Significance
Aedes aegypti is the principal vector of multiple arboviruses that significantly affect human health, including dengue, chikungunya, and zika. Development of tools for efficient genome engineering in this mosquito will not only lay the foundation for the application of genetic control strategies, but will also accelerate basic research on key biological processes involved in disease transmission. Here, we report the development of a transgenic CRISPR approach for rapid gene disruption in this organism. Given their high editing efficiencies, the Cas9 strains we developed can be used to quickly generate genome modifications, allowing for high-throughput gene targeting, and can possibly facilitate the development of gene drives, thereby accelerating comprehensive functional annotation and development of innovative population control strategies.
Keywords: Aedes aegypti, germline, cas9, CRISPR, mutagenesis
Abstract
The development of CRISPR/Cas9 technologies has dramatically increased the accessibility and efficiency of genome editing in many organisms. In general, in vivo germline expression of Cas9 results in substantially higher activity than embryonic injection. However, no transgenic lines expressing Cas9 have been developed for the major mosquito disease vector Aedes aegypti. Here, we describe the generation of multiple stable, transgenic Ae. aegypti strains expressing Cas9 in the germline, resulting in dramatic improvements in both the consistency and efficiency of genome modifications using CRISPR. Using these strains, we disrupted numerous genes important for normal morphological development, and even generated triple mutants from a single injection. We have also managed to increase the rates of homology-directed repair by more than an order of magnitude. Given the exceptional mutagenic efficiency and specificity of the Cas9 strains we engineered, they can be used for high-throughput reverse genetic screens to help functionally annotate the Ae. aegypti genome. Additionally, these strains represent a step toward the development of novel population control technologies targeting Ae. aegypti that rely on Cas9-based gene drives.
The yellow fever mosquito, Aedes aegypti, is the principal vector of many arboviruses, such as dengue, chikungunya, yellow fever, and zika. These pathogens are globally widespread and pose significant epidemiological burdens on infected populations, resulting in hundreds of millions infections and over 50,000 deaths per year (1–4). Due to the hazards they impose, many methods for controlling Ae. aegypti populations have been implemented, with the most common being chemical insecticides. However, chemical control has proven incapable of stopping the spread of Ae. aegypti, primarily due to the mosquito's ability to rapidly adapt to new climates, tendency to oviposit in minimal water sources, desiccation-tolerant eggs, and quick development of insecticide resistance (5, 6). Therefore, significant efforts are currently underway to discern the underlying molecular and genetic mechanisms important for arboviral vector competence, with the overall aim of developing insecticide-free ways to disrupt viral disease cycles (7). Importantly, uncovering these mechanisms hinges on the ability to stably insert and disrupt specific genes-of-interest through tailored genome engineering in a target-specific manner. Fortunately, several tools have been successfully employed in mosquitoes for targeted genome engineering that rely on either zinc finger nucleases (8–10), transcription activator-like effector nucleases (TALENs) (11, 12), or even homing endonuclease genes (13). However, because they rely on context-sensitive modular protein–DNA-binding interactions, each of these designer nucleases are time-consuming and complicated to engineer and validate, making them onerous for routine use in most laboratories.
To overcome the significant limitations posed by previous genome-editing tools, the clustered regularly interspaced short palindromic repeats/CRISPR-associated sequence 9 (CRISPR/Cas9) system, originally discovered in bacteria and archaea (14–20), has been adapted as a programmable (20, 21) precision genome-editing tool in a diversity of organisms (22–29), including mosquitoes (30–34). Briefly, the CRISPR/Cas9 system is guided by a chimeric programmable synthetic short-guide RNA (sgRNA) (20) that binds Cas9, directing it to a user-specified genomic DNA target sequence, via Watson–Crick base pairing between the sgRNA and the target DNA sequence, thereby generating site-specific double-strand (ds) DNA breaks. This system can be easily reprogrammed to modify virtually any desired genomic sequence by recoding the specificity-determining sequence of the sgRNA. Recoding constraints are minimal and simply require that the target sequence is unique, compared with the rest of the genome, and is located just upstream of a protospacer adjacent motif (PAM sequence), typically consisting of the three nucleotide motif NGG (20, 21). Both the minimal constraints and ease of use make CRISPR/Cas9 a powerful tool for genome-engineering applications.
Importantly, CRISPR-mediated genome engineering of organisms has been achieved in a variety of different ways. For example, direct injection of in vitro-purified sgRNAs combined with either purified Cas9 RNA, recombinant Cas9 protein, or even with Cas9 expression plasmids, have all been successful for a variety of organisms (29, 32, 35–40), including mosquitoes (30–34). However, the rate of mutagenesis and lethality have varied widely, both within and among these different studies, and such discrepancies are likely due to either the methods used to introduce the editing components, the variability in sgRNA functionality against target sites, or even unavoidable variability from manual injection of the components (41). To overcome these significant limitations, previous studies in other organisms have shown that stably expressing a transgenic provision of Cas9 in the germline can decrease toxicity to injected embryos, increase the rates of mutagenesis generated by both nonhomologous end joining (NHEJ) and homology-directed repair (HDR), and can also increase the rates of germline transmission of the disrupted allele to offspring (27, 41–46).
Germline expression of Cas9 is also essential for developing innovative technologies that rely on “gene drive,” which is a novel strategy proposed for the control of vector-borne diseases by rapidly spreading alleles in a population through super-Mendelian inheritance (47, 48). In mosquitoes, gene drives could potentially be used to rapidly disseminate a genetic payload that reduces pathogen transmission throughout a population, thereby suppressing vector competence and human disease transmission. Other possible applications include the suppression of the population by spreading alleles that impair fertility or viability (49, 50). A CRISPR homing-based gene-drive element consists of only a few components, such as an sgRNA and a germline-expressed Cas9 endonuclease that is positioned opposite its target site in the genome. The drive encodes the editing machinery (i.e., Cas9 and sgRNA), allowing it to cut the opposite allele and copy itself into this disrupted allele via HDR, thereby converting a heterozygote into a homozygote, enabling rapid invasion of the drive into a population. In fact, Cas9 has been used to develop highly promising homing-based gene drives in a number of organisms, including yeast (51), Drosophila (52), and Anopheles mosquitoes (52, 53); however, such a system has yet to be developed in Ae. aegypti.
We therefore aimed to develop a CRISPR/Cas9 transgenic system that would enable more robust and widespread Ae. aegypti genome-engineering applications, including disrupting genes important for vector competence, while also laying the foundation for the future development of gene drives. To do so, we utilized several previously described transcriptional regulatory elements (54, 55), many of which are active in the germline (56), to drive expression of Streptococcus pyogenes Cas9 in Ae. aegypti. In total, we developed six independent Cas9 expressing strains, and herein characterize and demonstrate their effectiveness for gene disruption and gene insertion via HDR. We demonstrate the efficiency of using these strains by disrupting six genes in Ae. aegypti, giving rise to severe phenotypes, such as having an extra eye (triple eyes), an extra maxillary palp (triple maxillary palps), a nonfunctional curved proboscis, malformed wings, eye pigmentation deficiencies, and pronounced whole-animal cuticle coloration defects. Furthermore, we also demonstrate the ease of generating double- and triple-mutant strains simultaneously from a single injection, a technique that will facilitate the ease of gene-function study in this nonmodel organism. Overall, gene-disruption efficiencies, survival rates, germline-transmission frequencies, and HDR rates were all significantly improved using the Cas9 strains we develop here. These strains should be highly valuable for facilitating the development of innovative control methods in this organism in the future.
Results
Construction of a Simple Transgenic CRISPR/Cas9 System for Ae. aegypti Mutagenesis.
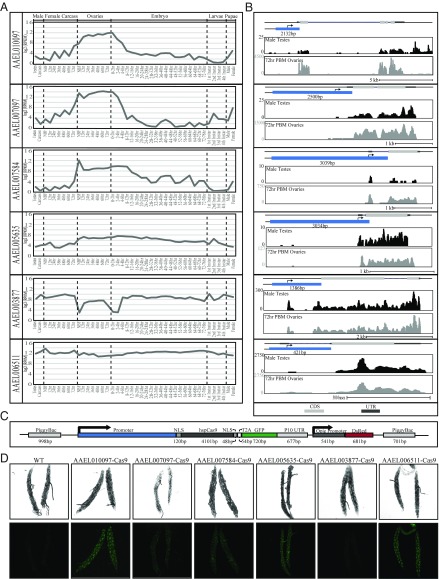
To express Cas9 in the germline of Ae. aegypti, we established transgenic mosquitoes harboring genomic sources of Cas9. To promote robust expression of Cas9, we utilized promoters from six genes including AAEL010097 (Exuperentia), AAEL007097 (4-nitrophenylphosphatase), AAEL007584 (trunk), AAEL005635 (Nup50), AAEL003877 (polyubiquitin), and AAEL006511 (ubiquitin L40), due to their constitutive high levels of expression during many developmental life stages (AAEL003877, AAEL006511, AAEL005635), or high levels of expression in the ovary triggered by uptake of a blood meal (AAEL010097, AAEL007097, AAEL007584), as evidenced from previous promoter characterization experiments or developmental transcriptional profiling (54–56) (Fig. 1 A and B). We inserted these various promoter fragments into a piggybac transposon upstream of the coding sequence for spCas9. Downstream to the promoter-driven Cas9 we included a self-cleaving T2A peptide and eGFP coding sequence, together serving as a visual indicator of promoter activity. We also included a baculovirus-derived Opie2 promoter (57) driving dsRed expression serving as a transgenesis marker (Fig. 1C). The piggybac transgenes were injected into the germline of wild-type embryos (Ae. aegypti genome sequence strain Liverpool) (58), and transgenic mosquitoes harboring these transgenes were readily identified by bright dsRed fluorescent expression in the abdomen (SI Appendix, Fig. S1). We established stable transgenic strains and dissected the germline tissues from each strain to assess promoter activity, and found moderate levels of eGFP present in the 0- to 24-h postblood meal ovary of the AAEL006511-Cas9 and AAEL010097-Cas9 line, and very-weak yet detectable levels of eGFP present in lines AAEL007097-Cas9, AAEL007584-Cas9, and AAEL005635-Cas9. The AAEL010097-Cas9 and AAEL006511-Cas9 lines also exhibited very-weak expression in the 4-d-old testes. Finally, the AAEL003877-Cas9 line exhibited no detectable eGFP signal, consistent with previous work indicating this promoter does not express in the germline (55) (Fig. 1D). Importantly, we were able to establish homozygous stocks for each line, demonstrating that no significant toxicity is associated with high transgenic expression of Cas9.
Fig. 1.
Rationally chosen, native promoters drive strong expression of Cas9. Log2 RPKM expression values for AAEL010097, AAEL007097, AAEL007584, AAEL005635, AAEL003877, and AAEL006511 were plotted across development. Samples include, from left to right: testis; male carcasses (lacking testes); carcasses of females before blood feeding (NBF), and at 12-, 24-, 36-, 48-, and 72-h postblood meal (PBM); ovaries from NBF females and at 12-, 24-, 36-, 48-, and 72-h PBM; embryos from 0–2 h through 72–76 h; whole larvae from first, second, third, and fourth instar; male pupae; female pupae (A). Genome browser snapshots of AAEL010097, AAEL007097, AAEL007584, AAEL005635, AAEL003877, and AAEL006511, including expression tracks for both 72-h PBM ovaries and testes. The light gray box indicates coding sequence; blue box represents promoter element with length indicated in bp; y axis shows the expression level based on raw read counts (B). Schematic representation of the piggybac-mediated Cas9 construct including the S. pyogenes Cas9-T2A-eGFP gene driven by selected promoters (blue), dsRed expressed under the control of the Opie2 promoter, which serves as a transgenesis marker. NLS represent nuclear localization signal (C). Confocal images using white light or GFP-filtering of transgenic Cas9 line ovaries (D). (Magnification, 200×.)
Germline Expression of Cas9 in Ae. aegypti Increases Targeting Efficiency.
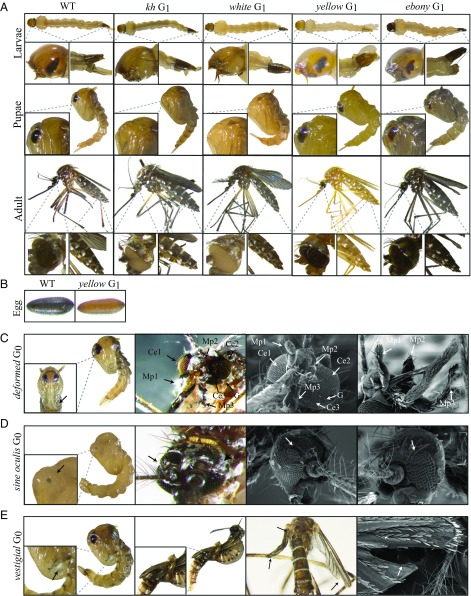
To test the efficacy of our transgenic Ae. aegypti Cas9 strains, we first attempted to mutate the kynurenine hydroxylase (kh) gene (AAEL008879), which was previously characterized as having an easily detectable dominant mutant eye phenotype, as the loss of kh gene function leads to severe eye pigmentation defects (31). To do so, we designed a sgRNA (Kh-sgRNA) targeting exon 4 of the kh gene, and to test its functionality we coinjected this in vitro-transcribed Kh-sgRNA into 450 wild-type embryos [referred to as generation 0 (G0)] along with purified recombinant Cas9 protein. Following injection, complete white-eye and partial mosaic white-eye mutants were readily observed in adults (SI Appendix, Fig. S2). The G0s had a survival rate of 35 ± 7% and a mutagenesis efficiency of 17 ± 3%, and a mutant heritability rate of 33 ± 7% in the G1 generation (Table 1). Once we confirmed functionality of the Kh-sgRNA using purified recombinant Cas9 protein, we then injected this Kh-sgRNA separately into 450 embryos (three injections of 150 embryos in biological triplicate) collected from each of our transgenic Ae. aegypti Cas9 strains without including recombinant Cas9 protein ;in the injection mix. Complete white-eye and mosaic white-eye mutants were readily observed in injected G0 mosquitoes from five of six lines, including AAEL010097-Cas9, AAEL007097-Cas9, AAEL007584-Cas9, AAEL005635-Cas9, and AAEL006511-Cas9. Notably, directly injecting Kh-sgRNAs into these transgenic Cas9 expression lines resulted in significantly higher G0 survival rates (61 ± 7%, 53 ± 4%, 63 ± 4%, 64 ± 6%, and 63 ± 4%, respectively), increased G0 mutagenesis efficiencies (85 ± 5%, 27 ± 6%, 52 ± 7%, 47 ± 3%, and 66 ± 4%, respectively), and increased overall heritable G1 mutation rates (60 ± 9%, 59 ± 6%, 59 ± 5%, 60 ± 6%, and 66 ± 3%, respectively) compared with when Kh-sgRNA was coinjected with recombinant Cas9 protein into wild-type embryos (Table 1). As reported previously, homozygous viable kh mutants have a dramatic eye pigmentation defect that can be visualized in larvae, pupae, and adults (31) (Fig. 2A). To confirm the phenotypic defects described above were due to mutagenesis of the kh gene, genomic DNA spanning the target site was amplified from homozygous mutant G1s (Fig. 2A) and sequenced, confirming the presence of insertion/deletions (indels) in the Kh-sgRNA genomic DNA target site (SI Appendix, Fig. S3). In contrast, Kh-sgRNA injected directly into the AAEL003877-Cas9 strain failed to show any visible mutant phenotypes in the G0 generation, presumably due to lack of germline expression by this promoter (55).
Table 1.
Summary of the injection, survival, and mutagenesis rates mediated by Kh-sgRNA, W1-sgRNA, and Y-sgRNA in wild-type and Cas9-expressing lines
| Ae. aegypti line | Injected component* | No. injected G0s | G0 adult survivors, % | G0 mosaic, % | G1 mutant adult†, % |
| Liverpool (wild-type) | Kh-sgRNA,W1-sgRNA,Y-sgRNA | 450, 450, 450 | 67 ± 4,63 ± 5,61 ± 4 | 0, 0, 0 | 0, 0, 0 |
| Liverpool (wild-type) | Cas9 protein | 450, 450, 450 | 39 ± 4,41 ± 4,44 ± 7 | 0, 0, 0 | 0, 0, 0 |
| Liverpool (wild-type) | Kh-sgRNA,W1-sgRNA,Y-sgRNA /Cas9 protein | 450, 450, 450 | 35 ± 7,40 ± 6,30 ± 8 | 17 ± 3,21 ± 3,18 ± 6 | 33 ± 7,32 ± 5,30 ± 6 |
| AAEL010097-Cas9 | Kh-sgRNA,W1-sgRNA,Y-sgRNA | 450, 450, 450 | 61 ± 7,71 ± 6,65 ± 7 | 85 ± 5,87 ± 6,90 ± 5 | 60 ± 9,69 ± 4,62 ± 4 |
| AAEL007097-Cas9 | Kh-sgRNA,W1-sgRNA,Y-sgRNA | 450, 450, 450 | 53 ± 4,58 ± 4,62 ± 9 | 27 ± 6,33 ± 4,31 ± 5 | 59 ± 6,56 ± 6,62 ± 5 |
| AAEL007584-Cas9 | Kh-sgRNA,W1-sgRNA,Y-sgRNA | 450, 450, 450 | 63 ± 4,55 ± 7,61 ± 5 | 52 ± 7,63 ± 8,57 ± 5 | 59 ± 5,51 ± 7,57 ± 9 |
| AAEL005635-Cas9 | Kh-sgRNA,W1-sgRNA,Y-sgRNA | 450, 450, 450 | 64 ± 6,67 ± 3,55 ± 4 | 47 ± 3,62 ± 7,49 ± 4 | 60 ± 6,50 ± 6,54 ± 4 |
| AAEL003877-Cas9 | Kh-sgRNA,W1-sgRNA,Y-sgRNA | 450, 450, 450 | 51 ± 9,61 ± 8,61 ± 7 | 0, 0, 0 | 0, 0, 0 |
| AAEL006511-Cas9 | Kh-sgRNA,W1-sgRNA,Y-sgRNA | 450, 450, 450 | 63 ± 4,57 ± 5,59 ± 6 | 66 ± 4,73 ± 6,70 ± 7 | 66 ± 3,65 ± 3,67 ± 4 |
Biological triplicate consisting of three independent injections (each injection replicate with 150 embryos total 450 embryos) were performed and the results are shown as the mean ± SE throughout table.
sgRNA 100 ng/μL; Cas9 protein 300 ng/μL.
The overall heritable mutation rate was calculated as the number of mutant G1s divided by the number of all G1s observed.
Fig. 2.
Severe mutant phenotypes caused by CRISPR/Cas9 mediated disruption. Larva, pupae, and adult phenotypes of wild-type, kh, white, yellow, and ebony mutant G1s, respectively, with clearly distinguishable eye (kh and white) and cuticle pigment (yellow and ebony) defects (A). Embryo phenotypes of wild-type (Left) and yellow mutants (Right) (B). Pupae and adult scanning electron microscopy images of the head of the deformed G0 mutants with three compound eyes (Ce), three maxillary palps (Mp), furrowed eyes, and deformed mouthparts (arrows) (C). Pupae and adult scanning electron microscopy images of the head of the sine oculis G0 mutants. Arrows point to the ectopic eyes (D). Images of pupae and adult wings of vestigial G0 mutants. Arrows point to pronounced wing, halteres, and forked wing defects (E). (Magnifications: whole-body images, 20×; Insets, 100×; SEM images, 150×.)
Given that CRISPR/Cas9 targeting efficiency varies significantly between loci and even between target sites within the same locus (26, 27, 30, 36), we wanted to further investigate the efficacy of these transgenic Cas9 expressing strains. We therefore designed two additional sgRNAs targeting uncharacterized conserved genes that may be useful for developing future control methodologies Ae. aegypti. One sgRNA (W1-sgRNA) was designed to target exon 3 of the white gene (AAEL016999) (SI Appendix, Fig. S4A), which is the Aedes 1:1 ortholog to Drosophilia melanogaster white and functions as an ATP-binding cassette transporter important for red and brown eye-color pigmentation (59, 60). Another sgRNA was designed to target exon 2 of the yellow gene (AAEL006830) (SI Appendix, Fig. S5A), which is the Aedes 1:1 ortholog to D. melanogaster yellow, a gene important for melanization of the cuticle (61, 62). To initially test their functionality, we coinjected either in vitro-transcribed W-sgRNA or Y-sgRNA, separately, into 450 wild-type embryos (three injections of 150 embryos in biological triplicate), each along with purified recombinant Cas9 protein. Following injection, complete white-eye and partial mosaic white-eye, or complete yellow-body and partial mosaic yellow-body mutants were readily observed in G0s, indicating that the sgRNAs are functional (Table 1 and SI Appendix, Fig. S2A). Once sgRNA functionality was confirmed using recombinant Cas9 protein, we then injected these sgRNAs separately into 450 embryos (three injections of 150 embryos in biological triplicate) collected from each of our transgenic Ae. aegypti Cas9-expressing strains without including recombinant Cas9 protein in the injection mix. Similar to the Kh-sgRNA results described above, remarkably higher survival rates, mutagenesis efficiencies, and heritable mutation rates were also achieved for both W-sgRNA and Y-sgRNA when injected into all transgenic Cas9 lines, except AAEL003877-Cas9 (Table 1). As in other Dipterans, homozygous G1 mutants for white have dramatic eye pigmentation defects, similar to the kh mutants, which can be visualized in larvae, pupa, and adults (Fig. 2A). Additionally, homozygous G1 mutants for yellow have a striking yellow cuticle pigmentation defect that can be visualized in, larvae, pupae, adults, and even eggs (Fig. 2 A and B). To confirm the phenotypic defects from these sgRNAs were due to mutagenesis of the white and yellow genes, genomic DNA from homozygous mutant G1s (Fig. 2A) was amplified spanning the target sites and sequenced confirming the presence of indels in both the W-sgRNA and Y-sgRNA genomic DNA target sites (SI Appendix, Figs. S4 and S5). Altogether, these results indicate that the Ae. aegypti transgenic Cas9 expression system that we generated can effectively express Cas9 protein, and direct injection of in vitro-transcribed sgRNAs is sufficient to rapidly disrupt function of target genes of interest and generate highly heritable mutations. Of all of the transgenic lines, the AAEL010097-Cas9 line consistently showed the highest survival rate and efficiency of mutagenesis. Therefore, we performed all subsequent experiments using this strain.
Precise Gene Disruption in Ae. aegypti.
To further measure the efficacy of the AAEL010097-Cas9 line, we decided to target additional genes causing readily visible phenotypes in related organisms that have yet to be studied in Ae. aegypti. Specifically, we chose genes AAEL005793, AAEL009950, AAEL009170, and AAEL003240, which are conserved 1:1 orthologs with the D. melanogaster genes ebony, deformed, vestigial, and sine oculis, respectively. Ebony encodes the enzyme N-β-alanyl dopamine (NBAD) synthetase that converts dopamine to NBAD. Loss of ebony function increases black cuticle pigment in D. melanogaster (63). Deformed is a homeobox-containing (Hox) transcription factor, loss-of-function of this gene results in dramatic defects in derivatives of the maxillary segments, mandibular segments, and anterior segments in D. melanogaster (64). Vestigial encodes a nuclear protein that plays a central role in the development of the wings and loss of vestigial results in the failure of proper wing development in D. melanogaster (65, 66). The sine oculis locus encodes a homeodomain-containing protein, which plays an essential role in eye development and loss of sine oculis function leads disruption of the eye disk formation in D. melanogaster (67, 68).
To disrupt these genes, a single sgRNA was designed (E-sgRNA, D-sgRNA, So-sgRNA, V-sgRNA) to target an exon in each of these genes (SI Appendix, Figs. S6–S9), and then individually injected into 200 AEL010097-Cas9 embryos. Injection of E-sgRNA resulted in 82% of G0s showing pronounced dark coloration of the adult cuticle (SI Appendix, Fig. S2). These mutations were heritable and we established homozygous viable stocks by pairwise mating G1s (Fig. 2A and Table 2). D-sgRNA injection resulted in the death of more than 72% of G0 mosquitoes before the pupal stage, indicating that this gene is critical for proper development. However, 87% of the mosquitoes that reached adulthood exhibited severe mutant phenotypes including an extra eye (triple eyes), mal-shaped proboscis, and even an extra maxillary palp (triple maxillary palps) (Fig. 2C and Table 2). Unsurprisingly, all surviving G0 mutant females failed to blood feed, preventing us from establishing homozygous mutant lines. In So-sgRNA injected mosquitoes, 61% of the surviving G0s had clearly visible eye defects do to cell death in the primordium region (Fig. 2D and Table 2). As with the deformed mutants, we were unable to establish homozygous sine oculis mutant lines because these mutations were homozygous-lethal. Finally, 70% of V-sgRNA-injected G0s died at the pupal stage, with many of the dead pupae showing deformed rudimentary wing appendages (Fig. 2E). Of the few individuals that survived to adulthood, 75% had undeveloped wings or halteres (Fig. 2E and Table 2), inhibiting their ability to fly or mate, making it impossible to establish homozygous mutant lines for this gene. To confirm that the phenotypes described above were caused by disruption of the specific genes targeted by the sgRNAs (E-sgRNA, D-sgRNA, So-sgRNA, V-sgRNA), genomic DNA from the mutants (Fig. 2 C–E) was amplified spanning the target sites and sequenced confirming a selection of indels in both the genomic DNA target sites (SI Appendix, Figs. S6–S9).
Table 2.
Summary of the injection, survival, and mutagenesis rates mediated by E-sgRNA, D-sgRNA, So-sgRNA, and V-sgRNA in AAEL010097-Cas9 line
| Ae. aegypti line | Injected component* | No. injected G0s | G0 larva survivors (%) | G0 adult survivors (%) | G0 mutant adult (%) | G1s mutant adult† (%) |
| AAEL010097-Cas9 | E-sgRNA | 200 | 115 (58) | 112 (56) | 92 (82) | 751 (67) |
| AAEL010097-Cas9 | D-sgRNA | 200 | 122 (61) | 33 (28) | 29 (87) | Lethal |
| AAEL010097-Cas9 | So-sgRNA | 200 | 104 (52) | 98 (52) | 59 (61) | Lethal |
| AAEL010097-Cas9 | V-sgRNA | 200 | 134 (67) | 40 (30) | 22 (75) | Lethal |
sgRNA 100 ng/μL.
The overall heritable mutation rate was calculated as the number of mutant G1s divided by the number of all G1s observed.
Multiple sgRNAs Dramatically Improve Mutagenesis Rates.
To determine if simultaneous injection of two sgRNAs into the embryos of the AAEL010097-Cas9 line could induce higher mutagenesis efficiencies and germline transmission rates, we designed another sgRNA (W2-sgRNA) also targeting exon 3 of the white gene (SI Appendix, Fig. S10). Initially we tested functionality of the W2-sgRNA by coinjecting this guide with recombinant Cas9 protein into wild-type embryos. We found that this W2-sgRNA is efficient at generating mutations as 41 ± 7% of injected embryos survived to adult, 19 ± 6% of adult survivors showed the mosaic and white-eye mutant phenotypes, and 27 ± 5% G1s showed the mutant white-eye phenotype (Table 3). Similar to results described above, directly injecting the W2-sgRNA into the embryos of the AAEL010097-Cas9 line, resulted in higher survival rates (62 ± 6%), greater mutagenesis efficiencies (67 ± 9%), and higher germline transmission rates (56 ± 9%) (Table 3). Importantly, when we coinjected W1-sgRNA (described above) and W2-sgRNA together into embryos of the AAEL010097-Cas9 line, white mutants were obtained in the G0s at an extremely higher frequency of 94 ± 3%, with a remarkable G1 transmission frequency of 96 ± 3% (Table 3). Furthermore, genomic sequencing confirmed that some of the injections generated large deletions spanning the genomic distance (∼350 bp) between the W1-sgRNA and W2-sgRNA target sites (SI Appendix, Fig. S10). Together, these results indicate that by simultaneous injection of multiple sgRNAs targeting the same gene, higher mutagenesis rates and large deletions can be readily achieved.
Table 3.
Summary of the injection, survival, and mutagenesis rates mediated by W1-sgRNA and W2-sgRNA in wild-type and AAEL010097-Cas9 line
| Ae. aegypti line | Injected component* | No. injected G0s | G0 adult survivors, % | G0 mosaic, % | G1s mutant adult† % |
| Liverpool (wild-type) | W2-sgRNA /Cas9 protein | 450 | 41 ± 7 | 19 ± 6 | 27 ± 5 |
| Liverpool (wild-type) | W1-sgRNA/W2-sgRNA /Cas9 protein | 450 | 37 ± 5 | 34 ± 5 | 37 ± 9 |
| AAEL010097-Cas9 | W2-sgRNA | 450 | 62 ± 6 | 67 ± 9 | 56 ± 9 |
| AAEL010097-Cas9 | W1-sgRNA/W2-sgRNA | 450 | 61 ± 8 | 94 ± 3 | 96 ± 3 |
Biological triplicate consisting of three independent injections (each injection replicate with 150 embryos total 450 embryos) were performed and the results are shown as the mean ± SE throughout table.
sgRNA 100 ng/μL; Cas9 protein 300 ng/μL.
The overall heritable mutation rate was calculated as the number of mutant G1s divided by the number of all G1s observed.
Highly Efficient Multiplex Gene Disruption from a Single Injection.
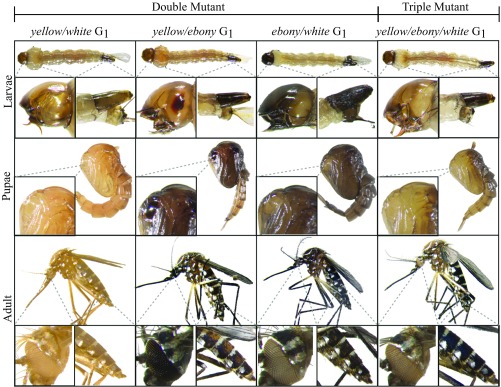
Given the laborious and time-consuming crossing required to generate Ae. aegypti lines with mutations at multiple genes, we explored whether double- or triple-gene knockout strains could quickly be generated in one step by coinjecting a multiplexed combination of sgRNAs targeting different genes. To do so, we made four mixes, including three combinations of two sgRNAs, including: W1-sgRNA and Y-sgRNA (mix 1); W1-sgRNA and E-sgRNA (mix 2); Y-sgRNA and E-sgRNA (mix 3); and one combination of three sgRNAs, W1-sgRNA, Y-sgRNA and E-sgRNA (mix 4). We injected these four mixes separately into 200 embryos from the AAEL010097-Cas9 line. We found that 90%, 87%, 93%, and 94% of the G0-injected survivors contained at least one mutant phenotype for the injection sets one to four, respectively (Table 4). Furthermore, 90%, 88%, and 92% of the G0 injected survivors had double-mutants when injected with sets one to three, respectively (Table 4 and SI Appendix, Fig. S11). When we coinjected a combination of three sgRNA (mix 4), we found that 67% of the surviving G0 adults contained mutations in all three genes targeted (Table 4 and SI Appendix, Fig. S11). Furthermore, these mutations were heritable as homozygous viable mutant stocks following pairwise mating of G1 multimutants (Fig. 3). Together, these results demonstrate that simultaneous and heritable multiple gene disruptions can be efficiently achieved by using transgenic expression of Cas9 and this simple technique should facilitate the rapid understanding of gene function and help tease apart gene networks in this nonmodel organism.
Table 4.
Single injection of multiple sgRNAs into the embryos of AAEL010097-Cas9 line results in highly efficient rates of multigene disruption
| Injected sgRNAs | Injected embryos | G0 adult survivors (%) | G0 mosaic (%) | Single mutant G0s (%); G1s* (%) | Double-mutant G0s (%); G1s† (%) | Triple-mutant G0s (%); G1s‡ (%) | ||||
| W | Y | E | W/Y | W/E | Y/E | W/Y/E | ||||
| W1-sgRNA/Y-sgRNA | 200 | 122 (61) | 110 (90) | 4 (4); 170 (15) | 7 (6); 238 (21) | N/A, N/A | 99 (90); 488 (43) | N/A; N/A | N/A; N/A | N/A; N/A |
| W1-sgRNA/E-sgRNA | 200 | 113 (56) | 98 (87) | 7 (7); 141 (17) | N/A; N/A | 5 (5); 224 (27) | N/A; N/A | 86 (88); 390 (47) | N/A; N/A | N/A; N/A |
| Y-sgRNA/E-sgRNA | 200 | 102 (51) | 95 (93) | N/A; N/A | 3 (3); 343(25) | 5 (5); 261 (15) | N/A; N/A | N/A; N/A | 87 (92); 536 (39) | N/A; N/A |
| W1-sgRNA/Y-sgRNA/E-sgRNA | 200 | 86 (43) | 81 (94) | 3 (4); 85 (11) | 3 (4); 54(7) | 1 (1); 23 (3) | 5 (6); 131 (17) | 9 (11); 101 (13) | 6 (7); 162 (21) | 54 (67); 85 (11) |
N/A, not applicable.
Single-mutant phenotype, W: white eye; Y: yellow body; E: dark body.
Double-mutant phenotypes, W/Y: white eye and yellow body, W/E: white eye and dark body; Y/E yellow body and dark body.
Triple-mutant phenotypes, W/Y/E: white eye, yellow body, and dark body.
Fig. 3.
Single injections of multiplexed sgRNAa robustly generate double- and triple-mutant mosquitoes. Larva, pupae, and adult G1 phenotypes for double-mutants, including: yellow body and white eyes (yellow/white), a mixture of yellow and dark body (yellow/ebony), dark body and white eyes (ebony/white), and one triple-mutant, which is a phenotypic mixture of yellow and dark body and white eyes (yellow/ebony/white). The striking differences between wild-type and mutant larva, pupae and adult are highlighted. (Magnifications: whole-body images, 20×; Insets, 100×.)
Increased Rates of HDR with dsDNA Donors.
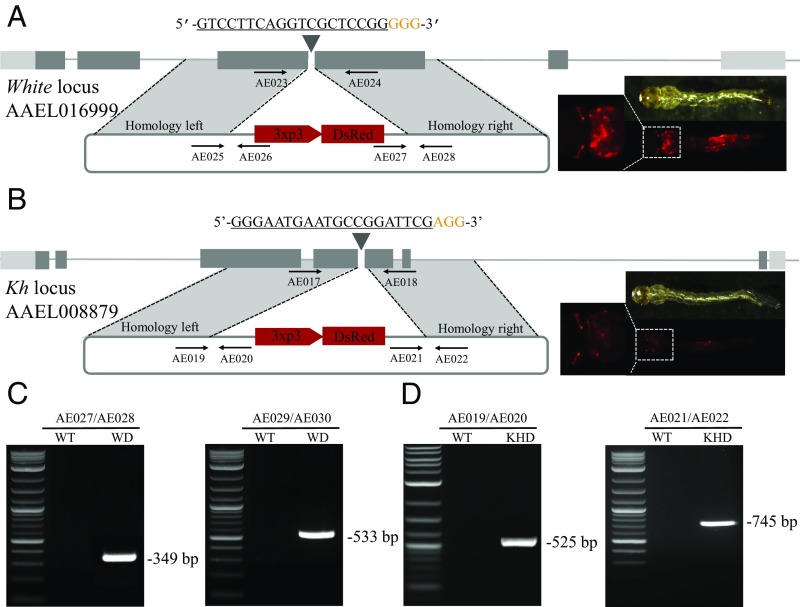
Given the exceedingly high rate of NHEJ-induced mutation when using theAAEL010097-Cas9 line, we wanted to assess knock-in rates mediated by HDR via dsDNA donors in this line. Our strategy was to employ two donor plasmids, a white-donor plasmid based on the W1-sgRNA and a kh-donor plasmid based on the Kh-sgRNA. Each of these two donor plasmids was designed to contain a dominant fluorescent marker consisting of 3xP3-dsRed, expressing in the larvae and adult photoreceptors, flanked by ∼1-kb homology arms that were derived from the genomic sequence immediately flanking the target cleavage sites (Fig. 4 A and B). We directly compared two approaches: (i) coinjection of dsDNA donor combined with in vitro transcribed sgRNA and purified recombinant Cas9 protein into wild-type embryos; and (ii) coinjection of circular dsDNA donor combined with in vitro transcribed sgRNA injected into AAEL010097-Cas9 line embryos. For the first approach, a total of 600 embryos were separately injected for both the white-donor and kh-donor, with a HDR rate of 0.15% and 0.14% (Table 5), respectively. For the second approach, a total of 600 embryos from the Ae. aegypti AAEL010097-Cas9 line were separately injected for both the white-donor and kh-donor, with a HDR rate of 2.36% and 2.48% (Table 5), respectively. Overall, we see a dramatic, yet consistent, 15- to 17-fold increase in rates of HDR when using the AAEL010097-Cas9 line compared with the nontransgenic method of supplying Cas9. Gene-specific insertion of our donor cassettes into the intended target sites was confirmed by subsequent genomic PCR and sequencing (Fig. 4 A–C and SI Appendix, Fig. S12).
Fig. 4.
Highly efficient site-specific integration via CRISPR-mediated HDR. Schematic representations of the white locus and white-donor construct (A), and the kh locus and kh-donor construct (B). Exons are shown as boxes, coding regions are depicted in black and the 5′ and 3′ UTRs in gray. Locations and sequences of the sgRNA targets are indicated with the PAM shown in yellow. Black arrows indicate approximate positions and directions of the oligonucleotide primers used in the study. The donor plasmids (blue) express fluorescent eye marker (3xp3-DsRed) inserted between regions of homology from the white and kh locus, respectively (A and B). Gene amplification analysis confirms site-specific integration of the white-donor construct into the white locus using combinations of genomic and plasmid donor-specific primers (933Cms3/933Cms4 expected 349 bp, and 933Cms5/933Cms6 expected 533 bp) (C), and also confirms the integration of the kh-donor construct into the kh locus using combinations of genomic and plasmid donor specific primers (924ms3/924ms4 expected 525 bp, and 924ms5/924ms6 expected 745 bp) with no amplification in wild-type (D). WT represents wild-type, WD represents knockin with white-donor, KHD represents knockin with kh-donor. (Magnification: 20×.)
Table 5.
Transgenic source of Cas9 results in increased rates of HDR with dsDNA donors
| Ae. aegypti strain | Injected component* | Injected embryos | Adult G0 survivors (%) | HDR event | |
| No. group founders/total group (%) | No. HDR G1/total G1 (%) | ||||
| Liverpool (wild-type) | W1-sgRNA/Cas9/white-donor | 600 | 251(41.83) | 1/20 (5) | 9/5,371 (0.15) |
| Liverpool (wild-type) | Kh-sgRNA/Cas9/kh-donor | 600 | 273 (45.50) | 1/20 (5) | 7/4,773 (0.14) |
| AAEL010097-Cas9 | W1-sgRNA/white-donor | 600 | 308 (51.33) | 4/20 (20) | 118/4,993 (2.36) |
| AAEL010097-Cas9 | Kh-sgRNA/kh-donor | 600 | 298 (49.67) | 5/20 (25) | 149/6,017 (2.48) |
The concentration of sgRNA is 100 ng/μL; Cas9 protein 300 ng/μL. white-donor and kh-donor plasmids 100 ng/μL.
Discussion
Previous studies have demonstrated effective CRISPR/Cas9 genome editing in the mosquito Ae. aegypti (30–34); however, these studies utilized a nontransgenic source of Cas9, limiting both survivorship and editing efficiencies. To overcome these previous limitations, and to reduce the complexity of injecting multiple components (i.e., Cas9 and sgRNA), herein we developed a simplified transgenic Cas9 expression system in Ae. aegypti, similarly to what is routinely used in other organisms such as D. melanogaster (27, 41, 42). Importantly, to achieve highly specific and consistent genome modifications, we demonstrate that embryos from these Cas9 strains need only be injected with easy-to-make sgRNAs. By using these Cas9 strains, we disrupted multiple genes that were either homozygous viable (kh, white, yellow, and ebony), or homozygous lethal (deformed, sine oculis, vestigial), resulting in dramatic phenotypes affecting viability, vision, flight, and blood feeding, and therefore may be useful for developing control strategies and genetic sexing techniques in the future. For example, in a yellow mutant background, the endogenous gene encoding yellow could be linked to the male-determining locus (32), using CRISPR-mediated HDR, to generate a robust genetic sexing system by which male embryos/larvae/adults would be dark and female embryos/larvae/adults would be yellow.
An appealing advantage of our Cas9 transgenic system is the ability to efficiently disrupt multiple genes simultaneously. We have demonstrated that we can efficiently generate large deletions, or even double- (yellow-white; ebony-white; yellow-ebony), or triple- (yellow-ebony-white) mutants. Importantly, these multimutants can rapidly be generated in a single-step approach by injecting multiple sgRNAs into the embryos of the transgenic Cas9 strains, significantly reducing downstream efforts. This rapid multiplex gene knockout approach will be instrumental for dissecting gene networks in this nonmodel organism. While the Cas9 strains were generated in the Liverpool genetic background, these strains can be introgressed into other genetic backgrounds if desired.
The germline Cas9 strains developed here may also bring us one step closer to engineering an effective CRISPR/Cas9-homing–based gene drive system (47, 48) in this organism. Homing-based drive systems rely on HDR to convert heterozygous alleles into homozygous alleles in the germline, and have recently been successfully engineered in two Anopheline mosquito species (51, 69). While these studies were fruitful at significantly biasing rates of Mendelian inheritance rates of the drive containing alleles, they were severely limited by the rapid evolution of resistance alleles generated by NHEJ, and are therefore not predicted to spread into diverse wild populations (70). It was hypothesized that these resistance alleles formed due to high levels of maternal deposition of Cas9 in the embryo, and by restricting Cas9 expression to the germline may subsequently increase rates of HDR and reduce rates of NHEJ. In addition to restricting expression to the germline, multiplexing of sgRNAs in the drive, and designing the drive to target a critical gene have also been proposed as innovative strategies to increase rates of HDR and reduce resistance caused by NHEJ (47, 48, 70); however, these hypotheses remain to be demonstrated. Notwithstanding, it will be interesting to determine if our Ae. aegypti Cas9 strains, each with varying expression in the germline, will be effective in a gene drive system designed for Ae. aegypti. This would be straightforward to test in a molecularly confined safe split-gene drive design where the Cas9 and the drive are positioned at different genomic loci. In this split-design, the Cas9 strains we developed can be directly tested without further modification by simply genetically crossing with a split gRNA-drive component and measuring rates of inheritance (49, 71–73).
While the CRISPR/Cas9 transgenic system developed here is quite effective, it would be useful to have the ability to supply the sgRNAs transgenically. In D. melanogaster, polymerase-3 promoters have been utilized to express sgRNAs, and through genetic crosses with Cas9 strains mutation efficiency could be increased up to 100% (42, 43).
Furthermore, it should be noted, that a slight disadvantage of these Cas9 strains for some groups may result from the fact that these strains were generated in the Liverpool genetic background, which restricts genome modifications to only this background. However, this can easily be overcome by simply using genetics to introgress these promoter-Cas9 transgenes into other genetic backgrounds, or by generating new transgenic strains in the desired background using the plasmids generated here.
Overall, our results demonstrate that our simplified transgenic Cas9 system has improved capacity to rapidly induce highly efficient and specific targeted genome modifications, including gene disruptions, deletions, and insertions. Given their high efficiencies, these Cas9 strains can be used to quickly generate genome modifications allowing for high-throughput gene targeting, thereby accelerating comprehensive functional annotation of the Ae. aegypti genome.
Materials and Methods
Insect Rearing.
Mosquitoes used in all experiments were derived from of the Ae. aegypti Liverpool strain, which was the source strain for the reference genome sequence (58).
Generation of Ae. aegypti Cas9 Transgenic Lines.
Transgenic Ae. aegypti Cas9 mosquitoes were created by injecting 0- to 1-h-old preblastoderm-stage embryos with a mixture of piggybac vector containing the Cas9 expressing plasmid designed above (200 ng/μL) and a source of piggyBac transposase [phsp-Pbac, (200 ng/μL)] (74–76).
Characterization of AAEL010097-Cas9 Insertion Site.
To characterize the Cas9 insertion site for AAEL010097-Cas9, we utilized a previously described inverse PCR protocol (77).
CRISPR Mediated Microinjections.
Embryonic collection and CRISPR microinjections were performed following previously established procedures (30, 78).
Supplementary Material
Acknowledgments
We thank Robert Harrell for providing the pBac-3xP3-dsRed and phsp-Pbac plasmids; Kate M. O’Connor-Giles for providing the p3xP3-EGFP/vasa-3xFLAG-NLS-Cas9-NLS plasmid and sequence; Alexander Knyshov for assisting with SEM imaging; and Anthony A. James for reading over the manuscript and providing insightful comments and edits. This work was supported by a private donation from https://www.maxmind.com/ (to O.S.A.); US NIH K22 Grant 5K22AI113060 (to O.S.A.); NIH R21 Grant 1R21AI123937 (to O.S.A.); NIH R21 Grant 1R21AI115271 (to B.J.W.); and Defense Advanced Research Project Agency Safe Genes Program Grant HR0011-17-2-0047 (to O.S.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All Cas9 plasmids and sequence maps have been deposited and made available for order and download at www.addgene.org/ [Addgene IDs 100580 (AAEL006511-Cas9), 100581 (AAEL003877-Cas9), 100608 (AAEL005635-Cas9), 100705 (AAEL007097-Cas9), 100706 (AAEL007584-Cas9) and 100707 (AAEL010097-Cas9)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711538114/-/DCSupplemental.
References
- 1.Barrett ADT, Higgs S. Yellow fever: A disease that has yet to be conquered. Annu Rev Entomol. 2007;52:209–229. doi: 10.1146/annurev.ento.52.110405.091454. [DOI] [PubMed] [Google Scholar]
- 2.Halstead SB. Dengue virus-mosquito interactions. Annu Rev Entomol. 2008;53:273–291. doi: 10.1146/annurev.ento.53.103106.093326. [DOI] [PubMed] [Google Scholar]
- 3.Weaver SC, Barrett ADT. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol. 2004;2:789–801. doi: 10.1038/nrmicro1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral Res. 2010;85:328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carvalho FD, Moreira LA. Why is Aedes aegypti linnaeus so successful as a species? Neotrop Entomol. 2017;46:243–255. doi: 10.1007/s13744-017-0520-4. [DOI] [PubMed] [Google Scholar]
- 6.Hemingway J, Field L, Vontas J. An overview of insecticide resistance. Science. 2002;298:96–97. doi: 10.1126/science.1078052. [DOI] [PubMed] [Google Scholar]
- 7.Franz AWE, Clem RJ, Passarelli AL. Novel genetic and molecular tools for the investigation and control of dengue virus transmission by mosquitoes. Curr Trop Med Rep. 2014;1:21–31. doi: 10.1007/s40475-013-0007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeGennaro M, et al. orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature. 2013;498:487–491. doi: 10.1038/nature12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liesch J, Bellani LL, Vosshall LB. Functional and genetic characterization of neuropeptide Y-like receptors in Aedes aegypti. PLoS Negl Trop Dis. 2013;7:e2486. doi: 10.1371/journal.pntd.0002486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMeniman CJ, Corfas RA, Matthews BJ, Ritchie SA, Vosshall LB. Multimodal integration of carbon dioxide and other sensory cues drives mosquito attraction to humans. Cell. 2014;156:1060–1071. doi: 10.1016/j.cell.2013.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aryan A, Anderson MAE, Myles KM, Adelman ZN. TALEN-based gene disruption in the dengue vector Aedes aegypti. PLoS One. 2013;8:e60082. doi: 10.1371/journal.pone.0060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smidler AL, Terenzi O, Soichot J, Levashina EA, Marois E. Targeted mutagenesis in the malaria mosquito using TALE nucleases. PLoS One. 2013;8:e74511. doi: 10.1371/journal.pone.0074511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aryan A, Anderson MAE, Myles KM, Adelman ZN. Germline excision of transgenes in Aedes aegypti by homing endonucleases. Sci Rep. 2013;3:1603. doi: 10.1038/srep01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 15.Brouns SJJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Held NL, Whitaker RJ. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ Microbiol. 2009;11:457–466. doi: 10.1111/j.1462-2920.2008.01784.x. [DOI] [PubMed] [Google Scholar]
- 17.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mojica FJ, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of archaea, bacteria and mitochondria. Mol Microbiol. 2000;36:244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 19.Mojica FJ, Juez G, Rodríguez-Valera F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol Microbiol. 1993;9:613–621. doi: 10.1111/j.1365-2958.1993.tb01721.x. [DOI] [PubMed] [Google Scholar]
- 20.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue W, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bassett AR, Tibbit C, Ponting CP, Liu J-L. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren X, et al. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci USA. 2013;110:19012–19017. doi: 10.1073/pnas.1318481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang N, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M, et al. Generation of heritable germline mutations in the jewel wasp Nasonia vitripennis using CRISPR/Cas9. Sci Rep. 2017;7:901. doi: 10.1038/s41598-017-00990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kistler KE, Vosshall LB, Matthews BJ. Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep. 2015;11:51–60. doi: 10.1016/j.celrep.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basu S, et al. Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Proc Natl Acad Sci USA. 2015;112:4038–4043. doi: 10.1073/pnas.1502370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall AB, et al. SEX DETERMINATION. A male-determining factor in the mosquito Aedes aegypti. Science. 2015;348:1268–1270. doi: 10.1126/science.aaa2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, et al. microRNA-309 targets the Homeobox gene SIX4 and controls ovarian development in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2016;113:E4828–E4836. doi: 10.1073/pnas.1609792113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong S, et al. Heritable CRISPR/Cas9-mediated genome editing in the yellow fever mosquito, Aedes aegypti. PLoS One. 2015;10:e0122353. doi: 10.1371/journal.pone.0122353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Z, et al. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics. 2013;195:289–291. doi: 10.1534/genetics.113.153825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gratz SJ, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei W, et al. Heritable genome editing with CRISPR/Cas9 in the silkworm, Bombyx mori. PLoS One. 2014;9:e101210. doi: 10.1371/journal.pone.0101210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma S, et al. CRISPR/Cas9 mediated multiplex genome editing and heritable mutagenesis of BmKu70 in Bombyx mori. Sci Rep. 2014;4:4489. doi: 10.1038/srep04489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilles AF, Schinko JB, Averof M. Efficient CRISPR-mediated gene targeting and transgene replacement in the beetle Tribolium castaneum. Development. 2015;142:2832–2839. doi: 10.1242/dev.125054. [DOI] [PubMed] [Google Scholar]
- 40.Sharma A, et al. Male sex in houseflies is determined by Mdmd, a paralog of the generic splice factor gene CWC22. Science. 2017;356:642–645. doi: 10.1126/science.aam5498. [DOI] [PubMed] [Google Scholar]
- 41.Sebo ZL, Lee HB, Peng Y, Guo Y. A simplified and efficient germline-specific CRISPR/Cas9 system for Drosophila genomic engineering. Fly (Austin) 2014;8:52–57. doi: 10.4161/fly.26828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Port F, Chen H-M, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA. 2014;111:E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kondo S, Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013;195:715–721. doi: 10.1534/genetics.113.156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gratz SJ, et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196:961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedland AE, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ablain J, Durand EM, Yang S, Zhou Y, Zon LI. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev Cell. 2015;32:756–764. doi: 10.1016/j.devcel.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Champer J, Buchman A, Akbari OS. Cheating evolution: Engineering gene drives to manipulate the fate of wild populations. Nat Rev Genet. 2016;17:146–159. doi: 10.1038/nrg.2015.34. [DOI] [PubMed] [Google Scholar]
- 48.Esvelt KM, Smidler AL, Catteruccia F, Church GM. Concerning RNA-guided gene drives for the alteration of wild populations. eLife. 2014;3:e03401. doi: 10.7554/eLife.03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burt A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc R Soc Lond B Biol Sci. 2003;270:921–928. doi: 10.1098/rspb.2002.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deredec A, Burt A, Godfray HC. The population genetics of using homing endonuclease genes in vector and pest management. Genetics. 2008;179:2013–2026. doi: 10.1534/genetics.108.089037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DiCarlo JE, Chavez A, Dietz SL, Esvelt KM, Church GM. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat Biotechnol. 2015;33:1250–1255. doi: 10.1038/nbt.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gantz VM, Bier E. Genome editing. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science. 2015;348:442–444. doi: 10.1126/science.aaa5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hammond A, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. 2016;34:78–83. doi: 10.1038/nbt.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akbari OS, Papathanos PA, Sandler JE, Kennedy K, Hay BA. Identification of germline transcriptional regulatory elements in Aedes aegypti. Sci Rep. 2014;4:3954. doi: 10.1038/srep03954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson MAE, Gross TL, Myles KM, Adelman ZN. Validation of novel promoter sequences derived from two endogenous ubiquitin genes in transgenic Aedes aegypti. Insect Mol Biol. 2010;19:441–449. doi: 10.1111/j.1365-2583.2010.01005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akbari OS, et al. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3 (Bethesda) 2013;3:1493–1509. doi: 10.1534/g3.113.006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ren L, et al. Comparative analysis of the activity of two promoters in insect cells. Afr J Biotechnol. 2011;10:8930–8941. [Google Scholar]
- 58.Nene V, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cosens D, Briscoe D. A switch phenomenon in the compound eye of the white-eyed mutant of Drosophila melanogaster. J Insect Physiol. 1972;18:627–632. [Google Scholar]
- 60.Klemenz R, Weber U, Gehring WJ. The white gene as a marker in a new P-element vector for gene transfer in Drosophila. Nucleic Acids Res. 1987;15:3947–3959. doi: 10.1093/nar/15.10.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Biessmann H. Molecular analysis of the yellow gene (y) region of Drosophila melanogaster. Proc Natl Acad Sci USA. 1985;82:7369–7373. doi: 10.1073/pnas.82.21.7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wittkopp PJ, Vaccaro K, Carroll SB. Evolution of yellow gene regulation and pigmentation in Drosophila. Curr Biol. 2002;12:1547–1556. doi: 10.1016/s0960-9822(02)01113-2. [DOI] [PubMed] [Google Scholar]
- 63.Wittkopp PJ, True JR, Carroll SB. Reciprocal functions of the Drosophila yellow and ebony proteins in the development and evolution of pigment patterns. Development. 2002;129:1849–1858. doi: 10.1242/dev.129.8.1849. [DOI] [PubMed] [Google Scholar]
- 64.Merrill VK, Turner FR, Kaufman TC. A genetic and developmental analysis of mutations in the deformed locus in Drosophila melanogaster. Dev Biol. 1987;122:379–395. doi: 10.1016/0012-1606(87)90303-4. [DOI] [PubMed] [Google Scholar]
- 65.Kim J, et al. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- 66.Klein T, Arias AM. The vestigial gene product provides a molecular context for the interpretation of signals during the development of the wing in Drosophila. Development. 1999;126:913–925. doi: 10.1242/dev.126.5.913. [DOI] [PubMed] [Google Scholar]
- 67.Cheyette BN, et al. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 68.Jusiak B, et al. Genome-wide DNA binding pattern of the homeodomain transcription factor Sine oculis (So) in the developing eye of Drosophila melanogaster. Genom Data. 2014;2:153–155. doi: 10.1016/j.gdata.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gantz VM, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci USA. 2015;112:E6736–E6743. doi: 10.1073/pnas.1521077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marshall JM, Buchman A, Sánchez C HM, Akbari OS. Overcoming evolved resistance to population-suppressing homing-based gene drives. Sci Rep. 2017;7:3776. doi: 10.1038/s41598-017-02744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Akbari OS, et al. BIOSAFETY. Safeguarding gene drive experiments in the laboratory. Science. 2015;349:927–929. doi: 10.1126/science.aac7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 73.Pfeiffer BD, Truman JW, Rubin GM. Using translational enhancers to increase transgene expression in Drosophila. Proc Natl Acad Sci USA. 2012;109:6626–6631. doi: 10.1073/pnas.1204520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Handler AM, Harrell RA., 2nd Germline transformation of Drosophila melanogaster with the piggyBac transposon vector. Insect Mol Biol. 1999;8:449–457. doi: 10.1046/j.1365-2583.1999.00139.x. [DOI] [PubMed] [Google Scholar]
- 75.Kokoza V, Ahmed A, Wimmer EA, Raikhel AS. Efficient transformation of the yellow fever mosquito Aedes aegypti using the piggyBac transposable element vector pBac[3xP3-EGFP afm] Insect Biochem Mol Biol. 2001;31:1137–1143. doi: 10.1016/s0965-1748(01)00120-5. [DOI] [PubMed] [Google Scholar]
- 76.Lobo NF, Hua-Van A, Li X, Nolen BM, Fraser MJ., Jr Germ line transformation of the yellow fever mosquito, Aedes aegypti, mediated by transpositional insertion of a piggyBac vector. Insect Mol Biol. 2002;11:133–139. doi: 10.1046/j.1365-2583.2002.00317.x. [DOI] [PubMed] [Google Scholar]
- 77.Huang AM, Rehm EJ, Rubin GM. Recovery of DNA sequences flanking P-element insertions in Drosophila: Inverse PCR and plasmid rescue. Cold Spring Harb Protoc. 2009;2009:pdb.prot5199. doi: 10.1101/pdb.prot5199. [DOI] [PubMed] [Google Scholar]
- 78.Aryan A, Myles KM, Adelman ZN. Targeted genome editing in Aedes aegypti using TALENs. Methods. 2014;69:38–45. doi: 10.1016/j.ymeth.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.