Significance
Herpesviruses have an extraordinarily complex relationship with their host, persisting for the lifetime of the host by way of a latent infection. Reactivation of replication is associated with significant disease risk, particularly in immunocompromised individuals. We characterize in depth transcriptional profiles of human cytomegalovirus latency. We show that a broad and concordant viral transcriptome is found in both an experimental model of latency and in asymptomatically infected individuals. We further define genes that are differentially regulated during latent and replicative states: candidates for key regulators controlling the switch between latency and reactivation. This work will help understand the persistence of complex DNA viruses and provides a path toward developing antiviral strategies to control herpesvirus entry into and exit from latency.
Keywords: cytomegalovirus, herpesvirus, transcriptome, latency, kernel density estimation
Abstract
The transcriptional program associated with herpesvirus latency and the viral genes regulating entry into and exit from latency are poorly understood and controversial. Here, we developed and validated a targeted enrichment platform and conducted large-scale transcriptome analyses of human cytomegalovirus (HCMV) infection. We used both an experimental hematopoietic cell model of latency and cells from naturally infected, healthy human subjects (clinical) to define the breadth of viral genes expressed. The viral transcriptome derived from experimental infection was highly correlated with that from clinical infection, validating our experimental latency model. These transcriptomes revealed a broader profile of gene expression during infection in hematopoietic cells than previously appreciated. Further, using recombinant viruses that establish a nonreactivating, latent-like or a replicative infection in CD34+ hematopoietic progenitor cells, we defined classes of low to moderately expressed genes that are differentially regulated in latent vs. replicative states of infection. Most of these genes have yet to be studied in depth. By contrast, genes that were highly expressed, were expressed similarly in both latent and replicative infection. From these findings, a model emerges whereby low or moderately expressed genes may have the greatest impact on regulating the switch between viral latency and replication. The core set of viral genes expressed in natural infection and differentially regulated depending on the pattern of infection provides insight into the HCMV transcriptome associated with latency in the host and a resource for investigating virus–host interactions underlying persistence.
All herpesviruses persist, in part, through the establishment of a latent infection. A central gap in our knowledge of herpesvirus biology is the extent of viral gene expression that occurs during latency. Human cytomegalovirus (HCMV), a member of the β-herpesvirus family, has the largest genome of any known human virus, at approximately 230-kbp in size (1–3) and encodes at least 170, and potentially as many as 754 unique ORFs (4). HCMV establishes latency in hematopoietic progenitor cells (HPCs) and myeloid lineage cells (5, 6). The latent state permits life-long persistence of the viral genome marked by sporadic bouts of reactivation, which allows for periods of typically subclinical virus shedding (7). In contrast to productive infection, viral genomes are maintained at low levels and viral gene expression is thought to be restricted during the latent infection and is rarely detected in the latent host. Therefore, how the programs of viral gene expression differ in various cell types or states of persistence in the host remains elusive (5, 6, 8). Understanding the cytomegalovirus transcriptome as part of the molecular basis of persistence in the healthy host is an important step toward developing strategies to control viral latency and reactivation. Reactivation of HCMV in the absence of adequate T-cell immunity results in life-threatening disease in solid organ and stem cell transplant recipients (9, 10), and HCMV is the leading infectious cause of congenital birth defects (11, 12).
Until recently, HCMV transcriptome analysis was predominantly restricted to productively infected fibroblasts due to technical challenges posed by HCMV infection and persistence (13, 14). The strict species restriction of HCMV has constrained latency studies to primary human cell models in CD34+ hematopoietic progenitor cells (HPCs) or CD14+ monocytes infected in vitro, where viral transcripts account for an exceptionally minor proportion of the RNA pool (15–20). Further, in the human host, it is estimated that only 1 in 104 to 105 mononuclear cells harbor viral genomes in healthy latently infected individuals (21), posing significant challenges for the bona fide detection of viral transcripts amid the overwhelming host transcriptome. To address these challenges, we developed a targeted enrichment platform to capture low-abundance viral transcripts from CD34+ HPCs infected in vitro and from peripheral blood mononuclear cells (PBMCs) isolated from asymptomatically infected individuals (clinical). These studies validate our enrichment platform and define the gene expression across the HCMV genome in CD34+ HPCs infected in vitro and in naturally infected PBMCs. Further, to explore the regulation of HCMV gene expression in the context of latency, we compared viral gene expression in CD34+ HPCs during infection with recombinant viruses that either (i) replicated and did not establish latency or (ii) maintained the viral genome but could not reactivate (22, 23). While highly expressed genes were highly expressed in all contexts of infection, genes differentially regulated in the context of latent and replicative states of infection in CD34+ HPCs were expressed at low to moderate levels. As many of the genes identified as expressed or regulated in these contexts do not have well-ascribed functions, they represent an important class of genes for studies aimed at understanding the regulation of latent vs. replicative states of infection.
Results
Robust and Reproducible Enrichment of HCMV Libraries from Infection of CD34+ HPCs.
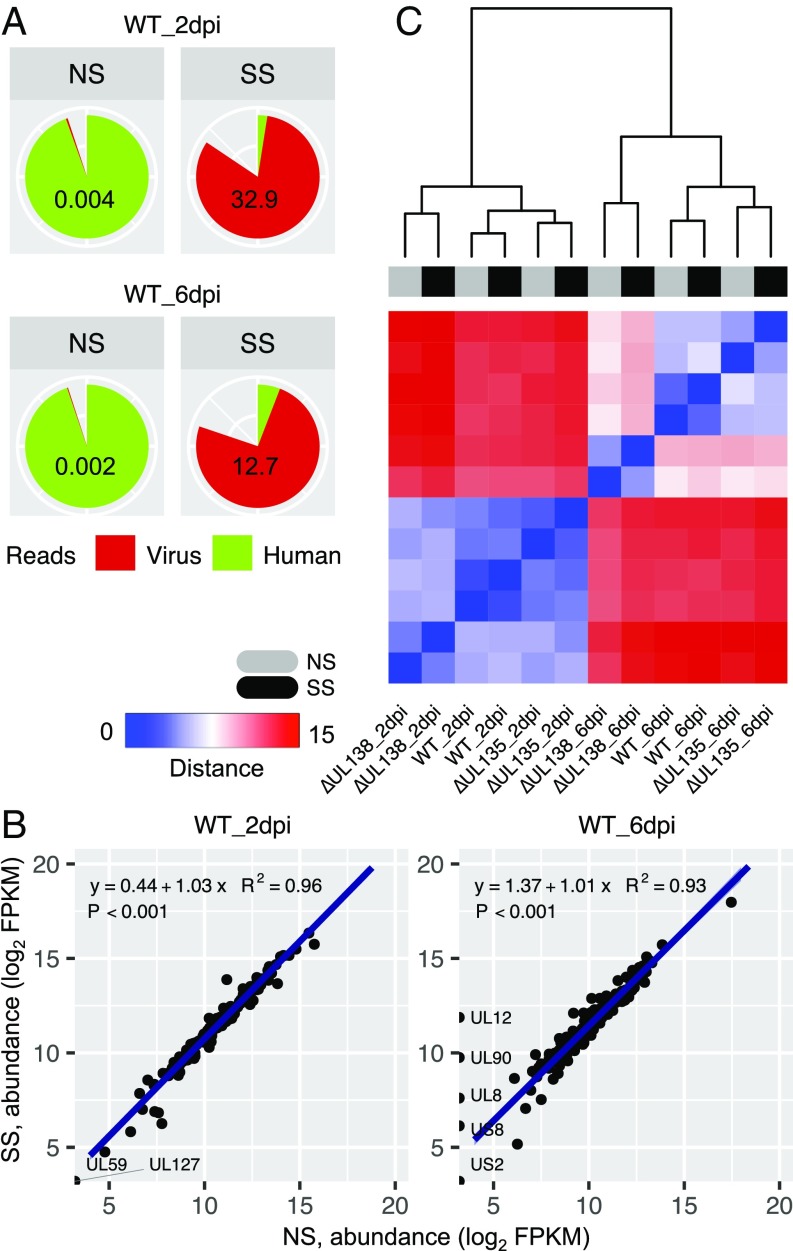
To obtain deep sequencing for rare HCMV sequence reads in CD34+ HPCs latently infected in vitro, we developed targeted probes (SureSelect, Agilent) to capture and enrich HCMV sequences from complex human samples for strand-specific RNA sequencing. This approach was used previously to enrich viral genomes from clinical/human libraries (24, 25). The enrichment probes were tiled along the HCMV TB40/E genome, excluding the internal repeat long (IRL) and terminal repeat long (TRL) regions and the viral long noncoding RNAs (4.9 kb, 2.7 kb, and 1.2 kb), which are expressed at high levels during infection in multiple contexts (13, 26, 27). Any sequences sharing identity to the human genome were also masked (excluded). We first examined the extent of the enrichment by comparing the ratio of virus-to-human reads (V/H) mapped in samples from TB40/E wild-type (WT)-infected CD34+ HPCs without [nonselected (NS)] vs. with [SureSelect enriched (SS)] enrichment. The V/H ratio accounts for both enrichment efficiency of virus reads and depletion efficiency of human reads in a SS sample. The V/H ratio increased 8,225-fold at 2 days postinfection (dpi) and 6,350-fold at 6 dpi in SS relative to NS (Fig. 1A). To confirm that samples did not acquire laboratory contamination during processing of NS samples, mock-infected samples were sequenced and the mean V/H ratio was 0.000089 (SI Appendix, Fig. S1A, NS, mock). The low availability of viral reads in NS CD34+ HPC libraries, despite all cells being infected, demonstrates the challenge of reconstructing HCMV transcriptomes, especially from clinical samples which harbor virus in a small proportion of cells. Notably, SureSelect enrichment increased viral reads to 81.92% (2 dpi) and 74.35% (6 dpi) of the total quality reads in the two samples (SI Appendix, Dataset S1), such that virus becomes a predominant species in the virus–host metatranscriptome.
Fig. 1.
Enrichment of HCMV libraries is an efficient and unbiased method for defining the transcriptome in samples where transcript abundance is low. CD34+ HPCs were infected with WT, ∆UL135, or ∆UL138 [multiplicity of infection (MOI) = 2] and cDNA libraries were prepared at 2 and 6 dpi with or without SureSelect enrichment and sequencing. (A) Pie charts illustrate the differences in the proportions of HCMV (red) and human (green) reads mapped between NS and SS samples from CD34+ HPCs infected with WT HCMV. The ratio of virus-to-human reads (V/H) is shown for each sample. For numbers of viral and human reads refer to SI Appendix, Dataset S1. (B) Gene abundance (FPKM) comparisons between NS and SS samples in A. Pearson’s correlation coefficient is shown. Level of confidence intervals for predictions of a linear model is 0.95. Genes absent in one sample are labeled. FPKM, fragments per kilobase of transcript per million mapped reads. (C) Heatmap displaying hierarchical clustering of the sample-to-sample distance matrix. NS (gray) and SS (black) libraries for six biological samples: WT, ∆UL135, and ∆UL138, each at 2 and 6 dpi, are included.
To identify possible sequence bias introduced by SS enrichment, we used Cufflinks-based quantification of viral gene expression, fragments per kilobase of transcript per million mapped reads (FPKM) (28) to correlate NS and SS samples in Fig. 1A (Upper and Lower). Linear regression indicated a high correlation between SS and NS samples at both 2 and 6 dpi (2 dpi: slope of 1.03 and R2 = 0.96; 6 dpi: slope of 1.01 and R2 = 0.93) (Fig. 1B). Despite the difference in viral genome coverage, only four genes (UL12, UL90, UL8, and US8) were detected at 6 dpi in the SS library but not the NS library. These may be genes expressed at levels not detected without enrichment or may indicate an enrichment bias. Collectively, these data indicate that the SS platform offers an efficient and reproducible enrichment without introducing substantial bias.
In addition to our analysis of in vitro infection of CD34+ HPCs with the TB40/E WT, we included two recombinant viruses containing disruptions in the ULb′ genes, UL135 and UL138. These genes have an antagonistic relationship that is important to regulating the transition between latent and reactivated states (22, 23, 29, 30) (SI Appendix, Fig. S1B). UL138 is suppressive to viral replication and recombinant viruses lacking UL138 (∆UL138) fail to establish a latent infection and instead productively replicate in CD34+ HPCs in the absence of a reactivation stimulus (22, 29, 30). In contrast, UL135 overcomes UL138-mediated suppression for reactivation (23). Recombinant viruses lacking UL135 (∆UL135) maintain viral genomes but fail to reactivate. These recombinant viruses represent powerful tools to distinguish the viral transcriptomes associated with latent-like vs. replicative states in CD34+ HPCs. Of note, these mutant viruses were generated by the substitution of stop codons for 5′ translational start codons to abrogate protein synthesis without disrupting the transcript.
We then calculated the Euclidean distance of viral gene expression with DESeq2 (31) between WT, ∆UL135, and ∆UL138 infection at 2 and 6 dpi to validate the enrichment for all samples. The heatmap shows that each SS biological sample was most closely related to its NS counterpart (Fig. 1C), independent of sequencing depth. Furthermore, the transcriptome of ∆UL135 infection was closely related to that of WT infection, but more different from ∆UL138 infection at both 2 and 6 dpi. At 6 dpi, greater diversity between the different viral transcriptomes was observed compared with 2 dpi.
Functional Antagonism Between UL135 and UL138 Reflected by Differential Expression of Low and Moderately Expressed Viral Genes in CD34+ HPCs.
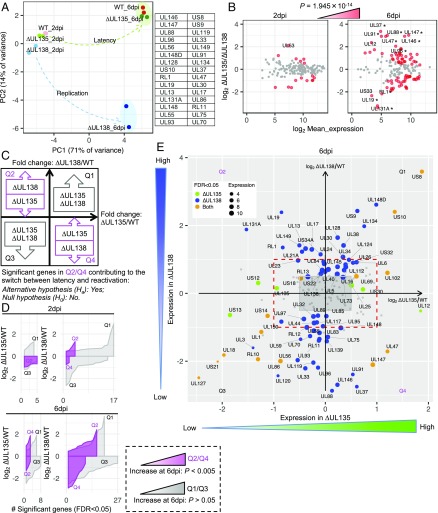
We applied several independent methods to assess the viral transcriptional program in WT and ∆UL135- and ∆UL138-infected CD34+ HPCs to further differentiate patterns of infection. Principal component analysis (PCA) revealed high similarity between WT and ∆UL135- and ∆UL138-mutant virus infections at 2 dpi; however, by 6 dpi when latency is being established, WT and ∆UL135 clustered tightly but ∆UL138 was segregated with respect to the first two principal components (PCs) (Fig. 2A). The difference between WT or ∆UL135 and ∆UL138 infections at 6 dpi was associated with PC2, accounting for 14% of the total variance (see SI Appendix, Fig. S1C for the scree plot and SI Appendix, Fig. S1D for the score plot of PC2 vs. PC3). The top 30 genes contributing to PC2 are listed (Fig. 2A, and see SI Appendix, Fig. S1E for the loadings). These data reflect a progressive transcriptional difference between the two patterns of infection (latent-like vs. replicative) associated with these viruses and is consistent with the opposing functions of UL135 and UL138 (23). Differential gene expression analysis between the two mutant viruses further indicated that differences in gene expression increased significantly from 2 to 6 dpi (P = 1.945 × 10−14, Fisher’s exact test). At 6 dpi, eight genes, UL12, UL37, UL47, UL88, UL91, UL96, UL146, and UL147, were considerably up-regulated in ∆UL135 while four genes, RL1, UL19, UL131A, and US33, were considerably up-regulated in ∆UL138 [more than fourfold change and false discovery rate (FDR) < 0.05, Fig. 2B]. Not surprisingly, 83% of these genes contribute to the variance of PC2.
Fig. 2.
Viral gene expression is differentially regulated in CD34+ HPCs infected with UL135- and UL138-mutant viruses. (A) PCA for infected CD34+ HPC samples (Fig. 1C) revealed two mutant viruses partitioning over time from 2 to 6 dpi. The green arrow represents a trajectory to the latent state of WT and ∆UL135 (green oval), while the blue arrow represents a trajectory to the replicative state of ∆UL138 (blue oval). This separation is associated with PC2 and the genes with the 30 highest absolute loadings are listed. (B) MA plot comparing two mutant viruses at 2 and 6 dpi. Genes are colored red if the FDR is <0.05, and they increase significantly (P = 1.945 × 10−14, Fisher’s exact test) from 2 to 6 dpi. Genes with more than fourfold change are indicated. An asterisk marks genes that are among the PC2 top loading genes in A. (C) Schematic illustration of a model of differential gene expression regulating the switch between latency and reactivation in CD34+ HPCs using two mutant viruses, ∆UL135 and ∆UL138. Two-dimensional differential expression for the fold change between ∆UL135 and WT is on the x axis and the fold change between ∆UL138 and WT is on the y axis. This analysis identifies subsets of genes that are concordantly (Q1/Q3) or antagonistically (Q2/Q4, highlighted by magenta) regulated in ∆UL135 and ∆UL138 transcriptomes. We hypothesize that the switch between latent and reactivation states requires a significant number of antagonistically regulated viral genes. (D) Quadrant-specific expression pattern using ribbon plot of fold change vs. significant gene counts. A significant increase (P < 0.005, Fisher’s exact test) of genes in Q2/Q4 (magenta), but not in Q1/Q3, at 6 dpi indicates null hypothesis is rejected. (E) Corresponding significant genes residing in quadrants Q2/Q4 or Q1/Q3 at 6 dpi. Red dashed rectangle highlights the twofold change. Green and blue dot size is proportional to the mean expression of individual genes in all ∆UL135 and ∆UL138 infections, respectively. Orange dot size is proportional to the mean expression of individual genes in all ∆UL135 and ∆UL138 infections. For corresponding significant genes at 2 dpi refer to SI Appendix, Fig. S2.
To further classify viral genes as concordantly or antagonistically expressed in ∆UL135 and ∆UL138 infections, we established a model examining 2D differential expression of viral genes in ∆UL135 and ∆UL138 infections each relative to WT. Genes antagonistically regulated are in quadrant Q2 or Q4, whereas genes concordantly regulated are in Q1 or Q3 (schematically illustrated in Fig. 2C). For example, Q2 genes are up-regulated in ∆UL138 infection and down-regulated in ∆UL135 infection. Analysis of significantly regulated genes (FDR < 0.05) using log2 fold change (FC) as a function of gene counts, revealed that over time postinfection the number of antagonistically (Q2/Q4), but not concordantly (Q1/Q3) regulated genes significantly increased (P < 0.005, Fisher’s exact test) (Fig. 2D). The differential expression of individual viral genes is shown in Fig. 2E at 6 dpi (see SI Appendix, Fig. S2 A and B for a log2 FC vs. log2 mean expression (MA) plot of each comparison and SI Appendix, Fig. S2C for 2 dpi). The correlation (Q1/Q3) and anticorrelation (Q2/Q4) of differential expression of all genes between the two comparisons are shown in SI Appendix, Fig. S2D. These data support a model whereby these antagonistically regulated genes (Q2/Q4) may contribute to the switch between latent and replicative states and reject the null hypothesis in Fig. 2C.
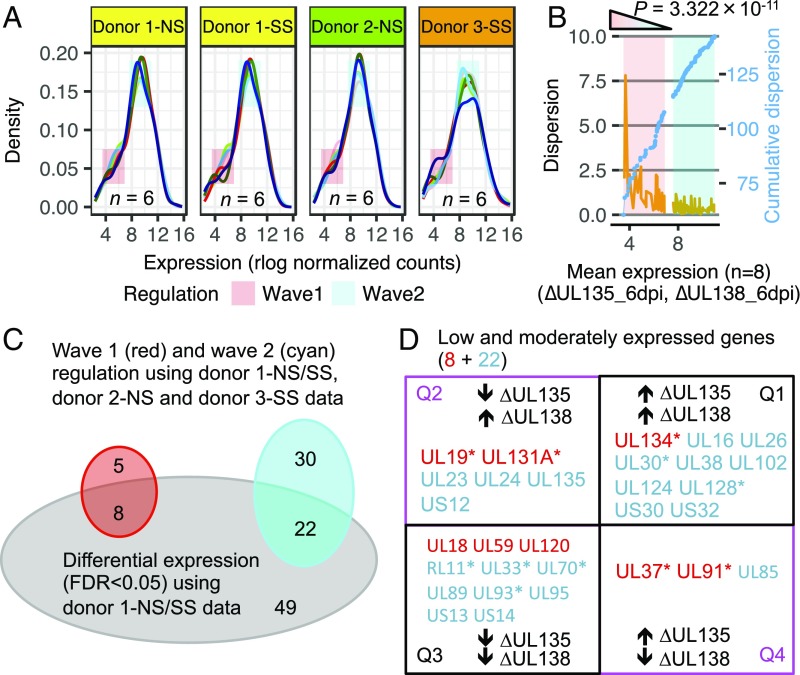
Given that infection of independent human CD34+ HPC donors resulted in high viral transcriptome variability between the biological replicates, we used kernel density estimation to investigate the viral gene expression in WT, ∆UL135, and ∆UL138 infections across two additional human donors (biological replicates). Kernel density estimation makes no assumption regarding distribution of the data, which allows for the unbiased classification of expression levels across samples (32). In Fig. 3A, six curves in each panel for WT, ∆UL135, and ∆UL138 infections at 2 and 6 dpi are shown for NS and SS samples from donor 1 (yellow), NS samples from donor 2 (green), and SS samples from donor 3 (orange). Strikingly, the six curves across different donors and virus infections were tightly aligned except for two distinct “waves,” one at low and the other at moderate expression levels (Fig. 3A). Examining different bandwidth settings in kernel density estimates validates that the wave 1 and wave 2 patterns are independent of this key parameter (SI Appendix, Fig. S3A). Using our data, 24 random samples generated showed tight overlap with one another, almost as a single curve, confirming the heterogeneity of gene expression within wave 1 and wave 2 between the real samples (SI Appendix, Fig. S3B). To quantify wave 1 and wave 2 variation in gene expression of two mutant viruses across biological replicates, we calculated genewise dispersion estimates in DESeq2 (31) for the genes whose expression fell within wave 1 or wave 2 across all available ∆UL135_6dpi and ∆UL138_6dpi transcriptomes. We found that the dispersion of wave 1 genes was significantly higher than that of wave 2 genes (Fig. 3B, P = 3.322 × 10−11, Wilcoxon rank sum test), indicating that antagonism between mutant virus infections was induced by two regulatory patterns. This was supported by the notion that the natural dimensionality of gene expression is determined not by individual genes, but by genes coregulated within transcriptional modules (33).
Fig. 3.
Low and moderately expressed genes exhibit high variability across different infections and cell donors. (A) Optimal kernel density estimates of expression levels of six samples (line colors correspond to sample colors in Fig. 2A) across three cell donors (yellow, green, and orange). Two regions of low and moderate expression (termed wave 1 and wave 2) exhibit high variation. (B) Dispersion (orange, Left y axis) and cumulative dispersion (blue, Right y axis) measurement for within wave 1 and wave 2 genes in ∆UL135_6dpi and ∆UL138_6dpi transcriptomes (n = 8). (C) Venn diagram displaying overlap between genes whose expression fell within wave 1 or wave 2 in all eight mutant viruses and genes from DE in Fig. 2E. (D) Thirty low and moderately expressed genes derived from shared kernel/dispersion workflow and DE metrics are organized in the four-quadrant model. * marks genes that are among the PC2 top loading genes in Fig. 2A.
To provide a robust profile of genes differentially expressed between ∆UL135 and ∆UL138 at 6 dpi across all donors, we aligned these two regulatory modules (13 wave 1 genes plus 52 wave 2 genes) to those that were differentially expressed between ∆UL135 and ∆UL138 infections each relative to WT (donor 1 data, see Fig. 2E). Thirty regulatory genes (8 in wave 1 and 22 in wave 2) were common between the two-distinct metrics [kernel/dispersion workflow vs. differential expression (DE)] using two partially overlapping datasets (Fig. 3C). The distribution of these 30 genes across the four quadrants is shown in Fig. 3D. The distribution of these 30 genes based on the metrics of dispersion vs. fold change is shown in SI Appendix, Fig. S4 and their functional annotation is shown in Tables S1 and S2. These combined methods identified genes in the two mutant virus infections that were differentially expressed across multiple cell donors (biological replicates).
Low Heterogeneity in the Viral Transcriptomes of ∆UL135- and ∆UL138-Infected Fibroblasts.
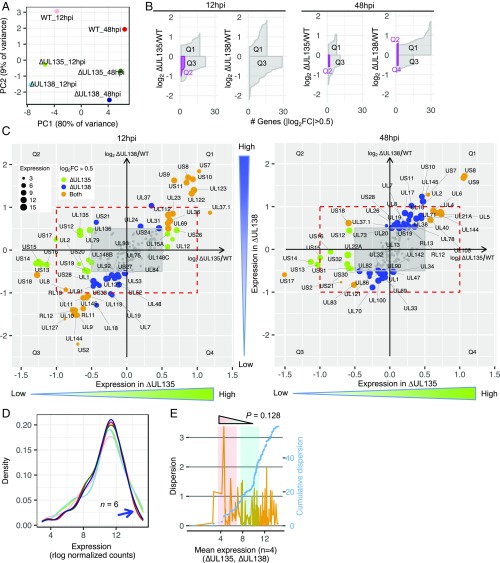
To further distinguish viral transcriptomes associated with infection in hematopoietic cells, we applied the same analysis pipeline to the dataset of fibroblast infection, a model of productive replication. We sequenced 12 samples infected with WT, ∆UL135, or ∆UL138 at 12, 24, 48, and 72 hpi. For brevity, 12 and 48 hpi results are shown in the main text and additional time points are in SI Appendix. In contrast to infection in CD34+ HPCs (SI Appendix, Fig. S1A), the ratio of virus-to-human reads increased over time during infection in fibroblasts (SI Appendix, Fig. S5A), indicative of a productive infection. The proportion of viral reads was similar between our samples (SI Appendix, Dataset S1) and those previously reported for fibroblast infection (34). By PCA, none of the six samples clustered and the greatest separation was based on time postinfection associated with PC1 (Fig. 4A and SI Appendix, Fig. S5B). This is in contrast to infection in CD34+ HPCs, where samples clustered at 2 dpi, and ∆UL138 separated from WT and ∆UL135 over time (Fig. 2A).
Fig. 4.
Viral gene expression is not antagonistically regulated in ∆UL135- and ∆UL138-infected fibroblasts. (A) PCA of six samples at 12 and 48 hpi. (B) Quadrant-specific expression pattern using ribbon plot of fold change vs. counts of genes with their absolute log2 FC >0.5. (C) Corresponding genes residing in quadrants Q2/Q4 or Q1/Q3 are shown. Red dashed rectangles highlight the twofold change. Green and blue dot size is proportional to the expression of individual genes in ∆UL135 and ∆UL138 infection, respectively. Orange dot size is proportional to the mean expression of individual genes in both ∆UL135 and ∆UL138 infection. For corresponding genes at the other two time points, refer to SI Appendix, Fig. S5. (D) Optimal kernel density estimates of expression levels of six samples (line colors match samples in A). Blue arrow indicates the variation caused by highly expressed genes. (E) Dispersion (orange, Left y axis) and cumulative dispersion (blue, Right y axis) measurement for ∆UL135 and ∆UL138 transcriptomes at 12 and 48 hpi (n = 4). Wave 1 (red shading) and wave 2 (cyan shading) from Fig. 3B are also highlighted in this dataset.
We next explored how viral gene expression might be differentially regulated in fibroblasts during ∆UL135 and ∆UL138 infection relative to WT. Analysis of log2 fold change as a function of gene counts revealed that the majority of genes were concordantly regulated (Q1/Q3) (Fig. 4B; see SI Appendix, Fig. S5C for 24 and 72 hpi). The maximal antagonistic expression was observed at 12 hpi with five genes (UL135, UL136, US12, US17, and US21) in Q2 (Fig. 4 B and C); however, these genes do not reach a twofold change (Fig. 4C, red rectangles in each panel). MA plots across the four time points comparing the mutant viruses or each mutant virus to WT show that expression of only a few genes was significantly different (P < 0.05, SI Appendix, Fig. S6). This is again in contrast to the antagonistic relationship between ∆UL135 and ∆UL138 that increases over time in CD34+ HPCs (Fig. 2D).
Kernel density estimation for the distribution of viral gene expression in fibroblasts revealed that the heterogeneity between viral transcriptomes was distributed similarly across the expression range such that the wave 1 and wave 2 regulatory patterns observed in CD34+ HPCs were lost (Fig. 4D). Using different bandwidth settings in kernel density estimates (SI Appendix, Fig. S7A) and comparing between 12 real and random samples (SI Appendix, Fig. S7B), we confirmed a similar level of variation across the expression range, including the high expression region (Fig. 4D, blue arrow). We then calculated the genewise dispersion estimates for two mutant virus infections at 12 and 48 hpi and found that the expression variability between them was low (Fig. 4E). Furthermore, there were no significant changes in viral gene expression within the wave 1 vs. wave 2 regions in the fibroblast dataset (P = 0.128, Wilcoxon rank sum test). Taken together, these results indicate that the UL138-mutant virus transcriptome is not substantially different from that of ∆UL135 or WT infection in fibroblasts, and instead three transcriptomes converge over time as indicated by PCA.
Analysis of HCMV Transcriptome in Clinical Latency.
Our targeted enrichment platform provided us with the sensitivity to analyze the HCMV transcriptome associated with latency in 12 healthy individuals (clinical latency). None of these donors were supporting active viral replication since no viral cytopathic effect or immediate early gene expression could be detected following incubation of fibroblasts with plasma from each of these donors (SI Appendix, Fig. S8A). Further, as HCMV genomes are maintained in PBMCs at or below the limit of quantitation of quantitative real-time PCR (qPCR), viral genomes were only detected in three of the donors and at a frequency well below that of a host gene (SI Appendix, Fig. S8B).
RNA isolated from PBMCs of the HCMV-seropositive subjects was pooled and viral cDNAs were enriched using our SureSelect platform for RNA sequencing. Given the low number of HCMV reads mapped (SI Appendix, Dataset S1), we analyzed read diversity by determining the percent identity of HCMV reads from clinical or in vitro infection samples to the TB40/E reference sequence (blastn, e-value < 1e-5) (SI Appendix, Fig. S9A). The clinical reads share significantly lower similarity to the TB40/E reference than in vitro infections (P < 0.01, Wilcoxon rank sum test). Further, clusters of those pooled HCMV reads and Shannon entropy estimates indicated that intrasample diversity was greatest for clinical reads (SI Appendix, Fig. S9B). Finally, comparison of variants using SAMtools (35) and GATK workflow (36) revealed high-confidence single nucleotide polymorphisms (SNPs) that were present in all in vitro samples, but not detected in clinical samples (SI Appendix, Fig. S9C). Collectively, these analyses demonstrate that the viral reads obtained from clinical samples represent bona fide natural infection.
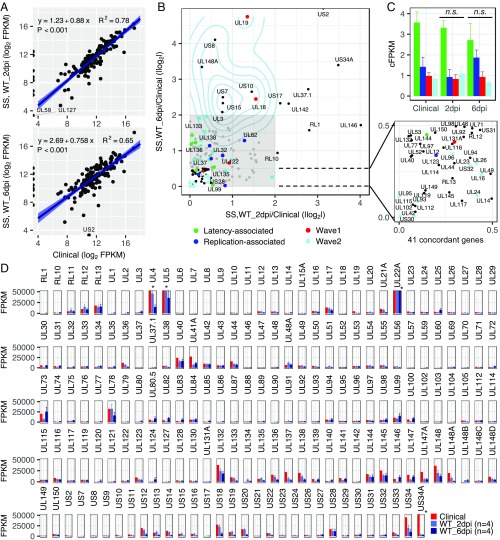
The enriched clinical transcriptome shared high correlation with the enriched viral transcriptomes from CD34+ HPCs infected in vitro at 2 dpi (R2 = 0.78) and 6 dpi (R2 = 0.65) (Fig. 5A). This indicates conservation of the transcriptomes associated with infection of hematopoietic cells. Nine genes (UL1, UL2, UL8, UL59, UL90, UL120, UL127, UL134, and UL148B) detected in in vitro transcriptomes were absent in the clinical samples, the majority of which encode putative membrane proteins or uncharacterized proteins.
Fig. 5.
Comparison of in vitro infection in CD34+ HPCs to clinical latency. Enriched HCMV transcriptomes from CD34+ HPCs infected with WT in vitro or from PBMCs isolated from seropositive individuals were compared. (A) Gene abundance (FPKM) comparisons between in vitro WT infection at 2 (Top) and 6 (Bottom) dpi vs. clinical latency. Pearson’s correlation coefficient is calculated. Level of confidence intervals for predictions of a linear model is 0.95. (B) Comparison of individual viral genes expressed at 2 or 6 dpi in vitro vs. clinical latency using absolute log fold change (ALFC). A total of 41 concordant genes (ALFC < 0.5, zoom) were identified. Similarly expressed (0.5 < ALFC < 2) genes are in the shaded area. Differences in viral gene expression (AFLC > 2) between 6 dpi in vitro and clinical latency are indicated by density plot. Latency- and replication-associated genes are indicated by green and blue dots, respectively. Wave 1 and wave 2 genes (Fig. 3D) are indicated by red and cyan dots, respectively. (C) FPKM of genes in the four groups in B was normalized by the geometric mean of 41 concordant genes (cFPKM). All error bars are SEM. n.s., no significant difference in the expression of each gene group between clinical and 2 or 6 dpi samples (P > 0.1, Wilcoxon test). (D) HCMV gene abundance across genome in clinical and in vitro (2 and 6 dpi) infections from four samples with three donors. All error bars are SEM; * indicates FPKM values >50,000 (see SI Appendix, Dataset S2 for FPKM values).
Comparing gene expression in clinical and in vitro infection samples revealed that 41 genes were concordantly expressed at 2 and 6 dpi in vitro relative to the clinical sample, defined as an absolute log2 fold change (ALFC) <0.5 (Fig. 5B, zoom). Half of these concordantly expressed genes are conserved among all herpesviruses or β-herpesviruses. We then examined latency-associated (UL133, UL135, UL136, UL138, UL144, and US28; Fig. 5B, green) and replication-associated genes (UL32, UL82, UL99, UL122, UL123; Fig. 4B, blue) in in vitro and clinical latency. These genes differed by ALFC <2. We also specifically examined the 30 genes in Fig. 3D that were differentially expressed during ∆UL135 and ∆UL138 infection across all replicates and found that the majority also differed by ALFC <2 (Fig. 5B, red and cyan dots). In Fig. 5C, FPKM was normalized to the 41 concordant genes (cFPKM) to facilitate intergroup comparisons between latency-associated (green), replication-associated (blue), wave 1 (red), and wave 2 (cyan) genes. There were no significant differences in the expression of each gene group between clinical and 2 or 6 dpi samples (P > 0.1, Wilcoxon test). These comparisons indicate a high level of conservation in transcriptomes between clinical samples and this experimental model.
To provide a landscape of HCMV gene expression associated with HCMV latency, mean expression from our in vitro (n = 4, three unique donor pools) and clinical samples is shown in Fig. 5D (see SI Appendix, Dataset S2 for FPKM values per sample). UL4, UL5, and UL22A were highly expressed across all samples; however, low levels of expression were detected from many regions across the genome. We also show a heatmap for expression of the 100 most highly expressed genes across all biological samples (SI Appendix, Fig. S10). These data further demonstrate consistency of gene expression between natural infection in PBMCs and CD34+ HPCs infected in vitro.
The latent transcriptome defined for in vitro infection and clinical samples includes a larger number of genes than anticipated based on our current understanding of HCMV latency. One concern is that the transcriptome may be influenced by a small number of cells supporting lytic replication. To exclude this possibility, we analyzed the transcriptomes from CD34+ HPCs infected for 10 d in the presence or absence of ganciclovir (GCV). GCV is a nucleoside analog that is toxic to cells replicating viral DNA and will kill cells undergoing lytic replication. While viral gene expression was generally decreased at 10 dpi compared with 2 or 6 dpi (SI Appendix, Fig. S11 vs. Fig. 5D), GCV treatment did not substantially alter the profile of gene expression in CD34+ HPCs infected with WT virus (SI Appendix, Fig. S11). From these data, we conclude that the profile of gene expression is indicative of the latent transcriptome and is not heavily influenced by cells undergoing lytic replication.
Discussion
Understanding the patterns of viral gene expression associated with HCMV persistence is an important goal toward defining the molecular underpinnings of latency and its associated health risks. This is a challenging question to address because CMV genomes are maintained and genes are expressed at exceedingly low levels in hematopoietic sites of latency. The development of a custom targeted enrichment platform was essential for transcriptome-wide characterization in latently infected human samples and in detecting viral transcripts expressed at low levels during in vitro infection of CD34+ HPCs. Targeted enrichment provides an efficient and robust method for recovering the HCMV transcriptome in the context of natural infection where HCMV transcripts typically comprise less than 0.0001% of the total transcriptome. Enrichment yielded a >6,000-fold increase in the ratio of virus-to-human reads without skewing the transcriptome (Fig. 1). Previous work by Rossetto et al. (27) reported transcripts in natural infection in the absence of enrichment; however, we found that the number of viral reads mapped in human samples was too low for accurate transcriptome quantification. Even following enrichment, three samples were pooled to provide sufficient reads for robust computational analysis. Our computational analysis defines the breadth of viral gene expression in the latently infected host (Fig. 5D) and identifies 141 genes [absolute log fold change (AFLC) < 2] that are similarly expressed in natural infection and experimental models of latency (Fig. 5B), providing targets of study to better understand HCMV latency in the host. Further, using viruses that differ in their ability to establish latency or reactivate in CD34+ HPCs extends our understanding of the viral transcriptome associated with the establishment of latent or replicative states in hematopoietic reservoirs (Figs. 2 and 3). Importantly, many genes identified by this study have been understudied and do not have well-understood functions in infection. Therefore, this study provides an initial road map to advance our understanding of HCMV latency and persistence.
Heterogeneity inherent to hematopoietic sites of HCMV latency highlights the likelihood that the transcriptome associated with these cell populations reflects not a single transcriptome, but an aggregate of many associated with specific subpopulations within the larger PBMC or CD34+ populations. As such, our study defines the breadth of viral gene expression in the host and in experimental models for latency rather than a single transcriptome. The transcriptome derived from clinical samples or CD34+ HPCs infected in vitro contains transcripts from all classically defined kinetic classes (14) (Fig. 5D). A common criticism of HCMV latent transcriptome studies is the likelihood that the latent transcriptome may be skewed by a disproportionate contribution of transcripts from a minority of cells undergoing lytic replication; a tenable argument given that HCMV reactivation is intimately linked to hematopoietic cell differentiation. We have addressed this possible caveat by defining the transcriptome in CD34+ HPCs infected in vitro and treated with ganciclovir to eliminate cells undergoing lytic replication. Under this treatment, the transcriptome defined in CD34+ HPCs was stable (SI Appendix, Fig. S11), indicating that our transcriptomes are not overwhelmingly influenced by cells productively replicating virus. We also detected low to no viral genomes or infectious virus in clinical samples (SI Appendix, Fig. S8), providing further evidence that the donors were not supporting detectable virus replication and were not viremic. Defining individual transcriptomes present within this aggregate transcriptome can only be addressed through single cell sequencing, which is challenging in the case of natural infection due to the low frequency of cells harboring HCMV genomes and expressing HCMV genes. Further, it is difficult to capture robust sequencing data for low abundance transcripts using single cell sequencing. While the data presented here represent unprecedented depth for the natural infection and computational analysis for HCMV, the landscape of viral gene expression is broadly consistent with other genome-wide studies in hematopoietic cells infected with HCMV (15, 16, 20, 26, 27, 37). The similarity between the HCMV transcriptome from clinical samples (PBMCs) and CD34+ HPCs infected in vitro (Fig. 5A) provides validation of our experimental CD34+ HPC model for the study of infection and latency in hematopoietic cells and suggests some conservation of the viral transcriptome across hematopoietic cell subpopulations.
The use of recombinant viruses that serve to shift infection to a predominantly nonreactivating, latent-like (∆UL135) or replicative (∆UL138) state is a powerful tool to identify genes important for latency or replication in CD34+ HPCs. While WT and ∆UL135 infections were strikingly similar, a number of HCMV genes were antagonistically expressed in the context of a ∆UL135 vs. a ∆UL138 infection in CD34+ HPCs (Fig. 2), a phenomenon not observed in fibroblasts that only support productive replication (Fig. 4). We identified 30 genes that were expressed at low to moderate levels across all biological replicates (Fig. 3) and were also detected in clinical samples (Fig. 5B). While we anticipate that genes important to infection and persistence in hematopoietic cells extend beyond these 30 genes, this represents a robust core group of gene candidates that may contribute to distinct patterns of infection. For example, higher expression of UL135, which promotes reactivation and replication, in ∆UL138 infection relative to WT fits within existing models (23, 38).
Genes antagonistically regulated in the context of ∆UL135 and ∆UL138 infections reflect the antagonistic functional relationship described for UL135 and UL138 (23, 38) and may be important to the switch between latent and replicative states. The UL135 and UL138 proteins are membrane bound and associated with cytoplasmic secretory membranes. As such, the mechanisms by which UL135 and UL138 impact viral gene expression may be indirect through their opposing effects on host signaling (38) or through an effect of UL138 in suppressing IE gene expression (39). By contrast, concordantly regulated genes suggest a partnership between UL135 and UL138, such that the loss of either partner produces a similar effect on viral gene expression. Consistent with this possibility, we previously demonstrated interaction between UL135 and UL138 proteins (38). The functional relevance of these differentially regulated viral genes to the outcomes associated ∆UL135 and ∆UL138 infection awaits further investigation to understand their role in regulating latency and the switch between latent and replicative states of infection.
In comparison with the genes differentially regulated during ∆UL135 and ∆UL138 infection in CD34+ HPCs, genes that are highly expressed in CD34+ HPCs and natural infection in PBMCs (e.g., UL4, UL5, UL22A, and UL132) were also highly expressed in fibroblasts and were not differentially impacted by ∆UL135 and ∆UL138 infection (Figs. 2, 3, and 5). These genes have been previously reported as being highly expressed in monocyte-derived cell types (26) and fibroblasts (13). While lncRNAs were excluded from our analyses, HCMV lncRNAs are also known to be highly expressed across cell types (13, 26, 27, 37). These highly expressed genes may play a fundamental role in infection regardless of cell type, but may be less likely to have substantial impact on the switch between latent and productive states because of their uniform expression across cell types and infection states. A comparison of the 100 most highly expressed genes between clinical latency, CD34+ HPCs infected in vitro, and fibroblasts infected in vitro, is provided in SI Appendix, Fig. S10. From our analysis, we propose a model whereby viral genes expressed at low to moderate levels and differentially regulated in ∆UL135 or ∆UL138 infection, although representing a minor subset of virus–host metatranscriptome, may have a greater impact on directing the pattern of infection than more abundantly expressed viral genes.
As nucleic acid detection approaches have exponentially increased in depth and sensitivity, the breadth of gene expression detected in the context of latency has also increased. As such, the notion of strict quiescence during latency is being challenged for all herpesviruses. Broad gene expression has also been reported in the context of herpes simplex virus type 1 (HSV-1) and varicella zoster virus (VZV) latency (40–44). In latently infected mice, single cell sequencing revealed HSV-1 lytic gene expression in the majority of infected dorsal root ganglia, which was accompanied by detectable protein expression (43, 45). Together these studies indicate that herpesvirus latency may be a more dynamic state than previously appreciated with regard to viral gene expression and highlights the risk in defining latency as the presence of a single latency transcript in the absence of a single lytic transcript. This study paves the way to establish paradigms of HCMV latency through enhanced definition of the viral transcriptome associated with natural infection in the host and latent vs. replicative states in an experimental model of latency.
Materials and Methods
Cells and Viruses.
For details on the culture and TB40/E strain infection of CD34+ HPCs and fibroblasts, see SI Appendix, SI Materials and Methods.
Clinical Latency Human Samples.
Healthy donors gave consent and PBMCs were collected using a consent form and protocol approved by the Institutional Review Board at the University of Arizona (IRB 1510182734). Human peripheral blood samples were obtained and PBMCs and plasma were cryopreserved from 12 healthy individuals known to be CMV seropositive. Human subjects ranged in age from 24 to 78 y old, with a mean ± SD age of 49 ± 21 y. Eleven of the 12 subjects self-identified as Caucasian, 1 as Asian; 2 subjects self-identified as Hispanic, and 10 as non-Hispanic. The presence of Abs against CMV was determined by ELISA on frozen plasma samples, as previously described (46). CMV titer was determined utilizing a standard curve from a confirmed clinical positive control as the reference with a negative control cutoff of 1:30. Additionally, a clinically verified negative CMV control was run with every plate. CMV serological titer ranged from 117 to 6,281, with a mean of 1,258 ± 1,698. Cryopreserved PBMC samples were thawed, cells were counted and immediately lysed in RNA/DNA lysis buffer (Zymo Research) and stored at −80 °C until nucleic acid was isolated. Three libraries were prepared for next-generation sequencing (NGS) from a pool of four donors.
SureSelect Enrichment RNA Bait Design.
SureSelect enrichment probes were designed in collaboration with the bait design team at Agilent Technologies. Enrichment probes were designed as overlapping (2× tiling), 120-mer RNA baits spanning the positive strand of the HCMV TB40/E reference genome (GenBank accession EF999921.1). Regions of the HCMV genome with more than 70% identity to the human genome were masked to avoid enrichment of nonviral sequences. The HCMV long noncoding RNAs (4.9 kb, 2.7 kb, and 1.2 kb) were also excluded, as they have been shown to be expressed to high levels during HCMV infection and may prevent adequate enrichment of rare transcripts expressed during HCMV latency (13, 27). Bait libraries were synthesized by Agilent. All bait designs are available in Agilent’s eArray software and from the corresponding author. Control baits, including sequences for TATA binding protein (Entrez gene accession no. 25833) and POU2F3 (accession NM014352) were incorporated to use in quantification of virus-sequence enrichment and human-sequence depletion.
NGS Library Preparation and Sequencing.
Samples frozen at −80 °C in RNA/DNA lysis buffer were thawed and RNA was isolated using the ZR-Duet Kit (Zymo Research), following manufacturer instructions. Isolated RNA was DNase I treated off column and repurified using the Machery-Nagel RNA II Kit. Following this additional DNaseI treatment, no sequences are amplified by PCR in the absence of reverse transcriptase, indicating the absence of contaminating DNA. RNA quality was assessed using the Agilent Bioanalyzer; RNA preparations used for subsequent NGS library preparation had an RNA integrity number (RIN) of ≥9. NGS library preparation was performed using manufacturer guidelines, including recommended quality control steps using either Agilent’s SureSelect Strand-Specific RNA Library Preparation Kit or Kapa Biosystems KAPA Stranded mRNA-Seq Kit. Briefly, 500 ng–1 μg of total RNA from CD34 cells or 4 µg of total RNA from fibroblasts was poly-A selected and chemically sheared, reverse transcribed, end repaired, adapter ligated, and PCR amplified. For samples processed without SureSelect enrichment, barcodes were added to adapters in a final, low cycle number PCR. All libraries were analyzed on the Agilent Bioanalyzer before SureSelect and the Advanced Analytical Technologies, Inc. (AATI) Fragmentation Analyzer (with AATI NGS High-Sensitivity Kit) before HiSeq loading for assessing library size and DNA contamination. For SureSelect enrichment, 100 ng of each library was hybridized to RNA enrichment probes (described above). After purification of enriched viral sequences, barcodes were added in a final PCR amplification step. Samples were multiplexed and sequenced using either HiSeq or the MiSeq. Raw sequencing data were demultiplexed and fastq files generated using either built-in software (MiSeq) or CASAVA (HiSeq). All project sequence reads are available at the National Center for Biotechnology Information (NCBI) under accession number GSE99823.
Quality Reads and Alignments.
RNAseq datasets refer to the following categories: (i) CD34+ HPC samples (2 and 6 dpi) were sequenced yielding a total of ∼269/143 million, uniform 101-bp paired-end reads for donor 1-NS/donor 2-NS samples, and ∼12/15 million, 151 bp (see SI Appendix, Fig. S12 for read-length distribution) paired-end reads for donor 1-SS/donor 3-SS samples; CD34+ HPC samples (10 dpi, SS) were sequenced yielding a total of ∼47 million, uniform 101-bp paired-end reads. (ii) Three pooled clinical samples contain ∼6 million, 151 bp (see SI Appendix, Fig. S12 for read length distribution) paired-end reads. (iii) Fibroblast samples were sequenced yielding a total of ∼204 million, 101-bp paired-end reads. (iv) Mock samples in CD34+ HPCs and fibroblasts were also sequenced (SI Appendix, Dataset S1). Raw sequence data were first evaluated using FastQC (v0.11.3, www.bioinformatics.babraham.ac.uk/projects/fastqc/) and preprocessed for quality through a combination of trimming and filtering using Trim Galore (v0.4.0, 15 bases were trimmed off from the 5′ end of the reads and five bases were trimmed off from the 3′ end; Phred score threshold of 20 and minimum length of 50 bp, paired, www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Quality reads were then uniquely mapped to HCMV (strain TB40/E, GenBank: EF999921.1) and human (GRCh38) genomes using Tophat2 (v2.1.1) (47) with strand-specific alignment of fr-firststrand. For HCMV alignment, the maximum intron size was set to 5 kb as described in the Tophat2 manual, and the uniquely mapped HCMV, but not human, reads were used for all subsequent analyses.
Differential Expression.
Raw read counts for each sample were obtained by mapping reads at the gene-level using HTSeq-count tool from the Python package HTSeq (48), with a stranded setting (reverse). DESeq2 R package (31) (v1.8.2) was then used to perform DE and statistical analysis, with a biological sample from SS and NS libraries of the same cell donor, grouped. We combined two-dimensional DE of ∆UL135/WT and ∆UL138/WT as Cartesian coordinates to form an antagonistic regulation model, where antagonistically and significantly (FDR < 0.05) regulatory genes reside at quadrants 2 and 4. Those genes were quantified by counts and the difference across time of postinfection was accessed using Fisher’s exact test.
Rlog-Based Kernel Density Variation.
Raw read counts for genes over a group of six biological samples (WT, ∆UL135, and ∆UL138, each at 2 and 6 dpi) of CD34+ HPC infection were normalized through a regularized logarithm transformation (rlog) implemented in DESeq2 (31), and kernel density estimates (KDEs) were then obtained using the density R function with default parameters. The six density curves were overlaid on one plot. The featured density variation was further evaluated using an extended group composed of all NS/SS replicates from different cell donors (the same six biological samples in each panel, four panels, for a total of 24 samples). Density variation across samples was accessed by different bandwidth settings (0.5, 0.75, 1, and 1.25), which determine the degree of smoothing in the estimate of the density function. In addition, density variation across samples was assessed by the comparison between real and random samples, where raw read count for each gene in each sample was the mean value of 100 permutations of involved real samples and rlog normalization was performed. The same pipeline is applied to the fibroblast dataset.
For details on additional computational analysis, see SI Appendix, SI Materials and Methods.
Statistical Tests.
Statistical tests were performed and Benjamini–Hochberg adjusted P values were calculated using R (https://www.r-project.org/).
Supplementary Material
Acknowledgments
We thank Nat Moorman (University of North Carolina-Chapel Hill) for helpful discussions and insight in designing the SureSelect enrichment platform; Dr. Jeff Frelinger, Dr. Joanne Berghout, Sebastian Zeltzer, and Mike Rak (University of Arizona) for helpful discussion and critical reading of the manuscript; Suzu Igarashi and Sebastian Zeltzer for assistance in analyzing donor plasma; Donna Collins-McMillen for assistance with library preparation; Ryan Sprissler and Jonathan Galina-Mehlman and the Arizona Research Laboratories Division of Biotechnology, University of Arizona Genetics Core, for expertise in library, preparation, analysis, and sequencing; Paula Campbell and Mark Curry (Arizona Cancer Center/Arizona Research Laboratories Division of Biotechnology Cytometry Core Facility) for expertise and assistance in flow cytometry; and special thanks to Terry Fox Laboratory for providing the M2-104 and Sl/Sl cells. This work was supported by Public Health Service Grants AI079059 and AI105062 (to F.G.) and AG048021 (to J.N.-Z.) from the National Institute of Allergy and Infectious Diseases and the National Institute on Aging, respectively. This work was also supported in part by the Cytometry Shared Resource, University of Arizona Cancer Center (P30CA023074). J.B. was supported by a National Cancer Institute Training Grant T32 (CA009213) and a fellowship from the American Cancer Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE99823).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710522114/-/DCSupplemental.
References
- 1.Davison AJ, et al. The human cytomegalovirus genome revisited: Comparison with the chimpanzee cytomegalovirus genome. J Gen Virol. 2003;84:17–28. doi: 10.1099/vir.0.18606-0. [DOI] [PubMed] [Google Scholar]
- 2.Murphy E, et al. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci USA. 2003;100:14976–14981. doi: 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy E, Rigoutsos I, Shibuya T, Shenk TE. Reevaluation of human cytomegalovirus coding potential. Proc Natl Acad Sci USA. 2003;100:13585–13590. doi: 10.1073/pnas.1735466100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stern-Ginossar N, et al. Decoding human cytomegalovirus. Science. 2012;338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reeves M, Sinclair J. Aspects of human cytomegalovirus latency and reactivation. Curr Top Microbiol Immunol. 2008;325:297–313. doi: 10.1007/978-3-540-77349-8_17. [DOI] [PubMed] [Google Scholar]
- 6.Goodrum F. Human cytomegalovirus latency: Approaching the Gordian knot. Annu Rev Virol. 2016;3:333–357. doi: 10.1146/annurev-virology-110615-042422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britt W. Manifestations of human cytomegalovirus infection: Proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol. 2008;325:417–470. doi: 10.1007/978-3-540-77349-8_23. [DOI] [PubMed] [Google Scholar]
- 8.Mocarski ES, Shenk T, Pass RF. Cytomegaloviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Lippincott, Williams & Wilkins; Philadelphia: 2007. pp. 2701–2673. [Google Scholar]
- 9.Razonable RR, Humar A. AST Infectious Diseases Community of Practice Cytomegalovirus in solid organ transplantation. Am J Transplant. 2013;13:93–106. doi: 10.1111/ajt.12103. [DOI] [PubMed] [Google Scholar]
- 10.Ariza-Heredia EJ, Nesher L, Chemaly RF. Cytomegalovirus diseases after hematopoietic stem cell transplantation: A mini-review. Cancer Lett. 2014;342:1–8. doi: 10.1016/j.canlet.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Cannon MJ. Congenital cytomegalovirus (CMV) epidemiology and awareness. J Clin Virol. 2009;46(Suppl 4):S6–S10. doi: 10.1016/j.jcv.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Syggelou A, Iacovidou N, Kloudas S, Christoni Z, Papaevangelou V. Congenital cytomegalovirus infection. Ann N Y Acad Sci. 2010;1205:144–147. doi: 10.1111/j.1749-6632.2010.05649.x. [DOI] [PubMed] [Google Scholar]
- 13.Gatherer D, et al. High-resolution human cytomegalovirus transcriptome. Proc Natl Acad Sci USA. 2011;108:19755–19760. doi: 10.1073/pnas.1115861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weekes MP, et al. Quantitative temporal viromics: An approach to investigate host-pathogen interaction. Cell. 2014;157:1460–1472. doi: 10.1016/j.cell.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodrum FD, Jordan CT, High K, Shenk T. Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: A model for latency. Proc Natl Acad Sci USA. 2002;99:16255–16260. doi: 10.1073/pnas.252630899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodrum F, Jordan CT, Terhune SS, High K, Shenk T. Differential outcomes of human cytomegalovirus infection in primitive hematopoietic cell subpopulations. Blood. 2004;104:687–695. doi: 10.1182/blood-2003-12-4344. [DOI] [PubMed] [Google Scholar]
- 17.Hargett D, Shenk TE. Experimental human cytomegalovirus latency in CD14+ monocytes. Proc Natl Acad Sci USA. 2010;107:20039–20044. doi: 10.1073/pnas.1014509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reeves MB, Sinclair JH. Analysis of latent viral gene expression in natural and experimental latency models of human cytomegalovirus and its correlation with histone modifications at a latent promoter. J Gen Virol. 2010;91:599–604. doi: 10.1099/vir.0.015602-0. [DOI] [PubMed] [Google Scholar]
- 19.Reeves M, Sinclair J. Regulation of human cytomegalovirus transcription in latency: Beyond the major immediate-early promoter. Viruses. 2013;5:1395–1413. doi: 10.3390/v5061395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung AK, Abendroth A, Cunningham AL, Slobedman B. Viral gene expression during the establishment of human cytomegalovirus latent infection in myeloid progenitor cells. Blood. 2006;108:3691–3699. doi: 10.1182/blood-2005-12-026682. [DOI] [PubMed] [Google Scholar]
- 21.Slobedman B, Mocarski ES. Quantitative analysis of latent human cytomegalovirus. J Virol. 1999;73:4806–4812. doi: 10.1128/jvi.73.6.4806-4812.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Umashankar M, et al. A novel human cytomegalovirus locus modulates cell type-specific outcomes of infection. PLoS Pathog. 2011;7:e1002444. doi: 10.1371/journal.ppat.1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umashankar M, et al. Antagonistic determinants controlling replicative and latent states of human cytomegalovirus infection. J Virol. 2014;88:5987–6002. doi: 10.1128/JVI.03506-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain V, et al. A toolbox for herpesvirus miRNA research: Construction of a complete set of KSHV miRNA deletion mutants. Viruses. 2016;8:E54. doi: 10.3390/v8020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Depledge DP, et al. Specific capture and whole-genome sequencing of viruses from clinical samples. PLoS One. 2011;6:e27805. doi: 10.1371/journal.pone.0027805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Damme E, et al. HCMV displays a unique transcriptome of immunomodulatory genes in primary monocyte-derived cell types. PLoS One. 2016;11:e0164843. doi: 10.1371/journal.pone.0164843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossetto CC, Tarrant-Elorza M, Pari GS. Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14 (+) monocytes and CD34 (+) cells. PLoS Pathog. 2013;9:e1003366. doi: 10.1371/journal.ppat.1003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodrum F, Reeves M, Sinclair J, High K, Shenk T. Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood. 2007;110:937–945. doi: 10.1182/blood-2007-01-070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrucelli A, Rak M, Grainger L, Goodrum F. Characterization of a novel Golgi apparatus-localized latency determinant encoded by human cytomegalovirus. J Virol. 2009;83:5615–5629. doi: 10.1128/JVI.01989-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hebenstreit D, et al. RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol Syst Biol. 2011;7:497. doi: 10.1038/msb.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heimberg G, Bhatnagar R, El-Samad H, Thomson M. Low dimensionality in gene expression data enables the accurate extraction of transcriptional programs from shallow sequencing. Cell Syst. 2016;2:239–250. doi: 10.1016/j.cels.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tirosh O, et al. The transcription and translation landscapes during human cytomegalovirus infection reveal novel host-pathogen interactions. PLoS Pathog. 2015;11:e1005288. doi: 10.1371/journal.ppat.1005288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raftery MJ, et al. Unravelling the interaction of human cytomegalovirus with dendritic cells by using SuperSAGE. J Gen Virol. 2009;90:2221–2233. doi: 10.1099/vir.0.010538-0. [DOI] [PubMed] [Google Scholar]
- 38.Buehler J, et al. Opposing regulation of the EGF receptor: A molecular switch controlling cytomegalovirus latency and replication. PLoS Pathog. 2016;12:e1005655. doi: 10.1371/journal.ppat.1005655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SH, Albright ER, Lee JH, Jacobs D, Kalejta RF. Cellular defense against latent colonization foiled by human cytomegalovirus UL138 protein. Sci Adv. 2015;1:e1501164. doi: 10.1126/sciadv.1501164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baird NL, Bowlin JL, Cohrs RJ, Gilden D, Jones KL. Comparison of varicella-zoster virus RNA sequences in human neurons and fibroblasts. J Virol. 2014;88:5877–5880. doi: 10.1128/JVI.00476-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giordani NV, et al. During herpes simplex virus type 1 infection of rabbits, the ability to express the latency-associated transcript increases latent-phase transcription of lytic genes. J Virol. 2008;82:6056–6060. doi: 10.1128/JVI.02661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kramer MF, Chen SH, Knipe DM, Coen DM. Accumulation of viral transcripts and DNA during establishment of latency by herpes simplex virus. J Virol. 1998;72:1177–1185. doi: 10.1128/jvi.72.2.1177-1185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma JZ, Russell TA, Spelman T, Carbone FR, Tscharke DC. Lytic gene expression is frequent in HSV-1 latent infection and correlates with the engagement of a cell-intrinsic transcriptional response. PLoS Pathog. 2014;10:e1004237. doi: 10.1371/journal.ppat.1004237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagel MA, et al. Varicella-zoster virus transcriptome in latently infected human ganglia. J Virol. 2011;85:2276–2287. doi: 10.1128/JVI.01862-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russell TA, Tscharke DC. Lytic promoters express protein during herpes simplex virus latency. PLoS Pathog. 2016;12:e1005729. doi: 10.1371/journal.ppat.1005729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wertheimer AM, et al. Aging and cytomegalovirus infection differentially and jointly affect distinct circulating T cell subsets in humans. J Immunol. 2014;192:2143–2155. doi: 10.4049/jimmunol.1301721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anders S, Pyl PT, Huber W. HTSeq: A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.