Abstract
Systemic acquired resistance (SAR) is an inducible defense response in plants that provides enhanced resistance against a variety of pathogens. In this regard, SAR marker gene PR1 (pathogenesis-related gene 1) was isolated from Brassica juncea and was named as BjPR1. The amino acid sequence of BjPR1 protein showed 99, 92, and 78% similarity with known PR1 proteins of Brassica rapa, Brassica napus, and Arabidopsis thaliana, respectively. Quantitative real-time PCR (qRT-PCR) analysis showed increased expression of BjPR1 gene both in local (infected) and distal (non-infected) leaves of B. juncea after Alternaria brassicae infection, whereas mechanical wounding showed expression only in local (wounded) leaves but not in distal (unwounded) leaves. Moreover, BjPR1 gene was strongly induced by salicylic acid (SA), whereas no such induction was observed following jasmonic acid (JA) or abscisic acid (ABA) treatments. To further elucidate gene regulation pattern of BjPR1, 2 kb promoter region of BjPR1 was isolated and subjected to in silico analysis which identified many potential cis-regulatory elements associated with plant defense as well as signaling pathways. The transient GUS expression analysis showed strong expression of GUS gene driven by BjPR1 promoter after SA treatment, while as ABA and JA downregulates GUS gene expression compared to control. In addition, BjPR1 promoter was significantly induced by wounding at local tissues. Hence, these results highlight the multiple role of BjPR1 gene in response to biotic and abiotic stresses. In addition, the present study also reported BjPR1 promoter as stress-specific inducible promoter that can be ideal candidate for controlling the expression of biotic stress response genes in transgenic plants.
Electronic supplementary material
The online version of this article (10.1007/s13205-017-1027-8) contains supplementary material, which is available to authorized users.
Keywords: Abscisic acid, Alternaria brassicae, Brassica juncea, Jasmonic acid, Pathogenesis-related proteins, Salicylic acid, Systemic acquired resistance
Introduction
Indian mustard (B. juncea L.) belonging to the family Brassicaeae is the second most important edible oilseed after groundnut in India. It contributes 28.6% in the total oilseeds production and sharing 27.8% in the India’s oilseed economy (Shekhawat et al. 2012). There are various factors including both biotic and abiotic stresses that lower the productivity of this crop, among which Alternaria blight caused by A. brassicae is a major constraint (Kolte 1985). Alternaria blight appears every year (endemic) and causes up to 36.88% loss of yield in mustard due Alternaria blight (Bal and Kumar 2014). In addition to direct losses, Alternaria blight also affects the quality of the seed by reducing size, causing seed discolouration, and reduced oil content (Kaushik et al. 1984). Improving Alternaria disease resistance through the conventional breeding in B. juncea has been challenged by complex nature of disease resistance, costly and less precise phenotyping, low heritability, environment sensitiveness, and limited availability of resistant germplasm. Second, present protective measures rely heavily on fungicides, generating adverse environmental consequences (Chen and Zhou 2009). The advancement in proteomics, genomics, and transcriptomics techniques has impressively accelerated the research in plant–pathogen interactions. In this regard, transgenic technology and molecular-assisted breeding will provide an alternative approach to develop disease-resistant varieties, which can overcome the problems related to the conventional breeding and fungicides. Furthermore, a better understanding of molecular mechanism of plant–pathogen interactions and identification of novel disease-resistant genes such as pathogen-related (PRs) genes will be an essential tool for crop improvement.
Pathogenesis-related (PR) proteins are of great interest for engineering plants not only to disease resistance but also for developing pest resistant varieties. Currently, PRs have been classified into 17 families, based on their amino acid sequence similarities, enzymatic activities and other biological properties. They have been numbered in sequence of discovery (Sels et al. 2008; Sinha et al. 2014). During host–pathogen interactions, PRs do not only accumulate locally in the infected leaf but also induced Systemically (Hamamouch et al. 2011). Among the classes of PR proteins, PR1 is one of the important pathogen-related proteins, which has been studied mostly in model plants (Arabidopsis and tobacco), but its molecular function remains unknown. A number of PR1 like proteins have been identified in many plant species including both mono and dicotyledonous plants (Mitsuhara et al. 2008; Li et al. 2011). PR1 group is found to be the most abundant group in PR gene families and is classified into two groups (acidic and basic proteins) based on the isoelectric point (Van Loon and Van Strien 1999). PR1 is universally known as a molecular indicator of induced plant immune system such as hypersensitivity response (HR) and systemic acquired resistance (SAR) (Jung and Hwang 2000; Jung et al. 2009). SAR is an inducible defense response in plants that provides enhanced resistance against broad range of pathogens. The previous reports have shown that transcript levels of PR1 gene increase significantly in plants exposed to biotic and abiotic stresses (Brederode et al. 1991; Mitsuhara et al. 2008), which suggests that they play an important role in combating these challenges. The PR1 gene induction following pathogen infection has been well documented in a number of crop species, viz., Paeonia suffruticosa, Nicotiana tobacum, Oryza sativa, etc. (Agrawal et al. 2001; Sujon et al. 2005; Yang et al. 2013). Overexpression of PR1 proteins in different crop systems has generally resulted in enhancing disease resistance against many pathogens (Alexander et al. 1993; Lawton et al. 1993; Niderman et al. 1995; Sarowar et al. 2005).
Plant immunity strongly relies on two important defense signaling regulatory pathways like JA and SA which act synergistically or antagonistically (Glazebrook 2005). Interestingly, exogenous treatment with defense hormonal stimulators salicylic acid (SA), jasmonate (JA), and ethylene (ET) has been reported earlier to regulate PR1 transcript accumulation which varies among plant species (Reymond and Farmer 1998; Kim and Hwang 2000; Zhang et al. 2010). JA- or an ethylene-dependent signaling pathway was found to induce the expression of basic PR1 genes strongly, whereas an SA-dependent pathway was found to increase the expression of acidic PR1 genes (Ward et al. 1991; Eyal et al. 1992; Niki et al. 1998).
One of the major challenges in plant genetic engineering is to find highly specific promoter which could drive the expression of target gene in transgenic crops (Hernandez-Garcia and Finer 2014). In general, constitutive promoters of both viral and plant origins have been commonly used to drive gene expression in many disease-resistant transgenic crops. These promoters cause a number of problems such as homology-dependent gene silencing, leading a fitness penalty in plant growth and development (Zheng et al. 2007). To solve this problem, spatially and temporally inducible promoters that are less exhaustive are needed to develop transgenic plants resistant to pathogens. Identification of ideal pathogen-inducible promoter mainly relies on the discovery of disease-resistant genes. The best feature of the pathogen-inducible promoter is the early and rapid activation in response to multiple phytopathogens. Till date, very few pathogen-inducible promoters have been isolated and characterized mainly from model plants. Therefore, it is very pertinent to isolate and characterize pathogen-inducible promoters for driving the expression of genes responsible for conferring disease resistance. Pathogen-inducible promoters usually possess many potential cis-regulatory elements based on their interaction with defense signaling molecules such as SA, JA, and ET or signals (Mazarei et al. 2008). Two important cis-acting elements, the GCC-like elements (Ohme-Tagaki et al. 2000) and the W-box (Eulgem et al. 2000) elements, have been well studied in pathogen-inducible promoters.
The aim of this study was to isolate and functionally characterize SAR marker gene BjPR1 and its promoter after Alternaria infection, wounding, SA and JA treatment in B. juncea. These results will provide novel insights into the Brassica-Alternaria pathosystem and their signaling cascades which are largely unknown.
Materials and methods
Plant material
Brassica juncea var. varuna and Nicotinia benthamiana plants were grown in a growth chamber at 22–24 °C under 12-h light and 12-h dark cycle. 40-day-old B. juncea plants were used for constructing a cDNA library and for endogenous gene expression assays. For transient assay, 1-month-old N. benthamiana plants were used.
Isolation and phylogenetic analysis of BjPR1
Brassica juncea cDNA library was constructed from total RNA of SA-treated leaf samples using BD SMART cDNA library construction kit following the manufacturer’s instructions (Clontech Inc., USA). The full-length sequence of BjPR1 gene was isolated from SA library as described by (Taweel et al. 2011) and sequenced. The bioinformatic tool GENSCAN (http://genes.mit.edu/GENSCAN.html accessed 17 June 2016) was used to predict the open reading frame of BjPR1 and its deduced amino acid sequence, respectively. Protein sequence similarity analysis of B. juncea PR1 protein was performed using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/blast), and for multiple sequence alignment, ClustalX was used. Phylogenetic relationships of B. juncea PR1 homologs were constructed using the neighbour-joining method with bootstrapping (1000 replicates) using MEGA 7.0 program (Kumar et al. 2016). The structural features of BjPR1 protein were analysed using the expert protein analysis system (http://www.expasy.org/). Conserved domain structure of BjPR1 protein was analysed by Pfam database (http://pfam.xfam.org/).
Culture of A. brassicae and inoculation
Alternaria brassicae stain was obtained from Indian Type Culture Collection (I.D. No. 81651) Division of Plant Pathology, IARI, New Delhi and cultured on radish dextrose agar at 22 °C for 20 days. The conidia of well-grown A. brassicae were suspended in sterile distilled water, filtered with two layers of muslin cloth, and diluted to 5 × 103 conidia/ml. Forty-five-day-old B. juncea plants were inoculated with 4–6 drops of spore suspension of A. brassicae (5 × 103 spores cm−3) on four different selected spots of the leaf surface and then incubated in a chamber at 25 °C, with 100% relative humidity. Control B. juncea plants were inoculated with 10 µl sterile distilled water. The leaf samples were collected from both local (infected) and distal (non-infected) leaves of B. juncea plants at 0-, 2-, 4-, 8-, 12-, 24-, 48-, 72-, and 96-h post-inoculation (hpi), stored at − 80 °C after being flash frozen in liquid nitrogen. Three different B. juncea plants were infected with A. brassicae on separate occasions to provide biological replicates for qRT-PCR analysis.
Wounding and hormonal treatments
Leaves of 40-day-old B. juncea plants were effectively wounded using sterile syringe needle and samples were harvested at different time intervals. For hormonal treatments, 40-day-old B. juncea plants were sprayed with 2-mM salicylic acid pH 7.0 (Chengguo et al. 2012), 100-μM jasmonic acid (Zhao and Chye 1999), and 50-µM ABA individually, kept separately in dark chamber to prevent cross talk signaling, evaporation, and light-induced degradation. Control plants were sprayed with sterile distilled water. Leaf samples for RNA isolation were harvested from control, SA, JA, and ABA-treated plants after 0-, 2-, 4-, 8-, 12-, 24-, 48-, and 72-h post-treatment.
RNA isolation and qRT-PCR
Total RNA was isolated from the control and treated leaf samples using the protocol of PureLink RNA Mini Kit (Ambion Life Technologies, USA). The purity and concentration of total RNA was determined by Nanodrop spectrophotometer (NanoDrop 2000c; Thermo Scientific, Wilmington, DE). First-strand cDNA was generated from 2-µg of DNase-treated total RNA by reverse transcriptase in 20-µl reaction volume containing oligo (dT) 18 primers, 10-mM dNTPS, and water following the manufacturer’s protocol (Fermentas, Canada). cDNA sample was 10 times diluted and kept at − 80 °C for further expression studies. qRT-PCR reaction mixture contains 2 µl of cDNA, 5 µl of SYBR green qRT-PCR master mix (Takara, Japan), and 0.5 µl (10 picomol) of PR1 forward and reverse primers (Table 1). The reactions were performed in triplicates and program of the qRT-PCR was; 95 °C for 5 min; followed by 40 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s. The relative levels of BjPR1 mRNA were evaluated against the A. thaliana housekeeping gene α-tubulin (GenBank accession no-NM_100360.4) amplified with specific primer pairs (Table 1). The relative expression levels of BjPR1 mRNA in all the treated samples were quantified by 2−∆∆CT method (Livak and Schmittgen 2001).
Table 1.
List of primers used in this study
| Gene | Sequence |
|---|---|
| PR1 | F-5′GAACACGTGCAATGGAGAATG3′ R- 5′CCATTGTTACACCTCGCTTTG3′ |
| Alpha tubulin | F-5′TGC TTT CGT TCA CTG GTA TGT3′ R-5′ CAG CAC CGA CCT CTT CAT AAT C3′ |
| AP1 | F-5′ GTAATACGACTCACTATAGGGC3′ R-5‘GTA ATA CGA CTC ACT ATA GGG C 3 |
| AP2-F | F-5′-ACTATAGGGCACGCGTGGT-3′ R-5′ACT ATA GGG CAC GCG TGG 3′ |
| GSP1 | 5′TATTTTTGTGTGTTCCCCGGCCGTAATGG3 |
| GSP2 | 5′CAAGAGCTCCCACAAGGGCAGCCAAAATTA3′ |
Isolation of BjPR1 promoter by genome walking
BjPR1 promoter was isolated from the B. juncea genome by PCR walking using Universal Genome Walker kit (Clontech, CA, USA). Briefly, high-quality genomic DNA was extracted from B. juncea using a DNeasy Plant Midi Kit (Qiagen, Valencia, CA). Purified DNA (2.5 μg) was digested at 37 °C with EcoRV, DraI, PvuII, and StuI restriction enzymes supplied with the GenomeWalker™ kit (Clontech, CA, USA). These restriction digestions generate blunt ends of the genomic DNA. Short adaptor DNA sequence provided with genome walker kit (Clontech laboratories Inc. http://www.clontech.com) was ligated to blunt end digested genomic DNA fragments, thus generating four Genome walker libraries (Table 1). These libraries were used as template for the isolation of promoters by two-step PCR (primary and secondary PCR) using adaptor-specific and gene-specific primers. The adaptor-specific primers provided in the kit and BjPR1 gene-specific primers for primary PCR and GSP2 for secondary PCR were designed within the 5′end of the BjPR1 sequence deposited in Genbank (accession no DQ359128) using primer 3.0 software (Table 1). The PCR purified product (BjPR1 promoter) approximately 2 kb was isolated from Stu1 library and cloned into pGEMT Easy vector for sequencing. To investigate the presence of cis-regulatory elements such as TATA box, CAAT box, and stress regulatory cis-acting element in BjPR1 promoter, sequence was analysed by PLACE (Higo et al. 1999) and PlantCARE (Lescot et al. 2002) databases of plant cis- regulatory DNA elements. The BjPR1 promoter sequence was submitted to the GenBank nucleotide sequence databases with accession number KC865598.
Vector construction and transient expression assays
The binary vector pORE-R2 (promoter less GUS reporter vector) was used in this study. The BjPR1 promoter was cloned in this binary vector at Pst1 and BamH1 site, respectively. The cloning of the BjPR1 promoter was confirmed by colony PCR and restriction analysis. The recombinant (BjPR1promoter-pORER2) GUS vector was further transformed into Agrobacterium tumefaciens strain (EHA105) by freeze thaw method and confirmed by colony PCR using promoter-specific primers. To investigate the BjPR1 promoter activity, transient assays were carried out in tobacco leaves. Agrobacterium strain EHA105 containing BjPR1-pORER2:GUS construct was grown in Luria–Bertani broth (LB) containing antibiotics (Rifampcin 25 µg mL−1and Kanamycin 50 µg mL−1) at 28 °C until the culture reached OD600 = 0.8. The culture was centrifuged at 7000 g for 10 min and resuspended in infiltration media containing (10-mM MES (pH 5.5), 10-mM MgCl2, and 100-µM acetosyringone) incubated at 28 °C for 3 h before agroinfiltration. The two young expanded leaves were infiltrated gently with the bacterial culture using 1-ml needleless syringe and kept in a growth chamber at 22 °C for 24 h. To examine the inducibility of PR1 promoter, 24 h of initial agroinfiltrated tobacco leaves were further infiltrated on the same spot with 2-mM SA, 100-µM MeJA, and 50-µM ABA samples were harvested after 24 h for GUS staining. For control, leaves were infiltrated with sterile distilled water. Leaf discs from both control- (negative and positive) and hormonal-treated samples were collected in small petridishes (Himedia), and immersed in GUS staining solution containing (0.1-M NaHPO4 pH 7.0, 0.5-mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.001-M EDTA pH 8.0, 20% methanol, 0.1% Triton X-100, and 0.5-mM X-gluc), and incubated at 37 °C for 16 h. After staining, samples were bleached by 75% ethanol 2–3 times and photographed. A. tumefaciens strain with promoter less GUS reporter vector was used as a negative control.
Results
Cloning and sequencing of BjPR1 gene
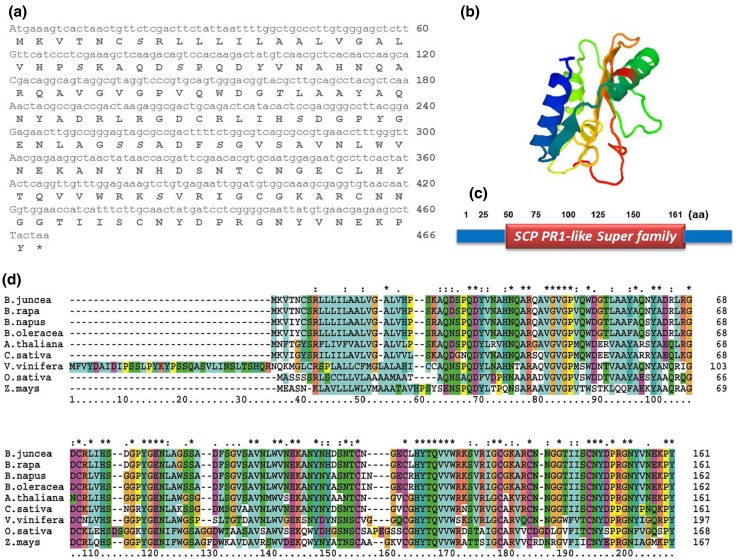
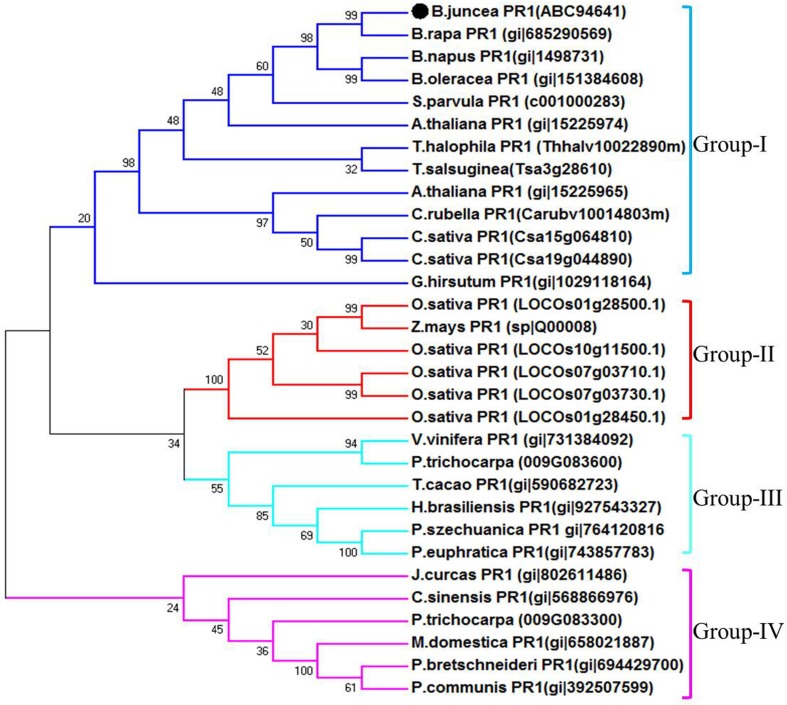
The full-length cDNA sequence of PR1 gene was isolated from SA-treated B. juncea library. The clone designated as BjPR1 (GenBank accession no. KM506762) was structurally and functionally characterised. Sequence analysis of BjPR1 revealed that it is comprised of 672 bp with an open reading frame of 486 nucleotides, encoding a protein of 161 amino acids with a calculated molecular mass of 17.53 kDa and a theoretical PI of 7.07. The nucleotide sequence of this gene also showed a 5′ untranslated region (5′ UTR) of 63 nucleotides and a 3′ UTR of 126 nucleotides. In silico subcellular localisation revealed that BjPR1 is expressed in Vacuole. The software NetPhos (http://www.cbs.dtu.dk/services/NetPhos) predicted that BjPR1 had six serines and one threonine as potential phosphorylation sites (Fig. 1a). The 3D structure of the BjPR1 protein is shown in (Fig. 1b).The predicted BjPR1 protein contained a conserved motif at residues 30–161 aa that belonged to the SCP-like super family (Fig. 1c). Its deduced amino acid sequence revealed highest similarity with PR1 proteins of its close relative B. rapa (99%) followed by A. thaliana (78%), Eutrema japonicum (78%), and Heve brassiliensis (66%), respectively (Fig. 1d). The phylogenetic relationships of BjPR1 with its homologs from both monocots and dicot plants were constructed through the neighbour-joining method using MEGA 7.1 program, which resulted four major clusters viz., I, II, III, and IV. The BjPR1 as well as PR1 genes of all Cruciferae members are grouped together in cluster I, and BjPR1 was nearest to PR1 of B. rapa followed by B. napus, B. oleracea, and S. parvula. However, BjPR1 was most diverged from PR1 of Gramineae family that are clustered in separate and distinct cluster IV (Fig. 2).
Fig. 1.
Structural analysis of BjPR1 nucleotide and amino acid sequences a Nucleotide and amino acid sequences of BjPR1 with putative phosphorylation sites like serine and tyrosine are shown in bold italics b 3D structure of BjPR1 protein c Conserved domain of the BjPR1 protein. The predicted BjPR1 protein contained a conserved motif at residues 30–161 aa that belonged to the SCP-PR1 like super family d Multiple sequence alignment of the BjPR1 protein sequence with other plant PR1 proteins. Comparison of deduced amino acid sequence of BjPR1 with other plant PR1 s from B. rapa, B.napus, B. oleracea, A. thaliana, C. sativa, O. sativa, and Z. mays. Conserved residues are shown with shaded colours
Fig. 2.
Phylogenetic relationship of BjPR1 with homologs of other plant species, constructed using the MEGA 7.0 program. Bootstrap values denote the divergence of each branch and the scale indicates branch length. BjPR1 is highlighted as black colour circular marker
BjPR1 expression in response to Alternaria infection
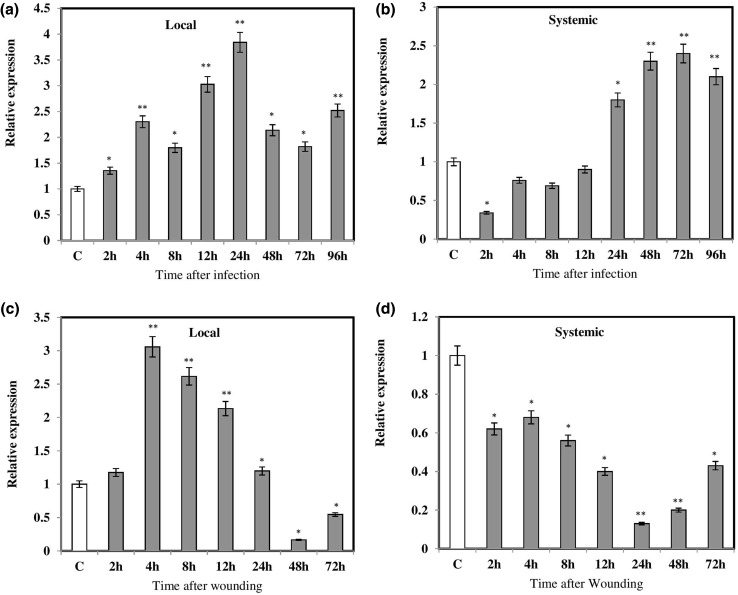
An accurate monitoring of disease progression in crop plants is very important to evaluate the role of pathogen-related genes. Therefore, we first studied the disease development in B. juncea during Alternaria infection. Pure culture of A. brassicae and its spore morphology are shown in (Fig. 3a‒b). After Alternaria infection, necrotic lesions appeared as grey circular areas at the site of inoculation on the infected leaves of B. juncea, while as no symptoms appeared on non-infected leaves (Fig. 3c‒d). These results showed the compatible interaction between Alternaria and B. juncea pathosystem. In the present study, we examine the transcriptional changes of BjPR1 gene in local (infected) and distal (non-infected) leaf sample of B. juncea in response to Alternaria infection. The qRT-PCR results showed that transcript levels of BjPR1 gene increase significantly in local infected tissue at 4 h (2.3 fold) and reached maximum at 24 h (3.8-fold) of post-inoculation as compared to control, but decreased sharply to a relatively low expression from 48 to 96 h (Fig. 4a). In comparison, the expression of BjPR1 gene in distal leaves (non-infected) was significantly lower at early stages but was detected after 48 hpi and reached to peak at 72 h (Fig. 4b). In general, the BjPR1 genes showed higher expression in local leaves (infected) than that of distal leaves (non-infected). These results, therefore, revealed that BjPR1 gene is induced both locally as well as systematically in B. juncea followed A. brassicae infection.
Fig. 3.
In vivo infection of B. juncea with A. brassiace a A. brassiace culture grown on root radish medium b Microscopic identification of A. brassicae fungus (Conidia under 100X microscope) c Uninfected B. juncea leaf as control d‒e B. juncea leaves after Alternaria infection
Fig. 4.
Local and systemic expression of BjPR1 gene after Alternaria infection and wounding: a expression of BjPR1 in local (infected) leaves at various time points; b expression of BjPR1 in distal (non-infected) leaves; c relative expression of BjPR1 in local (wounded) leaves; d relative expression of BjPR1 in distal (unwounded) leaves. SE for each bar is shown. The relative expression was calculated using ΔΔCt method. The asterisks indicate statistically significant differences between the control and treated B. juncea plants (*P < 0.05; **P < 0.01)
BjPR1 expression in response to wounding
Plants respond to wounding through induction of variety of genes both locally or systematically that contribute in healing of damaged tissues and further invasion of pathogens (Durrant et al. 2000). Therefore, in the present study, we examined the local and systemic expression of BjPR1 gene after wounding at early and late time intervals. Our results revealed that transcript levels of BjPR1 gene increases at 2 h and reached maximum at 4 h (threefold induction) followed by decline at later 8 h and 12 h in local tissues (Fig. 4c). In contrast, no systemic induction of BjPR1 was observed in leaves (unwound) when compared to control (Fig. 4d).
BjPR1 expression in response to hormonal treatments
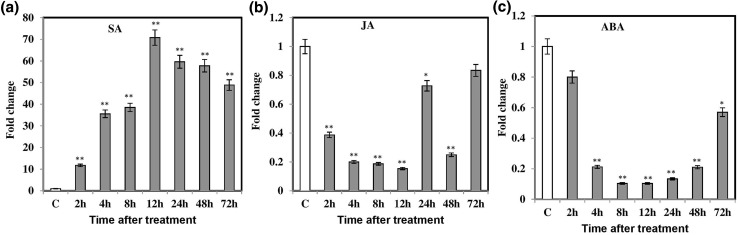
SA and JA are the plant hormones that are well-known modulators for plant defense system in plants. However, role of ABA in plant defense is not fully understood. To further examine the expression of BjPR1 gene in response to above hormones, B. juncea plants were treated with 2-mM SA, 100-µM JA, and 50-µM ABA, and are profiled over various time points. The transcript levels of BjPR1 gene were significantly increased at 2 h, reached the peak at 12 hpi rapidly after SA treatment, and then declined at later time points (Fig. 5a). In contrast, no significant transcript accumulation of BjPR1 was reported after JA treatment or ABA treatment at any time points (Fig. 5b‒c). SA-induced expression of BjPR1 further provides the evidence that it can be used as SA or SAR marker in B. juncea.
Fig. 5.
Transcriptional studies of BjPR1 under various hormonal stresses by qRT-PCR analysis. B. juncea plants were treated with 2-mM SA, 100-μM MeJA, and 50-μM ABA, respectively. Housekeeping gene alpha tubulin was used as internal control. All data are represented as means of three replicates (n = 3) ± SE. The asterisks indicate statistically significant differences between the control and hormone treated B. juncea plants (*P < 0.05; **P < 0.01)
Isolation and In silico analysis of BjPR1 promoter
The 2 kb upstream sequence of BjPR1 gene was isolated from B. juncea using genome walking approach and submitted into gene bank (accession no. KC865598.1). BjPR1 promoter sequence was scanned using PlantCARE and PLACE promoter databases for identifying putative cis-acting regulatory elements and are classified into three groups based on their function. The first group corresponds to basal regulatory elements that consist of two copies of TATA box and 7 copies of CAAT box (PlantCARE). The second group is related to pathogen and defense signaling-responsive cis-elements and comprise of GT-1 element (GAAAAA) required for rapid response to pathogen attack and salicylic acid inducible gene expression, WBOXATNPR-1 element required for salicylic acid response (PLACE), TCA element (CAGAAAAGGA) for SA response, TGACG motif for MeJA response, TC-rich motifs (ATTTTCTTCA) for defense response, and AT-rich sequence (TAAAATACT) for maximal elicitor-mediated activation (PlantCARE). Third group includes abiotic stress-related cis-elements such as ABREs motif (ACGT) for ABA-dependent expression, MYC-motif (CACATG), and MYB (GGATA) involved in early response to drought inducible gene induction, HSE element (AGAAAATTCG) involved in heat stress, Circadian motif (CAANNNNATC) involved in circadian control, AE-box (AGAAACT), G-box (CACGTT) involved in light response, and RY-element (CATGCATG) seed-specific regulation, (Fig. S1). A complete list of all predicted cis-elements present in the BjPR1 promoter was shown in Table 2. Hence, in silico analysis of BjPR1 promoter revealed that it could be induced by both biotic as well as abiotic stresses.
Table 2.
Putative cis-regulatory elements in BjPR1 promoter sequence identified by PlantCARE and PLACE promoter databases
| Motif | Copies | sequence | Function |
|---|---|---|---|
| TATA box | 2 | TTATA | Core promoter element |
| CAAT box | 10 | CAAT | Cis-acting regulatory element related to meristem (mesophyll) expression |
| TCA element | 1 | CAGAAAAGGA | Cis-acting element involved in salicylic acid responsiveness |
| GT-1 | 2 | GAAAAA | Cis-acting regulatory element required for rapid response to pathogen attack, salinity, and salicylic acid inducible gene expression |
| I-box | 1 | GATAGGG | Part of a light responsive element |
| MYC | 1 | CACATG | Cis-acting regulatory element involved in early response to drought and abscisic acid induction |
| HSE | 1 | AGAAAATTCG | Cis-acting element involved in heat stress responsiveness |
| Pollen specific | 4 | AGAAA | Required for pollen expression |
| MeJA motif | 1 | TGACG | Cis-acting regulatory element involved in methyl jasmonates responsiveness |
| RY-element | 1 | CATGCATG | Cis-acting regulatory element involved in seed-specific regulation |
| W-box | 3 | TTGAC, TGAC | Cis-acting regulatory element involved in direct fungal elicitor stimulated transcription of defense genes and activation of genes involved in response to wounding |
| MYB | 1 | GGATA | Involved in regulation of drought inducible gene expression |
| G-box | 2 | CACGTT, CACATGG | Cis-acting regulatory element involved in light responsiveness |
| AE-box | 1 | AGAAACTT | Part of a module for light response |
| Erd1 | 1 | ACGT | Cis-acting regulatory element required for early response to dehydration |
| TC-rich repeats | 2 | ATTTTCTTCA, | Cis-acting element involved in defense and stress responsiveness |
| GATA | 7 | GATA | Cis-acting regulatory element required for high level light regulated and tissue specific expression |
| AT-rich sequence | 1 | TAAAATACT | Element for maximal elicitor-mediated activation |
| ERE-motif | 1 | AATTCAAA | Ethylene-responsive element |
Hormonal response of BjPR1 promoter
To examine the inducibility of the BjPR1 promoter, we generated a construct containing the whole promoter region (1800-bp) fused with the GUS reporter gene in pORER2 vector (Fig. 6a). We further studied GUS expression levels driven by BjPR1 promoter using agrobacterium-mediated transient expression assay in N. benthamiana leaves (Fig. 6b). This technique is simple and widely used for the quantitative analysis of plant promoters in vivo (Li et al. 2011). We examined the GUS activity upon hormonal treatments such as (ABA, JA, and SA) after the Agrobacterium strains were infiltrated into N. benthamiana leaves. BjPR1 promoter showed basal expression under control conditions, while as no GUS activity was observed in negative control (Fig. 6c‒d). Interestingly, histochemical GUS analysis revealed that BjPR1 promoter showed weak expression after ABA or JA treatment as compared to untreated tobacco leaves (Fig. 6e‒f). As expected, BjPR1 promoter drives strong GUS gene expression in tobacco leaves after SA treatment, suggesting that the promoter is strongly and rapidly induced (Fig. 6g). We also observed moderate GUS activity driven by BjPR1 promoter in wounded B. juncea leaves (Fig. 6h).
Fig. 6.
Transient expression analysis of BjPR1 promoter in tobacco leaves: a schematic representation of BjPR1 promoter cloned in pORER2 vector (promoter less GUS reporter vector) at Pst1 and BamH1 sites for studying promoter inducibility; b healthy N. benthamiana plants for transient expression analysis. After 24 h of agroinfiltration, plants were treated with sterile water (control), ABA, JA, and SA, respectively. Wounding was carried out with sterile needle. Leaf samples were harvested from control and treated plants after 24 h of treatment for GUS staining; c Promoter less GUS reporter vector as negative control; d GUS gene expression driven by BjPR1 promoter without treatment; e effect of ABA on the expression of GUS gene driven by BjPR1 promoter; f effect of JA on the expression of GUS gene driven by BjPR1 promoter in tobacco leaf; g Effect of SA on the expression of GUS gene driven by BjPR1 promoter; h wound-induced GUS accumulation in tobacco leaf
Discussion
Plant pathogens are unquestionably the most versatile for ecological adaption and in the devastation of plant growth. However, different strategies have been carried out to increase disease resistance in plants like overexpression of pathogenesis-related (PR) proteins or antimicrobial peptides, modifying the resistance signaling pathway and even pyramiding the cloned resistance (R) genes (Grover and Gowthaman 2005). Genetic engineering has become an imperative approach to develop new varieties with high disease resistance. To date, a limited number of disease-resistant genes have been identified and characterized in B. juncea. Therefore, in the present study, one of the key genes of plant disease resistance PR1 and its promoter was studied for its expression pattern in response to Alternaria infection, wounding, and defense stimulators (SA and JA). The gene predicted (BjPR1) belongs to SCF family that is known to be antifungal and cell wall degrading proteins. Phylogenetic analysis of the predicted BjPR1 protein with other known PR1-like sequences revealed that they are grouped into distinct clades. However, BjPR1 fall within the same clade as other Brassica genus PR1 proteins (Fig. 2). Many studies have shown that PR1 plays multifaceted roles in plant defense and also are considered as signaling indicators of SA pathways, but the molecular function is still unknown.
Plant defense system is modulated by different PR proteins, which are the key players of plant immune system. The previous studies showed that PR1 expression is activated by various biotic and abiotic stresses (Mitsuhara et al. 2008; Thierry et al. 1995). Hou et al. (2012) reported that V. vinifera PR1 gene was highly induced after Plasmopara viticola inoculation and expression level of VvPR1 reached the highest peak at 24 hpi, suggesting that VvPR1 might be related to disease resistance. Similarly, transcript levels of PR1 gene isolated from Paeonia suffruticos increases significantly after Cylindrocladium canadense inoculation, and its expression peaked in 24 hpi, which implicated that PsPR1 might be involved in the disease defense (Yang et al. 2013). Our results showed that the expression of BjPR1 gene was significantly induced by A. brassiace (compatible interaction) in local as well as distal leaves; however, we also observed the expression of PR1 reached peak at 24 h after inoculation (Fig. 4a-b). The expression profiles of BjPR1 were similar to the previous studies, though transcript levels were relatively different, which may be due to nature or type of plant–pathogen interactions. These findings are supported by the similar expression pattern of PR1 gene in Arabidopsis and Solanum lycopersicoides inoculated with A. brassicola and Botrytis cinerea, respectively (Schenk et al. 2003; Smith et al. 2014). Interestingly, BjPR1 was induced both locally and systemically in B. juncea, which further provides the evidence for the role of BjPR1 in SAR. Furthermore, upregulation of SA marker gene PR1 upon SAR activation might directly contribute to resistance execution following fungal and oomycete pathogen assault, because PR1 proteins isolated from tobacco and tomato possess in vitro antifungal activity (Niderman et al. 1995). SA signaling generally regulates plant resistance against biotrophic pathogens, whereas JA/ET pathways are commonly associated with resistance to necrotrophic pathogens, and to herbivorous pests (Glazebrook 2005). The present study revealed that BjPR1 is induced by necrotrophic pathogen (A. brassicae) which suggests hormonal crosstalk in B. juncea. Mazumder et al. (2013) also reported the similar expression of PR1 gene and higher accumulation of salicylic acid (SA) after A. brassicicola inoculation in B. juncea. Therefore, it seems that Alternaria infection increases SA accumulation which acts as mobile signal that are transported to distal leaves from the local infected leaves to participate in SAR by activating defenses systemically (Hammond-Kosack and Jones 1996). SAR results in a heightened state of preparedness in the uninfected organs against subsequent infections. The induction of the SA and JA pathways is highly coordinated; with induction of one pathway occur at the expense of the other.
Wounding triggers the activation of many PR genes, thus understanding the expression pattern of BjPR1 under wounding treatment in B. juncea will help us to uncover its detailed role under different stresses. In the present study, BjPR1 was upregulated in local (wounded) leaves and reaches to maximum fold change at 4 h followed by decline at latter time points (Fig. 4c). Thus, BjPR1 seems to be early wound inducible gene as there are reports of early and late wounding responsive genes in plants (Scranton et al. 2013). Moreover, the defense response and wounding have been reported to share a number of components in their signaling pathways which includes SA, JA and ET (Maleck and Dietrich 1999). It was interesting to note that wounding did not increase the systemic expression of BjPR1 in distal (unwounded) leaves. This could be because of high accumulation of JA and ABA which might suppress the SA signaling pathways responsible for the establishment of SAR. Second, wound-induced expression of PR1 seems to be SA or JA independent in B. juncea as we could not observe the induction of BjPR1 in distal leaves (SAR) or after JA treatment. Therefore, further studies needs to be carried out to investigate the role of other hormones (especially ethylene) which might regulate PR genes during wounding.
Two important key players in plant defense response are the classical defense hormones SA and JA, both of which have been well described in model plant (Arabidopsis). However, the role of ABA is largely unknown. SA has received particular attention, because it activates many PR genes and SAR in plants (Yin and Hou 2007). Our results showed that SA leads rapid and strong induction of BjPR1 gene in B. juncea and can be used as SA signaling marker gene in B. juncea (Fig. 5a). In contrast, transcript levels of BjPR1 gene were not increased by JA in B. juncea at any time points which was upregulated in rice and tobacco plants (Xu et al. 1994; Agarwal 2000) (Fig. 5b). Interestingly, A. thaliana belonging to the same family as B. juncea (Crucifeciae) also displays similar results, as ATPR1 has been shown to be induced only by SA but not by JA (Thomma et al. 1998; Durrant and Dong 2004). Hence, increased SA levels in plants lead to the onset of systemic acquired resistance (SAR), an inducible defense response against broad spectrum of pathogens, and also promote PR gene induction (Durrant and Dong, 2004). ABA is a positive regulator mostly involved in abiotic stress responses, but has been also known to play positive or negative roles in plant defense through cross-interaction with SA, JA, and ET signaling transduction pathway. Our studies showed that BjPR1 was significantly downregulated by ABA (Fig. 5c), which was different with the previous studies (Hou et al. 2012; Gao et al. 2015). However, our results are consistent with the previous reports, indicating that ABA downregulates SAR marker gene (PR1) in Arabidopsis. These findings further provide the evidence that ABA plays negative role in B. juncea plant defense response by suppressing SAR marker gene (PR1). Collectively, ABA and JA showed antagonistic interaction with SA signaling pathway in B. juncea.
Despite the emergence of new techniques and years of study, one of the greatest challenges in the development of transgenic disease-resistant crop plants was the identification of inducible promoters which should replace exhaustive constitutive promoter (35S promoter). The use of constitutive promoters in plant genetic engineering is not always desirable, because constitutive overexpression of transgenes may compete for the building blocks that are required for plant growth under normal conditions. Therefore, stress or pathogen-inducible promoters are expected to be optimal for driving transgenes. In many crop species, variety of potential resistant genes for both biotic and abiotic stresses has been isolated, but using them in practical transgenic breeding has failed due to lack of stress-specific inducible promoters. The best stress-inducible promoter is the one induced by a wide array of stresses and must be inactive under stress-free conditions. In this regard, BjPR1 promoter regulating the expression of BjPR1 gene in B. juncea was isolated and subjected to in silico and GUS analysis to know whether it is an inducible or constitutive promoter. Expression of pathogen-related genes usually occurs either by SA- or JA-dependent pathway which is conferred by the presence of single or multiple copies of salicylic acid-responsive elements (SARE-motifs) or jasmonic acid-responsive elements (JAR motifs), respectively. Insilco analysis showed that BjPR1 promoter contains many SA-responsive cis-regulatory DNA elements such as TCA element (2 copies), GT1 motifs (3 copies), and W-box which might be responsible for the induction of BjPR1 after SA treatments. TCA, a cis-acting regulatory element involved in salicylic acid responsiveness, is known to be present in the non-translated regions of many monocot and dicot plant genes which are induced by one or more forms of stress (Goldsbrough et al. 1993). Similar location of TCA element was also observed in the untranslated region of BjPR1 promoter. On the other hand, GT1-elements are cis-acting regulatory elements required for rapid response to pathogen attack, salinity, and salicylic acid inducible gene expression. Our results have also identified the sequence element similar to GT1 motif in the promoter region of BjPR1 gene which further reveals that BjPR1 could be induced by SA. In addition, BjPR1 promoter also contains 2 copies of W-box (T)TGAC (C/T), another important cis-acting DNA element have been found in the promoters of a number of SA-inducible genes and have been shown to be essential for the full expression of SA-responsive gene SFR2. A well-known pathogen-related motif TC-rich repeats was also found in BjPR1 promoter which mediates biotic stress responses in plants. In addition to pathogen-responsive motifs, PR1 promoter also contains abiotic stress-related motifs, viz., drought (MYB-motif GGATA; MYC-motif-CACATG), heat (HSF-motif AGAAAATTCG), salt (GT1-motif GAAAAA, DRE-motif A/GCCGAC), and also W-box which mediates abiotic stress responses in plants like freezing, wounding, oxidative stress, drought, salinity, cold, and heat by binding with various WRKY TFs (Fig S1). Collectively, these findings indicated that transcription of BjPR1 promoter might be complex and regulated by a variety of biotic and abiotic stresses.
To further investigate the BjPR1 promoter activity, a qualitative GUS activity assay was conducted by Agrobacterium-mediated transient assay in tobacco leaves (Fig. 6c–h). Agrobacterium-mediated transient expression has been widely used for both qualitative and quantitative analysis of plant promoter and also for cis-element/trans-factor interactions. Moreover, the use of this technique is growing at an accelerated rate being fast as well as has no environmental risks associated with the production of stable transgenic plants, and hence have opened up new strategies for transgenic studies (Omidvar et al. 2008). Since PR gene expression has been reported to be induced by various factors such as pathogen attack, SA, JA, and wounding treatments. In addition, PR genes are considered as molecular indicators of the activation of SA and JA signaling pathways, and can be termed as signatures of these pathways. In this study, BjPR1 induction was monitored after treatment with defense stimulators (SA, JA, and ABA) as well as wounding. Transient assay revealed that BjPR1 promoter driven GUS gene activity shows basal expression under control conditions but increases significantly after SA treatment. On the other hand, BjPR1 driven GUS activity was decreased after ABA or JA treatments compared to control. In general, SA and JA predominantly have an antagonistic relationship (Pieterse et al. 2009), similar to that observed in our study. This further confirms that BPR1 is predominantly SA-dependent and can be used as a molecular indicator of SA signaling in B. juncea. The presence of potential SA-responsive cis-elements in BjPR1 promoter might be responsible for rapid and strong GUS gene expression in tobacco leaves after SA treatment. Mechanical wounding as a result of abiotic or biotic factors not only physically damages plant tissue, but it also provides entry for microbial and fungal pathogen invasion. Interestingly, BjPR1 promoter was also induced by wounding. Therefore, wound-induced expression of pathogenesis-related gene (BjPR1 and its promoter) further provides the evidence that they might play an important role in combating the detrimental effects of pathogen attack as well as wounding in B. juncea. Our results also identified many wound-responsive elements in the BjPR1 promoter such as W-box, G-box, and ethylene-responsive element which might be responsible for wound-induced expression of BjPR1 promoter.
Conclusion
In this study, we have isolated and functionally characterised SAR marker gene PR1 after Alternaria infection, wounding, and defense hormonal treatments in B. juncea. Hence, from the BjPR1 gene expression study, it is evident that BjPR1 gene is predominantly SA-dependent. However, the induction of BjPR1 gene by A. brassicae not by JA further provides the evidence of hormonal crosstalk in B. juncea. The present study also identified BjPR1 promoter as stress-inducible promoter which can be successfully and effectively used for the development of transgenic crops for fungal resistance.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We gratefully acknowledge the Project Director, National Research Centre on Plant Biotechnology Pusa Campus New Delhi for providing all the facilities required to complete this work.
Author contributions
AG has conceived and designed the work. SA performed all the experiments and wrote the manuscript. ZAM, JAB, AT & NC helped in bioinfromatic and statistical analysis. ZAM and PY helped in infection assays, SR & MS helped in manuscript editing. AG has given the final shape of manuscript and all authors aproved the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13205-017-1027-8) contains supplementary material, which is available to authorized users.
References
- Agrawal GK, Jwa NS, Rakwal R. A novel rice (Oryza sativa L.) acidic PR1 gene highly responsive to cut, phytohormones, and protein phosphatase inhibitors. Biochem Biophys Res Commun. 2000;274:157–165. doi: 10.1006/bbrc.2000.3114. [DOI] [PubMed] [Google Scholar]
- Agrawal GK, Rakwal R, Jwa NS, Agrawal VP. Signaling molecules and blast pathogen attack activates rice OsPR1a and OsPR1b genes: a model illustrating components participating during defense/stress response. Plant Physiol Biochem. 2001;39:1095–1103. doi: 10.1016/S0981-9428(01)01333-X. [DOI] [Google Scholar]
- Alexander D, Goodman RM, Gut-Rella M. Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc Natl Acad Sci USA. 1993;90:7327–7331. doi: 10.1073/pnas.90.15.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Taweel K, Fernando WGD, Brule-Babel AL. Construction and characterization of a Cdna library from wheat Infected with Fusarium graminearum Fg 2. Int J Mol Sci. 2011;12:613–626. doi: 10.3390/ijms12010613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal RS, Kumar A. Studies on the epidemiology of white rust and Alternaria leaf blight and their effect on the yield of Indian mustard. Afr J Agric Res. 2014;9:302–306. doi: 10.5897/AJAR2013.7352. [DOI] [Google Scholar]
- Brederode FT, Linthorst HJM, Bol JF. Differential induction of acquired resistance and PR gene expression in tobacco by virus infection, ethephon treatment, UV light and wounding. Plant Mol Biol. 1991;17:1117–1125. doi: 10.1007/BF00028729. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhou MG. Characterization of Fusarium graminearum isolates resistant to both carbendazim and a new fungicide JS399-19. Phytopathology. 2009;99:441–446. doi: 10.1094/PHYTO-99-4-0441. [DOI] [PubMed] [Google Scholar]
- Chengguo J, Liping Z, Lihong L, Jiansheng W, Chuanyou L, Qiaomei W. Multiple phytohormones signaling pathways modulate susceptibility of tomato plants to Alternaria alternate f. sp. Lycopersici. J Exp Bot. 2012;64(2):637–650. doi: 10.1093/jxb/ers360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X. Systemic acquired resistance. Ann Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE, Jones JDG. cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell. 2000;12:963–977. doi: 10.1105/tpc.12.6.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY super family of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Eyal Y, Sagee O, Fluhr R. Dark-induced accumulation of a basic pathogenesis-related (PR-1) transcript and a light requirement for its induction by ethylene. Plant Mol Biol. 1992;19:589–599. doi: 10.1007/BF00026785. [DOI] [PubMed] [Google Scholar]
- Gao L, Wang S, Li XY, Wei XY, Zhang YJ, Wang HY, Liu DQ. Expression and functional analysis of a pathogenesis-related protein 1 gene, TcLr19PR1, involved in wheat resistance against leaf rust fungus. Plant Mol Biol Report. 2015;33:797–805. doi: 10.1007/s11105-014-0790-5. [DOI] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Goldsbrough AP, Albrecht H, Stratford R. Salicylic acid-inducible binding of a tobacco nuclear protein to a 10 bp sequence which is strongly conserved amongst stress-inducible genes. Plant J. 1993;3:563–571. doi: 10.1046/j.1365-313X.1993.03040563.x. [DOI] [PubMed] [Google Scholar]
- Grover A, Gowthaman R. Strategies for development of fungus resistant transgenic plants. Curr Sci. 2005;84:330–340. [Google Scholar]
- Hamamouch N, Li CY, Seo PJ, Park CM, Davis EL. Expression of Arabidopsis pathogenesis-related genes during nematode infection. Mol Plant Pathol. 2011;12:355–364. doi: 10.1111/j.1364-3703.2010.00675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Silverman P, Raskin I, Jones JDG. Race-specific elicitors of cladosporium fulvum induce changes in cell morphology and the synthesis of ethylene and salicylic acid in tomato plants carrying the corresponding Cf disease resistance gene. Plant Physiol. 1996;110(4):1381–1394. doi: 10.1104/pp.110.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Garcia CM, Finer JJ. Identification and validation of promoters and cis- acting regulator elements. Plant Sci. 2014;217(218):109–119. doi: 10.1016/j.plantsci.2013.12.007. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis–acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou LX, Gao C, Che YM, Zhao FG, Liu X. Gene cloning and expression analysis of pathogenesis-related protein 1 in Vitis vinifera L. Plant Physiol J. 2012;48:57–62. [Google Scholar]
- Jung HW, Hwang BK. Isolation, partial sequencing, and expression of pathogenesis-related cDNA genes from pepper leaves infected by Xanthomonas campestris pv. vesicatoria. Mol Plant-Microbe Interact. 2000;13:136–142. doi: 10.1094/MPMI.2000.13.1.136. [DOI] [PubMed] [Google Scholar]
- Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT. Priming in systemic plant immunity. Science. 2009;324:89–91. doi: 10.1126/science.1170025. [DOI] [PubMed] [Google Scholar]
- Kaushik CD, Saharan GS, Kaushik JC (1984) Magnitude of losses in yield and management of Alternaria blight in rapeseed-mustard. Indian Phytopath 37:398 (Abstr.)
- Kim YJ, Hwang BK. Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiol Plant. 2000;108:51–60. doi: 10.1034/j.1399-3054.2000.108001051.x. [DOI] [Google Scholar]
- Kolte SJ. Diseases of annual edible oilseed crops. Boca Raton: CRC Press; 1985. p. 135. [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton K, Uknes S, Friedrich L, Gaffney T, Alexander D, Goodman R, Métraux JP, Kessmann H, Ahl Goy P, Gut-Ruella M, Ward E, Ryals J. The molecular biology of systemic acquired resistance. In: Fritig B, Legrand M, editors. Mechanisms of defence responses in plants. Dordrecht: Kluwer Academic Press; 1993. pp. 410–420. [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van De Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucl Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ilarslan H, Brachova L, Qian HR, Li L, Che P, Wurtele ES, Nikolau BJ. Reverse-genetic analysis of the two biotin-containing subunit genes of the heteromeric acetyl-coenzyme A carboxylase in Arabidopsis indicates a unidirectional functional redundancy. Plant Physiol. 2011;155:293–314. doi: 10.1104/pp.110.165910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maleck K, Dietrich RA. Defense on multiple fronts: how do plants cope with diverse enemies? Trends Plant Sci. 1999;4:215–219. doi: 10.1016/S1360-1385(99)01415-6. [DOI] [PubMed] [Google Scholar]
- Mazarei M, Teplova I, Hajimorad MR, Stewart CN. Pathogen phytosensing: plants to report plant pathogens. Sensors. 2008;8:2628–2641. doi: 10.3390/s8042628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder M, Das SU, Saha M, Chatterjee K, Bannerjee DB. Salicylic acid-mediated establishment of the compatibility between Alternaria brassicicola and Brassica juncea is mitigated by abscisic acid in Sinapis alba. Plant Physiol Biochem. 2013;70:43–51. doi: 10.1016/j.plaphy.2013.04.025. [DOI] [PubMed] [Google Scholar]
- Mitsuhara I, Iwai T, Seo S, Yanagawa Y, Kawahigasi H, Hirose S. Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180) Mol Genet Genom. 2008;279:415–427. doi: 10.1007/s00438-008-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niderman T, Genetet I, Bruyère T, Gees R, Stinzi A, Legrand M, Fritig B, Mösinger E. Pathogenesis-related PR1-proteins are antifungal. Isolation and characterization of three 14 kilo dalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol. 1995;108:17–27. doi: 10.1104/pp.108.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki T, Mitsuhara I, Seo S, Ohtsubo N, Ohashi Y. Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis-related (PR) protein genes in wounded mature tobacco leaves. Plant Cell Physiol. 1998;39:500–507. doi: 10.1093/oxfordjournals.pcp.a029397. [DOI] [Google Scholar]
- Ohme-Tagaki M, Suzuki K, Shinshi H. Regulation of ethylene-induced transcription of defense genes. Plant Cell Physiol. 2000;41:1187–1192. doi: 10.1093/pcp/pcd057. [DOI] [PubMed] [Google Scholar]
- Omidvar V, Siti AA, Marziah M, Maheran AA. A transient assay to evaluate the expression of polyhydroxybutyrate genes regulated by oil palm mesocarp-specific promoter. Plant Cell Rep. 2008;27:1451–1459. doi: 10.1007/s00299-008-0565-2. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Leon-Reyes A, Van der ES, Van Wees SCM. Networking by small-molecules hormones in plant immunity. Nat Chem Biol. 2009;5:308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- Reymond EE, Farmer P. Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol. 1998;1:404–411. doi: 10.1016/S1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- Sarowar S, Sujon S, Young JK, Eui NK. Overexpression of a pepper basic pathogenesis-related protein 1 gene in tobacco plants enhances resistance to heavy metal pathogen stresses. Plant Cell Rep. 2005;24:216–224. doi: 10.1007/s00299-005-0928-x. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Manners JM, Anderson JP, Simpson RS, Wilson IW, Somerville SC, Maclean DJ. Systemic gene expression in Arabidopsis during an incompatible interaction with Alternaria brassicicola. Plant Physiol. 2003;132:999–1010. doi: 10.1104/pp.103.021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scranton MA, Fowler JH, Girke T, Walling LL. Microarray Analysis of tomato’s early and late wound response reveals new regulatory targets for leucine aminopeptidase A. PLoS One. 2013;8(10):e77889. doi: 10.1371/journal.pone.0077889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sels J, Mathys J, De Coninck BM, Cammue BP, De Bolle MF. Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol Biochem. 2008;46:941–950. doi: 10.1016/j.plaphy.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Shekhawat K, Rathore SS, Premi OP, Kandpal BK, Chauhan JS. Advances in agronomic management of Indian mustard (Brassica juncea (L.) Czernj. Cosson): an overview. Int. J Agron. 2012;14:408284. [Google Scholar]
- Sinha M, Singh RP, Kushwaha GS, Iqbal N, Singh A, Kaushik S, et al. Current overview of allergens of plant pathogenesis related protein families. Sci World J. 2014;2014:19. doi: 10.1155/2014/543195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JE, Mengesha B, Tang H, Mengiste T, Bluhm BH. Resistance to Botrytis cinerea in Solanum lycopersicoides involves widespread transcriptional reprograming. BMC Genom. 2014;15:334. doi: 10.1186/1471-2164-15-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujon s, Sujon s, oung JK, Eui NK. Overexpression of a pepper basic pathogenesis-related protein gene in tobacco plants enhances resistance to heavy metal pathogen stresses. Plant Cell Rep. 2005;24:216–224. doi: 10.1007/s00299-005-0928-x. [DOI] [PubMed] [Google Scholar]
- Thierry N, lsabelle G, Thierry B, Rene G, Annick S, Michel L, Bernard F, Egon M. Pathogenesis relatedPR1 proteins are antifungal. Plant Physiol. 1995;108:17–27. doi: 10.1104/pp.108.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch Mani B, Vogelsang R, Cammue BPA, Broekaert WF. Separate jasmonate-dependent and salicylate dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon LC, Van Strien EA. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol. 1999;55:85–97. doi: 10.1006/pmpp.1999.0213. [DOI] [Google Scholar]
- Ward E, Uknes S, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Metraux JP, Ryals J. Coordinate gene activity in response to that induce systemic acquired resistance. Plant Cell. 1991;3:1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Chang PL, Liu D, Narasimhan ML, Raghothama KG, Hasegawa PM, Bressan RA. Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell. 1994;6:1077–1085. doi: 10.1105/tpc.6.8.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Zhang YX, Zheng GS. Gene cloning and expression analysis of pathogenesis-related protein 1 of Paeonia suffruticosa. Acta Hortic Sin. 2013;40:1583–1590. [Google Scholar]
- Yin LL, Hou XJ. The recent advances of salicylic acid as signal molecules of resistance in plant. Chin Agric Sci Bull. 2007;12:338–342. [Google Scholar]
- Zhang Y, Xu S, Ding P, Wang D, Cheng YT, Jing H. Control of salicylic acid synthesis And systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci USA. 2010;107:18220–18225. doi: 10.1073/pnas.1005225107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao KJ, Chye ML. Methyl jasmonate induces expression of a novel Brassica juncea chitinase with two chitin-binding domains. Plant Mol Biol. 1999;40:1009–1018. doi: 10.1023/A:1006266407368. [DOI] [PubMed] [Google Scholar]
- Zheng X, Deng W, Luo K, Duan H, Chen Y, McAvoy R, Song S, Pei Y, Li Y. The cauliflower mosaic virus (CaMV) 35S promoter sequence alters the level and patterns of activity of adjacent tissue and organ-specific gene promoters. Plant Cell Rep. 2007;26:1195–1203. doi: 10.1007/s00299-007-0307-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.