Abstract
Protein complexes involved in respiration, ATP synthesis, and protein import reside in the mitochondrial inner membrane; thus, proper regulation of these proteins is essential for cell viability. The m-AAA protease, a conserved hetero-hexameric AAA (ATPase associated with diverse cellular activities) protease, composed of the Yta10 and Yta12 proteins, regulates mitochondrial proteostasis by mediating protein maturation and degradation. It also recognizes and mediates the dislocation of membrane-embedded substrates, including foreign transmembrane (TM) segments, but the molecular mechanism involved in these processes remains elusive. This study investigated the role of the TM domains in the m-AAA protease by systematic replacement of one TM domain at a time in yeast. Our data indicated that replacement of the Yta10 TM2 domain abolishes membrane dislocation for only a subset of substrates, whereas replacement of the Yta12 TM2 domain impairs membrane dislocation for all tested substrates, suggesting different roles of the TM domains in each m-AAA protease subunit. Furthermore, m-AAA protease–mediated membrane dislocation was impaired in the presence of a large downstream hydrophilic moiety in a membrane substrate. This finding suggested that the m-AAA protease cannot dislocate large hydrophilic domains across the membrane, indicating that the membrane dislocation probably occurs in a lipid environment. In summary, this study highlights previously underappreciated biological roles of TM domains of the m-AAA proteases in mediating the recognition and dislocation of membrane-embedded substrates.
Keywords: ATPase associated with diverse cellular activities (AAA), membrane, mitochondria, transmembrane domain, yeast, Yta10, Yta12
Introduction
The mitochondrial inner membrane (IM)3 is the site for many membrane protein complexes functioning in various cellular processes: respiration, ATP synthesis, and protein import. Maintaining protein homeostasis at the mitochondrial IM is thus essential and critical for cellular fitness.
The m-AAA protease and the i-AAA protease form a major quality control system in the mitochondrial IM and mediate the degradation of misfolded or unassembled mitochondrial IM proteins (1–4). The i-AAA protease is a homo-oligomeric complex formed by Yme1, and the catalytic domain is exposed to the intermembrane space (IMS) (5, 6). The m-AAA protease is a heterohexameric complex composed of Yta10 and Yta12; each contains two N-terminal TMs, a soluble AAA+ domain, and a proteolytic domain exposed to the matrix (7, 8).
The function of the m-AAA proteases is not limited to quality control, but it also assists with protein folding of a mitochondrial ribosome subunit, MrpL32 (9), and maturation of cytochrome c peroxidase (Ccp1) (10). In the case of Ccp1 maturation, the m-AAA protease acts as an ATPase by pulling the Ccp1 TM to the matrix prior to processing by a rhomboid protease, Pcp1, in the IM (10, 11). Furthermore, an earlier study has shown that moderately hydrophobic TM segments can be dislocated by the m-AAA protease (12).
It has been reported that deletion of TMs of Yta10 or Yta12 causes severe defects in the degradation of integral membrane proteins but only mild defects in the degradation of peripherally associated membrane proteins, implicating the importance of TMs of the m-AAA protease in extracting the substrate TM segments from the membrane (13–15). However, it is poorly understood whether the m-AAA protease TMs are actively involved in the dislocation process or simply act to anchor the catalytic domain near the membrane.
To better understand the molecular mechanism of the m-AAA protease–mediated TM helix dislocation, this study focused on the role of TMs of the m-AAA protease. Individual TMs of Yta10 and Yta12 were replaced one at a time, and the capacity of these m-AAA protease variants to dislocate various membrane substrates was assessed. Our data show that replacement of TM2 of Yta10 impairs recognition of a subset of substrates, whereas replacement of Yta12 TM2 impairs membrane dislocation of all tested substrates, illustrating different roles of TMs in each subunit. Additionally, we have investigated the length of a soluble moiety in the IMS of the dislocation substrate the m-AAA protease is capable of extracting from the membrane.
Results
Yta10 or Yta12 variants with a replaced TM complement the respiratory growth defect of the corresponding deletion strain
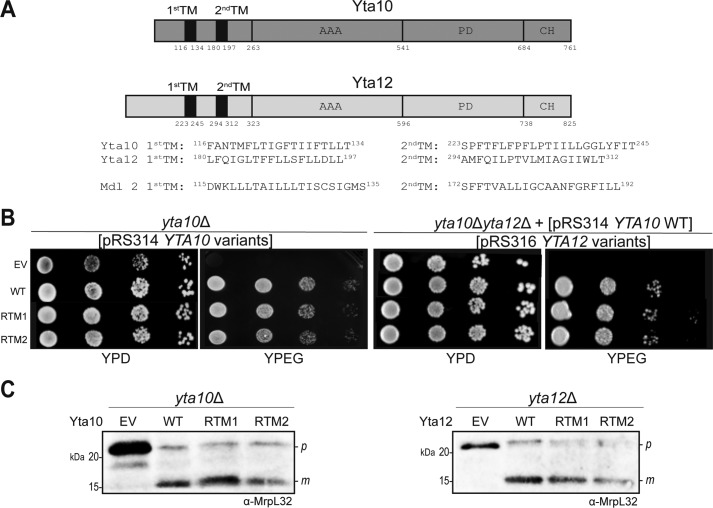
To elucidate the role of individual TMs of the m-AAA protease subunits, each TM of Yta10 and Yta12 was replaced with that of another mitochondrial IM protein, Mdl2, one at a time (Fig. 1A). These constructs were transformed into the yta10Δ or yta12Δ strain. As reported previously, the yta10Δ and yta12Δ strains were viable on fermentable carbon medium but could not grow on non-fermentable carbon medium, as the m-AAA protease is required for the maturation of the mitochondrial ribosome subunit MrpL32 (16).
Figure 1.
Yta10 and Yta12 variants with a replaced TM are functional. A, schematics of Yta10 and Yta12 with TMs highlighted in black. AAA, AAA domain; PD, proteolytic domain; CH, C-terminal helical domain. TM sequences are shown at the bottom. B, growth complementation assay. Yta10 or Yta12 variants were expressed in yta10Δ (left panel) or yta10Δ yta12Δ [pRS314 YTA10 WT] (right panel), respectively, under the endogenous promoter. The transformants were cultured in glucose-containing liquid medium prior to spotting on YPD or YPEG plates. The plates were incubated at 30 °C for 2 days prior to imaging. EV, empty vector; RTM1, replaced TM1; RTM2, replaced TM2. C, maturation of MrpL32. yta10Δ or yta10Δ yta12Δ [pRS314 YTA10 WT] cells expressing Yta10 or Yta12 variants were cultured overnight and lysed in sample buffer. The lysates were analyzed by SDS-PAGE and Western blotting. The blots were immunodecorated with anti-MrpL32 antibody. P, precursor; m, mature form.
To examine whether Yta10 and Yta12 variants are functional, a growth complementation assay was performed (Fig. 1B). All of the constructs rescued the growth defect of the corresponding deletion on non-fermentable medium. Furthermore, MrpL32 maturation in the Yta10 and Yta12 variants were unaffected (Fig. 1C). These results suggest that Yta10 or Yta12 variants with a replaced TM are correctly targeted, assembled into the m-AAA protease complex, and processed by MrpL32 in the mitochondria.
Dislocation substrates for the m-AAA protease
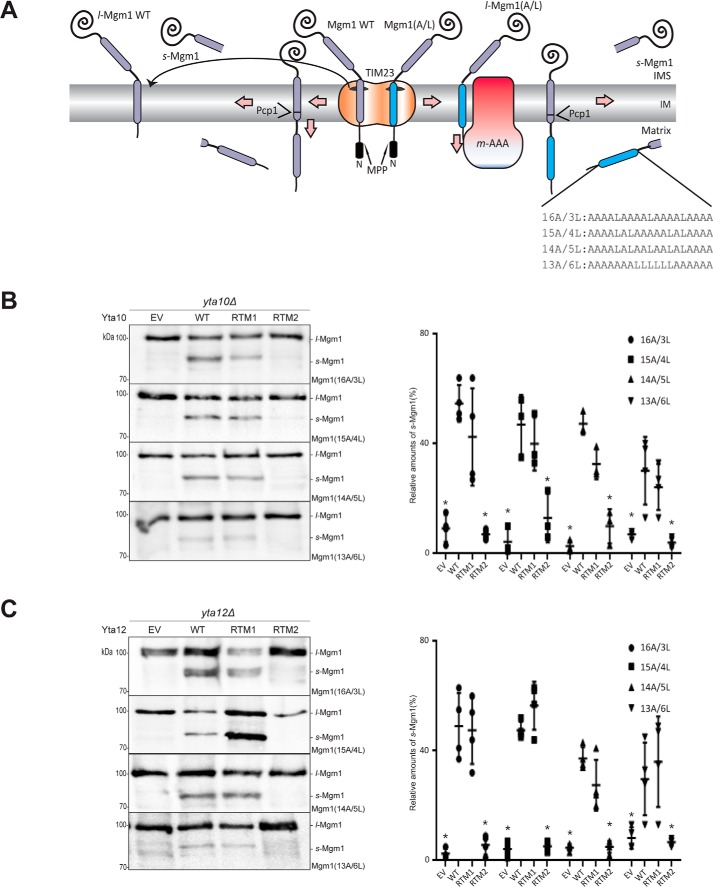
Mgm1 is an IM protein that naturally exists in two forms: l-Mgm1 and s-Mgm1. It carries an N-terminal presequence that is processed by mitochondrial processing peptidase (MPP) and two putative TMs. A recent report has shown that Mgm1 is co-translationally imported into mitochondria (17). Uniquely, the insertion efficiency of the TM1 into the IM is roughly 50% (18–20). When the TM1 is inserted into the IM, it becomes l-Mgm1. On the other hand, when the TM1 of Mgm1 is translocated to the matrix, the TM2 subsequently enters the IM, where it is further processed by Pcp1, and s-Mgm1 is formed (Fig. 2A, left). Here, the sorting of the TM1 of Mgm1 occurs at the level of the TIM23 complex.
Figure 2.
Dislocation of Mgm1(A/L) from the IM requires TM2 of Yta10 and Yta12. A, alternative topogenesis of Mgm1 WT and processing of Mgm1 (A/L). The sequences of the TM1 segment of Mgm1, Mgm1(16A/3L, 15A/4L, 14A/5L, 13A/6L), are shown. B, dislocation of Mgm1(A/L). Yeast transformants expressing the indicated Yta10 variant and Mgm1(A/L) were lysed in the presence of 25% TCA (final concentration). The lysates were analyzed by SDS-PAGE and Western blotting. The blots were immunodecorated with anti-HA antibody, which detects the HA tag at the C terminus of Mgm1(A/L) (left panel). Relative amounts of s-Mgm1 were quantified from three independent experiments and plotted with standard deviations. A t test was performed to examine the statistical significance of the observed results (right panel). C, Yta12 variants were tested in the same manner as described in B. The differences in dislocation efficiency were statistically significant between WT and EV/RTM2 (*, p < 0.05).
However, when the TM1 of Mgm1 is replaced with the model TM segment composed of varying number of alanines and leucines (19-n alanines and n leucines), this segment is inserted into the IM by the TIM23 complex, resulting in l-Mgm1 (21). Subsequently, l-Mgm1 can be dislocated by the m-AAA protease (12) (Fig. 2A, right). The dislocation efficiency has been shown to be dependent on the hydrophobicity; moderately hydrophobic TM segments are more readily dislocated than sufficiently hydrophobic ones (12). Thus, for the Mgm1 constructs carrying the engineered TM segments (Mgm1(A/L) constructs), the generation of s-Mgm1 is dependent on the dislocation activity of the m-AAA protease, as the cleavage of the TM2 by Pcp1 only occurs after dislocation of the TM1 by the m-AAA protease.
Replacement of TM2 of Yta10 and Yta12 impairs membrane dislocation of model TM segments in Mgm1
To assess the membrane dislocation efficiency of Yta10 and Yta12 TM mutants, the yta10Δ or yta12Δ strain carrying each Yta10 and Yta12 variant was transformed with a set of Mgm1(A/L) constructs. We reasoned that, if the role of Yta10 or Yta12 TMs is limited to anchoring the catalytic domain (ATPase domain and the metalloprotease domain) near the membrane, then substitution of TMs would have a minimal effect. In contrast, if the TMs of Yta10 or Yta12 are required either for the recognition or active membrane dislocation, then substitution of Yta10 TMs would compromise the membrane dislocation activity of m-AAA protease.
Replacement of TM1 (RTM1) of Yta10 or Yta12 did not have significant effects on the dislocation of the tested TM segments, as the ratio of l-Mgm1 to s-Mgm1 was comparable with the Yta10 or Yta12 WT strain (Fig. 2, B and C). Although subtle, a decrease in membrane dislocation of a sufficiently hydrophobic TM (Mgm1(14A/5L)) has been observed in both the Yta10 and Yta12 RTM1 variants (Fig. 2, B and C). In comparison, replacement of TM2 (RTM2) of Yta10 or Yta12 significantly reduced generation of the s-Mgm1 form, indicating that the membrane dislocation activity is severely impaired upon swapping of TM2 of the m-AAA protease (Fig. 2, B and C).
Replacement of TM2 of Yta10 selectively impairs membrane dislocation for only a subset of substrates
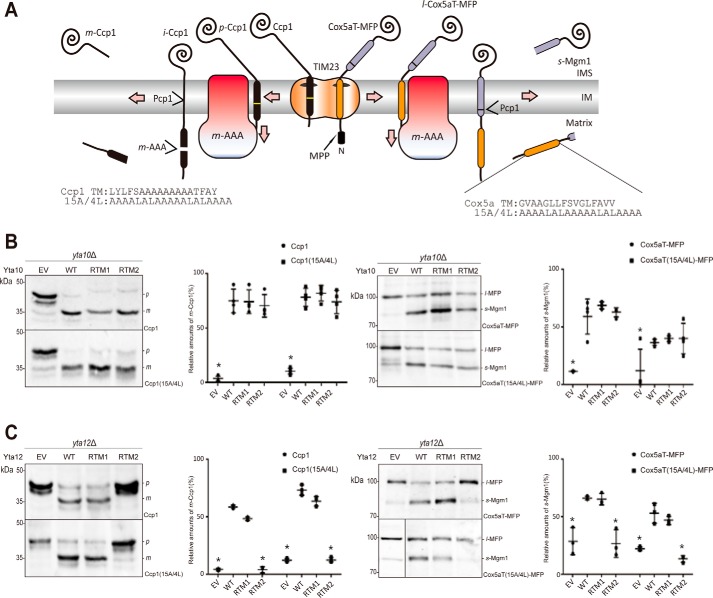
To distinguish whether the replacement of Yta10 or Yta12 TM2 causes a general defect in membrane dislocation activity or in recognizing substrates, other previously characterized m-AAA protease substrates were tested. Ccp1 is a natural substrate of the m-AAA protease. For maturation of Ccp1, precursor Ccp1 undergoes two-step cleavage by the m-AAA protease in the matrix and Pcp1 in the IM (Fig. 3A). Replacement of its TM1 with the model TM segment composed of 15 alanines and four leucines (Ccp1(15A/4L)) does not affect its maturation (12). For the Ccp1 constructs, membrane dislocation efficiency is measured by the production of mature Ccp1(m-Ccp1) (Fig. 3A, left).
Figure 3.
Dislocation activity is unaffected in Yta10 RTM2 but defective in Yta12 RTM2. A, schematic of processing of Ccp1, Ccp1(15A/4L), Cox5aT-MFP, and Cox5aT(15A/4L)-MFP by the m-AAA protease. The sequences of the TM1 segment of Ccp1 or Cox5a are shown, along with the 15A/4L segment sequence. B and C, expression of Ccp1, Ccp1(15A/4L), Cox5aT-MFP, or Cox5aT(15A/4L)-MFP in yta10Δ carrying the indicated Yta10 variants (B) or in yta10Δ yta12Δ [pRS314 YTA10 WT] carrying the indicated Yta12 variants (C). Lysates were prepared and analyzed as described in Fig. 2B (left panel). Relative amounts of m-Ccp1 or s-Mgm1 were quantified with standard deviations. A t test was performed to examine the statistical significance of the observed results (right panel). The differences in dislocation efficiency for Yta12 were statistically significant between WT and EV/RTM2 (*, p < 0.05). l-MFP, long Mgm1 fusion protein.
Cox5aT-MFP (MFP, Mgm1 fusion protein) is a chimera protein where Cox5a protein is truncated at residue 128 and fused to the C terminus of Mgm1 (22). Previous studies have shown that this C-terminal truncation at the IMS side converts Cox5a into the m-AAA protease substrate. Another m-AAA protease substrate, Cox5aT(15A/4L)-MFP, was prepared by replacing the Cox5a TM with the TM segment composed of 15A/4L in the background of Cox5aT-MFP. The membrane dislocation efficiency of Cox5aT-MFP variants was judged by the formation of s-Mgm1 (Fig. 3A, right).
First, the membrane dislocation efficiency of Ccp1 and Cox5aT-MFP variants in cells carrying the Yta10 variants was assessed. In contrast to the results with Mgm1(A/L) variants, the generation of m-Ccp1 or s-Mgm1 in Yta10 RTM2 strain was comparable with that of the Yta10 WT strain, indicating that dislocation of the Ccp1 and Cox5aT-MFP variants was unaffected by the TM2 replacement of Yta10 (Fig. 3B). These results thus implicate that replacement of Yta10 TM2 does not impair the general membrane dislocation activity of the m-AAA protease.
Replacement of TM2 of Yta12 causes a general defect in membrane dislocation activity of the m-AAA protease
Next, the dislocation efficiency of the Ccp1 and Cox5aT-MFP variants was tested in the Yta12 strains. Here, the generation of m-Ccp1 or s-Mgm1 was significantly reduced compared with the WT strain, indicating that the membrane dislocation of tested substrates was severely impaired (Fig. 3C). This suggests that replacement of TM2 of Yta12 causes a general defect in the membrane dislocation activity of the m-AAA protease, different from the effects of the TM2 replacement of Yta10.
Dislocation of a TM segment with a hydrophilic IMS moiety is inhibited in a length-dependent manner
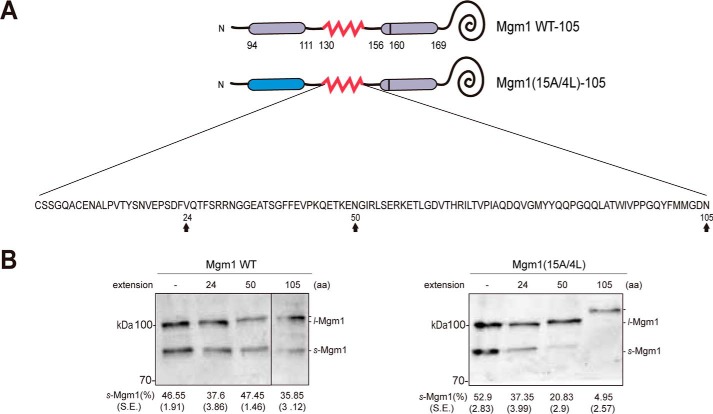
When the m-AAA protease mediates degradation of a membrane protein, not only the TMs but also the loop regions in the IMS should be pulled across the IM. How large can the soluble domain be for the m-AAA protease to handle? To determine the dislocation capacity of the m-AAA protease, a hydrophilic segment containing many polar and charged residues from the Escherichia coli leader peptidase periplasmic (P2) domain was introduced in between the two TMs of Mgm1(15A/4L) at various lengths (Fig. 4A).
Figure 4.
Dislocation of a TM segment with a large hydrophilic IMS moiety is impaired. A, schematic of Mgm1 WT and Mgm1(15A/4L) with a hydrophilic stretch. A soluble periplasmic (P2) domain from E. coli was added in between residues 130 and 131 of Mgm1 at the lengths of 24, 50, and 105 amino acids. The inserted amino acid sequence is shown, with arrows pointing toward the site of truncation. The Pcp1 cleavage site of Mgm1 (residue 160) is indicated by a black line in TM2. B, dislocation efficiency of Mgm1WT and Mgm1(15A/4L) with 24-, 50-, or 105-amino acid extensions. Yeast transformants expressing Mgm1WT (left panel) or Mgm1(15A/4L) (right panel) with 24-, 50-, or 105-amino acid extensions were lysed and analyzed as described in Fig. 2B. Relative amounts of s-Mgm1 quantified from three independent experiments are indicated at the bottom with standard errors.
Dislocation of TM1 of Mgm1(15A/4L) was gradually decreased by 24- and 50-amino acid-long extensions and completely inhibited by 105-amino acid-long extensions (Fig. 4B). An earlier study has shown that a tightly folded domain in downstream of the TM segment prevents membrane protein degradation by the m-AAA protease (15, 23). To check whether the hydrophilic segments used in our experiment form a tightly folded structure, the structure of the P2 domain was examined (PDB code 1T7D) (24). 24- and 50-amino acid-long segments are mostly unstructured, and a 105-amino acid segment consists of some β strands and unstructured loops. Thus, it is unlikely that membrane dislocation was prevented because of the folding structure of the hydrophilic extension. These results suggest that the m-AAA protease has limited capacity to dislocate a large IMS moiety.
Drastic dislocation impairment in the presence of a hydrophilic linker led us to think about the molecular environment from which the proteins are dislocated. We reasoned that, if dislocation occurs through the hydrophobic core of the lipid bilayer, then a large sum of energy is required; therefore, a long hydrophilic extension would prevent membrane dislocation, whereas, if it occurs through a proteinaceous pore, then the energy barrier would be lower, and the extension length would influence the dislocation efficiency less. To test the translocation efficiency of the P2 domain through the proteinaceous channel, increasing lengths of hydrophilic extensions were added in between the TM1 and the downstream Pcp1 cleavage site in Mgm1 WT (Fig. 4A). For the generation of s-Mgm1 in Mgm1 WT, the TM1 is translocated through a pore formed by the TIM23 complex (Fig. 2A); thus, Mgm1 WT with the hydrophilic domain extension could serve as a control for membrane dislocation through a proteinaceous environment. Here, the relative ratio of s-Mgm1 to l-Mgm1 was unaffected by the presence of a 24-, 50-, or 105-amino acid-long hydrophilic segment, demonstrating that addition of a hydrophilic moiety between the two TMs did not interfere with the translocation/insertion efficiency of Mgm1 (Fig. 4B). Further, this result showed that the presence of a long hydrophilic moiety upstream of the rhomboid cleavage site did not impair the cleavage efficiency of Pcp1. Taken together, these results suggest that the m-AAA protease is incapable of dislocating a large hydrophilic domain across the IM, subsequently implicating that membrane dislocation probably occurs through the lipid environment.
Discussion
The m-AAA TMs have been shown to be indispensable for integral membrane protein degradation (13), but their roles in extracting proteins out of the membrane remain poorly understood. By swapping each TM one at a time, we present data showing that replacement of the TM2s of m-AAA protease causes defects in the membrane dislocation of substrate proteins. Interestingly, replacement of TM2 of Yta10 and Yta12 exhibited different defects in substrate dislocation. Replacement of TM2 of Yta10 impaired the dislocation of Mgm1(A/L) variants only, whereas replacement of TM2 of Yta12 impaired the dislocation of all tested substrates.
The general dislocation impairment observed with TM2 replacement of Yta12 was unexpected because Ccp1 dislocation/maturation was shown to be unaffected in a yeast strain expressing a TM deletion mutant of Yta12 (13). Therefore, it is less likely that TM2 of Yta12 is indispensable for membrane protein dislocation or substrate recognition. Why would TM2 replacement cause a more severe defect than TM deletion in membrane dislocation? Augustin et al. (25) have shown that coordinated intersubunit signaling between AAA+ domains of Yta12 and Yta10 is especially critical for membrane dislocation. TM2 of Yta12 is located upstream of the AAA+ domain (Fig. 1A). If the TM2 replacement causes subtle misalignment between the AAA+ domains of Yta12 and Yta10, then it could impair efficient ATP hydrolysis relay and subsequent membrane dislocation. However, in the complete absence of a TM, the AAA+ domain of Yta12 could be better positioned via interaction with the AAA+ domain of Yta10.
Compared with Yta12, TM2 replacement of Yta10 impaired the membrane dislocation of Mgm1(A/L) variants only. Because dislocation of the Ccp1 and Cox5aT-MFP variants is normal, the dislocation function of the m-AAA protease is not abolished. Thus, these results imply that replacement of TM2 of Yta10 causes impairment in the process of selective substrate recognition.
Because Yta10 and Yta12 share high sequence homology (supplemental Fig. S1), at present we cannot exclude the possibility that TM2 of Yta10 and Yta12 has a redundant function in the recognition of membrane substrates. Although most substrates need one or the other, for some substrates, such as Mgm1(A/L), require both. In the case of the i-AAA protease Yme1, it has been shown that both N- and C-terminal helical domains serve to recognize different degradation substrates (26, 27). Given that the m-AAA protease plays a diverse role in mitochondrial proteostasis, it would not be surprising if it has multiple modes of substrate recognition. Further study is needed to clarify this issue.
Next, we probed the length of the hydrophilic polypeptides the m-AAA protease can dislocate through the membrane by introducing different lengths of hydrophilic linker in the m-AAA protease substrate. Our results show that the dislocation efficiency is decreased significantly as the hydrophilic extension lengthens. We reason that, if the dislocation occurs in a proteinaceous pore, then it would be less sensitive to the length of the hydrophilic moiety; however, if the dislocation occurs in the lipid bilayer, then the presence of a long hydrophilic segment would be energetically too costly. Hence, these results implicate that the m-AAA protease dislocates a TM segment from the lipid environment and that it would be incapable of dislocating a large IMS domain across the membrane. Subsequently, it can be predicted that, for the m-AAA protease to extract IM proteins for degradation in the matrix, either the IMS domain should be small or the large IMS domain should be cleaved off by corroborating with other proteases prior to extraction from the membrane.
Experimental procedures
Yeast strains
W303-1a (MATa, ade2, can1, his3, leu2, trp1, and ura3) was used as a parental strain in this study. yta10Δ (MATa, ade2, can1, his3, leu2, trp1, ura3, yta10::HIS3MX6) was made by standard homologous recombination, substituting YTA10 with an amplified HIS3MX6 (28). The primers used for the amplification of the HIS3MX6 cassette were 5′-CAGCGTTTGCAGACGTTATCTCGGATCCCCGGGTTAATTAA-3′ and 5′-TTGGGTAGAACGGTGTATTGTGTTGAATTCGAGCTCGTTTAAAC-3′. yta10Δ was used to make yta10Δyta12Δ (MATa, ade2, can1, his3, leu2, trp1, ura3, yta10::HIS3MX6, yta12::KanMX6) by substituting YTA12 with KanMX6. The primers used for the amplification of the KanMX6 cassette were 5′-TATCGGTTCGTTCAATAAGAAAGTC-3′ and 5′-GCCCTTAAGATGACCTACGTTTATT-3′. These yeast strains were cultured at 30 °C in this study.
Plasmid construction
YTA10 was amplified from the genomic DNA using the set of primers 5′-ATAGGGCGAATTGGAGCTCCACCGCGGTGGCGTTGTACATATATCTGCT-3′ and 5′-TATCGATAAGCTTGATATCGAATTCCTGCAGGATTTAATAAATGAAGGTGTT-3′. YTA12 was amplified from the genomic DNA using the set of primers 5′-GGTGGCGGCCGCTCTAGAACTAGTGGATCCACAGCGCGATACAATTTTC-3′ and 5′-ATCGATAAGCTTGATATCGAATTCCTGCAGAGGGAGTAGATTTGAAGTCTC-3′.
The set of primers amplified YTA10 and YTA12 with 1 kb upstream sequences and 500 bp downstream sequences to include endogenous promoters and potential transcriptional regulators and contained nucleotides for cloning the amplified product into pRS314 or pRS316, respectively, by homologous recombination ([pRS314 YTA10 WT], [pRS316 YTA12 WT]).
The TMs of Yta10 were replaced with a TM of Mdl2 using [pRS314 YTA10 WT] as template for site-directed mutagenesis. Two sets of primers used to replace TMs of Yta10 were 5′-TTAACCATATCATGTTCCATAGGCATGTCTTCCAGTAACAACTCAGGAGAC-3′ with 5′-AAGAAGTATGGCTGTAAGAAGCAGTTTCCAATCTTCCTTAGATCTGAAGTATTCTGATAA-3′ and 5′-GCTGCTAATTTTGGTAGATTTATATTATTGAGAAAAATAAATAGTTCACCACCA-3′ with 5′-ACAACCAATCAGTAAAGCAACAGTAAAAAAGGAAGAAGATCTCTCAATGTATTTGATG-3′, each used to replace TM1 and TM2 of Yta10 with a TM of Mdl2, respectively. In the same manner, 5′-TTAACCATATCATGTTCCATAGGCATGTCTAACAGTTTGGAAGAGCAAAGTG-3′ with 5′-AAGAAGTATGGCTGTAAGAAGCAGTTTCCAATCGTTAACATTTTTCGATACAGGATTACT-3′ and 5′-GCTGCTAATTTTGGTAGATTTATATTATTGCAAGAAGATCGGCACAAGC-3′ with 5′-ACAACCAATCAGTAAAGCAACAGTAAAAAAGGATTTAGCCCAATTGCCTTCTTG-3′ were used to replace TM1 and TM2 of Yta12 with a TM of Mdl2 using [pRS316 YTA12 WT] as a template for site-directed mutagenesis.
The Mgm1, Ccp1, and Cox5aT model proteins used in previous studies were adopted (12, 29). To make Mgm1 with a P2 domain inserted between residues 130 and 131 of Mgm1 WT and Mgm1(A/L), overlap PCR was performed (30). First, the N-terminal part of Mgm1 WT and Mgm1(A/L) was amplified using 5′-TGTTACGCATGCAAGCTTGATATCGAAATGAGTAATTCTACTTCATTAAGG-3′ and 5′-TAAATCCTTGATTCGATCTAGTTT-3′. The C-terminal part of Mgm1WT was amplified using 5′-GGTGAATCGATGAAGGAAAAG-3′ and 5′-TTAGAGAGCGTAATCTGGAAC-3′.
The P2 domain was amplified from LepH2 protein (31) using the primer 5′-AAACTAGATCGAATCAAGGATTTATGCAGTTCCGGCCAG3-′ in combination with 5′-CTTTTCCTTCATCGATTCACCAACGAAATCGCTCGGTTC-3′or 5′-CTTTTCCTTCATCGATTCACCATTTTCTTTGGTTTCCTGTTT-3′ or 5′-CTTTTCCTTCATCGATTCACCGTTGTCGCCCATCATGAA-3′ to generate different lengths of hydrophilic P2 stretches.
The N-terminal part of Mgm1 WT and Mgm1(15A/4L) was each fused to the P2 domain by overlap PCR, and the stitched products were further annealed to the C-terminal part of Mgm1 WT. The resulting PCR product was cloned into the pHP84HA plasmid (32) by homologous recombination.
Growth assay
yta10Δ expressing [pRS314 YTA10 variants] were cultured in -Trp(glucose) medium overnight at 30 °C. The overnight culture was diluted to 0.1 A600 and cultured to 0.5 A600 at 30 °C. 10 μl of culture was spotted on YPD (yeast extract peptone dextrose) or YPEG (yeast extract peptone ethanol, glycerol) plate and further incubated for 2 days at 30 °C. Photos were taken on a Chemi-doc-XRS+ system using Epi Light (Bio-Rad).
yta10Δ/yta12Δ [pRS314 YTA10 WT] expressing [pRS316 YTA12 variants] were cultured in -Trp -Ura (glucose) medium overnight at 30 °C. The overnight culture was diluted to 0.1 A600 and cultured to 0.5 A600 at 30 °C. 10 μl of culture was spotted on YPD or YPEG (ethanol/glycerol) plates and further incubated for 2 days at 30 °C. Photos were taken on a Chemi-doc-XRS+ system using Epi Light (Bio-Rad).
Protein preparation and Western blotting
Preparation of lysates, SDS-PAGE, and Western blotting were conducted as described previously (22). Briefly, yeast transformants were grown overnight in 5 ml of synthetic complete medium at 30 °C. Proteins were precipitated from 1 A600 unit of yeast cells by addition of trichloroacetic acid (TCA) (Sigma). Precipitated proteins were resuspended in 40 μl of sample buffer and incubated for 5 min at 95 °C prior to SDS-PAGE. The samples were separated on 6.5% or 12.5% Tris-HCl gels (Bio-Rad) and followed by Western blotting. Membranes were immunodecorated with an anti-HA antibody (Covance) and developed with the Amersham Bioscience advanced ECL kit on a Chemi-doc-XRS+ system (Bio-Rad). Quantification of detected bands was done using Image Lab 5.0 (Bio-Rad).
MrpL32 processing assay
From overnight cell cultures of yta10Δ expressing [pRS314 YTA10 variants] and yta10Δ/yta12Δ [pRS314 YTA10 WT] expressing [pRS316 YTA12 variants], whole-cell lysates were prepared by resuspending cells in sample buffer (50 mm Tris-HCl, 5% SDS, 5% glycerol, 50 mm DTT, 5 mm EDTA, 2 μg/ml leupeptin, 2 μg/ml pepstatin A, 1 μg/ml chymostatin, 0.15 mg/ml benzamidine, 0.1 mg/ml Pefabloc, 8.8 μg/ml aprotinin, 3 μg/ml Anitpain and bromphenol blue) and heating for 15 min at 60 °C. Prior to gel loading, the samples were centrifuged at 14,000 rpm for 5 min, and the supernatant fractions were loaded onto 15% Tris-HCl gels (Bio-Rad). Western blotting was followed with anti-MrpL32 antiserum (a kind gift from Drs. Langer and Tatsuta from the University of Cologne).
Author contributions
H. L. and H. K. conceived the project. H. L., S. L., S. Y., and H. K. designed the study, analyzed data, and wrote the manuscript. H. L., S. L., and S. Y. conducted the experiments.
Supplementary Material
Acknowledgments
We thank YeEun Kim, Hyeonseong Kim, Kwangjin Park, and Kyungyeun Song for initiating the project, the current laboratory members for critical reading of the manuscript, and Drs. Thomas Langer and Takashi Tatsuta (University of Cologne) for providing the anti-MrpL antibody.
The research was funded by National Research Foundation of Korea Grant NRF-2016R1A2B2013459 and a grant from the Promising-Pioneering Researcher Program through Seoul National University (to H. K.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Fig. S1.
- IM
- inner membrane
- AAA
- ATPase associated with diverse cellular activities
- IMS
- intermembrane space
- TM
- transmembrane
- MFP
- Mgm1 fusion protein.
References
- 1. Gerdes F., Tatsuta T., and Langer T. (2012) Mitochondrial AAA proteases: towards a molecular understanding of membrane-bound proteolytic machines. Biochim. Biophys. Acta 1823, 49–55 [DOI] [PubMed] [Google Scholar]
- 2. Tatsuta T. (2009) Protein quality control in mitochondria. J. Biochem. 146, 455–461 [DOI] [PubMed] [Google Scholar]
- 3. Baker M. J., Tatsuta T., and Langer T. (2011) Quality control of mitochondrial proteostasis. Cold Spring Harb. Perspect. Biol. 3, a007559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Janska H., Kwasniak M., and Szczepanowska J. (2013) Protein quality control in organelles: AAA/FtsH story. Biochim. Biophys. Acta 1833, 381–387 [DOI] [PubMed] [Google Scholar]
- 5. Leonhard K., Herrmann J. M., Stuart R. A, Mannhaupt G., Neupert W., and Langer T. (1996) AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 15, 4218–4229 [PMC free article] [PubMed] [Google Scholar]
- 6. Shah Z. H., Hakkaart G. A., Arku B., de Jong L., van der Spek H., Grivell L. A., and Jacobs H. T. (2000) The human homologue of the yeast mitochondrial AAA metalloprotease Yme1p complements a yeast yme1 disruptant. FEBS Lett. 478, 267–270 [DOI] [PubMed] [Google Scholar]
- 7. Lee S., Augustin S., Tatsuta T., Gerdes F., Langer T., and Tsai F. T. (2011) Electron cryomicroscopy structure of a membrane-anchored mitochondrial AAA protease. J. Biol. Chem. 286, 4404–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arlt H., Tauer R., Feldmann H., Neupert W., and Langer T. (1996) The YTA10–12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85, 875–885 [DOI] [PubMed] [Google Scholar]
- 9. Bonn F., Tatsuta T., Petrungaro C., Riemer J., and Langer T. (2011) Presequence-dependent folding ensures MrpL32 processing by the m-AAA protease in mitochondria. EMBO J. 30, 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tatsuta T., Augustin S., Nolden M., Friedrichs B., and Langer T. (2007) m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J. 26, 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Esser K., Tursun B., Ingenhoven M., Michaelis G., and Pratje E. (2002) A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1. J. Mol. Biol. 323, 835–843 [DOI] [PubMed] [Google Scholar]
- 12. Botelho S. C., Tatsuta T., von Heijne G., and Kim H. (2013) Dislocation by the m-AAA protease increases the threshold hydrophobicity for retention of transmembrane helices in the inner membrane of yeast mitochondria. J. Biol. Chem. 288, 4792–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Korbel D., Wurth S., Käser M., and Langer T. (2004) Membrane protein turnover by the m-AAA protease in mitochondria depends on the transmembrane domains of its subunits. EMBO Rep 5, 698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kihara A., Akiyama Y., and Ito K. (1999) Dislocation of membrane proteins in FtsH-mediated proteolysis. EMBO J. 18, 2970–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leonhard K., Guiard B., Pellecchia G., Tzagoloff A., Neupert W., and Langer T. (2000) Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol. Cell 5, 629–638 [DOI] [PubMed] [Google Scholar]
- 16. Nolden M., Ehses S., Koppen M., Bernacchia A., Rugarli E. I., and Langer T. (2005) The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 123, 277–289 [DOI] [PubMed] [Google Scholar]
- 17. Williams C. C., Jan C. H., and Weissman J. S. (2014) Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 346, 748–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Herlan M., Bornhövd C., Hell K., Neupert W., and Reichert A. S. (2004) Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J. Cell Biol. 165, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Herlan M., Vogel F., Bornhovd C., Neupert W., and Reichert A. S. (2003) Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 278, 27781–27788 [DOI] [PubMed] [Google Scholar]
- 20. Schäfer A., Zick M., Kief J., Steger M., Heide H., Duvezin-Caubet S., Neupert W., and Reichert A. S. (2010) Intramembrane proteolysis of Mgm1 by the mitochondrial rhomboid protease is highly promiscuous regarding the sequence of the cleaved hydrophobic segment. J. Mol. Biol. 401, 182–193 [DOI] [PubMed] [Google Scholar]
- 21. Botelho S. C., Osterberg M., Reichert A. S., Yamano K., Björkholm P., Endo T., von Heijne G., and Kim H. (2011) TIM23-mediated insertion of transmembrane α-helices into the mitochondrial inner membrane. EMBO J. 30, 1003–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park K., Botelho S. C., Hong J., Österberg M., and Kim H. (2013) Dissecting stop transfer versus conservative sorting pathways for mitochondrial inner membrane proteins in vivo. J. Biol. Chem. 288, 1521–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leonhard K., Stiegler A., Neupert W., and Langer T. (1999) Chaperone-like activity of the AAA domain of the yeast Yme1 AAA protease. Nature 398, 348–351 [DOI] [PubMed] [Google Scholar]
- 24. Paetzel M., Goodall J. J., Kania M., Dalbey R. E., and Page M. G. (2004) Crystallographic and biophysical analysis of a bacterial signal peptidase in complex with a lipopeptide-based inhibitor. J. Biol. Chem. 279, 30781–30790 [DOI] [PubMed] [Google Scholar]
- 25. Augustin S., Gerdes F., Lee S., Tsai F. T., Langer T., and Tatsuta T. (2009) An intersubunit signaling network coordinates ATP hydrolysis by m-AAA proteases. Mol. Cell 35, 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Graef M., Seewald G., and Langer T. (2007) Substrate recognition by AAA+ ATPases: distinct substrate binding modes in ATP-dependent protease Yme1 of the mitochondrial intermembrane space. Mol. Cell. Biol. 27, 2476–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rampello A. J., and Glynn S. E. (2017) Identification of a degradation signal sequence within substrates of the mitochondrial i-AAA protease. J. Mol. Biol. 429, 873–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 29. Kim H., Botelho S. C., Park K., and Kim H. (2015) Use of carbonate extraction in analyzing moderately hydrophobic transmembrane proteins in the mitochondrial inner membrane. Protein Sci. 24, 2063–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oldenburg K. R., Vo K. T., Michaelis S., and Paddon C. (1997) Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25, 451–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee H., Min J., von Heijne G., and Kim H. (2012) Glycosylatable GFP as a compartment-specific membrane topology reporter. Biochem. Biophys. Res. Commun. 427, 780–784 [DOI] [PubMed] [Google Scholar]
- 32. Kim H., Park H., Montalvo L., and Lennarz W. J. (2000) Studies on the role of the hydrophobic domain of Ost4p in interactions with other subunits of yeast oligosaccharyl transferase. Proc. Natl. Acad. Sci. U.S.A. 97, 1516–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.