB) and a following incubation at 37 °C for 30 min. cDNA was purified using silane be ads (Invitrogen) and then ligated to the 5′ adaptor (5Phos- NNNNNNNNNNAGATCGGAAGAGCACACGTCTG-3SpC3). Ligation was performed with T4 RNA ligase 1, high concentration (NEB) at 25 °C overnight. The ligated cDNA was purified using silane beads, and then subjected to PCR amplification with the following primers: 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGAC GCTCTTCCGATCT-3′, 5′-CAAGCAGAAGACGGCATACGAGATXXXXXXGTGACTGGAGTTCAGACGTGTGCTCTTCC GATC-3′ (XXXXXX represents index sequence). PCR product was purified by 8% TBE gel. The libraries were sequenced on Illumina Hiseq X10 with paired-end 2×150 bp read length.
Locus-specific m1A detection
We developed two different strategies for locus-specific detection of m1A sites at the first nucleotide of mRNA transcripts and those located internally, respectively.
For cap+1 m1A detection, 2 μg polyA+ RNA was fragmented to ~150 nt and then equally divided into two parts. One part of RNA sample was subjected to the AlkB demethylation treatment [“(+) AlkB”] and the other part was untreated [“(−) AlkB”]. Demethylation reaction was performed as described above. (+) AlkB and (−) AlkB sample were ligated to 3′ RNA linker (5′rAPP-AGATCGGAAGAGCGTCGTG-3SpC3) and then reverse transcription was carried out using TGIRT with the same RT condition as that of m1A-MAP. The 5′ cap-adaptor (5Phos- GTCTCAGCGTCCAGTGTCATC-3SpC3) was ligated to the reverse transcribed cDNAs. The regions harboring m1A were amplified by PCR using a universal forward primer (5′-GATGACACTGGACGCTGAGAC-3′) and gene specific reverse primers (listed in Table S5). The amplicons from different m1A-containing regions were mixed together for (+) AlkB and (−) AlkB sample, respectively and then subjected to DNA library construction using NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® (E7645) according to the manufacturer’s instructions. Deep sequencing for the libraries was performed on Illumina Hiseq X10 with paired-end 2×150 bp read length. Thus, cap+1 m1A to be validated can be covered with sufficient reads for the reliable examination of mutation rate.
For internal m1A detection, 200 ng polyA+ RNA was isolated from TRMT6/61A stable knock-down cells and mock control cells respectively, and then was fragmented to 150 nt using magnesium RNA fragmentation buffer (NEB). Fragmented RNA was ligated to 3′ RNA linker (5′rAPP-AGATCGGAAGAGCGTCGTG-3SpC3) and then reverse transcription was carried out using TGIRT with the same RT condition as that of m1A-MAP. The regions harboring m1A were amplified by PCR using specific primers. The amplicons from different m1A-containing regions were mixed together for each sample, and then subjected to DNA library construction using NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® (E7645) according to the manufacturer’s instructions. Deep sequencing for the libraries was performed on Illumina Hiseq X10 with paired-end 2×150 bp read length. Thus, the position of m1A to be validated can be covered with sufficient reads for the examination of mutation rate. Specific PCR primers are listed below:
NM_025099-m1A5643-FWD: AAAAAGCTCGGTCCGGGTTC;
NM_025099-m1A5643-RVS: TTAGCCGCAAAATCACGCTG;
NM_001193375-m1A978-FWD: ACACCTGTCCAAGCCCTAAT;
NM_001193375-m1A978-RVS: CTGAGGGGCCCTTATTCCCA;
NR_026951-m1A659-FWD: ACGTCGGCTCGTTGGTCTAG;
NR_026951-m1A659-RVS: ACAGTCAAGCCTCCTGCAGC;
NM_001256443-m1A486-FWD: CAAGGTTCCAGGCGAAGGG;
NM_001256443-m1A486-RVS: AGCCGGGGTCTCTGTGG.
Purification of Mitochondrial 12S mt-rRNA
Total RNA was isolated from cells using TRIzol, according to the manufacturer’s instructions (Invitrogen). An additional DNase I treatment step was performed to avoid DNA contamination. Bring 200 μg DNAse I treated total RNA to volume of 160 μL in nuclease-free water, and then add 1 μL 12S mt-rRNA probe mix (20 μM each). The probe mix contains 4 3′-biotin labeled oligoes that specifically target 12S mt-rRNA and the sequences were listed as follows: 12S-probe-1: GAGCTAATAGAAAGGCTAGGAC; 12S-probe-2: TCTATTGACTTGGGTTAATC; 12S-probe-3: TTATCGATTACAGAACAGGCT; 12S-probe-4: TCTACTCTTAGTTTACTGCTAA. The RNA mixture was heated at 65 °C for 10 min and followed by chilling on the ice. 40 μL 5 × Binding buffer (1.5 M NaCl, 10 mM EDTA, 100 mM Tris, pH 7.4) was added into the RNA mixture and mixed well by pipetting. The RNA mixture was incubated at room temperature for about 30 minutes. 200 μL Streptavidin C1 Dynabeads (Invitrogen) was resuspended in 200 μL Binding Buffer (300 mM NaCl, 2 mM EDTA, 20 mM Tris, pH 7.4) and then added into the RNA mixture. The RNA beads mixture was incubated at room temperature for 40 minutes by gently rotating. The beads was washed with Washing Buffer (30 mM NaCl, 1 mM EDTA, 10 mM Tris, pH 7.4, 1% SDS) at 40 °C for four times. RNA was eluted from beads by adding 200 μL Elution buffer (5 mM EDTA, 10 mM Tris, pH 7.4, 1% SDS) and incubating at 70 °C for 10 minutes. Isolated RNA was recovered by ethanol precipitation and 12S mt-rRNA was further selected using 4% Urea-PAGE. The purity of isolated 12S mt-rRNA was checked using RNA 6000 Pico Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies). The m1A methylation level of 12S mt-rRNA was analyzed by quantitative LC-MS/MS.
Primer extension
The primer extension was performed as previously reported (Bar-Yaacov et al., 2016; Chujo and Suzuki, 2012) with several modifications.
To detect m1A1374 in MT-ND5, 2 μg mRNA was mixed with 10 fmol 5′ biotin labeled primer (ND5- m1A1374-primer: CCTGCGAATAGGCTTCC) in a 5 μL solution containing 10 mM Tris, pH 7.5 and 1 mM EDTA. The RNA mixture was then heated at 80 °C for 2 min and slowly cooled to 25 °C over 55 min. The RT mixture (containing 2 μL 5× First-Strand Buffer, 1.5 μL 25 mM MgCl2, 0.5 μL 0.1 M DTT, 0.5 μL Superscript III, 0.25 μL RNase inhibitor, 0.25 μL 1.5 mM ddATP, dTTP, dCTP, and dGTP mix) was then added into the RNA mixture and mixed well. The reverse transcription was carried out at 55 °C for 1 h and then the RNA template was digested with RNase H. The reverse transcribed products were separated by 20% Urea-PAGE and then transferred to Nylon membrane, detected by chemiluminescent nucleic acid detection module (Thermo, 89880).
To detect m1A378 and m1A575 in 12S mt-rRNA, 50 ng fragmented total RNA or 20 ng immunoprecipitated RNA was mixed with 5 fmol 5′ biotin labeled primer (12S-m1A378-primer: TGTGTTCAGATATGTTAAAG; 12S-m1A575-primer: GAGGTGGTGAGGTTGATCG) and then subjected to reverse transcription as described above. The d/ddNTP mix consisted of dATP, dTTP, dCTP and ddGTP for m1A378 in 12S mt-rRNA; ddATP, dTTP, dCTP and dGTP for m1A575 in 12S mt-rRNA. The reverse transcription was carried out at 49 °C for 12S-m 1A378 and 55 °C for 12S-m1A575. The reverse transcribed products were analyzed as described above.
siRNA Knock-down and overexpression of TRMT61B
Two synthesized duplex RNAi oligoes targeting TRMT61B mRNA sequences were used: 5′-GGAUAUCAACCCAGGUGAUTT-3′ and 5′-GCGUGAUUCAUGGAAAUUATT-3′; a scrambled duplex RNAi oligo (5′-UCCUCCGAACGUGUCACGUTT) was used as the mock control. The siRNA oligo was transfected into HEK293T by Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions, and cells were harvested 48 h later after transfection. Knock-down efficiency was examined by qPCR. qPCR primers were listed as follows:
TRMT61B-qFWD: CAGGAGCAACCGAAGACAT;
TRMT61B-qRVS:ATATACAGCACATACACCACCAT;
For the construction of overexpression plasmid, TRMT61B was cloned into pcDNA3.1 using the following primers: FWD: TCGCGAAACACTATGCTAATGGC; RVS: GTTAAGTTGTGGTTTGACCTTCCTC. The empty pcDNA3.1 vector was used as the mock control. The plasmid was transfected into HEK293T by Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions and cells were harvested 48 h later after transfection.
Ribosome profiling
2 plates of 15-cm HEK293T cells were grown to 90% confluency; CHX was then added to the medium at a final concentration of 100μg/mL. Cells were treated in the CHX-medium for 7 min before they were harvested and lysed with 1 mL lysis buffer (10 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 100 mM KCl, 1% Triton X-100, 2 mM DTT, 100 μg/mL CHX, 0.5 U/μL RNase inhibitor, 1×complete protease inhibitor). The cell lysates were centrifuged at 15,000g for 15min, and the supernatant was collected and sent for the OD260 measuring. 100 μL lysates were kept as input sample and 1 mL TRIzol was added to purify the RNA. 1 μL Micrococcal Nuclease (NEB) per 25OD was added to the remaining cell lysates and incubated at 25 °C for 20 min. The digested cell lysates were used for performing ribosome foot-printing. The lysates were fractioned on 10/50% w/v sucrose gradients using the SW-40Ti rotor at 27,500rpm for 4h. 80S monosome fraction was collected, followed by the addition of equal volume of extraction buffer (1% SDS, 40 mM EDTA). RNA was isolated by phenol-chloroform extraction. RNA fragments between 28–30 nt were selected using 15% Urea-PAGE. Recovered RNA fragments were subjected to library construction. In brief, RNA samples were dephosphorylated with PNK (NEB) and ligated to 3′ RP linker (5′rAPP-CTGTAGGCACCATCAAT-3SpC3) using T4 RNA ligase2, truncated KQ (NEB). Reverse transcription was carried out using Superscript III (Invitrogen) with RP-RT primer (5Phos-AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGC-SpC18-C ACTCA-SpC18-TTCAGACGTGTGCTCTTCCGATCTATTGATGGTGCCTACAG). cDNA was circ-ligated with CircLigase II (Epicentre) and then amplified by PCR with primers (5′-AATGATACGGCGACCACCGAGATCTACAC-3′; 5′-CAAGCAGAAGACGGCATACGAGATXXXXXXGTGACTGGAGTTCAGACGTGTGCTCTTCC GATC-3′, XXXXXX represents index sequence). PCR products were purified by 8% TBE gel and sequenced on Illumina Hiseq 2500 with single end reads (50 bp).
qPCR-based m1A level evaluation
20 μg polyA+ RNA was isolated from HEK293T cells with TRMT61B overexpression, knock-down and the corresponding mock controls, respectively. RNA was fragmented into ~150 nt using magnesium RNA fragmentation buffer (NEB) and concentrated by ethanol precipitation. Fragmented RNA (as Input) was denatured and incubated with 2 μg anti-m1A antibody in IPP buffer (150 mM NaCl, 0.1% NP-40, 10 mM Tris, pH 7.4) at 4°C overnight. 20 μL Protein A/G UltraLink Resin (Pierce) was added to the RNA-antibody mixture and incubated at 4°C for additional 3 h. Resins were washed for twice with IPP buffer, once with low salt buffer (75 mM NaCl, 0.1% NP-40, 10 mM Tris, pH 7.4), once with high salt buffer (200 mM NaCl, 0.1% NP-40, 10 mM Tris, pH 7.4) and twice with TEN buffer (10 mM Tris, pH 7.4, 1 mM EDTA, pH 8.0, 0.05% NP-40). m1A-containing RNA was eluted from resins with 3 mg/mL N1-methyladenosine (Berry&Associates) in IPP buffer and purified by phenol chloroform extraction and ethanol precipitation. Input and immunoprecipitated samples were reverse transcribed into cDNA using Superscript III (Invitrogen) and quantified by qPCR using SYBR GREEN mix (Takara) on Roche Lightcycler 96 real-time PCR system. The m1A-IP/Input ratios of the targeted regions were calculated for the TRMT61B overexpression, knock-down and the corresponding control samples, respectively. The primers used for qPCR were listed below:
MT-CO1-qFWD: CCTATCATCTGTAGGCTCATTC;
MT-CO1-qRVS: GGAGGGTTCTTCTACTATTAGGAC;
MT-CO2-qFWD: ACAGATGCAATTCCCGGACG;
MT-CO2-qRVS: CCACAGATTTCAGAGCATTGACC;
MT-CO3-qFWD: CGCCTGATACTGGCATTTTG;
MT-CO3-qRVS: GACCCTCATCAATAGATGGAGAC;
MT-CYB-qFWD: CAACCCCCTAGGAATCACCTC;
MT-CYB-qRVS: GAGGGCGTCTTTGATTGTGTAG;
16S rRNA-qFWD: ATGAATGGCTCCACGAGGG;
16S rRNA-qRVS: CTTGCTGTGTTATGCCCGC.
Reductive dimethylation labeling
Mitochondrion was isolated from TRMT61B overexpression and mock control HEK293T cells according to the manufacturer’s (Thermo Fisher) and lysed with RIPA buffer followed by sonication. The extracted proteins from different samples were quantified with the BCA protein assay kit (Thermo Fisher). Equal amount of proteins from two samples were digested by trypsin on-column in 100 mM TEAB buffer and subjected to reductive dimethylation labeling. 4 μL of 4% (w/w) light or heavy formaldehyde was added to 100 μL of trypsin digested samples prepared from TRMT61B overexpression and mock control HEK293T cells, respectively. In the meantime, 4 μL of 0.6 M sodium cyanoborohydride was added and the samples were incubated at room temperature for 1h. The dimethylation labeling reaction was quenched by the addition of 1% (w/w) ammonia and 5% (w/w) formic acid. Finally, light and heavy labeled peptide samples were mixed, concentrated by vacuum, and analyzed on a Q Exactive mass spectrometer (Thermo Fisher).
LC-MS/MS and data analysis
The peptides were analyzed on a Q Exactive mass spectrometer (Thermo Fisher). Under the positive-ion mode, full-scan mass spectra was acquired over the m/z range from 350 to 1800 using the Orbitrap mass analyzer with mass resolution setting of 70000. MS/MS fragmentation was performed in a data-dependent mode, of which the 20 most intense ions were selected from each full-scan mass spectrum for high-energy collision induced dissociation (HCD) and MS2 analysis. MS2 spectra were acquired with a resolution setting of 17500 using the Orbitrap analyzer. Some other parameters in the centroid format: isolation window, 2.0 m/z units; default charge, 2+; normalized collision energy, 28%; maximum IT, 50 ms; dynamic exclusion, 20.0 s.
LC-MS/MS data was analyzed by ProLuCID (Xu et al., 2015) with static modification of cysteine (+57.0215 Da) and variable oxidation of methionine (+15.9949 Da). The searching results were filtered by DTASelect (Tabb et al., 2002) and peptides were also restricted to fully tryptic with a defined peptide false positive rate of 1%. The ratios of reductive dimethylation were quantified by the CIMAGE software as described before (Weerapana et al., 2010).
Pre-processing of raw sequencing data
A random barcode of 10 nt was included in the adapter that ligates to the 3′ end of cDNA and it cannot be precisely located in Read 1. Hence, only Read 2 data of m1A-MAP was used for subsequent analyses. Raw sequencing reads produced from m1A-MAP were firstly subjected to Trim_galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) for quality control and adaptor trimming. The minimum quality threshold was set to 20, and the minimum length required for reads after trimming was 30 nt. The remaining reads were further processed by removing the first 10 nt random barcode in the 5′ end. As for the ribosome profiling (Ribo-seq) data and the corresponding RNA-seq data, reads with a quality lower than 20 were discarded, and the adapter in 3′ end was trimmed. Processed reads with a length ranging from 25 nt to 35 nt in the ribosome profiling sample were kept for further analysis.
Reads mapping and PCR duplication removing
Processed reads were mapped to human transcriptome or mitochondrial genome using BWA-MEM with default parameters (version 0.7.15-r1140) (Li and Durbin, 2009). Reference transcriptome was prepared based on the Refseq annotation of human (hg19) downloaded from the table browser of UCSC database. The redundant sequences with the same Refseq id were removed. Transfer RNA (tRNA) sequences were also downloaded from UCSC table browser and integrated into the transcriptome. Mitochondrial genome and corresponding annotation were downloaded from NCBI (NC_012920.1). Reads mapping to an identical position of reference were considered as PCR duplications if their 10 nt random barcodes were the same, and only one of these reads was kept. Performances related to the processing of sam/bam file were done with the help of SAMtools (Li et al., 2009) (http://samtools.sourceforge.net/).
Identification of m1A sites
Mismatch rate of each nucleotide in the reference sequences was calculated for both (−) and (+) demethylase samples. Two parameters were defined to evaluate the dynamic change of mismatch rate in the (−) demethylase and (+) demethylase samples: difference of mismatch rate (Diff) and fold change of mismatch rate (FC). Diff was calculated by subtracting the mismatch rate in the (+) demethylase from that in the (−) demethylase sample, while FC was calculated by dividing the mismatch rate in the (−) demethylase by that in the (+) demethylase sample. FC was artificially set to “1000” if the mismatch rate in the (+) demethylase sample was “0”. A position was identified as an m1A site when the following criteria were met: a) FC>=3; b) Diff>=10%; c) the number of reads with a mismatch at the position was no less than 5; d) criteria (a–c) were all fulfilled in both replicates.
In order to evaluate the frequency of potential false positives caused by m1A-independent mismatch, we employed a reverse calling procedure. Specifically, the “reverse calculation” of Diff and FC values for each position were performed following the equations below:
Under such circumstance, only 17 sites passed the above-mentioned threshold, suggesting the m1A-independent mismatch should minimally interfere with the identification of true m1A sites.
Identification of TRMT61B targets in mitochondrial-encoded transcripts
For the identification of TRMT61B targeting sites in mitochondrial-encoded RNA, the mismatch rate in the Input sample was firstly calculated for mock control sample and TRMT61B overexpression sample, respectively. Two parameters were defined to evaluate the increase of mismatch rate from the mock to TRMT61B o/e sample: difference of mismatch rate (Diff) and fold change of mismatch rate (FC). Diff was calculated by subtracting the mismatch rate in the mock sample from that in the TRMT61B o/e sample, while FC was calculated by dividing the mismatch rate in the TRMT61B o/e by that in the mock sample. FC was artificially set to “1000” if the mismatch rate in the mock sample was “0”. A position was identified as TRMT61B target when the following criteria were met: a) FC>=5; b) Diff>=5%; c) the site was covered by more than 500 sequencing reads; d) criteria (a–c) were all fulfilled in both replicates.
Motif discovery and GO enrichment analysis
For the analysis of sequence consensus, 15 nt of sequence neighboring each m1A site was retrieved. These sequences were then subjected to DREME algorithm in MEME suite (Version 4.12.0) for the discovery of enriched motifs (Bailey et al., 2009). The shuffled input sequences were used as the background to eliminate potential false positives caused by the nucleotide composition.
Gene Ontology (GO) enrichment analyses were performed using DAVID web-based tool (http://david.abcc.ncifcrf.gov/) (Huang da et al., 2009).
Secondary structure and minimum free energy analysis
12 nucleotides of the 5′ end and 10 nucleotides of the 3′ end of each m1A site (hence 23 nt sequence in total) were retrieved for local structure analysis. RNAfold (v2.3.4) (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) was used to predict the secondary structure and calculate the corresponding minimum free energy (MFE). The length of the loop where m1A resides was determined based on the predicted structure; for an m1A site that is not located in a loop, this value was set to “0”. The significance test of MFE between m1A sites within the GUUCRA motif and other m1A sites was performed using Mann-Whitney U-test.
Ribosome profiling data analysis and TE calculation
Ribo-seq and corresponding RNA-seq reads were aligned to the transcriptome, and RPKM (Reads Per Kilobase per Million mapped reads) of each transcript was calculated. Translation efficiency (TE) was defined for each transcript as the ratio of the RPKM in Ribo-seq to the RPKM in RNA-seq.
For the analysis of influence of m1A on mitochondrial gene translation, mitochondrial ribosome profiling data were downloaded from the GEO Datasets (GSE48933) (Rooijers et al., 2013). The depth of reads covered for each nucleotide along the mitochondrial transcripts was retrieved using Samtools depth tool.
miRNA target analysis
The predicted miRNA targeting sites on mitochondrial coding genes were downloaded from the miRWalk database (v2.0) (Dweep and Gretz, 2015), which depends on the base-paring between “seed region” and gene sequence. The minimum length required for the match of seed region was set to 7 nt. The experimentally identified miRNA targeting sites were retrieved from the published CLASH results (Helwak et al., 2013).
Detection of mitochondrial nascent protein synthesis
The TRMT61B knock-down cells and mock control cells were washed with pre-warmed methionine-free medium and incubated in methionine-free medium for 45 min. Then Cells were treated with 50 μg/mL emetine in methionine-free medium for 15 min to inhibit cytoplasmic protein synthesis. Further the medium was replaced with methionine-free medium containing 1mM methionine analog L-azidohomoalanine (AHA) (Click Chemistry Tools) and 50 μg/mL emetine. After incubation for 1 h, cells were harvested using clod PBS. Total proteins from different cell lines were quantified with the BCA protein assay kit (Thermo Fisher). Equal amount of AHA-containing proteins were labeled with Cy3 (Click Chemistry Tool) via Click Chemistry. The click reaction was performed in RIPA buffer containing 550 μM Cy3-alkyne, 4mM BTTAA-CuSO4 complex and 151.43mM sodium ascorbate. The reaction was incubated for 2 h at 25 °C with gentle vortex. The labeled proteins were purified by chloroform/methanol precipitation and separated in a 8%–16% gradient SDS-PAGE, and the SDS-PAGE gel was visualized by Typhoon 9500 scan. The gel was also subjected to coomassie blue staining as loading control.
Enrichment of nascent protein
The TRMT61B overexpression cell lines and mock control cell lines were washed with pre-warmed methionine-free medium and starved of methionine in the methionine-free medium for 1 h. Then the medium was replaced with methionine-free medium containing 1 mM L-azidohomoalanine (AHA). After incubation for 2 h, the cells were harvested using cold PBS. Equal amount of AHA-containing proteins were labeled by biotin-alkyne (Click Chemistry Tool) via click reaction. The purified proteins were incubated with 200 μL Streptavidin Agarose Resin (Thermo) in RIPA buffer for 3 h at 29 °C. Resins we re washed 3 times with PBS and 3 additional times with water. Proteins were reduced by incubated at 37 °C in the presence of 10mM DTT in 6M Urea in PBS for 30min and later alkylated by adding iodoacetamide (Sigma) to the final concentration of 20 mM for another 30min. Resins were sequentially washed once with PBS and once with 100nM Triethylammonium bicarbonate (TEAB) buffer. Proteins were digested “on bead” with trypsin in 100 mM TEAB and 1 mM CaCl2 buffer overnight at 37 °C. The peptide solution was performed reductive dimethylation labeling and analyzed on a Q Exactive mass spectrometer.
QUANTIFICATION AND STATISTICAL ANALYSIS
p-values were calculated using unpaired two-sided Mann-Whitney U-test. *p < 0.05, **p < 0.01. N.S. stands for not significant. Error bars represent mean ± SD.
DATA AND SOFTWARE AVAILABILITY
Sequencing data have been deposited into the Gene Expression Omnibus (GEO) under the accession number: GSE102040. Original imaging data have been deposited to Mendeley Data and are available at http://dx.doi.org/10.17632/bf8cshtzmx.1
ADDITIONAL RESOURCES
A detailed protocol describing m1A-MAP used in this study is provided in Methods S1.
Supplementary Material
Methods S1, Detailed protocols for m1A-MAP, Related to STAR METHODS.
Table S1. List of m1A sites in the cytosolic tRNA identified by m1A-MAP, Related to Figure 1 and 2.
Table S2. List of m1A sites in the nuclear-encoded transcripts identified by m1A-MAP, Related to Figure 2 and 3.
Table S3. List of m1A sites in the mitochondrial-encoded transcripts identified by m1A-MAP, Related to Figure 4 and 5.
Table S4. List of m1A sites in the mitochondrial-encoded transcripts targeted by TRMT61B, Related to Figure 5.
Table S5. Sequences of gene-specific reverse primers used in locus-specific m1A detection, Related to STAR METHODS.
Highlights.
We report “m1A-MAP”, a misincorporation-based method, to map m1A at base resolution
m1A-MAP detects m1A methylation at mRNA cap and 5′-UTR
m1A-MAP reveals prevalent m1A modification in mitochondrial-encoded transcripts
m1A in different mRNA regions differentially impacts translation
Acknowledgments
The authors would like to thank Prof. Ying Liu for advice on methyltransferase, Prof. Yongliang Zhao for sharing antibody, Prof. Xinxiang Zhang and Mr. Xiaohui Zhang for quantitative MS measurements, Prof. Xiaorong Zhang and Mr. Kuanxing Gao for metabolic labelling assay and the Core Facilities at the School of Life Sciences, Peking University. This work was supported by the National Basic Research Foundation of China (nos. 2016YFC0900301 and 2014CB964900 to C.Y.), the National Natural Science Foundation of China (nos. 21522201 and 91740112 to C.Y.), US National Institutes of Health (R01 AG042400 and R01 GM122814 to S.-B.Q) and HHMI Faculty Scholar (55108556 to S.-B.Q).
Footnotes
AUTHOR CONTRIBUTIONS
X.L., X.X., and C.Y. conceived the base-resolution method and performed sequencing; Y.C., M.Z., K.W., J.L., and D.Y. performed MS analysis and cell biology experiments, under the guidance of C.Y., C.W., and X.-W.C. X.L., K.W., and J.Z. performed ribosome profiling and translation experiments. X.X. designed and performed the bioinformatics analysis, with the help of Y.M. C.Y. and S.-B.Q. supervised the project. All authors commented on and approved the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson JT, Droogmans L. Biosynthesis and function of 1-methyladenosine in transfer RNA. In: Grosjean H, editor. Fine-Tuning of RNA Functions by Modification and Editing. Berlin, Heidelberg: Springer Berlin Heidelberg; 2005. pp. 121–139. [Google Scholar]
- Antonicka H, Choquet K, Lin ZY, Gingras AC, Kleinman CL, Shoubridge EA. A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. Embo Rep. 2017;18:28–38. doi: 10.15252/embr.201643391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Yaacov D, Frumkin I, Yashiro Y, Chujo T, Ishigami Y, Chemla Y, Blumberg A, Schlesinger O, Bieri P, Greber B, et al. Mitochondrial 16S rRNA Is Methylated by tRNA Methyltransferase TRMT61B in All Vertebrates. PLoS Biol. 2016;14:e1002557. doi: 10.1371/journal.pbio.1002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Suzuki T. Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. RNA. 2012;18:2269–2276. doi: 10.1261/rna.035600.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark WC, Evans ME, Dominissini D, Zheng G, Pan T. tRNA base methylation identification and quantification via high-throughput sequencing. RNA. 2016;22:1771–1784. doi: 10.1261/rna.056531.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods. 2015;12:879–884. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn DB. The occurrence of 1-methyladenine in ribonucleic acid. Biochim Biophys Acta. 1961;46:198–200. doi: 10.1016/0006-3002(61)90668-0. [DOI] [PubMed] [Google Scholar]
- Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- Finer-Moore J, Czudnochowski N, O’Connell JD, 3rd, Wang AL, Stroud RM. Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA(3)(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target. J Mol Biol. 2015;427:3862–3876. doi: 10.1016/j.jmb.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Jaffrey SR, Pan T, Rechavi G, Suzuki T. RNA modifications: what have we learned and where are we headed? Nat Rev Genet. 2016;17:365–372. doi: 10.1038/nrg.2016.47. [DOI] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- Hauenschild R, Tserovski L, Schmid K, Thuring K, Winz ML, Sharma S, Entian KD, Wacheul L, Lafontaine DL, Anderson J, et al. The reverse transcription signature of N-1-methyladenosine in RNA-Seq is sequence dependent. Nucleic Acids Res. 2015;43:9950–9964. doi: 10.1093/nar/gkv895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- Helm M, Motorin Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat Rev Genet. 2017;18:275–291. doi: 10.1038/nrg.2016.169. [DOI] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kondo A, Yamamoto S, Nakaki R, Shimamura T, Hamakubo T, Sakai J, Kodama T, Yoshida T, Aburatani H, Osawa T. Extracellular Acidic pH Activates the Sterol Regulatory Element-Binding Protein 2 to Promote Tumor Progression. Cell Rep. 2017;18:2228–2242. doi: 10.1016/j.celrep.2017.02.006. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol. 2016a;12:311–316. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- Li X, Xiong X, Yi C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat Methods. 2016b;14:23–31. doi: 10.1038/nmeth.4110. [DOI] [PubMed] [Google Scholar]
- Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol. 2015;11:592–597. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, Wang X, Hao Z, Dai Q, Zheng G, et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell. 2016 doi: 10.1016/j.cell.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One. 2014;9:e110799. doi: 10.1371/journal.pone.0110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res. 2013;41:D262–267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanick S, Krecic A, Andersland J, Anderson JT. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA. 2005;11:1281–1290. doi: 10.1261/rna.5040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros PM, Prado MA, Zamboni N, D’Amico D, Williams RW, Finley D, Gygi SP, Auwerx J. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol. 2017;216:2027–2045. doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter-Dennerlein R, Dennerlein S, Rehling P. Integrating mitochondrial translation into the cellular context. Nat Rev Mol Cell Biol. 2015;16:586–592. doi: 10.1038/nrm4051. [DOI] [PubMed] [Google Scholar]
- Rooijers K, Loayza-Puch F, Nijtmans LG, Agami R. Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nat Commun. 2013;4:2886. doi: 10.1038/ncomms3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorbach J, Minczuk M. The post-transcriptional life of mammalian mitochondrial RNA. Biochem J. 2012;444:357–373. doi: 10.1042/BJ20112208. [DOI] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M, Nir R, Farouq D, Vainberg Slutskin I, Schwartz S. TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res. 2017;27:393–406. doi: 10.1101/gr.207613.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M, Fu Y, Pavon-Eternod M, He C, Pan T. Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA. 2010;16:1317–1327. doi: 10.1261/rna.2057810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saletore Y, Meyer K, Korlach J, Vilfan ID, Jaffrey S, Mason CE. The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 2012;13:175. doi: 10.1186/gb-2012-13-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014a;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014b;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation and degradation of human mitochondrial RNA: the prokaryotic past leaves its mark. Mol Cell Biol. 2005;25:6427–6435. doi: 10.1128/MCB.25.15.6427-6435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits P, Smeitink J, van den Heuvel L. Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J Biomed Biotechnol. 2010;2010:737385. doi: 10.1155/2010/737385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet. 2011;45:299–329. doi: 10.1146/annurev-genet-110410-132531. [DOI] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZM. Human mitochondrial mRNAs--like members of all families, similar but different. Biochim Biophys Acta. 2010;1797:1081–1085. doi: 10.1016/j.bbabio.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardo E, Nachbagauer C, Buzet A, Taschner A, Holzmann J, Rossmanith W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase--extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012;40:11583–11593. doi: 10.1093/nar/gks910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waku T, Nakajima Y, Yokoyama W, Nomura N, Kako K, Kobayashi A, Shimizu T, Fukamizu A. NML-mediated rRNA base methylation links ribosomal subunit formation to cell proliferation in a p53-dependent manner. J Cell Sci. 2016;129:2382–2393. doi: 10.1242/jcs.183723. [DOI] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D, Lu B, Liao L, Hewel J, Han X, et al. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteomics. 2015;129:16–24. doi: 10.1016/j.jprot.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You C, Dai X, Wang Y. Position-dependent effects of regioisomeric methylated adenine and guanine ribonucleosides on translation. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods. 2015;12:835–837. doi: 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubradt M, Gupta P, Persad S, Lambowitz AM, Weissman JS, Rouskin S. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat Methods. 2017;14:75–82. doi: 10.1038/nmeth.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods S1, Detailed protocols for m1A-MAP, Related to STAR METHODS.
Table S1. List of m1A sites in the cytosolic tRNA identified by m1A-MAP, Related to Figure 1 and 2.
Table S2. List of m1A sites in the nuclear-encoded transcripts identified by m1A-MAP, Related to Figure 2 and 3.
Table S3. List of m1A sites in the mitochondrial-encoded transcripts identified by m1A-MAP, Related to Figure 4 and 5.
Table S4. List of m1A sites in the mitochondrial-encoded transcripts targeted by TRMT61B, Related to Figure 5.
Table S5. Sequences of gene-specific reverse primers used in locus-specific m1A detection, Related to STAR METHODS.