Abstract
Vasodilator-stimulated phosphoprotein (VASP) is a processive actin polymerase with roles in the control of cell shape and cell migration. Through interaction with the cytoskeletal adaptor protein Zyxin, VASP can localize to damaged stress fibers where it serves to repair and reinforce these structures. VASP localization is mediated by its N-terminal Ena/VASP homology (EVH1) domain, which binds to the (W/F)PxφP motif (most commonly occurring as FPPPP) found in cytoskeletal proteins such as vinculin, lamellipodin, and Zyxin. Sequentially close clusters of four or five of these motifs frequently occur, as in the proline rich region of Zyxin with four such motifs. This suggests that tetrameric VASP might bind very tightly to Zyxin through avidity, with all four EVH1 domains binding to a single Zyxin molecule. Here, quantitative nuclear magnetic resonance titration analysis reveals a dominant bivalent 1:1 (Zyxin:EVH1) interaction between the Zyxin proline rich region and the VASP EVH1 domain that utilizes the EVH1 canonical binding site and a novel secondary binding site on the opposite face of the EVH1 domain. We further show that binding to the secondary binding site is specifically inhibited by mutation of VASP EVH1 domain residue Y39 to E, which mimics Abl-induced phosphorylation of Y39. On the basis of these findings, we propose a model in which phosphorylation of Y39 acts as a stoichiometry switch that governs binding partner selection by the constitutive VASP tetramer. These results have broader implications for other multivalent VASP EVH1 domain binding partners and for furthering our understanding of the role of Y39 phosphorylation in regulating VASP localization and cellular function.
Graphical abstract
The processive actin polymerase VASP (vasodilator-stimulated phosphoprotein) is an important regulator of the organization of the actin cytoskeleton.1–3 VASP and its homologues, Mena and Evl, are involved in the formation of lamellipodia4 and filopodia,5,6 as well as the cellular processes of phagocytosis,7 cell migration,3,4,8 and neuron axon guidance.9 VASP functions both as an actin polymerase and as an anti-capping protein,4,10,11 by protecting the barbed end of actin filaments from capping proteins. VASP and its homologues localize to specific sites in the cell such as lamellipodia and filopodia as well as focal adhesions and sites of stress fiber damage12–16 through binding to proline rich sequences with its EVH1 (Ena/VASP homology 1) domain.17,18
In response to cell mechanical stress, VASP is recruited to actin stress fibers, where it serves to reinforce and repair these highly regulated contractile structures.16 This is partially mediated by an interaction with Zyxin, a cytoskeletal protein involved in the mechanical stress response.15,16 Zyxin is phosphorylated at S142 in response to mechanical stimulation, which disrupts an inhibitory head–tail intramolecular interaction.19,20 Zyxin is then recruited to damaged stress fibers through an interaction between its LIM domains and unknown factors.21 Simultaneously, S142-phosphorylated Zyxin binds via multiple binding motifs in its disordered N-terminus to the proteins VASP and α-actinin.19
The interaction between VASP and Zyxin is mediated by both interactions between the N-terminal proline rich region of Zyxin and the EVH1 domain of VASP and an interaction between the LIM domain region of Zyxin and the proline rich region of VASP.20 This allows the interaction to be regulated by phosphorylation at multiple sites. First, Zyxin can adopt an inhibited conformation in which its LIM domains obstruct its proline rich region, which can be destabilized by phosphorylation of S142 by Akt.19,20,22 Second, although the LIM–VASP interaction can overcome this inhibitory conformation, phosphorylation of VASP at S157 by PKA prevents this.20 Third, phosphorylation of Y39 in the EVH1 domain of VASP by Abl inhibits its interaction with Zyxin,23 though it is not clear why given the placement of Y39 far from the EVH1 canonical binding site (here termed the primary binding site) known to bind the Listeria monocytogenes protein ActA.24
The VASP EVH1 domain binds the (W/F)PxφP motif, where x is any amino acid and φ is aliphatic.17,18 This motif most frequently occurs as FPPPP and very often occurs in clusters of four or five motifs, as in the cytoskeletal proteins lamellipodin and Zyxin, and ActA. This is thought to be a mechanism for enhancing binding between VASP and its binding partners, by combining the individual affinities of each of the four EVH1 domains in a VASP tetramer.25 Indeed, deletions in the C-terminal region of the EVH2 domain in Ena (Ena-VASP homology 2 domain) inhibit tetramerization and impair its ability to bind Zyxin in cell lysates26 and to localize VASP to focal adhesions.27 Inconsistent with this, however, are the facts that the EVH2 domain is dispensable for Listeria motility28 and that there seems to be a linear dependence of the rate of Listeria movement on the number of VASP binding sites in ActA.29 It has been proposed alternatively that a ligand molecule containing multiple FPPPP sites can be bound by multiple VASP tetramers, which would allow VASP to noncovalently cross-link its binding partners.25
Here, we provide evidence that a secondary binding site is present in the VASP EVH1 domain. This secondary binding site interacts weakly with short peptides containing a single (W/F)PxφP motif but displays enhanced binding to a Zyxin fragment containing all four (W/F)PxφP motifs. This enhancement could be created by Zyxin wrapping around the EVH1 domain in analogy to the structures of the homologous WASP-EVH1/WIP30,31 and RanBP2-RanBD1/Ran32 complexes. Analysis of nuclear magnetic resonance (NMR) titrations and of an EVH1–Zyxin chimera reveals a predominant orientation of the interaction, where the second FPPPP motif of Zyxin binds to the canonical binding site and a downstream region binds to the novel secondary binding site. This orientation was not observed when titrations were performed using the phosphomimetic EVH1 mutant Y39E, which demonstrates weakened or abolished binding at the secondary site. The secondary binding site influences the overall stoichiometry of the interaction between Zyxin and VASP, which provides insights regarding the effect of Y39 phosphorylation on the cellular localization of VASP.
MATERIALS AND METHODS
Protein Purification
EVH1 protein was expressed from a pMW172 vector with the human VASP EVH1 domain encoded (gift from M. Way, London Research Institute, London, U.K.) to produce a cleaved product consisting of EVH1 residues 2–115 with the non-native sequence GPGGRMS at the N-terminus. All proteins were expressed as previously described.33 The Y39E EVH1 mutants were produced by using site-directed mutagenesis. A pDEST17 plasmid (ampicillin resistance) encoding human Zyxin residues 41–140, with an N-terminal six-His fusion and a TEV cleavage site, was produced using the GATEWAY cloning system (Thermo Fisher Scientific). The Zyxin41–140 sequence was amplified from a RFP-Zyxin expression vector purchased from Addgene (Addgene plasmid 2672034). All were purified using native preparations as described previously33 and dialyzed into NMR buffer [20 mM KHPO4 and 50 mM KCl (pH 6.7)] following cleavage and removal of the His tag. NMR samples also contained 5 mM NaN3, 0.05% protease inhibitor cocktail (EDTA-free, Sigma), and 7% D2O. Unlabeled Zyxin41–140 protein used in the [15N]EVH1 titrations was concentrated to a volume of 600 μL, and a sample of 300 μL with 10 mM DSS was subjected to a one-dimensional NMR experiment to determine by comparison to DSS the stock solution concentration. NMR samples were concentrated with 3000 MWC centrifugal concentrators. The protein concentration was determined by UV absorption at 280 nm, using the theoretical extinction coefficients of 21000 cm−1 M−1 for VASP EVH1 and 19500 cm−1 M−1 for VASP EVH1 Y39E.
The EVH1 fusion protein with Zyxin was obtained by enzymatic digestion of the pMW172-EVH1 vector with EcoRI and BLP1. Polymerase chain reaction amplification from pDEST17-Zyxin41–140 was performed to obtain the SSGSG-GGG linker followed by Zyxin82–124. After enzyme digestion, ligation to pMW172-EVH1 was performed using the ThermoScientific rapid ligation kit. Expression and purification were performed in the same way as they were for [15N]EVH1 but using media enriched with [13C]glucose.
Peptide Production
Synthetic peptides encompassing the individual motif in Zyxin, Zyxin69–80, Zyxin91–102, Zyxin102–113, and Zyxin112–123 were purchased from Genscript (Piscataway, NJ). Peptides were dissolved in NMR buffer, and their pHs were adjusted using NaOH. The peptide concentration was determined using the theoretical extinction coefficient of 195 cm−1 M−1 at 257 nm.
NMR Data Collection and Processing
NMR experiments were performed at 25 °C using a Varian Inova 600 MHz spectrometer with a (H, C, N) Z-axis gradient probe. Spectra were processed with nmrPipe and analyzed with Sparky. Prior to Fourier transformation, [15N]EVH1 data were processed with an exponential window function. Peak positions and volumes were measured using the Sparky peak detection function.
Resonance Assignments
Resonance assignments for the EVH1 domain were obtained from the BioMagResBank, accession number 18569,18 and incongruent assignments or residues near the N-terminus were clarified by three-dimensional (3D) TOCSY and NOESY spectra of [15N]EVH1. Resonance assignments for the Y39E EVH1 mutant, and EVH1-Zyxin82–124, were obtained by 3D TOCSY and NOESY.
NMR Titration Experiments
Five [15N]EVH1-perspective HSQC and five [15N]Y39E EVH1-perspective HSQC titration experiments were performed. In each experiment, the sample with the highest concentration of Zyxin construct was produced first, and subsequent samples were produced by mixing part of the previous sample with a part of a stock of [15N]EVH1 or [15N]Y39E EVH1. The experiments were titrations of the [15N]EVH1 domain with Zyxin41–140, Zyxin69–80, Zyxin91–102, Zyxin102–113, and Zyxin112–123. In the Zyxin41–140 titration, the [15N]EVH1 concentration was held to 0.21 mM and the Zyxin41–140 concentrations were 0, 0.02, 0.03, 0.06, 0.13, 0.26, 0.52, and 1.03 mM. In the individual motif peptide titrations, the [15N]EVH1 concentration was held at 0.18 mM for all of them. Concentrations of Zyxin69–80 were 0, 0.15, 0.29, 0.59, 1.18, 2.35, 4.71, and 9.41 mM. In the Zyxin91–102 titration, the peptide concentrations were 0, 0.08, 0.17, 0.34, 0.67, 1.34, 2.68, and 5.36 mM. For Zyxin102–113, the concentrations were 0, 0.14, 0.28, 0.56, 1.11, 2.22, and 4.45 mM, and for Zyxin112–123, they were 0, 0.25, 0.51, 1.02, 2.03, and 4.07 mM. The experiments for the phosphomimetic mutant were titration of [15N]Y39E EVH1 with Zyxin41–140, in which the [15N]Y39E EVH1 concentration was held to 0.23 mM and the Zyxin41–140 concentrations were 0, 0.01, 0.03, 0.06, 0.12, 0.24, 0.49, and 0.97 mM. For the individual motif titrations, the [15N]Y39E EVH1 concentration was held at 0.18 mM and concentrations of Zyxin69–80 were 0, 0.04, 0.08, 0.15, 0.31, 0.61, 1.23, 2.46, and 4.92 mM. The concentrations of Zyxin91–102 were 0, 0.10, 0.20, 0.40, 0.80, 1.60, 3.19, and 6.38 mM; those of Zyxin102–113 were 0, 0.04, 0.08, 0.16, 0.32, 0.64, 1.28, 2.55, and 5.11 mM, and those of Zyxin112–123 were 0, 0.09, 0.19, 0.37, 0.74, 1.49, 2.97, and 5.95 mM.
The [15N]EVH1 and [15N]Y39E EVH1 spectra were recorded with spectral widths of 8 kHz in the proton dimension (2048 complex data points) and 1.8 kHz in the nitrogen dimension (256 complex data points).
Quantification of Motif Binding in the Context of Zyxin41–140
TITAN offers a wide range of binding models that can be used for two-dimensional line shape analysis.35 For [15N]Y39E EVH1 titrated with individual motif peptides, we used the simple two-state interaction available in TITAN and simultaneously fit 10 peak profiles. Additionally, TITAN offers the flexibility of including personalized binding models. For the [15N]EVH1 wild type titrated with individual motif peptides, we used the four-state binding model described in detail elsewhere36 and fit 10 peak profiles in the primary binding site and six in the secondary binding site. Peaks included in these analyses were selected on the basis of trajectories that displayed maximum Δδ values that exceeded the corresponding error in Δδ by a factor of at least 10. The error in Δδ was determined on the basis of peaks that were insensitive to ligand addition, where the average standard deviations of positions of these “invariant” peaks in each of the proton and nitrogen dimensions were taken as the chemical shift uncertainty for all peaks, and standard error propagation was applied to obtain the corresponding error in Δδ. TITAN also has the option for a model with flexible stoichiometry, which we used to analyze the titration of [15N]EVH1 (WT/Y39E) titrated with Zyxin41–140. An error analysis was performed using 200 repeats to determine the uncertainties of the fitting.
To approximate the distribution of bound motif populations at the primary site for the [15N]EVH1 and [15N]Y39E EVH1 cases at the highest Zyxin41–140 concentration, an analysis of the observed chemical shift relative to the bound chemical shifts for each individual motif peptide was performed. An approximating assumption is that chemical shift perturbations in EVH1 due to binding of a given Zyxin motif are the same in the context of Zyxin41–140 versus the individual motif peptide. For this analysis, the observed chemical shifts (Δδobsmax i) for 10 residues (separately analyzed in nitrogen and proton dimensions), for which the peak trajectory directions of individual motif titrations differed the most, were used to solve for the bound motif populations using the following equation:
where P1–P4 represent the populations of the EVH1 domain bound to motifs M1P–M4P, respectively, P5 is the population of free EVH1, ΔδboundMji represents the fitted chemical shift of the bound state for given residue i determined for titrations with individual motif peptides (j = 1–4), and Δδapo,i is the chemical shift of residue i in the primary site of free EVH1. With the additional constraint that the sum of Pi terms equals 1, Matlab was used to repeatedly solve the system of equations for eight residue-specific data sets selected randomly from a set of 20 data sets corresponding to the N and HN dimensions of 10 primary binding site residues. The average and standard deviation were obtained from 500 such runs. The same process was performed to approximate the bound motif populations in the most Zyxin41–140 -saturated [15N]Y39E EVH1 sample.
RESULTS
The Zyxin Proline Rich Region Induces Chemical Shift Changes at Expected and Unexpected Surfaces on VASP EVH1
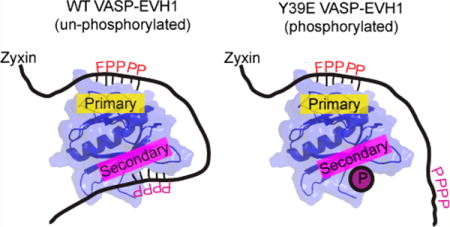
The proline rich region of Zyxin that harbors its four VASP EVH1 binding motifs [Zyxin residues 41–140, Zyxin41–140 (Figure 1A)] was expressed and purified, and its interaction with VASP EVH1 was investigated using NMR spectroscopy. As a first step, the [15N]EVH1 domain was titrated with unlabeled Zyxin41–140 (with the His tag cleaved) and a series of HSQC spectra were recorded. As expected, chemical shift perturbations were observed for residues previously characterized as being important for (W/F)PxφP ligand binding defined by the crystal structure of the Mena EVH1 domain/ActA complex and previous NMR studies of EVH1 titrated with (W/F)PxφP ligands.18,24,33 For example, residues W23, Y72, Q74, A75, W82, and N92 displayed large ligand-induced chemical shift changes (Figure 1B,C). Remarkably, chemical shift perturbations were also observed for residues remote from this well-characterized binding site. Namely, residues Ser2, Val5, Ile6, Arg48, Val50, and Ala65 showed perturbations similar in magnitude to those of residues in the canonical binding site (Figure 1B,D). Mapping these chemical shift changes onto the EVH1 structure shows localization of these noncanonical binding residues on the opposite side of the domain (Figure 1E), suggesting the presence of a secondary binding site. The possibility of slight pH changes being responsible for this observation was ruled out by pH titration (Figure S1). This region has previously been implicated in function, as mutation of the nearby Y39 altered the co-localization of and interaction between VASP and Zyxin.23
Figure 1.
Zyxin41–140 titration provides evidence of a secondary binding site in the EVH1 domain of VASP. (A) Amino acid sequence of the Zyxin41–140 construct, highlighting the four VASP binding motifs (red, bold type). (B) Composite chemical shift changes in [15N]EVH1 induced by addition of Zyxin41–140, calculated using the equation . Extracted regions of overlays of 15N–1H HSQC spectra of [15N]EVH1 titrated with Zyxin41–140 show trajectories for residues in (C) the canonical binding site and (D) a novel secondary site, all of which show maximum Δδ values in excess of 10 times the error in Δδ (determined to be 0.005). (E) Chemical shift changes mapped onto the structure of VASP EVH1 (Protein Data Bank entry 1EGX) show that Zyxin41–140 induces chemical shift perturbations for residues in the primary binding site (far right) but also for residues on the opposite face of the EVH1 domain (far left). Red denotes the largest chemical shift perturbation and purple no perturbation.
Quantitative Analysis of the Interaction of [15N]EVH1 with the Zyxin Proline Rich Region Reveals a Bivalent Binding Mechanism
The detection of two binding sites on EVH1 that potentially interact with the multiple binding motifs on Zyxin41–140 significantly complicates the quantitative interpretation of the Zyxin41–140 titration data. To begin to decipher this interaction, four 12-residue synthetic peptides (M1P, Zyxin69–80; M2P, Zyxin91–102; M3P, Zyxin102–113; M4P, Zyxin112–123) were used in titrations of wild-type [15N]EVH1 to determine the intrinsic affinities of each motif under the same sample conditions and to determine the chemical shifts of residues in the EVH1 primary site when bound to each of the four Zyxin motifs. As expected, chemical shift perturbations at the primary binding site were observed. Additionally, chemical shift perturbations at the novel secondary binding site were induced by titration with all four peptides. Therefore, the binding affinity was determined using a four-state model (free [15N]EVH1, only primary site bound, only secondary site bound, and both primary and secondary sites bound) as previously described.36 For all four peptides, this analysis yielded relatively weak affinities for the primary binding site and considerably weaker affinities for the secondary site (Table 1). These affinities are mostly weaker than the previously reported values for the same M1P–M4P peptides (74 μM, 61 μM, 197 μM, and no binding observed, respectively) determined by Trp fluorescence spectroscopy and fitted to a two-state model (free and 1:1 bound).18 It is possible that the affinity differences might be due to the lower pH (6.0) used in the Trp fluorescence measurements.18 Importantly, the NMR titration analysis yields motif-specific bound-state chemical shift values for residues in the primary binding site that are critical for characterizing the complex interaction between [15N]EVH1 and Zyxin41–140.
Table 1.
Dissociation Constants Calculated for EVH1 Primary and Secondary Binding Sites Titrated with Different Zyxin Peptides
| (μM) | (mM) |
|
||
|---|---|---|---|---|
| M1P | 237 ± 2 | 4.5 ± 0.1 | 0.82 | |
| M2P | 134 ± 2 | 4.4 ± 0.2 | 0.96 | |
| M3P | 235 ± 3 | 9.8 ± 0.3 | 1.50 | |
| M4P | 263 ± 3 | 2.1 ± 0.1 | 1.26 |
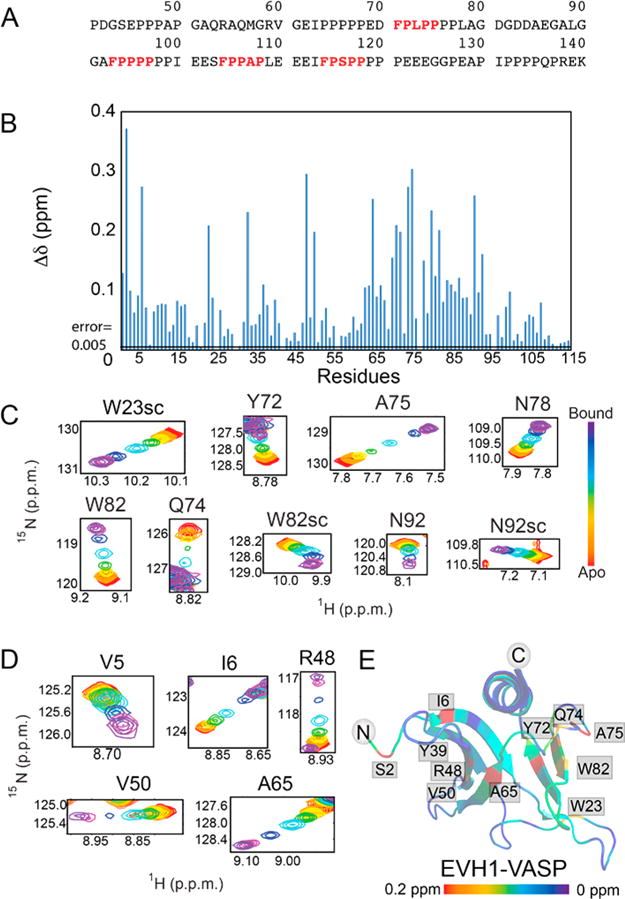
The amide proton and nitrogen chemical shifts of a given amino acid residue in a system at equilibrium are a useful readout that can be used to determine the population distribution over multiple states for which distinct chemical shift values are known. With multiple residues serving as observers of the same set of states, complex equilibria can be characterized if the chemical shifts of the different states sampled by a given residue are sufficiently distinct. For the case at hand, several residues in the [15N]EVH1 primary binding site show distinct peak trajectories upon being titrated with M1P–M4P (Figure 2A), enabling populations of the bound Zyxin motifs in the context of Zyxin41–140 to be approximated by assuming the observed [15N]EVH1–Zyxin41–140 chemical shift is a population-weighted average of the apo and individual peptide-bound chemical shifts. This analysis shows a strong preference for binding of M2 in Zyxin41–140 to the primary site [∼6.4, 73.4, 9.3, 6.5, and 4.3% occupancy for motifs M1–M4 and apo, respectively (Figure 2B)]. For comparison, Virtual Cell Modeling (VCell) software37 was used to simulate the expected population distribution for the most saturated [15N]EVH1–Zyxin41–140 sample condition if the four individual motifs in Zyxin were fully accessible and had the same intrinsic affinities as the corresponding short peptides (i.e., a 4:1 EVH1–Zyxin model). These simulations predict a significantly weaker preference of M2 binding [21, 35, 21, 19, and 4% occupancy for motifs M1–M4 and apo, respectively (Figure 2B)], indicating a significant impact of linking the four motifs together. Even if the simultaneous binding of EVH1 to nearest neighbor motifs is prohibited (i.e., all states with M1 and M2, M2 and M3, or M3 and M4 simultaneously bound are eliminated from the VCell model), the results are very similar (21, 35, 20, 19, and 5% occupancy for motifs M1–M4 and apo, respectively).
Figure 2.
Secondary binding site that creates a larger population of motif 2 bound to the canonical binding site. (A) Titrations show distinct chemical shift peak trajectories upon addition of each motif peptide (yellow, M1P; red, M2P; green, M3P; blue, M4P). The magenta peak shows titration with Zyxin41–140 at the highest Zyxin concentration. Purple shows the chemical shift of chimeric EVH1-Zyxin. (B) Distribution of Zyxin41–140 motifs bound at the canonical site of EVH1 deduced from chemical shifts (filled circles) compared with the expected distribution based on Kd values of individual peptides (stars). (C) Binding curves from average normalized chemical shift changes of 10 residues in the canonical binding site of EVH1 for individual peptide titrations and for Zyxin41–140. The yellow binding curve indicates titration with M1P, the red binding curve titration of EVH1 with M2P, the green binding curve titration of EVH1 with M3P, the blue binding curve titration of EVH1 with M4P, and the purple binding curve titration of EVH1 with Zyxin41–140. (D) Like panel C, for six residues at the novel secondary binding site of EVH1. (E) Binding curve from average normalized chemical shift changes for the primary binding site (blue dots) and for the secondary binding site (red dots) and a global fit to both curves (Kd = 32.2 μM). (F) Chimera of EVH1 and Zyxin (EVH1–Zyxin82–124) containing M2 (red), M3 (green), and M4 (blue) motifs. (G) Overlays of 15N–1H HSQC spectra for residues in the noncanonical site, titrated with M2P (red), M3P (green), M4P (green), or Zyxin41–140 (magenta) or in the context of EVH1–Zyxin82–124 (purple).
Taken together, the analyses described above demonstrate that Zyxin41–140 binds preferentially via M2 to the primary site, and that this binding mode is stabilized by interaction of another motif on the same Zyxin molecule with the secondary site to generate a bivalent bound state. This hypothesis is supported by an apparent enhancement of the primary site molar affinity compared to that seen with individual peptide titrations (Figure 2C), and an even more pronounced affinity enhancement for the secondary binding site (Figure 2D). Bivalent additivity has been widely studied36,38,39 and, in this case, could significantly stabilize the 1:1 interaction between Zyxin and the VASP EVH1 domain.
Quantitative interpretation of the binding curves obtained from titration of [15N]EVH1 with Zyxin41–140 is approached by considering a highly simplified two-state model, in which a single Zyxin molecule interacts with a single EVH1 molecule, and this interaction involves both primary and secondary sites in one unique Zyxin orientation. In this model, the apparent affinity enhancement of Zyxin41–140 relative to those of individual motif peptides would be due to the combined binding energies of both primary and secondary surfaces. Fitting of the binding data for six residues in the primary and six residues in the secondary binding surface to this model yields an apparent Kd value of 32.2 ± 1.7 μM (Figure 2E). The goodness of fit, evaluated by a χ2 analysis as described in detail elsewhere,36 yielded a reduced χ2 value of 4.19. This χ2 value is consistent with the two-state bivalent model being an oversimplification, as expected because the population distribution determined above indicates that multiple bound species are sampled. Separate fitting of just the primary or secondary site residues to the simple two-state model yields apparent Kd values of 21.8 ± 1.3 and 133.3 ± 5.8 μM for the primary and secondary sites, respectively, with corresponding χ2 values of 0.25 and 1.55 indicating good fits. The significant increases in affinity of both independently fitted primary and secondary site Kd values relative to the individual motif peptides, together with the overall affinity enhancement reflected in the highly simplified two-state bivalent fitting, are consistent with a more complex multistate model in which a dominantly populated bivalent state contributes to a significant enhancement of affinity. Additionally, the population distribution deduced from chemical shift perturbations supports a bivalently bound state with M2 bound to the primary site and a downstream epitope bound to the secondary site.
EVH1–Zyxin82–124 Fusion Protein Captures the Bivalently Bound State
To confirm that both primary and secondary sites are dominantly bound in a 1:1 stoichiometry in a bivalent manner, a fusion protein was constructed in which the C-terminus of EVH1 is fused to the proline rich region of Zyxin containing motifs M2–M4 [EVH1–Zyxin82–124 (Figure 2F)]. Importantly, no significant changes in [15N]EVH1 peak line widths were observed relative to WT EVH1, indicating the chimera is a monomer and not a domain-swapped dimer. Moreover, no NOESY crosspeaks were detected between the Zyxin and EVH1 components of the chimera, consistent with highly dynamic interfaces as expected from the weak intrinsic affinities characterized above. Resonance assignments for the EVH1 domain segment of the chimera were confirmed by standard triple-resonance NMR experiments. The 15N–1H HSQC spectrum of the 15N- and 13C-labeled EVH1–Zyxin82–124 chimera shows peak positions in both primary and secondary binding sites that are consistent with a more saturated state of the same binding trajectory as observed during titration with Zyxin41–140 (Figure 2A,G). This coincidence of the forced 1:1 stoichiometry in the chimera and the titration with Zyxin41–140 further justifies the bivalent model employed above for quantitative analysis of the Zyxin41–140 titration data. While the primary site shows a clear preference for M2 based on chemical shifts, it is not possible to distinguish a motif preference at the secondary site from the [15N]EVH1 perspective due to the lack of distinct chemical shift trajectories and the weak affinities of the individual motif peptides that prevented observation of larger populations of bound states (Figure 2G).
The Phosphomimetic EVH1 Y39E Mutation Induces Changes in the Remote Primary Binding Site and Abolishes Binding to the Secondary Site
It has been reported that phosphorylation of Y39 by Abl reduced the level of VASP accumulation in focal adhesions, and that mutation of Y39 to aspartic acid reduced the level of binding of VASP to Zyxin and altered cellular localization.23 Because Y39 is located within the secondary binding site described above (Figure 1E), we wondered whether placement of a negative charge at the Y39 position would inhibit binding at the EVH1 secondary site, thereby eliminating bivalent binding. Bacterial expression of EVH1 Y39D was poor (unpublished data), while EVH1 Y39E was robustly expressed and stable in aqueous solution. Therefore, EVH1 Y39E was used for phosphomimetic investigations, and backbone resonance assignments for this mutant (BioMagResBank entry 27193) were determined using a standard suite of 3D NMR experiments.40
To investigate whether the Y39E mutation alters the interaction of EVH1 with the Zyxin proline rich region, we first performed titrations with each of the four individual Zyxin motif peptides. Notably, significant chemical shift perturbations were observed for only residues of [15N]EVH1 Y39E in the primary binding site and not in the secondary site, indicating that indeed placement of a negative charge at residue 39 inhibits binding at the secondary site. Primary site binding affinities for each peptide, determined using a simple two-state binding model, are again relatively weak as was found in wild-type EVH1 (Table 2).
Table 2.
Dissociation Constants for the Primary Binding Site in Y39E EVH1
| Kdprimary (μM) | Xred2 | |
|---|---|---|
| M1P | 176 ± 2 | 0.91 |
| M2P | 126 ± 2 | 0.99 |
| M3P | 182 ± 2 | 0.84 |
| M4P | 240 ± 1 | 0.54 |
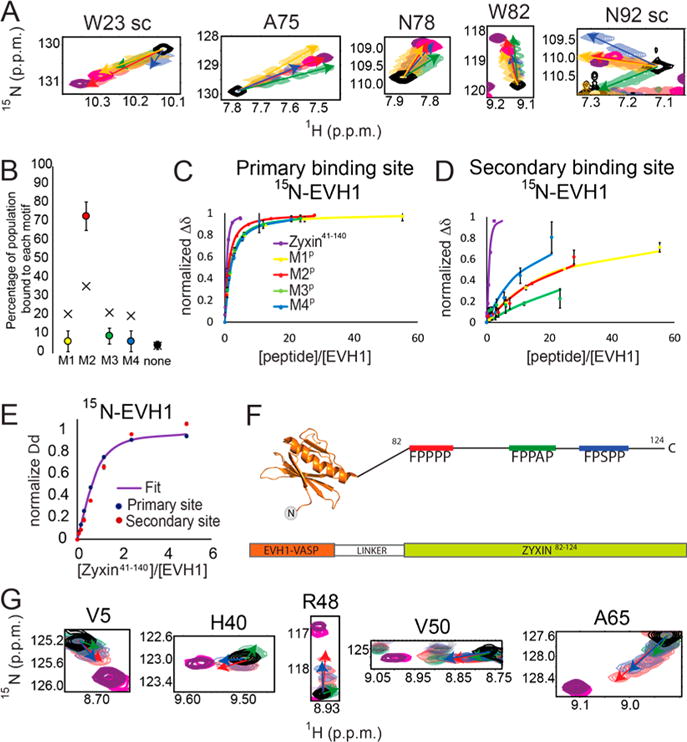
Next, Zyxin41–140 was titrated into [15N]EVH1 Y39E. While significant chemical shift changes in the primary binding site were observed, only minimal chemical shift perturbations were observed for residues at the secondary binding site, indicating a loss of the bivalent bound state (Figure 3). Consistent with this, the resulting binding curves for residues in the primary site could not be adequately fit to a simple two-state (1:1 stoichiometry) model (reduced χ2 value of 36.47), suggesting two or more EVH1 domains may bind to a single Zyxin molecule if a dominant 1:1 state is not achievable.
Figure 3.
Phosphomimetic mutation abolishes secondary site binding. (A) Bar plot showing differences between composite chemical shift perturbations of the 1H and 15N resonances upon addition of Zyxin41–140 to EVH1 WT and EVH1 Y39E. Those residues with the largest perturbation differences are located in the novel secondary binding site. Overlays of 15N–1H HSQC spectra for residues in (B) the canonical binding site and (C) the secondary site show binding for the primary but not secondary sites. (D) Mapping chemical shift changes induced by titration of [15N]EVH1 Y39E with Zyxin41–140 onto the structure of the VASP EVH1 domain (Protein Data Bank entry 1EGX) shows large perturbations for residues in the primary binding site (far right) but almost no perturbations for residues on the opposite face of the EVH1 Y39E domain (far left). Red indicates the largest chemical shift perturbation and purple no perturbation.
A key difference between the titrations of [15N]EVH1 WT and [15N]EVH1 Y39E is that the chemical shift trajectories differ (Figure 3B compared to Figure 1C). This difference can be explained by an altered population distribution of bound motifs. As was observed for WT EVH1, several residues in the [15N]EVH1 Y39E primary binding site show distinct peak trajectories upon being titrated with M1P–M4P, again enabling populations of the bound Zyxin motifs in the context of [15N]EVH1 Y39E with the most concentrated Zyxin41–140 condition to be approximated (Figure 4A). Strikingly, the populations were 16, 44, 20, 14, and 6% occupancy for motifs M1–M4 and apo, respectively, far more similar to the VCell-simulated population distribution for EVH1 Y39E based on individual motif peptide Kd values (simulation values of 22, 32, 23, 18, and 5% occupied by motifs M1–M4 and apo, respectively). Moreover, chemical shift perturbations induced by titration with Zyxin41–140 indicate binding at the primary but not at the secondary binding sites (Figures 3B–D and 4B,C). Together, these results demonstrate that the Y39E phosphomimetic mutation effectively abolishes the bivalent binding mode and enables the four motifs in Zyxin to interact with the primary binding site of EVH1 in a more equal population distribution. These results suggest that Y39 phosphorylation might act as a binding mode switch, where EVH1 domains with unphosphorylated Y39 bind to Zyxin predominantly in a bivalent 1:1 stoichiometry, and upon phosphorylation of tyrosine, up to four EVH1 domains could bind to one molecule of Zyxin.
Figure 4.
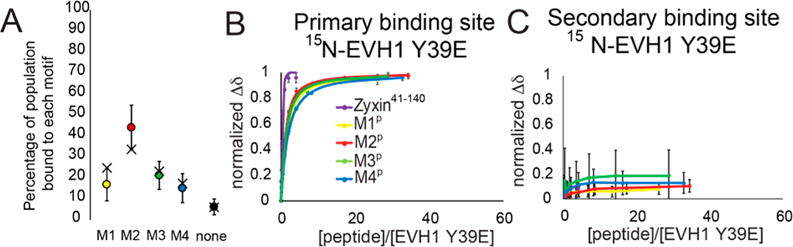
Population and binding curves for [15N]EVH1 Y39E. (A) Distribution of Zyxin41–140 motifs bound at the canonical site of EVH1 Y39E deduced from chemical shifts (filled circles) compared with the expected distribution based on Kd values of individual peptides (black crosses). (B) Binding curves from average normalized chemical shift changes of 10 residues in the canonical binding site of EVH1 Y39E for individual peptide titrations and for Zyxin41–140. The yellow binding curve indicates titration with M1P, the red binding curve titration of EVH1 Y39E with M2P, the green binding curve titration of EVH1 Y39E with M3P, the blue binding curve titration of EVH1 Y39E with M4P, and the purple binding curve titration of EVH1 Y39E with Zyxin41–140. (C) Like panel B, for six residues at the novel secondary binding site of EVH1 Y39E.
To investigate potential phosphorylation-induced allostery, we focused our attention on the backbone chemical shift assignments of the phosphomimetic EVH1 Y39E mutant. The peaks corresponding to N-terminal residues S2–C7 move in response to the Y39E phosphomimetic mutation, consistent with the structural interaction between Y39 and this region revealed in the NMR structure of human VASP EVH118 (1EGX and its homologues). Other residues in sequential or spatial proximity to Y39 also display new peak positions, such as H40, F47, and V49 (Figure 5), as expected. However, there are several residues more than 14 Å from the mutation site that display altered backbone and/or side chain NH chemical shifts. Striking examples include residues that reside in the primary binding site of EVH1, such as W82, A85, and W89 (Figure 5A). Mapping of the backbone NH chemical shift differences between WT and Y39E EVH1 onto the structure reveals a clear perturbation pathway from the mutation site to these residues (Figure 5B). These long-range perturbations of chemical environment induced by the phosphomimetic Y39E mutation suggest that phosphorylation of Tyr39 could similarly trigger a conformational rearrangement that alters the primary binding site of EVH1.
Figure 5.
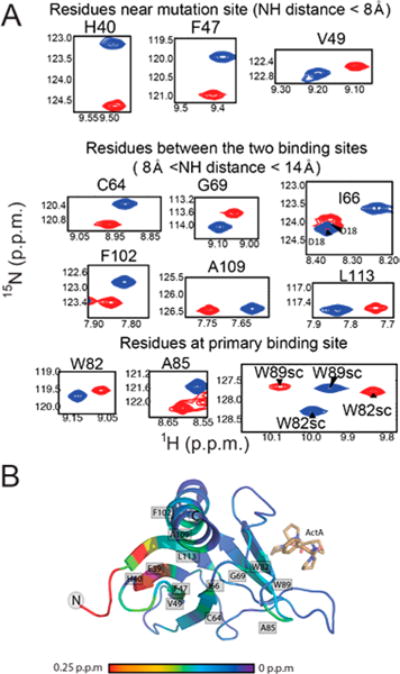
1H–15N chemical shift differences between wild-type EVH1 and mutant Y39E. (A) Chemical shift differences between wild-type EVH1 (blue spectra) and EVH1 Y39E (red spectra). Both spectra were recorded under identical buffer conditions and with the same parameters. All highlighted residues display differences in chemical shift that exceed the error in chemical shift value by a factor of at least 10. (B) Mapping of 1H–15N backbone chemical shift differences to the EVH1 structure. Significant changes (>0.25 ppm) are represented by red, and no change is shown as purple.
DISCUSSION
Understanding protein–protein interaction mechanisms that fine-tune signaling pathways is an important endeavor in biology. Here we studied the interaction between the EVH1 domain of VASP and the proline rich region of Zyxin. This interaction plays a critical role in the localization of VASP in focal adhesions.41 Interestingly, VASP mislocalization is achieved only when all four FPxφP motifs of Zyxin are compromised,42 showing that indeed these motifs mediate this interaction. VASP differs from MENA and EVL in its regulation by phosphorylation at Y39.23 Here, through application of NMR, we found a secondary binding site near Y39 in the VASP EVH1 domain that, along with the canonical EVH1 binding site, contributes to the interaction with the proline rich region of Zyxin. This secondary binding site is abolished by phosphomimetic mutation of Tyr39 to Glu. These studies suggest that Y39 phosphorylation acts as a stoichiometry switch, with one molecule of Zyxin binding to one EVH1 domain in the unphosphorylated state, and one molecule of Zyxin potentially binding to multiple EVH1 domains in the phosphorylated state.
Support for the biological relevance of the secondary binding interaction described here comes from the reported impact of phosphorylation of VASP EVH1 by Abl tyrosine kinase.23 In Bcr-Abl-positive leukemic cells, Abl phosphorylates Y39 of VASP, which is centrally located in the binding site we describe (Figure 1E). This may regulate the interaction between VASP and Zyxin, because the phosphomimetic mutant Y39D has a weakened ability to bind Zyxin in pull downs. Furthermore, overexpression of the Y39D or Y39F mutant, but not WT VASP, impairs adhesion of K562 cells to fibronectin, implying that a cycle of phosphorylation and dephosphorization is important for the proper regulation of VASP.23 Here, we provide evidence that the Y39E mutant of VASP EVH1 retains WT binding affinity for its primary site but is incapable of binding in the secondary site. Importantly, we show that this phosphomimetic mutation switches the stoichiometry of the interaction. Taken together, these data suggest that the secondary binding site could play a key role in the phosphorylation and dephosphorization cycle of VASP to regulate interactions with Zyxin and other proline rich binding partners.
EVH1 domains all share a conserved primary binding site that binds proline rich sequences adopting a polyproline type II (PPII) structure. The canonical (primary) binding site is a concave surface formed by one of the two β-sheets that form the domain’s β-sandwich fold (Figure 6, right). The secondary binding site we define here is a similar concave surface formed by the other β-sheet on the opposite side of the domain. The residues most sensitive to the secondary interaction include the very N-terminal sequence of EVH1 (M0-C7) as well as the side chains of R48, and N63, which are all near each other (Figure 6, right). Although interactions beyond the primary site have been reported,18,30 these additional EVH1 sites are highly distinct from the secondary binding surface described here.
Figure 6.
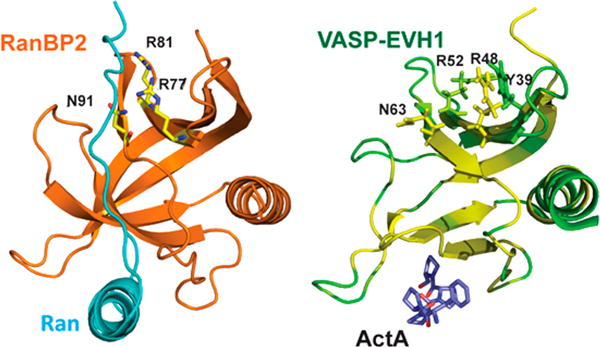
Tyr39 is located centrally in the secondary EVH1 binding site. In the left panel, the first Ran binding domain of RanBP2 (orange) binds Ran (cyan) using a secondary binding surface similar to the site we describe, employing residues R77, R81, and N91 that correspond to R48, R52, and N63, respectively, in VASP EVH1 (Protein Data Bank entry 1RRP). In the right panel, VASP EVH1 residue Y39 is located centrally in the secondary binding site. EVH1 is colored according to chemical shift perturbations induced by Zyxin41–140 (green, no perturbation; yellow, largest perturbation). Key residues are shown as sticks. The ActA sequence FPPPP (yellow) is shown bound in the primary site.
EVH1 domains are homologous in structure and sequence with RanBDs.43 The crystal structure of the first RanBD (denoted RanBD1) in RanBP2 bound to the nuclear transport protein Ran demonstrates that key features of the secondary binding site in VASP EVH1 are conserved in RanBDs.32 In this structure, a segment of Ran binds to the face of RanBD1 that corresponds to the secondary binding site we have described (Figure 6, left). The side chains of RanBP2 residues R77, R81, and N91 (corresponding to VASP EVH1 residues R48, R52, and N63, respectively) hydrogen bond with the backbone carbonyl groups of residues M179, A181, and L182 in Ran that are part of a PPII segment. Alignment of EVH1 and Ran binding domains shows that R48 and N63 are highly conserved,43 suggesting even broader utilization of this secondary binding site (e.g., by EVH1 domains in WASP and Homer). Importantly, the binding of a PPII structure to the corresponding “secondary site” in RanBD1 and the conservation of key interaction residues in EVH1 domains suggest that the EVH1 secondary site identified here might bind to sequences that adopt the PPII structure without the requirement of the W/FP motif.
VASP EVH1 has several binding partners that contain multiple proline rich motifs, some of which lack the canonical W/FPxφP primary site recognition motif. For instance, the Lipoma preferred partner (LPP), which is implicated in cell shape and motility, has two proline rich motifs (66DFLPPPP-PPLD76 and 87NFPPPPPLD95, only one of which has the canonical motif) separated by 12 residues, and direct binding of VASP EVH1 was previously reported.44 Similarly, Vinculin, a known EVH1 binding partner,45 has only one FPxφP motif (842FPPPPP847) but has another proline rich motif nearby (859APPKPPLP866). Even more surprising is Migfilin, a component in cell-matrix adhesions that offers a link with the actin cytoskeleton.46 Migfilin regulates localization of VASP to the cell-matrix adhesions via its proline rich region that interacts directly with the VASP EVH1 domain. Deletion of residues 84–112 in Migfillin eliminates VASP binding.47 Interestingly, this deletion contains two proline rich motifs without W or F (85CPPPPP90 and 104LPPPPPPPP112) separated by 14 residues. All of these proline rich motifs are expected to adopt PPII structure, which potentially could bind to the secondary binding site. Notably, the presence of neighboring proline rich motifs could enhance dramatically the affinity through bivalent binding as observed here for Zyxin.
It is well established that VASP functions in the cell as a constitutive tetramer.48 Four VASP polypeptide chains associate via the single-helix tetramerization domain to form a parallel right-handed four-helix coiled coil, causing VASP to form a “bouquet-like” tetramer.49 Truncation of the C-terminal region of VASP that contains the tetramerization domain causes a decrease in the VASP concentration at focal adhesions, indicating a tetramerization dependence for focal adhesion localization.27 Moreover, the possibility of VASP cross-linking several protein partners has been previously discussed.25 Notably, the work presented here suggests that when VASP residue Y39 is not phosphorylated, the bivalent interaction between one EVH1 domain and one multivalent protein ligand could allow a VASP tetramer to tightly localize as many as four different binding partners, effectively achieving reversible cross-linking of these partners (Figure 7A). Moreover, when VASP is phosphorylated at Y39, the resulting loss of secondary site binding would promote interaction of the VASP tetramer with a single multivalent protein partner due to the enhancement of avidity (Figure 7B). We propose a model in which Y39 phosphorylation acts as a molecular switch that governs the stoichiometry of interactions between the VASP tetramer and its binding partners that harbor multiple FPxφP motifs, where the unphosphorylated state acts as a cross-linker between multiple partners while the phosphorylated state binds to a single partner (Figure 7).
Figure 7.
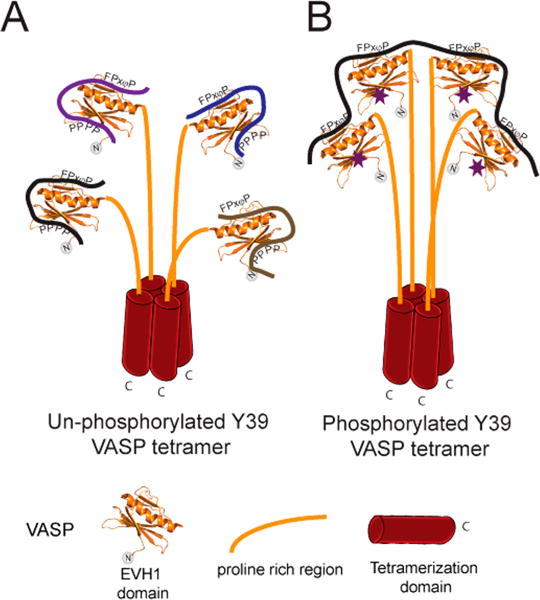
Proposed model for the stoichiometry switch controlled by phosphorylation of Y39 in VASP tetramers. (A) In the extreme case where all Y39 residues are unphosphorylated, each EVH1 domain in the VASP tetramer could interact with a molecule of Zyxin or any other binding partner. Bivalent interactions could form due to the presence of nearby proline rich motifs. (B) In the extreme case where each Y39 residue is phosphorylated (purple stars), one molecule with multiple FPxφP motifs (like Zyxin, ActA, or lammellipodin) could bind to one tetramer of VASP. Phosphorylation of Y39 could recruit VASP exclusively to a specific binding partner with multiple canonical binding sites, while dephosphorylation could cause cross-linking between multiple binding partners.
In summary, a secondary binding site in the VASP EVH1 domain was identified and characterized through NMR investigations of the interactions between the proline rich region of Zyxin and [15N]EVH1. By analysis of population distributions over bound states, we determined that this secondary binding site directs the orientation of Zyxin binding through formation of a bivalent interaction between Zyxin and EVH1. In this bivalently bound state, M2 in Zyxin interacts with the primary site and downstream Zyxin regions interact with the secondary site in EVH1. Additionally, we observed that the secondary binding site is abolished by phosphomimetic mutation of Y39 to E. Phosphorylation dependence at Y39 for the interaction between VASP and Zyxin has been previously reported.23 On the basis of our findings, we hypothesize that phosphorylation of Y39 acts as a stoichiometry switch in the interaction between the VASP tetramer and Zyxin. These results have important implications for the regulation of VASP interactions with its other binding partners that contain multiple neighboring proline rich motifs, and more broadly for potential regulation of interactions involving other EVH1 domains in which residues implicated in secondary site function are conserved.
Supplementary Material
Acknowledgments
Funding
This investigation was supported by the National Science Foundation (MCB-1157806 and MCB-1615350) and through graduate student training grant support to L.A.A. by the National Institutes of Health (2T32GM008267).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.7b00618.
Comparison of 15N–1H HSQC chemical shift perturbations of [15N]EVH1 residue H40 induced by pH titration versus by Zyxin41–140 titration (PDF)
ORCID
Linda K. Nicholson: 0000-0002-2701-1530
Notes
Theauthors declare no competing financial interest.
References
- 1.Breitsprecher D, Kiesewetter AK, Linkner J, Urbanke C, Resch GP, Small JV, Faix J. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 2008;27:2943–2954. doi: 10.1038/emboj.2008.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breitsprecher D, Kiesewetter AK, Linkner J, Vinzenz M, Stradal TE, Small JV, Curth U, Dickinson RB, Faix J. Molecular mechanism of Ena/VASP-mediated actin-filament elongation. EMBO J. 2011;30:456–467. doi: 10.1038/emboj.2010.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- 4.Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, Gertler FB. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 5.Lebrand C, Dent EW, Strasser GA, Lanier LM, Krause M, Svitkina TM, Borisy GG, Gertler FB. Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of netrin-1. Neuron. 2004;42:37–49. doi: 10.1016/s0896-6273(04)00108-4. [DOI] [PubMed] [Google Scholar]
- 6.Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 7.Coppolino MG, Krause M, Hagendorff P, Monner DA, Trimble W, Grinstein S, Wehland J, Sechi AS. Evidence for a molecular complex consisting of Fyb/SLAP, SLP-76, Nck, VASP and WASP that links the actin cytoskeleton to Fcgamma receptor signalling during phagocytosis. J Cell Sci. 2001;114:4307–4318. doi: 10.1242/jcs.114.23.4307. [DOI] [PubMed] [Google Scholar]
- 8.Bear JE, Loureiro JJ, Libova I, Fassler R, Wehland J, Gertler FB. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 2000;101:717–728. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- 9.Goh KL, Cai L, Cepko CL, Gertler FB. Ena/VASP proteins regulate cortical neuronal positioning. Curr Biol. 2002;12:565–569. doi: 10.1016/s0960-9822(02)00725-x. [DOI] [PubMed] [Google Scholar]
- 10.Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, Schafer DA. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem. 2005;280:28653–28662. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bear JE, Gertler FB. Ena/VASP: towards resolving a pointed controversy at the barbed end. J Cell Sci. 2009;122:1947–1953. doi: 10.1242/jcs.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rottner K, Behrendt B, Small JV, Wehland J. VASP dynamics during lamellipodia protrusion. Nat Cell Biol. 1999;1:321–322. doi: 10.1038/13040. [DOI] [PubMed] [Google Scholar]
- 13.Reinhard M, Halbrugge M, Scheer U, Wiegand C, Jockusch BM, Walter U. The 46/50 kDa phosphoprotein VASP purified from human platelets is a novel protein associated with actin filaments and focal contacts. EMBO J. 1992;11:2063–2070. doi: 10.1002/j.1460-2075.1992.tb05264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tokuo H, Ikebe M. Myosin X transports Mena/VASP to the tip of filopodia. Biochem Biophys Res Commun. 2004;319:214–220. doi: 10.1016/j.bbrc.2004.04.167. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman LM, Jensen CC, Chaturvedi A, Yoshigi M, Beckerle MC. Stretch-induced actin remodeling requires targeting of zyxin to stress fibers and recruitment of actin regulators. Mol Biol Cell. 2012;23:1846–1859. doi: 10.1091/mbc.E11-12-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith MA, Blankman E, Gardel ML, Luettjohann L, Waterman CM, Beckerle MC. A zyxin-mediated mechanism for actin stress fiber maintenance and repair. Dev Cell. 2010;19:365–376. doi: 10.1016/j.devcel.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niebuhr K, Ebel F, Frank R, Reinhard M, Domann E, Carl UD, Walter U, Gertler FB, Wehland J, Chakraborty T. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J. 1997;16:5433–5444. doi: 10.1093/emboj/16.17.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ball LJ, Kuhne R, Hoffmann B, Hafner A, Schmieder P, Volkmer-Engert R, Hof M, Wahl M, Schneider-Mergener J, Walter U, Oschkinat H, Jarchau T. Dual epitope recognition by the VASP EVH1 domain modulates polyproline ligand specificity and binding affinity. EMBO J. 2000;19:4903–4914. doi: 10.1093/emboj/19.18.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Call GS, Chung JY, Davis JA, Price BD, Primavera TS, Thomson NC, Wagner MV, Hansen MD. Zyxin phosphorylation at serine 142 modulates the zyxin head-tail interaction to alter cell-cell adhesion. Biochem Biophys Res Commun. 2011;404:780–784. doi: 10.1016/j.bbrc.2010.12.058. [DOI] [PubMed] [Google Scholar]
- 20.Moody JD, Grange J, Ascione MP, Boothe D, Bushnell E, Hansen MD. A zyxin head-tail interaction regulates zyxin-VASP complex formation. Biochem Biophys Res Commun. 2009;378:625–628. doi: 10.1016/j.bbrc.2008.11.100. [DOI] [PubMed] [Google Scholar]
- 21.Smith MA, Blankman E, Deakin NO, Hoffman LM, Jensen CC, Turner CE, Beckerle MC. LIM domains target actin regulators paxillin and zyxin to sites of stress fiber strain. PLoS One. 2013;8:e69378. doi: 10.1371/journal.pone.0069378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan CB, Liu X, Tang X, Fu H, Ye K. Akt phosphorylation of zyxin mediates its interaction with acinus-S and prevents acinus-triggered chromatin condensation. Cell Death Differ. 2007;14:1688–1699. doi: 10.1038/sj.cdd.4402179. [DOI] [PubMed] [Google Scholar]
- 23.Maruoka M, Sato M, Yuan Y, Ichiba M, Fujii R, Ogawa T, Ishida-Kitagawa N, Takeya T, Watanabe N. Abl-1-bridged tyrosine phosphorylation of VASP by Abelson kinase impairs association of VASP to focal adhesions and regulates leukaemic cell adhesion. Biochem J. 2012;441:889–899. doi: 10.1042/BJ20110951. [DOI] [PubMed] [Google Scholar]
- 24.Prehoda KE, Lee DJ, Lim WA. Structure of the enabled/VASP homology 1 domain-peptide complex: a key component in the spatial control of actin assembly. Cell. 1999;97:471–480. doi: 10.1016/s0092-8674(00)80757-6. [DOI] [PubMed] [Google Scholar]
- 25.Zimmermann J, Labudde D, Jarchau T, Walter U, Oschkinat H, Ball LJ. Relaxation, Equilibrium Oligomerization, and Molecular Symmetry of the VASP (336–380) EVH2 Tetramer. Biochemistry. 2002;41:11143–11151. doi: 10.1021/bi020379x. [DOI] [PubMed] [Google Scholar]
- 26.Ahern-Djamali SM, Comer AR, Bachmann C, Kastenmeier AS, Reddy SK, Beckerle MC, Walter U, Hoffmann FM. Mutations in Drosophila enabled and rescue by human vasodilator-stimulated phosphoprotein (VASP) indicate important functional roles for Ena/VASP homology domain 1 (EVH1) and EVH2 domains. Mol Biol Cell. 1998;9:2157–2171. doi: 10.1091/mbc.9.8.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haffner C, Jarchau T, Reinhard M, Hoppe J, Lohmann SM, Walter U. Molecular cloning, structural analysis and functional expression of the proline-rich focal adhesion and microfilament-associated protein VASP. EMBO J. 1995;14:19–27. doi: 10.1002/j.1460-2075.1995.tb06971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geese M, Loureiro JJ, Bear JE, Wehland J, Gertler FB, Sechi AS. Contribution of Ena/VASP proteins to intracellular motility of listeria requires phosphorylation and proline-rich core but not F-actin binding or multimerization. Mol Biol Cell. 2002;13:2383–2396. doi: 10.1091/mbc.E02-01-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith GA, Theriot JA, Portnoy DA. The tandem repeat domain in the Listeria monocytogenes ActA protein controls the rate of actin-based motility, the percentage of moving bacteria, and the localization of vasodilator-stimulated phosphoprotein and profilin. J Cell Biol. 1996;135:647–660. doi: 10.1083/jcb.135.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volkman BF, Prehoda KE, Scott JA, Peterson FC, Lim WA. Structure of the N-WASP EVH1 domain-WIP complex: insight into the molecular basis of Wiskott-Aldrich Syndrome. Cell. 2002;111:565–576. doi: 10.1016/s0092-8674(02)01076-0. [DOI] [PubMed] [Google Scholar]
- 31.Peterson FC, Deng Q, Zettl M, Prehoda KE, Lim WA, Way M, Volkman BF. Multiple WASP-interacting protein recognition motifs are required for a functional interaction with N-WASP. J Biol Chem. 2007;282:8446–8453. doi: 10.1074/jbc.M609902200. [DOI] [PubMed] [Google Scholar]
- 32.Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature. 1999;398:39–46. doi: 10.1038/17969. [DOI] [PubMed] [Google Scholar]
- 33.Greenwood AI, Kwon J, Nicholson LK. Isomerase-Catalyzed Binding of Interleukin-1 Receptor-Associated Kinase 1 to the EVH1 Domain of Vasodilator-Stimulated Phosphoprotein. Biochemistry. 2014;53:3593–3607. doi: 10.1021/bi500031e. [DOI] [PubMed] [Google Scholar]
- 34.Bhatt A, Kaverina I, Otey C, Huttenlocher A. Regulation of focal complex composition and disassembly by the calcium-dependent protease calpain. J Cell Sci. 2002;115:3415–3425. doi: 10.1242/jcs.115.17.3415. [DOI] [PubMed] [Google Scholar]
- 35.Waudby CA, Ramos A, Cabrita LD, Christodoulou J. Two-Dimensional NMR Lineshape Analysis. Sci Rep. 2016;6:24826. doi: 10.1038/srep24826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogals MJ, Greenwood AI, Kwon J, Lu KP, Nicholson LK. Neighboring phosphoSer-Pro motifs in the undefined domain of IRAK1 impart bivalent advantage for Pin1 binding. FEBS J. 2016;283:4528–4548. doi: 10.1111/febs.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaff J, Fink CC, Slepchenko B, Carson JH, Loew LM. A general computational framework for modeling cellular structure and function. Biophys J. 1997;73:1135–1146. doi: 10.1016/S0006-3495(97)78146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daum S, Lucke C, Wildemann D, Schiene-Fischer C. On the benefit of bivalency in peptide ligand/pin1 interactions. J Mol Biol. 2007;374:147–161. doi: 10.1016/j.jmb.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 39.Jencks WP. On the attribution and additivity of binding energies. Proc Natl Acad Sci U S A. 1981;78:4046–4050. doi: 10.1073/pnas.78.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leopold MF, Urbauer JL, Wand AJ. Resonance assignment strategies for the analysis of nmr spectra of proteins. Mol Biotechnol. 1994;2:61–93. doi: 10.1007/BF02789290. [DOI] [PubMed] [Google Scholar]
- 41.Nix DA, Fradelizi J, Bockholt S, Menichi B, Louvard D, Friederich E, Beckerle MC. Targeting of Zyxin to Sites of Actin Membrane Interaction and to the Nucleus. J Biol Chem. 2001;276:34759–34767. doi: 10.1074/jbc.M102820200. [DOI] [PubMed] [Google Scholar]
- 42.Drees B, Friederich E, Fradelizi J, Louvard D, Beckerle MC, Golsteyn RM. Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. J Biol Chem. 2000;275:22503–22511. doi: 10.1074/jbc.M001698200. [DOI] [PubMed] [Google Scholar]
- 43.Callebaut I, Cossart P, Dehoux P. EVH1/WH1 domains of VASP and WASP proteins belong to a large family including Ran-binding domains of the RanBP1 family. FEBS Lett. 1998;441:181–185. doi: 10.1016/s0014-5793(98)01541-5. [DOI] [PubMed] [Google Scholar]
- 44.Petit MM, Fradelizi J, Golsteyn RM, Ayoubi TA, Menichi B, Louvard D, Van de Ven WJ, Friederich E. LPP, an actin cytoskeleton protein related to zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol Biol Cell. 2000;11:117–129. doi: 10.1091/mbc.11.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harbeck B, Huttelmaier S, Schluter K, Jockusch BM, Illenberger S. Phosphorylation of the vasodilator-stimulated phosphoprotein regulates its interaction with actin. J Biol Chem. 2000;275:30817–30825. doi: 10.1074/jbc.M005066200. [DOI] [PubMed] [Google Scholar]
- 46.Tu Y, Wu S, Shi X, Chen K, Wu C. Migfilin and Mig-2 link focal adhesions to filamin and the actin cytoskeleton and function in cell shape modulation. Cell. 2003;113:37–47. doi: 10.1016/s0092-8674(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Tu Y, Gkretsi V, Wu C. Migfilin interacts with vasodilator-stimulated phosphoprotein (VASP) and regulates VASP localization to cell-matrix adhesions and migration. J Biol Chem. 2006;281:12397–12407. doi: 10.1074/jbc.M512107200. [DOI] [PubMed] [Google Scholar]
- 48.Bachmann C, Fischer L, Walter U, Reinhard M. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J Biol Chem. 1999;274:23549–23557. doi: 10.1074/jbc.274.33.23549. [DOI] [PubMed] [Google Scholar]
- 49.Kühnel K, Jarchau T, Wolf E, Schlichting I, Walter U, Wittinghofer A, Strelkov SV. The VASP tetramerization domain is a right-handed coiled coil based on a 15-residue repeat. Proc Natl Acad Sci U S A. 2004;101:17027–17032. doi: 10.1073/pnas.0403069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


