Abstract
Molecular self-assembly is the dominant form of chemical reaction in living systems, yet efforts at systems biology modeling are only beginning to appreciate the need for and challenges to accurate quantitative modeling of self-assembly. Self-assembly reactions are essential to nearly every important process in cell and molecular biology and handling them is thus a necessary step in building comprehensive models of complex cellular systems. They present exceptional challenges, however, to standard methods for simulating complex systems. While the general systems biology world is just beginning to deal with these challenges, there is an extensive literature dealing with them for more specialized self-assembly modeling. This review will examine the challenges of self-assembly modeling, nascent efforts to deal with these challenges in the systems modeling community, and some of the solutions offered in prior work on self-assembly specifically. The review concludes with some consideration of the likely role of self-assembly in the future of complex biological system models more generally.
Keywords: Biomolecular aggregates, Biomolecular dynamics, Self-organized systems, Computer modeling
1. Introduction
Self-assembly reactions account for the overwhelming majority of the reaction events occurring in the cell. Most eukaryotic proteins function normally in complexes and self-assembly of these complexes is a key step in nearly all major cellular functions [8]. Examples of processes critically dependent on self-assembly include genome replication [147, 195, 172, 19]; gene transcription and transcript degradation [19, 111, 127]; protein synthesis and degredation [112, 53]; cell movement and shape control [200, 45, 34, 81]; cell-to-cell communication including gap-junction assembly and regulation [188]; formation of membrane complexes such as pore-forming toxins [12]; and mechanotransduction [198, 202, 9]. Through these processes, the assembly and disassembly of molecular complexes and machines plays a crucial role in essentially all regulatory processes in cell biology. Given the centrality of self-assembly to cell biology, one cannot hope to develop truly comprehensive quantitative models of systems biology without tackling self-assembly. Yet self-assembly has until recently been largely absent from major efforts at developing general systems biology modeling tools (e.g., [146, 82, 60, 65, 182, 181, 108, 160, 185]) or handled only with one-off special cases for particular systems of importance (e.g.,[59, 199, 96]). Even the most ambitious efforts at large-scale biochemical modeling largely focus on traditional enzymatic chemistry or transcriptional dynamics and only implicitly model the self-assembly reactions involved in those processes (e.g., recent comprehensive models of whole-cell or whole-organism transcriptional and metabolomic modeling [189, 18]). This situation is beginning to change as some major systems biology tools (e.g., [69, 68, 55]) and modeling efforts [96] have begun to incorporate methods suitable to complex self-assembly, but major challenges remain.
These challenges of self-assembly modeling largely arise from the extremely large space of possible pathways accessible to the intermediate species of a self-assembly reaction network. The number of possible reaction trajectories by which a set of free monomers can assemble into a complex grows in general exponentially in the complex size, leading to an enormous combinatorial explosion in pathway space for even moderate-sized assemblies and astronomical numbers for large complexes, such as virus capsids or cytoskeletal networks. This is problematic for experimental study of assembly systems, as it is rarely possible to discriminate experimentally among these pathways except at a coarse level, particularly for highly symmetric or repetitive structures. It likewise creates problems for the most popular modeling methods. Mass action differential equation (DE) models are generally unsuitable for non-trivial assemblies because they require either extensive simplifications [70, 56, 125] or enormous numbers of equations and variables to account for the many possible intermediates [90]. Brownian dynamics (BD) models, even highly coarse-grained [167, 17, 51, 10], are likewise challenged by the large numbers of reactants and long timescales typical of self-assembly systems, requiring themselves great simplifications of reaction processes that generally make them unsuitable for accurate quantitative modeling [58]. Methods based on Gillespie’s stochastic simulation algorithm (SSA) can provide an effective balance between DE and BD, but face their own challenges because the underlying reaction networks are too large to model explicitly [26, 59, 64]. For similar reasons, self-assembly networks are extremely challenging for experimental characterization [27, 29, 93, 100, 141, 222, 223] and model inference as well [104, 210]. For example, the high computational cost and large numbers of intermediate species make it computationally infeasible to learn models via prevailing Bayesian parameter inference schemes [67], which require large numbers of simulation trajectories.
Over the recent decades, however, a specialized literature on self assembly modeling has grown for handling a number of challenging systems of independent importance. Cytoskeletal assembly (i.e., actin and microtubule assembly) has been the subject of extensive modeling work, leading to many seminal results in the basic biophysics of molecular assembly processes. Viral capsid assembly [70] has a long history as one of the primary model systems for macromolecular self assembly, both from an experimental and a computational perspective. Another key model system is amyloid aggregation, the basis for many major public health threats, including Alzheimers disease, Huntington’s disease, Parkinsons disease, prion disease, and type II diabetes. Fig. 1 shows a few examples of important model systems for self-assembly and models through which they have been studied. The practical importance of these and other systems has led them to attract their own modeling communities to find solutions to the special challenges of molecular self-assembly to computational modeling. In these fields, one can find studies both anticipating the challenges beginning to face broader systems biology efforts and often offering at least partial solutions to these challenges.
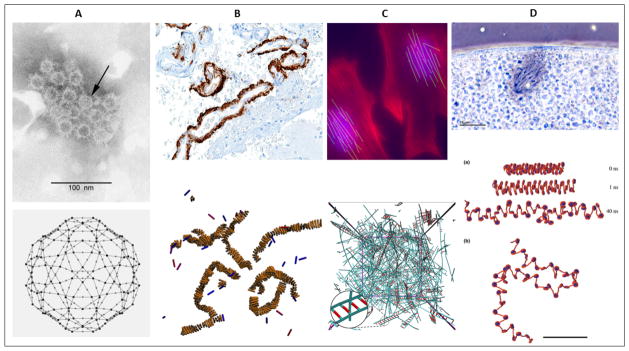
Figure 1.
Example model systems for self-assembly simulation. (A) Viral capsid assembly. Top: Hepatitis virus [77], Bottom: Coarse-grained SSA simulation of HPV assembly [211]. (B) Amyloid aggregation. Top: High magnification micrograph of cerebral amyloid angiopathy with senile plaques in the cerebral cortex (amyloid beta, as seen in Alzheimer disease) [23], Bottom: Coarse-grained Monte Carlo simulation of amyloid aggregation with two state monomer model [11]. (C) Cytoskeletal assembly and disassembly. Top: Highly oriented actin fibers in shear stress cultivated rat cells [2], Bottom: BD simulation of actin cytoskeleton composed mainly of actin and actin crosslinking proteins [98]. (D) Genome organization. Top: Chromatin fibers during mitosis, Xenopus egg [22], Bottom: BD simulations of nucleosome structure and dynamics [151]. For more on nucleosome assembly, see [19].
The remainder of this review will consider in more detail both the special diffculty of self-assembly modeling and the literature addressing it. It will first discuss some of the important roles of self-assembly in cellular biochemistry as well as the role of systems modeling methods in understanding these systems. It will then discuss some of the successful approaches to self-assembly modeling that have emerged through this literature, as well as continuing challenges. It will conclude with consideration of how quantitative self-assembly modeling may shape future efforts in modeling biological systems more generally.
2. Why does self-assembly (SA) matter?
2.1. The role of self-assembly in general cell biology
Self-assembly is everywhere in biology, beginning with the most fundamental processes of molecular biology, all of which depend on the self-assembly of specialized complexes, structures, or molecular machines. Examples of self-assembled molecular machines fundamental to molecular biology include DNA polymerases (replication), RNA polymerases (transcription), the spliceasome (splicing), the ribosome (translation), and the proteasome (protein degradation). Each of these processes is critical in different ways to the regulation of complex biological systems and thus has been the focus of specialized modeling efforts. For example, the transcription complex is one of the most well studied systems in molecular biology, with experimental work on the interaction of classic 1D and 3D diffusion of transcription factors [74] inspiring kinetic models of the recruitment process [95]. More specialized examples of self-assembly continue to be elucidated, with prominent recent examples including the RISC complex involved in miRNA [168, 116, 137, 88] and the Cas9-gRNA complex [33] implementing the CRISPR/Cas system [138, 186].
Within eukaryotic biology specifically, a more specialized set of self-assembly systems have evolved critical roles. The cytoskeleton is an unusually large, dynamic, and complicated molecular assembly, making it a crucial target of modeling efforts. The cytoskeleton itself is essential to intracellular transport [150, 152], cell movement and shape control [7, 149], mechanotransduction [201], and cell division [79], among many other functions. Furthermore, the dynamic process of assembly and disassembly is central to each of these functions. Actin and microtubule assembly and disassembly have been key model systems for self-assembly from the early days of molecular biology [91, 134, 61, 49, 170, 124, 204, 20, 62] and have inspired numerous computational models (e.g., [159, 130, 52, 179, 57, 161]). Transport processes in the eukaryotic cell frequently depend on other kinds of specialized self-assemblies, in addition to the cytoskeleton. For example, much eukaryotic transport involves the assembly of specialized machinery for construction and scission of cargo-carrying vesicles, such as the clathrin and COPI/COP-II coat systems [136, 50], which have inspired their own modeling literature (e.g., [37, 117, 87]).
Beyond its role in general cell and molecular biology, self-assembly is crucial to a number of disease-specific processes. Amyloid diseases are perhaps the prime example of a disease specifically of self-assembly, where aberrant assembly is the mechanism of illness. Numerous such diseases are known, including many major public health threats. Perhaps best known are Alzheimer’s disease (characterized by aggregates of the Aβ peptide and the Tau protein[122, 94]), Huntington’s disease (characterized by aggregates of the Huntingtin protein [113]), Parkinson’s disease [169], amytrophic lateral sclerosis [197], type II diabetes, and a variety of known prion diseases such as Creutzfeldt-Jakob [133, 28]. Alzheimer’s and dementia, for example, are strongly associated with aging and affected roughly 36 million people in 2010 [208, 209]. It is becoming increasingly clear that the ability to form the amyloid state is a widespread, generic property of proteins [102] making the process of amyloidogenesis an important topic of theoretical study. From a physical perspective, the main question is what forces stabilize the aggregates into the oligomer (small soluble disordered clusters) and fibrillar (long, many-chain highly structured -sheet-containing aggregates) states associated with neurotoxicity [165]. For a broader discussion of these forces, see [109, 110, 139]. From a computational perspective, the focus is both on identifying the structure of oligomeric intermediates and fibers but also elucidating the kinds of assembly pathways available. This is an especially challenging computational problem due to the intrinsic disorder in the system.
Viral illnesses form another broad class of self-assembly-driven illness, in which assembly of large complexes (i.e., the viruses themselves) are the mechanism of the disease process. Virus assembly is of obvious medical importance, given the millions who die each year from viral illnesses, e.g., 1.5 million from AIDS alone [38]. A fundamental understanding of this crucial aspect of the viral life cycle and infectivity may offer avenues for therapeutics or vaccines [223]. Additionally, there many factors making viruses appealing to the modeling community, including the deep experimental literature on their assembly and a high degree of symmetry in the final structure that allows for large complexes to be produced from small numbers of distinct subunit types. Viral assembly modeling has thus become a subfield in itself. Virus assembly has been a crucial platform for many basic advances in self-assembly modeling, including the use of DE [220], BD [167, 144, 71, 128], and SSA [218, 78, 97] methods. It has likewise been a platform for developing a variety of specialized versions of these modeling methods, such as rule-based approaches to simulating extremely large reaction networks [89] and derivative-free optimization approaches to model inference [104, 210]. Viral capsids have been a focus of intense theoretical study into the basic biophysics of self-assembly [70, 71, 42, 187, 24] as well as for identifying potential new avenues for assembly-mediated treatment [105, 31, 106, 187, 203, 215, 101, 43, 86, 171, 143].
3. Self-assembly modeling and simulation
3.1. The challenge of quantitative modeling of self-assembly reaction networks
At the root of much of the diffculty of modeling self-assembly is the extraordinarily large number of intermediates and pathways potentially accessible to a self-assembly system. Large number of reactants present problems in different ways to most conventional modeling and model inference methods (see Section 3.2 below). They likewise present a challenge to experimental characterization of such systems, as there is no practical way to monitor huge numbers of distinct molecular species. While details vary by geometry, in general the number of possible intermediates (partially built structures) one might encounter on the way to a complete assembly will blow up exponentially in the assembly size. This problem has probably been most intensively studied in the virus assembly literature, as it is particularly pronounced for large, highly symmetric structures, of which viral capsids are a prime example. Even a coarse-grained model of an icosahedral virus capsid, consisting of just twelve subunits, has 750 possible intermediate structures [120]. For real viral structures, which typically have several hundred proteins, the numbers of potential intermediates will be astronomical. Similar problems will arise to a lesser degree with large, asymmetric assemblies (e.g., the ribosome [103, 126]) as well as with larger but less symmetric assemblies such as the cytoskeleton. While the number of species possible for a linear filament is small, once one allows for branching [216], numbers of possible branched filaments or networks can blow up exponentially in the structure size as well. Note that this is not a unique challenge of self-assembly, as similar issues arise in other combinatorially explosive systems, such as signaling networks [16, 80].
A related concern for modeling, particularly with respect to self-assembly in cell biology, is the issue of small copy numbers [63, 194] resulting in an inherently discrete and stochastic reaction system. The issue occurs for many cellular systems involving reactants that occur in just a few copies per cell, but is especially an issue for self-assembly because the large number of intermediates guarantees that most are present in zero or one copies at any given time [135]. The issue is exacerbated by the fact that self-assembly reactions are frequently nucleation-limited, meaning that they are characterized by slow and relatively rare nucleation events followed by comparatively rapid polymerization. Nucleation-limited growth is well established for several of the major model systems in self-assembly, such as virus capsids [141, 217], amyloids [107], and actin and tubulin fibers [14]. A large body of theory suggests the nucleation-limited growth is crucial to their robust operation [156, 142, 141, 44, 191, 140]. In nucleation-limited systems, nearly every species is unpopulated at most times. Small copy numbers are problematic computationally in part because they mean that discretization errors inherent to efficient continuum models became substantial. In part, they are problematic because they mean that self-assembly must be treated as a stochastic system, forcing the use of less efficient simulation methods than the continuum approximations usable when all species are well populated [63, 194, 41] (see Section 3.4).
A second major challenge of self-assembly reactions is their long timescales (see Figure 2), and in particular the large gap between timescales of the full assembly reaction and the individual polymerization steps of assembly. Full assembly reactions of large complexes in vitro may have timescales measured in minutes to days (although assembly in vivo may be substantially faster [115, 36, 175]) while individual reaction steps are typically many orders of magnitude faster [180, 222]. In part, this is a side effect of nucleation-limited growth mentioned above: nucleation reactions are necessarily much slower than the subsequent elongation reactions [167, 221]. Furthermore, the nucleation reactions themselves may in fact require extensive trial-and-error involving much faster formation and breakdown of transient partial intermediates [221, 217, 184]. Large timescales, and a large dynamic range of timescales, are challenges for essentially all standard modeling methods, whether that manifests in a need for large numbers of timesteps in a continuum method or large numbers of discrete events for a stochastic simulation.
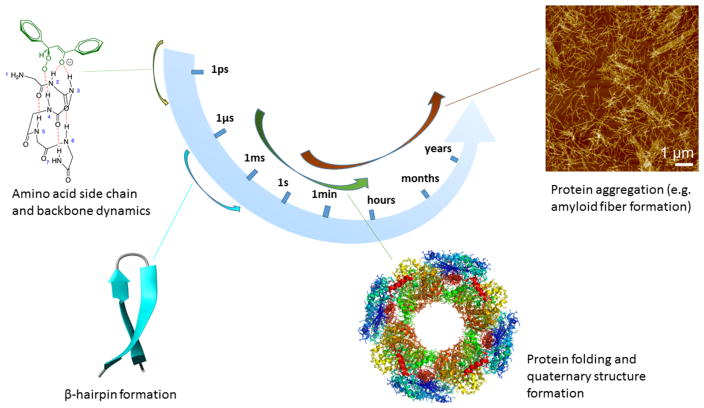
Figure 2.
Timescales for protein dynamics and aggregation. The figure illustrates some of the basic biological processes that go into self-assembly and their relevant timescales, illustrated by the specific example of amyloid aggregation. Figure based on material from [6, 21, 207, 206]
A third class of challenge arises from the fact that self-assembly reactions are unusually sensitive to the many ways in which the physical biology of the cell differs from that of in vitro models. For example, physical confinement — by the cell membrane, subcellular compartments, or other large structures such as the cytoskeleton or genome — is commonly neglected in modeling reaction systems yet cannot be ignored when dealing with reactions that result in products comparable in size to the spaces in which they form. A related issue is that self-assembly processes are also well known to be unusually sensitive to macromolecular crowding [148, 119, 75], a key distinguishing feature of the cellular environment. Numerous theoretical and experimental studies have suggested both the need for and the challenge to correcting simulation methods to account for the effects of crowding on assembly processes (e.g., [129, 166]). Examples include the effects on several aspects of DNA replication such as helicase activity and the sensitivity of DNA polymerase to salt [1], on protein-protein binding affinity and specificity [99], on the kinetics and morphology of amyloid self-assembly [115], on the stochasticity of gene expression machinery [76], and on viral capsid assembly [175, 36].
3.2. Modeling methodologies
Despite the difficulties they present to modelers, a variety of modeling methods have proven valuable for self-assembly. Table 1 describes a few of the primary methods that have emerged for self-assembly modeling. While most are drawn from older techniques for more general reaction chemistry modeling, in the self-assembly context they often present novel challenges or require specialized adaptations. This section covers three of the most successful methodologies that have been developed for self-assembly, some of the particular challenges they have faced in the self-assembly context, and how they have been adapted to meet those challenges.
Table 1.
Common modeling methodologies for self-assembly. The table lists principal techniques for self-assembly modeling, some systems biology software packages implementing them, and some notable applications in self-assembly modeling.
| Reaction Representation | Description | Software Packages | Applications |
|---|---|---|---|
| Law of Mass Action (Deterministic) | Expresses any well-mixed chemical system as a collection of coupled non-linear first order differential equations which typically must be numerically integrated. PDEs must be used when space is explicitly included | BioNetGen [15], COPASI [82], VCell [146], DBSolve [66] | Virus Assembly: [125, 173, 27, 70], Metabolomic Networks[90] |
| SSA/Gillespie Approaches | Provides a way to simulate kinetically correct trajectories consistent with the Chemical Master Equation | Moleculizer [114], BioNetGen [15], VCell [146], DESSA [218] | Virus assembly: [184, 97] |
| Spatial Stochastic | Usually combine Gillespie or Stochastic Langevin with diffusion or subunit geometry | MCell [182], StochSim [108], VCell [146], Smoldyn [4], SRSim [69] | Geometric Constraints with Diffusion: [68], Amyloid-Beta: [192] |
| Rule-Based | Primarly network-free rule-based methods which may incorporate stochasticity and spatial modeling | RuleMonkey [32], BioNetGen, ML-Space [13], VCell [164], SRSim [69] | Multivalent ligand-receptor interactions: [213], Prion Aggregation [154], Virus Assembly: [167, 218] |
| Brownian Dynamics | An explicitly spatial model where Brownian motion is computed with the Langevin equation | Smoldyn [4], MCell [182] | Multiscale Reaction-Diffusion [58], Virus Assembly: [167, 71, 128, 46, 47, 17], Crowding/Amyloids: [205], Clathrin Cage Formation: [87] |
3.2.1. Mass action differential equation (DE) models
Much modeling of reaction systems classically has arisen, at least initially, from DE models based on the chemical Law of Mass Action. Such models represent any generic chemical reaction network
in terms of a system of differential equations of the form
Accumulating these contributions across a full set of reactions and reactant species defines a system of differential equations modeling the time evolution of all reactants in the system. Such DE models were the basis of many of the earliest cell simulation systems, such as E-cell [193], ProMoT/Diva [65], Virtual Cell [162], GEPASI [118] and others. Later extensions of these models allowed for consideration of spatial heterogeneity via partial differential equation (PDE) reaction-diffusion models:
for reactant-specific diffusion coefficients di.
DE models provided a basis for some of the first approaches to modeling many self-assembly systems. Classic results on molecular assembly of polymers derived from such models include [132, 131] and they were integral to seminal models of microtubule polymerization [39]. They likewise were used for early attempts at more complex systems, such as the first dynamic models of viral self-assembly [223, 221], where they provided early insights into the parameter space of self-assembly [221]. They continue to prove valuable in that context for such problems as interpreting complex experimental data [70, 173, 27]
The most substantial challenge to DE models on self-assembly systems is computational tractability, as such models need to keep explicit track of all species that might be present in a given simulation. While that number grows only linearly in assembly size for linear polymers, it blows up exponentially in size for more complex structures such as viruses. In practice, the solution to that problem has typically been to simplify: either manually via simplified versions of structures or conflation of subsets of structures [220, 221] or through automated methods for pruning low-usage pathways [48]. While there is good empirical evidence that such strategies can yield quantitatively accurate models [223, 48], degrees of accuracy can be sensitive to structure and pathways used [120]. DE models further provide no good solution for the problem of modeling discretization of small copy number reactions.
3.3. Brownian dynamics (BD) models
The challenges self-assembly modeling presents to DEs led to an alternative approach based on Brownian dynamics (BD) particle models. In a BD approach, we explicitly model a finite set of assembly subunits in three dimensional space. These subunits diffuse through space under a model of Brownian motion, implemented by a variant of damped Langevin dynamics [51]. Models of binding dynamics can be implemented either by discrete reactions occuring upon particle collisions or via short-range binding forces, leading to gradual agglomeration of particles over the course of a simulation. BD models have the considerable advantage over DE models that one need not devote computational resources to any species not present at a specific instant in time. Run time thus depends on the number of particles modeled, not the number of species they might in principle form.
Such models have perhaps been most pronounced in their use with viral capsid systems, perhaps because their exceptionally large space of intermediates makes them especially challenging for DE models. Through viral capsid work, they have been the basis of numerous important insights into the basic biophysics of self-assembly. BD models were introduced to capsid studies nearly two decades ago [167], have seen a series of important methodological advances since [144, 71, 128] and continue to be the basis of new approaches and applications (e.g., [10, 51, 17]). They have also seen important roles in modeling various other challenging assembly systems, such as clathrin [87]. Insights arising from BD models include understanding the importance of nucleation limited growth to ensuring robust assembly and preventing kinetic trapping [73], the sensitivity to numerous parameter variations [46], and the potential sources of misassembly [46, 71]. In more recent years, these models have been extended to issues diffcult to model with other methods, such as understanding the role of the genome in RNA virus assembly [47].
The advantages of BD methods, however, come with some significant tradeoffs. First, the large size and long timescale of assembly reactions generally requires substantial structural simplifications. Second, such models typically can accommodate only modest numbers of particles, ranging up to a few thousand per simulation for state-of-the-art methods [17, 72, 155]. For relatively large structures, that may be too few to capture more than a small fraction of possible assembly trajectories. Third, they generally cannot produce quantitatively correct assembly rates, because of the large gap between diffusion rates and assembly rates. Effectively, systems need to be shifted into domains of extremely rapid assembly, through unrealistically high binding rates or concentrations, in order to yield computationally tractable simulations of the complete assembly process. Some more advanced versions of this approach can somewhat mitigate these issues, for example the use of Green’s function reaction dynamics (GFRD) to reduce the computational time needed to compute trajectories of particles between collision events [196].
3.4. Stochastic simulation algorithm (SSA) methods
Just as BD models were introduced to self-assembly modeling to address the weaknesses of DE models, so have models based on the stochastic simulation algorithm (SSA) [63] (also known as “Gillespie models” after their inventor) been adopted to address the weaknesses of BD models. In an SSA model, we represent a system at an instant in time by discrete counts of molecular species (monomers or partial assemblies). Simulation progress proceeds via reaction events, which for a self-assembly system will largely consist of single binding or dissociation reactions. Classically, one assumes a uniform, well-mixed system, in which reaction times can be approximated with exponential waiting time distributions [63]. The SSA approach can also accommodate spatial heterogeneity through modeling as an array of well-mixed, discretized spatial compartments, a variant known as spatial SSA, e.g. [183, 5].
SSA models offer considerable advantages but also involve important tradeoffs with the previously considered methods. They can be implemented to have run times independent of the number of potential species, unlike DE models, and can thus handle arbitrarily large reaction networks [64]. However, their run time does depend on the number of discrete particles present, limiting them to finite numbers of protein copies, as do BD models. They are, however, typically much more efficient than comparable BD models since they do not need to model diffusion explicitly [64] and are practical over a much broader range of parameter domains [184]. In addition, they provide an explicit quantitative model yielding kinetically correct samples from a set of reactions and associated rate constants. However, because they do not explicitly model space, they do not easily handle steric constraints that are important to such processes as aberrant assembly [167], interaction of proteins with a flexible genome [47, 214], or any form of continuous flexibility in proteins or complexes [153, 81].
SSA methods have needed some special adaptations to deal with the challenges of self-assembly. Probably the most important advance is the use of rule-based modeling (e.g. [177]), a strategy independently developed for the self-assembly field under the name local rule modeling [167] and later introduced to SSA models under that name [218]. Rule-based models allow one to avoid explicitly constructing the reaction network, an infeasible task for all but trivial self-assembly systems, and rather represent only the current state of the system and its immediate neighbors [54, 218]. This reduces run time from dependence on the size of the network to dependence only on the number of species and reactions present at any instant in time. While steric constraints are a challenge for rule-base models, that challenge has been overcome for some systems, e.g., in modeling multivalent ligand-receptor interactions [123]. Further improvements to queuing methods for discrete event implementations of SSA [89, 40] made it possible to accelerate run time by eliminating quadratic time/memory dependencies in the standard algorithms. Additionally, a set of more specialized theory has been developed to deal with the problem of extreme divergence between timescales of monomeric reactions versus the complete assembly process. Generic methods for accelerating SSA can be helpful, e.g., [145, 25], as well as more specialized variants specifically for self-assembly [121]. Other improvements include hybrid methods combining SSA with ideas from agent-based modeling [3].
While SSA methods have not yet seen as wide use as BD in the self-assembly field, they have proven to have important applications for which neither DE nor BD methods are suitable. Because of their ability to handle complex geometries and long time scales, SSA models have proven valuable for exploring parameter dependence of assembly systems by making it practical to sample large numbers of trajectories over long time scales [211] and to sample trajectories from particularly complex geometries or pathway sets [97]. They have also become a valuable platform for fitting models to experimental data, where their ability to fit an explicit timescale, to function over wide parameter ranges, and to model complex geometries are all crucial features [211, 210, 175, 176].
4. Self-assembly in broader systems biology modeling
In recent years, efforts at systems biology modeling have begun increasingly to recognize the importance of self-assembly to comprehensive modeling of complex biochemical systems. For example, a number of general systems biology simulation tools have begun to incorporate handling of self-assembly in various ways. An early example was Moleculizer [114], which incorporated basic models of assembly reactions via a rule-based SSA model with special purpose corrections accounting for altered diffusion rates of growing species. Similar kinds of models have become important more generally in modeling tools, such as RuleBender [212], which have made it possible to integrate similar rule-based SSA models into other tools for systems biology modeling. The Virtual Cell [163] has recently added handling of self-assembly reactions, using a special-purpose extension based on a form of coarse-grained BD models of self-assembly [4, 35], as well as explicit handling of rule-based modeling [164]. The most recent version of the E-Cell [193] simulation environment (ECell4) has also been updated to include capabilities for modeling self-assembly such as a network-free rule model [54] and a spatial SSA method [183]. While none of these systems yet incorporates all of the specializations found in such methods in self-assembly specific contexts, they represent important steps towards generic tools for modeling complex reaction networks that include but are not specific to self-assembly.
This need for handling the kind of combinatorial explosive reaction network that characterizes self-assembly is also beginning to be reflected in systems biology language design. For example, the Systems Biology Markup Language (SBML) [85, 84], which has become the de facto standard for specifying models in systems biology, has been updated in more recent versions to accommodate the kind of network-free rule-based models needed for self-assembly work [83]. While it has long been possible to generate SBML from a rule specification through external tools, such as BioNetGen [55], native support of the modeling language is necessary to achieve the benefits of network-free modeling needed to make complicated self-assembly modeling tractable. Handling of steric constraints that become imporant in formation of more complicated assemblies remains a hard problem for the field, however, and is so far handled only in more specialized self-assembly simulation languages [218].
Recent years have also seen claims of the first true whole-cell simulations [96, 157], an effort that necessarily involves modeling numerous processes that depend on self-assembly. In practice, such efforts have not relied on a general-purpose simulation engine suitable to both self-assembly and more conventional reaction chemistry, favoring instead general purpose methods ill-suited to self-assembly coupled to special-purpose handling of particular kinds of self-assembly. The landmark work of Karr et al. [96] establishing a comprehensive simulation of M. genitalium biochemistry, relied on a series of special-purpose modules, several of which involved ad hoc methods for specific examples of self-assembly, such as macromolecular complexation and ribosome assembly. Nonetheless, even this kind of special-purpose handling remains the exception in similar efforts at comprehensive modeling of whole-cell reaction networks (e.g., [189, 18]).
5. Conclusions
Self-assembly is a greatly important but long neglected issue in the quantitative modeling of biological systems. While it is conventionally seen as a specialized form of chemistry, it is in fact the dominant form of reaction in living systems. It poses distinctive challenges for modeling methods, though, that prevailing methods in systems biology cannot handle. Self-assembly modeling has, however, been studied intensively in many more specialized contexts, leading to an appreciation of these challenges and a variety of ways they can be addressed. As more general systems biology efforts are beginning to embrace the necessity of accommodating self-assembly, this specialized literature can provide guidance and at least partial answers to some of the biggest obstacles these efforts will encounter. This review was intended to provide a brief overview of the particular challenges of self-assembly modeling, how they have been approached to date, and how these methods have been used in the past and are beginning to be incorporated into comprehensive models of systems biology. The hope behind this review is that better awareness of obstacles and solutions already identified by self-assembly modelers can assist the broader systems modeling community in anticipating and navigating the same issues.
An appreciation for the past literature allows us to predict some of the future paths comprehensive systems modeling efforts are likely to follow. For the most part, where general efforts systems biology modeling has considered self-assembly, it has been as special cases with special-purpose methods for specific systems (e.g., [17, 47, 211, 174, 158, 92, 99, 178, 155, 10, 219, 210]). Given the many examples of self-assembly in cell biology, it is safe to say this is not a sustainable solution; rather general systems biology efforts will need to start to think of self-assembly as the normal case that must be accommodated and integrated into simulation design via both model specifications and simulation algorithms. Modeling methods that will work for both self-assembly and for other kinds of chemistry exist [13, 199, 3, 146, 15, 114, 185, 32], but will need to become the standard for modeling tools and languages. More foresighted efforts in a variety of systems modeling contexts can help point the way (e.g., [114, 193, 4, 96, 35, 164]), although most remain behind the state-of-the-art in modeling of self-assembly specifically.
At the same time, there are many challenges for which good solutions do not yet exist. For example model inference [30] remains an extremely difficult problem for self-assembly systems [223, 104, 211, 210], where the Bayesian methods usually favored by the field [67] are unusable in practice, and it is likely advances in both biotechnology and inference algorithms will be needed to address it. The field is beginning to tackle this challenge, e.g., with BioNetFit [190], which uses a genetic algorithm to provide curve fitting capabilities compatible with ODE (BioNetGen) and Network Free (NFSim) model specifications and has proven successful in fitting to steady-state and time-series oligomerization data. There are also, as yet, no universally good methods for modeling hard self-assembly systems. Each of the major approaches covered here — SSA [64], BD [167], and DE [220] — has tradeoffs that make them unsuitable for some questions. It remains to be seen whether more general solutions might arise from advances in one or more of these methods, clever hybrid approaches, or some wholly new ideas. It is also worth noting that self-assembly systems are challenging to characterize experimentally, for similar reasons to their challenge to modelers. The solutions to that issue, as well, are likely to lie in pooled efforts by experimentalists and computational researchers to advance experimental biotechnology and model-fitting algorithms in complementary ways. Indeed, self-assembly may be a particularly valuable test case for addressing the hard problems in building detailed and predictive quantitative models of complex biological systems, where the field can begin to think of modelers and experimentalists not as two communities but as two inseparable pieces of the future practice of biological discovery.
Acknowledgments
M.T. was supported in part by U.S. National Institutes of Health award T32EB009403. R.S. was supported in part by U.S. National Institutes of Health award R01GM117425, U.S. Air Force Office of Scientific Research Award FA9550-13-1-0108, Commonwealth of Pennsylvania CURE grant 1042545, and an award from the University of Pittsburgh Medical Center (UPMC). The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations or conclusions.
References
- 1.Akabayov B, Akabayov SR, Lee SJ, Wagner G, Richardson CC. Impact of macromolecular crowding on dna replication. Nature communications. 2013;4:1615. doi: 10.1038/ncomms2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alioscha-Perez M, Benadiba C, Goossens K, Kasas S, Dietler G, Willaert R, Sahli H. A robust actin filaments image analysis framework. PLoS Comput Biol. 2016;12(8):e1005063. doi: 10.1371/journal.pcbi.1005063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amar P, Paulevé L. Hsim: A hybrid stochastic simulation system for systems biology. Electronic Notes in Theoretical Computer Science. 2015;313:3–21. [Google Scholar]
- 4.Andrews SS, Addy NJ, Brent R, Arkin AP. Detailed simulations of cell biology with smoldyn 2.1. PLoS Comput Biol. 2010;6(3):e1000705. doi: 10.1371/journal.pcbi.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews SS, Bray D. Stochastic simulation of chemical reactions with spatial resolution and single molecule detail. Physical biology. 2004;1(3):137. doi: 10.1088/1478-3967/1/3/001. [DOI] [PubMed] [Google Scholar]
- 6.Apetri MM, Harkes R, Subramaniam V, Canters GW, Schmidt T, Aartsma TJ. Direct observation of α-synuclein amyloid aggregates in endocytic vesicles of neuroblastoma cells -licenced under cc by 4.0. PloS one. 2016;11(4):e0153020. doi: 10.1371/journal.pone.0153020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115(6):705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- 8.Bai Y, Luo Q, Liu J. Protein self-assembly via supramolecular strategies. Chemical Society Reviews. 2016;45(10):2756–2767. doi: 10.1039/c6cs00004e. [DOI] [PubMed] [Google Scholar]
- 9.Bailey J, Critser P, Whittington C, Kuske J, Yoder MC, Voytik-Harbin S. Collagen oligomers modulate physical and biological properties of three-dimensional self-assembled matrices. Biopolymers. 2011;95(2):77–93. doi: 10.1002/bip.21537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baschek JE, Klein HC, Schwarz US. Stochastic dynamics of virus capsid formation: direct versus hierarchical self-assembly. BMC biophysics. 2012;5(1):22. doi: 10.1186/2046-1682-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bieler NS, Knowles TP, Frenkel D, Vácha R. Connecting macroscopic observables and microscopic assembly events in amyloid formation using coarse grained simulations. PLoS Comput Biol. 2012;8(10):e1002692. doi: 10.1371/journal.pcbi.1002692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bischofberger M, Iacovache I, Boss D, Naef F, van der Goot FG, Molina N. Revealing assembly of a pore-forming complex using single-cell kinetic analysis and modeling. Biophysical journal. 2016;110(7):1574–1581. doi: 10.1016/j.bpj.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bittig A, Uhrmacher A. Ml-space: Hybrid spatial gillespie and particle simulation of multilevel rule-based models in cell biology. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2016 doi: 10.1109/TCBB.2016.2598162. [DOI] [PubMed] [Google Scholar]
- 14.Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by arp2/3 complex and wasp/scar proteins. Nature. 2000;404(6781):1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- 15.Blinov ML, Faeder JR, Goldstein B, Hlavacek WS. Bionetgen: software for rule-based modeling of signal transduction based on the interactions of molecular domains. Bioinformatics. 2004;20(17):3289–3291. doi: 10.1093/bioinformatics/bth378. [DOI] [PubMed] [Google Scholar]
- 16.Blinov ML, Faeder JR, Goldstein B, Hlavacek WS. A network model of early events in epidermal growth factor receptor signaling that accounts for combinatorial complexity. Biosystems. 2006;83(2):136–151. doi: 10.1016/j.biosystems.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 17.Boettcher MA, Klein HC, Schwarz US. Role of dynamic capsomere supply for viral capsid self-assembly. Physical biology. 2015;12(1):016014. doi: 10.1088/1478-3975/12/1/016014. [DOI] [PubMed] [Google Scholar]
- 18.Bordbar A, Monk JM, King ZA, Palsson BO. Constraint-based models predict metabolic and associated cellular functions. Nature Reviews Genetics. 2014;15(2):107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- 19.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nature structural & molecular biology. 2013;20(1):14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buxbaum R, Heidemann S. A thermodynamic model for force integration and microtubule assembly during axonal elongation. Journal of theoretical biology. 1988;134(3):379–390. doi: 10.1016/s0022-5193(88)80068-7. [DOI] [PubMed] [Google Scholar]
- 21.By Awapuhi Lee (Own work). [CC BY-SA 4.0 (http://creativecommons.org/licenses/by-sa/4.0)], via Wikimedia Commons. 1x9j. 2015
- 22.By Doc. RNDr. Josef Reischig, CSc. (Author’s archive) [CC BY-SA 3.0 (http://creativecommons.org/licenses/by-sa/3.0)], via Wikimedia Commons. Chromatin fibers (261 19) mitosis; xenopus egg.
- 23.By Nephron (Own work). [CC BY-SA 3.0 (http://creativecommons.org/licenses/by-sa/3.0) or GFDL (http://www.gnu.org/copyleft/fdl.html)], via Wikimedia Commons. Cerebral amyloid angiopathy -2b- amyloid beta - very high mag
- 24.Cadena-Nava RD, Comas-Garcia M, Garmann RF, Rao A, Knobler CM, Gelbart WM. Self-assembly of viral capsid protein and rna molecules of different sizes: requirement for a specific high protein/rna mass ratio. Journal of virology. 2012;86(6):3318–3326. doi: 10.1128/JVI.06566-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao Y, Gillespie DT, Petzold LR. Efficient step size selection for the tau-leaping simulation method. The Journal of chemical physics. 2006;124(4):044109. doi: 10.1063/1.2159468. [DOI] [PubMed] [Google Scholar]
- 26.Cao Y, Li H, Petzold L. Efficient formulation of the stochastic simulation algorithm for chemically reacting systems. The journal of chemical physics. 2004;121(9):4059–4067. doi: 10.1063/1.1778376. [DOI] [PubMed] [Google Scholar]
- 27.Casini GL, Graham D, Heine D, Garcea RL, Wu DT. In vitro papillomavirus capsid assembly analyzed by light scattering. Virology. 2004;325(2):320–327. doi: 10.1016/j.virol.2004.04.034. [DOI] [PubMed] [Google Scholar]
- 28.Cescatti M, Saverioni D, Capellari S, Tagliavini F, Kitamoto T, Ironside J, Giese A, Parchi P. Analysis of conformational stability of abnormal prion protein aggregates across the spectrum of creutzfeldt-jakob disease prions. Journal of virology. 2016:JVI–00144. doi: 10.1128/JVI.00144-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C, Kao CC, Dragnea B. Self-assembly of brome mosaic virus capsids: Insights from shorter time-scale experiments. The Journal of Physical Chemistry A. 2008;112(39):9405–9412. doi: 10.1021/jp802498z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou IC, Voit EO. Recent developments in parameter estimation and structure identification of biochemical and genomic systems. Mathematical biosciences. 2009;219(2):57–83. doi: 10.1016/j.mbs.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cole AG. Modulators of hbv capsid assembly as an approach to treating hepatitis b virus infection. Current Opinion in Pharmacology. 2016;30:131–137. doi: 10.1016/j.coph.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Colvin J, Monine MI, Gutenkunst RN, Hlavacek WS, Von Hoff DD, Posner RG. Rulemonkey: software for stochastic simulation of rule-based models. BMC bioinformatics. 2010;11(1):1. doi: 10.1186/1471-2105-11-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using crispr/cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cosgrove DJ. Re-constructing our models of cellulose and primary cell wall assembly. Current opinion in plant biology. 2014;22:122–131. doi: 10.1016/j.pbi.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cowan AE, Moraru II, Schaff JC, Slepchenko BM, Loew LM. Spatial modeling of cell signaling networks. Methods in cell biology. 2012;110:195. doi: 10.1016/B978-0-12-388403-9.00008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.del Álamo M, Rivas G, Mateu MG. Effect of macromolecular crowding agents on human immunodeficiency virus type 1 capsid protein assembly in vitro. Journal of virology. 2005;79(22):14271–14281. doi: 10.1128/JVI.79.22.14271-14281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Den Otter WK, Renes MR, Briels W. Self-assembly of three-legged patchy particles into polyhedral cages. Journal of Physics: Condensed Matter. 2010;22(10):104103. doi: 10.1088/0953-8984/22/10/104103. [DOI] [PubMed] [Google Scholar]
- 38.Denning DW. Minimizing fungal disease deaths will allow the unaids target of reducing annual aids deaths below 500 000 by 2020 to be realized. Phil Trans R Soc B. 2016;371(1709):20150468. doi: 10.1098/rstb.2015.0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annual review of cell and developmental biology. 1997;13(1):83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 40.Diamond B, Krahl D, Nastasi A, Tag P. Extendsim advanced techology: integrated simulation database. Proceedings of the Winter Simulation Conference; Winter Simulation Conference; 2010. pp. 32–39. [Google Scholar]
- 41.Duncan A, Erban R, Zygalakis K. Hybrid framework for the simulation of stochastic chemical kinetics arXiv preprint arXiv: 151203988. 2015 [Google Scholar]
- 42.Dykeman EC, Stockley PG, Twarock R. Building a viral capsid in the presence of genomic rna. Physical Review E. 2013;87(2):022717. doi: 10.1103/PhysRevE.87.022717. [DOI] [PubMed] [Google Scholar]
- 43.Dykeman EC, Stockley PG, Twarock R. Solving a levinthal’s paradox for virus assembly identifies a unique antiviral strategy. Proceedings of the National Academy of Sciences. 2014;111(14):5361–5366. doi: 10.1073/pnas.1319479111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of wave1-induced actin nucleation by rac1 and nck. Nature. 2002;418(6899):790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 45.Edwards M, Zwolak A, Schafer DA, Sept D, Dominguez R, Cooper JA. Capping protein regulators fine-tune actin assembly dynamics. Nature Reviews Molecular Cell Biology. 2014;15(10):677–689. doi: 10.1038/nrm3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elrad OM, Hagan MF. Mechanisms of size control and polymorphism in viral capsid assembly. Nano letters. 2008;8(11):3850–3857. doi: 10.1021/nl802269a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elrad OM, Hagan MF. Encapsulation of a polymer by an icosahedral virus. Physical biology. 2010;7(4):045003. doi: 10.1088/1478-3975/7/4/045003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Endres D, Miyahara M, Moisant P, Zlotnick A. A reaction landscape identifies the intermediates critical for self-assembly of virus capsids and other polyhedral structures. Protein science. 2005;14(6):1518–1525. doi: 10.1110/ps.041314405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Engqvist-Goldstein ÅE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annual review of cell and developmental biology. 2003;19(1):287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 50.Engqvist-Goldstein ÅE, Warren RA, Kessels MM, Keen JH, Heuser J, Drubin DG. The actin-binding protein hip1r associates with clathrin during early stages of endocytosis and promotes clathrin assembly in vitro. The Journal of cell biology. 2001;154(6):1209–1224. doi: 10.1083/jcb.200106089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Erban R. Proc R Soc A. 2167. Vol. 470. The Royal Society; 2014. From molecular dynamics to brownian dynamics; p. 20140036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Erban R, Flegg MB, Papoian GA. Multiscale stochastic reaction–diffusion modeling: application to actin dynamics in filopodia. Bulletin of mathematical biology. 2014;76(4):799–818. doi: 10.1007/s11538-013-9844-3. [DOI] [PubMed] [Google Scholar]
- 53.Errington WJ, Khan MQ, Bueler SA, Rubinstein JL, Chakrabartty A, Privé GG. Adaptor protein self-assembly drives the control of a cullin-ring ubiquitin ligase. Structure. 2012;20(7):1141–1153. doi: 10.1016/j.str.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 54.Faeder JR, Blinov ML, Goldstein B, Hlavacek WS. Rule-based modeling of biochemical networks. Complexity. 2005;10(4):22–41. [Google Scholar]
- 55.Faeder JR, Blinov ML, Hlavacek WS. Rule-based modeling of biochemical systems with bionetgen. Systems biology. 2009:113–167. doi: 10.1007/978-1-59745-525-1_5. [DOI] [PubMed] [Google Scholar]
- 56.Fallahi-Sichani M, Flynn JL, Linderman JJ, Kirschner DE. Differential risk of tuberculosis reactivation among anti-tnf therapies is due to drug binding kinetics and permeability. The Journal of Immunology. 2012;188(7):3169–3178. doi: 10.4049/jimmunol.1103298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fass J, Pak C, Bamburg J, Mogilner A. Stochastic simulation of actin dynamics reveals the role of annealing and fragmentation. Journal of theoretical biology. 2008;252(1):173–183. doi: 10.1016/j.jtbi.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franz B, Flegg MB, Chapman SJ, Erban R. Multiscale reaction-diffusion algorithms: Pde-assisted brownian dynamics. SIAM Journal on Applied Mathematics. 2013;73(3):1224–1247. [Google Scholar]
- 59.Frazier JM, Chushak Y, Foy B. Stochastic simulation and analysis of biomolecular reaction networks. BMC systems biology. 2009;3(1):1. doi: 10.1186/1752-0509-3-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Funahashi A, Matsuoka Y, Jouraku A, Morohashi M, Kikuchi N, Kitano H. Celldesigner 3.5: a versatile modeling tool for biochemical networks. Proceedings of the IEEE. 2008;96(8):1254–1265. [Google Scholar]
- 61.Gardner K, Bennett V. Modulation of spectrin–actin assembly by erythrocyte adducin. Nature. 1987;328(6128):359–362. doi: 10.1038/328359a0. [DOI] [PubMed] [Google Scholar]
- 62.Gardner MK, Hunt AJ, Goodson HV, Odde DJ. Microtubule assembly dynamics: new insights at the nanoscale. Current opinion in cell biology. 2008;20(1):64–70. doi: 10.1016/j.ceb.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gillespie DT. Exact stochastic simulation of coupled chemical reactions. The journal of physical chemistry. 1977;81(25):2340–2361. [Google Scholar]
- 64.Gillespie DT. Stochastic simulation of chemical kinetics. Annu Rev Phys Chem. 2007;58:35–55. doi: 10.1146/annurev.physchem.58.032806.104637. [DOI] [PubMed] [Google Scholar]
- 65.Ginkel M, Kremling A, Nutsch T, Rehner R, Gilles ED. Modular modeling of cellular systems with promot/diva. Bioinformatics. 2003;19(9):1169–1176. doi: 10.1093/bioinformatics/btg128. [DOI] [PubMed] [Google Scholar]
- 66.Gizzatkulov NM, Goryanin II, Metelkin EA, Mogilevskaya EA, Peskov KV, Demin OV. Dbsolve optimum: a software package for kinetic modeling which allows dynamic visualization of simulation results. BMC systems biology. 2010;4(1):109. doi: 10.1186/1752-0509-4-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Golightly A, Wilkinson DJ. Bayesian parameter inference for stochastic biochemical network models using particle markov chain monte carlo. Interface focus. 2011:rsfs20110047. doi: 10.1098/rsfs.2011.0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gruenert G, Ibrahim B, Lenser T, Lohel M, Hinze T, Dittrich P. Rule-based spatial modeling with diffusing, geometrically constrained molecules. BMC bioinformatics. 2010;11(1):1. doi: 10.1186/1471-2105-11-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grünert G, Dittrich P. Using the srsim software for spatial and rule-based modeling of combinatorially complex biochemical reaction systems. International Conference on Membrane Computing; Springer; 2010. pp. 240–256. [Google Scholar]
- 70.Hagan MF. Modeling viral capsid assembly. Advances in chemical physics. 2014;155:1. doi: 10.1002/9781118755815.ch01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hagan MF, Chandler D. Dynamic pathways for viral capsid assembly. Biophysical journal. 2006;91(1):42–54. doi: 10.1529/biophysj.105.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hagan MF, Elrad OM. Understanding the concentration dependence of viral capsid assembly kineticsthe origin of the lag time and identifying the critical nucleus size. Biophysical journal. 2010;98(6):1065–1074. doi: 10.1016/j.bpj.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hagan MF, Elrad OM, Jack RL. Mechanisms of kinetic trapping in self-assembly and phase transformation. The Journal of chemical physics. 2011;135(10):104115. doi: 10.1063/1.3635775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hammar P, Leroy P, Mahmutovic A, Marklund EG, Berg OG, Elf J. The lac repressor displays facilitated diffusion in living cells. Science. 2012;336(6088):1595–1598. doi: 10.1126/science.1221648. [DOI] [PubMed] [Google Scholar]
- 75.Hancock R. A role for macromolecular crowding effects in the assembly and function of compartments in the nucleus. Journal of structural biology. 2004;146(3):281–290. doi: 10.1016/j.jsb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 76.Hansen MM, Meijer LH, Spruijt E, Maas RJ, Rosquelles MV, Groen J, Heus HA, Huck WT. Macromolecular crowding creates heterogeneous environments of gene expression in picolitre droplets. Nature nanotechnology. 2016;11(2):191–197. doi: 10.1038/nnano.2015.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He B, Fan Q, Yang F, Hu T, Qiu W, Feng Y, Li Z, Li Y, Zhang F, Guo H, Zou X, Tu C. Hepatitis virus in long-fingered bats, Myanmar. Emerg Infect Dis. 2013;19(4):638–640. doi: 10.3201/eid1904.121655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hemberg M, Yaliraki SN, Barahona M. Stochastic kinetics of viral capsid assembly based on detailed protein structures. Biophysical journal. 2006;90(9):3029–3042. doi: 10.1529/biophysj.105.076737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heng YW, Koh CG. Actin cytoskeleton dynamics and the cell division cycle. The international journal of biochemistry & cell biology. 2010;42(10):1622–1633. doi: 10.1016/j.biocel.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 80.Hlavacek WS, Faeder JR, Blinov ML, Perelson AS, Goldstein B. The complexity of complexes in signal transduction. Biotechnology and bioengineering. 2003;84(7):783–794. doi: 10.1002/bit.10842. [DOI] [PubMed] [Google Scholar]
- 81.Holmes NA, Walshaw J, Leggett RM, Thibessard A, Dalton KA, Gillespie MD, Hemmings AM, Gust B, Kelemen GH. Coiled-coil protein scy is a key component of a multiprotein assembly controlling polarized growth in streptomyces. Proceedings of the National Academy of Sciences. 2013;110(5):E397–E406. doi: 10.1073/pnas.1210657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U. Copasia complex pathway simulator. Bioinformatics. 2006;22(24):3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 83.Hucka M, Bergmann FT, Hoops S, Keating SM, Sahle S, Schaff JC, Smith LP, Wilkinson DJ. The systems biology markup language (sbml): language specification for level 3 version 1 core. J Integr Bioinforma. 2015;12(2):266. doi: 10.2390/biecoll-jib-2015-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hucka M, Finney A, Bornstein BJ, Keating SM, Shapiro BE, Matthews J, Kovitz BL, Schilstra MJ, Funahashi A, Doyle JC, et al. Evolving a lingua franca and associated software infrastructure for computational systems biology: the systems biology markup language (sbml) project. Systems biology. 2004;1(1):41–53. doi: 10.1049/sb:20045008. [DOI] [PubMed] [Google Scholar]
- 85.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. The systems biology markup language (sbml): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19(4):524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 86.Hueging K, Doepke M, Vieyres G, Bankwitz D, Frentzen A, Doerrbecker J, Gumz F, Haid S, Wölk B, Kaderali L, et al. Apolipoprotein e codetermines tissue tropism of hepatitis c virus and is crucial for viral cell-to-cell transmission by contributing to a postenvelopment step of assembly. Journal of virology. 2014;88(3):1433–1446. doi: 10.1128/JVI.01815-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ilie IM, den Otter WK, Briels WJ. Rotational brownian dynamics simulations of clathrin cage formation. The Journal of chemical physics. 2014;141(6):065101. doi: 10.1063/1.4891306. [DOI] [PubMed] [Google Scholar]
- 88.Iwasaki S, Sasaki HM, Sakaguchi Y, Suzuki T, Tadakuma H, Tomari Y. Defining fundamental steps in the assembly of the drosophila rnai enzyme complex. Nature. 2015;521(7553):533–536. doi: 10.1038/nature14254. [DOI] [PubMed] [Google Scholar]
- 89.Jamalyaria F, Rohlfs R, Schwartz R. Queue-based method for efficient simulation of biological self-assembly systems. Journal of Computational Physics. 2005;204(1):100–120. [Google Scholar]
- 90.Jamshidi N, Palsson BØ. Mass action stoichiometric simulation models: incorporating kinetics and regulation into stoichiometric models. Biophysical journal. 2010;98(2):175–185. doi: 10.1016/j.bpj.2009.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Janmey PA. Phosphoinositides and calcium as regulators of cellular actin assembly and disassembly. Annual Review of Physiology. 1994;56(1):169–191. doi: 10.1146/annurev.ph.56.030194.001125. [DOI] [PubMed] [Google Scholar]
- 92.Jiang W, Luo J, Nangia S. Multiscale approach to investigate self-assembly of telodendrimer based nanocarriers for anticancer drug delivery. Langmuir. 2015;31(14):4270–4280. doi: 10.1021/la503949b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnson JM, Tang J, Nyame Y, Willits D, Young MJ, Zlotnick A. Regulating self-assembly of spherical oligomers. Nano letters. 2005;5(4):765–770. doi: 10.1021/nl050274q. [DOI] [PubMed] [Google Scholar]
- 94.Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert J-C, Chung J, Naj AC, Kunkle BW, Wang L-S, Bis JC, Bellenguez C, et al. A novel alzheimer disease locus located near the gene encoding tau protein. Molecular psychiatry. 2015 doi: 10.1038/mp.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kalay Z. Kinetics of self-assembly via facilitated diffusion: Formation of the transcription complex. Physical Review E. 2015;92(4):042716. doi: 10.1103/PhysRevE.92.042716. [DOI] [PubMed] [Google Scholar]
- 96.Karr JR, Sanghvi JC, Macklin DN, Gutschow MV, Jacobs JM, Bolival B, Assad-Garcia N, Glass JI, Covert MW. A whole-cell computational model predicts phenotype from genotype. Cell. 2012;150(2):389–401. doi: 10.1016/j.cell.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Keef T, Micheletti C, Twarock R. Master equation approach to the assembly of viral capsids. Journal of theoretical biology. 2006;242(3):713–721. doi: 10.1016/j.jtbi.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 98.Kim T, Hwang W, Lee H, Kamm RD. Computational analysis of viscoelastic properties of crosslinked actin networks. PLoS Comput Biol. 2009;5(7):e1000439. doi: 10.1371/journal.pcbi.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim YC, Best RB, Mittal J. Macromolecular crowding effects on protein–protein binding affinity and specificity. The Journal of chemical physics. 2010;133(20):205101. doi: 10.1063/1.3516589. [DOI] [PubMed] [Google Scholar]
- 100.Kler S, Asor R, Li C, Ginsburg A, Harries D, Oppenheim A, Zlotnick A, Raviv U. Rna encapsidation by sv40-derived nanoparticles follows a rapid two-state mechanism. Journal of the American Chemical Society. 2012;134(21):8823–8830. doi: 10.1021/ja2110703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Klumpp K, Crépin T. Capsid proteins of enveloped viruses as antiviral drug targets. Current opinion in virology. 2014;5:63–71. doi: 10.1016/j.coviro.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 102.Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nature reviews Molecular cell biology. 2014;15(6):384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 103.Kressler D, Hurt E, Ba J. βler. Driving ribosome assembly. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2010;1803(6):673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 104.Kumar MS, Schwartz R. A parameter estimation technique for stochastic self-assembly systems and its application to human papillomavirus self-assembly. Physical biology. 2010;7(4):045005. doi: 10.1088/1478-3975/7/4/045005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lamorte L, Titolo S, Lemke CT, Goudreau N, Mercier JF, Wardrop E, Shah VB, von Schwedler UK, Langelier C, Banik SS, et al. Discovery of novel small-molecule hiv-1 replication inhibitors that stabilize capsid complexes. Antimicrobial agents and chemotherapy. 2013;57(10):4622–4631. doi: 10.1128/AAC.00985-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lampel A, Bram Y, Ezer A, Shaltiel-Kario R, Saad JS, Bacharach E, Gazit E. Targeting the early step of building block organization in viral capsid assembly. ACS chemical biology. 2015;10(8):1785–1790. doi: 10.1021/acschembio.5b00347. [DOI] [PubMed] [Google Scholar]
- 107.Lazo ND, Grant MA, Condron MC, Rigby AC, Teplow DB. On the nucleation of amyloid β-protein monomer folding. Protein Science. 2005;14(6):1581–1596. doi: 10.1110/ps.041292205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Le Novere N, Shimizu TS. Stochsim: modelling of stochastic biomolecular processes. Bioinformatics. 2001;17(6):575–576. doi: 10.1093/bioinformatics/17.6.575. [DOI] [PubMed] [Google Scholar]
- 109.Leckband D, Israelachvili J. Intermolecular forces in biology. Quarterly reviews of biophysics. 2001;34(02):105–267. doi: 10.1017/s0033583501003687. [DOI] [PubMed] [Google Scholar]
- 110.Leckband D, Sivasankar S. Forces controlling protein interactions: theory and experiment. Colloids and surfaces B: Biointerfaces. 1999;14(1):83–97. [Google Scholar]
- 111.Lee NN, Chalamcharla VR, Reyes-Turcu F, Mehta S, Zofall M, Balachandran V, Dhakshnamoorthy J, Taneja N, Yamanaka S, Zhou M, et al. Mtr4-like protein coordinates nuclear rna processing for heterochromatin assembly and for telomere maintenance. Cell. 2013;155(5):1061–1074. doi: 10.1016/j.cell.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157(3):624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li SH, Li XJ. Huntingtin–protein interactions and the pathogenesis of huntington’s disease. TRENDS in Genetics. 2004;20(3):146–154. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 114.Lok L, Brent R. Automatic generation of cellular reaction networks with moleculizer 1.0. Nature biotechnology. 2005;23(1):131–136. doi: 10.1038/nbt1054. [DOI] [PubMed] [Google Scholar]
- 115.Ma B, Xie J, Wei L, Li W. Macromolecular crowding modulates the kinetics and morphology of amyloid self-assembly by β-lactoglobulin. International journal of biological macromolecules. 2013;53:82–87. doi: 10.1016/j.ijbiomac.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 116.Makhnevych T, Houry WA. The role of hsp90 in protein complex assembly. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2012;1823(3):674–682. doi: 10.1016/j.bbamcr.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 117.Matthews R, Likos CN. Structures and pathways for clathrin self-assembly in the bulk and on membranes. Soft Matter. 2013;9(24):5794–5806. [Google Scholar]
- 118.Mendes P. Gepasi: a software package for modelling the dynamics, steady states and control of biochemical and other systems. Computer applications in the biosciences: CABIOS. 1993;9(5):563–571. doi: 10.1093/bioinformatics/9.5.563. [DOI] [PubMed] [Google Scholar]
- 119.Minton AP. Implications of macromolecular crowding for protein assembly. Current opinion in structural biology. 2000;10(1):34–39. doi: 10.1016/s0959-440x(99)00045-7. [DOI] [PubMed] [Google Scholar]
- 120.Misra N, Lees D, Zhang T, Schwartz R. Pathway complexity of model virus capsid assembly systems. Computational and Mathematical Methods in Medicine. 2008;9(3–4):277–293. [Google Scholar]
- 121.Misra N, Schwartz R. Efficient stochastic sampling of first-passage times with applications to self-assembly simulations. The Journal of chemical physics. 2008;129(20):204109. doi: 10.1063/1.3026595. [DOI] [PubMed] [Google Scholar]
- 122.Mondragón-Rodríguez S, Perry G, Luna-Muñoz J, Acevedo-Aquino M, Williams S. Phosphorylation of tau protein at sites ser396–404 is one of the earliest events in alzheimer’s disease and down syndrome. Neuropathology and applied neurobiology. 2014;40(2):121–135. doi: 10.1111/nan.12084. [DOI] [PubMed] [Google Scholar]
- 123.Monine MI, Posner RG, Savage PB, Faeder JR, Hlavacek WS. Modeling multivalent ligand-receptor interactions with steric constraints on configurations of cell-surface receptor aggregates. Biophysical journal. 2010;98(1):48–56. doi: 10.1016/j.bpj.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Biochemistry. 2012;81(1):661. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- 125.Morozov AY, Bruinsma RF, Rudnick J. Assembly of viruses and the pseudo-law of mass action. The Journal of chemical physics. 2009;131(15):155101. doi: 10.1063/1.3212694. [DOI] [PubMed] [Google Scholar]
- 126.Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. Visualizing ribosome biogenesis: parallel assembly pathways for the 30s subunit. Science. 2010;330(6004):673–677. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Müller-McNicoll M, Neugebauer KM. How cells get the message: dynamic assembly and function of mrna-protein complexes. Nature Reviews Genetics. 2013;14(4):275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 128.Nguyen HD, Reddy VS, Brooks CL. Deciphering the kinetic mechanism of spontaneous self-assembly of icosahedral capsids. Nano Letters. 2007;7(2):338–344. doi: 10.1021/nl062449h. [DOI] [PubMed] [Google Scholar]
- 129.Nicolau DV, Burrage K. Stochastic simulation of chemical reactions in spatially complex media. Computers & Mathematics with Applications. 2008;55(5):1007–1018. [Google Scholar]
- 130.Okazaki K-i, Sato T, Takano M. Temperature-enhanced association of proteins due to electrostatic interaction: A coarse-grained simulation of actin–myosin binding. Journal of the American Chemical Society. 2012;134(21):8918–8925. doi: 10.1021/ja301447j. [DOI] [PubMed] [Google Scholar]
- 131.Oosawa F. Size distribution of protein polymers. Journal of theoretical biology. 1970;27(1):69–86. doi: 10.1016/0022-5193(70)90129-3. [DOI] [PubMed] [Google Scholar]
- 132.Oosawa F, Higashi S. Statistical thermodynamics of polymerization and polymorphism of protein. Progress in theoretical biology. 1967;1:79–164. [Google Scholar]
- 133.Palmer MS, Dryden AJ, Hughes JT, Collinge J. Homozygous prion protein genotype predisposes to sporadic creutzfeldt–jakob disease. Nature. 1991;352(6333):340–342. doi: 10.1038/352340a0. [DOI] [PubMed] [Google Scholar]
- 134.Pantaloni D, Carlier MF. How profilin promotes actin filament assembly in the presence of thymosin β4. Cell. 1993;75(5):1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- 135.Paulsson J. Models of stochastic gene expression. Physics of life reviews. 2005;2(2):157–175. [Google Scholar]
- 136.Pfeffer SR, Rothman JE. Biosynthetic protein transport and sorting by the endoplasmic reticulum and golgi. Annual review of biochemistry. 1987;56(1):829–852. doi: 10.1146/annurev.bi.56.070187.004145. [DOI] [PubMed] [Google Scholar]
- 137.Pham JW, Sontheimer EJ. Molecular requirements for rna-induced silencing complex assembly in the drosophila rna interference pathway. Journal of Biological Chemistry. 2005;280(47):39278–39283. doi: 10.1074/jbc.M509202200. [DOI] [PubMed] [Google Scholar]
- 138.Plagens A, Tripp V, Daume M, Sharma K, Klingl A, Hrle A, Conti E, Urlaub H, Randau L. In vitro assembly and activity of an archaeal crispr-cas type ia cascade interference complex. Nucleic acids research. 2014;42(8):5125–5138. doi: 10.1093/nar/gku120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Podgornik R, Aksoyoglu MA, Yasar S, Svensek D, Parsegian VA. Dna equation of state: In vitro vs in viro. The Journal of Physical Chemistry B. 2016;120(26):6051–6060. doi: 10.1021/acs.jpcb.6b02017. [DOI] [PubMed] [Google Scholar]
- 140.Pollard TD, Cooper JA. Actinand actin-binding proteins, a critical evaluation of mechanisms and functions. Annual review of biochemistry. 1986;55(1):987–1035. doi: 10.1146/annurev.bi.55.070186.005011. [DOI] [PubMed] [Google Scholar]
- 141.Prevelige PE, Jr, Thomas D, King J. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophysical journal. 1993;64(3):824. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pring M, Evangelista M, Boone C, Yang C, Zigmond SH. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 2003;42(2):486–496. doi: 10.1021/bi026520j. [DOI] [PubMed] [Google Scholar]
- 143.Lingappa VR, Hurt CR, Garvey E. Capsid assembly as a point of intervention for novel anti-viral therapeutics. Current pharmaceutical biotechnology. 2013;14(5):513–523. doi: 10.2174/13892010113149990201. [DOI] [PubMed] [Google Scholar]
- 144.Rapaport D, Johnson J, Skolnick J. Supramolecular self-assembly: molecular dynamics modeling of polyhedral shell formation. Computer physics communications. 1999;121:231–235. [Google Scholar]
- 145.Rathinam M, Petzold LR, Cao Y, Gillespie DT. Stiffness in stochastic chemically reacting systems: The implicit tau-leaping method. The Journal of Chemical Physics. 2003;119(24):12784–12794. [Google Scholar]
- 146.Resasco DC, Gao F, Morgan F, Novak IL, Schaff JC, Slepchenko BM. Virtual cell: computational tools for modeling in cell biology. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2012;4(2):129–140. doi: 10.1002/wsbm.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ritzi M, Knippers R. Initiation of genome replication: assembly and disassembly of replication-competent chromatin. Gene. 2000;245(1):13–20. doi: 10.1016/s0378-1119(00)00020-2. [DOI] [PubMed] [Google Scholar]
- 148.Rivas G, Fernández JA, Minton AP. Direct observation of the enhancement of noncooperative protein self-assembly by macromolecular crowding: indefinite linear self-association of bacterial cell division protein ftsz. Proceedings of the National Academy of Sciences. 2001;98(6):3150–3155. doi: 10.1073/pnas.051634398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rivero F, Koppel B, Peracino B, Bozzaro S, Siegert F, Weijer CJ, Schleicher M, Albrecht R, Noegel AA. The role of the cortical cytoskeleton: F-actin crosslinking proteins protect against osmotic stress, ensure cell size, cell shape and motility, and contribute to phagocytosis and development. Journal of Cell Science. 1996;109(11):2679–2691. doi: 10.1242/jcs.109.11.2679. [DOI] [PubMed] [Google Scholar]
- 150.Rogers SL, Gelfand VI. Membrane trafficking, organelle transport, and the cytoskeleton. Current opinion in cell biology. 2000;12(1):57–62. doi: 10.1016/s0955-0674(99)00057-5. [DOI] [PubMed] [Google Scholar]
- 151.Rosa A, Everaers R. Structure and dynamics of interphase chromosomes. PLoS Comput Biol. 2008;4(8):e1000153. doi: 10.1371/journal.pcbi.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ross JL, Ali MY, Warshaw DM. Cargo transport: molecular motors navigate a complex cytoskeleton. Current opinion in cell biology. 2008;20(1):41–47. doi: 10.1016/j.ceb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Roychaudhuri R, Yang M, Hoshi MM, Teplow DB. Amyloid β-protein assembly and alzheimer disease. Journal of Biological Chemistry. 2009;284(8):4749–4753. doi: 10.1074/jbc.R800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rubenstein R, Gray P, Cleland T, Piltch M, Hlavacek W, Roberts R, Ambrosiano J, Kim JI. Dynamics of the nucleated polymerization model of prion replication. Biophysical chemistry. 2007;125(2):360–367. doi: 10.1016/j.bpc.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 155.Ruiz-Herrero T, Hagan MF. Simulations show that virus assembly and budding are facilitated by membrane microdomains. Biophysical journal. 2015;108(3):585–595. doi: 10.1016/j.bpj.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by bni1 and profilin. Nature cell biology. 2002;4(8):626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- 157.Sanghvi JC, Regot S, Carrasco S, Karr JR, Gutschow MV, Bolival B, Jr, Covert MW. Accelerated discovery via a whole-cell model. Nature methods. 2013;10(12):1192–1195. doi: 10.1038/nmeth.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Saric A, Chebaro YC, Knowles TP, Frenkel D. Crucial role of nonspecific interactions in amyloid nucleation. Proceedings of the National Academy of Sciences. 2014;111(50):17869–17874. doi: 10.1073/pnas.1410159111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Saunders MG, Voth GA. Comparison between actin filament models: coarse-graining reveals essential differences. Structure. 2012;20(4):641–653. doi: 10.1016/j.str.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 160.Sauro HM, Hucka M, Finney A, Wellock C, Bolouri H, Doyle J, Kitano H. Next generation simulation tools: the systems biology workbench and biospice integration. Omics A Journal of Integrative Biology. 2003;7(4):355–372. doi: 10.1089/153623103322637670. [DOI] [PubMed] [Google Scholar]
- 161.Schafer DA, Jennings PB, Cooper JA. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. The Journal of Cell Biology. 1996;135(1):169–179. doi: 10.1083/jcb.135.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Schaff J, Fink CC, Slepchenko B, Carson JH, Loew LM. A general computational framework for modeling cellular structure and function. Biophysical journal. 1997;73(3):1135. doi: 10.1016/S0006-3495(97)78146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schaff JC, Loew LM. The virtual cell. Pacific Symposium on Biocomputing; Citeseer. 1999. pp. 228–239. [DOI] [PubMed] [Google Scholar]
- 164.Schaff JC, Vasilescu D, Moraru II, Loew LM, Blinov ML. Rule-based modeling with virtual cell. Bioinformatics. 2016:btw353. doi: 10.1093/bioinformatics/btw353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Schmit JD, Ghosh K, Dill K. What drives amyloid molecules to assemble into oligomers and fibrils? Biophysical journal. 2011;100(2):450–458. doi: 10.1016/j.bpj.2010.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Schnell S, Turner T. Reaction kinetics in intracellular environments with macromolecular crowding: simulations and rate laws. Progress in biophysics and molecular biology. 2004;85(2):235–260. doi: 10.1016/j.pbiomolbio.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 167.Schwartz R, Shor PW, Prevelige PE, Berger B. Local rules simulation of the kinetics of virus capsid self-assembly. Biophysical journal. 1998;75(6):2626–2636. doi: 10.1016/S0006-3495(98)77708-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the rnai enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 169.Selkoe DJ. Cell biology of protein misfolding: the examples of alzheimer’s and parkinson’s diseases. Nature cell biology. 2004;6(11):1054–1061. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- 170.Sheetz MP, Wayne DB, Pearlman AL. Extension of filopodia by motor-dependent actin assembly. Cell motility and the cytoskeleton. 1992;22(3):160–169. doi: 10.1002/cm.970220303. [DOI] [PubMed] [Google Scholar]
- 171.Shi J, Friedman DB, Aiken C. Retrovirus restriction by trim5 proteins requires recognition of only a small fraction of viral capsid subunits. Journal of virology. 2013;87(16):9271–9278. doi: 10.1128/JVI.00713-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shin J, Jardine P, Noireaux V. Genome replication, synthesis, and assembly of the bacteriophage t7 in a single cell-free reaction. ACS synthetic biology. 2012;1(9):408–413. doi: 10.1021/sb300049p. [DOI] [PubMed] [Google Scholar]
- 173.Singh S, Zlotnick A. Observed hysteresis of virus capsid disassembly is implicit in kinetic models of assembly. Journal of Biological Chemistry. 2003;278(20):18249–18255. doi: 10.1074/jbc.M211408200. [DOI] [PubMed] [Google Scholar]
- 174.Slyngborg M, Fojan P. A computational study of the self-assembly of the rfffr peptide. Physical Chemistry Chemical Physics. 2015;17(44):30023–30036. doi: 10.1039/c5cp01324k. [DOI] [PubMed] [Google Scholar]
- 175.Smith GR, Xie L, Lee B, Schwartz R. Applying molecular crowding models to simulations of virus capsid assembly in vitro. Biophysical journal. 2014;106(1):310–320. doi: 10.1016/j.bpj.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Smith GR, Xie L, Schwartz R. Modeling effects of rna on capsid assembly pathways via coarse-grained stochastic simulation. PloS one. 2016;11(5):e0156547. doi: 10.1371/journal.pone.0156547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sneddon MW, Faeder JR, Emonet T. Efficient modeling, simulation and coarse-graining of biological complexity with nfsim. Nature methods. 2011;8(2):177–183. doi: 10.1038/nmeth.1546. [DOI] [PubMed] [Google Scholar]
- 178.Spiriti JM, Zuckerman DM. Tabulation as a high-resolution alternative to coarse-graining protein interactions: Initial application to virus capsid subunits. Biophysical Journal. 2016;110(3):495a. doi: 10.1063/1.4938479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Splettstoesser T, Holmes KC, Noé F, Smith JC. Structural modeling and molecular dynamics simulation of the actin filament. Proteins: Structure, Function, and Bioinformatics. 2011;79(7):2033–2043. doi: 10.1002/prot.23017. [DOI] [PubMed] [Google Scholar]
- 180.Stanley CB, Perevozchikova T, Berthelier V. Structural formation of huntingtin exon 1 aggregates probed by small-angle neutron scattering. Biophysical journal. 2011;100(10):2504–2512. doi: 10.1016/j.bpj.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Stiles J, Van Helden D, Bartol T, SALPETER M. Miniature endplate current rise times¿ 100 mu s from improved dual recordgs can be modified with passive acetylcholine diffusion from a synaptic ves icle. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(12):5747–5752. doi: 10.1073/pnas.93.12.5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Stiles JR, Bartol TM, et al. Monte carlo methods for simulating realistic synaptic microphysiology using mcell. Computational neuroscience: realistic modeling for experimentalists. 2001:87–127. [Google Scholar]
- 183.Stundzia AB, Lumsden CJ. Stochastic simulation of coupled reaction–diffusion processes. Journal of computational physics. 1996;127(1):196–207. [Google Scholar]
- 184.Sweeney B, Zhang T, Schwartz R. Exploring the parameter space of complex self-assembly through virus capsid models. Biophysical journal. 2008;94(3):772–783. doi: 10.1529/biophysj.107.107284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Takahashi K, Ishikawa N, Sadamoto Y, Sasamoto H, Ohta S, Shiozawa A, Miyoshi F, Naito Y, Nakayama Y, Tomita M. E-cell 2: multi-platform e-cell simulation system. Bioinformatics. 2003;19(13):1727–1729. doi: 10.1093/bioinformatics/btg221. [DOI] [PubMed] [Google Scholar]
- 186.Tamulaitis G, Venclovas Č, Siksnys V. Type iii crispr-cas immunity: Major differences brushed aside. Trends in Microbiology. 2016 doi: 10.1016/j.tim.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 187.Tan Z, Maguire ML, Loeb DD, Zlotnick A. Genetically altering the thermodynamics and kinetics of hepatitis b virus capsid assembly has profound effects on virus replication in cell culture. Journal of virology. 2013;87(6):3208–3216. doi: 10.1128/JVI.03014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Thévenin AF, Kowal TJ, Fong JT, Kells RM, Fisher CG, Falk MM. Proteins and mechanisms regulating gap-junction assembly, internalization, and degradation. Physiology. 2013;28(2):93–116. doi: 10.1152/physiol.00038.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Thiele I, Swainston N, Fleming RM, Hoppe A, Sahoo S, Aurich MK, Haraldsdottir H, Mo ML, Rolfsson O, Stobbe MD, et al. A community-driven global reconstruction of human metabolism. Nature biotechnology. 2013;31(5):419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Thomas BR, Chylek LA, Colvin J, Sirimulla S, Clayton AH, Hlavacek WS, Posner RG. Bionetfit: a fitting tool compatible with bionetgen, nfsim, and distributed computing environments. Bioinformatics. 2015:btv655. doi: 10.1093/bioinformatics/btv655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tobacman LS, Korn ED. The kinetics of actin nucleation and polymerization. Journal of Biological Chemistry. 1983;258(5):3207–3214. [PubMed] [Google Scholar]
- 192.Toivari E, Manninen T, Nahata AK, Jalonen TO, Linne ML. Effects of transmitters and amyloid-beta peptide on calcium signals in rat cortical astrocytes: Fura-2am measurements and stochastic model simulations. PloS one. 2011;6(3):e17914. doi: 10.1371/journal.pone.0017914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Tomita M, Hashimoto K, Takahashi K, Shimizu TS, Matsuzaki Y, Miyoshi F, Saito K, Tanida S, Yugi K, Venter JC, et al. E-cell: software environment for whole-cell simulation. Bioinformatics. 1999;15(1):72–84. doi: 10.1093/bioinformatics/15.1.72. [DOI] [PubMed] [Google Scholar]
- 194.Turner TE, Schnell S, Burrage K. Stochastic approaches for modelling in vivo reactions. Computational biology and chemistry. 2004;28(3):165–178. doi: 10.1016/j.compbiolchem.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 195.Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The rcaf complex mediates chromatin assembly during dna replication and repair. Nature. 1999;402(6761):555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 196.van Zon JS, Ten Wolde PR. Greens-function reaction dynamics: a particle-based approach for simulating biochemical networks in time and space. The Journal of chemical physics. 2005;123(23):234910. doi: 10.1063/1.2137716. [DOI] [PubMed] [Google Scholar]
- 197.Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. Mutations in fus, an rna processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Vogel V. Mechanotransduction involving multimodular proteins: converting force into biochemical signals. Annu Rev Biophys Biomol Struct. 2006;35:459–488. doi: 10.1146/annurev.biophys.35.040405.102013. [DOI] [PubMed] [Google Scholar]
- 199.Voliotis M, Thomas P, Grima R, Bowsher CG. Stochastic simulation of biomolecular networks in dynamic environments. PLoS Comput Biol. 2016;12(6):e1004923. doi: 10.1371/journal.pcbi.1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang H, Lacoche S, Huang L, Xue B, Muthuswamy SK. Rotational motion during three-dimensional morphogenesis of mammary epithelial acini relates to laminin matrix assembly. Proceedings of the National Academy of Sciences. 2013;110(1):163–168. doi: 10.1073/pnas.1201141110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang N, Butler JP, Ingber DE, et al. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260(5111):1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 202.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nature reviews Molecular cell biology. 2009;10(1):75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 203.Wang YJ, Lu D, Xu YB, Xing WQ, Tong XK, Wang GF, Feng CL, He PL, Yang L, Tang W, et al. A novel pyridazinone derivative inhibits hepatitis b virus replication by inducing genome-free capsid formation. Antimicrobial agents and chemotherapy. 2015;59(11):7061–7072. doi: 10.1128/AAC.01558-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW. A protein factor essential for microtubule assembly. Proceedings of the National Academy of Sciences. 1975;72(5):1858–1862. doi: 10.1073/pnas.72.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wieczorek G, Zielenkiewicz P. Influence of macromolecular crowding on protein-protein association ratesa brownian dynamics study. Biophysical journal. 2008;95(11):5030–5036. doi: 10.1529/biophysj.108.136291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wikimedia Commons/C.rose.kennedy. Julia-colonna active site structure1. 2010 [Google Scholar]
- 207.Wikimedia Commons/Isabella Daidone. Beta hairpin. 2006 [Google Scholar]
- 208.Wimo A, Jönsson L, Bond J, Prince M, Winblad B, International AD. The worldwide economic impact of dementia 2010. Alzheimer’s & Dementia. 2013;9(1):1–11. doi: 10.1016/j.jalz.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 209.Wortmann M. Dementia: a global health priority-highlights from an adi and world health organization report. Alzheimer’s research & therapy. 2012;4(5):1. doi: 10.1186/alzrt143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Xie L, Smith G, Schwartz R. Applying derivative-free optimization to fit kinetic parameters of viral capsid self-assembly models from multi-source bulk in vitro data. Biophysical Journal. 2015;108(2):470a–471a. [Google Scholar]
- 211.Xie L, Smith GR, Feng X, Schwartz R. Surveying capsid assembly pathways through simulation-based data fitting. Biophysical journal. 2012;103(7):1545–1554. doi: 10.1016/j.bpj.2012.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Xu W, Smith AM, Faeder JR, Marai GE. Rulebender: a visual interface for rule-based modeling. Bioinformatics. 2011;27(12):1721–1722. doi: 10.1093/bioinformatics/btr197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yang J, Monine MI, Faeder JR, Hlavacek WS. Kinetic monte carlo method for rule-based modeling of biochemical networks. Physical Review E. 2008;78(3):031910. doi: 10.1103/PhysRevE.78.031910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Yang L, Gal J, Chen J, Zhu H. Self-assembled fus binds active chromatin and regulates gene transcription. Proceedings of the National Academy of Sciences. 2014;111(50):17809–17814. doi: 10.1073/pnas.1414004111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yang Z, Reeves M, Ye J, Trang P, Zhu L, Sheng J, Wang Y, Zen K, Wu J, Liu F. Rnase p ribozymes inhibit the replication of human cytomegalovirus by targeting essential viral capsid proteins. Viruses. 2015;7(7):3345–3360. doi: 10.3390/v7072775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ydenberg CA, Smith BA, Breitsprecher D, Gelles J, Goode BL. Cease-fire at the leading edge: New perspectives on actin filament branching, debranching, and cross-linking. Cytoskeleton. 2011;68(11):596–602. doi: 10.1002/cm.20543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zandi R, van der Schoot P, Reguera D, Kegel W, Reiss H. Classical nucleation theory of virus capsids. Biophysical journal. 2006;90(6):1939–1948. doi: 10.1529/biophysj.105.072975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Zhang T, Rohlfs R, Schwartz R. Implementation of a discrete event simulator for biological self-assembly systems. Proceedings of the Winter Simulation Conference; 2005; IEEE; 2005. p. 9. [Google Scholar]
- 219.Zhang T, Schwartz R. Simulation study of the contribution of oligomer/oligomer binding to capsid assembly kinetics. Biophysical journal. 2006;90(1):57–64. doi: 10.1529/biophysj.105.072207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zlotnick A. To build a virus capsid: an equilibrium model of the self assembly of polyhedral protein complexes. Journal of molecular biology. 1994;241(1):59–67. doi: 10.1006/jmbi.1994.1473. [DOI] [PubMed] [Google Scholar]
- 221.Zlotnick A. Theoretical aspects of virus capsid assembly. Journal of Molecular Recognition. 2005;18(6):479–490. doi: 10.1002/jmr.754. [DOI] [PubMed] [Google Scholar]
- 222.Zlotnick A, Aldrich R, Johnson JM, Ceres P, Young MJ. Mechanism of capsid assembly for an icosahedral plant virus. Virology. 2000;277(2):450–456. doi: 10.1006/viro.2000.0619. [DOI] [PubMed] [Google Scholar]
- 223.Zlotnick A, Johnson JM, Wingfield PW, Stahl SJ, Endres D. A theoretical model successfully identifies features of hepatitis b virus capsid assembly. Biochemistry. 1999;38(44):14644–14652. doi: 10.1021/bi991611a. [DOI] [PubMed] [Google Scholar]