Abstract
Transcriptional regulation involves both positive and negative regulatory elements. The Dig1 negative regulators are part of a fungal-specific module that includes a transcription factor (a Ste12 family member) and a Dig1 family member. In S. cerevisiae the post-genome-duplication Dig1/Dig2 proteins regulate MAP kinase controlled signaling pathways involved in mating and filamentous growth. We have identified the single Dig1 ortholog in the fungal pathogen Candida albicans. Genetic studies and transcriptional profiling experiments show that this single protein is implicated in regulation of MAP kinase-controlled processes involved in mating, filamentous growth and biofilm formation, and also influences cAMP-regulated processes. This suggests that the multiple cellular roles of the Dig1 protein are ancestral, and predate the sub-functionalization apparent in S. cerevisiae after the genome duplication. Intriguingly, even though loss of Dig1 function in C. albicans enhances filamentous growth and biofilm formation, colonization of the murine gastrointestinal tract is reduced in the mutant. The complexity of the processes influenced by Dig1 in C. albicans, and the observation that Dig1 is one of the few regulatory proteins that were retained in the duplicated state after the whole genome duplication event in yeast, emphasizes the important role of these negative regulators in fungal transcriptional control.
Graphical abstract
Introduction
Tight control of transcriptional activity permits microorganisms to tolerate and adapt to environmental changes and survive complex challenges from sources such as host immune defenses. Saccharomyces cerevisiae Ste12p is the founding member of the Ste12-like protein family that contains fungal-specific transcription factors typically involved in the positive regulation of filamentation and mating. Activation of the S. cerevisiae Ste12p transcription factor is controlled both by the inhibitory proteins Dig1/2p and by post-translational modification (phosphorylation), while in the wider fungal kingdom, alternative splicing also plays a role in the control of some family members (reviewed in (1)) reflecting the need for proper regulation of this important class of transcription factors for normal cell function.
In addition to S. cerevisiae, several closely related species are predicted to have maintained two Dig paralogs after the whole genome duplication event. These include Saccharomyces paradoxus, Saccharomyces mikatae and Saccharomyces bayanus, suggesting a central role for the Dig1p family members as transcription factor regulators. Interestingly, S. cerevisiae Dig1p and Dig2p appear to have subfunctionalized, as they form two distinct complexes; Tec1p-Ste12p-Dig1p which is responsible for filamentous growth through the activation of genes containing a Tec1-binding site (TCS), and Dig2p-Ste12p-Dig1p which regulates mating genes (2).
Candida albicans is a versatile opportunistic fungal pathogen that is capable of causing superficial to severe systemic infections in humans. C. albicans can reversibly switch between yeast and filamentous morphologies such as hyphae and pseudohyphae in response to environmental cues and this adaptability is thought to contribute to its virulence. C. albicans also undergoes an epigenetic white-opaque switch that is regulated in part by the Mating Type-Like (MTL) locus (3–5). Switching is heritable and stochastic between the two phenotypic states in both MTLa and MTLα cells. In contrast, MTLa/α cells are sterile and the switch to the opaque-phase is inhibited by a transcriptional complex comprised of the homeodomain proteins Mtla1p and Mtlα2p (5).
Regulation of filamentation in white cells is largely controlled by two main signaling pathways; the cyclic-AMP/Protein kinase A (cAMP/PKA) pathway and a mitogen-activated protein kinase (MAPK) pathway (6–11). The cAMP/PKA pathway activates the transcription factor Efg1p, which is a major regulator of the yeast-hyphal transition and a repressor of the white-opaque switch (12–14). MAPK pathway signals converge on two transcription factors, Cph1p and Tec1p, which are required for hyphal growth, biofilm formation and invasive growth (6, 15–17). Several components of the filamentous MAPK signaling cascade are also members of the mating MAPK pathway, and pheromone-induced signals are transduced to activate Cph1p and subsequently turn on genes required for mating (18).
Here we identify and characterize the single C. albicans orthologue of ScDig1p and ScDig2p and show that CaDig1p is a negative regulator of filamentous growth and colonization of the murine gastrointestinal tract. These studies therefore establish that Dig1p function contributes significantly to the commensalism of C. albicans. Furthermore, we provide evidence that in the absence of CaDig1p, MTLa cells are constitutively activated for the pheromone response and can mate with both MTLa and MTLα cell types, showing that CaDig1p regulates both filamentation and mating through the MAPK pathway. We also identified a genetic interaction between CaDIG1 and the cAMP-PKA pathway, raising the intriguing possibility that CaDig1p is a key component of two separate signaling cascades in C. albicans, and that in the process of duplication and subfunctionalization the resulting S. cerevisiae protein set lost this function. It thus appears that the single Dig1 protein of C. albicans connects to more regulatory circuits than the two orthologs in S. cerevisiae.
Results
Deletion of ORF19.1666 in Candida albicans results in invasive growth
In order to identify possible C. albicans orthologs of S. cerevisiae Dig1p and Dig2p, protein sequence data was obtained from the Saccharomyces Genome Database (SGD) (http://www.yeastgenome.org/), and compared with the genomes of various species available in the SGD Fungal BLAST tool. We identified a previously uncharacterized C. albicans open reading frame designated ORF19.1666, which is syntenic with ScDIG2. Global alignment of the Orf19.1666 protein sequence revealed 17 % percentage identity with ScDig1p and 18 % with ScDig2p. We proceeded to construct a homozygous deletion mutant of ORF19.1666 that we refer to here as dig1Δ/dig1Δ. We used an MTLa/α strain, and replaced one allele of DIG1 with the ARG4 selectable marker and the second allele with HIS1 to create strain HJR8 (Strains used in the study are listed in Supplementary Table S1). Correct integrations of the various disruption cassettes at the DIG1 locus were confirmed by colony PCR. The oligos used in these experiments are listed in Supplementary Table S2, and the plasmids used are listed in Supplementary Table S3.
dig1Δ/dig1Δ colonies from HJR8 were similar to those of Scdig1Δ/dig2Δ double mutants described in S. cerevisiae (19, 20) in that they were crenulated and hard to the touch, with numerous hyphal-like projections emanating from the border. We also observed that dig1Δ/dig1Δ colonies were invasive on YPD-agar. Invasive strains are often associated with filamentous growth (8), which is in part regulated by the MAPK pathway in C. albicans. As shown in the right panel of Figure 1A, when grown to stationary phase in liquid YPD medium at 30°C, cells of C. albicans dig1Δ/dig1Δ exhibited a mixed morphology resembling opaque-phase C. albicans cells and a combination of pseudohyphal and highly-polarized cells that are similar in appearance to both mating and hyphal projections based on the lack of non-constricted, parallel-sided cell walls, in contrast to the wild type cells in the left panel of Figure 1A (11, 21, 22). In order to determine if the dig1Δ/dig1Δ cells also exhibit the characteristic pimpled cell-surface of opaque cells (4), we used scanning electron microscopy (SEM) to visualize the cells at high resolution (Figure 1B). Wild-type white and opaque phase cells were included for comparison. Cells of the dig1Δ/dig1Δ strain exhibited a slightly larger, more elongated cellular morphology than that of white cells but lacked the pimpled surface associated with true opaque phase cells suggesting that the morphology of these cells is neither classically white nor opaque. We confirmed that the cell and colony morphology changes were the result of the dig1 mutation by reintroducing the DIG1 gene into a disruption strain as shown in supplementary figure 1.
Figure 1. Imaging.
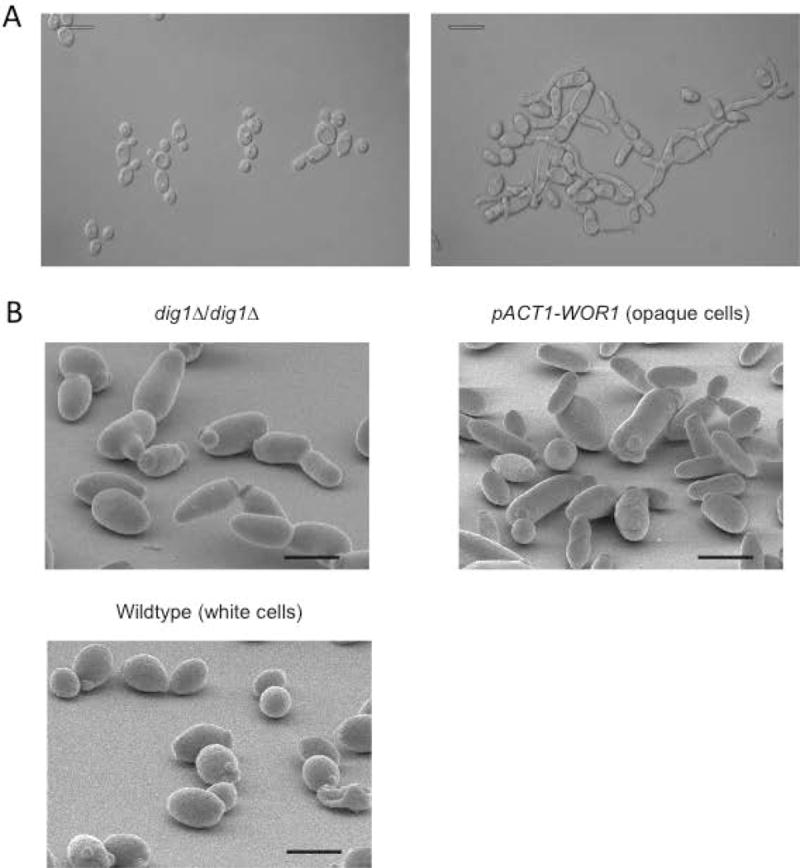
(A) Overnight cultures of wild-type SN148 (right panel) or HJR8 dig1Δ/dig1Δ (left panel) imaged by Differential Interference Contrast (DIC) microscopy show mixed morphology of the mutant cells. Scale bar 10 μm.
(B) C. albicans strain HJR8 dig1Δ/dig1Δ was inoculated onto YPD agar and incubated at 30 °C for 24 hours before processing for scanning electron microscopy; comparison of the dig1Δ strain with opaque (pACT1-WOR1) and white phase cells. Scale bar 15 μm.
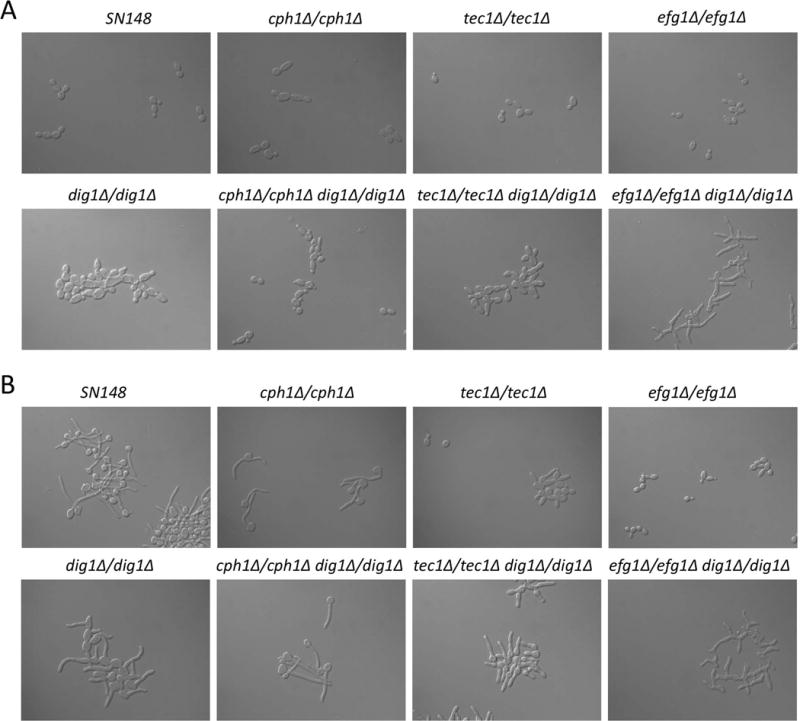
Depending on the environmental cues and cell type, the MAPK signal activates filamentation programs transduced by the transcription factors Cph1p and Tec1p (6, 15). A third key transcription factor, Efg1p, is activated by a second signaling cascade, designated the cAMP-PKA pathway (12). In order to determine if Dig1p is influencing these pathways during invasive growth, we constructed double homozygous deletions of CPH1, TEC1 or EFG1 in wild type and homozygous dig1Δ mutant backgrounds. The PCR-confirmed double knockouts were used in plate-washing assays to test for invasive growth in both YPD and Spider medium using single knockout mutants as controls (Figure 2). After 5 days of growth at 30 °C or 37 °C the homozygous dig1Δ and homozygous double mutant dig1Δ/cph1Δ colonies were unperturbed by washing and thus highly invasive, while the dig1Δ/efg1Δ and dig1Δ/tec1Δ homozygous double mutant colonies were somewhat eroded. The wild type and single mutant deletions of cph1Δ, tec1Δ and efg1Δ all behaved in a similar manner, showing a low level of resistance to the plate-washing assay, and thus a low level of invasiveness. The extreme resistance of the dig1Δ/cph1Δ mutant to the plate-washing assay suggests that Cph1p is not needed for invasive growth in the absence of Dig1p.
Figure 2. Invasive growth assays.
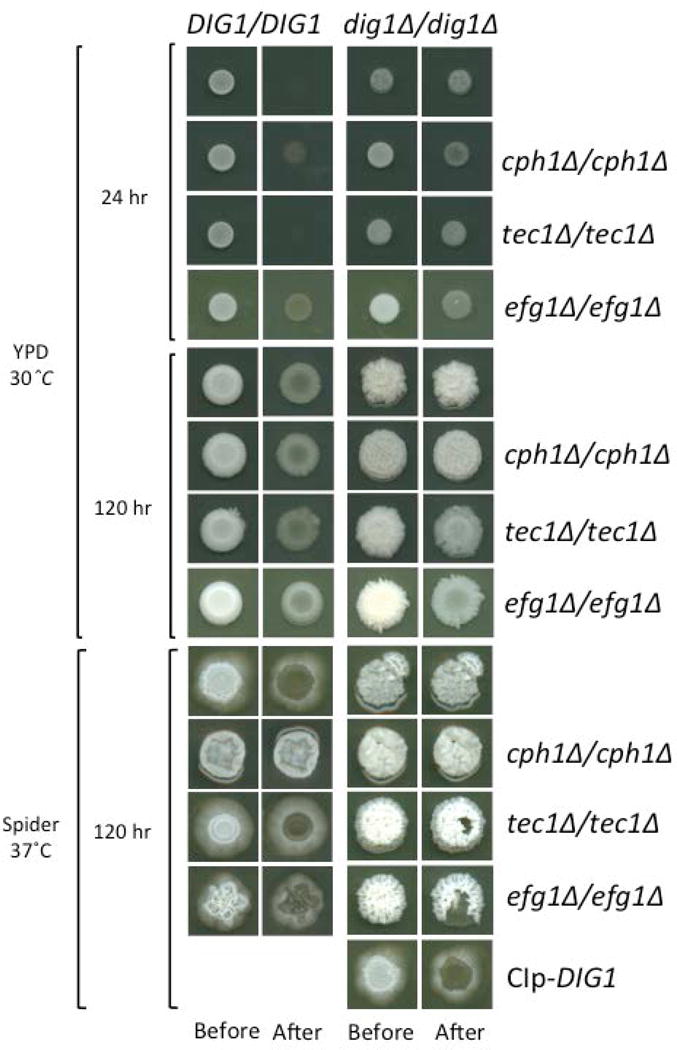
Strains were spotted on YPD and Spider plates and grown for the specified times (before) prior to washing with a stream of water for 15 seconds (after). After 120 h the dig1 mutant and the dig1Δ/cph1Δ double mutant were highly invasive and particularly resistant to washing. All strains are MTLa/α.
Dig1p is a negative regulator of biofilm formation
C. albicans forms biofilms on implanted medical devices such as in-dwelling catheters (for a review see (23)). Biofilms provide a controlled microenvironment that can promote cell-cell signaling and allow circumvention of host immune responses (24, 25). In addition to their invasive properties, dig1Δ cells have a tendency to flocculate and stick to the walls of glass and polypropylene vessels routinely used for culturing C. albicans in the laboratory. This characteristic suggested that Dig1p might negatively regulate biofilm formation, and biofilm formation is influenced by both Cph1p and Tec1p (16, 26). To investigate this further, we first tested the ability of strains deleted for DIG1 to form biofilms; a wild type MTLa strain and two independently constructed MTLa dig1Δ strains, CAY3570 and CAY3571, were induced to form biofilms for 24–48 hours in the presence or absence of pheromone (27) (Figure 3A and B). Under these conditions both dig1Δ strains formed significantly more robust biofilms when compared with the wild type regardless of the presence or absence of pheromone induction, confirming a role for Dig1p in the modulation of biofilm formation.
Figure 3. Enhanced biofilm formation in dig1Δ mutants.
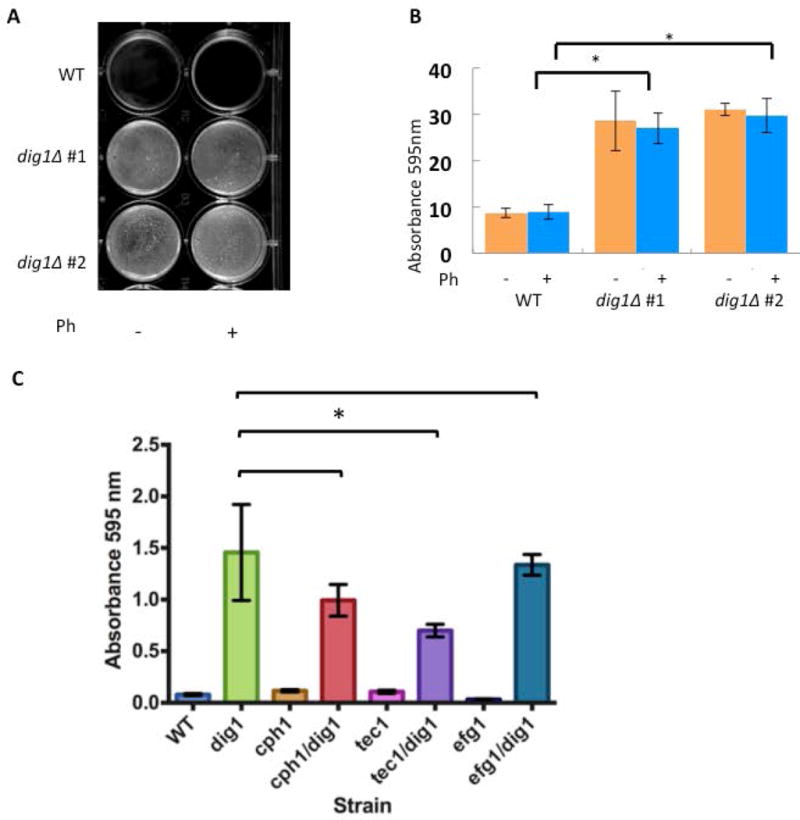
(A) Representative image of biofilm formation in two independent MTLa dig1Δ mutants (CAY3570 and CAY3571) and the wild type control (RBY1132). Cells were grown as pheromone-stimulated biofilms in SCD media for 48 hours at room temperature without shaking. Ph = 10 μM α-pheromone. Reports (16, 26) both describe increased sexual biofilm in the SC5314 homozygote MTLa control compared to the regular biofilm. We believe the lack of difference in our data might be due to the fact we are using a different wild type control.
(B) Cells from strains CAY3570 and CAY3571were washed with PBS and adherence quantified by crystal violet staining. *= p <.01, Student’s t-test. error bars = standard deviation. n= 3 biological replicates.
(C) To determine the role of each transcription factor (Cph1p, Tec1p, Efg1p) in the formation of dig1∆/dig1∆-enhanced biofilms, wild type, single and double homozygous deletion mutants were challenged to form biofilms in the absence of pheromone. Tec1p is the major contributor to the formation of enhanced dig1∆/dig1∆ biofilms. All strains are MTLa/α. Cells were incubated in 6 well plates in SCD media for 24–48 hours at room temperature without shaking, washed with PBS and stained with crystal violet. Absorbance at 595 nm was recorded. Student’s unpaired t-test. Standard deviation indicated by error bars, n=3. *=p < 0.05.
To better understand the role of Dig1p as a negative regulator of biofilm formation and to determine which transcription factor(s) are responsible for the biofilm program, we assayed biofilm formation in the homozygous disruption double mutant strains cph1Δ/dig1Δ, efg1Δ/dig1Δ and tec1Δ/dig1Δ, together with the single null mutant controls. We observed a significant increase in biofilm formation in the three double knock-outs compared with the respective single transcription factor null mutants, confirming that loss of Dig1p is sufficient to induce biofilm formation independently of these three transcription factors (Figure 3C). While none of the double mutant combinations prevented biofilm formation, each caused an overall reduction compared to the dig1Δ homozygous null, suggesting the redundancy was not absolute.
dig1Δ cells mate homothallically and with increased efficiency
We next investigated the effects of deleting DIG1 on aspects of the mating MAPK pathway. Previous studies in S. cerevisiae have described no discernable effect on the efficacy of mating in dig1/2Δ double or single mutants (19, 20). We performed a series of quantitative mating assays whereby the dig1Δ MTLa strain was crossed with white and opaque mating-testers from wild type MTLa and MTLα cells (Figure 4). We carried out the mating assays with MTLa dig1Δ cells, which we had previously observed to be pheromone-responsive without the need to specifically select for opaque form cells. The quantitative mating assay revealed an increased frequency of mating products in each cross when one of the mating partners was deleted for DIG1, indicating that Dig1p negatively regulates mating in C. albicans. Intriguingly, we also observed prototrophic mating products arising from MTLa-a crosses between MTLa dig1Δ cells and opaque cells of the wild type MTLa mating tester strain, suggesting that Dig1p is also a negative regulator of homothallic, same-sex mating in C. albicans.
Figure 4. Quantitative mating in the dig1Δ mutant.
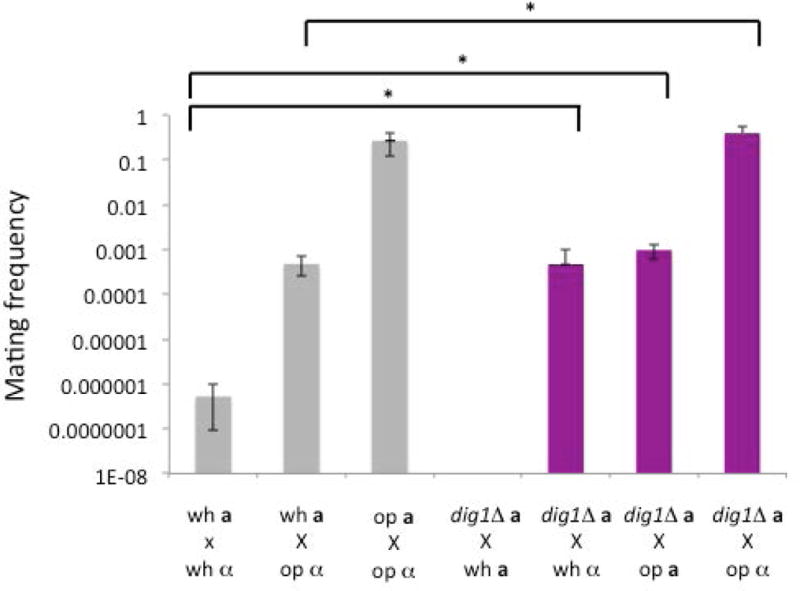
Quantitative mating assays of crosses between WT (SN148) or the dig1Δ MTLa (HJR6) strain and a white or opaque MTLα (3315) or MTLa (3745) mating tester. The crosses involving the dig1 mutants show an increase in mating frequency compared with the wild type white-opaque cross. The assay also identifies detectable homothallic mating between the dig1Δ MTLa strain and the MTLa mating tester strain; no mating is detected between normal MTLa strains mating with MTLa testers. wh, white cells. op, op cells. * = p <.05, Kruskal- Wallis. error bars, s.e.m. n= 3–6 biological replicates per cross.
Multiple signals promote dig1Δ cell mating
In S. cerevisiae, full activation of Ste12p is dependent on two mechanisms; the release of Dig1p/Dig2p from its activation domain and/or DNA binding domain, and the phosphorylation of critical residues on Ste12p by MAPKs Fus3p or Kss1p (28–31). To test if deletion of DIG1 alone is enough to activate Dig1p-related transcription factors, or if upstream signaling components also play a role, we deleted two members of the heterotrimeric G-protein responsible for transducing the pheromone signal through the MAPK cascade (32). Cag1p and Ste4p are the respective Gα and Gβ subunits of the G protein. We constructed MTLa dig1Δ/cag1Δ and dig1Δ/ste4Δ double mutants and performed halo and qualitative cross-patch mating assays. Neither MTLa dig1Δ/cag1Δ nor MTLa dig1Δ/ste4Δ strains generated a halo in response to α-pheromone (Figure 5A), and both were sterile in the mating assay (Figure 5B), suggesting that in addition to inactivation of repression from Dig1p, signaling through the mating MAPK pathway is essential for activation of the underlying transcription factors.
Figure 5. Halo and mating assays.
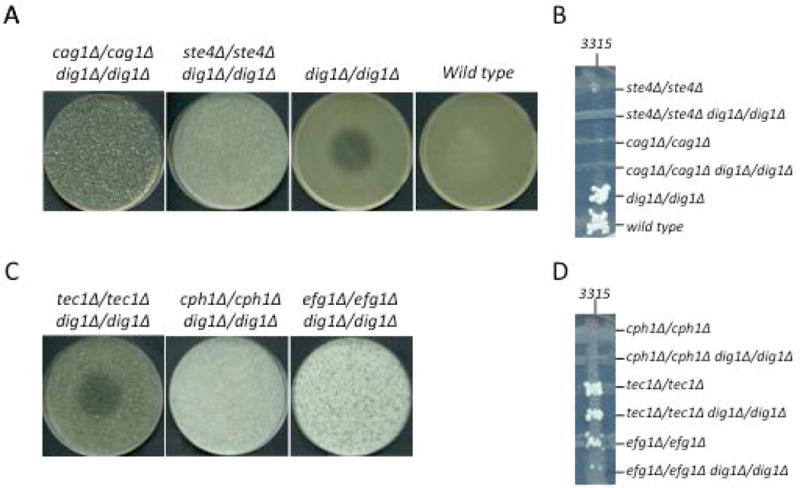
Each of the G-protein (Cag1p and Ste4p) and transcription factor (Tec1p, Cph1p, Efg1p) double and/or single mutants were assayed for their response to alpha-factor peptide (A, C) and their ability to mate with MTLα tester strain 3315 (B, D). Only dig1Δ/dig1Δ (row A) and tec1Δ/tec1Δ dig1Δ/dig1Δ (row B) were capable of arresting the cell cycle in response to pheromone, indicating that Tec1p is not required for transduction of the pheromone signal in the absence of Dig1p while Cag1p, Ste4p, Cph1p and Efg1p are. Cross-patch mating assays reveal that efg1Δ/efg1Δ dig1Δ/dig1Δ, tec1Δ/tec1Δ dig1Δ/dig1Δ and their respective single mutants are mating-competent, in addition to dig1Δ/dig1Δ and the SN148 wild type. All strains other than 3315 are MTLa.
Cph1p can complement the mating defect of S. cerevisiae ste12 mutants and is the main transcription factor controlling mating in C. albicans (6, 33, 34). Since the roles of Cph1p, Tec1p and Efg1p were interrelated in the invasive growth and biofilm assays, we endeavored to separate the function of these transcription factors by asking which of the three are required for the pheromone response and mating. MTLa/α strains are inherently sterile so MTLa versions of dig1Δ/cph1Δ, dig1Δ/efg1Δ and dig1Δ/tec1Δ were isolated for these experiments. When challenged with α-pheromone, MTLa dig1Δ/tec1Δ generated a halo whereas MTLa dig1Δ/cph1Δ and MTLa dig1Δ/efg1Δ did not (Figure 5C), indicating that both Cph1p and Efg1p are critical for the pheromone response and thus that signal transduction through the cAMP-PKA pathway is required in addition to MAPK signaling. Cross-patch mating assays revealed that MTLa dig1Δ/cph1Δ is sterile while MTLa dig1Δ/tec1Δ and MTLa dig1Δ/efg1Δ generated prototrophic mating products, although the latter exhibited a lower mating frequency than MTLa dig1Δ cells alone (Figure 5D). This suggests that while Tec1p and Efg1p are not essential for mating they help to promote it, at least in the dig1Δ mutant background. Taken together, these data suggest that MAPK and Efg1p-mediated signals work co-operatively to facilitate pheromone response and mating in a Dig1p-dependent manner.
dig1Δ strains are defective in colonization of the murine GI tract
Candida species effectively colonize healthy individuals and rates of carriage are estimated in the range of 53 to 75 % of the population (35, 36). That C. albicans has the capacity to adapt and undergo the yeast-to-hypha transition and/or a phenotypic switch in response to environmental cues is thought to contribute to its ability to persist within a host. Based on the mixed phenotype of the dig1Δ cell morphology and the propensity of the cells to form biofilms in vitro, we asked if C. albicans strains lacking Dig1p were capable of efficient colonization of the murine gastrointestinal tract. Mice were inoculated with either wild type or dig1Δ C. albicans cells, and colonization was measured in genome equivalents per gram of cecal contents one, three or six days post-inoculation (Figure 6). Initially, colonization of the murine GI tract by dig1Δ strains was generally greater than that of wild type cells, however this trend was not sustained and the number of dig1Δ genome equivalents/gram decreased considerably over time to the point where the fungal genomic DNA content of three out of five samples were below the limit of detection. In contrast, levels of genome equivalents/gram in mice inoculated with wild type C. albicans strains generally increased over time by several orders of magnitude. This data indicates that the dig1Δ strains are defective in the colonization of the mouse GI tract.
Figure 6. dig1Δ/dig1Δ is defective in colonization of the murine GI tract.
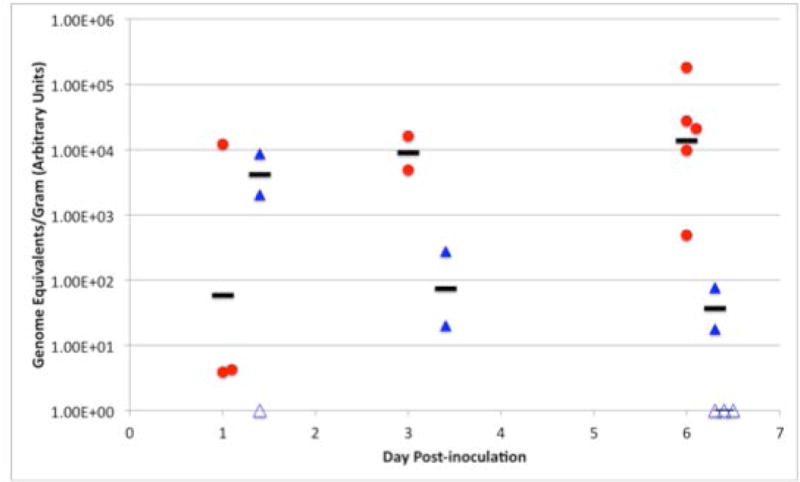
Mice were inoculated by oral gavage with wild type (JDM001, leu2−ura3−) or dig1Δ/dig1Δ (HJR6, leu2−ura3−) C. albicans strains and sacrificed after 1, 3 or 6 days before harvesting, extraction and quantification of genomic fungal DNA content. Each symbol represents cecal fungal burden for individual mice. Black bars indicate the geometric mean for samples that were positive for the C. albicans genomes. Open symbols indicate samples that were below the limit of detection. The Y-axis shows Genome equivalents per gram (measured in arbitrary units) as detected by real time PCR normalized to the wet weight of pelleted cecum contents. The X-axis shows Day post-inoculation. WT (red) and mutant (blue) samples were collected on the same day and are offset for clarity.
Gene expression analysis of Dig1
The phenotypic consequences of dig1Δ mutants and the genetic interactions with transcription factor mutants suggested that dig1 was influencing gene expression. We used microarray analysis to assess the direct consequences of dig1 deletion on gene expression patterns implicated in mating and filamentous growth using MTLa dig1Δ and MTLa/α dig1Δ strains. Of the top 50 genes expressed in each condition, 15 were in common (many encoding hyphal specific surface proteins, but also genes like CEK1 that are implicated in mating). This suggests that a significant component of the genes upregulated in the absence of Dig1 were independent of the status of the MTL locus (Table 1). In the 35 genes specific for MTLa dig1 cells, there were 10 that were associated with pheromone induction, including FUS3 and STE2, while in the 35 specific genes for the MTLa/α dig1 strain there were many uncharacterized genes, as well as a mix of pheromone-responsive genes such as FAV1, FAV2 and FAV3 and biofilm-induced genes like LDG8. Analysis using the Candida Genome Database Gene Ontology Slim Mapper, showed that the top 20% of significantly upregulated genes of in MTLa/α and MTLa strains were enriched for a variety of general biological processes including conjugation, cell wall organization, biofilm formation, pathogenesis, response to drugs, cell adhesion, filamentous growth, cell cycle and interspecies interaction between organisms (Supplementary Table S4).
Table 1.
Top 50 up regulated genes in dig1 mutant MTLa/alpha and MTLa strains. The top 50 genes in the a/α set were all greater than 10 fold above their respective controls, those in the MTLa cells were all greater than 2.5 fold above their respective controls. All genes selected had a P value of 0.1 or less, and are ranked by their fold induction.
| a cell specific | common | a/alpha cell specific | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gene | name | pheromone | filaments | biofilm | gene | name | pheromone | filaments | biofilm | gene | name | pheromone | filaments | biofilm |
| orf19.2164.1 | MFA1 | Y | orf19.3384 | Y | Y | orf19.7167 | ||||||||
| orf19.1616 | FGR23 | Y | Y | Y | orf19.1321 | HWP1 | Y | Y | Y | orf19.5401 | FLO9 | |||
| orf19.4481 | MFALPHA | Y | orf19.1327 | Y | Y | orf19.1401 | EAP1 | Y | Y | |||||
| orf19.36.1 | orf19.5302 | PGA31 | orf19.5404 | |||||||||||
| orf19.3736 | KAR4 | Y | orf19.1725 | orf19.5 | ||||||||||
| orf19.460 | CEK2 | Y | Y | orf19.4688 | DAG7 | Y | orf19.5402 | |||||||
| orf19.3829 | PHR1 | Y | orf19.2990 | XOG1 | Y | orf19.701 | CFL11 | |||||||
| orf19.720 | GST3 | Y | Y | orf19.6202 | RBT4 | Y | orf19.2131 | |||||||
| orf19.1156 | FUS1 | Y | orf19.2886 | CEK1 | Y | Y | orf19.5212 | |||||||
| orf19.669 | PRM1 | Y | orf19.756 | SAP7 | orf19.7305 | |||||||||
| orf19.4377 | KRE1 | Y | Y | orf19.4082 | DDR48 | Y | orf19.301 | PGA18 | ||||||
| orf19.6420 | PGA13 | Y | orf19.6274 | PBR1 | Y | Y | orf19.1562 | Y | ||||||
| orf19.842 | ASR3 | Y | orf19.1415 | FRE10 | Y | orf19.1914 | FAV3 | Y | ||||||
| orf19.1618 | GFA1 | orf19.7606 | orf19.2431 | Y | Y | |||||||||
| orf19.1486 | Y | orf19.3740 | PGA23 | orf19.4509 | LPF28 | |||||||||
| orf19.6287 | AAT21 | Y | orf19.3499 | LDG8 | Y | |||||||||
| orf19.4593 | RGA2 | Y | orf19.1844 | |||||||||||
| orf19.5517 | ADH7 | orf19.3369 | MOH1 | Y | Y | |||||||||
| orf19.7111.1 | SOD3 | Y | orf19.1600 | |||||||||||
| orf19.2444 | CHS7 | Y | orf19.5635 | PGA7 | Y | |||||||||
| orf19.1827 | Y | Y | orf19.1120 | FAV2 | Y | Y | ||||||||
| orf19.1779 | MP65 | Y | Y | orf19.5400 | ||||||||||
| orf19.5285 | PST3 | Y | orf19.3380 | HWP2 | Y | Y | ||||||||
| orf19.2843 | RHO1 | orf19.3444 | SGE13 | |||||||||||
| orf19.2685 | PGA54 | Y | Y | orf19.7609 | PGA11 | |||||||||
| orf19.7304 | orf19.4215 | FET34 | Y | |||||||||||
| orf19.4651 | PGA53 | orf19.7170 | ||||||||||||
| orf19.696 | STE2 | Y | orf19.6859 | |||||||||||
| orf19.1799 | GAP5 | orf19.5760 | IHD1 | Y | Y | |||||||||
| orf19.6563 | KCH1 | Y | Y | orf19.5645 | MET15 | Y | ||||||||
| orf19.3974 | PUT2 | Y | orf19.1367.1 | |||||||||||
| orf19.1690 | TOS1 | Y | orf19.3801 | FAV1 | Y | |||||||||
| orf19.6527 | PRM10 | Y | Y | orf19.1636 | STE50 | Y | ||||||||
| orf19.2480.1 | AUT7 | Y | orf19.7550 | IFA14 | Y | |||||||||
| orf19.5307 | JEN2 | Y | orf19.5305 | RHD3 | ||||||||||
To further investigate the effect of dig1 deletion on aspects of mating, we identified 48 genes that were designated as being pheromone responsive in at least two independent studies to define a pheromone regulon (37, 38). We then compared these genes with our data set and found that half of these pheromone-response genes were upregulated in dig1Δ MTLa cells. A further 21 genes that were induced in these dig1Δ cells were also induced in one but not both of the other studies, suggesting that the pheromone response is inherently activated in the absence of DIG1 (Figure S3).
As our results also suggest that Dig1 is involved in biofilm formation, we examined our dig1Δ data sets for enrichment of genes that are known to be targets of Efg1, Tec1 or Cph1 regulation during this process (Supplementary Table S5). We compiled two gene regulons using data from Nobile et al (39) that identified genes that are downregulated in efg1Δ or tec1Δ mutants during biofilm formation and are also binding targets of Efg1 or Tec1, respectively, as determined by ChIP-chip experiments. The Efg1 regulon consisted of 179 genes in total, 60 of which were upregulated in dig1Δ MTLa/α cells, while the Tec1 regulon comprised 36 genes, 18 of which were also upregulated in dig1Δ MTLa/α cells. Similarly we used transcriptional profiling data from Lin et al (16) to find genes that are up-regulated during pheromone-stimulated biofilm formation, which is principally controlled by Cph1, and also to identify downregulated genes in a cph1Δ mutant. Lin et al (16) demonstrate that deletion of CPH1 eliminates the transcriptional response to biofilm formation and as such only eight genes are downregulated more than 2-fold under pheromone-stimulated biofilm-forming conditions, all of which are uncharacterized or dubious open reading frames (19.1821, 19.1302, 19.6450, 19.1450, 19.4094, 19.6970, 19.6115 and 19.50) and four of which are upregulated in MTLa/α cells 19.1302, 19.6450, 19.4094, 19.6115). However in wild-type cells, 52 genes are significantly up regulated, 24 of which are also induced in dig1Δ MTLa cells and 43 in dig1Δ MTLa/α cells. Overall, this data provides good transcriptional evidence of a role for Dig1 in the negative regulation of filamentous growth, pheromone response, and processes such as biofilm formation in concert with the transcription factors Tec1, Efg1 and Cph1.
Discussion
Candida albicans is an opportunistic fungal pathogen capable of causing superficial and systemic infections in humans. The ability of C. albicans to switch between various morphological forms depending on its host environment is thought to contribute to its virulence as filamentous growth states are associated with tissue invasion, biofilm formation, evasion of innate host defences and mating. Although the mechanisms of activation of filamentous growth pathways have been extensively studied, less is known about which factors control the negative regulation of filamentation. In this study, we have identified a previously uncharacterized Orf that shares sequence similarity with Saccharomyces cerevisiae Dig1p and Dig2p. Deletion of the gene encoding this Orf triggers invasive growth in C. albicans and so we have retained the yeast designation of Dig1 (for Down-regulation of Invasive Growth). Mutants lacking CaDIG1 form cultures of hyperpolarized cells, form robust biofilms, are highly invasive in vitro but defective at colonization in vivo and are de-repressed for both heterothallic and homothallic mating. Furthermore, when we delete key transcription factors thought to act downstream of Dig1p we find evidence to suggest that CaDig1 influences filamentation and mating through multiple signaling pathways including those controlled by MAPK and by cAMP-PKA pathways. This reveals a previously unappreciated level of complexity and plasticity between the two major signaling pathways and suggests a potential genetic link between the two through DIG1-EFG1 interactions. A genetic interaction between ScDIG1/2 and the cAMP-PKA-modulated transcription factor Efg1 ortholog Sok2 has not been observed in S. cerevisiae (19, 20). It is possible the DIG1-EFG1 interaction represents an ancestral program that was lost at some point in the S. cerevisiae lineage; alternatively, CaDig1 may have gained this extra function after divergence from a common ancestor. Further genetic analyses of multiple species would be required to test this hypothesis.
In C. albicans, Cph1p, Tec1p and Efg1p all contribute to activation of filamentous growth (8, 9, 11). Intriguingly, our data reveals that co-deletion of CPH1 with DIG1 has no effect on the invasive phenotype of the dig1Δ/dig1Δ mutant, while loss of EFG1 or TEC1 considerably reduces it. This is inconsistent with S. cerevisiae models where deletion of TEC1 or STE12, but not the EFG1 ortholog PHD1, blocks dig1Δ/dig2Δ invasive properties (19, 40, 41). Therefore we propose that in C. albicans dig1Δ/dig1Δ strains, Tec1p can regulate the invasive growth program independently of Cph1p and instead acts together with Efg1p, whereas in S. cerevisiae Ste12p and Tec1p exhibit a co-dependent MAPK response. The paralog of PHD1, SOK2, is so far untested for interactions with DIG1/2.
One surprising observation from our study was that in the absence of CaDig1p, Efg1p is essential for signal transduction in response to pheromone and environmental cues such as temperature, pH and the presence of serum (Supplementary Figure S2, Figure 7), indicating a potential role for CaDig1p outside of the traditional MAPK transcription factors and a new connection to the regulation of the cAMP-PKA signaling pathway. Furthermore, individually both the efg1Δ and dig1Δ single mutants would be expected to bolster mating in C. albicans (MTLa efg1Δ mutants are locked in the opaque-phase and dig1Δ/dig1Δ mutants are mating-proficient) yet the combinatorial effect actually compromises mating as the dig1Δ/efg1Δ double mutant shows reduced numbers of mating products resulting from cross-patch mating assays and a lack of a halo in response to the α-pheromone challenge. Taken together, these results provide evidence to suggest that the cAMP-PKA and MAPK signaling pathways are linked to an unexpected degree and may work synergistically to facilitate mating in C. albicans, a function that is not present in S. cerevisiae where MAPK signals inhibit cAMP-PKA in response to pheromone (42), or alternatively that the Efg1 protein may function independently of the cAMP pathway in C. albicans mating.
Figure 7. Transcription factor response to yeast or hyphae-inducing conditions in dig1Δ/dig1Δ MTLa/α background.
Each of the single and double transcription factor mutants (Dig1p, Tec1p, Cph1p, Efg1p) were assayed for their response to strong (A) yeast (pH 4.0, 25 °C) or (B) hyphae (pH 7.0, 37°C, 20 % v/v foetal calf serum) inducing conditions for 180 or 90 minutes respectively. Only the efg1Δ/efg1Δ dig1Δ/dig1Δ mutant completely fails to respond to either condition. All strains are MTLa/α.
Our investigation revealed that as well as constitutive activation of the filamentous growth pathway, cells deleted for CaDIG1 show increased activation of the pheromone-signaling pathway. This was evident by their being able to undergo homothallic mating and form clear haloes in response to a pheromone challenge. Such homothallic mating could be explained by the relative expression levels of BAR1 (orthologue of ScBAR1) and MFA1. Under normal circumstances, an opaque MTLa cell expresses both a- and α-pheromones (encoded by MFa and MFα respectively) when stimulated by an external source of α-pheromone (37, 43). In these cells BAR1 is also significantly induced and regulates the degradation of α-pheromone to prevent self-mating (44). However, if barrier activity (Bar1p) is compromised or if a unisexual population is exposed to high levels of pheromone from an external source, same-sex mating can occur (45). Comparison of relative gene expression levels in dig1Δ and wild type C. albicans reveals that the expression of MFα is six times greater than that of BAR1 in the mutant while in the wild type this differential is reduced to only 1.5 times (data not shown). This suggests that the increased expression level of α-pheromone in the MTLa dig1Δ cells generates sufficient pheromone to escape barrier activity and promote a-a mating. Taken together with the efficient mating frequency of a-α mating between dig1Δ and wild type cells, this study provides key evidence for CaDig1p as a negative regulator of mating signaling C. albicans.
Intriguingly, while C. albicans (and likely the common ancestor) encodes only one DIG gene, S. cerevisiae and several Saccharomyces species that arose after the whole genome duplication event, have maintained two Dig paralogs. This is both unusual as the most common fate of duplicated genes is gene loss (46), and interesting as retention of both paralogs serves to highlight the importance of the DIG family of negative regulators in fungal transcriptional control. Intriguingly, the single Dig protein at the focus of this study appears involved in a broader range of activities than the combinatorial effect of its S. cerevisiae orthologs; namely genetic interaction with the cAMP-PKA pathway, host colonization, biofilm formation, cellular morphology, mating, pheromone response and invasive growth compared to just the latter three in S. cerevisiae. So why does S. cerevisiae retain two paralogs whose gene products perform fewer biological functions than the ancestral protein? Although the evolutionary decisions behind the process of gene loss are not fully understood, a major influence on the maintenance of duplicate genes is the number of posttranslational modifications that the resulting proteins undergo; genes that encode highly phosphorylated proteins are much more likely to be retained as paralogs than those that do not (47) and the same is true of other types of posttranslationally modified proteins. Amoutzias et al. suggest that this could be due, at least in part, to the need to maintain transcriptional control during the period of genomic instability observed after a WGD event, where numerous duplicate genes are assimilated into a genome. Mutations at various phosphorylation sites would then be an efficient mechanism to promote neo- or sub-functionalization of paralogs making them more likely to be retained and facilitating transcriptional re-wiring to accommodate duplicated genes and maintain control of biological networks. Since ScDig1 and ScDig2 are predicted to contain 36 and 11 phospho-sites respectively (http://www.yeastgenome.org/), it is possible that in this scenario, the ancestral Dig protein was involved in several important biological processes and was functionally controlled by posttranslational modifications. It is possible that as S. cerevisiae diverged, certain of these functions and transcription factor-TF-regulator interactions that promote C. albicans pathogenesis such as host colonization, regulation of biofilms and cellular morphology were lost as they were not required for the non-pathogenic yeast resulting in highly phosphorylated but subfunctionalized paralogs. However, functional analyses of more members of the family will be necessary to establish the actual evolutionary trajectory of the Dig1 orthologs.
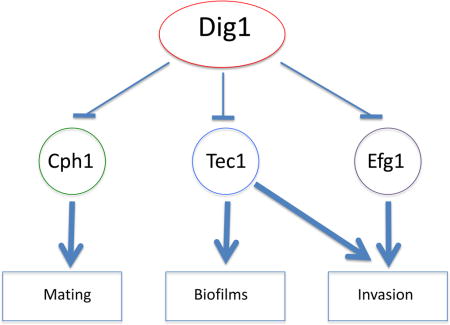
Overall, this work implicates a role for the Dig1 transcription factor regulator in cellular pathways involved in mating, biofilm formation and invasive growth. Intriguingly, although mutants are activated for these processes when assessed in vitro, in vivo colonization of the murine GI tract is defective. In these pathways the Dig1 function appears to play its role in conjunction with the transcription factors Efg1, Tec1 and Cph1, but the relative importance of the TF/Dig1 module varies for each process. As summarized in Figure 8, Dig1 regulation of Cph1 has a major role in mating, with Tec1 and Efg1 effects relatively minor, while Dig1-repressed invasive growth has Cph1 playing a minor role, with Efg1 and Tec1 having major effects. In the case of biofilm formation Efg1 and Cph1 play a less significant role than Tec1. This suggests that the single Dig1 orthlog in C. albicans plays a more extensive role in signal regulation than the two orthologs of S. cerevisiae. Further work will be required to establish the specific molecular role of Dig1 in these various pathways.
Figure 8. Schematic representing Dig1p-regulated transcription factors.
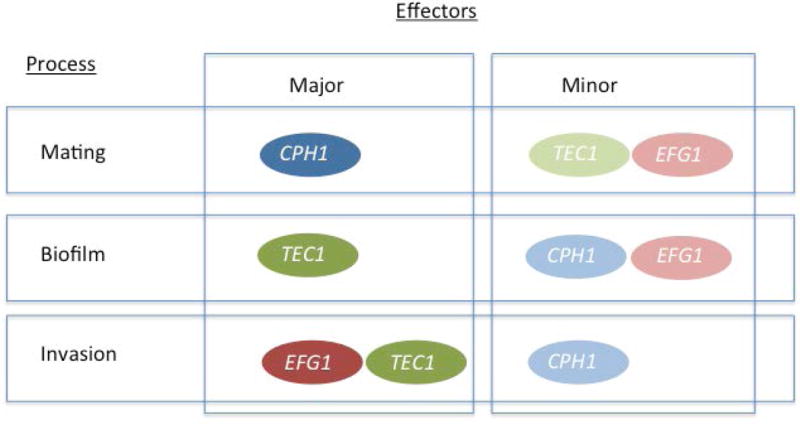
Genetic interactions between DIG1 and CPH1, EFG1 or TEC1 reveal that while all three contribute to mating, biofilm formation and invasive growth, the impact of transcription factor activity varies depending on the biological process. The major contributor(s) to each process are shown here in bold colours while the minor contributor(s) are lightly shaded.
Materials and Methods
Media and Strains
C. albicans strains were routinely cultured in YPD medium (1 % w/v Bacto-yeast extract, 2 % w/v Bacto-peptone, 2 % w/v glucose) supplemented with 50 mg L−1 uridine plus 2 % w/v Bacto-agar for plates. For selection of opaque-phase colonies, strains were grown on Synthetic Dextrose (SD) (0.67 % yeast nitrogen base lacking amino acids, 2 % w/v glucose, 0.15 % w/v complete amino acid mixture, 0.005 % w/v uridine, pH adjusted to 6.0) supplemented with 5 μg mL−1 phloxine B (4) and incubated at 24 °C. Putative transformants and mating products were selected for on SD minus the relevant amino acids at 30 °C and 24 °C respectively. Ura− strains were selected for on 5-FOA media (SD plus 1 g L−1 of 5-fluoroorotic acid) and grown at 30 °C.
All strains used in this study are listed in the supplementary data in Table S1 and are derived from SN148 unless otherwise stated. Oligonucleotides and plasmids used in this study are listed in Table S2. Strain construction was performed using the lithium acetate method of transformation (48) and homologous recombination of PCR-generated pFA-cassettes at the desired locus (49, 50). Putative transformants were selected for on SD media minus uridine, histidine, arginine or leucine. DIG1 was deleted from strains CAY3570, CAY3571, CAY4366, and CAY4437 in a similar manner to above, except PCR using oligos BL1757-1760 generated homologous cassettes from pSN52 or pSN40 (49). Deletion was confirmed by using primers internal to the LEU2 or HIS1 markers (49) in combination with BL1763 or BL1764 and by using primers internal to the DIG1 ORF (BL1761/62). CPH1 (CAY4436) and TEC1 (CAY4437) deletion constructs were created by PCR amplification of approximately 500bp homologous flanking regions and subsequent cloning into plasmid psfs2a (50). Transformants were selected on YPD+ nourseothricin (200ug/ml). Heterozygous CPH1 and DIG1 strains were incubated on maltose media for 3–4 days in order to excise the nourseothricin marker (50) and allow for a second round of transformation. Deletion was confirmed by PCR with primers internal to the CPH1 or TEC1 ORFs (BL1341/1432 and BL1173/74, respectively) and by junction check using primers BL1339/BL70 and BL1340/BL71 (CPH1) or BL1171/BL70 and BL1172/BL71.
Plate-washing assay
A single colony of the relevant C. albicans strain was inoculated into 5 mL of YPD and incubated shaking overnight at 30 °C. The OD600 of each culture was measured and approximately equal cell numbers of each strain were spotted on to YPD or Spider medium plates and incubated at 30 °C for 1, 2 or 5 days before washing under a constant stream of water for 15 seconds. Plates were scanned immediately before and after washing using an Epson Perfection V500 Photo colour scanner.
Biofilm assays
The pheromone-induced biofilm assay was performed as described in (16) with the following modifications. Strains were inoculated in 3 mL of SD and grown overnight at 25°C. Approximately 5.0 × 107 cells were added to 12 well dishes (Costar, Corning, inc) containing 1 mL of SD media (dig1Δ/dig1Δ cells) or Lee’s + glucose media (dig1Δ/pMET3 cells) in the presence of 0.01% DMSO or 10 μM C. albicans synthetic α-pheromone. Cells were incubated 24–48 hours, then decanted and each well washed twice with 1 mL of phosphate-buffered saline (PBS). Crystal violet staining was used to quantify adherence as described in (54). The results for each strain were averaged and plotted on a bar chart with error bars representing the standard deviation. Statistical significance was calculated using a student’s t-test (dig1Δ/dig1Δ experiments) or one-way ANOVA (dig1Δ/pMET3 experiments).
The pheromone-independent biofilm assay was performed in triplicate in a similar manner but with the following changes: Briefly, a single colony of the relevant C. albicans strain was inoculated into 5 mL of SD and grown overnight at room temperature. After 24 hours, the absorbance of each culture was measured and the equivalent of 1 mL of cells at OD600 2.5 was added to 1 mL of SD in a 24-well polystyrene plate. Plates were swirled to distribute cells evenly and then incubated statically for 48 hours at room temperature. The SD media was then decanted and remaining cells were washed three times in 700 μL of PBS. Samples were allowed to air-dry for 45 minutes before staining with 385 μL of 0.4 % w/v aqueous crystal violet for a further 45 minutes. Wells were washed 4 times with 1 mL of sterile distilled water and de-stained by addition of 700 μL of 95 % v/v ethanol. After 45 minutes at room temperature, the de-stain solution was diluted 1 in 10 with 95 % ethanol and the absorbance at OD495 was measured and averaged.
Murine Colonization
Female Swiss Webster mice (18–20 g) received antibiotics in their drinking water (tetracycline (1 mg mL−1), streptomycin (2 mg mL−1), gentamycin (0.1 mg mL−1) beginning four days prior to inoculation. C. albicans cells were grown in YPD medium at 37 °C for 24 hours. After washing twice in PBS, cells were counted with a hemocytometer; cells that were not separated because of the dig1Δ/dig1Δ mutant morphology defect were still counted as separate cells. Cultures were adjusted to 5 × 108 cells mL−1 and equal volumes of the two strains (Ura- and His-) were mixed, creating a 1:1 mixture of the two auxotrophs. Mice were inoculated by oral gavage with 5 × 107 C. albicans cells. Mice were sacrificed on day 1, 3 or 6 post-inoculation.
Cecum contents collected in PBS were centrifuged at 3600 g for 7 min to pellet particulate material, including fungal cells. DNA was extracted from 70 mg of contents by bead beating in phenol-chloroform (55), followed by chloroform extraction, ethanol precipitation, and Qiagen DNeasy purification with RNase treatment. qPCR was performed with primers RDNAF1 and RDNAR1 (Table S2). Numbers of genomes were measured using a standard curve and are expressed in arbitrary units. Composite results from two experiments are shown.
Ethics statement
All mouse protocols were approved by Tufts University’s Institutional Animal Care and Use Committee.
Microarrays
To isolate total RNA from C. albicans strains, a single colony was inoculated in 20 mL of YPD and incubated overnight shaking at 30 °C. Cultures were then diluted to OD600 0.1 in 100 mL of fresh YPD and grown at 30 °C until they reached OD600 0.6–0.7. Total RNA was extracted using an RNeasy Mini Kit (Qiagen) and quantified. RNA was reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) and the resulting cDNA was directly labeled with Cy3 or Cy5 monoreactive dyes and purified using a QIAquick PCR clean-up column (Qiagen) before hybridization to microarrays (56). A minimum two biological replicates with dye-swaps were performed for each strain and analysed with a P-value set at <0.05 for statistical significance. Data submitted to GEO, file GSE70085.
Halo assay
Halo assays based on (57) were modified for this study: A single colony of the relevant C. albicans strain was inoculated into 1 mL of YPD and mixed with 25 mL of molten YPD containing 1 % w/v agarose and pre-cooled to 45 °C. The mixture was allowed to set in a petri dish and 10 μL of 1 mg mL−1 alpha-factor pheromone were spotted onto the plates before incubation at 24 °C for 2–3 days.
Cross-patch Mating assay
To determine their mating capacity, relevant C. albicans strains were crossed with opaque tester strains 3315 (MTLα) or 3745 (MTLa) both of which harbour complementary auxotrophies for Lys and Trp. A single colony of each tester strain or the strain of interest was streaked onto YPD plates and incubated at 24 °C for 24–48 hrs. Strains were then crossed by replica plating on YPD and incubated for a further 24 hrs at 24 °C. Mating products were selected for by replica plating onto SD minus five amino acids and incubating at 24 °C for up to 5 days.
Scanning Electron Microscopy
Cells were grown on YPD agar for 24 hours at 30 °C, and then fixed with 2.5% (w/v) glutaraldehyde in 0.1 M sodium cacodylate buffer. Cells were then postfixed with 1% aqueous osmium tetroxide. Following fixation, cells were dehydrated gradually using a 15% gradient ethanol series and subsequently dried using a critical point dryer. The samples were then coated with 20nm of gold palladium (60:40) in an Emitech K550 sputter coater. Cells were imaged with a Hitachi S-2700 scanning electronic microscopy and collected with Quartz PCI software.
Quantitative Mating Assays
Quantitative mating assays were performed as described in (43) with the following modifications. C. albicans strains were incubated in liquid SCD medium overnight at 24 °C. Aliquots of 2×107 cells of each mating type were mixed and spotted onto 0.8 μm filters on Spider medium (1.35 % agar, 1 % nutrient broth, 0.4 % potassium phosphate, and 2 % mannitol, pH 7.2) at 24 °C for 4–5 days. Cells were scraped off, resuspended in H2O, and plated on SD dropout media for selection of parents as well as mating products. Percentage of mating products = 100 × (no. of colonies on double-selection medium/no. minimal parents on single selection medium). Double-selection medium was SCD–arginine –histidine, whereas single-selection medium was SCD–arginine and SCD–histidine. Experiments were repeated in three to six biological replicates.
Supplementary Material
Supplementary Figure S1. Colony morphology from the wild type strain SN148, the dig1∆/dig1∆ strain and the revertant strain dig1∆/dig1∆ (CIp-DIG1), in the MTLa/α and MTLa/a backgrounds, cultured on YPD agar at 30˚C for 48h, and cellular morphology after 3h of culture in liquid YPD at 30˚C 220 rpm, from a starting OD600 of 0.3. The cells were visualized using DIC at 1000× magnification with a LEICA DM6000 microscope.
Each of the single and double G protein mutants (Dig1p, Cag1, Ste4p) were assayed for their response to strong (A) yeast (pH 4.0, 25 °C) or (B) hyphae (pH 7.0, 37°C, 20 % v/v foetal calf serum) inducing conditions for 180 or 90 minutes respectively. The dig1Δ/dig1Δ mutant fails to respond to yeast conditions. All strains are MTLa.
Supplementary Figure S3. Venn diagrams illustrating two sets of up-regulated genes obtained from microarray analysis of dig1∆/dig1∆ MTLa and dig1∆/dig1∆ MTLa/α and their overlap with previously described hyphal-specific genes (Nantel et al., 2002), pheromone-specific genes (Bennett et al., 2003 and Bennett & Johnson, 2006), Efg1 regulon (Nobile et al, 2012) and Tec1 regulon (Nobile et al, 2012). For the microarray analysis we included the fifty genes with highest up-regulation for each set studied – higher than 2.5-fold for dig1∆/dig1∆ MTLa and higher than 10-fold for dig1∆/dig1∆ MTLa/α. The lists of genes in each intersection are listed in supplemental table S6.
Acknowledgments
We are grateful to Cécile Beaurepaire and André Nantel of the BRI Microarray Lab, National Research Council, Canada and Baharul Choudhury and Faïza Tebbji for their help with microarrays and data normalization, and Susan Sillaots for help with the complementation constructs. We also thank Dannie Durand for her insight and helpful discussions.
Footnotes
Author Contributions
Conceived and designed the experiments: HR, CMS, MPH, KTWG, CAK, MW, RJB
Performed the experiments: HR, YC, CMS, MPH, KTWG, CAK, TOCM
Analyzed the data: HR, YS, CMS, MPH, KG, CAK,TOCM
Contributed reagents/materials/analysis tools: CAK, MW, RJB
Wrote the paper: HR, MW
References
- 1.Wong Sak Hoi J, Dumas B. Ste12 and Ste12-like proteins, fungal transcription factors regulating development and pathogenicity. Eukaryotic cell. 2010;9(4):480–5. doi: 10.1128/EC.00333-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chou S, Lane S, Liu H. Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Molecular and cellular biology. 2006;26(13):4794–805. doi: 10.1128/MCB.02053-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. “White-opaque transition”: a second high-frequency switching system in Candida albicans. Journal of bacteriology. 1987;169(1):189–97. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson JM, Soll DR. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. Journal of bacteriology. 1987;169(12):5579–88. doi: 10.1128/jb.169.12.5579-5588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110(3):293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266(5191):1723–6. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 7.Kohler JR, Fink GR. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(23):13223–8. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90(5):939–49. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 9.Stoldt VR, Sonneborn A, Leuker CE, Ernst JF. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. The EMBO journal. 1997;16(8):1982–91. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonneborn A, Bockmuhl DP, Gerads M, Kurpanek K, Sanglard D, Ernst JF. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Molecular microbiology. 2000;35(2):386–96. doi: 10.1046/j.1365-2958.2000.01705.x. [DOI] [PubMed] [Google Scholar]
- 11.Si H, Hernday AD, Hirakawa MP, Johnson AD, Bennett RJ. Candida albicans white and opaque cells undergo distinct programs of filamentous growth. PLoS pathogens. 2013;9(3):e1003210. doi: 10.1371/journal.ppat.1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bockmuhl DP, Ernst JF. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics. 2001;157(4):1523–30. doi: 10.1093/genetics/157.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonneborn A, Tebarth B, Ernst JF. Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infection and immunity. 1999;67(9):4655–60. doi: 10.1128/iai.67.9.4655-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS biology. 2007;5(10):e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schweizer A, Rupp S, Taylor BN, Rollinghoff M, Schroppel K. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Molecular microbiology. 2000;38(3):435–45. doi: 10.1046/j.1365-2958.2000.02132.x. [DOI] [PubMed] [Google Scholar]
- 16.Lin CH, Kabrawala S, Fox EP, Nobile CJ, Johnson AD, Bennett RJ. Genetic control of conventional and pheromone-stimulated biofilm formation in Candida albicans. PLoS pathogens. 2013;9(4):e1003305. doi: 10.1371/journal.ppat.1003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahni N, Yi S, Daniels KJ, Huang G, Srikantha T, Soll DR. Tec1 mediates the pheromone response of the white phenotype of Candida albicans: insights into the evolution of new signal transduction pathways. PLoS biology. 2010;8(5):e1000363. doi: 10.1371/journal.pbio.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Chen J, Lane S, Liu H. A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Molecular microbiology. 2002;46(5):1335–44. doi: 10.1046/j.1365-2958.2002.03249.x. [DOI] [PubMed] [Google Scholar]
- 19.Cook JG, Bardwell L, Kron SJ, Thorner J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes & development. 1996;10(22):2831–48. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- 20.Tedford K, Kim S, Sa D, Stevens K, Tyers M. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Current biology: CB. 1997;7(4):228–38. doi: 10.1016/s0960-9822(06)00118-7. [DOI] [PubMed] [Google Scholar]
- 21.Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends in microbiology. 2004;12(7):317–24. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Chapa YLB, Lee S, Regan H, Sudbery P. The mating projections of Saccharomyces cerevisiae and Candida albicans show key characteristics of hyphal growth. Fungal biology. 2011;115(6):547–56. doi: 10.1016/j.funbio.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Ganguly S, Mitchell AP. Mucosal biofilms of Candida albicans. Current opinion in microbiology. 2011;14(4):380–5. doi: 10.1016/j.mib.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Z, Thompson A, Sobue T, Kashleva H, Xu H, Vasilakos J, et al. Candida albicans biofilms do not trigger reactive oxygen species and evade neutrophil killing. The Journal of infectious diseases. 2012;206(12):1936–45. doi: 10.1093/infdis/jis607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganguly S, Bishop AC, Xu W, Ghosh S, Nickerson KW, Lanni F, et al. Zap1 control of cell-cell signaling in Candida albicans biofilms. Eukaryotic cell. 2011;10(11):1448–54. doi: 10.1128/EC.05196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi S, Sahni N, Daniels KJ, Lu KL, Srikantha T, Huang G, et al. Alternative mating type configurations (a/alpha versus a/a or alpha/alpha) of Candida albicans result in alternative biofilms regulated by different pathways. PLoS biology. 2011;9(8):e1001117. doi: 10.1371/journal.pbio.1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daniels KJ, Srikantha T, Lockhart SR, Pujol C, Soll DR. Opaque cells signal white cells to form biofilms in Candida albicans. The EMBO journal. 2006;25(10):2240–52. doi: 10.1038/sj.emboj.7601099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bardwell L, Cook JG, Voora D, Baggott DM, Martinez AR, Thorner J. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes & development. 1998;12(18):2887–98. doi: 10.1101/gad.12.18.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung W, Olson KA, Breitkreutz A, Sadowski I. Characterization of the basal and pheromone-stimulated phosphorylation states of Ste12p. European journal of biochemistry/FEBS. 1997;245(2):241–51. doi: 10.1111/j.1432-1033.1997.00241.x. [DOI] [PubMed] [Google Scholar]
- 30.Bardwell L, Cook JG, Zhu-Shimoni JX, Voora D, Thorner J. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(26):15400–5. doi: 10.1073/pnas.95.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson KA, Nelson C, Tai G, Hung W, Yong C, Astell C, et al. Two regulators of Ste12p inhibit pheromone-responsive transcription by separate mechanisms. Molecular and cellular biology. 2000;20(12):4199–209. doi: 10.1128/mcb.20.12.4199-4209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dignard D, Andre D, Whiteway M. Heterotrimeric G-protein subunit function in Candida albicans: both the alpha and beta subunits of the pheromone response G protein are required for mating. Eukaryotic cell. 2008;7(9):1591–9. doi: 10.1128/EC.00077-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malathi K, Ganesan K, Datta A. Identification of a putative transcription factor in Candida albicans that can complement the mating defect of Saccharomyces cerevisiae ste12 mutants. The Journal of biological chemistry. 1994;269(37):22945–51. [PubMed] [Google Scholar]
- 34.Magee BB, Legrand M, Alarco AM, Raymond M, Magee PT. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Molecular microbiology. 2002;46(5):1345–51. doi: 10.1046/j.1365-2958.2002.03263.x. [DOI] [PubMed] [Google Scholar]
- 35.Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS pathogens. 2010;6(1):e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bougnoux ME, Diogo D, Francois N, Sendid B, Veirmeire S, Colombel JF, et al. Multilocus sequence typing reveals intrafamilial transmission and microevolutions of Candida albicans isolates from the human digestive tract. Journal of clinical microbiology. 2006;44(5):1810–20. doi: 10.1128/JCM.44.5.1810-1820.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennett RJ, Johnson AD. The role of nutrient regulation and the Gpa2 protein in the mating pheromone response of C. albicans. Molecular microbiology. 2006;62(1):100–19. doi: 10.1111/j.1365-2958.2006.05367.x. [DOI] [PubMed] [Google Scholar]
- 38.Bennett RJ, Uhl MA, Miller MG, Johnson AD. Identification and characterization of a Candida albicans mating pheromone. Molecular and cellular biology. 2003;23(22):8189–201. doi: 10.1128/MCB.23.22.8189-8201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148(1–2):126–38. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan O, Shapiro RS, Kurat CF, Mayhew D, Baryshnikova A, Chin B, et al. Global gene deletion analysis exploring yeast filamentous growth. Science. 2012;337(6100):1353–6. doi: 10.1126/science.1224339. [DOI] [PubMed] [Google Scholar]
- 41.Breitkreutz A, Boucher L, Breitkreutz BJ, Sultan M, Jurisica I, Tyers M. Phenotypic and transcriptional plasticity directed by a yeast mitogen-activated protein kinase network. Genetics. 2003;165(3):997–1015. doi: 10.1093/genetics/165.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arkinstall SJ, Papasavvas SG, Payton MA. Yeast alpha-mating factor receptor-linked G-protein signal transduction suppresses Ras-dependent activity. FEBS letters. 1991;284(1):123–8. doi: 10.1016/0014-5793(91)80777-z. [DOI] [PubMed] [Google Scholar]
- 43.Alby K, Schaefer D, Bennett RJ. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature. 2009;460(7257):890–3. doi: 10.1038/nature08252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaefer D, Cote P, Whiteway M, Bennett RJ. Barrier activity in Candida albicans mediates pheromone degradation and promotes mating. Eukaryotic cell. 2007;6(6):907–18. doi: 10.1128/EC.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alby K, Bennett RJ. Interspecies pheromone signaling promotes biofilm formation and same-sex mating in Candida albicans. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(6):2510–5. doi: 10.1073/pnas.1017234108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–5. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 47.Amoutzias GD, He Y, Gordon J, Mossialos D, Oliver SG, Van de Peer Y. Posttranslational regulation impacts the fate of duplicated genes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(7):2967–71. doi: 10.1073/pnas.0911603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11(4):355–60. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 49.Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryotic cell. 2005;4(2):298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–27. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 51.Gola S, Martin R, Walther A, Dunkler A, Wendland J. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast. 2003;20(16):1339–47. doi: 10.1002/yea.1044. [DOI] [PubMed] [Google Scholar]
- 52.Schaub Y, Dunkler A, Walther A, Wendland J. New pFA-cassettes for PCR-based gene manipulation in Candida albicans. Journal of basic microbiology. 2006;46(5):416–29. doi: 10.1002/jobm.200510133. [DOI] [PubMed] [Google Scholar]
- 53.Al-Rawi N, Laforce-Nesbitt SS, Bliss JM. Deletion of Candida albicans SPT6 is not lethal but results in defective hyphal growth. Fungal genetics and biology: FG & B. 2010;47(4):288–96. doi: 10.1016/j.fgb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Porman AM, Hirakawa MP, Jones SK, Wang N, Bennett RJ. MTL-independent phenotypic switching in Candida tropicalis and a dual role for Wor1 in regulating switching and filamentation. PLoS genetics. 2013;9(3):e1003369. doi: 10.1371/journal.pgen.1003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: Greene Publishing and Wiley Interscience; 1989. [Google Scholar]
- 56.Nantel A, Dignard D, Bachewich C, Harcus D, Marcil A, Bouin AP, et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Molecular biology of the cell. 2002;13(10):3452–65. doi: 10.1091/mbc.E02-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Achstetter T. Regulation of alpha-factor production in Saccharomyces cerevisiae: a-factor pheromone-induced expression of the MF alpha 1 and STE13 genes. Molecular and cellular biology. 1989;9(10):4507–14. doi: 10.1128/mcb.9.10.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Colony morphology from the wild type strain SN148, the dig1∆/dig1∆ strain and the revertant strain dig1∆/dig1∆ (CIp-DIG1), in the MTLa/α and MTLa/a backgrounds, cultured on YPD agar at 30˚C for 48h, and cellular morphology after 3h of culture in liquid YPD at 30˚C 220 rpm, from a starting OD600 of 0.3. The cells were visualized using DIC at 1000× magnification with a LEICA DM6000 microscope.
Each of the single and double G protein mutants (Dig1p, Cag1, Ste4p) were assayed for their response to strong (A) yeast (pH 4.0, 25 °C) or (B) hyphae (pH 7.0, 37°C, 20 % v/v foetal calf serum) inducing conditions for 180 or 90 minutes respectively. The dig1Δ/dig1Δ mutant fails to respond to yeast conditions. All strains are MTLa.
Supplementary Figure S3. Venn diagrams illustrating two sets of up-regulated genes obtained from microarray analysis of dig1∆/dig1∆ MTLa and dig1∆/dig1∆ MTLa/α and their overlap with previously described hyphal-specific genes (Nantel et al., 2002), pheromone-specific genes (Bennett et al., 2003 and Bennett & Johnson, 2006), Efg1 regulon (Nobile et al, 2012) and Tec1 regulon (Nobile et al, 2012). For the microarray analysis we included the fifty genes with highest up-regulation for each set studied – higher than 2.5-fold for dig1∆/dig1∆ MTLa and higher than 10-fold for dig1∆/dig1∆ MTLa/α. The lists of genes in each intersection are listed in supplemental table S6.


