Abstract
In the past fifteen years, the proteasome has been validated as an anticancer drug target and 20S proteasome inhibitors (such as bortezomib and carfilzomib) have been approved by the FDA for the treatment of multiple myeloma and some other liquid tumors. However, there are shortcomings of clinical proteasome inhibitors, including severe toxicity, drug resistance and no effect in solid tumors. At the same time, extensive research has been conducted in the areas of natural compounds and old drug repositioning toward the goal of discovering effective, economical, low toxicity proteasome-inhibitory anticancer drugs. A variety of dietary polyphenols, medicinal molecules, metallic complexes and metal-binding compounds have been found to be able to selectively inhibit tumor cellular proteasomes and induce apoptotic cell death in vitro and in vivo, supporting the clinical success of specific 20S proteasome inhibitors bortezomib and carfilzomib. Therefore, the discovery of natural proteasome inhibitors and researching old drugs with proteasome inhibitory properties may provide an alternative strategy for improving the current status of cancer treatment and even prevention.
Keywords: 26S proteasome, protein degradation, proteasome inhibitor, natural compounds, drug repurposing, tea polyphenols, metal complexes, cancer therapy and prevention
1 Introduction
The 26S proteasome is part of a cellular protein degradation pathway that plays a vital part in many cellular functions. Research into the inhibition of the proteasome for the purpose of anticancer therapy, particularly by chemical inhibitors, has yielded distinct and encouraging outcomes that are changing the status of cancer treatment.
1.1 The ubiquitin-proteasome system
The ubiquitin-proteasome system (UPS) (Fig. 1) is a vital proteolysis mechanism that is responsible for 80–90% of protein degradation in the cell [1–4]. As a result, the UPS is essential to cellular homeostasis. The UPS is a protein-based cascade that is responsible for degrading proteins that are involved in many cellular events, including cell cycle progression, apoptosis, DNA damage/repair, endocytosis, drug resistance, angiogenesis and cell differentiation [5–11]. In addition, research has shown that the UPS plays an important role in tumorigenesis and tumor survival [11–14].
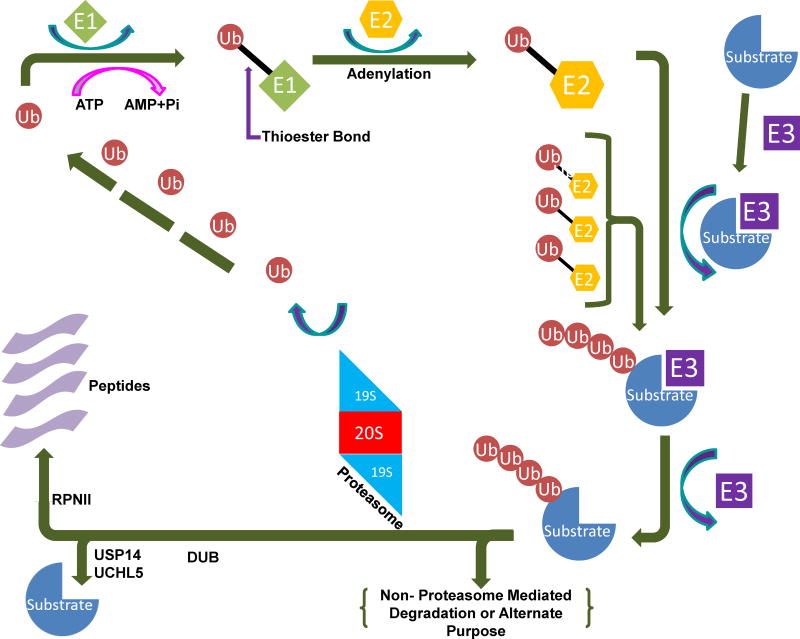
Fig. 1.
The ubiquitin-proteasome system (UPS). The pathway begins with the conjugation of ubiquitin molecules to a substrate protein, forming a polyubiquitin chain through E1, E2 and E3 enzymes. The chain is then recognized and removed by the 19S lid of the 26S proteasome, followed by either release or degradation of the protein by the 20S catalytic core.
The UPS contains two major steps. The first step is the covalent attachment of ubiquitin(s) to a protein substrate—a process called ubiquitination—and the second is the degradation of the substrate by the 20S proteasome [15] (Figure 1). The UPS has become a promising target for anticancer therapies in the last twenty years, and continuing research strengthens its potential as a target for cancer treatment and even prevention.
1.2 Ubiquitin conjugation
Ubiquitination, the first step in the UPS, is the conjugation of ubiquitin to a protein substrate. Ubiquitin is a small protein consisting of 76 amino acids. Ubiquitination is conducted by a series of enzymes belonging to three classes, known as E1, E2, and E3 (Figure 1). This cascade starts with the activation of ubiquitin. Using ATP to fuel the process, E1 adenylates ubiquitin to create a high energy E1-ubiquitin thionene [16]. There are only two types of E1 enzymes, but each type can interact with one of the approximately three dozen E2 enzymes to continue the cascade [17]. This is important to help maintain the precise regulation necessary for the UPS to function.
There are ~35 E2 enzymes [17]. They conjugate ubiquitin through transthioesterification [18]. This prepares ubiquitin for the E3 ligase. It is at this point that the activated, conjugated ubiquitin is attached to a substrate protein at a lysine residue. The substrate can receive either just one ubiquitin (mono-ubiquitination), or multiple ubiquitin molecules. Monoubiquitinated substrates are used in a variety of cellular processes, including cell signaling and histone modification [19,20]. Typically, monoubiquitinated substrates are not thought to be degraded by the proteasome, although there is some suggestion that proteins shorter than 150 amino acids may be degraded after monoubiquitination [21]. Multi-monoubiquitination occurs when the substrate protein is monoubiquitinated at several different lysine residues [22]. Polyubiquitination, in contrast, is the result of additional ubiquitin molecules becoming attached to one of seven lysine residues on an ubiquitin monomer bound to the substrate. Proteins tagged with polyubiquitin chains, usually involving four or more ubiquitin molecules, may then go to the 20S proteasome for degradation [23] (Figure 1).
1.3 Deubiquitinases
Deubiquitinases (DUBs) are enzymes that identify and then remove attached ubiquitins (deubiquitination) from substrate proteins and therefore help regulate the degradation of the substrates by proteasomes [24]. There are five types of DUBs: (i) ubiquitin-specific proteases (USP), (ii) ubiquitin carboxyl-terminal hydrolases (UCH), (iii) ovarian tumor-like, (iv) Machado-Joseph Disease (MJD), and (v) JAMM (metallo-protease family) [25]. There are three DUBs present in the 19S proteasome: UCHL5 (UCH37) and USP14 are two cysteine proteases associated with some 19S subunits, while POH1 (RPN11) is a Zn-dependent metalloprotease and a permanent member of 19S. UCHL5 and USP14 mediate a stepwise removal of ubiquitin from the protein by disassembling the chain from its distal tip, whereas POH1 is the only essential DUB of 26S proteasome (Fig. 2a). Since there are several outstanding, comprehensive reviews on DUBs and their inhibitors in this special issue, we will not review this topic in detail.
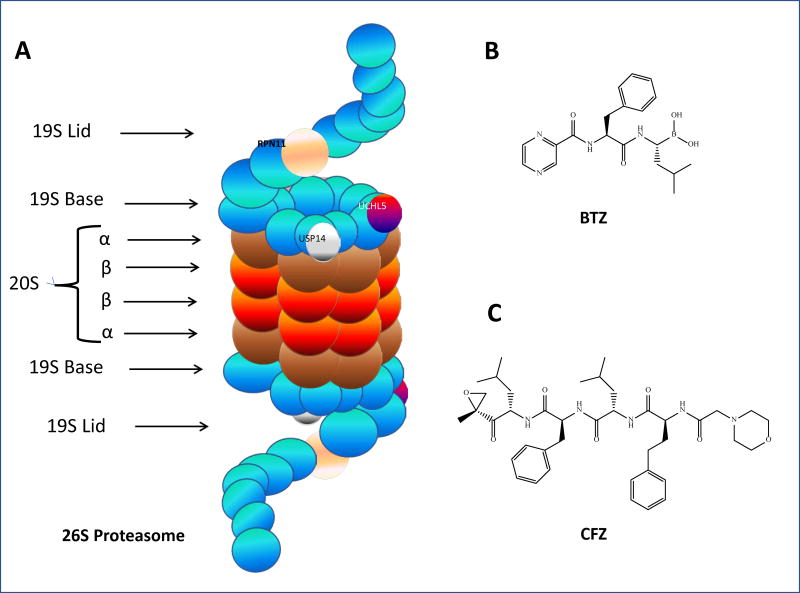
Fig. 2.
The 26S proteasome and chemical structures of BTZ and CFZ. A. 26S proteasome: the 19S–associated DUBs (Rpn11, USP14, and UCHL5) are pictured, as are the alpha and beta subunits which make up 20S core particle; B. Bortezomib; C. carfilzomib
1.4 The 26S proteasome
The 26S proteasome is a multi-subunit protease complex, with 44 polypeptides and a molecular weight of ~2,400 kDa [1–3,26]. This ATP-dependent holocomplex is responsible for protein degradation, the second step of the UPS [27]. Typically, only polyubiquitinated substrates with chains of four or more ubiquitin molecules attached to the lysine sites (not K-63) without branching ultimately end up in the proteasome [28]. The 26S proteasome itself is a closely regulated multi-protein machine that is important to protein integrity, as it functions to degrade misfolded and damaged proteins [27]. Aside from its function in protein quality control, it is also pivotal to metabolic regulation and cell cycle-controlled antigen presentation. The 26S proteasome also degrades the proteins responsible for cellular regulation and apoptosis as well.
The proteasome has varied forms. It can be constitutive (20S) or immunological (20Si); both forms have the same beginning. The expression of the 26S proteasome is directly linked to transcription factor Nrf1, and inhibition of Nrf1 was recently shown to suppress the 26S proteasome by Weyburne et al. [29]. The assembly of the proteasome is overseen mainly by two pairs of proteasome-assembling chaperone (PAC) facilitators, PAC1-PAC2 and PAC3-PAC4. These facilitators form the α-ring of the 20S core particle first. After the α-ring is created, the PACs use the α-ring to build the β-ring attached to it. At this point PAC3-4 is degraded. This half-assembled core particle is called the 15S. Eventually two 15S particles will bind at their β-rings and form the 20S core particle, the type of β-subunits determining which form of proteasome will be made [30]. In the immunological 20Si, the active β-subunits β1, β2, and β5 are replaced with LMP2, MECL-1, and LMP7, respectively [31]. While little is understood about the assembly of the 19S lid and base, it is believed they are formed at this point, using the α-ring of the core particle as an anchor and possibly PAC1-PAC2 as well [32].
The 26S proteasome, once its assembly is completed, is arranged so that the 19S ring proteins form a lid and a base for the 20S core particle (Fig. 2A). The lid is arranged in helical bundles that form a horseshoe shape, with its spiral shapes made by aromatic, hydrophobic “pore-loops.” The 19S is the gateway to proteolytic activity in the 26S [30]. The 19S lid is comprised of nine unique Rpn subunits [33]. It has three identified deubiquitinating enzymes (DUBs), which serve to cleave ubiquitin molecules from incoming ubiquitinated protein substrates: two cysteine DUBs, UCHL5/UCH37 and USP14/USP6, both of which act on the distal end of ubiquitin chains, and the metalloprotease RPN11, which engages in complete cleavage of the ubiquitin chain at the proximal end [34]. Other functions of the lid remain to be determined. The 19S base consists of ten subunits, six of which are AAA-ATPases. The base functions in recognition of ubiquitin chains on incoming protein substrates, as well as unfolding the proteins in preparation for proteolysis and opening a “gate” formed by the 20S core particle’s α-rings, thus allowing the protein to enter the core particle for degradation [33].
The α-rings of the 20S admit substrates into the proteolytic core. Following ubiquitin tag recognition and the unfolding of the substrate protein, the protein passes through the α-ring of the 20S. A mature 26S has six proteolytic sites with three types of activities: trypsin (T)-like activity (cleavage after basic residue), chymotrypsin (CT)-like activity (cleavage after hydrophobic residue), and peptidyl glutamyl peptide-hydrolyzing (PGPH)-like activity (cleavage after acidic residue) [30]. Here in the core, proteins are degraded via nucleophilic attack from the oxygen atom on the side chain of the N-terminal threonine (Thr1Oγ) of each subunit. Proteins are cleaved into oligopeptides ranging in length from three to fifteen amino acids [33] (Figure 2). Some proteasome inhibitors, such as the FDA-approved bortezomib, work by binding to one of these catalytic subunits and impairing the proteasome’s ability to degrade proteins [35,36].
1.5 Proteasomes and cancer
Cancer cells are generally distinguished by one or both of two homeostatic instabilities: they can have rampant cellular proliferation, and/or a distinct lack of apoptosis [37]. The proteasome is closely involved in many regulatory pathways within the cell, including proliferation and apoptosis. Elevated levels of 26S proteasome, as well as unusually high levels of proteasome activity, have been detected in several different types of cancer [38–40]. High proteasome activity seems to be important for cancer cell survival, as it likely aids in protection against apoptosis pathways and ridding the cell of damaged proteins [40]. Therefore, inhibition of the proteasome is an area of interest in cancer research. Proteasome inhibition has been shown to induce apoptosis in a variety of cancers, and some studies have shown cancer cells to be more sensitive to proteasome inhibition than normal cells [41–44]. The molecular mechanisms by which proteasome inhibitors could selectively target tumor cells have been widely studied, including those below.
1.5.1 Increasing oxidative stress in cancer cells via proteasome inhibitors
Enhanced proliferation can result in higher levels of reactive oxygen species (ROS) in cancer cells, leading to oxidative stress. Oxidized proteins can become toxic to cells and must be degraded. Proteasome inhibition may in some cases further increase oxidative stress in cancer cells, leading to cell death [45,46]. Interestingly, the 20S proteasome is unusually resistant to oxidative stress compared to the 26S proteasome, allowing it to function in degradation of oxidation-damaged proteins even in conditions of high oxidative stress [47]. This resilience would be useful in cancer cells exhibiting higher levels of oxidative stress, and could make the proteasome a good target for anticancer therapy.
1.5.2 Selective apoptosis induction in cancer cells via proteasome inhibitors
Apoptosis, a highly conserved function, is a two-stage process (commitment and execution) that is committed by proteases of the caspase class. The UPS is important to this process because it affects a cell’s sensitivity to apoptosis through regulation of the levels of proteins that are involved in the control of apoptosis. Apoptosis dysregulation is a factor in many human diseases, including cancer, as mentioned before. It has been shown that proteasome inhibitors induce apoptosis in a variety of cancer cells, sometimes in a cancer cell-specific manner [48–50]. Proteasome inhibition has also been shown to cause cell cycle arrest [51,52]. In some cases, proteasome inhibition may even induce apoptosis in proliferative cells while protecting quiescent cells [53]. The mechanism through which proteasome inhibitors induce apoptosis is an area of study, and studies have suggested these mechanisms may involve an increase in levels of tumor suppressor p53 and an increase in cell cycle regulator p27, as well as a decrease in anti-apoptotic protein Bcl-2 [54–56]. The apparent preference for inducing apoptosis in cancer cells makes proteasome inhibition potentially less toxic to non-malignant cells than other cancer therapies, and is therefore an attractive solution.
2 Proteasome inhibitors in clinical studies
There are five main types of proteasome inhibitors, classified by how they interact with the active site threonine residue. The five types are: (i) peptide aldehydes, (ii) peptide boronates, (iii) peptide vinyl sulfones, (iv)peptide epoxyketones, and (v) β-lactone. All of the “peptide” proteasome inhibitors’ moieties are required for binding to the substrate “pockets” in the 20S core particle. Peptide aldehydes are quite unstable and easily oxidized into inactive acids, thus limiting their capability to specifically inhibit the 20S proteasome (though they sometimes also inhibited serine and cysteine proteases). Peptide boronates have experienced more success, especially as small molecule 20S proteasome inhibitors [57].
Some early clinical UPS inhibitors include immunomodulatory imide drugs (IMiDs). Cereblon, the original target for the IMiDs, is a cullin-RING E3 ligase. By controlling which proteins are tagged and transferred to the proteasome, the function of the 26S proteasome is consequently inhibited [58,59]. E3 ligases fall into four classes: (i) Really Interesting New Gene (RINGs) (which target proteins and mediate the transfer of ubiquitin from an E2), (ii) E6-AP Carboxyl-Terminus (HECTs (which are homologous to E6 and transfer ubiquitin to their own cysteine residues and then to a substrate), (iii) the U-boxes (which behave like RINGs), and (iv) the RING-Inbetween-RINGs (which behave like hybrids of RINGs and HECTs). Some IMiDs, thalidomide, lenalidomide, and pomalidomide are used in combination with proteasome inhibitors, resulting in enhanced therapeutic effects [60,14].
Bortezomib (BTZ, Fig. 2B) was the first FDA-approved proteasome inhibitor. Approved in 2003 for the treatment of multiple myeloma, it falls under the peptide boronate category of proteasome inhibitors and binds reversibly to the 20S proteasome via its boron atom. It has been shown to inhibit the β5 subunit of the 20S core particle in the 26S, thus reducing its CT-like activity [57]. Importantly, BTZ can also increase sensitivity to other chemotherapeutic drugs and radiation, useful in light of the problem of drug resistance in cancer [61]. However, BTZ has a number of side effects, including thrombocytopenia and peripheral neuropathy [62]. Additionally, resistance to BTZ, whether initial or after treatment with the drug, is also a major issue in the clinic [63].
In 2012, carfilzomib (CFZ, Fig. 2C), a second generation proteasome inhibitor, was approved . Different from BTZ, CFZ is an irreversible proteasome inhibitor that binds to the 20S proteasome and falls into the peptide epoxyketone class [14,60]. It was designed in an effort to address resistance to BTZ as well as to try to avoid some of its side effects, though CFZ does have side effects of its own. Indeed, carfilzomib does show efficacy in BTZ-refractory multiple myeloma patients [64]. However, a recent trial suggests that CFZ may not be as effective when used alone compared to when used in combination with dexamethasone [65]. Patients who exhibited resistance to BTZ may also develop eventual resistance to CFZ as well, highlighting a need for more therapeutic agents for these patients [66]. It should also be noted that BTZ and CFZ are mainly used for the treatment of hematological tumors, not solid tumors. Therefore, new proteasome inhibitors are needed for the treatment of solid cancers.
3 Natural compounds and repurposed drugs with proteasome-inhibitory activity
Traditional chemotherapeutic agents, though effective, have some problems. They are often expensive to develop and result in high costs per dose. Due to toxicity, they can have a variety of side effects, some of which can be severe and affect quality of life. Additionally, many patients develop resistance to chemotherapy. Therefore, there is an urgent need for the development of new drugs to either overcome resistance or enhance sensitivity to existing drugs. In light of the issues posed by traditional anticancer drugs, many researchers are interested in finding ways to develop safer, more economical therapeutic agents.
One area of drug development that has garnered great interest recently is through the use of natural compounds. These come from a variety of sources, including traditional Chinese medicine, herbs, and even diet. Often, these natural compounds have been in use already for centuries, especially in the case of traditional Chinese medicine, meaning their safety profile can be well known. Additionally, they are sometimes less toxic than current chemotherapy drugs and radiation. These factors, along with their wide availability around the world, suggest that potentially these natural compounds may be used as a part of combination treatment therapies. Natural compounds can be used synergistically to enhance the effect of current anticancer therapies and aid in overcoming chemo-drug resistance, and they can also be used as a starting point for the design of new drugs via the development of modified prodrug or synthetic analog versions of their natural counterparts.
In addition, one creative way researchers avoid high cost and the time-consuming issues of new drug discovery/ development is through old drug repurposing, in which a drug that is already approved for the treatment of another disease or condition is studied for treatment of a different disease, such as cancer. This is often a faster, more economical way of bringing a drug to new treatment –drugs that are already approved come with an established safety profile, and effective dosage is already known. This cuts development time (and therefore cost) down greatly, and can potentially identify drugs that may be safer to use than traditional chemotherapy. Here, promising research on natural compounds and some repurposed drugs as anticancer therapies is reviewed comprehensively, with a focus on their molecular targets and potentials for cancer treatment and prevention.
3.1 Flavonoids
It is well known that a diet rich in fruits and vegetables is healthier for a variety of reasons. One such reason is the enriched levels of flavonoids present in fruits, vegetables, and other plant materials, a class of molecules characterized by polyphenol rings. In recent years, flavonoids have garnered attention for their various beneficial effects in humans, including antioxidant, anti-inflammatory, and anticancer properties. Many flavonoids have been researched in preclinical and clinical studies as potential drugs for the prevention and treatment of various diseases, including cancer.
3.1.1 Tea polyphenols
One area of great interest is tea polyphenols. Tea is an extremely popular beverage worldwide, and studies have shown that drinking tea can be beneficial for cancer prevention. Available in a variety of different leaf types, tea is inexpensive, easy to prepare, and rarely has any negative effect on the drinker – in fact, drinking tea boasts a plethora of beneficial health effects [67]. This, combined with the efficacy of tea polyphenols in research studies involving cancer models, makes them of great interest as potential drug candidates for anticancer therapy.
One such study examined several similar tea polyphenols. These included (−)-epigallocatechin-3-gallate (EGCG) (Fig. 3a), (−)-epigallocatechin (EGC), (−)-epicatechin-3-gallate (ECG), (−)-gallocatechin-3-gallate (GCG), (−)-catechin-3-gallate (CG), (−)-epicatechin (EC), (−)-gallocatechin (GC), and (−)-catechin (C). It was hypothesized that polyphenols that included an ester bond would be able to inhibit the proteasome, but polyphenols without the ester bond would not. To test this, the CT-like activity of purified 20S proteasome in the presence of each polyphenol was determined first. As predicted, the ester bond-containing polyphenols (EGCG, EGC, GCG, and CG) were able to potently inhibit CT-like activity in vitro (EGCG being by far the most potent), while the compounds that did not contain an ester bond (EC, GC, and C) were not. This indicated that the ester bond was essential for proteasome inhibition by the tea polyphenols [68]. Next, the compounds were analyzed in silico for their susceptibility for nucleophilic attack by the proteasome, a factor used to predict the ability of the compound to bind with and subsequently inhibit the proteasome. EGCG was found to be the most susceptible to nucleophilic attack, though the other ester bond-containing compounds were highly susceptible as well. The non-ester bond-containing polyphenols, on the other hand, had low susceptibility to nucleophilic attack. Following this, EGCG and EGC were tested in Jurkat T cell extract. EGCG was able to inhibit the 26S proteasome, while EGC was not. Furthermore, EGCG affected only the CT-like activity and PGPH activity of the 20S proteasome, indicating that it was a specific inhibitor [68]. Finally, EGCG and EGC were tested in intact Jurkat T cells, where again, EGCG was a potent inhibitor of the 26S proteasome and EGC was not (as measured by inhibition of chymotrypsin-like activity). It was also tested in MCF-7 breast cancer cells and PC-3 and LNCaP prostate cancer cells, where similar results were observed [68]. Additionally, based on observed accumulation of proteasome target proteins p27(Kip1) and IKB-α in response to EGCG treatment in Jurkat T cells, it was hypothesized that EGCG should cause G1 phase cell cycle arrest due to accumulation of these proteins in treated cells, since overexpression of the proteins can suppress the transition from G1 to S phase. Indeed, a 12 hour EGCG treatment was able to induce G1 arrest in Jurkat T cells and LNCaP cells, which correlated with an increase in p27(Kip1) and IKB-α. Finally, EGCG-induced G1 arrest was tested in an SV40-transformed human fibroblast line (VA-13) alongside the normal, untransformed line (WI-38). The accumulation of p27(Kip1) and IKB-α was observed in VA-13 cells, along with an increased population of cells in G1 phase, indicating G1 arrest. In contrast, the WI-38 untransformed cells showed no increase in p27(Kip1) and only low levels of IKB-α, and they were much less sensitive to G1 arrest [68]. Overall, this study showed that the ester bond is an essential factor for EGCG’s inhibitory properties, and that it is a potent and specific proteasome inhibitor when tested with purified proteasomes and in cultured cells. Furthermore, EGCG seemed to have little effect on normal, untransformed cells, indicating that it could potentially be a safe and relatively nontoxic treatment for cancer [68].
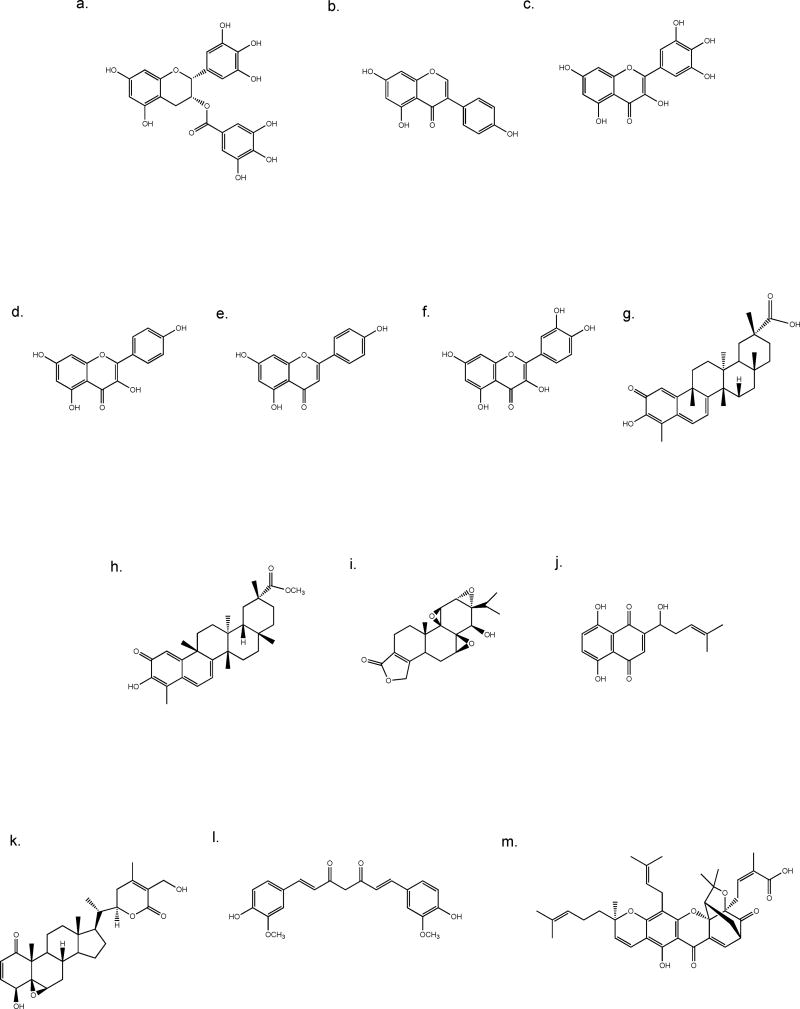
Fig. 3.
Chemical structures of dietary flavonoids (a–f) and medicinal compounds (g–m). a. EGCG, b. genestein, c. myricetin, d. kaempferol, e. apigenin, f. quercetin, g. celastrol, h. pristimerin, i. triptolide, j. shikonin, k. withaferin A, l. curcumin, m. gambogic acid.
Though potent both with purified 20S proteasome and in a variety of cell lines as described above, one drawback to EGCG is its poor bioavailability in humans. This is due to both EGCG’s instability at physiological pH and its susceptibility to modification by catechol-O-methyltransferase (COMT), which is known to methylate EGCG. In humans, the level of COMT activity is determined by a polymorphism in the gene for COMT, which results in three phenotypes: high COMT activity, moderate COMT activity, or low COMT activity [69,70]. A 2010 study hypothesized that breast cancer cells with high COMT activity would be less sensitive to EGCG treatment, but that COMT inhibition could reverse this [71]. To test this idea, MDA-MB-231 breast cancer cells with high COMT expression were treated with EGCG alone, a COMT inhibitor dinitrocatechol (DNC) alone, or a combination of the two. Both EGCG alone and DNC alone decreased COMT activity (providing further evidence that EGCG is a COMT substrate), but together further decreased COMT activity. The combination treatment also enhanced EGCG’s proteasome inhibitory effect (DNC alone had no effect in terms of proteasome inhibition). Combination treatment also increased caspases 3 and 7, both biomarkers of apoptosis, and induced apoptotic cellular morphological change. Finally, when normal, nontransformed cell lines were treated in comparison to tumor cell lines, very little effect was seen with combination treatment, while tumor cells showed a decrease in viability. Hence, COMT is likely responsible for EGCG’s low bioavailability in humans, since COMT inhibition can enhance the effects of EGCG in cancer cells [71].
One method for circumventing the effects of COMT methylation is through the design of synthetic analogs of EGCG. Another 2010 study investigated several synthetic analogs: 5, a symmetrical analog of EGCG; 7, similar to 5; and a prodrug of each (6 and 8, respectively) [72]. (Please note that the numbers of the analogs were originally from the publication [72]; for chemical structures of these compounds, please see the original paper). The prodrugs were acetylated versions of each analog. An acetylated version of EGCG, called 4, was also designed. First, the compounds were tested to examine their ability to inhibit purified 20S proteasome via a CT-like activity assay. None of the peracetates were able to inhibit the 20S proteasome, but EGCG’s analog, compound 5, caused inhibition. 5’s activity was the best, followed by EGCG and compound 7. Next, compounds 5 and 7 were tested alongside EGCG, which was used as a comparison control in MDA-MB-231 cells with high COMT activity. In this case, compound 7 most potently inhibited the 26S proteasome, followed by 5 and then EGCG. This suggested that EGCG and compound 5 are substrates of COMT. To confirm this, the researchers co-treated the cells with a COMT inhibitor (DNC), and found that the effects of EGCG and compound 5 were improved, while that of compound 7 remained the same. This indicated that compound 7 was more resistant to COMT methylation than the other two. Next, the compounds and their peracetate prodrugs were tested in cells to investigate whether any of them caused growth inhibition. It was found that the peracetates (6 and 8) did indeed cause growth inhibition. When compared with EGCG’s peracetate, 4, both 6 and 8 outperformed it, with 8 being the most potent. When this experiment was repeated in the presence of DNC, the growth inhibition caused by compounds 4 and 6 improved, but 8 did not, indicating that compound 8 is the least susceptible to COMT methylation in cells [72].
To further understand the mechanism behind the reduced susceptibility of EGCG’s synthetic analogs to COMT methylation, another study was done using compounds 5 and 7, in addition to compounds 16 and 21 (based off of 5 and 7, respectively, but each missing a gallate group) [73]. First, each compound was analyzed to determine how many sites for nucleophilic attack it possessed. Compounds 16 and 21 only had one site for nucleophilic attack each, but compounds 5 and 7 had two, suggesting they may be more susceptible to nucleophilic attack by the β5 subunit of the 20S proteasome (and hence more likely to bind tightly). To further investigate this, docking studies to the β5 subunit (which is responsible for CT-like activity) were performed for each compound. Due to its equally-spaced hydroxyl and carbonyl groups, compound 7 was able to take on a very stable binding conformation, and also had the lowest free energy. It was followed by compound 5, which had slightly higher free energy, likely due to its two extra hydroxyl groups, which could hinder its ability to bind as well as 7. Compounds 16 and 21 adopted a docking conformation that was very different from compounds 5 and 7, due to each missing a gallate group, and this resulted in much higher free energy. Therefore, these compounds were less likely to bind to the β5 subunit. Ultimately this study suggested that compound 7, which was proven to be more resistant to COMT methylation than EGCG in the previous study, is also more likely to bind tightly to the β5 subunit of the 20S proteasome, matching its performance in in vivo studies. Therefore, design and in silico study of synthetic EGCG analogs could prove to be a good route for circumventing EGCG’s shortcomings in terms of COMT methylation [73].
3.1.2 Soy isoflavones
In addition to tea polyphenols, there are many other flavonoids being studied for the treatment and prevention of cancer, including some that may also target the 26S proteasome. One well known category of flavonoids is soy isoflavones. These are found in products made from the soybean plant, which includes soymilk, tofu, vegetarian meat imitation products, and of course, soybeans themselves, which are commonly eaten as snacks like edamame [74]. One particular soy isoflavone, genistein (Fig. 3b), is currently of interest because of its ability to prevent the growth of tumor cells. Genistein’s mechanism in this regard was relatively unknown, but one study found that it is a potent inhibitor of the proteasome and may induce apoptosis via this route [75]. In silico studies showed that genestein can indeed bind to the β5 subunit of the 20S proteasome, suggesting that it could inhibit CT-like activity. When used in LNCaP prostate cancer and MCF-7 breast cancer cells, genistein was able to inhibit the proteasome as predicted. This was evidenced by the accumulation of ubiquitinated proteins along with proteasomal target proteins p27(Kip1) and IKB-α, an indication of proteasome inhibition. Increased levels of Bax indicated induction of apoptosis in the genistein-treated cells. Interestingly, genistein showed selectivity in simian virus-40 transformed fibroblasts over normal human fibroblasts, causing proteasome inhibition only in the transformed cells [75].
A second study showed that genestein and some other flavonoids, all extracted from the stem of Spatholobus suberectus, were able to inhibit the CT-like activity of the 20S proteasome. Seven compounds were tested, and of those, three were found to have good inhibitory activity in purified 20S proteasome. Those compounds were genestein, isoliquiritigen, and 7-hydroxyflavanone, and they showed a dose-dependent inhibitory effect when tested on the 20S proteasome using a proteasome inhibitor, epoximicin, as a positive control [76].
3.1.3 Other dietary flavonoids
Dietary flavonoids can come from a variety of sources, mainly plant products. The roots, barks, leaves, seeds, and flowers of many plants can be eaten raw, used as a tea, or made into other food products. Some people may get dietary flavonoids via a dietary supplement, though a common source of dietary flavonoids for people around the world is simply through the consumption of fruits and vegetables, which are part of a healthy diet [77]. One study examined four different dietary flavonoids to determine whether they could inhibit the proteasome and induce apoptosis in cancer cells [78]. The dietary flavonoids included myricetin, kaempferol, apigenin, and quercetin (Fig. 3c–f), all of which can be found in a variety of fruits and vegetables (though apigenin can also be found in particular abundance in the flowers of the chamomile plant, which is often prepared as a tea [79]). The compounds were first studied to determine whether they could inhibit purified 20S proteasome, followed by experiments with Jurkat T cells to determine whether they could inhibit the 26S proteasome in intact cells. CT-like activity was measured in both cases. Next, levels of proteasome target proteins Bax and IKB-α were measured in intact Jurkat T cells as an indicator of proteasome inhibition, as well as apoptotic proteins caspase-3 and PARP. In all experiments, apigenin and quercetin were far more potent than the other two dietary flavonoids in terms of proteasome inhibition and induction of apoptosis. When docked to the β5 subunit using docking software, apigenin and quercetin were much more likely to bind than kaempferol or myricetin as well, indicating that the 20S proteasome is likely their target. Lastly, when apigenin was tested in normal, non-transformed cells, no proteasome inhibition or apoptosis induction was observed as it was with the cancer cells, suggesting that apigenin is selective towards cancer cells [78]. Apigenin has also been studied in breast cancer cells, where it showed similar effects, as well as a mouse model, where mice were given MDA-MB-231 breast cell tumors and then treated with apigenin. In this in vivo experiment, when the tumors reached sizes of ~120 mm3 on average, the mice were randomly grouped and were treated by daily subcutaneously injection with 25 or 50 mg/kg apigenin, or vehicle for 29 days, resulting in 22% growth inhibition in the 25 mg/kg apigenin-treated tumors (P < 0.05) and to 43% growth inhibition in the 50 mg/kg apigenin-treated tumors (P < 0.01), respectively [80]. Apigenin not only resulted in significantly smaller tumors in the mice, but also inhibited proteasomal chymotrypsin-like activity in tumors grown in the mice. Furthermore, apigenin treatment caused no apparent toxicity issues in the mice, indicating that it could be a safe and selective treatment to develop for cancer therapy [80].
3.2 Medicinal compounds
Traditional Chinese medicine has been a unique source for compounds being studied for the treatment and prevention of many diseases, including cancer. The medicinal compounds used in traditional medicine are usually isolated from herbal plants native to China and have been used throughout history for the treatment of a variety of ailments. Chinese herbal medicine is unique in that it is typically prepared as a combination of a variety of different herbs that may work together, rather than a single formulation, perhaps foreshadowing their ability to work synergistically with existing pharmaceutical drugs [81]. While natural compounds including herbs are safe to consume, they can have side effects (like any other drugs), which are often the result of improper use or dosage [82]. Thus, it is important that these traditional medicines are studied further. Though many of these medicines have been used for centuries, their mechanisms in vitro and in vivo are now under investigation in order to shed more light on their anticancer properties and molecular targets (such as 19S or 20S proteasomes).
3.2.1 Celastrol
Celastrol (Fig. 3g), a compound from the root bark of a Chinese plant known as the “Thunder of God Vine,” is one such herb that has been associated with anticancer properties, and has been investigated for its mechanism of action in a variety of research studies [83–85]. Celastrol has been identified as a proteasome inhibitor in one such study [86]. Traditionally used for the treatment of many inflammatory conditions, this study first noted that celastrol is similar in structure to quercetin, a dietary flavonoid mentioned above. It was tested in comparison to oridonin, a compound extracted from another Chinese plant that was predicted to have low proteasome inhibitory activity based on computational docking studies. In purified 20S proteasome, celastrol selectively inhibited CT-like activity, while oridonin had very little inhibitory activity even at high concentrations. The same inhibitory effects from Celastrol, but not oridonin, were observed in intact PC-3 prostate cancer cells, along with increased level of proteasome target proteins. Caspase-3 levels also increased in addition to PARP cleavage, indicating induction of apoptosis by celastrol. Next, celastrol was tested in AR-positive LNCaP prostate cancer cells, where it was able to inhibit the proteasome and caused a decrease in AR levels [86]. This finding was significant due to AR’s role in driving prostate cancer cell growth [87]. In mice growing PC-3 prostate tumors, celastrol was also able to decrease cancer growth [86]. In these experiments, celastrol, injected subcutaneously, was used at 1–3 mg/kg/day for up to 31 days, resulting in 65% to 82% inhibition.
Celastrol has also been shown to work synergistically with existing proteasome inhibitors, a factor which could make it very useful clinically. A 2009 study showed that celastrol, along with pristimerin, could enhance the effects of temozolomide in several melanoma lines [88]. In a temozolomide-resistant line, SK-MEL-173, both combination therapies were more effective in inhibiting cell proliferation than temozolomide alone. When tested in four other resistant melanoma lines, the celastrol/temozolomide combination resulted in a decrease in the IC50 of temozolomide in each line. Resistant SK-MEL-173 cells were also tested to determine whether celastrol could inhibit the proteasome. To do this, the cells were treated with celastrol and then studied via Western blot analysis to look for accumulation of ubiquitinated proteins. Indeed, an accumulation of ubiquitinated proteins was observed, along with an accumulation of proteasomal substrate IKB, indicating that the proteasome was inhibited. Since IKB phosphorylation is the precursor to NFKB activation, which regulates gene expression in the nucleus, this study also examined NFKB’s role in celastrol’s mechanism of action. It was found that when melanoma cells were pretreated with celastrol, followed by temozolomide treatment shortly thereafter, IKB phosphorylation was inhibited, thus inhibiting the activation of NFKB. Though NFKB expression levels did not decrease, it was found that its translocation to the nucleus, an essential step for NFKB to affect gene expression, was blocked. This could further explain celastrol’s mechanism of action for decreasing cell proliferation in otherwise resistant melanoma lines [88].
3.2.2 Pristimerin
Pristimerin (Fig. 3h), the methyl ester of celastrol, is another natural Chinese compound that has been studied for use in cancer treatment. Also used traditionally to treat inflammation, pristimerin was found to bind tightly to the β5 subunit in docking studies and successfully inhibited purified 20S proteasome and 26S proteasome. Like celastrol, it was able to decrease AR levels in PC-3 prostate cancer cells, in addition to several other AR-positive lines. Overall, pristimerin was found to inhibit proteasome activity and cause apoptosis via AR inhibition [89].
3.2.3 Triptolide
Triptolide (Fig. 3i), yet another compound isolated from the Chinese “Thunder of God Vine,” also shows proteasome inhibitory activity. When used in PC-3 prostate cancer cells and MDA-MB-231 breast cancer cells, triptolide potently inhibited cell growth, with the breast cancer line being slightly more responsive to treatment than the PC-3 cells [90]. Unlike celastrol and pristimerin, triptolide did not show any activity towards purified 20S proteasome, an outcome that was predicted by docking studies. However, triptolide was able to potently inhibit the CT-like activity of the proteasome in intact PC-3 cells in both a dose-dependent and time-dependent manner. Furthermore, it was able to induce apoptosis in both PC-3 cells and MDA-MB-231 cells --again, the breast cancer cells were more responsive. The authors of the study propose two possible explanations for why triptolide does not inhibit purified proteasome, but can inhibit the proteasome in intact cells. First, some bonds within the triptolide molecule could hinder its ability to bind to the β5 subunit of the 20S proteasome. Second, triptolide could inhibit other subunits of the 26S proteasome besides the 20S subunit. The authors suggest that modified forms of triptolide could prove to be very effective anticancer drugs [90].
3.2.4 Shikonin
Shikonin (Fig. 3j) is a medicinal compound extracted from the root of Lithospermum erythrorhizon, an herb which has been used for centuries in Chinese medicine for the treatment of burns, cuts, and lesions caused by disease. In research today, it is known to cause apoptosis in a variety of cancer cell lines. Though shikonin advanced to clinical trials, its mechanism for inducing apoptosis was unknown. A 2009 study indicated that shikonin could inhibit the 26S proteasome in both human PC-3 prostate cancer cells and murine hepatoma H22 lines in a dose-dependent manner [91]. Shikonin also induced apoptosis in these cell lines. It was also shown that proteasome inhibition precedes cell death. Finally, in an in vivo experiment with a mouse model, mice were injected with P388 leukemia cells. While all control mice died within 23 days, all but one shikonin-treated mouse survived to the end of the experiment at 28 days [91].
3.2.5 Withaferin A
Withaferin A (Fig. 3k) is a steroidal lactone extracted from the leaves of the Withania somnifera plant, also known as the Indian winter cherry. The plant has been used in Southeast Asia for millennia towards a vast variety of applications in traditional medicine, ranging from tumor and wound care to the promotion of youth [92]. Like the other compounds discussed, withaferin A has been investigated for its antitumor effect in an effort to elucidate its effectiveness and mechanism of action. A 2007 study found that the compound potently inhibits the 20S proteasome, with the β5 subunit as its target [93]. Initial computer docking studies indicated that withaferin A could bind to the β5 subunit of the 20S proteasome, thus inhibiting its CT-like activity. Indeed, when tested in vitro with purified 20S proteasome, inhibition of CT-like activity was observed. A similar effect was seen in vivo in PC-3 prostate cancer xenografts treated with withaferin A. Induction of apoptosis was also observed in these cells, as distinguished by western blotting for apoptotic biomarkers. Following treatment with withaferin A, an increase in caspase-3 and PARP was observed following proteasome inhibition, with morphological changes occurring in the cells as well. Similar results were observed in LNCaP cells, along with a time-dependent increase in ubiquitinated proteins and a decrease in androgen receptor expression. Lastly, a human PC-3 xenograft was created in mice. Mice treated with withaferin A had much smaller tumors when compared to the control mice, with one treated mouse becoming tumor-free. Tumor samples were analyzed following the experiment, and decreased chymotrypsin-like activity was observed along with increased ubiquitination of proteasome target proteins Bax, IκB-α, and p27. Caspase-3 was found to be increased, and immunostaining of the tumor samples detected apoptotic cells [93].
3.2.6 Curcumin
Curcumin (Fig. 3l), a compound found in turmeric, is of particular interest in cancer research currently. Used in both Asian cuisine and medicine for centuries, it is a spice used around the world today. Recently, interest in curcumin has spiked as its many beneficial effects are becoming more apparent in research studies, and it is still widely used in a medicinal context in India [94]. Curcumin has shown good anticancer properties, especially in colon cancer models. The mechanism of curcumin’s action against colon cancer is a prominent area of research, and it has been reported that the proteasome is a target of curcumin.
A study on curcumin and its involvement with the proteasome found that curcumin was susceptible to nucleophilic attack at two carbonyl carbons, and was therefore able to bind to the β5 subunit in an in silico model [95]. In purified 20S proteasome, it was able to inhibit not only CT-like activity, but also trypsin-like and PGPH-like activity (though to a lesser extent than chymotrypsin-like activity). When tested in two human colon cancer lines, HCT-116 and SW480, it was found that curcumin was able to inhibit CT-like activity in a dose-dependent manner, and could induce apoptosis in the cells, as evidenced by accumulation of apoptotic proteins. The study also examined whether curcumin causes G1 arrest, but did not find any significant evidence of this, which indicated that proteasome inhibition does indeed cause cells to undergo apoptosis. Lastly, a mouse model injected with HCT-116 cells was found to have smaller tumors with diminished chymotrypsin-like activity and signs of apoptosis. In this in vivo experiment, curcumin was used at 500 mg/kg body weight given intragastrically every day for 3 weeks, resulting in 40% inhibition of tumor growth. Interestingly, curcumin also showed a decrease in biomarkers for cell proliferation in the tumor cells, indicating that curcumin was able to slow the growth of tumor cells [95].
Like EGCG (section 3.1.1), curcumin does have some issues with bioavailability in humans, in addition to having poor solubility in water. In light of this, a 2010 study attempted to circumvent these issues through the design of curcumin analogs and water-soluble amino acid conjugates [96]. A monoacetate and a diacetate of curcumin were designed first, and their effects on HCT-116 and SW480 colon cancer cell proliferation were tested alongside curcumin. In the HCT-116 cells, all compounds had similar effects, but in the SW480 cells, curcumin monoacetate was less effective than the other two. Measurement of CT-like activity showed that the acetates were much less potent in terms of proteasome inhibition. Next, four different amino acid conjugates of curcumin (Bisglycinoylcurcumin dihydrochloride salt, 10; bisalaninoylcurcumin dihydrochloride salt, 11; bisvalinoylcurcumin dihydrochloride salt, 12; and a poly-L-glutamic acid conjugate of curcumin, 13) were tested alongside curcumin for their ability to inhibit purified 20S proteasome. All of the amino acid conjugates proved to be more potent inhibitors of the 20S proteasome than unmodified curcumin, 13 being the most potent. Next, the conjugates were tested in LNCaP prostate cancer cells to determine their effect on cell proliferation. In contrast to the in vitro experiment, compound 13 was the least potent inhibitor. All of the other conjugates, which are water-soluble, outperformed both 13 and unmodified curcumin. Following this, conjugates 10–12 were tested in PC-3 and LNCaP cells, where they were found to inhibit CT-like activity. As an explanation for why 13 performs well in vitro but not in vivo, the authors suggest that this may be due to 13’s high molecular weight, which could make it difficult for the conjugate to enter the cells. Overall, the data from this study suggests that water-soluble amino acid conjugates of curcumin could be a good way to harness its anticancer effects for human treatment [96].
Similarly, a recent study showed that 4-arylidene analogs of curcumin could have potent inhibitory effects on the proteasome [97]. A particularly potent analog, named 33, was able to specifically inhibit two of the 19S DUBs (USP14 and, to a lesser extent, UCHL5). IC50 data for the DUBs was significantly lower than that of the 20S core particle when treated with 33. As a result, the proteasome was inhibited, as shown by elevated levels of ubiquitinated proteins in cells. Cell studies using A549 cancer cells and normal HLF cells showed that 33 induced apoptosis in both lines, but to a lesser extent in the normal HLF line, suggesting some selectivity. Like celastrol, 33 was also found to inhibit the nuclear translocation of NFKB via an increase in ubiquitinated IKB [97]. Deubiquitination of IKB and its subsequent proteasomal degradation is necessary for the translocation of NFKB to the nucleus, and thus its ability to regulate gene expression [98]. This could further explain 33’s mechanism of proteasome inhibition and its consequences relating to apoptosis in cancer cells [97].
3.2.7 Gambogic acid
Gambogic acid (GA) (Fig. 3m) is the principal pigment of gamboge resin of several Garcinia species, which has been used as traditional Chinese medicine for the treatment of human diseases including cancer for many years [99]. GA has been approved by the Chinese Food and Drug Administration for the treatment of multiple cancers in clinical trials [100]. Even though several potential molecular targets of GA have been reported, its molecular targets have not been clearly elucidated. It has been reported that GA inhibits tumor proteasome activity, with potency comparable to bortezomib, but with less toxicity [101]. It was further confirmed that GA gains the proteasome inhibitory function after being metabolized by intracellular CYP2E1 [101]. The expression of the CYP2E1 gene is very high in tumor tissues but low in many normal tissues, thus producing tissue-specific proteasome inhibition and tumor-specific toxicity. These have been demonstrated in other cancer cell models [102,103].
3.3 Metal complexes
Metallic complexes are of growing interest in the field of cancer therapy. Metal ions have been shown to bind with nucleic acids or proteins, altering their conformation and biological functions [104]. Since the approval of the platinum complex cisplatin in 1978, metals have become an attractive tool for potential anticancer drugs. However, cisplatin and its subsequent platinum derivatives (e.g., carboplatin and oxiplatin) have shown severe side effects such as neuron- and nephrotoxicity, ototoxicity, and electrolyte disturbances [105]. Therefore, novel metallic complexes are needed.
3.3.1 Gold complexes
The use of gold in anticancer therapy arose from the anti-inflammatory properties attributed to gold compounds, as well as the structural similarity of Au(III) to cisplatin –the centers of both Au(III) and Pt(II) share a d8 electronic configuration, and therefore they both give rise to square planar complexes [105–107]. Au(III) dithiocarbamates of simple amines and oligopeptides inhibit both the proliferation of breast cancer cells in vitro and tumor growth in vivo by inducing apoptosis [13,107]. Notably, the treatment of human xenografts with this class of metal complexes resulted in inhibition of tumor proteasome in vivo [107].
Auranofin (Aur) is a gold(I)-containing compound, which has been used clinically to treat rheumatic arthritis for more than 30 years. Since Aur also has anti-cancer effects with less toxicity compared to currently used chemotherapeutic agents, Aur was recently approved by FDA for Phase II clinical trial in cancer therapy [108,109]. Even though several potential molecular targets for the anti-inflammatory and anti-cancer activities of Aur have been reported, only recently it was found that Aur inhibits the 26S proteasome but not 20S proteasome peptidases , a mechanism different from the FDA-approved proteasome inhibitor bortezomib [110–112]. Molecularly, Aur inhibits 19S-associated DUBs USP14 and UCHL5 [111, 112]. A computational study predicted the docking between Aur and the 19S-associated DUBs [111]. It was found that compound L2 (an active metabolite of Aur) could bind to the active site of UCHL5 with relatively high CDOCKER Energy of −14.51 kcal/mol; the binding model shows that the side chains of His164, Phe165 and Asp179 of UCHL% active site can coordinate to Au+ with distances of 3.181 Å, 2.537 Å and 2.776Å, respectively. Moreover, two ethyl groups stretch towards hydrophobic side chains of Phe79 and Leu179. When compound L2 was docked to the active site of USP14, there were three ligand-poses produced, suggesting that compound L2 could also inhibit USP14 activity but relatively less than UCHL5. Furthermore, Aur showed selective toxicity and accumulation of ubiquitinated proteins to the monocytes from AML patients over monocytes from healthy volunteers [111]. These results demonstrate that low toxicity nature of Aur could be related to its tumor-selective effects in terms of DUB inhibition and growth suppression [111–112]. Please see another review on “Metal-based 19S proteasome-associated deubiquitinase inhibitors as potential anticancer agents” in the same special issue for details. Additionally, it was recently found that that the proteasome inhibition is required for Aur-induced cytotoxicity, unveiling a novel mechanism for the anti-cancer effects of Aur [111,112].
3.3.2 Organic tin complexes
Organotin compounds have been well-documented in their biocidal uses, but have also become of interest as cancer therapy [113–115]. A 2009 study examined the effect of two organotin compounds, triphenyltin (TPT) and tributyltin (TBT), on breast cancer cells [116]. The compounds were first studied using purified 20S proteasome to determine whether they could inhibit proteasomal activity in vitro. It was found that both compounds could inhibit CT-like activity, but TPT was far more potent. Computer docking studies further validated this claim. Next, the compounds were tested in MDA-MB-231 breast cancer cells, where both TBT and TPT were found to inhibit the CT-like activity of the proteasome in these cells, TPT again being the most potent. Consistent with the conclusion of TPT as a potent 20S proteasome inhibitor and apoptosis inducer, after TPT treatment of Jurkat T cells, elevated levels of ubiquitinated proteins (indicating proteasome inhibition) and apoptosis (PARP cleavage and morphological changes) were observed. Finally, immunoprecipitation of proteasome complexes from MDA-MB-231 cells treated with TPT showed Sn only in cells treated with TPT but not the control, indicating that Sn does indeed bind to the proteasome and that TPT’s target is the proteasome [116].
3.3.3 Copper complexes
Copper is an essential trace element for humans; however, higher levels of copper within tumor cells have been observed in several cancers, and copper has been shown to play a role in angiogenesis [117–120]. Therefore, copper chelating compounds are of great interest in cancer therapy due to their ability to bind copper (see section 3.4). Copper-containing compounds are also of interest as proteasome inhibitors. Copper complexes have been shown to result in proteasome inhibition and apoptosis in leukemia cells, whereas copper salts [i.e., CuCl2, leading to the presence of the exa-aquo copper (II) ion in water solution] do not [121]. Pyrithione, which has metal chelating properties when combined with copper (CuPt), has been shown to inhibit the 19S DUBs and the 20S proteasome [122]. Several tested Cu(II) complexes (such as those of 8-OHQ, DSF, DDTC, CQ, etc.) have been shown to inhibit CT-like activity in several prostate cancer lines, inhibiting the 26S proteasome and inducing apoptosis. Interestingly, proteasome inhibition was not seen in normal human breast cell line MCF-10A when tested in comparison, indicating that these complexes may be selective towards cancer cells [123].
3.3.4 Other metal complexes
Zn(II) complexes have also been tested to examine their anticancer effects alongside Cu(II). A Zn(II) complex, along with a zinc salt, showed CT-like inhibitory effects when studied in vitro with purified 20S proteasome. Additionally, the zinc complex caused proteasome inhibition and apoptosis in C4-2B cancer cells, and also showed specificity towards cancer cells over normal MCF-10A breast cells [124]. Gallium(III)-containing compounds are also apoptosis inducers and can inhibit CT-like activity in prostate cancer cells; in one study, they demonstrated a 70% reduction in tumor growth [125]. Complexes of cadmium, though known as contaminants and carcinogens, have demonstrated inhibition against CT-like activity and have also been shown to induce cancer cell-specific apoptosis in breast cancer lines [126]. Based on computer docking studies, these cadmium complexes are believed to inhibit the β5 subunit [126].
3.4 Metal-binding compounds
The discovery of cisplatin in 1965 sparked great interest towards other metal complexes for the treatment of cancer. Though some were hesitant at first towards the idea of using a compound containing a heavy metal in humans, cisplatin was eventually approved for clinical use in the 1970’s. While toxic at high doses, it is widely used as a chemotherapy drug for the successful treatment of many cancers at doses safe for human patients [127]. High serum or tissue levels of copper were found in many types of human cancers including breast, prostate, colon, lung, and brain , suggesting that copper could be used as a novel selective target for cancer therapies [118,128–133]. Today, research on copper (metal) binding compounds continues in an effort to find safer and more effective treatments for cancer. Metal-binding compounds currently being studied are coordinated to a variety of different metals, such as copper and zinc. The resulting complexes have shown effectiveness at proteasome inhibition.
3.4.1 8-hydroxyquinoline and clioquinol
8-hydroxyquinoline (Fig. 4a) is an example of one such compound. It has been studied alongside its derivative, clioquinol, for use as an anticancer agent. Clioquinol (Fig. 4b) was used from the 1950’s through the 1970’s as an antifungal agent. Though it is rarely used now due to toxicity problems, it is of interest for cancer therapy, perhaps as a starting point for the design of safer analogs [134]. It is known that both 8-hydroxyquinoline and clioquinol can bind to copper (in situ complexation), and that both agents exhibit potent anticancer effects ranging from growth suppression to apoptosis. However, the role of copper ions and copper chelation by both compounds was unclear. Therefore, one study compared 8-hydroxyquinoline and clioquinol to four analogs, named 2, 3, 5, and 6, which were highly similar in structure but unable to bind to copper [135]. Analogs 2 and 3 were similar to 8-hydroxyquinoline, and analogs 5 and 6 were similar to clioquinol. First, all of the compounds were given the opportunity to form copper complexes or for in situ complexation; as expected, the analogs did not. Next, using DCIS human breast cancer cells, it was found that 8-hydroxyquinoline and clioquinol were able to facilitate cellular copper uptake, but the non-copper binding analogs could not. The cells showed much higher intercellular copper levels after treatment with 8-hydroxyquinoline and clioquinol than with their non-copper binding analogs, though membrane permeability of each compound was very similar. Next, DCIS cells were treated with 8-hydroxyquinoline, clioquinol, and their analogs to measure their effects on cell proliferation. The non-copper binding compounds had little to no effect, but the copper-binding compounds were able to potently inhibit cell proliferation. Furthermore, both 8-hydroxyquinoline and clioquinol were found to cause proteasome inhibition and subsequent induction of apoptosis, but their non-copper binding analogs could not. Therefore, the data from this study suggests that copper binding is essential for the anticancer effects of 8-hydroxyquinoline and clioquinol [135]. A similar study investigating the role of copper complexing to clioquinol and pyrrolidine dithiocarbamate found that, when complexed to copper, these metal-binding compounds could cause proteasome inhibition and decreased cell proliferation in breast cancer cell lines, but were unable to do so when not complexed to copper [134].
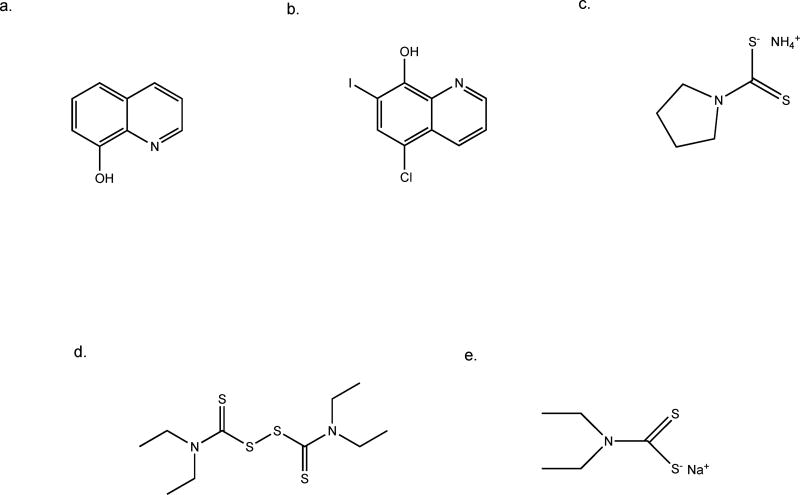
Fig. 4.
Chemical structures of selected metal binding compounds. a. 8-hydroxyquinoline, b. clioquinol, c. pyrrolidine dithiocarbamate, d. disulfiram, e. sodium diethyldithiocarbamate.
3.4.2 Dithiocarbamates
Disulfiram (Fig. 4d), an aldehyde dehydrogenase inhibitor, is another member of the dithiocarbamate family that is of interest as an anticancer therapy. Able to bind to copper, disulfiram (DSF) has a long history as an FDA-approved anti-alcoholism drug. As mentioned before, this makes disulfiram a particularly attractive inhibitor as it could be used as a repurposed drug. Disulfiram gained FDA-approval for its original use in patients with alcoholism in 1951 [136]. Due to its many years in use, its dosage and safety profile are well-established, meaning development of disulfiram as a cancer drug would be much faster and cheaper than developing a novel compound.
When bound to copper, the DSF-Cu complex is capable of inhibiting the 20S proteasome in vitro, while unbound DSF had very little effect [137]. This effect was replicated in MDA-MB-231 cells, where DSF-Cu was able to potently inhibit the CT-like activity of the proteasome in vivo. DSF-Cu also inhibited cell proliferation and caused apoptosis in MDA-MB-231 cells. It was determined that proteasome inhibition occurs before apoptosis in DSF-Cu treated cells. Lastly, two breast cell lines (one was a tumor cell line and the other was a normal, untransformed breast cell line) were tested together with DSF-Cu treatment to determine specificity and toxicity of the DSF-Cu complex. Again, DSF-Cu (but not DSF and Cu alone) potently inhibited the proteasome and caused apoptosis in the malignant line, but not in the untransformed cells. Since the cultured cells had relatively low cellular copper levels in comparison to cancer cells in a physiological setting, another experiment was done in which MDA-MB-231 cells were cultured in media containing copper prior to treatment. Then, the cells were treated with DSF or a control. The DSF-treated cells still underwent proteasome inhibition and apoptosis, indicating that in the presence of high intracellular copper levels, DSF can cause proteasome inhibition and apoptosis. Finally, MDA-MB-231 cells were injected into a mouse model in order to study the effects of DSF alone in a physiological setting. Knowing that cellular copper levels are higher in tumor cells in a physiological setting, it was hypothesized based on the last piece of data that DSF should have an effect on these tumors. DSF did indeed slow tumor growth dramatically in treated mice compared to control mice. Additionally, harvested tumor cells showed inhibition of CT-like activity and accumulation of apoptotic biomarkers. Therefore, DSF seems to be a potent proteasome inhibitor both in vitro and in vivo. This, in addition to its selectivity between malignant and normal cells, makes it an attractive drug candidate. Another important consideration is the fact that DSF is already FDA-approved and has a well-established safety profile, which could make development of DSF as an anticancer therapy much easier [137].
Another member of the dithiocarbamate family, sodium diethyldithiocarbamate (or DDTC) (Fig. 4e), is also a known chelator of copper. Previously studied for the treatment of AIDS, it was also studied as a proteasome inhibitor for anticancer therapy. First, DDTC and copper were mixed and allowed to form a complex. As predicted, DDTC was able to bind to copper. Next, DDTC-Cu was tested in two prostate cancer cell lines, LNCaP and C4-2B. It was able to significantly inhibit the CT-like activity in both lines, while DDTC and copper alone showed little to no effect. Additionally, AR levels dropped in these lines, and induction of apoptosis occurred. CT-like activity could also be inhibited in MDA-MB-231 and MCF-7 breast cancer lines by DDTC-Cu. Visible morphological changes were observed in these cells when treated by DDTC-Cu, but not when treated with DDTC or Cu alone. Again, this data showed that copper binding is an essential part of DDTC’s role in cancer cells [138].
Pyrrolidine dithiocarbamate (Fig. 4c) itself is also of interest as a proteasome inhibitor. A member of the dithiocarbamate family, which is both metal chelator and antioxidant, pyrrolidine dithiocarbamate has been shown to be a good potential anticancer agent when bound to copper, but its effects when bound to other metals, such as zinc, were unknown. To test this, pyrrolidine dithiocarbamate was mixed with zinc (II) chloride or copper (II) chloride, and the mixtures were first tested for their ability to inhibit purified 20S proteasome alongside zinc (II) chloride and copper (II) chloride alone. All were found to inhibit the 20S proteasome, with copper (II) chloride performing best for the metals alone, and the pyrrolidine dithiocarbamate-copper mixture performing best for the latter. Next, it was found that the pyrrolidine dithiocarbamate-zinc mixture was indeed able to inhibit proteasome activity in MDA-MB-231 breast cancer cells in a dose-dependent manner. Both mixtures were found to induce apoptosis in these breast cancer cells, as well as MCF-7 breast cancer cells, DCIS cells, and PC-3 cells. When cells were pretreated with a calpain inhibitor, however, the mixtures did not achieve such potent induction of apoptosis, indicating that calpain is likely involved in apoptosis caused by zinc and copper mixtures with pyrrolidine dithiocarbamate [139]. Also, a series of complexes with diethyldithiocarbamate ligand and three different metals (Ni, Cu, Zn) was tested in human breast cancer MDA-MB-231 cells. Zinc and copper complexes, but not nickel complex, were found to be more active against cellular 26S proteasome than against purified 20S proteasome core particle. One of the possible explanations is inhibition of JAMM domain in the 19S proteasome lid [140]. It is also possible that they inhibit other 19S proteasome associated-DUBs (see below section). There is a most recent report in which the authors report on the synthesis, physico-chemical characterization, and solution behavior of two gold(III) pyrrolidine dithiocarbamates (PDT), namely [Au(III)Br2(PDT)] and [Au(III)Cl2(PDT)]. They found that the bromide derivative was more effective than the chloride one in inducing cell death for several cancer cell lines. [Au(III)Br2(PDT)] elicited oxidative stress with effects on the permeability transition pore, a mitochondrial channel whose opening leads to cell death [141].
3.4.3 Pyrithione
It has been reported that metal-containing compounds could induce cytotoxicity in many cancer cells via targeting the ubiquitin-proteasome system. Similar to disulfiram, pyrithione (PT) also has excellent metal chelating properties, and the zinc complex of pyrithione, for example, has been found to have significant anticancer effects. Several studies by using pyrithione as a metal-binding compound have found that besides the inhibitory effect of PT itself on proteasome function, different metal pyrithione compounds (copper, zinc, nickel, platinum) exhibit a dramatic difference on proteasome function and cytotoxicity, demonstrating the metals’ contributing effect. For example, at effective doses, only CuPT but not NiPT, ZnPT and PtPT inhibits the 20S proteasome peptidases; all of them inhibit the 19S-DUBs [142–145].
4 Conclusions and Perspectives
In 2003, BTZ was approved by the FDA as the first 20S proteasome inhibitor anticancer drug, for the treatment of multiple myeloma (MM). Since then, BTZ-based therapies have become a staple for treating patients with multiple myeloma at all stages. Thanks to BTZ and other immunomodulatory drugs, the survival rate of MM patients has significantly increased. However, BTZ has several shortcomings, including dose-limiting peripheral neuropathy and relapse or development of resistance in many patients. To overcome these limitations, several second generation proteasome inhibitors have been developed, and tested first pre-clinically and then clinically, including the FDA-approved CFZ. In addition, researchers have looked into other targets of the UPS, such as E3 ligases and 19S DUBs, in order to discover new, novel anticancer drugs and/or to overcome BTZ resistance.
Similar effort has been put in the areas of natural compounds and old drug repositioning. Given the high cost of new drug discovery, it is not surprising that natural compounds and repurposed drugs have become such popular research subjects recently. In addition to their cost effectiveness, they also have a multitude of benefits over their current chemotherapy counterparts, often including good selectivity, lower toxicity, and more widespread availability. Natural compounds, especially traditional Chinese herbs, have been in use for centuries for treatment of an astoundingly large variety of ailments. Though their effects and benefits are well known among local communities, it is important that these compounds are characterized officially through scientific research so that they can be used safely in the clinical as well. Also, derivatives, prodrugs, and synthetic analogs can be designed based on natural compounds in order to improve their safety profile, circumvent bioavailability issues, and increase potency. In this article, we have reviewed many natural compounds and old drugs that have proteasome-inhibitory activity, providing alternative approaches for developing proteasome inhibitor anticancer drugs at low cost.
Importantly, natural proteasome inhibitors (unlike BTZ) can be used for cancer prevention. Given the availability and lack of the adverse effects seen in other anti-cancer therapies, it is not surprising that the bioactive molecules found in foods are being explored as cancer preventatives. These molecules, such as green tea polyphenol EGCG (which has potent proteasome-inhibitory activity), have been shown to cause cell cycle arrest and apoptosis, which can interfere with cancer formation at multiple stages. Curcumin serves as another example of a natural proteasome inhibitor compound. Curcumin, in combination with resveratrol in liposome encapsulation, decreased the occurrence of prostate cancer.
Many bioactive molecules can induce anti-oxidative reactions and/or detoxifying enzymes which can prevent tumor development due to DNA damage and remove carcinogens. This encourages their use as cancer preventatives. Sulforaphane and EGCG, which are inhibitors of 19S DUBs and the 20S proteasome, respectively, are effective anti-oxidants and activate phase II detoxifying enzymes. Flavonoids, many of which are 20S proteasome inhibitors as reviewed, specifically show an inverse relationship with the risk of colon, breast and prostate cancers. The more flavonoid- rich food consumed, the lower the risk.
Researchers are currently working to utilize food derived cancer preventatives. They must overcome poor solubility, and low bioavailability related to varying absorption rates, chemical/biochemical instabilities, and transformational behaviors of the active molecules. Such hurtles have also made it difficult to determine dose amounts, which in turn hinders the use of bioactive molecules from food. As more is understood about these bioactive molecules, they may become pivotal preventative measures for numerous types of cancer.
Continuing research with polyphenols and flavonoids supports their use as cancer therapy treatments, as they demonstrate potent proteasome inhibitory activity, low toxicity and tumor selectivity. Their abundance in nature and the body’s ability to recognize and use dietary medicinal compounds lends further greater study. Selective inhibition of tumor proteasome could lead to induction of apoptotic activities in many varieties of cancer and help increase the efficacy of existing and approved drugs. In summary, proteasome inhibition is a promising area for the search for safe and effective cancer treatments and prevention, and natural compounds may prove to be an invaluable source for the next generation of successful proteasome inhibitors. It should be mentioned that in addition to targeting 20S and 19S proteasomes for treating cancers as reviewed here, researchers have been searching for compounds that could selectively inhibit upstream events of the ubiquitin-proteasome pathway, including E1, E2 and E3 enzymes as well as ubiquitin-binding proteins, for improving the current status of cancer therapies [146–149].
Acknowledgments
We apologize to all whose relevant contributions were not cited in our review due to space limitations. We thank Michael Miotto for his assistance in Figure 1 drawing. This work was partially supported by National Cancer Institute grant R21CA184788 (to Q. Ping Dou) and National Institutes of Health grant P30 CA022453 (to the Karmanos Cancer Institute at Wayne State University).
References
- 1.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79(1):13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 2.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7(2):215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg AL, Akopian TN, Kisselev AF, Lee DH, Rohrwild M. New insights into the mechanisms and importance of the proteasome in intracellular protein degradation. Biol Chem. 1997;378(3–4):131–140. [PubMed] [Google Scholar]
- 4.Dou QP, Zonder JA. Overview of Proteasome Inhibitor-Based Anti-cancer Therapies: Perspective on Bortezomib and Second Generation Proteasome Inhibitors versus Future Generation Inhibitors of Ubiquitin-Proteasome System. Current cancer drug targets. 2014;14(6):517–536. doi: 10.2174/1568009614666140804154511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mocciaro A, Rape M. Emerging regulatory mechanisms in ubiquitin-dependent cell cycle control. J Cell Sci. 2012;125(Pt 2):255–263. doi: 10.1242/jcs.091199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orlowski RZ. The role of the ubiquitin-proteasome pathway in apoptosis. [Article] Cell Death & Differentiation. 1999;6(4):303. doi: 10.1038/sj.cdd.4400505. [DOI] [PubMed] [Google Scholar]
- 7.Daulny A, Tansey WP. Damage control: DNA repair, transcription, and the ubiquitin-proteasome system. DNA Repair. 2009;8(4):444–448. doi: 10.1016/j.dnarep.2009.01.017. doi: https://doi.org/10.1016/j.dnarep.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Strous GJ, Govers R. The ubiquitin-proteasome system and endocytosis. J Cell Sci. 1999;112(10):1417. doi: 10.1242/jcs.112.10.1417. [DOI] [PubMed] [Google Scholar]
- 9.Ruckrich T, Kraus M, Gogel J, Beck A, Ovaa H, Verdoes M, et al. Characterization of the ubiquitin-proteasome system in bortezomib-adapted cells. Leukemia. 2009;23(6):1098–1105. doi: 10.1038/leu.2009.8. [DOI] [PubMed] [Google Scholar]
- 10.Rahimi N. The ubiquitin-proteasome system meets angiogenesis. Mol Cancer Ther. 2012;11(3):538–548. doi: 10.1158/1535-7163.mct-11-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie P, Fan Y, Zhang H, Zhang Y, She M, Gu D, et al. CHIP represses myocardin-induced smooth muscle cell differentiation via ubiquitin-mediated proteasomal degradation. Mol Cell Biol. 2009;29(9):2398–2408. doi: 10.1128/mcb.01737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tu Y, Chen C, Pan J, Xu J, Zhou Z-G, Wang C-Y. The Ubiquitin Proteasome Pathway (UPP) in the regulation of cell cycle control and DNA damage repair and its implication in tumorigenesis. International Journal of Clinical and Experimental Pathology. 2012;5(8):726–738. [PMC free article] [PubMed] [Google Scholar]
- 13.Milacic V, Chen D, Ronconi L, Landis-Piwowar KR, Fregona D, Dou QP. A novel anticancer gold(III) dithiocarbamate compound inhibits the activity of a purified 20S proteasome and 26S proteasome in human breast cancer cell cultures and xenografts. Cancer Res. 2006;66(21):10478–10486. doi: 10.1158/0008-5472.can-06-3017. [DOI] [PubMed] [Google Scholar]
- 14.Dalla Via L, Nardon C, Fregona D. Targeting the ubiquitin-proteasome pathway with inorganic compounds to fight cancer: a challenge for the future. Future Med Chem. 2012;4(4):525–543. doi: 10.4155/fmc.11.187. [DOI] [PubMed] [Google Scholar]
- 15.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 16.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 17.Chaugule Viduth K, Walden H. Specificity and disease in the ubiquitin system. Biochemical Society Transactions. 2016;44(1):212–227. doi: 10.1042/BST20150209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee I, Schindelin H. Structural Insights into E1-Catalyzed Ubiquitin Activation and Transfer to Conjugating Enzymes. Cell. 2008;134(2):268–278. doi: 10.1016/j.cell.2008.05.046. doi: https://doi.org/10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
- 19.Sigismund S, Polo S, Di Fiore PP. Signaling Through Monoubiquitination. In: Madshus IH, editor. Signalling from Internalized Growth Factor Receptors. Berlin, Heidelberg: Springer Berlin Heidelberg; 2004. pp. 149–185. [DOI] [PubMed] [Google Scholar]
- 20.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 Complex Is Essential for Histone Monoubiquitination by the Rad6-Bre1 Complex, Which Signals for Histone Methylation by COMPASS and Dot1p. Journal of Biological Chemistry. 2003;278(37):34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 21.Braten O, Livneh I, Ziv T, Admon A, Kehat I, Caspi LH, et al. Numerous proteins with unique characteristics are degraded by the 26S proteasome following monoubiquitination. Proceedings of the National Academy of Sciences. 2016;113(32):E4639–E4647. doi: 10.1073/pnas.1608644113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadowski M, Suryadinata R, Tan AR, Roesley SNA, Sarcevic B. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life. 2012;64(2):136–142. doi: 10.1002/iub.589. [DOI] [PubMed] [Google Scholar]
- 23.Ristic G, Tsou W-L, Todi SV. An optimal ubiquitin-proteasome pathway in the nervous system: the role of deubiquitinating enzymes. Frontiers in Molecular Neuroscience. 2014;7:72. doi: 10.3389/fnmol.2014.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dambacher CM, Worden EJ, Herzik MA, Martin A, Lander GC. Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. eLife. 2016;5:e13027. doi: 10.7554/eLife.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isono E, Nagel M-K. Deubiquitylating enzymes and their emerging role in plant biology. Frontiers in Plant Science. 2014;5:56. doi: 10.3389/fpls.2014.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Bortezomib as the First Proteasome Inhibitor Anticancer Drug: Current Status and Future Perspectives. Current cancer drug targets. 2011;11(3):239–253. doi: 10.2174/156800911794519752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreas Schweitzer AA, Till Rudack, Florian Beck, Gunter Pfeifer, Plitzko Jurgen M, Eri Sakata, Klaus Schulten, Friedrich Forster, Wolfgang Baumeister. Structure of the human 26S proteasome at a resolution of 3.9 A°. Proceedings of the National Academy of Sciences of the United States. 2016 doi: 10.1073/pnas.1608050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paraskevopoulos K, Paraskevopoulos K, Kriegenburg F, Tatham Michael H, Rösner Heike I, Medina B, Larsen Ida B, et al. Dss1 Is a 26S Proteasome Ubiquitin Receptor. Molecular Cell. 2014;56(3):453–461. doi: 10.1016/j.molcel.2014.09.008. doi: http://dx.doi.org/10.1016/j.molcel.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weyburne ES, Wilkins OM, Sha Z, Williams DA, Pletnev AA, de Bruin G, et al. Inhibition of the Proteasome β2 Site Sensitizes Triple-Negative Breast Cancer Cells to β5 Inhibitors and Suppresses Nrf1 Activation. Cell Chemical Biology. 2017;24(2):218–230. doi: 10.1016/j.chembiol.2016.12.016. doi: http://dx.doi.org/10.1016/j.chembiol.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livneh I, Cohen-Kaplan V, Cohen-Rosenzweig C, Avni N, Ciechanover A. The life cycle of the 26S proteasome: from birth, through regulation and function, and onto its death. [Review] Cell Res. 2016;26(8):869–885. doi: 10.1038/cr.2016.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrington DA, Gregerson DS. Immunoproteasomes: Structure, Function, and Antigen Presentation. Progress in molecular biology and translational science. 2012;109:75–112. doi: 10.1016/B978-0-12-397863-9.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saeki Y, Toh-e A, Kudo T, Kawamura H, Tanaka K. Multiple Proteasome-Interacting Proteins Assist the Assembly of the Yeast 19S Regulatory Particle. Cell. 2009;137(5):900–913. doi: 10.1016/j.cell.2009.05.005. doi: https://doi.org/10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka K. The proteasome: Overview of structure and functions. Proceedings of the Japan Academy. Series B, Physical and Biological Sciences. 2009;85(1):12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Arcy P, Wang X, Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacology & Therapeutics. 2015;147:32–54. doi: 10.1016/j.pharmthera.2014.11.002. doi: http://dx.doi.org/10.1016/j.pharmthera.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Sridhar S, Bhat G, Guruprasad K. Analysis of bortezomib inhibitor docked within the catalytic subunits of the Plasmodium falciparum 20S proteasome. SpringerPlus. 2013;2:566. doi: 10.1186/2193-1801-2-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams J, Kauffman M. Development of the proteasome inhibitor Velcade (Bortezomib) Cancer Invest. 2004;22(2):304–311. doi: 10.1081/cnv-120030218. [DOI] [PubMed] [Google Scholar]
- 37.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 38.Chen L, Madura K. Increased Proteasome Activity, Ubiquitin-Conjugating Enzymes, and eEF1A Translation Factor Detected in Breast Cancer Tissue. [10.1158/0008-5472.CAN-05-0201] Cancer research. 2005;65(13):5599. doi: 10.1158/0008-5472.CAN-05-0201. [DOI] [PubMed] [Google Scholar]
- 39.Kumatori A, Tanaka K, Inamura N, Sone S, Ogura T, Matsumoto T, et al. Abnormally high expression of proteasomes in human leukemic cells. Proc Natl Acad Sci U S A. 1990;87(18):7071–7075. doi: 10.1073/pnas.87.18.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arlt A, Bauer I, Schafmayer C, Tepel J, Muerkoster SS, Brosch M, et al. Increased proteasome subunit protein expression and proteasome activity in colon cancer relate to an enhanced activation of nuclear factor E2-related factor 2 (Nrf2) Oncogene. 2009;28(45):3983–3996. doi: 10.1038/onc.2009.264. [DOI] [PubMed] [Google Scholar]
- 41.Adams J. The proteasome: structure, function, and role in the cell. Cancer Treatment Reviews. 2003;29:3–9. doi: 10.1016/s0305-7372(03)00081-1. doi: http://dx.doi.org/10.1016/S0305-7372(03)00081-1. [DOI] [PubMed] [Google Scholar]
- 42.Delic J, Masdehors P, Omura S, Cosset JM, Dumont J, Binet JL, et al. The proteasome inhibitor lactacystin induces apoptosis and sensitizes chemo- and radioresistant human chronic lymphocytic leukaemia lymphocytes to TNF-alpha-initiated apoptosis. British Journal of Cancer. 1998;77(7):1103–1107. doi: 10.1038/bjc.1998.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LeBlanc R, Catley LP, Hideshima T, Lentzsch S, Mitsiades CS, Mitsiades N, et al. Proteasome inhibitor PS-341 inhibits human myeloma cell growth in vivo and prolongs survival in a murine model. Cancer Res. 2002;62(17):4996–5000. [PubMed] [Google Scholar]
- 44.An B, Goldfarb RH, Siman R, Dou QP. Novel dipeptidyl proteasome inhibitors overcome Bcl-2 protective function and selectively accumulate the cyclin-dependent kinase inhibitor p27 and induce apoptosis in transformed, but not normal, human fibroblasts. Cell Death Differ. 1998 Dec;5(12):1062–75. doi: 10.1038/sj.cdd.4400436. [DOI] [PubMed] [Google Scholar]
- 45.Lipchick BC, Fink EE, Nikiforov MA. Oxidative Stress and Proteasome Inhibitors in Multiple Myeloma. Pharmacological research. 2016;105:210–215. doi: 10.1016/j.phrs.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Yen J, Kaiser P, Huang L. Regulation of the 26S Proteasome Complex During Oxidative Stress. Science signaling. 2010;3(151):ra88–ra88. doi: 10.1126/scisignal.2001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aiken CT, Kaake RM, Wang X, Huang L. Oxidative Stress-Mediated Regulation of Proteasome Complexes. Molecular & Cellular Proteomics : MCP. 2011;10(5) doi: 10.1074/mcp.M110.006924. R110.006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crawford LJA, Walker B, Ovaa H, Chauhan D, Anderson KC, Morris TCM, et al. Comparative Selectivity and Specificity of the Proteasome Inhibitors BzLLLCOCHO, PS-341, and MG-132. [10.1158/0008-5472.CAN-06-0605] Cancer research. 2006;66(12):6379. doi: 10.1158/0008-5472.CAN-06-0605. [DOI] [PubMed] [Google Scholar]
- 49.Kazi A, Lawrence H, Guida WC, McLaughlin ML, Springett GM, Berndt N, et al. Discovery of a novel proteasome inhibitor selective for cancer cells over non-transformed cells. Cell Cycle. 2009;8(12):1940–1951. doi: 10.4161/cc.8.12.8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding W-X, Ni H-M, Gao W, Chen X, Kang JH, Stolz DB, et al. Oncogenic transformation confers a selective susceptibility to the combined suppression of the proteasome and autophagy. Molecular cancer therapeutics. 2009;8(7):2036–2045. doi: 10.1158/1535-7163.MCT-08-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Y, Wang K, Zhen S, Wang R, Luo W. Carfilzomib induces G2/M cell cycle arrest in human endometrial cancer cells via upregulation of p21Waf1/Cip1 and p27Kip1. Taiwan J Obstet Gynecol. 2016;55(6):847–851. doi: 10.1016/j.tjog.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 52.Huang H, Liu N, Liao Y, Liu N, Cai J, Xia X, et al. Platinum-containing compound platinum pyrithione suppresses ovarian tumor proliferation through proteasome inhibition. Journal of Experimental & Clinical Cancer Research : CR. 2017;36:79. doi: 10.1186/s13046-017-0547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drexler HCA. Activation of the cell death program by inhibition of proteasome function. Proc Natl Acad Sci U S A. 1997;94(3):855–860. doi: 10.1073/pnas.94.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pandit B, Gartel AL. Proteasome Inhibitors Induce p53-Independent Apoptosis in Human Cancer Cells. The American Journal of Pathology. 2011;178(1):355–360. doi: 10.1016/j.ajpath.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nam S, Smith DM, Dou QP. Tannic Acid Potently Inhibits Tumor Cell Proteasome Activity, Increases p27 and Bax Expression, and Induces G<sub>1</sub> Arrest and Apoptosis. Cancer Epidemiology Biomarkers &amp; Prevention. 2001;10(10):1083. [PubMed] [Google Scholar]
- 56.Mortenson MM, Schlieman MG, Virudachalam S, Lara PN, Gandara DG, Davies AM, et al. Reduction in BCL-2 levels by 26S proteasome inhibition with bortezomib is associated with induction of apoptosis in small cell lung cancer. Lung Cancer. 2005;49(2):163–170. doi: 10.1016/j.lungcan.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 57.Adams J. Potential for proteasome inhibition in the treatment of cancer. Drug Discov Today. 2003;8(7):307–315. doi: 10.1016/s1359-6446(03)02647-3. [DOI] [PubMed] [Google Scholar]
- 58.Liu Y, Huang X, He X, Zhou Y, Jiang X, Chen-Kiang S, et al. A novel effect of thalidomide and its analogs: suppression of cereblon ubiquitination enhances ubiquitin ligase function. Faseb j. 2015;29(12):4829–4839. doi: 10.1096/fj.15-274050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lopez-Girona A, Mendy D, Ito T, Miller K, Gandhi AK, Kang J, et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012;26(11):2326–2335. doi: 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kubiczkova L, Pour L, Sedlarikova L, Hajek R, Sevcikova S. Proteasome inhibitors - molecular basis and current perspectives in multiple myeloma. Journal of Cellular and Molecular Medicine. 2014;18(6):947–961. doi: 10.1111/jcmm.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buac D, Shen M, Schmitt S, Kona FR, Deshmukh R, Zhang Z, et al. From Bortezomib to other Inhibitors of the Proteasome and Beyond. Current pharmaceutical design. 2013;19(22):4025–4038. doi: 10.2174/1381612811319220012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moreau P, Pylypenko H, Grosicki S, Karamanesht I, Leleu X, Grishunina M, et al. Subcutaneous versus intravenous administration of bortezomib in patients with relapsed multiple myeloma: a randomised, phase 3, non-inferiority study. The Lancet Oncology. 2011;12(5):431–440. doi: 10.1016/S1470-2045(11)70081-X. doi: http://dx.doi.org/10.1016/S1470-2045(11)70081-X. [DOI] [PubMed] [Google Scholar]
- 63.Lü S, Wang J. The resistance mechanisms of proteasome inhibitor bortezomib. Biomarker Research. 2013;1:13–13. doi: 10.1186/2050-7771-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steiner RE, Manasanch EE. Carfilzomib boosted combination therapy for relapsed multiple myeloma. Onco Targets Ther. 2017;10:895–907. doi: 10.2147/ott.s102756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sonneveld P, Broijl A. Treatment of relapsed and refractory multiple myeloma. Haematologica. 2016;101(4):396–406. doi: 10.3324/haematol.2015.129189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Riz I, Hawley TS, Hawley RG. KLF4-SQSTM1/p62-associated prosurvival autophagy contributes to carfilzomib resistance in multiple myeloma models. Oncotarget. 2015;6(17):14814–14831. doi: 10.18632/oncotarget.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang CS, Landau JM. Effects of tea consumption on nutrition and health. J Nutr. 2000;130(10):2409–2412. doi: 10.1093/jn/130.10.2409. [DOI] [PubMed] [Google Scholar]
- 68.Nam S, Smith DM, Dou QP. Ester bond-containing tea polyphenols potently inhibit proteasome activity in vitro and in vivo. J Biol Chem. 2001;276(16):13322–13330. doi: 10.1074/jbc.M004209200. [DOI] [PubMed] [Google Scholar]
- 69.Lachman HM, Papolos DF, Saito T, Yu YM, Szumlanski CL, Weinshilboum RM. Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics. 1996;6(3):243–250. doi: 10.1097/00008571-199606000-00007. [DOI] [PubMed] [Google Scholar]
- 70.Weinshilboum RM, Otterness DM, Szumlanski CL. Methylation pharmacogenetics: catechol O-methyltransferase, thiopurine methyltransferase, and histamine N-methyltransferase. Annu Rev Pharmacol Toxicol. 1999;39:19–52. doi: 10.1146/annurev.pharmtox.39.1.19. [DOI] [PubMed] [Google Scholar]
- 71.Landis-Piwowar K, Chen D, Chan TH, Dou QP. Inhibition of catechol-O-methyltransferase activity in human breast cancer cells enhances the biological effect of the green tea polyphenol (−)-EGCG. Oncol Rep. 2010;24(2):563–569. doi: 10.3892/or_00000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huo C, Yang H, Cindy Cui Q, Dou QP, Chan TH. Proteasome Inhibition in Human Breast Cancer Cells with High Catechol-O-methyltransferase Activity by Green tea polyphenol EGCG analogs. Bioorganic & medicinal chemistry. 2010;18(3):1252. doi: 10.1016/j.bmc.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanwar J, Mohammad I, Yang H, Huo C, Chan TH, Dou QP. Computational modeling of the potential interactions of the proteasome β5 subunit and catechol-O-methyltransferase-resistant EGCG analogs. International Journal of Molecular Medicine. 2010;26(2):209–215. doi: 10.3892/ijmm_00000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barrett JR. The Science of Soy: What Do We Really Know? Environmental Health Perspectives. 2006;114(6):A352–A358. doi: 10.1289/ehp.114-a352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kazi A, Daniel KG, Smith DM, Kumar NB, Dou QP. Inhibition of the proteasome activity, a novel mechanism associated with the tumor cell apoptosis-inducing ability of genistein. Biochem Pharmacol. 2003;66(6):965–976. doi: 10.1016/s0006-2952(03)00414-3. [DOI] [PubMed] [Google Scholar]
- 76.Shim SH. 20S proteasome inhibitory activity of flavonoids isolated from Spatholobus suberectus. Phytother Res. 2011;25(4):615–618. doi: 10.1002/ptr.3342. [DOI] [PubMed] [Google Scholar]
- 77.Egert S, Rimbach G. Which sources of flavonoids: complex diets or dietary supplements? Adv Nutr. 2011;2(1):8–14. doi: 10.3945/an.110.000026.10.3945/an.110.000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen D, Daniel KG, Chen MS, Kuhn DJ, Landis-Piwowar KR, Dou QP. Dietary flavonoids as proteasome inhibitors and apoptosis inducers in human leukemia cells. Biochem Pharmacol. 2005;69(10):1421–1432. doi: 10.1016/j.bcp.2005.02.022. doi: http://dx.doi.org/10.1016/j.bcp.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 79.Srivastava JK, Shankar E, Gupta S. Chamomile: A herbal medicine of the past with bright future. Molecular medicine reports. 2010;3(6):895–901. doi: 10.3892/mmr.2010.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen D, Landis-Piwowar KR, Chen MS, Dou QP. Inhibition of proteasome activity by the dietary flavonoid apigenin is associated with growth inhibition in cultured breast cancer cells and xenografts. Breast Cancer Res. 2007;9(6):R80. doi: 10.1186/bcr1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yuan R, Lin Y. Traditional Chinese medicine. Pharmacology & Therapeutics. 2000;86(2):191–198. doi: 10.1016/s0163-7258(00)00039-5. doi: http://dx.doi.org/10.1016/S0163-7258(00)00039-5. [DOI] [PubMed] [Google Scholar]
- 82.Ergil KV, Kramer EJ, Ng AT. Chinese herbal medicines. Western Journal of Medicine. 2002;176(4):275–279. [PMC free article] [PubMed] [Google Scholar]
- 83.Dai Y, DeSano J, Tang W, Meng X, Meng Y, Burstein E, et al. Natural Proteasome Inhibitor Celastrol Suppresses Androgen-Independent Prostate Cancer Progression by Modulating Apoptotic Proteins and NF-kappaB. PLOS ONE. 2010;5(12):e14153. doi: 10.1371/journal.pone.0014153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pang X, Yi Z, Zhang J, Lu B, Sung B, Qu W, et al. Celastrol Suppresses Angiogenesis-Mediated Tumor Growth through Inhibition of AKT/Mammalian Target of Rapamycin Pathway. Cancer research. 2010;70(5):1951–1959. doi: 10.1158/0008-5472.CAN-09-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Raja SM, Clubb RJ, Ortega-Cava C, Williams SH, Bailey TA, Duan L, et al. Anticancer activity of celastrol in combination with ErbB2-targeted therapeutics for treatment of ErbB2-overexpressing breast cancers. Cancer Biology & Therapy. 2011;11(2):263–276. doi: 10.4161/cbt.11.2.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang H, Chen D, Cui QC, Yuan X, Dou QP. Celastrol, a triterpene extracted from the Chinese "Thunder of God Vine," is a potent proteasome inhibitor and suppresses human prostate cancer growth in nude mice. Cancer Res. 2006;66(9):4758–4765. doi: 10.1158/0008-5472.can-05-4529. [DOI] [PubMed] [Google Scholar]
- 87.Mahajan K, Malla P, Lawrence HR, Chen Z, Kumar-Sinha C, Malik R, et al. ACK1/TNK2 Regulates Histone H4 Tyr88-phosphorylation and AR Gene Expression in Castration-Resistant Prostate Cancer. Cancer Cell. 2017;31(6):790–803. e798. doi: 10.1016/j.ccell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen M, Rose AE, Doudican N, Osman I, Orlow SJ. Celastrol synergistically enhances temozolomide cytotoxicity in melanoma cells. Mol Cancer Res. 2009;7(12):1946–1953. doi: 10.1158/1541-7786.mcr-09-0243. [DOI] [PubMed] [Google Scholar]
- 89.Yang H, Landis-Piwowar KR, Lu D, Yuan P, Li L, Reddy GP, et al. Pristimerin induces apoptosis by targeting the proteasome in prostate cancer cells. J Cell Biochem. 2008;103(1):234–244. doi: 10.1002/jcb.21399. [DOI] [PubMed] [Google Scholar]
- 90.Lu L, Kanwar J, Schmitt S, Cui QC, Zhang C, Zhao C, et al. Inhibition of tumor cellular proteasome activity by triptolide extracted from the Chinese medicinal plant ‘thunder god vine’. Anticancer Res. 2011;31(1):1–10. [PMC free article] [PubMed] [Google Scholar]
- 91.Yang H, Zhou P, Huang H, Chen D, Ma N, Cui QC, et al. Shikonin exerts antitumor activity via proteasome inhibition and cell death induction in vitro and in vivo. Int J Cancer. 2009;124(10):2450–2459. doi: 10.1002/ijc.24195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mirjalili MH, Moyano E, Bonfill M, Cusido RM, Palazon J. Steroidal lactones from Withania somnifera, an ancient plant for novel medicine. Molecules. 2009;14(7):2373–2393. doi: 10.3390/molecules14072373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang H, Shi G, Dou QP. The tumor proteasome is a primary target for the natural anticancer compound Withaferin A isolated from "Indian winter cherry". Mol Pharmacol. 2007;71(2):426–437. doi: 10.1124/mol.106.030015. [DOI] [PubMed] [Google Scholar]
- 94.Hatcher H, Planalp R, Cho J, Torti FM, Torti SV. Curcumin: From ancient medicine to current clinical trials. Cellular and molecular life sciences : CMLS. 2008;65(11):1631–1652. doi: 10.1007/s00018-008-7452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Milacic V, Banerjee S, Landis-Piwowar KR, Sarkar FH, Majumdar APN, Dou QP. Curcumin inhibits the proteasome activity in human colon cancer cells in vitro and in vivo. Cancer research. 2008;68(18):7283–7292. doi: 10.1158/0008-5472.CAN-07-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wan SB, Yang H, Zhou Z, Cui QC, Chen D, Kanwar J, et al. Evaluation of curcumin acetates and amino acid conjugates as proteasome inhibitors. Int J Mol Med. 2010;26(4):447–455. doi: 10.3892/ijmm_00000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yue X, Zuo Y, Ke H, Luo J, Lou L, Qin W, et al. Identification of 4-arylidene curcumin analogues as novel proteasome inhibitors for potential anticancer agents targeting 19S regulatory particle associated deubiquitinase. Biochem Pharmacol. 2017;137:29–50. doi: 10.1016/j.bcp.2017.04.032. [DOI] [PubMed] [Google Scholar]
- 98.Henkel T, Machleidt T, Alkalay I, Kronke M, Ben-Neriah Y, Baeuerle PA. Rapid proteolysis of I[kappa]B-[alpha] is necessary for activation of transcription factor NF-[kappa]B. [10.1038/365182a0] Nature. 1993;365(6442):182–185. doi: 10.1038/365182a0. [DOI] [PubMed] [Google Scholar]
- 99.PDR for Herbal Medicines. Second. Montvale, NJ: Medical Economics Company; 2000. [Google Scholar]
- 100.Zhou ZTWJ. Phase I human tolerability trial of gambogic acid. Chin J New Drugs. 2007;16(1):679–682. [Google Scholar]
- 101.Li X, Liu S, Huang H, Liu N, Zhao C, Liao S, et al. Gambogic acid is a tissue-specific proteasome inhibitor in vitro and in vivo. Cell Rep. 2013;3(1):211–222. doi: 10.1016/j.celrep.2012.11.023. [DOI] [PubMed] [Google Scholar]
- 102.Shi X, Chen X, Li X, Lan X, Zhao C, Liu S, et al. Gambogic acid induces apoptosis in imatinib-resistant chronic myeloid leukemia cells via inducing proteasome inhibition and caspase-dependent Bcr-Abl downregulation. Clin Cancer Res. 2014;20(1):151–163. doi: 10.1158/1078-0432.ccr-13-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shi X, Lan X, Chen X, Zhao C, Li X, Liu S, et al. Gambogic acid induces apoptosis in diffuse large B-cell lymphoma cells via inducing proteasome inhibition. Scientific Reports. 2015;5:9694. doi: 10.1038/srep09694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Barry NPE, Sadler PJ. Exploration of the medical periodic table: towards new targets. [10.1039/C3CC41143E] Chemical Communications. 2013;49(45):5106–5131. doi: 10.1039/C3CC41143E. [DOI] [PubMed] [Google Scholar]
- 105.Dilruba S, Kalayda GV. Platinum-based drugs: past, present and future. [journal article] Cancer Chemotherapy and Pharmacology. 2016;77(6):1103–1124. doi: 10.1007/s00280-016-2976-z. [DOI] [PubMed] [Google Scholar]
- 106.Tiekink ERT. Anti-cancer potential of gold complexes. [journal article] Inflammopharmacology. 2008;16(3):138–142. doi: 10.1007/s10787-007-0018-5. [DOI] [PubMed] [Google Scholar]
- 107.Nardon C, Schmitt SM, Yang H, Zuo J, Fregona D, Dou QP. Gold(III)-Dithiocarbamato Peptidomimetics in the Forefront of the Targeted Anticancer Therapy: Preclinical Studies against Human Breast Neoplasia. PLOS ONE. 2014;9(1):e84248. doi: 10.1371/journal.pone.0084248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fiskus W, Saba N, Shen M, Ghias M, Liu J, Gupta SD, et al. Auranofin induces lethal oxidative and endoplasmic reticulum stress and exerts potent preclinical activity against chronic lymphocytic leukemia. Cancer Res. 2014;74(9):2520–2532. doi: 10.1158/0008-5472.can-13-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Phase I and II Study of Auranofin in Chronic Lymphocytic Leukemia (CLL) https://clinicaltrials.gov/ct2/show/NCT01419691.
- 110.Madeira JM, Gibson DL, Kean WF, Klegeris A. The biological activity of auranofin: implications for novel treatment of diseases. Inflammopharmacology. 2012;20(6):297–306. doi: 10.1007/s10787-012-0149-1. [DOI] [PubMed] [Google Scholar]
- 111.Liu N, Li X, Huang H, Zhao C, Liao S, Yang C, et al. Clinically used antirheumatic agent auranofin is a proteasomal deubiquitinase inhibitor and inhibits tumor growth. Oncotarget. 2014;5(14):5453–5471. doi: 10.18632/oncotarget.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen X, Shi X, Zhao C, Li X, Lan X, Liu S, et al. Anti-rheumatic agent auranofin induced apoptosis in chronic myeloid leukemia cells resistant to imatinib through both Bcr/Abl-dependent and -independent mechanisms. Oncotarget. 2014;5(19):9118–9132. doi: 10.18632/oncotarget.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Varela-Ramirez A, Costanzo M, Carrasco YP, Pannell KH, Aguilera RJ. Cytotoxic effects of two organotin compounds and their mode of inflicting cell death on four mammalian cancer cells. Cell biology and toxicology. 2011;27(3):159–168. doi: 10.1007/s10565-010-9178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pellerito L, Nagy L. Organotin(IV)n+ complexes formed with biologically active ligands: equilibrium and structural studies, and some biological aspects. Coordination Chemistry Reviews. 2002;224(1):111–150. doi: http://dx.doi.org/10.1016/S0010-8545(01)00399-X. [Google Scholar]
- 115.Nath M, Yadav R, Eng G, Musingarimi P. Characteristic spectral studies and in vitro Antimicrobial and in vivo multi-infection antifungal activities in mice of new organotin(IV) derivatives of heterocyclic amino acids. Applied Organometallic Chemistry. 1999;13(1):29–37. doi: 10.1002/(SICI)1099-0739(199901)13:1<29::AID-AOC809>3.0.CO;2-D. [DOI] [Google Scholar]
- 116.Shi G, Chen D, Zhai G, Chen MS, Cui QC, Zhou Q, et al. The Proteasome Is a Molecular Target of Environmental Toxic Organotins. Environmental Health Perspectives. 2009;117(3):379–386. doi: 10.1289/ehp.11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gupte A, Mumper RJ. Elevated copper and oxidative stress in cancer cells as a target for cancer treatment. Cancer Treatment Reviews. 2009;35(1):32–46. doi: 10.1016/j.ctrv.2008.07.004. doi: http://dx.doi.org/10.1016/j.ctrv.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 118.Kuo HW, Chen SF, Wu CC, Chen DR, Lee JH. Serum and tissue trace elements in patients with breast cancer in Taiwan. Biol Trace Elem Res. 2002;89(1):1–11. doi: 10.1385/bter:89:1:1. [DOI] [PubMed] [Google Scholar]
- 119.Zuo XL, Chen JM, Zhou X, Li XZ, Mei GY. Levels of selenium, zinc, copper, and antioxidant enzyme activity in patients with leukemia. Biol Trace Elem Res. 2006;114(1–3):41–53. doi: 10.1385/bter:114:1:41. [DOI] [PubMed] [Google Scholar]
- 120.Brem SS, Zagzag D, Tsanaclis AM, Gately S, Elkouby MP, Brien SE. Inhibition of angiogenesis and tumor growth in the brain. Suppression of endothelial cell turnover by penicillamine and the depletion of copper, an angiogenic cofactor. Am J Pathol. 1990;137(5):1121–1142. [PMC free article] [PubMed] [Google Scholar]
- 121.Daniel KG, Gupta P, Harbach RH, Guida WC, Dou QP. Organic copper complexes as a new class of proteasome inhibitors and apoptosis inducers in human cancer cells. Biochem Pharmacol. 2004;67(6):1139–1151. doi: 10.1016/j.bcp.2003.10.031. doi: http://dx.doi.org/10.1016/j.bcp.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 122.Liu N, Liu C, Li X, Liao S, Song W, Yang C, et al. A novel proteasome inhibitor suppresses tumor growth via targeting both 19S proteasome deubiquitinases and 20S proteolytic peptidases. Scientific Reports. 2014;4:5240. doi: 10.1038/srep05240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hindo SS, Frezza M, Tomco D, Heeg MJ, Hryhorczuk L, McGarvey BR, et al. Metals in Anticancer Therapy: Copper(II) Complexes as Inhibitors of the 20S Proteasome. European journal of medicinal chemistry. 2009;44(11):4353–4361. doi: 10.1016/j.ejmech.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Frezza M, Hindo SS, Tomco D, Allard M, Cui QC, Heeg MJ, et al. Comparative Activities of Nickel(II) and Zinc(II) Complexes of Asymmetric [NN’O] Ligands as 26S Proteasome Inhibitors. Inorganic chemistry. 2009;48(13):5928–5937. doi: 10.1021/ic900276g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Verani CN. Metal complexes as inhibitors of the 26S proteasome in tumor cells. Journal of Inorganic Biochemistry. 2012;106(1):59–67. doi: 10.1016/j.jinorgbio.2011.09.003. doi: http://dx.doi.org/10.1016/j.jinorgbio.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 126.Zhang Z, Bi C, Buac D, Fan Y, Zhang X, Zuo J, et al. Organic cadmium complexes as proteasome inhibitors and apoptosis inducers in human breast cancer cells. Journal of Inorganic Biochemistry. 2013;0:1–10. doi: 10.1016/j.jinorgbio.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kelland L. The resurgence of platinum-based cancer chemotherapy. [10.1038/nrc2167] Nat Rev Cancer. 2007;7(8):573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 128.Rizk SL, Sky-Peck HH. Comparison between concentrations of trace elements in normal and neoplastic human breast tissue. Cancer Res. 1984;44(11):5390–5394. [PubMed] [Google Scholar]
- 129.Huang YL, Sheu JY, Lin TH. Association between oxidative stress and changes of trace elements in patients with breast cancer. Clin Biochem. 1999;32(2):131–136. doi: 10.1016/s0009-9120(98)00096-4. [DOI] [PubMed] [Google Scholar]
- 130.Habib FK, Dembinski TC, Stitch SR. The zinc and copper content of blood leucocytes and plasma from patients with benign and malignant prostates. Clin Chim Acta. 1980;104(3):329–335. doi: 10.1016/0009-8981(80)90390-3. [DOI] [PubMed] [Google Scholar]
- 131.Nayak SB, Bhat VR, Upadhyay D, Udupa SL. Copper and ceruloplasmin status in serum of prostate and colon cancer patients. Indian J Physiol Pharmacol. 2003;47(1):108–110. [PubMed] [Google Scholar]
- 132.Diez M, Arroyo M, Cerdan FJ, Munoz M, Martin MA, Balibrea JL. Serum and tissue trace metal levels in lung cancer. Oncology. 1989;46(4):230–234. doi: 10.1159/000226722. [DOI] [PubMed] [Google Scholar]
- 133.Turecky L, Kalina P, Uhlikova E, Namerova S, Krizko J. Serum ceruloplasmin and copper levels in patients with primary brain tumors. Klin Wochenschr. 1984;62(4):187–189. doi: 10.1007/BF01731643. [DOI] [PubMed] [Google Scholar]
- 134.Daniel KG, Chen D, Orlu S, Cui QC, Miller FR, Dou QP. Clioquinol and pyrrolidine dithiocarbamate complex with copper to form proteasome inhibitors and apoptosis inducers in human breast cancer cells. Breast Cancer Research. 2005;7(6):R897–R908. doi: 10.1186/bcr1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhai S, Yang L, Cui QC, Sun Y, Dou QP, Yan B. Tumor cellular proteasome inhibition and growth suppression by 8-hydroxyquinoline and clioquinol requires their capabilities to bind copper and transport copper into cells. J Biol Inorg Chem. 2010;15(2):259–269. doi: 10.1007/s00775-009-0594-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.De Sousa A. The Pharmacotherapy of Alcohol Dependence: A State of the Art Review. Mens Sana Monographs. 2010;8(1):69–82. doi: 10.4103/0973-1229.58820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen D, Cui QC, Yang H, Dou QP. Disulfiram, a clinically used anti-alcoholism drug and copper-binding agent, induces apoptotic cell death in breast cancer cultures and xenografts via inhibition of the proteasome activity. Cancer Res. 2006;66(21):10425–10433. doi: 10.1158/0008-5472.can-06-2126. [DOI] [PubMed] [Google Scholar]
- 138.Pang H, Chen D, Cui QC, Dou QP. Sodium diethyldithiocarbamate, an AIDS progression inhibitor and a copper-binding compound, has proteasome-inhibitory and apoptosis-inducing activities in cancer cells. Int J Mol Med. 2007;19(5):809–816. [PubMed] [Google Scholar]
- 139.Milacic V, Chen D, Giovagnini L, Diez A, Fregona D, Dou QP. Pyrrolidine dithiocarbamate-zinc(II) and -copper(II) complexes induce apoptosis in tumor cells by inhibiting the proteasomal activity. Toxicology and Applied Pharmacology. 2008;231(1):24–33. doi: 10.1016/j.taap.2008.03.009. doi: http://dx.doi.org/10.1016/j.taap.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cvek B, Milacic V, Taraba J, Dou QP. Ni(II), Cu(II), and Zn(II) diethyldithiocarbamate complexes show various activities against the proteasome in breast cancer cells. J Med Chem. 2008;51(20):6256–6258. doi: 10.1021/jm8007807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nardon C, Chiara F, Brustolin L, Gambalunga A, Ciscato F, Rasola A, et al. Gold(III)-pyrrolidinedithiocarbamato Derivatives as Antineoplastic Agents. ChemistryOpen. 2015;4(2):183–191. doi: 10.1002/open.201402091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu N, Liu C, Li X, Liao S, Song W, Yang C, et al. A novel proteasome inhibitor suppresses tumor growth via targeting both 19S proteasome deubiquitinases and 20S proteolytic peptidases. Sci Rep. 2014;4:5240. doi: 10.1038/srep05240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao C, Chen X, Zang D, Lan X, Liao S, Yang C, et al. A novel nickel complex works as a proteasomal deubiquitinase inhibitor for cancer therapy. Oncogene. 2016;35(45):5916–5927. doi: 10.1038/onc.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhao C, Chen X, Yang C, Zang D, Lan X, Liao S, et al. Repurposing an antidandruff agent to treating cancer: zinc pyrithione inhibits tumor growth via targeting proteasome-associated deubiquitinases. Oncotarget. 2017;8(8):13942–13956. doi: 10.18632/oncotarget.14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhao C, Chen X, Zang D, Lan X, Liao S, Yang C, et al. Platinum-containing compound platinum pyrithione is stronger and safer than cisplatin in cancer therapy. Biochem Pharmacol. 2016;116:22–38. doi: 10.1016/j.bcp.2016.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tomasello MF, Nardon C, Lanza V, Di Natale G, Pettenuzzo N, Salmaso S, et al. New comprehensive studies of a gold(III) Dithiocarbamate complex with proven anticancer properties: Aqueous dissolution with cyclodextrins, pharmacokinetics and upstream inhibition of the ubiquitin-proteasome pathway. Eur J Med Chem. 2017;138:115–127. doi: 10.1016/j.ejmech.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 147.An H, Statsyuk AV. An inhibitor of ubiquitin conjugation and aggresome formation. Chem Sci. 2015;6(9):5235–5245. doi: 10.1039/c5sc01351h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yang Y, Kitagaki J, Dai RM, Tsai YC, Lorick KL, Ludwig RL, et al. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007;67(19):9472–9481. doi: 10.1158/0008-5472.can-07-0568. [DOI] [PubMed] [Google Scholar]
- 149.Xu GW, Ali M, Wood TE, Wong D, Maclean N, Wang X, et al. The ubiquitin-activating enzyme E1 as a therapeutic target for the treatment of leukemia and multiple myeloma. Blood. 2010;115(11):2251–2259. doi: 10.1182/blood-2009-07-231191. [DOI] [PMC free article] [PubMed] [Google Scholar]